Abstract
The microbiome is now considered our ‘second genome’ with potentially comparable importance to the genome in determining human health. There is, however, a relatively limited understanding of the broader environmental factors, particularly social conditions, that shape variation in human microbial communities. Fulfilling the promise of microbiome research — particularly the microbiome’s potential for modification — will require collaboration between biologists and social and population scientists. For life scientists, the plasticity and adaptiveness of the microbiome calls for an agenda to understand the sensitivity of the microbiome to broader social environments already known to be powerful predictors of morbidity and mortality. For social and population scientists, attention to the microbiome may help answer nagging questions about the underlying biological mechanisms that link social conditions to health. We outline key substantive and methodological advances that can be made if collaborations between social and population health scientists and life scientists are strategically pursued.
We are an amalgamation of cells, both human and microbial, and there is growing evidence that the trillions of microbes that inhabit the human body — collectively referred to as the human microbiota — have profound implications for human health1. This complex human ecosystem belies the traditional dichotomy between ‘good’ and ‘bad’ bacteria, giving way to a more nuanced consideration of changing and interacting networks of microbes. The microbiome is now considered our ‘second genome’ with potentially larger importance than the genome in shaping human health2. What makes the microbiome potentially so pivotal for shaping health — and potentially of significant interest to social science and population health researchers — is its plasticity, or ability to be altered, and, specifically, its responsiveness to the environment3. Yet, despite rapid technological progress in description and sequencing, gaps remain in our understanding of the broader environmental factors that shape inter-individual variation in these microbial communities4, particularly in regards to how social conditions may influence this variation.
We argue that fulfilling the promise ofmicrobiome research — particularly the human microbiome’s potential for modification — will require closer collaboration between life scientists and social and population health scientists who can consider the interaction of multiple levels of environmental exposures. The very nature of the microbiome, particularly its plasticity and adaptiveness to the environment, opens the door to a broader research agenda focused on how social conditions influence the microbiome. For analytic purposes, we focus on early-life conditions, socioeconomic resources and social relationships as case studies for the possibilities these collaborations may hold. Decades of experimental and observational evidence demonstrate that these social conditions influence morbidity and mortality at levels far exceeding individual behaviours such as obesity or even medical interventions such as anti-hypertensives. For social and population scientists, attention to the microbiome may help answer nagging questions about the underlying biological mechanisms that link social conditions to health, and promote attention to policy and other upstream factors that drive changes in the microbiome at the population level.
In this Perspective, we first detail existing research on how social environments influence the gut microbiome, focusing particularly on early-life conditions, socioeconomic factors and social relationships. We then outline potential interdisciplinary collaborations across these three substantive areas, detailing ways in which existing population-based studies and methods could be employed to test novel hypotheses about the human gut microbiome. In sum, we detail key substantive and methodological advances that can be made if collaborations between the social and population sciences and life sciences are strategically pursued, with a particular, though not exclusive, emphasis on the gut microbiome, where the largest number of microbial communities in the human body can be found.
Current research on social environments and gut microbiome
The gastrointestinal tract is estimated to harbour roughly 90% of our indigenous microbes5. There is increasing empirical evidence, both from animal and human population studies, that distal gut community patterns play an important role in a broad range of physiological functions of their host, including immune system maturation, metabolic and inflammatory processes, and even the brain and behaviour via the ‘gut-brain axis’6. Indeed, the gut microbiota is now implicated in a wide array of chronic diseases, including type 1 and type 2 diabetes, inflammatory bowel disease, obesity, cardiovascular disease and cancer, which remain among the leading causes of morbidity and mortality in the developed — and increasingly developing — world1,2. Moreover, there is a robust body of research demonstrating how diet and nutritional factors influence the gut microbiome, which may prove a key pathway linking diet to health7. The rapid acceleration of research — including successful clinical interventions involving fecal transplants — points to the general consensus that the gut microbiome could radically transform research and interventions to improve human health.
Despite large advances, however, scientific knowledge of the gut microbiome — especially the way broader environments shape its variability over the life course — remains in its infancy. Research is still limited regarding how broader social environments and conditions shape exposures and ultimately influence its composition, especially research employing human subjects. This limitation is important because recent evidence showed that the environment — rather than genetics — predominantly shapes human gut microbial composition8. How this ‘environment’ is defined and measured has not yet been well developed. Yet, a small, but growing body of literature is looking at how our social environment shapes acquisition and exchange of microbes — from the people we interact with to the environments in which we work and live9–13. In the sections below, we explore existing research that touches on how our social environments, specifically early-life conditions, social relationships and socioeconomic conditions, may shape the human gut microbiome9. Figure 1 outlines a general conceptual framework for potential pathways linking social environments and the microbiome over the life course, some of which we highlight below.
Fig. 1 |. Proposed relationships between social conditions, the gut microbiome, and morbidity and mortality.
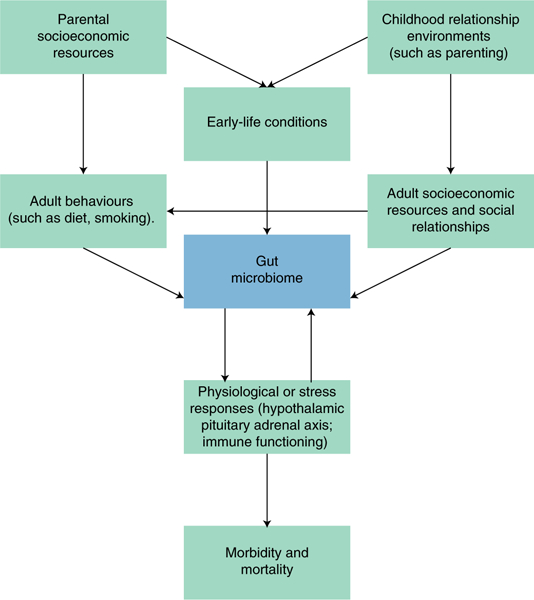
Note: this figure is not intended to be a comprehensive overview of all possible causal relationships. It suggests where social and population health scientists are best positioned to contribute to microbiome research, focusing particularly on the possible pathways between social conditions and the gut microbiome.
Early-life conditions.
It is well known that birth and early life are critical periods for the acquisition and development of an individual’s microbiome. Broadly, prenatal and early-life environments play an important role in developmental trajectories for both the immune-and stress-response systems, with implications for developing microbiota as well14–16. Since humans are born mostly (albeit not fully) microbially sterile, it is necessarily through interaction with the social and physical environment around them that subsequent microbial colonization takes place. Specifically, exposures such as mode of delivery and initiation and duration of breastfeeding influence its composition, and these early-life exposures are in turn shaped by one’s social status17–19. The quality of fetal environments — such as proper nutrition — is strongly patterned by income and education, including in the developed world20–22. This remains true after birth. While 91% of mothers with a college degree breast feed in the United States, that rate falls to 69% for mothers with a high-school degree. These rates fall to 28% and 14%, respectively, for six months of exclusive breastfeeding23.
We are beginning to see evidence that the lack of healthy microbiome development in children can have severe and lasting consequences24. For example, evidence from fecal transplant studies in mice show that the microbiomes of undernourished children, drawn from a sample in Malawi, negatively affect physical and cognitive development23,25. Despite this evidence of how extreme malnutrition in early life in the developing world alters the developing microbiome, we have little evidence from the developed world regarding how variation in early-life socioeconomic resources influences the developing gut microbiome.
Social relationships.
Social relationships have long been linked to health and mortality, including inflammation and immune response22. Indeed, the evidence is that the influence of social relationships on health and mortality exceeds medical interventions such as quitting smoking and the use of anti-hypertensives, like statins26. Given the robustness of this relationship, and the links between social relationships and inflammation and immune functioning, it is not surprising that research is now beginning to explore whether and how social interactions shape the microbiome.
In terms of existing research, studies using primate models suggest that social relationships impact the composition of the gut microbiota through direct microbial sharing between individuals27–30. Tung and colleagues[29], for example, found that social network and social group predicted the species in the gut microbiome of 48 wild baboons, even after adjusting for other shared factors including kinship, diet and shared environments, suggesting the importance of direct physical contact during social interactions in transmitting gut microbiomes. A few human studies have begun to document these relationships. For example, two recent studies found that individuals living together had more similar gut18 and skin18,19 microbiota than did those living apart. Some even hypothesize that microbes can help explain the evolution of social behaviours31; the effect of microbiota on the hosts’ central nervous system could operate via chemical signals that are used as social communication. This manipulation could benefit fitness of bacteria, such as reproduction and transmission, and food cravings and preferences32,33.
But despite some tantalizing evidence of the influence of social relationships on the gut microbiome, studies in human populations remain relatively small in number. Nonetheless, there is related evidence to support further exploration of connections between social relationships and the gut microbiome. There is growing empirical support for links between social and physical environments. Humans sharing homes have more similar skin microbiomes compared with those not sharing a home, probably due to skin shedding, respiratory activity and skin-surface contact34. When families moved, their microbial signature followed them to the new home, and individuals who left the home for several days saw a decline in their contribution to the home microbiome34. These findings suggest a mechanism for social transmission of bacterial communities through the built environment, which could apply to socially shared spaces such as schools, work and public transportation. In light of this, as well as recent primate evidence that ‘immigrant’ males share gut microbiome characteristics from both their birth and adult communities35, life-course residential and migration histories could play an important role in human microbiome dynamics.
Socioeconomic conditions.
Extensive research shows that adult socioeconomic resources, in addition to early-life socioeconomic resources, influence morbidity and mortality. Life expectancy difference at age 25 can differ by as much as 16 years between those with the lowest and highest levels of educational attainment36. While there is little direct existing evidence linking adult socioeconomic resources to the gut microbiome, the link is highly plausible12. The gut microbiome is strongly implicated in metabolic and inflammatory disorders. Adult socioeconomic resources, in turn, pattern chronic inflammatory diseases and metabolic disorders, ranging from diabetes to heart disease. The prevalence of diabetes is twice as high among those with lower compared with higher educational attainment. Among those with diabetes and myocardial infarction, the mortality risk is substantially greater for those with lower educational attainment and incomes37. Moreover, there is a robust literature linking socioeconomic status to inflammatory markers, such C-reactive protein, more generally. To some extent, this pathway is linked via behaviours. In the United States, obesity prevalence is 28% for women with a college degree compared with 45% for women without a high-school degree38. Given the already robust evidentiary body linking diet to the gut microbiome, dietary behaviours may be a key path linking socioeconomic status to the gut microbiome.
In addition to possible behavioural pathways that could link adult socioeconomic resources to the gut microbiome, there is emerging evidence of the role of psychosocial stress in modulating the microbiome33,39,40. This is important because psychosocial stress is a key pathway between many social environments (such as limited socioeconomic resources and social relationships) and morbidity and mortality outcomes41. Perhaps most striking is a study of bees that demonstrated that position in their social hierarchies influenced the gut microbiome, with both diet and stress mediating these relationships42. Rodent models have convincingly shown that exposure to psychological stressors can alter the gut microbiome, through neuroendocrine response, the integrity of barrier defences and the internalization of microbes43. In mice, exposure to social stressors has been shown to alter homeostatic interactions between the intestinal microbiota and the immune system, leading to increased susceptibility to enteric infection, and overproduction of inflammatory mediators that induce anxiety-like behaviour44. States of isolation, such as maternal neglect, appear to influence the gut microbial composition in animal models45 at least in part through stress43,46. For example, in captive rhesus monkeys, maternal separation stress induced reductions in lactobacilli in intestinal microflora and higher rates of opportunistic enteric infection45. Prenatal stress in mothers has also been shown to impact the microbiota of offspring in mice, which in turn decreased the abundance of Lactobacillus in the gut microbiota of their offspring47. More generally, there is some evidence that the effect of maternal stress on child anxiety and mental health disorders may be modulated by the gut microbiome47. These alterations were subsequently related to changes in the offspring’s metabolite profiles involved in energy balance, as well as with disruptions of amino acid profiles in the developing brain47,48.
We note that we have largely highlighted influences of social conditions on the microbiome. There is, however, empirical evidence supporting the idea that the composition of the microbiome may modulate individual behaviours, preferences and choices, and thus potentially shape individuals’ social interactions and environments. If confirmed, this bacterial ‘manipulation’ of the human host has perplexing implications for the evolution of phenotypical traits. This is an active area of study, mostly with animal models, and has already sparked controversy regarding the likely (or unlikely) sustainability of a strategy involving bacterial exploitation of their hosts33.
The need for better data
Overall, the existing evidence provides a strong rationale for the potential importance of social conditions for the dynamics of gut microbial composition across the life course, but thus far, population-based evidence to confirm these relationships is limited. To move forward, the significant challenge of data availability needs to be addressed. While animal models have been invaluable in understanding the mechanisms underlying gut microbial composition and function, including the possible influence of social conditions, they are constrained by some important limitations — some of which are general issues with animal models and some of which are specific to the study of the gut microbiome. First, the basic biological variance between mice (and some primates) and human models may limit the potential to translate what we learn about mice to humans49,50. Second, even if we overcome this challenge, animal studies are constrained in their ability to examine how more complex social phenomena, such as human social relationships and networks, influence the gut microbiome. While we can draw useful parallels, similar to basic biological differences, the social and cognitive differences between humans and animals constrain comparisons.
Existing human studies also have some important limitations, especially if the goal is to explore how social environments influence the gut microbiome. Early human gut microbiome studies were constrained by small, non-randomly selected, samples. For example, the Human Microbiome Project (HMP), directed and funded by the National Institutes of Health to map the healthy human microbiota in 2012, was conducted on a single non-random sample of 256 individuals from St. Louis and Houston, most of whom were researchers and students51. Despite the small number of non-Whites in the HMP sample, comparisons were made across race and ethnicity (Asian, Black, Mexican, Puerto Rican, White), with the investigators reporting that a “wide variety of taxa, gene families and metabolic pathways were differentially distributed with subject ethnicity at every body habitat, representing the phenotype with the greatest number … of total associations with the microbiome”51. These incidental findings of strong associations with race and ethnicity suggest the need to characterize the microbiome in diverse, population-based samples. Yatsuneko et al. also highlighted the need for diverse samples, showing strong geographical differences in microbiome structure and function for residents of the United States compared with the Amazon in Venezuela and rural Malawi52.
More recently, there have been attempts to collect much larger samples53, but these attempts fall far short both of population representativeness and measurement of the ‘macroenvironment’. Voluntary crowd-sourcing models such as The American Gut Project (http://americangut.org) or UK-based MapMyGut (https://mapmygut.com) have been shown to be particularly non-representative. For example, only 6% of respondents in the American Gut Project are obese compared with a 37% adult obesity rate overall in the United States54. As interest in the microbiome grows, larger studies are including microbiome collection. Key examples include the Belgian Flemish Gut Flora Project and the Dutch LifeLines Study53,55. Both studies include rich detail in regards to biological, anthropomorphic and general health data; however, they contain more limited data on social environments — socioeconomic, family, work and community — compared with social and demographic-based population health studies. Moreover, participants were not randomly selected, but rather were recruited through media campaigns, thus introducing selection and sample bias. The TwinsUK Study, one of most prolific human studies of the gut microbiome thus far, was designed with a specific biomedical focus on the heritability of common diseases, with only superficial attention to the social environment. The disproportionately female and White volunteer sample does not reflect the race, ethnic and socioeconomic diversity of the overall UK population56–63. Importantly, the highly selected nature of these samples limits variation in both the microbial exposures and phenotypic outcomes of interest, reducing their analytical potential and ultimately their scientific generalizability.
What are the implications of non-representative samples in this emerging science? One related cautionary tale comes from the neuroscience of brain development. This research has largely been conducted on non-representative ‘convenience’ samples of volunteers, leading some to argue that these study findings may be skewed64. A recent study supports this contention. Researchers compared representative and non-representative samples of children in an imaging study focused on brain development; findings in data that better represented the population were markedly different to those in the unrepresentative sample, showing a very different pattern of how differing regions of the brain develop as children age65. While this scenario may or may not repeat itself for existing microbiome and health research, it will be important to be aware of the heterogeneity of associations across different populations and how and why this variation may arise.
Collaborations with population health sciences
Given existing limitations, in this section, we detail ways in which existing population-based studies and methods could be employed to test novel hypotheses about social environments and the gut microbiome. But to start, we want to emphasize the broader potential methodological contributions that population health scientists might make to this field. One of the central challenges of human microbiome research, much like basic social science research, is how to demonstrate causal relationships. While animal models provide a straight-forward platform, in and of themselves, they are not sufficient to fully explore the social determinants of the gut microbiome. Observational human microbiome studies, however, can suffer from familiar issues related to unobserved confounding and causal inference.
Collaborations between life science and social and population health researchers, however, can draw on a long history in the social sciences of methods to improve causal inference in observational data, including family-based designs, natural or ‘quasi-experiments’ as well as population-based field experiments66. Opportunities for such research designs require microbiome data from ongoing, preferably longitudinal, studies with rich social, environmental and phenotypic data on participants. As the largest cost in obtaining data from a large representative population is drawing the sample frame and the initial enrolment of participants, we argue that adding the microbiome to existing population surveys is the most cost-effective approach relative to designing new studies from scratch. Many long-running population-based studies (for example, the Panel Study of Income Dynamics) follow families, which would allow for testing for intergenerational effects. Most large longitudinal population-based surveys also already collect biological data, ranging from blood to saliva. This would allow microbiome analysis in conjunction with high-quality health data (including genetic and epigenetic data), while also leveraging the experience of these studies in getting their participants to provide these types of more sensitive data67.
An ideal study design to investigate some of the pathways alluded to above should satisfy three conditions. First, as most population studies aspire to be, it should be representative of a target metapopulation rather than based on highly selected samples that preclude more than modest generalization of inferences. Second, it should be flexible enough to maximize opportunities to make genuine causal statements rather than being a source of association measures, which, in most cases, cannot be elevated to the status of estimates of causal effects, which are uncontaminated by omitted variable biases and confounding or selection mechanisms of various types. Third, because many of the relations portrayed in Fig. 1 are a function of lags and delayed impacts, the ideal study should be longitudinal and a source of information on events that unfold over multiple stages in the life course of individuals.
Early-life conditions.
Over the past 10 to 15 years, research on the Developmental Origins of Adult Health and Disease (DOHaD) has produced robust empirical evidence suggesting that prenatal and early postnatal exposures have a strong influence on early growth and development and, under some conditions, have significant delayed impacts on adult health outcomes68–72. The first three years of life are crucial for colonization of the gut microbiome52,73. This points to the configuration of the microbiome as one pathway through which early conditions may operate, calling for social and population health scientists working under the DOHaD paradigm to explicitly include investigation of the microbiome over the life course.
Both bodies of research confront remarkably similar problems, such as the existence of critical and sensitive periods, accumulation of damage and synergies over the life course, path dependencies, and reversibility properties41,74–77. It is likely that these problems, to which DOHaD and microbiome researchers have arrived independently, might have common solutions. Moreover, both groups are pointing to epigenetic modifications as an important mechanism through which embryonic and prenatal exposures, on one hand, and composition of the microbiome, on the other, may sometimes operate72,78–81.
While there are many examples of potentially fruitful joint research, an area that offers promise of very immediate rewards concerns the effects of early nutritional status and nutritional shocks on growth, development and adult health outcomes. Barker’s seminal research68 implicated fetal nutritional impairments as an important determinant of adult chronic conditions, including obesity, coronary heart disease and type 2 diabetes, whereas a large literature in population health sciences investigates the effects of infant and child nutrition on diseases in adult mortality and disability (for a review see ref. 82). Until recently, this body of research made no reference to the relation between nutrition and the microbiome. New research suggests that a paradigmatic shift is in order. A study of Malawian malnourished infants showed, via fecal transplantation in mice, that “gut microbial immaturity is causally related to child malnutrition” as “immature microbiota transmit impaired growth, altered bone morphology, and metabolic abnormalities”83. This could be a smoking gun, as it shows that the microbiome is one mechanism that mediates the association of prenatal or neonatal malnutrition and later morbidity and mortality. If replicated, it provides a heretofore unknown and modifiable pathway.
Establishing relations involving the microbiome may also be required to fully understand other processes identified by DOHaD (and variants), such as those relating early exposure to acute stress and later mental health84–86, childhood poverty and adversity to late onset of chronic illnesses87–89, exposure to shocks such as influenza, natural disasters, wars to a broad array of chronic ailments90, or recurrent childhood or adolescent infectious diseases, sustained inflammation, and later heart and circulatory disorders91–94.
But how can we explore these questions? One approach involves exploiting quasi-experimental conditions generated by famines, including the Dutch Famine95 and the Great Chinese Famine96, which produce quasi-randomly selected subpopulations exposed and unexposed to a ‘treatment’. Studies of the Dutch Famine have uncovered, in samples of mid-life and older adults, that those exposed to the famine in utero, compared with those in utero in the months just preceding the famine, have everything from higher mortality rates from cardiovascular diseases to differences in DNA methylation97–99 linked to metabolic health and transgenerational impacts on metabolic health. Differences in DNA methylation explained a significant portion of the differences in metabolic health between those with and without exposure to the famine in utero. Data on the gut microbiome could be added to these existing cohorts, with people currently in their 60s (Great Chinese Famine) and 70s (Dutch Famine). This could elucidate relations conjectured by DOHaD.
Though less common, randomized control trials, or field experiments are also being conducted to measure the impact of social interventions on health outcomes in larger population-based samples. For example, a newly launched, randomized (conditional) cash-transfer experiment is enrolling 1,000 infants, to track how a randomly assigned increase in income ($333 a month) affects cognitive development in poor children100. A wealth of longitudinal and quasi-experimental research points strongly to the influence of poverty on maternal stress and mental health, as well as the infants’ cognitive development in early life. The addition of data on the gut microbiome would allow for the testing of the role of the gut microbiome as a mediator in these relationships. As already previously detailed, studies showing connections between the gut microbiome, maternal stress and cognitive development would suggest these are pathways worth exploring.
Furthermore, there are a growing number of studies that involve controlled interventions that monitor large populations over extended periods of time. Thus, studies built around cash-transfer programmes, such as Progresa in Mexico, have become an ideal study design that many other low-to middle-income countries are following, thus generating massive datasets on a multiplicity of conditions and outcomes101,102. In addition, international organizations such as the World Bank, the International Food Policy Research Institute, the Inter-American Development Bank and the World Health Organization periodically initiate studies that involve a multiplicity of social protection interventions and are designed to facilitate causal inference. Experiments involving cash transfers or those based on nutritional interventions could add a module to collect information on pregnant women, maternal health and nutrition, peri-and postnatal birth exposures, microbiota composition and various childhood outcomes.
Lastly, although the empirical evidence is still too fragile to confirm them, DOHaD and related theories pose conjectures involving transgenerational effects of some significance. To the extent that these may be contingent or directly influenced by changes in the microbiome, there will be ample room to test evolutionary biology hypotheses about the development (and disappearance) of phenotypical traits with strong impacts on reproduction and longevity. Thus, large population studies with the characteristics described above will not only be useful to population health scientists, but also could have potential large spill-over effects benefitting the growth of other disciplines.
Socioeconomic conditions.
Social and population health scientists have spent decades gathering and analysing data that demonstrate that socioeconomic markers, such as education, income and wealth, matter a great deal for adult health and mortality, as we detailed earlier103. Evidence linking behavioural factors, such as obesity, and psychosocial stress to both the gut microbiome and socioeconomic health disparities, provide a plausible basis for testing whether adult socioeconomic resources influence the gut microbiome14,15,39,101. To date, however, there is virtually no work exploring the potential of gut microbial composition as a biological mechanism linking adult socioeconomic status to morbidity and mortality outcomes.
Existing population-based longitudinal studies that have documented the influence of income and educational attainment on morbidity and mortality are large in number and increasingly include a wide array of more basic biological data collected from saliva, blood and even urine, hair and nail samples104. The addition of microbial data, then, would be a natural extension. These studies follow large populations of cohorts for extended periods of time and are a source of very rich information. Examples include the Health and Retirement Study (HRS), and its sister studies around the world, the National Study of Adult and Adolescent Health (AddHealth) and the British Cohort Studies.
Although these studies are not designed to be experimental, changing exogenous conditions sometimes induce a quasi-experimental set-up that can be exploited. For example, the HRS in the United States has provided rich information on the effects of the Great Recession on individual health status changes105. The HRS, in addition to many other population-based studies, has been used to test the influence of changes in schooling laws, as a source of exogenous change, on health, with outcomes ranging from diabetes and strokes to later life cognitive decline and mortality104,105. Many studies, including the HRS and AddHealth, now include genetic data, which allow for a Mendelian randomization approach — drawn from genetic variants linked to educational attainment — to test the causal influence of educational attainment on health106–108. Many of these studies could be replicated to test the influence of these exogenous changes on microbial composition, for example.
Combining animal experiments with insights from the kinds of observational human data detailed above may offer an especially unique methodological strategy to strengthen causal findings. There is, of course, already precedence for this. For example, a well-known study found that the transplantation of gut microbiota from obese humans to lean, germ-free recipient mice transfers an increased adiposity phenotype relative to transplants from lean donors79. This approach relies on hybrid human-animal studies to produce a robust causal design. They first find associations between phentoypes (like obesity in this case) and the composition of the gut microbiome, and then they use animal models (with human fecal samples) to further test the causality of that associational relationship. For example, we know the chronic inflammatory conditions, like diabetes, are more deadly for those with low compared with high educational attainment, even after accounting for body mass index37. Fecal transplants drawn from those with type 2 diabetes, but who varied in their educational attainment and were comparable on characteristics like body mass index, could then be transplanted into mice to see how the gut microbiota influenced morbidity and mortality outcomes in these models. Is the gut microbiome a biological mechanism that can help clarify why lower levels of educational attainment are so harmful for health?
Social relationships.
Social and behavioural scientists have also shown that quality, quantity and duration of intimate contact and social relations are important for health, potentially as a buffer from stress (for reviews see refs 109,110). Across a wide array of empirical designs, ranging from animal models to human longitudinal studies and randomized controlled trials, the relationship between social relationships and health and mortality is the most well documented among specific social conditions that influence health and mortality111. Indeed, a recent review found that social relationships are a stronger predictor of mortality than smoking111. Limited social interactions may contribute to reduced diversity in gut microbial communities among older persons or the socially isolated, another plausible biological mechanism for the strong associations of social relationships and health. As already detailed, there is some evidence from primate models that social relationships, perhaps in part via direct microbial sharing, exerts an influence on gut microbial composition. Indeed, a recent study demonstrated that the oral microbiome infiltrates the gut microbiome, supporting evidence for microbial exchange via salivary mechanisms112. This area is ripe for study in human models to further test and elucidate pathways between social interactions, gut microbial composition, and morbidity and mortality outcomes.
Our recent data collection in the Wisconsin Longitudinal Study (WLS) took a step in the direction of integrating social and biological approaches to the gut microbiome. This cohort of nearly 10,317 1957 Wisconsin high-school graduates, their spouses and a subsample of siblings113 has been followed for the past 60 years and includes extensive social and phenotypic measurement of everything from high-school records, education and occupation histories, to childhood conditions, health status, cognition, disability, mortality and genetic data. Fecal samples from a randomly selected subsample of 436 participants were recently collected, targeting a mixture of sibling and spousal pairs in their mid-70s67.
Because the subsample includes siblings as well as spouses, most of whom have been married and lived together for nearly their entire adult lives, we were able to compare microbiome composition among those who share environments due to living arrangements across most of their adult lives to those who share only early upbringing conditions. We found114 that shared environments in adult life had a stronger influence on microbial composition in later life than did shared early-life environments. This provided a mechanism to test the relatively plasticity of the gut microbiome in population-based data. We also found that the similarity between spouses, compared with unrelated individuals, was entirely driven by married couples who reported they had a very close relationship; in short, the shared gut microbial composition of married couples who rated their relationship as ‘somewhat’ good was similar to that of unrelated individuals.
Modest as it may be, this finding alone is important, as it generates new questions and problems. First, it is somewhat unexpected given extant empirical evidence that points to the early colonization and stable character of the microbiome3. Second, it confirms other findings in epidemiology according to which environments shared by siblings explain only a small fraction of adult outcomes115. Finally, one puzzle to resolve is whether or not the impact of shared environments by spouses on the microbiome is part of a chain of events that accounts for within-couple similarity in chronic illness116.
Conclusion
We believe these early days of microbiome research offer exciting opportunities for collaborations between the life sciences and the social and population health sciences, a time when theory and measurement in both realms is still developing and before disciplinary conventional wisdom has the chance to solidify. For social and population health scientists, the study of the microbiome may help elucidate currently unknown biological pathways that link social conditions to health and mortality, and provide a target for intervention.
For biologists, collaborations with social and population health scientists can enhance knowledge of a new range of environmental factors that may influence microbial composition and, in turn, health. Insights from the social and behavioural health research can also help contextualize existing findings. For example, the robust evidentiary base linking diet and the gut microbiome should be considered in the context of broader population health research that documents how structural conditions, ranging from economic resources to neighbourhood environments, constrain nutritional choices individuals make.
Both fields will benefit from the joining of social science and basic biological methodological approaches. Embedding animal models in the context of population-based studies provides a novel approach to improving causal methods for social and population scientists. Having access to large, population representative, longitudinal-based studies with high-quality phenotypic measures can vastly expand the quality of research produced by biologists. Success will ultimately be measured by the ability of scientists across all disciplines to understand how the microbiome influences human health and social trajectories (and vice versa), and how social and medical interventions may use this knowledge to improve both individual well-being and population health.
Acknowledgements
The authors thank members of the Rey Laboratory for their insights and support. This work was supported by the National Institute of Food and Agriculture, US Department of Agriculture (2016-67017-24416, FER), the National Institute on Aging, National Institutes of Health (NIH) (AG041868, PH), the Center for the Demography of Health and Aging (AG017266, PH), and the Clinical and Translational Science Award (CTSA), NIH National Center for Advancing Translational Sciences (NCATS) (UL1TR000427, ZZT). Additional support was provided by the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Clemente JC, Ursell LK, Parfrey LW & Knight R The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice EA & Segre JA The human microbiome: our second genome. Annu. Rev. Genomics Hum. Genet. 13, 151–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK & Knight R Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho I & Blaser MJ The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sender R, Fuchs S & Milo R Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster J & Neufeld KA Gut-brain axis: how the microbiome influences anxiety and depression. Int. J. Neuropsychopharmacol. 17, 27–27 (2014). [Google Scholar]
- 7.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothschild D et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Adams RI, Bateman AC, Bik HM & Meadow JF Microbiota of the indoor environment: a meta-analysis. Microbiome 3, 49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stilling RM, Bordenstein SR, Dinan TG & Cryan JF Friends with social benefits: host-microbe interactions as a driver of brain evolution and development? Front. Cell Infect. Microbiol. 4, 147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamper CE et al. The microbiome of the built environment and human behavior: implications for emotional health and well-being in postmodern Western societies. Int. Rev. Neurobiol. 131, 289–323 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Rook GA, Raison CL & Lowry CA Microbial ‘old friends, immunoregulation and socioeconomic status. Clin. Exp. Immunol. 177, 1–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay BB & Arrietta MC Let Them Eat Dirt: Saving Our Children from an Oversanitized World 304 (Greystone Books, New York, 2016). [Google Scholar]
- 14.McDade TW The ecologies of human immune function. Annu. Rev. Anthropol. 21, 495–521 (2005). [Google Scholar]
- 15.Coe CL & Laudenslager ML Psychosocial influences on immunity, including effects on immune maturation and senescence. Brain Behav. Immun. 21, 1000–1008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagundes CP, Glaser R & Kiecolt-Glaser JK Stressful early life experiences and immune dysregulation across the lifespan. Brain Behav. Immun. 27, 8–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennis CL Breastfeeding initiation and duration: a 1990–2000 literature review. J. Obstet. Gynecol. Neonatal Nurs. 31, 12–32 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Dominguez-Bello MG et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller NT et al. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. Int. J. Obes. 39, 665–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer MS, Sequin L, Lydon J & Goulet L Socio-economic disparities in pregnancy outcome: why do the poor fare so poorly? Paediatr. Perinat. Epidemiol. 14, 194–210 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Joseph KS, Liston RM, Dodds L, Dahlgren L & Allen AC Socioeconomic status and perinatal outcomes in a setting with universal access to essential health care services. CMAJ 177, 583–590 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Berg G, van Eijsden M, Vrijkotte TG & Gemke RJ Educational inequalities in perinatal outcomes: the mediating effect of smoking and environmental tobacco exposure. PLoS ONE 7, e37002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anstey EH, Chen J, Elam-Evans LD & Perrine CG Racial and geographic differences in breastfeeding — United States, 2011–2015. MMWR Morb. Mortal. Wkly Rep. 66, 723–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Codagnone MG et al. Programming bugs: microbiota and the developmental origins of brain health and disease. Biol. Psychiatry https://doi.org/10.10167j.biopsych.2018.06.014 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Blanton LV et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang YC et al. Social relationships and physiological determinants of longevity across the human life span. Proc. Natl Acad. Sci. USA 113, 578–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moeller AH et al. Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2, e1500997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett G et al. Host age, social group, and habitat type influence the gut microbiota of wild ring-tailed lemurs (Lemur catta). Am. J. Primatol. 78, 883–892 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Tung J et al. Social networks predict gut microbiome composition in wild baboons. Elife 4, e05224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amaral WZ et al. Social influences on Prevotella and the gut microbiome of young monkeys. Psychosom. Med. 79, 888–897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewin-Epstein O, Aharonov R & Hadany L Microbes can help explain the evolution of host altruism. Nat. Commun. 8, 14040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Archie EA & Tung J Social behavior and the microbiome. Curr. Opin. Behav. Sci. 6, 28–34 (2015). [Google Scholar]
- 33.Johnson KV & Foster KR Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 16, 647–655 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Lax S et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grieneisen LE, Livermore J, Alberts S, Tung J & Archie EA Group living and male dispersal predict the core gut microbiome in wild baboons. Integr. Comp. Biol. 57, 770–785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rostron BL, Boies JL & Arias E Education reporting and classification on death certificates in the United States. Vital Health Stat. 2, 1–21 (2010). [PubMed] [Google Scholar]
- 37.Perna L, Thien-Seitz U, Ladwig KH, Meisinger C & Mielck A Socio-economic differences in life expectancy among persons with diabetes mellitus or myocardial infarction: results from the German MONICA/ KORA study. BMC Public Health 10, 135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogden CL et al. Prevalence of obesity among adults, by household income and education — United States, 2011–2014. MMWR Morb. Mortal. Wkly Rep. 66, 1369–1373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen AP, Dinan TG, Clarke G & Cryan JF A psychology of the human brain-gut-microbiome axis. Soc. Personal. Psychol. Compass 11, e12309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lach G, Schellekens H, Dinan TG & Cryan JF Anxiety, depression, and the microbiome: a role for gut peptides. Neurotherapeutics 15, 36–59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marmot M & Wilkinson RG Psychosocial and material pathways in the relation between income and health: a response to Lynch. et al. BMJ 322, 1233–1236 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwong WK & Moran NA Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey MT et al. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav. Immun. 25, 397–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey MT Influence of stressor-induced nervous system activation on the intestinal microbiota and the importance for immunomodulation. Adv. Exp. Med. Biol. 817, 255–276 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Bailey MT & Coe CL Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev. Psychobiol. 35, 146–155 (1999). [PubMed] [Google Scholar]
- 46.O’Mahony SM et al. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol. Psychiatry 65, 263–267 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Jasarevic E, Howerton CL, Howard CD & Bale TL Alterations in the vaginal microbiome by maternal stress are associated with metabolic reprogramming of the offspring gut and brain. Endocrinology 156, 3265–3276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goyal MS, Venkatesh S, Milbrandt J, Gordon JI & Raichle ME Feeding the brain and nurturing the mind: linking nutrition and the gut microbiota to brain development. Proc. Natl Acad. Sci. USA 112, 14105–14112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z & Knight R Dietary effects on human gut microbiome diversity. Br. J. Nutr. 113, S1–S5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen TL, Vieira-Silva S, Liston A & Raes J How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhernakova A et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Preliminary Characterization of the American Gut Population (American Gut Project, 2014); http://americangut.org/wp-content/uploads/2016/02/mod1_main.pdf [Google Scholar]
- 55.Falony G et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Moayyeri A, Hammond CJ, Hart DJ & Spector TD The UK Adult Twin Registry (TwinsUK Resource). Twin Res. Hum. Genet. 16, 144–149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodrich JK et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson MA et al. Signatures of early frailty in the gut microbiota. Genome Med. 8, 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beaumont M et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 17, 189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie H et al. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. 3, 572–584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodrich JK et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menni C et al. Gut microbiome diversity and high-fibre intake are related to lower long-term weight gain. Int. J. Obes. 41, 1099–1105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu J et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ. Res. 117, 817–824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falk EB et al. What is a representative brain? Neuroscience meets population science. Proc. Natl Acad. Sci. USA 110, 17615–17622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.LeWinn KZ, Sheridan MA, Keyes KM, Hamilton A & McLaughlin KA Sample composition alters associations between age and brain structure. Nat. Commun. 8, 874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morgan SL & Winship C Counterfactuals and Causal Inference: Analytical Methods for Social Research Social Research, 1st edn (Cambridge Univ. Press, Cambridge, 2014). [Google Scholar]
- 67.Herd P et al. The influence of social conditions across the life course on the human gut microbiota: a pilot project with the Wisconsin Longitudinal Study. J. Gerontol. B Psychol. Sci. Soc. Sci. 73, 124–133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barker DJP Mothers, Babies, and Health in Later Life. 2nd edn (Vol. ix, Churchill Livingstone, Edinburgh, 1998). [Google Scholar]
- 69.Gluckman P & Hanson M The Fetal Matrix: Evolution, Development and Disease (Cambridge Univ. Press, Cambridge, 2004). [Google Scholar]
- 70.Langley-Evans SC Fetal Nutrition and Adult Disease: Programming of Chronic Disease Through Fetal Exposure to Undernutrition (CABI Publishing, Wallingford, 2004). [Google Scholar]
- 71.Bateson P & Gluckman P Plasticity, Robustness, Development and Evolution (Cambridge Univ. Press, Cambridge, 2011). [Google Scholar]
- 72.Gluckman P, Beedle A, Buklijas T, Low F & Hanson M Principles of Evolutionary Medicine 2nd edn, 400 (Oxford Univ. Press, Oxford, 2016). [Google Scholar]
- 73.Faith JJ et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wamala SP, Lynch J & Kaplan GA Women’s exposure to early and later life socioeconomic disadvantage and coronary heart disease risk: the Stockholm Female Coronary Risk Study. Int. J. Epidemiol. 30, 275–284 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Pensola TH & Martikainen P Cumulative social class and mortality from various causes of adult men. J. Epidemiol. Community Health 57, 745–751 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo Y & Waite LJ The impact of childhood and adult SES on physical, mental, and cognitive well-being in later life. J. Gerontol. B Psychol. Sci. Soc. Sci. 60, S93–S101 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lynch JW et al. Childhood and adult socioeconomic status as predictors of mortality in Finland. Lancet 343, 524–527 (1994). [DOI] [PubMed] [Google Scholar]
- 78.Cortese R, Lu L, Yu Y, Ruden D & Claud EC Epigenome-microbiome crosstalk: a potential new paradigm influencing neonatal susceptibility to disease. Epigenetics 11, 205–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harris RA et al. Colonic mucosal epigenome and microbiome development in children and adolescents. J. Immunol. Res. 2016, 9170162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Indrio F et al. Epigenetic matters: the link between early nutrition, microbiome, and long-term health development. Front. Pediatr. 5, 178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monk C, Spicer J & Champagne FA Linking prenatal maternal adversity to developmental outcomes in infants: the role of epigenetic pathways. Dev. Psychopathol. 24, 1361–1376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barker DJ, Eriksson JG, Forsen T & Osmond C Fetal origins of adult disease: strength of effects and biological basis. Int. J. Epidemiol. 31, 1235–1239 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Smith MI et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meaney MJ Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu. Rev. Neurosci. 24, 1161–1192 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Mcewen BS Protective and damaging effects of stress mediators: allostasis and allostatic load. N. Engl. J. Med. 338, 171–179 (1998). [DOI] [PubMed] [Google Scholar]
- 86.Knudsen EI, Heckman JJ, Cameron JL & Shonkoff JP Economic, neurobiological, and behavioral perspectives on building America’s future workforce. Proc. Natl Acad. Sci. USA 103, 10155–10162 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Forsdahl A Commentary: childhood deprivation and adult mortality. Int. J. Epidemiol. 31, 308–308 (2002). [Google Scholar]
- 88.Hayward MD & Gorman BK The long arm of childhood: the influence of early-life social conditions on men’s mortality. Demography 41, 87–107 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Bengtsson T & Lindstrom M Childhood misery and disease in later life: the effects on mortality in old age of hazards experienced in early life, southern Sweden, 1760–1894. Popul. Stud. 54, 263–277 (2000). [DOI] [PubMed] [Google Scholar]
- 90.Almond D & Currie J Killing me softly: the fetal origins hypothesis. J. Econ. Perspect. 25, 153–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finch C The Biology of Human Longevity: Inflammation, Nutrition, and Aging in the Evolution of Lifespans 1st edn (Academic Press, San Diego, 2007). [Google Scholar]
- 92.Fong IW Emerging relations between infectious diseases and coronary artery disease and atherosclerosis. CMAJ 163, 49–56 (2000). [PMC free article] [PubMed] [Google Scholar]
- 93.Kermack WO & McKendrick AG A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. A: Math. Phys. Eng. Sci. 115, 700–721 (1927). [Google Scholar]
- 94.McDade TW, Rutherford J, Adair L & Kuzawa CW Early origins of inflammation: microbial exposures in infancy predict lower levels of C-reactive protein in adulthood. Proc. Biol. Sci. 277, 1129–1137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lumey LH et al. Cohort profile: the Dutch Hunger Winter families study. Int. J. Epidemiol. 36, 1196–1204 (2007). [DOI] [PubMed] [Google Scholar]
- 96.Li C & Lumey LH Exposure to the Chinese famine of 1959–61 in early life and long-term health conditions: a systematic review and meta-analysis. Int. J. Epidemiol. 46, 1157–1170 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Tobi EW et al. DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci. Adv. 4, eaao4364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roseboom T, de Rooij S & Painter R The Dutch famine and its long-term consequences for adult health. Early Hum. Dev. 82, 485–491 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Painter RC et al. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 115, 1243–1249 (2008). [DOI] [PubMed] [Google Scholar]
- 100.Mother’s money. The Economist (3 May 2018). [Google Scholar]
- 101.Rivera JA, Sotres-Alvarez D, Habicht JP, Shamah T & Villalpando S Impact of the Mexican program for education, health, and nutrition (Progresa) on rates of growth and anemia in infants and young children: a randomized effectiveness study. JAMA 291, 2563–2570 (2004). [DOI] [PubMed] [Google Scholar]
- 102.Behrman JR & Todd PE Randomness in the Experimental Samples of Progresa Working Paper 38. (International Food Policy Research Institute, Washington DC, 1999). [Google Scholar]
- 103.Chetty R et al. The association between income and life expectancy in the United States, 2001–2014. JAMA 315, 1750–1766 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Crimmins E, Jung Ki, K. & Sarinnapha V Biodemography: new approaches to understanding trends and differences in population health and mortality. Demography 47, S41–S64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McInerney M, Mellor JM & Nicholas LH Recession depression: mental health effects of the 2008 stock market crash. J. Health Econ. 32, 1090–1104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Glymour MM, Kawachi I, Jencks CS & Berkman LF Does childhood schooling affect old age memory or mental status? Using state schooling laws as natural experiments. J. Epidemiol. Community Health 62, 532–537 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davies NM, Dickson M, Davey Smith G, van den Berg G & Windmeijer F Preprint at bioRxiv 10.1101/074815 (2016). [DOI] [Google Scholar]
- 108.Tillmann T et al. Education and coronary heart disease: Mendelian randomisation study. BMJ 358, j3542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.House JS, Landis KR & Umberson D Social relationships and health. Science 241, 540–545 (1988). [DOI] [PubMed] [Google Scholar]
- 110.Umberson D, Crosnoe R & Reczek C Social relationships and health behavior across life course. Annu. Rev. Sociol. 36, 139–157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holt-Lunstad J, Smith TB & Layton JB Social relationships and mortality risk: a meta-analytic review. PLoS Med. 7, e1000316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cao X Intestinal inflammation induced by oral bacteria. Science 358, 308–309 (2017). [DOI] [PubMed] [Google Scholar]
- 113.Herd P, Carr D & Roan C Cohort profile: Wisconsin Longitudinal Study (WLS). Int. J. Epidemiol. 43, 34–41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dill-McFarland KA et al. Social relationships, social isolation, and the human gut microbiota. Preprint at bioRxiv https://doi.org/10.110¼28938 (2018). [Google Scholar]
- 115.Lawlor DA, Clark H, Davey Smith G & Leon DA Childhood intelligence, educational attainment and adult body mass index: findings from a prospective cohort and within sibling-pairs analysis. Int. J. Obes. 30, 1758–1765 (2006). [DOI] [PubMed] [Google Scholar]
- 116.Meyler D, Stimpson JP & Peek MK Health concordance within couples: a systematic review. Soc. Sci. Med. 64, 2297–2310 (2007). [DOI] [PubMed] [Google Scholar]


