The discovery of a methanogen that can conserve energy to support growth solely from the oxidation of organic carbon coupled to the reduction of an extracellular electron acceptor expands the possible environments in which methanogens might thrive. The potential importance of c-type cytochromes for extracellular electron transfer to syntrophic bacterial partners and/or Fe(III) minerals in some Archaea was previously proposed, but these studies with Methanosarcina acetivorans provide the first genetic evidence for cytochrome-based extracellular electron transfer in Archaea. The results suggest parallels with Gram-negative bacteria, such as Shewanella and Geobacter species, in which multiheme outer-surface c-type cytochromes are an essential component for electrical communication with the extracellular environment. M. acetivorans offers an unprecedented opportunity to study mechanisms for energy conservation from the anaerobic oxidation of one-carbon organic compounds coupled to extracellular electron transfer in Archaea with implications not only for methanogens but possibly also for Archaea that anaerobically oxidize methane.
KEYWORDS: AQDS reduction, Methanosarcina, c-type cytochrome, extracellular electron transfer, genetics, transcriptome
ABSTRACT
Extracellular electron exchange in Methanosarcina species and closely related Archaea plays an important role in the global carbon cycle and enhances the speed and stability of anaerobic digestion by facilitating efficient syntrophic interactions. Here, we grew Methanosarcina acetivorans with methanol provided as the electron donor and the humic analogue, anthraquione-2,6-disulfonate (AQDS), provided as the electron acceptor when methane production was inhibited with bromoethanesulfonate. AQDS was reduced with simultaneous methane production in the absence of bromoethanesulfonate. Transcriptomics revealed that expression of the gene for the transmembrane, multiheme, c-type cytochrome MmcA was higher in AQDS-respiring cells than in cells performing methylotrophic methanogenesis. A strain in which the gene for MmcA was deleted failed to grow via AQDS reduction but grew with the conversion of methanol or acetate to methane, suggesting that MmcA has a specialized role as a conduit for extracellular electron transfer. Enhanced expression of genes for methanol conversion to methyl-coenzyme M and the Rnf complex suggested that methanol is oxidized to carbon dioxide in AQDS-respiring cells through a pathway that is similar to methyl-coenzyme M oxidation in methanogenic cells. However, during AQDS respiration the Rnf complex and reduced methanophenazine probably transfer electrons to MmcA, which functions as the terminal reductase for AQDS reduction. Extracellular electron transfer may enable the survival of methanogens in dynamic environments in which oxidized humic substances and Fe(III) oxides are intermittently available. The availability of tools for genetic manipulation of M. acetivorans makes it an excellent model microbe for evaluating c-type cytochrome-dependent extracellular electron transfer in Archaea.
INTRODUCTION
Extracellular electron exchange is central to the environmental function of diverse Archaea that oxidize and/or produce methane. Some methanogens can divert electrons from methane production to the reduction of extracellular electron carriers such as Fe(III), U(VI), V(IV), and anthraquinone-2,6-disulfonate (AQDS), a humic acid analog (1–9). Diversion of electron flux from methane production to extracellular electron transfer may influence the extent of methane production and metal geochemistry in anaerobic soils and sediments. Methanogens such as Methanothrix (formerly Methanosaeta) and Methanosarcina species can accept electrons via direct interspecies electron transfer from electron-donating partners, such as Geobacter species, in important methanogenic environments such as anaerobic digesters and rice paddy soils (10–12). Anaerobic methane oxidation also plays an important role in the global carbon cycle and diverse anaerobic methane-oxidizing archaea (ANME) transfer electrons derived from methane oxidation to extracellular electron acceptors, such as other microbial species, Fe(III), or extracellular quinones (13–20). The electrical contacts for extracellular electron exchange have yet to be definitively identified in any of these Archaea.
It has been hypothesized that outer-surface cytochromes enable electron transfer to electron-accepting microbial partners or Fe(III) in some ANME (13–19). Genes for multiheme c-type cytochromes that are present in ANME genomes can be highly expressed and in some instances the encoded proteins have been detected (14, 19). The putative function of outer-surface cytochromes is terminal electron transfer to extracellular electron acceptors, similar to the role that outer surface c-type cytochromes play in extracellular electron transfer in Gram-negative bacteria such as Shewanella and Geobacter species (21–23). Similar c-type cytochrome electrical contacts have been proposed for Fe(III)-reducing Archaea, such as Ferroglobus and Geoglobus species (24–26). However, the study of the mechanisms for extracellular electron transfer in these archaea has been stymied by the lack of microorganisms available in pure culture that can grow via extracellular electron transfer and are genetically tractable.
Tools are available for genetic manipulation of the methanogen Methanosarcina acetivorans (27–29). A methyl‐coenzyme M reductase from an uncultured ANME was introduced into M. acetivorans to generate a strain that could convert methane to acetate with simultaneous reduction of Fe(III) (30). Most of the electrons from the methane consumed were recovered in acetate (30), and it was not shown that energy was conserved from Fe(III) reduction. In vitro reactions catalyzed by membrane vesicles of wild-type M. acetivorans suggested that the membrane-bound heterodisulfide reductase HdrDE reduced Fe(III)-citrate and AQDS and that an outer-surface multiheme c-type cytochrome might also function as a potential electron donor for Fe(III)-citrate reduction (31). However, in vitro assays with cell components are not a definitive approach for determining the physiologically relevant mechanisms involved in the reduction of Fe(III) and AQDS. This is because constituents that do have access to extracellular electron acceptors in vivo are exposed to extracellular electron acceptors in vitro and many reduced cofactors and redox-proteins, including c-type cytochromes, can nonspecifically reduce these electron acceptors (32). Analysis of the phenotypes of intact cells that result from specific gene deletions can provide more conclusive evidence.
We report here that M. acetivorans can be grown without methane production with AQDS as the sole electron acceptor. Analysis of gene expression patterns and phenotypes of gene deletion strains suggest a mechanism for energy conservation during extracellular electron transfer.
RESULTS AND DISCUSSION
Growth of M. acetivorans with AQDS as the sole terminal electron acceptor.
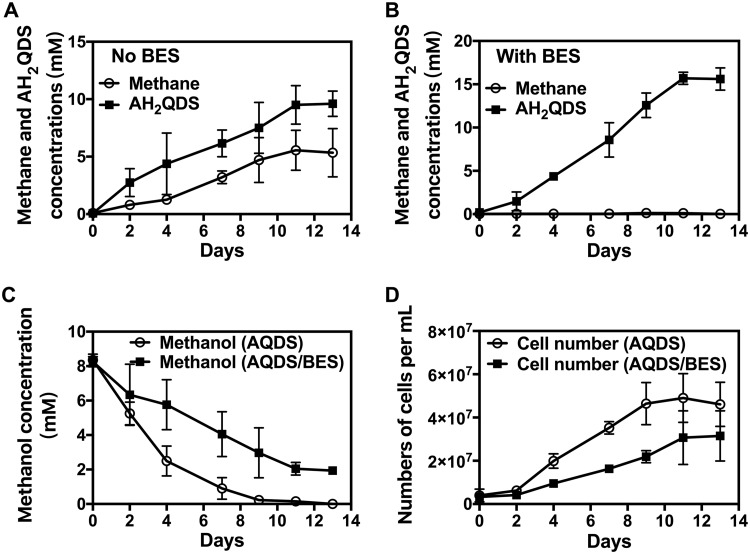
In medium with methanol provided as the electron donor and AQDS as a potential electron acceptor, M. acetivorans simultaneously produced methane and reduced AQDS (Fig. 1a). The addition of bromoethanesulfonate (BES) inhibited methane production and increased the extent of AQDS reduction (Fig. 1B; Fig. S1). The metabolism of methanol (Fig. 1c) was accompanied by an increase in cell numbers (Fig. 1D). In the BES-amended cultures, 6.3 ± 0.43 mM (mean of triplicate cultures ± the standard deviation) methanol was consumed with the reduction of 15.7 ± 0.61 mM AQDS. When the need to divert some of the methanol metabolized to cell biomass is considered, this stoichiometry is consistent with the oxidation of methanol to carbon dioxide, with AQDS serving as the sole electron acceptor: CH3OH + 3AQDS + H2O → 3AH2QDS + CO2. Methanol consumption stopped once all the AQDS was reduced in the BES-amended cultures (Fig. 1C). However, in the absence of BES, all of the methanol could be consumed because methanol was also converted to methane.
FIG 1.
Growth of M. acetivorans with methanol provided as an electron donor and AQDS as an electron acceptor in the presence or absence of BES. (A) Methane and AH2QDS concentrations generated by cultures grown without BES. (B) Methane and AH2QDS concentrations generated by cultures grown with BES. (C and D) Methanol concentrations (C) and cell numbers (D) from cultures grown in the presence or absence of BES. The complete inhibition of methane production in the presence of BES is also shown on an expanded scale in Fig. S1.
Methane concentrations detected in Methanosarcina acetivorans cultures grown with methanol as the electron donor and AQDS as the electron acceptor in the presence of the methanogenic inhibitor BES. Each point and error bars represent the average and standard deviation of triplicate measurements. Download FIG S1, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The growth of M. acetivorans with AQDS as the sole electron acceptor (Fig. 1) is the first example of a methanogen conserving energy to support growth with electron transfer to an external electron acceptor. The ability of M. acetivorans to grow in this manner and the availability of tools for genetic manipulation (27–29) provide an opportunity for functional analysis of extracellular electron transfer by an archaeon.
Transcriptomics and gene deletion studies demonstrate that the multiheme c-type cytochrome MmcA is involved in AQDS reduction.
In order to obtain insight into potential electron carriers involved in AQDS reduction, the transcriptome of cells grown with AQDS as the sole electron acceptor in the presence of BES was compared to the transcriptome of cells grown with methanol in the absence of AQDS or BES, so that methane production was the sole route of electron flux. The generation time (0.69 ± 0.13 days) of the cells grown via methanogenesis (Fig. S2) was longer than previously reported (generation time, 6.3 h; [33]). The lower growth rate in our study might be due to the omission of cysteine and lower sulfide content in our medium (0.3 mM compared to 0.5 mM) in order to reduce medium constituents that might abiotically reduce AQDS. However, methanogenic growth was ∼4-fold faster than growth via AQDS respiration in the presence of BES (generation time, 2.9 ± 0.18 days). Consistent with the lower growth rate, most of the genes related to cell growth (amino acid biosynthesis; protein synthesis; biosynthesis of purines, pyrimidines, nucleosides, and nucleotides; and transcription) had greater expression in methanogenic cells than cells grown via AQDS respiration (see Table S1B in the supplemental material).
Methanol (A), methane (B), AQDS concentrations (C), and cell numbers (D) in control cultures. All points and error bars represent the average and standard deviation for triplicate samples. For methanogenic conditions, Methanosarcina acetivorans cells were grown with methanol as the substrate. For the cells/BES/methanol condition, methanogenesis was inhibited with BES, and no electron acceptor (AQDS) was provided. For the cells/BES/AQDS condition, cells were provided with electron acceptor (AQDS), methanogenesis was inhibited with BES, and no electron donor (methanol) was provided. For the no cells/BES/AQDS condition, BES and AQDS were added to the medium but no inoculum was added. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Differential expression of genes from M. acetivorans cells grown with methanol provided as electron donor and AQDS (with BES) as electron acceptor (experimental condition) versus cells grown via methylotrophic methanogenesis (control condition). Values were obtained with EdgeR software. Only genes with P values of ≥0.05 are included in this table. Positive log fold change values indicate that the gene was more highly transcribed in AQDS-respiring cells; negative log fold change values indicate that the gene was more highly transcribed in cells grown via methylotrophic methanogenesis. (B) Number of growth-related genes that were more highly expressed in M. acetivorans cells grown with methanol as the electron donor and AQDS as the electron acceptor in the presence of BES (experimental condition) or in M. acetivorans cells grown via methylotrophic methanogenesis (control condition). These results are a summary of data presented in more detail in Table S1A. Download Table S1, XLSX file, 0.3 MB (366.2KB, xlsx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Remarkably, despite the lower growth rate on AQDS, the gene MA0658, which encodes a seven-heme, outer-surface c-type cytochrome, was 4.5-fold more highly expressed in AQDS-reducing versus methanogenic cells (Table 1). For future reference, this cytochrome was designated MmcA (membrane multiheme cytochrome A). Multiheme c-type cytochromes are of particular interest as potential electron carriers in extracellular electron transport because of the well-documented role of multiheme c-type cytochromes in bacteria such as Shewanella and Geobacter species that are highly effective in extracellular electron transfer (21–23). MA3739, a gene coding for a five-heme c-type cytochrome, was transcribed at similar levels as mmcA, and 4.1-fold-higher expression was detected in AQDS-reducing than methanogenic cells (Table 1).
TABLE 1.
Differential expression of genes encoding c-type cytochrome proteins in M. acetivorans cellsa
| Locus | No. of: |
Predicted localization | Fold upregulationb |
P | FDR | |
|---|---|---|---|---|---|---|
| Heme groups |
Transmembrane helices |
|||||
| MA0658 | 7 | 1 | Membrane | 4.53 | 0.002 | 0.006 |
| MA3739 | 5 | 0 | Unknown | 4.14 | 0.047 | 0.031 |
| MA0167 | 1 | 1 | Membrane | 5.97 | 0.018 | 0.037 |
| MA2925 | 2 | 1 | Membrane | NS | ||
| MA2908 | 2 | 1 | Membrane | NS | ||
Cells were grown with methanol provided as the electron donor and AQDS as the electron acceptor in the presence of BES or were grown via methanogenesis with methanol as the substrate. Genes were only considered differentially expressed if the P value and FDR (false discovery rate) were ≤0.05. NS, no significant difference in read abundance between conditions.
That is, in AQDS/BES versus methanogenesis.
There are three other putative c-type cytochrome genes in the M. acetivorans genome (26). MA0167, which encodes a monoheme cytochrome with predicted localization in the cell membrane, was six times more highly expressed in cells grown via AQDS respiration (Table 1). Functional analysis of the outer membrane of G. sulfurreducens has suggested that a monoheme c-type cytochrome may play a role in regulating the expression of multiheme c-type cytochromes, possibly by providing a sensor function (34, 35). It is possible that the protein encoded by MA0167 plays a similar role in M. acetivorans. The expressions of MA2925 and MA2908, both of which encode two-heme c-type cytochromes, were comparable in AQDS-reducing versus methanogenic cells (Table 1). These cytochromes are homologous to methylamine utilization protein G (MauG) and the diheme cytochrome c peroxidase (CcpA). MauG is required for aerobic methylamine metabolism (36–38), and CcpA proteins reduce hydrogen peroxide to water and protect the cell from reactive oxygen species (39, 40). Thus, it seems unlikely that either of these cytochromes is involved in extracellular electron transfer.
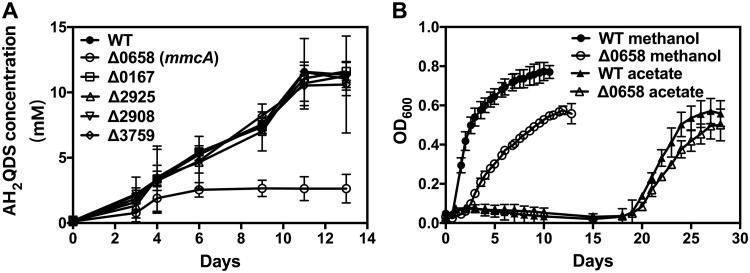
In order to evaluate the potential role of c-type cytochromes in AQDS reduction, deletion mutant strains were constructed in M. acetivorans for each c-type cytochrome gene in the genome (Table 1). Only the deletion of mmcA inhibited AQDS reduction (Fig. 2A). Deletion of mmcA had a slight impact on methanogenic growth with methanol (Fig. 2B). These results suggest that MmcA is a major component for extracellular electron transfer to AQDS but not for the conversion of methanol to methane.
FIG 2.
Impact of deletion of c-type cytochrome genes on growth of M. acetivorans under different conditions. (A) AH2QDS production during growth with methanol as the electron donor and AQDS as the acceptor in the presence of BES. The locus for the deleted cytochrome gene is designated next to the corresponding symbol. (B) Growth of wild-type and ΔMA0658 strains under methanogenic conditions as measured as the A600 with methanol or acetate provided as the substrates.
Previous studies have suggested that MmcA is part of the Rnf complex, which is required for acetoclastic methanogenesis (41), and that mmcA is cotranscribed with Rnf genes located in the same region of the chromosome (42). However, deletion of the MmcA gene did not substantially impact growth on acetate (Fig. 2B) or transcription of other genes from the Rnf complex (Fig. S2). Furthermore, the expression profiles of mmcA and genes for the Rnf complex were also different (Tables 1 and 2).
TABLE 2.
Comparison of transcripts from genes coding for components of the Rnf and Mrp complexes in M. acetivorans cellsa
| Locus | Description | Gene | Fold upregulationb |
P | FDR |
|---|---|---|---|---|---|
| MA0659 | Electron transport complex protein RnfC | rnfC | 1.52 | 0.02 | 0.04 |
| MA0660 | Electron transport complex protein RnfD | rnfD | NS | ||
| MA0661 | Electron transport complex protein RnfG | rnfG | 1.66 | 0.006 | 0.01 |
| MA0662 | Electron transport complex protein RnfE | rnfE | 1.45 | 0.02 | 0.05 |
| MA0663 | Electron transport complex protein RnfA | rnfA | 1.66 | 0.006 | 0.01 |
| MA0664 | Electron transport complex protein RnfB | rnfB | 1.57 | 0.008 | 0.01 |
| MA4572 | Multisubunit sodium/proton antiporter, MrpA subunit | mrpA | 5.44 | 5.77 × 10–8 | 5.07 × 10–6 |
| MA4665 | Multisubunit sodium/proton antiporter, MrpB subunit | mrpB | 5.41 | 8.99 × 10–8 | 6.06 × 10–6 |
| MA4570 | Multisubunit sodium/proton antiporter, MrpC subunit | mrpC | 6.50 | 7.25 × 10–8 | 5.71 × 10–6 |
| MA4569 | Multisubunit sodium/proton antiporter, MrpD subunit | mrpD | 4.84 | 1.38 × 10–7 | 7.21 × 10–6 |
| MA4568 | Multisubunit sodium/proton antiporter, MrpE subunit | mrpE | 3.70 | 3.79 × 10–6 | 5.56 × 10–5 |
| MA4567 | Multisubunit sodium/proton antiporter, MrpF subunit | mrpF | 4.79 | 3.78 × 10–7 | 1.28 × 10–5 |
| MA4566 | Multisubunit sodium/proton antiporter, MrpG subunit | mrpG | 4.57 | 3.39 × 10–7 | 1.20 × 10–5 |
Cells were grown with methanol and AQDS in the presence of BES or were grown via methanogenesis with methanol as the substrate. Genes were only considered differentially expressed if the P value and FDR were ≤0.05. NS, no significant difference in read abundance.
That is, in AQDS/BES versus methanogenesis.
Model for electron transport to AQDS via MmcA.
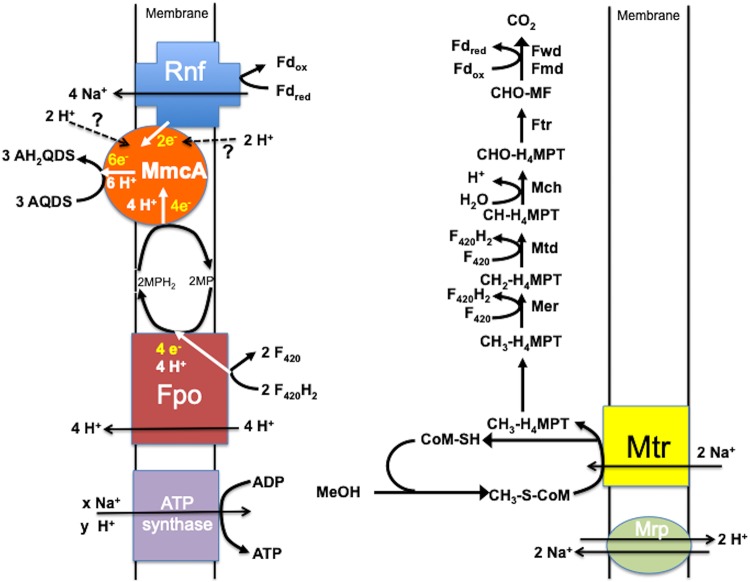
MmcA is a strong candidate for the terminal AQDS reductase because its localization in the cell membrane (42) is likely to provide access to AQDS and because of the well-known role of outer-membrane multiheme c-type cytochromes in reduction of AQDS and various forms of Fe(III) in Gram-negative bacteria such as Shewanella and Geobacter species (21–23, 43). It was previously suggested that MmcA could be a terminal reductase for the reduction of soluble Fe(III)-citrate, based on the in vitro oxidation of MmcA in membrane vesicles upon addition of Fe(III)-citrate (31). Such in vitro assays can be poor predictors of in vivo activity because Fe(III)-citrate typically oxidizes c-type cytochromes in vitro, regardless of physiological function, due to its very positive redox potential. However, as detailed below, multiple lines of evidence support a model in which energy can be conserved when MmcA serves as the terminal reductase during in vivo methanol oxidation coupled to AQDS reduction (Fig. 3).
FIG 3.
Proposed model for extracellular electron transport to AQDS by M. acetivorans when methanol is provided as the electron donor and methanogenesis is prevented by the addition of BES.
During methane production from methanol, methanol is converted to CH3-CoM by the activity of three enzymes, methyltransferase 1 (MtaB), methyltransferase 2 (MtaA), and methanol corrinoid protein (MtaC) (44–46). The oxidation of one molecule of CH3-CoM to CO2 generates the reducing equivalents necessary to reduce three molecules of CH3-CoM to methane. During methanol oxidation coupled to AQDS reduction in the presence of BES, the step that reduces CH3-CoM to methane is blocked, but the option for CH3-CoM oxidation remains (Fig. 3). Genes coding for enzymes involved in the oxidation of CH3-CoM to carbon dioxide were more highly expressed in methanogenic cells, consistent with increased transcription of growth-related genes in methanogenic cells and the need for this pathway to generate reductants to support methanogenesis (Table S2).
Differential expression of genes coding for proteins from the oxidative branch of the methylotrophic methanogenesis pathway and methyl-coenzyme M reductase (Mcr) in M. acetivorans cells grown with methanol and AQDS in the presence of BES or cells grown via methanogenesis with methanol as the substrate. Negative values indicate that genes were more significantly expressed in methanogenic cells. Genes were only considered differentially expressed if the P value and FDR (False Discovery Rate) were ≤0.05. NS: no significant difference in read abundance between conditions Download Table S2, DOCX file, 0.1 MB (131.7KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential expression of genes encoding isomers of MtaB, MtaA, and MtaC suggested that there might be some differences in the route for methanol conversion to CH3-CoM (Table 3). The genes for the isomers MtaB1, MtaA1, and MtaC1 were more highly transcribed in methanogenic cells, whereas AQDS-respiring cells had greater expression of genes coding for the alternative MtaB, MtaA, and MtaC isomers (Table 3). Differences in the activity of these isomers are unknown, but in previous studies mtaA1, mtaB1, and mtaC1 genes were highly transcribed during methanogenesis from methanol and MtaA1 was required for growth on methanol, whereas MtaA2 was dispensable (46).
TABLE 3.
Differential expression of genes encoding methanol methyltransferase enzymes in M. acetivorans cellsa
| Locus | Annotation | Gene | Fold upregulationb |
P | FDR |
|---|---|---|---|---|---|
| MA4379 | Co-methyl-5-hydroxybenzimi- dazolylcobamide:2-mercapto-ethanesulphonic acid methyltransferase, isozyme 1 |
mtaA1 | –1.68 | 0.01 | 0.02 |
| MA0455 | Methanol:5-hydroxybenzimidazolyl-cobamide methyltransferase, isozyme 1 |
mtaB1 | –6.84 | 0.02 | 0.04 |
| MA0456 | Corrinoid-containing methyl-accepting protein, isozyme 1 |
mtaC1 | –7.95 | 0.01 | 0.03 |
| MA4392 | Methanol:5-hydroxybenzimidazolylcobamide methyltransferase, isozyme 2 |
mtaB2 | 68.55 | 5.70 × 10–11 | 2.56 × 10–7 |
| MA4391 | Corrinoid-containing methyl-accepting protein, isozyme 2 |
mtaC2 | 48.28 | 3.27 × 10–10 | 5.54 × 10–7 |
| MA1615 | Co-methyl-5-hydroxybenzimidazolylcobamide:2- mercapto-ethanesulphonic acid methyltransferase, isozyme 2 |
mtaA2 | 5.39 | 1.77 × 10–7 | 8.04 × 10–6 |
| MA1616 | Methanol:5-hydroxybenzimidazolylcobamide methyltransferase, isozyme 3 |
mtaB3 | 9.66 | 5.24 × 10–8 | 4.89 × 10–6 |
| MA1617 | Corrinoid-containing methyl-accepting protein, isozyme 3 |
mtaC3 | 8.49 | 2.52 × 10–7 | 1.00 × 10–5 |
Cells were grown with methanol provided as an electron donor and AQDS provided as an electron acceptor in the presence of BES or cells grown via methanogenesis with methanol as the substrate. Negative values indicate that genes were more significantly expressed in methanogenic cells. Genes were only considered differentially expressed if the P value and FDR were ≤0.05.
That is, in AQDS/BES versus methanogenesis.
Oxidation of methanol to carbon dioxide is expected to yield reduced ferredoxin and reduced F420 (F420H2). It is likely that the Rnf complex oxidizes reduced ferredoxin with electron transfer to MmcA (47). Transcripts for genes coding for components of the Rnf complex were slightly higher (∼1.5-fold) than those in methanogenic cells (Table 2), suggesting an important role for the Rnf complex in energy conservation from methanol oxidation coupled to AQDS reduction.
In methanogenic cells, the membrane-bound Fpo complex (F420:methanophenazine oxidoreductase) oxidizes F420H2 derived from methanol oxidation with the reduction of methanophenazine and proton translocation (48–52). Transcription of all Fpo subunit genes was higher in methanogenic cells than AQDS-reducing cells, as expected because of the importance of Fpo in oxidizing F420H2 in cells producing methane (Table S3). However, all of the Fpo complex genes were also being actively expressed in AQDS-respiring cells, suggesting that Fpo is important for the oxidation of F420H2 generated in methanol-oxidizing, AQDS-reducing cells. The reduced methanophenazine that Fpo generates from F420H2 oxidation could transfer electrons to MmcA (41, 42, 47, 53). Although it has also been proposed that reduced methanophenazine may be able to directly transfer electrons to extracellular electron carriers in M. acetivorans (31), the requirement for MmcA for growth via AQDS reduction indicates that this is an unlikely route for AQDS reduction.
Differential expression of genes coding for proteins from the proton-translocating F420:methanophenazine oxidoreductase (Fpo) complex in M. acetivorans cells grown with methanol as the electron donor and AQDS (in the presence of BES) as the acceptor versus cells grown via methylotrophic methanogenesis with methanol as the substrate. Negative values indicate that genes were more significantly expressed in methanogenic cells. Genes were only considered differentially expressed if the P value and FDR (false discovery rate) were <0.05. Download Table S3, DOCX file, 0.1 MB (108.7KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In methanogenic cells, reduced methanophenazine could also donate electrons to the membrane-bound heterodisulfide reductase HdrDE (52, 54–59). In vitro evidence with membrane vesicles suggested that HdrDE can reduce AQDS with CoM-SH and CoB-SH oxidation to form CoM-S-S-CoB (31). However, the redox-active components of HdrE and HdrD responsible for electron transfer to an electron acceptor are embedded in the membrane and the cytoplasm, respectively (52), and thus unlikely to access extracellular AQDS in vivo. The relative expressions of hdrD and hdrE were slightly lower in AQDS-reducing cells than in methanogenic cells (Table S2). Furthermore, the inability of the MmcA-deficient strain to grow via AQDS reduction indicates that HdrDE is not capable of functioning as the sole AQDS reductase to support growth. Thus, based on the lack of strong evidence for a role for HdrDE, the likely simpler and more direct route for AQDS-dependent oxidation of reduced methanophenazine is electron transfer to MmcA.
Based on these considerations and current understanding of the function of the redox proteins involved (52, 60, 61), it is apparent that a net positive export of Na+ and H+ outside the cell membrane during AQDS respiration that can support the generation of ATP is feasible (Fig. 3). In this model, two Na+ must be translocated into the cell for the initial oxidation of CH3-S-CoM (62–64). Two moles of F420H2 and one mole of reduced ferrodoxin are generated per mole of CH3-S-CoM oxidized to carbon dioxide. Fpo oxidizes the F420H2 with H+ translocation and the reduction of methanophenazine (49–51). The reduced methanophenazine transfers electrons to MmcA, which reduces AQDS. The Rnf complex oxidizes the reduced ferredoxin coupled with Na+ translocation (41, 47, 65) and the reduction of MmcA. MmcA may transfer protons, as well as electrons during AQDS reduction, as observed in other c-type cytochromes (66–71). The ATP synthase couples both Na+ and H+ transport to ATP synthesis (72), but the H+/Na+ antiporter complex Mrp can be important for balancing external Na+/H+ ratios (73). Genes for Mrp were highly expressed in AQDS-reducing cells (Table 2).
Uncertainties in the stoichiometry of Na+/H+ transport per ATP synthesized and the total amount of H+ translocated prevent an accurate estimate of the theoretical ATP yield per mole of methanol oxidized with the reduction of AQDS. However, the proposed metabolic route suggests a likely mechanism for net ATP synthesis to support the observed growth of M. acetivorans with methanol oxidation coupled to AQDS reduction.
Implications.
The discovery that M. acetivorans can conserve energy to support growth from the oxidation of a one-carbon compound coupled to the reduction of an extracellular electron acceptor has important implications for the biogeochemistry of anaerobic soils and sediments and provides a genetically tractable model microbe for further analysis of the mechanisms of extracellular electron transfer in Archaea. Humic substances and Fe(III) are often abundant extracellular electron acceptors in a wide variety of anaerobic soils and sediments, and their availability for microbial respiration can reduce the extent of methane production (74–77). Competition for electron donors between methanogens and Fe(III)- and humic-reducing microorganisms is one factor (78–80). However, the finding that some methanogens may conserve energy by reducing extracellular electron acceptors suggests a mechanism for methanogens to survive in environments in which Fe(III) and oxidized forms of humic substances are abundant and then rapidly switch to methane production as these extracellular electron acceptors are depleted.
A comprehensive survey of the ability of diverse methanogens to conserve energy to support growth from electron transport to extracellular electron acceptors is warranted. Most methanogens, including other Methanosarcina species, lack membrane-bound multiheme cytochromes like MmcA and would need other mechanisms for extracellular electron transfer. The findings that MmcA is not essential for methane production and that expression of mmcA was increased when AQDS served as an electron acceptor suggest that the primary role of MmcA is extracellular electron transfer. If so, the presence of MmcA further suggests that there are environments in which the capacity for extracellular electron transfer substantially benefits M. acetivorans.
A wide diversity of archaea are capable of extracellular electron transfer (81). For archaea such as Ferroglobus placidus (24), Geoglobus ahangari (25), and diverse ANME (13–19), it has been proposed that outer-membrane cytochromes are the terminal reductase. It also appears that methanogens have evolved efficient means of extracellular electron transport; however, the mechanisms are poorly understood. The rapid nonphysiological reduction of extracellular electron acceptors by a range of redox-active proteins and cofactors in vitro necessitates genetically tractable model organisms for physiologically relevant functional studies. Thus, M. acetivorans may serve as an important model organism for better understanding cytochrome-based extracellular electron transfer in Archaea.
MATERIALS AND METHODS
Strains and growth conditions.
Methanosarcina acetivorans strains were routinely cultured under strict anaerobic conditions at 37°C in a previously described (27) medium with either 8.5 mM methanol or 40 mM acetate provided as the substrates.
M. acetivorans mutant strains were constructed with M. acetivorans WWM1 (Δhpt) (82) as the parent strain, as described previously (28). For the construction of MA0658, MA3739, MA2908, MA0167, and MA2925 deletion strains, genes were replaced with the pac gene (puromycin resistance gene). First, regions 500 to 1,000 bp upstream and downstream from the target genes were amplified by PCR (see Table S4 and Fig. S4 in the supplemental material). The DNA fragments of the upstream and downstream regions of MA0658 were digested with SacI/XbaI and EcoRI/XhoI. Upstream and downstream regions of MA3739 were digested with SalI/XbaI and SacI/NotI. Upstream and downstream regions of MA2908, MA0167, and MA2925 were digested with XhoI/HindIII and BamHI/NotI. The upstream fragment was ligated into the pJK3 plasmid (27). The downstream fragment was ligated into the pJK3 plasmid already containing the upstream fragment. This recombinant plasmid was then linearized and used for transformation. The deletion and replacement of all genes with pac was verified with primers (Table S4). All transformants were selected on medium supplemented with puromycin (2 μM final concentration), as previously described (27).
Reverse-transcription PCR with primers targeting two other genes from the Rnf complex in the Methanosarcina acetivorans genome, MA0659 and MA0664. RNA was extracted from cells of the MA0658 deletion mutant strain of M. acetivorans grown with acetate provided as the substrate for growth. Download FIG S3, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Steps involved in construction of the mutant strain ΔMA0658. The coding region from amino acid residues 54Cys to 457Arg was deleted. P1, P2, and P3 were primers utilized for verification of the mutant construct. Primer sequences used for construction and verification of this mutant strain and other strains included in this study are provided in Table S4. Download FIG S4, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used to construct various deletion mutant strains (restriction sites are underlined). Genetically recombinant plasmids were checked for accuracy by PCR with verification primers. Download Table S4, DOCX file, 0.1 MB (124.3KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additions of anthraquinone-2,6,-disulphonate (AQDS) were made from a concentrated stock to provide a final concentration of 16 mM. Cysteine was omitted from all cultures. When noted, 2-bromoethanesulfonate (BES) was added from a concentrated stock to provide a final concentration of 15 mM. Growth with AQDS was measured by determining numbers of cells stained with acridine orange with epifluorescence microscopy (83). For comparing methanogenic growth in wild-type and mutant cells, growth was monitored by spectrometry at an absorbance of 600 nm (84).
Analytical techniques.
Methanol concentrations were monitored with a gas chromatograph equipped with a headspace sampler and a flame ionization detector (Clarus 600; Perkin-Elmer, Inc., San Jose, CA). Methane in the headspace was measured by gas chromatography with a flame ionization detector (Shimadzu, GC-8A) as previously described (85). The production of reduced AQDS reduction was monitored by spectrophotometry at an absorbance of 450 nm as previously described (86).
RNA extraction.
Cells were harvested from triplicate 50 ml cultures of M. acetivorans grown with methanol (10 mM) provided as the electron donor and AQDS (16 mM) in the presence of the methanogenesis inhibitor BES (15 mM) or via methanogenesis with 40 mM methanol provided as the substrate. Cells were harvested when AQDS-respiring cultures had reduced ∼8 mM AQDS (midexponential phase) and when methanogenic cells reached an optical density at 600 nm of 0.5.
Cells were split into 50-ml conical tubes (BD Sciences), mixed with RNAProtect (Qiagen) in a 1:1 ratio, and pelleted by centrifugation at 3,000 × g for 15 min at 4°C. Pellets were then immediately frozen in liquid nitrogen and stored at –80°C. Total RNA was extracted from all six cell pellets according to the previously described protocol (87) and cleaned using an RNeasy minikit (Qiagen). All six RNA samples (three AQDS-respiring and three methanogenic) were then treated with Turbo DNA-free DNase (Ambion, Austin, TX). In order to ensure that samples were not contaminated with genomic DNA, PCR with primers targeting the 16S rRNA gene was done with RNA that had not been reverse transcribed. Further enrichment of mRNA was done with the MICROBExpress kit (Ambion), according to the manufacturer’s instructions.
RT-PCR analysis.
Total RNA was prepared from M. acetivorans hpt and ΔMA0658 strains grown methanogenically with acetate (40 mM). Complementary DNA (cDNA) was prepared by reverse transcription with AMV reverse transcriptase (New England Biolabs, Ipswich, MA) with the primers TCAGCATGCCTCATTCCAAC (MA0659) or TCGCAGACAGCCTTAACGTC (MA0664) according to the manufacturer’s specifications. This cDNA was then used as a template for PCR with the following primers: CAGTGACCTCGCTTATGTCC/TCAGCATGCCTCATTCCAAC (MA0695) or TGTGGAGGTTGCGGATTTGC/TCGCAGACAGCCTTAACGTC (MA0664). The amplified fragments were analyzed by agarose gel electrophoresis.
Illumina sequencing and data analysis.
Directional multiplex libraries were prepared with the ScriptSeq v2 RNA-Seq Library preparation kit (Epicentre), and paired-end sequencing was performed on a Hi-Seq 2000 platform at the Deep Sequencing Core Facility at the University of Massachusetts Medical School in Worchester, MA.
All raw data generated by Illumina sequencing were quality checked by visualization of base quality scores and nucleotide distributions with FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Initial raw nonfiltered forward and reverse sequencing libraries contained an average of 134,187,478 ± 20,358,059 reads that were ∼100 bp in length (Table S5). Sequences from all of the libraries were trimmed and filtered with Trimmomatic (88), with the sliding window approach set to trim bases with quality scores lower than 3, strings of 3+N’s, and reads with a mean quality score lower than 20. Bases were also cut from the start and end of reads that fell below a threshold quality of 3, and any reads smaller than 50 bp were eliminated from the library. These parameters yielded an average of 90,596,717 ± 23,433,670 quality reads per RNA-Seq library.
(A) Summary of statistics from RNAseq libraries assembled from RNA extracted from Methanosarcina acetivorans cells grown via methylotrophic methanogenesis with methanol provided as the substrate (3 biological replicates). (B) Summary of statistics from RNAseq libraries assembled from RNA extracted from Methanosarcina acetivorans cells grown with methanol provided as the electron donor and AQDS as the electron acceptor in the presence of BES (3 biological replicates). (C) The glmQLFit() function was used to fit the negative binomial generalized log-linear model for each tag. Fitted values for each gene and biological replicate (three AQDS/BES [AQB] and three methanol [Met] samples) are provided in the table below. For differential expression analysis of these genes, the glmQLFTest() function was used, which applies empirical Bayes quasi-likelihood (QL) F tests to the fitted values. Download Table S5, DOCX file, 0.9 MB (952.3KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All paired-end reads were then merged with FLASH (89), resulting in 40,312,494 ± 8,686,910 reads with an average read length of 145 bp. After merging the QC-filtered reads, SortMeRNA (90) was used to separate all rRNA reads from nonribosomal reads, and this resulted in 30,679,551 ± 6,275,120 mRNA reads.
Mapping of mRNA reads.
Trimmed and filtered mRNA reads from the triplicate samples for the two different culture conditions were mapped against the M. acetivorans strain C2A genome (NC_003552) downloaded from IMG/MER (img.jgi.doe.gov) using ArrayStar software (DNAStar). Analyses of reads from all three biological replicates for each condition demonstrated that the results were highly reproducible (Table S5 and Fig. S5).
Figure showing QuarterRoot mean deviance versus average log2 cpm (counts per million). The raw QL dispersion estimates are represented by black dots. The trend line is the mean dependent trend that was fitted to the raw QL dispersion estimates. The raw estimates were then squeezed toward this trend, called EB estimates (red dots). EBs were used in place of raw values for downstream hypothesis testing because the EB strategy reduces the uncertainty of the estimates and improves testing power. Download FIG S5, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reads were normalized and processed for differential expression studies using the edgeR package in Bioconductor (91), with AQDS/BES considered the experimental condition and methanol the control. Genes with P values of ≤0.05 were considered differentially expressed. Using these criteria, 1,188 genes were downregulated, 2,121 genes were not differentially expressed, and 1,182 genes were upregulated (Table S1).
Genome data analysis.
Gene sequence data for M. acetivorans C2A was acquired from the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov) or from GenBank at the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov). Initial analyses were done with tools available on the Integrated Microbial Genomes (IMG) website (img.jgi.doe.gov). Some protein domains were identified with NCBI conserved domain search (92) and Pfam search (93) functions. Transmembrane helices were predicted with TMpred (94), TMHMM (95), and HMMTOP (96), and signal peptides were identified with PSORTb v3.0.2 (97) and Signal P v4.1 (98).
Data availability.
Illumina sequence reads have been submitted to the SRA NCBI database under BioProject PRJNA509433 and Biosample SAMN10580613 (SRX5113605 to SRX5113610).
ACKNOWLEDGMENTS
This research was supported by the Army Research Office and was accomplished under grant W911NF-17-1-0345.
The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the Army Research Office or the U.S. Government.
Footnotes
Citation Holmes DE, Ueki T, Tang H-Y, Zhou J, Smith JA, Chaput G, Lovley DR. 2019. A membrane-bound cytochrome enables Methanosarcina acetivorans to conserve energy from extracellular electron transfer. mBio 10:e00789-19. https://doi.org/10.1128/mBio.00789-19.
REFERENCES
- 1.Vargas M, Kashefi K, Blunt-Harris EL, Lovley DR. 1998. Microbiological evidence for Fe(III) reduction on early Earth. Nature 395:65–67. doi: 10.1038/25720. [DOI] [PubMed] [Google Scholar]
- 2.Bond DR, Lovley DR. 2002. Reduction of Fe(III) oxide by methanogens in the presence and absence of extracellular quinones. Environ Microbiol 4:115–124. doi: 10.1046/j.1462-2920.2002.00279.x. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes FJ, de Bok FAM, Tuan DD, Stams AJM, Lettinga G, Field JA. 2002. Reduction of humic substances by halorespiring, sulphate-reducing and methanogenic microorganisms. Environ Microbiol 4:51–57. doi: 10.1046/j.1462-2920.2002.00258.x. [DOI] [PubMed] [Google Scholar]
- 4.Bodegom PM, Scholten JC, Stams AJ. 2004. Direct inhibition of methanogenesis by ferric iron. FEMS Microbiol Ecol 49:261–268. doi: 10.1016/j.femsec.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, Dong HL, Bishop ME, Wang HM, Agrawal A, Tritschler S, Eberl DD, Xie SC. 2011. Reduction of structural Fe(III) in nontronite by methanogen Methanosarcina barkeri. Geochim Cosmochim Acta 75:1057–1071. doi: 10.1016/j.gca.2010.11.009. [DOI] [Google Scholar]
- 6.Zhang J, Dong HL, Liu D, Fischer TB, Wang S, Huang LQ. 2012. Microbial reduction of Fe(III) in illite-smectite minerals by methanogen Methanosarcina mazei. Chem Geol 292:35–44. doi: 10.1016/j.chemgeo.2011.11.003. [DOI] [Google Scholar]
- 7.Zhang J, Dong HL, Zhao LD, McCarrick R, Agrawal A. 2014. Microbial reduction and precipitation of vanadium by mesophilic and thermophilic methanogens. Chem Geol 370:29–39. doi: 10.1016/j.chemgeo.2014.01.014. [DOI] [Google Scholar]
- 8.Sivan O, Shusta SS, Valentine DL. 2016. Methanogens rapidly transition from methane production to iron reduction. Geobiology 14:190–203. doi: 10.1111/gbi.12172. [DOI] [PubMed] [Google Scholar]
- 9.Holmes DE, Orelana R, Giloteaux L, Wang LY, Shrestha P, Williams K, Lovley DR, Rotaru AE. 2018. Potential for Methanosarcina to contribute to uranium reduction during acetate-promoted groundwater bioremediation. Microb Ecol 76:660–667. doi: 10.1007/s00248-018-1165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotaru AE, Shrestha PM, Liu F, Markovaite B, Chen S, Nevin KP, Lovley DR. 2014. Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri. Appl Environ Microbiol 80:4599–4605. doi: 10.1128/AEM.00895-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rotaru AE, Shrestha PM, Liu FH, Shrestha M, Shrestha D, Embree M, Zengler K, Wardman C, Nevin KP, Lovley DR. 2014. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci 7:408–415. doi: 10.1039/C3EE42189A. [DOI] [Google Scholar]
- 12.Holmes DE, Shrestha PM, Walker DJF, Dang Y, Nevin KP, Woodard TL, Lovley DR. 2017. Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in methanogenic rice paddy soils. Appl Environ Microbiol 83:e00223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glöckner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol 12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 14.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. 2015. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526:531. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 15.Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. 2015. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- 16.McGlynn SE. 2017. Energy metabolism during anaerobic methane oxidation in ANME Archaea. Microbes Environ 32:5–13. doi: 10.1264/jsme2.ME16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmers PHA, Welte CU, Koehorst JJ, Plugge CM, Jetten MSM, Stams A. 2017. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1. doi: 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai C, Leu AO, Xie G-J, Guo J, Feng Y, Zhao J-X, Tyson GW, Yuan Z, Hu S. 2018. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J 12:1929–1939. doi: 10.1038/s41396-018-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krukenberg V, Riedel D, Gruber-Vodicka HR, Buttigieg PL, Tegetmeyer HE, Boetius A, Wegener G. 2018. Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia. Environ Microbiol 20:1651–1666. doi: 10.1111/1462-2920.14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MSM, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi L, Dong H, Reguera G, Beyenal H, Lu A, Liu J, Yu H-Q, Fredrickson JK. 2016. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol 14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- 22.Ueki T, DiDonato LN, Lovley DR. 2017. Toward establishing minimum requirements for extracellular electron transfer in Geobacter sulfurreducens. FEMS Microbiol Lett 364:fnx093. [DOI] [PubMed] [Google Scholar]
- 23.Aklujkar M, Coppi MV, Leang C, Kim BC, Chavan MA, Perpetua LA, Giloteaux L, Liu A, Holmes DE. 2013. Proteins involved in electron transfer to Fe(III) and Mn(IV) oxides by Geobacter sulfurreducens and Geobacter uraniireducens. Microbiology 159:515–535. doi: 10.1099/mic.0.064089-0. [DOI] [PubMed] [Google Scholar]
- 24.Smith JA, Aklujkar M, Risso C, Leang C, Giloteaux L, Holmes DE. 2015. Mechanisms involved in Fe(III) respiration by the hyperthermophilic archaeon Ferroglobus placidus. Appl Environ Microbiol 81:2735–2744. doi: 10.1128/AEM.04038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manzella MP, Holmes DE, Rocheleau JM, Chung A, Reguera G, Kashefi K. 2015. The complete genome sequence and emendation of the hyperthermophilic, obligate iron-reducing archaeon “Geoglobus ahangari” strain 234T. Stand Genomic Sci 10:77. doi: 10.1186/s40793-015-0035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kletzin A, Heimerl T, Flechsler J, van Niftrik L, Rachel R, Klingl A. 2015. Cytochromes c in Archaea: distribution, maturation, cell architecture, and the special case of Ignicoccus hospitalis. Front Microbiol 6 doi: 10.3389/fmicb.2015.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A 94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buan N, Kulkarni G, Metcalf W. 2011. Genetic methods for Methanosarcina species. Methods Enzymol 494:23–42. doi: 10.1016/B978-0-12-385112-3.00002-0. [DOI] [PubMed] [Google Scholar]
- 29.Nayak DD, Metcalf WW. 2017. Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc Natl Acad Sci U S A 114:2976–2981. doi: 10.1073/pnas.1618596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soo VWC, McAnulty MJ, Tripathi A, Zhu F, Zhang L, Hatzakis E, Smith PB, Agrawal S, Nazem-Bokaee H, Gopalakrishnan S, Salis HM, Ferry JG, Maranas CD, Patterson AD, Wood TK. 2016. Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb Cell Fact 15:11. doi: 10.1186/s12934-015-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Z, Joshi P, Gorski CA, Ferry JG. 2018. A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nature Comm 9:1642. doi: 10.1038/s41467-018-04097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coppi MV, O’Neil RA, Leang C, Kaufmann F, Methe BA, Nevin KP, Woodard TL, Liu A, Lovley DR. 2007. Involvement of Geobacter sulfurreducens SfrAB in acetate metabolism rather than intracellular Fe(III) reduction. Microbiology 153:3572–3585. doi: 10.1099/mic.0.2007/006478-0. [DOI] [PubMed] [Google Scholar]
- 33.Buan NR, Metcalf WW. 2010. Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol Microbiol 75:843–853. doi: 10.1111/j.1365-2958.2009.06990.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim B-C, Leang C, Ding YR, Glaven RH, Coppi MV, Lovley DR. 2005. OmcF, a putative c-type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochrome in Geobacter sulfurreducens. J Bacteriol 187:4505–4513. doi: 10.1128/JB.187.13.4505-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim B-C, Postier BL, DiDonato RJ, Chaudhuri SK, Nevin KP, Lovley DR. 2008. Insights into genes involved in electricity generation in Geobacter sulfurreducens via whole genome microarray analysis of the OmcF-deficient mutant. Bioelectrochemistry 73:70–75. doi: 10.1016/j.bioelechem.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Jones LH, Pearson AR, Wilmot CM, Davidson VL. 2006. Mechanistic possibilities in MauG-dependent tryptophan tryptophylquinone biosynthesis. Biochem 45:13276–13283. doi: 10.1021/bi061497d. [DOI] [PubMed] [Google Scholar]
- 37.Pearson AR, Jones LH, Higgins L, Ashcroft AE, Wilmot CM, Davidson VL. 2003. Understanding quinone cofactor biogenesis in methylamine dehydrogenase through novel cofactor generation. Biochemistry 42:3224–3230. doi: 10.1021/bi027073a. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Graichen ME, Liu A, Pearson AR, Wilmot CM, Davidson VL. 2003. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry 42:7318–7325. doi: 10.1021/bi034243q. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann M, Seidel J, Einsle O. 2009. CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase. J Mol Biol 393:951–965. doi: 10.1016/j.jmb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Atack JM, Kelly DJ. 2007. Structure, mechanism and physiological roles of bacterial cytochrome c peroxidases. Adv Microb Physiol 52:73–106. doi: 10.1016/S0065-2911(06)52002-8. [DOI] [PubMed] [Google Scholar]
- 41.Schlegel K, Welte C, Deppenmeier U, Muller V. 2012. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J 279:4444–4452. doi: 10.1111/febs.12031. [DOI] [PubMed] [Google Scholar]
- 42.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, Ferry JG. 2006. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J Bacteriol 188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voordeckers JW, Kim BC, Izallalen M, Lovley DR. 2010. Role of Geobacter sulfurreducens outer surface c-type cytochromes in reduction of soil humic acid and anthraquinone-2,6-disulfonate. Appl Environ Microbiol 76:2371–2375. doi: 10.1128/AEM.02250-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bose A, Pritchett MA, Rother M, Metcalf WW. 2006. Differential regulation of the three methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. J Bacteriol 188:7274–7283. doi: 10.1128/JB.00535-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway de Macario E, Ferry JG, Jarrell KF, Jing H, Macario AJL, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res 12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bose A, Pritchett MA, Metcalf WW. 2008. Genetic analysis of the methanol- and methylamine-specific methyltransferase 2 genes of Methanosarcina acetivorans C2A. J Bacteriol 190:4017–4026. doi: 10.1128/JB.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M, Tomb JF, Ferry JG. 2011. Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol 11:165. doi: 10.1186/1471-2180-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulkarni G, Kridelbaugh DM, Guss AM, Metcalf WW. 2009. Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc Natl Acad Sci U S A 106:15915–15920. doi: 10.1073/pnas.0905914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abken HJ, Deppenmeier U. 1997. Purification and properties of an F420H2 dehydrogenase from Methanosarcina mazei Go1. FEMS Microbiol Lett 154:231–237. doi: 10.1016/S0378-1097(97)00330-3. [DOI] [Google Scholar]
- 50.Baumer S, Ide T, Jacobi C, Johann A, Gottschalk G, Deppenmeier U. 2000. The F420H2 dehydrogenase from Methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases. J Biol Chem 275:17968–17973. doi: 10.1074/jbc.M000650200. [DOI] [PubMed] [Google Scholar]
- 51.Welte C, Deppenmeier U. 2011. Re-evaluation of the function of the F-420 dehydrogenase in electron transport of Methanosarcina mazei. FEBS J 278:1277–1287. doi: 10.1111/j.1742-4658.2011.08048.x. [DOI] [PubMed] [Google Scholar]
- 52.Welte C, Deppenmeier U. 2014. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta Bioenerg 1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Suharti S, Wang M, de Vries S, Ferry JG. 2014. Characterization of the RnfB and RnfG subunits of the Rnf complex from the archaeon Methanosarcina acetivorans. PLoS One 9:e97966. doi: 10.1371/journal.pone.0097966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ide T, Baumer S, Deppenmeier U. 1999. Energy conservation by the H2:heterodisulfide oxidoreductase from Methanosarcina mazei Go1: identification of two proton-translocating segments. J Bacteriol 181:4076–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heiden S, Hedderich R, Setzke E, Thauer RK. 1993. Purification of a cytochrome b containing H2:heterodisulfide oxidoreductase complex from membranes of Methanosarcina barkeri. Eur J Biochem 213:529–535. doi: 10.1111/j.1432-1033.1993.tb17791.x. [DOI] [PubMed] [Google Scholar]
- 56.Heiden S, Hedderich R, Setzke E, Thauer RK. 1994. Purification of a 2-subunit cytochrome-b-containing heterodisulfide reductase from methanol-grown Methanosarcina barkeri. Eur J Biochem 221:855–861. doi: 10.1111/j.1432-1033.1994.tb18800.x. [DOI] [PubMed] [Google Scholar]
- 57.Hedderich R, Hamann N, Bennati M. 2005. Heterodisulfide reductase from methanogenic archaea: a new catalytic role for an iron-sulfur cluster. Biol Chem 386:961–970. doi: 10.1515/BC.2005.112. [DOI] [PubMed] [Google Scholar]
- 58.Abken HJ, Tietze M, Brodersen J, Baumer S, Beifuss U, Deppenmeier U. 1998. Isolation and characterization of methanophenazine and function of phenazines in membrane-bound electron transport of Methanosarcina mazei Gol. J Bacteriol 180:2027–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duszenko N, Buan NR. 2017. Physiological evidence for isopotential tunneling in the electron transport chain of methane-producing Archaea. Appl Environ Microbiol 83:e00950-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar VS, Ferry JG, Maranas CD. 2011. Metabolic reconstruction of the archaeon methanogen Methanosarcina acetivorans. BMC Syst Biol 5:28. doi: 10.1186/1752-0509-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benedict MN, Gonnerman MC, Metcalf WW, Price ND. 2012. Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A. J Bacteriol 194:855–865. doi: 10.1128/JB.06040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gottschalk G, Thauer RK. 2001. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim Biophys Acta 1505:28–36. doi: 10.1016/s0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- 63.Becher B, Muller V, Gottschalk G. 1992. N5-methyl-tetrahydromethanopterin:coenzyme M methyltransferase of Methanosarcina strain Go1 is an Na+-translocating membrane protein. J Bacteriol 174:7656–7660. doi: 10.1128/jb.174.23.7656-7660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lienard T, Becher B, Marschall M, Bowien S, Gottschalk G. 1996. Sodium ion translocation by N5-methyltetrahydromethanopterin: coenzyme M methyltransferase from Methanosarcina mazei Go1 reconstituted in ether lipid liposomes. Eur J Biochem 239:857–864. doi: 10.1111/j.1432-1033.1996.0857u.x. [DOI] [PubMed] [Google Scholar]
- 65.Buckel W, Thauer RK. 2018. Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: a historical review. Front Microbiol 9:401. doi: 10.3389/fmicb.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshikawa S, Shimada A. 2015. Reaction mechanism of cytochrome c oxidase. Chem Rev 115:1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 67.Morgado L, Dantas JM, Bruix M, Londer YY, Salgueiro CA. 2012. Fine tuning of redox networks on multiheme cytochromes from Geobacter sulfurreducens drives physiological electron/proton energy transduction. Bioinorg Chem Appl 2012:298739. doi: 10.1155/2012/298739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Louro RO, Catarino T, Turner DL, Picarra-Pereira MA, Pacheco I, LeGall J, Xavier AV. 1998. Functional and mechanistic studies of cytochrome c3 from Desulfovibrio gigas: thermodynamics of a “proton thruster. Biochemistry 37:15808–15815. doi: 10.1021/bi981505t. [DOI] [PubMed] [Google Scholar]
- 69.Morgado L, Bruix M, Pessanha M, Londer YY, Salgueiro CA. 2010. Thermodynamic characterization of a triheme cytochrome family from Geobacter sulfurreducens reveals mechanistic and functional diversity. Biophys J 99:293–301. doi: 10.1016/j.bpj.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gunner MR, Mao J, Song Y, Kim J. 2006. Factors influencing the energetics of electron and proton transfers in proteins: what can be learned from calculations. Biochim Biophys Acta 1757:942–968. doi: 10.1016/j.bbabio.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dantas JM, Morgado L, Aklujkar M, Bruix M, Londer YY, Schiffer M, Pokkuluri PR, Salgueiro CA. 2015. Rational engineering of Geobacter sulfurreducens electron transfer components: a foundation for building improved Geobacter-based bioelectrochemical technologies. Front Microbiol 6:752. doi: 10.3389/fmicb.2015.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlegel K, Leone V, Faraldo-Gómez JD, Müller V. 2012. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc Natl Acad Sci U S A 109:947–952. doi: 10.1073/pnas.1115796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jasso-Chavez R, Diaz-Perez C, Rodriguez-Zavala JS, Ferry JG. 2017. Functional role of MrpA in the MrpABCDEFG Na+/H+ antiporter complex from the archaeon Methanosarcina acetivorans. J Bacteriol 199:e00662-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klupfel L, Piepenbrock A, Kappler A, Sander M. 2014. Humic substances as fully regenerable electron acceptors in recurrently anoxic environments. Nature Geosci 7:195–200. doi: 10.1038/ngeo2084. [DOI] [Google Scholar]
- 75.Thamdrup B. 2000. Bacterial manganese and iron reduction in aquatic sediments. Adv Microb Ecol 16:41–84. doi: 10.1007/978-1-4615-4187-5_2. [DOI] [Google Scholar]
- 76.Lovley DR. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev 55:259–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keller J, Weisenhorn P, Megonigal J. 2009. Humic acids as electron acceptors in wetland decomposition. Soil Biol Biochem 41:1518–1522. doi: 10.1016/j.soilbio.2009.04.008. [DOI] [Google Scholar]
- 78.Roden EE, Wetzel RG. 2003. Competition between Fe(III)-reducing and methanogenic bacteria for acetate in iron-rich freshwater sediments. Microb Ecol 45:252–258. doi: 10.1007/s00248-002-1037-9. [DOI] [PubMed] [Google Scholar]
- 79.Lovley DR, Phillips EJP. 1987. Competitive mechanisms for inhibition of sulfate reduction and methane production in the zone of ferric iron reduction in sediments. Appl Environ Microbiol 53:2636–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cervantes FJ, Velde S, Lettinga G, Field JA. 2000. Competition between methanogenesis and quinone respiration for ecologically important substrates in anaerobic consortia. FEMS Microbiol Ecol 34:161–171. doi: 10.1111/j.1574-6941.2000.tb00766.x. [DOI] [PubMed] [Google Scholar]
- 81.Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol 49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- 82.Pritchett MA, Zhang JK, Metcalf WW. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl Environ Microbiol 70:1425–1433. doi: 10.1128/aem.70.3.1425-1433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lovley DR, Phillips EJ. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54:1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mouser PJ, Holmes DE, Perpetua LA, DiDonato R, Postier B, Liu A, Lovley DR. 2009. Quantifying expression of Geobacter spp. oxidative stress genes in pure culture and during in situ uranium bioremediation. ISME J 3:454–465. doi: 10.1038/ismej.2008.126. [DOI] [PubMed] [Google Scholar]
- 85.Holmes DE, Giloteaux L, Orellana R, Williams KH, Robbins MJ, Lovley DR. 2014. Methane production from protozoan endosymbionts following stimulation of microbial metabolism within subsurface sediments. Front Microbiol 5:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445–448. doi: 10.1038/382445a0. [DOI] [Google Scholar]
- 87.Holmes DE, Risso C, Smith JA, Lovley DR. 2012. Genome-scale analysis of anaerobic benzoate and phenol metabolism in the hyperthermophilic archaeon Ferroglobus placidus. ISME J 6:146–157. doi: 10.1038/ismej.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kopylova E, Noe L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 91.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hofmann K, Stoffel W. 1993. TMBASE: a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler 374:166. [Google Scholar]
- 95.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 96.Tusnady GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 97.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methane concentrations detected in Methanosarcina acetivorans cultures grown with methanol as the electron donor and AQDS as the electron acceptor in the presence of the methanogenic inhibitor BES. Each point and error bars represent the average and standard deviation of triplicate measurements. Download FIG S1, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Methanol (A), methane (B), AQDS concentrations (C), and cell numbers (D) in control cultures. All points and error bars represent the average and standard deviation for triplicate samples. For methanogenic conditions, Methanosarcina acetivorans cells were grown with methanol as the substrate. For the cells/BES/methanol condition, methanogenesis was inhibited with BES, and no electron acceptor (AQDS) was provided. For the cells/BES/AQDS condition, cells were provided with electron acceptor (AQDS), methanogenesis was inhibited with BES, and no electron donor (methanol) was provided. For the no cells/BES/AQDS condition, BES and AQDS were added to the medium but no inoculum was added. Download FIG S2, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Differential expression of genes from M. acetivorans cells grown with methanol provided as electron donor and AQDS (with BES) as electron acceptor (experimental condition) versus cells grown via methylotrophic methanogenesis (control condition). Values were obtained with EdgeR software. Only genes with P values of ≥0.05 are included in this table. Positive log fold change values indicate that the gene was more highly transcribed in AQDS-respiring cells; negative log fold change values indicate that the gene was more highly transcribed in cells grown via methylotrophic methanogenesis. (B) Number of growth-related genes that were more highly expressed in M. acetivorans cells grown with methanol as the electron donor and AQDS as the electron acceptor in the presence of BES (experimental condition) or in M. acetivorans cells grown via methylotrophic methanogenesis (control condition). These results are a summary of data presented in more detail in Table S1A. Download Table S1, XLSX file, 0.3 MB (366.2KB, xlsx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential expression of genes coding for proteins from the oxidative branch of the methylotrophic methanogenesis pathway and methyl-coenzyme M reductase (Mcr) in M. acetivorans cells grown with methanol and AQDS in the presence of BES or cells grown via methanogenesis with methanol as the substrate. Negative values indicate that genes were more significantly expressed in methanogenic cells. Genes were only considered differentially expressed if the P value and FDR (False Discovery Rate) were ≤0.05. NS: no significant difference in read abundance between conditions Download Table S2, DOCX file, 0.1 MB (131.7KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential expression of genes coding for proteins from the proton-translocating F420:methanophenazine oxidoreductase (Fpo) complex in M. acetivorans cells grown with methanol as the electron donor and AQDS (in the presence of BES) as the acceptor versus cells grown via methylotrophic methanogenesis with methanol as the substrate. Negative values indicate that genes were more significantly expressed in methanogenic cells. Genes were only considered differentially expressed if the P value and FDR (false discovery rate) were <0.05. Download Table S3, DOCX file, 0.1 MB (108.7KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reverse-transcription PCR with primers targeting two other genes from the Rnf complex in the Methanosarcina acetivorans genome, MA0659 and MA0664. RNA was extracted from cells of the MA0658 deletion mutant strain of M. acetivorans grown with acetate provided as the substrate for growth. Download FIG S3, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Steps involved in construction of the mutant strain ΔMA0658. The coding region from amino acid residues 54Cys to 457Arg was deleted. P1, P2, and P3 were primers utilized for verification of the mutant construct. Primer sequences used for construction and verification of this mutant strain and other strains included in this study are provided in Table S4. Download FIG S4, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used to construct various deletion mutant strains (restriction sites are underlined). Genetically recombinant plasmids were checked for accuracy by PCR with verification primers. Download Table S4, DOCX file, 0.1 MB (124.3KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Summary of statistics from RNAseq libraries assembled from RNA extracted from Methanosarcina acetivorans cells grown via methylotrophic methanogenesis with methanol provided as the substrate (3 biological replicates). (B) Summary of statistics from RNAseq libraries assembled from RNA extracted from Methanosarcina acetivorans cells grown with methanol provided as the electron donor and AQDS as the electron acceptor in the presence of BES (3 biological replicates). (C) The glmQLFit() function was used to fit the negative binomial generalized log-linear model for each tag. Fitted values for each gene and biological replicate (three AQDS/BES [AQB] and three methanol [Met] samples) are provided in the table below. For differential expression analysis of these genes, the glmQLFTest() function was used, which applies empirical Bayes quasi-likelihood (QL) F tests to the fitted values. Download Table S5, DOCX file, 0.9 MB (952.3KB, docx) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Figure showing QuarterRoot mean deviance versus average log2 cpm (counts per million). The raw QL dispersion estimates are represented by black dots. The trend line is the mean dependent trend that was fitted to the raw QL dispersion estimates. The raw estimates were then squeezed toward this trend, called EB estimates (red dots). EBs were used in place of raw values for downstream hypothesis testing because the EB strategy reduces the uncertainty of the estimates and improves testing power. Download FIG S5, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2019 Holmes et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Illumina sequence reads have been submitted to the SRA NCBI database under BioProject PRJNA509433 and Biosample SAMN10580613 (SRX5113605 to SRX5113610).