Abstract
Myo‐inositol oxygenase (MIOX) is the first enzyme in the inositol route to ascorbate (L‐ascorbic acid, AsA, vitamin C). We have previously shown that Arabidopsis plants constitutively expressing MIOX have elevated foliar AsA content and displayed enhanced growth rate, biomass accumulation, and increased tolerance to multiple abiotic stresses. In this work, we used a combination of transcriptomics, chromatography, microscopy, and physiological measurements to gain a deeper understanding of the underlying mechanisms mediating the phenotype of the AtMIOX4 line. Transcriptomic analysis revealed increased expression of genes involved in auxin synthesis, hydrolysis, transport, and metabolism, which are supported by elevated auxin levels both in vitro and in vivo, and confirmed by assays demonstrating their effect on epidermal cell elongation in the AtMIOX4 over‐expressers. Additionally, we detected up‐regulation of transcripts involved in photosynthesis and this was validated by increased efficiency of the photosystem II and proton motive force. We also found increased expression of amylase leading to higher intracellular glucose levels. Multiple gene families conferring plants tolerance/expressed in response to cold, water limitation, and heat stresses were found to be elevated in the AtMIOX4 line. Interestingly, the high AsA plants also displayed up‐regulation of transcripts and hormones involved in defense including jasmonates, defensin, glucosinolates, and transcription factors that are known to be important for biotic stress tolerance. These results overall indicate that elevated levels of auxin and glucose, and enhanced photosynthetic efficiency in combination with up‐regulation of abiotic stresses response genes underly the higher growth rate and abiotic stresses tolerance phenotype of the AtMIOX4 over‐expressers.
Keywords: abiotic stress, ascorbate, ascorbic acid, redox biology, vitamin C
1. INTRODUCTION
L‐Ascorbic acid (AsA) also known as vitamin C is important for human, animal, and plant health. In animals, vitamin C helps in collagen formation, decreases cholesterol content, boosts immune function, and improves wound healing. Vitamin C deficiency on the other hand leads to scurvy. Humans and other primates are unable to produce vitamin C, so they depend on fruits and vegetables to satisfy their need for this vitamin (Figueroa‐Mendez & Rivas‐Arancibia, 2015; Leong, Liu, Oey, & Burritt, 2017; Weaver et al., 2014). In plants, ascorbate acts as a redox buffer and a cofactor of enzymes involved in various biochemical processes. Ascorbate also participates in vitamin E regeneration and acts as a precursor of organic acids including threonic and tartaric acids (Debolt, Cook, & Ford, 2006; Gallie, 2013; Gest, Gautier, & Stevens, 2013). Ascorbate modulates the cell cycle, flowering time, and signal transduction affecting plant growth and development (Ortiz‐Espín, Sanchez‐Guerrero, Sevilla, & Jiménez, 2017). Vitamin C‐deficient (vtc) mutants have lower AsA, slower growth, and show early senescence (Conklin, Saracco, Norris, & Last, 1999; Dowdle, Ishikawa, Gatzek, Rolinski, & Smirnoff, 2007; Kerchev et al., 2011; Pastori et al., 2003).
Despite the importance of vitamin C for human health, it was until 1998 when a pathway for its biosynthesis in plants was revealed (Wheeler, Jones, & Smirnoff, 1998). Two additional routes contributing to the formation of AsA, the L‐gulose and D‐galacturonate pathways, were proposed in 2003 (Agius et al., 2003; Wolucka & Montagu, 2003) and an additional route that uses myo‐inositol as a precursor was proposed by Lorence, Chevone, Mendes, and Nessler (2004) (Figure 1).
Figure 1.
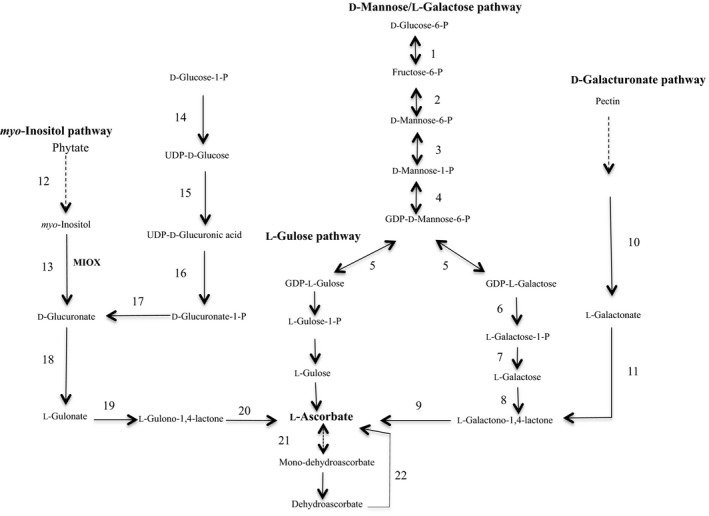
The ascorbate metabolic network in plants and animals. The D‐mannose/L‐galactose route, the L‐gulose shunt, the D‐galacturonate route, and the myo‐inositol pathway. Enzymes: 1 phosphoglucose isomerase: 2 phosphomannose isomerase; 3 phosphomannose mutase; 4 GDP‐mannose pyrophosphorylase; 5 GDP‐mannose, 3′5′ epimerase; 6 GDP‐galactose phosphorylase; 7 L‐galactose‐1‐P‐phosphatase; 8 L‐galactose dehydrogenase; 9 L‐galactono‐1,4‐lactone dehydrogenase; 10 galacturonate reductase; 11 aldonolactonase; 12 myo‐inositol 1‐P phosphatase; 13 myo‐inositol oxygenase; 14 UDP‐glucose pyrophosphorylase; 15 UDP‐glucose dehydrogenase; 16 UDP‐glucuronate pyrophosphorylase; 17 glucuronate 1‐kinase; 18 glucuronate reductase; 19 glucuronolactonase; 20 L‐gulono‐1,4‐lactone oxidase; 21 monodehydroascorbate reductase; 22 dehydroascorbate reductase. Broken arrows indicate multiple reactions
Ascorbate is the most abundant antioxidant in plants present at concentrations orders of magnitude higher than others. Ascorbate scavenges reactive oxygen species (ROS) and protects plants from abiotic and biotic stresses by maintaining ROS homeostasis (Foyer & Noctor, 2011). Plants with elevated AsA content have been developed by over‐expressing biosynthetic genes and master regulators (reviewed in Yactayo‐Chang, Acosta‐Gamboa, Nepal, & Lorence, 2018). Plants with lower vitamin C (vtc mutants) are sensitive to high light, heat, osmotic, and oxidative stresses (Cho et al., 2016; Conklin, DePaolo, Wintle, Schatz, & Buckenmeyer, 2013; Tóth, Nagy, Puthur, Kovacs, & Garab, 2011), but are resistant to Pseudomonas syringae infection due to increased expression of pathogen‐related (PR) genes via reduced glutathione accumulation (Pavet et al., 2005). Contrastingly, enhanced resistance of low ascorbate Arabidopsis thaliana to Pseudomonas syringae is due to higher ROS priming with increased salicylic acid (SA) and enhanced expression of SA induced PR proteins (Mukherjee et al., 2010). Plants with elevated AsA on the other hand are tolerant to salt, cold, heat, osmotic, oxidative, high light, and water limitation stresses (reviewed in Yactayo‐Chang et al., 2018).
Ascorbate acts as cofactor for the thioglucosidase degradation enzyme, myrosinase that produces toxic chemicals against herbivores and pathogens (Burmeister, Cottaz, Rollin, Vasella, & Henrissat, 2000), and there are reports indicating that AsA can also act as an antioxidant agent and protect caterpillars from ROS generated from the oxidation of ingested polyphenols (Barbehenn, 2002). Other work reported that when the peach potato aphid (Myzus persicae) was allowed to feed on potato leaves with normal AsA content, the aphid grew twice the size of its counterpart growing on leaves with low AsA content, indicating that elevated AsA may aid in aphid growth (Kerchev et al., 2013). When Arabidopsis roots were infected with the nematode Heterodera schachtii, MIOX played a significant role in the development of syncytium and female nematodes. The study of metabolites in roots containing syncytia showed that a decrease in the level of myo‐inositol was responsible for syncytium development (Siddique et al., 2009, 2014). These examples highlight the species‐specific and context‐dependent versatile role of AsA during biotic stress.
Arabidopsis plants constitutively expressing enzymes in the myo‐inositol pathway: myo‐inositol oxygenase, glucuronate reductase, or L‐gulono‐1,4‐lactone oxidase (GLOase, a.k.a. GulLO) have 1.5‐fold to threefold elevated AsA content compared to wild‐type (WT) control (Lorence et al., 2004; Radzio, Lorence, Chevone, & Nessler, 2003; Tóth et al., 2011; Yactayo‐Chang, 2011). Using manual phenotyping, we and others have showed that Arabidopsis MIOX4 and GulLO over‐expressers also display enhanced biomass accumulation of both aerial and underground tissues and increased tolerance to cold, heat, salt, and pyrene, an environmental pollutant (Lisko et al., 2013; Tóth et al., 2011). Using high‐throughput phenotyping approaches, we have quantified the enhanced growth rate and biomass accumulation of the high AsA line (Acosta‐Gamboa, 2019; Lisko, Aboobucker, Torres, & Lorence, 2014; Lisko et al., 2013; Yactayo‐Chang et al., 2018). In this study, we report our findings on the diverse mechanisms underlying the enhanced growth rate, biomass accumulation, and broad abiotic stress tolerance of the high AsA line using a combination of transcriptomics (RNA‐seq), RT‐qPCR, liquid chromatography–tandem mass spectroscopy (LC‐MS/MS), cell biology, confocal microscopy, and physiological measurements.
2. MATERIALS AND METHODS
2.1. Plants and growth conditions
In this study, we used Arabidopsis thaliana (A. thaliana) wild‐type accession Columbia (ABRC stock CS‐60000), single‐insertion homozygous line over‐expressing AtMIOX4 (AtMIOX4 L21, Yactayo‐Chang, 2011), a vitamin C defective mutant (vtc1‐1) (Conklin et al., 1999), and the auxin sensor line (R2D2) (Liao et al., 2015). Seeds were sterilized with 70% (v/v) ethanol for 5 min followed by 50% (v/v) sodium hypochlorite with 0.05% (v/v) Tween‐20 for 10 min. Then, seeds were washed 5–6 times with sterile distilled water. Plants were grown in Murashige and Skoog (MS) media pH 5.6 for 12 days after stratification for 3 days at 4°C. Seedlings from MS plates were transferred to soil (Promix PGX; Premier Horticulture) and grown in a growth chamber at 23°C, 65% humidity, 165–210 μmol m− 2 s− 1 light intensity, and 10:14‐hr photoperiod.
2.2. Construction of transgenic plants
The MIOX4 ORF was subcloned into the KpnI and SacI sites of the pBIB‐Kan binary vector placing it under the control of the CaMV 35S promoter with duplicated enhancer. The AtMIOX4:pBIB‐Kan construct was inserted into Agrobacterium tumefaciens strain GV3101 via heat shock. Arabidopsis thaliana var. Columbia plants were transformed with AtMIOX4:pBIB‐Kan construct via the floral dip method (Clough & Bent, 1998). Seedlings were selected on MS media plates containing 50 mg/L kanamycin. After 2 weeks, antibiotic‐resistant seedlings were transferred to soil and plants were grown until maturity. Leaves were collected for AsA measurements, and PCR‐based screens to confirm the presence of the transgene. This process was repeated until homozygous lines (plants with a 100% germination score in the presence of the selection marker) were found. A single‐insertion line was identified via Southern blot as described next.
2.3. Southern blot
Genomic DNA of the AtMIOX4 OE line was isolated using the CTAB method. Briefly, 4–5 g of fresh tissue was ground in a mortar and pestle‐cooled with liquid nitrogen. Then, 20 ml of CTAB buffer (1 M Tris‐HCl pH 8.0, 5 M NaCl, 0.5 M EDTA, 2% CTAB, and 1% (v/v) β‐mercaptoethanol) was added, vortexed, and incubated at 65°C for 20 min. After incubation, 20 ml of chloroform: isoamyl alcohol 24:1 (v/v) was added. The lysates were gently mixed and centrifuged at 4,000 g at 4°C for 20 min. The upper aqueous phase was transferred to new tube, and total DNA was extracted and precipitated with 3M NaAc pH 5.2. The precipitated DNA was dissolved in TE buffer. Five micrograms of extracted DNA was digested with SacI or KpnI. Digested DNA was separated on 0.8% (w/v) agarose 1× TAE gel and electroblotted onto a positively charged nylon membrane (Roche), using 1× TBE as transfer buffer. The UV cross‐linked membrane was then pre‐hybridized at 65°C for 15 min in Church buffer (sodium phosphate buffer containing 0.5 M Na2HPO4, pH 7.2, 20% (w/v) SDS, 1 mM EDTA, and 1% BSA) and hybridized overnight at 65°C with 50 ng of DNA probe each labeled with 50 μCi[32P]‐dCTP. The probes were prepared using Prime‐It® Rm T Random Primer Labeling kit (Stratagene). The probe for AtMIOX4:pBIB‐Kan was amplified with npt‐II forward (5′‐AGA GGC TAT TCG GCT ATG AC‐3′) and npt‐II reverse (5′‐AGC TCT TCA GCA ATA TCA CG‐3′) primers at 55°C annealing temperature. After hybridization, the blots were washed with phosphate buffer (0.5 M Na2HPO4, 20% (w/v) SDS, 1 mM EDTA, and 33 mM NaCl pH 7.2 at 65°C. Photographs were taken by exposing membranes to film for 2–3 days at −80°C (Biomax XAR, Kodak).
2.4. Measurement of plant growth
Growth of AtMIOX4 OE line and WT control was quantified using a Scanalyzer HTS phenotyping platform. Seeds were sterilized and germinated on MS media; 13 days after sowing robust seedlings were transferred to 85 × 73 mm Quick Pot 15 RW trays (HerkuPlast Kubern) containing Arabidopsis plant growing media (Lehle Seeds). Blue mesh was placed on top of the soil to reduce algal growth and improve segmentation during image analysis. Images of Arabidopsis plants were captured every 2 days, from seedling establishment to full vegetative growth (developmental stage 6.1 as defined by Boyes et al., 2001) using a LemnaTec Scanalyzer HTS system controlled using the LemnaControl software. Images were captured during a specific time window each day (3.5 hr after onset of daylight ±30 min). Visible (RGB) images were taken using a piA2400‐17gcCCD camera (Basler, Ahrensburg, Germany) with a resolution of 2,454 × 2056 pixels. Images of A. thaliana plants (128 images = 16 biological replicates × 8 time points) were analyzed using the LemnaGrid software. Analysis of the images acquired with the VIS camera was done as previously described (Acosta‐Gamboa et al., 2017). Once all the rosette leaves were identified, projected leaf surface area (cm2) was calculated for each plant. One‐way ANOVA was used for statistical analysis of growth rate throughout the vegetative growth at 0.05 significance level.
2.5. RNA extraction and sequencing
Three biological replicates of leaf samples were collected when plants completed the vegetative growth stage (stage 5.0 according to Boyes et al., 2001). Total RNA was purified using the RNeasy kit (Qiagen) following the manufacturer's instructions. RNA quality and quantity were assessed with an Experion system using the Experion™ RNA HighSens chips (Bio‐Rad). RNA samples were submitted to the Core Facility at Michigan State University for sequencing. Six libraries were prepared and sequenced using manufacturer recommendations with a Hi‐Seq Illumina 2500 system. Libraries were prepared using the Illumina TruSeq Stranded mRNA Library Preparation Kit. After QC and quantification, the libraries were combined into a single pool for multiplexed sequencing. The pool was loaded on two lanes of an Illumina HiSeq 2500 Rapid Run flow cell (v2). Sequencing was performed in a 1 × 50 bp single read format. Base calling was done by Illumina Real Time Analysis (RTA) v1.18.64, and output of RTA was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v1.8.4. Sequencing data used in this study (#SRA accession: PRJNA515221) will be available upon publication.
2.6. Bioinformatics analysis of raw RNA‐Seq data
Quality analysis of raw data was performed using FastQC v0.11.7 (Andrews, 2014). Trim‐Galore v0.4.3.1 was used for trimming the Illumina universal adapter sequences and removal of reads less than 40 bp in length (Krueger, 2015). TopHat v 2.1.1 was used for aligning the reads with the whole‐genome sequence of A. thaliana genome (TAIR10) (https://www.arabidopsis.org/) using maximum edit distance 2.0, library type unstranded, maximum intron length 10,000 bp, minimum intron length 50 bp and default parameters (Kim, Pertea, et al., 2013; Kim, Baek, et al., 2013). Cufflinks (v2.2.1) and Cuffmerge (v3.0) algorithms were used next for transcript compilation and gene identification using A. thaliana gene annotation TAIR10_gff3/TAIR10_GFF3_genes_transposons.gff (https://www.arabidopsis.org/). Differential expression analysis was performed using the CuffDiff (v7.0) algorithm. The CummeRbund algorithm R package (v2.22.0) was used for data visualization (Trapnell et al., 2012). Differentially expressed (DE) transcripts were selected based on an absolute value of log2 ratio ≥ 1.5 with FDR adjusted p‐value ≤ 0.05). False discovery rate correction (FDR) was performed using the Benjamini–Hochberg method. Blast2GO (Conesa & Gotz, 2008) and DAVID v6.8 (Huang, Sherman, & Lempicki, 2009) were used for gene ontology and functional enrichment analysis with a false discovery rate correction using the Benjamini–Hochberg method with FDR adjusted p‐value ≤ 0.05. MapMan 3.5.1R2 (Usadel et al., 2012) was employed for visualizing up‐ and down‐regulated genes affecting metabolism in plants mapping with Ath_AGI_LOCUS_TAIR10_Aug2012 (https://www.arabidopsis.org). The gene enrichment analysis was performed using PageMan module of MapMan 3.5.1R2 mapping with Ath_AGI_LOCUS_TAIR10_Aug2012 (https://www.arabidopsis.org/). Binwise Wilcoxon tests, no multiple corrections and over representation cutoff value of 3 were used for the analysis (Usadel et al., 2006).
2.7. RNA‐seq, assembly, and annotation
Foliar tissue from Arabidopsis AtMIOX4 L21 plants and wild‐type controls was collected at developmental stage 5.0. High‐quality RNA was used to make six cDNA libraries, three from each genotype. These libraries were sequenced using the Illumina HiSeq 2500 Rapid Run flow cell with 1x50 bp paired end format. A total of 200,736,389 raw reads were obtained. The percentage of adapters in samples was up to 3.1%. The Trim‐Galore script was used to remove the reads <40 bp in all samples. After removal of adapters and low‐quality reads, 185,235,265 bp of high‐quality reads were obtained. Reads that achieved a high Phred score (>38) indicating good quality sequences were used for the assembly. To assess the sequencing quality, the reads were mapped to the A. thaliana genome (TAIR10) using TopHat. The functional gene ontology annotation of transcripts was performed using the Blast algorithm. The quality and mapping of the RNA‐Seq data obtained from Illumina sequencing were good for transcriptome analysis of the AtMIOX4 OE line.
2.8. Real‐time quantitative PCR (RT‐qPCR) for gene expression analysis
Real‐time quantitative PCR (RT‐qPCR) was used for biological validation of the RNA‐seq data using reference genes previously published and following MIQE guideline (Bustin et al., 2009; Czechowski, Stitt, Altmann, Udvardi, & Scheible, 2005). Three biological replicates of leaf samples were collected when plants completed the vegetative growth stage. Total RNA was purified using the Purelink™ RNA mini kit (Ambion, Life Technologies) following the manufacturer's instructions. The RNA quality and quantity were assessed with an Experion system using the Experion™ RNA HighSens chips (Bio‐Rad). Residual DNA in the sample was removed using DNA‐free™ DNA removal kit (Invitrogen). Complimentary DNA (cDNA) were prepared by following manufacturer's instruction (iScript select cDNA synthesis kit, Bio‐Rad). Validated primers were obtained from AtRTPrimer database (Han & Kim, 2006). The primers for photosystem II (PSII) related center transcripts and glutathione reductase (GRX480) were validated using the guidelines by Bustin et al. (2009). RT‐qPCR was performed following manufacturer's instruction (SsoFastTM EvaGreen® supermixes, Bio‐Rad) in a CFX384TM real‐time system (Bio‐Rad). Normalization of transcripts counts was obtained using ACTIN2 and UBQ10 as internal controls. The relative expression of the genes was calculated using 2−ΔΔt method (Livak & Schmittgen, 2001). Three biological replicates and two technical replicates were used for RT‐qPCR. Two‐way Student's t test was performed at 0.05 significance level for normalized expression value.
2.9. Di‐nitro salicylic acid (DNS) assay for reducing sugars estimation
Six biological replicates of leaf samples were collected when plants completed the vegetative growth stage. Reducing sugars were estimated using di‐nitro salicylic assay (DNS). Plant samples were ground and extracted twice in 1 ml of 0.5 mM sodium acetate buffer (pH 5.0) using Precellys 24 lyzer (Bertin Technologies), and 100 μl of DNS reagent (Sigma) was added and incubated in boiling water bath (95°C) for 20 min. The reaction was stopped by adding 300 μl ddH2O. The 125 μl of reaction mixture was transferred into a 96‐well clear plastic reading plate with 2 technical replicates and absorbance was measured at 540 nm in a 96‐well plate reader (BioTek). Two‐way Student's t test was performed at a 0.05 significance level. The experiment was replicated twice with similar results.
2.10. Glucose oxidase–glucose peroxidase (GOD‐POD) assay for estimation of glucose
Five biological replicates of leaf samples were collected when plants completed the vegetative growth stage. Plant samples were ground and extracted twice in 1 ml of 0.5 mM sodium acetate buffer pH 4.5 using Precellys 24 lyzer (Bertin Technologies). Sixty microliters of sample, 54 μl of GOD‐POD enzyme mix (Sigma)) and 6 μl of O‐dianisidine was mixed and incubated at 37°C for 30 min. The reaction was stopped by adding 60 μl of 10 M HCl. Absorbance was recorded at 540 nm in a 96‐well clear plastic reading plate with two technical replicates in a 96‐well plate reader (BioTek). Two‐way Student's t test was performed at 0.05 significance level. The experiments were replicated twice with similar results.
2.11. Photosynthetic efficiency measurements
Plants were grown on MS media in 23°C, 65% humidity, 165–210 μmol m− 2 s− 1 light intensity, and 10:14‐hr photoperiod after stratification for 3 days. Twelve days after germination plants were transferred to Arabidopsis soil (Promix PGX; Premier Horticulture LTEE). MultispeQ v1.0 (Kuhlgert et al., 2016) was used to measure the photosystem II efficiency (ΦII) and proton motive force (vH+) 21, 23, 26, and 28 days after germination. Fifteen biological replicates for each genotype were measured. Experiment was replicated twice with similar results. Data shown correspond to one of the experiments. Two‐way Student's t test was performed at a 0.05 significance level for each day of measurement.
2.12. Hormone and metabolite quantification using LC‐MS/MS
Leaf samples from 10 biological replicates for each genotype were collected and lyophilized in liquid nitrogen when plants completed the vegetative growth stage. Hormones extraction and analysis were performed at the University of Nebraska, Lincoln, with modifications of Lan, Krosse, Achard, Dam, and Bede (2014) and Westfalla et al. (2016 ). Briefly, lyophilized samples were ground using a TissueLyser (Qiagen). Samples were spiked with deuterium‐labeled internal standards and extracted in ice‐cold methanol: acetonitrile (MeOH:CAN,1:1,v/v) for 2 min at 15 Hzs‐1 then centrifuged at 16,000 g for 5 min at 4⁰C. Supernatants were pooled and evaporated in a Labconco SpeedVac. The pellets after evaporation were redissolved in 200 μl of 30% MeOH. For LC separation, a ZORBAX Eclipse Plus C18 column (2.1 × 100 mm, Agilent) was used flowing at 0.45 ml/min. The gradient of the mobile phases A (0.1% acetic acid) and B (0.1% acetic acid/90% acetonitrile) was as follows: 5% B for 1 min, to 60% B in 4 min, to 100% B in 2 min, hold at 100% B for 3 min, to 5% B in 0.5 min. The LC system was interfaced with a Sciex QTRAP 6,500+ mass spectrometer equipped with a TurboIonSpray (TIS) electrospray ion source. Analyst software (version 1.6.3) was used to control sample acquisition and data analysis. The QTRAP 6,500+ mass spectrometer was tuned and calibrated according to the manufacturer's recommendations. All hormones were detected using MRM transitions that were previously optimized using standards. The instrument was set up to acquire in positive and negative ion switching. For quantification, an external standard curve was prepared using a series of standard samples containing different concentrations of unlabeled hormones and fixed concentrations of deuterium‐labeled standards (D5‐IAA, D5‐tZ, D5‐tZR, D2‐JA, D6‐ABA, D4‐SA). Two‐way Student's t test was performed at 0.05 significance level.
2.13. Semi‐quantification of in vivo auxin
Crosses between AtMIOX4 L21 (♂) and auxin sensor (R2D2) (♀) were used for visualization and in vivo semi‐quantitative analysis of auxin. Plants were grown on MS media in 23°C, 65% humidity, 165–210 μmol m− 2 s− 1 light intensity, and 10:14‐hr photoperiod after stratification for 3 days. Twelve days after germination, plants were transferred to Arabidopsis soil (Promix PGX; Premier Horticulture). AtMIOX4 L21 and R2D2 line were crossed and F1 generation were selected using kanamycin as a selectable marker and PCR using primers for MIOX4 and kanamycin resistance genes. The positive crosses were taken for the in vivo auxin quantification. The positive crosses were grown on MS media horizontally and vertically for in vivo auxin quantification of shoots and roots, respectively. The plants were grown at 23°C, 65% humidity, 165–210 μmol m− 2 s− 1 light intensity, and 10:14‐hr photoperiod for 7 days. The seedlings were mounted in propidium iodide (10 μg/ml) for root scanning and in PBS for shoots scanning. The image acquisition and analysis of roots and shots were performed in Cytation™ 5 cell imaging multimode reader and Gen3.03 software (BioTek). Venus was excited at 488 nm and detected at 498–530 nm, ntdTomato was excited at 561 nm and detected at 571–630, propidium iodide was excited at 561 nm and detected at 571–700 nm (Liao et al., 2015).
2.14. Root length, hypocotyl length, and hypocotyl cell length measurement
Plants were grown on plates positioned vertically on MS media in 23°C, 65% humidity, 165–210 μmol m− 2 s− 1 light intensity, and 10:14‐hr photoperiod after stratification for 3 days. For primary root length measurement, plants were grown on 150–210 μmol m− 2 s− 1 light intensity, whereas for hypocotyl measurement plants were grown on dark condition in same growth chamber. Nine days after germination the primary root length and the hypocotyl length were measured with a caliper. Fifteen biological replicates for each genotype were used in this experiment and a two‐way Student's t test was performed at 0.05 significance level.
For cell length measurements, hypocotyl of plants grown vertically on MS media on dark conditions was mounted in 1× PBS. The image acquisition and analysis were performed using Cytation™ 5 cell imaging multimode reader (BioTek) and cell length was measured using the Gen3.03 software (BioTek). Hypocotyl epidermal cell length was the average of 6 biological replicates for each genotype. The length of five cells was measured from each sample. Two‐way Student's t test was performed at a 0.05 significance level (Das, St‐Onge, Voesenek, Pierik, & Sasidharan, 2016).
2.15. Ascorbate measurements
Foliar tissue samples (40–60 mg) were collected at developmental stage 5.0 (Boyes et al., 2001) and immediately frozen in liquid nitrogen. The samples were ground in a cold mortar and pestle using liquid nitrogen and adding 0.75 ml of fresh meta‐phosphoric acid (6%) to the sample. The plant extracts were immediately transferred to a 1.5‐ml amber tube and centrifuged at 14,000 g for 15 min at 4°C. For the measurement of reduced ascorbate, the supernatants were transferred to 1.5‐ml amber tubes and kept on ice. There 0.95 ml of the meta‐phosphoric acid buffer was added to a quartz cuvette, and 50 μl of supernatant was added and mixed by inverting 2–3 times. The absorbance was recorded at 265 nm. Twenty microliters of ascorbate oxidase was added and mixed, and absorbance was recorded at 265 nm. For the measurement of oxidized ascorbate, 0.95 ml of the meta‐phosphoric buffer was added into a quartz cuvette and 50 μl of supernatant was added and mixed, and absorbance was recorded at 265 nm. The mix was recovered in a 1.5‐ml amber tube and 1 μl DTT (200 mM) was added mixed and incubated at dark for 20 min. The final absorbance was recorded at 260 nm. The reduced and oxidized ascorbate content was calculated using the extinction coefficient as [(Total change in absorbance)/14.3]*20*0.75(1/sample weights in grams). The total ascorbate content was calculated as the sum of reduced and oxidized ascorbate pools.
2.16. Quantification of H2O2 accumulation
Foliar tissue samples (40–60 mg) were collected at developmental stage 5.0 and immediately frozen in liquid nitrogen. H2O2 was quantified using Amplex Red (Molecular Probes, Invitrogen) as previously described (Katano, Kataoka, Fujii, & Suzuki, 2017; Suzuki et al., 2015). Briefly, the plant samples were ground using liquid nitrogen. The Amplex red reagent (50 mM sodium phosphate buffer pH 7.4, 50 μM Amplex red and 0.05 U/ml horseradish peroxidase) was added to ground tissues and centrifuged at 13,000 g for 12 min at 4°C. After centrifugation, 450 μl of supernatant was transferred to a 1.5‐ml amber tube and incubated for 30 min in the dark at room temperature. The absorbance was recorded at 560 nm. The concentration of H2O2 was calculated using a standard curve obtained from 0, 0.5, 1, 3, 6, and 9 μM H2O2.
2.17. Methionine‐derived aliphatic glucosinolate HPLC analysis
When plants completed the vegetative growth stage, leaf samples (~100 mg of DW) from ten biological replicates per genotype were collected and frozen in liquid nitrogen. Glucosinolates extraction was carried out with 80% MeOH and aryl sulfatase (Sigma). Recovered desulfoglucosinolates were lyophilized in SpeedVac and redissolved in 500 μl of ultrapure water. Samples were analyzed in a reverse phase HPLC system (Dionex; Ultimate 3,000) equipped with an AcclaimTM 300 C18 column (4.6 × 150 mm, 3 μm particle size, 300A°). The mobile phase was composed of acetonitrile (A) and 0.1% TFA in water (B). The gradient program was: 0–1 min, 1.5% B; 1–6 min, 1.5%–5% B; 6–8 min, 5%–7% B; 8–18 min, 7%–21% B; 18–23 min, 21%–29% B; 23–23.1 min, 29%–100% B; 23.1–24 min 100% B, and 24.1–28 min 1.5% B; flow 1.0 ml/min. Glucoerucin (Nacalai Tesque), 4‐methylthiobutil glucosinolate (4MTB), a methionine‐derived aliphatic glucosinolate, was analyzed. Detection was performed at 229 nm using a photodiode array. Glucoerucin UV spectra acquisition and analysis were carried following methods described by Grosser and Dam (2017) and Madsen, Kunert, Reichelt, Gershenzon, and Halkier (2015 ).
2.18. Pathogen infection assay
High ascorbate (AtMIOX4 L21), low ascorbate (vtc2‐1), and WT (CS‐60000) A. thaliana plants were challenged with the virulent pathogen Pst DC3000 (Pseudomonas syringae pv tomato DC3000). Pst DC3000 was grown on King's B medium for 2 to 3 days and resuspended on MgCl2 (10 mM) and Silwet/L77 (0.02%) in 105 colony‐forming unit (cfu)/ml. Two‐week‐old seedlings grown on MS media were dip‐inoculated with bacteria and covered for 1 hr and uncovered slowly. The amount of bacteria present in plants was analyzed 1 hr after dipping (Day 0) and 3 d after dipping (Day 3). Three biological replicates were pooled and measured for bacterial growth as cfu for each genotype (Zou, Yang, Shi, Dong, & Hua, 2014). The experiment was replicated three times with similar results.
3. RESULTS
After years of work, we have detected gene silencing in the AtMIOX4 lines we initially published (Lorence et al., 2004). To continue studying the possible roles of constitutive expression of AtMIOX4 in plants, we decided to develop new transgenic lines. The line used in this study was one of the new ones we developed (Figure 2a). AtMIOX4 line 21 was chosen for this study because it is homozygous, harbors a single copy of the MIOX4 ORF (Figure 2c), and recapitulates the phenotype of the lines previously reported (Lorence et al., 2004) that includes elevated levels of AsA (Figure 2b), enhanced biomass (Figure 2d) and tolerance to multiple abiotic stresses (Acosta‐Gamboa, 2019) relative to the wild‐type control (Lisko et al., 2014; Yactayo‐Chang, 2011; Yactayo‐Chang et al., 2018).
Figure 2.
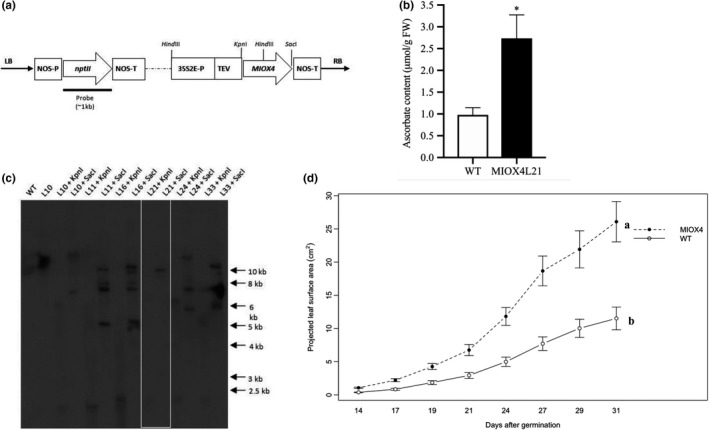
AtMIOX4 OE line accumulates high ascorbate and enhanced growth. (a) The AtMIOX4:pBIB‐Kan construct containing the myo‐inositol oxygenase (AtMIOX4) and the neomycin phosphotransferase II (nptII) ORFs. 35S2E‐P: cauliflower mosaic virus double‐enhanced promoter, TEV, tobacco etch virus translational enhancer; NOS‐P, promoter, NOS‐T terminator. (b) Total foliar AsA content of A. thaliana MIOX4 plants. Values are means ± SEM (n = 6), *p < .05 at 0.05 significance level. (c) Southern blot, illustrating the number of copies of the transgene in multiple AtMIOX4 lines. DNA was digested with either KpnI or SacI; WT: wild type (negative control); arrows indicate molecular weight marker. (d) The projected leaf surface area of AtMIOX4 L21 line is significantly higher compared to WT control. Data are means ± SEM (n = 8), p < .001 at 0.05 significance level
3.1. MIOX4 is highly expressed in the AtMIOX4 line
The MIOX4 (At4g26260) gene was significantly over‐expressed in the AtMIOX4 transcriptome compared to the WT control, as shown by RT‐qPCR (Figure 3b).
Figure 3.
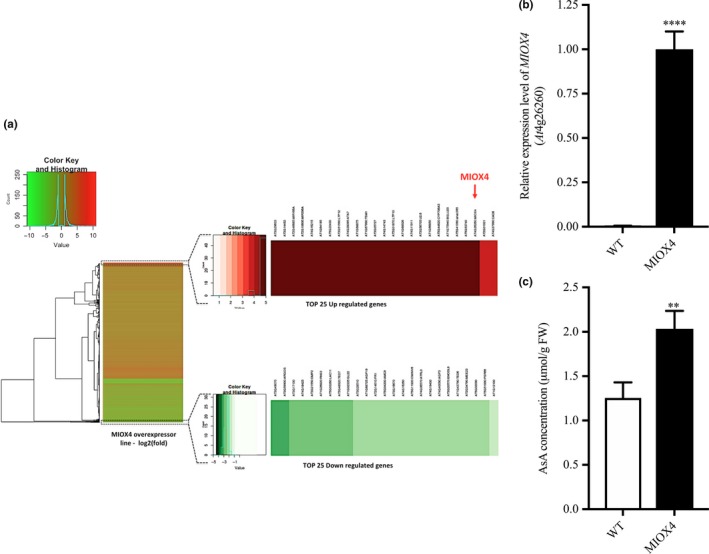
Global effects of the over‐expression of AtMIOX4 at the transcriptomic level. (a) MIOX4 was highly up‐regulated among differentially expressed transcripts shown in the heat map. (b) Validation of MIOX4 expression via RT‐qPCR. Data are means ± SEM (n = 3). (c) L‐Ascorbate concentration was significantly higher in MIOX4 OE line compared to WT control. Data are means ± SEM (n = 15). **Indicates p < .01, ****indicates p‐value < .0001 at 0.05 significance level
Differentially expressed transcripts (DETs) in the AtMIOX4 OE line were identified by comparison with the transcriptome of WT control. Genes with an adjusted p‐value FDR < 0.05 and a log2 fold change value of 2 were assigned as differentially expressed. As a result, 947 up‐regulated transcripts and 357 down‐regulated transcripts were differentially expressed in the AtMIOX4 OE line compared to the control (Figure 3a). Gene enrichment analysis has shown that photosynthesis, lipid metabolism, specialized metabolism, and hormone metabolism‐related genes are up‐regulated, whereas transcripts enriched in cell wall metabolism were down‐regulated in AtMIOX4 OE line compared to control (Figure S6). According to the functional annotation of DETs, most of the differential transcripts have been proven to be correlated to different biological processes related to developmental regulation and response to stresses in plants. The majority of up‐regulated transcripts fall under the response to stimulus biological process. These include response to external stimulus, response to organic substance, response to endogenous stimulus, response to auxin stimulus, response to abscisic acid stimulus, response to jasmonic acid stimulus, response to stress, response to water deprivation, response to cold stress, and response to salt stress (Table S4). On the other hand, the majority of down‐regulated transcripts were grouped under biological processes as response to chemical stimulus, response to stimulus and response to stresses (Table S5). We identified transcripts belonging to the precursors of three microRNAs (At3g63375, At2g46685, and At2g10606), a transcript related with TSM1 (At1g67990), TRNG.1 (AtCg00100), and a snoRNA (At1g32385, At1g20015) involved in cell regulation that were exclusively expressed in the AtMIOX4 OE line. Similarly, transcripts related to stress responses such as lipid transfer proteins related to pathogenesis (At4g22513, At3g51590, At4g28395, At1g66850, At5g01870), an extensin family protein (At5g22430), a protease inhibitor (At4g22513), a guard cell‐specific transmembrane protein (At3g14452), a defensing‐like family member protein (At1g64195, At3g05727), a stay‐green like protein (At4g11911), a cytochrome P450 family protein (At5g44620), a β‐glucosidase 20 (At1g75940), a NAC domain containing protein (At5g41090), and hypothetical proteins (At3g29633, At5g53740, At1g68526, At1g68875, At4g16240, At4g16215, At3g15534) were among the top 25 up‐regulated genes in the AtMIOX OE line (Table 1). On the other hand, development‐related genes such as a plant self‐compatibility protein S1 family (At3g24065), an aspartyl protease family protein (At5g48430), a putative non‐LTR retroelement reverse transcriptase (At2g07767; At3g49330), a plant invertase/pectin methylesterase inhibitor family protein (At5g12880; At3g17130), transmembrane proteins (At3g49070; At4g18425; At3g21550), GASA10 (At5g59845), ARGOs (At3g59900), IDL3 (At5g09805), RNS3 (At1g26820), LAC11 (At5g03260), AGP19 (At1g68725) and stress response genes such as PR1 (At2g14610), GATA19 (At4g36620), and MRD1 (At1g53480) were down‐regulated in the AtMIOX4 OE line compared to the control (Table 2). Additionally, amino acid metabolism genes lysine‐ketoglutarate reductase/saccharopine dehydrogenase functional protein (At4g33150), involved in lysine degradation and induced by jasmonate (JA), and arginase (At4g08870), involved in defense response and induced by methyl jasmonate (MeJA), were abundant in AtMIOX4 OE line compared to WT control (Tables S1–S3; Figure 3a).
Table 1.
List of top 25 down‐regulated genes in AtMIOX4 OE line compared to WT control
| Gene id | Gene | Log2 (fold_change) | TAIR information |
|---|---|---|---|
| At3g24065 | – | −5.13702 | Plant self‐incompatibility protein S1 family |
| At5g48430 | – | −4.01838 | Eukaryotic aspartyl protease family protein |
| At1g53480 | MRD1 | −3.79043 | Arabidopsis mto 1 responding down 1 |
| At2g07767 | – | −3.61645 | Putative non‐LTR retroelement reverse transcriptase |
| At3g49330 | – | −3.3819 | Putative non‐LTR retroelement reverse transcriptase |
| At5g12880 | – | −3.36218 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| At3g49070 | – | −3.27256 | Transmembrane protein |
| At5g59845 | GASA10 | −3.2326 | Gibberellic acid stimulated Arabidopsis 10 |
| At3g59900 | ARGOs | −3.22481 | Auxin‐regulated gene involved in organ size |
| At5g09805 | IDL3 | −2.89875 | Inflorescence deficient in abscission (IDA)‐like 3 |
| At3g17130 | – | −2.86408 | Plant invertase/pectin methylesterase inhibitor superfamily protein |
| At4g18425 | DMP4 | −2.83296 | DUF679 domain membrane protein 4 |
| At3g22070 | – | −2.82244 | Proline‐rich family protein |
| At3g21550 | DMP2 | −2.71775 | Arabidopsis thaliana DUF679 domain membrane protein 2 |
| At1g26820 | RNS3 | −2.64418 | Ribonuclease 3 |
| At5g03260 | LAC11 | −2.63596 | Laccase 11 |
| At5g48920 | TED7 | −2.61501 | Tracheary element differentiation‐related 7 |
| At2g31940 | – | −2.60708 | Oxidoreductase/transition metal ion‐binding protein |
| At1g02335 | GL22 | −2.59085 | Germin‐like protein subfamily 2 member 2 precursor |
| At3g28510 | ‐ | −2.49273 | P‐loop containing nucleoside triphosphate hydrolases superfamily protein |
| At1g68725 | AGP19 | −2.44671 | Arabinogalactan protein 19 |
| At2g14610 | PR1 | −2.40485 | Pathogenesis‐related gene 1 |
| At4g36620 | GATA19 | −2.39598 | GATA transcription factor 19 |
| At5g04200 | AMC9 | −2.3909 | Metacaspase 9 |
| At3g16670 | – | −2.38297 | Pollen O allergen extensin family protein |
Table 2.
Defense response genes up‐regulated in AtMIOX4 OE line relative to WT plants
| Gene Id | Gene | Log2 (fold change) | TAIR information |
|---|---|---|---|
| At1g67990 | TSM1 | Inf | Encodes tapetum specific O‐methyltransferase |
| At4g37990 | CAD‐B2 | 3.85 | Encodes an aromatic alcohol:NADP+ oxidoreductase |
| At1g56650 | MYB 75 | 1.65 | Encodes MYB domain containing transcription factor |
| At2g38240 | JAO4 | 2.26 | Encodes 2‐oxoglutarate/Fe(II)‐dependent oxygenases |
| At3g55970 | JAO3 | 3.04 | Encodes 2‐oxoglutarate/Fe(II)‐dependent oxygenases |
| At5g05600 | JAO2 | 2.38 | Encodes 2‐oxoglutarate/Fe(II)‐dependent oxygenases |
| At5g61168 | AACT1 | −1.75 | Encodes anthocyanin 5‐aromatic acyltransferase 1 |
| At1g49530 | GGPS6 | 1.57 | Encodes mitochondrial‐targeted geranylgeranyl pyrophosphate synthase 6 |
| At2g24210 | TPS10 | 3.39 | Encodes terpene synthase 10 |
| At3g45310 | LAS1 | 2.01 | Encodes cysteine protease superfamily protein |
| At4g16740 | ATPSO3 | 2.79 | Encodes an (E,E)_‐alpha‐farnese synthase |
| At2g06050 | OPR3 | 1.76 | Encodes 12‐oxophytodienoate reductase |
| At3g16470 | JR1 | 2.04 | Encodes jasmonate‐responsive 1 |
| At4g37150 | MES9 | 2.04 | Encodes methylesterase 9 |
| At1g05680 | UGT74E2 | 3.70 | Encodes uridine diphosphate glycosyltransferase 74E2 |
| At3g11480 | BSMT1 | 2.33 | Encodes SABATH methyltransferase |
| At4g11740 | VSP2 | 3.12 | Encodes vegetative storage protein 2 |
| At5g24780 | VSP1 | 3.15 | Encodes vegetative storage protein 1 |
| At1g19610 | PDF1.4 | 2.03 | Predicted to encode plant defensin family protein |
| At1g43160 | RAP2.6 | 1.77 | Encodes ethylene response factor subfamily B‐4 of ERF/AP2 transcription factor family |
| At5g13330 | RAP 2.6L | 2.28 | Encodes ethylene response factor subfamily B‐4 of ERF/AP2 transcription factor family |
| At4g36900 | RAP2.10 | 1.46 | Encodes member of the DREB subfamily A‐5 of ERF/AP2 transcription factor family |
We measured elevated foliar AsA content in the AtMIOX4 OE at the time leaf samples was taken for transcriptome analysis (1.63‐fold) compared to the control (Figure 3c), confirming our previous reports that over‐expression of AtMIOX4 leads to higher AsA content in Arabidopsis both in the absence and in response to abiotic stresses (Lisko et al., 2013; Lorence et al., 2004; Yactayo‐Chang, 2011). The transcripts only expressed in the AtMIOX4 OE line might have multiple roles in plant physiology, such as development, the oxidation–reduction process, carbohydrate and glucosinolate metabolism, and defense response against fungus, which could be vital for development and the stress tolerance/resistance phenotype observed in the AtMIOX4 OE line. A selected number of the DET from each group were validated through RT‐qPCR and those results are presented in the following sections. We have categorized the DETs in different physiological processes in plants. We have screened some of the hallmark genes functionally important to particular physiological process and validated their expression using RT‐qPCR.
3.2. Primary metabolism is up‐regulated in MIOX4 OE
The DETs were classified into different categories using the MapMan software (Usadel et al., 2009). Based on our results, the DETs were associated with primary metabolism, photosynthesis, biotic and abiotic stress, oxidative stress and secondary metabolism. Among the significantly over‐expressed genes in the AtMIOX4 OE line involved in primary metabolism, we identified transcripts related with sugar metabolism such as hexokinase like protein 1 (HKL1: At1g50460), α‐amylase‐like (AMY1: At4g25000), and cellulose synthase (At4g24000). In addition, transcripts related to sugar and starch metabolism were significantly up‐regulated in AtMIOX4 OE line compared to the control (Figure 4a). Interestingly, a starch degrading enzyme, α‐amylase (At4g25000), was significantly up‐regulated in the AtMIOX4 transcriptome which was validated using RT‐qPCR (Figure 4b). α‐Amylase catabolizes starch and releases free glucose and dextrin. Therefore, we proceeded to measure the intracellular pool of free glucose in the high AsA line using two different methods. As shown in Figure 4c, we found that the intracellular concentration of reducing sugars was significantly higher in the AtMIOX4 OE line (41.64 ± 7.61 μM/g FW) compared to controls (23.13 ± 2.02 μM/g FW). In support of this, the intracellular concentration of glucose in the AtMIOX4 OE line (31.92 ± 0.95 μM/g FW) was significantly higher compared to the one found in the control (19.61 ± 0.99 μM/g FW), as shown by a glucose‐specific enzymatic assay (Figure 4d). In summary, intracellular glucose concentration was 1.62‐fold higher in AtMIOX4 OE line compared to WT control. This indicates that the AtMIOX4 OE line has elevated expression of α‐amylase which leads to higher available intracellular glucose in these plants.
Figure 4.
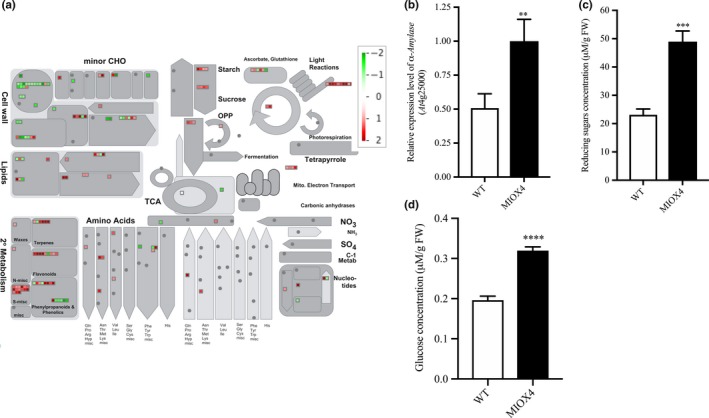
Carbohydrate metabolism is enhanced in the high ascorbate line. (a) The overall effect of MIOX4 OE in primary and secondary metabolisms. Figure was obtained using MapMan 3.5.1 R2. Up‐regulated transcripts are indicated by red, while down‐regulated transcripts are indicated in green. (b) α‐Amylase was significantly up‐regulated in MIOX4 OE compared to WT as shown by RT‐qPCR. Data are means ± SEM (n = 3). (c) Reducing sugars concentration was significantly higher in MIOX4 OE line compared to WT as shown by DNS assay. D) Glucose concentration was significantly higher in MIOX4 OE line compared to controls as shown by GOD‐POD assay. **Indicates p < .01, ***indicates p < .001 and ****indicates p < .0001 at 0.05 significance level. DNS = Di‐nitrosalicylic acid, GOD‐POD = glucose oxidase–glucose peroxidase. Data are means ± SEM of three replicates for RT‐qPCR and six replicates for DNS and GOD‐POD assays
3.3. MIOX4 over‐expression positively influences photosynthetic efficiency
Transcripts involved in light reactions of photosynthesis, as photosystem II‐related transcripts (PSBA‐ATCG00020, PSBC‐ATCG00280, PSBB‐ATCG00680) and photosystem I‐related transcripts (PSAJ‐ATCG00630, PSAC‐ATCG01060, NDHG‐ATCG01080) were significantly up‐regulated in the AtMIOX4 OE line (Figure 5a). In support of this, over‐expression of photosystem II center‐related transcript (PSBB‐ AtCg00680) was validated with RT‐qPCR (Figure 5b). To determine whether the increased gene expression we observed is physiologically significant, we measured photosynthetic efficiency using a handheld device (Kuhlgert et al., 2016). As illustrated in Figure 5c,d, photosystem II efficiency and proton motive force (vH+) were significantly higher in the AtMIOX4 OE line compared to WT control at 21, 23, 26, and 28 days after germination. Taking together these results show that the AtMIOX4 OE line is photosynthetically more efficient than its WT counterpart.
Figure 5.
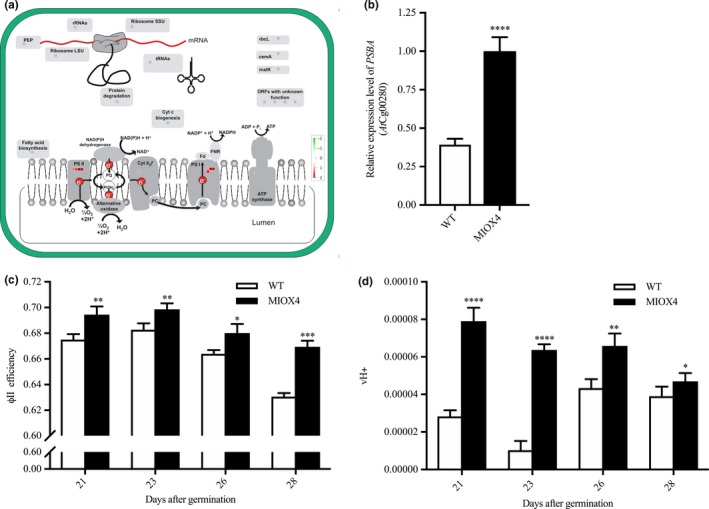
The high ascorbate MIOX4 line is photosynthetically more efficient. (a) Transcripts related to photosystem I and II (PSI, PSII) were up‐regulated (indicated by red squares) in MIOX4 OE line compared to WT. Figure was obtained using MapMan V3.5.1R2. (b) Over‐expression of transcripts related to PSII center was validated using RT‐qPCR. Data are means ± SEM (n = 3). (c,d) Photosystem II efficiency (ΦII efficiency) and proton motive force (vH+) were significantly higher in MIOX4 OE line compared to WT from 21 to 28 days after germination. Data are means ± SEM (n = 15). **indicates p < .01, ***indicates p < .001, and ****indicates p < .0001 at 0.05 significance level
3.4. MIOX4 over‐expression leads to increase auxin
Next, we investigated the role of phytohormones in the enhanced growth and abiotic stress tolerance phenotype recorded for the AtMIOX4 OE line. Analysis of the transcriptome data indicates that genes involved in auxin biosynthesis (YUCCA6: At5g25620, NIT2: At3g44300), auxin–amino acid conjugate hydrolase (ILL6: At1g44350), auxin transport (auxin efflux carrier family protein), and small auxin up RNAs (SAUR2: At4g34780, SAUR4: At4g34800, SAUR36: At2g45210) were up‐regulated in AtMIOX4 OE line compared to WT controls, whereas auxin response small RNA (SAUR53: At1g19840) and auxin‐regulated organ size transcript (ARGOs: At3g59900) were down‐regulated in AtMIOX4 OE line compared to WT (Figure 6a). The differential expression of auxin biosynthesis (YUCCA6, NIT2), auxin–amino acid conjugate hydrolase (ILL6), and transcripts involved in regulation of organ size (ARGOs) was confirmed via RT‐qPCR (Figure 6b).
Figure 6.
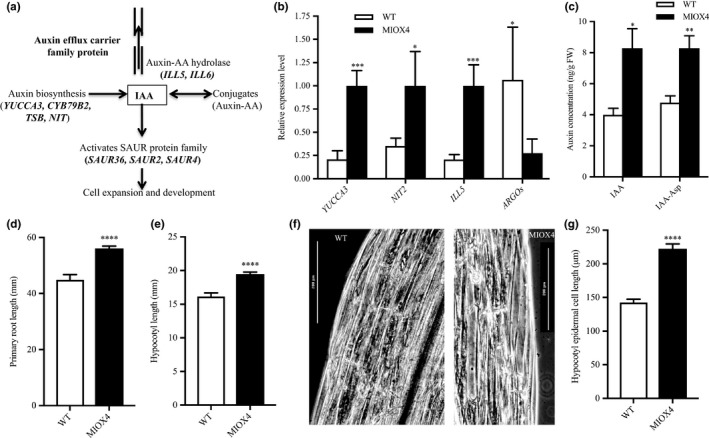
Increased auxin metabolism in high ascorbate line. (a) Transcripts related to auxin biosynthesis, auxin–amino acid conjugate hydrolases, auxin transport, and auxin response small RNA were up‐regulated in the MIOX4 transcriptome compared to WT. (b) Expression of transcripts related to auxin metabolism was validated using RT‐qPCR. Data are means ± SEM (n = 3). (c) Active auxin and inactive auxin (IAA‐Asp) were significantly higher in MIOX4 OE compared to WT as shown by LC‐MS/MS. Data are means ± SEM (n = 5). (d,e) Hypocotyl length of seedlings grown in dark conditions and primary root length of seedlings grown in light conditions were significantly longer in MIOX4 compared to WT 9 days after germination. Data are means ± SEM (n = 20). (f,g) Hypocotyl epidermal cell length was significantly higher in MIOX4 OE compared to WT. Data are means ± SEM (n = 30). **Indicates p < .01, ***indicates p < .001, and ****indicates p < .0001 at 0.05 significance level
Next, we determined the intracellular level of auxin and its conjugated form using LC‐MS/MS. The intracellular concentrations of the active auxin (IAA) and inactive auxin (IAA‐Ile) were significantly higher in AtMIOX4 OE line compared to WT (Figure 6c). To understand the effect of elevated auxin in AtMIOX4 OE line, next we measured the elongation of the primary root in light conditions and the hypocotyl length/hypocotyl epidermal cell length in dark conditions in the AtMIOX4 and controls. As shown in Figure d–e, the primary root of the high AsA line was longer than the control when plants grew under light conditions, and their hypocotyl was longer when plants grew in the dark. Next, we investigated whether the hypocotyl elongation in the AtMIOX4 OE line was due to increase in cell number or cell length using phase‐contrast microscopy. We found that hypocotyl epidermal cell length was significantly elongated in the AtMIOX4 OE line compared to WT controls (Figure 6f,g).
Subsequently, we took advantage of the auxin sensor line R2D2 developed by Liao et al. (2015) to monitor in vivo auxin levels in our high AsA line. For this, crosses between the R2D2 and the AtMIOX4 line (both lines present in the A. thaliana Col‐0 ecotype) were made. Positive crosses were selected using kanamycin as a selectable marker and PCR to confirm the presence of the transgenes of interest. Two positive crosses and controls were used for the in vivo auxin visualization and semi‐quantification. The R2D2 line expresses lower ntdTomato fluorescence (red) in the presence of normal auxin level. When there is a higher level of auxin, there is a higher level of ntdTomato fluorescence, compared to n3 X DII‐Venus (green) expression which was prominent in both shoots (Figure 7a,b) and roots (Figure 7d,e). As illustrated in Figure 7, the ratio of ntdTomato fluorescence to n3X DII‐Venus fluorescence was significantly higher in the crosses compared to R2D2 control in both shoots (Figure 7c) and roots (Figure 7f).
Figure 7.
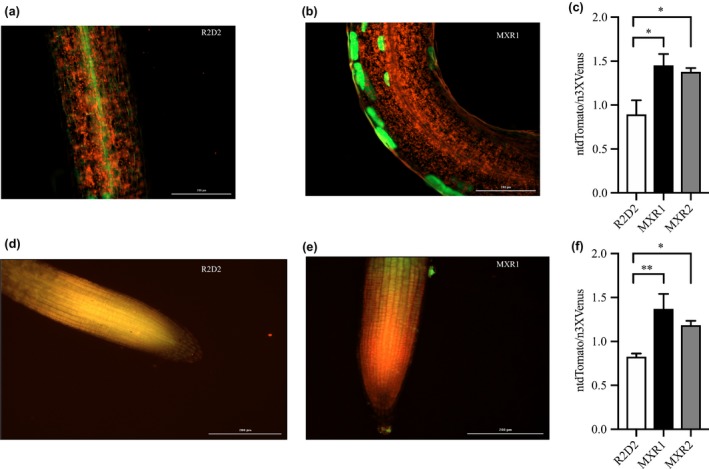
In vivo visualization and semi‐quantification of auxin response in AtMIOX4 × R2D2 crosses. (a–c) The ntdTomato expression was significantly higher in MIOX4 × R2D2 crosses compared to R2D2 control in shoots. (d–f) The ntdTomato expression was significantly higher in MIOX4 × R2D2 crosses compared to R2D2 control in roots. ntdTomato = red, DII‐venus = green. Data are means ± SEM (n = 4). Scale bars = 200 μm. Data were analyzed using one‐way ANOVA and multiple comparisons were performed comparing crosses with controls using Dunnett's multiple comparisons test at 0.05 significance level. *Indicates p < .05 and **indicates p < .01
We did not find evidence of differentially regulated transcripts involved in cytokinin metabolism in the AtMIOX4 OE line. The level of active cytokinin (cis‐zeatin), isomeric and relatively less active cytokinin (trans‐zeatin), and cytokinin conjugate (zeatin‐riboside) levels were similar in AtMIOX4 OE line compared to WT control as data from LC‐MS/MS show (Figure 8a–c). Taken all together, these results indicate that there are elevated levels of auxin due to the transcriptional up‐regulation of auxin metabolism in the AtMIOX4 OE line and that the elevated auxin levels observed in the AtMIOX4 OE line are functionally relevant. Our observations support that there is a positive association between high ascorbate level, elevated auxin level, and cell elongation in the AtMIOX4 OE line.
Figure 8.
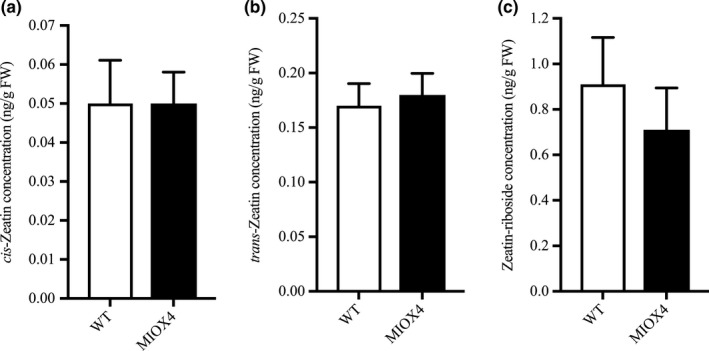
Cytokinin metabolism was unaffected by MIOX4 over‐expression. (a) Trans‐zeatin, (b) cis‐zeatin, and (c) zeatin‐riboside levels were similar in MIOX4 OE compared to WT as shown by LC‐MS/MS. Data are means ± SEM (n = 10) at 0.05 significance level
3.5. MIOX4 over‐expressers tolerate abiotic stresses independently of abscisic acid up‐regulation
We also examined some of the possible molecular mechanisms mediating the abiotic stress tolerance phenotype in AtMIOX4 OE line. We identified abiotic stress response‐related based on the expression profiles of the DETs transcripts using MapMan. Among the statistically significant transcripts, we found that heat stress response transcripts 17.8KDa class I heat‐shock protein (HSP17.8‐CI‐ At1g07400), 17.4 KDa class III heat‐shock protein (HSP17.4‐CIII‐ At1g54050), cold‐regulated protein 15a (COR15A‐At2g42540), and the drought‐induced protein (ATDI21‐At4g15910) were significantly up‐regulated in AtMIOX4 OE line compared to WT control (Figure 9a, Table S3) The expression of cold stress tolerance transcript (COR15A) and salt stress‐tolerance transcript (NAM) were validated using RT‐qPCR (Figure 9b).
Figure 9.
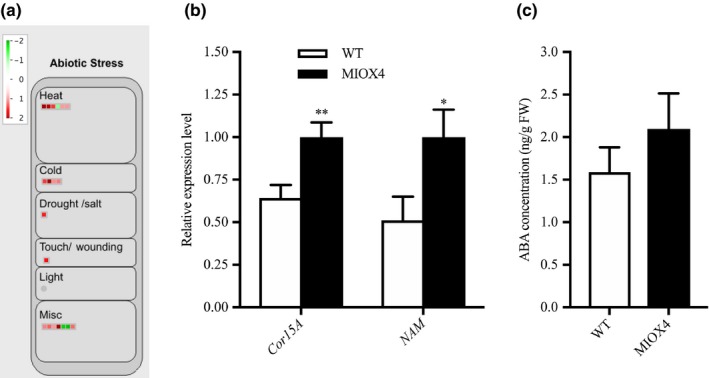
Abiotic stress response genes were differentially expressed in high ascorbate line. (a) Heat, cold, and drought stress response transcripts were up‐regulated in MIOX4 OE compared to WT. (b) Validation of over‐expression of cold‐regulated protein (CoR15a) and NAM transcription factor using RT‐qPCR. Data are means ± SEM (n = 3). (c) Abscisic acid levels remained similar in MIOX4 OE and WT as shown by LC‐MS/MS. Data are means ± SEM (n = 10); *indicates p < .05 at 0.05 significance level
We also looked at whether levels of abscisic acid were altered in the AtMIOX4 OE line compared to controls using LC‐MS/MS. Abscisic acid levels were similar in AtMIOX4 OE and the WT line (Figure 9c). We then used Genevestigator to find signature genes that were over‐expressed in publically available abiotic stresses experiments. Genevestigator analysis of the top 200 up‐regulated (log2 fold > 1.75) and down‐regulated transcripts (log2 fold < −1.25) was compared against signature expression transcripts from 50 similar perturbations. We have found that cinnamyl alcohol dehydrogenase (AtCAD8: At4g37990), N‐acetyl transferase activity 1 (At2g39030), lactoylglutathione lyase (GLYI7:At1g80160), bHLH family protein (At1g10585), and NAC domain containing protein 19 (NAC019:At1g52890) were up‐regulated in both, the set of abiotic stress perturbation experiments and in the AtMIOX4 OE line (Figure S4). Taken together, these results point out to an important role for a conserved module of stress response genes in the abiotic stress tolerance phenotype observed in AtMIOX4 OE line (Acosta‐Gamboa, 2019).
3.6. High AsA exerts a contrasting regulation on glutathione reductase and ascorbate peroxidase genes
Antioxidants maintain the homeostasis of ROS in plants. We looked for genes involved in the antioxidant cycle network in the high AsA line to identify possible crosstalk within the redox network. We found that superoxide dismutase expression was similar in AtMIOX4 OE line compared to WT control (Figure 10a). Glutathione reductase 480 (GRX480: At1g28480), which regulates the protein redox state, and dehydroascorbate reductase 1 (DHAR1: At1g19570), involved in recycling of reduced ascorbate, were up‐regulated while peroxisomal ascorbate peroxidase 5 (pAPX5: At4g35970), involved in scavenging ROS, was down‐regulated in the AtMIOX4 OE line compared to WT control. The expression of APX5 and GRX480 was validated using RT‐qPCR (Figure 10b). Interestingly, expression of genes involved in multiple pathways of L‐ascorbate biosynthesis was similar in the AtMIOX4 OE and the control, while expression of DHAR1, involved in recycling of reduced ascorbate, was up‐regulated (Figure S1). Down‐regulation of peroxisomal APX5 leads to significantly higher reactive oxygen species (H2O2) in AtMIOX4 OE compared to its WT counterpart (Figure 10c). Taking together, these results indicate that high AtMIOX4 OE leads to higher ROS concentration due to down‐regulation of peroxisomal APX gene. Additionally, high AsA leads to up‐regulation of glutathione peroxidase and DHAR1 genes which maintain the ROS homeostasis in AtMIOX4 OE line.
Figure 10.
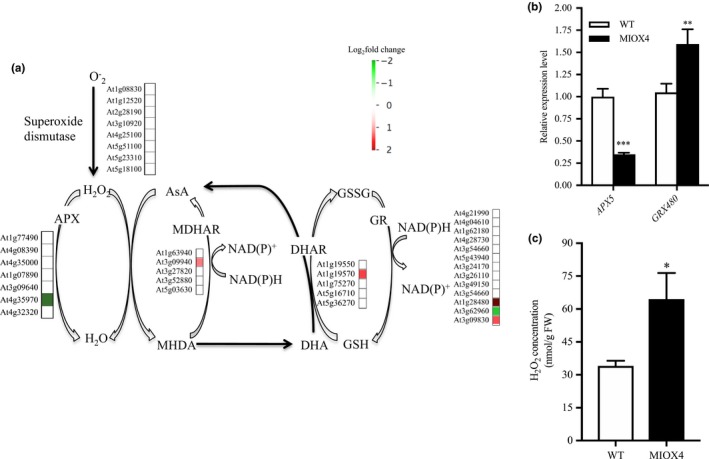
Antioxidant crosstalk in MIOX4 OE. (a) The ascorbate recycling enzyme DHAR1 and glutathione reductase (GRX480) were significantly up‐regulated, whereas ascorbate peroxidase 5 (APX5) was significantly down‐regulated in the MIOX4 OE transcriptome compared to WT. (b) RT‐qPCR validation of up‐regulated and down‐regulated transcripts involved in ascorbate–antioxidant crosstalk. Data are means ± SEM (n = 3). (c) H2O2 accumulation is higher in foliar tissue of MIOX4 OE line compared to WT. Data are means ± SEM (n = 4). *Indicates p < .05, **indicates p < .01, and ***indicates p < .001 at 0.05 significance level. APX, Ascorbate peroxidase; AsA, ascorbate; GR, glutathione reductase; GSH, reduced glutathione; GSSG, oxidized glutathione; H2O2, hydrogen peroxide
3.7. High ascorbate plants are transcriptionally primed for herbivore defense
Genes related to cell wall metabolism, specialized metabolism, and defense hormone biosynthesis are important for the biotic stress response of plants. We found that genes involved in arabinogalactan metabolism and xyloglucan metabolism were down‐regulated, whereas cellulose synthesis was up‐regulated in AtMIOX4 OE line compared to controls. Three members of the arabinogalactan gene family, AGP19: At1g68725, AGP3 (At4g40090), and AGP14 (At5g56540) were down‐regulated. Similarly, xyloglucan metabolism genes XTH17 (At1g65310), XTH25 (At5g57550), and XTH10 (At2g14620) were down‐regulated, whereas genes related to cellulose synthesis, CSLA 10 (At1g24070), CSLA 1 (At4g16590), and CSLG2 (At4g24000) were up‐regulated in AtMIOX4 OE line compared to WT control.
Specialized metabolites are also an important line of defense in plants to combat stresses. Genes related to phenolic biosynthesis, O‐methyl transferase family 2 protein (At1g76790), were up‐regulated, while laccase biosynthesis genes, LAC11 (At5g03260) and LAC17 (At5g60020), were down‐regulated in the AtMIOX4 OE line compared to WT. The phenylpropanoid biosynthesis gene, TSM1 (At1g67990), was only expressed in the AtMIOX4 OE line, while A. thaliana CAD‐B2 (At4g37990) was up‐regulated in AtMIOX4 OE line compared to controls. Flavonoid metabolism‐related transcription MYB 75 (At1g56650), which is involved in radical scavenging activity and anthocyanin metabolism, 2OG‐Fe(II) oxygenase family proteins (At2g38240, At3g55970, At5g05600), and AACT1 (At5g61168) were up‐regulated in AtMIOX4 OE line compared to controls. Similarly, terpene biosynthesis genes, GGPS6 (At1g49530), TPS10 (At2g24210), LAS1 (At3g45310), and ATTPS3 (At4g16740), were up‐regulated in AtMIOX4 OE line compared to WT.
Defense hormones, such as jasmonic acid (JA), salicylic acid (SA), and ethylene, are critical to induce the systemic acquired defense response in plants. Genes related to JA biosynthesis and metabolism, OPR3 (At2g06050) and JR1 (At3g16470), were up‐regulated in the AtMIOX4 OE line compared to WT control. Similarly, genes related to SA metabolism, MES9 (At4g37150), involved in conversion of methyl salicylate to active SA, UDP‐glucosyl transferase family protein (At1g05680), and BSMT1 (At3g11480) were also up‐regulated in the AtMIOX4 OE line. JA response transcripts, VSP2 (At4g11740), VSP1 (At5g24780), and PDF1.4 (At1g19610) were significantly up‐regulated in the AtMIOX4 OE line compared to WT. Similarly, we noted that ethylene response factors as RAP2.6 (At1g43160), RAP 2.6L (At5g1330), and RAP2.10 (At4g36900) were significantly up‐regulated in the AtMIOX4 OE line compared to WT (Figure 11a; Table 2). The expression of the signature genes that play a significant role in biotic stress response (MYB27: At3g53200, OPR3, VSP2: At4g11740 and RAP2.6: At1g43160) was validated using RT‐qPCR (Figure 11b). OPR3 regulates the biosynthesis of JA in plants. The over‐expression of the OPR3 leads to higher levels of JA and JA‐Ile, while the metabolite intermediate 12‐oxophytodienoate (OPDA) was similar in AtMIOX4 OE line compared to WT control (Figure 11c–e). The over‐expression of the genes related to specialized metabolites, such as phenylpropanoid, flavonoid, and terpene biosynthesis; up‐regulation of genes involved in JA and SA biosynthesis; elevated level of JA and JA‐Ile hormones; and expression of JA response genes, such as VSP1, VSP2, and PDF 1.4, suggests that AtMIOX4 OE plants are ready to defend themselves from herbivores attack.
Figure 11.
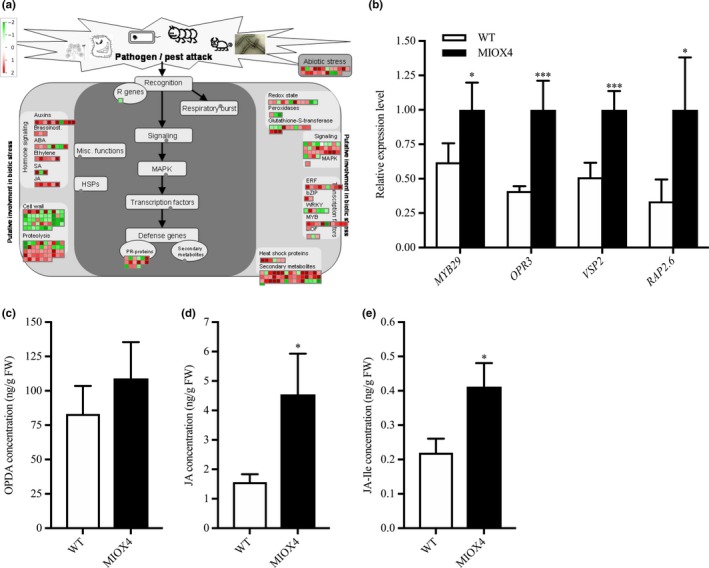
Biotic stress response transcripts are differentially expressed in high ascorbate line. (a) Transcripts related to defense hormone biosynthesis, biotic stress response transcription factors, and specialized metabolite biosynthesis were up‐regulated while some transcripts related to cell wall metabolism were down‐regulated in MIOX4 OE compared to WT. (b) The up‐regulation of transcripts involved in biotic stress response was validated using RT‐qPCR. Data are means ± SEM (n = 3). (c) Defense hormone jasmonates (JA) and (d) jasmonic acid‐Ile (JA‐Ile) were significantly higher in MIOX4 OE line compared to WT. Data are means ± SEM (n = 5), *indicates p < .05, **indicates p < .01, and ***indicates p < .001 at 0.05 significance level
3.8. Genes from the glucosinolate biosynthetic pathway are up‐regulated in MIOX4 over‐expressers
Glucosinolates are specialized metabolites present mostly in plants of the Brassicaceae family. It is well known that glucosinolates and their hydrolysis products act as defense compounds against herbivores and pathogens. Glucosinolates are derived from amino acids and are grouped into aliphatic, aromatic, and indolic glucosinolates based on their different side chain structures. We have found that genes involved in the glucosinolate ‐myrosinase system, flavin monooxygenase glucosinolate S‐oxygenase 2 (FMOGS‐OX2: At1g62540), alkenyl hydroxyalkyl producing 2 (AOP2: At4g03060), MYB domain protein 76 (MYB76: At5g07700), myrosinase binding protein 1 (MBP1: At1g52040), cytochrome P450 CYP83B1 (SUR2: At4g31500), and epithiospecifier protein (ESP: At1g54040) were elevated in the AtMIOX4 OE line compared to WT control (Figure 12a). The CYP83B1 gene is involved in the indol‐ and benzyl‐glucosinolates biosynthesis which has tryptophan and phenylalanine as precursors (Sønderby, Burow, Rowe, Kliebenstein, & Halkier, 2010). The MYB 76 is transcriptional activator of methionine‐derived aliphatic glucosinolates in A. thaliana (Col‐0) (Gigolashvili, Engqvist, Yatusevich, & Muller, 2008; Sønderby et al., 2007). We found that the level of glucoerucin, a methionine‐derived aliphatic glucosinolate, was elevated ~2.5‐fold in AtMIOX4 OE line compared to WT control (Figure 12b). Interestingly, we found that negative regulator of methionine biosynthesis Arabidopsis MTO 1 responding down 1 (At5g53480) was significantly down‐regulated in AtMIOX4 OE line (Table S2).
Figure 12.
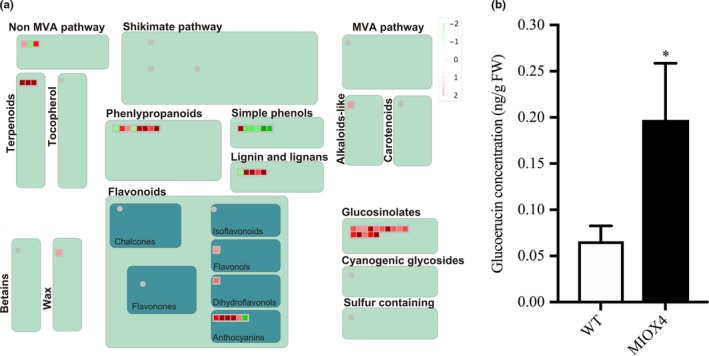
Glucosinolate metabolism was enhanced by higher ascorbate in MIOX4 plants. (a) Glucosinolate metabolism‐related transcripts were up‐regulated in MIOX4 OE compared to WT as indicated by red squares in the figure obtained from MapMan v3.5.1R2. (b) Glucoerucin concentration was higher in MIOX4 OE compared to WT as shown by HPLC. Data are means ± SEM (n = 6); *indicates p < .05 at 0.05 significance level
3.9. Severity of down‐regulation of pathogen‐resistant genes is compensated by increased SA level
Surprisingly, we found that genes related to the pathogen response as pathogenesis‐related proteins (At2g14610), glycosyl hydrolase superfamily protein (At4g16260), peroxidase family proteins (At5g51890), germin (GLP3: At5g51890), leucine‐rich repeat lipid family protein (LRR: At5g23400), and disease resistance responsive family protein (At1g6580) were down‐regulated in the AtMIOX4 OE line compared to WT controls (Figure 13a). To test whether down‐regulation of these genes affected the pathogen defense, we challenged AtMIOX4 OE (high AsA), vtc 1‐1 (low AsA), and WT line (moderate AsA) with Pseudomonas syringae. One‐way ANOVA was performed, and Tukey's post hoc test was used to find out mean difference of bacterial count between different genotypes at 0.05 significant level. Initially, the bacterial count did not differ significantly in all genotypes. Later, 3 days after infection, the vtc1‐1 (low AsA) lines significantly decrease the number of bacterial count compared to other lines and resistant to P. syringae attack, whereas the bacterial count in AtMIOX4 OE line does not differ significantly compared in WT lines after 3 days of infection, indicating AtMIOX4 does not seem to compromised to defend against P. syringae attack (Figure 13b). We evaluated the level of SA in AtMIOX4 because SA is responsible for protecting the plants against pathogens by provoking the systemic defense response. The foliar level of SA was higher in AtMIOX4 OE line compared to controls, as shown by LC‐MS/MS, without the challenge of biotic stress (Figure 13c). Despite the down‐regulated pathogen defense response transcripts, the AtMIOX4 line was not compromised by the pathogen challenge. This can be due to the presence of high level of SA, which could compensate for the down‐regulation of pathogen response genes by expressing lipid transfer proteins related to pathogenesis (At3g51590, At4g28395, At1g66850, At5g01870) and PROTEASE INHIBITOR (At4g22513) genes only in the AtMIOX4 OE line.
Figure 13.
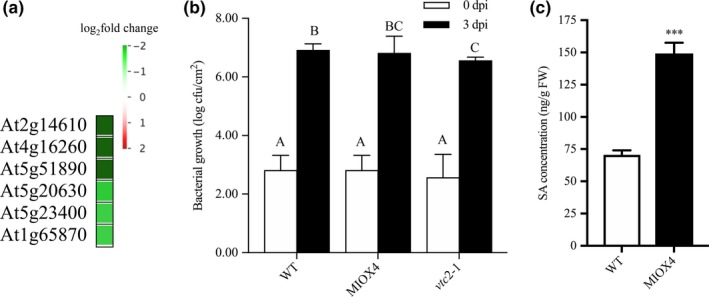
Differentially expressed genes involved in pathogen defense response in high ascorbate line. (a) List of genes that were down‐regulated and important for pathogen defense response in MIOX4 OE compared to WT. (b) Pseudomonas syringae infection did not alter the susceptibility of lines with high ascorbate level. MIOX4 OE response was similar to WT when challenged with P. syringae, whereas low ascorbate line vtc2‐1 was resistant to P. syringae infection. Data are means ± SD (n = 3). (c) Defense hormone salicylic acid (SA) was higher in MIOX4 OE line compared to WT l. Data are means ± SEM (n = 5), *indicates p < .05, and ****indicates p < .0001 at 0.05 significance level
4. DISCUSSION
Ascorbate is fundamental for proper development and response to biotic and abiotic stress in plants (Conklin et al., 1999; DeBolt et al., 2006; Dowdle et al., 2007; Gallie, 2013; Gest et al., 2013; Kerchev et al., 2011; Ortiz‐Espín et al., 2017; Pastori et al., 2003). The D‐mannose/L‐galactose pathway (Wheeler et al., 1998) is well characterized compared to other ascorbate routes. Other pathways also called “alternative routes” are also functional in different plants species, tissues and developmental stage‐specific manner (Lorence et al., 2004; Yactayo‐Chang, 2011; Yactayo‐Chang et al., 2018). The MIOX4 transcript is expressed during reproductive stage in leaf stipules and was found predominantly in cytoplasm and in nucleus (Alford, Rangarajan, Williams, & Gillaspy, 2012; Endres & Tenhaken, 2011; Kanter et al., 2005). Over‐expression of enzymes involved in the myo‐inositol pathway leads to increased AsA content in Arabidopsis (Lorence et al., 2004; Radzio et al., 2003; Yactayo‐Chang, 2011; Yactayo‐Chang et al., 2018). This has also been confirmed by other laboratories (Tóth et al., 2011). Additionally, a study in a T‐DNA insertion mutant of the vtc4 enzyme showed that this proteins is involved in myo‐inositol synthesis resulting in decreased AsA in kiwifruit (Actinidia deliciosa) in mutants compared to WT plants (Torabinejad, Donahue, Gunesekera, Allen‐Daniels, & Gillaspy, 2009). A recent study reports no conversion of myo‐inositol into AsA in the AtMIOX4 lines in which we have detected gene silencing (the ones reported in the Lorence et al., 2004 paper) (Kavkova, Blochl, & Tenhaken, 2018). Because of the gene silencing issue, we developed new transgenic lines. The AtMIOX4 line 21 used in this transcriptomic paper is homozygous, contains a single copy of the transgene and consistently has shown elevated AsA in more than 10 generations we have grown these plants.
The rosettes of the low ascorbate A. thaliana vtc2 mutant are smaller compared to WT due to intracellular structural deformity similar to programmed cell death (Olmos, Kiddle, Pellny, Kumar, & Foyer, 2006; Pavet et al., 2005), whereas plants over‐expressing Arabidopsis myo‐inositol oxygenase (MIOX4) (Lorence et al., 2004) have significantly higher (1.5 to 2‐fold) ascorbate. Higher AsA results in enhanced biomass, and increased tolerance to abiotic stresses including cold, heat, salt, and pyrene, an environmental pollutant (Lisko et al., 2013). In this study, we report the physiological and molecular mechanisms likely underlying the enhanced biomass and abiotic stresses tolerance phenotype in AtMIOX4 OE using a comprehensive approach combining transcriptomic analysis, with LC‐MS/MS, cell biology, and physiological measurements.
We found that an α‐amylase‐like (At4g25000) transcript was over‐expressed and resulted in higher intracellular concentration of reducing sugars and glucose in AtMIOX4 OE line compared to WT using RNA‐Seq, RT‐qPCR, biochemical assay, and enzymatic assay (Figure 4a–d). Glucose acts as a primary carbon and energy source in plants and plays a crucial role in plant growth, development, and physiology by coordinating with phytohormone signals. Enhanced production of glucose leads to manipulation of root architecture for maximum development through interactions with auxin metabolism genes (Li et al., 2007; Moore et al., 2003; Peng et al., 2018). Furthermore, increased concentration of UDP‐glucose leads to accumulation of higher biomass in plants and helps in vegetative phase change by repressing the miR156A/miR156C (Christopher, Rensburg, & Ende, 2017; Yang, Xu, Koo, He, & Poethig, 2013). Additionally, glucose induces expression of MYB family transcription factors and biosynthetic genes in the aliphatic glucosinolate pathway and in the presence of JA acts synergistically regulating glucosinolates accumulation (Guo et al., 2013; Miao et al., 2016). Enhanced intracellular glucose accumulation indicates the higher pool of primary carbon and energy source, and signaling molecule for stresses tolerance, which might contribute to higher biomass and abiotic stresses tolerance phenotype in AtMIOX4 OE line compared to WT.
We have shown that photosynthesis light reaction‐associated transcripts were up‐regulated and that photosynthetic efficiency and proton motive force were higher in AtMIOX4 OE line compared to WT (Figure 5a–d). The AtMIOX4 OE line has higher non‐photochemical quenching and energy‐dependent quenching due to the presence of elevated ascorbate in the lumen. Additionally, in heat‐stressed leaves of high AsA plants, ascorbate acts as PSII donor by retarding photo‐inactivation (Tóth et al., 2011). The electrons are passed down the photosynthetic electron transfer chain and carbon be ultimately used to generate reducing power that can be used to fix carbon in the dark reactions (Johnson, 2016). Higher rate of photosynthesis is associated with higher biomass production and greater yield in plants (Evans, 2013; Taylaran, Adachi, Ookawa, Usuda, & Hirasawa, 2011). Hence, increase in photosynthesis efficiency is observed in AtMIOX4 OE plant.
The growth and development of plants are regulated by the interaction between growth and defense hormones. We have shown that expression of auxin biosynthesis, auxin transport, auxin–amino acid conjugate hydrolase, and auxin response signaling factors was up‐regulated in AtMIOX4 OE line compared to the control (Figure 6a,b). The concentration of intracellular active auxin (IAA) and conjugated auxin (IAA‐Asp) was higher in AtMIOX4 OE line compared to WT (Figure 6c). The functional response of elevated auxin in these plants is evidenced by increased primary root length when grown in light conditions, elongated hypocotyl and hypocotyl epidermal cell when grown in dark conditions and both visualization and semi‐quantification of auxin levels using confocal microscopy. Auxin concentration was higher in both roots and shoots of AtMIOX4 OE line harboring the auxin sensor (R2D2) relative to the R2D2 control (Figure 7a‐f). Auxin tightly regulates growth and development of plants. Tryptophan‐dependent auxin biosynthesis transcripts were over‐expressed in AtMIOX4 OE line as nitrilase family gene converts indole‐3‐acetonitrile to IAA, whereas YUCCA family proteins convert indole‐3‐pyruvic acid to IAA which is the rate limiting step for tryptophan‐dependent auxin biosynthesis. Over‐expression of both branches of auxin biosynthesis pathway leads to high IAA and IAA‐Asp level (Korasick, Enders, & Strader, 2013; Zhao et al., 2001). YUCCA6 over‐expression results in high auxin phenotype such as delayed senescence, narrow and downward curled rosette leaves and tolerance to drought stress (Kim et al., 2011; Kim, Pertea, et al., 2013; Kim, Baek, et al., 2013). The over‐expression of YUCCA6, YUCCA1, and external application of auxin derivative delayed senescence and drought stress tolerance in plants due to elevated auxin (Kim et al., 2011; Lee et al., 2011; Mueller‐Roeber & Balazadeh, 2013). We have found YUCCA3 over‐expression, delayed senescence, and abiotic stress tolerance phenotype in the AtMIOX4 OE line compared to WT control. Additionally, auxin promotes plant organ elongation (hypocotyl, roots, and coleoptiles) by enhancing the proton extrusion through the membrane H+ ATPase. Auxin lowers apoplastic pH through activation of membrane H+ ATPase, which induces cell wall loosening proteins facilitating inward movement of K+ and water (Claussen, Luthe, Blatt, & Bottger, 1997; Philippar et al., 2006; Takahashi, Hayashi, & Kinoshita, 2012; Tyburski, Dunajska‐Ordak, Skorupa, & Tretyn, 2012), whereas cytokinins are involved in cell division and morphogenesis regulation, and abiotic/biotic stresses tolerance (Zalabak et al., 2013). However, constitutive expression of AtMIOX4 OE did not seem to lead to changes in cytokinin (cis‐zeatin, trans‐zeatin, and zeatin‐riboside) levels in A. thaliana foliar tissue (Figure 8a–c), which can be observed in both RNA‐Seq and LC‐MS/MS data.
Analysis of our transcriptomics results and published evidence indicates that abundance of intracellular primary carbon source, increased photosynthetic efficiency, and elevated auxin contribute to higher biomass phenotype in AtMIOX4 OE line compared to WT control. Additionally, delayed senescence in plants was indicated by the presence of high free auxin which is consistent with our results.
4.1. Cell wall metabolism is differentially regulated in AtMIOX4
The cell wall is important to maintain the structural integrity and stresses tolerance of plants. We found that cell wall metabolism genes were differentially modulated in AtMIOX4 OE line compared to WT control (Figure 11a; Table S3). Lignin maintains the structure of the secondary cell wall. We found that laccase biosynthesis genes (LAC11 and LAC17) were down‐regulated in AtMIOX4 OE line compared to WT control. Knockout of laccase biosynthesis (LAC1 and LAC17) genes in A. thaliana results in reduced plant growth and vascular development, decreased root diameter due to reduced lignification and increased saccharification (Berthet et al., 2011; Zhao et al., 2013) which contrasts with our results. Auxin decreases the expression of AGP3 whereas AGP19 is involved in plant growth, cell division and expansion, leaf development and reproduction (Goda et al., 2004; Yang, Sardar, McGovern, Zhang, & Showalter, 2007). Interestingly, A. thaliana arabinogalactan protein (AtAGP17) over‐expression results in resistance to infection by Agrobacterium (Gaspar et al., 2004). The expression of the gene longofolia is involved in polar cell expansion by turgor‐driven approach by controlling XTH17 and XTH24 expression (Lee et al., 2018). In contrast, CSLA/CSLA‐like gene family members that are involved in mannan and xyloglucan backbones biosynthesis (Liepman & Cavalier, 2012) were up‐regulated in AtMIOX4 OE line compared to the WT control.
4.2. Abiotic stress tolerance of high ascorbate line was independent of increased abscisic acid level
Prior studies have shown that low ascorbate A. thaliana lines were sensitive to abiotic stresses such as heat, cold, light, and oxidative stress (Conklin et al., 2013; Pavet et al., 2005). On the other hand, plants with elevated ascorbate were tolerant to abiotic stresses as heat, cold, drought, salt, and oxidative stress (Eltayeb et al., 2006; Eltayeb, Fujikawa, & Esaka, 2012; Lisko et al., 2013; Tóth et al., 2011; Yactayo‐Chang et al., 2018). We found that heat stress response genes (HSP17.8‐CI; HSP17.4‐CIII), cold stress response genes (COR15A), and drought stress response genes (ATDI21, NAC019) were up‐regulated in the AtMIOX4 OE line compared to WT (Figure 9a,b; Figure S3), whereas intracellular concentration of ABA was similar in AtMIOX4 OE line and WT (Figure 9c). Over‐expression of class I and class II HSPs is important for basal thermotolerance by interacting with translation factor (eEF1B), and knockdown of class II HSPs by RNAi results in severe heat sensitivity (McLoughlin et al., 2016). Moreover, over‐expression of HSP 17.8 aggregates the A. thaliana citrate synthase enzyme and protects it from heat at 43°C (Liu, Huang, Li, & Wu, 2005). Additionally, HSP 17.8 over‐expression results in freezing tolerance in maize (Shou et al., 2004). Similarly, over‐expression of HSP 17.4‐CII results in heat stress tolerance in tomato through coregulation of heat stress‐induced heat‐shock transcription factor A2 (HsfA2) (Port et al., 2004). Constitutive expression of Cor15A enhances freezing tolerance of protoplast and chloroplast in A. thaliana. This is due to decrease incidence of lamellar to hexagonal II phase transitions which occur in the close proximity of chloroplast membrane and plasma membrane due to dehydration caused by freezing (Artus et al., 1996; Steponkus, Uemura, Joseph, Gilmour, & Thomashow, 1998; Thalhammer, Bryant, Sulpice, & Hincha, 2014). Interestingly, over‐expression of NAC family transcription factors (ATAF, CUC, and NAM) in rice enhances drought resistance and salt tolerance by transactivating activity (Hu et al., 2006). In addition to this, over‐expression of ANAC019 increases heat tolerance and drought tolerance in A. thaliana (Guan, Yue, Zeng, & Zhu, 2014; Tran et al., 2004).
Peroxisomal ascorbate peroxidase 5 (pAPX5) is involved in scavenging ROS, which is down‐regulated in the AtMIOX4 OE line compared to WT (Figure 9a,b). High amount of ROS are produced in peroxisomes due to peroxisomal specific enzymes as acyl coA oxidase, glycolate oxidase, sulfite oxidase, superoxide dismutase, sarcosine oxidase, and polyamine oxidase in different plant species (Arent, Pye, & Henriksen, 2008; del Rio, Sandalio, Corpas, Palma, & Barroso, 2006; Douce & Neuburger, 1999; Goyer et al., 2004; Hansch et al., 2006; Kamada‐Nobusuda, Hayashi, Fukazawa, Sakakibara, & Nishimura, 2008; Planas‐Portell, Gallart, Tiburcio, & Altabella, 2013). Reactive oxygen species produced in peroxisomes are scavenged by pAPX or catalase (CAT). Spinach pAPX has high affinity for H2O2 compared to catalase (Ishikawa et al., 1998). Reduction in expression of pAPX results in higher level of H2O2. Enhanced level of H2O2 in peroxisomes results in up‐regulated expression of genes involved in oxidative stress response as small heat‐shock family proteins, DNAJ heat‐shock family protein, aromatic alcohol: NADPH reductase, and bHLH transcription factor. It was previously observed that silencing of pAPX results in oxidative stress tolerance in rice (del Rio et al., 2006; Sousa et al., 2015).
Based on a combination of the results shown here and published evidence, we propose a model where elevated AsA in A. thaliana reduces the expression of pAPX resulting in high level of peroxisomal ROS. High ROS in peroxisomes may act as signaling molecules and elevate the expression of abiotic stresses tolerance genes as heat response genes (HSP17.8‐CI; HSP17.4‐CIII), DNAJ heat‐shock protein family, bHLH transcription factor, and aromatic alcohol: NADPH reductase. This will result in plants tolerant to oxidative stress. In addition to this, cold stress response gene (COR15A), drought stress response genes (ATDI21, NAC019) that were up‐regulated in AtMIOX4 OE line could lead to cold and drought stress tolerance.
4.3. High ascorbate plants are transcriptionally primed for herbivore defense
Genes related to specialized metabolism were differentially regulated in AtMIOX4 OE line compared to WT control. Phenolics biosynthesis gene o‐methyl transferase family 2 protein was up‐regulated. Phenylpropanoid biosynthesis genes (TSM1, CAD‐B2), flavonoid biosynthesis (MYB75), the JAO2, JAO3, JAO4, AACT1 genes, and terpene biosynthesis genes (GGPS6, LAS1, TPS10) were up‐regulated in the high AsA line compared to controls (Figure 11a). Over‐expression of MYB75 results in accumulation of anthocyanin and protects A. thaliana from high light intensity (Gonzalez, Zhao, Leavitt, & Lloyd, 2008; Li et al., 2016). Glucosinolates‐myrosinase system genes such as flavin monooxygenase glucosinolate s‐oxygenase 2, alkenyl hydroxyalkyl producing 2, MYB domain protein 76, myrosinase binding protein 1, and epithiospecifier protein were up‐regulated in AtMIOX4 OE line compared to WT (Figure 12a). MYB76 transcription factor positively regulates the biosynthesis of methionine‐derived aliphatic glucosinolates (Gigolashvili et al., 2008; Sønderby et al., 2007). MYB76 is involved in synthesis of methylsulfinylalkyl glucosinolates from methylthioalkyl glucosinolates catalyzed by FMOGSOXS enzyme family (Hansen, Kliebenstein, & Halkier, 2007; Li, Hansen, Ober, Kliebenstein, & Halkier, 2008). Epithiospecifier protein is involved in nitrile formation upon glucosinolates hydrolysis and making Arabidopsis less appealing to specialist herbivores (Burow, Halkier, & Kliebenstein, 2010; Lambrix, Reichelt, Mitchell, Klibenstein, & Gershenzon, 2001). Methionine‐derived glucosinolates acts as a chemoprotective compounds in plant defense against biotic stress (Gigolashvili, Yatusevich, Berger, Muller, & Flugge, 2007). Increased levels of glucoerucin observed in AtMIOX4 OE might indicate that high AsA plants could be less susceptible to herbivore attack.
Further analysis on other glucosinolates present in AtMIOX4 OE line is now in progress and will help us to understand the effect of glucosinolates in this plant.
Jasmonic acid biosynthesis gene and JA‐responsive genes were up‐regulated in AtMIOX4 OE line compared to WT. JA biosynthesis intermediate metabolites (OPDA) were similar but JA and JA‐Ile levels were significantly higher in AtMIOX4 OE line compared to controls (Figure 11a–e). Over‐expression of OPR3 results in increased level of JA and JA‐Ile in A. thaliana and plants become resistance to insects and fungi (Suza & Staswick, 2008) which is similar to our results. Interestingly, JA is also involved in plant development, growth, reproduction and protects plants against abiotic stresses as salinity, drought, and UV radiation (Kerchev et al., 2011; Wasterneck & Hause, 2013). Salicylic acid metabolism genes and SA level were up‐regulated in AtMIOX4 OE line compared to WT control. In contrast, pathogenesis‐related proteins were down‐regulated in AtMIOX4 OE line compared to WT control (Figures 11a, 13a, c). The expression of pathogenesis‐related proteins was significantly down‐regulated in AtMIOX4 OE line. Surprisingly, the response of AtMIOX4 OE line was similar to WT control when challenged with P. syringae. This may be due to exclusive expression of pathogen response genes in AtMIOX4 line (Figure 3a; Table 3). Salicylate protects the plants from biotrophic and hemibiotrophic pathogens by interacting with NPR1 (Palmer et al., 2017). Salicylic acid not only protect plants against biotic stress, it is also involved in growth, development of plants and increases tolerance against abiotic stresses as cold, heat, drought, metal by increasing antioxidant capacity (Bartoli, Casalongué, Simontacchi, Marquez‐Garcia, & Foyer, 2013; Gimenez et al., 2017; Horváth et al., 2015). Furthermore, ethylene‐responsive element binding factor 13 induces the over‐expression of RAP transcriptions factor. Over‐expression of RAP2.6 resulted in increased resistance to nematode (Heterodera schachttii) in A. thaliana by enhanced callose deposition (Ali, Abbas, Kreil, & Bohlmann, 2013).
Table 3.
List of top 25 up‐regulated genes in AtMIOX4 OE line compared to WT control
| Gene id | Gene | Log2 (fold_change) | TAIR information |
|---|---|---|---|
| At3g29633 | – | inf | Encodes hypothetical protein |
| Atcg00100 | TRNG.1 | inf | Encodes tRNA‐gly |
| At1g32385 | – | inf | snoRNA‐rRNA modification |
| At3g14452 | – | inf | Encodes hypothetical transmembrane protein |
| At1g20015 | – | inf | snoRNA‐rRNA modification |
| At3g63375 | MIR167B | inf | Targets ARF6 and ARF8 (auxin response factors—ARF) |
| At2g46685 | MIR166A | inf | Targets several HD‐ZIPIII family members |
| At2g10606 | MIR396A | inf | Targets several GRF family members |
| At4g16240 | – | inf | Encodes hypothetical protein |
| At4g16215 | – | inf | Encodes hypothetical protein |
| At1g64195 | – | inf | Encodes a defensin‐like (DEFL) family protein.. |
| At3g15534 | – | inf | Encodes hypothetical protein |
| At5g22430 | – | inf | Encodes pollen Ole e1 allergen and extensin family protein. |
| At4g22513 | ‐ | inf | Encodes a protease inhibitor/seed storage/LTP family protein |
| At3g51590 | LTP12 | inf | Encodes lipid transfer protein 12 |
| At4g28395 | ATA7 | inf | Encodes lipid transfer protein |
| At1g68875 | – | inf | Encodes hypothetical protein |
| At1g67990 | TSM1 | inf | Tapetum specific methyl transferase |
| At3g05727 | – | inf | Encodes a defensin‐like (DEFL) family protein |
| At4g14743 | – | inf | Encodes reverse transcriptase‐related family protein |
| At5g01870 | LTP10 | inf | Predicted to encode a pathogenesis‐related protein |
| At1g68526 | – | inf | Encodes hypothetical protein |
| At4g11911 | – | inf | Encodes stay‐green‐like protein |
| At3g56705 | U2.6 | inf | Encodes U2 small nucleolar RNA6 |
| At1g66850 | – | inf | Encodes bifunctional inhibitor/lipid transfer protein |
4.4. Crosstalk between high AsA, SA and JA
Glutathione reductase 480 (GRX480) and DHAR1 which is involved in recycling of reduced ascorbate were both up‐regulated while peroxisomal pAPX5 was down‐regulated in AtMIOX4 OE line compared to WT control (Figure 9a,b). It was reported that, under high light stress, elevated H2O2 is associated with up‐regulation of small heat‐shock proteins and transcription factors related to anthocyanin biosynthesis (Vanderauwera et al., 2005). Similarly, anthocyanins protect plants against photo‐protection and photo‐oxidative damage. Mutation of thylakoid APX results in elevated H2O2 under high light stress which increases the expression of genes involved in anthocyanin biosynthesis regulation (Maruta et al., 2014). We found that the positive regulator of anthocyanin biosynthesis MYB75 and the anthocyanin biosynthesis 2OG‐Fe(II) oxygenase family proteins were up‐regulated in AtMIOX4 OE line compared to control (Table S2). These results suggest that high AsA A. thaliana are ready to protect themselves from photo‐oxidative damage by regulating anthocyanin biosynthesis.
Salicylic acid negatively regulates the JA biosynthesis genes and decreases expression of JA‐responsive genes (PDF1.2 and VSP2) (Chehab et al., 2011; Leon‐Reyes et al., 2010; Vlot, Dempsey, & Klessig, 2009). We found that JA and SA were both elevated and JA‐responsive defensin proteins were highly expressed in AtMIOX4 OE line compared to control. GRX480 was inducible by SA and suppresses the JA‐responsive defensin (PDF1.2) transcription in a NPR1‐dependent manner. GRX480 interacts with TGA2 transcription factor and suppresses the ethylene response factor 59 (ERF59) resulting in suppression of JA response defensin (Ndamukong et al., 2007). Published literature indicates that higher levels of endogenous ethylene diminish the SA and JA antagonism (Leon‐Reyes et al., 2010; Leon‐Reyes et al., 2010). Ethylene response factors as RAP2.6, RAP2.6L, RAP 2.10 were up‐regulated in AtMIOX4 OE line compared to WT control (Figure 11a). The over‐expression of the ethylene response factors might contribute to diminish the SA and JA antagonism, hence increased level of both SA and JA in high AsA A. thaliana.
5. CONCLUSIONS AND FUTURE PERSPECTIVES
We have here demonstrated the association between high AsA, increased intracellular glucose, enhanced photosynthetic efficiency, and increased auxin level in AtMIOX4 OE line. The enhanced glucose, auxin, and photosynthetic efficiency are likely explanations for the enhanced biomass phenotype of the high AsA line. Furthermore, the up‐regulated expression of signature genes and transcription factors for heat stress, cold stress, water limitation stress, and salt stress tolerance over‐expression likely lead to abiotic stresses tolerance in this high AsA AtMIOX4 OE line. The fact that abscisic acid was similar in AtMIOX4 and controls indicates that abiotic stresses tolerance in this line is most likely independent of up‐regulation of ABA. Interestingly, biotic stress response genes related to JA, SA, specialized metabolism, and glucosinolates biosynthesis and metabolism genes were up‐regulated in the high AsA line. The defense hormones JA, JA‐Ile, and SA were elevated in the high AsA line compared to its counterparts. These defense hormones and elevated expression of biotic stress response signature genes might confer the resistance phenotype of AtMIOX4 in response to biotic stresses. Furthermore, the up‐regulated expression of ethylene response factors in the high AsA line strongly suggests the presence of elevated levels of ethylene. Elevated ethylene might abolish the SA and JA antagonism in high AsA line. These results not only shed light into the molecular mechanisms underlying the enhanced biomass and abiotic stress tolerance phenotype of the high AsA line but also point to a conserved module of biotic stress response genes. Future studies are necessary to test the ability of high AsA lines to respond when challenged with herbivores. Next, steps should also include the expression of AtMIOX4 in crops of commercial value to fully exploit the potential biotechnological value of this knowledge.
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
NN performed the experiments, prepared figures, and wrote initial draft of the manuscript; JPYC generated the MIOX4 homozygous line; JPYC and LMAG prepared RNA samples for sequencing; MEGR contributed to HPLC analysis; KMJ and MAV analyzed the RNA‐Seq data; and AL conceived the experiments, secured funding, and wrote final draft of the manuscript.
Supporting information
ACKNOWLEDGMENTS
We thank Dr. Fabricio Medina‐Bolivar (A‐State) for providing access to the HPLC, Dr. Sophie Alvarez (UNL) for technical assistance in LC‐MS/MS measurements, Dr. Jian Hua (Cornell) for assistance with pathogens assays, Krishna Deo Sharma (A‐State) for assistance in microscopy, Zachary Campbell (A‐State) for technical assistance, and Kimberly Lee (A‐State) for plant care. We thank Dr. Fiona Goggin (UAF) for insightful suggestions to the project. NN thanks the Molecular Biosciences PhD Program for providing stipend support. We also thank the Arkansas Biosciences Institute (ABI, A‐State) for providing facilities to conduct experiments. This research was funded by the Arkansas Center for Plant Powered Production (Arkansas EPSCoR Track 1, fund EPS 0701890), the Plant Imaging Consortium (PIC, AR EPSCoR Track 2, fund IIA 1430427) funded by the National Science Foundation and a grant to AL from the Arkansas Research Alliance.
Nepal N, Yactayo‐Chang JP, Medina‐Jiménez K, et al. Mechanisms underlying the enhanced biomass and abiotic stress tolerance phenotype of an Arabidopsis MIOX over‐expresser. Plant Direct. 2019;3:1–27. 10.1002/pld3.165
REFERENCES
- Acosta‐Gamboa, L. (2019) Assessing the contribution of multiple ascorbate pathways to abiotic stress tolerance using phenomic approaches. PhD Dissertation, Arkansas State University, AR. Retrieved from ProQuest Dissertations and Thesis https://search.proquest.com/docview/2217758146/A800619EC5DF4DBFPQ/1?accountxml:id=8363
- Acosta‐Gamboa, L. M. , Liu, S. , Langley, E. , Campbell, Z. C. , Castro‐Guerrero, N. , Mendoza‐Cózatl, D. , & Lorence, A. (2017). Moderate to severe water limitation differentially affects the phenome and ionome of Arabidopsis. Functional Plant Biology, 44, 94–106. [DOI] [PubMed] [Google Scholar]
- Agius, F. , Gonzalez‐Lamothe, R. , Caballero, L. J. , Munoz‐Blanco, J. , Botella, A. M. , & Valpuesta, V. (2003). Engineering increased vitamin C levels in plants by over‐expression of a D‐galacturonic acid reductase. Nature Biotechnology, 21, 177–181. 10.1038/nbt777 [DOI] [PubMed] [Google Scholar]
- Alford, S. R. , Rangarajan, P. , Williams, P. , & Gillaspy, G. E. (2012). myo‐Inositol oxygenase is required for responses to low energy conditions in Arabidopsis thaliana . Frontiers in Plant Science, 3, 69 10.3389/fpls.2012.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, M. A. , Abbas, A. , Kreil, D. P. , & Bohlmann, H. (2013). Over‐expression of the transcription factor RAP2.6 leads to enhanced callose deposition in syncytia and enhanced resistance against the beet cyst nematode Heterodera schachtii in Arabidopsis roots. BMC Plant Biology, 13, 1–13. 10.1186/1471-2229-13-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2014). FastQC: A quality control tool for high throughput sequence data. Version 0.11. Cambridge, UK: 2. Babraham Institute. [Google Scholar]
- Arent, S. , Pye, V. E. , & Henriksen, A. (2008). Structure and function of plant acyl‐CoA oxidases. Plant Physiology and Biochemistry, 46, 292–301. 10.1016/j.plaphy.2007.12.014 [DOI] [PubMed] [Google Scholar]
- Artus, N. N. , Uemura, M. , Steponkus, P. L. , Gilmour, S. J. , Lin, C. , & Thomashow, M. F. (1996). Constitutive expression of the cold‐regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proceedings of the National Academy of Sciences of the United States of America, 93, 13404–13409. 10.1073/pnas.93.23.13404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbehenn, R. V. (2002). Gut‐based antioxidant enzymes in a polyphagous and a graminivorous grasshopper. Journal of Chemical Ecology, 28, 1329–1347. [DOI] [PubMed] [Google Scholar]
- Bartoli, C. G. , Casalongué, C. A. , Simontacchi, M. , Marquez‐Garcia, B. , & Foyer, C. H. (2013). Interactions between hormone and redox signaling pathways in the control of growth and cross tolerance to stress. Environmental and Experimental Botany, 94, 73–88. [Google Scholar]
- Berthet, S. , Demont‐Caulet, N. , Pollet, B. , Bidzinski, P. , Cezard, L. , Le Bris, P. , … Jouanin, L. (2011). Disruption of LACCASE4 and 17 results in tissue‐specific alterations to lignification of Arabidopsis thaliana stems. The Plant Cell, 23, 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D. C. , Zayed, A. M. , Ascenzi, R. , McCaskill, A. J. , Hoffman, N. E. , Davis, K. R. , & Görlach, J. (2001). Growth stage–based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. The Plant Cell, 13, 1499–1510. 10.1105/TPC.010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister, W. P. , Cottaz, S. , Rollin, P. , Vasella, A. , & Henrissat, B. (2000). High resolution X‐ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base. Journal of Biological Chemistry, 275, 39385–39393. 10.1074/jbc.M006796200 [DOI] [PubMed] [Google Scholar]
- Burow, M. , Halkier, B. A. , & Kliebenstein, D. J. (2010). Regulatory networks of glucosinolates shape Arabidopsis thaliana fitness. Current Opinion in Plant Biology, 13, 348–353. 10.1016/j.pbi.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Bustin, S. A. , Benes, V. , Garson, J. A. , Hellemans, J. , Huggett, J. , Kubista, M. , … Wittwer, C. T. (2009). The MIQE guideline: Minimum information for publication of quantitative real‐time PCR experiments. Clinical Chemistry, 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Chehab, E. W. , Kim, S. , Savchenko, T. , Kliebenstein, D. , Dehesh, K. , & Braam, J. (2011). Intronic T‐DNA insertion renders Arabidopsis OPR3 a conditional jasmonic acid‐producing mutant. Plant Physiology, 156, 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, K. M. , Nguyen, H. T. , Kim, S. Y. , Shin, J. S. , Cho, D. H. , Hong, S. B. , … Ok, S. H. (2016). CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytologist, 209, 664–678. 10.1111/nph.13612 [DOI] [PubMed] [Google Scholar]
- Christopher, H. , Rensburg, J. A. , & den Ende, W. V. (2017). UDP‐Glucose: A potential signaling molecule in plants? Frontiers in Plant Science, 8, 2230 10.3389/fpls.2017.02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen, M. , Luthe, H. , Blatt, M. , & Bottger, M. (1997). Auxin‐induced growth and its linkage to potassium channels. Planta, 201, 227–234. 10.1007/BF01007708 [DOI] [Google Scholar]
- Clough, S. J. , & Bent, A. F. (1998). Floral dip: A simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conesa, A. , & Gotz, S. (2008). Blast2Go: A comprehensive suite for functional analysis in plant genomics. International Journal of Plant Genomics, 2008, 1–13. 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin, P. L. , DePaolo, D. , Wintle, B. , Schatz, C. , & Buckenmeyer, G. (2013). Identification of Arabidopsis VTC3 as a putative and unique dual function protein kinase: Protein phosphatase involved in the regulation of the ascorbic acid pool in plants. Journal of Experimental Botany, 64, 2793–2804. 10.1093/jxb/ert140 [DOI] [PubMed] [Google Scholar]
- Conklin, P. L. , Saracco, S. A. , Norris, S. R. , & Last, R. L. (1999). Identification of ascorbic acid‐deficient Arabidopsis thaliana mutants. Genetics, 154, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M. K. , & Scheible, W. R. (2005). Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology, 139, 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, D. , St‐Onge, K. R. , Voesenek, L. A. , Pierik, R. , & Sasidharan, R. (2016). Ethylene‐ and shade‐induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiology, 172, 718–733. 10.1104/pp.16.00725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt, S. , Cook, D. R. , & Ford, C. M. (2006). L‐Tartaric acid synthesis from vitamin C in higher plants. Proceedings of the National Academy of Sciences of the United States of America, 103, 5608–5613. 10.1073/pnas.0510864103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio, L. A. , Sandalio, L. M. , Corpas, F. J. , Palma, J. M. , & Barroso, J. B. (2006). Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiology, 141, 330–335. 10.1104/pp.106.078204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce, R. , & Neuburger, M. (1999). Biochemical dissection of photorespiration. Current Opinion in Plant Biology, 2, 214–222. 10.1016/S1369-5266(99)80038-7 [DOI] [PubMed] [Google Scholar]
- Dowdle, J. , Ishikawa, T. , Gatzek, S. , Rolinski, S. , & Smirnoff, N. (2007). Two genes in Arabidopsis thaliana encoding GDP‐L‐galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. The Plant Journal, 52, 673–689. 10.1111/j.1365-313X.2007.03266.x [DOI] [PubMed] [Google Scholar]
- Eltayeb, A. E. , Kawano, N. , Badawi, G. H. , Kaminaka, H. , Sanekata, T. , Morishima, I. , … Tanaka, K. (2006). Enhanced tolerance to ozone and drought stresses in transgenic tobacco over‐expressing dehydroascorbate reductase in cytosol. Physiol Planta, 127, 57–65. 10.1111/j.1399-3054.2006.00624.x [DOI] [Google Scholar]
- Eltayeb, H. A. , Fujikawa, Y. , & Esaka, M. (2012). Overexpression of the acerola (Malpighia glabra) monodehydroascorbate reductase gene in transgenic tobacco plants results in increased ascorbate levels and enhanced tolerance to salt stress. South African Journal of Botany, 78, 295–301. 10.1016/j.sajb.2011.08.005 [DOI] [Google Scholar]
- Endres, S. , & Tenhaken, R. (2011). Down‐regulation of the myo‐inositol oxygenase gene family has no effect on cell wall composition in Arabidopsis. Planta, 234, 157–169. 10.1007/s00425-011-1394-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J. R. (2013). Improving photosynthesis. Plant Physiology, 162, 1780–1793. 10.1104/pp.113.219006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa‐Mendez, R. , & Rivas‐Arancibia, S. (2015). Vitamin C in health and disease: Its role in the metabolism of cells and redox state in the brain. Frontiers in Physiology, 6, 397 10.3389/fphys.2015.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer, C. H. , & Noctor, G. (2011). Ascorbate and glutathione: The heart of the redox hub. Plant Physiology, 155, 2–18. 10.1104/pp.110.167569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie, D. R. (2013). L‐Ascorbic acid: A multifunctional molecule supporting plant growth and development. Journal of Experimental Botany, 64, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar, Y. M. , Nam, J. , Schultz, C. J. , Lee, L. Y. , Gilson, P. R. , Gelvin, S. B. , & Bacic, A. (2004). Characterization of the Arabidopsis lysine‐rich arabinogalactan protein AtAGP17 mutant (rat1) that results in a decreased efficiency of Agrobacterium transformation. Plant Physiology, 135, 2162–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gest, N. , Gautier, H. , & Stevens, R. (2013). Ascorbate as seen through plant evolution: The rise of a successful molecule? Journal of Experimental Botany, 64, 33–53. 10.1093/jxb/ers297 [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T. , Engqvist, M. , Yatusevich, R. , Muller, C. & Flugge, U. C. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytology 177: 627–642. [DOI] [PubMed] [Google Scholar]
- Gigolashvili, T. , Yatusevich, R. , Berger, B. , Muller, C. , & Flugge, U. I. (2007). The R2R3‐MYB transcription factor HAG1/MYB28 is a regulator of methionine‐derived glucosinolate biosynthesis in Arabidopsis thaliana . The Plant Journal, 51, 247–261. [DOI] [PubMed] [Google Scholar]
- Gimenez, M. J. , Serrano, M. , Valvarde, J. M. , Martinez, R. D. , Castillo, S. , Valero, D. , & Guillen, F. (2017). Preharvest salicylic acid and acetylsalicylic acid treatments preserve quality and enhance antioxidants systems during postharvest storage of sweet cherry cultivars. Journal of the Science of Food and Agriculture, 97, 1220–12228. [DOI] [PubMed] [Google Scholar]
- Goda, H. , Sawa, S. , Asami, T. , Fujioka, S. , Shimada, Y. , & Yoshida, S. (2004). Comprehensive comparison of auxin‐regulated and brassinosteroid‐regulated genes in Arabidopsis. Plant Physiology, 134, 1555–1573. 10.1104/pp.103.034736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, A. , Zhao, M. , Leavitt, J. M. , & Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal, 53, 814–827. 10.1111/j.1365-313X.2007.03373.x [DOI] [PubMed] [Google Scholar]
- Goyer, A. , Johnson, T. L. , Olsen, L. J. , Collakova, E. , Shachar‐Hill, Y. , Rhodes, D. , & Hanson, A. D. (2004). Characterization and metabolic function of a peroxisomal sarcosine and pipecolate oxidase from Arabidopsis. Journal of Biological Chemistry, 279, 16947–16953. [DOI] [PubMed] [Google Scholar]
- Grosser, K. , & van Dam, N. M. (2017). A straightforward method for glucosinolate extraction and analysis with high‐pressure liquid chromatography (HPLC). Journal of Visualized Experiments, 121, e55425 10.3791/55425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Q. , Yue, X. , Zeng, H. , & Zhu, J. (2014). The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress‐responsive gene regulation and thermotolerance in Arabidopsis. The Plant Cell, 26, 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, R. , Shen, W. , Qian, H. , Zhang, M. , Liu, L. , & Wang, Q. (2013). Jasmonic acid and glucose synergistically modulate the accumulation of glucosinolates in Arabidopsis thaliana . Journal of Experimental Botany, 64, 5707–5719. 10.1093/jxb/ert348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S. , & Kim, D. (2006). AtRTPrimer: Database for Arabidopsis genome‐wide homogeneous and specific RT‐PCR primer‐pairs. BMC Bioinfo, 7, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansch, R. , Lang, C. , Riebeseel, E. , Lindigkeit, R. , Gessler, A. , Rennenberg, H. , & Mendel, R. R. (2006). Plant sulfite oxidase as novel producer of H2O2: Combination of enzyme catalysis with a subsequent non‐enzymatic reaction step. Journal of Biological Chemistry, 281, 6884–6888. [DOI] [PubMed] [Google Scholar]
- Hansen, B. G. , Kliebenstein, D. J. , & Halkier, B. A. (2007) Identification of a flavin‐ monooxygenase as the S‐oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant Journal 50: 902–910. [DOI] [PubMed] [Google Scholar]
- Horváth, E. , Brunner, S. Z. , Bela, K. , Papdi, C. S. , Szabados, L. , Tari, I. , & Csiszár, J. (2015). Exogenous salicylic acid‐triggered changes in the glutathione transferases and peroxidases are key factors in the successful salt stress acclimation of Arabidopsis thaliana . Functional Plant Biology, 42, 1129–1140. 10.1071/FP15119 [DOI] [PubMed] [Google Scholar]
- Hu, H. , Dai, M. , Yao, J. , Xiao, B. , Li, X. , Zhang, Q. , & Xiong, L. (2006). Over‐expressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the United States of America, 103, 12987–12992. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. W. , Sherman, B. T. , & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protocols, 4, 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ishikawa, T. , Yoshimura, K. , Sakai, K. , Tamoi, M. , Takeda, T. , & Shigeoka, S. (1998). Molecular characterization and physiological role of a glyoxysome‐bound ascorbate peroxidase from spinach. Plant and Cell Physiology, 39, 23–34. 10.1093/oxfordjournals.pcp.a029285 [DOI] [PubMed] [Google Scholar]
- Johnson, M. P. (2016). Photosynthesis. Essays in Biochemistry, 60, 255–273. 10.1042/EBC20160016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada‐Nobusuda, T. , Hayashi, M. , Fukazawa, M. , Sakakibara, H. , & Nishimura, M. (2008). A putative peroxisomal polyamine oxidase, AtPAO4, is involved in polyamine catabolism in Arabidopsis thaliana . Plant and Cell Physiology, 49, 1272–1282. 10.1093/pcp/pcn114 [DOI] [PubMed] [Google Scholar]
- Kanter, U. , Usadel, B. , Guerineau, F. , Li, Y. , Pauly, M. , & Tenhaken, R. (2005). The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell‐wall matrix polysaccharides. Planta, 221, 243–254. 10.1007/s00425-004-1441-0 [DOI] [PubMed] [Google Scholar]
- Katano, K. , Kataoka, R. , Fujii, M. , & Suzuki, N. (2017). Differences between seedlings and flowers in anti‐ROS based heat responses of Arabidopsis plants deficient in cyclic nucleotide gated channel 2. Plant Physiology and Biochemistry, 123, 288–296. 10.1016/j.plaphy.2017.12.021 [DOI] [PubMed] [Google Scholar]
- Kavkova, I. , Blochl, E. C. , & Tenhaken, R. (2018). The myo‐inositol pathway does not contribute to ascorbic acid synthesis. Plant Biology, 25, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev, P. I. , Karpinska, B. , Morris, J. A. , Hussain, A. , Verrall, S. R. , Hedley, P. E. , … Hancock, R. D. (2013). Vitamin C and the abscisic acid‐insensitive 4 transcription factor are important determinants of aphid resistance in Arabidopsis. Antioxidants & Redox Signaling, 18, 2091–2105. [DOI] [PubMed] [Google Scholar]
- Kerchev, P. I. , Pellny, T. K. , Vivancos, P. D. , Kiddle, G. , Hedden, P. , Driscoll, S. , … Foyer, C. H. (2011). The transcription factor ABI4 is required for the ascorbic acid‐dependent regulation of growth and regulation of jasmonate‐dependent defense signaling pathways in Arabidopsis. The Plant Cell, 23, 3319–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. , & Salzberg, S. L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. H. , Xu, Z. Y. , Na, Y. J. , Yoo, Y. J. , Lee, J. , Sohn, E. J. , & Hwang, I. (2011). Small heat shock protein Hsp17.8 functions as an AKR2A cofactor in the targeting of chloroplast outer membrane proteins in Arabidopsis. Plant Physiology, 157, 132–146. 10.1104/pp.111.178681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. I. , Baek, D. , Park, H. C. , Chun, H. J. , Oh, D. H. , Lee, M. K. , … Yun, D. J. (2013). Over‐expression of Arabidopsis YUCCA6 in potato results in high‐auxin developmental phenotypes and enhanced resistance to water deficit. Molecular Plant, 6, 337–349. 10.1093/mp/sss100 [DOI] [PubMed] [Google Scholar]
- Korasick, D. A. , Enders, T. A. , & Strader, L. C. (2013). Auxin biosynthesis and storage forms. Journal of Experimental Botany, 64, 2541–2555. 10.1093/jxb/ert080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger, F. (2015). Trim galore. A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. v0.4.3.1. Cambridge, UK: Babraham Institute. [Google Scholar]
- Kuhlgert, S. , Austic, G. , Zegarac, R. , Osei‐Bonsu, I. , Hoh, D. , Chilvers, M. I. , … Kramer, D. M. (2016). MultispeQ Beta: A tool for large‐scale plant phenotyping connected to the open PhotosynQ network. Royal Society Open Science, 3, 160592 10.1098/rsos.160592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrix, V. , Reichelt, M. , Mitchell, O. T. , Klibenstein, D. J. , & Gershenzon, J. (2001). The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia in herbivory. The Plant Cell, 13, 2793–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, Z. , Krosse, S. , Achard, P. , van Dam, N. M. , & Bede, J. C. (2014). DELLA proteins modulate Arabidopsis defenses induced in response to caterpillar herbivory. Journal of Experimental Botany, 65, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. , Jung, J. H. , Han, D. Y. , Seo, P. J. , Park, W. J. , & Park, C. M. (2011) Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 235: 923–938. [DOI] [PubMed] [Google Scholar]
- Lee, Y. K. , Rhee, J. Y. , Lee, S. H. , Chung, G. C. , Park, S. J. , Segami, S. , … Choi, G. (2018). Functionally redundant LNG3 and LNG4 genes regulate turgor‐driven polar cell elongation through activation of XTH17 and XTH24 . Plant Molecular Biology, 97, 23–36. 10.1007/s11103-018-0722-0 [DOI] [PubMed] [Google Scholar]
- Leong, S. Y. , Liu, T. , Oey, I. , & Burritt, D. J. (2017). Ascorbic acid in processed plant‐based foods In Munne‐Bosch S., Burrit D. J., Diaz‐Vivancos P., Fujita M., & Lorence A. Ascorbic acid in plant growth, Development and stress tolerance (pp. 431–469). Hossain, MA: Springer. [Google Scholar]
- Leon‐Reyes, A. , Does, D. V. , De Lange, E. S. , Delker, C. , Wasternack, C. , Van Wees, S. C. M. , … Pieterse, C. M. J. (2010). Salicylate‐mediated suppression of jasmonate‐responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta, 232, 1423–1432. 10.1007/s00425-010-1265-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Hansen, B. G. , Ober, J. A. , Kliebenstein, D. J. , & Halkier, B. A. (2008). Subclade of flavin‐monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiology, 148, 1721–1733. 10.1104/pp.108.125757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Wang, W. , Gao, J. , Yin, K. , Wang, R. , Wang, C. , … Qiu, J. L. (2016). MYB75 phosphorylation by MPK4 is required for light‐induced anthocyanin accumulation in Arabidopsis. The Plant Cell, 28, 2866–2883. 10.1105/tpc.16.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Smith, C. , Corke, F. , Zheng, L. , Merali, Z. , Ryden, P. , … Bevan, M. W. (2007). Signaling from an altered cell wall to the nucleus mediates sugar‐responsive growth and development in Arabidopsis thaliana . The Plant Cell, 19, 2500–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, C. Y. , Smet, W. , Brunoud, G. , Yoshida, S. , Vernoux, T. , & Weijers, D. (2015). Reporters for sensitive and quantitative measurement of auxin response. Nature Methods, 12, 207–210. 10.1038/nmeth.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman, A. H. , & Cavalier, D. M. (2012). The CELLULOSE SYNTHASE‐LIKE A and CELLULOSE SYNTHASE‐LIKE C families: Recent advances and future perspectives. Froniers in Plant Science, 3, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisko, K. A. , Aboobucker, S. I. , Torres, R. , & Lorence, A. (2014) Engineering elevated vitamin C in plants to improve their nutritional content, growth, and tolerance to abiotic stress. In Phytochemicals – Biosynthesis, Function and Application” R Jetter (ed). Recent Advances in Phytochemistry 44, 109–128. [Google Scholar]
- Lisko, K. A. , Torres, R. , Harris, R. S. , Belisle, M. , Vaughan, M. M. , Jullian, B. , … Lorence, A. (2013). Elevating vitamin C content via overexpression of myo‐inositol oxygenase and L‐gulono‐1,4‐lactone oxidase in Arabidopsis leads to enhanced biomass and tolerance to abiotic stresses. In Vitro Cellular & Developmental Biology – Plant, 49, 643–655. 10.1007/s11627-013-9568-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Huang, W. , Li, M. , & Wu, Q. (2005). Purification and characterization of two small heat shock proteins from Anabaena sp. PCC 7120. IUBMB Life, 57, 449–454. 10.1080/15216540500138402 [DOI] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorence, A. , Chevone, B. I. , Mendes, P. , & Nessler, C. L. (2004). myo‐Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiology, 134, 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, S. R. , Kunert, G. , Reichelt, M. , Gershenzon, J. , & Halkier, B. A. (2015). Feeding on leaves of the glucosinolate transporter mutant gtr1gtr2 reduces fitness of Myzus persicae . Journal of Chemical Ecology, 41, 975–984. 10.1007/s10886-015-0641-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta, T. , Noshi, M. , Nakamura, M. , Matsuda, S. , Tamoi, M. , Ishikawa, T. , & Shigeoka, S. (2014). Ferulic acid 5‐hydroxylase 1 is essential for expression of anthocyanin biosynthesis‐associated genes and anthocyanin accumulation under photo‐oxidative stress in Arabidopsis. Plant Science, 219–220, 61–68. 10.1016/j.plantsci.2014.01.003 [DOI] [PubMed] [Google Scholar]
- McLoughlin, F. , Basha, E. , Fowler, M. E. , Kim, M. , Bordowitz, J. , Katiyar‐Agarwal, S. , & Vierling, E. (2016). Class I and II small heat shock proteins together with HSP101 protect protein translation factors during heat stress. Plant Physiology, 172, 1221–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, H. , Cai, C. , Wei, J. , Huang, J. , Chang, J. , Qian, H. , … Wang, Q. (2016). Glucose enhances indolic glucosinolate biosynthesis without reducing primary sulfur assimilation. Scientific Reports, 6, 31854 10.1038/srep31854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, B. , Zhou, L. , Rolland, F. , Hall, Q. , Cheng, W. H. , Liu, Y. X. , … Sheen, J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science, 300, 332–336. 10.1126/science.1080585 [DOI] [PubMed] [Google Scholar]
- Mueller‐Roeber, B. , & Balazadeh, S. (2013). Auxin and its role in plant senescence. Journal of Plant Growth Regulation, 33, 21–33. 10.1007/s00344-013-9398-5 [DOI] [Google Scholar]
- Mukherjee, M. , Larrimore, K. E. , Ahmed, N. J. , Bedick, T. S. , Barghouthi, N. T. , Traw, M. B. , & Barth, C. (2010). Ascorbic acid deficiency in Arabidopsis induces constitutive priming that is dependent on hydrogen peroxide, salicylic acid, and the NPR1 gene. Molecular Plant Microbe Interactions, 3, 340–351. [DOI] [PubMed] [Google Scholar]
- Ndamukong, I. , Abdallat, A. A. , Thurow, C. , Fode, B. , Zander, M. , Weigel, R. , & Gatz, C. (2007). SA‐inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA‐responsive PDF1.2 transcription. The Plant Journal, 50, 128–139. 10.1111/j.1365-313X.2007.03039.x [DOI] [PubMed] [Google Scholar]
- Olmos, E. , Kiddle, G. , Pellny, T. K. , Kumar, S. , & Foyer, C. H. (2006). Modulation of plant morphology, root architecture, and cell structure by low vitamin C in Arabidopsis thaliana . Journal of Experimental Botany, 57, 1645–1655. 10.1093/jxb/erl010 [DOI] [PubMed] [Google Scholar]
- Ortiz‐Espín, A. , Sanchez‐Guerrero, A. , Sevilla, F. , & Jiménez, A. (2017). The role of ascorbate in plant growth and development In Burrit D. J., Diaz‐Vivancos P., Fujita M., & Lorence A. Ascorbic acid in plant growth, development and stress tolerance (pp. 25–46). New York: Springer. [Google Scholar]
- Pastori, G. M. , Kiddle, G. , Antoniw, J. , Bernard, S. , Veljovic‐Jovanovic, S. , Verrier, P. J. , … Foyer, C. H. (2003). Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. The Plant Cell, 15, 939–951. 10.1105/tpc.010538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet, V. , Olmos, E. , Kiddle, G. , Mowla, S. , Kumar, S. , Antoniw, J. , … Foyer, C. H. (2005). Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiology, 139, 1291–1303. 10.1104/pp.105.067686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Chen, L. , Li, S. , Zhang, Y. , Xu, R. , Liu, R. , … Li, Y. (2018). BRI1 and BAK1 interact with G proteins and regulate sugar‐responsive growth and development in Arabidopsis. Nature Communications, 9, 1522 10.1038/s41467-018-03884-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar, K. , Buchsenschutz, K. , Edwards, D. , Loffler, J. , Luthen, H. , Kranz, E. , … Hedrich, R. (2006). The auxin‐induced K(+) channel gene Zmk1 in maize functions in coleoptile growth and is required for embryo development. Plant Molecular Biology, 61, 757–768. 10.1007/s11103-006-0047-2 [DOI] [PubMed] [Google Scholar]
- Planas‐Portell, J. , Gallart, M. , Tiburcio, A. F. , & Altabella, T. (2013). Copper‐containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana . BMC Plant Biology, 13, 1–13. 10.1186/1471-2229-13-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port, M. , Tripp, J. , Zielinski, D. , Weber, C. , Heerklotz, D. , Winkelhaus, S. , … Scharf, K. D. (2004). Role of Hsp17.4‐CII as co‐regulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiology, 135, 1457–1470. 10.1104/pp.104.042820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzio, J. A. , Lorence, A. , Chevone, B. I. , & Nessler, C. L. (2003). L‐Gulono‐1,4‐lactone oxidase expression rescues vitamin C‐deficient Arabidopsis (vtc) mutants. Plant Molecular Biology, 53, 837–844. 10.1023/B:PLAN.0000023671.99451.1d [DOI] [PubMed] [Google Scholar]
- Shou, H. , Bordallo, P. , Fan, J. B. , Yeakley, J. M. , Bibikova, M. , Sheen, J. , & Wang, K. (2004). Expression of an active tobacco mitogen‐activated protein kinase kinase kinase enhances freezing tolerance in transgenic maize. Proceedings of the National Academy of Sciences of the United States of America, 101, 3298–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique, S. , Endres, S. , Atkins, J. M. , Szakasits, D. , Wieczorek, K. , Hoffman, J. , … Bohlmann, H. (2009). Myo‐inositol oxygenase genes are involved in the development of syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytologist, 184, 457–472. [DOI] [PubMed] [Google Scholar]
- Siddique, S. , Endres, S. , Sobczak, M. , Radakovic, Z. S. , Fragner, L. , Grundler, F. M. W. , … Bohlmann, H. (2014). Myo‐inositol oxygenase is important for the removal of excess myo‐inositol from syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytologist, 201, 476–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby, I. E. , Burow, M. , Rowe, C. H. , Kliebenstein, D. J. , & Halkier, A. B. (2010). A complex interplay of three R2R3 MYB transcription factors determine the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiology, 153, 348–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby, I. E. , Hansen, B. G. , Bjarnholt, N. , Ticconi, C. , Halkier, B. A. , & Kliebenstein, D. J. (2007) A systems biology approach identifies a R2R3MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2: e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa, R. H. V. , Carvalho, F. E. L. , Ribeiro, C. W. , Passaia, G. , Cunha, J. R. , Lima‐Melo, Y. , … Silveira, J. A. (2015). Peroxisomal APX knockdown triggers antioxidant mechanisms favorable for coping with high photorespiratory H2O2 induced by CAT deficiency in rice. Plant, Cell and Environment, 38, 499–513. [DOI] [PubMed] [Google Scholar]
- Steponkus, P. L. , Uemura, M. , Joseph, R. A. , Gilmour, S. J. , & Thomashow, M. F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 95, 14570–14575. 10.1073/pnas.95.24.14570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suza, W. P. , & Staswick, P. E. (2008). The role of JAR1 in jasmonoyl‐L‐isoleucine production during Arabidopsis wound response. Planta, 227, 1221–1232. 10.1007/s00425-008-0694-4 [DOI] [PubMed] [Google Scholar]
- Suzuki, N. , Devireddy, A. R. , Inupakutika, M. A. , Baxter, A. , Miller, G. , Song, L. , … Mittler, R. (2015). Ultra‐fast alterations in mRNA levels uncover multiple players in light stress acclimation in plants. The Plant Journal, 84, 760–772. 10.1111/tpj.13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K. , Hayashi, K. , & Kinoshita, T. (2012). Auxin activates the plasma membrane H+‐ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiology, 159, 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylaran, R. D. , Adachi, S. , Ookawa, T. , Usuda, H. , & Hirasawa, T. (2011). Hydraulic conductance as well as nitrogen accumulation plays a role in the higher rate of leaf photosynthesis of the most productive variety of rice in Japan. Journal of Experimental Botany, 62, 4067–4077. 10.1093/jxb/err126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer, A. , Bryant, G. , Sulpice, R. , & Hincha, D. K. (2014). Disordered cold regulated 15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo . Plant Physiology, 166, 190–201. 10.1104/pp.114.245399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabinejad, J. , Donahue, J. L. , Gunesekera, B. N. , Allen‐Daniels, M. J. , & Gillaspy, G. E. (2009). VTC4 is a bifunctional enzyme that affects myo‐inositol and ascorbate biosynthesis in plants. Plant Physiology, 150, 951–961. 10.1104/pp.108.135129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth, S. Z. , Nagy, V. , Puthur, J. T. , Kovacs, L. , & Garab, G. (2011). The physiological role of ascorbate as photosystem II electron donor: Protection against photo‐inactivation in heat‐stressed leaves. Plant Physiology, 156, 382–392. 10.1104/pp.110.171918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, L. S. , Nakashima, K. , Sakuma, Y. , Simpson, S. D. , Fujita, Y. , Maruyama, K. , … Yamaguchi‐Shinozaki, K. (2004). Isolation and functional analysis of Arabidopsis stress‐inducible NAC transcription factors that bind to a drought‐responsive cis‐element in the early responsive to dehydration stress 1 promoter. The Plant Cell, 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D. R. , … Pachter, L. (2012). Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyburski, J. , Dunajska‐Ordak, K. , Skorupa, M. , & Tretyn, A. (2012). Role of ascorbate in the regulation of the Arabidopsis thaliana root growth by phosphate availability. Journal of Experimental Botany, 2012, 1–11. [Google Scholar]
- Usadel, B. , Nagel, A. , Steinhauser, D. , Gibon, Y. , Bläsing, O. E. , Redestig, H. , … Stitt, M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics, 7, 535 10.1186/1471-2105-7-535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel, B. , Poree, F. , Nagel, A. , Lohse, M. , Czedik‐Eysenberg, A. , & Stitt, M. (2009). A guide to using MapMan to visualize and compare omics data in plants: A case study in the crop species, Maize. Plant, Cell and Environment, 32, 1211–1229. 10.1111/j.1365-3040.2009.01978.x [DOI] [PubMed] [Google Scholar]
- Vanderauwera, S. , Zimmermann, P. , Rombauts, S. , Vandenabeele, S. , Langebartels, C. , Gruissem, W. , … Van Breusegem, F. (2005). Genome‐wide analysis of hydrogen peroxide‐regulated gene expression in Arabidopsis reveals a high light‐induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiology, 139, 806–821. 10.1104/pp.105.065896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A. C. , Dempsey, D. A. , & Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology, 47, 177–206. 10.1146/annurev.phyto.050908.135202 [DOI] [PubMed] [Google Scholar]
- Wasternack, C. , & Hause, B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Annals of Botany, 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, C. M. , Dwyer, J. T. , Fulgoni, V. L. , King, J. , Leveille, G. , MacDonald, R. , … Schnakenberg, D. (2014). Processed foods: Contributions to nutrition. American Journal of Clinical Nutrition, 99, 1525–1542. 10.3945/ajcn.114.089284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfalla, C. S. , Zubieta, C. , Alvarez, S. , Schraft, E. , Marcellin, R. , Ramirez, L. , & Jez, J. M. (2016). Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 113, 13917–13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, G. L. , Jones, M. A. , & Smirnoff, N. (1998). The biosynthetic pathway of vitamin C in higher plants. Nature, 393, 365–369. 10.1038/30728 [DOI] [PubMed] [Google Scholar]
- Wolucka, B. A. , & Montagu, M. V. (2003). GDP‐mannose 3',5'‐epimerase forms GDP‐L‐gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. Journal of Biological Chemistry, 278, 47483–47490. [DOI] [PubMed] [Google Scholar]
- Yactayo‐Chang, J. P. (2011) Stable co‐expression of vitamin C enhancing genes for improved production of a recombinant therapeutic protein, hIL‐12, in Arabidopsis thaliana . Master Thesis, Arkansas State University. Retrieved from ProQuest Dissertations and Theses, https://search.proquest.com/docview/912371383?pq-origsite=gscholar.
- Yactayo‐Chang, J. P. , Acosta‐Gamboa, L. M. , Nepal, N. , & Lorence, A. (2018). The role of plant high‐throughput phenotyping in the characterization of the response of high ascorbate plants to abiotic stresses In Hossain M. A., Munné‐Bosch S., Burritt D. J., Diaz‐Vivancos P., Fujita M., & Lorence A. (Eds.), Ascorbic acid in plant growth, development and stress tolerance (pp. 321–354). New York: Springer. [Google Scholar]
- Yang, J. , Sardar, H. S. , McGovern, K. R. , Zhang, Y. , & Showalter, A. M. (2007). A lysine‐rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. The Plant Journal, 49, 629–640. 10.1111/j.1365-313X.2006.02985.x [DOI] [PubMed] [Google Scholar]
- Yang, L. , Xu, M. , Koo, Y. , He, J. , & Poethig, R. S. (2013). Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife, 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalabak, D. , Pospisilova, H. , Smehilova, M. , Mrizova, K. , Frebort, I. , & Galuszka, P. (2013). Genetic engineering of cytokinin metabolism: Prospective way to improve agricultural traits of crop plants. Biotechnology Advances, 31, 97–117. 10.1016/j.biotechadv.2011.12.003 [DOI] [PubMed] [Google Scholar]
- Zhao, Q. , Nakashima, J. , Chen, F. , Yin, Y. , Fu, C. , Yun, J. , … Dixon, R. A. (2013). Laccase is necessary and non‐redundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. The Plant Cell, 25, 3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Christensen, S. K. , Fankhauser, C. , Cashman, J. R. , Cohen, J. D. , Weigel, D. , & Chory, J. (2001). A role for flavin monooxygenase‐like enzymes in auxin biosynthesis. Science, 291, 306–309. 10.1126/science.291.5502.306 [DOI] [PubMed] [Google Scholar]
- Zou, B. H. , Yang, D. L. , Shi, Z. Y. , Dong, H. S. , & Hua, J. (2014). Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis . Plant Physiology, 165, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


