Abstract
Objective:
Mutational signatures provide insights into the biological processes shaping tumor genomes and may inform patient therapy. We sought to define the mutational signatures of i) endometrioid and serous endometrial carcinomas (ECs), stratified into the four molecular subtypes, ii) uterine carcinosarcomas, and iii) matched primary and metastatic ECs.
Methods:
Whole-exome sequencing MC3 data from primary endometrioid and serous carcinomas (n=232) and uterine carcinosarcomas (n=57) from The Cancer Genome Atlas (TCGA), and matched primary and metastatic ECs (n=61, 26 patients) were reanalyzed, subjected to mutational signature analysis using deconstructSigs, and correlated with clinicopathologic and genomic data.
Results:
POLE (ultramutated) and MSI (hypermutated) molecular subtypes displayed dominant mutational signatures associated with POLE mutations (15/17 cases) and microsatellite instability (55/65 cases), respectively. Most endometrioid and serous carcinomas of copy-number low (endometrioid) and copy-number high (serous-like) molecular subtypes, and carcinosarcomas displayed a dominant aging-associated signature 1. Only 15% (9/60) of copy-number high (serous-like) ECs had a dominant signature 3 (homologous recombination DNA repair deficiency (HRD)-related), a prevalence significantly lower than that found in high-grade serous ovarian carcinomas (54%, p<0.001) or basal-like breast cancers (46%, p<0.001). Shifts from aging- or POLE- to MSI-related mutational processes were observed in the progression from primary to metastatic ECs in a subset of cases.
Conclusions:
The mutational processes underpinning ECs vary even among tumors of the same TCGA molecular subtype and in the progression from primary to metastatic ECs. Only a minority of copy-number high (serous-like) ECs display genomics features of HRD and would likely benefit from HRD-directed therapies.
1. INTRODUCTION
Endometrial cancer (EC) is the fourth most common female malignancy in the United States[1]. Since 2000, both the incidence and death rates in EC have been steadily increasing, and despite advances in therapy, when compared to 1975, the overall 5-year relative survival for patients with EC has decreased[2]. Hence, it is imperative to gain more insights into the biology of EC to improve management of the disease and advance the use of targeted therapies. In 2013, The Cancer Genome Atlas (TCGA) identified four distinct EC molecular subtypes, which carry prognostic and predictive information, including the POLE (ultramutated) subtype characterized by polymerase epsilon (POLE) exonuclease domain mutations (EDMs) and very high mutation rates, the MSI (hypermutated) subtype of microsatellite unstable tumors with high mutation rates, the copy-number low (endometrioid) subtype characterized by CTNNB1 mutations, and the copy-number high (serous-like) subtype characterized by high levels of copy number alterations and recurrent TP53 mutations[3]. Heterogeneity in the clinical behavior of tumors of the same molecular subtype has been documented. For instance, patients with ECs of POLE subtype are reported to have a favorable prognosis, however POLE ECs with recurrent disease are on record, suggesting that molecular heterogeneity may be present in this seemingly homogenous subtype[4]. A formal classification of uterine carcinosarcomas into the molecular subtypes has not been reported to date, however extensive copy-number alterations and recurrent somatic mutations akin to those observed in both endometrioid and serous ECs have been found[5]. Whilst TCGA focused on primary untreated ECs, whole-exome sequencing (WES) analysis of primary ECs and matched abdominopelvic metastases revealed novel recurrent alterations in metastatic ECs, temporal heterogeneity of driver genetic alterations, and demonstrated that <50% of somatic mutations were conserved from primary to metastasis within ECs[6].
Shortly after the publication of the EC TCGA study, Alexandrov and colleagues derived ‘mutational signatures’ from the analysis of cancer genomes[7], based on the principle that the type of nucleotide substitution and their context (i.e. the bases immediately before and after the substitution) may provide important information about the oncogenic processes operative in the development and progression of a cancer. By applying mathematical algorithms to the aggregate of somatic mutations present in individual cancers, 30 mutational signatures have been identified. These include signatures that can be indicative of specific forms of DNA repair defects in cancer cells (e.g. homologous recombination (HR) DNA repair defects (signature 3), DNA mismatch repair (MMR) defects (signatures 6, 15, 20 and 26) and POLE EDMs (signatures 10 and 14)), exposure to mutagenic/carcinogenic stimuli (e.g. UV light (signature 7) and tobacco (signatures 4 and 29)) and other forms of genotoxic insults (e.g. activity of the APOBEC enzymes (signatures 2 and 13))[8,9]. These phenomena leave characteristic imprints/scars in the cancer genome in the form of specific patterns of mutations (i.e. mutational signatures)[8,9]. With variable levels of understanding and interpretation of genomic test results among clinicians[10], mutational signatures may help clarify the driving biological processes and assist in guiding therapy[9], even in the absence of a targetable mutation.
In a recent study of gynecologic and breast cancers by TCGA, MSI-high ECs were found to often display DNA MMR mutational signatures[11], however a detailed systematic evaluation of the mutational signatures in ECs has not been performed to date. In addition, in other cancer types, there is evidence to suggest that the dominant mutational signatures present in the primary tumors may differ from those identified in the metastatic lesions from the same patients[12,13], a phenomenon that has yet to be investigated in EC. Hence, in this study, we sought to define the mutational signatures of i) endometrioid and serous ECs stratified into the four molecular subtypes, ii) uterine carcinosarcomas, a histologic EC subtype associated with poor outcome, and iii) primary ECs matched to their metastatic lesions.
2. MATERIAL AND METHODS
2.1. Whole-exome sequencing (WES) data from TCGA
The recently updated WES-derived somatic mutation MC3 data[14] of the 232 endometrioid and serous ECs with molecular subtype classification[3], uterine carcinosarcomas (n=57)[5], high-grade serous ovarian cancers (HGSOC; n=233; n=225 with ≥20 somatic single nucleotide variants, SNVs)[15] and basal-like breast cancers (n=147; n=146 with ≥20 somatic SNVs)[16] from TCGA were retrieved from the Genomic Data Commons (GDC; https://gdc.cancer.gov/about-data/publications/mc3-2017)[14].
2.2. Mutational signatures
Mutational signatures were defined by deconstructsigs using all SNVs[17] at default parameters, as previously described[18], for samples with ≥20 somatic SNVs[19].
2.3. MSIsensor and MLH1 promoter methylation scores
For the quantification of microsatellite instability (MSI), MSIsensor was employed, as described by Niu et al.[20], and samples with an MSIsensor score ≥3.5 were deemed MSI-high. Level 3 Illumina HumanMethylation450K methylation data for endometrioid and serous ECs with molecular subtype classification (n=232)[3] and uterine carcinosarcomas (n=57)[5] were obtained from the TCGA GDC Legacy Archive. A beta-value threshold of <0.3 was used for unmethylated DNA/CpG locus, as described[21].
2.4. Large-scale state transitions (LSTs), mutations affecting homologous recombination (HR) DNA repair genes and deletion length
LST scores, a genomic feature of HR deficiency (cut-off ≥15)[22], and information on bi-allelic somatic and germ line pathogenic mutations in a curated list of 102 HR-related DNA repair genes in copy-number high (serous-like) ECs, HGSOCs and basal-like breast cancers were obtained from Riaz et al.[23]. In addition, the length of small deletions (indels) was assessed in these tumors, as HR-defective cancers have been shown to have an enrichment for deletions ≥5bp [24].
2.5. WES analysis of primary and matched metastatic ECs
WES data in the form of Binary Sequence Alignment Map (BAM) files from primary ECs and matched metastatic lesions from 26 EC patients described by Gibson et al.[6] were obtained from dbGaP (accession phs001127.v1.p1). Sequencing data analysis was performed as previously described[18](Supplementary Methods). Only somatic mutations with ≥20 reads in the respective normal samples were considered. Somatic copy number alterations and loss of heterozygosity (LOH) were defined using FACETS[25], as previously described[18]. The cancer cell fractions (CCFs) of all mutations were computed using ABSOLUTE (v1.0.6)[26], as previously described[18].
2.6. Phylogenetic tree construction
A given mutation was considered “shared” if it was present in both the primary and metastatic lesion. We defined mutations “private to the primary lesion” and “private to the metastatic lesion” as those present only in the primary tumor or only in the metastasis, respectively. Mutational signatures were defined for shared and private mutations separately using deconstructSigs[17] as described above, for cases with ≥20 shared and/or private SNVs. To reconstruct the phylogeny of the primary and matched metastatic ECs we used Treeomics[27] based on all synonymous and non-synonymous mutations identified, as previously described[18].
2.7. Statistical analyses
Comparisons of LST scores and indel length between different tumor types were performed using the Mann-Whitney U test. Associations between specific clinicopathologic features/molecular features/molecular signatures and molecular subtypes were tested using Fisher’s exact tests. Progression-free survival curves were calculated using the Kaplan–Meier method with the log-rank test. P-values of <0.05 were considered statistically significant. Statistical analyses were performed using IBM SPSS v24.0.0.0 and Prism 7.
3. RESULTS
3.1. Mutational signatures of primary endometrioid and serous ECs stratified according to molecular subtypes
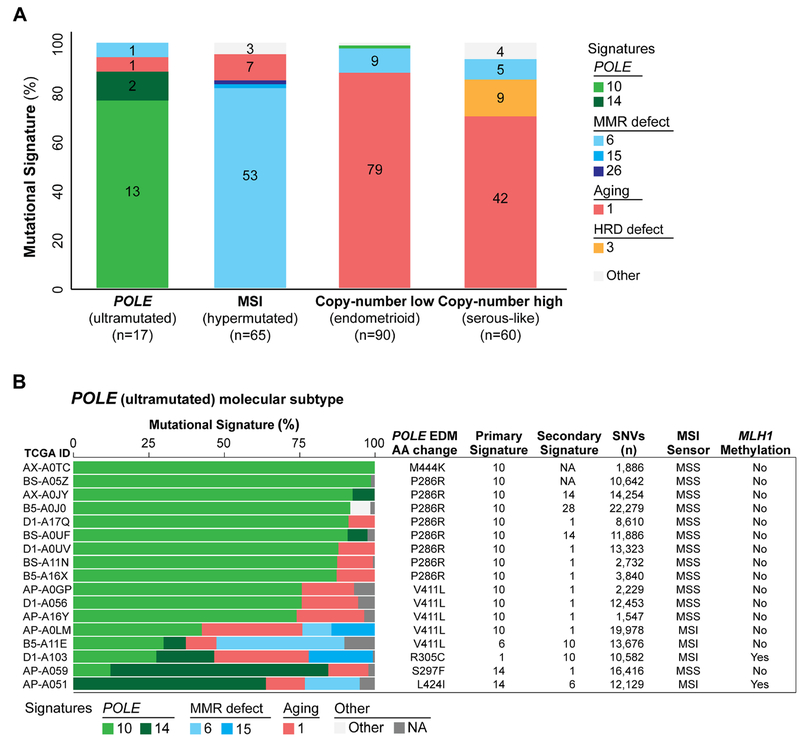
We first defined whether primary treatment-naïve endometrioid and serous ECs from TCGA stratified according to the molecular subtypes would have distinct mutational signatures. This analysis revealed that 76% (13/17) of the ECs of POLE (ultramutated) subtype had a dominant signature 10 associated with POLE mutations[28]. The majority of ECs of MSI (hypermutated) subtype (85%; 55/65) had a dominant mutational signature associated with defective DNA MMR (53/65, signature 6; 1/65, signature 15; 1/65, signature 26; Fig. 1a; Supplementary Figs. S1 and S2). Conversely, the majority of ECs of copy-number low (endometrioid) and copy-number high (serous-like) subtypes had the aging-related signature 1 as the most prevalent dominant mutational signature (88%, 79/90 and 70%, 42/60, respectively; Figs. 1 and 2, Supplementary Figs. S1 and S3).
Figure 1. Mutational signatures of primary endometrioid and serous endometrial cancers stratified according to molecular subtype.
A) The dominant mutational signatures of 232 endometrioid and serous endometrial carcinomas from The Cancer Genome Atlas (TCGA) [3] stratified according to molecular subtype. Mutational signatures are color-coded according to the legend on the right, and the number of endometrial cancers displaying a specific dominant mutational signature is shown in each bar. B) The mutational signatures of the 17 endometrial cancers of the 232 endometrioid and serous endometrial cancers studied by TCGA [3] classified as of POLE (ultramutated) molecular subtype, harboring POLE exonuclease domain mutations, are shown. Cases are sorted by their proportion of signature 10 associated with POLE alterations, and molecular signatures are color-coded according to the legend. Information on the exact POLE exonuclease domain mutation (amino acid), the primary and secondary molecular signatures, the number of non-synonymous single nucleotide variants (SNVs), microsatellite instability (MSIsensor score) and MLH1 promoter methylation is provided for each case on the right. AA, amino acid change; EDM, exonuclease domain mutation; MSI, microsatellite instability; MSS, microsatellite stable.
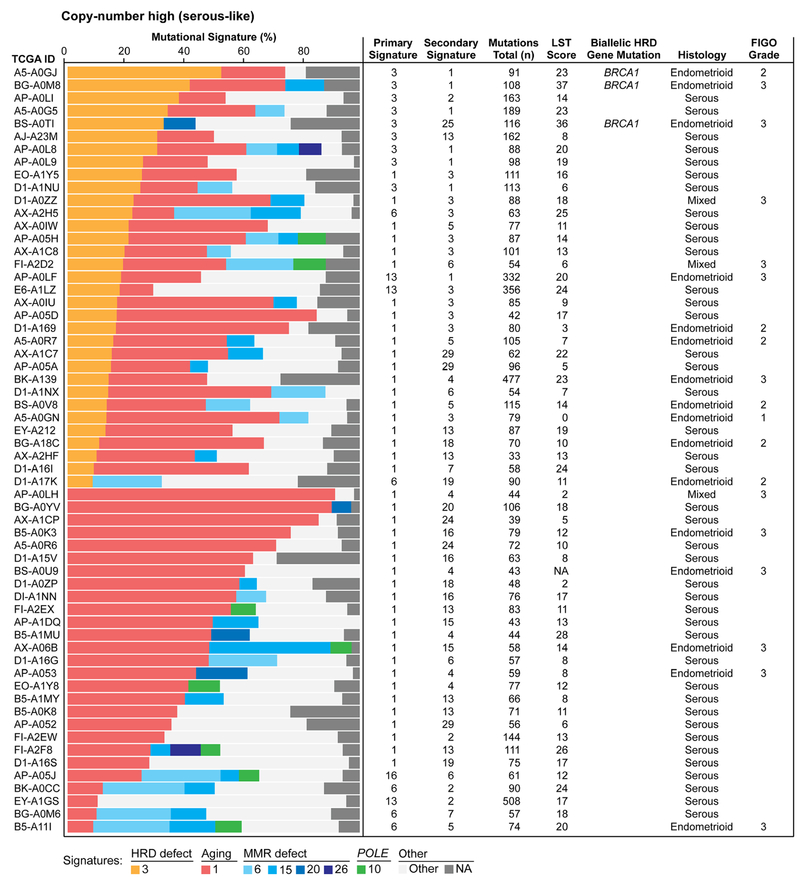
Figure 2. Mutational signatures and homologous recombination DNA repair defects in endometrial cancers of copy-number high (serous-like) molecular subtype.
Mutational signatures of 60 endometrioid and serous endometrial carcinomas of copy-number high (serous-like) molecular subtype from The Cancer Genome Atlas (TCGA), sorted according to the proportion of mutational signatures 3 and 1. Mutational signatures are color-coded according to the legend on the bottom. Information on the primary and secondary signatures, total number of somatic mutations, large-scale state transition (LST) scores, bi-allelic homologous recombination DNA repair (HRD) gene mutations, histology and FIGO grade for endometrioid and mixed endometrial carcinomas are provided for each case on the right.
In the TCGA study, POLE ECs were found to have the best and copy-number high (serous-like) ECs the worst progression-free survival[3]. As a hypothesis-generating analysis, we used the dominant mutational signatures to categorize ECs into i) dominant POLE signatures 10 or 14, ii) dominant MSI signatures 6, 15, 20 or 26, iii) dominant aging signature 1, copy-number low and iv) dominant aging signature 1, copy-number high. This stratification was found to be significantly associated with progression-free survival (p=0.045, log-rank) and recapitulated the survival curves based on the molecular subtypes reported by TCGA[3](Supplementary Fig. 1b).
3.2. POLE EDM type and POLE mutational signatures
Of the 17 cases within the POLE (ultramutated) molecular subtype, all of which harbored a POLE EDM, 13 ECs displayed a dominant signature 10 associated with POLE mutations, one a dominant aging-related signature 1, one a dominant DNA MMR signature 6, and two ECs had a dominant signature 14 (Fig. 1b). In the latter two cases, co-occurrence of POLE EDMs with either MLH1 gene promoter methylation or MSH6 mutations were detected, consistent with the notion that signature 14 is present in tumors with concurrent loss of POLE and MMR function[29,30]. We also observed that the percentage of signature 10 present in a given EC varied according to the specific POLE EDM. ECs with M444K or P286R POLE hotspot EDMs had the highest percentage of mutational signature 10 (median 91%, range 87-100%), followed by ECs with V411L mutations (median 74%, range 30-76%). By contrast, the three ECs with POLE EDMs not affecting known hotspots either had a dominant aging-related signature 1 (R305C) or a dominant signature 14 (i.e. S297F and L424I; Fig. 1b). Furthermore, POLE ECs with a dominant signature 10 were significantly less likely to be MSI-high, as defined by MSI Sensor, than those with dominant signatures 1, 6 or 14 (8% vs 75% MSI-high, p=0.02, Fisher’s exact test; Fig. 1b)[30]. Of note, we identified one copy-number low (endometrioid) EC with a dominant molecular signature 10 (Fig. 1a; Supplementary Figs. S1 and S3); this case harbored a POLE L424V EDM, a known pathogenic mutation associated with familial colonic polyposis and colorectal cancer[31], but not considered to constitute a POLE hotspot mutation in ECs.
3.3. HR DNA repair defects in copy-number high (serous-like) ECs
Nine copy-number high (serous-like) cancers (15%) had a dominant mutational signature 3 associated with defective homologous recombination DNA repair (HRD), and were of endometrioid (n=3) and serous (n=6) histologic subtypes. Of the remaining 51 copy-number high (serous-like) ECs, ten (10/51; 20%) had a secondary signature 3; of these, seven were of serous, two of endometrioid and one of mixed histologic subtype (Fig. 2).
Of the nine ECs with a dominant signature 3, six (67%) displayed high LST scores[22], another genomic feature associated with HR deficiency. Furthermore, three of these nine ECs, all with high LST scores, harbored bi-allelic pathogenic germline BRCA1 mutations, and were of endometrioid histology (Fig. 2). Also, five ECs with a secondary signature 3 (50%) had high LST scores, however no bi-allelic genetic alterations affecting HRD genes were found.
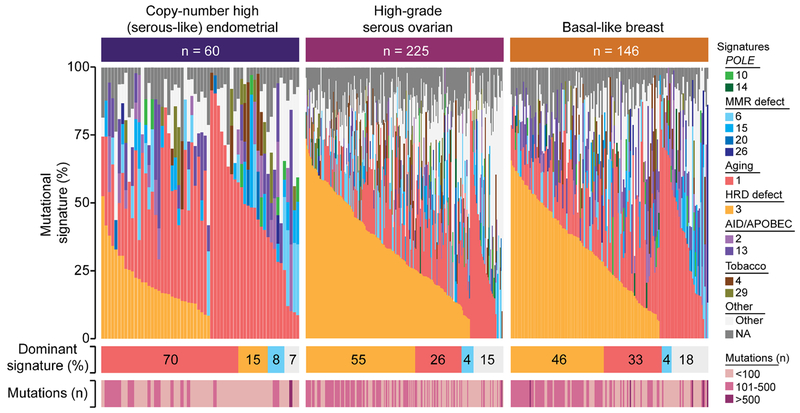
In the EC study by TCGA[3], it was reported that copy-number high (serous-like) ECs shared many molecular features with both HGSOCs and basal-like breast cancers, and, therefore, some of the treatment paradigms applicable to these cancers could also be extended to copy-number high (serous-like) ECs. HGSOCs and basal-like breast cancers frequently harbor alterations affecting the HRD pathway[15,16,23]. Mutational signature analysis performed here revealed that 55% of HGSOCs (132/225)[15] and 46% of basal-like breast cancers (67/146)[16] harbored a dominant HRD-related signature 3, as compared to 15% of copy-number high (serous-like) ECs (p<0.001 and p=0.007, respectively, Fisher’s exact test; Fig. 3). Consistent with this observation, other features of defective HRD, including LST scores, the average length of small deletions[24] and bi-allelic genetic alterations in HRD genes were significantly lower in ECs of copy-number high (serous-like) molecular subtype than in HGSOCs and basal-like breast cancers (Supplementary Fig. S4). These data suggest that despite the high prevalence of TP53 mutations and high levels of copy number alterations in copy-number high (serous-like) ECs, the DNA repair defects and mutational processes involved in the tumorigenesis of the majority of copy-number high (serous-like) ECs may differ from those of HGSOCs and basal-like breast cancers.
Figure 3. Mutational signature 3 associated with homologous recombination DNA repair defects in copy-number high (serous-like) endometrial cancers, high-grade serous ovarian cancers and basal-like breast cancers from The Cancer Genome Atlas.
Mutational signatures were defined for copy-number high (serous-like) endometrial cancers (n=60), high-grade serous ovarian cancers (n=225) and basal-like breast cancers (n=146) with ≥20 single nucleotide variants from The Cancer Genome Atlas (TCGA) [3,15,16], color-coded according to the legend. Cases are sorted by the proportion of signatures 3 and 1. The percentage of each dominant signature and the total number of non-synonymous somatic mutations for each case are shown below. HRD, homologous recombination DNA repair deficiency; MSI, microsatellite instability.
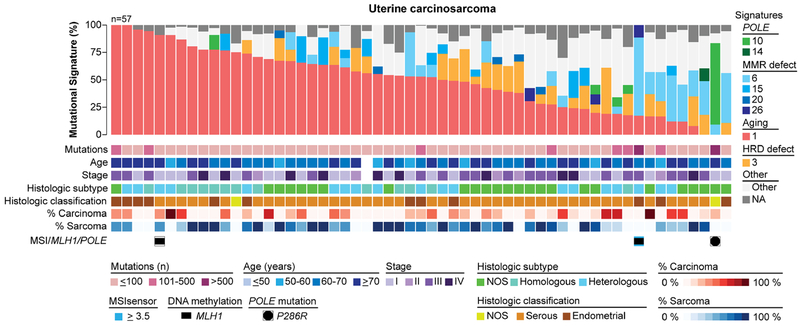
3.4. Mutational signatures of uterine carcinosarcoma
Uterine carcinosarcomas are aggressive biphasic neoplasms with high-grade malignant epithelial and mesenchymal elements. We sought to define whether the mutational processes underlying uterine carcinosarcomas would differ from those of endometrioid and serous carcinomas. Mutational signature analysis of the 57 uterine carcinosarcomas from TCGA[5] revealed a dominant aging-related signature 1 in 77% (44/57) of cases (Fig. 4). In addition, 4/57 (7%) uterine carcinosarcomas had a dominant mutational signature 3 associated with defective HRD, and 12/57 (21%) had a secondary signature 3; however, none of these harbored bi-allelic genomic alterations affecting HR-related genes[23]. One uterine carcinosarcoma had a dominant POLE-related signature 10 and a P286R POLE hotspot EDM, and six cases had a dominant defective DNA MMR signature (signatures 6 and 15; Fig. 4). No associations between histologic type (e.g. homologous/heterologous), histologic classification (e.g. endometrioid/serous), percentage of carcinoma/sarcoma present in the frozen tissue subjected to WES and the mutational signatures were found (Fig. 4).
Figure 4. Mutational signatures of uterine carcinosarcomas.
Mutational signatures for 57 primary treatment-naïve uterine carcinosarcomas from The Cancer Genome Atlas (TCGA) [5], sorted by the proportion of signature 1, and color-coded according to the legend. The number of non-synonymous somatic mutations per case, age at diagnosis, stage, histologic subtype, histologic classification, percentage of carcinoma in the frozen tissue subjected to whole-exome sequencing, percentage of sarcoma in the frozen tissue subjected to whole-exome sequencing, MSIsensor score, MLH1 promoter methylation and POLE mutations are provided below the signatures. HRD, homologous recombination DNA repair deficiency; MSI, microsatellite instability; NOS, not otherwise specified.
Taken together, uterine carcinosarcomas not only harbor mutations found in both endometrioid and serous carcinomas, but also the mutational processes that shaped the genomes of uterine carcinosarcomas were similar to those found in endometrioid and serous carcinomas of copy-number low (endometrioid) and copy-number high (serous-like) subtypes.
3.5. Mutational signatures in the progression from primary to metastatic EC
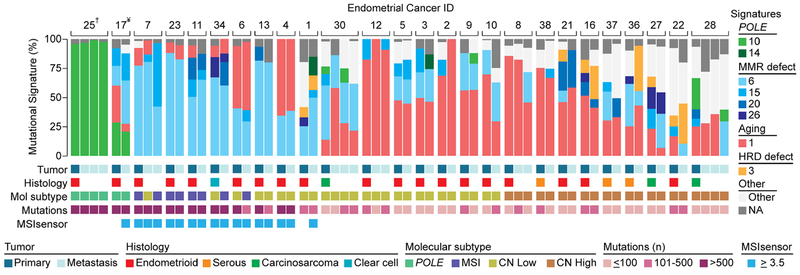
To define whether the mutational spectra of ECs change with the progression from primary to metastatic disease, we retrieved WES data from 26 primary ECs (19 endometrioid, three serous, three carcinosarcomas and one clear cell carcinoma) and 35 matched metastases from Gibson et al.[6]. We performed mutational signature analysis of i) all mutations in a given primary tumor/metastasis, ii) mutations shared between primary tumors and metastases, iii) mutations private to the primary tumor or iv) mutations private to the metastasis of a given case. We reasoned that these private mutations may stem from mutational processes playing a role in tumor maintenance and progression. When assessing all mutations present in a given tumor/metastasis, we observed changes in the dominant mutational signature from primary tumor to metastasis in 8 of the 26 ECs (31%; Fig. 5); these included changes from a dominant aging-related signature 1 in the primary to a dominant DNA MMR-related signature 6 in the metastasis (e.g. EC10, EC27) or changes from a dominant POLE-related signature 10 in the primary to dominant signatures 1 and 6 in two and one metastases of the same case, respectively (EC28; Fig. 5).
Figure 5. Mutational signatures of primary endometrial cancers and matched metastases.
Mutational signatures of primary and metastatic endometrial cancers from 26 patients from Gibson et al [6], sorted according to molecular subtype, color-coded according to the legend. Information on histology and molecular subtype was obtained from Gibson et al [6] and is displayed below each case along with the number of non-synonym ous somatic mutations and MSIsensor scores. HRD, homologous recombination DNA repair deficiency; MSI, microsatellite instability, †, P286R POLE somatic mutation; ¥, V411L POLE somatic mutation.
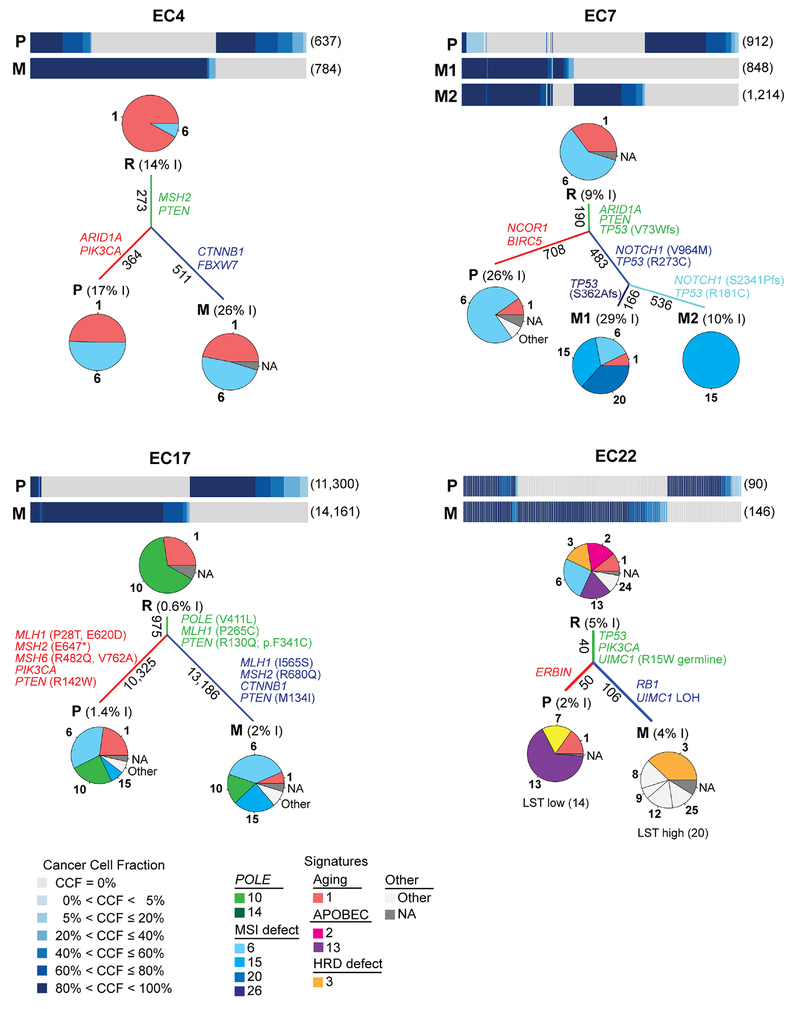
Signature analysis of shared vs private mutations was performed in the 13 cases with ≥20 shared/private mutations (n=12 endometrioid, n=1 clear cell histology), which revealed that distinct DNA repair mechanisms might play a role in the progression from primary EC to metastatic disease in a subset of cases. In the V411L POLE-mutant EC17, the mutations shared between the primary tumor and metastasis had a dominant POLE signature 10, whereas the dominant mutational processes underpinning the primary tumor and metastasis itself were MSI-related (signatures 6/15; Fig. 6). In this case EC17, a somatic MLH1 missense mutation not associated with loss of heterozygosity (LOH) of the wild-type allele was found in the shared mutations (root), whereas two additional MLH1, two MSH6 and one MSH2 mutations were found in the primary tumor only and additional MLH1 and MSH2 mutations in the metastasis only, suggesting that the somatic DNA MMR gene mutations in this tumor may be secondary to the ultramutator POLE EDM[30,32]. Furthermore, we observed a dominant aging mutational process (signature 1) in the shared mutations of EC4 and EC6 but the mutations private to the primary and matched metastatic tumors had dominant MSI-related signatures (signatures 6/15/20) and an increase in the fraction of small indels (Fig. 6, Supplementary Fig. S5). No genetic alterations in DNA MMR genes restricted to the mutations unique to the primary or metastatic tumors were found in these two cases; it is plausible that the MSI and shift in mutational signature may be due to methylation of MLH1 in these cases, a common event in endometrioid ECs[3]. EC7, EC13, EC23 and EC34 displayed a dominant MSI signature 6 and a secondary signature 1 when assessing the shared mutations; in these cases, an even further increase in the proportion of different MSI signatures (6/15/20) and in the fraction of small indels was observed in the mutations private in the primary tumor and the metastases (Fig. 6, Supplementary Fig. S5).
Figure 6. Mutational signatures of shared and private mutations identified in primary endometrial cancers and matched metastases.
Cancer cell fractions of non-synonymous somatic mutations identified in primary endometrial cancers and metastases using ABSOLUTE [26], color-coded according to the legend. The number of non-synonymous somatic mutations identified in a given lesion is shown in parentheses on the right. Pie charts represent the mutational signatures identified in mutations shared between primary tumors and metastases (root, R) in mutations private to the primary lesion (P) and in mutations private to the metastases (M), color-coded according to the legend. The length of the branches is proportional to the number of somatic mutations that are shared/ unique to a given lesion, and the number of mutations is shown alongside the branches. The percentage of small insertions and deletions (indels, I) are included within parentheses next the R, P or M labels. Selected pathogenic somatic mutations are shown along their corresponding branches. For EC22, the LST scores are shown below the pie charts. EC, endometrial cancer; LST, large-scale state transition; M, metastasis; P, primary tumor; R, root. Additional plots are shown in Supplementary Fig. S5.
Finally, the shared mutations of EC22 displayed a dominant signature 6, as well as APOBEC signatures 2 and 13 and the HRD-related signature 3. Whilst a shift in the mutational signatures was observed and the primary tumor mutations displayed a dominant APOBEC-related signature 13, the mutations private in the metastasis had a dominant HRD-related signature 3. Both the primary tumor and its metastasis harbored a p.R15W germline mutation affecting UIMC1, which encodes the BRCA1-interacting protein RAP80 and plays a role in BRCA1-mediated DNA damage responses by recruiting BRCA1 to DNA double-strand breaks[33]. Importantly, bi-allelic inactivation of UIMC1, through LOH of the wild-type allele, was only detected in the metastasis. Consistent with this finding, we found that the LST score was only high in the metastasis of EC22 (LST score 20), but not in the primary tumor (LST score 14), providing evidence to suggest that defective HR, likely driven by UIMC1 bi-allelic inactivation, may have contributed to the progression to metastatic disease in this EC.
Taken together, the acquisition of additional genetic instability, through defects in DNA repair mechanisms including DNA MMR, APOBEC and HR, may play a role in the progression from primary EC to metastatic disease and may have therapeutic implications.
4. DISCUSSION
Here we demonstrate that the genomes of endometrioid ECs of POLE (ultramutated) and MSI (hypermutated) molecular subtypes are shaped by specific mutational processes (i.e. loss of polymerase proofreading and MSI), whilst in endometrioid and serous ECs of copy-number low/high subtypes and uterine carcinosarcomas aging-related mutational processes are the most dominant. The mutational signature analysis performed here expands on the TCGA molecular classification, demonstrating that there is heterogeneity in the mutational processes even among tumors of the same molecular subtype. In fact, MSI-high tumors were found to lack a dominant MSI mutational signature in 15% of cases and only 15% of copy-number high (serous-like) cancers had an HRD signature. In addition, we have observed that whilst the TCGA molecular subtype of an EC is generally stable from primary tumor to metastasis, mutational signature shifts from primary to metastatic endometrial cancer take place in >25% of ECs, suggesting that additional defects in DNA repair mechanisms may drive or be acquired in the progression of ECs. These findings support the notion that the biological processes that drive the development of ECs may differ from those that result in their progression. Furthermore, these results impact on the delivery of precision medicine approaches for EC patients, given that mutational signatures and a clear understanding of the underlying mutagenic processes/pathways present in a given cancer may inform treatment options, in particular in the absence of a targetable mutation or gene copy number alteration (e.g. genomics features of HRD even in the absence of bi-allelic BRCA1/BRCA2 inactivation). Importantly, given the shifts observed in the mutational signatures between the primary tumor and metastasis in a subset of cases, one could speculate that in the advanced setting, genomics analyses should be performed on the metastasis rather than the primary tumor.
Specific POLE and POLD1 mutations have been shown to lead to ultramutation[29]. Here we observed that specific POLE EDMs might have distinct impacts on the mutational processes, given that the mutational signature 10 was more abundant in ECs harboring M444K or P286R EDMs than in those harboring V411L or other EDMs. This is consistent with recent observations that the V411L mutation reduces exonuclease activity without completely abolishing it, whereas the P286R mutation essentially inactivates proofreading[29]. Mutational signature analysis also identified a copy-number low (endometrioid) EC with a POLE L424V EDM mutation that was not classified as of POLE subtype by TCGA, and a uterine carcinosarcoma with a POLE P286R hotspot mutation. On the other hand, we identified two MSI (hypermutated) and one copy-number high (serous-like) EC with POLE EDM mutations not affecting hotspot residues lacking POLE mutational signatures. In the process of translating the molecular classification into the clinical setting[34]. POLE gene sequencing is required. With increasing evidence that POLE mutations are an early event in endometrial and colorectal cancers[28,30] and that ECs harboring POLE hotspot EDMs may be sensitive to nucleoside analogs[35] or immune checkpoint inhibitors[28,36], prospective clinical trials focusing on the treatment of advanced POLE ECs are warranted. In this setting, mutational signature analysis may help to determine whether the POLE mutation not only shaped the primary tumor genome but also continues to act as the tumor driver itself, as we observed that metastases from primary POLE-mutant ECs were potentially driven by DNA MMR-related mutational processes.
Defining the mutational signatures of ECs may assist in refining the classification and therapy of EC patients. With the approval of the immune checkpoint inhibitor pembrolizumab for advanced MSI-high/DNA MMR-deficient cancers[37], accurate identification of MSI-driven tumors is paramount. MSI immunohistochemistry in EC has a sensitivity of 86-100% and specificity of 48-81%[38]. Using a combination of MSIsensor and MLH1 promoter hypermethylation analysis, we identified all ECs classified by TCGA within the MSI (hypermutated) molecular subtype. In ten of the MSI (hypermutated) tumors, however, MSI-related mutational processes accounted only for a minority of the somatic mutations detected (Supplementary Fig. S2). Further studies are warranted to assess whether MSI-high ECs with a dominant aging mutational signature are less sensitive to immune checkpoint inhibitors than those with a dominant MSI mutational signature.
HGSOCs and basal-like and/or triple-negative breast cancers often show defects in HRD and due to genetic alterations affecting this pathway, including germline and somatic BRCA1 and BRCA2 mutations, and are sensitive to platinum-based chemotherapy and poly(ADP-ribose) polymerase (PARP) inhibitors[39]. Based on the similarity in copy number alterations between HGSOCs, basal-like breast cancers and copy-number high (serous-like) ECs, it has been suggested that copy-number high (serous-like) ECs may also be candidates for therapies currently employed for the management of HGSOCs and basal-like breast cancers[3]. We noted, however, that despite similar levels of gene copy number alterations, the underlying DNA repair defects that shaped the majority of the genomes of copy-number high (serous-like) ECs are likely distinct from those of HGSOCs and basal-like breast cancers, given their lower levels of mutational signature 3, LST scores, average indel length and number of cases harboring bi-allelic HR-related gene alterations. This is further supported by the relative reduced response rates to platinum agents of uterine serous carcinomas as compared to HGSOCs[40]. Importantly, however, we did identify three endometrioid ECs with a dominant signature 3, high LSTs and bi-allelic BRCA1 alterations, features consistent with HRD and potential benefit from therapies targeting this type of DNA repair defects.
Our study has several limitations. This is a retrospective analysis of publicly available data. The sequencing depth of the primary and metastatic samples was on average 77x[6], thus we cannot rule out that subclonal mutations were missed. In addition, we could perform mutational signature analysis separately only in the subset of the matched primary and metastatic ECs with sufficient shared and private mutations. More comprehensive analyses including whole-genome sequencing may better define the mutational processes that are driving the progression from primary to metastatic disease in EC.
Despite these limitations, our study demonstrated that EC is a heterogeneous disease with multiple mutational processes shaping the mutational spectra of these lesions. Although the molecular subtype classification proposed by TCGA for endometrioid and serous carcinomas captures some of this heterogeneity, our mutational signature analysis identified examples that would result in their reclassification. We also observed a shift from aging- or POLE- to MSI-related mutational processes in the progression from primary to metastatic ECs in a subset of cases. With the decreasing costs of genomic sequencing, routine whole-genome or even multiple-site sequencing of tumors is rapidly becoming reality in the clinical setting. In the era of precision medicine, information on the mutational processes shaping the genome of a given EC may provide important information in addition to the specific mutations and gene copy number alteration.
Supplementary Material
HIGHLIGHTS.
MSI and POLE endometrial cancers (ECs) show specific mutational signatures
The majority of ECs and carcinosarcomas display the aging-associated signature 1
POLE mutations have an allele-specific impact on mutational processes
15% of serous-like ECs have the defective homologous recombination DNA repair mutational signature 3
Shifts in the mutational signatures take place in the progression of ECs
ACKNOWLEDGEMENTS
We thank S.H. Kim and L.A Moukarzel for critical reading of the manuscript.
Funding support
J.S. Reis-Filho is funded in part by the Breast Cancer Research Foundation. Research reported in this publication was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No. P30CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
N. Riaz reports research support from Pfizer and Bristol-Myers Squibb, and personal fees from the Illumina Speakers’ Bureau, outside the submitted work. J.S. Reis-Filho reports personal/consultancy fees from VolitionRx, Page.AI and Goldman Sachs, outside the submitted work. The remaining authors have no conflicts of interest to declare.
REFERENCES
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 68 (2018) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson B, et al. Annual Report to the Nation on the Status of Cancer, 1975-2014, Featuring Survival. J Natl Cancer Inst. 109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 497 (2013) 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cosgrove CM, Tritchler DL, Cohn DE, Mutch DG, Rush CM, Lankes HA, et al. An NRG Oncology/GOG study of molecular classification for risk prediction in endometrioid endometrial cancer. Gynecol Oncol. 148 (2018) 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cherniack AD, Shen H, Walter V, Stewart C, Murray BA, Bowlby R, et al. Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell. 31 (2017) 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gibson WJ, Hoivik EA, Halle MK, Taylor-Weiner A, Cherniack AD, Berg A, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet. 48 (2016) 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 500 (2013) 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nik-Zainal S, Morganella S. Mutational Signatures in Breast Cancer: The Problem at the DNA Level. Clin Cancer Res. 23 (2017) 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 15 (2014) 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brusco LL, Wathoo C, Mills Shaw KR, Holla VR, Bailey AM, Johnson AM, et al. Physician interpretation of genomic test results and treatment selection. Cancer. 124 (2018) 966–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 33 (2018) 690–705 e699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 346 (2014) 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ng CKY, Bidard FC, Piscuoglio S, Geyer FC, Lim RS, de Bruijn I, et al. Genetic Heterogeneity in Therapy-Naive Synchronous Primary Breast Cancers and Their Metastases. Clin Cancer Res. 23 (2017) 4402–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 173 (2018) 371–385 e318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 474 (2011) 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 490 (2012) 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17 (2016) 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weigelt B, Bi R, Kumar R, Blecua P, Mandelker DL, Geyer FC, et al. The Landscape of Somatic Genetic Alterations in Breast Cancers From ATM Germline Mutation Carriers. J Natl Cancer Inst. 110 (2018) 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mueller JJ, Schlappe BA, Kumar R, Olvera N, Dao F, Abu-Rustum N, et al. Massively parallel sequencing analysis of mucinous ovarian carcinomas: genomic profiling and differential diagnoses. Gynecol Oncol. 150 (2018) 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 30 (2014) 1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 173 (2018) 291–304 e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Popova T, Manie E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 72 (2012) 5454–5462. [DOI] [PubMed] [Google Scholar]
- [23].Riaz N, Blecua P, Lim RS, Shen R, Higginson DS, Weinhold N, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. 8 (2017) 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alexandrov L, Kim J, Haradhvala NJ, Huang MN, Ng AWT, Boot A, et al. The Repertoire of Mutational Signatures in Human Cancer. BioRxiv 322859, 2018. (doi: 10.1101/322859). [DOI] [Google Scholar]
- [25].Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44 (2016) e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 30 (2012) 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Reiter JG, Makohon-Moore AP, Gerold JM, Bozic I, Chatterjee K, Iacobuzio-Donahue CA, et al. Reconstructing metastatic seeding patterns of human cancers. Nat Commun. 8 (2017) 14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Temko D, Van Gool IC, Rayner E, Glaire M, Makino S, Brown M, et al. Somatic POLE exonuclease domain mutations are early events in sporadic endometrial and colorectal carcinogenesis, determining driver mutational landscape, clonal neoantigen burden and immune response. J Pathol. 245 (2018) 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Andrianova MA, Bazykin GA, Nikolaev SI, Seplyarskiy VB. Human mismatch repair system balances mutation rates between strands by removing more mismatches from the lagging strand. Genome Res. 27 (2017) 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Haradhvala NJ, Kim J, Maruvka YE, Polak P, Rosebrock D, Livitz D, et al. Distinct mutational signatures characterize concurrent loss of polymerase proofreading and mismatch repair. Nat Commun. 9 (2018) 1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bellido F, Pineda M, Aiza G, Valdes-Mas R, Navarro M, Puente DA, et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med. 18 (2016) 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Billingsley CC, Cohn DE, Mutch DG, Stephens JA, Suarez AA, Goodfellow PJ. Polymerase varepsilon (POLE) mutations in endometrial cancer: clinical outcomes and implications for Lynch syndrome testing. Cancer. 121 (2015) 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nikkila J, Coleman KA, Morrissey D, Pylkas K, Erkko H, Messick TE, et al. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 28 (2009) 1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].McAlpine J, Leon-Castillo A, Bosse T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J Pathol. 244 (2018) 538–549. [DOI] [PubMed] [Google Scholar]
- [35].Van Gool IC, Rayner E, Osse EM, Nout RA, Creutzberg CL, Tomlinson IPM, et al. Adjuvant Treatment for POLE Proofreading Domain-Mutant Cancers: Sensitivity to Radiotherapy, Chemotherapy, and Nucleoside Analogues. Clin. Cancer Res 24, 2018, 3197–3203. [DOI] [PubMed] [Google Scholar]
- [36].Mittica G, Ghisoni E, Giannone G, Aglietta M, Genta S, Valabrega G. Checkpoint inhibitors in endometrial cancer: preclinical rationale and clinical activity. Oncotarget. 8 (2017) 90532–90544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lemery S, Keegan P, Pazdur R. First FDA Approval Agnostic of Cancer Site - When a Biomarker Defines the Indication. N Engl J Med. 377 (2017) 1409–1412. [DOI] [PubMed] [Google Scholar]
- [38].McConechy MK, Talhouk A, Li-Chang HH, Leung S, Huntsman DG, Gilks CB, et al. Detection of DNA mismatch repair (MMR) deficiencies by immunohistochemistry can effectively diagnose the microsatellite instability (MSI) phenotype in endometrial carcinomas. Gynecol Oncol. 137 (2015) 306–310. [DOI] [PubMed] [Google Scholar]
- [39].Mylavarapu S, Das A, Roy M. Role of BRCA Mutations in the Modulation of Response to Platinum Therapy. Front Oncol. 8 (2018) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bestvina CM, Fleming GF. Chemotherapy for Endometrial Cancer in Adjuvant and Advanced Disease Settings. Oncologist. 21 (2016) 1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.