Abstract
Sepsis is a multiorgan disease affecting the ileum and jejunum (small intestine), liver, skeletal muscle, and lung clinically. The specific metabolic changes in the ileum, jejunum, liver, skeletal muscle, and lung have not previously been investigated. Live Pseudomonas aeruginosa, isolated from a patient, was given via i.v. catheter to pigs to induce severe sepsis. Eighteen hours later, ileum, jejunum, medial gastrocnemius skeletal muscle, liver, and lung were analyzed by nontargeted metabolomics analysis using gas chromatography/mass spectrometry. The ileum and the liver demonstrated significant changes in metabolites involved in linoleic acid metabolism: the ileum and lung had significant changes in the metabolism of valine/leucine/isoleucine; the jejunum, skeletal muscle, and liver had significant changes in arginine/proline metabolism; and the skeletal muscle and lung had significant changes in aminoacyl-tRNA biosynthesis, as analyzed by pathway analysis. Pathway analysis also identified changes in metabolic pathways unique for different tissues, including changes in the citric acid cycle (jejunum), β-alanine metabolism (skeletal muscle), and purine metabolism (liver). These findings demonstrate both overlapping metabolic pathways affected in different tissues and those that are unique to others and provide insight into the metabolic changes in sepsis leading to organ dysfunction. This may allow therapeutic interventions that focus on multiple tissues or single tissues once the relationship of the altered metabolites/metabolism to the underlying pathogenesis of sepsis is determined.
The idea that sepsis is linked to metabolic alterations goes back to observations made in animals challenged with endotoxin, in which distorted mitochondria were observed.1 Alterations in mitochondrial respiration, mitochondrial complex activities, and nucleotide levels have been observed in cells, tissue homogenates, and isolated mitochondria.2 The manner in which protein metabolism is affected after 24 hours of endotoxemia in pigs has found a net protein synthesis in the liver, along with increased amino acid transamination in muscle, with glutamine and alanine effluxes serving as substrates for the liver.3 Recently, the application of metabolomics analysis to circulating plasma or urine of patients with pneumonia and/or sepsis has been used to identify metabolites with prognostic and predictive value in specific clinical settings.4, 5, 6, 7
Sepsis is a multiorgan disease, affecting the gut, liver, skeletal muscle, and kidney. The severity of sepsis in patients is classified clinically according to the presence of evidence of inflammation, multiple organ failure, or organ failure with hypotension. To date, studies of metabolic alterations have primarily focused on either circulating metabolic changes in serum or specifically in a single organ (eg, skeletal muscle). Given that cellular metabolic dysregulation is implicated in disease,8 the contribution of metabolic changes in an organ-specific manner is clinically relevant to the variable effects on organ failure linked to survival.9 In the present study, an organ systems approach was taken to identify the metabolic changes occurring in the jejunum, ileum, skeletal muscle, liver, and lung using a nontargeted metabolomics approach to identify metabolic pathways altered in a porcine model of sepsis induced by 18 hours of continuous i.v. infusion of Pseudomonas aeruginosa.
Materials and Methods
Compliance with Ethical Standards
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Animals
Twenty-seven female Yorkshire crossbred pigs (Sus domesticus) from multiple litters (Metz Farms, Russellville, AR), weighing 20 to 25 kg, were used in the present study. A dedicated veterinarian of the Arkansas Children's Hospital Research Institute (Little Rock, AR; American Association for Laboratory Animal Care certified) assessed the general health status of all animals. Pigs were individually housed in steel pens (2 × 3 m) located in a controlled housing biosafety level 2 facility, within a large animal cubicle with a plastic-coated mesh-style grid floor. The facilities were temperature controlled (22°C to 24°C) with a 12-hour light-dark cycle and standardized food intake (1 kg/day) (Teklad; Envigo, Alice, TX) and water available ad libitum. Pens were enriched with toys to stimulate natural pig rooting behavior and to prevent stereotypic behavior. A heating pad was added to the pen to ensure natural body temperature control after surgery. Clinical examination was performed on arrival, before surgery (7 to 10 days after arrival), and after surgery (twice daily on the first 3/4 days after surgery and then every 2/3 days until the sepsis study). The present animal study, including its justification for number, breed, and use of single sex, was approved by the animal experiment ethics committee (Institutional Animal Care and Use Committee) of University of Arkansas Medical Sciences (Little Rock, AR).
Presurgical Catheter Placement, Anesthesia Induction, and Surgical Procedure
Catheters were first implanted in the abdominal aorta, inferior vena cava, splenic vein, an extracaval vein, a portal vein, and the hepatic vein to surgically sample blood, monitor pressures, administer medications and infusions, and sample blood from multiple organs, while a Swan Ganz catheter was placed in the right jugular vein to monitor pulmonary pressures, as previously described.10, 11 Briefly, animals were acclimatized for 7 to 10 days and underwent a presurgery clinical examination under anesthesia by an assigned veterinarian. Anesthesia was induced with i.m. injection of a mixture of tiletamine hydrochloride and zolazepam hydrochloride (Telazol; Zoetis, Parsippany, NJ). The animal was then intubated and put under isoflurane general anesthesia maintained at 2% with oxygen. Flunixin meglumine (Flunixamine; Zoetis; 2 mg/kg BW) was administered as analgesic, and lincomycin-spectinomycin (Linco-Spectin; Zoetis; lincomycin, 5 mg/kg BW, and spectinomycin, 10 mg/kg BW) was administered as a surgical antibiotic prophylaxis via ear vein cannula. During surgery, 500 to 1000 mL Ringer's lactate solution was given to maintain blood pressure and fluid balance. ECG, CO2, and oxygen saturation were monitored continuously.
A midline laparotomy was performed to place the abdominal aorta catheter for blood sampling over time and to monitor mean arterial pressure. An inferior caval vein catheter was then placed to allow post-surgical administration of medications. A second arterial catheter, just before the bifurcation, and a splenic vein catheter were placed, allowing infusion of para-aminohippuric acid for plasma flow measurements. A Swan Ganz catheter (5 Fr; number 132F5; Edwards Life Sciences, Irvine, CA) was inserted via the right jugular vein to monitor mean pulmonary arterial pressure. The position of the tip of the Swan Ganz catheter was determined after the pressure readings when the catheter was pushed through the right atrium and ventricle. In addition, an extracaval vein, a portal vein, and a hepatic vein catheter were placed for multiorgan blood sampling. The duration of the surgery was approximately 5 hours. Postoperative care was standardized, as described in detail previously,10, 11 and monitored by the assigned veterinarian. After surgery, ECG, pulse oximetry, and ventilation were continued until the animal responded with multiple swallow reflexes, performed eye blinking, demonstrated evidence of fighting the ventilator, and had attempts to rise. A second dose of lincomycin-spectinomycin was then given intramuscularly. Breathing and heart rate were observed until the animal was able to sit sternally. During the first 4 days after surgery, animals were clinically examined twice daily (body temperature, appearance, and behavior), and buprenorphine (0.03 mg/kg BW) and lincomycin-spectinomycin were administered via the implanted central vein catheter. Animals were fed half their normal food ration during the first 2 days after surgery. Catheters were checked daily for the first 3 days, and their patency was observed by antimicrobial and anticoagulant catheter filling, using 0.5 mL of 20 mg/mL gentamicin and 225 U/mL alfa-chymotrypsin, which was set in the catheter, not injected, as previously described.10 Subsequently, animals were examined, and catheters were checked for patency every 2 to 3 days. Animals were acclimated to a small movable cage (0.9 × 0.5 × 0.3 m) to ensure that the P. aeruginosa experiments could be performed in the cage in conscious animals in free standing, sitting, or lying positions both before and after surgery and during the recovery period (7 to 10 days). Of the 27 animals, 5 were lost because of intestinal complications (ileus, ascites, or peritonitis) during the recovery phase from the catheter implantation surgery. However, no animals were lost during the surgery and the P. aeruginosa experiment. At the time of the sepsis experiment, animals had been acclimated at least 10 times in this movable cage.
Septic Response to P. aeruginosa Porcine Study Design
Animals were randomized to either experimental or control group 7 to 10 days after surgery. Basal blood pressures, plasma flow, blood gas, clinical chemistry, and inflammatory data (preseptic period; time = −2 to 0 hours) measurements were started 4 to 6 hours after the last food intake (half of the daily amount: 0.5 kg). No food was administered for the duration of the subsequent 18-hour experimental period. The experimental period began with the introduction of bacteremia (time = 0 hours) by i.v. continuous infusion of P. aeruginosa (IRS 12-44; Shriners Burns Institute, University of Texas Medical Branch, Galveston, TX; original from a burn patient at Brook Army Medical Center, San Antonio, TX; 109 colony-forming units/hour in 1 mL 0.9% NaCl solution). The control group received 0.9% NaCl solution in the same volume. The P. aeruginosa dose was based on the method of Rimmele et al,12 which was tested for virulence and adjusted in a pilot study (three animals). Fluid resuscitation (30 mL/kg BW/hour 0.9% NaCl, intravenously) was started at the same time as the P. aeruginosa infusion. Continuous hemodynamic monitoring ensured that the hyperdynamic state was kept in the expected ranges for severe sepsis and not shock, including maximal body temperature ±2°C (normal range, 37°C to 39.6°C),4 maximal mean pulmonary arterial pressure of 35 mm Hg (normal range, 11 to 24 mm Hg),4 heart rate of 200 beats per minute (normal range, 90 to 107 beats per minute),4 and mean arterial pressure >60 mm Hg (normal range, 86 to 123 mm Hg)4 using the arterial and Swan Ganz catheter for invasive blood pressure measurement [monitor: Propaq Encore (Welch Allyn, Skaneateles Falls, NY); transducer: TruWave (Edwards Lifesciences, Irvine, CA)]. If the heart rate was >200 beats per minute and/or mean pulmonary arterial pressure was >35 mm Hg, the P. aeruginosa infusion was stopped to limit the increase of the right ventricular afterload3 until heart rate or mean pulmonary arterial pressure was <200 beats per minute or <35 mm Hg, respectively. Animals were euthanized 18 hours (time = 18) after bacterial infusion with 125 mg/kg pentobarbital sodium and 16 mg/kg phenytoin sodium (Euthanasia-III Solution; Med-Pharmex, Pomona, CA), administered via the central vein catheter. Immediately after euthanasia, approximately 200-mg fragments of skeletal muscle (medial gastrocnemius, right hind leg, and middle section), jejunum, ileum, liver, and lung were collected from 9 P. aeruginosa and 13 control animals, placed on dry ice, and stored at −80°C.
Nontargeted Metabolomics Analysis by GC/MS Instrumentation
Samples were then processed and analyzed by gas chromatography/mass spectrometry (GC/MS), as previously described.13, 14 A 20- to 50-mg sample was cut from the flash-frozen tissue and immediately weighed, followed by quickly mincing the tissue and addition of extraction solution (50% acetonitrile, 50% water, and 0.3% formic acid, made fresh daily) prepared at a concentration of 25 mg/475 μL solution. The sample was then immediately homogenized on ice for 10 to 25 seconds, placed on dry ice, and stored at – 80°C. The samples were crash deproteinated by methanol precipitation and spiked with D27-deuterated myristic acid as an internal standard for retention time (RT) locking and dried. The derivatized trimethylsilyl–D27-deuterated myristic acid standard RT was set at 16.727 minutes. Reactive carbonyls were stabilized at 50°C with methoxyamine hydrochloride in dry pyridine. Metabolites were made volatile with trimethylsilyl groups using N-methyl-N-(trimethylsilyl) trifluoroacetamide or N-methyl-N-trimethylsilyltrifluoroacetamide with catalytic trimethylchlorosilane at 50°C. GC/MS methods follow previous studies using a 6890N GC connected to a 5975 inert single quadrupole MS (Agilent Technologies, Santa Clara, CA).15, 16 The two wall-coated, open-tubular GC columns connected in series were both from J&W/Agilent (part number 122-5512; Agilent Technologies), DB5-MS, 15 m in length, 0.25 mm in diameter, with a 0.25-μm luminal film. Positive ions, generated with conventional electron ionization at 70 eV, were scanned broadly from 600 to 50 m/z in the detector throughout the 45-minute cycle time. Data were acquired using MSD ChemStation (Agilent Technologies) by identifying metabolites on the basis of their mass fragmentation patterns and RT. Rawdata formatted files were exported for further analysis in Automatic Mass Spectral Deconvolution and Identification Software (freeware developed by Drs. Steve Stein, W. Gary Mallard, and their coworkers at the National Institute of Standards and Technology, Gaithersburg, MD17, 18, 19). Deconvoluted spectra were annotated as metabolites, to the extent possible, using an orthogonal approach that incorporated both RT from GC and the fragmentation pattern observed in electron ionization mass spectrometer. Peak annotation was based primarily on our own RT-locked spectral library of metabolites, built on the Fiehn GC/MS Metabolomics RTL Library (a gift from Agilent, part number G1676-9000020, 21), the Wiley 9th–National Institute of Standards and Technology 2011 commercial library (Agilent G1730-64000), and other spectral libraries. Once annotation was complete, a cross-tabulated spreadsheet was generated, listing the integrated peak area for each metabolite versus sample identity. This was accomplished using a custom Visual Basic program in Microsoft Excel (Microsoft Corp., Redmond, WA) that grouped peaks across samples on the basis of identical metabolite annotation and RT proximity. Peak alignment across samples was further confirmed using SpectConnect22 to assess similarity in spectral fragmentation patterns and by manual curation.
Metabolomic Statistical Analysis
The raw, transformed, and sorted data used for each of the three comparisons in the metabolomics analyses can be found in Supplemental Tables S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, and S15. Up to three missing values per group were imputed using the lowest value in the same group, with groups missing four or more values excluded from the analysis. The raw data from this study are accessible through the NIH Common Fund's Data Repository and Coordinating Center (supported by NIH grant U01-DK097430) Metabolomics Workbench (https://www.metabolomicsworkbench.org/data; Project ID = PR000802). The processed data used in the analyses described herein are found in Supplemental Tables S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14, and S15. t-Test analysis was performed on metabolite peak areas (as representative of concentration) using Metaboanalyst version 3.5 using a Pareto scaling feature.23, 24, 25 Heat maps were generated using Metaboanalyst. Post-hoc graphs were generated in Prism 7.0 (GraphPad Software, Inc., La Jolla, CA). All data are shown as means ± SEM, unless otherwise indicated.
Pathway Analysis
Pathway analyses in Metabolomic Pathway Analysis (MetPA) were conducted through three routes.26 Pathway enrichment analysis supports both overrepresentation analysis as well as Gene Set Enrichment Analysis–based approaches. The available algorithms include Fishers exact test, the hypergeometric test, the global test,27 and GlobalAncova.28 MetPA's pathway topological analysis is based on the centrality measures of a metabolite in a given metabolic network. Centrality is a local quantitative measure of the position of a node relative to the other nodes and is often used to estimate a node's relative importance or role in network organization.29 Because metabolic networks are directed graphs, MetPA uses relative betweenness centrality and out degree centrality measures to calculate compound importance. The pathway impact was calculated as the sum of the importance measures of the matched metabolites normalized by the sum of the importance measures of all metabolites in each pathway. Finally, MetPA provides several univariate analyses performed at the compound level to provide a more detailed view of the distribution of individual metabolite concentrations with regard to phenotypes. They include the t-test, one-way analysis of variance, and linear regression. The pathway data used in MetPA were downloaded as KEGG Markup Language (KGML) files from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.30 Chemical compounds and pathway topology information were parsed into graph models using the KEGGgraph package.31 The current library contains 874 metabolic pathways from 11 model organisms, including humans, mouse, Drosophila, Arabidopsis, and Escherichia coli. Metabolic pathways are presented as a network of chemical compounds with metabolites as nodes and reactions as edges. The graph generation and manipulation were implemented using Graphviz version 2.40.1 (AT&T Lab Research, Atlanta, Georgia; http://www.graphviz.org, last accessed December 8, 2018) and ImageMagick version 6.9.10 (ImageMagick Studio LLC, Landerberg, PA; http://www.imagemagick.org, last accessed December 8, 2018). This visualization system supports lossless zooming, dragging, and linking operations based on Ajax (Asynchronous JavaScript with XML) technology.32 All relevant information was obtained by clicking on the corresponding graphical elements.26
Results
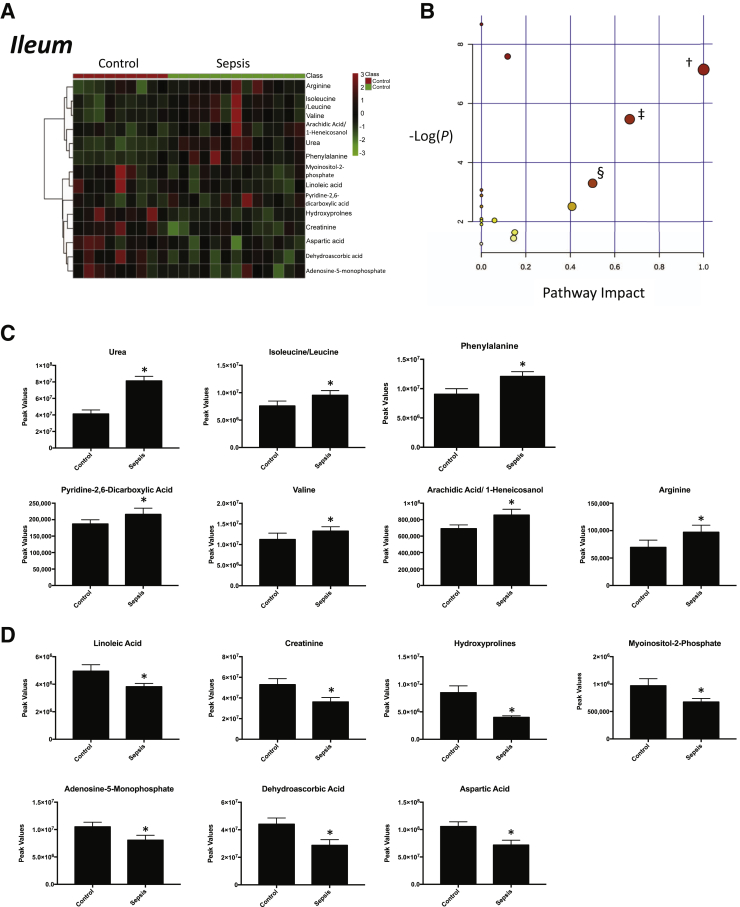
Nontargeted analysis of the ileum from septic pigs collected 18 hours after the induction of sepsis with i.v. P. aeruginosa identified 97 metabolites that were present (Supplemental Figure S1). Of these metabolites, 14 were significantly different in septic ileum compared with control animal ileum (Figure 1A). Of these metabolites, seven were significantly increased in ileum from septic animals (Figure 1C) and seven were significantly decreased (Figure 1D). Enrichment analysis of these metabolites using a pathway-associated metabolite set, a disease-associated metabolite set, a predicted metabolite set, and a location-associated metabolite set identified a >10-fold enrichment for urea cycle metabolites (Supplemental Figure S2A); approximately 10-fold enrichment in metabolites associated with epilepsy, acute seizures, and phenylketonuria (Supplemental Figure S2B); metabolites involved in the transport into the mitochondria/carnitine exchange (Supplemental Figure S2C); and mitochondria (Supplemental Figure S2D). Analysis of these 14 significantly altered metabolites using pathway enrichment analysis revealed significant enrichment for linoleic acid metabolism, valine/leucine/isoleucine biosynthesis, and phenylalanine/tyrosine/tryptophan biosynthesis (Figure 1B). After 18 hours of P. aeruginosa sepsis, the ileum demonstrated alterations in fatty acid (linoleic), branched-chain amino acid (valine/leucine), and phenylalanine metabolism (Table 1).
Figure 1.
Significantly altered metabolites in ileum obtained from sepsis-challenged pigs compared with controls. A: Heat map of significantly altered metabolites using t-test results. B: Pathway enrichment analysis of significantly altered metabolites. Metabolic pathway analysis shows all metabolic pathways arranged according to the scores from enrichment analysis (y axis) and from topology analysis (x axis). C and D: Significantly increased (C) or decreased (D) metabolite peak signal in sepsis-challenged ileum compared with vehicle-control ileum. n = 9 control ileum samples (C and D); n = 13 septic ileum samples (C and D). ∗P < 0.05 versus control, false-discovery rate (FDR) < 0.33; †linoleic acid metabolism, P = 7.9 × 10−4, FDR = 0.02 (metabolites: linoleic acid and phosphatidylcholine); ‡valine, leucine, and isoleucine biosynthesis, P = 4.2 × 10−3, FDR = 0.08 (metabolites: l-leucine and l-valine); §phenylalanine, tyrosine, and tryptophan biosynthesis, P = 0.3.69 × 10−2, FDR = 0.59 (metabolites: phenylalanine).
Table 1.
Summary of Significant Metabolites Identified by Nontargeted Metabolomics by Tissue
| Ileum | Jejunum | Gastrocnemius skeletal muscle | Liver | Lung |
|---|---|---|---|---|
| Urea | Urea | Urea | Urea | Urea |
| Hydroxyprolines | Hydroxyprolines | Hydroxyprolines | Proline | |
| Phenylalanine | Phenylalanine | Phenylalanine | Phenylalanine | |
| Dehydroascorbic acid | Dehydroascorbic acid | |||
| Isoleucine/leucine | Isoleucine/leucine | |||
| Aspartic acid | Aspartic acid | Aspartic acid | ||
| Arachidic acid/1-heneicosanol | Arachidic acid/1-heneicosanol | |||
| Myoinositol-2-phosphate | ||||
| Arginine | ||||
| Valine | Valine | |||
| Creatinine | Creatinine | |||
| Pyridine-2,6-dicarboxylic acid | ||||
| Adenosine-5-monophosphate | ||||
| Linoleic acid | Linoleic acid | |||
| Aminomalonic acid | ||||
| Uracil | ||||
| Fumaric acid | Fumaric acid | |||
| Spermidine | ||||
| 1-Monomyristin | ||||
| 2-Hydroxybutyric acid | 2-Hydroxybutyric acid | 2-Hydroxybutyric acid | ||
| 1,2-Propanediol | ||||
| 6-Hydroxynicotinic acid | 6-Hydroxynicotinic acid | |||
| Succinic acid | ||||
| Malic acid | ||||
| Methionine | ||||
| Glutamic acid | ||||
| Methylglutamic acid | Methylglutamic acid | |||
| Threonine | ||||
| 1,2-Propanediol | ||||
| 2-Hydroxybutyric acid | ||||
| Anserine | ||||
| 2-Hydroxyglutaric acid | ||||
| Glycine | ||||
| 2-Hydroxypyridine | ||||
| Carnosine | ||||
| Pantothenic acid | Pantothenic acid | |||
| Glycerol 1-phosphate | ||||
| Sarcosine | ||||
| Threonine | Threonine | |||
| 5′-Deoxy-5′-methylthioadenosine | ||||
| Adenosine | ||||
| Inosine | ||||
| Cysteinesulfinic acid | ||||
| β-Glycerolphosphate | ||||
| N-methylalanine/2-aminobutanoic acid/N-ethylglycine | ||||
| Ornithine | ||||
| Glucose and other aldohexoses | ||||
| Fructose or similar ketohexose | ||||
| Nicotinamide | ||||
| Glycolic acid | ||||
| Palmitoleic acid | ||||
| Maltose or similar disaccharide | ||||
| Guanine | ||||
| Ribulose-5-phosphate | ||||
| Cholesterol | ||||
| O-methylphosphate | ||||
| 2-Aminoadipic acid | ||||
| β-Alanine | ||||
| 3-Phosphoglyceric acid | ||||
| Putrescine | ||||
| Oleic acid | ||||
| Alanine |
Significant metabolites found in sepsis and named in pathway enrichment analysis (versus tissue control) are shown in bold. Italics indicates multiple tissues.
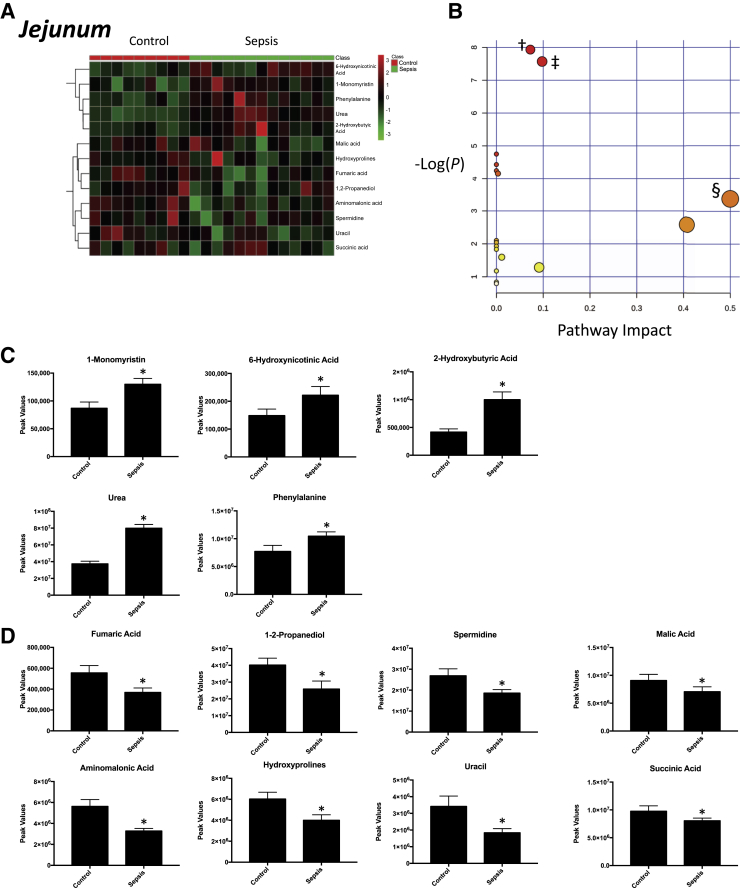
Nontargeted analysis of the jejunum detected 99 metabolites present in septic animals compared with controls (Supplemental Figure S3). Of the metabolites detected (ie, identified, named, and consistently measured across samples), 13 were significantly altered septic jejunum compared with controls (Figure 2A); 5 were significantly increased in jejunum from septic animals (Figure 2C); and 8 were significantly decreased (Figure 2D). Enrichment analysis using a pathway-associated metabolite set, a disease-associated metabolite set, a predicted metabolite set, and a location-associated metabolite set identified a greater than eightfold enrichment in metabolites involved in the citric acid cycle and phenylalanine/tyrosine metabolism (Supplemental Figure S4A), hyperphenylalaninemia (Supplemental Figure S4B), 2-hydroxybutyrate exchange (Supplemental Figure S4C), and metabolism characteristic of the epidermis and testes (Supplemental Figure S4D). Analysis of these 13 significantly altered metabolites using pathway enrichment analysis revealed significant enrichment for arginine/proline metabolism, citric acid cycle, and phenylalanine/tyrosine/tryptophan biosynthesis (Figure 2B). After 18 hours of P. aeruginosa sepsis, the jejunum demonstrated 3 overlapping metabolite alterations found in the ileum (urea, hydroxyprolines, and phenylalanine) and 10 additional uniquely altered metabolites (Table 1).
Figure 2.
Significantly altered metabolites in jejunum obtained from sepsis-challenged pigs compared with controls. A: Heat map of significantly altered metabolites using t-test significant metabolites. B: Pathway enrichment analysis of significantly altered metabolites. Metabolic pathway analysis shows all metabolic pathways arranged according to the scores from enrichment analysis (y axis) and from topology analysis (x axis). C and D: Significantly increased (C) or decreased (D) metabolite peak signal in sepsis-challenged jejunum compared with vehicle-control jejunum. n = 9 control jejunum samples (C and D); n = 13 septic jejunum samples (C and D). ∗P < 0.05 versus control, false-discovery rate (FDR) < 0.38; †arginine and proline metabolism, P = 3.5904 × 10−4, FDR = 0.02 (metabolites: hydroxyproline, spermidine, fumaric acid, and urea); ‡citric acid cycle, P = 5.1567 × 10−4, FDR = 0.02 (metabolites: succinic acid, malic acid, and fumaric acid); §phenylalanine, tyrosine, and tryptophan biosynthesis, P = 0.03, FDR = 0.39 (metabolite: phenylalanine).
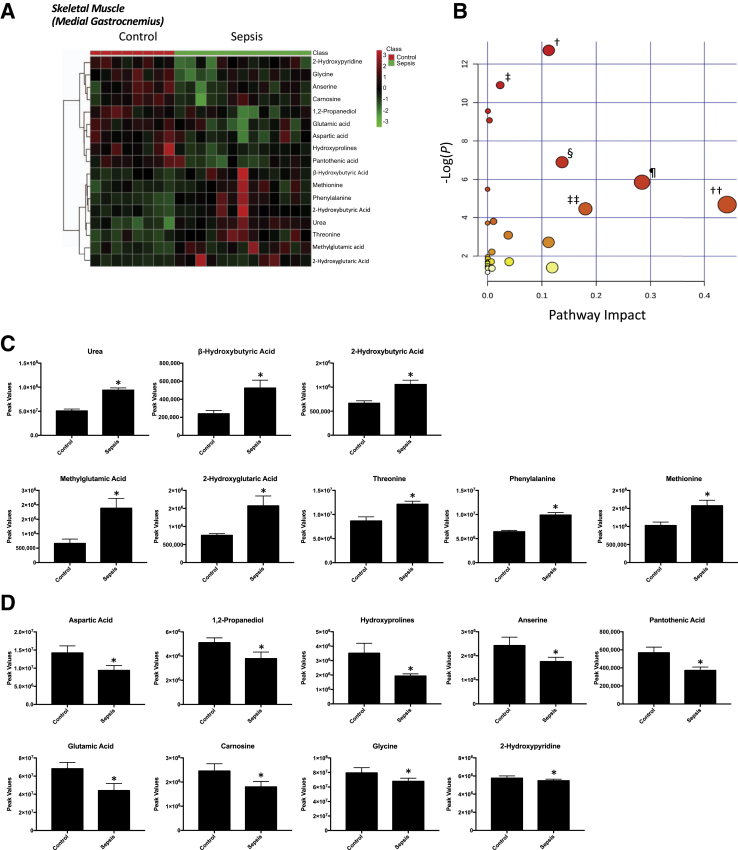
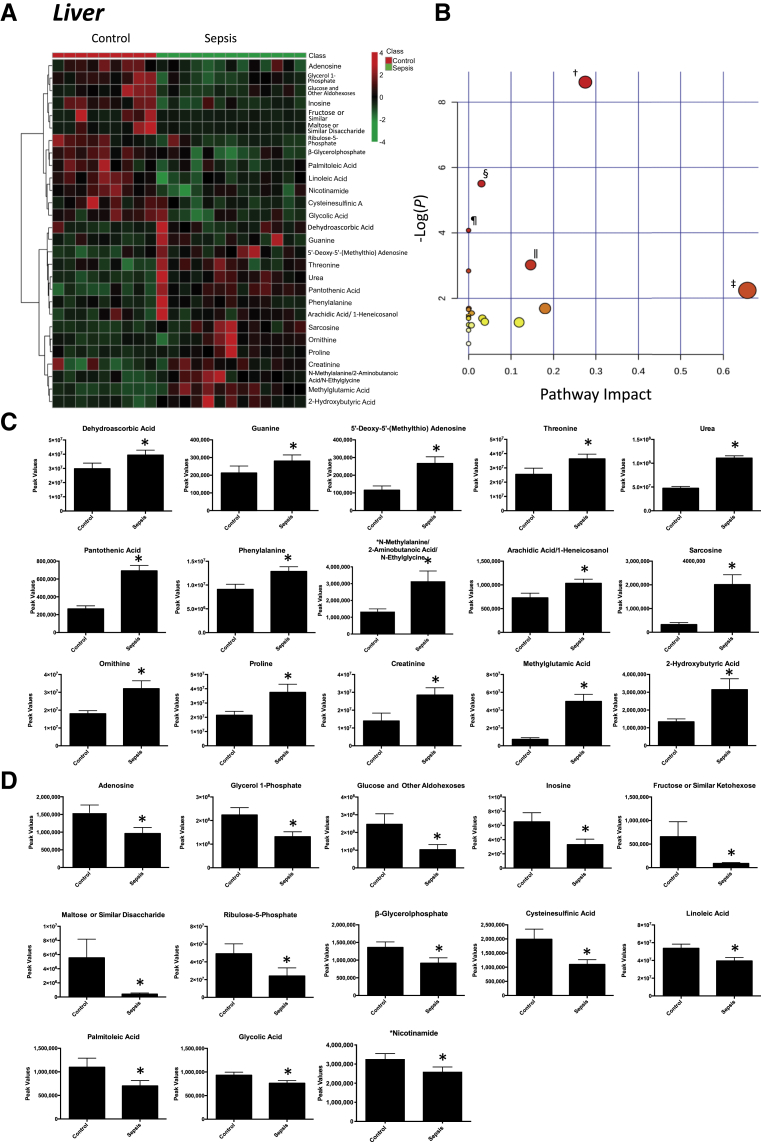
Nontargeted analysis of the medial gastrocnemius skeletal muscle identified 91 metabolites in septic animals compared with controls (Supplemental Figure S5). Of these metabolites, 17 were altered in the gastrocnemius muscle compared with control animal gastrocnemius muscle (Figure 3A); 8 were significantly increased in septic animals (Figure 3C); and 9 were significantly decreased (Figure 3D). Enrichment analysis of these metabolites using a pathway-associated metabolite set, a disease-associated metabolite set, a predicted metabolite set, and a location-associated metabolite set identified a >14-fold enrichment in metabolites involved in β-alanine metabolism and ammonia recycling metabolism (Supplemental Figure S6A), aseptic meningitis (Supplemental Figure S6B), l-threonine deaminase (Supplemental Figure S6C), and metabolism characteristic of the muscle, mitochondria, and lysosome locations (Supplemental Figure S6D). Pathway enrichment analysis of the 17 significantly altered metabolites revealed enrichment for arginine/proline metabolism, propionate metabolism, alanine/aspartate/glutamine metabolism, and phenylalanine/tyrosine/tryptophan biosynthesis (Figure 3B). After 18 hours of P. aeruginosa sepsis, the gastrocnemius skeletal muscle demonstrated 13 significant alterations compared with nonseptic parallel controls, including five metabolites overlapping with changes seen in the ileum and jejunum (Table 1). Nontargeted analysis of the liver identified 114 metabolites that were present in septic animals compared with controls (Supplemental Figure S7). Of these metabolites, 28 were significantly different in the liver compared with controls (Figure 4A); 15 were increased (Figure 4C); and 13 were decreased (Figure 4D). Further enrichment analysis of these metabolites using a pathway-associated metabolite set identified enrichment for arginine/proline metabolism, starch/sucrose metabolism, and phenylacetate metabolism (Supplemental Figure S8). Pathway enrichment analysis revealed significant enrichment for arginine and proline metabolism, linoleic acid metabolism, aminoacyl-tRNA biosynthesis, and glycine, serine, and threonine metabolism (Figure 4B). After 18 hours of P. aeruginosa sepsis, the liver had the highest number of significantly altered metabolites (n = 28) compared with the other tissues tested. Eleven of these significant metabolites overlapped with the other four tissues, whereas 17 were unique to the liver (Table 1). The metabolic pathways identified parallel changes in the jejunum and skeletal muscle (arginine and proline metabolism) and the ileum (linoleic acid). The liver uniquely demonstrated significant changes in metabolites involved in purine metabolism and aminoacyl-tRNA biosynthesis.
Figure 3.
Significantly altered metabolites in medial gastrocnemius muscle obtained from sepsis-challenged pigs compared with controls. A: Heat map of significantly altered metabolites using t-test results. B: Pathway enrichment analysis of significantly altered metabolites. Metabolic pathway analysis shows all metabolic pathways arranged according to the scores from enrichment analysis (y axis) and from topology analysis (x axis). C and D: Significantly increased (C) or decreased (D) metabolite peak signal in sepsis-challenged skeletal muscle (medial gastrocnemius) compared with vehicle-control skeletal muscle (medial gastrocnemius). n = 9 control gastrocnemius samples (C and D); n = 13 septic gastrocnemius samples (C and D). ∗P < 0.05 versus control, false discovery rate (FDR) < 0.26; †aminoacyl-tRNA biosynthesis, P = 3.00 × 10−06, FDR = 0.00024 (metabolites: phenylalanine, glycine, methionine, threonine, glutamic acid, and aspartic acid); ‡β-alanine metabolism, P = 1.83 × 10−05, FDR = 0.00073364 (metabolites: aspartic acid, pantothenic acid, carnosine, and anserine); §arginine and proline metabolism, P = 0.0010111, FDR = 0.00073364 (metabolites: glutamic acid, aspartic acid, urea, and hydroxyproline); ¶glycine, serine, and threonine metabolism, P = 0.0028628, FDR = 0.00073364 (metabolites: aspartic acid, threonine, and glycine); ††cyanoamino acid metabolism, P = 0.0041371, FDR = 0.047282 (metabolites: glycine and aspartic acid); ‡‡pantothenate and CoA biosynthesis, P = 0.011631, FDR = 0.011631 (metabolites: aspartic acid and pantothenic acid).
Figure 4.
Significantly altered metabolites in liver obtained from sepsis-challenged pigs compared with controls. A: Heat map of significantly altered metabolites using t-test significant results. B: Pathway enrichment analysis of significantly altered metabolites. Metabolic pathway analysis shows all metabolic pathways arranged according to the scores from enrichment analysis (y axis) and from topology analysis (x axis). C and D: Significantly increased (C) or decreased (D) metabolite peak signal in sepsis-challenged liver compared with vehicle-control liver. n = 9 vehicle controls (C and D); n = 9 control liver samples (C and D); n = 13 septic liver samples (C and D). ∗P < 0.05 versus control, false discovery rate (FDR) < 0.18; †arginine and proline metabolism, P = 1.81 × 10−4, FDR = 0.014 (metabolites: l-ornithine, proline, creatinine, sarcosine, and urea); ‡linoleic acid metabolism, P = 0.10, FDR = 1 (linoleic acid); §purine metabolism, P = 4.1 × 10−3, FDR = 0.162 (metabolites: guanine, inosine, adenosine, and urea); ¶aminoacyl-tRNA biosynthesis, P = 0.016, FDR = 0.451 (metabolites: phenylalanine, threonine, and proline); ‖glycine, serine, and threonine metabolism, P = 0.048, FDR = 0.93 (metabolites: threonine and sarcosine).
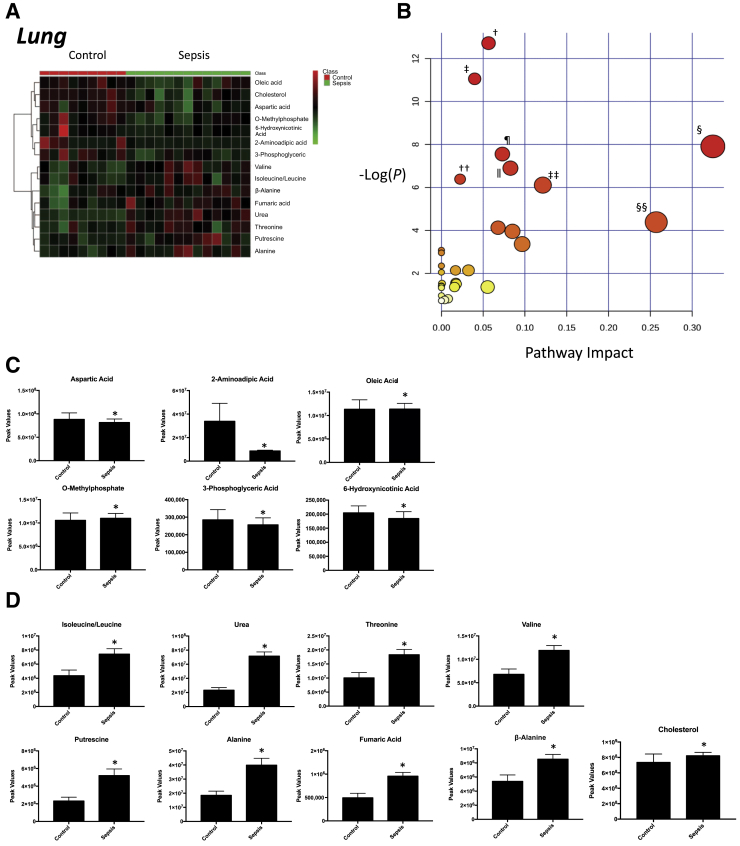
Nontargeted analysis of the lung identified 91 metabolites in septic animals compared with controls (Supplemental Figure S9). Of these metabolites, 15 were significantly different in the septic lungs compared with control lungs (Figure 5A). Of these metabolites, nine were significantly increased in the lung from septic animals (Figure 5C) and six were significantly decreased (Figure 5D). Enrichment analysis of these significant metabolites using a pathway-associated metabolite set and a location-associated metabolite set identified a >10-fold enrichment in metabolites involved in the urea cycle, aspartate metabolism, arginine/proline metabolism (Supplemental Figure S10A), and metabolism linked to placenta and mitochondrial locations (Supplemental Figure S10B). Analysis of these 15 significantly altered metabolites using pathway enrichment analysis revealed significant enrichment for arginine/proline metabolism, alanine/aspartate/glutamine metabolism, valine/leucine/isoleucine metabolism, and β alanine metabolism (Figure 5B). After 18 hours of P. aeruginosa sepsis, seven of the significantly altered metabolites overlapped with the other four tissues, with eight unique to the liver (Table 1). The significantly altered lung metabolites were enriched for metabolites involved in arginine and proline metabolism (like the jejunum, skeletal muscle, and liver) and alanine/aspartate/glutamine metabolism (like skeletal muscle).
Figure 5.
Significantly altered metabolites in lung obtained from sepsis-challenged pigs compared with controls. A: Heat map of significantly altered metabolites using t-test significant results. B: Pathway enrichment analysis of significantly altered metabolites. Metabolic pathway analysis shows all metabolic pathways arranged according to the scores from enrichment analysis (y axis) and from topology analysis (x axis). C and D: Significantly increased (C) or decreased (D) metabolite peak signal in sepsis-challenged lung compared with vehicle-control lung. n = 9 control lung samples (C and D); n = 13 septic lung samples (C and D). ∗P < 0.05 versus control, false discovery rate (FDR) < 0.3; †aminoacyl-tRNA biosynthesis, P = 3.00 × 105, FDR = 0.00024 (metabolites: aspartic acid, valine, alanine, isoleucine, leucine, and threonine); ‡valine, leucine, and isoleucine biosynthesis, P = 1.58 × 10−05, FDR = 0.00063115 (metabolites: valine, leucine, isoleucine, and threonine); §alanine, aspartate, and glutamate metabolism, P = 3.667 × 10−4, FDR = 0.00036665 (metabolites: aspartic acid, alanine, fumaric acid, and valine); ¶pantothenate and CoA biosynthesis, P = 5.24 × 10−4, FDR = 0.010479 (metabolites: aspartic acid and β-alanine); ‖arginine and proline metabolism, P = 1.01 × 10−2, FDR = 0.016178 (metabolites: aspartic acid, fumaric acid, urea, and putrescine); ††valine, leucine, and isoleucine degradation, P = 1.68 × 10−3, FDR = 0.022 (metabolites: leucine, isoleucine, and valine); ‡‡nicotinate and nicotinamide metabolism, P = 2.22 × 10−3, FDR = 0.025 (metabolites: aspartic acid, 6-hydroxynicotinate, and fumaric acid); §§β-alanine metabolism, P = 1.24 × 10−3, FDR = 0.124 (metabolites: aspartic acid and β-Alanine).
Discussion
Most, if not all, disease states exhibit alterations in the metabolic profile of tissues affected. In several of our previous studies, we have characterized the metabolome network of several physiological perturbations across organ systems that has demonstrated the diverse and complementary changes in organ metabolism.33, 34, 35, 36 Noteworthy are our observations across these studies that the least affected system with respect to metabolites is the circulation, suggesting a full panel analysis of organs is necessary to gain a full profile of the underlying pathologic mechanisms driving disease. As sepsis is a multiorgan disease with evidence for involvement of mitochondrial dysfunction and metabolic alterations, the metabolic profiles of several tissues were investigated in our pig model of sepsis using a nontargeted metabolomics approach. Sepsis is a multiorgan disease, and our work has the metabolic changes that occur in several disease states.33, 34, 35, 36 Exercise-induced alterations in skeletal muscle, heart, liver, and serum metabolome33; the effects of the kinase inhibitor sorafenib/sunitinib/erlotinib on heart, muscle, liver, and plasma metabolism34, 35; and how human amylin impairs protein biosynthesis and alters major cellular signaling pathways in the heart, brain, and liver of humanized diabetic rat model in vivo were identified.36 Overall, the fewest significant metabolic alterations in the serum/plasma have been identified compared with other tissues. These fewer changes in serum/plasma compared with complimentary tissues suggest that only a fraction of the diverse and sometimes complementary changes in organ metabolism are reflected in the serum/plasma, which was the driving rationale for the present study focusing on metabolic changes in organs affected in sepsis.
Ileum and Jejunum (Gut)
The systemic evaluation of the metabolic changes in the gut, including the ileum and jejunum, has not been previously reported. Alterations in the ileum involved linoleic metabolism, branched-chain amino acids (valine/leucine/isoleucine), and phenylalanine (Figure 1D). The jejunum had alterations in arginine metabolism, citric acid cycle intermediates, and phenylalanine (Figure 2D). Recent studies have linked the gut immune system with critical care disease outcomes using immune-enhancing diets to combat gut failure in critical care37, 38 and sepsis.3, 39, 40, 41, 42, 43, 44, 45, 46 Immune-enhancing diets include linoleic acid and arginine, in addition to glutamine, fish oil (ω-3 fatty acids), and nucleotides.47, 48, 49 The semiessential amino acid arginine is beneficial under stress conditions,50 promoting nitrogen retention and mucosal barrier integrity,51 along with the release of anabolic hormones and improved wound healing.52 Enteral nutrition supplementation of arginine53, 54 with glutamine has been reported to be more effective in protecting the intestinal mucosa in cases of endotoxemia.55, 56, 57, 58 The findings in the present study raise the possibility that the benefits of supplementing with linoleic acid and arginine may be due to metabolic defects seen in the small intestine (ileum and jejunum). Furthermore, the metabolic deficits may be greater than previously recognized, and nutritional supplementation should also include sufficient protein levels that contain branched-chain amino acids and phenylalanine as they were also altered in the present study. It is clear that all elements of the intestine are impacted by sepsis and critical illness, including the epithelium, the immune system, and the microbiome.59 The metabolic changes herein are a compilation of all of these components, including sepsis-associated changes in intestinal permeability60, 61 and enterocyte cell death.62
Skeletal Muscle
Sepsis induces skeletal muscle atrophy and the proteolysis of the sarcomere.63 Interestingly, although the gastrocnemius muscles in our sepsis model showed a mild influx of inflammatory cells between the muscle fibers in all of the septic animals (and also in two of the control animals), no histologic signs of muscle atrophy were noted.64 A previous study identified the effects of endotoxemia on pig muscle protein metabolism and identified an increase in amino acid transamination from skeletal muscle (hindquarter) and portal-drained viscera (reflecting the intestinal tract), leading to glutamine and alanine effluxes thought to serve as substrates for the liver.3 The changes in skeletal muscle metabolism that occur after sepsis have not been detailed to the extent identified in the present study. Highly enriched pathways affected in the gastrocnemius skeletal muscle in sepsis in the current study included the following: i) amino-acyl-tRNA biosynthesis, ii) β-alanine metabolism, and iii) arginine and proline metabolism (Figure 3D). The altered metabolites related to aminoacyl-tRNA biosynthesis in skeletal muscle after sepsis in the current study include phenylalanine (+), methionine (+), threonine (+), glycine (−), glutamate (−), and aspartate (−) (Figure 3, B and C). Given the critical role of aminoacyl-tRNA biosynthesis in protein synthesis, which is suppressed in catabolic states like sepsis- and unloading-induced muscle atrophy, this is an interesting observation.65
The significant metabolites identified in the gastrocnemius skeletal muscle after 18 hours of sepsis that are involved in β-alanine metabolism include aspartic acid (−), pantothenic acid (−), carnosine (−), and anserine (−) (Figure 3C). Of these metabolites, a link between carnosine concentrations and sepsis has been reported previously.66, 67 Carnosine is a muscle biomolecule first identified at the beginning of the 20th Century.66, 68 Carnosine is a dipeptide that can scavenge α, β unsaturated aldehydes formed during oxidative stress–induced lipid peroxidation and reactive oxygen species formation.66, 69, 70, 71 In preclinical sepsis models, supplementing carnosine suppresses inflammatory cytokines (macrophage migration inhibitory factor, IL-8, and tumor necrosis factor-α), increases the NF-κB inhibitory factor IκBα, and decreases phosphorylated p65 and phosphorylated IIKα/β.67 Other work using a single dose of carnosine at the time of cecal ligation and puncture demonstrated that carnosine supplementation decreased serum and tissue malondialdehyde levels compared with control animals.66 Chronic β-alanine supplementation can significantly augment muscle carnosine concentrations,72, 73 presenting a viable therapeutic approach. The role of decreased β-alanine in the current model has not been tested directly with skeletal muscle function; direct supplementation of these metabolites will offer valuable information in future studies.
Metabolites significantly altered in gastrocnemius skeletal muscle after 18 hours of sepsis are involved in arginine and proline metabolism and include glutamic acid (−), aspartic acid (−), hydroxyproline (−), and urea (+) (Figure 3, B and C). Significantly decreased plasma glutamate was reported in intensive care unit patients with sepsis compared with healthy control subjects74 and has been hypothesized to be an underlying contributor to the sepsis state.46, 75 The metabolic response in catabolic states, including sepsis, is characterized by a negative nitrogen balance, with accelerated proteolysis of amino acids from the peripheral muscle.76, 77, 78, 79, 80 The catabolic response is mediated, in part, by glucocorticoids,79 which release amino acids in skeletal muscle for use by other tissues, such as the liver conversion of amino acids to glucose.76, 81 When rats are given the glucocorticoid dexamethasone, skeletal muscle glutamic acid and aspartic acid levels decrease significantly,82 as in the present study. Glutamate is generated and metabolized throughout the body, being most abundant in skeletal muscle, brain, kidneys, and liver.83
Liver
The present study is the first organ-level analysis, to our knowledge, investigating the metabolic changes in the liver during sepsis. The liver is a target of sepsis-related injury, including hypoxic hepatitis secondary to ischemia due to shock, cholestasis resulting from altered bile metabolism, and hepatocellular injury caused by inflammation.84 In a healthy state, bile salts are taken up by hepatocytes via the basolateral sodium taurocholate cotransporter and organic anion transporting proteins. The phospholipid export pump facilitates excretion of phosphatidylcholine, which forms mixed micelles in bile together with bile salts and cholesterol. Bile salts are reabsorbed in cholangiocytes as well as hepatocytes. Similar mechanisms for bile acid resorption exist in the enterocytes of the terminal ileum.85 During microbial infection, the liver switches from tolerogenic responses to immunogenic responses, while initiating the production of acute-phase proteins.86, 87 Changes in the liver's metabolic roles appear to be down-regulated, including the production and transport (reabsorption) of bile, as indicated by changes in plasma.84, 85 Studies of plasma in nonhuman primates (Macaca fascicularis) inoculated with E. coli also suggest altered liver metabolic states,88 and increased bile acids (taurine) and conjugated bile acids (eg, taurocholate and taurolithocholate 3-sulfate) were interpreted as sensitive measures of sepsis-induced cholestasis in a mouse model of sepsis.89
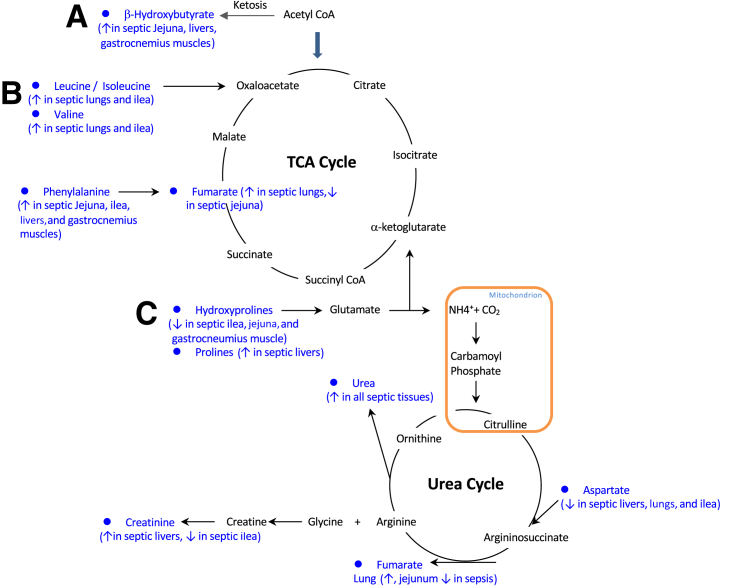
The significant metabolites identified in the liver after 18 hours of sepsis, which are involved with arginine and proline metabolism, include significant increases in ornithine, proline, creatinine, sarcosine, and urea (Figure 4D). In this pathway, arginine is metabolized to urea and ornithine (via arginase) or creatine.90 Creatine is metabolized to sarcosine or creatinine (via the intermediate creatine phosphate), and ornithine is metabolized to proline (via ornithine aminotransferase).90 The accumulation of serum ornithine, proline, creatinine, sarcosine, and urea has been reported to be significantly elevated in systemic inflammatory response syndrome and sepsis patients, in addition to accumulation of taurine.91 The significant decrease in liver linoleic acid and significant alterations in purine metabolism metabolites (increased guanine and urea and decreased inosine and adenosine) have not been reported previously, to our knowledge.
Lung
Metabolic changes in the lung after cecal ligation and puncture sepsis have been reported to include alterations in amino acid metabolism, purine metabolism, lipid metabolism, and energy regulation.92 Metabolites identified in the lung with the present study are involved with aminoacyl-tRNA biosynthesis, valine/leucine/isoleucine biosynthesis, alanine/aspartate/glutamate metabolism, and pantothenate and CoA biosynthesis, including significantly increased valine, alanine, isoleucine, leucine, and threonine and significantly decreased aspartic acid (Figure 5D). In addition, metabolites involved in arginine and proline metabolism included significantly increased fumaric acid, urea, and putrescine and significantly decreased aspartic acid (Figure 5D). The significant alterations identified in the present study did not overlap with those in the previous mouse study92 and may represent differences in the timing of the sepsis (18 hours versus 1 week), the model used (pig versus mouse), or the infection (i.v. P. aeruginosa versus cecal ligation and puncture).
The Urea Cycle and Sepsis
Urea nitrogen is present in all water pools [including plasma (blood urea nitrogen)] in the body and reflects the balance between urea production via the urea cycle as a by-product of protein degradation and urinary excretion. The urea cycle converts toxic ammonia to urea for excretion and occurs primarily in liver and, to a lesser extent, the kidneys.93, 94, 95 In the current study, urea as the only metabolite significantly increased in all tissues tested and indicates that urea is present in all water pools (intracellular and extracellular) (Table 1 and Figure 6). Because ammonia is absorbed by the lungs, muscle, and large intestine96 and is not produced in the lung or muscle (with smaller amounts made in the small intestines),93 the elevated urea concentrations likely represent a combination of increased production in relation to increased protein degradation and possibly reduced kidney excretion. In sepsis, blood urea nitrogen levels are elevated, reflecting the complex balance between urea production, metabolism, and excretion.97 Factors that contribute to elevated blood urea nitrogen include glomerular filtration and tubular reabsorption in the kidney, dietary protein intake, endogenous protein catabolism (eg, muscle atrophy), glucocorticoid-dependent catabolism, volume status, and gastrointestinal tract bleeding.97 The decrease in urea clearance in kidneys during endotoxemia involves the cytokine down-regulation of urea transporters,98 contributing to the buildup of systemic urea and accumulation in the ileum, jejunum, skeletal muscle, liver, and lung identified in the present study.
Figure 6.
Major metabolic pathways affected by sepsis, determined by nontargeted metabolomic analysis. Alterations in amino acids feeding into ketosis (A), the tricarboxylic acid (TCA) cycle (B), and the urea cycle (C) were observed in two or more tissues in the present study. Blue indicates metabolites significantly altered in two or more tissues (detailed in parentheses).
Conclusions
Herein, we present a multiorgan approach to identify tissue-specific metabolic changes during sepsis, identifying both unique and systematic alterations that may prove to be therapeutic targets that support the tissue-specific changes in metabolism (Table 1). After 18 hours of sepsis, the ileum and the liver had significant changes in linoleic acid metabolism; the ileum and lung had significant changes in valine/leucine/isoleucine metabolism; the jejunum, skeletal muscle, and liver had significant changes in arginine/proline metabolism; and the skeletal muscle and lung had significant changes in aminoacyl-tRNA biosynthesis, as determined by pathway analysis (Figures 1D, 2D, 3D, 4D, 5D, and Figure 6). Pathway analysis also identified tissue-specific changes in metabolic pathways, including changes in the citric acid cycle (jejunum), β-alanine metabolism (skeletal muscle), and purine metabolism (liver). The overlapping and tissue-specific changes in metabolism provide insight into the metabolic changes in sepsis that may contribute to subsequent organ dysfunction. This knowledge can potentially allow the future development of therapeutic interventions. These interventions could focus on treating multiple tissues (eg, supplementing deficits in arginine/proline metabolism in jejunum, skeletal muscle, and liver) or specific susceptible organs and their symptoms (eg, supplementing deficits in carnosine found only in skeletal muscle). Future studies can then determine the clinical significance of these organ metabolite deficits and determine their relationship to the underlying pathogenesis of sepsis.
Acknowledgments
We thank the assigned veterinarians and animal caretakers of the Arkansas Children's Hospital Research Institute for dedication in providing optimal care for the pigs in our experiment; and Joshua Spore, Bea Zoer, Eva M.C. Oosterlaken, and Sonya R. Mehta for skilled technical assistance in performing the animal experiments and/or sample processing. The RT-locked spectral library of metabolites was built on the Fiehn GC/MS Metabolomics RTL Library, kindly provided by Agilent.
Footnotes
Supported by the NIH grants R01GM084447 (N.E.D.), S10RR027047 (N.E.D.) and R01HL104129 (M.S.W.).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2019.05.021.
Supplemental Data
Heat map of metabolites identified using nontargeted metabolomics of ileum samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control ileum samples; n = 13 septic ileum samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of ileum samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A), disease-associated (B), predicted (C), or location (D) metabolite set using t-test significant metabolites. Insets: scales for P values.
Heat map of metabolites identified using nontargeted metabolomics of jejunum samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control jejunum samples; n = 13 septic jejunum samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of jejunum samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A), disease-associated (B), predicted (C), and location (D) metabolite set using t-test significant metabolites. Insets: scales for P values.
Heat map of metabolites identified using nontargeted metabolomics of gastrocnemius skeletal muscle samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control gastrocnemius samples; n = 13 septic gastrocnemius samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of gastrocnemius skeletal muscle samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A), disease-associated (B), predicted (C), and location (D) metabolite set using t-test significant metabolites. n = 9 vehicle controls; n = 13 sepsis challenged.
Heat map of metabolites identified using nontargeted metabolomics of liver samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control liver samples; n = 13 septic liver samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of liver samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated metabolites set using t-test significant metabolites. Inset: scale for P values.
Heat map of metabolites identified using nontargeted metabolomics of lung samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control lung samples; n = 13 septic lung samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of lung samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A) and location (B) metabolite set using significantly altered metabolites by t-test results. Insets: scales for P values.
References
- 1.Levy E., Slusser R.J., Ruebner B.H. Hepatic changes produced by a single dose of endotoxin in the mouse: electron microscopy. Am J Pathol. 1968;52:477–502. [PMC free article] [PubMed] [Google Scholar]
- 2.Singer M., Brealey D. Mitochondrial dysfunction in sepsis. Biochem Soc Symp. 1999;66:149–166. doi: 10.1042/bss0660149. [DOI] [PubMed] [Google Scholar]
- 3.Bruins M.J., Soeters P.B., Deutz N.E. Endotoxemia affects organ protein metabolism differently during prolonged feeding in pigs. J Nutr. 2000;130:3003–3013. doi: 10.1093/jn/130.12.3003. [DOI] [PubMed] [Google Scholar]
- 4.Seymour C.W., Yende S., Scott M.J., Pribis J., Mohney R.P., Bell L.N., Chen Y.F., Zuckerbraun B.S., Bigbee W.L., Yealy D.M., Weissfeld L., Kellum J.A., Angus D.C. Metabolomics in pneumonia and sepsis: an analysis of the GenIMS cohort study. Intensive Care Med. 2013;39:1423–1434. doi: 10.1007/s00134-013-2935-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Simon M., Morales J.M., Modesto-Alapont V., Gonzalez-Marrachelli V., Vento-Rehues R., Jorda-Minana A., Blanquer-Olivas J., Monleon D. Prognosis biomarkers of severe sepsis and septic shock by 1H NMR urine metabolomics in the intensive care unit. PLoS One. 2015;10:e0140993. doi: 10.1371/journal.pone.0140993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers A.J., McGeachie M., Baron R.M., Gazourian L., Haspel J.A., Nakahira K., Fredenburgh L.E., Hunninghake G.M., Raby B.A., Matthay M.A., Otero R.M., Fowler V.G., Rivers E.P., Woods C.W., Kingsmore S., Langley R.J., Choi A.M. Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS One. 2014;9:e87538. doi: 10.1371/journal.pone.0087538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neugebauer S., Giamarellos-Bourboulis E.J., Pelekanou A., Marioli A., Baziaka F., Tsangaris I., Bauer M., Kiehntopf M. Metabolite profiles in sepsis: developing prognostic tools based on the type of infection. Crit Care Med. 2016;44:1649–1662. doi: 10.1097/CCM.0000000000001740. [DOI] [PubMed] [Google Scholar]
- 8.Carre J.E., Singer M. Cellular energetic metabolism in sepsis: the need for a systems approach. Biochim Biophys Acta. 2008;1777:763–771. doi: 10.1016/j.bbabio.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Singer M., De Santis V., Vitale D., Jeffcoate W. Multiorgan failure is an adaptive, endocrine-mediated, metabolic response to overwhelming systemic inflammation. Lancet. 2004;364:545–548. doi: 10.1016/S0140-6736(04)16815-3. [DOI] [PubMed] [Google Scholar]
- 10.Ten Have G.A., Bost M.C., Suyk-Wierts J.C., van den Bogaard A.E., Deutz N.E. Simultaneous measurement of metabolic flux in portally-drained viscera, liver, spleen, kidney and hindquarter in the conscious pig. Lab Anim. 1996;30:347–358. doi: 10.1258/002367796780739862. [DOI] [PubMed] [Google Scholar]
- 11.Poeze M., Bruins M.J., Kessels F., Luiking Y.C., Lamers W.H., Deutz N.E. Effects of L-arginine pretreatment on nitric oxide metabolism and hepatosplanchnic perfusion during porcine endotoxemia. Am J Clin Nutr. 2011;93:1237–1247. doi: 10.3945/ajcn.110.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimmele T., Assadi A., Benatir F., Boselli E., Kaminski C., Arnal F., Lambert C., Goudable J., Chassard D., Allaouchiche B. Validation of a Pseudomonas aeruginosa porcine model of septic shock. J Infect. 2006;53:199–205. doi: 10.1016/j.jinf.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee R., Bultman S.J., Holley D., Hillhouse C., Bain J.R., Newgard C.B., Muehlbauer M.J., Willis M.S. Non-targeted metabolomics of Brg1/Brm double-mutant cardiomyocytes reveals a novel role for SWI/SNF complexes in metabolic homeostasis. Metabolomics. 2015;11:1287–1301. doi: 10.1007/s11306-015-0786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee R., He J., Spaniel C., Quintana M.T., Wang Z., Bain J., Newgard C.B., Muehlbauer M.J., Willis M.S. Non-targeted metabolomics analysis of cardiac Muscle Ring Finger-1 (MuRF1), MuRF2, and MuRF3 in vivo reveals novel and redundant metabolic changes. Metabolomics. 2015;11:312–322. doi: 10.1007/s11306-014-0695-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiehn O., Wohlgemuth G., Scholz M., Kind T., Lee D.Y., Lu Y., Moon S., Nikolau B. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 16.Roessner U., Wagner C., Kopka J., Trethewey R.N., Willmitzer L. Technical advance: simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- 17.Halket J.M., Przyborowska A., Stein S.E., Mallard W.G., Down S., Chalmers R.A. Deconvolution gas chromatography/mass spectrometry of urinary organic acids: potential for pattern recognition and automated identification of metabolic disorders. Rapid Commun Mass Spectrom. 1999;13:279–284. doi: 10.1002/(SICI)1097-0231(19990228)13:4<279::AID-RCM478>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 18.Mallard W.G., Reed J. US Department of Commerce, National Institute of Standards and Technology (NIST) Standard Reference Data Program; Gaithersburg, MD: 1997. Automated Mass Spectral Deconvolution and Identification System: AMDIS User Guide. [Google Scholar]
- 19.Stein S.E. An integrated method for spectrum extraction and compound identification from GC/MS data. J Am Soc Mass Spectrom. 1999;10:770–781. [Google Scholar]
- 20.Kind T., Wohlgemuth G., Lee D.Y., Lu Y., Palazoglu M., Shahbaz S., Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopka J., Schauer N., Krueger S., Birkemeyer C., Usadel B., Bergmuller E., Dormann P., Weckwerth W., Gibon Y., Stitt M., Willmitzer L., Fernie A.R., Steinhauser D. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 22.Styczynski M.P., Moxley J.F., Tong L.V., Walther J.L., Jensen K.L., Stephanopoulos G.N. Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Anal Chem. 2007;79:966–973. doi: 10.1021/ac0614846. [DOI] [PubMed] [Google Scholar]
- 23.Xia J., Wishart D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics. 2016;55:14.10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- 24.Xia J., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0: making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goeman J.J., van de Geer S.A., de Kort F., van Houwelingen H.C. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 27.Hummel M., Meister R., Mansmann U. GlobalANCOVA: exploration and assessment of gene group effects. Bioinformatics. 2008;24:78–85. doi: 10.1093/bioinformatics/btm531. [DOI] [PubMed] [Google Scholar]
- 28.Aittokallio T., Schwikowski B. Graph-based methods for analysing networks in cell biology. Brief Bioinform. 2006;7:243–255. doi: 10.1093/bib/bbl022. [DOI] [PubMed] [Google Scholar]
- 29.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J.D., Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25:1470–1471. doi: 10.1093/bioinformatics/btp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger S.I., Iyengar R., Ma'ayan A. AVIS: AJAX viewer of interactive signaling networks. Bioinformatics. 2007;23:2803–2805. doi: 10.1093/bioinformatics/btm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia J., Wishart D.S. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–2344. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 33.Starnes J.W., Parry T.L., O'Neal S.K., Bain J.R., Muehlbauer M.J., Honcoop A., Ilaiwy A., Christopher P.M., Patterson C., Willis M.S. Exercise-induced alterations in skeletal muscle, heart, liver, and serum metabolome identified by non-targeted metabolomics analysis. Metabolites. 2017;7:E40. doi: 10.3390/metabo7030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen B.C., Parry T.L., Huang W., Beak J.Y., Ilaiwy A., Bain J.R., Newgard C.B., Muehlbauer M.J., Patterson C., Johnson G.L., Willis M.S. Effects of the kinase inhibitor sorafenib on heart, muscle, liver and plasma metabolism in vivo using non-targeted metabolomics analysis. Br J Pharmacol. 2017;174:4797–4811. doi: 10.1111/bph.14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen B.C., Parry T.L., Huang W., Ilaiwy A., Bain J.R., Muehlbauer M.J., O'Neal S.K., Patterson C., Johnson G.L., Willis M.S. Non-targeted metabolomics analysis of the effects of tyrosine kinase inhibitors sunitinib and erlotinib on heart, muscle, liver and serum metabolism in vivo. Metabolites. 2017;7:E31. doi: 10.3390/metabo7030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ilaiwy A., Liu M., Parry T.L., Bain J.R., Newgard C.B., Schisler J.C., Muehlbauer M.J., Despa F., Willis M.S. Human amylin proteotoxicity impairs protein biosynthesis, and alters major cellular signaling pathways in the heart, brain and liver of humanized diabetic rat model in vivo. Metabolomics. 2016;12:95. doi: 10.1007/s11306-016-1022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sertaridou E., Papaioannou V., Kolios G., Pneumatikos I. Gut failure in critical care: old school versus new school. Ann Gastroenterol. 2015;28:309–322. [PMC free article] [PubMed] [Google Scholar]
- 38.Musso G., Gambino R., Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 39.Bruins M.J., Deutz N.E., Soeters P.B. Aspects of organ protein, amino acid and glucose metabolism in a porcine model of hypermetabolic sepsis. Clin Sci (Lond) 2003;104:127–141. doi: 10.1042/CS20020275. [DOI] [PubMed] [Google Scholar]
- 40.Bruins M.J., Lamers W.H., Meijer A.J., Soeters P.B., Deutz N.E. In vivo measurement of nitric oxide production in porcine gut, liver and muscle during hyperdynamic endotoxaemia. Br J Pharmacol. 2002;137:1225–1236. doi: 10.1038/sj.bjp.0704993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruins M.J., Luiking Y.C., Soeters P.B., Akkermans L.M., Deutz N.E. Effect of prolonged hyperdynamic endotoxemia on jejunal motility in fasted and enterally fed pigs. Ann Surg. 2003;237:44–51. doi: 10.1097/00000658-200301000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruins M.J., Soeters P.B., Lamers W.H., Deutz N.E. L-arginine supplementation in pigs decreases liver protein turnover and increases hindquarter protein turnover both during and after endotoxemia. Am J Clin Nutr. 2002;75:1031–1044. doi: 10.1093/ajcn/75.6.1031. [DOI] [PubMed] [Google Scholar]
- 43.Bruins M.J., Soeters P.B., Lamers W.H., Meijer A.J., Deutz N.E. L-arginine supplementation in hyperdynamic endotoxemic pigs: effect on nitric oxide synthesis by the different organs. Crit Care Med. 2002;30:508–517. doi: 10.1097/00003246-200203000-00003. [DOI] [PubMed] [Google Scholar]
- 44.Preiser J.C., Luiking Y., Deutz N. Arginine and sepsis: a question of the right balance? Crit Care Med. 2011;39:1569–1570. doi: 10.1097/CCM.0b013e318215c1ea. [DOI] [PubMed] [Google Scholar]
- 45.Vissers Y.L., Debats I.B., Luiking Y.C., Jalan R., van der Hulst R.R., Dejong C.H., Deutz N.E. Pros and cons of L-arginine supplementation in disease. Nutr Res Rev. 2004;17:193–210. doi: 10.1079/NRR200490. [DOI] [PubMed] [Google Scholar]
- 46.Luiking Y.C., Poeze M., Dejong C.H., Ramsay G., Deutz N.E. Sepsis: an arginine deficiency state? Crit Care Med. 2004;32:2135–2145. doi: 10.1097/01.ccm.0000142939.81045.a0. [DOI] [PubMed] [Google Scholar]
- 47.Gumus M., Tekin R., Firat U., Onder A., Kapan M., Boyuk A., Aldemir M., Kilinc C. The effects of pomegranate on bacterial translocation in rats with obstructive jaundice. Eur Rev Med Pharmacol Sci. 2013;17:1488–1494. [PubMed] [Google Scholar]
- 48.Marik P.E., Zaloga G.P. Immunonutrition in high-risk surgical patients: a systematic review and analysis of the literature. JPEN J Parenter Enteral Nutr. 2010;34:378–386. doi: 10.1177/0148607110362692. [DOI] [PubMed] [Google Scholar]
- 49.Martindale R.G., McClave S.A., Vanek V.W., McCarthy M., Roberts P., Taylor B., Ochoa J.B., Napolitano L., Cresci G., American College of Critical Care Medicine. A.S.P.E.N. Board of Directors Guidelines for the provision and assessment of nutrition support therapy in the adult critically ill patient: Society of Critical Care Medicine and American Society for Parenteral and Enteral Nutrition: executive summary. Crit Care Med. 2009;37:1757–1761. doi: 10.1097/CCM.0b013e3181a40116. [DOI] [PubMed] [Google Scholar]
- 50.Zhou M., Martindale R.G. Arginine in the critical care setting. J Nutr. 2007;137:1687S–1692S. doi: 10.1093/jn/137.6.1687S. [DOI] [PubMed] [Google Scholar]
- 51.Wang G., Wen J., Xu L., Zhou S., Gong M., Wen P., Xiao X. Effect of enteral nutrition and ecoimmunonutrition on bacterial translocation and cytokine production in patients with severe acute pancreatitis. J Surg Res. 2013;183:592–597. doi: 10.1016/j.jss.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 52.Mizock B.A. Immunonutrition and critical illness: an update. Nutrition. 2010;26:701–707. doi: 10.1016/j.nut.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Viana M.L., Santos R.G., Generoso S.V., Arantes R.M., Correia M.I., Cardoso V.N. Pretreatment with arginine preserves intestinal barrier integrity and reduces bacterial translocation in mice. Nutrition. 2010;26:218–223. doi: 10.1016/j.nut.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 54.Quirino I.E., Correia M.I., Cardoso V.N. The impact of arginine on bacterial translocation in an intestinal obstruction model in rats. Clin Nutr. 2007;26:335–340. doi: 10.1016/j.clnu.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Zhou X., Wu X., Yin Y., Zhang C., He L. Preventive oral supplementation with glutamine and arginine has beneficial effects on the intestinal mucosa and inflammatory cytokines in endotoxemic rats. Amino Acids. 2012;43:813–821. doi: 10.1007/s00726-011-1137-2. [DOI] [PubMed] [Google Scholar]
- 56.Coeffier M., Dechelotte P. Combined infusion of glutamine and arginine: does it make sense? Curr Opin Clin Nutr Metab Care. 2010;13:70–74. doi: 10.1097/MCO.0b013e328333c27f. [DOI] [PubMed] [Google Scholar]
- 57.Chamorro S., de Blas C., Grant G., Badiola I., Menoyo D., Carabano R. Effect of dietary supplementation with glutamine and a combination of glutamine-arginine on intestinal health in twenty-five-day-old weaned rabbits. J Anim Sci. 2010;88:170–180. doi: 10.2527/jas.2008-1698. [DOI] [PubMed] [Google Scholar]
- 58.Kul M., Vurucu S., Demirkaya E., Tunc T., Aydinoz S., Meral C., Kesik V., Alpay F. Enteral glutamine and/or arginine supplementation have favorable effects on oxidative stress parameters in neonatal rat intestine. J Pediatr Gastroenterol Nutr. 2009;49:85–89. doi: 10.1097/MPG.0b013e318198cd36. [DOI] [PubMed] [Google Scholar]
- 59.Klingensmith N.J., Coopersmith C.M. The gut as the motor of multiple organ dysfunction in critical illness. Crit Care Clin. 2016;32:203–212. doi: 10.1016/j.ccc.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fredenburgh L.E., Velandia M.M., Ma J., Olszak T., Cernadas M., Englert J.A., Chung S.W., Liu X., Begay C., Padera R.F., Blumberg R.S., Walsh S.R., Baron R.M., Perrella M.A. Cyclooxygenase-2 deficiency leads to intestinal barrier dysfunction and increased mortality during polymicrobial sepsis. J Immunol. 2011;187:5255–5267. doi: 10.4049/jimmunol.1101186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fink M.P. Intestinal epithelial hyperpermeability: update on the pathogenesis of gut mucosal barrier dysfunction in critical illness. Curr Opin Crit Care. 2003;9:143–151. doi: 10.1097/00075198-200304000-00011. [DOI] [PubMed] [Google Scholar]
- 62.Piton G., Belon F., Cypriani B., Regnard J., Puyraveau M., Manzon C., Navellou J.C., Capellier G. Enterocyte damage in critically ill patients is associated with shock condition and 28-day mortality. Crit Care Med. 2013;41:2169–2176. doi: 10.1097/CCM.0b013e31828c26b5. [DOI] [PubMed] [Google Scholar]
- 63.Kim H.C., Cho H.Y., Hah Y.S. Role of IL-15 in sepsis-induced skeletal muscle atrophy and proteolysis. Tuberc Respir Dis (Seoul) 2012;73:312–319. doi: 10.4046/trd.2012.73.6.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ten Have G.A.M., Deutz R.C.I., Engelen M., Wolfe R.R., Deutz N.E.P. Characteristics of a Pseudomonas aeruginosa induced porcine sepsis model for multi-organ metabolic flux measurements. Lab Anim. 2018;52:163–175. doi: 10.1177/0023677217718003. [DOI] [PubMed] [Google Scholar]
- 65.Gordon B.S., Kelleher A.R., Kimball S.R. Regulation of muscle protein synthesis and the effects of catabolic states. Int J Biochem Cell Biol. 2013;45:2147–2157. doi: 10.1016/j.biocel.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahin S., Oter S., Burukoglu D., Sutken E. The effects of carnosine in an experimental rat model of septic shock. Med Sci Monit Basic Res. 2013;19:54–61. doi: 10.12659/MSMBR.883758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun C., Wu Q., Zhang X., He Q., Zhao H. Mechanistic evaluation of the protective effect of carnosine on acute lung injury in sepsis rats. Pharmacology. 2017;100:292–300. doi: 10.1159/000479879. [DOI] [PubMed] [Google Scholar]
- 68.Kim M.Y., Kim E.J., Kim Y.N., Choi C., Lee B.H. Effects of alpha-lipoic acid and L-carnosine supplementation on antioxidant activities and lipid profiles in rats. Nutr Res Pract. 2011;5:421–428. doi: 10.4162/nrp.2011.5.5.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boldyrev A.A., Stvolinsky S.L., Fedorova T.N., Suslina Z.A. Carnosine as a natural antioxidant and geroprotector: from molecular mechanisms to clinical trials. Rejuvenation Res. 2010;13:156–158. doi: 10.1089/rej.2009.0923. [DOI] [PubMed] [Google Scholar]
- 70.Nikolic J., Stojanovic I., Pavlovic R., Sokolovic D., Bjelakovic G., Beninati S. The role of L-arginine in toxic liver failure: interrelation of arginase, polyamine catabolic enzymes and nitric oxide synthase. Amino Acids. 2007;32:127–131. doi: 10.1007/s00726-006-0309-y. [DOI] [PubMed] [Google Scholar]
- 71.Guliaeva N.V., Dupin A.M., Levshina I.P., Obidin A.B., Boldyrev A.A. Carnosine prevents the activation of free-radical lipid oxidation during stressBiull Eksp Biol Med. 1989;107:144–147. Russian. [PubMed] [Google Scholar]
- 72.Stegen S., Bex T., Vervaet C., Vanhee L., Achten E., Derave W. beta-Alanine dose for maintaining moderately elevated muscle carnosine levels. Med Sci Sports Exerc. 2014;46:1426–1432. doi: 10.1249/MSS.0000000000000248. [DOI] [PubMed] [Google Scholar]
- 73.Stellingwerff T., Decombaz J., Harris R.C., Boesch C. Optimizing human in vivo dosing and delivery of beta-alanine supplements for muscle carnosine synthesis. Amino Acids. 2012;43:57–65. doi: 10.1007/s00726-012-1245-7. [DOI] [PubMed] [Google Scholar]
- 74.Luiking Y.C., Poeze M., Ramsay G., Deutz N.E. Reduced citrulline production in sepsis is related to diminished de novo arginine and nitric oxide production. Am J Clin Nutr. 2009;89:142–152. doi: 10.3945/ajcn.2007.25765. [DOI] [PubMed] [Google Scholar]
- 75.Poeze M., Luiking Y.C., Breedveld P., Manders S., Deutz N.E. Decreased plasma glutamate in early phases of septic shock with acute liver dysfunction is an independent predictor of survival. Clin Nutr. 2008;27:523–530. doi: 10.1016/j.clnu.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 76.Douglas R.G., Shaw J.H. Metabolic response to sepsis and trauma. Br J Surg. 1989;76:115–122. doi: 10.1002/bjs.1800760205. [DOI] [PubMed] [Google Scholar]
- 77.Wernerman J., Vinnars E. The effect of trauma and surgery on interorgan fluxes of amino acids in man. Clin Sci (Lond) 1987;73:129–133. doi: 10.1042/cs0730129. [DOI] [PubMed] [Google Scholar]
- 78.Rennie M.J. Muscle protein turnover and the wasting due to injury and disease. Br Med Bull. 1985;41:257–264. doi: 10.1093/oxfordjournals.bmb.a072060. [DOI] [PubMed] [Google Scholar]
- 79.Kagan R.J., Matsuda T., Hanumadass M., Castillo B., Jonasson O. The effect of burn wound size on ureagenesis and nitrogen balance. Ann Surg. 1982;195:70–74. doi: 10.1097/00000658-198201001-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitch W.E. Amino acid release from the hindquarter and urea appearance in acute uremia. Am J Physiol. 1981;241:E415–E419. doi: 10.1152/ajpendo.1981.241.6.E415. [DOI] [PubMed] [Google Scholar]
- 81.Frayn K.N. Hormonal control of metabolism in trauma and sepsis. Clin Endocrinol (Oxf) 1986;24:577–599. doi: 10.1111/j.1365-2265.1986.tb03288.x. [DOI] [PubMed] [Google Scholar]
- 82.Ardawi M.S., Jamal Y.S. Glutamine metabolism in skeletal muscle of glucocorticoid-treated rats. Clin Sci (Lond) 1990;79:139–147. doi: 10.1042/cs0790139. [DOI] [PubMed] [Google Scholar]
- 83.Taylor L., Curthoys N.P. Glutamine metabolism: role in acid-base balance∗. Biochem Mol Biol Educ. 2004;32:291–304. doi: 10.1002/bmb.2004.494032050388. [DOI] [PubMed] [Google Scholar]
- 84.Strnad P., Tacke F., Koch A., Trautwein C. Liver: guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- 85.Geier A., Fickert P., Trauner M. Mechanisms of disease: mechanisms and clinical implications of cholestasis in sepsis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:574–585. doi: 10.1038/ncpgasthep0602. [DOI] [PubMed] [Google Scholar]
- 86.Heymann F., Tacke F. Immunology in the liver: from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13:88–110. doi: 10.1038/nrgastro.2015.200. [DOI] [PubMed] [Google Scholar]
- 87.Bauer M., Press A.T., Trauner M. The liver in sepsis: patterns of response and injury. Curr Opin Crit Care. 2013;19:123–127. doi: 10.1097/MCC.0b013e32835eba6d. [DOI] [PubMed] [Google Scholar]
- 88.Langley R.J., Tipper J.L., Bruse S., Baron R.M., Tsalik E.L., Huntley J., Rogers A.J., Jaramillo R.J., O'Donnell D., Mega W.M., Keaton M., Kensicki E., Gazourian L., Fredenburgh L.E., Massaro A.F., Otero R.M., Fowler V.G., Jr., Rivers E.P., Woods C.W., Kingsmore S.F., Sopori M.L., Perrella M.A., Choi A.M., Harrod K.S. Integrative “omic” analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes. Am J Respir Crit Care Med. 2014;190:445–455. doi: 10.1164/rccm.201404-0624OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Recknagel P., Gonnert F.A., Westermann M., Lambeck S., Lupp A., Rudiger A., Dyson A., Carre J.E., Kortgen A., Krafft C., Popp J., Sponholz C., Fuhrmann V., Hilger I., Claus R.A., Riedemann N.C., Wetzker R., Singer M., Trauner M., Bauer M. Liver dysfunction and phosphatidylinositol-3-kinase signalling in early sepsis: experimental studies in rodent models of peritonitis. PLoS Med. 2012;9:e1001338. doi: 10.1371/journal.pmed.1001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morris S.M., Jr. Arginine metabolism: boundaries of our knowledge. J Nutr. 2007;137:1602S–1609S. doi: 10.1093/jn/137.6.1602S. [DOI] [PubMed] [Google Scholar]
- 91.Su L., Li H., Xie A., Liu D., Rao W., Lan L., Li X., Li F., Xiao K., Wang H., Yan P., Li X., Xie L. Dynamic changes in amino acid concentration profiles in patients with sepsis. PLoS One. 2015;10:e0121933. doi: 10.1371/journal.pone.0121933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y., Liu Y., Cao Q., Shi X., Lu H., Gao S., Yang R. Metabolomic analysis for the protective effects of mangiferin on sepsis-induced lung injury in mice. Biomed Chromatogr. 2018;32:e4208. doi: 10.1002/bmc.4208. [DOI] [PubMed] [Google Scholar]
- 93.Huizenga J.R., Gips C.H., Tangerman A. The contribution of various organs to ammonia formation: a review of factors determining the arterial ammonia concentration. Ann Clin Biochem. 1996;33:23–30. doi: 10.1177/000456329603300103. [DOI] [PubMed] [Google Scholar]
- 94.Emmanuel B. Urea cycle enzymes in tissues (liver, rumen epithelium, heart, kidney, lung, and spleen) of sheet (Ovis aries) Comp Biochem Physiol. 1980;65B:693–697. [Google Scholar]
- 95.Hoagland R., Mansfield C.M. Glycolytic properties of muscular tissue. J Biol Chem. 1917;31:501–517. [Google Scholar]
- 96.Klisiecki A.J. The absorption of ammonia by the lungs and heart. Exp Physiol. 1934;24:225–235. [Google Scholar]
- 97.Beier K., Eppanapally S., Bazick H.S., Chang D., Mahadevappa K., Gibbons F.K., Christopher K.B. Elevation of blood urea nitrogen is predictive of long-term mortality in critically ill patients independent of “normal” creatinine. Crit Care Med. 2011;39:305–313. doi: 10.1097/CCM.0b013e3181ffe22a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmidt C., Hocherl K., Bucher M. Cytokine-mediated regulation of urea transporters during experimental endotoxemia. Am J Physiol Renal Physiol. 2007;292:F1479–F1489. doi: 10.1152/ajprenal.00460.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heat map of metabolites identified using nontargeted metabolomics of ileum samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control ileum samples; n = 13 septic ileum samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of ileum samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A), disease-associated (B), predicted (C), or location (D) metabolite set using t-test significant metabolites. Insets: scales for P values.
Heat map of metabolites identified using nontargeted metabolomics of jejunum samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control jejunum samples; n = 13 septic jejunum samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of jejunum samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A), disease-associated (B), predicted (C), and location (D) metabolite set using t-test significant metabolites. Insets: scales for P values.
Heat map of metabolites identified using nontargeted metabolomics of gastrocnemius skeletal muscle samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control gastrocnemius samples; n = 13 septic gastrocnemius samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of gastrocnemius skeletal muscle samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A), disease-associated (B), predicted (C), and location (D) metabolite set using t-test significant metabolites. n = 9 vehicle controls; n = 13 sepsis challenged.
Heat map of metabolites identified using nontargeted metabolomics of liver samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control liver samples; n = 13 septic liver samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of liver samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated metabolites set using t-test significant metabolites. Inset: scale for P values.
Heat map of metabolites identified using nontargeted metabolomics of lung samples obtained from sepsis-challenged pigs compared with vehicle controls. n = 9 control lung samples; n = 13 septic lung samples.
Enrichment analysis results obtained from results of nontargeted metabolomics of lung samples obtained from sepsis-challenged pigs compared with vehicle controls. Enrichment by pathway-associated (A) and location (B) metabolite set using significantly altered metabolites by t-test results. Insets: scales for P values.