The gut microbiome is critical in providing resistance against colonization by exogenous microorganisms. The mechanisms via which the gut microbiota provide colonization resistance (CR) have not been fully elucidated, but they include secretion of antimicrobial products, nutrient competition, support of gut barrier integrity, and bacteriophage deployment.
KEYWORDS: bacterial enteric infection, bacteriocins, bile acids, colonization resistance, enteric pathogens, gut microbiota, microbiome, mucus layer, nutrient competition, short-chain fatty acids, bacteriophages
SUMMARY
The gut microbiome is critical in providing resistance against colonization by exogenous microorganisms. The mechanisms via which the gut microbiota provide colonization resistance (CR) have not been fully elucidated, but they include secretion of antimicrobial products, nutrient competition, support of gut barrier integrity, and bacteriophage deployment. However, bacterial enteric infections are an important cause of disease globally, indicating that microbiota-mediated CR can be disturbed and become ineffective. Changes in microbiota composition, and potential subsequent disruption of CR, can be caused by various drugs, such as antibiotics, proton pump inhibitors, antidiabetics, and antipsychotics, thereby providing opportunities for exogenous pathogens to colonize the gut and ultimately cause infection. In addition, the most prevalent bacterial enteropathogens, including Clostridioides difficile, Salmonella enterica serovar Typhimurium, enterohemorrhagic Escherichia coli, Shigella flexneri, Campylobacter jejuni, Vibrio cholerae, Yersinia enterocolitica, and Listeria monocytogenes, can employ a wide array of mechanisms to overcome colonization resistance. This review aims to summarize current knowledge on how the gut microbiota can mediate colonization resistance against bacterial enteric infection and on how bacterial enteropathogens can overcome this resistance.
INTRODUCTION
The human gastrointestinal (GI) tract is colonized by an enormous number of microbes, collectively termed gut microbiota, including bacteria, viruses, fungi, archaea, and protozoa. Bacteria achieve the highest cell density, estimated to be approximately 1011 bacteria/ml in the colon (1). Research has long focused on the pathogenicity of microbes and not on their potential beneficial roles in human health. These beneficial roles include aiding immune system maturation, production of short-chain fatty acids (SCFAs), vitamin synthesis, and providing a barrier against colonization with potential pathogens (2). Additionally, the gut microbiota have extensive interactions with our immune system, and they have been associated with many immune-mediated diseases both in and outside the gut (3–5). Over the last 10 years, there has been increased interest in elucidating the bidirectional relationship between the gut microbiota and human health and disease. This has been partly propelled by improved sequencing technologies, allowing the profiling of entire microbial communities at high efficiency and low cost (6).
Hundreds of different bacterial species inhabiting the healthy human gut have been identified (7, 8). Initial studies seeking to elucidate the relationship between the human microbiota and health and disease were largely observational; the gut microbiota compositions of diseased and healthy groups would be compared and subsequently associated with clinical markers (9). Currently, the field is moving toward more functional and mechanistic studies by including other omics techniques.
In healthy individuals, the gut microbiota provide protection against infection by deploying multiple mechanisms, including secretion of antimicrobial products, nutrient competition, support of epithelial barrier integrity, bacteriophage deployment, and immune activation. Together, these mechanisms contribute to resistance against colonization by exogenous microorganisms (colonization resistance [CR]) (10). Also, however, in the absence of a fully functional immune system, the gut microbiota can provide crucial and nonredundant protection against a potentially lethal pathogen (11). This review discusses the mechanisms used by the gut microbiota to provide CR, the impacts of various drugs on the gut microbiota and thereby on CR, and the strategies of specific bacterial pathogens to overcome CR and ultimately cause enteric infection.
MECHANISMS PROVIDING COLONIZATION RESISTANCE
The gut microbiota produce various products with antimicrobial effects, including SCFAs, secondary bile acids and bacteriocins. Each of these contributes to CR in a product-specific manner. Their general mechanisms of action are described below. The contribution of the immune system in conferring CR has been extensively reviewed previously and is outside the scope of this review (12, 13).
Short-Chain Fatty Acids
SCFAs are mainly produced by bacteria through fermentation of nondigestible carbohydrates (Fig. 1) (14). The three main SCFAs are acetate, propionate, and butyrate, constituting 90% to 95% of the total SCFA pool (15). Under homeostatic conditions, butyrate is the main nutrient for enterocytes and is metabolized through β-oxidation. Thereby, an anaerobic milieu inside the gut can be maintained (16). SCFAs can impair bacterial growth by affecting intracellular pH and metabolic functioning. SCFA concentrations have been shown to be inversely related to pH throughout different regions of the gut (17). At lower pH, SCFAs are more prevalent in their nonionized forms, and these nonionized acids can diffuse across the bacterial membrane into the cytoplasm. Within the cytoplasm, they dissociate, resulting in a buildup of anions and protons, leading to a lower intracellular pH (18).
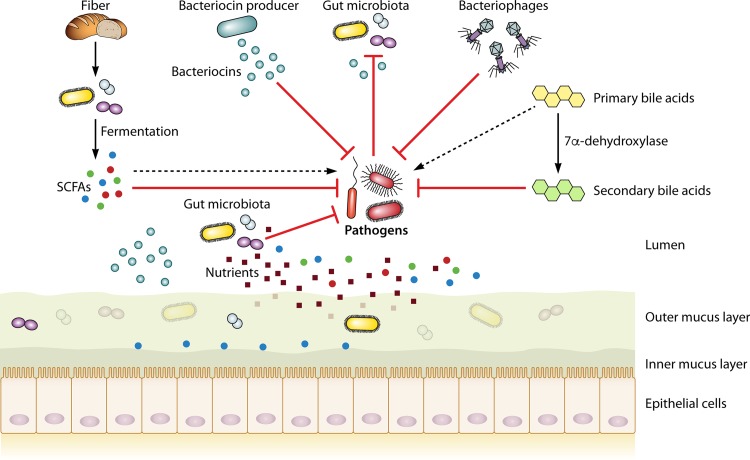
FIG 1.
Outline of gut microbiota-mediated colonization resistance mechanisms. Fiber obtained from the diet is fermented by the gut microbiota into SCFAs. Bacteriocin producers produce bacteriocins capable of targeting a specific pathogen. Primary bile acids can be converted by a very select group of gut microbiota into secondary bile acids, which generally have properties antagonistic to pathogens. Nutrient competition of native microbiota can limit access to nutrients for a pathogen. Specific organisms can use SCFAs, bacteriocins, and primary bile acids to increase their virulence, as discussed in the text.
In the presence of acetate, metabolic functioning of Escherichia coli could be impaired by preventing the biosynthesis of methionine, leading to accumulation of toxic homocysteine and growth inhibition. Growth inhibition was partly relieved by supplementing the growth medium with methionine, showing that this metabolic dysfunction is one of the factors by which SCFAs impair bacterial growth (19).
Bile Acids
Bile acids, which possess antimicrobial properties, are produced by the liver and excreted in the intestinal tract to aid in the digestion of dietary lipids. After production of primary bile acids in the liver, they are subsequently conjugated with glycine or taurine to increase solubility (20). They are then stored in the gallbladder and, upon food intake, are released into the duodenum to increase solubilization of ingested lipids. A large part of the conjugated primary bile acids (50% to 90%) is reabsorbed in the distal ileum, while the remainder can be subjected to bacterial metabolism in the colon (20). There, conjugated bile acids can be deconjugated by bile salt hydrolases (BSH), which are abundantly present in the gut microbiome (21). Deconjugated primary bile acids can subsequently be converted into the two main secondary bile acids, deoxycholic acid and lithocholic acid, by a few bacteria, mostly Clostridium species, via 7α-dehydroxylation through a complex biochemical pathway (21–23) (Fig. 1). A crucial step during the conversion is encoded by the baiCD genes, which are found in several Clostridium strains, including Clostridium scindens (24). Deoxycholic acid is bactericidal to many bacteria, including Staphylococcus aureus, Bacteroides thetaiotaomicron, Clostridioides difficile, bifidobacteria, and lactobacilli, by membrane disruption and subsequent leakage of cellular content (25–28).
The importance of bacteria for conversion of primary bile acids was demonstrated by investigating bile acid profiles in germ-free mice, where no secondary bile acids could be measured (29). Very few colonic bacteria, less than 0.025% of the total gut microbiota, are capable of performing 7α-dehydroxylation (23, 30). One of these bacteria, C. scindens, is associated with colonization resistance against C. difficile through secondary-bile-acid production (22, 31). A follow-up in vivo study demonstrated that C. scindens provided CR in the first day postinfection (p.i.), but protection and secondary-bile-acid production were lost at 72 days p.i. (32). C. scindens on its own was also not sufficient to inhibit C. difficile outgrowth in humans (33). Together, these studies suggest either that C. scindens requires cooperation with other secondary-bile-acid-producing bacteria or that other mechanisms were involved in providing CR. The secondary bile acid lithocholic acid may exert its antimicrobial effects, and potentially its effects on CR, in an indirect manner. Lithocholic acid has been shown to enhance transcription for the antimicrobial peptide LL-37 in gut epithelium using an HT-29 cell line (34). However, no increased mRNA transcription or protein translation of LL-37 was observed in another study using a Caco2 cell line (35).
Bacteriocins
Bacteriocins are short, toxic peptides produced by specific bacterial species that can inhibit the colonization and growth of other species (36) (Fig. 1). Their mechanisms of action are multiple and include disturbing RNA and DNA metabolism and killing cells through pore formation in the cell membrane (37–40). Bacteriocins can be divided into those produced by Gram-positive bacteria and those produced by Gram-negative bacteria. Further classification of bacteriocins has been extensively discussed previously (41, 42). The bacteriocins of Gram-positive bacteria are mostly produced by lactic acid bacteria (e.g., Lactococcus and Lactobacillus) and some Streptococcus species and are further subdivided into three major classes on the basis of the molecular weight of the bacteriocins and the presence of posttranslational modifications (42). Bacteriocins produced by Gram-negative bacteria, mostly by Enterobacteriaceae, can be broadly divided into high-molecular-weight proteins (colicins) and lower-molecular-weight peptides (microcins) (41).
The lantibiotic nisin is the best-studied bacteriocin and is produced by Lactococcus lactis strains. It has potent activity against many Gram-positive bacteria but has much less intrinsic activity against Gram-negative organisms (43–45). By itself, nisin does not induce growth inhibition of Gram-negative bacteria, since binding to lipid II—the main target—is prevented by the outer bacterial membrane (46). Therefore, studies have used different methods to overcome this problem by combining nisin with chelating agents, like EDTA, antibiotics, and engineered nisin peptides (47–52). These compounds can destabilize the outer membrane, allowing nisin to exert its damaging effect (53, 54).
Several in vivo models have confirmed the potency of bacteriocins in providing CR.
Lactobacillus salivarius UCC 118, which produces the bacteriocin Abp118, was able to significantly protect mice from infection by direct killing of Listeria monocytogenes, while a UCC 118 mutant could not, confirming the protective role of Abp118 against the foodborne pathogen (55).
Another example is Bacillus thuringiensis DPC 6431, which produces the bacteriocin thuricin (36). Thuricin targets several C. difficile strains, including the highly virulent PCR ribotype 027. In vitro, its activity was more potent than that of metronidazole, the common treatment for C. difficile infection (56). In a colon model system, metronidazole, vancomycin, and thuricin all effectively reduced C. difficile levels. However, thuricin has the advantage of conserving the gut microbiota composition. This is highly relevant, as a disturbed microbiota is associated with increased susceptibility to infection (57, 58).
Enterobacteriaceae members can produce specific bacteriocins called colicins, and one example, colicin FY, is encoded by the Yersinia frederiksenii Y27601 plasmid. Recombinant E. coli strains capable of producing colicin FY were shown to be highly effective against Yersinia enterocolitica in vitro (59). In vivo experiments were performed by first administering the recombinant E. coli strains, after which mice were infected with Y. enterocolitica. In mice with normal gut microbiota, the recombinant strains did not inhibit Y. enterocolitica infection, while infection was effectively reduced in mice pretreated with streptomycin (59). This was most probably the result of increased colonization capacity of recombinant E. coli in the inflamed gut, while the normal gut microbiota provided sufficient CR to prevent E. coli colonization (59).
Microcins are also produced by Enterobacteriaceae but differ from colicins in several ways (60). For example, microcins are much smaller (<10 kDa), and microcin production is not lethal to the producing bacterium, in contrast to colicin production (60). E. coli Nissle 1917, capable of producing microcin M and microcin H47, could significantly inhibit Salmonella enterica serovar Typhimurium in vitro and in vivo (61). This inhibition, however, was seen only during intestinal inflammation, when S. Typhimurium expresses siderophores to scavenge iron from an iron-depleted environment. As microcins are able to conjugate to siderophores and S. Typhimurium takes up the siderophore during iron scavenging, microcins are introduced into the bacterial cell in a Trojan-horse-like manner (62).
In silico identification of bacteriocin gene clusters shows that much remains to be discovered in this area, as 74 clusters were identified in the gut microbiota (63). Not all of these clusters may be active in vivo, but it illustrates the potential relevance of bacteriocin production by the gut microbiota to providing colonization resistance.
Nutrient Competition
Bacteria have to compete for nutrients present in the gut. This is especially relevant for bacterial strains belonging to the same species, as they often require similar nutrients. The importance of nutrient competition in providing CR has been shown in multiple studies using multiple E. coli strains (64–67). Indigenous E. coli strains compete with pathogenic E. coli O157:H7 for the amino acid proline (64). In fecal suspensions, depletion of the proline pool by high-proline-utilizing E. coli strains inhibited growth of pathogenic E. coli. This inhibition could be reversed by adding proline to the medium, thereby confirming nutrient competition between the strains (64). In addition to amino acids, different E. coli strains use distinct sugars present in the intestinal mucus (65). When two commensal E. coli strains that together utilize the same sugars as E. coli O157:H7 were present in the mouse gut, E. coli O157:H7 was unable to colonize after it was administered to the mice. However, E. coli O157:H7 successfully colonized when only one of the commensals was present. This indicated that the two commensals complement each other to sufficiently deplete all the sugars used by the pathogenic E. coli strain (66). Nutrient competition is not limited to macronutrients but can extend to micronutrients, such as iron. S. Typhimurium is known to take up large amounts of iron from the inflamed gut during infection (67). Upon a single administration of the probiotic E. coli Nissle 1917, which was proposed to scavenge iron very efficiently, S. Typhimurium levels were reduced by more than 2 log units during infection via the limitation of iron availability. Administration of E. coli Nissle 1917 prior to infection with S. Typhimurium led to 445-fold lower colonization (67).
Finally, genome scale metabolic models have been used to reconstruct microbiome-wide metabolic networks, which could partly predict which species utilize specific compounds from their environment (68). These models have been used to study nutrient utilization by C. difficile, which is described below.
Together, these studies show that colonization resistance through nutrient competition is most effective when the microbiota take up key nutrients that are required by the pathogen (Fig. 1). Future strategies, therefore, could aim at administering probiotic strains that are able to outcompete pathogens for specific nutrients. This is especially relevant at times of gut microbiota disturbance, e.g., during and following antibiotic treatment, as this is the time window in which it is easiest for exogenous bacteria to colonize the GI tract.
Mucus Layers
The gut barrier consists of the inner and outer mucus layers, the epithelial barrier, and its related immune barrier. It is beyond the scope of this review to discuss the full immunological characteristics of the epithelial barrier, the highly complex host-microbe interactions occurring at the mucus layer, and host-associated genetic polymorphisms associated with mucus layer composition, as these have been extensively described previously (12, 13, 69, 70). Instead, a general description with various examples of how the mucus layer provides CR is given.
The inner mucus layer is impenetrable and firmly attached to the epithelium, forming a physical barrier for bacteria, thereby preventing direct interaction with the epithelial layer and a potential inflammatory response (71, 72). Commensal gut microbes reside and metabolize nutrients in the nonattached outer mucus layer. Thinning of the mucus layer leads to increased susceptibility to pathogen colonization, which can result from a Western-style diet deficient in microbiota-accessible carbohydrates (MACs) (58). When MACs were scarce, mucus-degrading bacteria (Akkermansia muciniphila and Bacteroides caccae) fed on the outer mucus layer in a gnotobiotic-mouse model, resulting in closer proximity of bacteria to the epithelial layer (58). The host adapted by increasing muc2 expression, the main producer of intestinal mucin glycans, but failed to do so sufficiently. Inner-mucus-layer damage, however, could be reversed by administration of Bifidobacterium longum, perhaps due to stimulation of mucus generation (73).
The composition of the microbiota is thus a contributing factor to the integrity of the mucus barrier. Genetically identical mice housed in different rooms at the same facility showed distinct microbiota compositions, with one group of mice showing a more penetrable barrier (74). When fecal-microbiota transplant (FMT) was performed on germ-free mice, they displayed the same barrier function as their respective donors. No specific microbes were identified as responsible for the change in observed barrier function (74).
In conclusion, the mucus layers provide a first barrier of defense against colonization by exogenous microorganisms. Diet has been shown to be an important factor in the proper functioning of the layers, suggesting that dietary intervention, or specific pro- and prebiotics, may be a future therapeutic option.
Bacteriophages
Bacteriophages are the most abundant microorganisms on our planet and are also present in high numbers in the human gut (75, 76). Bacteriophages have been proposed as potential alternatives to antibiotics, as they are highly specific, targeting only a single or a few bacterial strains, thereby minimizing the impact on commensal members of the microbiota (75, 77) (Fig. 1). Their complex interactions in the intestine with both host immunity and bacterial inhabitants are starting to be explored, but much remains to be elucidated (76). Here, we focus on their relationship with bacterial enteropathogens.
Vibrio cholerae infection could be controlled using a prophylactic phage cocktail in mice and rabbits (78). The prophylactic cocktail killed V. cholerae in vitro, reduced colonization by V. cholerae in the mouse gut, and prevented cholera-like diarrhea in rabbits. Importantly, the authors suggest that the concentration of phages in the gut is an important criterion for successful prevention of infection, as the time between phage cocktail administration and V. cholerae inoculation was associated with treatment outcome (78). Similar findings have been demonstrated for Campylobacter jejuni colonization in chickens, where a phage cocktail reduced C. jejuni levels by several orders of magnitude (79).
Bacteriophages can also confer a competitive advantage on commensals. Enterococcus faecalis V583 harbors phages that infect and kill other E. faecalis strains, thereby creating a niche for E. faecalis V583 (80).
Phages play an important role in excluding specific gut bacteria and can thereby contribute to CR. Therapeutic use in humans is not yet performed on a wide scale in the Western world, as sufficient evidence for their safety and efficacy is still lacking (81). However, recent case reports indicate that bacteriophage treatment has definite future potential for treating multidrug-resistant bacteria (82, 83).
EFFECTS OF VARIOUS NONANTIBIOTIC DRUGS ON GUT COLONIZATION RESISTANCE
Antibiotics have long been known for their deleterious effect on the gut microbiota. Recently, various other drugs have come to our attention due to their impact on our microbial ecosystem. As the effects of antibiotics have been extensively reviewed previously (84, 85), the focus in the current review is on nonantibiotic drugs, namely, proton pump inhibitors (PPIs), antidiabetics, and antipsychotics.
Proton Pump Inhibitors
PPIs inhibit gastric acid production and are among the most prescribed drugs in Western countries (86). A significant association between long-term use of PPIs and the risk of several bacterial enteric infections has been demonstrated in multiple systematic reviews (87–90).
Several studies have associated PPI use with microbiota alterations that may specifically predispose to C. difficile infection and to small-intestinal bacterial outgrowth (91–95). Especially taxa prevalent in the oral microbiota (e.g., Streptococcus) were associated with PPI use, likely resulting from increased gastric pH, allowing colonization by the bacteria further down the gastrointestinal tract (91–94). Administering PPIs to 12 healthy volunteers for 4 weeks did not result in changes in diversity or in the overall microbiota composition. However, the abundances of specific taxa associated with C. difficile infection and gastrointestinal bacterial overgrowth increased, thereby potentially lowering colonization resistance against C. difficile (91).
The results of two mouse studies suggest that reduced bactericidal effect, due to increased stomach pH, may be the most important factor for increased enteric infection risk. Mice received PPIs 7 days prior to infection with the murine pathogen Citrobacter rodentium, which resulted in increased numbers of C. rodentium organisms in the cecum 1 h postinoculation compared to control mice (96). Similar results were observed in another study, where treatment of mice with PPIs led to increased colonization by vancomycin-resistant enterococci and Klebsiella pneumoniae (97). In spite of their general acceptance as a model for gut disturbances, it is important to note that the mice were pretreated with clindamycin, which may limit generalizability (97). This is an important issue when studying the effects of PPIs, as the combined use of medications in the human population complicates the study of the effects of PPIs on the microbiota and CR. Even though large-scale studies have adjusted for confounders to filter out the effect of PPIs on the gut microbiota, this does not represent a mechanistic study, where only PPIs would be administered (92, 98).
Therefore, more mechanistic studies investigating how PPIs increase the risk for enteric infection are required. These studies should exclusively administer PPIs to healthy human volunteers or animals.
Antidiabetics
Metformin is the primary drug prescribed for treatment of type II diabetes mellitus (T2DM) and mainly acts by reducing hepatic glucose production, thereby lowering blood glucose levels (99). The current increase in the number of T2DM patients is unprecedented, and it is therefore crucial to evaluate metformin’s effect on the gut microbiota and colonization resistance (100).
The microbiota of T2DM patients are, among other changes, characterized by depletion of butyrate-producing bacteria (101, 102). Metformin administration increases both the abundance of butyrate and other SCFA-producing bacteria and fecal SCFA levels and may thus contribute to colonization resistance. The underlying mechanisms remain unknown (101, 103).
Another effect of metformin has been studied in an in vitro model, where it was found to reduce tight-junction dysfunction of the gut barrier by preventing tumor necrosis factor alpha (TNF-α)-induced damage to tight junctions (104). Similar findings of improvement of tight-junction dysfunction were demonstrated using two in vivo models, one using interleukin-10-deficient mice and one using a mouse colitis model (105, 106). As tight junctions are a critical part of epithelial barrier integrity, alleviating their impaired functioning likely improves CR.
In conclusion, metformin may have beneficial effects on CR, as its ability to raise SCFA concentrations and improve tight-junction function suggests. The effects of metformin on the gut microbiota and CR in healthy organisms need further evaluation.
Antipsychotics
The interest in whether antipsychotics affect gut microbiota composition and colonization resistance may surge after a recent publication demonstrating that antipsychotics target microbes based on their structural composition (107). This led to the suggestion that antibacterial activity may not simply be a side effect of antipsychotics but can be part of their mechanism of action (107). Various antipsychotics have been investigated for their antibacterial effects, several of which are highlighted here.
In an in vitro model, olanzapine has been demonstrated to completely inhibit the growth of two potentially pathogenic bacteria, E. coli and E. faecalis (108). Pimozide has been shown to inhibit the internalization of several bacteria, including L. monocytogenes (109). An in vitro screening test evaluated the effects of fluphenazine on 482 bacterial strains belonging to 10 different genera. Growth inhibition was demonstrated in multiple species, including five out of six Bacillus spp., 95 out of 164 staphylococci, 138 out of 153 V. cholerae strains, and Salmonella serovars Typhi and Typhimurium. Significant protection by administering fluphenazine was shown in a mouse model infected with S. Typhimurium, as the number of viable cells in several organs was lower and overall survival was higher than for controls (110).
Antipsychotics can also be used in combination with antibiotics to exert a synergistic antibacterial effect. Flupenthixol dihydrochloride (FD) was demonstrated to have antibacterial activity both in vitro and in vivo (111). Coadministration of FD and penicillin yielded extra protection against S. Typhimurium compared to singular administration of either drug (111). As antipsychotics have only recently been recognized to have potential antimicrobial effects, studies have looked only at the effects on pathogens. It is likely that gut commensals are also affected by these drugs, but future studies will have to confirm this hypothesis.
Apart from their potential antibacterial effects, several antipsychotics were shown to increase intestinal permeability in the distal ileum in rats and therefore showed a possible detrimental effect on CR (112). Curiously enough, use of antidepressants was associated with increased risk of C. difficile infection development, although no underlying mechanism has yet been elucidated (113).
In conclusion, antipsychotics have definite antibacterial effects, but to our knowledge, no studies have yet been performed regarding their effects on colonization resistance and bacterial enteric infection in vivo.
COLONIZATION RESISTANCE AGAINST SPECIFIC BACTERIAL ENTERIC PATHOGENS
Other than antibiotic resistance acquisition, enteric pathogens possess multiple virulence factors to overcome CR and cause infection. Some of these factors are common and apply to many bacterial species, while others are organism specific. Mechanisms implicated in antibiotic resistance development include horizontal gene transfer, mutational resistance, and altering the structure and thereby the efficacy of the antibiotic molecule. Full reviews describing these mechanisms in depth can be found elsewhere (114, 115). Here, the main focus is on how several of the most prevalent and dangerous bacterial enteropathogens overcome the mechanisms providing CR described here, namely, secretion of antimicrobial products, nutrient competition, mucus barrier integrity, and bacteriophage deployment. As insufficient knowledge is available on how each specific enteropathogen overcomes CR by rendering bacteriophages ineffective, apart from the well-known and conserved CRISPR (clustered regularly interspaced short palindromic repeat)-Cas, an overview of the currently known bacterial defense mechanisms is provided at the end of this review.
C. difficile
C. difficile-associated diarrhea is the most common hospital-acquired infection, causing more than 450,000 diarrheal cases per year in the United States alone (116). Clinical symptoms can range from self-limiting diarrhea to bloody diarrhea, pseudomembranous colitis, and ultimately death (117). However, in healthy individuals as well, CR is not always successful against this opportunistic pathogen, resulting in asymptomatic colonization in 2% to 15% of the healthy population (118). The reason why some asymptomatically colonized patients do not develop infection while others do may well be found in the gut microbiome, although no mechanisms have yet been elucidated. C. difficile contains a pathogenicity locus with the information to produce its two major toxins, TcdA and TcdB. The significance of a third toxin, called binary toxin, is less clear. Toxin production in the colon is facilitated by disruption of the native gut microbiota, for instance, through antibiotic use (119).
The effects of SCFAs on C. difficile throughout its life cycle are currently unclear (120–122). In an antibiotic-treated-mouse model, decreased SCFA levels were associated with impaired CR against C. difficile (120). CR was subsequently restored 6 weeks after ending antibiotic treatment, with a concomitant increase in SCFAs, probably resulting from restoration of the fermentative activity of the microbiota (120). Restoration of SCFA levels is also seen as an effect after fecal-microbiota transplantations in humans (122). However, SCFA supplementation could not induce a significant decrease in C. difficile shedding levels up to 6 weeks postinfection (121). No study has yet investigated whether C. difficile possesses any mechanisms by which it becomes resistant to the effects of SCFAs, which warrants further research.
Compared to the effects of SCFAs, there is more clarity on the effects of bile acids on C. difficile. Secondary bile acids are toxic to both C. difficile spores and vegetative cells, while primary bile acids generally stimulate growth and spore germination (123–125). During antibiotic treatment, conversion of primary into secondary bile acids is suppressed, and the reduction of secondary bile acids leads to a more favorable environment for C. difficile (120). In addition, C. difficile isolates causing the most severe disease in mice were also the isolates that showed the highest resistance to lithocholic acid in vitro (126). A relationship between the disease score and deoxycholic acid could not be shown (126). Secondary bile acid resistance may be strain dependent, but further research is warranted to draw this conclusion with certainty.
Intrinsic antibacteriocin properties have been described for C. difficile (127, 128). Nisin can inhibit the growth of vegetative cells and prevent C. difficile spore germination in vitro (44). However, this does not hold for all C. difficile strains, as the mutant strain MC119 had normal growth in sublethal concentrations. It was demonstrated that this resistance was at least partly due to export of nisin by an ABC transporter (127). Another identified mechanism was a net positive charge on the bacterial cell surface, resulting in lower efficacy of nisin, since nisin is attracted to a low negative charge on the cell surface (128).
Using genome scale metabolic models in antibiotic-treated mice, it was demonstrated that C. difficile does not necessarily compete for specific nutrients against specialized bacteria but that it adapts to utilize a wide array of nutrients. This allows colonization of diverse microbiomes where C. difficile is not limited to a specific nutrient niche (129). A follow-up study, also using a multiomics approach, showed that C. difficile alters the transcriptional activity of low-abundance taxa especially. The main genes showing decreased transcription in these low-abundance taxa during infection compared to mock-infected mice were carbohydrate acquisition and utilization genes. A possible reason for this could be that C. difficile attempts to create its own nutrient niche to facilitate colonization (130).
However, others have found specific nutrients that may be important for C. difficile colonization and/or outgrowth. Three highly virulent ribotypes (RT), RT017, RT027, and RT078, have recently been demonstrated to utilize trehalose as a nutrient source (131, 132). This was confirmed in a mouse model, where mice were challenged with spores of either RT027 or a non-trehalose-metabolizing ribotype. After trehalose administration, RT027 mice showed higher mortality in a dose-dependent manner (131).
C. difficile postantibiotic outgrowth depends partly on the production of succinate and sialic acid by commensals. B. thetaiotaomicron is capable of metabolizing polysaccharides and thereby produces sialic acid. Upon inoculation with C. difficile, mice monocolonized by B. thetaiotaomicron had an approximately five times higher density of C. difficile in their feces than germ-free mice (133). Expression levels of genes involved in sialic acid metabolism were increased in the B. thetaiotaomicron model, and as expected, a sialidase-deficient B. thetaiotaomicron mutant led to highly reduced production of sialic acid and lower C. difficile density (133). The density of C. difficile was higher in B. thetaiotaomicron-infected mice fed a polysaccharide-rich diet than in mice fed a chow diet (134). The succinate-to-butyrate pathway was crucial for C. difficile expansion in B. thetaiotaomicron-infected mice, as wild- type (WT) C. difficile was more effective in establishing infection than a succinate-transporter-deficient C. difficile strain (134).
Micronutrient availability can affect the virulence of C. difficile. High zinc levels have been demonstrated to exacerbate C. difficile infection in mouse models (135). Mice fed a high-zinc diet had higher toxin levels, higher proinflammatory cytokine levels, and increased loss of barrier function. Furthermore, it was shown that calprotectin, a zinc-binding protein, was important for limiting zinc availability to C. difficile during infection (135).
Together, these studies demonstrate the importance of specific nutrients used by C. difficile to establish colonization and infection.
Efficient colonization of the epithelial barrier is made possible by flagella and pili (136, 137). When mice were inoculated with flagellated or nonflagellated C. difficile strains, higher levels of flagellated C. difficile were found in mouse ceca (136). The exact destination of nonflagellated C. difficile remained unknown, as levels were not measured in feces or in sections of the small intestine. Regarding pili, it has been shown that type IV pili did not play a role in initial colonization but were crucial for epithelial adherence and long-lasting infection (137).
S. Typhimurium
S. Typhimurium is a nontyphoidal Salmonella serovar and an important cause of gastroenteritis in humans. It has been estimated that globally 3.4 million invasive nontyphoidal Salmonella infections occur each year, 65.2% of which are attributable to serovar Typhimurium (138). It mostly causes self-limiting, nonbloody diarrhea in otherwise healthy individuals. However, it can lead to bloodstream infections and metastatic spread with eventual death, especially in infants and immunocompromised individuals (138, 139). S. Typhimurium contains two pathogenicity islands, SPI1 and SPI2. SPI1 contains mostly information for causing intestinal disease and cell invasion, while SPI2 is necessary for intracellular survival (140).
The effects of SCFAs on S. Typhimurium are not yet well defined. Butyrate and propionate have been demonstrated to reduce the expression of invasion genes, while acetate increased their expression in S. Typhimurium (141, 142). However, conflicting results exist. An S. Typhimurium knockout mutant unable to metabolize butyrate caused less inflammation than a WT S. Typhimurium strain, suggesting that butyrate is crucial for S. Typhimurium virulence (143). Furthermore, the study demonstrated that butyrate was necessary for the expression of invasion genes in mouse models. In contrast, propionate inhibited S. Typhimurium in a dose-dependent manner in vitro, probably due to disturbance of intracellular pH (144). In an in vivo setting, it was demonstrated that a cocktail of propionate-producing Bacteroides species was sufficient to mediate CR against S. Typhimurium (144).
S. Typhimurium has developed mechanisms to overcome bile acids encountered in the gut. When exposed to individual bile acids at sublethal levels in vitro, it can become resistant to originally lethal levels by changing the gene and protein expression of several virulence regulators (145, 146). In addition, it has been demonstrated that a mixture of cholate and deoxycholate confers synergistic inhibition on invasion gene expression in S. Typhimurium (147).
Innate resistance of S. Typhimurium to bacteriocins produced by Gram-positive bacteria is naturally conferred through its Gram-negative outer membrane (148).
Usage of nutrients produced by gut microbiota is believed to facilitate S. Typhimurium outgrowth. By causing inflammation and thereby altering the microbiota composition, S. Typhimurium provides itself with a competitive advantage (149, 150).
Metabolic profiling in mice showed increased luminal lactate levels in the inflamed gut during S. Typhimurium infection, which could result from depletion of butyrate-producing bacteria (149). When butyrate is scarce, enterocytes switch to glycolysis, with lactate as the end product. Lactate is an important nutrient for S. Typhimurium, as indicated by decreased colonization of cecal and colonic lumen by an S. Typhimurium mutant lacking two lactate dehydrogenases (149). As explained in the introduction, an anaerobic milieu is maintained in the gut under homeostatic conditions. However, diffusion of oxygen from the tissue to the lumen is enabled by inflammation caused by S. Typhimurium, which alters enterocyte metabolism (151). Oxygen can then be used by S. Typhimurium to ferment several carbohydrates through respiration (152–155). In conclusion, these findings suggest that S. Typhimurium creates its own niche in the gut by causing inflammation, subsequently shifting the microbiota composition and thereby nutrient availability so that it can optimally colonize and expand.
An intact and well-functioning mucus layer is crucial for protection against S. Typhimurium infection. WT mice infected with the attenuated ΔaroA strain, which causes severe colitis, showed increased muc2 gene expression and MUC2 production (156). Mortality and morbidity were high in Δmuc2 mice, and higher numbers of the pathogen were found in their livers and ceca and close to the epithelial layer (156).
S. Typhimurium may profit from mucin-degrading commensal microbiota. In a gnotobiotic mouse model, complementation with mucin-degrading A. muciniphila during S. Typhimurium infection allowed S. Typhimurium to dominate the bacterial community 5 days p.i. (157). This was not caused by an absolute increase in cell numbers but by a decrease in other microbiota members. In addition, the complementation with A. muciniphila led to increased inflammation, as indicated by increased histopathology scores and protein and mRNA levels of proinflammatory cytokines. Although generally considered a beneficial bacterium, A. muciniphila exacerbated S. Typhimurium infection by thinning the mucus layer, thereby promoting translocation of the pathogen to the epithelial layer (157).
Enterohemorrhagic E. coli
Shiga-toxin-producing E. coli (STEC) comprises a group of E. coli strains capable of producing Shiga toxins. Enterohemorrhagic E. coli (EHEC) is a subgroup of STEC whose members cause more severe disease, often with complications. Each year, approximately 100,000 people are infected by the most common EHEC serotype, O157:H7 (158). Clinical presentation includes abdominal pain and bloody diarrhea, which can progress to toxin-mediated hemolytic uremic syndrome (159). The virulence of EHEC strains is mostly encoded by Shiga toxin genes, stx1 and stx2, and by locus of enterocyte effacement (lee) genes, which are imperative for initial attachment to epithelial cells (160).
At present, outcomes regarding the effects of SCFAs on EHEC are mixed (161–165). LEE protein and gene expression was already enhanced at 1.25 mM butyrate, while for acetate and propionate, only minor changes were detected at 20 mM, with acetate producing a repressive effect. In a separate growth experiment, acetate was more efficient in inhibiting growth of EHEC than butyrate and propionate (162). Acetate was observed to have small repressive effects on EHEC in a study by Nakanishi et al., and this was also found by Fukuda et al. (162, 165). Mice fed acetylated starch prior to infection showed higher fecal acetate levels and improved survival rates compared to starch-fed mice (165). Acetate also prevented gut barrier dysfunction as measured by transepithelial electrical resistance and prevented translocation of the Shiga toxin to the basolateral side of the epithelial cells (165). In Caco2 cells, EHEC epithelial adherence was 10-fold higher when grown on butyrate than when grown on acetate or propionate (162). These results indicate that butyrate may be less effective in inhibiting EHEC growth, and potentially colonization, than acetate and propionate, for which the exact pathways and genes involved have been elucidated (162, 163). In contrast, butyrate was found to be effective against EHEC in a pig model (161). Piglets given sodium butyrate 2 days prior to being infected with EHEC showed no symptoms 24 h p.i., while the control group developed multiple signs of disease, e.g., histopathological signs of kidney damage. The sodium butyrate group did not show any signs of inflammation and shed fewer viable cells than the control group within 48 h (161). In vitro assays demonstrated that butyrate enhanced bacterial clearance, ultimately leading the authors to suggest that butyrate could be developed as a new drug to treat EHEC infection (161).
EHEC has multiple traits to fight against the potentially deleterious effects of bile acids. Bile acid mixtures upregulated gene expression of the AcrAB efflux pump and downregulated ompF, a gene encoding an outer membrane porin (166). In addition, other genes responsible for limiting penetration of bile acids through the membrane (basR and basS) were upregulated, and this effect was concentration dependent. Interestingly, the bile acid mixtures did slightly downregulate stx2 subunit genes, encoding Shiga toxin production (166).
EHEC possesses natural resistance to bacteriocins, especially nisin, through its Gram-negative outer membrane, as described above. Three EHEC strains were screened for, among others, potential resistance to several colicinogenic E. coli strains (167). In vitro, resistance to E. coli strains producing a single colicin was observed, but resistance to multiple colicins was rarely observed and could never be linked to acquiring a specific plasmid (167).
Nutrient competition for proline and several sugars between EHEC and commensal E. coli strains is described in the introduction. In addition, ethanolamine (EA), a source of carbon, nitrogen, and energy for EHEC, has been investigated. It was demonstrated that EA could diffuse across the bacterial membrane and that the eut genes were crucial for metabolizing EA. eut sequences were absent in native bacterial genomes in the bovine gut, apart from commensal E. coli, indicating that EA provides a nutrient niche for E. coli. When the eutB gene was knocked out in E. coli strain EDL933, it was outcompeted by commensal E. coli due to its inability to utilize EA, indicating its critical importance for colonization (168). During further transcriptomic investigations of EA utilization, it was noticed that genes involved in gluconeogenesis were upregulated if no glucose was supplemented. Knockout of two genes within the gluconeogenesis pathway led to a growth defect in a coculture with the wild type (169). This is in line with a previous finding that optimal usage of gluconeogenic substrates by EDL933 is important for colonization (170). Since this effect was seen in medium consisting of bovine small-intestinal contents, the relevance for the human gut remains unclear (169).
Coculturing of EHEC with B. thetaiotaomicron led to upregulation of genes involved in nutrient competition in EHEC compared to culturing EHEC alone (171). In addition, the presence of B. thetaiotaomicron resulted in upregulation of multiple virulence genes, including lee, likely due to regulation of a transcription factor involved in sensing carbon metabolite concentrations in the environment (171). Using a combination of in vitro and in vivo methods, Pacheco et al. showed that fucose cleaved from mucins by B. thetaiotaomicron could be an important nutrient for upregulating virulence and intestinal colonization by EHEC (172). Interestingly, fucose sensing and subsequent regulation of virulence genes were more important for successful colonization than utilization of fucose for energy. This example indicates not only that nutrients can be utilized for energy, but that they can be important environmental signals for properly regulating the timing of virulence (172).
Human colonoid monolayers were used to study the initial colonization mechanisms of EHEC (173). This study showed that EHEC disturbs the tight junctions, preferentially attaches to mucus-producing cells, and subsequently impairs the mucus layer (173). In addition, by using various in vitro models, it was demonstrated that the metalloprotease StcE, produced by EHEC, enables degradation of MUC2 in the inner mucus layer, which may pave the way to the epithelial surface (174).
S. flexneri
Shigella infections mostly occur in developing countries, with S. flexneri the species most frequently found (175). Annually, an estimated 164,000 people die from shigellosis worldwide (176). Clinical presentation includes a wide variety of symptoms, including severe diarrhea, possibly containing blood and mucus, and abdominal pain (160). S. flexneri contains a virulence plasmid (pINV) that is necessary for invasion of epithelial cells and intracellular survival (160).
No studies seem to have investigated the resistance mechanisms of S. flexneri against SCFAs. Butyrate has been investigated as a potential therapeutic agent, as it counteracts a putative virulence mechanism of S. flexneri, namely, decreasing LL-37 expression in the gut (177, 178). By suppressing LL-37 expression, S. flexneri is able to colonize more deeply into intestinal crypts (178). Butyrate was able to increase rectal LL-37 expression in a subgroup of patients, which was associated with less inflammation in rectal mucosa and lower levels of proinflammatory cytokines (177). However, butyrate treatment did not seem to impact clinical recovery (177).
The type 3 secretion system (T3SS), which is able to directly inject bacterial protein into host cells and cause infection, is considered a key virulence factor. The S. flexneri T3SS can sense and bind the secondary bile acid deoxycholate, which leads to colocalization of protein translocators at the needle tip (179, 180). In S. flexneri mutants lacking the needle structure, the deoxycholate-associated adhesion to and invasion of host epithelial cells by S. flexneri were diminished (181). At physiological levels of bile salts, S. flexneri is able to grow normally in vitro, but at increased concentrations, growth is significantly reduced (182). Transcriptomics showed that during exposure to physiological bile salt levels, genes involved in drug resistance and virulence were upregulated, which was subsequently confirmed using reverse transcription-quantitative PCR (RT-qPCR). Deletion of a multidrug efflux pump led to sensitivity to bile salts and growth inability, confirming the importance of the pump to bile salt resistance (182).
Bacteriocin resistance has not been well studied in S. flexneri, but downregulating antimicrobial peptide production in the gut has been suggested to be an important virulence mechanism (183). The downregulation of LL-37 early in infection was demonstrated both in gut biopsy specimens from patients and in cell lines (183). Since protein and gene expression were not downregulated to the same degree, the authors speculated that there is an interference mechanism during active transcription of LL-37. Transcription of other antimicrobial peptides was also downregulated, especially in the human β-defensin hBD family (178, 183). It was demonstrated that S. flexneri shows high sensitivity to LL-37 and hBD-3 peptides in vitro (178). This suggests that by downregulating expression of antimicrobial peptides, S. flexneri creates an environment in which it can survive and ultimately cause severe disease.
It is unknown how S. flexneri competes and utilizes nutrients in the luminal side of the gut. Therefore, a short description of how the bacterium rewires host cell metabolism to support its survival after entering host cells is provided. These findings might be translatable and can at least provide insight into potential nutrient usage by S. flexneri in the lumen. Using a combination of metabolomics and proteomics, it was demonstrated that S. flexneri does not alter host cell metabolism in HeLa cells but that it captures the majority of the pyruvate output (184). Pyruvate was demonstrated to be a crucial carbon source for S. flexneri cultured on a HeLa cell derivative, using metabolomics, transcriptomics, and bacterial mutants (185). S. flexneri converts pyruvate into acetate via a very quick but energy-inefficient pathway, allowing rapid expansion of the bacterium intracellularly without rapid destruction of the host cell (184).
S. flexneri possesses special systems to alter mucus composition. Human colonoid monolayers infected with S. flexneri showed increased extracellular release of mucins (186). The increased extracellular mucins were trapped at the cell surface, which surprisingly favored access of S. flexneri to the apical surface, subsequently promoting cell invasion and cell-to-cell spread (186). Furthermore, expression of several genes encoding production of mucins and mucin glycosylation patterns were altered (186). Together, these results suggest that S. flexneri can alter the mucus environment so that it can promote its own virulence.
C. jejuni
C. jejuni is associated with foodborne gastroenteritis and is estimated to cause more than 800,000 infections annually in the United States alone (187). Major clinical symptoms include diarrhea (both with and without blood), fever, and abdominal cramping (160). In rare cases, it can give rise to Guillain-Barré syndrome and reactive arthritis (187). It is a commensal bacterium in avian species, and it is not yet well understood why it causes disease in humans (188).
There is a distinct lack of research on the resistance mechanisms of C. jejuni against SCFAs, but one study found that SCFAs are important for colonization in chickens (189). Acetinogenesis, the conversion of pyruvate to acetate, is a crucial metabolic pathway for optimal colonization by C. jejuni. Mutants unable to use this pathway show impaired colonization and decreased expression of acetinogenesis genes. Upon encountering a mixture of SCFAs at physiological levels, the mutant was surprisingly able to restore acetinogenesis gene expression to WT levels. Therefore, it was investigated whether expression of acetinogenic genes differs throughout the intestinal tract, as SCFAs are most abundant in distal parts of the intestine. It was observed that both gene expression and C. jejuni levels were highest in the cecum. The authors suggested that C. jejuni can monitor SCFA levels in the gut so that, in response, it can express colonization factors (189). As this is the only study suggesting this hypothesis, further research is required for validation.
Results regarding bile acid resistance in C. jejuni are mixed, which may stem from using different animal models or bile acids. A specific multidrug efflux pump, CmeABC, was important for bile resistance in chickens (190). ΔcmeABC mutants showed impaired growth in vitro and unsuccessful colonization in chickens upon cholate administration, while cholate did not affect growth and colonization by the WT (190). This suggests that the efflux pump is critical for proper colonization by C. jejuni by mediating bile acid resistance. Another study elucidated the effects of secondary bile acids on C. jejuni (191). Upon administration of deoxycholate prior to and during infection, mice showed decreased colitis. Unexpectedly, C. jejuni luminal colonization levels were not affected (191). In conclusion, C. jejuni colonization seems not to be affected by bile acids, but they may be important in limiting disease progression.
Bacteriocin resistance is not common in C. jejuni. Multiple C. jejuni isolates (n = 137) were screened for resistance to two anti-Campylobacter bacteriocins, OR-7 and E-760, produced by the gut inhabitants L. salivarius and Enterococcus faecium. However, no isolates were found to harbor resistance (192). In a follow-up study, chickens were successfully colonized with a C. jejuni strain prior to bacteriocin treatment, with the aim of studying bacteriocin resistance. Resistance developed in most chickens but was lost upon ending bacteriocin administration, suggesting resistance instability in vivo (193).
In contrast to most other enteric pathogens, C. jejuni does not metabolize carbohydrates as its main energy source. It is unable to oxidize glucose, fructose, galactose, and several disaccharides, including lactose, maltose, and trehalose, resulting from the absence of 6-phosphofructokinase (194–197). Fucose could be metabolized by some C. jejuni strains due to the occurrence of an extra genomic island (197). The main energy sources for C. jejuni are organic acids, including acetate, and a limited number of amino acids (198–200). It is currently unclear what these metabolic adaptations mean for its colonization potential, but it is possible that C. jejuni occupies a unique macronutrient niche.
Iron regulation systems are critical for colonization by and persistence of C. jejuni. In the presence of sufficient iron, transporter and acquisition genes are downregulated (201). Mutants lacking genes involved in either iron acquisition or transport were severely impaired in colonizing the chick gut (201). Free-iron concentrations are extremely low in the gut, which forces C. jejuni to utilize other iron sources. It was demonstrated that lactoferrin and transferrin can also be used for this purpose, and molecular pathways have been described (202). In short, transferrin-bound iron can be utilized only if it is in close proximity to the bacterial cell surface. Thereafter, it is most likely that iron is freed from the bacterial cell surface proteins, transported across the outer membrane, and subsequently internalized by an ABC transporter (202). Additionally, both in an in vitro setting and in a controlled human infection model with C. jejuni, the most upregulated genes were involved in iron acquisition (188, 203). These results suggest that iron regulation is maintained extremely well and that C. jejuni can obtain sufficient iron even in a harsh environment, such as the gut.
C. jejuni resides in the mucus layer prior to invading the epithelial cell. It can cross and reside there because of its powerful flagellum, which can change in conformation or rotation upon being challenged by higher viscosity (204, 205). C. jejuni can thus cross the mucus layer at speeds that cannot be met by other enteric pathogens, and the flagellum can subsequently be used as an adhesin (205, 206).
Another characteristic important for C. jejuni’s success in crossing the mucus layer is its helix shape. In a mouse model, a WT strain or either of two rod-shaped C. jejuni bacteria, a Δpgp1 or Δpgp2 mutant, was administered to cause infection (207). The rod-shaped mutants were demonstrated to be mostly nonpathogenic, whereas the WT strain caused severe inflammation. The mutants were to some extent able to colonize the mucus layer but could not cross it, explaining their nonpathogenicity (207).
V. cholerae
V. cholerae is one of the first bacterial pathogens for which the microbiota has been considered to play an important role against infection (208). It is mainly prevalent in contaminated brackish or salt water and can cause outbreaks, particularly during wars and after natural disasters. In the first 2 years following the 2010 earthquake in Haiti, more than 600,000 people were infected with V. cholerae serogroup O1 biotype Ogawa, resulting in more than 7,000 deaths (209). The clinical course is characterized by watery diarrhea, which can be so severe that it can result in dehydration, hypovolemic shock, and death (210). V. cholerae colonizes the small intestine by employing the toxin-coregulated pilus, after which it can cause severe infection and clinical symptoms through cholera enterotoxin production (210).
V. cholerae is able to utilize its acetate switch, the shift from elimination to assimilation of acetate, to increase its own virulence (211). In a Drosophila model, it was demonstrated that crbRS controlled the acetate switch, while acs1 was required for acetate assimilation (211). When either of the genes was knocked out, mortality decreased. Competition experiments demonstrated that WT V. cholerae had a growth advantage over the ΔcrbS strain when the ΔcrbS and WT V. cholerae strains were administered together in a 9:1 ratio. This led the authors to suggest that acetate utilization may be important early in infection, when low levels of V. cholerae cells are present (211). Furthermore, acetate consumption led to dysregulation of host insulin signaling pathways, ultimately leading to intestinal steatosis and increased mortality. Dysregulation of host insulin signaling was not observed in the ΔcrbS or Δacs1 strain, further confirming the role of acetate in V. cholerae virulence (211).
V. cholerae has a master regulator, toxT, which can directly activate several virulence factors, including toxin production. Cholera toxin production was reduced by 97% when V. cholerae was grown in the presence of bile, which could be reversed after growing the same cells in bile-free medium for a few hours (212). ctx and tcpA, encoding cholera toxin and the major structural unit of the toxin-coregulated pilus (respectively) and regulated by toxR and toxT, were highly repressed during bile exposure (212). Additionally, motility was increased approximately 1.6-fold in the presence of bile (212). To elucidate which exact components of bile acids were responsible for the repression of these virulence genes, bile was fractionated. It was found that several unsaturated fatty acids strongly repressed ctx and tcpA and that they upregulated expression of flrA, leading to increased motility (213). The reason for the upregulation of flrA and downregulation of tcpA could be that the flagellum increases the speed of passing through the mucus layer, while the pilus would only slow it down. When lower concentrations of bile at the epithelial surface are encountered, expression can be reversed (214).
Two outer membrane porins, OmpU and OmpT, are directly regulated by the master regulator toxR. Upon encountering bile acids, ompU and ompT are regulated in such a way that bile acid entrance is prevented (215, 216). Furthermore, ΔtoxR mutants are more sensitive to bile acids due to changed outer membrane composition (215). Recently, it was shown that toxR also regulates leuO (217). leuO was demonstrated to confer bile resistance independently of the two porins, although its exact resistance mechanism is not yet elucidated (217).
Bacteriocin resistance in V. cholerae, to our knowledge, has not been studied, and future studies will have to reveal whether any resistance is present.
An important nutrient through which V. cholerae gains a competitive advantage is sialic acid, a component of the mucus layer. Using streptomycin-pretreated mice that were given a mutant strain defective in sialic acid transport (ΔsiaM), it was shown that sialic acid is not required for initial colonization but that it is important for persistent colonization (218). Competition assays of the two mutant strains in the mouse intestine (small intestine, cecum, and large intestine) showed that the ΔsiaM strain was less fit to compete in each environment, further indicating the necessity of sialic acid utilization for niche expansion of V. cholerae (218).
The El Tor strain may have a competitive advantage over “classical” strains due to its differential carbohydrate metabolism (219). When grown in a glucose-rich medium, classical strains display a growth defect compared to El Tor. It was observed that this was due to production of organic acids through glucose metabolism, leading to acidification of the medium. El Tor biotypes were found to produce acetoin, a neutral compound, and to decrease organic acid production. This prevented acidification of the medium, leading to better growth. El Tor strains were also more successful in colonizing mice, especially when extra glucose was administered. The classical types were shown to be able to produce acetoin, but glucose led to only a minor increase in the transcription of genes necessary for acetoin production (219). These studies have shown that specific metabolic pathways are used by V. cholerae to successfully colonize the gut.
One of the first studies on how the mucus layer can potentially be crossed by V. cholerae was reported almost 50 years ago (220). There, motile and nonmotile strains were compared for pathogenicity after administration to mice. It was observed that motile strains were almost always deadly 36 h p.i., while most nonmotile strains had a mortality rate of under 35% (220). One hypothesis offered by the authors was that, together with mucinase, the flagellum could effectively pass the mucus barrier (220). Specific mucin degradation mechanisms employed by V. cholerae have been identified since, with hemagglutinin/protease (Hap), and TagA being the major ones (221–225). The presence of mucins and limitation of carbon sources and bile acids maximized production of Hap, while glucose could partly reverse this effect (221). This may indicate that under the conditions encountered in the gut, V. cholerae quickly aims to cross the mucus layer and be in close contact with the epithelial cells. TagA, which is similar to StcE as described for EHEC, is also capable of degrading mucin (222). In conclusion, V. cholerae has developed a way of sensing environmental conditions and, in response to them, is able to upregulate virulence factors that can degrade mucins. A simplified overview of V. cholerae virulence factors opposing CR can be found in Fig. 2.
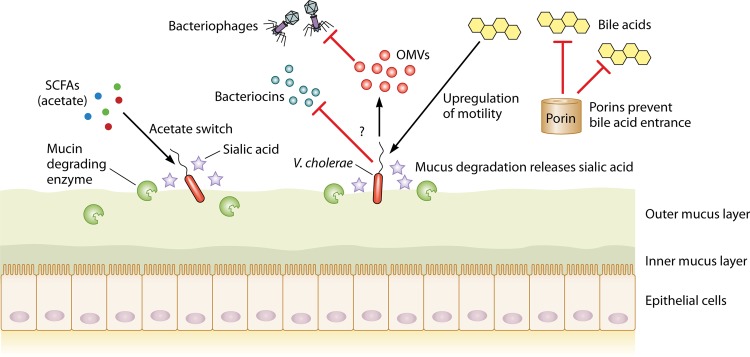
FIG 2.
V. cholerae uses a wide array of mechanisms to overcome CR. First, it employs its acetate switch to use acetate to upregulate its own virulence. Nothing about potential bacteriocin resistance is presently known, and this subject remains to be studied. To protect itself from bacteriophages, V. cholerae produces outer membrane vesicles (OMVs) that act as a decoy binding site for the attacking phages (see Bacterial Defense Mechanisms against Bacteriophages). Regulation of outer membrane porins allows them to prevent entry of bile acids when they are encountered. By employing specific mucin-degrading enzymes, V. cholerae releases sialic acid and subsequently metabolizes it.
Y. enterocolitica
Yersiniosis is mostly contracted through food or water contaminated with Y. enterocolitica, and its prevalence is much higher in developing countries than in high-income nations (160, 226). It is characterized by mild gastroenteritis and abdominal pain and is usually self-limiting, though pseudoappendicitis illnesses can occur (160). Virulence is mostly conferred through the presence of a 64- to 75-kb plasmid on which several virulence genes are present, including yadA, which is crucial for epithelial adherence (227).
The resistance of Y. enterocolitica to antibacterial compounds has not been much studied. One study investigated the effects of SCFAs, including acetic acid and propionic acid, on Y. enterocolitica at 4°C. Y. enterocolitica was less sensitive to acetic acid when cultured anaerobically than under aerobic culturing. Propionic acid was similarly effective in inhibiting growth with both culture methods (228). Even though conditions like 4°C are not representative of the intestinal environment, this study might provide some initial clues to the effects of SCFAs on Y. enterocolitica. It is clear that more research is required to further elucidate potential resistance mechanisms.
ompR, a transcriptional regulator in Y. enterocolitica, is probably able to upregulate expression of the AcrAB-TolC efflux pump, which in turn is regulated by two components of the efflux pump, acrR and acrAB (229). A mixture of bile acids, but not the secondary bile acid deoxycholate, was found to be the strongest inducer of acR and acrAB (229). Whether the upregulation of these efflux pump components contributes to bile acid resistance remains to be elucidated.
Bacteriocin resistance is so far mostly unknown in Y. enterocolitica. WA-314 and 8081 are both 1B:O8 strains that are highly infective in murine models (230). WA-314 possesses a putative colicin cluster for colicin production, but no expression was observed in a spot-on-lawn assay with 8081 and the colicin-sensitive E. coli K12 (230). It is likely that no specific resistance to colicin is present, as colicin has been shown to effectively inhibit Y. enterocolitica infections in vivo (59).
Like most other enteric pathogens, Y. enterocolitica has sophisticated systems to acquire sufficient iron. Using these systems, Y. enterocolitica may be more efficient at scavenging iron than commensal members, thereby providing itself with a competitive advantage. Y. enterocolitica expresses yersiniabactin, encoded by ybt, a highly efficient siderophore and a crucial component for lethality in mouse models (231, 232). The exact mechanisms of iron uptake and transport have been extensively reviewed previously (233). Proteomics analysis revealed that Y. enterocolitica serovar 1A, whose pathogenic role is unclear, uses different proteins to successfully scavenge iron, as it lacks the Ybt protein (234).
Y. enterocolitica is the only pathogenic Yersinia species that can metabolize sucrose, cellobiose, indole, sorbose, and inositol (235). Additionally, it can degrade EA and 1,2-propanediol (1,2-PD) by using tetrathionate as a terminal electron acceptor (235).
Mucus layer invasion and adherence of Y. enterocolitica were elucidated in great detail several decades ago (236–240). The YadA protein is used for initial attachment to the mucus (240). The preferential binding site on mucins is their carbohydrate moiety, but binding to mucin proteins is also possible under specific conditions (238). Y. enterocolitica uses a plasmid, pYV, with mucin degradation enzymes to thin the mucus layer, facilitating crossing of the mucus layer (237, 240). Y. enterocolitica containing the pYV plasmid is not only able to successfully invade and degrade the mucus layer, but is also highly efficient in multiplying in this environment (240). After interacting with the mucus layer, its bacterial cell surface was altered so that Y. enterocolitica became less efficient in colonizing the brush border (240). This may be a host response mechanism to prevent Y. enterocolitica invasion into deeper tissues. In a rabbit infection model, persistent goblet cell hyperplasia and increased mucin secretion were observed throughout the small intestine over 14 days (236). The extent of hyperplasia was associated with the severity of mucosal damage, indicating a compensatory mechanism. The mucin composition changed in infected rabbits, with a decrease in sialic acid and an increase in sulfate (236).
L. monocytogenes
L. monocytogenes causes listeriosis, a foodborne disease. Listeriosis is not highly prevalent, with an estimated 23,150 people infected in 2010 worldwide, but it has a high mortality rate of 20% to 30% (241). The most common syndrome is febrile gastroenteritis, but complications can develop, such as bacterial sepsis and meningitis (241). This is especially relevant for vulnerable patient groups, such as immunocompromised individuals, neonates, and fetuses (242). Virulence genes are present on an 8.2-kb pathogenicity island, which includes internalin genes necessary for invading host cells (243).
Culturing L. monocytogenes in the presence of high levels of butyrate leads to incorporation of more straight-chain fatty acids in the membrane (244, 245). This is not a natural state for L. monocytogenes, as normally its membrane consists of a very high percentage of branched-chain fatty acids. When subsequently exposed to LL-37, it displays a survival defect compared to bacteria not grown in the presence of butyrate (244). It was not elucidated whether this survival defect was due to increased stress, altered membrane composition, or differentially regulated virulence factors. The effects of propionate on L. monocytogenes growth, metabolism, and virulence factor expression are dependent on temperature, oxygen availability, and pH (246). Therefore, it is not possible to ascribe a general function to propionate in relation to L. monocytogenes.
L. monocytogenes possesses several bile acid resistance mechanisms, and in vitro transcriptome and proteome analyses have provided insight into them. Transcriptomics analysis revealed that in response to cholic acid, among others, two efflux pumps were upregulated, mdrM and mdrT (247). BrtA was shown to regulate the expression of the efflux pumps and to be able to sense bile acid levels. Bacterial abundance was determined in multiple organs of mice infected with knockout strains of either efflux pump, but not in the intestine (247). Proteomic analyses found many changes in response to bile salts and included proteins associated with efflux pumps, metabolism, and DNA repair (248).
Bile salt hydrolases (BSH) are another way of combatting encountered bile acids. It was demonstrated that all Listeria species that infect mammals showed BSH enzyme activity. BSH was crucial during infection of guinea pigs, as demonstrated by the decreased ability of a Δbsh strain to cause a persistent infection (249). At decreased pH levels, e.g., in the duodenum, bile salts are more acidic and show higher toxicity (250). However, this toxicity seems to be strain dependent (251). The strain responsible for a 2011 outbreak even displayed higher bile resistance at pH 5.5 than at pH 7.0, further indicating that bile susceptibility may be strain dependent (251).
As discussed in Bacteriocins above, the Abp118 bacteriocin produced by L. salivarius protected mice from L. monocytogenes infection (55).
However, several bacteriocins have been shown to be ineffective against L. monocytogenes, and the responsible mechanisms have been partly elucidated. Innate nisin resistance has been associated with multiple loci (252). One crucial gene was anrB, encoding a permease in an ABC transporter. Loss of the gene resulted in high sensitivity, not only to nisin, but also to several other bacteriocins (252). The mannose phosphotransferase system (Man-PTS), encoded by mptACD, is a main sugar uptake system, and two of its outer membrane proteins, IIC and IID, can serve as class II bacteriocin receptors (253). In naturally resistant and spontaneously resistant strains, reduced expression of mptC and mptD was observed, although this could not be linked to receptor mutations (254). The mpt operon is partly regulated by manR, and a manR mutant did not show any activation of the mpt operon (255). Development of bacteriocin resistance was to some extent dependent on available carbohydrates (256). Several sugar sources impaired growth of L. monocytogenes when exposed to the bacteriocin leucocin A. Increased sensitivity to leucocin A was hypothesized to be related to sugar uptake by the Man-PTS. When specific sugars are present, cells may not downregulate the system even in the presence of bacteriocins, which possibly allows leucocin A to use the Man-PTS as a docking molecule (256). Not only does L. monocytogenes display bacteriocin resistance, it also produces a bacteriocin, listeriolysin S, that modifies the gut microbiota so that intestinal colonization is promoted (257). Allobaculum and Alloprevotella, genera known to contain SCFA-producing strains, were significantly decreased in mice treated with listeriolysin S. L. monocytogenes strains unable to produce listeriolysin S were impaired in competing with the native gut microbiota and colonized less efficiently (257).
Most reports about metabolic adaptations of L. monocytogenes have logically described intracytosolic adaptations, as L. monocytogenes replicates intracellularly (258). Limited information is available on nutrient competition of L. monocytogenes inside the lumen. Comparison of genome sequences between colonizing Listeria and noncolonizing Listeria led to identification of, among others, a vitamin B12-dependent 1,2-PD degradation pathway in colonizing Listeria, dependent on the pduD gene (259). Mice were coinfected with a ΔpduD strain and a WT strain. Within 3 h after feeding, a large number of the ΔpduD bacteria were shed in feces, and 21 h later, the number of viable cells had decreased significantly. At 10 days p.i., the ΔpduD strain was completely cleared, while the WT strain was shed for up to 4 more days. This indicates that the ability to degrade 1,2-PD offers L. monocytogenes a distinct competitive advantage (259).
Multiple adhesins and internalins that facilitate L. monocytogenes retention in the mucus layer have been characterized (260–263). InlB, InlC, InlL, and InlJ were demonstrated to bind to MUC2, but not to epithelial cell surface MUC1 (262, 263). Histopathological analysis of a listeriosis rat model revealed that L. monocytogenes was present in the mucus layer less than 3 h p.i. (261). At that time point, very few L. monocytogenes bacteria were present on the epithelial cells (261).
BACTERIAL DEFENSE MECHANISMS AGAINST BACTERIOPHAGES
As research investigating how each enteric pathogen overcomes CR by rendering bacteriophages ineffective is still in its infancy, here, we describe the most widely employed resistance mechanisms. The bacteriophage infectious cycle involves a lytic cycle and a lysogenic cycle. Phages have to bind to a receptor on the bacterial surface to be able to insert their genomic material, usually DNA, into the bacterial cytoplasm and subsequently circularize their DNA (264). Here, lysogenic and lytic bacteriophage mechanisms start to branch (Fig. 3). Lytic phages start DNA replication, assemble their proteins, and pack their DNA into the typical bacteriophage shape with a capsid head and tail. After sufficient replication, the phages use lytic enzymes to form holes in the bacterial cell membrane, eventually leading to lysis of the cell and phage spreading. Lysogenic phages integrate their DNA in the bacterial chromosome and become prophages. Reproduction is then ensured through vertical transmission, and upon induction, prophages can also enter the lytic cycle (265) (Fig. 3). In general, factors that induce the lytic phase are compounds or conditions with bactericidal effects, e.g., a DNA-damaging agent (266).
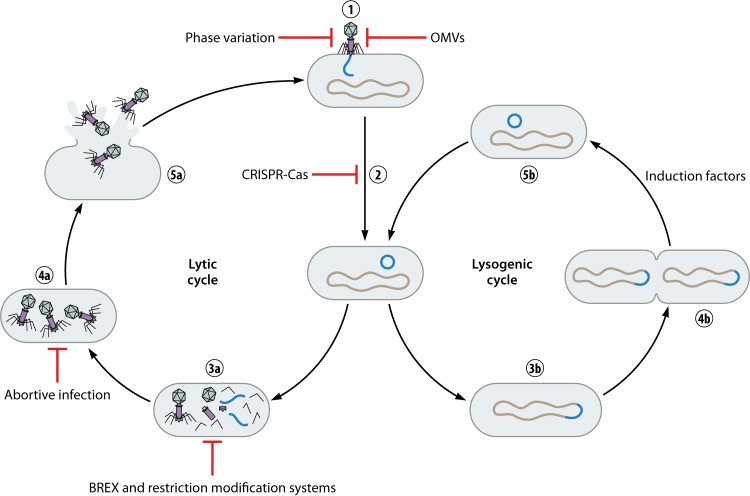
FIG 3.
Lytic and lysogenic bacteriophage infection cycles with bacterial defense mechanisms. The first two steps of infection are identical for the lytic and lysogenic cycles, namely, phage binding followed by DNA insertion and DNA circularization (1 and 2). The lysogenic cycle then branches off by integrating its DNA into the bacterial chromosome and becoming a prophage, thereby ensuring its replication (3b). Only upon encountering induction factors will the prophage leave the bacterial chromosome, after which it can enter the lytic cycle (4b and 5b). In the lytic cycle, phage DNA and protein are replicated and subsequently assembled into full phages (3a and 4a). The phages then lyse the bacterial cell, are released, and can infect other bacteria (5a). Bacteria possess multiple mechanisms to prevent killing by bacteriophages, starting with blocking attachment. This can be achieved through phase variation or production of OMVs. After phage DNA entry, CRISPR-Cas can recognize this foreign DNA and degrade it. Phage DNA and protein replication can be prevented by BREX and restriction-modification systems, while full phage assembly can be prevented by abortive infection.
The first step in preventing bacteriophage infection is to prevent surface receptor recognition. Outer membrane vesicles are produced by Gram-negative bacteria and have several functions, including interbacterial communication (267). They have a surface composition highly similar to that of the bacterium and may thereby serve as decoys for attacking phages (268) (Fig. 3). Indeed, V. cholerae outer membrane vesicles were shown to neutralize a V. cholerae-specific phage in a dose-dependent manner (Fig. 2) (268). This effect was seen only when the O1 antigen, the bacteriophage target on V. cholerae, was included in the outer membrane vesicle structure (268).
V. cholerae possesses another mechanism to prevent O1 phage receptor recognition (269) (Fig. 3). Two genes necessary for O1 biosynthesis were shown to use phase variation to induce variation in the O1 antigen composition (269). Mutants using phase variation were resistant to the O1 antigen phage but displayed impaired colonization in a mouse model (269). As the O1 antigen is an important virulence factor, e.g., for immune evasion, this demonstrates that enteric pathogens constantly have to deal with multiple CR mechanisms (269).
The second step in phage infection is injection of its DNA, and this can be prevented by superinfection exclusion systems, which are mostly encoded by prophages (Fig. 3). The E. coli prophage HK97 encodes gp15, a probable inner transmembrane protein (270). Remarkably, HK97 gp15 has putative homologues resembling the YebO protein family in many Enterobacteriaceae (270). GP15 prevented DNA injection into the bacterial cytoplasm by preventing proper formation of a complex consisting of an inner membrane glucose transporter and part of the tape measure protein (270, 271). This example illustrates how bacteria can incorporate phage DNA to protect themselves against future phage attacks.
DNA replication can be prevented by restriction-modification systems (Fig. 3). These systems consist of a methyltransferase and a restriction endonuclease. Exogenous DNA is not tagged by the methyltransferase, while “self”’ DNA is tagged (272, 273). Subsequently, nontagged DNA can be cleaved. This system is viewed as a primitive innate bacterial defense system. However, it was found that the system is not perfect, as restriction-modification systems can also attack self-DNA (274).
Currently, many groups are actively investigating the adaptive bacterial immune system CRISPR-Cas, and this has been extensively reviewed previously (275, 276). CRISPR-Cas is present in about 45% of sequenced bacterial genomes, although it is unknown if its prevalence is similar in gut bacteria (277, 278). In short, it consists of CRISPR arrays (sets of short repetitive DNA elements with variable DNA sequences [spacers] separating the repetitive DNA sets) and an operon of CRISPR-associated genes (Cas). The spacers are pieces of foreign DNA derived from bacteriophage DNA or other mobile genetic elements, such as plasmids. The defense mechanism consists of adaptation followed by expression and interference. During adaptation, Cas proteins can recognize foreign phage DNA and integrate a piece of this DNA as a new spacer into the CRISPR array. This allows the bacterium to build an immunological memory of all the phages it previously encountered. The expression response entails transcription of the CRISPR array, followed by processing into smaller RNA pieces (crRNAs). crRNAs consist of two outer parts of repeated DNA sequences with a spacer in between. To form the eventual Cas-crRNA complex, crRNAs are combined with at least one Cas protein. This complex then travels through the bacterial cell, and when it identifies a cDNA sequence representative of the previously encountered bacteriophage, it cleaves and degrades the foreign DNA.
In 2015, a novel phage resistance system, called bacteriophage exclusion (BREX), was discovered (279). BREX is able to block DNA replication but does not prevent bacteriophage attachment to the bacterium (Fig. 3). It also uses methylation as guidance to identify self and exogenous DNA but is different from restriction-modification systems, as it does not cleave the exogenous DNA (279). Almost 10% of all bacterial genomes sequenced were found to have BREX, suggesting that it is a well-conserved defense mechanism against bacteriophages (279). In spite of this promise as a defense mechanism, no further papers have been published regarding BREX functioning in, e.g., pathogenic bacteria.
Bacterial cells can perform an apoptosis-like action called abortive infection, resulting in death of the infected cell and thereby protecting surrounding bacterial cells (280) (Fig. 3). These systems have not been much elucidated for enteric pathogens at a molecular level, though the relevance of the system has been shown for the gut bacteria Shigella dysenteriae and E. coli (281, 282). The abortive infection systems are best studied in L. lactis, a bacterium widely used in production of fermented foods (283).
CONCLUDING REMARKS
Currently, bacterial enteric infections still cause a heavy disease burden worldwide. For many bacterial pathogens, the virulence factors involved in infection are understood, but less is known concerning the failure of the gut microbiota to provide colonization resistance against these enteropathogens. A more comprehensive understanding of why the microbiota fail to confer sufficient CR could lead to development of specific therapies aiming to restore CR. It is likely that no single bacterium will be used as the “holy grail” to restore CR but that a bacterial consortium with complementary functions will be used instead. This would be preferable to the currently often used FMT, where it is not well known what exact components are transferred to the patient. One could imagine not only that these consortia could be used to treat existing infections, but that they could also be administered prophylactically in susceptible patient groups. In addition, more attention has recently been given to several drugs that were previously not linked to gut health due to their potentially disturbing effect on the gut microbiota and perhaps CR. In conclusion, we reviewed many of the latest insights in the rapidly evolving fields of gut microbiota, colonization resistance, and bacterial enteric infection. We are looking forward to the coming years, when more knowledge on gut microbiota and CR will undoubtedly be gained, ultimately leading to more microbiota-based therapies.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. B.V.H.H. and E.J.K. are supported by an unrestricted grant from Vedanta Biosciences Inc. W.V.S. is supported by a Royal Society Wolfson Research Merit Award (WM160092). V.B.Y. is supported by grant AI124255 from the National Institutes of Health (United States) and is a consultant for Vedanta Biosciences Inc.
Q.R.D., R.D.Z., B.V.H.H., and E.J.K. designed the structure and content of the review and performed literature research. Q.R.D. wrote the manuscript with guidance from R.D.Z. and B.V.H.H. We all critically reviewed and revised the manuscript and read and approved the final manuscript.
REFERENCES
- 1.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer F, Backhed F. 2013. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 3.Round JL, Palm NW. 2018. Causal effects of the microbiota on immune-mediated diseases. Sci Immunol 3:eaao1603. doi: 10.1126/sciimmunol.aao1603. [DOI] [PubMed] [Google Scholar]
- 4.Opazo MC, Ortega-Rocha EM, Coronado-Arrazola I, Bonifaz LC, Boudin H, Neunlist M, Bueno SM, Kalergis AM, Riedel CA. 2018. Intestinal microbiota influences non-intestinal related autoimmune diseases. Front Microbiol 9:432. doi: 10.3389/fmicb.2018.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Gao Z, Wang H, Wu H, Liu Y, Yang Y, Han L, Wang X, Zhao L, Tong X. 2018. Intestinal immunomodulatory cells (T lymphocytes): a bridge between gut microbiota and diabetes. Mediators Inflamm 2018:9830939. doi: 10.1155/2018/9830939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadowsky MJ, Staley C, Heiner C, Hall R, Kelly CR, Brandt L, Horuts KA. 2017. Analysis of gut microbiota—an ever changing landscape. Gut Microbes 8:268–275. doi: 10.1080/19490976.2016.1277313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. 2019. A new genomic blueprint of the human gut microbiota. Nature 568:499–504. doi: 10.1038/s41586-019-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vollaard EJ, Clasener HA. 1994. Colonization resistance. Antimicrob Agents Chemother 38:409–414. doi: 10.1128/AAC.38.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becattini S, Littmann ER, Carter RA, Kim SG, Morjaria SM, Ling L, Gyaltshen Y, Fontana E, Taur Y, Leiner IM, Pamer EG. 2017. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med 214:1973–1989. doi: 10.1084/jem.20170495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martens EC, Neumann M, Desai MS. 2018. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol 16:457–470. doi: 10.1038/s41579-018-0036-x. [DOI] [PubMed] [Google Scholar]
- 13.Pickard JM, Zeng MY, Caruso R, Nunez G. 2017. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis P, Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 15.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Litvak Y, Byndloss MX, Baumler AJ. 2018. Colonocyte metabolism shapes the gut microbiota. Science 362:eaat9076. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cummings JH, Pomare EW, Branch WJ, Naylor CPE, Macfarlane GT. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Repaske DR, Adler J. 1981. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol 145:1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Byrne C, Booth IR, Roe AJ, McLaggan D. 2002. Inhibition of Escherichia coli growth by acetic acid a problem with methione byosynthesis and homocysteine toxicity. Microbiology 148:2215–2222. doi: 10.1099/00221287-148-7-2215. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann AF. 1999. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med 159:2647–2658. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 21.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano S, Nakam R, Tamaki M, Masuda N, Oda H. 1981. Isolation and characterization of thirteen intestinal microorganisms capable of 7 alpha-dehydroxylating bile acids. Appl Environ Microbiol 41:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang DJ, Ridlon JM, Moore DR II, Barnes S, Hylemon PB. 2008. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim Biophys Acta 1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sannasiddappa TH, Lund PA, Clarke SR. 2017. In vitro antibacterial activity of unconjugated and conjugated bile salts on Staphylococcus aureus. Front Microbiol 8:1581. doi: 10.3389/fmicb.2017.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurdi P, Kawanishi K, Mizutani K, Yokota A. 2006. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol 188:1979–1986. doi: 10.1128/JB.188.5.1979-1986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe M, Fukiya S, Yokota A. 2017. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J Lipid Res 58:1143–1152. doi: 10.1194/jlr.M075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thanissery R, Winston JA, Theriot CM. 2017. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Wells JE, Berr F, Thomas LA, Dowling RH, Hylemon PB. 2000. Isolation and characterization of cholic acid 7α-dehydroxylating fecal bacteria from cholesterol gallstone patients. J Hepatol 32:4–10. doi: 10.1016/S0168-8278(00)80183-X. [DOI] [PubMed] [Google Scholar]
- 31.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, Schurch CM, McCoy KD, Kuehne SA, Minton NP, Stecher B, Bernier-Latmani R, Hapfelmeier S. 2016. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front Cell Infect Microbiol 6:191. doi: 10.3389/fcimb.2016.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amrane S, Bachar D, Lagier JC, Raoult D. 2018. Clostridium scindens is present in the gut microbiota during Clostridium difficile infection: a metagenomic and culturomic analysis. J Clin Microbiol 56:e01663-17. doi: 10.1128/JCM.01663-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Termen S, Tollin M, Rodriguez E, Sveinsdottir SH, Johannesson B, Cederlund A, Sjovall J, Agerberth B, Gudmundsson GH. 2008. PU.1 and bacterial metabolites regulate the human gene CAMP encoding antimicrobial peptide LL-37 in colon epithelial cells. Mol Immunol 45:3947–3955. doi: 10.1016/j.molimm.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 35.Peric M, Koglin S, Dombrowski Y, Gross K, Bradac E, Ruzicka T, Schauber J. 2009. VDR and MEK-ERK dependent induction of the antimicrobial peptide cathelicidin in keratinocytes by lithocholic acid. Mol Immunol 46:3183–3187. doi: 10.1016/j.molimm.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Rea MC, Sit CS, Clayton E, O'Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. 2010. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A 107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks WM, Bottrill AR, Pierrat OA, Durrant MC, Maxwell A. 2007. The action of the bacterial toxin, microcin B17, on DNA gyrase. Biochimie 89:500–507. doi: 10.1016/j.biochi.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 38.Destoumieux-Garzon D, Peduzzi J, Thomas X, Djediat C, Rebuffat S. 2006. Parasitism of iron-siderophore receptors of Escherichia coli by the siderophore-peptide microcin E492m and its unmodified counterpart. Biometals 19:181–191. doi: 10.1007/s10534-005-4452-9. [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. 2004. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol Cell 14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 41.Rebuffat S. 2011. Bacteriocins from Gram-negative bacteria: a classification?, p 55–72. In Drider D, Rebuffat S (ed), Prokaryotic antimicrobial peptides: from genes to applications. Springer, New York, NY. [Google Scholar]
- 42.Rea MC, Ross RP, Cotter PD, Hill C. 2011. Classification of bacteriocins from Gram-positive bacteria, p 29–53. In Drider D, Rebuffat S (ed), Prokaryotic antimicrobial peptides: from genes to applications. Springer, New York, New York, NY. [Google Scholar]
- 43.Severina E, Severin A, Tomasz A. 1998. Antibacterial efficacy of nisin against multidrug-resistant Gram-positive pathogens. J Antimicrob Chemother 41:341–347. doi: 10.1093/jac/41.3.341. [DOI] [PubMed] [Google Scholar]
- 44.Le Lay C, Dridi L, Bergeron MG, Ouellette M, Fliss IL. 2016. Nisin is an effective inhibitor of Clostridium difficile vegetative cells and spore germination. J Med Microbiol 65:169–175. doi: 10.1099/jmm.0.000202. [DOI] [PubMed] [Google Scholar]
- 45.Benkerroum N, Sandine WE. 1988. Inhibitory action of nisin against Listeria monocytogenes. J Dairy Sci 71:3237–3245. doi: 10.3168/jds.S0022-0302(88)79929-4. [DOI] [PubMed] [Google Scholar]
- 46.Stevens KA, Sheldon BW, Klapes NA, Klaenhammer TR. 1991. Nisin treatment for inactivation of Salmonella species and other gram-negative bacteria. Appl Environ Microbiol 57:3613–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prudencio CV, Mantovani HC, Cecon PR, Vanetti MC. 2015. Differences in the antibacterial activity of nisin and bovicin HC5 against Salmonella Typhimurium under different temperature and pH conditions. J Appl Microbiol 118:18–26. doi: 10.1111/jam.12680. [DOI] [PubMed] [Google Scholar]
- 48.Singh AP, Prabha V, Rishi P. 2013. Value addition in the efficacy of conventional antibiotics by nisin against Salmonella. PLoS One 8:e76844. doi: 10.1371/journal.pone.0076844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh AP, Preet S, Rishi P. 2014. Nisin/beta–lactam adjunct therapy against Salmonella enterica serovar Typhimurium: a mechanistic approach. J Antimicrob Chemother 69:1877–1887. doi: 10.1093/jac/dku049. [DOI] [PubMed] [Google Scholar]
- 50.Li Q, Montalban-Lopez M, Kuipers OP. 2018. Increasing the antimicrobial activity of nisin-based lantibiotics against gram-negative pathogens. Appl Environ Microbiol 84:e00052-18. doi: 10.1128/AEM.00052-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritchie JM, Greenwich JL, Davis BM, Bronson RT, Gebhart D, Williams SR, Martin D, Scholl D, Waldor MK. 2011. An Escherichia coli O157-specific engineered pyocin prevents and ameliorates infection by E. coli O157:H7 in an animal model of diarrheal disease. Antimicrob Agents Chemother 55:5469–5474. doi: 10.1128/AAC.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gillor O, Nigro LM, Riley MA. 2005. Genetically engineered bacteriocins and their potential as the next generation of antimicrobials. Curr Pharm Des 11:1067–1075. doi: 10.2174/1381612053381666. [DOI] [PubMed] [Google Scholar]
- 53.Hasper HE, Kramer NE, Smith JL, Hillman JD, Zachariah C, Kuipers OP, de Kruijff B, Breukink E. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636–1637. doi: 10.1126/science.1129818. [DOI] [PubMed] [Google Scholar]
- 54.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. doi: 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 55.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CGM. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A 104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rea MC, Dobson A, O'Sullivan O, Crispie F, Fouhy F, Cotter PD, Shanahan F, Kiely B, Hill C, Ross RP. 2011. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A 108:4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rea MC, Alemayehu D, Casey PG, O'Connor PM, Lawlor PG, Walsh M, Shanahan F, Kiely B, Ross RP, Hill C. 2014. Bioavailability of the anti-clostridial bacteriocin thuricin CD in gastrointestinal tract. Microbiology 160:439–445. doi: 10.1099/mic.0.068767-0. [DOI] [PubMed] [Google Scholar]
- 58.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, Martens EC. 2016. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosák J, Micenková L, Hrala M, Pomorská K, Kunova Bosakova M, Krejci P, Göpfert E, Faldyna M, Šmajs D. 2018. Colicin FY inhibits pathogenic Yersinia enterocolitica in mice. Sci Rep 8:12242. doi: 10.1038/s41598-018-30729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillor O, Kirkup BC, Riley MA. 2004. Colicins and microcins: the next generation antimicrobials. Adv Appl Microbiol 54:129–146. doi: 10.1016/S0065-2164(04)54005-4. [DOI] [PubMed] [Google Scholar]
- 61.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. 2016. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rebuffat S. 2012. Microcins in action: amazing defence strategies of Enterobacteria. Biochem Soc Trans 40:1456–1462. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
- 63.Walsh CJ, Guinane CM, Hill C, Ross RP, O'Toole PW, Cotter PD. 2015. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project's reference genome database. BMC Microbiol 15:183. doi: 10.1186/s12866-015-0515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Momose Y, Hirayama K, Itoh K. 2008. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek 94:165–171. doi: 10.1007/s10482-008-9222-6. [DOI] [PubMed] [Google Scholar]
- 65.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun 76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, Conway T. 2013. Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS One 8:e53957. doi: 10.1371/journal.pone.0053957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. 2013. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Borenstein E, Kupiec M, Feldman MW, Ruppin E. 2008. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci U S A 105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 70.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. 2013. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A 110:17059–17064. doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansson ME, Larsson JM, Hansson GC. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A 108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Atuma C, Strugala V, Allen A, Holm L. 2001. The adherent gastrointestinal mucus gel layer thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol 280:G922–G929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 73.Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, Backhed F. 2018. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe 23:27–40.e7. doi: 10.1016/j.chom.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jakobsson HE, Rodriguez-Pineiro AM, Schutte A, Ermund A, Boysen P, Bemark M, Sommer F, Backhed F, Hansson GC, Johansson ME. 2015. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keen EC. 2015. A century of phage research: bacteriophages and the shaping of modern biology. Bioessays 37:6–9. doi: 10.1002/bies.201400152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mirzaei MK, Maurice CF. 2017. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol 15:397–408. doi: 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 77.Wittebole X, De Roock S, Opal SM. 2014. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5:226–235. doi: 10.4161/viru.25991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yen M, Cairns LS, Camilli A. 2017. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat Commun 8:14187. doi: 10.1038/ncomms14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fischer S, Kittler S, Klein G, Glunder G. 2013. Impact of a single phage and a phage cocktail application in broilers on reduction of Campylobacter jejuni and development of resistance. PLoS One 8:e78543. doi: 10.1371/journal.pone.0078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. 2012. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A 109:17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henein A. 2013. What are the limitations on the wider therapeutic use of phage? Bacteriophage 3:e24872. doi: 10.4161/bact.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T. 2017. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61:e00954-17. doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. 2018. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018:60–66. doi: 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. 2014. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 85.Becattini S, Taur Y, Pamer EG. 2016. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Forgacs I, Loganayagam A. 2008. Overprescribing proton pump inhibitors. BMJ 336:2–3. doi: 10.1136/bmj.39406.449456.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leonard J, Marshall JK, Moayyedi P. 2007. Systematic review of the risk of enteric infection in patients taking acid suppression. Am J Gastroenterol 102:2047–2056. doi: 10.1111/j.1572-0241.2007.01275.x. [DOI] [PubMed] [Google Scholar]
- 88.Wei L, Ratnayake L, Phillips G, McGuigan CC, Morant SV, Flynn RW, Mackenzie IS, MacDonald TM. 2017. Acid-suppression medications and bacterial gastroenteritis: a population-based cohort study. Br J Clin Pharmacol 83:1298–1308. doi: 10.1111/bcp.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hafiz RA, Wong C, Paynter S, David M, Peeters G. 2018. The risk of community-acquired enteric infection in proton pump inhibitor therapy: systematic review and meta-analysis. Ann Pharmacother 52:613–622. doi: 10.1177/1060028018760569. [DOI] [PubMed] [Google Scholar]
- 90.Bavishi C, Dupont HL. 2011. Systematic review: the use of proton pump inhibitors and increased susceptibility to enteric infection. Aliment Pharmacol Ther 34:1269–1281. doi: 10.1111/j.1365-2036.2011.04874.x. [DOI] [PubMed] [Google Scholar]
- 91.Freedberg DE, Toussaint NC, Chen SP, Ratner AJ, Whittier S, Wang TC, Wang HH, Abrams JA. 2015. Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial. Gastroenterology 149:883–885.e9. doi: 10.1053/j.gastro.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJ, Dijkstra G, Franke L, Xavier RJ, Jonkers D, Wijmenga C, Weersma RK, Zhernakova A. 2016. Proton pump inhibitors affect the gut microbiome. Gut 65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jackson MA, Goodrich JK, Maxan M-E, Freedberg DE, Abrams JA, Poole AC, Sutter JL, Welter D, Ley RE, Bell JT, Spector TD, Steves CJ. 2016. Proton pump inhibitors alter the composition of the gut microbiota. Gut 65:749–756. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takagi T, Naito Y, Inoue R, Kashiwagi S, Uchiyama K, Mizushima K, Tsuchiya S, Okayama T, Dohi O, Yoshida N, Kamada K, Ishikawa T, Handa O, Konishi H, Okuda K, Tsujimoto Y, Ohnogi H, Itoh Y. 2018. The influence of long-term use of proton pump inhibitors on the gut microbiota: an age-sex-matched case-control study. J Clin Biochem Nutr 62:100–105. doi: 10.3164/jcbn.17-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sieczkowska A, Landowski P, Gibas A, Kamińska B, Lifschitz C. 2018. Long-term proton pump inhibitor therapy leads to small bowel bacterial overgrowth as determined by breath hydrogen and methane excretion. J Breath Res 12:036006. doi: 10.1088/1752-7163/aa9dcf. [DOI] [PubMed] [Google Scholar]
- 96.Yasutomi E, Hoshi N, Adachi S, Otsuka T, Kong L, Ku Y, Yamairi H, Inoue J, Ishida T, Watanabe D, Ooi M, Yoshida M, Tsukimi T, Fukuda S, Azuma T. 2018. Proton pump inhibitors increase the susceptibility of mice to oral infection with enteropathogenic bacteria. Dig Dis Sci 63:881–889. doi: 10.1007/s10620-017-4905-3. [DOI] [PubMed] [Google Scholar]
- 97.Stiefel U, Rao A, Pultz MJ, Jump RL, Aron DC, Donskey CJ. 2006. Suppression of gastric acid production by proton pump inhibitor treatment facilitates colonization of the large intestine by vancomycin-resistant Enterococcus spp. and Klebsiella pneumoniae in clindamycin-treated mice. Antimicrob Agents Chemother 50:3905–3907. doi: 10.1128/AAC.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Imhann F, Vich Vila A, Bonder MJ, Lopez Manosalva AG, Koonen DPY, Fu J, Wijmenga C, Zhernakova A, Weersma RK. 2017. The influence of proton pump inhibitors and other commonly used medication on the gut microbiota. Gut Microbes 8:351–358. doi: 10.1080/19490976.2017.1284732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Foretz M, Guigas B, Bertrand L, Pollak M, Viollet B. 2014. Metformin: from mechanisms of action to therapies. Cell Metab 20:953–966. doi: 10.1016/j.cmet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 100.Smyth S, Heron A. 2006. Diabetes and obesity: the twin epidemics. Nat Med 12:75–80. doi: 10.1038/nm0106-75. [DOI] [PubMed] [Google Scholar]
- 101.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jorgensen T, Levenez F, Dore J, Meta HITC, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O. 2015. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 103.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, Ståhlman M, Olsson LM, Serino M, Planas-Fèlix M, Xifra G, Mercader JM, Torrents D, Burcelin R, Ricart W, Perkins R, Fernàndez-Real JM, Bäckhed F. 2017. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 104.Zhou HY, Zhu H, Yao XM, Qian JP, Yang J, Pan XD, Chen XD. 2017. Metformin regulates tight junction of intestinal epithelial cells via MLCK-MLC. Eur Rev Med Pharmacol Sci 21:5239–5246. doi: 10.26355/eurrev_201711_13847. [DOI] [PubMed] [Google Scholar]
- 105.Deng J, Zeng L, Lai X, Li J, Liu L, Lin Q, Chen Y. 2018. Metformin protects against intestinal barrier dysfunction via AMPKalpha1-dependent inhibition of JNK signalling activation. J Cell Mol Med 22:546–557. doi: 10.1111/jcmm.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xue Y, Zhang H, Sun X, Zhu MJ. 2016. Metformin improves ileal epithelial barrier function in interleukin-10 deficient mice. PLoS One 11:e0168670. doi: 10.1371/journal.pone.0168670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morgan AP, Crowley JJ, Nonneman RJ, Quackenbush CR, Miller CN, Ryan AK, Bogue MA, Paredes SH, Yourstone S, Carroll IM, Kawula TH, Bower MA, Sartor RB, Sullivan PF. 2014. The antipsychotic olanzapine interacts with the gut microbiome to cause weight gain in mouse. PLoS One 9:e115225. doi: 10.1371/journal.pone.0115225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lieberman LA, Higgins DE. 2009. A small–molecule screen identifies the antipsychotic drug pimozide as an inhibitor of Listeria monocytogenes infection. Antimicrob Agents Chemother 53:756–764. doi: 10.1128/AAC.00607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dastidar SG, Chaudhury A, Annadurai S, Roy S, Mookerjee M, Chakrabarty AN. 1995. In vitro and in vivo antimicrobial action of fluphenazine. J Chemother 7:201–206. doi: 10.1179/joc.1995.7.3.201. [DOI] [PubMed] [Google Scholar]
- 111.Jeyaseeli L, Dasgupta A, Dastidar SG, Molnar J, Amaral L. 2012. Evidence of significant synergism between antibiotics and the antipsychotic, antimicrobial drug flupenthixol. Eur J Clin Microbiol Infect Dis 31:1243–1250. doi: 10.1007/s10096-011-1435-3. [DOI] [PubMed] [Google Scholar]
- 112.Cussotto S, Strain CR, Fouhy F, Strain RG, Peterson VL, Clarke G, Stanton C, Dinan TG, Cryan JF. 2018. Differential effects of psychotropic drugs on microbiome composition and gastrointestinal function. Psychopharmacology 235:1–15. doi: 10.1007/s00213-018-5006-5. [DOI] [PubMed] [Google Scholar]
- 113.Rogers MA, Greene MT, Young VB, Saint S, Langa KM, Kao JY, Aronoff DM. 2013. Depression, antidepressant medications, and risk of Clostridium difficile infection. BMC Med 11:121. doi: 10.1186/1741-7015-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Munita JM, Arias CA. 2016. Mechanisms of antibiotic resistance. Microbiol Spectr 4:VMBF-0016-2015. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. 2016. Clostridium difficile infection. Nat Rev Dis Primers 2:16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Crobach MJT, Vernon JJ, Loo VG, Kong LY, Pechine S, Wilcox MH, Kuijper EJ. 2018. Understanding Clostridium difficile colonization. Clin Microbiol Rev 31:e00021-17. doi: 10.1128/CMR.00021-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Keller JJ, Kuijper EJ. 2015. Treatment of recurrent and severe Clostridium difficile infection. Annu Rev Med 66:373–386. doi: 10.1146/annurev-med-070813-114317. [DOI] [PubMed] [Google Scholar]
- 120.Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Seekatz AM, Theriot CM, Rao K, Chang YM, Freeman AE, Kao JY, Young VB. 2018. Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe 53:64–73. doi: 10.1016/j.anaerobe.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sorg JA, Sonenshein AL. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol 191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wilson KH. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 18:1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lewis BB, Carter RA, Pamer EG. 2016. Bile acid sensitivity and in vivo virulence of clinical Clostridium difficile isolates. Anaerobe 41:32–36. doi: 10.1016/j.anaerobe.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McBride SM, Sonenshein AL. 2011. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun 79:167–176. doi: 10.1128/IAI.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McBride SM, Sonenshein AL. 2011. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology 157:1457–1465. doi: 10.1099/mic.0.045997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jenior ML, Leslie JL, Young VB, Schloss PD. 2017. Clostridium difficile colonizes alternative nutrient niches during infection across distinct murine gut microbiomes. mSystems 2:e00063-17. doi: 10.1128/mSystems.00063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jenior ML, Leslie JL, Young VB, Schloss PD. 2018. Clostridium difficile alters the structure and metabolism of distinct cecal microbiomes during initial infection to promote sustained colonization. mSphere 3:e00261-18. doi: 10.1128/mSphere.00261-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Collins J, Robinson C, Danhof H, Knetsch CW, van Leeuwen HC, Lawley TD, Auchtung JM, Britton RA. 2018. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553:291–294. doi: 10.1038/nature25178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Collins J, Danhof H, Britton RA. 2018. The role of trehalose in the global spread of epidemic Clostridium difficile. Gut Microbes :1–6. doi: 10.1080/19490976.2018.1491266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota–liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. 2014. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zackular JP, Moore JL, Jordan AT, Juttukonda LJ, Noto MJ, Nicholson MR, Crews JD, Semler MW, Zhang Y, Ware LB, Washington MK, Chazin WJ, Caprioli RM, Skaar EP. 2016. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med 22:1330–1334. doi: 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun 69:7937–7940. doi: 10.1128/IAI.69.12.7937-7940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McKee RW, Aleksanyan N, Garret EM, Tamayo R. 2018. Type IV pili promote Clostridium difficile adherence and persistence in a mouse model of infection. Infect Immun 86:e00943-17. doi: 10.1128/IAI.00943-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. 2015. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg Infect Dis 21:941–949. doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kwon YM, Ricke SC. 1998. Induction of acid resistance of Salmonella Typhimurium by exposure to short-chain fatty acids. Appl Environ Microbiol 64:3458–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lawhon S, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short‐chain fatty acids alter Salmonella Typhimurium invasion gene expression and virulence through BarASirA. Mol Microbiol 46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 143.Bronner DN, Faber F, Olsan EE, Byndloss MX, Sayed NA, Xu G, Yoo W, Kim D, Ryu S, Lebrilla CB, Baumler AJ. 2018. Genetic ablation of butyrate utilization attenuates gastrointestinal Salmonella disease. Cell Host Microbe 23:266–273.e4. doi: 10.1016/j.chom.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. 2018. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe 24:296–307.e7. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.van Velkinburgh JC, Gunn JS. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance. Infect Immun 67:1614–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hernandez SB, Cota I, Ducret A, Aussel L, Casadesus J. 2012. Adaptation and preadaptation of Salmonella enterica to bile. PLoS Genet 8:e1002459. doi: 10.1371/journal.pgen.1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Eade CR, Hung CC, Bullard B, Gonzalez-Escobedo G, Gunn JS, Altier C. 2016. Bile acids function synergistically to repress invasion gene expression in Salmonella by destabilizing the invasion regulator hilD. Infect Immun 84:2198–2208. doi: 10.1128/IAI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gänzle MG, Hertel C, Hammes WP. 1999. Resistance of Escherichia coli and Salmonella against nisin and curvacin A. Int J Food Microbiol 48:37–50. doi: 10.1016/S0168-1605(99)00026-4. [DOI] [PubMed] [Google Scholar]
- 149.Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, Chanin RB, Behrendt CL, Hooper LV, Santos RL, Winter SE. 2018. Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23:54–64.e6. doi: 10.1016/j.chom.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Baumler AJ. 2016. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Spiga L, Winter MG, Furtado de Carvalho T, Zhu W, Hughes ER, Gillis CC, Behrendt CL, Kim J, Chessa D, Andrews-Polymenis HL, Beiting DP, Santos RL, Hooper LV, Winter SE. 2017. An oxidative central metabolism enables Salmonella to utilize microbiota-derived succinate. Cell Host Microbe 22:291–301.e6. doi: 10.1016/j.chom.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, Andrews-Polymenis HL, Winter SE, Bäumler AJ. 2017. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13:e1006129. doi: 10.1371/journal.ppat.1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Faber F, Tran L, Byndloss MX, Lopez CA, Velazquez EM, Kerrinnes T, Nuccio SP, Wangdi T, Fiehn O, Tsolis RM, Baumler AJ. 2016. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534:697–699. doi: 10.1038/nature18597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zarepour M, Bhullar K, Montero M, Ma C, Huang T, Velcich A, Xia L, Vallance BA. 2013. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun 81:3672–3683. doi: 10.1128/IAI.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ganesh BP, Klopfleisch R, Loh G, Blaut M. 2013. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One 8:e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Eppinger M, Cebula TA. 2015. Future perspectives, applications and challenges of genomic epidemiology studies for food-borne pathogens: a case study of enterohemorrhagic Escherichia coli (EHEC) of the O157:H7 serotype. Gut Microbes 6:194–201. doi: 10.4161/19490976.2014.969979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pai CH, Ahmed N, Lior H, Johnson WM, Sims HV, Woods DE. 1988. Epidemiology of sporadic diarrhea due to verocytotoxin-producing Escherichia coli: a two-year prospective study. J Infect Dis 157:1054–1057. doi: 10.1093/infdis/157.5.1054. [DOI] [PubMed] [Google Scholar]
- 160.Carroll KC, Pfaller MA, Landry ML, McAdam AJ, Patel R, Richter SS, Warnock DW. 2019. Manual of clinical microbiology, 12th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 161.Xiong H, Guo B, Gan Z, Song D, Lu Z, Yi H, Wu Y, Wang Y, Du H. 2016. Butyrate upregulates endogenous host defense peptides to enhance disease resistance in piglets via histone deacetylase inhibition. Sci Rep 6:27070. doi: 10.1038/srep27070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. 2009. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 163.Takao M, Yen H, Tobe T. 2014. LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol Microbiol 93:1302–1313. doi: 10.1111/mmi.12737. [DOI] [PubMed] [Google Scholar]
- 164.Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DS, O'Brien AD. 2013. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proc Natl Acad Sci USA 110:E2126–2133. doi: 10.1073/pnas.1222014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 166.Kus JV, Gebremedhin A, Dang V, Tran SL, Serbanescu A, Barnett Foster D. 2011. Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 193:4509–4515. doi: 10.1128/JB.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schamberger GP, Diez-Gonzalez F. 2005. Assessment of resistance to colicinogenic Escherichia coli by E. coli O157:H7 strains. J Appl Microbiol 98:245–252. doi: 10.1111/j.1365-2672.2004.02452.x. [DOI] [PubMed] [Google Scholar]
- 168.Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol 13:365–377. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- 169.Bertin Y, Deval C, de la Foye A, Masson L, Gannon V, Harel J, Martin C, Desvaux M, Forano E. 2014. The gluconeogenesis pathway is involved in maintenance of enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. PLoS One 9:e98367. doi: 10.1371/journal.pone.0098367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K–12 (MG1655) in the mouse intestine. Infect Immun 72:1666–1676. doi: 10.1128/IAI.72.3.1666-1676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. 2014. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.In J, Foulke-Abel J, Zachos NC, Hansen AM, Kaper JB, Bernstein HD, Halushka M, Blutt S, Estes MK, Donowitz M, Kovbasnjuk O. 2016. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol 2:48–62.e3. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hews CL, Tran SL, Wegmann U, Brett B, Walsham ADS, Kavanaugh D, Ward NJ, Juge N, Schuller S. 2017. The StcE metalloprotease of enterohaemorrhagic Escherichia coli reduces the inner mucus layer and promotes adherence to human colonic epithelium ex vivo. Cell Microbiol 19:e12717. doi: 10.1111/cmi.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM. 1999. Global burden of Shigella infections implications for vaccine development. Bull World Health Organ 77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 176.Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi A. 2018. Shigellosis. Lancet 391:801–812. doi: 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 177.Raqib R, Sarker P, Mily A, Alam NH, Arifuzzaman ASM, Rekha RS, Andersson J, Gudmundsson GH, Cravioto A, Agerberth B. 2012. Efficacy of sodium butyrate adjunct therapy in shigellosis a randomized, double-blind, placebo-controlled clinical trial. BMC Infect Dis 12:111. doi: 10.1186/1471-2334-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sperandio B, Regnault B, Guo J, Zhang Z, Stanley SL Jr, Sansonetti PJ, Pedron T. 2008. Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J Exp Med 205:1121–1132. doi: 10.1084/jem.20071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. 2007. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun 75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. 2008. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem 283:18646–18654. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Brotcke Zumsteg A, Goosmann C, Brinkmann V, Morona R, Zychlinsky A. 2014. IcsA is a Shigella flexneri adhesin regulated by the type III secretion system and required for pathogenesis. Cell Host Microbe 15:435–445. doi: 10.1016/j.chom.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 182.Nickerson KP, Chanin RB, Sistrunk JR, Rasko DA, Fink PJ, Barry EM, Nataro JP, Faherty CS. 2017. Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect Immun 85:e01067-16. doi: 10.1128/IAI.01067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, Agerberth B, Gudmundsson GH. 2001. Downregulation of bactericidal peptides in enteric infections a novel immune escape mechanism. Nat Med 7:180–185. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- 184.Kentner D, Martano G, Callon M, Chiquet P, Brodmann M, Burton O, Wahlander A, Nanni P, Delmotte N, Grossmann J, Limenitakis J, Schlapbach R, Kiefer P, Vorholt JA, Hiller S, Bumann D. 2014. Shigella reroutes host cell central metabolism to obtain high-flux nutrient supply for vigorous intracellular growth. Proc Natl Acad Sci U S A 111:9929–9934. doi: 10.1073/pnas.1406694111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Waligora EA, Fisher CR, Hanovice NJ, Rodou A, Wyckoff EE, Payne SM. 2014. Role of intracellular carbon metabolism pathways in Shigella flexneri virulence. Infect Immun 82:2746–2755. doi: 10.1128/IAI.01575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sperandio B, Fischer N, Joncquel Chevalier-Curt M, Rossez Y, Roux P, Robbe Masselot C, Sansonetti PA. 2013. Virulent Shigella flexneri affects secretion, expression, and glycosylation of gel-forming mucins in mucus-producing cells. Infect Immun 81:3632–3643. doi: 10.1128/IAI.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kaakoush NO, Castano-Rodriguez N, Mitchell HM, Man SM. 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Crofts AA, Poly FM, Ewing CP, Kuroiwa JM, Rimmer JE, Harro C, Sack D, Talaat KR, Porter CK, Gutierrez RL, DeNearing B, Brubaker J, Laird RM, Maue AC, Jaep K, Alcala A, Tribble DR, Riddle MS, Ramakrishnan A, McCoy AJ, Davies BW, Guerry P, Trent MS. 2018. Campylobacter jejuni transcriptional and genetic adaptation during human infection. Nat Microbiol 3:494–502. doi: 10.1038/s41564-018-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Luethy PM, Huynh S, Ribardo DA, Winter SE, Parker CT, Hendrixson DR. 2017. Microbiota-derived short-chain fatty acids modulate expression of Campylobacter jejuni determinants required for commensalism and virulence. mBio 8:e00407-17. doi: 10.1128/mBio.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun X, Winglee K, Gharaibeh RZ, Gauthier J, He Z, Tripathi P, Avram D, Bruner S, Fodor A, Jobin C. 2018. Microbiota-derived metabolic factors reduce campylobacteriosis in mice. Gastroenterology 154:1751–1763.e2. doi: 10.1053/j.gastro.2018.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hoang KV, Stern NJ, Saxton AM, Xu F, Zeng X, Lin J. 2011. Prevalence, development, and molecular mechanisms of bacteriocin resistance in Campylobacter. Appl Environ Microbiol 77:2309–2316. doi: 10.1128/AEM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hoang KV, Stern NJ, Lin J. 2011. Development and stability of bacteriocin resistance in Campylobacter spp. J Appl Microbiol 111:1544–1550. doi: 10.1111/j.1365-2672.2011.05163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Velayudhan J, Kelly DJ. 2002. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni an essential role for phosphoenolpyruvate carboxykinase. Microbiology 148:685–694. doi: 10.1099/00221287-148-3-685. [DOI] [PubMed] [Google Scholar]
- 195.Line JE, Hiett KL, Guard-Bouldin J, Seal BS. 2010. Differential carbon source utilization by Campylobacter jejuni 11168 in response to growth temperature variation. J Microbiol Methods 80:198–202. doi: 10.1016/j.mimet.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 196.Gripp E, Hlahla D, Didelot X, Kops F, Maurischat S, Tedin K, Alter T, Ellerbroek L, Schreiber K, Schomburg D, Janssen T, Bartholomäus P, Hofreuter D, Woltemate S, Uhr M, Brenneke B, Grüning P, Gerlach G, Wieler L, Suerbaum S, Josenhans C. 2011. Closely related Campylobacter jejuni strains from different sources reveal a generalist rather than a specialist lifestyle. BMC Genomics 12:584. doi: 10.1186/1471-2164-12-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Muraoka WT, Zhang Q. 2011. Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J Bacteriol 193:1065–1075. doi: 10.1128/JB.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Velayudhan J, Jones MA, Barrow PA, Kelly DJ. 2004. L-serine catabolism via an oxygen-labile l-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect Immun 72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wright JA, Grant AJ, Hurd D, Harrison M, Guccione EJ, Kelly DJ, Maskell DJ. 2009. Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155:80–94. doi: 10.1099/mic.0.021790-0. [DOI] [PubMed] [Google Scholar]
- 200.Leach S, Harvey P, Wait R. 1997. Changes with growth rate in the membrane lipid composition of and amino acid utilization by continuous cultures of Campylobacter jejuni. J Appl Microbiol 82:631–640. doi: 10.1111/j.1365-2672.1997.tb02873.x. [DOI] [PubMed] [Google Scholar]
- 201.Palyada K, Threadgill D, Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol 186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Miller CE, Rock JD, Ridley KA, Williams PH, Ketley JM. 2008. Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J Bacteriol 190:1900–1911. doi: 10.1128/JB.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu MM, Boinett CJ, Chan ACK, Parkhill J, Murphy MEP, Gaynor EC. 2018. Investigating the Campylobacter jejuni transcriptional response to host intestinal extracts reveals the involvement of a widely conserved iron uptake system. mBio 9:e01347-18. doi: 10.1128/mBio.01347-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ferrero R, Lee A. 1988. Motility of Campylobacter jejuni in a viscous environment comparison with conventional rod-shaped bacteria. J Gen Microbiol 134:53–59. doi: 10.1099/00221287-134-1-53. [DOI] [PubMed] [Google Scholar]
- 205.Szymanski CM, King M, Haardt M, Armstrong GD. 1995. Campylobacter jejuni motility and invasion of Caco-2 cells. Infect Immun 63:4295–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.McSweegan E, Walker RI. 1986. Identification and characterization of two Campylobacter jejuni adhesins for cellular and mucous substrates. Infect Immun 53:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Stahl M, Frirdich E, Vermeulen J, Badayeva Y, Li X, Vallance BA, Gaynor EC. 2016. The helical shape of Campylobacter jejuni promotes in vivo pathogenesis by aiding transit through intestinal mucus and colonization of crypts. Infect Immun 84:3399–3407. doi: 10.1128/IAI.00751-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Freter R. 1955. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J Infect Dis 97:57–65. doi: 10.1093/infdis/97.1.57. [DOI] [PubMed] [Google Scholar]
- 209.Barzilay EJ, Schaad N, Magloire R, Mung KS, Boncy J, Dahourou GA, Mintz ED, Steenland MW, Vertefeuille JF, Tappero JW. 2013. Cholera surveillance during the Haiti epidemic—the first 2 years. N Engl J Med 368:599–609. doi: 10.1056/NEJMoa1204927. [DOI] [PubMed] [Google Scholar]
- 210.Kaper JB, Morris JG Jr, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. doi: 10.1128/CMR.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, Mekalanos JJ, Watnick PI. 2014. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16:592–604. doi: 10.1016/j.chom.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gupta S, Chowdhury R. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect Immun 65:1131–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chatterjee A, Dutta PK, Chowdhury R. 2007. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect Immun 75:1946–1953. doi: 10.1128/IAI.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sengupta C, Ray S, Chowdhury R. 2014. Fine tuning of virulence regulatory pathways in enteric bacteria in response to varying bile and oxygen concentrations in the gastrointestinal tract. Gut Pathog 6:38. doi: 10.1186/s13099-014-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun 68:1491–1497. doi: 10.1128/IAI.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Duret G, Delcour AH. 2010. Size and dynamics of the Vibrio cholerae porins OmpU and OmpT probed by polymer exclusion. Biophys J 98:1820–1829. doi: 10.1016/j.bpj.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, Bina JE. 2015. Vibrio cholerae leuO transcription is positively regulated by toxR and contributes to bile resistance. J Bacteriol 197:3499–3510. doi: 10.1128/JB.00419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.McDonald ND, Lubin JB, Chowdhury N, Boyd EF. 2016. Host-derived sialic acids are an important nutrient source required for optimal bacterial fitness in vivo. mBio 7:e02237-15. doi: 10.1128/mBio.02237-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yoon SS, Mekalanos JJ. 2006. 2,3-butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect Immun 74:6547–6556. doi: 10.1128/IAI.00695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Guentzel MN, Berry LJ. 1975. Motility as a virulence factor for Vibrio cholerae. Infect Immun 11:890–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Silva AJ, Pham K, Benitez JA. 2003. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149:1883–1891. doi: 10.1099/mic.0.26086-0. [DOI] [PubMed] [Google Scholar]
- 222.Szabady RL, Yanta JH, Halladin DK, Schofield MJ, Welch RA. 2011. TagA is a secreted protease of Vibrio cholerae that specifically cleaves mucin glycoproteins. Microbiology 157:516–525. doi: 10.1099/mic.0.044529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Benitez JA, Spelbrink RG, Silva A, Philips TE, Stanley CM, Boesman-Finkelstein M, Finkelstein RA. 1997. Adherence of Vibrio cholerae to cultured differentiated human intestinal cells an in vitro colonization model. Infect Immun 65:3474–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Booth BA, Boesman–Finkelstein M, Finkelstein RA. 1984. Vibrio cholerae hemagglutinin protease nicks cholera enterotoxin. Infect Immun 45:558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Finkelstein RA, Hanne LF. 1982. Purification and characterization of the soluble hemagglutinin (cholera lectin) (produced by Vibrio cholerae. Infect Immun 36:1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rahman A, Bonny TS, Stonsaovapak S, Ananchaipattana C. 2011. Yersinia enterocolitica: Epidemiological studies and outbreaks. J Pathog 2011:239391. doi: 10.4061/2011/239391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bancerz-Kisiel A, Pieczywek M, Łada P, Szweda W. 2018. The most important virulence markers of Yersinia enterocolitica and their role during infection. Genes 9:235. doi: 10.3390/genes9050235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.El-Ziney MG, De MH, Debeve Re JM. 1997. Growth and survival kinetics of Yersinia enterocolitica IP 383 O:9 as affected by equimolar concentrations of undissociated short-chain organic acids. Int J Food Microbiol 34:233–247. doi: 10.1016/S0168-1605(96)01194-4. [DOI] [PubMed] [Google Scholar]
- 229.Raczkowska A, Trzos J, Lewandowska O, Nieckarz M, Brzostek K. 2015. Expression of the AcrAB components of the AcrAB-TolC multidrug efflux pump of Yersinia enterocolitica is subject to dual regulation by OmpR. PLoS One 10:e0124248. doi: 10.1371/journal.pone.0124248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Garzetti D, Bouabe H, Heesemann J, Rakin A. 2012. Tracing genomic variations in two highly virulent Y. enterocolitica strains with unequal ability. BMC Genomics 13:467. doi: 10.1186/1471-2164-13-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Carniel E, Mazigh D, Mollaret HH. 1987. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun 55:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. 1993. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65 000 Da and pesiticin sensitivity. Mol Microbiol 8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 233.Perry RD, Fetherston JD. 2011. Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect 13:808–817. doi: 10.1016/j.micinf.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kanaujia PK, Bajaj P, Kumar S, Singhal N, Virdi JS. 2015. Proteomic analysis of Yersinia enterocolitica biovar 1A under iron-rich and iron-poor conditions indicate existence of efficiently regulated mechanisms of iron homeostasis. J Proteomics 124:39–49. doi: 10.1016/j.jprot.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 235.Reuter S, Connor TR, Barquist L, Walker D, Feltwell T, Harris SR, Fookes M, Hall ME, Petty NK, Fuchs TM, Corander J, Dufour M, Ringwood T, Savin C, Bouchier C, Martin L, Miettinen M, Shubin M, Riehm JM, Laukkanen-Ninios R, Sihvonen LM, Siitonen A, Skurnik M, Falcao JP, Fukushima H, Scholz HC, Prentice MB, Wren BW, Parkhill J, Carniel E, Achtman M, McNally A, Thomson NR. 2014. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc Natl Acad Sci U S A 111:6768–6773. doi: 10.1073/pnas.1317161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mantle M, Atkins E, Kelly J, Thakore E, Buret A, Gall DG. 1991. Effects of Yersinia enterocolitica infection on rabbit intestinal and colonic goblet cells and mucin morphometrics, histochemistry, and biochemistry. Gut 32:1131–1138. doi: 10.1136/gut.32.10.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mantle M, Basaraba L, Peacock SC, Gall DG. 1989. Binding of Yersinia enterocolitica to rabbit intestinal brush border membranes, mucus, and mucin. Infect Immun 57:3292–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mantle M, Husar SD. 1994. Binding of Yersinia enterocolitica to purified, native small intestinal mucins from rabbits and humans involves interactions with the mucin carbohydrate moiety. Infect Immun 62:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mantle M, Rombough C. 1993. Growth in and breakdown of purified rabbit small intestinal mucin by Yersinia enterocolitica. Infect Immun 61:4131–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Paerregaard A, Espersen F, Jensen OM, Skurnik M. 1991. Interactions between Yersinia enterocolitica and rabbit ileal mucus growth, adhesion, penetration, and subsequent changes in surface hydrophobicity. Infect Immun 59:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.de Noordhout CM, Devleesschauwer B, Angulo FJ, Verbeke G, Haagsma J, Kirk M, Havelaar A, Speybroeck N. 2014. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect Dis 14:1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.McLauchlin J. 1990. Human listeriosis in Britain, 1967-85, a summary of 722 cases. Epidemiol Infect 104:181–189. doi: 10.1017/S0950268800059343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Roberts AJ, Wiedmann M. 2003. Pathogen, host and environmental factors contributing to the pathogenesis of listeriosis. Cell Mol Life Sci 60:904–918. doi: 10.1007/s00018-003-2225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sun Y, Wilkinson BJ, Standiford TJ, Akinbi HT, O'Riordan MX. 2012. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J Bacteriol 194:5274–5284. doi: 10.1128/JB.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Julotok M, Singh AK, Gatto C, Wilkinson BJ. 2010. Influence of fatty acid precursors, including food preservatives, on the growth and fatty acid composition of Listeria monocytogenes at 37 and 10°C. Appl Environ Microbiol 76:1423–1432. doi: 10.1128/AEM.01592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rinehart E, Newton E, Marasco MA, Beemiller K, Zani A, Muratore MK, Weis J, Steinbicker N, Wallace N, Sun Y. 2018. Listeria monocytogenes response to propionate is differentially modulated by anaerobicity. Pathogens 7:60. doi: 10.3390/pathogens7030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Quillin SJ, Schwartz KT, Leber JH. 2011. The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol Microbiol 81:129–142. doi: 10.1111/j.1365-2958.2011.07683.x. [DOI] [PubMed] [Google Scholar]
- 248.Payne A, Schmidt TB, Nanduri B, Pendarvis K, Pittman JR, Thornton JA, Grissett J, Donaldson JR. 2013. Proteomic analysis of the response of Listeria monocytogenes to bile salts under anaerobic conditions. J Med Microbiol 62:25–35. doi: 10.1099/jmm.0.049742-0. [DOI] [PubMed] [Google Scholar]
- 249.Dussurget O, Cabanes D, Dehoux P, Lecuit M, European Listeria Genome Consortium, Buchrieser C, Glaser P, Cossart P. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol 45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 250.Begley M, Gahan CGM, Hill C. 2002. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl Environ Microbiol 68:6005–6012. doi: 10.1128/AEM.68.12.6005-6012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.White SJ, McClung DM, Wilson JG, Roberts BN, Donaldson JR. 2015. Influence of pH on bile sensitivity amongst various strains of Listeria monocytogenes under aerobic and anaerobic conditions. J Med Microbiol 64:1287–1296. doi: 10.1099/jmm.0.000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Collins B, Curtis N, Cotter PD, Hill C, Ross RP. 2010. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various lactam antibiotics. Antimicrob Agents Chemother 54:4416–4423. doi: 10.1128/AAC.00503-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci U S A 104:2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kjos M, Nes IF, Diep DB. 2011. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol 77:3335–3342. doi: 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Dalet K, Cenatiempo Y, Cossart P, European Listeria Genome Consortium, Héchard Y. 2001. A sigma(54)-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263–3269. doi: 10.1099/00221287-147-12-3263. [DOI] [PubMed] [Google Scholar]
- 256.Balay DR, Ganzle MG, McMullen LM. 2018. The effect of carbohydrates and bacteriocins on the growth kinetics and resistance of Listeria monocytogenes. Front Microbiol 9:347. doi: 10.3389/fmicb.2018.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Quereda JJ, Dussurget O, Nahori MA, Ghozlane A, Volant S, Dillies MA, Regnault B, Kennedy S, Mondot S, Villoing B, Cossart P, Pizarro–Cerda J. 2016. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci U S A 113:5706–5711. doi: 10.1073/pnas.1523899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Chen GY, Pensinger DA, Sauer JD. 2017. Listeria monocytogenes cytosolic metabolism promotes replication, survival, and evasion of innate immunity. Cell Microbiol 19:e12762. doi: 10.1111/cmi.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Schardt J, Jones G, Muller-Herbst S, Schauer K, D'Orazio SEF, Fuchs TM. 2017. Comparison between Listeria sensu stricto and Listeria sensu lato strains identifies novel determinants involved in infection. Sci Rep 7:17821. doi: 10.1038/s41598-017-17570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mariscotti JF, Quereda JJ, Garcia-Del Portillo F, Pucciarelli MG. 2014. The Listeria monocytogenes LPXTG surface protein Lmo1413 is an invasin with capacity to bind mucin. Int J Med Microbiol 304:393–404. doi: 10.1016/j.ijmm.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 261.Pron B, Boumaila C, Jaubert F, Sarnacki S, Monnet J, Berche P, Gaillard J. 1998. Comprehensive study of the intestinal stage of listeriosis in a rat ligated ileal loop system. Infect Immun 66:747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Linden SK, Bierne H, Sabet C, Png CW, Florin TH, McGuckin MA, Cossart P. 2008. Listeria monocytogenes internalins bind to the human intestinal mucin MUC2. Arch Microbiol 190:101–104. doi: 10.1007/s00203-008-0358-6. [DOI] [PubMed] [Google Scholar]
- 263.Popowska M, Krawczyk-Balska A, Ostrowski R, Desvaux M. 2017. InlL from Listeria monocytogenes is involved in biofilm formation and adhesion to mucin. Front Microbiol 8:660. doi: 10.3389/fmicb.2017.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Segev N, Laub A, Cohen G. 1980. A circular form of bacteriophage P1 DNA made in lytically infected cells of Escherichia coli. Virology 101:261–271. doi: 10.1016/0042-6822(80)90501-2. [DOI] [PubMed] [Google Scholar]
- 265.Davies EV, Winstanley C, Fothergill JL, James CE. 2016. The role of temperate bacteriophages in bacterial infection. FEMS Microbiol Lett 363:fnw015. doi: 10.1093/femsle/fnw015. [DOI] [PubMed] [Google Scholar]
- 266.Refardt D, Rainey PB. 2010. Tuning a genetic switch: experimental evolution and natural variation of prophage induction. Evolution 64:1086–1097. doi: 10.1111/j.1558-5646.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- 267.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Reyes-Robles T, Dillard RS, Cairns LS, Silva-Valenzuela CA, Housman M, Ali A, Wright ER, Camilli A. 2018. Vibrio cholerae outer membrane vesicles inhibit bacteriophage infection. J Bacteriol 200:e00792-17. doi: 10.1128/JB.00792-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Seed KD, Faruque SM, Mekalanos JJ, Calderwood SB, Qadri F, Camilli A. 2012. Phase variable O antigen biosynthetic genes control expression of the major protective antigen and bacteriophage receptor in Vibrio cholerae O1. PLoS Pathog 8:e1002917. doi: 10.1371/journal.ppat.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Cumby N, Edwards AM, Davidson AR, Maxwell KL. 2012. The bacteriophage HK97 gp15 moron element encodes a novel superinfection exclusion protein. J Bacteriol 194:5012–5019. doi: 10.1128/JB.00843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Cumby N, Reimer K, Mengin-Lecreulx D, Davidson AR, Maxwell KL. 2015. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol Microbiol 96:437–447. doi: 10.1111/mmi.12918. [DOI] [PubMed] [Google Scholar]
- 272.Haberman A, Heywood J, Meselson M. 1972. DNA modification methylase activity of Escherichia coli restriction endonucleases K and P. Proc Natl Acad Sci U S A 69:3138–3141. doi: 10.1073/pnas.69.11.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Brockes JP, Brown PR, Murray K. 1972. The deoxyribonucleic acid modification enzyme of bacteriophage P1. Biochem J 127:1–10. doi: 10.1042/bj1270001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pleska M, Qian L, Okura R, Bergmiller T, Wakamoto Y, Kussell E, Guet CC. 2016. Bacterial autoimmunity due to a restriction-modification system. Curr Biol 26:404–409. doi: 10.1016/j.cub.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 275.Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. 2016. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 276.Amitai G, Sorek R. 2016. CRISPR-Cas adaptation: insights into the mechanism of action. Nat Rev Microbiol 14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 277.Touchon M, Bernheim A, Rocha EP. 2016. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J 10:2744–2754. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a Web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Gross SR. 1954. Abortive Infection of a strain of Escherichia coli by colipphage T2. J Bacteriol 68:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Fields KL. 1969. Comparison of the action of colicins El and K on Escherichia coli with the effects of abortive infection by virulent bacteriophages. J Bacteriol 97:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Smith HS, Pizer LI, Pylkas L, Lederberg S. 1969. Abortive infection of Shigella dysenteriae P2 by T2 bacteriophage. J Virol 4:162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ainsworth S, Stockdale S, Bottacini F, Mahony J, van Sinderen D. 2014. The Lactococcus lactis plasmidome: much learnt, yet still lots to discover. FEMS Microbiol Rev 38:1066–1088. doi: 10.1111/1574-6976.12074. [DOI] [PubMed] [Google Scholar]