Abstract
Organogenesis involves integration of myriad cell types, and dysregulation of cellular gene networks results in birth defects, affecting 5 per cent of live births. Congenital heart defects (CHD) are the most common malformations and result from disruption of discrete subsets of cardiac progenitor cells1, yet the transcriptional changes in individual progenitors that lead to organ-level defects remain unknown. Here, we employed single-cell RNA sequencing (scRNA-seq) to interrogate early cardiac progenitor cells as they become specified during normal and abnormal cardiogenesis, revealing how dysregulation of specific cellular sub-populations has catastrophic consequences. A network-based computational method for scRNA-seq that predicts lineage-specifying transcription factors2,3 identified Hand2 as a specifier of outflow tract cells but not right ventricular cells, despite failure of right ventricular formation in Hand2-null mice4. Temporal single-cell transcriptome analysis of Hand2-null embryos revealed failure of outflow tract myocardium specification, whereas right ventricular myocardium was specified but failed to properly differentiate and migrate. Loss of Hand2 also led to dysregulation of retinoic acid signaling and disruption of anterior-posterior patterning of cardiac progenitors. This work reveals transcriptional determinants that specify fate and differentiation in individual cardiac progenitor cells, and exposes mechanisms of disrupted cardiac development at single-cell resolution, providing a framework to investigate congenital heart defects.
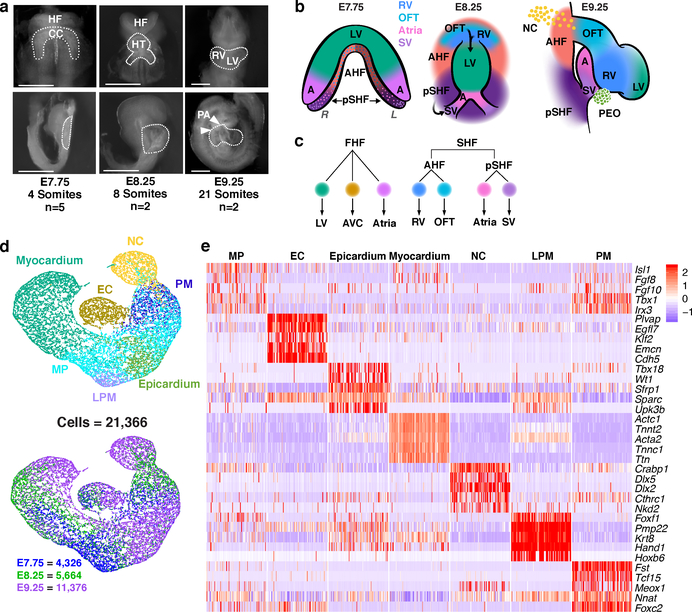
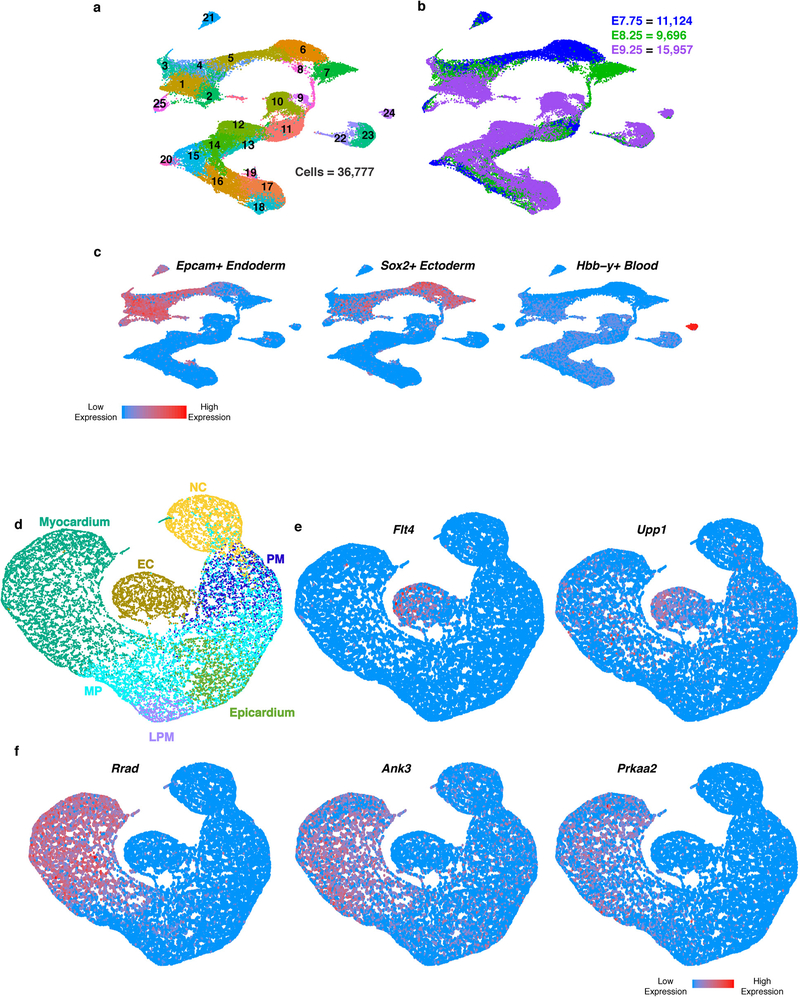
The heart develops from diverse cell lineages specified from two pools of mesodermally-derived cardiac progenitor cells (CPCs), the first and second heart fields (FHF, SHF), and from multipotent neural crest cells1. Genetic analyses, are revealing mutations that contribute to CHD5, and the next challenge is to identify specific cell types affected by such mutations. To address this challenge, we identified transcriptional features of cardiac cell specification and morphogenesis by sequencing over 36,000 individual cells collected from the cardiogenic region of mouse embryos at three developmental stages: 1) as CPCs begin to differentiate in the late cardiac crescent at embryonic day (E) 7.75; 2) as the FHF forms a linear heart tube and the SHF migrates into the anterior and posterior poles of the tube (E8.25); and 3) as the heart tube loops and incorporates the SHF-derived outflow tract (OFT), right ventricle (RV), sinus venosus (SV) and atrial cells with the FHF-derived left ventricle (LV), atrial and atrioventricular canal (AVC) cells (E9.25) (Fig. 1a–c; Extended Data Fig. 1a-c; Supplementary Table 1). Among these, transcriptomes of 21,366 mesoderm and neural crest cells were partitioned into 7 broadly defined populations6: multipotent Isl1+ progenitors, endothelial/endocardial cells, epicardium, myocardium, neural crest, paraxial mesoderm and lateral plate mesoderm (Fig. 1d, e; Supplementary Table 2). Several genes with de novo mutations identified through whole-exome sequencing of CHD probands and parents5 had expression specific to or enriched in one of these populations (Extended Data Fig. 1d–f).
Figure 1: Single-cell RNA-seq reveals heterogeneity of cardiogenic regions during early embryonic development.
a, Representative images of mouse embryos at E7.75, E8.25 and E9.25 used for cell collection, with micro-dissected regions indicated, in frontal view (top) and right sagittal view (bottom). Scale bar, 200 μm. Single-cell experiments were repeated with n=5 biologically independent embryos at E7.75, and n=2 biologically independent embryos at E8.25 and E9.25; similar results were obtained for embryos collected at the same developmental stage. b, Spatial organization of captured cardiac cell populations at each stage: frontal view at E7.75 and E8.25; right sagittal view at E9.25. Darker shaded region on left side of SHF at E7.75 indicates left-right asymmetric patterning. c, Lineage relationships between myocardial subtypes and progenitor domains d, UMAP plot of all captured mesodermal and neural crest populations colored by cluster identity and embryonic stage of collection. e, Expression heatmap of 5 marker genes of broadly defined populations. Statistics for differential gene expression tests were applied to n = 21, 366 cells. Data are shown for 100 cells subsampled from each population. Scale indicates Z-scored expression values. HF, head folds; CC, cardiac crescent; HT heart tube; RV, right ventricle; LV, left ventricle; PA, pharyngeal arches; FHF, first heart field; SHF, second heart field; AHF, anterior heart field; pSHF, posterior second heart field; NC, neural crest cells; OFT, outflow tract; AVC, atrioventricular canal; SV, sinus venosus; A, Atria; PEO, proepicardial organ containing epicardial cells. MP, multipotent progenitors; EC, endocardium/endothelial cells; PM, paraxial mesoderm; LPM, lateral plate mesoderm.
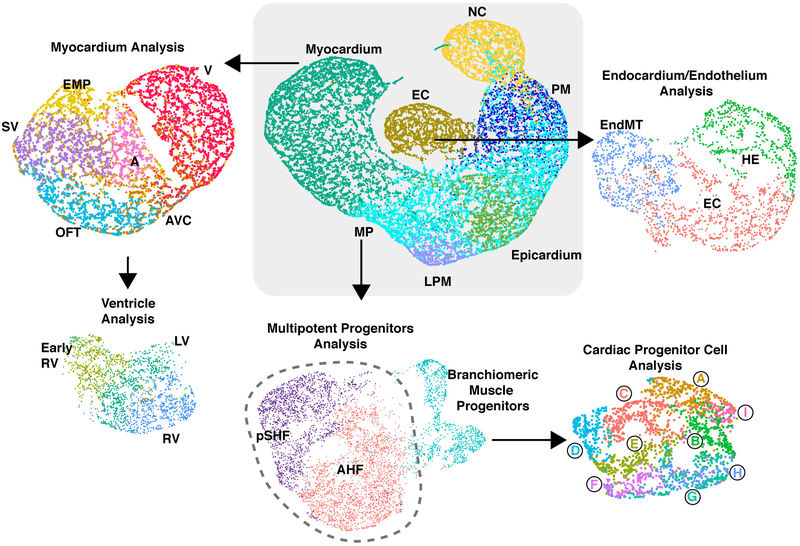
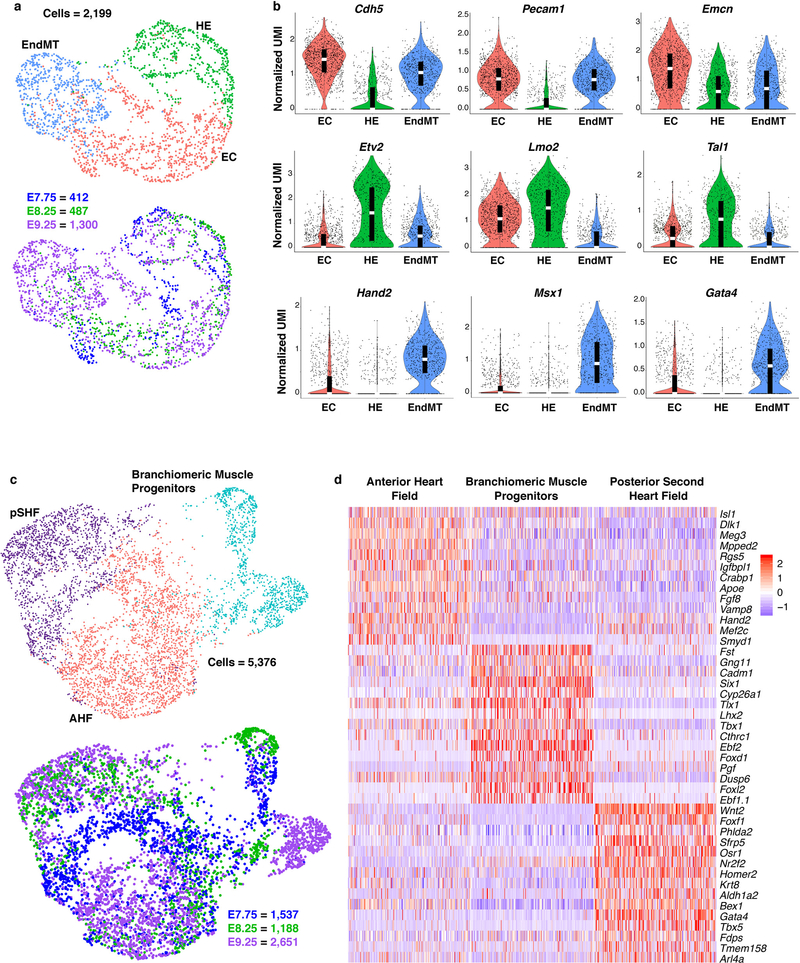
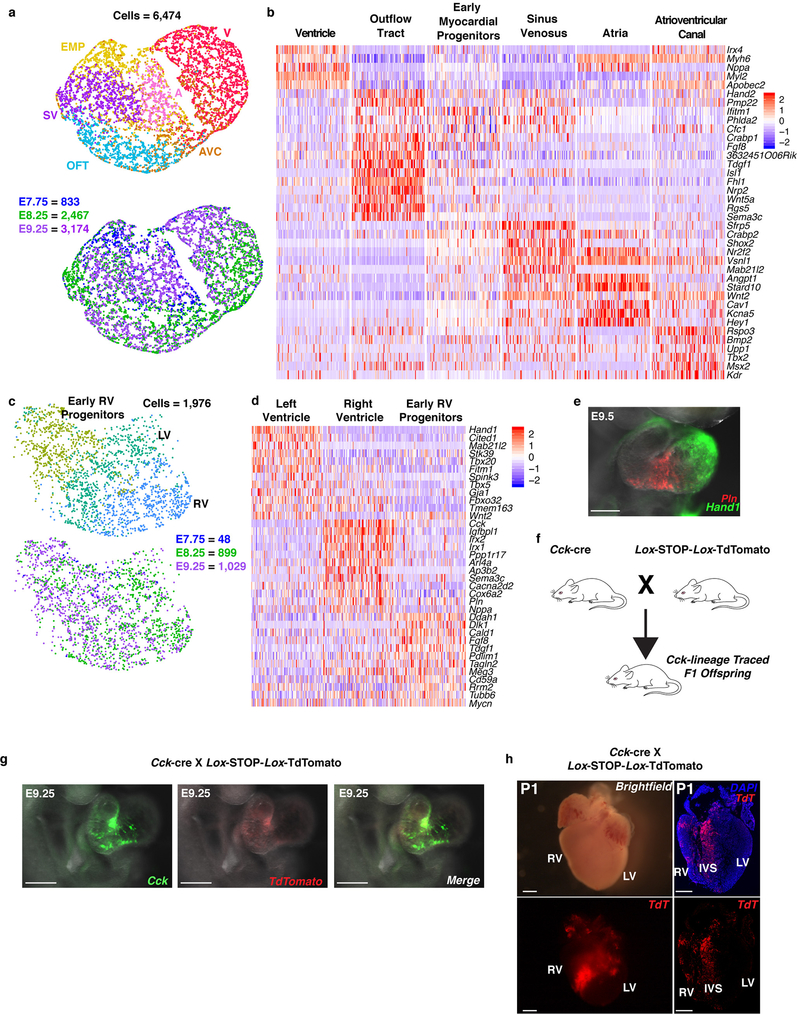
Within each broad population, further transcriptome analyses revealed distinct cell types characteristic of unique progenitor pools (Extended Data Fig. 2), which we validated and resolved spatially by in situ hybridization of specific marker genes (Extended Data Fig. 3). Three subpopulations of the endocardial/endothelial lineage emerged: hematoendothelial progenitors, specified endothelial/endocardial cells and endocardial cells initiating an endothelial-to-mesenchymal transition program typical of valve development (Extended Data Fig. 4a, b). The Isl1+ multipotent progenitor population segregated into the anterior and posterior domains of the SHF (Anterior Heart Field [AHF] and posterior Second Heart Field [pSHF], respectively), and the branchiomeric muscle progenitors that share a common origin with SHF cells in the cardiopharyngeal mesoderm (Extended Data Fig. 4c, d). Moreover, we identified transcriptomes representative of ventricular, atrial, SV, AVC and OFT myocardium (Extended Data Fig 5a, b). This dataset therefore represents a catalog of cardiac cell states that arise during embryonic development, providing a foundation to study the transcriptional dynamics underlying specification of multiple cardiac subtypes, and to uncover novel subtype-specific genes (Supplementary Table 2).
While the left and right ventricles perform similar functions, they arise from distinct progenitors, and dysregulation of genes uniquely enriched in each population leads to chamber-specific congenital malformations1. Focusing on ventricular (V) progenitors identified LV-fated cells, an early, less differentiated RV progenitor, and a more differentiated RV pool of cells (Extended Data Fig. 5c, d). Several genes were enriched in only one chamber, consistent with the ventricles’ distinct origins and physiology (Supplementary Table 2). Surprisingly, phospholamban (Pln), a critical regulator of calcium handling, and cholecystokinin (Cck), an intestinal hormone, were predominantly expressed in RV cells, which we confirmed with in-situ hybridization (Extended Data Fig. 5d–g). Lineage tracing of Cck-expressing cells and their progeny using a constitutive Cck-ires-Cre transgenic mouse crossed with a floxed TdTomato reporter mouse established their specific contribution to the RV, particularly the trabecular myocardium and interventricular septum (IVS) post-natally (Extended Data Fig. 5f–h).
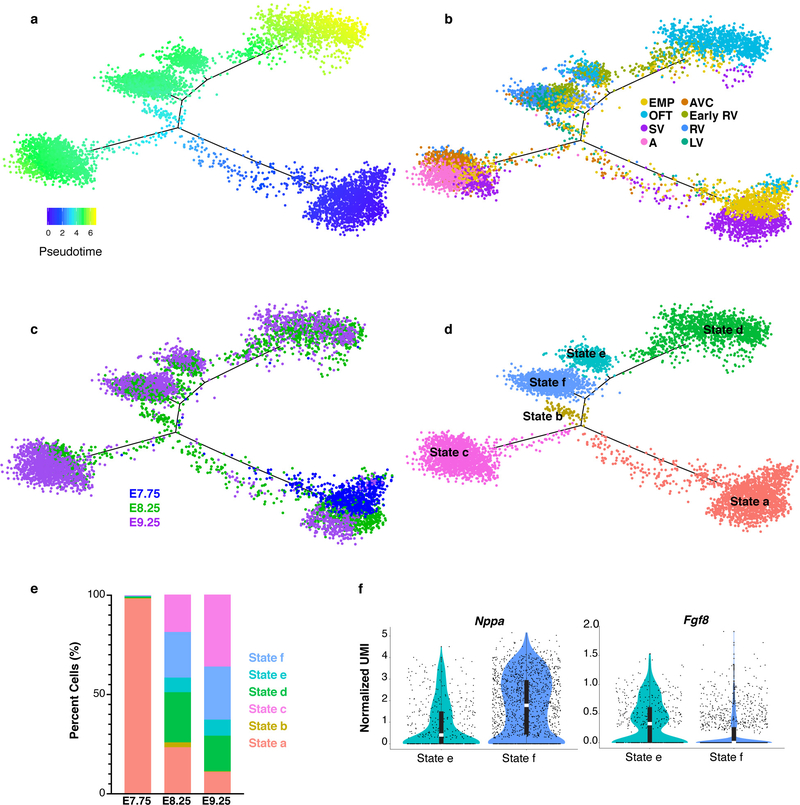
A myocardial population that we could not ascribe to a known cell type comprised a large proportion of E7.75 cells and expressed genes of multiple subtypes. We used pseudotemporal ordering to test the hypothesis that this cluster represents a mixed population of early myocardium progenitors (EMP) (Extended Data Fig. 6a–e)7. This analysis placed the majority of EMP cells at the start of the pseudotime trajectory, while EMP cells from later time points were represented in all other myocardial cell states, consistent with our hypothesis. Additionally, the early RV progenitor cells segregated with the OFT state (State d), differentiated LV/RV state (State f) and an intermediate state (State e) that expressed lower levels of differentiated ventricle genes, such as Nppa and higher levels of Fgf8, supporting their designation as early RV progenitors (Extended Data Fig. 6b, d, f; Supplementary Table 2).
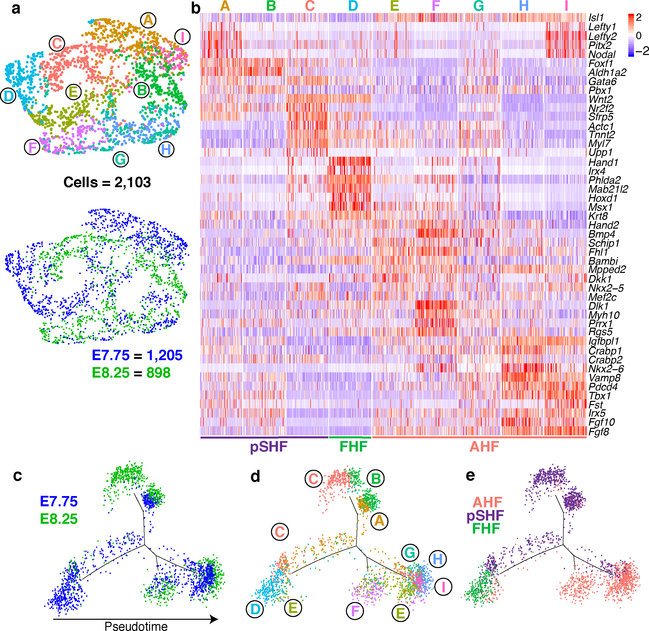
The large number of early CPCs in our dataset compared to previous single-cell CPC analyses8–11, allowed us to characterize the heterogeneity of this compartment. AHF and pSHF populations captured at E7.75 and E8.25 segregated into nine subpopulations (Fig. 2a, b; Supplementary Table 2). Populations A-C were of pSHF origin, while E-I were of AHF origin. Clusters A and I were derived from E7.75 and the majority of cells in each population co-expressed the left-sided genes Nodal, Lefty2 and Pitx2, revealing transcriptomes of left-right asymmetric cells of the AHF and pSHF (Supplementary Table 2). Population D had enriched expression of FHF, LV and sarcomeric genes with minimal Isl1 expression, features indicative of early FHF progenitors, while population F appears to represent the earliest outflow tract cells (Fig. 2b). Pseudotime analysis suggests that pSHF clusters A and B, AHF clusters E and F, and AHF clusters H and I, represent sequential stages of three differentiating populations (Fig. 2c–e). In contrast, pSHF cluster C encompasses a continuum of cells that are differentiating to myocardium (Fig. 2b, d).
Figure 2: Analysis of cardiac progenitor cell populations reveals early specification dynamics of myocardial subtypes.
a, UMAP plot of AHF and pSHF subclusters colored by cluster identity and embryonic stage of collection. b, Expression of indicated highly and uniquely expressed genes in subpopulations from (a). Scale indicates Z-scored expression values. Pseudotime trajectory of CPCs colored by c, embryonic stage of collection, d, population identity and e, AHF, pSHF or FHF origin.
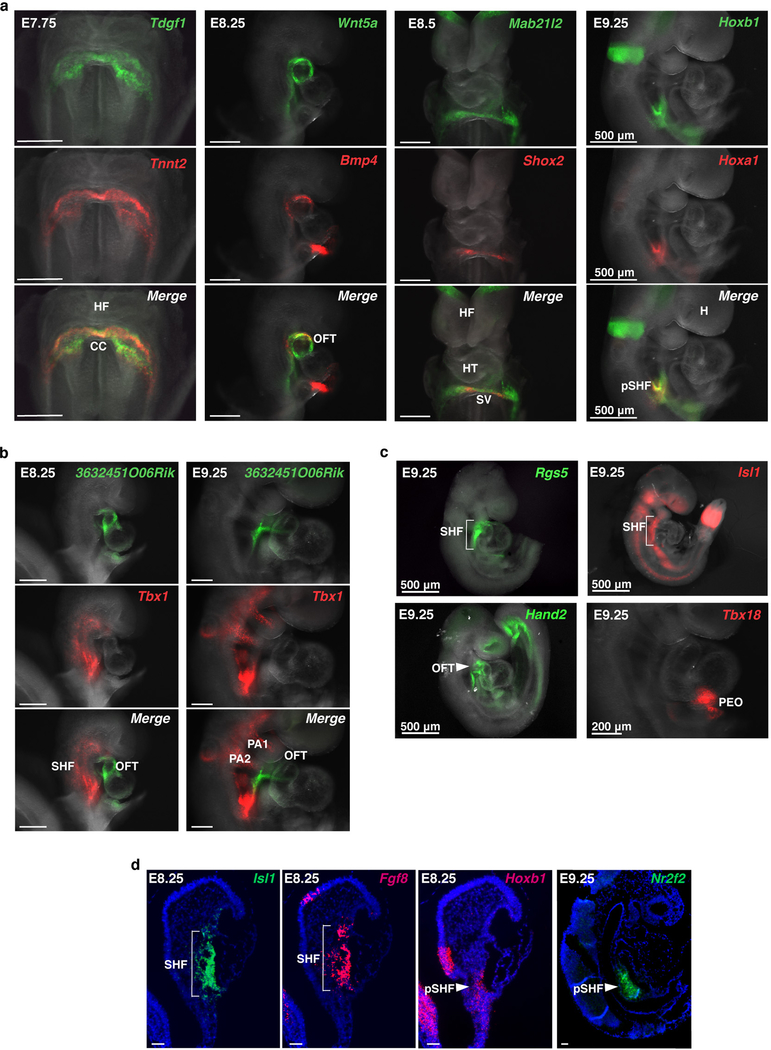
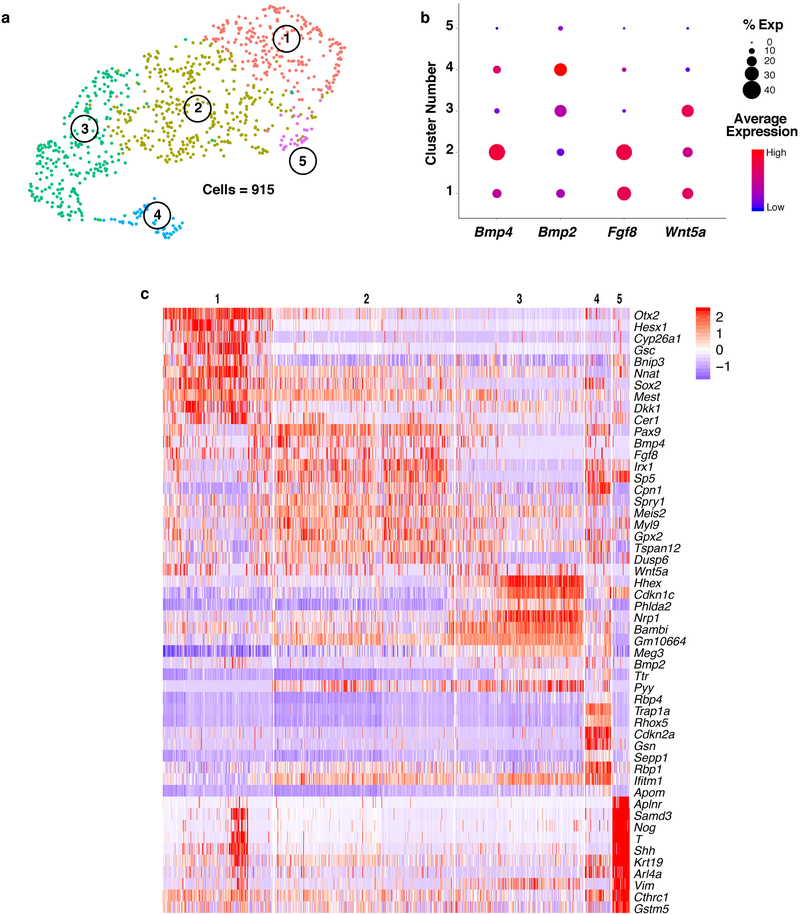
As CPCs form the cardiac crescent, their cardiac gene expression is driven by FGF and BMP signals secreted from the adjacent anterior endoderm1. Analysis of definitive endoderm cells captured at E7.75 revealed enriched expression of cardiac inducers Bmp2, Bmp4 and Fgf8 in specific subpopulations, suggesting CPCs receive combinatorial inductive signals from distinct endodermal domains (Extended Data Fig. 7; Supplementary Table 2). One endodermal cluster (Cluster 5) expressed genes characteristic of mesendoderm, while another (Cluster 1) may represent cardiogenic anterior visceral endoderm, revealing the previously undescribed transcriptomes of these specific endodermal populations. Notably, the gene Wnt5a was enriched in multiple clusters that co-expressed Bmp4, Bmp2 and Fgf8 (Extended Data Fig. 7b). It is intriguing to consider whether the spatial positioning of these endoderm populations relative to the adjacent cardiac mesoderm determines myocardium subtype specification.
The molecular regulators governing the progressive specification decisions of cardiac progenitors into distinct sub-lineages are largely unknown. We applied a Boolean network-based lineage specifier prediction method2,3 to AHF, RV and OFT cells captured at E7.75 and E8.25 and identified Irx4 and Plagl1 as specifiers of RV cells, and Hand2, Tead2 and Arid3b as OFT cell fate determinants. Irx4 is an established specifier of ventricular identity1, and Arid3b is important for SHF progenitor deployment in the OFT12, serving as positive controls for this analysis. Although Hand2 expression was enriched in the OFT myocardium (Extended Data Fig. 5b), it was curious that Hand2 was predicted as a lineage specifier for the OFT, but not RV, myocardium, as its global deletion causes severe RV hypoplasia4. This phenotype is recapitulated upon specific deletion of Hand2 in the SHF, underscoring its requirement in this progenitor compartment, although the mechanism has been elusive13.
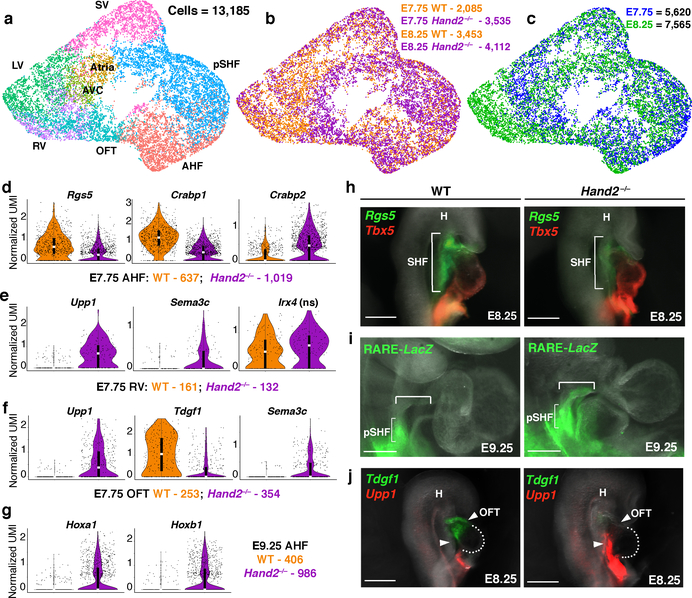
To resolve the discrepancy of the predicted lineage-specifying function of Hand2 in the OFT, but not RV, and the morphologic loss-of-function consequence, we analyzed single-cell transcriptomes of WT and Hand2-null cardiac progenitors captured at E7.75 and E8.25 (Fig. 3a–c; Supplementary Table 3). AHF, OFT and RV precursors in Hand2-null embryos were transcriptionally dysregulated as early as E7.75 (Fig. 3d–g; Extended Data Fig. 8a–c), well before any morphologic defect. For instance, the Rgs5 gene, which we identified as a marker of the AHF and OFT, was downregulated in Hand2-null AHF and OFT cells at E7.75 (Fig. 3d, h; Extended Data Fig. 8b, d). The chromatin remodeling gene Smyd1 was downregulated in Hand2-null AHF cells at E8.25, consistent with the observation that Smyd1 loss phenotypically mimics that of Hand2 loss14 (Extended Data Fig. 8e).
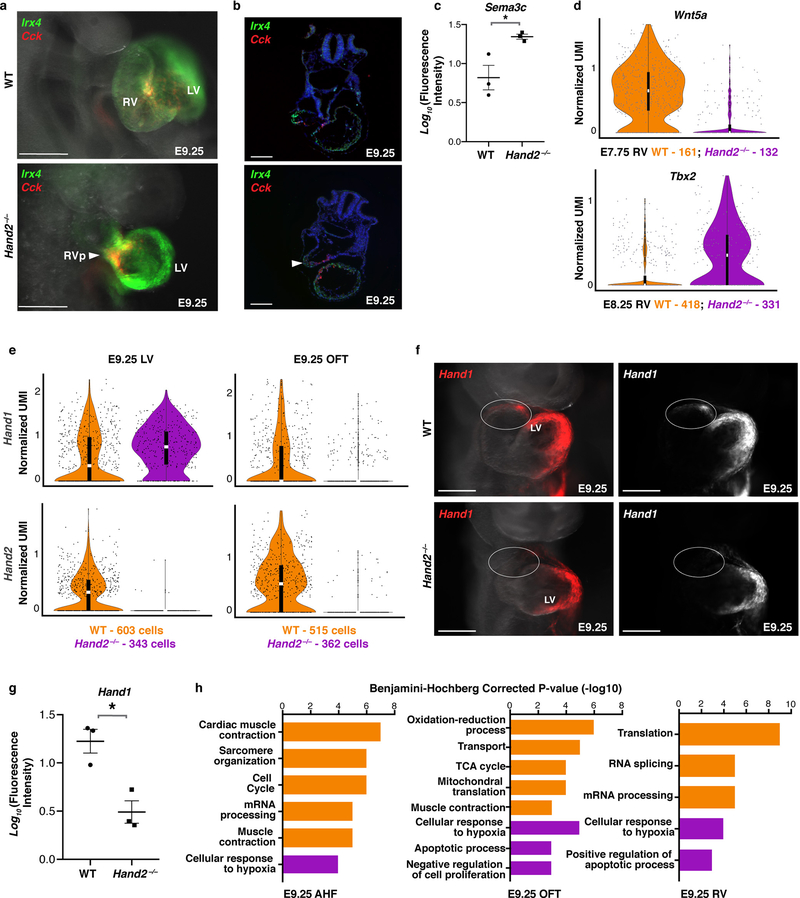
Figure 3: Transcriptional dysregulation in Hand2-null embryos reveals ectopic retinoic acid signaling with posteriorization of AHF-derivatives.
a, UMAP plot of WT and Hand2-null cells colored by cluster, b, genotype and c, embryonic stage of collection. Violin plots showing expression of genes in WT and Hand2-null AHF, RV and OFT cells at E7.75 (d-f) and AHF cells at E9.25 (g). ns, not significant. All genes represented have a Bonferroni correction adjusted p-value < 1×10–4 (Wilcoxon rank sum test, two-sided). h, mRNA in situ hybridization for expression of Rgs5 and Tbx5 at E8.25. i, Expression of lacZ transgene driven by retinoic acid response element (RARE) validated at E9.25 by whole-mount in situ hybridization focused on cardiac region. Bracketed region indicates ectopic RARE activity in Hand2-null embryos compared to WT. j, mRNA in situ hybridization for expression of Tdgf1 and Upp1 at E8.25. All scale bars, 200 μm. n=3 independent experiments with similar results for h, i and j, shown in right lateral views. pSHF, posterior second heart field; SHF, second heart field; H, head; OFT, outflow tract. Violin plot summary statistics: center white line represents median gene expression and central black rectangle spans the first quartile to the third quartile of the data distribution. The whiskers indicate value at 1.5x interquartile range above the third quartile or below the first quartile.
SHF cells undergo a binary decision to adopt an anterior or posterior lineage identity8, and we investigated whether this decision was disrupted upon Hand2 loss. Crabp1 and Crabp2 are opposing regulators of retinoic acid (RA) signaling, which is involved in posteriorization of SHF progenitors, resulting in an atrial-like fate15. Crabp1, normally high in the AHF, sequesters RA to facilitate its catabolism, whereas Crabp2, which promotes RA nuclear transport and subsequent transcriptional activation16, is low in AHF cells. In Hand2-null AHF cells, Crabp1 was downregulated, while Crabp2 was upregulated, which would be expected to cause ectopic RA signaling and AHF posteriorization (Fig. 3d). Hand2-null embryos expressing a LacZ reporter driven by an RA response element (RARE-LacZ) revealed ectopic LacZ expression in the AHF progenitors extending into the cardiac outflow tract region (Fig. 3i, Extended Data Fig. 8d). In agreement, Hoxa1 and Hoxb1, established RA transcriptional targets15 and pSHF markers8, were upregulated in Hand2-null AHF cells at E9.25 (Fig. 3g; Extended Data Fig. 8d, f). Upp1, a gene typically expressed in pSHF derivatives, was also ectopically expressed in Hand2-null OFT and RV cells (Fig 3e, f, j; Extended Data Fig. 8d). Notably, the proportions of AHF and pSHF cells captured at E7.75 were higher in Hand2-null mutants than in WT embryos, likely reflecting more proliferative, less differentiated SHF cells (Extended Data Fig. 8g).
In addition to this posteriorized gene signature, the AHF and its derivatives displayed broader dysregulation. Sema3c, which is expressed in later E8.5-E9.0 OFT myocardium to attract PlexinA2-positive neural crest cells17, was ectopically expressed in the Hand2-null AHF at E8.25, and OFT and RV progenitors at E7.75 (Fig. 3e, f; Extended Data Fig. 8e). The OFT marker gene Tdgf1 was downregulated in Hand2-null OFT cells at E7.75 and almost absent at E8.25 (Fig. 3f, j; Extended Data Fig. 8d). In contrast, WT and Hand2-null RV cells had equivalent expression levels of Irx4 at E7.75 suggesting Hand2 loss did not prevent acquisition of ventricular identity (Fig. 3e). However, markers of differentiation towards working myocardium were downregulated in Hand2-null RV cells at E8.25, indicating that these cells were in fact dysregulated after specification (Extended Data Fig. 8h).
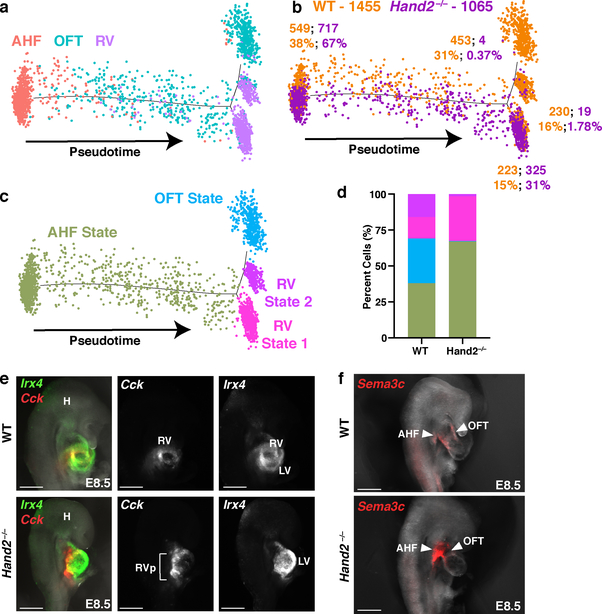
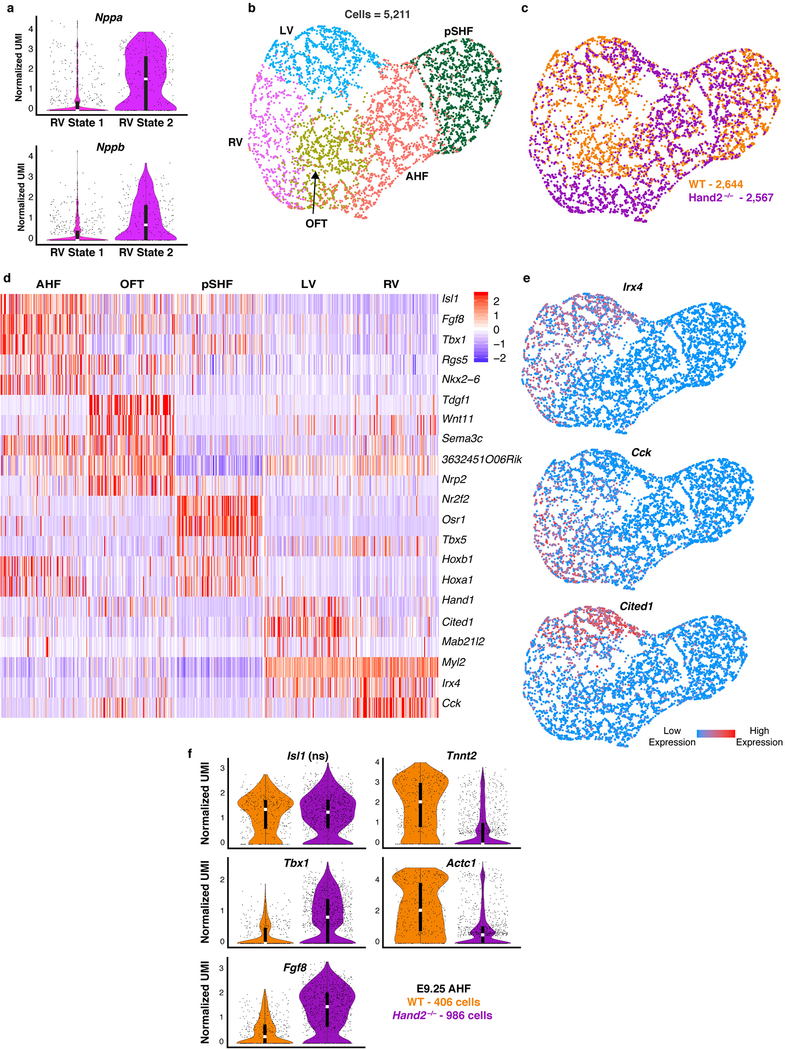
To test the prediction that Hand2 specifies OFT cells, we ordered WT and Hand2-null AHF, OFT and RV cells captured at E8.25 in pseudotime (Fig. 4a–c). The resulting trajectory began with AHF cells and split into three cell states: one OFT state, and two distinct RV states. Hand2-null cells were severely depleted in the OFT state, and while RV State 1 comprised both WT and Hand2-null cells in comparable numbers, WT cells predominated in RV State 2 (Fig. 4b, d). Differential expression analysis between the RV states indicated that maturation genes such as Nppa and Nppb were highly expressed in the WT-enriched State 2 (Extended Data Fig. 9a; Supplementary Table 3). These data suggest that Hand2-null OFT-fated cells had disrupted specification, while RV-fated cells were appropriately specified but had differentiation defects, consistent with the lineage-specifier analysis. In agreement, cardiac transcriptomes from E9.25 indicated the presence of an RV population that comprised Hand2-null cells in comparable numbers to WT cells (Extended Data Fig. 9b–e; Supplementary Table 3), despite the absence of the RV chamber in Hand2 mutants. Furthermore, Hand2-null AHF cells at E9.25 failed to upregulate differentiation genes and continued to express progenitor markers consistent with a differentiation defect (Extended Data Fig. 9f).
Figure 4: Hand2 loss disrupts outflow tract myocardial cell specification as well as right ventricle myocardial cell differentiation and migration.
a, Pseudotime trajectory of AHF, OFT and RV cells at E8.25 colored by cluster identity, b, genotype and c, cell state. n = 2 biologically independent embryos per genotype. Numbers in b indicate absolute number of cells of each genotype in each cell state. Percentages indicate proportions of cells per genotype in each state, graphically represented in (d). e, Expression of Cck and Irx4 by in situ hybridization in right lateral view to localize RV cells at E8.5 and corresponding fluorescence image for Cck or Irx4 expression alone. n=2 independent experiments with similar results. Scale bar, 200 μm. f, Expression of Sema3c to localize AHF cells and derivatives at E8.5 in right lateral view. n = 3 independent experiments with similar results. Scale bar, 200 μm. H, head; RV, right ventricle; RVp, right ventricle progenitors; LV, left ventricle; AHF, anterior heart field; OFT, outflow tract.
In-situ hybridization demonstrated that Irx4+/Cck+ RV cells were present in Hand2-null embryos but were located in the AHF area behind the LV at E8.5 and in the area of the OFT at E9.25 (Fig. 4e; Extended Data Fig. 10a, b). Moreover, in Hand2-null embryos at E8.5, Sema3c-positive RV cells accumulated in the pharyngeal mesoderm behind the heart instead of populating the cardiac outflow tract (Fig. 4f; Extended data Fig 10c; Supplementary Videos 1–4). Wnt5a, which promotes AHF cell migration18, was downregulated, while Tbx2 expression, which impairs AHF migration19, was upregulated in Hand2-null RV-specified cells (Extended Data Fig. 10d), in agreement with their arrested migration.
At E9.25, Hand1, partially redundant with Hand220, was selectively downregulated in Hand2-null OFT, but not LV, progenitors (Extended Data Fig. 10e–g). Thus, Hand2-null OFT progenitors lacked Hand1 and Hand2, which may explain the failure of OFT specification. Moreover, the AHF, OFT, and RV Hand2-null cells more highly expressed genes controlling apoptosis and the response to hypoxia (Extended Data Fig. 10h), despite no overt signs of heart failure such as cardiac edema.
In this study, we used scRNA-seq to reveal mechanisms of normal organogenesis and how regulatory defects in discrete cell subsets can lead to morphologic developmental defects. The ability to acquire quantitative and spatial resolution of large numbers of individual transcriptomes allowed us to dissect functional deficits such as improper differentiation and migration versus disrupted fate specification in distinct cell types in response to loss of a single transcription factor, Hand2. Single-cell transcriptomics therefore offers a potent strategy to more effectively determine the precise mechanisms and cell types underlying phenotypic presentation of developmental defects associated with genetic variation. Meticulous dissection of transcriptional landscapes during organogenesis is a prerequisite to defining preventive approaches for birth defects and potential post-natal intervention for ongoing sequelae of human malformations.
Methods
Animal models
Animal studies were conducted in strict compliance with all relevant ethical regulations in the animal use protocols, UCSF animal use guidelines and the NIH Guide for the care and Use of Laboratory Animals. All protocols concerning animal use were approved by the Institutional Animal Care and Use Committee (IACUC) at UCSF and were accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Transcriptomes were captured from wildtype (WT) and Hand2-null embryos from intercrossed C57BL/6 mice heterozygous for the Hand2-null allele4. The sexes of all embryos used for capture of single-cell transcriptomes are listed in Supplementary Table 1a. Lineage tracing of Cck expressing cells was performed using Cck-ires-Cre (JAX stock #012706)21 and Ai14 (JAX stock #007914)22 mice. Validation of ectopic RA signaling in the Hand2 mutant was done by crossing the mutant line to RARE- hsp68LacZ mice (JAX stock #008477)23.
Timed matings between male (8–10 weeks of age) and female (6–8 weeks of age) mice were set up where noon on the day of plug detection was considered E0.5. Pregnant females were identified by echocardiography performed at E6.5 and sacrificed to harvest embryos at E7.75, E8.25 and E9.25 for scRNA-seq and at E7.75, E8.25, E8.5 and E9.25 for whole mount and section-based in-situ hybridization experiments. Transcriptomes from at least 2 embryos were collected per embryonic stage, per genotype. The sample sizes of embryos used for single-cell transcriptome analysis at each time point was chosen to obtain cell numbers comparable to estimated cell numbers in the cardiogenic region at each embryonic stage. Embryos were developmentally matched at each time point by somite count (4, 8, and 21 somites for E7.75, E8.25, and E9.25, respectively).
Randomization was not implemented; experimental groups were determined by genotype i.e., Hand2 wild type embryos were compared to Hand2-null embryos. Covariates were not relevant to the analysis of the Hand2-null phenotype as the developmental defect is highly penetrant regardless of embryo sex, and the developmental stages analyzed were prior to the onset of overt heart failure. Investigators were not blinded to allocation of embryos during experiments. Blinding was not possible for the WT and Hand2-null embryo comparisons due to the need to match somite counts to control for developmental timing.
Embryo dissection and single-cell library generation
The entire cardiogenic region was dissected at each time point, including the SHF region that lies behind the cardiac crescent and heart tube, as well as the first and second pharyngeal arches at E9.25. Due to the small size of embryos at these stages, some surrounding tissue (indicated in Fig. 1a) encompassing the posterior lateral plate mesoderm (E7.75), head folds (E7.75 and E8.25), and endoderm, was microdissected to ensure complete retrieval of cardiac populations. Embryos were dissected in cold PBS (Life Technologies, CAT# 14190250), de-yolked and placed in PBS/1% FBS (ThermoFisher Scientific, CAT# 10439016) solution on ice until dissociation (approximately 3 hours). Yolk sac DNA was extracted (QuickExtract DNA Extraction Solution, Epicentre, CAT# QE09050) and used for genotyping to distinguish Hand2 WT and Hand2-null embryos before further microdissection of cardiac regions at each stage. Dissected cardiac tissue was incubated in 200 μl TrypLE (ThermoFisher Scientific, CAT# 12563029) for 5 min, triturated with a 200 μl pipette tip, and incubated for an additional 5 min. The TrypLE solution was quenched with 600 μl PBS/1% FBS. Cells were filtered through a 70 μm cell strainer (BD Falcon, CAT# 08–771-2), centrifuged at 150 rcf for 3 min, and resuspended in 35 μl PBS/1% FBS. Single-cell droplet libraries from this suspension were generated in the 10X Genomics Chromium controller according to the manufacturer’s instructions in the Chromium Single Cell 3′ Reagent Kit v2 User Guide. The cell capture efficiency of the Chromium controller is ~57%, thus, we loaded all cells dissected from embryos without pre-counting, to minimize cell loss and maximize the number of captured single cells. Additional components used for library preparation include the Chromium Single Cell 3′ Library and Gel Bead Kit v2 (PN-120237) and the Chromium Single Cell 3′ Chip kit v2 (PN-120236).
Single-cell transcriptome library preparation and sequencing
Libraries were prepared according to the manufacturer’s instructions using the Chromium Single Cell 3′ Library & Gel Bead Kit v2 (PN- 120237) and Chromium i7 Multiplex Kit (PN-120262). Final libraries were sequenced on the NextSeq 500 and Hiseq 4000. Somite-matched WT and Hand2-null replicate libraries from each litter were pooled and sequenced in the same lane. Sequencing parameters were selected according to the Chromium Single Cell v2 specifications. All libraries were sequenced to a mean read depth of at least 50,000 total aligned reads per cell.
Processing of raw sequencing reads
Raw sequencing reads were processed using the Cell Ranger v2.2.0 pipeline from 10X Genomics. Briefly, reads were demultiplexed, aligned to the mouse mm10 genome and UMI counts were quantified per gene per cell to generate a gene-barcode matrix. Data from multiple samples (WT only analysis, WT/Hand2-null E7.75/E8.25 and WT/Hand2-null E9.25 analyses) were aggregated and normalized to the same sequencing depth, resulting in a combined gene-barcode matrix of all samples.
Cell filtering and cell-type clustering analysis
We sequenced the transcriptomes of 36,777 cells captured from WT and 37,149 cells captured from Hand2-null embryos in total. Further filtering and clustering analyses of these cells were performed with the Seurat v2.2 R package, as described in the tutorials (http://satijalab.org/seurat/)6. For each aggregated dataset (WT only, WT/Hand2-null E7.75/E8.25, WT/Hand2-null E9.25), cells were normalized for genes expressed per cell and total expression, then multiplied by a scale factor of 10,000 and log-transformed. Cells that were of low quality or represented doublets were excluded from our analyses - this was achieved by filtering out cells with greater than 8000 and fewer than 1500 genes in Seurat. We then performed a linear regression on all genes to eliminate technical variability due to the number of genes detected, embryonic time point, embryo replicate and stage of the cell cycle (ScaleData Function). For the Hand2-null analyses, we also regressed out the Hand2 gene to eliminate its contribution to cell clustering. Highly variable genes in the dataset were computed and used as input for Principal Component Analysis. Significant PCs were used for downstream graph-based, semi-unsupervised clustering into distinct populations (FindClusters Function) and Uniform Manifold Approximation and Projection (UMAP)24 dimensionality reduction was used to project these populations in 2D. For clustering, the resolution parameter, which indirectly controls the number of clusters, was approximated based on the number of cells according to Seurat guidelines; a vector of resolution parameters was passed to the FindClusters function and the optimal resolution that established discernible clusters with distinct marker gene expression was selected. One or two cell clusters would emerge that expressed marker genes representing multiple populations; these contained cells with low UMI and gene counts that escaped the first filtering step. These cells were removed from the analyses. To identify marker genes, the clusters were compared pairwise for differential gene expression using the Wilcoxon rank sum test for single-cell gene expression (FindAllMarkers function, min.pct = 0.25, min.diff.pct = 0.1, return.thresh (p-value cut-off) = 1×10–4). To assign identities to these subpopulations, we cross-referenced their marker genes with known cardiac subtype markers and in situ hybridization data from the literature (Supplementary Table 2). We also validated several of these marker genes by fluorescence in situ hybridization (Extended Data Fig. 3). We removed blood, endoderm-, and ectoderm- derived clusters based on their expression of known blood markers such as the hemoglobin genes, endoderm markers such as Epcam and Foxa2, and ectoderm markers such as Pou3f1 and Sox2 (Extended Data Fig. 1c), retaining the cells of mesodermal or neural crest identity. The clustering approach was then repeated for these retained mesodermal and neural crest cells, beginning with the regression of technical variables, identification of highly variable genes, Principal Component Analysis, graph-based clustering, UMAP projection and marker analysis. Similarly, all reclustering analyses (Extended Data Fig. 2) were processed as described above. For computing differentially expressed genes between Pitx2-positive (normalized UMI >0.1) and Pitx2-negative (normalized UMI < 0.1) cells in clusters A and I (Fig. 2a) and for the Hand2-null analyses, the FindMarkers function was used on WT and Hand2-null cells from each cluster with the following parameters: Wilcoxon rank sum test, min.diff.pct = 0.1, min.pct = 0.25, logFC.threshold = 0.2. Additionally, the max.cells.per.ident argument was used to ensure that equivalent numbers of cells were considered in the analysis. For each population analyzed, the number of cells (n) given to this argument was set to the population/genotype that had the lower cell number. An adjusted p-value (Bonferroni Correction) cut-off < 1×10–4 was used to identify differentially expressed genes.
Prediction of cell fate determinants
Cell fate determinants for OFT and RV from the AHF were predicted using a modified version of the method that we previously developed2,3. This procedure was performed on E7.75 and E8.25 WT OFT, RV and AHF cells. One hundred cells were randomly selected from the AHF and from the OFT and RV daughter populations and the normalized ratio difference (NRD) was computed for all combinations of these 100 cells, yielding 10,000 parent-daughter cell combinations. The NRD was calculated for all pairs of differentially expressed TFs between OFT, RV and AHF cells and averaged over the 10,000 cell combinations. We minimized the impact of zero inflation by relying on the highly stringent adjusted (Bonferroni) p-value cutoff (< 1×10–4) and by filtering out very lowly expressed genes in defining differentially expressed TFs (average expression >= 0.5 normalized UMI). We filtered out genes that were detected in less than 25% of cells in populations being compared, and only tested genes that show a minimum difference of 0.1 in the fraction of detection between the two groups. TF pairs whose mean NRD was more than 0.05 in one lineage direction but less than 0.01 in the other lineage direction were selected. Finally, the TF pairs that resided in the strongly connected component of the GRN were kept as the final candidate cell fate determinants.
Cell trajectory analysis
Pseudotime analyses were performed using the Monocle 2 package, as described in the tutorials (http://cole-trapnell-lab.github.io/monocle-release/)7. Differentially expressed genes, as determined in Seurat using the FindAllMarkers function, between the myocardium, CPC and WT/Hand2-null AHF, OFT and RV cells were used as input for temporal ordering of these cells along the differentiation trajectory.
In Situ hybridization experiments
Each in situ hybridization experiment was replicated at least twice for identifying spatial expression of genes and three times for quantification of in situ signal for differentially expressed genes in the Hand2-null analysis. For whole-mount experiments: de-yolked whole embryos were fixed in a 4% formaldehyde solution (ThermoFisher Scientific, CAT# 28906) overnight at 4°C followed by 2X PBST washes and 5-minute incubations in a dehydration series of 25%, 50%, 75% and 100% methanol (Fisher Scientific, CAT# A454–1). At this point embryos were stored in 100% methanol at −20°C until the in situ protocol was initiated. Yolk sac DNA was used for genotyping. The whole-mount in situ assay was adapted from the protocol formulated for whole-mount zebrafish embryos25 using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics, CAT# 323100), with minor modifications (the air-drying step was excluded in our protocol, Protease Plus was used for embryo permeabilization, and the 0.2X SSCT wash step between reagent incubations was reduced to 3X 8 mins). Whole-mount embryos were imaged in 0.1% PBST using the Leica M165 fluorescent dissecting scope (FC/PLANAPO 1.0x #10450028; Camera – DFC3000G; ET GFP #10447408, ET mCHER #10450195, Acquisition Software – LAS V4.6). Quantification of transcript signal from whole-mount WT and Hand2-null embryos was performed using ImageJ v1.51m9. The mean grey value and integrated density of a defined area, that was kept consistent between WT and mutant embryos, as well as the background fluorescence level for the same defined area per embryo was measured. The corrected total fluorescence for each gene was calculated using the following formula: Integrated density – (Area x Background fluorescence). Corrected total fluorescence values were the log10 transformed before t-tests were conducted to satisfy the prerequisite assumptions of normality.
For in situ hybridization experiments performed on embryo and postnatal day 1 (p1) heart sections: embryos and p1 hearts were washed 3X in PBS after overnight fixation in 4% formaldehyde and stored in 70% ethanol (VWR, CAT# 89125–186) indefinitely until embedding. Embryos were embedded in Histogel (Thermo Scientific, CAT# R904012) and paraffin processed using standard protocols and embedded for transverse sectioning, while p1 hearts were directly processed in paraffin. Tissue slices were serially sectioned at 5 μm intervals, mounted on slides and stored at room temperature until initiation of the RNAscope protocol for paraffin embedded sections (User manual catalog number 322452-USM). Sections were imaged with a Zeiss Axio Observer.Z1 inverted epifluorescence microscope (Carl Zeiss Microscopy, Thornwood, NY) with Zeiss Axiocam MRm and PCO.edge sCMOS (PCO.Imaging, Kelheim, Germany) monochrome cameras run by Zeiss Zen imaging software. For 3D expression reconstruction, embryos were embedded in low melting agarose and imaged with a Zeiss lightsheet Z.1 selective plane illumination microscope (Carl Zeiss Microscopy, Thornwood, NY) with 488nm and 561nm lasers, tandem PCO.edge sCMOS cameras (PCO.Imaging, Kelheim, Germany) and Zeiss Zen imaging software. 3D reconstructions of multi-view images were performed using Bitplane Imaris software v.9.0.2 (Andor Technology PLC, Belfast, N.Ireland).
Catalog numbers for RNAscope probes used in this study: Cck, 402271-C3; Cited1, 432471; Hand1, 429651-C2; Irx4, 504831; Pln, 506241; Rgs5, 430181; Sema3c, 441441-C3; Tdgf1, 506411; Tbx5, 519581-C2 and Upp1, 504841-C2; Tbx1, 481911-C2; Nr2f2, 480301-C3; Hoxb1, 541861; LacZ, 313451; Tbx18, 515221-C2; Mab21l2, 456901; Wnt5a, 316791; Bmp4, 401301- C2; 3632451O06Rik, 502031; C130080G10Rik, 506051; Tnnt2, 418681-C4; Actc1, 510361-C2; Cacna2d2, 449221-C2; Tbx2, 448991-C2; Shox2, 554291-C3; TdTomato, 317041-C2.
Statistics and reproducibility
Standard statistical analyses were performed using GraphPad Prism 8. The number of replicates, statistical test used and test result are described in the figure legends. The level of significance in all graphs is represented as follow: *P<0.05 and **P<0.01. For fluorescence quantification, corrected total fluorescence values were log transformed before t-tests were conducted to satisfy the prerequisite assumptions of normality. For all quantifications, the mean +/− s.e.m is reported. No experimental samples were excluded from the statistical analyses. Sample size was not pre- determined through power calculations, and no randomization or investigator blinding approaches were implemented during the experiments and data analyses. When representative results are presented to indicate expression patterns of genes in WT embryos, at least two independent embryos were analyzed.
Data availability
All source data, including sequencing reads and single-cell expression matrices have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE126128. Data underlying each figure are available in the Source Data Files, Supplementary Information and on the UCSC cell browser at https://mouse-cardiac.cells.ucsc.edu. Users can use the cell browser to explore the data, view expression of genes of interest in each UMAP plot and download datasets for custom analysis.
Code Availability
All analyses were performed using standard protocols with previously described R packages6,7. R scripts available upon request.
Extended Data
Extended Data Figure 1: Novel genes associated with CHD are enriched in specific cardiac populations.
a, UMAP plot of all captured cell populations colored by cluster and b, embryonic stage of collection. c, UMAP feature plot showing expression of marker genes used to identify and remove endoderm (Epcam), ectoderm (Sox2) and blood (Hbb-y) cell populations. Statistics for differential gene expression tests were applied to n = 36, 777 cells. d, UMAP plot of all mesodermal and neural crest populations captured at E7.75, E8.25 and E9.25 colored by cluster identity from Fig. 1d. e, Expression of Flt4 and Upp1 in Endocardium/Endothelium population. f, Expression of Rrad, Ank3 and Prkaa2 in sub-populations of the Myocardium. Statistics for differential gene expression tests were applied to n = 21,366 cells. MP, multipotent progenitors; EC, endocardium/endothelial cells; PM, paraxial mesoderm; LPM, lateral plate mesoderm.
Extended Data Figure 2: Focused analyses of cardiac populations.
Schema of progressive subdivisions of broadly clustered cell populations from Fig. 1d that are discussed in the manuscript.
Extended Data Figure 3: Spatial validation of marker gene expression by in situ hybridization.
a, Ventral view of Tdgf1 and Tnnt2 expression in the cardiac crescent (CC), right lateral views of Wnt5a and Bmp4 in the outflow tract (OFT), Mab21l2 and Shox2 in the sinus venosus (SV), and Hoxa1 and Hoxb1 in the posterior second heart field (pSHF) by in situ hybridization at days indicated that informed assignment of population identities in Extended Data Figure 4d and 5a. b, Expression of Tbx1 in the second heart field (SHF) and pharyngeal arches (PA) and novel unannotated gene 3632451O06Rik in the OFT at E8.25 and E9.25. c, Expression of Rgs5 and Isl1 in the SHF, Hand2 in the SHF and OFT and Tbx18 in the proepicardial organ (PEO) of E9.25 embryos. Scale bars indicate 200 μm unless otherwise noted. d, In situ hybridization of mRNA expression of Isl1, Fgf8 and Hoxb1 at E8.25 and Nr2f2 at E9.25 in right lateral histologic sections. n=2 independent embryos per gene for all panels. Scale bars indicate 50 μm. HF, head fold; HT, heart tube; H, head.
Extended Data Figure 4: Heterogeneity in endocardium/endothelium and multipotent progenitor populations.
a, UMAP plot of reclustered Endocardium/Endothelium population colored by cluster and embryonic stage of collection. b, Violin plot of markers indicating distinct subpopulations of endocardium/endothelial cells. Summary statistics reported in violin plots: the center white line represents median gene expression and the central black rectangle spans the first quartile to the third quartile of the data distribution. The whiskers above or below the box indicate value at 1.5x interquartile range above the third quartile or below the first quartile. Statistics for differential gene expression tests were applied to n=2,199 cells. c, UMAP plot of reclustered multipotent progenitor populations colored by cluster and embryonic stage of collection. d, Heatmap showing curated list of marker genes that identify pSHF, AHF and branchiomeric muscle progenitors. Scale indicates Z-scored expression values. Statistics for differential gene expression tests were applied to n=5,376 cells. HE, hemato- endothelial progenitors; EC, endocardial/endothelial cells; EndMT, endothelial-mesenchymal transition cells; AHF, anterior heart field; pSHF, posterior second heart field.
Extended Data Figure 5: Focused analyses of myocardial populations and spatial validation of right ventricle markers.
a, UMAP plot of reclustered “Myocardium” population colored by cluster and embryonic stage of collection. b, Heatmap of highly and uniquely expressed genes in myocardial subpopulations. Scale indicates Z-scored expression values. Statistics for differential gene expression tests were applied to n=6,474 cells. c, UMAP plot of reclustered ventricle populations colored by cluster and embryonic stage of collection. d, Heatmap showing curated list of genes that identify left ventricle (LV), right ventricle (RV) and early RV progenitors. Scale indicates Z-scored expression values. Statistics for differential gene expression tests were applied to n=1,976 cells. e, mRNA expression of left ventricle marker Hand1 (green) and Pln (red) in frontal view of the E9.5 heart showing enrichment in right ventricle region by whole mount in situ hybridization. n=2 independent embryos per gene, Scale bar, 200 μm. f, Breeding scheme for lineage-tracing Cck expressing cells. g, mRNA expression of endogenous Cck and TdTomato driven by Cck-cre transgene at E9.25 in right oblique view of the heart. n=2 independent embryos per gene; scale bar, 200 μm. h, Expression of TdTomato in whole-mount and sectioned postnatal day 1 (P1) heart from Ai14xCck-cre lineage-traced mice showing location of progeny of Cck-expressing cells. Left panels show brightfield view (top) or TdTomato (bottom) of whole-mount P1 heart; right panels show sections of TdTomato and DAPI (top) or TdTomato alone (bottom) in P1 heart section. n=2 independent embryos. Scale bar, 100 μm. EMP, early myocardial progenitors; SV, sinus venosus; A, atria; AVC, atrioventricular canal; OFT, outflow tract; V, ventricle; RV, right ventricle; LV, left ventricle; IVS, interventricular septum.
Extended Data Figure 6: Pseudotemporal ordering of myocardium populations.
Pseudotime trajectory of myocardium populations colored by a, pseudotime value, b, cluster identity, c, embryonic stage of collection and d, cell state. Pseudotime trajectory analysis was applied to n=6,474 cells. e, Percentage of cells in each state that were captured at E7.75, E8.25 or E9.25. f, Violin plots showing expression of Nppa and Fgf8 in State e and State f from pseudotime trajectory in (d). Statistics for differential gene expression tests were applied to n = 455 cells from each state. Bonferroni correction adjusted p-value < 1×10–4 (Wilcoxon rank sum test, two-sided). Summary statistics reported in violin plots: the center white line represents median gene expression and the central black rectangle spans the first quartile to the third quartile of the data distribution. The whiskers above or below the box indicate value at 1.5x interquartile range above the third quartile or below the first quartile.
Extended Data Figure 7: Endoderm populations adjacent to the cardiac crescent.
a, UMAP plot of endoderm populations captured at E7.75 colored by cluster. b, DotPlot highlighting expression patterns of known and novel endodermal secreted factors, Fgf8, Bmp4, Bmp2, and Wnt5a. The size of the dot indicates the percentage of cells expressing that gene within a cluster (% exp), while the color encodes the average expression level of that gene within a cluster. c, Expression heatmap of the top ten marker genes of each endodermal population and secreted factors from (b). Scale indicates Z-scored expression values. Statistics for differential gene expression tests were applied to n = 915 cells.
Extended Data Figure 8: Transcriptional perturbation in Hand2-null embryos.
a, Heatmap of marker genes of populations from Fig. 3a. Statistics for differential gene expression tests were applied to n = 13,185 cells. b, Heatmap of differentially expressed genes between WT and Hand2-null OFT and c, RV cells captured at E7.75 and E8.25. Statistics were applied to n = 253 OFT cells/genotype at E7.75, n = 276 OFT cells/genotype at E8.25, n= 132 RV cells/genotype at E7.75, and n=331 RV cells/genotype at E8.25. Scale indicates Z-scored expression values. d, Quantification of fluorescence signal for indicated genes in Fig. 3h–j and Extended Data Fig. 8f. n=3 independent embryos per genotype. The mean +/− s.e.m is indicated. Two-tailed t-test. *P<0.05 and **P<0.01. e, Violin plots showing expression of Smyd1 and Sema3c in WT and Hand2-null E8.25 AHF cells. f, Expression of Hoxb1 in WT and Hand2-null embryos at E9.25 by whole mount in situ hybridization in right lateral view. Arrowheads indicate expanded anterior Hoxb1 expression in Hand2-null embryos. Scale bar, 500 μm. n=3 independent experiments with similar results. g, Proportion of WT and Hand2-null cells from each population captured at E7.75. n = 5 WT embryos and n = 3 Hand2-null embryos. The mean +/− s.e.m is indicated. Two-tailed t-test. *P<0.05 and **P<0.01. h, Violin plots showing expression of Nppa and Nppb in E8.25 WT and Hand2-null RV cells. All genes in a, b, c, e, h have a Bonferroni correction adjusted p-value < 1×10–4 (Wilcoxon rank sum test, two-sided). Violin plot summary statistics: center white line represents median gene expression and central black rectangle spans the first to third quartile of the data distribution. The whiskers indicate value at 1.5x interquartile range above the third quartile or below the first quartile.
Extended Data Figure 9: Right ventricle cells are present in Hand2-null embryos at E9.25.
a, Violin plots showing expression of Nppa and Nppb in RV State 1 and 2 from Fig. 4c. Bonferroni correction adjusted p-value < 1×10–4 (Wilcoxon rank sum test, two-sided). Statistics for differential gene expression tests were applied to n = 251 cells from each State b, UMAP plot of subset of cardiac populations captured at E9.25 colored by cluster and c, genotype. d, Curated list of highly and uniquely enriched genes in cardiac populations at E9.25. Scale indicates Z-scored expression values. e, UMAP feature plot showing expression domains of Irx4, Cited1, and Cck indicating presence of LV and RV at E9.25. Statistics for differential gene expression tests for d and e were applied to n=5,211 cells. f, Violin plots of genes differentially expressed in WT vs Hand2-null AHF cells captured at E9.25. Bonferroni correction adjusted p-value < 1×10–4 (Wilcoxon rank sum test, two-sided). Isl1 is shown to indicate equivalent expression, and thus AHF identity, in WT and Hand2-null cells. ns, not significant. Violin plot summary statistics: center white line represents median gene expression and the central black rectangle spans the first quartile to the third quartile of the data distribution. The whiskers indicate value at 1.5x interquartile range above the third quartile or below the first quartile. RV, right ventricle; LV, left ventricle; OFT, outflow tract; AHF, anterior heart field; pSHF, posterior second heart field.
Extended Data Figure 10: Right ventricle cell migration is impaired in Hand2-null embryos.
a, Whole-mount in situ hybridization for Irx4 and Cck in right lateral view and b, transverse sections at E9.25 indicating presence of RV cells in Hand2 mutants (arrowheads). n=2 independent experiments with similar results. Scale bar, 200 μm. c, Quantification of Sema3c fluorescence signal in Fig. 4h. n=3 replicate embryos per genotype. The mean +/− s.e.m indicated. Two-tailed t-test. *P<0.05. d, Violin plots of Wnt5a and Tbx2 expression in WT and Hand2-null RV cells at E7.75 and E8.25, respectively and e, Hand1 and Hand2 expression in LV and OFT cells at E9.25. All genes in d, e have a Bonferroni correction adjusted p-value < 1×10–4 (Wilcoxon rank sum test, two-sided). Violin plot summary statistics: center white line represents median gene expression and central black rectangle spans the first to third quartile of the data distribution. Whiskers indicate value at 1.5x interquartile range above the third quartile or below the first quartile. RV, right ventricle; RVp, right ventricle progenitors; LV, left ventricle; OFT, outflow tract; AHF, anterior heart field. f, In situ hybridization for Hand1 in WT and Hand2-null embryos at E9.25 in frontal view. n=3 independent experiments with similar results. g, Quantification of Hand1 fluorescent signal in the OFT. n=3 replicated embryos per genotype. The mean +/− s.e.m is indicated. Two-tailed t-test. *P<0.05. h, GO biological process terms of differentially expressed genes in WT and Hand2-null AHF (n = 406 cells per genotype), OFT (n = 362 cells per genotype) or RV (n = 227 cells per genotype) cells at E9.25, as determined with DAVID v6.8. Significant functional enrichment was statistically determined using a modified Fisher’s exact test (EASE score) followed by Benjamini-Hochberg correction for multiple comparisons, with 0.01 as a P-value cut-off.
Supplementary Material
Acknowledgements
The authors thank Dr. Benoit Bruneau and members of the Srivastava lab for helpful discussion and feedback and Dr. Cole Trapnell for guidance on scRNA-seq analysis. The authors acknowledge the Gladstone Histology and Light Microscopy Core, the Gladstone Genomics Core and the Gladstone Bioinformatics Core for their technical expertise and the Gladstone Animal Facility for support with mouse colony maintenance. We thank T. Marsh, Dr. J. Debnath, H. Yin, Dr. Françoise Chanut, B. Taylor, T. Roberts and G. Maki for their assistance with imaging, literature review, editing and graphics. We thank Dr. Maximilian Haeussler and Dr. Matthew Speir for formatting and hosting the processed datasets on the UCSC cell browser. Y.D.S was supported by the UCSF Chancellor’s Fellowship, Genentech Foundation Fellowship, Discovery Fellows Program and Phi Beta Kappa Graduate Scholarship. C.A.G. is a HHMI fellow of the Damon Runyon Cancer Research Foundation (DRG-2206–14). S.S.R is a Winslow Fellow. D.S. is supported by the National Heart Lung and Blood Institute (R01 HL057181, P01 HL089707, UM1HL098179, UM1HL128761), the California Institute for Regenerative Medicine (DISC2–09098), the Roddenberry Foundation, the L.K. Whittier Foundation, and the Younger Family Fund. S.O. is supported by an FNR CORE grant (C15/BM/10397420) and S.R. by the University of Luxembourg IRP Grant (R- AGR-3227–11). This work was also supported by NIH/NCRR grant C06 RR018928 to the Gladstone Institutes.
The authors declare no competing financial or non-financial interests as defined by Nature Research.
References
- 1.Srivastava D Making or breaking the heart: from lineage determination to morphogenesis. Cell 126, 1037–48 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Okawa S, Nicklas S, Zickenrott S, Schwamborn JC & del Sol A A generalized gene-regulatory network model of stem cell differentiation for predicting lineage specifiers. Stem Cell Reports 7, 307–315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okawa S & del Sol A A computational strategy for predicting lineage specifiers in stem cell subpopulations. Stem Cell Res. 15, 427–434 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Srivastava D et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHand. Nat. Genet 16, 154–160 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Jin SC et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet 49, 1593–1601 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu X et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lescroart F et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 359, 1177–1181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLaughter DM et al. Single-cell resolution of temporal gene expression during heart development. Dev. Cell 39, 480–490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G et al. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev. Cell 39, 491–507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia G et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uribe V, Bad C & Sanz-ezquerro JJ Arid3b is essential for second heart field cell deployment and heart patterning. 4168–4181 (2014). doi: 10.1242/dev.109918 [DOI] [PubMed] [Google Scholar]
- 13.Tsuchihashi T et al. Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev. Biol 351, 62–69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlieb PD et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat. Genet 31, 25–32 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Stefanovic S & Zaffran S Mechanisms of retinoic acid signaling during cardiogenesis. Mech. Dev 143, 9–19 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Napoli JL Cellular retinoid binding-proteins, CRBP, CRABP, FABP5 : Effects on retinoid metabolism, function and related diseases. Pharmacol. Ther 173, 19–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein JA, Aghajanian H & Singh MK Semaphorin signaling in cardiovascular development. Cell Metab. 21, 163–173 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha T et al. Loss of Wnt5a disrupts second heart field cell deployment and may contribute to OFT malformations in DiGeorge syndrome. Hum. Mol. Genet 24, 1704–1716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupays L, Kotecha S, Angst B & Mohun TJ Tbx2 misexpression impairs deployment of second heart field derived progenitor cells to the arterial pole of the embryonic heart. Dev. Biol 333, 121–131 (2009). [DOI] [PubMed] [Google Scholar]
- 20.George RM & Firulli AB Hand factors in cardiac development. Anat. Rec 107, 101–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi H et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossant J, Zirngibl R, Giguere V, Cado D & Shago M Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5, 1333–1344 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Gross-Thebing T, Paksa A & Raz E Simultaneous high-resolution detection of multiple transcripts combined with localization of proteins in whole-mount embryos. BMC Biol. 12, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data, including sequencing reads and single-cell expression matrices have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE126128. Data underlying each figure are available in the Source Data Files, Supplementary Information and on the UCSC cell browser at https://mouse-cardiac.cells.ucsc.edu. Users can use the cell browser to explore the data, view expression of genes of interest in each UMAP plot and download datasets for custom analysis.