Abstract
Background
The adaptation of a marine prokaryote to live in freshwater environments or vice versa is generally believed to be an unusual and evolutionary demanding process. However, the reasons are not obvious given the similarity of both kinds of habitats.
Results
We have found major differences at the level of the predicted metaproteomes of marine and freshwater habitats with more acidic values of the isoelectric points (pI) in marine microbes. Furthermore, by comparing genomes of marine-freshwater phylogenetic relatives, we have found higher pI values (basic shift) in the freshwater ones. This difference was sharper in secreted > cytoplasmic > membrane proteins. The changes are concentrated on the surface of soluble proteins. It is also detectable at the level of total amino acid composition and involves similarly core and flexible genome- encoded proteins.
Conclusions
The marked changes at the level of protein amino acid composition and pI provide a tool to predict the preferred habitat of a culture or a metagenome-assembled genome (MAG). The exact physiological explanation for such variations in the pIs and electrostatic surface potentials is not known yet. However, these changes might reflect differences in membrane bioenergetics derived from the absence of significant Na+ concentrations in most freshwater habitats. In any case, the changes in amino acid composition in most proteins imply that a long evolutionary time is required to adapt from one type of habitat to the other.
Electronic supplementary material
The online version of this article (10.1186/s40168-019-0731-5) contains supplementary material, which is available to authorized users.
Keywords: Isoelectric point, Marine-freshwater transition, Metaproteome, Basic shift, Electrostatic surface potential
Background
One classic conundrum of microbiology, or actually of biology at large, is the marked borderline that exists between freshwater and marine habitats [1]. Although aquatic environments share many features and ecological parameters, the microbes found throughout both systems have different characteristics at several levels. First, although the major microbial taxa have representation in both, the proportions of each are very different. For instance, the phylum Actinobacteria and the class Betaproteobacteria are two notorious examples of taxa that are more abundant in freshwater [2–4], while classes Alphaproteobacteria and Gammaproteobacteria are more abundant in marine waters [4, 5]. Second, although it might be an artifact of lack of coverage, there are lower-level taxa that appear to be altogether absent in one of the groups of habitats regardless of how abundant they are in the other. Some relevant examples are acI Actinobacteria [6], Limnohabitans, and Polynucleobacter Betaproteobacteria [7, 8], which dominate freshwater but are absent in marine habitats. LD12 Alphaproteobacteria, including Ca. Fonsibacter [9–11], have only been found also in freshwater and estuarine systems, although their relatives in the SAR11 clade are widespread in marine habitats. On the other hand, Prochlorococcus species [12] or Gammaproteobacteria groups such as SAR86 [13] are found only in marine ecosystems. The explanation for such differences is not obvious considering the similarity of aquatic pelagic habitats aside from the salinity and the influence of terrestrially derived organic matter [1]. On the other hand, there are reports of multiple marine clades being detected, albeit in small numbers, in freshwater habitats [14–16], and the opposite is true for marine regions neighboring the continents, particularly near large estuaries like the Amazon on the Atlantic coast of Brazil [17] or the Baltic Sea [18, 19]. Thus, the differences cannot be explained by physical isolation. Still, excluding some microbes that can survive and remain rare, such as Escherichia coli or Vibrio cholerae, there is no known example of microbes of the same species (with > 95% average nucleotide identity, ANI) being found in both types of aquatic environments.
One problem to understand the real differences between these two kinds of aquatic systems is the enormous diversity within each of them. Particularly, freshwater lakes vary in their trophic status (from oligotrophic to highly eutrophic) and other environmental parameters, all of them having profound implications in the taxonomic composition. Recently, we were involved in the first metagenomic study of Lake Baikal, Siberia, Russia [20]. This is the largest and deepest (max. 1600 m, average 1300 m) lake on Earth [21], ultraoligotrophic and with relatively little influence from terrestrial sources (all features that make it similar to marine off-shore waters) while having very low salt content (dominated by Ca2+ and HCO3-, being particularly low in Na+ and K+) [22–24]. Interestingly, we found some groups with close relatives among bona fide marine lineages, including the first freshwater Pelagibacter-like (SAR11 clade) metagenome-assembled genome (MAG) within the typically marine clade I [20]. In previous studies, we compared the pI patterns of this SAR11 MAG [20] and a freshwater Synechococcus [25] with their marine closest relatives and, in spite of their relative phylogenetic proximity, noticed significant differences in the global values of their predicted proteome pIs.
The variations in the global proteome pI plots of cells depend on the amino acid overall charge and have important implications on protein structure and properties [26]. It is generally accepted that prokaryotic genomes have a bimodal shape with two maxima [27], one at acidic pH corresponding largely to dissolved proteins (cytoplasmic or secreted) and one at basic pH of the membrane proteins that have a basic (positively charged) domain intracellularly to facilitate the generation of the proton motive force. In between these two peaks, there is a minimum at ca. neutral values that correspond to the intracellular pH at which proteins of equivalent pI value would be the least soluble. In salt-in halophiles, the alkaline peak nearly disappears because they have a large amount of intracellular K+ [28]. The adaptation to hypersaline environments (much more saline than seawater) leads to these changes in their inhabitants (halophiles) and has been known for long [29]. Thus, hyperhalophiles such as Haloquadratum walsbyi or Salinibacter ruber have their proteomes markedly displaced to acidic values [28]. However, marine bacteria and archaea are expected to be salt-out strategists, i.e., that keep most inorganic salts (particularly Na+) outside the cell, maintaining a relatively salt-free cytoplasm [28].
The large change in this value detected in the freshwater microbes mentioned above made us wonder if it could be a general phenomenon and what could be the underlying reason for such a broad deviation. There is a current database with pI calculations and amino acid properties for more than > 5000 bacteria and archaea [30], and prior studies identified a correlation between salinity and pIs of microbes [31]. However, there are no studies comparing bona fide freshwater and salt-adapted microbial predicted proteomes including hypothetical proteins derived from metagenomes. Here, filling this gap, we have analyzed in detail some specific cases when closely related microbes by whole genome comparisons have been retrieved from marine and freshwater habitats; furthermore, there is metagenomic evidence (by recruitment of metagenomic reads) showing that they are actually adapted to live in either one or the other environment. We have also dissected these pI values depending on the localization of the proteins and used available three-dimensional models to determine whether there was a preferential location of the charges. Our data confirm that indeed the predicted proteins, regardless of location in the cell, accumulate fewer negative charges in prokaryotes coming from freshwater environments, corresponding to a significant deviation in the amino acidic composition. This fact, among other consequences, implies a large sequence variation that requires long evolutionary times to carry out the transition between marine and freshwater habitats or vice versa.
Results
Global pIs of metaproteomes from different aquatic habitats
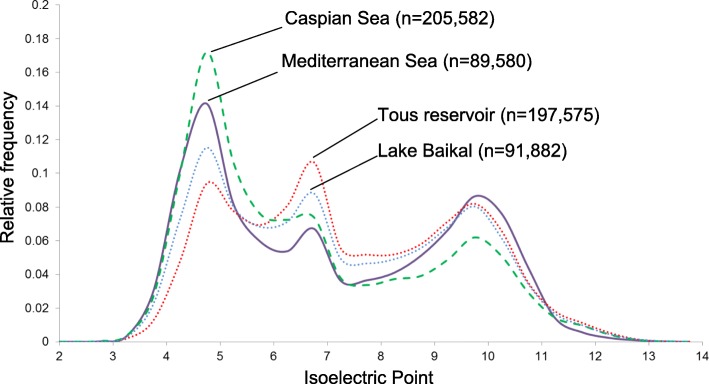
A global metagenomic approach was first used to assess if the changes in pI could be detected at the level of the microbial community as a whole. Specifically, we used metagenomic datasets from freshwater (Tous reservoir and Lake Baikal), brackish (Caspian Sea), and marine (Mediterranean Sea) environments from similar depths, which were assembled and annotated, taking all predicted and hypothetical proteins from contigs > 5 kb and obtaining sets of more than 85,000 proteins for each environment (Fig. 1). Interestingly, the three highest peaks were observed at nearly identical pI values (4.5, 6.8, and 9.8) for the different aquatic habitats in spite of the large differences in salinity (ca. 0.05 in freshwater habitats, 1.2 in the Caspian, and 3.8% in the Mediterranean) and community structure [20, 32, 33]. A major difference was observed in the acidic peak, with brackish and marine environments having higher relative frequencies of proteins with lower pIs, compared to their freshwater counterparts. On the other hand, a higher peak of neutral pIs was observable in freshwater systems. Finally, the segment of the plot corresponding to basic pIs (8–9.5 and 11–14) was also higher in freshwaters, although the relative frequencies of pIs from 9.5–11 were a little higher in the Mediterranean Sea. However, these changes could just reflect the variation in the community structure, i.e., very different taxonomic composition depending on the habitat. Therefore, we have analyzed the pI features of the proteomes of related microbes that are bona fide inhabitants of one or the other kind of habitat.
Fig. 1.
Metaproteome pI versus relative frequency plot of marine (Mediterranean Sea, 30 m deep), brackish (Caspian Sea, 15 m), and freshwater (Tous reservoir, 12–25 m, and Lake Baikal, 20 m) habitats. N indicates the number of proteins retrieved from each metaproteome and used in this comparison
Overall pI patterns within phyla
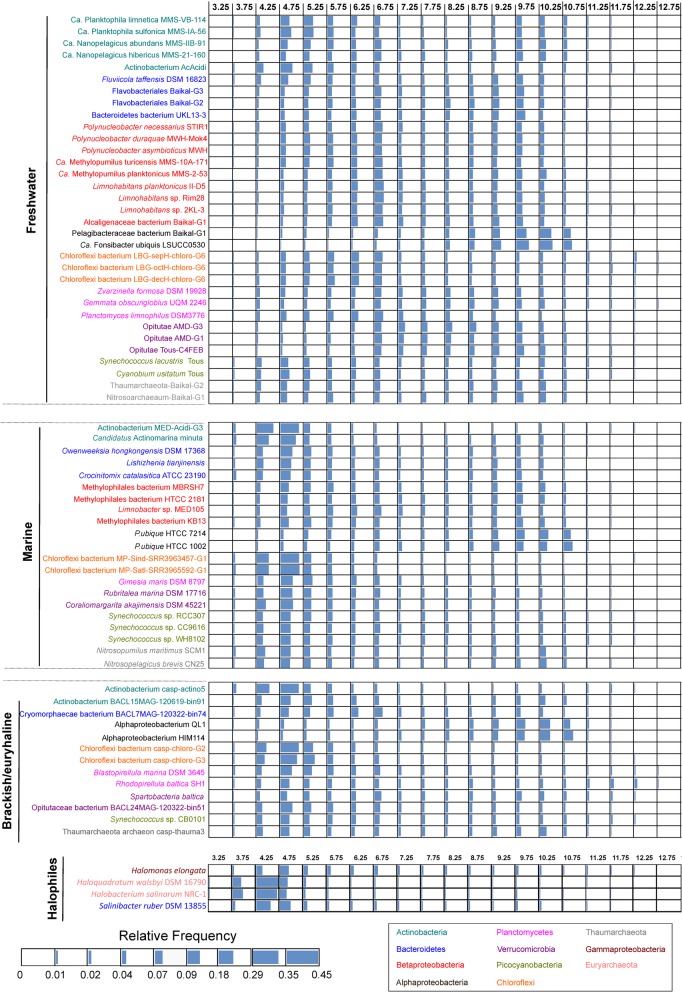
To assess if the differences in the global pI distributions were due to the habitat or taxonomic bias, we selected a total of 71 prokaryotes from public databases and compared their overall pI values (Fig. 2). We used examples of bona fide freshwater, brackish, and marine microbes, some of them retrieved as MAGs (> 70% of completeness) from the environments compared in Fig. 1, and others as pure cultures. We selected representatives from class Alphaproteobacteria (SAR11, Roseobacter, and Rhodospirillaceae), order Betaproteobacteriales, Chloroflexi, Planctomycetes, Verrucomicrobia, Cyanobacteria (Synechococcus/Cyanobium), phyla Actinobacteria, Bacteroidetes, and Thaumarchaeota (Fig. 2 and Additional file 1: Figsure S1–S6, see Additional file 2 for extra information on each selected microbe). The Bray-Curtis distances obtained between relative frequencies of pIs and the statistical analysis conducted with PERMANOVA allowed us to compare both habitat and taxonomic distance effects on the relative frequencies of pIs in the dataset of the selected microbes (see the “Methods” section and Additional file 2). We obtained an R2 of 0.336 and 0.45 for habitat and phyla variables respectively, confirming that there is a taxonomic bias, with an important influence in the pIs, as happens in SAR11 (see Additional file 1: Figure S1 and Additional file 2). However, both variables significantly explained the differences in pI. The principal component analysis (PCA) plot also showed the effect of taxonomic bias (SAR11) and a generally observed different distribution of freshwater and salt-adapted microbes (Additional file 2). In all cases (habitat-specific metaproteomes and phyla by phyla species comparison), we noticed differences between freshwater, brackish, and marine predicted proteome pI patterns (Figs. 1 and 2 and Additional file 1 Figures. S1–S6). Thus, these plots could help to identify in a relatively straightforward manner the salinity of origin of the different microbes without prior knowledge.
Fig. 2.
Whole proteome isoelectric point versus relative frequency plot of freshwater, marine, brackish, and halophilic selected prokaryotes. Genomes are color-coded according to their taxonomic affiliation and arranged according to their origin. Bar plots represent the increasing relative frequencies of each isoelectric point value (pI)
First, halophiles present a single acid peak at low pIs (highest among the microbes compared). Second, brackish and marine species tend to show bimodal patterns and display a higher peak of acidic proteins compared to freshwater ones. One exception was SAR11 (Additional file 1: Figure S1), which always presented a higher peak of basic proteins independently of the origin. Third, it is particularly remarkable the high peak of neutral proteins (with pIs ranging from 6 to 8) in some freshwater species, while this peak is very low or absent in salt-adapted species. This was the case of Flavobacteriales, Betaproteobacteriales, Verrucomicrobia, Planctomycetes, or Thaumarchaeota (Additional file 1: Figures. S2, S5, and S6). Our data could facilitate the prediction of a microbe natural ecosystem and it could be established as a rule of thumb to infer the preferred habitat of microbes, particularly useful in mixed systems such as estuaries.
Pairwise comparisons of close phylogenetic neighbors
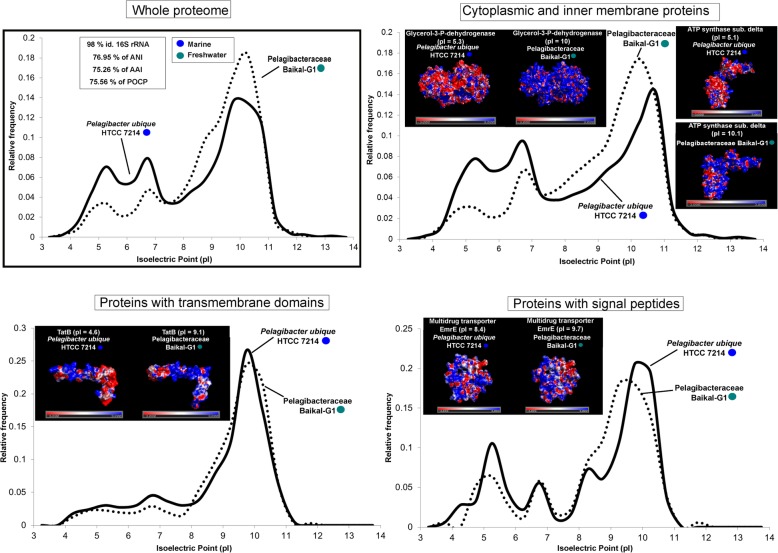
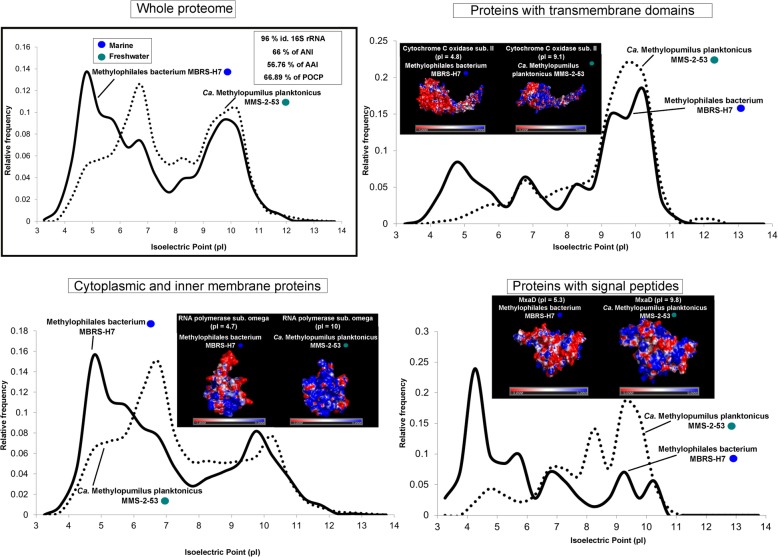
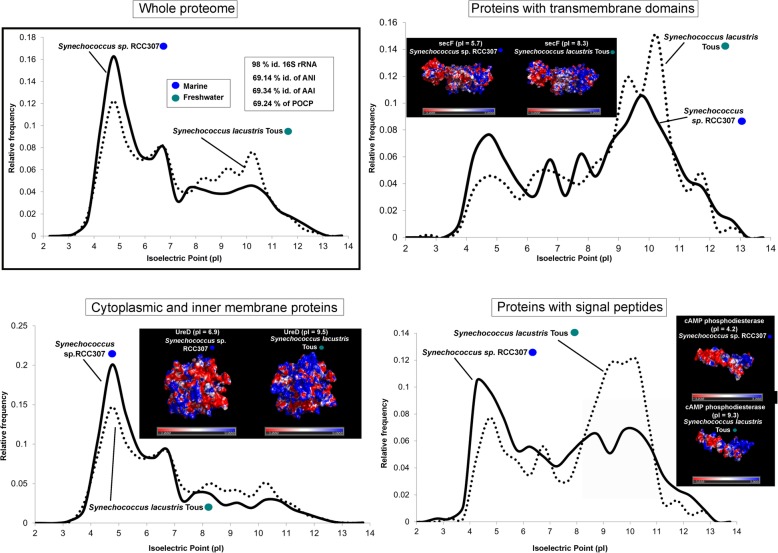
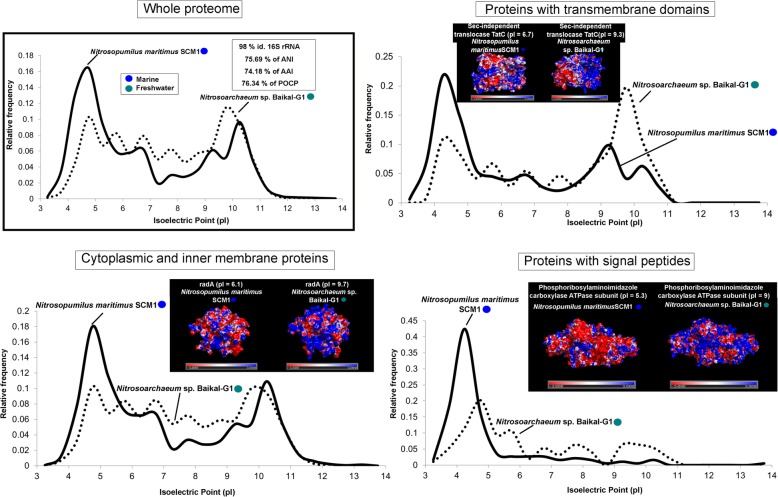
We selected pairs of microbes that are as closely associated phylogenetic neighbors as available (same family or genera, whenever possible), but one of them is freshwater and the other marine inhabitants. In these cases, the effect of the taxonomic distance was reduced to the minimum presently available in databases. Thus, we could compare two species from the family Nitrosopumilaceae (Nitrosoarchaeum sp. Baikal-G1, MAG, vs Nitrosopumilus maritimus SCM1, culture), two SAR11 members from family Pelagibacteraceae (Pelagibacteraceae bacterium Baikal-G1, MAG, vs Pelagibacter ubique HTCC7214, culture), two picocyanobacteria from the order Synechococcales (Synechococcus sp. RCC307, culture, vs Synechococcus lacustris Tous, culture), and finally two species from the family Methylophylaceae (Methylophilales bacterium MBRS-H7, culture vs Methylopumilus planktonicus MMS-2-53, culture). We chose pairs of microbes with similar cell and genome sizes displaying similar metabolic and ecological roles in the environment to reduce to the minimum other factors. Values of average nucleotide identity (ANI), average amino acid identity (AAI), 16S rRNA gene identity, and percentage of conserved proteins (POCP) were also calculated for each pair. We divided the proteome into three categories: cytoplasmic and inner membrane proteins that are submitted to the cytoplasmic environment, proteins with transmembrane domains, and secreted (with signal peptide), i.e., exposed to the extracellular environment. The average pIs were also calculated for these three categories. The differences between freshwater and marine microbes appear clear at all levels (Figs. 3, 4, 5, and 6). We found in these pairs of microbes whose ANI varied between 66 and 77% that the AAI values were similar (when not lower) than the nucleotide identity. This is contrary to the expectations in comparisons of phylogenetic neighbors that tend to have more similarity at the level of amino acids than nucleotides [34, 35]. This also indicated a major shift in the composition of amino acids of the core genome (shared genes, see below).
Fig. 3.
Proteome (whole and at different locations, secreted, cytoplasmic, and transmembrane) pI values versus relative frequency of P. ubique HTCC7214 (marine, culture) and Pelagibacteraceae bacterium Baikal-G1 (freshwater, MAG). Insets show tridimensional electrostatic surface potential 3D models of individual proteins selected for each location (secreted, cytoplasmic, and transmembrane). The potentials were colored from − 3 kcal mol−1(red) to + 3 kcal mol−1 (blue). Values of ANI, AAI, POCP, and percentage of identity of 16S rRNA for each couple of microbes are also indicated
Fig. 4.
Proteome (whole and at different locations, secreted, cytoplasmic, and transmembrane) pI values versus relative frequency of Ca. Methylopumilus planktonicus MMS-2-53 (freshwater, culture) and Methylophilales bacterium MBRS-H7 (marine, culture). Insets show tridimensional electrostatic surface potential 3D models of individual proteins selected for each location. The potentials were colored from − 3 kcal mol−1(red) to + 3 kcal mol−1 (blue). Values of ANI, AAI, POCP, and percentage of identity of 16S rRNA for each couple of microbes are also indicated
Fig. 5.
Proteome (whole and at different locations, secreted, cytoplasmic, and transmembrane) pI values versus relative frequency of Synechococcus lacustris Tous (freshwater, culture) and Synechococcus sp. RCC307 (marine, culture). Insets show tridimensional electrostatic surface potential 3D models of individual proteins selected for each location. The potentials were colored from − 3 kcal mol−1 (red) to + 3 kcal mol−1 (blue). Values of ANI, AAI, POCP, and percentage of identity of 16S rRNA for each couple of microbes are also indicated
Fig. 6.
Proteome (whole and at different locations, secreted, cytoplasmic, and transmembrane) pI values versus relative frequency of Nitrosopumilus maritimus SCM1 (marine, culture) and Nitrosoarchaeum sp. Baikal-G1 (freshwater, MAG). Insets show tridimensional electrostatic surface potential 3D models of individual proteins selected for each location (secreted, cytoplasmic, and transmembrane). The potentials were colored from − 3 kcal mol−1(red) to + 3 kcal mol−1 (blue). Values of ANI, AAI, POCP, and percentage of identity of 16S rRNA for each couple of microbes are also indicated
Changes in the amino acid composition
The variations in the amino acid composition (expressed as Mole% for each microbe) in the compared pairs are noteworthy (Additional file 1: Figure S7). A general trend that is conserved in the four cases is the higher percentage of acid amino acids (aspartic and glutamic) in the marine representatives (from 0.6 to 1.4% higher). This is in agreement with the overall higher peak of acidic proteome pIs in these salt-adapted microbes. Actually, the global percentage of charged amino acids comprising both acid and basic types (aspartic and glutamic acids, lysine, histidine, and arginine) is higher in marine microbes, i.e., the acid increase in marine is more accentuated than the basic amino acids in freshwater representatives. However, three out of four freshwater microbes analyzed here display higher percentages of basic amino acids compared with their marine relatives. The only exception observed is the case of S. lacustris Tous, which presents a nearly identical percentage of basic amino acids when compared with its marine relative Synechococcus RCC307. Nevertheless, there are noticeable differences, for instance, S. lacustris shows a higher percentage of lysine (K) residues on its whole proteome, while RCC307 compensates this decrease in lysine by having more arginine (R) residues, resulting in practically the same percentage of total basic amino acids in both genomes. On the other hand, the higher percentage of basic residues in Methylophilales, Pelagibacteraceae, and archaeal Nitrosopumilaceae genomes is significant. Nitrosoarchaeum sp. Baikal-G1 presents a higher percentage of all three basic amino acids, compared to its marine relative Nitrosopumilus maritimus SCM1. Ca. Methylopumilus planktonicus MMS-2-53 displays a higher percentage of arginine and histidine, but slightly lower lysine compared to its marine relative. Hence, a global picture contemplating these variations in the overall pI plots, electrostatic surface potential of proteins, and percentages of basic/acid amino acids could help predicting the freshwater or salt-adapted origin of a novel microbe of unknown source.
Changes are located at the surface of proteins
In salt-in halophiles, the changes in aminoacidic composition are concentrated at the level of the protein surface producing an even sharper change in the electrostatic surface potential [36]. Therefore, we analyzed the case of some individual protein homologs for which there were tridimensional models to assess if the difference in the predicted global pI was concentrated at the level of the protein surface. We chose homologs with a marked difference in the global pI between freshwater and marine representatives, and only proteins with well-established tertiary structures (retrieved from SWISS-MODEL) were considered. Indeed, we observed a higher accumulation of negative charges (acid amino acids) on the surface of proteins from marine microbes, while positive electrostatic potentials (with more basic amino acids) are seen in freshwater microbes (Figs. 3, 4, 5, and 6). We are showing only some individual protein examples for each category/prokaryote; however, many other protein homologs presented substantial differences in their pIs (see Additional file 3). Thus, it appears that the electrostatic surface potential between pairs of homologs differs significantly between marine and freshwater species. These differences are even more evident when introducing halophiles into the comparison (Additional file 1: Figure S8). These significant variations were apparent in all three categories: cytoplasmic, membrane, and secreted proteins. However, the differences in electrostatic surface potential were more marked in the secreted > cytoplasmic > membrane, which indicates a more radical change in the extracellular than in the intracellular environment between freshwater and marine microbes.
Pan-genome pI distribution
We also calculated the pIs for each category (core and flexible proteins) to assess if the change in the global proteome might be due to variations affecting homologous proteins as those shown above or it could be an extreme consequence of a change in the differential gene pool present in freshwater or marine microbes, i.e., it could be due to the flexible or core genes [37, 38]. Therefore, we analyzed the global pI plots of both components in the four close relative comparisons (Additional file 1: Figure S9). Indeed, marine representatives always had a higher peak of acidic proteins, independently if the coding genes belong to the core or flexible genome component. Actually, only the case of Ca. Methylopumilus displayed a different pattern between core and flexible genes (both being more displaced towards neutrality-basicity compared to their marine homolog). In the other cases, there were no differences altering significantly the patterns observed in whole genomes. The flexible genome had in three of the examples a higher basic peak, probably reflecting enrichment in membrane proteins, largely transporters, and sensors that are typical components of the flexible genome (involved in habitat-adaptation and niche-occupancy) [39].
Discussion
Previous studies showed that bacteria and archaea, independently of their origin, presented a bimodal pI pattern, while eukaryotes showed a trimodal pattern [27]. Others have shown a multi-modal distribution picture of the pIs in different organisms as a consequence of the chemical properties of the different amino acids (rather than sequence evolution) [26] or as a result of discrete pKr values for the amino acids [40]. Our observations indicate that the pI pattern varies significantly among microbes, having cases of unimodal (salt-in halophiles) and bimodal patterns (most of the microbes analyzed here) to multimodal (Thaumachaeaota, Verrucomicrobia, Planctomycetes, or Betaproteobacteriales).
The pI of a certain protein is a major indicator of the properties of the macromolecule. It determines the water solubility of the protein as well as the interactions with the chemical environment. Typically, intracellular pH of microbes (including alkaliphilic or acidophilic ones) is near neutrality [41, 42], and proteins are less soluble at pH values near their pI. Thus, cytoplasmic and secreted proteins, that mostly work in a soluble form, tend to have pIs far from neutrality, mostly acidic [41]. On the other hand, membrane proteins only interact with water in their exposed domains and tend to have alkaline pIs to compensate for the positive charge outside of the membrane created by the proton gradient [43]. The consistent difference that we have detected between freshwater and marine microbes indicates a significant change in one of these two aspects of cell biology, either the intracellular pH or the bioenergetics of the cell (perhaps both). Another factor that likely interacts with the protein charge is the presence of other solutes in the water phase at both sides of the membrane. Most cells maintain a significant concentration of K+ cations inside while keeping Na+ outside. Typically, marine microbes would need higher intracellular potassium concentrations in order to compensate the sodium ions abundant in the extracellular environment. Depending on the cation concentration, soluble proteins need to have more or less negative charges to maintain a proper hydration sphere [44]. This is why halophiles with salt-in strategies must have very acidic soluble proteins [28]. Freshwater must impose limitations to the accessibility to the main cellular cations, particularly Na+, that might be limiting in salt-poor environments like Lake Baikal [22]. These conditions could lead to adaptations consisting of less intracellular potassium. It is thus not surprising that a less acidic proteome might be favorable for freshwater microbes. Other predicted physiological differences between the two types of aquatic microbes include the preference for H+- over Na+-based electron or nutrient transport mechanisms [41], but this is unlikely to have effects over the global proteome as described here.
The kind of analysis that we have done in this work has previously been hampered by the lack of close relatives specialized in living in either freshwater or marine habitats. However, provided that most of the microbial examples that we have used are difficult to retrieve in pure culture and are only available as genomes, it is not feasible to carry out physiological or biophysical experiments that could clarify the meaning of the patterns that we have found. Hence, our hypotheses rely on predicted proteins from either pure cultures or MAGs. For instance, one crucial point would be determining the intracellular K+ concentration, which to date has been only done in E. coli [45], a marine Pseudomonas [46], and some halophiles [47]. Similarly, we need to understand how different microbes regulate their cytoplasmic pH in response to environmental changes. However, there is a significant difficulty in measuring the cytoplasmic pH of microbes under growth conditions [42]. Furthermore, some microbes undergo small variations in the pH of their cytoplasm (up to 0.1 units per pH unit change), while others such as E. coli or Coxiella burnetti are subjected to much wider changes [48, 49].
As could be expected, there is a taxonomic component in the pI patterns, for example, SAR11 clade members tend to have the pI plot displaced towards basic values (Additional file 1: Figure S1, see Additional file 2). That streamlined bacteria, independently of their origin, should have a tendency to basicity in their pIs is to be expected (Additional file 1: Figure S10) considering their higher surface/volume ratio, which leads to a higher membrane/cytoplasmic proteins ratio. Still, even in these cases, the differential value in the freshwater-marine comparison was detectable (i.e., regardless of the pI range always marine have more acidic average values). This general pattern was confirmed by the amino acid composition that shows common trends in organisms as phylogenetically distant as Pelagibacter and Thaumarchaeota. It was also remarkable that in closely related microbes but from different origin (marine or freshwater), AAI was similar and (in most cases) lower than ANI, i.e., amino acid similarity is lower than nucleotide identity. This was also observed in freshwater, euryhaline, and marine Synechococcus/Cyanobium genera [50]. This is the opposite of what we found when comparing similarly distant microbes but living in the same type of aquatic habitats, such as freshwater acI Actinobacteria [6]. The values are consistently AAI < ANI as could be expected from the existence of neutral changes due to the degeneration of the genetic code.
Our work underscores the important changes that a microbe must suffer to get adapted to freshwater from a marine habitat or vice versa. If many (or most) proteins change in their amino acidic composition, the number of changes, i.e., the evolutionary time involved, have to be large. Although several studies assured that marine-freshwater transitions tended to be infrequent [51, 52], it has been proven that some close relatives to marine microbes are found in freshwater habitats (SAR11 Pelagibacteraceae, Rhodobacteraceae, and Flavobacteria) [14–16, 20]. Furthermore, microbial transitions have also been recorded in brackish ecosystems such as Baltic Sea [18], in which some species of brackish origin transit to either marine or freshwater systems. Therefore, the transition, although demanding, could have happened at some stages in the long evolutionary history of microbes, but the adaptation to be freshwater or marine adapted is a crucial evolutionary decision that every species take.
Conclusions
There is a large change in amino acid composition among microbes depending on whether they live in marine or freshwater habitats. The change can already be detected by relatively low values of AAI (compared to ANI) and is reflected by a major shift in the pI pattern of the cell predicted proteome, with an increase in the acidic peak in the marine microbes and another (albeit more moderate) in the neutral and basic peaks for the freshwater ones. These changes occur also in closely related microbes, i.e., they do not reflect a taxonomic bias. Furthermore, we have been able to see changes in individual proteins with 3D models and their overall surface electrostatic potential, indicating that the changes tend to accumulate on the surface of the protein, particularly when they are soluble (cytoplasmic or secreted).
We propose that our results indicate an important change in cell physiology due to the absence of salts in the freshwater habitats. This absence might imply specific requirements of membrane characteristics (membranes could change in composition when exposed to the absence of salts in significant amounts since the stability of lipid bilayers could be affected), bioenergetics (differences in the electrochemical gradient across the membrane), intracellular pH (a change in the intracellular pH would modify the solubility of the proteins), or K+ concentration (requiring less acidity to compensate the positive charge of intracellular cations) or a combination of these or other components of cell biology which apply throughout the prokaryotic domain, bacteria, and archaea.
Methods
Metagenomic datasets and bacterial genomes used in this work
All metagenomic datasets used in this work are publicly available in the NCBI/SRA databases: Mediterranean Sea [33], Caspian Sea [32], Lake Baikal [20], Tous reservoir [25]. All bacterial and archaeal genomes used in this study, together with their accession/Genbank number (NCBI), habitat, isolation/origin, reference, type of genome, and phylum are shown in the Additional file 2. The eight genomes used in the protein-by protein-based comparison were previously published: Synechococcus lacustris Tous [50], Synechococcus sp. RCC307 [53], Methylopumilus planktonicus MMS-2-53 [54], Methylophilales bacterium MBRSH7 [55], Pelagibacteraceae bacterium Baikal-G1 [20], Pelagibacter ubique HTCC 7214 (ASM70138v1), Nitrosopumilus maritimus SCM1 [56], and Nitrosoarchaeum sp. Baikal-G1 [20].
Protein isoelectric point determination
The isoelectric point calculations and amino acid features of each predicted protein and microbe were calculated with the software Pepstats from the EMBOSS package [57]. To determine the pI distribution from metaproteomes, we obtained all proteins from the assembled contigs larger than 5 kb, which were representative of the different taxa inhabiting each ecosystem [20, 32, 33]. We used at least 85,000 proteins per metagenome (Mediterranean Sea 30 m, Lake Baikal 20 m, Caspian Sea 15 m, and Tous reservoir 12–25 m).
Statistical analysis of the different habitat adapted microbes
Bray-Curtis distances between the relative frequencies of the pIs were calculated to evaluate if the differences in pIs between the 71 habitat-adapted microbes (Fig. 2) were due to the habitats (freshwater, marine, brackish, halophile) or taxonomic classification (phyla). Then, we performed a normalization of the dataset and a PERMANOVA analysis with Vegan R-package [58]. We also obtained a two-dimensional principal component analysis plot with FactoMineR package [59] and calculated average and standard deviation values between all relative frequencies of different microbes for acid (3.25–6.25), neutral (6.75–7.25), and basic (7.75–13.75) pIs. All the statistical analyses are shown in Additional file 2.
Category classification of different proteins
Each protein was categorized into cytoplasmic/inner membrane, proteins with transmembrane domains, and proteins with signal peptides according to the SignalP [60] and Phobius [61] tools predictions. The pIs of the different proteins, their transmembrane domain topology, and presence/absence of signal peptides for the eight microbes used in this comparison are shown in Additional file 3.
Structure homology modeling and determination of the electrostatic surface potential of different proteins
The selected proteins for the pair-wise microbe comparison were first modeled for their tertiary structure with the SWISS-MODEL online tool [62–64]. The extracted PDB was then visualized with PYMOL [65], and the electrostatic surface potential was calculated with APBS tool [66]. The surface potentials were mapped from − 3 kcal mol−1 (red) to + 3 kcal mol−1 (blue).
Pan-genome analysis
The different freshwater and marine genomes used in the structural comparison were also subjected to a pan-genome analysis. Core and flexible genomes were determined with OrthoMCL and getHomologues software [67, 68].
Additional files
Figure S1. Whole proteome pI versus relative frequency plot of some representatives from different habitats of the class Alphaproteobacteria. A) Rhodospirillaceae and Roseobacter clades and B) SAR11 clade. Figure S2. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) phylum Bacteroidetes and B) Order Betaproteobacterales. Figure S3. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) phylum Chloroflexi and B) phylum Actinobacteria. Figure S4. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) Genera Synechococcus/Cyanobium and B) Assorted halophiles (bacteria and archaea). Figure S5. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) phylum Verrucomicrobia and B) phylum Planctomycetes. Figure S6. Whole proteome pI versus relative frequency plot of some representatives from different habitats of the phylum Thaumarchaeota. Figure S7. Star diagrams and amino acid composition of prokaryotic relatives from marine and freshwater origin. Figure S8. Structural model of proteins from different habitat-adapted archaea. Insets show electrostatic surface potential 3D models of N5-carboxyaminoimidazole ribonucleotide synthetase (secreted protein) and radA (cytoplasmic). The potentials were colored from -3 kcal mol-1(red) to +3 kcal mol-1 (blue). Whole proteome pI versus relative frequency plot of Haloquadratum walsbyi DSM 16790 (halophile, culture), Nitrosopumilus maritimus SCM1 (marine, culture), Nitrosoarchaeum sp. Baikal-G1 (freshwater, MAG). Figure S9. Isoelectric point versus relative frequency plot of the pan-genome (core and flexible genome) of freshwater and marine prokaryotes. N indicates the number of proteins in either core or flexible genomes. A) P.ubique HTCC 7214 and Pelagibacteraceae bacterium Baikal-G1. B) Ca. Methylopumilus planktonicus MMS-2-53 and Methylophilales bacterium MBRS-H7. C) Synechococcus sp. RCC307 and Synechococcus lacustris Tous. D) Nitrosopumilus maritimus SCM1 and Nitrosoarchaeum sp. Baikal-G1. Figure S10. Whole proteome pI versus relative frequency plot of some streamlined bacteria from different habitats. (PDF 4739 kb)
Prokaryotic genomes used in this study. Name of the genome, accession/Genbank number (NCBI), habitat, isolation/origin, reference, type of genome and taxonomy fields are provided. Habitat, phyla and Isoelectric point relative frequencies of each microbe. Statistical analysis of the compared dataset (Bray-Curtis+PERMANOVA, Principal Component Analysis, Average + Standard Deviation). (XLSX 60 kb)
Classification and pI values of the different types of proteins retrieved from the eight prokaryotes used in the electrostatic surface potential and pI comparisons. The pIs of the different proteins, their transmembrane domain topology and presence/absence of signal peptides are shown. (XLSX 784 kb)
Acknowledgements
This work was supported by grant “VIREVO” CGL2016-76273-P [AEI/FEDER, EU] (co-founded with FEDER funds).
Abbreviations
- AAI
Average amino acid identity
- ANI
Average nucleotide identity
- MAG
Metagenome-assembled genome
- pI
Isoelectric point
- POCP
Percentage of conserved proteins
Authors’ contributions
FRV and PJCY conceived the project. PJCY performed analyses. FRV and PJCY wrote the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
All metagenomic datasets and microbial genomes used in this work are publicly available in the NCBI/SRA databases.
Ethics approval and consent to participate
Ethics approval was not required for the study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hobbie JE. A comparison of the ecology of planktonic bacteria in fresh and salt water. Limnol Oceanogr. Wiley Online Library. 1988;33:750–764. [Google Scholar]
- 2.Ghai R, McMahon KD, Rodriguez-Valera F. Breaking a paradigm: cosmopolitan and abundant freshwater actinobacteria are low GC. Environ Microbiol Rep. 2012;4:29–35. doi: 10.1111/j.1758-2229.2011.00274.x. [DOI] [PubMed] [Google Scholar]
- 3.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. Key roles for freshwater Actinobacteria revealed by deep metagenomic sequencing. Mol Ecol. 2014;23:6073–6090. doi: 10.1111/mec.12985. [DOI] [PubMed] [Google Scholar]
- 4.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giovannoni S. J. Genome Streamlining in a Cosmopolitan Oceanic Bacterium. Science. 2005;309(5738):1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 6.Neuenschwander SM, Ghai R, Pernthaler J, Salcher MM. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2017. [DOI] [PMC free article] [PubMed]
- 7.Kasalický V, Jezbera J, Hahn MW, Šimek K. The diversity of the Limnohabitans genus, an important group of freshwater bacterioplankton, by characterization of 35 isolated strains. PLoS One. Public Library of Science. 2013;8:e58209. doi: 10.1371/journal.pone.0058209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn MW, Koll U, Jezberová J, Camacho A. Global phylogeography of pelagic Polynucleobacter bacteria: restricted geographic distribution of subgroups, isolation by distance and influence of climate. Environ Microbiol. 2015;17:829–840. doi: 10.1111/1462-2920.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salcher MM, Pernthaler J, Posch T. Seasonal bloom dynamics and ecophysiology of the freshwater sister clade of SAR11 bacteria ‘that rule the waves’(LD12) ISME J. 2011;5:1242. doi: 10.1038/ismej.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaremba-Niedzwiedzka K, Viklund J, Zhao W, Ast J, Sczyrba A, Woyke T, et al. Single-cell genomics reveal low recombination frequencies in freshwater bacteria of the SAR11 clade. Genome Biol. 2013;14:R130. doi: 10.1186/gb-2013-14-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henson MW, Lanclos VC, Faircloth BC, Thrash JC. Cultivation and genomics of the first freshwater SAR11 (LD12) isolate. ISME J. Nature Publishing Group. 2018;12:1846. doi: 10.1038/s41396-018-0092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, et al. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch Microbiol. Springer. 1992;157:297–300. doi: 10.1007/BF00245165. [DOI] [Google Scholar]
- 13.Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Richter RA, Valas R, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. Nature Publishing Group. 2012;6:1186. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Yoshizawa S, Sun Y, Huang Y, Chu X, González JM, et al. Repeated evolutionary transitions of Flavobacteria from marine to non-marine habitats. Environ Microbiol. Wiley Online Library. 2018. [DOI] [PubMed]
- 15.Paver SF, Muratore D, Newton RJ, Coleman ML. Reevaluating the salty divide: phylogenetic specificity of transitions between marine and freshwater systems. mSystems. Am Soc Microbiol. 2018;3:e00232–e00218. doi: 10.1128/mSystems.00232-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon M, Scheuner C, Meier-Kolthoff JP, Brinkhoff T, Wagner-Döbler I, Ulbrich M, et al. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. Nature Publishing Group. 2017;11:1483. doi: 10.1038/ismej.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satinsky Brandon M, Zielinski Brian L, Doherty Mary, Smith Christa B, Sharma Shalabh, Paul John H, Crump Byron C, Moran Mary. The Amazon continuum dataset: quantitative metagenomic and metatranscriptomic inventories of the Amazon River plume, June 2010. Microbiome. 2014;2(1):17. doi: 10.1186/2049-2618-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herlemann Daniel PR, Labrenz Matthias, Jürgens Klaus, Bertilsson Stefan, Waniek Joanna J, Andersson Anders F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME Journal. 2011;5(10):1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmfeldt Karin, Dziallas Claudia, Titelman Josefin, Pohlmann Kirsten, Grossart Hans-Peter, Riemann Lasse. Diversity and abundance of freshwaterActinobacteriaalong environmental gradients in the brackish northern Baltic Sea. Environmental Microbiology. 2009;11(8):2042–2054. doi: 10.1111/j.1462-2920.2009.01925.x. [DOI] [PubMed] [Google Scholar]
- 20.Cabello-Yeves PJ, Zemskaya TI, Rosselli R, Coutinho FH, Zakharenko AS, Blinov VV, et al. Genomes of novel microbial lineages assembled from the sub-ice waters of Lake Baikal. Appl Environ Microbiol. 2018;84:e02132–e02117. doi: 10.1128/AEM.02132-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galazy GI. Atlas of Lake Baikal. GUGK, Moscow (in Russ. 1993;
- 22.Falkner KK, Measures CI, Herbelin SE, Edmond JM, Weiss RF. The major and minor element geochemistry of Lake Baikal. Limnol Oceanogr. 1991;36:413–423. doi: 10.4319/lo.1991.36.3.0413. [DOI] [Google Scholar]
- 23.Votintsev KK, Mescheryakova AI, Popovskaya GI. Cycle of organic matter in Lake Baikal. Nauka, Novosibirsk; 1975.
- 24.Votinsev KK, Popovskaya GI. The peculiarity of the biotic cycle in Lake Baikal. Dokl Akad Nauk SSSR. 1979;216:666–669. [Google Scholar]
- 25.Cabello-Yeves PJ, Haro-Moreno JM, Martin-Cuadrado A-B, Ghai R, Picazo A, Camacho A, et al. Novel Synechococcus genomes reconstructed from freshwater reservoirs. Front Microbiol. 2017;8:1151. doi: 10.3389/fmicb.2017.01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiller Georg F., Caraux Gilles, Sylvester Nicole. The modal distribution of protein isoelectric points reflects amino acid properties rather than sequence evolution. PROTEOMICS. 2004;4(4):943–949. doi: 10.1002/pmic.200200648. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz R., Ting C. S., King J. Whole Proteome pI Values Correlate with Subcellular Localizations of Proteins for Organisms within the Three Domains of Life. Genome Research. 2001;11(5):703–709. doi: 10.1101/gr.GR-1587R. [DOI] [PubMed] [Google Scholar]
- 28.Bardavid RE, Oren A. Acid-shifted isoelectric point profiles of the proteins in a hypersaline microbial mat: an adaptation to life at high salt concentrations? Extremophiles. 2012;16:787–792. doi: 10.1007/s00792-012-0476-6. [DOI] [PubMed] [Google Scholar]
- 29.Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62:504–544. doi: 10.1128/mmbr.62.2.504-544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlowski LP. Proteome-pI: proteome isoelectric point database. Nucleic Acids Res. Oxford University Press; 2016;gkw978. [DOI] [PMC free article] [PubMed]
- 31.Kiraga Joanna, Mackiewicz Pawel, Mackiewicz Dorota, Kowalczuk Maria, Biecek Przemysław, Polak Natalia, Smolarczyk Kamila, Dudek Miroslaw R, Cebrat Stanislaw. The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms. BMC Genomics. 2007;8(1):163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehrshad M, Amoozegar MA, Ghai R, Fazeli SAS, Rodriguez-Valera F. Genome reconstruction from metagenomic datasets reveals novel microbes in the brackish waters of the Caspian Sea. Appl Environ Microbiol. 2016;AEM:03381–03315. doi: 10.1128/AEM.03381-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haro-Moreno JM, López-Pérez M, José R, Picazo A, Camacho A, Rodriguez-Valera F. Fine metagenomic profile of the Mediterranean stratified and mixed water columns revealed by assembly and recruitment. Microbiome. BioMed Central; 2018;6:128. [DOI] [PMC free article] [PubMed]
- 34.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 35.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madern D., Ebel C., Zaccai G. Halophilic adaptation of enzymes. Extremophiles. 2000;4(2):91–98. doi: 10.1007/s007920050142. [DOI] [PubMed] [Google Scholar]
- 37.Medini Duccio, Donati Claudio, Tettelin Hervé, Masignani Vega, Rappuoli Rino. The microbial pan-genome. Current Opinion in Genetics & Development. 2005;15(6):589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Mira A, Martín-Cuadrado AB, D’Auria G, Rodríguez-Valera F. The bacterial pan-genome: a new paradigm in microbiology. Int Microbiol. 2010;13:45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 40.Wu Songfeng, Wan Ping, Li Jianqi, Li Dong, Zhu Yunping, He Fuchu. Multi-modality of pI distribution in whole proteome. PROTEOMICS. 2006;6(2):449–455. doi: 10.1002/pmic.200500221. [DOI] [PubMed] [Google Scholar]
- 41.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol. Elsevier; 2009;55:1–317. [DOI] [PubMed]
- 42.Booth IR. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. American Society for Microbiology (ASM); 1985;49:359. [DOI] [PMC free article] [PubMed]
- 43.Boyd D, Beckwith J. The role of charged amino acids in the localization of secreted and membrane proteins. Cell. Cell Press. 1990;62:1031–1033. doi: 10.1016/0092-8674(90)90378-r. [DOI] [PubMed] [Google Scholar]
- 44.Gitlin Irina, Carbeck Jeffrey D., Whitesides George M. Why Are Proteins Charged? Networks of Charge–Charge Interactions in Proteins Measured by Charge Ladders and Capillary Electrophoresis. Angewandte Chemie International Edition. 2006;45(19):3022–3060. doi: 10.1002/anie.200502530. [DOI] [PubMed] [Google Scholar]
- 45.Schultz S. G. Cation Transport in Escherichia coli: I. Intracellular Na and K concentrations and net cation movement. The Journal of General Physiology. 1961;45(2):355–369. doi: 10.1085/jgp.45.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takacs FP, Matula TI, MacLeod RA. Nutrition and metabolism of marine bacteria XIII. Intracellular concentrations of sodium and potassium ions in a marine Pseudomonad. J Bacteriol. Am Soc Microbiol; 1964;87:510–518. [DOI] [PMC free article] [PubMed]
- 47.Deole Ratnakar, Challacombe Jean, Raiford Douglas W., Hoff Wouter D. An Extremely Halophilic Proteobacterium Combines a Highly Acidic Proteome with a Low Cytoplasmic Potassium Content. Journal of Biological Chemistry. 2012;288(1):581–588. doi: 10.1074/jbc.M112.420505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hackstadt T. Estimation of the cytoplasmic pH of Coxiella burnetii and effect of substrate oxidation on proton motive force. J Bacteriol. Am Soc Microbiol; 1983;154:591–597. [DOI] [PMC free article] [PubMed]
- 49.Ahmed S, Booth IR. The effect of-galactosides on the protonmotive force and growth of Escherichia coli. Microbiology. Microbiology Society. 1983;129:2521–2529. doi: 10.1099/00221287-129-8-2521. [DOI] [PubMed] [Google Scholar]
- 50.Cabello-Yeves PJ, Picazo A, Camacho A, Callieri C, Rosselli R, Roda-Garcia JJ, et al. Ecological and genomic features of two widespread freshwater picocyanobacteria. Environ Microbiol. Wiley Online Library; 2018; [DOI] [PubMed]
- 51.Logares Ramiro, Bråte Jon, Bertilsson Stefan, Clasen Jessica L., Shalchian-Tabrizi Kamran, Rengefors Karin. Infrequent marine–freshwater transitions in the microbial world. Trends in Microbiology. 2009;17(9):414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 52.Logares R, Bråte J, Heinrich F, Shalchian-Tabrizi K, Bertilsson S. Infrequent transitions between saline and fresh waters in one of the most abundant microbial lineages (SAR11). Mol Biol Evol. Oxford University Press; 2009;27:347–357. [DOI] [PubMed]
- 53.Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, et al. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salcher MM, Neuenschwander SM, Posch T, Pernthaler J. The ecology of pelagic freshwater methylotrophs assessed by a high-resolution monitoring and isolation campaign. ISME J. 2015;9:2442. doi: 10.1038/ismej.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jimenez-Infante Francy, Ngugi David Kamanda, Vinu Manikandan, Alam Intikhab, Kamau Allan Anthony, Blom Jochen, Bajic Vladimir B., Stingl Ulrich. Comprehensive Genomic Analyses of the OM43 Clade, Including a Novel Species from the Red Sea, Indicate Ecotype Differentiation among Marine Methylotrophs. Applied and Environmental Microbiology. 2015;82(4):1215–1226. doi: 10.1128/AEM.02852-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Könneke M, Bernhard AE, de La Torre JR, Walker CB. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 57.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 58.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. The vegan package. Community Ecol Packag. 2007;10:631–637. [Google Scholar]
- 59.Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 60.Petersen Thomas Nordahl, Brunak Søren, von Heijne Gunnar, Nielsen Henrik. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 61.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. Oxford University Press; 2007;35:W429–W432. [DOI] [PMC free article] [PubMed]
- 62.Guex Nicolas, Peitsch Manuel C., Schwede Torsten. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. ELECTROPHORESIS. 2009;30(S1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 63.Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L, et al. The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res. Oxford University Press; 2016;45:D313–D319. [DOI] [PMC free article] [PubMed]
- 64.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018. [DOI] [PMC free article] [PubMed]
- 65.DeLano WL. The PyMOL molecular graphics system. http//www pymol org. DeLano scientific; 2002;
- 66.Lerner MG, Carlson HA. APBS plugin for PyMOL. Ann Arbor Univ Michigan. 2006.
- 67.Li L. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Research. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Contreras-Moreira Bruno, Vinuesa Pablo. GET_HOMOLOGUES, a Versatile Software Package for Scalable and Robust Microbial Pangenome Analysis. Applied and Environmental Microbiology. 2013;79(24):7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Whole proteome pI versus relative frequency plot of some representatives from different habitats of the class Alphaproteobacteria. A) Rhodospirillaceae and Roseobacter clades and B) SAR11 clade. Figure S2. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) phylum Bacteroidetes and B) Order Betaproteobacterales. Figure S3. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) phylum Chloroflexi and B) phylum Actinobacteria. Figure S4. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) Genera Synechococcus/Cyanobium and B) Assorted halophiles (bacteria and archaea). Figure S5. Whole proteome pI versus relative frequency plot of some representatives from different habitats of A) phylum Verrucomicrobia and B) phylum Planctomycetes. Figure S6. Whole proteome pI versus relative frequency plot of some representatives from different habitats of the phylum Thaumarchaeota. Figure S7. Star diagrams and amino acid composition of prokaryotic relatives from marine and freshwater origin. Figure S8. Structural model of proteins from different habitat-adapted archaea. Insets show electrostatic surface potential 3D models of N5-carboxyaminoimidazole ribonucleotide synthetase (secreted protein) and radA (cytoplasmic). The potentials were colored from -3 kcal mol-1(red) to +3 kcal mol-1 (blue). Whole proteome pI versus relative frequency plot of Haloquadratum walsbyi DSM 16790 (halophile, culture), Nitrosopumilus maritimus SCM1 (marine, culture), Nitrosoarchaeum sp. Baikal-G1 (freshwater, MAG). Figure S9. Isoelectric point versus relative frequency plot of the pan-genome (core and flexible genome) of freshwater and marine prokaryotes. N indicates the number of proteins in either core or flexible genomes. A) P.ubique HTCC 7214 and Pelagibacteraceae bacterium Baikal-G1. B) Ca. Methylopumilus planktonicus MMS-2-53 and Methylophilales bacterium MBRS-H7. C) Synechococcus sp. RCC307 and Synechococcus lacustris Tous. D) Nitrosopumilus maritimus SCM1 and Nitrosoarchaeum sp. Baikal-G1. Figure S10. Whole proteome pI versus relative frequency plot of some streamlined bacteria from different habitats. (PDF 4739 kb)
Prokaryotic genomes used in this study. Name of the genome, accession/Genbank number (NCBI), habitat, isolation/origin, reference, type of genome and taxonomy fields are provided. Habitat, phyla and Isoelectric point relative frequencies of each microbe. Statistical analysis of the compared dataset (Bray-Curtis+PERMANOVA, Principal Component Analysis, Average + Standard Deviation). (XLSX 60 kb)
Classification and pI values of the different types of proteins retrieved from the eight prokaryotes used in the electrostatic surface potential and pI comparisons. The pIs of the different proteins, their transmembrane domain topology and presence/absence of signal peptides are shown. (XLSX 784 kb)
Data Availability Statement
All metagenomic datasets and microbial genomes used in this work are publicly available in the NCBI/SRA databases.