Abstract
Many long Drosophila introns are processed by an unusual recursive strategy.
The presence of ~200 adjacent splice acceptor and splice donor sites, termed ratchet points (RPs), were inferred to reflect “zero nucleotide exons” whose sequential processing subdivides removal of long host introns. We used CRISPR-Cas9 to disrupt several intronic RPs in the animal, and some recapitulated characteristic loss-of-function phenotypes. Unexpectedly, selective disruption of RP splice donors revealed constitutive retention of unannotated short exons. Functional minigene tests confirm that unannotated cryptic splice donor sites are critical for recognition of intronic RPs, demonstrating that recursive splicing involves the recognition of cryptic RP-exons. We generalize this mechanism, since canonical, conserved, splice donors are specifically enriched in a +40–80 nt window downstream of known and newly-annotated intronic RPs, and exhibit similar properties to a newly-recognized class of expressed RP-exons. Overall, these studies unify the mechanism of Drosophila recursive splicing with that in mammals.
Introduction
Large introns create challenges for accurate processing, due to seemingly modest information encoded by minimal splice donor (GU) and acceptor (AG) sites. One established concept is the splicing machinery defines the smallest available unit; thus, introns are defined when they are relatively small, but exon definition becomes critical when flanking introns are larger1. In this way, many cryptic splicing signals within intronic context might be avoided. Still, it is puzzling how proper junctions are decoded as introns increase from tens to hundreds of kilobases, even megabases in mammalian genomes, given that splicing rates of short and very long introns are similar2. Coupling of RNA Polymerase II, chromatin, and factors involved in splice site recognition and spliceosome assembly, may facilitate processing at long introns3. For example, factors involved in splice site pairing, including U1 snRNP and U2AF65, associate with Pol II. At the same time, exons can preferentially associate with nucleosomes, are marked by distinctive histone modifications, and associate with U2 snRNP. Thus, organization and scaffolding afforded by Pol II and chromatin can aid the specificity and efficiency of splicing across long distances3.
Another consideration is the process of recursive splicing, whereby splicing of long introns is achieved in stepwise fashion. This breaks up the daunting task of processing a larger intron into several smaller, more manageable segments. The Lopez lab first recognized this mechanism during processing of the 77kb intron of Drosophila Ultrabithorax (Ubx). This intronic space includes two short cassette exons (mI and mII) that can be present or absent in different isoforms. However, unlike typical alternative splicing reactions, careful analysis showed that processing of these Ubx microexons involves splicing that regenerates 5’ splice sites at their junctions4. The same Ubx intron was subsequently shown to contain a “ratchet point” (RP) without a recognizable microexon; thus, it is marked only by a juxtaposed AG:GU splice acceptor-donor pair5. This same study predicted 165 candidate RPs within long introns of >100 genes, suggesting that recursive splicing is utilized broadly to process long introns in Drosophila5.
Of note, the vast majority of predicted RPs (155/165) were not associated with known exons and therefore do not appear in mature mRNA; seven novel RPs were validated using rt-PCR assays5. It would take another decade, until the advent of deep RNA-sequencing surveys, for broad experimental confirmation of recursive splicing. In particular, total RNA-seq data from diverse Drosophila stages, tissues and cell types permitted de novo annotation of 197 “zero nucleotide exon” RPs from 130 introns of 115 genes6. Little is known about the recursive splicing mechanism, although the process is believed constitutive and appears especially sensitive to U2AF activity6. How an exon of zero nucleotides would be recognized by the splicing machinery is mysterious, and previous sequence analysis downstream of known intronic ratchet points did not reveal sequence motifs or compelling conserved regions6.
Mammals also utilize recursive splicing, but seem to harbor far fewer intronic recursive splice sites6,7. Notably, while exon signatures were not identified at Drosophila “0-nt exon” RPs6, functional studies in human cells provided evidence that exon definition via recursive splicing exons (“RS-exons”) is critical for recursive splicing7. Thus, mammalian RS-exon splicing is analogous to the strategy described for the initial Ubx microexons4.
Many fundamental questions regarding recursive splicing remain. For example, it is not resolved whether recursively spliced products represent obligate intermediates on the way to mature mRNAs (Supplementary Figure 1). The “sawtooth” pattern of total RNA-seq reads at certain loci, which dips characteristically at RP sites, is consistent with these being co-transcriptional splicing intermediates6 (Supplementary Figure 1). However, it is possible that recursive splicing is a pathway parallel to non-recursive splicing. The residence of RPs in the very longest transcription units does not render them amenable to direct mechanistic observation using in vitro splicing assays, and recursive splicing has otherwise been studied only with minigenes. Other basic questions include what effect recursive splicing has on gene expression, whether recursive splicing matters in vivo, and whether there truly are fundamental differences in recursive splicing mechanism between flies and mammals, as suggested by available literature8.
In this study, we use molecular genetic information from CRISPR engineering to reveal that recursive splicing in Drosophila proceeds via unannotated cryptic RP-exons. We validate this model using functional tests, and extend it genomewide, by showing that unannotated, high-scoring, conserved splice donors are present in a distinctive length window downstream of known and novel intronic RPs. Recursive splicing is utilized on a continuum, since beyond the hundreds of splicing events inferred to involve cryptic RP-exons, we also recognize scores of expressed RP-exons. These findings now unify the mechanism of recursive splicing in flies with mammals.
Results
In vivo mutagenesis of ratchet points in Drosophila
To our knowledge, no studies of recursive splicing have yet involved endogenous sites in intact animals4–7,9. Therefore, we exploited CRISPR-Cas9 to mutagenize ratchet points (RPs) in Drosophila and assess consequences on phenotypes and RNA processing. We successfully targeted intronic RPs in Beadex (Bx), Ultrabithorax (Ubx) and kuzbanian (kuz); the former lies between non-coding exons while the latter two reside between coding exons. In all three cases, we characterized alleles that selectively disrupted RP splice donor sites (Figure 1A, molecular details provided in Supplementary Figure 2). While the Bx[RP] mutant was viable and lacked overt defects, the Ubx[RP] and kuz[RP] mutants were lethal. Our RP mutants failed to complement known amorphic Ubx10 and kuz11 mutants, and we proceeded to detailed phenotypic analyses.
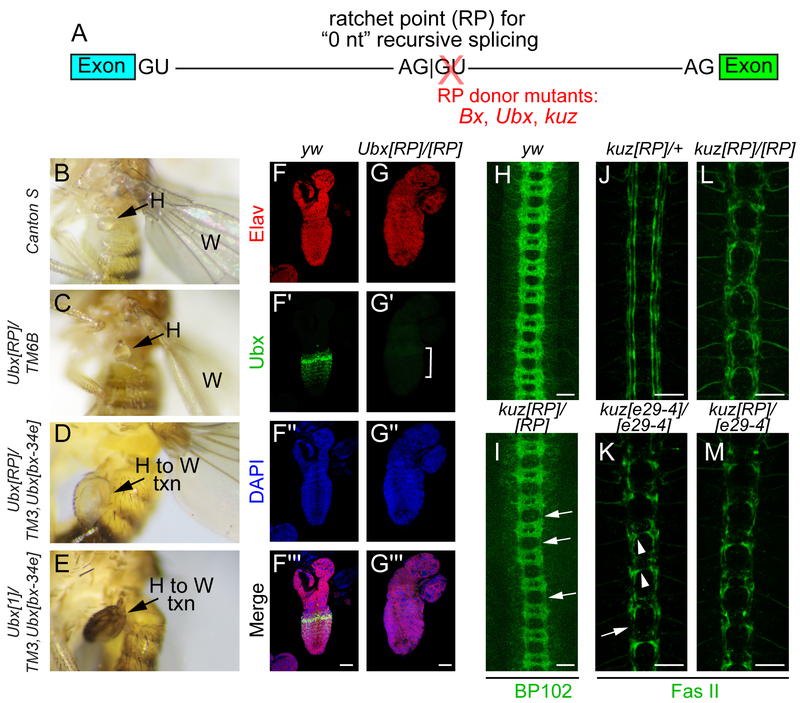
Figure 1. Ratchet point donor mutants of Ubx and kuz are strong loss-of-function alleles.
(A) Recursive splicing at long introns sequentially removes intron segments at paired splice acceptor-donor sites (AG:GU), also known as a ratchet point (RP), without leaving behind a mature exon. We used CRISPR-Cas9 to selectively mutagenize several Drosophila RP donor sites. (B-E) Lateral images of adult flies that illustrate phenotypes of Ubx[RP] mutants. (B) Wildtype (Canton S) fly with wing (W) and haltere (H) labeled. (C) Ubx[RP] heterozygote (in trans to TM6B balancer) shows mild enlargement of the haltere, indicative of Ubx haploinsufficiency. (D) Ubx[RP]/[bx-34e] (in trans to TM3 balancer) shows an overt haltere-to-wing transformation (H to W txn). (E) The phenotype of the RP mutant is similar to the known amorphic allele Ubx[1] over TM3. (F-G) Immunostaining of first instar larval CNS. (F) Control yw shows the normal segmental pattern of Ubx protein (green) in the ventral nerve cord, counterstained with pan-neuronal Elav (red) and DAPI (blue). (G) Ubx[RP] homozygote selectively lacks Ubx protein. (H-M) Ventral images of stage 16 embryos stained with α-BP102 (H, I) or α-Fas II (J, M) to reveal all CNS axons or subsets of ipsilateral axons, respectively. (H) BP102 exhibits a characteristic ladder-like staining pattern in control (yw) embryo. (I). kuz[RP] homozygote display thickening of the commissures and thinning of longitudinal connectives (arrows). (J) kuz[RP]/+ heterozygote exhibits a normal Fas II pattern of three bundles of longitudinal axons on either side of the ventral midline. All three kuz mutant combinations, kuz[e29–4] homozygotes (K), kuz[RP] homozygotes (L) and kuz[RP]/[e29–4] trans-heterozygotes (M) exhibit similar Fas II defects. These include failure to establish the longitudinal tracts and midline crossing defects. Scale bars in F-G indicate 40 μm and in H-M indicate 20 μm.
Ubx[RP]/+ animals exhibit mild haltere enlargement, consistent with partial conversion to wing identity (Figure 1B–C). As the dominant Ubx effect can be difficult to visualize, we sensitized the background using the Ubx hypomorphic allele on the TM3 balancer. Strikingly, viable Ubx[RP]/TM3, Ubx[bx-34e] animals exhibit overt transformation of halteres into wings equivalent in severity to amorphic Ubx[1] in trans to TM3 (Figure 1D–E). Moreover, immunostaining of larval CNS showed the normal pattern of Ubx protein in wildtype ventral nerve cord was basically undetectable in lethal Ubx[RP] homozygotes (Figure 1F–G). Thus, Ubx[RP] abrogates Ubx function and protein accumulation.
With kuz[RP], homozygous mutant embryos phenocopied previously described zygotic defects in CNS axonal patterning11. For example, BP102 staining showed reduction of longitudinal bundles and accumulation of commissural material in kuz[RP] homozygotes compared to controls (Figure 1H–I). Fasciclin II (Fas II) staining also recapitulated known kuz defects. kuz[RP]/+ heterozygotes exhibit characteristic Fas II patterns of three longitudinal axonal tracts on either side of the midline (Figure 1J), while amorphic kuz[e29–4] homozygotes fail to elaborate the longitudinal tracts and present midline crossing defects (Figure 1K). We find similar phenotypes for kuz[RP] homozygotes (Figure 1L) and kuz[RP]/[e29–4] trans-heterozygotes (Figure 1M), indicating kuz[RP] is a strong loss-of-function allele.
Altogether, these tests provide first evidence that altering recursive splicing in the animal can disrupt endogenous gene function and generate mutant phenotypes.
Molecular analysis of RP mutants reveals constitutive retention of cryptic exons
We analyzed molecular consequences of RP mutations on RNA processing. Of note, since previous mutational tests of recursive splicing were done with minigenes, it has been difficult to ascertain if this process generates obligate intermediates towards mRNA. Alternatively, there could be parallel processing pathways that skip recursive sites, and/or recursive splicing might theoretically generate dead-end products (Supplementary Figure 1).
rt-PCR analyses to detect an intermediate amplicon downstream of the ratchet point (Figure 2A) yielded specific products from each RP mutant (Figure 2B), indicating successful splicing into ratchet points. Moreover, we detected mature mRNA products from all three RP mutants. However, mature mRNA amplicons from RP mutants were longer in all three cases (Figure 2C). Interestingly, sequencing of these products showed mutant transcripts retained sequences that originate from immediately 3’ of the RP splice acceptor, and are spliced to cognate downstream exons through cryptic splice donor sites (Figure 2D and Supplementary Figure 2). Since the ectopic exon in Bx resides in its 5’ UTR, this does not affect protein output. However, the inclusion of novel exons in kuz and Ubx disrupts their reading frames. Of note, rt-PCR tests exclusively detected RP-mutant transcripts bearing the ectopic exons, whether coding or non-coding. Thus, splicing into these RP sites is constitutive, and they are obligate in vivo intermediates toward mature mRNAs.
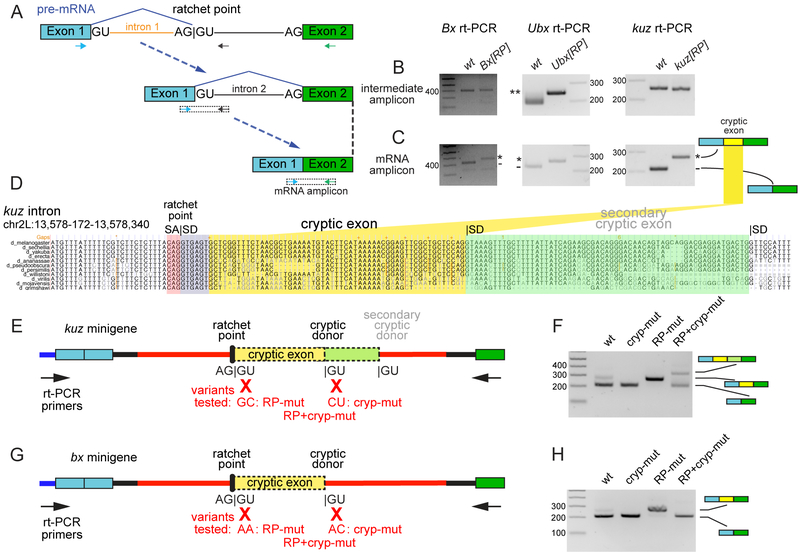
Figure 2. Molecular evaluation of RP mutants reveals existence of cryptic exons.
(A) Schematic of recursive splicing depicting removal of an intron in two steps. In the first step, a portion of the intron is removed (orange) resulting in intermediate pre-mRNA with a regenerated 5’ splice site. In the second step, the remainder of the intron (black) is removed, producing mRNA. Arrows are used to display primers to specifically amplify intermediate and mRNA transcripts. (B) Both wildtype and homozygous RP mutant animals produced intermediate amplicons, indicating that RP donor mutations did not disrupt recursive splicing. **Ubx[RP] mutants have a 38nt insertion that disrupts the RP and separates SA and SD by 38nt; thus, lengthening the intermediate amplicon. (C) Compared to wildtype, RP donor mutants consistently had larger mRNA amplicons that contained cryptic exon retention. (D) UCSC genome browser screenshot of the first ratchet point in kuz (kuz-RP1, with ratchet point splice acceptor [SA] in red and splice donor [SD] in purple; the SD was disrupted in kuz[RP] allele), along with highlighted regions showing cryptic exon (yellow) and a secondary cryptic exon (green) revealed in mutagenesis experiments. (E, G) Schematics of minigene constructs. The ~2.5 kb intronic RP locus used in the kuz (E) and the Bx (G) minigene is shown as a red line. Variants tested and primers used for rt-PCR are as indicated. (F, H) rt-PCR was used to evaluate spliced products from minigenes. (F) S2-R+ cells were transfected with WT kuz minigene (wt), or variants containing mutations in cryptic splice donor (cryp-mut), RP-splice donor (RP-mut), or both (RP+cryp-mut). (H) S2-R+ cells were transfected with WT Bx minigene and an analogous set of variants. In both cases, mutation of RP donor sites resulted in cryptic exon retention, while mutation of both RP+cryp donor sites lowered the efficiency of mature splicing. With kuz, the RP+cryp mut construct reveals usage of an extended cryptic exon. Uncropped gel images are shown in Supplementary Data Set 1.
Recursive splicing is mediated by short cryptic exons
The complete retention and characteristic size of retained cryptic exons implied their involvement in recursive splicing. In particular, we hypothesized that intronic ratchet points do not represent “0-nt exons” as originally suggested6, but actually proceed by an exon definition strategy involving unannotated cryptic exons, whose inclusion is subsequently suppressed. In this model, the strategy of Drosophila recursive splicing might resemble that of Ubx mini-exons (mI and mII) and mammalian recursive splicing7 (Supplementary Figure 1C). In some cases the putative splice donor site of the cryptic exon is conserved, as with Ubx-RP, but the putative splice donors of Bx-RP and kuz-RP cryptic exons are less conserved; none of these cryptic exons reflect coding constraint (Supplementary Figure 2C). By contrast, their companion RP sequences are perfectly constrained in the most distantly aligned Drosophilid genomes. If cryptic exons are integral to the recursive splicing reaction, they would represent an unusual case of RNA processing in which some cis signals are better conserved than others.
We utilized minigenes to test kuz-RP and Bx-RP intronic processing in S2-R+ cells, along with mutants in RP splice donors, cryptic splice donors, or both (Figure 2E, G). Both kuz and Bx wildtype minigenes produced expected spliced mRNA products (Figure 2F, H, wt lanes). Unexpectedly, a second band was observed for wildtype kuz and to a lesser extent, for wildtype Bx and confirmed by sequencing to be spliced mRNA that included the cryptic exon. This further suggests the presence of unannotated cryptic exons at intronic RPs. Indeed, total RNA-seq shows that the cryptic kuz exon is partially retained in S2-R+ but not in tissues (Supplementary Figure 3).
Consistent with our animal mutants, mutation of RP donor sites caused constitutive inclusion of cryptic exons from kuz-RP and Bx-RP minigenes (Figure 2F, H, RP-mut lanes, quantified in Supplementary Figure 4). Disruption of cryptic splice donors resulted in exclusion of cryptic exons for ectopic kuz and for ectopic Bx (Figure 2F, H, cryp-mut lanes). In these tests, cryptic exon skipping, whether through recursive splicing or loss of cryptic exon definition, would yield similar spliced products (see Supplementary Figure 1B: path 1 vs. path 2). To distinguish these possibilities, we examined constructs with both cryptic and RP splice donors mutated. If the reaction proceeded via recursive splicing, spliced mRNAs would include cryptic exons, whereas if recursive splicing were abolished due to loss of exon definition, spliced mRNAs would exclude cryptic exons.
The double Bx mutant predominantly yielded one spliced product that lacked the cryptic exon (Figure 2H, RP+cryp-mut lane). The kuz double mutant yielded two products – without cryptic exon and with a longer exon (Figure 2F, RP+cryp-mut lane). Sequencing revealed that retention of an extended cryptic exon in double mutants was due to usage of a downstream, poorly-conserved, secondary cryptic splice donor (Figure 2D). Cryptic exon skipping in Bx and kuz suggests loss of recursive splicing due to loss of exon definition, and the lower levels of spliced products in RP+cryp donor mutants indicates that recursive splicing contributes to effective processing. Moreover, activation of a novel cryptic exon in kuz double mutants suggests that fortuitous splice elements can easily compensate for disruptions in normally-recognized cryptic splice donor sequences. We return to the latter point in evolutionary analysis. Together, these data provide evidence that exon definition is an important step during recursive splicing.
Genomewide re-annotation of Drosophila intronic RPs and RP-exons
We sought to generalize the cryptic RP-exon model for intronic “0-nt” recursive splicing. Before doing so, we aimed to expand the catalog of Drosophila ratchet points. Recent efforts used ~11 billion paired end reads from ~100 Drosophila stages, tissues, and cell lines to annotate 197 intronic RPs from 130 introns of 115 genes6. However, as these are transient intermediates of co-transcriptionally processed RNA, total RNA-seq data are not optimal for their detection.
We collected data representing actively transcribed RNA (chromatin RNA-seq, nascent RNA-seq, GRO-seq) from S2 cells, embryos, heads and ovaries (Supplementary Data Set 2), and observed enrichment for intronic coverage and junction-spanning reads at known RPs, compared to total RNA and mRNA datasets (Supplementary Figure 5A–B). We then developed a pipeline to annotate recursive splicing events (Supplementary Note 1). As before6, we focused on intronic junction spanning reads with tandem splice acceptor and donor motifs at the 3’ end, and emphasized loci with sawtooth RNA-seq patterns. We observed strong enrichment of minimal paired splice acceptor and donor motifs (AG:GT) only when the junctions were located within introns >5 kb (Supplementary Figure 5C), confirming our pipeline identified genuine splicing events and validating our decision to triage other types of split reads with non-canonical junctions. However, bearing in mind that recursive intermediates are transient, we relaxed the requirement for sawtooth RNA-seq patterns if candidate RPs exhibited strong splice sites (see Methods). We manually examined all candidates to filter potential false positives.
Although we only analyzed 4 tissue types and many-fold fewer mapped RNA-seq reads than previously6, we substantially increase the scope of recursive splicing. Our pipeline recovered 187/197 previously annotated RPs6, and in total identified 304 unique RPs in 188 introns of 169 genes (Figure 3A and Supplementary Data Set 3). The newly recognized RPs share sequence and evolutionary properties of known recursive sites, as shown in partitioned analyses (Supplementary Figure 6). Notably, the 93 novel RPs with sawtooth RNA-seq evidence exhibit comparable phyloP conservation scores to known RPs, while 24 novel RPs lacking overt sawtooth RNA-seq patterns were only moderately less constrained (Supplementary Figure 6A). Overall, intronic RPs are (1) well-conserved across the Drosophilid phylogeny (Figure 3B), (2) share consensus splice motif characteristics including strong polypyrimidine tracts (Figure 3C), and (3) preferentially reside within especially long host introns (Supplementary Figure 6B).
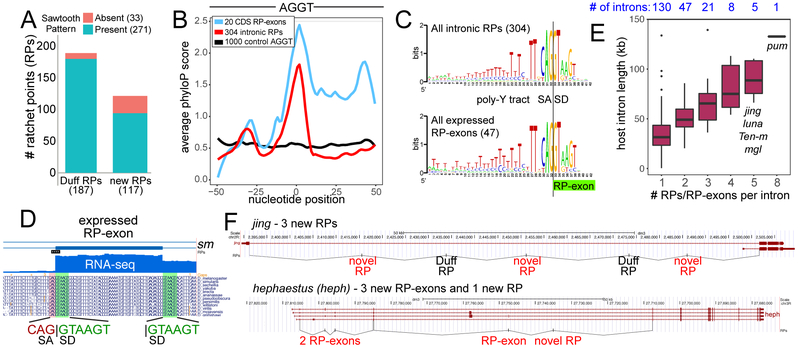
Figure 3. Genomewide annotation of novel intronic RPs and RP-exons.
(A) Summary of intronic RP annotations (i.e., with no evidence for an expressed exon) made in this study using nascent RNA-seq datasets, 187 of which were previously reported6 and 117 of which are novel. Presence of sawtooth RNA-seq patterns are noted. (B) Evolutionary conservation for all intronic RPs, RP-exons and control intronic AGGT sites. This was evaluated by averaging phyloP scores at each nucleotide position about the RP. (C) Sequence logos for the aggregate collections of intronic RPs and RP-exon splice junctions. (D) Example of RP-exon in the sm gene. It contains a perfectly conserved canonical splice donor (GTAAGT) at the beginning of an expressed cassette exon, which also uses a conserved GTAAGT splice donor. (E) Comparison of intron lengths and number of RPs per intron. Boxes represent the interquartile ranges and n for each group is indicated above plot. (F) Examples of novel intronic RPs and expressed RP-exons identified in jing and hephaestus (heph).
The first characterized cases of recursive splicing, Ubx microexons mI and mII4, resemble cryptic exon retention in [RP] mutant animals. Moreover, although usage of the kuz cryptic exon results in a nonsense product, we detect endogenous inclusion in S2-R+ (Supplementary Figure 3). This led us to suspect there might be a larger class of expressed, recursively-spliced exons beyond Ubx, which would exist on a continuum of alternative splicing in Drosophila with intronic RPs that putatively proceed via cryptic exons. For example, msi is a gene where we detect an intronic RP with sawtooth RNA-seq pattern and a cassette exon that regenerates a 5’ splice site (Supplementary Figure 7). We adapted our pipeline to annotate cassette exons with very high scoring splice donor sites at their precise 5’ ends. We identified 47 expressed RP-exons, nested within 42 introns of 41 genes (Supplementary Data Set 3). Although these exons are skipped in many libraries, all have spliced RNA-seq evidence for expression (Supplementary Data Set 4), and thus represent a class of alternative splicing. A majority of these reside within 5’ UTRs, although some specify coding sequences (CDS, 12) and alternative start codons (CDS:5’ UTR, 8). The 20 loci that include CDS content exhibit conserved aggregate phyloP profiles indicative of coding sequence (Figure 3B and Supplementary Figure 8). We illustrate an RP-exon from sm with highly conserved coding sequence and strikingly conserved RP-like tandem SA:SD sequence at its 5’ end (Figure 3D); others are in Supplementary Data Set 4.
Overall, the sequence content at tandem SA:SD sites between the aggregate intronic RPs and expressed “RP-exon” classes is nearly identical (Figure 3C), and their total host intron lengths are also similar (~55 kb for intronic RPs, ~43 kb for expressed RP-exons). This suggests these are mechanistically similar splicing processes. Interestingly, there is a linear relationship between intron length and total number of RPs per intron (Figure 3E), and intronic RPs and expressed RP-exons tend to be evenly distributed throughout their resident introns (Figure 3F). Together, these findings suggest that recursive splicing preferentially aids processing of long Drosophila introns.
Drosophila ratchet points are associated with cryptic exons genomewide
With our expanded annotation of recursive splice sites in hand, we assessed the breadth of the cryptic exon model for processing intronic RPs. We used NNSPLICE to score potential splice donors (SDs) in a 1 kb window downstream of intronic RP sites. Notably, within 100 nt of RPs, >1/3 of RPs had very high-scoring SDs (>0.8), and >1/2 of RPs scored >0.7 (Figure 4A). To investigate a potential positional bias of these SDs, we plotted their locations at various thresholds, and compared them against SDs downstream of 1000 control intronic AGGT sites. Amongst high scoring (>0.8) SDs, we observe a clear positional bias ~40–80 nt downstream of RP sites (Figure 4B), while background levels of high-scoring SDs were seen throughout the query window downstream of control AGGT sites. Analysis of other bins of SD scores showed similar positional bias, with modest enrichment even at the 0.5–0.6 range (Supplementary Figure 9A). Thus, while our main analyses focus on the top-scoring sites, we conclude the strong majority of recursive sites utilize a positionally constrained cryptic donor.
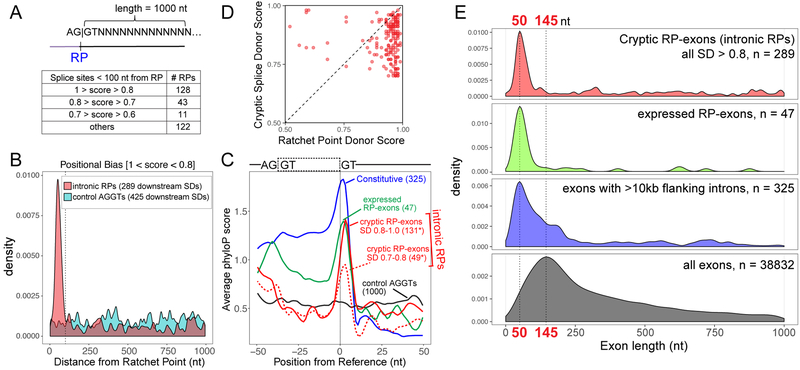
Figure 4. Genomewide identification of RP-associated cryptic exons.
(A) Schematic of strategy used to identify potential splice sites downstream of intronic RPs within a 1000nt window and a summary of non-overlapping RPs that were found to have high-scoring cryptic splice donors <100nt from the RPs. (B) Positional density of high-scoring splice donor sites downstream of intronic RPs (red) and control AGGT (cyan). A total of 289 SDs were found within a 1 kb region for all 304 RPs, whereas 485 SDs were found within a 1 kb region of 1000 control AGGT sites. Distance of splice site from RP is indicated on the x-axis and the dotted line marks 100nt from RPs. Similar positional bias was observed for lower-scoring bins of cryptic splice donors (Supplementary Figure 9). (C) Evolutionary conservation of splice donors from constitutive exons [with flanking intron length >10 kb] (blue), RP-exons (green) and potential cryptic exons [SDs <100nt from intronic RPs] (red line – high score splice sites, red dotted line – moderate score splice sites). *Note that some intronic RPs had >1 high scoring SD within 100nt. (D) Cryptic splice donors are generally weaker than their paired RP donors. Each dot represents the NNSPLICE scores predicted at a given intronic recursive site; only the highest-scoring (>0.7) cryptic splice donors <100 nt from RPs were considered in this analysis. (E) Preferred length of exon definition for cryptic RP-exons, RP-exon and constitutive exons within long intronic contexts. Plotted are exon lengths inferred from intronic RPs + cryptic donor (red), RP-exons (green), constitutive exons with flanking intron length >10 kb (blue) and all Drosophila exons with flanking introns (black). In the case of intronic RPs, all SDs (>0.8 score) found within 1 kb of RPs were used to generate positional density plots (B, E); however, only proximal SDs are predominantly likely to contribute to cryptic exon processing.
We used phyloP to assess conservation of cryptic exon splice donors. We emphasize that when centering such analysis on RPs themselves6, no positionally-biased conservation is apparent downstream (Figure 3B). However, bearing in mind that RP cryptic exons are heterogeneous in length, we reconfigured conservation analyses by centering on cryptic exon splice-site donors, segregated by distance from RPs (Figure 4C and Supplementary Figure 9B). Satisfyingly, we now observe that high-scoring cryptic exon donor splice sites are highly conserved if they are within 100 nts of RPs, while those located further away are not conserved. There was lesser constraint on lower-scoring bins of cryptic splice donors, but clear selection remained at the same position relative to RPs (Supplementary Figure 9). Thus, there is a strong evolutionary constraint on cryptic splice donors, even though their exons are ultimately not utilized in mature mRNAs. Overall, the strong sequence and positional constraint on cryptic splice donor sites, indicates their general importance for recursive splicing. We illustrate conservation of RP-cryptic donor regions from UCSC Genome Browser alignments in Supplementary Data Set 5. Nevertheless, these alignments show that cryptic donor sequences are often less constrained than their partner RP sequences, as reflected by phyloP analyses.
In mammals, usage of cryptic recursively-spliced exons is suppressed following exon definition by competition7. As mentioned, Drosophila differs from mammals in that intronic RPs comprise an extremely abundant class of recursive splicing events. To test if suppression of cryptic exons at RPs can be accommodated by splice donor competition, we compared relative NNSPLICE strengths of pairs of intronic RP and cryptic donors. Of course, we could only do so for identified cryptic SD sites, which progressively get more modest in strength (Supplementary Figure 9). We therefore focused this analysis on the very best cryptic splice donors, i.e., those with scores >0.7. Indeed, for the vast majority of cases, the cryptic donor is weaker than its partner RP donor (Figure 4D), consistent with the model for SD competition.
Finally, we plotted cryptic RP-exon sizes and found them to be tightly distributed around 40–80 nt in length. Notably, this matches the size range of our newly-recognized, broad class of recursive cassette exons (RP-exons). By comparison, the average length of Drosophila exons is about three times larger (Figure 4E). In light of this, we wondered whether recursive splicing exons utilize unique architectural properties. Broadly speaking, intron definition prevails when introns are short, while exon definition prevails when introns are long. However, to our knowledge, the correlation of flanking exon lengths has not been examined systematically in Drosophila. To evaluate this, we identified a set of constitutive exons that are embedded within long flanking introns (>10 kb flanking on either side). Interestingly, the length profiles of these exons mirror those of recursive exons, both cryptic and RP-exons (Figure 4E). The preferred length of constitutive and recursive exons, and their distinct size profile, unifies the strategy of exon definition within long introns in Drosophila.
Discussion
In a prescient discussion on the first case of recursive splicing, Lopez and colleagues eloquently stated that “The internal exons of Ubx are not simply small ships adrift in an intronic ocean, precarious recognition of their splice sites causing more or less frequent skipping in different cellular contexts”4. Instead, Ubx microexons proved to be recursively processed, thereby regenerating 5’ splice sites, such that these short exons could also be alternatively spliced in some isoforms. They concluded that “This mechanism has important implications not only for understanding alternative splicing regulation but also the processing of long introns in complex transcription units”4. Nearly twenty years later, we use genetic, molecular, and computational analyses that elaborate on the far-reaching implications of these statements.
In particular, following the recent molecular validation of ~200 ratchet point events in Drosophila6, many of which were computationally predicted5, we provide evidence that redefines the mechanism of “0-nucleotide” recursive splicing and broadly extends the scope of both constitutive and alternative recursive splicing in Drosophila. We perform the first in vivo RP mutagenesis to demonstrate that disruption of the second step of recursive splicing that is required to skip the cryptic RP-exon can interfere with endogenous gene function. Notably, we show that characteristically-sized, conserved, cryptic exons are critical for recursive splicing via exon definition. That is, Drosophila has a preponderance of introns for which inclusion of the cryptic exon is constitutively suppressed, even though its recognition is central to the recursive splicing process. Not only do we greatly expand the catalog of recursive splicing events that proceed via cryptic RP-exons, we annotate scores of recursive cassette exons in flies. Thus, Ubx recursive microexons are not a lone case, and these events represent a continuum of specialized alternative splicing. Importantly, our studies unify this mRNA processing strategy between flies and mammals, the latter of which may also have hundreds of recursive splice sites that are utilized under certain circumstances7.
Interestingly, the sequence content and length of cryptic exons are evolving, and their splice donor sites turn over more quickly than their companion RP sequences. Even when we experimentally manipulate a reasonably well-conserved cryptic splice donor site, the spliceosome can utilize a fortuitous, non-conserved, donor site. Therefore, RP cryptic exons harbor curious properties: they are functionally critical, yet relatively less conserved modules, in an otherwise well-conserved process of long intron splicing control. The evolutionary plasticity of recursive splicing could potentially lead to the interchange of cryptic and alternative RP-exons at long introns. We observe that progressively longer introns tend to have more recursive sites, that recursive sites are not randomly distributed within such introns but tend to subdivide them. Moreover, GO analysis indicates genes undergoing recursive splicing are enriched for developmental processes, especially neurogenesis and neuronal differentiation (Supplementary Data Set 6). Thus, recursive splicing may preferentially aid the processing of certain types of neural genes.
Online Methods
CRISPR-Cas9-mediated mutagenesis
kuz[RP] and Bx[RP]:
We used the transgenic Cas9-gRNA system12 to perform mutagenesis in the yw background. In the case of Bx, a single gRNA was directed at the recursive splice site using a PAM proximal to the AGGT sequence. In the case of kuz, two gRNAs were directed to flank the recursive splice site. In our typical transgenic CRISPR pipeline we establish 8 lines of candidate mutagenized chromosomes, and evaluate them by PCR and Sanger sequencing13. For Bx, 5/8 candidates contained mutations in the vicinity of the ratchet point, but only one mutant disrupted the site, and contained an alteration in the RP donor. For kuz, 7/8 candidates contained mutations in the vicinity of the ratchet point, but again only one mutant actually disrupted the site and contained an alteration in the RP donor.
Ubx[RP]:
We mutagenized the Ubx-RP using CRISPR-Cas9 and a single stranded oligo donor (ssODN) that abolished the ratchet point splice donor site. The gRNA was directed at the ratchet point and cloned into pCFD3. Injections were performed into yw; nos-Cas9[II-attP40] (BestGene Inc., Chino Hills) and the progeny of surviving animals were screened for site-specific incorporation of the ssODN. These experiments were far less efficient than the transgenic approach. We screened ~600 candidate lines, and recovered a single RP mutant.
All gRNA sequences, screening oligos and ssODN details are provided in Supplementary Table 1.
Immunostaining
To study kuz phenotypes, we balanced kuz[RP] and the amorphic allele kuz[e29–4] (BDSC#5804) over Cyo[Ubi-GFP]. We collected embryos from the homozygous and trans-heterozygous crosses as well as control Canton S animals at 25°C. Embryos were aged, fixed and stained using the following primary antibodies: chicken anti-GFP (1:1000, Abcam #ab13970), mouse anti-BP102 (1:10, DSHB) and anti mouse-Fas II (1:100, 1D4, DSHB). Secondary antibodies used were made in donkey and conjugated to Alexa-488, −568 or −647 (Jackson ImmunoResearch). Stacks of images were obtained using a Leica confocal microscope using a 40x oil immersion objective and maximum projections were generated using ImageJ-LOCI plugin. kuz mutant animals were identified by the lack of GFP staining.
To study Ubx phenotypes, we used the amorphic allele Ubx[1] (BDSC#2866) and the balancer chromosome TM3, Sb, Ser, which also carries the hypomorphic allele Ubx[bx-34e]. To stain for Ubx, Ubx[RP]/TM6B-[ubi-GFP] or yw flies were allowed to lay eggs in cages for 24 hrs at 25°C. After sufficient time, GFP-negative 1st instar larvae were hand-picked under a fluorescence microscope and dissected to obtain CNS. The samples were fixed and incubated with the following primary antibodies: rat anti-Elav (1:100, 7E8A10, DSHB) and mouse anti-Ubx (1:10, FP3.38, DSHB).
Constructs and cell culture
We created a minimal construct consisting of the following fragments stitched together from kuz: fragment of exon 2 (from start codon to end of exon 2), exon 3, a reduced version of intron 3 (131 nt of 5’ end and 290 nt of 3’ end), and 150 nt of exon 4. NotI and EcoRV restriction sites were added between the two intron 3 fragments to allow for further modifications. All fragments were cloned into pAC-5.1-V5-His using Gibson Assembly®.
To create pAC-kuzMG-kuzRI, a ~2.6 kb fragment surrounding kuz-RP1 was PCRed from wildtype animals and cloned into the minimal vector using NotI and EcoRV sites. Similarly, to create pAC-kuzMG-BxRI, we used ~2.5 kb fragment surrounding Bx-RP. To obtain ratchet point splice donor mutants, PCR was performed on RP mutant animals and cloned into the minimal vector. We used site directed mutagenesis to make all other mutants constructs. All primers used for cloning and mutagenesis can be found in Supplementary Table 1.
All transfections in this study were performed using S2-R+ cells cultured in Schneider Drosophila medium with 10% fetal Bovine serum. Cells were seeded in 6-well plates at a density of 1 million/mL and transfected with 100 ng of construct using the Effectene transfection kit [Qiagen]. Cells were harvested following three days of incubation.
rt-PCR of mRNA and recursive intermediates
Ubx[RP] and kuz[RP]:
mutant stocks were made with GFP balancers. Homozygous 1st instar larval mutants (GFP-) were hand-picked under a fluorescence microscope. Animals were homogenized and RNA was extracted using the standard Trizol protocol. 2 μg of RNA was treated with Turbo DNase [Ambion] for 45 min before cDNA synthesis using SuperScript III [Life Technology] with random hexamers. rt-PCRs were done using AccuPrime™ Pfx DNA polymerase [ThermoFisher Scientific] with standard protocol using 28 cycles for mRNA and 34 cycles for intermediates.
Bx[RP] - similar to Ubx[RP] and kuz[RP], except for the following differences: homozygous mutant adult flies were homogenized and RNA was extracted using Trizol. 5 μg of RNA was DNase treated and reverse transcribed using random hexamers. rt-PCRs were done using 35 cycles for mRNA and intermediates.
Cell culture: RNA was collected from transfected cells using Trizol. 5 μg of RNA was treated with Turbo DNase [Ambion] for 45 min before cDNA synthesis using SuperScript III [Life Technology] with random hexamers. rt-PCRs were done using AccuPrime™ Pfx DNA polymerase [ThermoFisher Scientific] with standard protocol using 26 cycles and primers that were specific to minigene construct. All primers with descriptions can be found in Supplementary Table 1.
Bioinformatic annotation of putative ratchet points from nascent RNA datasets
We used publicly available nascent RNA-seq and genomic run-on sequencing (GRO-seq) datasets from NCBI’s sequence read archive14–21, described in Supplementary Data Set 2. The datasets were mapped to the Drosophila melanogaster BDGP R5 (dm3) reference genome using TopHat2 under default settings22.
In theory, recursive splice sites should contain tandem splice acceptor and donor sequences (Supplementary Figure 1A). Therefore, to identify putative recursive splice sites, we first collated all junction-spanning loci (in the case of splice junctions, introns) and kept those that contained the AGGT tetranucleotide across the 3’ end (Supplementary Note 1). This ensures that the junctions have tandem minimal consensus splice acceptor (AG) and splice donor (GT) motifs. We then classified the 3’ ends of AGGT junctions based on where they occur, such as exon junctions, exon body, introns, cassette exon junctions or intergenic space, using RefSeq Gene annotations (Supplementary Note 1). Since recursive splice sites should occur within intronic regions of the transcriptome, we only further analyzed events that were unambiguously intronic (“0-nt” recursive splicing candidates) or mapping to cassette exon junctions (RP-exon candidates – see below). These loci were examined in depth for potential ratchet points.
Up to this point, to cast a wide net, we had qualified all AGGT junctions as candidates. However, to narrow down to a set of likely candidates, we employed the splice site prediction tool, NNSPLICE23, to quantify 3’ and regenerated 5’ splice site scores (Supplementary Note 1). Simultaneously, we merged and converted all nascent RNA and GRO sequencing datasets into the browser-friendly bigWig format, and manually inspected all intronic recursive splicing candidates for the characteristic saw-tooth pattern expected of ratchet points. We found 271 sites that had high splice site scores and were supported by clear saw-tooth patterns (Figure 3A, Supplementary Note 1).
However, we observed that the strength of the saw-tooth pattern was a continuum, which likely depends on library properties such as coverage and inherent host gene properties, such as recursive intermediate stability. Therefore, we reasoned that we were systematically removing potential RPs by requiring a saw-tooth pattern, and sought to acquire these by selecting sites with high splice site scores. We set splice score cutoffs to mirror the scores of RPs that were supported by saw-tooth patterns (< 0.75) and found a total of 33 additional intronic RP loci (Figure 3A, Supplementary Note 1).
Bioinformatic annotation of RP-exons from nascent RNA datasets
After identifying cryptic exons associated with intronic RPs through bioinformatics and experimentation, we inferred that some known cassette exons might also be processed by recursive splicing. We utilized an analogous strategy to identify a set of expressed RP-exons for further inspection (see above) and supplemented these with all annotated cassette exons that were not sampled due to sample or tissue type. Typically, saw-tooth patterns are used in the annotation of RPs. However, since expressed RP-exons are stable exons recovered in transcriptomic data, we had to rely on splice site score alone to predict potential recursive splicing. Therefore, we scored all 3’ and regenerated 5’ splice sites from cassette exon junctions using NNSPLICE, and employed a strict cutoff of 0.85, resulting in the annotation of 47 expressed RP-exons (Supplementary Note 1). These were generally alternatively spliced.
Identification of potential cryptic exon donor splice sites
Following the observation of intron retention in kuz[RP], Bx[RP] and Ubx[RP], we manually browsed other RPs and noticed a similar and regular occurrence of 5’ splice sites downstream of the AGGT site. To formalize this observation, we used NNSPLICE to search for splice sites of varying strengths within a 1 kb region downstream of all putative intronic RPs. As control, we used a set of AGGT sites from introns of matched length as the RPs, and likewise looked for splice sites.
Evaluation of recursive splice site and cryptic exon donor site conservation
We used phyloP scores from the UCSC Genome Browser to assess conservation. Briefly, potential ratchet points from intronic cryptic RP-exon and expressed RP-exon categories were anchored at position 0 and phyloP scores for all sites were summed and averaged at each position from −50 to +50.
To calculate conservation of cryptic exon splice donors, we split all cryptic splice sites into two categories: those occurring <100 nt of RPs and those occurring >100 nt from RP. For each set of cryptic sites, we anchored each cryptic splice site at position 0 and calculated phyloP scores for each nucleotide position from −50 to +50. To graph conservation, we averaged the phyloP scores at each position per set.
Statistics and Reproducibility
Adult fly phenotypes in Figure 1B–D were evaluated using >100 animals from each genotype, and the phenotypes were completely penetrant. For immunostaining experiments in Figure 1F–G, we imaged 3 yw and 2 Ubx[RP/RP] 1st instar larval CNS, and the normal Ubx pattern was observed in all wildtype CNS and completely absent in Ubx mutants. For immunostaining experiments in Figure 1H–I, we imaged 7 yw and 7 kuz[RP]/[RP] embryonic CNS, and the mutant phenotypes were completely penetrant. For Figure 1J–M, we imaged 7 kuz[RP]/+ (J), 8 kuz[e29–4]/[e29–4] (K), 9 kuz[RP]/[RP] (L) and 3 kuz[RP]/[e29–4] embryonic CNS. All control CNS were normal whereas all kuz mutant combinations exhibited the defects shown in the representative images.
For Figure 2, the rt-PCR experiments were performed in biological triplicates and the cell culture reporter experiments were performed in biological duplicates.
Data Availability and Code availability Statement
Source data for plots in Figures 3–4 are available with the Supplementary Data Sets online. Custom scripts used in this study are available from the authors upon request.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center and the Developmental Studies Hybridoma Bank for fly stocks and antibodies used in this study. S.K. supported by the Mochida Memorial Foundation for Medical and Pharmaceutical Research. Work in E.C.L.’s group was supported by the National Institutes of Health (R01-NS083833 and R01-GM083300) and MSK Core Grant P30-CA008748).
Footnotes
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.De Conti L, Baralle M & Buratti E Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA 4, 49–60 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Singh J & Padgett RA Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16, 1128–33 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollander D, Naftelberg S, Lev-Maor G, Kornblihtt AR & Ast G How Are Short Exons Flanked by Long Introns Defined and Committed to Splicing? Trends Genet 32, 596–606 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Hatton AR, Subramaniam V & Lopez AJ Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon-exon junctions. Mol Cell 2, 787–96 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J & Lopez AJ Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics 170, 661–74 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duff MO et al. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 521, 376–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sibley CR et al. Recursive splicing in long vertebrate genes. Nature 521, 371–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook-Andersen H & Wilkinson MF Molecular biology: Splicing does the two-step. Nature 521, 300–1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly S et al. Splicing of many human genes involves sites embedded within introns. Nucleic Acids Res 43, 4721–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bender W et al. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science 221, 23–9 (1983). [DOI] [PubMed] [Google Scholar]
- 11.Fambrough D, Pan D, Rubin GM & Goodman CS The cell surface metalloprotease/disintegrin Kuzbanian is required for axonal extension in Drosophila. Proc Natl Acad Sci U S A 93, 13233–8 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo S & Ueda R Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo S et al. New genes often acquire male-specific functions but rarely become essential in Drosophila. Genes Dev 31, 1841–1846 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohn F, Sienski G, Handler D & Brennecke J The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157, 1364–79 (2014). [DOI] [PubMed] [Google Scholar]
- 15.McMahon AC et al. TRIBE: Hijacking an RNA-Editing Enzyme to Identify Cell-Specific Targets of RNA-Binding Proteins. Cell 165, 742–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YA et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol Cell 63, 97–109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez J, Menet JS & Rosbash M Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol Cell 47, 27–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sienski G, Donertas D & Brennecke J Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozhkov NV, Hammell M & Hannon GJ Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27, 400–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrari F et al. “Jump start and gain” model for dosage compensation in Drosophila based on direct sequencing of nascent transcripts. Cell Rep 5, 629–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W et al. Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms. Mol Cell 59, 819–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reese MG, Eeckman FH, Kulp D & Haussler D Improved splice site detection in Genie. J Comput Biol 4, 311–23 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for plots in Figures 3–4 are available with the Supplementary Data Sets online. Custom scripts used in this study are available from the authors upon request.