Abstract
Fatty acid binding protein 4 (FABP4) is a leaderless lipid carrier protein primarily expressed by adipocytes and macrophages that not only functions intracellularly but is also secreted. The secretion is mediated via unconventional mechanism(s), and in a variety of species, metabolic dysfunction is correlated with elevated circulating FABP4 levels. In diabetic animals, neutralizing antibodies targeting serum FABP4 increase insulin sensitivity and attenuate hepatic glucose output, suggesting the functional importance of circulating FABP4. Using animal and cell-based models, we show that FABP4 is secreted from white, but not brown, adipose tissue in response to lipolytic stimulation in a sirtuin-1 (SIRT1)–dependent manner via a mechanism that requires some, but not all, autophagic components. Silencing of early autophagic genes such as Ulk1/2, Fip200, or Beclin-1 or chemical inhibition of ULK1/2 or VPS34 attenuated secretion, while Atg5 knockdown potentiated FABP4 release. Genetic knockout of Sirt1 diminished secretion, and serum FABP4 levels were undetectable in Sirt1 knockout mice. In addition, blocking SIRT1 by EX527 attenuated secretion while activating SIRT1 by resveratrol-potentiated secretion. These studies suggest that FABP4 secretion from adipocytes is regulated by SIRT1 and requires early autophagic components.
Introduction
Obesity-induced metabolic disease is linked to a chronic low-grade inflammatory state associated with adipose insulin resistance and altered adipokine release (1,2). Concomitant with insulin resistance is increased adipocyte lipolysis, leading to elevated circulating fatty acids and, ultimately, triglycerides. Fatty acid binding proteins (FABPs) belong to a supergene family of lipid-binding proteins that function in fatty acid storage, transport, and lipid signaling (3–5). Adipocytes express FABP4 (major form) and FABP5 (minor form), which serve to facilitate lipolysis and free fatty acid (FFA) release from the cell in response to adrenergic signaling (3). FABP4 forms a physical complex with both hormone-sensitive lipase and CGI-58 to facilitate lipid droplet hydrolysis, and in mouse models, deletion of FABP4 reduces fat cell lipolysis ∼70% (6).
Fabp4 null mice are protected from insulin resistance, asthma, atherosclerosis, and inflammation (7–9), and metabolic dysfunction linked to FABP4 has largely been attributed to its lipid shuttle activity in adipocytes and its ability to control macrophage polarization, endoplasmic reticulum stress, and inflammation (9,10). However, recent literature has shown FABP4 is also secreted from adipocytes and, to a much lesser extent, macrophages (11). Serum FABP4 levels are elevated in patients with obesity and metabolic syndrome (12) and implicated in disease progression in some cancers (13). Circulating FABP4 has been shown to regulate production of hepatic glucose (14) and release of insulin from pancreatic β-cells (15). As a result, circulating FABP4 is currently being evaluated as a potential clinical biomarker for metabolic and cardiovascular diseases (16–18).
FABP4 lacks the classical secretory signal sequence and is shown to be released by fat cells via an unconventional protein secretion (UPS) mechanism. FABP4 is secreted unconventionally in response to lipolytic stimuli, increased intracellular Ca2+ levels, or hypoxia (11,14,15,19,20) and is independent of apoptosis (21). Recently, Villeneuve et al. (22) demonstrated in 3T3-L1 adipocytes that a very small fraction of FABP4 secretion was mediated by multivesicular bodies/exosomes and that fat cells secrete FABP4 via an endosomal mechanism that requires secretory lysosomes, but does not require the autophagic protein ATG5. In this study, we provide additional insight into FABP4 secretion, implicating additional autophagic proteins and the deacetylase SIRT1 as major regulators of regulated FABP4 release from fat cells.
Research Design and Methods
Reagents and Chemicals
Forskolin (FSK), ATGListatin (ASTAT), TNF-α, 8-Br-cAMP, EX527, resveratrol, isoproterenol, 3-methyl adenine (3MA), N-acetyl d-sphingosine (NAS), puromycin, (−)-N6-(2-phenylisopropyl)-adenosine (PIA), adenosine deaminase, and insulin (I5500) were obtained from Sigma-Aldrich (St. Louis, MO). TNF-α was purchased from R&D Systems (Minneapolis, MN) and troglitazone from Cayman Chemical. PIK-III (VPS34 inhibitor) was obtained as a gift from the Novartis Institutes for BioMedical Research (Cambridge, MA), and the ULK 1/2 inhibitor MRT68921 was purchased from APExBIO (Houston, TX).
Cell Culture and Cell Lines
3T3-L1 fibroblasts were grown to confluence in six-well plates and differentiated to mature adipocytes as described previously (23). Differentiated adipocytes were maintained in DMEM (Invitrogen, Carlsbad, CA) with 10% FBS and used for experimentation from days 10 to 12. Atg5 and Fip200 knockdown 3T3-L1 cells were made as described (24) and maintained in 2 μg/mL puromycin. To obtain SirT1-silenced 3T3-L1 cells, preadipocytes were transduced with an shRNA lentivirus and selected with puromycin as described previously (25). The shRNA clones directed toward Sirt-1 were TRCN0000039294 and TRCN0000039296 from the University of Minnesota Genomic Center with sequences 5′-CCGGGCCATGTTTGATATTGAGTATCTCG AGATACTCAATATCAAACATGGCTTTG-3′ and 5′-CCGGGAGGGTAATCAATACCTGTTTCTCGAGAAACAGGTATTGATTACCCTCTTTTTG-3′, respectively.
Murine OP9 stromal cells were purchased from American Type Culture Collection (CRL-2749; Manassas, VA). OP9 cells were propagated in minimum essential medium-α (MEM-α) with 20% FBS. Upon confluence, the cells were differentiated with MEM-α supplemented with 15% Knockout Serum Replacement (10828-028; Invitrogen) for 4 days. Subsequently, the cells were further cultured in MEM-α with 20% FBS for 5–7 additional days (26).
Immortalized mouse embryonic fibroblasts (MEFs) were grown in DMEM containing 0.1% penicillin, 0.1% streptomycin, and 10% FBS. Cells were grown to confluence and differentiated 2 days postconfluence with media containing insulin, dexamethasone, 3-isobutyl-1-methylxanthine, and troglitazone as previously described (23).
Primary Fat Cells
Primary adipocytes were isolated from C57BL/6J mice maintained on a high-fat diet for 12 weeks (3282; Bio-Serv, Flemington, NJ). Epididymal and inguinal white adipose tissues (eWATs and iWATs, respectively) were dissected, minced, and digested with type I collagenase (1 mg/mL CLS-1; Worthington Biochemical Corporation) for 1 h at 37°C in Krebs–Ringer–HEPES (KRH; 118 mmol/L NaCl, 4.75 mmol/L KCl, 1.2 mmol/L KH2PO4, 2.44 mmol/L MgSO4, 25 mmol/L NaHCO3, 25 mmol/L HEPES, and 2.5 mmol/L CaCl2, pH 7.4) buffer supplemented with 0.5% fatty acid–free BSA and 5 mmol/L glucose. Adipocytes were filtered through a 100-μm membrane followed by three washes (centrifuged at 4,000g for 10 min) with BSA-supplemented KRH buffer. The floating adipocytes were recovered and treated with 1 µg/mL bovine insulin with 100 nmol/L PIA and 1 unit/mL adenosine deaminase or 10 μmol/L FSK plus 1 unit/mL adenosine deaminase (27). The cells were incubated in KRH with 0.5% fatty acid–free BSA with insulin or FSK for 2 h at 37°C with gentle shaking (100 rpm), followed by centrifugation at 4,000g for 10 min, allowing separation of the supernatant from the cells.
Adipose Tissue Explants
eWAT, iWAT, perigonadal WAT (pWAT), and brown adipose tissue (BAT) depots were collected from C57BL/6J mice maintained on a chow diet at 15 weeks of age. A total of 25–30 mg of tissue was washed thoroughly with KRH buffer and subsequently minced, washed in KRH, and incubated with KRH with 0.5% fatty acid–free BSA supplemented with insulin or FSK for 4 h. The KRH media was collected and centrifuged at 10,000g to remove cell debris.
Analysis of Protein Secretion
Differentiated adipocytes were washed with 37°C PBS and incubated in secretion medium (KRH containing 5 mmol/L glucose plus 0.5% fatty acid–free BSA) for up to 4 h with vehicle (VEH) or 20 μmol/L FSK. For chemical inhibitor treatments, the cells were preincubated with these inhibitors for 2 h in DMEM with 2% FBS prior to induction of secretion and during the process of secretion. After incubation for the indicated times, the medium was collected and centrifuged at 10,000g to remove cell debris. Cells were then lysed in buffer containing 50 mmol/L Tris, pH 7.5, 50 mmol/L NaCl, 1 mmol/L EDTA, 1% deoxycholate, 1% Triton X-100, 0.1% SDS supplemented with protease (Calbiochem, Darmstadt, Germany), and phosphatase inhibitors (Sigma-Aldrich). Protein content was quantified using the bicinchoninic acid assay (Sigma-Aldrich). A limited number of experiments were carried out using secretion medium containing DMEM with 0.5% BSA or DMEM containing 2% FBS with essentially identical results.
Immunoblotting
Protein samples (2% of the total sample from six wells, for both the media with the secreted proteins and the cellular protein extracts) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane (Li-Cor Biosciences, Lincoln, NE) for immunoblotting. The primary antibodies used were polyclonal antibodies to FABP4 and FABP5 (28), anti–β-actin (A5361), and anti-ULK1 (A7481) from Sigma-Aldrich and anti–galectin-3 (87985), anti–acetylated lysine (9441), anti-ATG14 (96752), anti–phospho-ATG14 (13155), anti-P62 (5114), and anti-LC3B (2775) from Cell Signaling Technology (Danvers, MA). Anti–retinol binding protein 4 (RBP4) (AF3476), anti–caspase-3 (AF-605), and anti-cleaved caspase-3 (MAB835) were obtained from R&D Systems. Anti-ATG5 (110-53818) and anti–nicotinamide phosphoribosyl transferase (NAMPT) (NB-100-594) were purchased from Novus Biologicals (Littleton, CO). Anti–beclin-1 (sc-11427) was obtained from Santa Cruz Biotechnology (Dallas, TX). The anti-SIRT1 antibody (07-131) was purchased from Millipore Sigma (Burlington, MA), and anti-FIP200 was made in the laboratory of D.-H. Kim as described (24).
Analytical Assays
Unesterified FFAs were measured using the HR series nonesterified fatty acid kit (Wako Chemicals, Richmond, VA) as per the manufacturer’s protocol. RNA for quantitative real-time PCR was obtained using TRIzol reagent (Invitrogen), and iScript (Bio-Rad, Hercules, CA) was used to make cDNA as per the manufacturer’s protocol. Quantitative RT-PCR amplification was performed on the Bio-Rad CFX 96 real-time system with SYBR Green Supermix.
Transcription factor II E (TFIIE) was used as the internal control to normalize expression. The primers used were TFIIE forward, 5′-CAAGGCTTTAGGGGACCAGATAC-3′, and reverse, 5′-CATCCATTGACTCCACAGTGACAC-3′, and SIRT1 forward, 5′-GGCTACCGAGACAACC TCCTG-3′, and reverse, 5′-AGTCCAGTCACTAGAGCTGGC-3′. For hypoxia experiments, a hypoxia chamber (BioSpherix Ltd, Parish, NY) containing 1% oxygen and 5% CO2 was used. FABP4 was detected in the serum samples of C57BL/6J (wild-type) and whole-body SIRT-deficient (SIRT1 KO) mice using a sandwich ELISA kit from Lifespan Biosciences Inc (LS-F11412-1; Seattle, WA) as per the manufacturer’s protocol.
Statistical Analysis
All of the results are expressed as SEM. For experiments performed with 3T3-L1 cells, the data presented have a sample size of three, and individual experiments were repeated at least three times. Statistical significance was determined using an unpaired two-tailed Student t test.
Results
FABP4 Is Secreted From White Fat, but Not Brown Fat, in Response to Lipolytic Stimulation
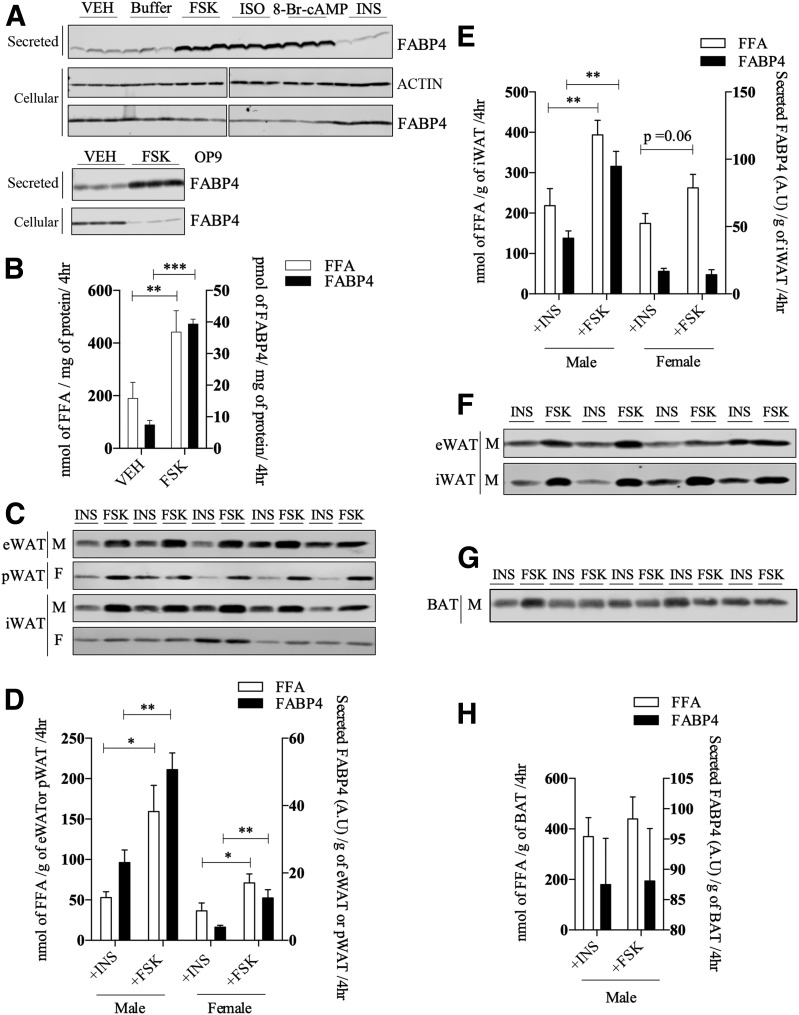
To better understand the mechanism of FABP4 secretion, we used adipose tissue and adipocytes from C57BL/6J mice as well as two differentiating adipocyte cell lines (3T3-L1 and OP9) to assess basic parameters of FABP4 secretion. Consistent with previously published studies (14,19,20,22), FABP4 is secreted by 3T3-L1 adipocytes in response to a variety of lipolytic agonists (Fig. 1A) and is independent of cell death, as evidenced by a lack of FSK-stimulated caspase-3 activation (Supplementary Fig. 1A). Because insulin is antilipolytic, it was used to establish the level of basal FABP4 secretion in many experiments and did not differ from either buffer or VEH (0.1% DMSO) controls. FABP4 secretion paralleled fatty acid efflux, although quantitatively, and the release of FFA exceeded that of FABP4 by three to four orders of magnitude (Fig. 1B) and was linear for at least 6 h (Supplementary Fig. 1B). Similarly, FSK stimulated FABP4 secretion by another murine adipocyte cell line, OP9, to approximately the same extent (Fig. 1A). Further, we assessed FABP4 secretion from adipose tissue explants of chow-fed mice. Male eWAT, male WAT, and female pWAT depots exhibited increased FABP4 secretion in response to FSK when compared with explants incubated with insulin; however, no appreciable regulation of FABP4 secretion was observed from the iWAT depot of female mice (Fig. 1C and E). Additionally, primary adipocytes from eWAT and iWAT of male mice on a high-fat diet exhibited FSK-stimulated increase in FABP4 secretion as compared with insulin treatment (Fig. 1F). Although BAT explants released FABP4 basally, the secretion was not regulated by FSK (Fig. 1G and H).
Figure 1.
FABP4 secretion in response to lipolytic stimuli. A (top): Western analysis of FABP4 secretion from differentiated 3T3-L1 adipocytes in response to a 4-h treatment with VEH, buffer, 20 μmol/L FSK, 10 μmol/L isoproterenol (ISO), 1 mmol/L 8-Br-cAMP, or 500 nmol/L insulin (INS). The intracellular levels of β-actin and FABP4 were determined immunochemically. Because these samples were analyzed on different gels, the indicated break in the image denotes the two separate gels used. A (bottom): Secreted and intracellular FABP4 in differentiated OP9 adipocytes in response to VEH or 20 μmol/L FSK. B: Levels of FABP4 (quantified from A) and fatty acids (FFA) secreted (as measured by the colorometric assay described in research design and methods) by 3T3-L1 adipocytes treated with VEH or 20 μmol/L FSK. C: Tissue explants from eWAT, pWAT, and iWAT were isolated and treated with 20 μmol/L FSK or 500 nmol/L INS for 4 h, and the secretion of FABP4 was evaluated immunochemically. D: Quantitation of FABP4 (from C) and FFA secretion by eWAT and pWAT explants. E: Quantitation of FABP4 and FFA secretion by iWAT explants from male or female C57BL/6J mice as shown in C. F: FABP4 secretion from primary adipocytes derived from eWAT and iWAT of high-fat–fed C57BL/6J mice in response to 2-h treatment with either 20 μmol/L FSK or 500 nmol/L INS. G: Explant tissue from interscapular BAT of male C57BL/6J mice was isolated as described and treated with 20 μmol/L FSK or 500 nmol/L INS for 4 h. The secretion of FABP4 was evaluated immunochemically. H: Quantitation of FABP4 and FFA secretion by BAT explants from male C57BL/6J mice as shown in G. *P < 0.05; **P < 0.01; ***P < 0.001. A.U., arbitrary units; F, female; hr, hours; M, male.
Work from Yoon et al. (29) has shown that NAMPT enzyme (eNAMPT), another protein lacking a conventional secretion signal, is also secreted in response to lipolytic conditions, as are other adipocyte proteins such as FABP5 and galectin-3 (30) (Supplementary Fig. 1C). Importantly, when release of the conventionally secreted RBP4 was analyzed under the same FSK-stimulated conditions, its secretion was not regulated (Supplementary Fig. 1C). Ertunc et al. (20) have reported that intracellular triglyceride hydrolysis is required for FABP4 secretion. Consistent with this, when the ATGL was inhibited with ATGListatin, there was a decrease in stimulated FABP4 secretion and FFA release (Supplementary Fig. 1D and E). Similar to the results of Wu et al. (15), we also observed that FABP4 and eNAMPT secretion is modestly increased under hypoxic conditions, albeit to a much lesser extent than more direct lipolytic stimuli (Supplementary Fig. 1F).
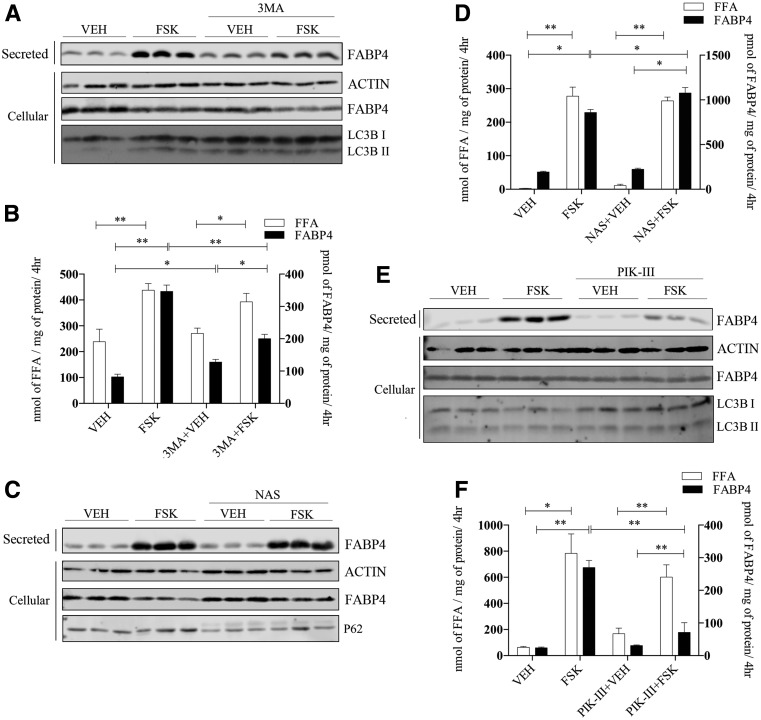
FABP4 Secretion Is Mediated by Some, but Not All, Autophagic Proteins
Unconventional secretion of FABP4 release from adipose tissue occurs via an endosomal mechanism involving lysosomes, but does not require the autophagic protein ATG5 (22) that is involved in vesicle elongation and autophagosome maturation (31). However, it has been demonstrated that autophagosomes and autolysosomes can be formed in an ATG5-independent manner that could be used to deliver cargo to lysosomes for secretion. As such, we expanded the evaluation of the role of autophagy in FABP4 secretion using chemical and molecular inhibitors. To this end, adipocytes were treated with the classical autophagic inhibitor 3MA (32) or the autophagic inducer NAS (33). 3MA treatment reduced autophagy as evidenced by the increase in LC3BI as compared with LC3BII and concurrently blunted FABP4 secretion (Fig. 2A), whereas FFA release was unaffected (Fig. 2B). In contrast, treatment with NAS modestly potentiated FABP4 secretion in the absence of an effect on fatty acid release (Fig. 2C and D). NAS treatment activated autophagy, as indicated by the decrease in P62 levels (Fig. 2C). Inhibiting VPS34, a class III phosphatidylinositol 3-kinase (PI3K) using the chemical inhibitor PIK-III (34), markedly abrogated FABP4 release from adipocytes without affecting FFA efflux (Fig. 2E and F).
Figure 2.
FABP4 secretion in response to chemical regulators of autophagy. A: Differentiated 3T3-L1 adipocytes were pretreated for 2 h with or without 5 mmol/L 3MA, washed, and then incubated in secretion media with VEH or 20 μmol/L FSK in the presence of 3MA for 4 h. Secreted FABP4 and cellular actin, FABP4, and LC3BI and -II levels were determined immunochemically. B: Quantification secreted FFA and FABP4 from A. C: Differentiated 3T3-L1 adipocytes were pretreated for 2 h with or without 30 μmol/L NAS, washed, and then incubated in secretion media with VEH or 20 μmol/L FSK in the presence of NAS for 4 h. Secreted FABP4 and cellular β-actin, FABP4, and P62 levels were determined immunochemically. D: Levels of secreted FFA and FABP4 from C. E: Differentiated 3T3-L1 adipocytes were pretreated for 2 h with or without 7.5 μmol/L of the PI3K inhibitor PIK-III, washed, and then incubated in secretion media with VEH or 20 μmol/L FSK in the presence of PIK-III for 4 h. Secreted FABP4 and cellular β-actin, FABP4, and LC3BI and -II levels were determined immunochemically. F: Quantitation of FFA and FABP4 secretion from E. *P < 0.05; **P < 0.01. hr, hours; kd, knockdown.
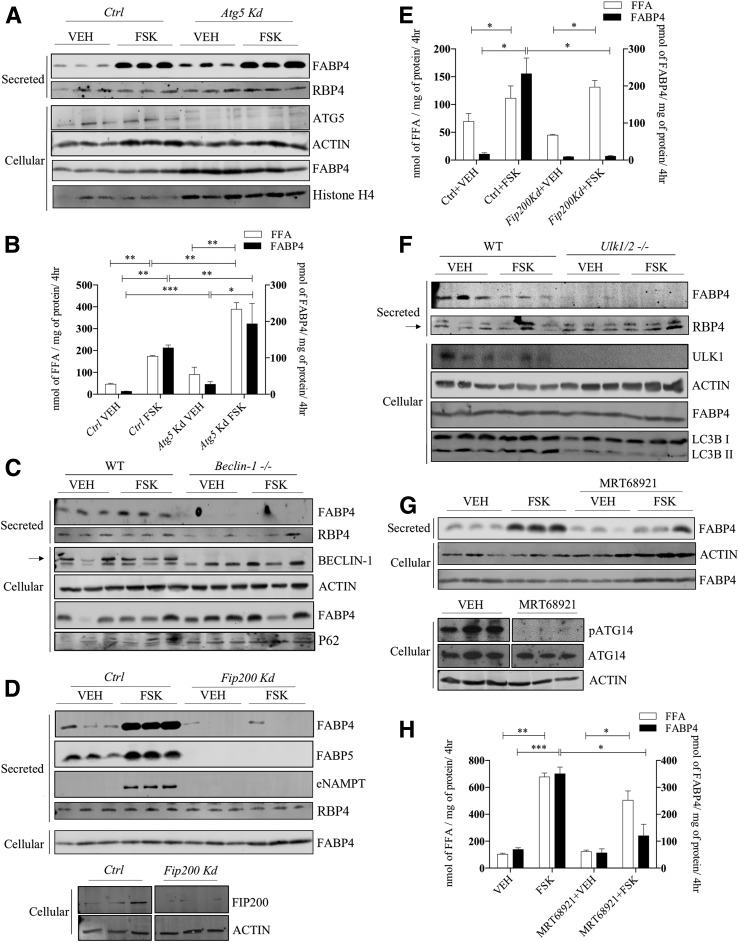
FABP4 Secretion Requires Early Components of Autophagy
Given the rather broad effects of both 3MA and NAS on autophagy, we then explored targeted analysis of autophagic components using gene silencing and knockout approaches. Similar to the results of Villeneuve et al. (22), silencing of Atg5 did not attenuate either FABP4 secretion or FFA efflux (Fig. 3A and B) but instead modestly potentiated both FABP4 secretion and FFA release from cultured 3T3-L1 adipocytes. Because ATG5 is a component of secretory autophagy (35), this result suggests that unconventional secretion of FABP4 is mediated by a fundamentally different mechanism. To parallel studies of ATG5 with other autophagic components, beclin-1, a core component of a PI3K complex involving VPS34–beclin-1–VPS15–ATG14L, was evaluated for its role in FABP4 secretion using differentiated MEF cells from wild-type and Beclin-1 null mice. Although FSK modestly stimulated FABP4 secretion from differentiated control cells, Beclin-1–null differentiated cells failed to secrete FABP4 under any conditions and accumulated P62 due to the block in autophagy (Fig. 3C). Extending these studies, FIP200 is a subunit of the ULK1–ATG13–ATG101–FIP200 complex and is crucial for autophagic initiation and activation of beclin–VPS34 complex (36). Figure 3D and E shows that similar to Beclin-null MEFs, Fip200-silenced 3T3-L1 adipocytes failed to secrete FABP4, whereas FFA efflux as well as secretion of the classically secreted protein RBP4 was unaffected. Similarly, FIP200 is also required for FABP5 and NAMPT release (Fig. 3D). Lastly, when ULK1, the serine/threonine protein kinase master regulator of autophagy, was evaluated for its influence on FABP4 secretion using differentiated Ulk1/2-deficient MEF cells, FABP4 release was undetectable, whereas secretion of RBP4 was unaffected (Fig. 3F). As an alternative to the genetic knockout model, we also evaluated the ULK1/2 kinase–specific inhibitor MRT68921 (37) and demonstrated its effectiveness via loss of phosphorylation of its target protein, ATG14 (Fig. 3G). FSK-stimulated FABP4 secretion in MRT68921-treated 3T3-L1 adipocytes was attenuated in the absence of an alteration on FFA efflux (Fig. 3G and H).
Figure 3.
FABP4 secretion in response to molecular regulators of early autophagic proteins. A: Lentiviral-infected control (Ctrl) or Atg5-silenced 3T3-L1 adipocytes were treated with VEH or 20 μmol/L FSK for 4 h, and the secretion of FABP4 and RBP4, as well as the cellular levels of ATG5, β-actin, FABP4, and histone H4, was evaluated immunochemically. B: Quantitative analysis of the levels of secreted FFA and FABP4 from A. C: MEFs from a wild-type C57BL/6J or beclin-1 null mouse were differentiated as described in research design and methods, and secreted FABP4 and RBP4 and cellular levels of beclin-1, β-actin, FABP4, and P62 were determined immunochemically. D (top): Lentiviral-infected control or Fip200-silenced 3T3-L1 adipocytes were treated with VEH or 20 μmol/L FSK for 4 h, and the secretion of FABP4, FABP5, eNAMPT, and RBP4 as well as cellular FABP4 was determined immunochemically. D (bottom): Cellular levels of FIP200 and β-actin in control or Fip200-silenced 3T3-L1 adipocytes. E: Quantitative analysis of FABP4 (from D) and FFAs secreted by control or Fip200-silenced 3T3-L1 adipocytes. F: MEFs from wild-type C57BL/6J or ULK1/2 null mice were differentiated and the expression of secreted FABP4 and RBP4 and cellular levels of ULK1/2, β-actin, FABP4, and LC3B were determined immunochemically. The arrow indicates specific RBP4 band. G: Differentiated 3T3-L1 adipocytes were treated with or without 2 μmol/L of the ULK1/2 inhibitor MRT68921 for 2 h followed by addition of either VEH or 20 μmol/L FSK for 4 h. The levels of secreted FABP4 and cellular β-actin and FABP4 were evaluated immunochemically. Additionally, the cellular levels of phosphorylated ATG14 (pATG14), ATG14, and β-actin were evaluated immunochemically. H: Quantitation of FABP4 and FFA secreted by 3T3-L1 adipocytes in response to VEH or 20 μmol/L FSK stimulation in the presence and absence of the ULK1/2 inhibitor MRT68921. *P < 0.05; **P < 0.01; ***P < 0.001. hr, hours; kd, knockdown.
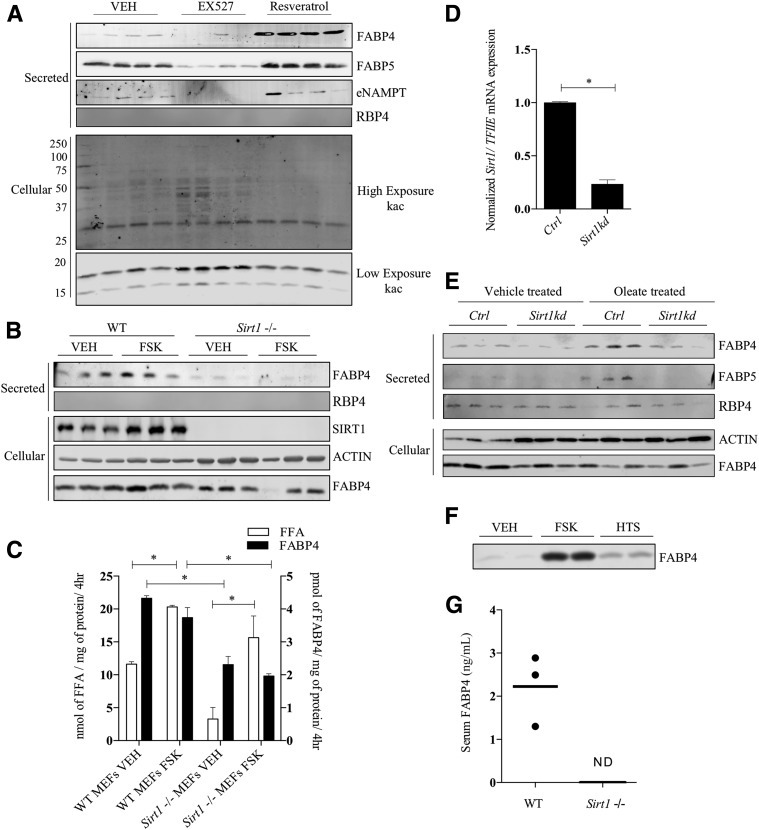
SIRT1 Is Required for FABP4 Secretion
The cAMP-dependent protein kinase A phosphorylates S434 of SIRT1 and activates its deacetylase activity to promote fatty acid oxidation and energy expenditure, as well as regulate the protein quality control system (38). A growing body of evidence shows that SIRT1 positively regulates autophagy (39), and Sathyanarayan et al. (40) demonstrated that SIRT1 activation results in part from ATGL-dependent lipolysis. Moreover, work from Yoon et al. (29) has shown that adipocyte secretion of NAMPT to form an extracellular pool of enzyme (eNAMPT) was SIRT1 dependent. To assess the role of SIRT1 on FABP4 secretion, the SIRT1 deacetylase inhibitor EX527 or the SIRT1 activator resveratrol was used. As shown in Fig. 4A, treatment of cultured 3T3-L1 adipocytes with EX527 reduced basal FABP4, FABP5, and eNAMPT secretion, whereas resveratrol treatment increased secretion of these proteins. Similarly, differentiated Sirt1−/− MEF cells also showed severe impairment in FABP4 secretion (Fig. 4B), even though FFA efflux in the presence of FSK was unaffected (Fig. 4C). In parallel, control and Sirt1 knockdown 3T3-L1 adipocytes (Fig. 4D) were profiled for their influence on FABP secretion. As shown in Fig. 4E, FABP4 and FABP5 were found secreted from control cells, but to a lesser extent from Sirt1-deficient cells. Treatment of Sirt1-deficient cells with FSK resulted in increased expression of SIRT1 (results not shown), even in the presence of the inactivating shRNA, due to protein kinase A activation that ultimately increases SIRT1 expression (38). As an alternative experimental approach to increase intracellular fatty acids, two strategies were used: oleate loading and FABP4 inhibition. When oleate was added to the extracellular medium, FABP4 and FABP5 secretion were potentiated in control, but not in Sirt1-silenced, 3T3-L1 adipocytes (Fig. 4E). Furthermore, the secretion of the classically secreted protein RBP4 was unaffected by Sirt1 knockdown or oleate treatment (Fig. 4E). Similarly, when the pan-specific FABP inhibitor HTS01037 (41) was used to displace bound FFA from their intracellular binding partners, basal FABP4 secretion was also increased (Fig. 4F), suggesting that intracellular FFAs are positive regulators of UPS. Consistent with a requirement for SIRT1, levels of FABP4 were undetectable in serum from Sirt1 null mice (Fig. 4G) compared with wild-type controls.
Figure 4.
FABP4 secretion in response to regulators of SIRT1. A: Differentiated 3T3-L1 adipocytes were treated with 10 μmol/L EX527 or 10 μmol/L resveratrol for 16 h, and the secretion of FABP4, FABP5, eNAMPT, and RBP4 was determined immunochemically. In parallel, the cellular extract was probed for acetylated lysine using the polyspecific anti-Kac antibody. The lysine acetylation results are from a single gel but are shown as different exposures to allow a better representation of the resulting signal across the entire blot. B: Differentiated Sirt1-deficient and control MEFs were treated with VEH or 20 μmol/L FSK for 4 h and secreted FABP4 and RBP4 and cellular SIRT1, β-actin, and FABP4 determined immunochemically. C: Quantitative analysis of the levels of FABP4 (from B) and FFA secreted by control and SIRT1-deficient adipocytes. D: Levels of Sirt1 mRNA relative to TFIIE measured in control and Sirt1 silenced 3T3-L1 adipocytes. E: Control and Sirt1-silenced 3T3-L1 adipocytes were preincubated for 18 h with 400 μmol/L potassium oleate complexed to 100 μmol/L BSA. The basal levels of FABP4, FABP5, and RBP4 secretion were determined immunochemically. F: Differentiated 3T3-L1 adipocytes were incubated with 50 μmol/L of the pan-FABP inhibitor HTS01037 (HTS) or 20 μmol/L FSK, and the levels of secreted FABP4 were determined immunochemically. G: Serum FABP4 levels in chow-fed wild-type (WT) and Sirt1 null C57BL/6J mice as evaluated by an ELISA (Lifespan Biosciences Inc.) conducted per the manufacturer’s protocol. *P < 0.05. hr, hour; kd, knockdown; ND, not detected.
Discussion
Increased circulating FABP4 as a consequence of obesity is correlated with a variety of pathophysiologies including type 2 diabetes, cardiac dysfunction, Cushing syndrome, end-stage renal disease, and ovarian and breast cancer (13,17,42,43). The circulating form of FABP4 increases approximately two- to fourfold in both murine models of metabolic dysfunction and in obese humans, while polymorphisms in the human FABP4 promoter that reduce its expression are protective for development of cardiovascular disease (9). Moreover, therapeutic monoclonal antibodies targeting the secreted form of FABP4 attenuate hepatic glucose output and may constitute a feasible approach to treatment for some forms of type 2 diabetes (44).
Previous work by Cao et al. (14), Wu et al. (15), and Villeneuve et al. (22) have shown that the leaderless protein FABP4 is secreted from adipocytes in an unconventional manner in a process distinct from the classical endoplasmic reticulum–Golgi system, as well as by processes unlinked to cellular apoptosis or necrosis. FABP4 release from adipose tissue is mediated in part by secretory lysosomes independently from vesicular or exosomal mechanisms and occurs largely by adipocytes rather than macrophages. Secretory FABP4 localizes to structures that are positive for late endosomal markers M6PR and Rab7 but does not require the function of ATG5 (22) (Fig. 3A), suggesting that conventional autophagic mechanisms may not play a role in FABP4 secretion. Recently, Flaherty et al. (45) reported a lipase-independent mechanism for fatty acid release by adipocytes that does result in a small amount of FABP4 secretion and defines a second mechanism for lipid efflux that does not require classical lipolytic machinery. Our studies in this paper demonstrate that the primary mechanism for FABP4 secretion does not involve multivesicular bodies but requires classical lipase activation or a source of increased cellular fatty acids and is markedly stimulated by agents that increase intracellular cAMP (Fig. 1). As such, while there are similarities in FABP4 secretion to the process described by Flaherty et al. (45), there are also clearly significant differences.
To evaluate if processes upstream of ATG5 would mediate FABP4 secretion, we used a variety of chemical and molecular inhibitors and demonstrated that components of the autophagy initiation process are required for UPS. Whereas the general autophagy inhibitor 3MA attenuated secretion and the autophagy activator N-acetyl sphingosine potentiated secretion, targeting specific complexes using gene silencing and knockout cell lines revealed that the canonical autophagy initiation proteins ULK1/2 and FIP200 of the ULK complex and VPS34–beclin-1 of the PI3KC3 complex mediate FABP4 secretion (Figs. 2 and 3). The role(s) for ULK1/2, FIP200, and VPS34 in UPS were recently highlighted by Goodwin et al. (46), who showed that ferritin and its binding partner nuclear receptor coactivator-4 are targeted to lysosomes by a form of selective autophagy that requires ULK1/2-FIP200 complex, ATG9A, and VPS34, but none of the other classical autophagy genes. Moreover, Nishida et al. (47) have shown that some phagophores receive membranes from the trans-Golgi or late endosomes in a noncanonical process that requires ULK1 and beclin-1 but is independent of ATG5 and that intermediate vesicles formed with composite membranes from the isolation membrane and endosomes can eventually fuse with lysosomes and release its cargo via lysosomal exocytosis. Via an analogous pathway, FABP4 may be initially recruited to a double-layered isolation membrane using the ULK1/2-FIP200 and beclin-1–VPS34 complexes without using classical autophagy proteins such as ATG5. This may provide an explanation for why FABP4 colocalizes with endosomal markers (22) while still requiring autophagy initiation proteins. It should be noted that although the focus of this work was on FABP4, the secretion of other proteins via unconventional mechanisms such as FABP5 and eNAMPT is also mediated by the ULK1/2-FIP200 and beclin-1–VPS34 complexes (Fig. 3 and Supplementary Fig. 1C), suggesting that this type of secretion may be more widespread than previously appreciated.
The second major finding from this work is the requirement for SIRT1. SIRT1 is an NAD-dependent deacetylase that participates in a number of regulatory processes linked to nutrient deprivation or depletion. Indeed, one such metabolic response to nutrient depletion is adipocyte lipolysis in which the liberated fatty acids can serve as an energy source by the muscle and/or liver. Importantly, as shown by Khan et al. (48), ATGL promotes autophagy via SIRT1 to control hepatic lipid droplet catabolism. Yoon et al. (29) have demonstrated that SIRT1 is required for unconventional secretion of eNAMPT from adipocytes via deacetylation of K53. Consistent with this, inhibition of SIRT1 attenuated not only eNAMPT secretion but also that of FABP4 and FABP5. Although acetylation of FABP4 or FABP5 has not been characterized, several components of the autophagy apparatus are controlled via acetylation–deacetylation cycles. SIRT1 mediates the deacetylation and activation of beclin-1 at K430 and K437, and the deacetylated state of VPS34 promotes autophagy initiation (49,50). Our results show that VPS34–beclin-1 activity is crucial for FABP4 release, suggesting that SIRT1 may function at multiple levels, deacetylating cargo proteins such as eNAMPT and possibly FABP4, as well as deacetylating the secretion machinery promoting UPS. Consistent with this, serum levels of FABP4 from Sirt1 null mice were undetectable, and although the sample size was not large (n = 3 for control and Sirt1 null), the results were unequivocal.
Because lipolytic agents increase FFA release from fat cells and FABP4 facilitates cytoplasmic solubilization and trafficking of fatty acids, it is possible that secreted FABP4 facilitates FFA efflux from fat cells. However, several lines of experimentation oppose this consideration. Firstly, the non–fatty acid binding mutant of FABP4 (R126Q) is secreted similarly to that of wild-type protein (20), and secondly, the amount of FABP4 secreted is only 0.1–0.01% that of the amount of fatty acid released (Fig. 1B). Moreover, as shown by analysis of FABP4 secretion in PIK-III–treated cells (Fig. 2E and F) or in cells lacking Fip200 (Fig. 3D and E), attenuation of FABP4 secretion occurred independently from FFA secretion. FFA binding to FABP4 is pH dependent (5), and lipids rapidly dissociate from the protein at the lower pH of the lysosome. Inhibition of lysosomal acid lipase with LAListat had no effect on FABP4 secretion (results not shown), suggesting that while fatty acids may be produced within the lysosome as part of lipophagy, they are not likely to be bound to FABP4 unless the pH of secretory lysosomes increases to near neutrality.
In sum, the findings presented in this study point toward unconventional secretion of FABP4 as being mediated by components of the ULK1/2–FIP200 complex in conjunction with beclin-1–VPS34. Secretion is regulated, in part, by nutrient deprivation that activates the classical cAMP–protein kinase A lipolytic cascade and the obligatory activation of SIRT1, resulting in the secretion of FABP4 and likely other proteins of fat cells via unconventional mechanisms. The molecular apparatus that facilitates such processes and defines unconventional secretion in terms of cargo selection and regulation is largely unexplored but may be novel loci with therapeutic potential for those with metabolic disease.
Supplementary Material
Article Information
Acknowledgments. The authors thank the members of the Bernlohr laboratory for helpful suggestions. PIK-III (VPS34 inhibitor) was generously provided by Dr. Leon O. Murphy, Novartis Institutes for BioMedical Research, Ulk1/2−/− MEFs were kindly provided by Dr. M. Kundu, St. Jude Children’s Research Hospital (Memphis, TN), and Becn1−/− MEFs were provided by Dr. Zhenyu Yue, Icahn School of Medicine at Mount Sinai (New York, NY) to D.-H.K.
Funding. This work was supported by National Institutes of Health grant DK-053189 and the Minnesota Agricultural Experiment Station (MN 70-015) to D.A.B. and National Institute on Aging grant AG-055452, National Institute of Diabetes and Digestive and Kidney Diseases grants DK-108790 and DK-114401, and the American Diabetes Association (1-16-IBS-203) to D.G.M.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. A.J., A.V.H., and D.A.B. developed hypotheses, carried out experiments, interpreted results, and wrote the manuscript. E.K.B. carried out experiments and interpreted results. M.W.M. developed molecular reagents, interpreted results, and edited the manuscript. S.-I.I. and D.G.M. developed hypotheses, interpreted results, and edited the manuscript. D.-H.K. developed hypotheses, interpreted results, and edited manuscript. D.A.B. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 77th Scientific Sessions of the American Diabetes Association, San Diego, CA, 9–13 June 2017.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-1367/-/DC1.
References
- 1.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest 2003;112:1785–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung UJ, Choi M-S. Obesity and its metabolic complications: the role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int J Mol Sci 2014;15:6184–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coe NR, Simpson MA, Bernlohr DA. Targeted disruption of the adipocyte lipid-binding protein (aP2 protein) gene impairs fat cell lipolysis and increases cellular fatty acid levels. J Lipid Res 1999;40:967–972 [PubMed] [Google Scholar]
- 4.LaLonde JM, Bernlohr DA, Banaszak LJ. The up-and-down beta-barrel proteins. FASEB J 1994;8:1240–1247 [DOI] [PubMed] [Google Scholar]
- 5.Sha RS, Kane CD, Xu Z, Banaszak LJ, Bernlohr DA. Modulation of ligand binding affinity of the adipocyte lipid-binding protein by selective mutation. Analysis in vitro and in situ. J Biol Chem 1993;268:7885–7892 [PubMed] [Google Scholar]
- 6.Hertzel AV, Smith LA, Berg AH, et al. . Lipid metabolism and adipokine levels in fatty acid-binding protein null and transgenic mice. Am J Physiol Endocrinol Metab 2006;290:E814–E823 [DOI] [PubMed] [Google Scholar]
- 7.Tuncman G, Erbay E, Hom X, et al. . A genetic variant at the fatty acid-binding protein aP2 locus reduces the risk for hypertriglyceridemia, type 2 diabetes, and cardiovascular disease. Proc Natl Acad Sci U S A 2006;103:6970–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Hertzel AV, Steen KA, Wang Q, Suttles J, Bernlohr DA. Uncoupling lipid metabolism from inflammation through fatty acid binding protein-dependent expression of UCP2. Mol Cell Biol 2015;35:1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steen KA, Xu H, Bernlohr DA. FABP4/aP2 regulates macrophage redox signaling and inflammasome activation via control of UCP2. Mol Cell Biol 2017;37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jahansouz C, Xu H, Hertzel AV, et al. . Partitioning of adipose lipid metabolism by altered expression and function of PPAR isoforms after bariatric surgery. Int J Obes 2018;42:139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlottmann I, Ehrhart-Bornstein M, Wabitsch M, Bornstein SR, Lamounier-Zepter V. Calcium-dependent release of adipocyte fatty acid binding protein from human adipocytes. Int J Obes 2014;38:1221–1227 [DOI] [PubMed] [Google Scholar]
- 12.Xu A, Wang Y, Xu JY, et al. . Adipocyte fatty acid-binding protein is a plasma biomarker closely associated with obesity and metabolic syndrome. Clin Chem 2006;52:405–413 [DOI] [PubMed] [Google Scholar]
- 13.Gharpure KM, Pradeep S, Sans M, et al. . FABP4 as a key determinant of metastatic potential of ovarian cancer. Nat Commun 2018;9:2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao H, Sekiya M, Ertunc ME, et al. . Adipocyte lipid chaperone AP2 is a secreted adipokine regulating hepatic glucose production. Cell Metab 2013;17:768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu LE, Samocha-Bonet D, Whitworth PT, et al. . Identification of fatty acid binding protein 4 as an adipokine that regulates insulin secretion during obesity. Mol Metab 2014;3:465–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stejskal D, Karpisek M. Adipocyte fatty acid binding protein in a Caucasian population: a new marker of metabolic syndrome? Eur J Clin Invest 2006;36:621–625 [DOI] [PubMed] [Google Scholar]
- 17.Tso AWK, Xu A, Sham PC, et al. . Serum adipocyte fatty acid binding protein as a new biomarker predicting the development of type 2 diabetes: a 10-year prospective study in a Chinese cohort. Diabetes Care 2007;30:2667–2672 [DOI] [PubMed] [Google Scholar]
- 18.Tu W-J, Zeng X-W, Deng A, et al. . Circulating FABP4 (fatty acid-binding protein 4) is a novel prognostic biomarker in patients with acute ischemic stroke. Stroke 2017;48:1531–1538 [DOI] [PubMed] [Google Scholar]
- 19.Mita T, Furuhashi M, Hiramitsu S, et al. . FABP4 is secreted from adipocytes by adenyl cyclase-PKA- and guanylyl cyclase-PKG-dependent lipolytic mechanisms. Obesity (Silver Spring) 2015;23:359–367 [DOI] [PubMed] [Google Scholar]
- 20.Ertunc ME, Sikkeland J, Fenaroli F, et al. . Secretion of fatty acid binding protein aP2 from adipocytes through a nonclassical pathway in response to adipocyte lipase activity. J Lipid Res 2015;56:423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf BB, Schuler M, Echeverri F, Green DR. Caspase-3 is the primary activator of apoptotic DNA fragmentation via DNA fragmentation factor-45/inhibitor of caspase-activated DNase inactivation. J Biol Chem 1999;274:30651–30656 [DOI] [PubMed] [Google Scholar]
- 22.Villeneuve J, Bassaganyas L, Lepreux S, et al. . Unconventional secretion of FABP4 by endosomes and secretory lysosomes. J Cell Biol 2018;217:649–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Student AK, Hsu RY, Lane MD. Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J Biol Chem 1980;255:4745–4750 [PubMed] [Google Scholar]
- 24.Ro SH, Jung CH, Hahn WS, et al. . Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy 2013;9:2103–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobo S, Wiczer BM, Smith AJ, Hall AM, Bernlohr DA. Fatty acid metabolism in adipocytes: functional analysis of fatty acid transport proteins 1 and 4. J Lipid Res 2007;48:609–620 [DOI] [PubMed] [Google Scholar]
- 26.Wolins NE, Quaynor BK, Skinner JR, et al. . OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J Lipid Res 2006;47:450–460 [DOI] [PubMed] [Google Scholar]
- 27.Viswanadha S, Londos C. Determination of lipolysis in isolated primary adipocytes. Methods Mol Biol 2008;456:299–306 [DOI] [PubMed] [Google Scholar]
- 28.Hertzel AV, Bennaars-Eiden A, Bernlohr DA. Increased lipolysis in transgenic animals overexpressing the epithelial fatty acid binding protein in adipose cells. J Lipid Res 2002;43:2105–2111 [DOI] [PubMed] [Google Scholar]
- 29.Yoon MJ, Yoshida M, Johnson S, et al. . SIRT1-mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab 2015;21:706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes RC. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim Biophys Acta 1999;1473:172–185 [DOI] [PubMed] [Google Scholar]
- 31.Hanada T, Noda NN, Satomi Y, et al. . The Atg12-Atg5 conjugate has a novel e3-like activity for protein lipidation in autophagy. J Biol Chem 2007;282:37298–37302 [DOI] [PubMed] [Google Scholar]
- 32.Heckmann BL, Yang X, Zhang X, Liu J. The autophagic inhibitor 3-methyladenine potently stimulates PKA-dependent lipolysis in adipocytes. Br J Pharmacol 2013;168:163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang YP, Hu LF, Zheng HF, et al. . Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin 2013;34:625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dowdle WE, Nyfeler B, Nagel J, et al. . Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 2014;16:1069–1079 [DOI] [PubMed] [Google Scholar]
- 35.Kimura T, Jia J, Claude-Taupin A, et al. . Cellular and molecular mechanism for secretory autophagy. Autophagy 2017;13:1084–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell RC, Tian Y, Yuan H, et al. . ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013;15:741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petherick KJ, Conway OJL, Mpamhanga C, et al. . Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem 2015;290:11376–11383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerhart-Hines Z, Dominy JE Jr., Blättler SM, et al. . The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 2011;44:851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 2014;32:1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sathyanarayan A, Mashek MT, Mashek DG. ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep 2017;19:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hertzel AV, Hellberg K, Reynolds JM, et al. . Identification and characterization of a small molecule inhibitor of fatty acid binding proteins. J Med Chem 2009;52:6024–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu A, Tso AWK, Cheung BMY, et al. . Circulating adipocyte-fatty acid binding protein levels predict the development of the metabolic syndrome: a 5-year prospective study. Circulation 2007;115:1537–1543 [DOI] [PubMed] [Google Scholar]
- 43.Ishimura S, Furuhashi M, Watanabe Y, et al. . Circulating levels of fatty acid-binding protein family and metabolic phenotype in the general population. PLoS One 2013;8:e81318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burak MF, Inouye KE, White A, et al. . Development of a therapeutic monoclonal antibody that targets secreted fatty acid-binding protein aP2 to treat type 2 diabetes. Sci Transl Med 2015;7:319ra205 [DOI] [PubMed] [Google Scholar]
- 45.Flaherty SE III, Grijalva A, Xu X, Ables E, Nomani A, Ferrante AW Jr. A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science 2019;363:989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodwin JM, Dowdle WE, DeJesus R, et al. . Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Rep 2017;20:2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishida Y, Arakawa S, Fujitani K, et al. . Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009;461:654–658 [DOI] [PubMed] [Google Scholar]
- 48.Khan SA, Sathyanarayan A, Mashek MT, Ong KT, Wollaston-Hayden EE, Mashek DG. ATGL-catalyzed lipolysis regulates SIRT1 to control PGC-1α/PPAR-α signaling. Diabetes 2015;64:418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun T, Li X, Zhang P, et al. . Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun 2015;6:7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su H, Yang F, Wang Q, et al. . VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol Cell 2017;67:907–921.e7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.