Abstract
Nurse scientists play an important role in studying complex relationships among human genetics, environmental factors, and the microbiome, all of which can contribute to human health and disease. Therefore, it is essential that they have the tools necessary to execute a successful microbiome research study. The purpose of this article is to highlight important methodological factors for nurse scientists to consider when designing a microbiome study. In addition to considering factors that influence host-associated microbiomes (i.e., microorganisms associated with organisms such as humans, mice, and rats), this manuscript highlights study designs and methods for microbiome analysis. Exemplars are presented from nurse scientists who have incorporated microbiome methods into their program of research. This review is intended to be a resource to guide nursing-focused microbiome research and highlights how study of the microbiome can be incorporated to answer research questions.
Keywords: microbiome, research methods, genetics
Nurse scientists have emerged as leaders in the effort to better understand the complex relationships among the human genome, the genomes of the microbiota, and the multitude of environmental forces that shape superorganisms and contribute to health outcomes (Brooks et al., 2017; Cong et al., 2017; Edwards, Cunningham, Dunlop, & Corwin, 2017; Fourie et al., 2016). Nurses offer a unique holistic perspective on patient health that spans areas as diverse as midwifery, cardiology, nephrology, neurology, and psychology. They are thus well suited to join the microbiome research revolution but require additional training to increase their “omics” literacy.
Over the past several decades, authors have widely cited the idea that the number of microbial cells in the human body outnumbers our mammalian cells by a ratio of 10:1 (Goodman & Gordon, 2010; Luckey, 1972; Savage, 1977). Given that there are 37.2 trillion mammalian cells in the human body, this idea suggests an enormous pool of cells whose effects on health were open to study (Bianconi et al., 2013). Although more recent calculations put the number closer to 1.3 microbial cells per 1 mammalian cell (Sender, Fuchs, & Milo, 2016), each of the thousands of microbial strains that comprise the microbiome contains a unique genome, and microbial genes do vastly outnumber mammalian genes. Furthermore, this vast reservoir of microbial genes plays a significant role in maintaining the homeostasis of the host (Gilbert et al., 2018; Khanna & Tosh, 2014; Schirmer et al., 2016; Stamper et al., 2016). Microorganisms perform vital functions, including (but not limited to) development of the immune system (Bartman, Chong, & Alegre, 2015; Iebba, Nicoletti, & Schippa, 2012; Wei et al., 2010), metabolism and digestion of food materials (David et al., 2014; Kau, Ahern, Griffin, Goodman, & Gordon, 2011; Sonnenburg & Backhed, 2016), and synthesis of vitamins, cytokines, and neurotransmitters (Dinan & Cryan, 2017; Gilbert et al., 2016; Griffin et al., 2017; Schirmer et al., 2016; Sonnenburg & Backhed, 2016).
Microbiomes are critical to health, and authors have suggested that they be considered a new endocrine organ (Baquero & Nombela, 2012; Clarke et al., 2014). The addition of microbiome analyses to research designs has provided valuable information related to physiologic and pathophysiologic processes and may have a role in the diagnosis and treatment of disease (Bartman et al., 2015; Tamboli, Neut, Desreumaux, & Colombel, 2004; Yang & Zubcevic, 2017). Even in neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases, microorganisms may play a role, either as potential causative or facilitating agents or as diagnostic responders to changing physiological conditions associated with disease progression (Keshavarzian et al., 2015; Sampson et al., 2016; Vogt et al., 2017). Understanding the role of the microbiome is additionally critical for interpreting results from animal studies.
The purpose of this article is to highlight important methodological factors for nurse scientists to consider when designing a microbiome study. In addition to considering factors that influence host-associated microbiomes (i.e., microorganisms associated with organisms such as humans, mice, and rats), this article highlights study designs and methods for microbiome analysis. We have provided a list of commonly used definitions in microbiome research in Table 1.
Table 1.
Common Terms in Microbiome Research.
| Term | Definition |
|---|---|
| 16S ribosomal RNA (rRNA) gene | The gene encoding for the small subunit of the prokaryotic ribosome, which contains nine variable regions that can be targeted with PCR (Janda & Abbott, 2007). |
| 16S rRNA gene amplicon sequencing | A PCR-based sequencing method targeting microbial rRNA genes, which is used to identify and determine the relative abundance of microorganisms present within a given sample and to compare microbial communities between samples (Janda & Abbott, 2007). |
| Alpha diversity | Microbiota diversity within an individual site or sample diversity; one value per sample (Jost, 2007). |
| Beta diversity | Between-sample changes in taxon diversity (Jost, 2007). |
| Dysbiosis | A breakdown in the homeostasis of a microbial community, leading to overgrowth of detrimental microorganisms (Tamboli et al., 2004). |
| Metabolomics | The comprehensive, qualitative, and quantitative study of all the small molecules in an organism that result from cellular metabolism and give insight to the network of biochemical reactions in a cell (Liu & Locasale, 2017). |
| Metagenome/metagenomics | The combination of microbial genomes and genes in an environment or sample/the direct genetic analysis of genomes contained within an environmental sample (Thomas, Gilbert, & Meyer, 2012). |
| Microbiome | The collection of microbial organisms and genetic information (i.e., genomes) at a given site (i.e., skin, oral, stool, others; Ursell et al., 2012). |
| Microbiota | The microbial taxa associated with multicellular organisms, from plants to humans (Ursell et al., 2012). |
| Operational taxonomic unit | Similar sequences are clustered together to represent a microbial taxon based on a predetermined similarity identity cutoff. Thresholds of 97–100% have been used to represent an approximate “species” level grouping (Edgar, 2018). |
| Shotgun metagenome sequencing | A DNA sequencing method that enables comprehensive sampling of all genes in all organisms in a given complex microbial sample instead of targeting a specific marker gene (Sharpton, 2014). |
| Shotgun metatranscriptome sequencing (metatranscriptomics) | Analysis of all messenger RNAs encoded by a group of microorganisms within a complex sample. Metatranscriptomics describes the pool of expressed genes at a given time point in a complex microbial sample (Warnecke & Hess, 2009). |
Note. PCR = polymerase chain reaction.
Factors Influencing the Human Microbiome
The human microbiome is affected by numerous factors that researchers must take into account when designing studies, including genetics, age, diet, antibiotics, and environment (Figure 1). These factors cause alterations in the microbiome, which may lead to negative or positive effects on health (Bokulich et al., 2016; Conlon & Bird, 2014; Cresci & Bawden, 2015). For instance, dietary intake and antibiotic use can cause dysbiosis, a pathologic alteration in the native microbial community, in the gut microbiota and contribute to diseases such as obesity, asthma, diabetes, inflammatory bowel disease, and cardiovascular disease (Bokulich et al., 2016; David et al., 2014; Zoetendal & de Vos, 2014). In turn, several factors can influence dietary intake and nutrition, including cultural or religious beliefs, geographical location, and socioeconomic status, adding further complexity to the research design and external validity of study results (Burkitt, Walker, & Painter, 1972; Darmon & Drewnowski, 2015; Griffin et al., 2017; McInerney et al., 2016). Conversely, breast milk and pre- and probiotics contribute to the abundance of microbes that overall benefit immune health and function (Conlon & Bird, 2014; Langdon, Crook, & Dantas, 2016). Because of their influence on microbial communities, it is important to take these factors into consideration when designing a research study.
Figure 1.
Some common factors affecting the microbiome: Humans and animals.
As with human studies, researchers must take into account several factors that influence the animal microbiome when designing animal studies. For example, animal age, species, diet, and housing considerations (such as grouped vs. single animal housing) are strong drivers of the microbiome and can actually have a stronger influence on research results than the main independent variable of interest (Hoy et al., 2015; Lees et al., 2014; McCord et al., 2014). Other factors, such as time of sample collection and accounting for circadian rhythmicity of microbial relative abundance, may not have as strong a microbiome effect as the previous factors but should still be taken into consideration and controlled during study design and interpretation of results (Liang, Bushman, & FitzGerald, 2015). Providing and maintaining a rationale for a consistent sampling time will account for potential variation in bacterial expression patterns throughout the circadian day.
Setting Up a Microbiome Research Project
Advances in human microbiome research depend on precise study execution, control, and reproducibility. Aside from the factors described above, researchers must consider a number of other elements to ensure high-quality data analysis and results. These elements include study design, sample collection methods, and processing and storage for downstream analysis. We provide a summary of the specific recommendations for human and animal microbiome studies that we describe in this article in Table 2.
Table 2.
Recommendations for Conducting a Microbiome Study.
| Recommendations for a Microbiome Study |
|---|
|
| Special considerations for human studies |
|
| Special considerations for animal studies |
|
Note. PCR = polymerase chain reaction; rRNA = ribosomal RNA; PERMANOVA = permutational analysis of variance.
Study Design
Because no standard methods have yet been established in microbiome research, effect sizes vary by disorders (Kelly et al., 2015; La Rosa et al., 2012). For singular microbiome studies, doing a pilot study first will help define adequate sample and effect sizes. Alternatively, large cohort studies like the Human Microbiome Project (HMP) and tools like “Evident” (https://github.com/biocore/Evident) have published methods to estimate sample sizes based on projected effect sizes (Goodrich et al., 2014).
Sample Collection Methods
Most biological sample collection protocols for human microbiome studies are noninvasive or minimally invasive and cause minimal risk to the participants (except for collection of hard tissues from the oral cavity and vagina and rectal swabs; Kuczynski et al., 2011; McInnes & Cutting, 2010). Although fecal sampling for microbiome studies is noninvasive, participants may view this route of sample collection as unpleasant, and researchers should consider and account for potential attrition in participants in longitudinal studies (Ricardo-Rodrigues et al., 2015). Devoting time to education on procedures for sample collection and return can increase compliance and will increase the sample yield (Wolf et al., 2001).
Researchers collect samples once or multiple times over a specific study period, and in some cases, favor longitudinal studies (Faust, Lahti, Gonze, de Vos, & Raes, 2015). Regardless of the number of collection time points, however, accurate collection techniques and sample handling are crucial for preventing sample contamination and postcollection microbial growth (Jordan et al., 2017; Vogtmann et al., 2017). Use of either repeated sampling or time series sampling gives insight into the volatility, resilience, composition, and relative abundance of microbial communities and can provide baseline data for genetically distinct individuals (David et al., 2014). It should be noted that because of genetics, diet, and study compliance, human microbiome studies can have much higher interindividual variability than animal studies. For these reasons, longitudinal studies can provide a more comprehensive perspective on microbiota diversity (within-subject and between-subject diversities) compared to single time point cross-sectional sampling. Although previously cost prohibitive for many researchers due to the historically high cost of microbiome sequencing, longitudinal microbiome studies have become more accessible to more investigators because of the reduced cost of next-generation sequencing (NGS) methods and increased automation for the DNA extraction and sequencing processes. Therefore, investigators should plan to perform replicate sampling from the same individual (Gohl et al., 2016). The numbers of recommended replicates vary based on the research question and statistical tests used, but generally two to three replicates are recommended per sample site. The Manual of Procedures for the HMP has described in detail the methods for sample collection, transport, and storage for each microbial habitat; the information is publicly accessible (McInnes & Cutting, 2010).
Sample Processing and Storage
Methodologies used for sample processing, storage, and transport can impact DNA and RNA recovery and the final observed microbial community structure. Because these steps occur at the beginning of the workflow, they affect all downstream stages. A standardized sample tracking system using electronic- and paper-based methods with an established labeling system will allow close monitoring of the samples through collection, processing, storage, and downstream analysis. The tracking system is also critical for long-term storage because it could provide precise locations of samples in an −80°C freezer.
Sample processing will generally follow the instructions from a commercial kit chosen based on the research question and previously published studies. Sample storage prior to nucleic acid extraction includes flash freezing either on dry ice or liquid nitrogen followed by long-term storage at −20°C or −80°C. Commercially available alternatives allow for medium-term storage at room temperature in stabilizing and inactivating reagents, allowing for home collection and postal shipment of oral, vaginal, and fecal samples (e.g., products from companies such as DNA Genotek, Norgen Biotek, Zymo, and others). DNA-based analyses are less sensitive to short-term environmental shifts than RNA-, metabolite-, or protein-based analysis; thus, DNA sampling is less onerous. Metabolomics or metatranscriptomics studies require more rigorous standardization of sample processing and storage methods for rapid stabilization of samples due to the faster nature of by-product degradation. For example, in metabolomics analysis protocols, a standardized weight of fecal sample is aliquoted to an Eppendorf tube, flash frozen in liquid nitrogen, and eventually freeze-dried by lyophilization before shipment for processing in order to ensure accuracy and reproducibility of sample results (Deda, Gika, & Theodoridis, 2018).
Maintaining constant environmental conditions is critical during storage and shipping. The number of freeze–thaw cycles, resulting from fluctuation of environmental conditions, can impact microbial composition and nucleic acid integrity (Sergeant, Constantinidou, Cogan, Penn, & Pallen, 2012). Investigators should thoroughly document any significant environmental changes that occur to samples and should then account for potential effects on the data with statistical modeling (Chen et al., 2012).
Microbiome Sequencing Methods
Rigorous study design, sample collection, and storage and controlling for environmental confounders are all important factors in planning microbiome research, but understanding and proper selection of appropriate sequencing methods is vital for a successful study. Despite the power of cultivation-independent molecular methods, all such methods contribute bias. Sequencing results, therefore, are a proxy for the “true” microbial community, and it is important to understand the limitations of different molecular approaches. In this section, we review some commonly used approaches for microbiome sequencing.
16S rRNA Sequencing Versus Shotgun Metagenomics Sequencing
Before the development of nucleic acid–based molecular tools, including DNA-based amplification techniques such as the polymerase chain reaction (PCR), characterization of the human microbiota was limited to microscopy and laboratory cultivation of organisms (Ursell et al., 2012). Consequently, the microorganisms that were studied in depth were those that could be successfully grown in vitro (such as Escherichia coli) but were not necessarily dominant organisms in the original sample. This limitation led to a sparse view of complex microbial communities, elsewhere labeled the “great plate count anomaly” (Staley & Konopka, 1985), and prevented scientists from comprehensively identifying microbial populations throughout the body. Although important developments in cultivation approaches have enabled recovery of previously “unculturable” organisms (Nichols et al., 2010), most organisms remain resistant to growth under laboratory conditions. To circumvent such difficulties, cultivation-independent methods have been widely employed to characterize complex microbial communities (Amann, Ludwig, & Schleifer, 1995).
The most commonly used methods today include 16S ribosomal RNA (rRNA) gene amplicon sequencing and “shotgun” metagenome sequencing. Both approaches are culture-independent techniques that use DNA sequence data to infer the presence and relative abundance of microorganisms and specific genes from those organisms. The use of molecular sequence data to identify microorganisms is necessary because microorganisms have a limited number of morphologies and look similar to one another, and many organisms share functional features or phenotypes (e.g., metabolic activity) even when they are not closely related. Prior studies in which investigators used morphology or metabolic activity as a guide to phylogeny were eventually revealed to be highly problematic (Fox et al., 1980). DNA-dependent analyses also allow for the identification and classification of many types of organisms that are historically laborious or currently impossible to grow in a controlled laboratory setting (Morgan & Huttenhower, 2012). In addition, these methods have been more accessible to a broad range of scientists due to the availability of highly reliable commercial kits and limited equipment needs. Less specific experience is needed to perform DNA extraction, PCR, and sequencing than to perform cultivation, and many of these tasks can be outsourced to external facilities. Thus, researchers require a basic understanding of these processes for properly interpreting results. Major advances in sequencing technology, bioinformatic algorithms, and computational capacity have facilitated the development of the throughput of these methods, concomitant with reduced cost.
16S rRNA gene amplicon sequencing
Until recently, the most common method for investigating the microbiome was 16S rRNA gene amplicon sequencing (Schmidt, DeLong, & Pace, 1991). The rRNAs were identified as phylogenetic markers very early on (Woese & Fox, 1977) due to a number of favorable features: (a) the presence of rRNA genes in all known organisms due to the integral role of rRNA as a component of the ribosome and, therefore, in messenger RNA (mRNA) translation (Birtel, Walser, Pichon, Burgmann, & Matthews, 2015); (b) the presence of highly conserved regions of the gene, allowing for the design of broad-range PCR primers; (c) the presence of highly variable regions, allowing for the use of the gene for phylogenetic analysis; (d) the low rate of rRNA lateral gene transfer between different microbial lineages (Kurland, Canback, & Berg, 2003); and (e) the absence of translation of the gene, thereby avoiding codon degeneracy issues that complicate PCR primer design in protein-coding genes.
Because the small-subunit rRNA gene (i.e., 16S or 18S rRNA gene) is present in the genomes of all microorganisms, the regions of the gene which are highly conserved (or unchanged) are ideal target sites for the so-called universal primers. Universal primers are used to target a wide range of microorganisms, including unknown organisms that have not been cultivated. Between these highly conserved regions in the 16S rRNA gene are nine hypervariable regions that have changed over the course of microbial evolution (Figure 2). These regions, which are more similar between closely related organisms and more different between more distantly related organisms, can thereby serve as a guide to microbial taxonomy. For taxonomic purposes, the full 16S rRNA gene (approximately 1,542 base pairs [bp] in E. coli; see Figure 2) is traditionally used (Brosius, Palmer, Kennedy, & Noller, 1978). When attempting to determine whether two microorganisms belong to the same species (itself a difficult concept in the field of microbiology; Rossello-Mora & Amann, 2001), researchers have traditionally used a threshold of 97% similarity for exclusion (i.e., two organisms with 16S rRNA genes with lower than 97% similarity are considered to be distinct species; Stackebrandt & Goebel, 1994). Although some of the newest sequencing technologies are able to sequence the entire 16S rRNA gene (e.g., Oxford Nanopore and Pacific Biosciences), historically, much shorter sequences have been employed for analysis of microbial communities. Even now, full gene sequencing is expensive and requires sophisticated sequencing pipelines (Yang, Wang, & Qian, 2016).
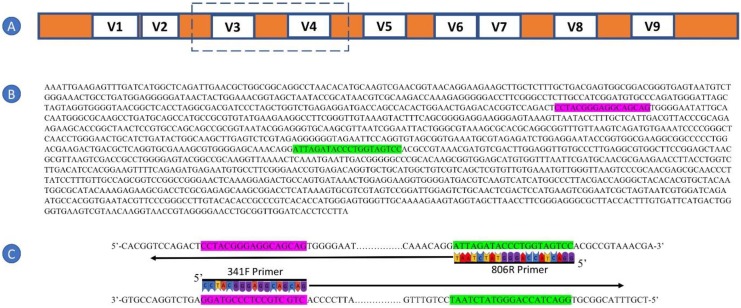
Figure 2.
Targeting the small-subunit (16S) ribosomal rRNA (rRNA) gene in microbial communities using polymerase chain reaction (PCR). (A) Schematic of the conserved and variable regions of the 16S rRNA gene and PCR primers. The 16S rRNA gene has nine variable regions that are dispersed throughout the conserved regions of the gene. Primers used for sequencing of broad categories of microorganisms (e.g., all bacteria) are designed in the conserved (orange) region, and the generated amplicons span one or more variable (white) regions. The box indicates the approximate region targeted by the commonly used primers 341F and 806 R. (B) A full-length sequence (1,541 bases) of the “top” strand of a 16S rRNA gene of Escherichia coli (GenBank Accession Number J01859). The forward primer location is highlighted in pink and the reverse primer location is highlighted in green. Note that the length of the 16S rRNA gene can vary substantially between different taxa, but conserved regions generally remain similar, even among distantly related taxa. The primer sites are targeted by the “forward” primer 341F (CCTACGGGAGGCAGCAG) and the “reverse” primer 806R (GGACTACNVGGGTWTCTAAT; where N = any base, V = A, C or G, and W = A or T). Note that only a single variant of the degenerate 806R primer matches the E. coli 16S rRNA gene and is shown here. (C) A visual of the annealing of 341F (targeting the pink sequence on the “bottom” strand) and 806R (targeting the green sequence on the “top” strand) primers to the E. coli 16S rRNA gene sequence. These primers can also anneal to most bacterial 16S rRNA genes and generate PCR amplicons containing the V3 and V4 variable regions that are used for annotation purposes. (figure modified from Del Chierico et al., 2015.)
The more traditional approach to 16S rRNA sequencing has been to choose one or two hypervariable regions to target via PCR for short-read NGS. The ability to acquire a very large number of short sequences (on the scale of 100–500 bp [bp]) necessitated the development of new techniques to allow for rapid analysis of large datasets. Prior to starting a microbiome study, it is important that researchers consult both the literature and experts in field; this practice can help them tailor the choice of the primer set to the goals of the study. Not all primer sets work for all organisms. For example, some hypervariable regions are suitable for distinguishing between closely related species from the same genus, but other regions may have no differences between the two species (Ionescu et al., 2016). Clearly, the ideal universal primers should match the rRNA genes of all organisms and amplify one or more hypervariable regions of the 16S rRNA gene to accurately categorize known and unknown organisms. No such perfect primer set exists, however, and the choice of primers may need to be adjusted for each microbiome study based on the goals of the study and the specific genera of interest (Kuczynski et al., 2011). After amplification and sequencing, the sequences in the variable regions of the 16S rRNA gene are used as genetic fingerprints to identify bacteria by phylum to species and below (Reyes-Lopez et al., 2003).
Because these NGS approaches to sequencing of the rRNA genes generate so many sequences (usually in the range of 1,000–100,000 sequences per sample), investigators employ data reduction strategies. One data reduction strategy is to group highly similar sequences together for taxonomic identification. In many cases, sequences are grouped together at 97% similarity (i.e., all sequences within any given group are at least 97% similar to every other sequence in the group). These groups of similar sequences are called operational taxonomic units (OTUs) and have been used as a proxy for microbial “species.” This clustering approach is an informatics strategy only, and the clustering does not depend on any external biological information. As a result, by using this method, it is possible to group sequences from distinct taxa into a single OTU as well as to split sequences from a single taxon into multiple OTUs. Researchers have developed solutions to control for the ambiguity of using representative DNA sequences as a proxy for a bacterial taxon (Callahan et al., 2016), but a full discussion is beyond the scope of this work.
After clustering sequences into OTUs, the researcher selects a representative sequence from each OTU for annotation and then assigns this annotation to all the sequences in the OTU. One common mechanism for annotation is to identify the sequences that are most closely related to the representative sequence and use the taxonomic information from those sequences to assign taxonomy to the unknown OTU. For this technique, investigators use 16S rRNA gene sequence databases such as SILVA (Pruesse et al., 2007) or Greengenes (DeSantis et al., 2006). Two software packages for the analysis of large amplicon data sets are used extensively and process data from raw sequences to annotated and tabulated OTU data: QIIME (Caporaso et al., 2010) and mothur (Schloss et al., 2009).
Overall, the 16S rRNA gene amplicon sequencing approach has both beneficial and detrimental features. The beneficial features include (a) highly robust PCR amplification, allowing the use of low amounts of DNA input; (b) a universal gene present in all organisms; (c) established bioinformatics pipelines; and (d) data amenable to straightforward statistical analysis. Conversely, the detrimental features include (a) the use of PCR, which is dependent on a priori knowledge for primer design and can introduce substantial bias (Green, Venkatramanan, & Naqib, 2015); (b) poor and inconsistent resolution at the taxonomic level of genus and species (Zeigler, 2003); (c) lack of information about the functional gene content of the identified organisms (see a possible solution to this issue in Langille et al., 2013); and (d) limited capacity for multidomain detection (i.e., bacteria, archaea, eukarya) within a single assay. These detrimental features have driven the development of shotgun sequencing approaches.
Shotgun metagenomics sequencing is a more comprehensive technique that potentially allows for sampling all genes from all organisms present in a given sample. In addition to taxonomic information, as derived from 16S rRNA gene amplicon sequencing, shotgun sequencing provides sequence data from other genes, including the so-called functional genes (i.e., not housekeeping genes) that not all organisms have. Functional genes include, but are not limited to, genes involved in nutrient metabolism, downstream signaling, antibiotic production, antibiotic resistance, motility, metabolism (including fermentation), respiration (aerobic/anaerobic), and secondary metabolite production (Cresci & Bawden, 2015; Kuczynski et al., 2011; Langdon et al., 2016). Furthermore, shotgun sequencing can target bacteria, archaea, fungi, microeukaryotes, and some viruses simultaneously. Although a number of approaches exist for preparing genomic DNA for shotgun sequencing, many protocols start by fragmenting the DNA into relatively small pieces (generally 250- to 600-bp fragments; Di Bella, Bao, Gloor, Burton, & Reid, 2013; Morgan & Huttenhower, 2012). This fragmentation is necessary for Illumina sequencing platforms, which do not tolerate very large DNA fragments. Other sequencing platforms, such as those from Oxford Nanopore and Pacific Biosciences, can use much larger pieces of genomic DNA (>10,000 bp), but currently have a relatively high error rate that has limited their use in sequence analysis of complex microbial communities. Improved sequencing strategies and sequencing analysis algorithms may increase the use of these platforms in the future (Frank et al., 2016; Huson et al., 2018). After fragmentation, genomic DNA is prepared to be loaded onto a sequencer through a process called “library preparation,” which involves a series of enzymatic steps, including repair of the ends of the DNA and ligation of sequencing adapters. The ligation step is used to incorporate known DNA sequences into the ends of unknown genomic DNA in a sequence-independent manner. Subsequently, the known sequences (called sequencing adapters) are used to manipulate the DNA by the way of PCR amplification (to increase the total amount of DNA but without selecting for any specific sequences) and to initiate the sequencing reaction, again without selection for any specific sequence from the source genomic DNA. In most cases, the sequencing adapters also contain sample-specific information (so-called barcodes), which allow the mixing of multiple samples on a single sequencing run. Fragmenting and barcoding the DNA sequences is performed to exploit fully the tremendous amount of data generated on a sequencing run by splitting the sequencing data yield into output from multiple samples, thereby decreasing the per-sample cost.
The data yield from shotgun metagenome sequencing can be daunting. Millions to hundreds of millions of short sequences (generally 150 bases, in pairs) are generated using Illumina sequencing platforms. These data can be analyzed in a variety of ways (Sharpton, 2014). Common strategies include (but are not limited to) (a) searching for lineage-specific marker genes to annotate sequences accurately (Segata et al., 2012), (b) high-throughput basic logical alignment search tool analysis of individual sequences against reference sequence databases such as the National Center for Biotechnology Information nonredundant database (Buchfink, Xie, & Huson, 2015; Huson et al., 2016; Meyer et al., 2008) to annotate data and assign sequences to taxonomic and functional gene groups, and (c) assembly of larger DNA sequences (“contigs”) from the short-read data (called de novo assembly) and subsequent mapping of sequence data back to the assembled contigs (Brown et al., 2011). Ultimately, the annotated data are used to characterize the gene content of microbial communities, measure diversity, and identify differences in the relative abundance of microbial features (i.e., taxa, genes, and pathways) between different groups of samples.
Although shotgun metagenome sequencing has many advantages, there are also challenges. In particular, analysis of the large data output is computationally demanding, the short sequences generated from Illumina sequencers can be difficult to annotate, and the relative abundance of sequences from each organism is affected by both the abundance of the organism and the size of its genome. In samples with a high amount of host DNA, shotgun sequencing can yield very little microbial DNA because there is no PCR selection of microbial genes. Shotgun metagenome data are richer in total numbers of sequences, in number of organisms targeted, and in regions of the genome targeted. Conversely, as the complexity is much greater, low-abundance features can be difficult to detect. Unlike 16S rRNA gene amplicon sequencing, the field has not settled on standard approaches for data analysis.
Pros and cons of 16S and SMS technology
Despite notable differences between 16S rRNA and shotgun sequencing approaches, there are several common factors for researchers to consider when they are starting microbiome research studies. Both approaches depend on workflows upstream of the library preparation and sequencing, including sampling, sample storage, and genomic DNA extraction (Brooks et al., 2015; Choo, Leong, & Rogers, 2015). Investigators should thus take great care to establish proper experimental design and sampling, storage, and library preparation procedures to reduce bias (Goodrich et al., 2014; Ionescu et al., 2016). Recently, cell- and DNA-based standards have been developed for microbiome studies that allow for the assessment of DNA extraction efficiency and bias associated with library preparation (Tighe et al., 2017). In addition to upstream sources of bias, library preparation protocols can differ greatly. For 16S rRNA gene amplicon sequencing, a large number of primer sets can be used as well as a wide range of PCR mastermixes and PCR conditions that can greatly influence the observed microbial community (Green et al., 2015). Similarly, for shotgun sequencing approaches, there are a number of different techniques for fragmenting genomic DNA as well as a wide range of kits for library preparation. These preparation protocols can also influence the observed microbial community. Finally, analysis can be influenced by the software used for data analysis, databases used for output classification, clustering strategies, and taxonomic level of analysis. Microbiome analysis by 16S rRNA amplification is still the most commonly used approach because it is relatively inexpensive (generally in the range of 15–40 USD per sample, depending on amplicon length and depth of sequencing), the data analysis can be performed using established pipelines, and there is a large body of archived data (Ranjan, Rani, Metwally, McGee, & Perkins, 2016). Additionally, as the technology has been around longer, many online tutorials are available on the bioinformatics and statistical analysis of this technology.
Shotgun metagenome sequencing, as described above, has many advantages over 16S rRNA gene amplicon sequencing but remains substantially more expensive (generally in the range of 100–400 USD per sample, depending on library preparation protocol, sequencing read length, and depth of sequencing) and is, itself, not immune from improper annotation of sequences. Although some software packages have included attempts to use 16S rRNA gene amplicon sequence data to infer the presence of specific functional genes (Langille et al., 2013), shotgun data provide a more rigorous option to generate taxonomic data, functional gene data, and coupled taxonomic information for functional genes (i.e., allow for the identification of the source organism for a gene of interest that is not a rRNA gene). Shotgun sequence data can be used for assembly of large genomic fragments and, in some cases, near complete genomes (Albertsen et al., 2013; Luo, Tsementzi, Kyrpides, & Konstantinidis, 2012; Sieber et al., 2018). In addition, shotgun data can be used to generate gene catalogs for specific environments, including host-associated environments such as skin or feces (Karlsson, Nookaew, & Nielsen, 2014; Qin et al., 2010).
Finally, it should be noted that 16S rRNA gene amplicon sequencing and shotgun metagenome sequencing can be used in tandem. 16S sequencing can be performed on very large sample sets, and the data generated from that initial sequencing can be used to guide the selection of samples for more in-depth interrogation using shotgun sequencing. In addition, comparing results from 16S and shotgun sequencing of human-associated microbial communities from many different body sites has revealed a wide range of different microbial communities in the same niche from different healthy individuals but much lower variation in the relative abundance of metabolic pathways at a high level (Huttenhower et al., 2012). Ultimately, the scientific questions being asked in each study should dictate the choice of method used.
Classifying Gene Activity: Metatranscriptomics
Shotgun sequencing techniques, in which nucleic acids are sheared and adapters are ligated in a sequence-independent manner, are also employed to target mRNAs in a method broadly termed RNAseq (Di Bella et al., 2013). When applied to a complex community of microorganisms, this technique is called metatranscriptomics. Conceptually, the approach is straightforward. Total RNA extracts are converted to double-stranded complementary DNA and subsequently prepared for high-throughput NGS. Unlike shotgun metagenome sequencing, however, the total RNA extract must be manipulated before sequencing because, in most cases, most (>80%) of the RNA is composed of ribosomes. While analysis of ribosomal RNA directly can be extremely useful, metatranscriptomics focuses on the analysis of mRNA. Thus, the mRNA must be enriched, either by removal of rRNAs (i.e., ribosomes) or by specific capture of mRNA. Microorganisms do not, however, polyadenylate their mRNAs; thus, poly(A) capture techniques that are standard in eukaryotic transcriptome sequencing cannot be used in the field of microbiology (Bashiardes, Zilberman-Schapira, & Elinav, 2016). rRNAs can be removed through a number of strategies (Stewart, Ottesen, & DeLong, 2010), but the most straightforward is to hybridize total RNA with anti-sense biotinylated DNA oligonucleotide probes targeting the rRNAs of a range of different microbial taxa (Xiong et al., 2012). These probes (and the RNA bound to them) are removed from the solution using streptavidin-coated magnetic beads (streptavidin has an extremely high affinity for biotin; Weber, Ohlendorf, Wendoloski, & Salemme, 1989). The RNA remaining in the solution is prepared for sequencing using standard RNA sequencing library preparation protocols (reviewed in Sarode, Parris, Ganesh, Seston, & Stewart, 2016). These libraries are then sequenced deeply, as with shotgun metagenome sequencing. In host–microbial ecosystems, host RNA (both rRNA and mRNA) can dominate microbial transcripts; thus, further RNA selection may be required.
Despite these difficulties, microbial metatranscriptome sequence data can be highly valuable. RNA-based sequencing approaches provide information regarding microorganisms active at the time of sampling and (because microorganisms alter transcript abundances rapidly) thus a snapshot of microbial gene expression patterns (Sarode et al., 2016). In addition, metatranscriptome sequencing provides quantitative data that can be used to characterize essential and condition-specific gene expression patterns. Thus, metatranscriptomics provides the added information of gene transcription and activity (compared to simply identifying presence or absence of a gene) and has the potential to address research questions related to external factors and their influence on microbiome activity in healthy and disease states (Helbling, Ackermann, Fenner, Kohler, & Johnson, 2012).
Quantifying Metabolic Output of the Microbiome: Metabolomics
Although shotgun metagenomic and metatranscriptome sequencing are critical tools for characterizing community structure and activity in microbial environments, these methods still serve as proxies for metabolic activity. Direct measurement of metabolites can be used in concert with molecular data to demonstrate that a shift in microbial community structure and in microbial gene expression patterns lead to a change in physiochemical conditions (Hale et al., 2018; Tankou et al., 2018; Yan et al., 2018). Metabolomics is the study of the metabolite composition within a cell type, tissue, or other sample and allows for an analysis of functional processes that are occurring as a result of cellular metabolism (Patti, Yanes, & Siuzdak, 2012). Such analyses can be more sensitive to changing conditions than shotgun metagenomics (Gowda & Djukovic, 2014), and recent research has shown that the microbial phenotype, or the functional result of metabolic pathways, is more suggestive of disease or health than is the presence or absence of bacteria alone (Gowda & Djukovic, 2014).
The two main techniques used for metabolomics analysis are nuclear magnetic resonance (NMR)- and mass spectrometry (MS)-based metabolomics, and both platforms are used to characterize the metabolic profiles of biological fluids such as urine, feces, serum, and saliva (Dettmer, Aronov, & Hammock, 2007; Lindon, Nicholson, Holmes, & Everett, 2000; Patti et al., 2012). MS-based metabolomics is usually coupled with gas chromatography, ion chromatography, or liquid chromatography to analyze specific metabolite components in the sample. Comparison of the NMR- and MS-based metabolomics is beyond the scope of this article, but each system has unique benefits and limitations that researchers should consider in light of the design of the microbiome study. An increase or decrease in relevant metabolites, such as short-chain fatty acids, can lead to hypothesis generation and pathway analysis of processes associated with microbiota pathology that may lead to disease (Romick-Rosendale et al., 2009). If shotgun sequencing is not performed, metabolite data can be paired with the 16S rRNA sequencing to identify associations between specific taxa and metabolites. The lack of associated metabolic data is a common criticism of 16S rRNA gene amplicon sequencing; thus, metabolomics can be an important tool in studying the processes involved in regulation of the gut microbiota (Ranjan et al., 2016). Likewise, even with the information-rich data from shotgun metagenomic and metatranscriptome sequencing, metabolite analysis provides a definitive link of microbial community structure and activity with microbiome function. The analysis of host metabolic profiles in the context of the host-specific microbiome has important potential in the study of personalized medicine and may allow greater understanding of how the microbiota is involved in human health and disease.
State and of the Science and Future Directions
To date, several nurse scientists have incorporated microbiome research into their research questions. Some areas studied by nurse scientists using microbiomics are feeding method (i.e. breastfed vs. formula fed) in preterm infants (Cong et al., 2017; Cong et al., 2016), irritable bowel syndrome (Fourie et al., 2017; Fourie et al., 2016), and the vaginal microbial community (Brooks et al., 2017). Although this list is not a comprehensive one of all the work done by nurse scientists, it provides exemplars of advancement in the field. Table 3 describes in detail various research designs and methodological techniques in which microbiome analysis can be incorporated to answer research questions relevant to nursing. Although there has been a large amount of progress in the field of microbiome research in the past several years, most work is associational. Associational studies narrow future research questions and are important starting points; however, more mechanistic studies are needed to understand the effect of microorganisms on health and disease. Nurse scientists’ training to view the patient as an interconnected unit can be harnessed to incorporate the microbiota into research questions aimed at improving patient outcomes, reducing symptom burden from chronic disease, and promoting long-term health.
Table 3.
Microbiome Research Performed by Nurse Scientists.
| Citation | Purpose/Objectives | Sample Site | Research Design | Microbiome Methods | Main Findings | Implications/Future Directions | Analysis Pipeline |
|---|---|---|---|---|---|---|---|
| Brooks et al. (2017) | Investigate the stability of microbiome profiles, clustered into CSTs, across different data sets. The investigators also sought to analyze whether CSTs can be used to assess dynamics in the microbiome. | Vaginal microbiome samples | A secondary analysis of five data sets that had two or more longitudinal vaginal samples per subject | Four of the data sets were derived from 16S rRNA amplicon-based surveys, and one was based on whole-metagenome shotgun sequencing. The amplicon-based data sets were generated by 454 pyrosequencing while the metagenomic sequence data were generated by Illumina sequencing. | Healthy subjects tended to persist in a CST profile for an average of 2–3 weeks or more, while subjects with dysbiosis had CST profiles that changed more often (often in response to medication). Changes in CST profiles occurred gradually in some subjects and as quickly as 1 day in others. The presence of Gardnerella vaginalis was a strong predictor of an upcoming CST change. | There is a lack of dense longitudinal measurements of the vaginal microbiome, and further work can be aimed at repeated measures of the vaginal microbiome with detailed accompanying clinical information. This would allow experiments to control for factors that can affect bacterial growth and/or pH, such as hormone levels, clothing, antibiotic/antifungal use, diet, douching practices, and presence of semen. | Statistical modeling of the five data sets was performed in R to calculate associational and predictive values of the CSTs in each data set. |
| Cong et al. (2017) | Explore the effect of feeding types on gut microbial colonization of preterm infants in the neonatal intensive care unit (NICU) and investigate the contribution of different feeding types on the development of gut microbial diversities over the first 30 days of life | Stool sample collected directly from diaper | Secondary analysis of Cong et al. (2016), with additional subjects added. Thirty-three stable preterm infants were followed for 30 days. Infants were classified into six groups based on feeding: mother’s own milk [MOM], human donated milk [HDM], formula, MOM + HDM, MOM + formula, and HDM + formula during days 0–10, 11–20, and 21–30 after birth. | DNA was extracted using the MoBio Power Soil kit (MoBio Laboratories, Inc.). The V4 variable region of the 16S rRNA gene was sequenced | The MOM group had the highest abundance of Clostridiales, Lactobacillales, and Bacillales, and the lowest abundance of Enterobacteriales. Stool samples of the infants in the HDM, formula, and HDM + formula groups had a high abundance of Enterobacteriales at all time points. Alpha diversity (α-diversity) was higher in the MOM group compared to other groups over the three 10-day intervals. Higher α-diversity of the microbial community was associated with older day of life (p < .001), fed MOM (p < .01), and female gender (p < .05). Beta diversity (β-diversity) was evaluated using PERMNOVA, and feeding type explained the greatest variance in the community structure of the factors tested (11%; p < .001). | Future work may explore the different mechanisms by which MOM affects the diversity and composition of the preterm infant’s gut microbiota. Further exploration of the influences of gender and feeding type on microbial community is suggested. Further work is also necessary on testing ways HDM can be preserved to maintain compositional integrity to promote a more diverse gut microbiota. | Using QIIME software, OTUs were determined by clustering reads to the Greengenes 16S reference data set (2013–08 release) with a 97% identity cutoff. Validity of the pipeline was tested using a mock bacterial community. |
| Cong et al. (2016) | Explore day-to-day gut microbiome patterns in preterm infants during their early life in the NICU and investigate the relationship between clinical factors (e.g., infant demographics, mode of delivery, feeding type, antibiotic use, and health conditions) and patterns of infant gut microbial colonization | Stool sample collected directly from diaper | Prospective longitudinal study. Three hundred and seventy-eight fecal samples were analyzed from 29 preterm infants. Factors hypothesized to contribute to OTU diversity in microbiome samples were tested using generalized linear mixed modeling. Gender difference in microbial community was tested. | DNA was extracted using the MoBio Power Soil kit (MoBio Laboratories, Inc.). The V4 region if the 16S rRNA gene was sequenced. | α-Diversity increased from .31 ± .32 at postnatal Day 5 of life to .49 ± .19 at Day 30, with an average increase of .008 per day (p < .001). Factors associated with microbial α-diversity were time (postnatal days), p < .01; feeding type (using MOM or not), p < .01; and gender, p < .05. Male infants began life with low α-diversity but normalized to female values by Day 20–30. Overall mean of α-diversity was higher in females (.58 ± .22) than males (.48 ± .26) during first 30 days of life (p < .05). Feeding type was also found to be a significant influencing factor in microbial diversity in the gut microbiome. | Feeding type in preterm infants may be an important driver in diversity of the gut microbiome. The authors state that future studies with larger sample sizes and a more geographically diverse population would be informative. | Using QIIME software, OTUs were determined by clustering reads to the Greengenes 16S reference data set (2013–08 release) with a 97% identity cutoff. |
| Fourie et al. (2016) | Assess whether the oral microbiome differs between participants with IBS and healthy controls, and explore whether the oral microbiome correlates with variation in symptom severity of visceral sensitivity and pain | Oral buccal mucosal membrane cells were collected using a Pytobrush® (Cooper Surgical, Berlin, Germany) | Patients with IBS (n = 20) and healthy controls (n = 20) were matched on age, gender, race, and weight. Visceral pain was assessed before and after ingestion of an intestinal permeability test sugar solution by the Gastrointestinal Pain Pointer (GIPP), an electronic self-report interface. | Amplification, purification, hybridization, and microarray analysis were completed by second genome. The 16S rRNA gene was sequenced using the 27F/1492 R primer set. Bacterial 16S rRNA gene amplicons were fragmented, biotin-labeled, and hybridized to the PhyloChip™ Array (version G3). | Pain severity was highest in overweight participants with IBS and correlated to the abundance of 60 OTUs, 4 genera, 5 families, and 4 orders of bacteria (r 2 > .4, p < .001). Analysis of β-diversity showed significant OTU separation of the overweight IBS patients from other groups. Having IBS and being overweight was the most significant predictor for the severity of visceral pain and variation in the microbiome. | This study was performed as a pilot for future research that will be aimed at expanding the literature on the oral microbiome in functional GI disorders and symptoms. The investigators plan future research analyzing both fecal and oral samples for further comparisons on if and how the microbiomes vary in IBS. | Data were captured using Affymetrix software (GeneChip® Microarray Analysis Suite). Fluorescent intensity was calculated and hybridization scores (HybScore) were derived. |
| Fourie et al. (2017) | Evaluate structural and functional changes in the colonic microbiome when persistent IBS-like symptoms were induced in rats by a repeated stressor (visceral hypersensitivity, increased colonic permeability, and increased fecal pellet output) | Distal colonic mucosa epithelial cells | 13 adult Sprague Dawley rats (200–220 g) were exposed to WA stress for 1 hr each day for 10 consecutive days. These were compared to a control, sham-stress arm (n = 13). Animals were sacrificed at the end of the protocol and 10 mm of distal colon was removed. | DNA was extracted using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) and the V4 region of the 16S rRNA gene was sequenced using primers F515/R806. | WA-stress animals exhibited higher α-diversity and moderately differed in community structure (β-diversity) compared with controls. The WA microbiome was enriched in proteobacteria and depleted in several beneficial taxa. Decreased energy and lipid metabolism and an increased capacity for fatty acid and sulfur metabolism were inferred for the WA microbiome. | Future experiments may target sulfur metabolites and sulfur-metabolizing bacteria because increased capacity for sulfur metabolism was inferred. These targets may successfully modulate sulfur production to prevent or alleviate symptoms and may be studied as future targets of intervention in patients with IBS. Additionally, restoration of beneficial microbes or functional domains (such as fatty acid metabolism dynamics) could be studied to translate these findings back to human patients with IBS. | SecondGenome was contracted to perform data processing. Using mothur’s Bayesian classifier, OTUs were determined by clustering reads to the Greengenes 16S reference data set with a 99% identity cutoff. |
Note. CSTs = community state types; GI = gastrointestinal; IBS = irritable bowel syndrome; miRNA = micro RNA; NICU = neonatal intensive care unit; OTUs = operational taxonomic units; QIIME = Quantitative Insights Into Microbial Ecology software; rRNA = ribosomal RNA; WA = water avoidance.
Conclusion
Nurse scientists have a unique lens based on their patient-focused training and are well equipped to study the complex relationships among the human genome, the genomes of the microbiota, the multitude of environmental forces that influence health, and disease. Ensuring that they have the tools and resources they need when incorporating microbiome methods into a study is vital to moving microbiomics research forward. Identifying important covariates, meticulously designing the study from research team to sample storage, and selecting the microbiome sequencing method that is most appropriate for the research question will support a scientifically sound microbiome study.
Acknowledgments
The authors would like to thank Drs. Joan Austin and Ann Cashion for their comments on this review and Kevin Grandfield, publication manager for the UIC Department of Biobehavioral Health Science, for editorial assistance.
Authors’ Note: The opinions expressed herein, and the interpretation and reporting of these data, are the responsibility of the author(s) and should not be seen as an official recommendation or interpretation of the National Institutes of Health.
Author Contributions: Katherine A. Maki, Ana F. Diallo, Mark B. Lockwood, Alexis T. Franks, Stefan J. Green, and Paule V. Joseph contributed equally in the conception, design and acquisition of data, drafting of the manuscript, and critical revision of the manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) declared the following financial support for the research, authorship, and/or publication of this article: Support provided by the National Institute of Nursing Research (to PVJ, and Intramural Research Training Award to ATF); Office of Workforce Diversity, National Institutes of Health (NIH) to PVJ; Sigma Theta Tau International, Alpha Lambda Chamber to KAM; and University of Illinois at Chicago, College of Nursing, to KAM.
ORCID iD: Alexis T. Franks  https://orcid.org/0000-0002-0550-334X
https://orcid.org/0000-0002-0550-334X
Stefan J. Green  https://orcid.org/0000-0003-2781-359X
https://orcid.org/0000-0003-2781-359X
Paule V. Joseph  https://orcid.org/0000-0002-1198-9622
https://orcid.org/0000-0002-1198-9622
References
- Albertsen M., Hugenholtz P., Skarshewski A., Nielsen K. L., Tyson G. W., Nielsen P. H. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nature Biotechnology, 31, 533–538. doi:10.1038/nbt.2579 [DOI] [PubMed] [Google Scholar]
- Amann R. I., Ludwig W., Schleifer K. H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews, 59, 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F., Nombela C. (2012). The microbiome as a human organ. Clinical Microbiology and Infection, 18, 2–4. doi:10.1111/j.1469-0691.2012.03916.x [DOI] [PubMed] [Google Scholar]
- Bartman C., Chong A. S., Alegre M. L. (2015). The influence of the microbiota on the immune response to transplantation. Current Opinion in Organ Transplantation, 20, 1–7. doi:10.1097/mot.0000000000000150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiardes S., Zilberman-Schapira G., Elinav E. (2016). Use of metatranscriptomics in microbiome research. Bioinformatics and Biology Insights, 10, 19–25. doi:10.4137/bbi.S34610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi E., Piovesan A., Facchin F., Beraudi A., Casadei R., Frabetti F.…Canaider S. (2013). An estimation of the number of cells in the human body. Annals of Human Biology, 40, 463–471. doi:10.3109/03014460.2013.807878 [DOI] [PubMed] [Google Scholar]
- Birtel J., Walser J. C., Pichon S., Burgmann H., Matthews B. (2015). Estimating bacterial diversity for ecological studies: Methods, metrics, and assumptions. PLoS One, 10, e0125356 doi:10.1371/journal.pone.0125356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N. A., Chung J., Battaglia T., Henderson N., Jay M., Li H.…Blaser M. J. (2016). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Science Translational Medicine, 8, 343ra82 doi:10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. P., Buck G. A., Chen G., Diao L., Edwards D. J., Fettweis J. M.…Zhou Y. H. (2017). Changes in vaginal community state types reflect major shifts in the microbiome. Microbial Ecology in Health and Disease, 28, 1303265 doi:10.1080/16512235.2017.1303265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. P., Edwards D. J., Harwich M. D., Jr, Rivera M. C., Fettweis J. M., Serrano M. G.…Buck G. A. (2015). The truth about metagenomics: Quantifying and counteracting bias in 16 S rRNA studies. BMC Microbiology, 15, 66 doi:10.1186/s12866-015-0351-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. (1978). Complete nucleotide sequence of a 16 S ribosomal RNA gene from Escherichia coli. Proceedings of the National Academy of Sciences of the USA, 75, 4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. T., Davis-Richardson A. G., Giongo A., Gano K. A., Crabb D. B., Mukherjee N.…Triplett E. W. (2011). Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for Type 1 diabetes. PLoS One, 6, e25792 doi:10.1371/journal.pone.0025792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nature Methods, 12, 59–60. doi:10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Burkitt D. P., Walker A. R., Painter N. S. (1972). Effect of dietary fibre on stools and the transit-times, and its role in the causation of disease. Lancet, 2, 1408–1412. [DOI] [PubMed] [Google Scholar]
- Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13, 581–583. doi:10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K.…Gordon J. I. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bittinger K., Charlson E. S., Hoffmann C., Lewis J., Wu G. D.…Li H. (2012). Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics, 28, 2106–2113. doi:10.1093/bioinformatics/bts342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo J. M., Leong L. E., Rogers G. B. (2015). Sample storage conditions significantly influence faecal microbiome profiles. Scientific Reports, 5, 16350 doi:10.1038/srep16350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G., Stilling R. M., Kennedy P. J., Stanton C., Cryan J. F., Dinan T. G. (2014). Minireview: Gut microbiota: The neglected endocrine organ. Molecular Endocrinology, 28, 1221–1238. doi:10.1210/me.2014-1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X., Judge M., Xu W., Diallo A., Janton S., Brownell E. A.…Graf J. (2017). Influence of feeding type on gut microbiome development in hospitalized preterm infants. Nursing Research, 66, 123–133. doi:10.1097/nnr.0000000000000208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X., Xu W., Janton S., Henderson W. A., Matson A., McGrath J. M.…Graf J. (2016). Gut microbiome developmental patterns in early life of preterm infants: Impacts of feeding and gender. PLoS One, 11, e0152751 doi:10.1371/journal.pone.0152751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon M. A., Bird A. R. (2014). The impact of diet and lifestyle on gut microbiota and human health. Nutrients, 7, 17–44. doi:10.3390/nu7010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresci G. A., Bawden E. (2015). Gut microbiome: What we do and don’t know. Nutrition in Clinical Practice, 30, 734–746. doi:10.1177/0884533615609899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon N., Drewnowski A. (2015). Contribution of food prices and diet cost to socioeconomic disparities in diet quality and health: A systematic review and analysis. Nutrition Reviews, 73, 643–660. doi:10.1093/nutrit/nuv027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E.…Turnbaugh P. J. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505, 559–563. doi:10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deda O., Gika H. G., Theodoridis G. A. (2018). Rat fecal metabolomics-based analysis In Theodoridis G., Gika H., W. I (Eds.), Metabolic profiling: Methods in molecular biology (Vol. 1738). New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- Del Chierico F., Ancora M., Marcacci M., Camma C., Putignani L., Conti S. (2015). Choice of next-generation sequencing pipelines. Methods in Molecular Biology, 1231, 31–47. doi:10.1007/978-1-4939-1720-4_3 [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K.…Andersen G. L. (2006). Greengenes, a chimera-checked 16 S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72, 5069–5072. doi:10.1128/aem.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer K., Aronov P. A., Hammock B. D. (2007). Mass spectrometry-based metabolomics. Mass Spectrometry Reviews, 26, 51–78. doi:10.1002/mas.20108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella J. M., Bao Y., Gloor G. B., Burton J. P., Reid G. (2013). High throughput sequencing methods and analysis for microbiome research. Journal of Microbiological Methods, 95, 401–414. doi:10.1016/j.mimet.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Dinan T. G., Cryan J. F. (2017). Microbes, immunity, and behavior: Psychoneuroimmunology meets the microbiome. Neuropsychopharmacology, 42, 178–192. doi:10.1038/npp.2016.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2018). Updating the 97% identity threshold for 16 S ribosomal RNA OTUs. Bioinformatics, 34, 2371–2375. doi:10.1093/bioinformatics/bty113 [DOI] [PubMed] [Google Scholar]
- Edwards S. M., Cunningham S. A., Dunlop A. L., Corwin E. J. (2017). The maternal gut microbiome during pregnancy. MCN: The American Journal of Maternal/Child Nursing, 42, 310–317. doi:10.1097/nmc.0000000000000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K., Lahti L., Gonze D., de Vos W. M., Raes J. (2015). Metagenomics meets time series analysis: Unraveling microbial community dynamics. Current Opinion in Microbiology, 25, 56–66. doi:10.1016/j.mib.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Field D., Garrity G., Gray T., Morrison N., Selengut J., Sterk P.…Wipat A. (2008). The minimum information about a genome sequence (MIGS) specification. Nature Biotechnology, 26, 541–547. doi:10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourie N. H., Wang D., Abey S. K., Creekmore A. L., Hong S., Martin C. G.…Henderson W. A. (2017). Structural and functional alterations in the colonic microbiome of the rat in a model of stress induced irritable bowel syndrome. Gut Microbes, 8, 33–45. doi:10.1080/19490976.2016.1273999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourie N. H., Wang D., Abey S. K., Sherwin L. B., Joseph P. V., Rahim-Williams B.…Henderson W. A. (2016). The microbiome of the oral mucosa in irritable bowel syndrome. Gut Microbes, 7, 286–301. doi:10.1080/19490976.2016.1162363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A.…Woese C. R. (1980). The phylogeny of prokaryotes. Science, 209, 457–463. [DOI] [PubMed] [Google Scholar]
- Frank J. A., Pan Y., Tooming-Klunderud A., Eijsink V. G., McHardy A. C., Nederbragt A. J., Pope P. B. (2016). Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Scientific Reports, 6, 25373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Blaser M. J., Caporaso J. G., Jansson J. K., Lynch S. V., Knight R. (2018). Current understanding of the human microbiome. Nature Medicine, 24, 392–400. doi:10.1038/nm.4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Quinn R. A., Debelius J., Xu Z. Z., Morton J., Garg N.…Knight R. (2016). Microbiome-wide association studies link dynamic microbial consortia to disease. Nature, 535, 94–103. doi:10.1038/nature18850 [DOI] [PubMed] [Google Scholar]
- Gohl D. M., Vangay P., Garbe J., MacLean A., Hauge A., Becker A.…Beckman K. B. (2016). Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nature Biotechnology, 34, 942–949. doi:10.1038/nbt.3601 [DOI] [PubMed] [Google Scholar]
- Goodman A. L., Gordon J. I. (2010). Our unindicted coconspirators: Human metabolism from a microbial perspective. Cell Metabolism, 12, 111–116. doi:10.1016/j.cmet.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. K., Di Rienzi S. C., Poole A. C., Koren O., Walters W. A., Caporaso J. G.…Ley R. E. (2014). Conducting a microbiome study. Cell, 158, 250–262. doi:10.1016/j.cell.2014.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda G. A., Djukovic D. (2014). Overview of mass spectrometry-based metabolomics: Opportunities and challenges. Methods in Molecular Biology, 1198, 3–12. doi:10.1007/978-1-4939-1258-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S. J., Venkatramanan R., Naqib A. (2015). Deconstructing the polymerase chain reaction: Understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS One, 10, e0128122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin N. W., Ahern P. P., Cheng J., Heath A. C., Ilkayeva O., Newgard C. B.…Gordon J. I. (2017). Prior dietary practices and connections to a human gut microbial metacommunity alter responses to diet interventions. Cell Host & Microbe, 21, 84–96. doi:10.1016/j.chom.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale V. L., Jeraldo P., Mundy M., Yao J., Keeney G., Scott N.…Chia N. (2018). Synthesis of multi-omic data and community metabolic models reveals insights into the role of hydrogen sulfide in colon cancer. Methods, 149, 59–68. doi:10.1016/j.ymeth.2018.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbling D. E., Ackermann M., Fenner K., Kohler H. P., Johnson D. R. (2012). The activity level of a microbial community function can be predicted from its metatranscriptome. The ISME Journal, 6, 902–904. doi:10.1038/ismej.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy Y. E., Bik E. M., Lawley T. D., Holmes S. P., Monack D. M., Theriot J. A., Relman D. A. (2015). Variation in taxonomic composition of the fecal microbiota in an inbred mouse strain across individuals and time. PLoS One, 10, e0142825 doi:10.1371/journal.pone.0142825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Albrecht B., Bağcı C., Bessarab I., Górska A., Jolic D., Williams R. B. (2018). MEGAN-LR: New algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biology Direct, 13, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H., Beier S., Flade I., Gorska A., El-Hadidi M., Mitra S.…Tappu R. (2016). MEGAN community edition—Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Computational Biology, 12, e1004957 doi:10.1371/journal.pcbi.1004957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C., Gevers D., Knight R., Abubucker S., Badger J. H., Chinwalla A. T.…Giglio M. G. (2012). Structure, function and diversity of the healthy human microbiome. Nature, 486, 207–214. doi:10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iebba V., Nicoletti M., Schippa S. (2012). Gut microbiota and the immune system: An intimate partnership in health and disease. International Journal of Immunopathology and Pharmacology, 25, 823–833. doi:10.1177/039463201202500401 [DOI] [PubMed] [Google Scholar]
- Ionescu D., Overholt W. A., Lynch M. D. J., Neufeld J. D., Naqib A., Green S. (2016). Microbial community analysis using high-throughput amplicon sequencing. In Yates M. V., Nakatsu C. H., Miller R. V., Pillai S. D. (Eds.), Manual of environmental microbiology (4th ed, pp. 2.4.2-1–2.4.2-26). Washington, DC: ASM Press. [Google Scholar]
- Janda J. M., Abbott S. L. (2007). 16 S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. Journal of Clinical Microbiology, 45, 2761–2764. doi:10.1128/jcm.01228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S., Baker B., Dunn A., Edwards S., Ferranti E., Mutic A. D.…Rodriguez J. (2017). Maternal-child microbiome: Specimen collection, storage, and implications for research and practice. Nursing Research, 66, 175–183. doi:10.1097/nnr.0000000000000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. (2007). Partitioning diversity into independent alpha and beta components. Ecology, 88, 2427–2439. [DOI] [PubMed] [Google Scholar]
- Karlsson F. H., Nookaew I., Nielsen J. (2014). Metagenomic data utilization and analysis (MEDUSA) and construction of a global gut microbial gene catalogue. PLoS Computational Biology, 10, e1003706 doi:10.1371/journal.pcbi.1003706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau A. L., Ahern P. P., Griffin N. W., Goodman A. L., Gordon J. I. (2011). Human nutrition, the gut microbiome and the immune system. Nature, 474, 327–336. doi:10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B. J., Gross R., Bittinger K., Sherrill-Mix S., Lewis J. D., Collman R. G.…Li H. (2015). Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics, 31, 2461–2468. doi:10.1093/bioinformatics/btv183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzian A., Green S. J., Engen P. A., Voigt R. M., Naqib A., Forsyth C. B.…Shannon K. M. (2015). Colonic bacterial composition in Parkinson’s disease. Movement Disorders, 30, 1351–1360. doi:10.1002/mds.26307 [DOI] [PubMed] [Google Scholar]
- Khanna S., Tosh P. K. (2014). A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clinic Proceedings, 89, 107–114. doi:10.1016/j.mayocp.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Kim D., Hofstaedter C. E., Zhao C., Mattei L., Tanes C., Clarke E.…Bittinger K. (2017). Optimizing methods and dodging pitfalls in microbiome research. Microbiome, 5, 52 doi:10.1186/s40168-017-0267-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J., Lauber C. L., Walters W. A., Parfrey L. W., Clemente J. C., Gevers D., Knight R. (2011). Experimental and analytical tools for studying the human microbiome. Nature Reviews Genetics, 13, 47–58. doi:10.1038/nrg3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C. G., Canback B., Berg O. G. (2003). Horizontal gene transfer: A critical view. Proceedings of the National Academy of Sciences of the USA, 100, 9658–9662. doi:10.1073/pnas.1632870100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa P. S., Brooks J. P., Deych E., Boone E. L., Edwards D. J., Wang Q.…Shannon W. D. (2012). Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS One, 7, e52078 doi:10.1371/journal.pone.0052078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon A., Crook N., Dantas G. (2016). The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Medicine, 8, 39 doi:10.1186/s13073-016-0294-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A.…Huttenhower C. (2013). Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nature Biotechnology, 31, 814–821. doi:10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees H., Swann J., Poucher S. M., Nicholson J. K., Holmes E., Wilson I. D., Marchesi J. R. (2014). Age and microenvironment outweigh genetic influence on the Zucker rat microbiome. PLoS One, 9, e100916 doi:10.1371/journal.pone.0100916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Bushman F. D., FitzGerald G. A. (2015). Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proceedings of the National Academy of Sciences of the U S A, 112, 10479–10484. doi:10.1073/pnas.1501305112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon J. C., Nicholson J. K., Holmes E., Everett J. R. (2000). Metabonomics: Metabolic processes studied by NMR spectroscopy of biofluids. Concepts in Magnetic Resonance, 12, 289–320. doi:10.1002/1099-0534(2000)12:5<289::AID-CMR3>3.0.CO;2-W [Google Scholar]
- Liu X., Locasale J. W. (2017). Metabolomics: A primer. Trends Biochem Sci, 42, 274–284. doi:10.1016/j.tibs.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey T. D. (1972). Introduction to intestinal microecology. American Journal of Clinical Nutrition, 25, 1292–1294. doi:10.1093/ajcn/25.12.1292 [DOI] [PubMed] [Google Scholar]
- Luo C., Tsementzi D., Kyrpides N. C., Konstantinidis K. T. (2012). Individual genome assembly from complex community short-read metagenomic datasets. ISME Journal, 6, 898–901. doi:10.1038/ismej.2011.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord A. I., Chapman C. A., Weny G., Tumukunde A., Hyeroba D., Klotz K.…Goldberg T. L. (2014). Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation. American Journal of Primatology, 76, 347–354. doi:10.1002/ajp.22238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M., Csizmadi I., Friedenreich C. M., Uribe F. A., Nettel-Aguirre A., McLaren L.…McCormack G. R. (2016). Associations between the neighbourhood food environment, neighbourhood socioeconomic status, and diet quality: An observational study. BMC Public Health, 16, 984 doi:10.1186/s12889-016-3631-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes P., Cutting M. (2010). Human Microbiome Project—Core microbiome sampling protocol A. National Institutes of Health Human Microbiome Project Initiative, 12.0 Retrieved from https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000228.v4.p1 [Google Scholar]
- Meyer F., Paarmann D., D’Souza M., Olson R., Glass E. M., Kubal M.…Edwards R. A. (2008). The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics, 9, 386 doi:10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan X. C., Huttenhower C. (2012). Chapter 12: Human microbiome analysis. PLoS Computational Biology, 8, e1002808 doi:10.1371/journal.pcbi.1002808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols D., Cahoon N., Trakhtenberg E. M., Pham L., Mehta A., Belanger A.…Epstein S. S. (2010). Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Applied and Environmental Microbiology, 76, 2445–2450. doi:10.1128/aem.01754-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti G. J., Yanes O., Siuzdak G. (2012). Innovation: Metabolomics: The apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13, 263–269. doi:10.1038/nrm3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., Glockner F. O. (2007). SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research, 35, 7188–7196. doi:10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C.…Wang J. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464, 59–65. doi:10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Rani A., Metwally A., McGee H. S., Perkins D. L. (2016). Analysis of the microbiome: Advantages of whole genome shotgun versus 16 S amplicon sequencing. Biochemical and Biophysical Research Communications, 469, 967–977. doi:10.1016/j.bbrc.2015.12.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lopez M. A., Mendez-Tenorio A., Maldonado-Rodriguez R., Doktycz M. J., Fleming J. T., Beattie K. L. (2003). Fingerprinting of prokaryotic 16 S rRNA genes using oligodeoxyribonucleotide microarrays and virtual hybridization. Nucleic Acids Research, 31, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Rodrigues I., Jimenez-Garcia R., Hernandez-Barrera V., Carrasco-Garrido P., Jimenez-Trujillo I., Lopez-de-Andres A. (2015). Adherence to and predictors of participation in colorectal cancer screening with faecal occult blood testing in Spain, 2009–2011. European Journal of Cancer Prevention, 24, 305–312. doi:10.1097/cej.0000000000000088 [DOI] [PubMed] [Google Scholar]
- Romick-Rosendale L. E., Goodpaster A. M., Hanwright P. J., Patel N. B., Wheeler E. T., Chona D. L., Kennedy M. A. (2009). NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril). Magnetic Resonance in Chemistry, 47, S36–S46. doi:10.1002/mrc.2511 [DOI] [PubMed] [Google Scholar]
- Rossello-Mora R., Amann R. (2001). The species concept for prokaryotes. FEMS Microbiological Reviews, 25, 39–67. [DOI] [PubMed] [Google Scholar]
- Sampson T. R., Debelius J. W., Thron T., Janssen S., Shastri G. G., Ilhan Z. E.…Mazmanian S. K. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell, 167, 1469–1480. e1412 doi:10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarode N., Parris D. J., Ganesh S., Seston S. L., Stewart F. J. (2016). Generation and analysis of microbial metatranscriptomes In Marylynn V. Yates, Cindy H. Nakatsu, Robert V. Miller, Suresh D. Pillai. (Eds.), Manual of environmental microbiology (4th ed; pp. 2.4.5-1–2.4.5-19). Washington, DC: ASM Press. [Google Scholar]
- Savage D. C. (1977). Microbial ecology of the gastrointestinal tract. Annual Review of Microbiology, 31, 107–133. doi:10.1146/annurev.mi.31.100177.000543 [DOI] [PubMed] [Google Scholar]
- Schirmer M., Smeekens S. P., Vlamakis H., Jaeger M., Oosting M., Franzosa E. A.…Xavier R. J. (2016). Linking the human gut microbiome to inflammatory cytokine production capacity. Cell, 167, 1125,–1136. e1128 doi:10.1016/j.cell.2016.10.020 [DOI] [PubMed] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B.…Weber C. F. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied Environmental Microbiology, 75, 7537–7541. doi:10.1128/aem.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. M., DeLong E. F., Pace N. R. (1991). Analysis of a marine picoplankton community by 16 S rRNA gene cloning and sequencing. Journal of Bacteriology, 173, 4371–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods, 9, 811–814. doi:10.1038/nmeth.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biology, 14, e1002533 doi:10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant M. J., Constantinidou C., Cogan T., Penn C. W., Pallen M. J. (2012). High-throughput sequencing of 16 S rRNA gene amplicons: Effects of extraction procedure, primer length and annealing temperature. PLoS One, 7, e38094 doi:10.1371/journal.pone.0038094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton T. J. (2014). An introduction to the analysis of shotgun metagenomic data. Frontiers in Plant Science, 5, 209 doi:10.3389/fpls.2014.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber C. M. K., Probst A. J., Sharrar A., Thomas B. C., Hess M., Tringe S. G., Banfield J. F. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nature Microbiology, 3, 836–843. doi:10.1038/s41564-018-0171 -1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J. L., Backhed F. (2016). Diet–microbiota interactions as moderators of human metabolism. Nature, 535, 56–64. doi:10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Goebel B. (1994). Taxonomic note: A place for DNA–DNA reassociation and 16 S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic and Evolutionary Microbiology, 44, 846–849. [Google Scholar]
- Staley J. T., Konopka A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annual Review of Microbiology, 39, 321–346. doi:10.1146/annurev.mi.39.100185.001541 [DOI] [PubMed] [Google Scholar]
- Stamper C. E., Hoisington A. J., Gomez O. M., Halweg-Edwards A. L., Smith D. G., Bates K. L.…Lowry C. A. (2016). The microbiome of the built environment and human behavior: Implications for emotional health and well-being in postmodern Western societies. International Review of Neurobiology, 131, 289–323. doi:10.1016/bs.irn.2016.07.006 [DOI] [PubMed] [Google Scholar]
- Stewart F. J., Ottesen E. A., DeLong E. F. (2010). Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME Journal, 4, 896–907. doi:10.1038/ismej.2010.18 [DOI] [PubMed] [Google Scholar]
- Tamboli C. P., Neut C., Desreumaux P., Colombel J. F. (2004). Dysbiosis in inflammatory bowel disease. Gut, 53, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tankou S. K., Regev K., Healy B. C., Tjon E., Laghi L., Cox L. M.…Weiner H. L. (2018). A probiotic modulates the microbiome and immunity in multiple sclerosis. Annals of Neurology, 83, 1147–1161. doi:10.1002/ana.25244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Gilbert J., Meyer F. (2012). Metagenomics—A guide from sampling to data analysis. Microbial Informatics and Experimentation, 2, 3 doi:10.1186/2042-5783-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe S., Afshinnekoo E., Rock T. M., McGrath K., Alexander N., McIntyre A.…Mason C. E. (2017). Genomic methods and microbiological technologies for profiling novel and extreme environments for the extreme microbiome project (XMP). Journal of Biomolecular Techniques, 28, 31–39. doi:10.7171/jbt.17-2801-004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell L. K., Metcalf J. L., Parfrey L. W., Knight R. (2012). Defining the human microbiome. Nutrition Reviews, 70, S38–S44. doi:10.1111/j.1753-4887.2012.00493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N. M., Kerby R. L., Dill-McFarland K. A., Harding S. J., Merluzzi A. P., Johnson S. C.…Rey F. E. (2017). Gut microbiome alterations in Alzheimer’s disease. Scientific Reports, 7, 13537 doi:10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtmann E., Chen J., Amir A., Shi J., Abnet C. C., Nelson H.…Sinha R. (2017). Comparison of collection methods for fecal samples in microbiome studies. American Journal of Epidemiology, 185, 115–123. doi:10.1093/aje/kww177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke F., Hess M. (2009). A perspective: Metatranscriptomics as a tool for the discovery of novel biocatalysts. Journal of Biotechnology, 142, 91–95. doi:10.1016/j.jbiotec.2009.03.022 [DOI] [PubMed] [Google Scholar]
- Weber P. C., Ohlendorf D. H., Wendoloski J. J., Salemme F. R. (1989). Structural origins of high-affinity biotin binding to streptavidin. Science, 243, 85–88. [DOI] [PubMed] [Google Scholar]
- Wei B., Wingender G., Fujiwara D., Chen D. Y., McPherson M., Brewer S.…Braun J. (2010). Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. Journal of Immunology, 184, 1218–1226. doi:10.4049/jimmunol.0902620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. (1977). Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proceedings of the National Academy of Sciences of the USA, 74, 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R. L., Zybert P., Brouse C. H., Neugut A. I., Shea S., Gibson G.…Basch C. E. (2001). Knowledge, beliefs, and barriers relevant to colorectal cancer screening in an urban population: A pilot study. Family & Community Health, 24, 34–47. [DOI] [PubMed] [Google Scholar]
- Xiong X., Frank D. N., Robertson C. E., Hung S. S., Markle J., Canty A. J.…Parkinson J. (2012). Generation and analysis of a mouse intestinal metatranscriptome through Illumina based RNA-sequencing. PLoS One, 7, e36009 doi:10.1371/journal.pone.0036009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Gao X. J., Li T., Wei B., Wang P., Yang Y., Yan R. (2018). Fecal microbiota transplantation in experimental ulcerative colitis reveals associated gut microbial and host metabolic reprogramming. Applied and Environmental Microbiology, 84, 1–11. doi:10.1128/aem.00434-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Wang Y., Qian P. Y. (2016). Sensitivity and correlation of hypervariable regions in 16 S rRNA genes in phylogenetic analysis. BMC Bioinformatics, 17, 135 doi:10.1186/s12859-016-0992-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Zubcevic J. (2017). Gut–brain axis in regulation of blood pressure. Frontiers in Physiology, 8, 845 doi:10.3389/fphys.2017.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler D. R. (2003). Gene sequences useful for predicting relatedness of whole genomes in bacteria. International Journal of Systemic and Evolutionary Microbiology, 53, 1893–1900. doi:10.1099/ijs.0.02713-0 [DOI] [PubMed] [Google Scholar]
- Zoetendal E. G., de Vos W. M. (2014). Effect of diet on the intestinal microbiota and its activity. Current Opinions in Gastroenterology, 30, 189–195. doi:10.1097/mog.0000000000000048 [DOI] [PubMed] [Google Scholar]