Abstract
Amplification of the epidermal growth factor receptor gene (EGFR) represents one of the most commonly observed genetic lesions in glioblastoma (GBM), however, therapies targeting this signaling pathway have failed clinically. Here, using human tumors, primary patient-derived xenografts (PDXs), and a murine model for GBM, we demonstrate that EGFR inhibition leads to increased invasion of tumor cells. Further, EGFR inhibitor-treated GBM demonstrate altered oxidative stress, with increased lipid peroxidation, and generation of toxic lipid peroxidation products. A tumor cell subpopulation with elevated aldehyde dehydrogenase (ALDH) levels were determined to comprise a significant proportion of the invasive cells observed in EGFR inhibitor-treated GBM. Our analysis of ALDH1A1 protein in newly diagnosed GBM revealed detectable ALDH1A1 expression in 69% (35/51) of the cases, but in relatively low percentages of tumor cells. Analysis of paired human GBM before and after EGFR inhibitor therapy showed an increase in ALDH1A1 expression in EGFR-amplified tumors (p<0.05, n=13 tumor pairs), and in murine GBM ALDH1A1-high clones were more resistant to EGFR inhibition than ALDH1A1-low clones. Our data identify ALDH levels as a biomarker of GBM cells with high invasive potential, altered oxidative stress, and resistance to EGFR inhibition, and reveal a therapeutic target whose inhibition should limit GBM invasion.
Keywords: EGFRvIII, therapeutic resistance, tumor heterogeneity, invasion, GBM, oxidative stress
INTRODUCTION
Glioblastoma, a malignant brain cancer, is characterized by diffuse invasion of tumor cells into the adjacent brain parenchyma and by the increased activity of receptor tyrosine kinase (RTK) signaling pathways. The most commonly altered RTK, the epidermal growth factor receptor (EGFR), demonstrates gene amplification in up to 45% of patients (1) and expression of a constitutively active EGFR variant, EGFRvIII, in approximately 19% (2–4). Despite the high frequency of EGFR alterations in GBM and their demonstrated roles in oncogenesis (5,6), chemotherapeutic modalities targeting EGFR signaling have had disappointing results in clinical trials, and have not increased the median survival for GBM patients, which remains at less than two years (7,8).
Several factors contribute to therapeutic resistance to EGFR inhibition (9). Such factors include: some EGFR inhibitors are sub-optimal in their receptor binding pocket occupancy (10); limited brain access of systemically administered drugs, especially to invading tumor cells outside of the enhancing tumor mass; tumor cell utilization of mechanisms that increase drug metabolism and detoxification, including expression of efflux pumps (11,12); and the molecularly heterogeneous nature of GBM, whose aggressive biology is driven by the summation of multiple signaling inputs (13,14). Thus, and despite effective on-target inhibition, tumor cells appear able to re-establish downstream PI3K/Akt/mTOR signaling pathway activity via suppression of key negative regulators and the activation of other RTKs (15–17), including MET or PDGFRB (18,19). In general, both acquired and innate resistance mechanisms contribute to therapeutic resistance.
Sustained proliferation, as observed in malignant tumors like GBM, requires changes in cellular metabolism that support rapid cell membrane turnover and the prevention of oxidative damage (20). EGFR signaling itself can increase the activity of such protective pathways, including de novo lipogenesis (21,22) and protection against oxidative stress-induced apoptosis (23). In this study we have investigated relationships between EGFR activity and tumor protective pathways, and how maintaining these protective pathways is associated with resistance to EGFR inhibitor therapy. In so doing we have identified potential points of therapeutic vulnerability that may be relevant to improving GBM treatment outcomes.
MATERIALS AND METHODS
Cell culture conditions & reagents.
EGFRvIII-expressing murine tumorspheres were isolated as previously described (24). GBM43 and GBM6 are primary cell cultures of patient-derived xenografts, established as previously described (25). All cell lines were analyzed before using and each subsequent year by short tandem repeat (STR) analysis to ensure the identity and validity of cells and confirmed to be negative by PCR for mycoplasma contamination. Tumorspheres were cultured and passaged on low-attachment plates under standard neurosphere conditions as previously described (26). Doubling time was calculated using an online calculator (Roth V. 2006 Doubling Time Computing: http://www.doubling-time.com/compute.php). For Western blotting the following antibodies were used: phospho-EGFR Y1173 (Cell Signaling 4407), EGFR (Santa Cruz sc-03), phospho-Akt S473 (Cell Signaling 4060), Akt (Cell Signaling 9272), phospho-p44/42 MAPK T202/Y204 (Cell Signaling 9101), p44/42 MAPK (Cell Signaling 9107), phospho-Src Family Y416 (Cell Signaling 2101), Src (Cell Signaling 2109), ALDH1A1 (Cell Signaling 12035), GAPDH (Millipore MAB374).
Inhibitors.
Erlotinib HCl (Selleckchem S1023), Mitomycin C (Santa Cruz sc-3514), dasatinib (LC Laboratories D3307), lapatinib (LC Laboratories L4804), DEAB (Sigma-Aldrich D86256), Tyrphostin AG1478 (Sigma-Aldrich T4182)(27), and disulfiram (Sigma-Aldrich T1132).
Cell viability assays MTT assay.
For MTT assay dissociated cells were plated as 4×104-8×104 cells per well in a 6 well plate with and without appropriate drug treatments. Cells were incubated for 3 days and assayed by MTT (Promega) or DyLight 800 NHS Ester and flow cytometry.
Human GBM.
We analyzed paired sets of surgical specimens from patients with glioblastoma before and after treatment with an EGFR inhibitor from three different institutions, the University of California, San Francisco (UCSF), the University of California, Los Angeles, and from a clinical trial cohort (15) for which tissue samples were acquired before or under treatment, 5 days. Cases were selected based on availability of paired tumor specimens from before and after EGFR inhibitor therapy and EGFR amplification. Unpaired human GBM from initial presentation were obtained through the Brain Tumor Research Center Biorepository at UCSF. All tumor specimens were obtained according to a protocols approved by the respective institutional review boards.
In vivo transplantation.
Tumor cells were transplanted orthotopically into the striatum of nude mice (Simonsen) as previously described (24). To assess invasion following EGFR inhibition tumors were allowed to propagate for 7 days prior to 5 days of treatment with erlotinib (150mg/kg per day via oral gavage). To assess tumor area in the clones, tumors were allowed to propagate for 2 days prior to 10 days of treatment daily with erlotinib or erlotinib plus disulfiram (100mg/kg and 150 mg/kg, respectively, via intraperitoneal injection). In both experiments, brains were examined at 12 days post transplant or at humane disease endpoint, if sooner. Control vehicle treated mice were included in each experiment. An image of each tumor was taken at a similar anterior-posterior position using a Zeiss Observer inverted microscope at 1.25x and 5x magnification. To assess invasion, results are maximum distance invaded, as measured by longest line between two tumor foci, normalized to total tumor area, calculated using ImageJ software. Tumor area was determined by hand tracing total intracerebral tumor area blinded to tumor cell and treatment arm, calculated using ImageJ software. All animal procedures and care was performed according to institutional animal care and use policies (IACUC # AN169893–02).
Invasion assays.
Two dimensional invasion assays were performed as previously described (28). Six images of invaded cells per insert were taken at 200x magnification on a Zeiss Axioimager MI microscope. The total number of invaded cells at 16 hours per image were counted using ImageJ Software and averaged for each insert. Cells were pre-treated for 2.5 days in 1μM erlotinib and remained exposed to 1μM of erlotinib during the invasion assay. Data was normalized to control in each experiment. Three dimensional invasion assays were performed as previously described (28). Images of each sphere were taken at 0 hours, 16 hours and 24 hours using a Zeiss Observer inverted microscope at 25x magnification. The total area of each sphere was calculated using ImageJ Software and invaded area for each sphere was normalized to its center sphere area. Appropriate drug concentrations were added to both the underlying matrigel coat and to the sphere containing media.
Immunohistochemistry and immunofluorescence.
Tumor-bearing mice were euthanized, perfused with 4% paraformaldehyde, and brains were fixed overnight in 4% PFA, rinsed in PBS and store in 70% ethanol until further processing. Human GBM formalin-fixed paraffin embedded sections and tissue microarrays were immunostained for ALDH1A1 (Abcam ab52492). Immunohistochemistry was performed according to standard protocols on the Ventana Medical Systems Benchmark XT. Images were taken using an Olympus BX41 microscope and Olympus DP72 camera. Immunostaining was quantified using ImageJ Software and percent DAB positive for each tumor was averaged across two images per tumor. Histological analysis was performed on hematoxylin and eosin stained sections. For immunofluorescence 2×103 cells were plated into each well of a Biocoat Poly-D-lysine 8 Well Culture Slide (Corning 354688) for 3 days, followed by 2 hours with appropriate drug treatments. Cells were stained with 1:50 dilution of 4-hydroxynonenal antibody (Abcam ab46545). Cells were then stained with 4ug/mL goat anti-rabbit AlexaFluor 488 IgG (Gibco A11008) and nuclei labeled with DAPI. Images were taken using the Zeiss LSM780 Confocal Microscope. The total number of cells and number of 4HNE positive cells per image were counted using ImageJ Software and averaged across three pictures per well.
ALDEFLUOR Assay.
The ALDEFLUOR assay measures the conversion of the Aldefluor substrate BODIPY aminoacetaldehyde (BAAA) to the negatively charged BODIPY aminoacetate (BAA-) (29). The ALDEFLUOR kit (STEMCELL Technologies 01700) was used as described to measure proportions of ALDH+ cells. Cells were ALDEFLUOR stained at a concentration of 5×105 cells/mL for 45 minutes at 37°C, followed by DyLight 800 NHS Ester (Life Technologies, Cat#46421) staining for 15 minutes at RT as a live/dead marker. DEAB or disulfiram were added to samples in ALDEFLUOR buffer for 10 minutes at RT before ALDEFLUOR staining. Flow cytometry was performed on a BD FACSAria using FACS DIVA software, followed by analysis with FlowJo. A minimum of 10000 events were collected for each sample. In instances when cells were sorted, the top 10% (ALDH High) and bottom 40% (ALDH High) were sorted into NBM supplemented with B-27 for use in invasion assays, or into RLT buffer for RNA extraction.
Image-IT Lipid Peroxidation.
The Image-IT Lipid Peroxidation kit for live cell analysis (Image-iT Lipid Peroxidation kit, Thermo Fisher Scientific Inc.) was used as described to measure degree of lipid peroxidation. Results are displayed as a ratio of oxidized BODIPY C11 reagent (~510nm) over unoxidized BODIPY C11 reagent (~591nm), as per manufacturer’s instructions. Cells were plated at 1×104 cells/mL for three days, followed by appropriate treatment. Cells were centrifuged then dissociated with Cell Dissociation Solution, Non-Enzymatic and replated in full media with 10uM Image-IT Lipid Peroxidation probe for 30 minutes at 37°C, followed by DyLight 800 NHS Ester (Life Technologies, Cat#46421) staining for 15 minutes at RT as a live/dead marker. 100uM cumene hydroperoxide induces lipid peroxidation and was added to cells for 2 hours at 37°C prior to staining as a positive control. Flow cytometry was performed on a BD FACSAria using FACS DIVA software, followed by analysis with FlowJo. A minimum of 10,000 events were collected for each sample.
Reverse Transcription and qPCR.
Total RNA was collected from cells using an RNeasy Mini Kit (Qiagen 74106). cDNA was synthesized using SuperScript III First-Strand Synthesis System for RT-PCR with Oligo(dT) primers (Invitrogen, Cat#18080–051). qPCR was performed with FastStart Universal SYBR Green Master (Rox) (Roche, cat# 049138500011) on the Applied Biosystems 7900HT Fast Real-Time PCR System. Ct values were normalized to GAPDH Ct values for each individual sample and then averaged across experiments.
Statistics.
All statistics were performed using GraphPad Prism 5.0 software. The one-way ANOVA with Tukey’s test for multiple comparisons, Mann-Whitney or the Student’s t-test were used to determine differences between experimental groups as appropriate.
RESULTS
Pro-invasive phenotype stimulated by EGFR inhibition.
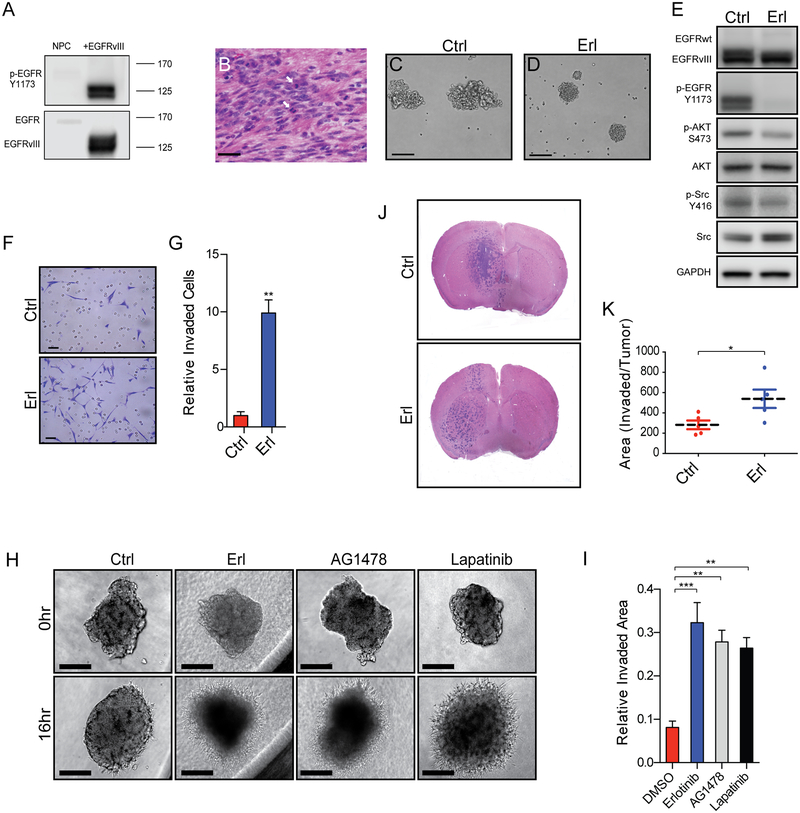
To explore mechanisms of EGFR inhibitor resistance in GBM, we utilized a murine model for GBM driven by constitutive activation of EGFR (EGFRvIII) (Figure 1A) (24,26). Tumor cells derived from orthotopic tumors that occur in these mice, can be maintained in vitro as tumorspheres that, in turn, generate highly proliferative and invasive tumors upon orthotopic engraftment (Figure 1B) with robust EGFR activation. We refer to these tumor-propagating cells as cancer stem cells (CSCs), To study the emergence of EGFR inhibitor resistance we cultured cells in the presence of the EGFR inhibitor erlotinib. Within 2 hours of EGFR inhibition changes in tumorsphere morphology were evident. Ragged, irregular spheres became compact, round spheres with a smooth outer surface upon EGFR inhibition (Figure 1C–D). Western blot analysis revealed decreased phosphorylation of Y1173 in treated cells, as well as decreased signaling downstream of EGFR, including decreased phosphorylation of AKT (S473) and Src (Y416) (Figure 1E). With respect to biologic properties, acute EGFR inhibition increased tumor cell invasion in matrigel boyden chambers (Figure 1F–G), and increased tumorsphere invasion in matrigel 3D assays (Figure 1H–I). The pro-invasive phenotype conferred by EGFR inhibition was not specific to erlotinib and was observed with two additional pharmacological EGFR inhibitors: AG1478 (27) and lapatinib. As expected, EGFR inhibition also decreased tumor cell proliferation (Supplemental Figure 1E). While EGFR inhibition decreased downstream signaling pathway activity (Figure 1E), inhibition of PI3K/AKT or Src signaling pathway alone was not sufficient to confer the pro-invasive phenotype (Supplemental Figure 1A-D).
Figure 1. EGFR inhibition in murine cancer stem cells (CSCs) drive increased tumor cell invasion in vitro and in vivo.
(A) Tumor-prone neural progenitor cells express high levels of phosphorylated EGFRvIII and (B) generate invasive, highly proliferative (arrows) tumors when transplanted orthotopically. (C-D) Cultured CSCs grow as irregular, ragged spheres but become compact and round upon inhibition of EGFR, erlotinib (Erl) versus control (Ctrl). (E) Representative Westerns demonstrating inhibition of activation of EGFR and several downstream effectors with Erl. Increased tumor cell invasion in (F-G) 2D invasion assays and (H-I) 3D spheroid invasion assays with Erl and in (H) with AG1478 and lapatinib. (J-K) Increased invasive area in orthotopic tumors with EGFR inhibition as compared to control. Representative images and summary quantifications are shown. Scale bars denote 30μm in (B), 100μm in (C-D, H), and 50μm in (F). Shown are mean ± SEM from biologic replicates or representative data from biologic replicates, (G) n=3, (I) n=3 performed twice, (K) n=5 per treatment. *p<0.05; ** p<0.005; ***p<0.0005.
To determine the in vivo relevance of EGFR inhibitor-stimulated invasiveness, mice harboring orthotopic tumors were treated with erlotinib (150mg/kg/d) for five days and tumors were examined in the brain of euthanized mice at this time point. Inhibition of EGFR conferred a 1.9-fold increase in area of invasive tumor as compared to control treated tumors (Figure 1J–K).
Pro-invasive phenotype associated with an ALDH high subpopulation.
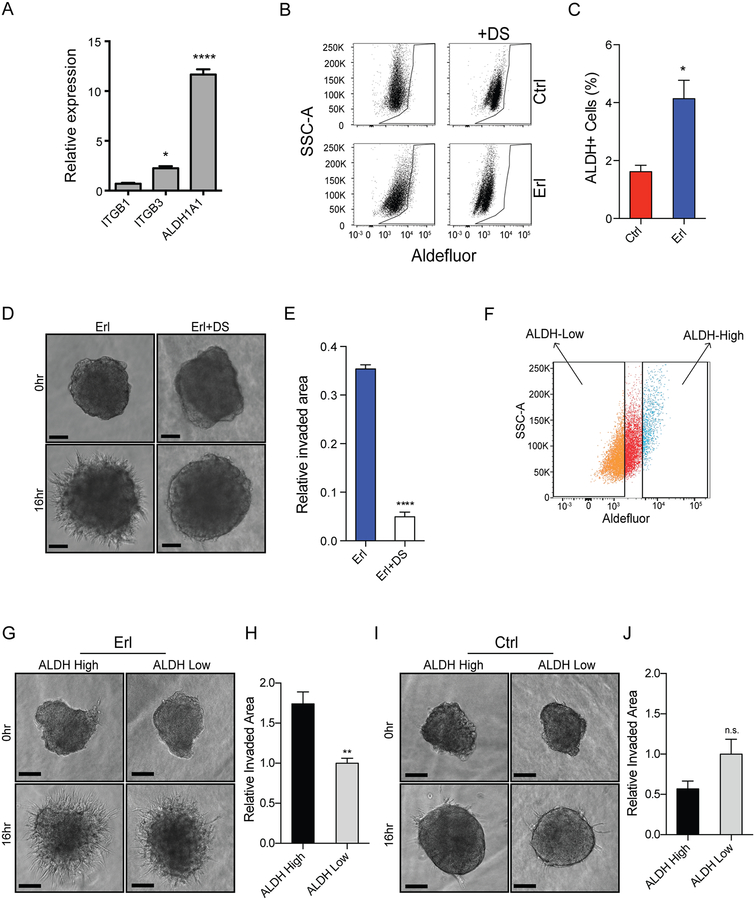
A gene family often implicated in chemotherapeutic resistance encode aldehyde dehydrogenases that detoxify reactive aldehyde species. EGFR inhibition in CSCs resulted in a striking 11-fold increase in aldehyde dehydrogenase 1A1 (ALDH1A1) expression relative to control (Figure 2A). The increase in ALDH1A1 expression conferred by erlotinib was also observed with AG1478 and lapatinib (Supplemental Figure 1F). EGFR inhibition also resulted in increased expression of several genes implicated in cell adhesion and motility, including integrin beta chain beta 3 (ITGB3), CXCR4, S100A4, and TIMP3 (Figure 2A and Supplemental Figure 1G). EGFR inhibition resulted in increased ALDH activity (Figure 2B–C) by ALDEFLUOR assay(29). Treatment of CSCs with the irreversible ALDH inhibitor disulfiram (DS) (Figure 2D–E) or a structurally distinct ALDH inhibitor N,N-diethylabenzaldehyde (DEAB) (Supplemental Figure 2A-B) blocked the pro-invasive phenotype stimulated by EGFR inhibition.
Figure 2. Expansion of invasive ALDH+ subpopulation in CSCs upon EGFR inhibition.
(A) Gene expression of ITGB1, ITGB3, and ALDH1A1 in CSCs resistant to EGFR inhibition relative to control treated CSCs. (B-C) Increased Aldefluor staining in erlotinib-treated cells (left, bottom) relative to control (left, top) by flow cytometry. Baseline fluorescence established by inhibiting ALDH with disulfiram (DS) without (right, top) and with (right, bottom) erlotinib. (D-E) DS inhibits erlotinib-induced invasion in spheroid invasion assay. (F) Schematic demonstrating Aldefluor sorting strategy. Erlotinib-treated cells (16 h) prior to staining were sorted by FACs and denoted as ALDH-high, top 10% of Aldefluor stained cells, or ALDH-low, bottom 40% of Aldefluor stained cells. (G) Increased erlotinib-induced invasion in sorted ALDH-high cells (left) vs. ALDH-low cells (right). (H) Quantification normalized to ALDH-low cells. (I-J) ALDH-high and ALDH-low cells exhibit similar invasion in the absence of EGFR inhibition. Representative images and quantification over biologic replicates, mean ± SEM (n=3). n.s. denotes not significant. *, p<0.05, **p<0.005, ****, p<0.0001.
To determine whether elevated ALDH activity was associated with invasive cells, we sorted bulk untreated tumor cells based on ALDH activity (Figure 2F). With EGFR inhibition, sorted ALDH-high cells proved more invasive than ALDH-low cells in 3D matrigel invasion assays (Figure 2G–H). Strikingly, this difference in invasion was dependent on EGFR inhibition (Figure 2I–J). Thus, a highly pro-invasive phenotype required both high ALDH activity and EGFR inhibition.
ALDH1A1-high CSCs show intrinsic EGFR inhibitor resistance.
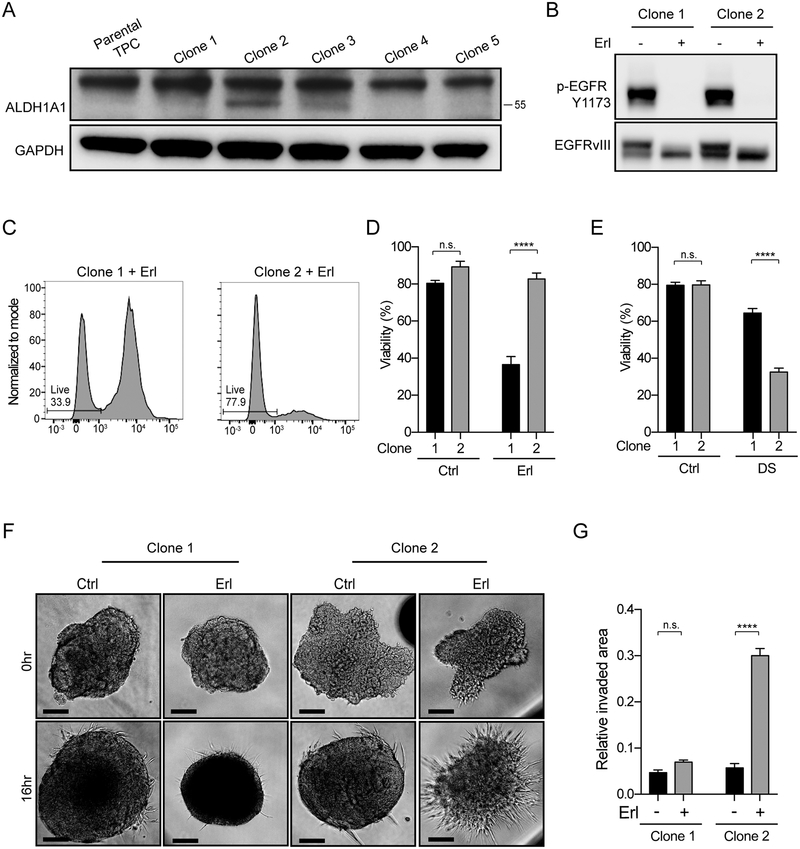
EGFR inhibition resulted in a rapid change in tumor invasive behavior dependent on ALDH-activity. We hypothesized that heterogeneity in ALDH1A1 expression could be an intrinsic mechanism of resistance to EGFR inhibition. In untreated bulk tumor cell cultures, ALDH1A1 protein was not apparent by western blotting (Figure 3A and Supplemental Figure 2C). Single cell-derived clonal analysis of bulk tumor, however, revealed a range of ALDH1A1 expression levels across clones (Figure 3A). Two clones with different ALDH1A1 protein expression, similar levels of EGFRvIII activation, and similar cell growth under control conditions, demonstrated a marked difference in sensitivity to EGFR inhibition (Figure 3A–D). The ALDH1A1-high clone 2, had a distinct survival advantage over the ALDH1A1-low clone 1 upon EGFR inhibitor treatment (Figure 3C–D). Moreover, EGFR inhibition conferred a pro-invasive phenotype in the ALDH1A1-high clone 2 (Figure 3F–G). Cell viability of clone 2 was markedly reduced by DS treatment (Figure 3E).
Figure 3. ALDH-high subpopulation exhibits increased resistance to erlotinib and increased sensitivity to ALDH inhibition.
(A) ALDH1A1 expression across clones derived from untreated bulk tumor cell cultures. GAPDH protein loading control. Note: ALDH1A1 (55 kDa) is distinct from higher molecular weight non-specific band in mouse. (B) Clones 1 and 2 have similar EGFRvIII phosphorylation levels under basal conditions and after erlotinib treatment. (C-D) Increased viability of clone 2 relative to clone 1 only in the presence of erlotinib. (E) Viability assay demonstrating selective vulnerability of clones to disulfiram (DS). (F-G) Pro-invasive phenotype in ALDH1A1-high clone 2 upon EGFR inhibition by erlotinib. Representative images and quantification of biologic triplicates are shown, mean ± SEM. ****p<0.0001, n.s. denotes not significant.
ALDH1A1-high CSCs are protected from drug-induced lipid peroxidation.
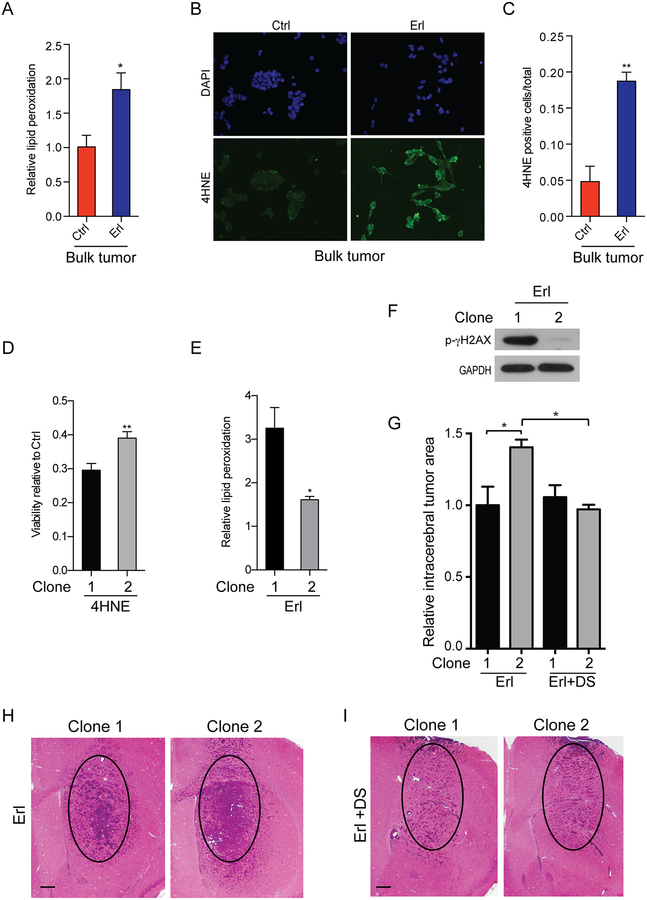
Oncogenic activation of EGFR alters cellular metabolism and up-regulates several pro-survival pathways (30). Inhibition of EGFR can decrease the activity of these pathways and lead to increased oxidative stress (31). Increased oxidative stress can promote lipid membrane peroxidation and the generation of toxic aldehyde species. To investigate changes in lipid membrane peroxidation, in association with EGFR inhibitor treatment, we examined the levels of these metabolites following EGFR inhibition of bulk tumor cells. Inhibition of EGFR nearly doubled the lipid membrane peroxidation levels as demonstrated by a shift in peak fluorescence emission upon oxidation of C11-BODIPY581/591 (32), (Figure 4A). In addition, the cellular accumulation of toxic protein adducts containing 4-HNE, a destructive aldehyde generated by lipid peroxidation, were readily observed upon EGFR inhibition (Figure 4B–C).
Figure 4. ALDH-high CSCs exhibit reduced levels of erlotinib-induced lipid peroxidation, reduced sensitivity to toxic lipid peroxidation products, and increased resistance to EGFR inhibition in vivo.
(A) Increase in lipid peroxidation in bulk tumor cells with acute erlotinib treatment detected by FACs. (B-C) Increased levels of the lipid peroxidation product 4-hydroxynonenal (4-HNE) in erlotinib-treated bulk tumor cells. (D) Differential sensitivity of clones to exogenous 4-HNE. (E) ALDH1A1-low clone 1 with increased levels of erlotinib-induced lipid peroxidation as compared to ALDH1A1-high clone 2. Normalized to respective vehicle control. (F) Increased p-gammaH2AX in DNA damage in ALDH1A1-low clone 1 treated with erlotinib. Representative images and quantification, mean ± SEM from biologic triplicate. (G) ALDH-high clone 2 generated larger intracerebral tumors than ALDH-low clone 1 when EGFR was inhibited (Erl). Inhibition of ALDH by disulfiram (DS) in the context of EGFR inhibition abolished this effect (Erl+DS). (H,I) Representative images of ALDH-high clone 2 treated with erlotinib (Erl) (H) or erlotinib plus disulfiram (Erl+DS) (I) as compared to ALDH-low clone 1. Circle denotes same area for comparison. Scale bar 300μm. *p<0.05; **p<0.005.
Aldehyde dehydrogenases, including ALDH1A1, metabolize and remove harmful reactive aldehydes produced by lipid peroxidation, including 4-HNE (33,34). To determine whether CSCs with elevated ALDH levels were more resistant to lipid peroxidation, we applied exogenous 4-HNE to clones 1 and 2, with results showing increased viability of ALDHA1-high clone 2 cells, relative to ALDHA1-low clone 1 cells (Figure 4D). In relation to ALDHA1-low cells, ALDHA1-high clone 2 cells showed decreased levels of lipid peroxidation upon EGFR inhibition (Figure 4E). High ALDH1A1 levels were also associated with greater protection from DNA damage induced by EGFR inhibition, as demonstrated by decreased phospho-gammaH2AX levels in these cells (Figure 4F).
ALDH1A1-high CSCs show decreased sensitivity to EGFR inhibition in vivo.
EGFR inhibitor treatment of orthotopic tumors demonstrated that ALDH1A1–high clone 2 cells were more resistant to therapy and developed larger tumors than ALDH1A1-low clone 1 cells. When ALDH1A1 was inhibited by DS there was no difference in tumor area in ALDH1A1-high and -low tumors (Figure 4G). In the absence of any therapy ALDH1A1-high and -low tumors were indistinguishable.
ALDH activity in human GBM PDX confers a pro-invasive phenotype.
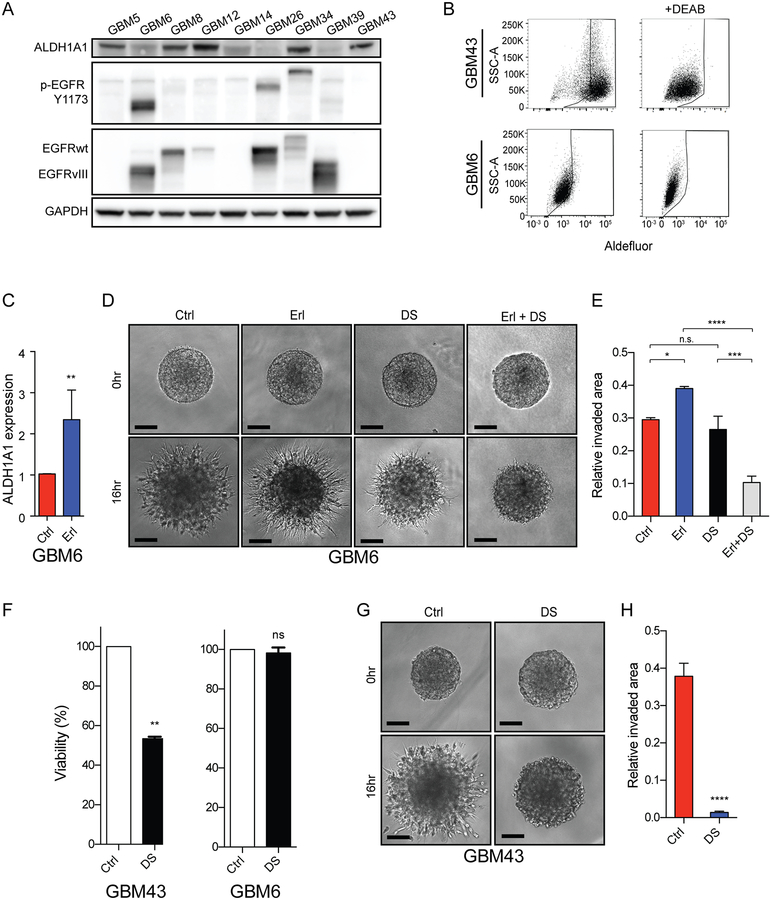
To investigate potential functions for ALDH activity in human GBM we analyzed patient-derived xenografts (PDXs). ALDH1A1 could be detected in five of nine PDX lines, with variable levels of expression evident among the five positive lines (Figure 5A). GBM6, with amplified and expressed EGFRvIII, expressed low level ALDH1A1, and showed low ALDH activity (Figure 5A, 5B). In ALDH1A1-low GBM6 inhibition of EGFR increased ALDH1A1 expression 2.3-fold (Figure 5C) and increased tumor cell invasion (Figure 5D–E). Erlotinib-induced invasion could be blocked by ALDH inhibition by DS. In contrast to GBM6, GBM43 had high endogenous levels of ALDH1A1 protein and high ALDH activity (Figure 5A–B). Inhibition of ALDH by DS in GBM43 resulted in decreased cell viability relative to ALDH-low GBM6 and nearly abolished tumor cell invasion (Figure 5F–H).
Figure 5. Induction of ALDH1A1 expression and a pro-invasive phenotype upon EGFR inhibition in an EGFR-activated patient-derived GBM xenograft.
(A) ALDH1A1 expression and EGFR expression and phosphorylation across a cohort of human GBM xenografts. GAPDH protein loading control. (B) Aldefluor activity (left panels) in GBM43 (top) and GBM6 (bottom) relative to baseline fluorescence established by inhibiting ALDH with DEAB (right panels). (C) Erlotinib-induced increase in ALDH1A1 mRNA expression in GBM6. (D-E) Erlotinib-induced and disulfiram-sensitive increase in invasion in GBM6. (F) ALDH1A1-high GBM43 demonstrate increased sensitivity to disulfiram treatment than GBM6 and (G-H) GBM43 invasion is inhibited by disulfiram treatment. Representative images and quantification, mean ± SEM of biologic triplicate. *p<0.05; **p<0.01; ***p<0.0005; ****p<0.0001.
ALDH1A1 expression in patient GBM and increases following EGFR inhibitor therapy.
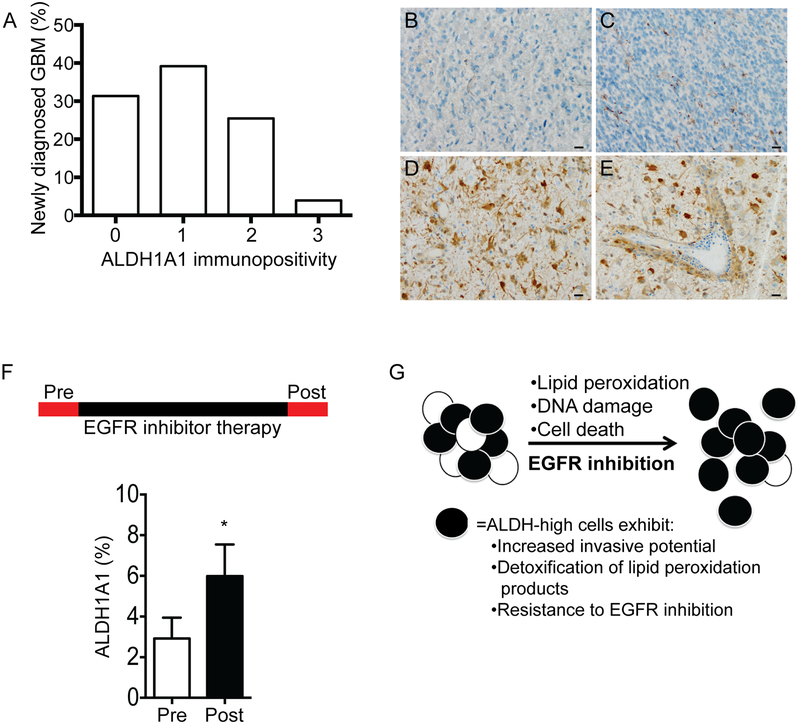
ALDH1A1 protein is expressed in the majority of human GBM (69%, 35 of 51 GBM at initial diagnosis), however, the fraction of positive tumor cells is relatively low with a mean immunopositivity of 8.0% ± 12.5% among positive tumors (Figure 6A). ALDH1A1-positive tumor cells were observed in perivascular regions and as individual infiltrating cells (Figure 6B–E). While ALDH1A1-positive cells were frequently present at the infiltrative tumor edge, they were also present in central tumor regions. Given the elevated ALDH1A1 expression in our murine tumor cells upon EGFR inhibition, we assessed ALDH1A1 levels in paired human GBM from before and after EGFR inhibitor therapy (Figure 6F). In tumors with EGFR amplification, there was a significant increase in ALDH1A1-positive tumor cells following EGFR inhibitor therapy (p<0.05, n=13 tumor pairs). In contrast, there was no increase in ALDH1A1 in tumors without EGFR amplification (n=9 tumor pairs).
Figure 6. ALDH1A1 expression in human GBM and in paired tumors before and after EGFR inhibitor therapy.
(A) The majority of treatment naïve human GBM express ALDH1A1 protein but the fraction of positive tumor cells is low as denoted by the frequency of tumors with immunohistochemistry scores < 3 (having <25% positivity). Tumors with robust immunopositivity for ALDH1A1 often had prominent perivascular collections of ALDH1A1-positive tumor cells. (B-E) Representative immunostaining for ALDH1A1, including (B) absent, score 0; (C) <5% tumor cells, score 1; and (D-E) >25% tumor cells positive, score 3. (F) ALDH1A1 protein levels in paired human GBM samples obtained before (Pre-) and after (Post-) EGFR inhibitor therapy with EGFR amplification. (G) Schematic representation of EGFR inhibition and enrichment of an ALDH-high population of glioma cells.
DISCUSSION
Alterations in EGFR signaling are common in GBM, yet strategies to target EGFR-associated signaling abnormalities have had limited therapeutic efficacy. Here, we identify a subpopulation of tumor cells with elevated levels of the detoxifying enzyme ALDH that resist EGFR inhibition. We showed that ALDH expression was associated with protection from EGFR inhibitor-mediated lipid peroxidation and a pro-invasive phenotype. Pharmacologic inhibition of ALDH selectively targeted the EGFR inhibitor resistant subpopulation, and in so doing caused decreased viability and invasion of these cells. Our data suggest therapeutic strategies targeting ALDH-high tumor cell populations, that have greater resistance to oxidative stress, may be effective when used in combination with EGFR inhibition.
The molecular biology of GBM includes increased lipid peroxidation and generation of toxic end products, such as 4-HNE, that can lead to DNA damage and cell death (35–37). To offset this anti-tumor characteristic, GBM employ several protective mechanisms. To decrease production of peroxidized lipids, aggressive tumors often maintain relatively high levels of saturated fatty acids, which are less susceptible to lipid peroxidation. Indeed, oncogenic activation of receptor tyrosine kinases, including EGFR, drives de novo lipogenesis and the generation of highly saturated phospholipids (22,35). The inhibition of EGFR can suppress several of these protective mechanisms, including decreased de novo lipogenesis and increased oxidative phosphorylation (31,38). Using a murine model for GBM, we demonstrate EGFR inhibition results in increased lipid peroxidation and increased production of toxic 4-HNE adducts.
Aldehyde dehydrogenases, a family of nineteen enzymes, detoxify endogenous and exogenous aldehydes in a NAD(P)+-dependent manner, and have been implicated in EGFR inhibitor resistance in lung, breast and gastric cancer (31,39). Consistent with their role in the detoxification of lipid peroxidation products, we have shown that ALDH-high GBM cells exhibit decreased lipid peroxidation upon EGFR inhibition and increased resistance to 4-HNE associated cell toxicity.
In murine cancer stem cells (CSCs), increased aldehyde dehydrogenase (ALDH) activity following EGFR inhibition may be due, in part, to a selective survival of ALDH-high tumor cells (Figure 6G). Elevated ALDH activity has previously been associated with increased tumor cell invasion, self-renewal, and cancer stem-like properties in several cancers including GBM (29,40–43). Our data suggest an association between high ALDH activity and increased invasive phenotype. It is not clear if increased invasive phenotype is due to increased ALDH activity or, rather, elevated ALDH is simply an associated, but not causative, molecular characteristic of invasive GBM cells. Interestingly, tumor metabolism and oxidative phosphorylation may differ in highly invasive versus less invasive regions of GBM (44). Increased ALDH1A1 may be one mechanism to manage high metabolic stress at the invasive edge. In non-neoplastic brain, astrocytes also exhibit metabolic heterogeneity as evidenced by an ALDH1A1-high subpopulation of astrocytes that reside in close proximity to blood vessels (45).
ALDH activity has also been used to define tumor cell populations with increased resistance to chemotherapy, including agents targeting EGFR (31,39,46–48). Given the importance of oxidative stress and lipid peroxidation in aggressive cancers, it is not surprising that ALDH-high tumor cell subpopulations may play important roles in resistance to several chemotherapeutics. In general, drug resistance is ascribed to a cell population with increased ALDH activity rather than the function of a single ALDH enzyme. Due to the large number of ALDH enzymes and increased expression of several ALDH enzymes in cancer, it has been difficult to assign causality to a specific ALDH (31). Our study indicates increased aldehyde dehydrogenase activity mediated by ALDH1A1 contributes to increased detoxification of reactive aldehydes downstream of lipid peroxidation and increased resistance to EGFR inhibition.
Both human (GBM43) and murine (CSC clone 2) GBM cells with elevated expression of ALDH1A1 were sensitive to the irreversible ALDH inhibitor disulfiram. Based on data from several studies suggesting its cytotoxic effect on tumor cells (49,50) disulfiram is currently in clinical trials for the treatment of GBM (, , , ; clinicaltrials.gov). Our data confirm a cytotoxic effect of disulfiram, but additionally suggest that combination therapy with both ALDH and EGFR inhibitors may be especially effective in treating a subset of GBM with abnormal and elevated EGFR activity.
Supplementary Material
Acknowledgements:
We are grateful and acknowledge the UCSF Brain Tumor SPORE Tissue Core (P50CA097257) for providing histology services and the UCSF Helen Diller Family Comprehensive Cancer Center Laboratory for Cell Analysis Shared Resource Facility (P30CA082103) for microscopy services.
Financial support:
This study was funded by NIH/NINDS R01 NS081117 to J.J.P., NIH/NCI U01 CA168878 to J.J.P., NIH/NCI U01 CA229345 to J.J.P., NIH/NCI P50CA221747 to C.D.J., the Oncosuisse (OCS-01680-02-2005 to M.E.H.), NIH/NCI R01 CA179071 to A.L., NIH/NCI P50-CA211015 to A.L., and resources were provided by the UCSF Brain Tumor SPORE Biorepository NIH/NCI P50CA097257 to J.J.P.. We also acknowledge and thank the T.J. Martell Foundation, the Gerson and Barbara Bakar Philanthropic Fund, and the Sence Foundation for their support of this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of interest:
The authors declare that they have no conflicts of interest with the contents of this article
REFERENCES
- 1.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89:2965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89:4309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boockvar JA, Kapitonov D, Kapoor G, Schouten J, Counelis GJ, Bogler O, et al. Constitutive EGFR signaling confers a motile phenotype to neural stem cells. Mol Cell Neurosci. 2003;24:1116–30. [DOI] [PubMed] [Google Scholar]
- 6.Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, et al. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–77. [DOI] [PubMed] [Google Scholar]
- 7.Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaeckle KA, Uhm JH, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:5603–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MCM, Kros JM, Carpentier AF, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol Off J Am Soc Clin Oncol. 2009;27:1268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkovich KJ, Hariono S, Garske AL, Zhang J, Blair JA, Fan Q-W, et al. Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer Discov. 2012;2:450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y-J, Huang W-C, Wei Y-L, Hsu S-C, Yuan P, Lin HY, et al. Elevated BCRP/ABCG2 expression confers acquired resistance to gefitinib in wild-type EGFR-expressing cells. PloS One. 2011;6:e21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pick A, Wiese M. Tyrosine kinase inhibitors influence ABCG2 expression in EGFR-positive MDCK BCRP cells via the PI3K/Akt signaling pathway. ChemMedChem. 2012;7:650–62. [DOI] [PubMed] [Google Scholar]
- 13.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–7. [DOI] [PubMed] [Google Scholar]
- 15.Hegi ME, Diserens A-C, Bady P, Kamoshima Y, Kouwenhoven MCM, Delorenzi M, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol Cancer Ther. 2011;10:1102–12. [DOI] [PubMed] [Google Scholar]
- 16.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. [DOI] [PubMed] [Google Scholar]
- 17.Fenton TR, Nathanson D, de Albuquerque Ponte C, Kuga D, Iwanami A, Dang J, et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc Natl Acad Sci U S A. 2012;109:14164–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jun HJ, Acquaviva J, Chi D, Lessard J, Zhu H, Woolfenden S, et al. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I, et al. De-repression of PDGFRβ transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3:534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. [DOI] [PubMed] [Google Scholar]
- 21.Guo D, Reinitz F, Youssef M, Hong C, Nathanson D, Akhavan D, et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 2011;1:442–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo D, Prins RM, Dang J, Kuga D, Iwanami A, Soto H, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal. 2009;2:ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orcutt KP, Parsons AD, Sibenaller ZA, Scarbrough PM, Zhu Y, Sobhakumari A, et al. Erlotinib-mediated inhibition of EGFR signaling induces metabolic oxidative stress through NOX4. Cancer Res. 2011;71:3932–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips JJ, Huillard E, Robinson AE, Ward A, Lum DH, Polley M-Y, et al. Heparan sulfate sulfatase SULF2 regulates PDGFRα signaling and growth in human and mouse malignant glioma. J Clin Invest. 2012;122:911–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res Off J Am Assoc Cancer Res. 2006;12:2264–71. [DOI] [PubMed] [Google Scholar]
- 26.Lindberg OR, McKinney A, Engler JR, Koshkakaryan G, Gong H, Robinson AE, et al. GBM heterogeneity as a function of variable epidermal growth factor receptor variant III activity. Oncotarget. 2016; 19(Suppl6): vi52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Z, Parmar S, Peng X-X, Shen T, Robey RW, Bates SE, et al. The epidermal growth factor tyrosine kinase inhibitor AG1478 and erlotinib reverse ABCG2-mediated drug resistance. Oncol Rep. 2009;21:483–9. [PMC free article] [PubMed] [Google Scholar]
- 28.Wade A, Engler JR, Tran VM, Phillips JJ. Measuring sulfatase expression and invasion in glioblastoma. Methods Mol Biol Clifton NJ. 2015;1229:507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B, et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17:1000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raha D, Wilson TR, Peng J, Peterson D, Yue P, Evangelista M, et al. The Cancer Stem Cell Marker Aldehyde Dehydrogenase Is Required to Maintain a Drug-Tolerant Tumor Cell Subpopulation. Cancer Res. 2014;74:3579–90. [DOI] [PubMed] [Google Scholar]
- 32.Pap EH, Drummen GP, Winter VJ, Kooij TW, Rijken P, Wirtz KW, et al. Ratio-fluorescence microscopy of lipid oxidation in living cells using C11-BODIPY(581/591). FEBS Lett. 1999;453:278–82. [DOI] [PubMed] [Google Scholar]
- 33.Makia NL, Bojang P, Falkner KC, Conklin DJ, Prough RA. Murine hepatic aldehyde dehydrogenase 1a1 is a major contributor to oxidation of aldehydes formed by lipid peroxidation. Chem Biol Interact. 2011;191:278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, et al. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem. 2007;282:25668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rysman E, Brusselmans K, Scheys K, Timmermans L, Derua R, Munck S, et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010;70:8117–26. [DOI] [PubMed] [Google Scholar]
- 36.Cheeseman KH, Burton GW, Ingold KU, Slater TF. Lipid peroxidation and lipid antioxidants in normal and tumor cells. Toxicol Pathol. 1984;12:235–9. [DOI] [PubMed] [Google Scholar]
- 37.Germain E, Chajès V, Cognault S, Lhuillery C, Bougnoux P. Enhancement of doxorubicin cytotoxicity by polyunsaturated fatty acids in the human breast tumor cell line MDA-MB-231: relationship to lipid peroxidation. Int J Cancer. 1998;75:578–83. [DOI] [PubMed] [Google Scholar]
- 38.Martin MJ, Eberlein C, Taylor M, Ashton S, Robinson D, Cross D. Inhibition of oxidative phosphorylation suppresses the development of osimertinib resistance in a preclinical model of EGFR-driven lung adenocarcinoma. Oncotarget. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corominas-Faja B, Oliveras-Ferraros C, Cuyàs E, Segura-Carretero A, Joven J, Martin-Castillo B, et al. Stem cell-like ALDH(bright) cellular states in EGFR-mutant non-small cell lung cancer: a novel mechanism of acquired resistance to erlotinib targetable with the natural polyphenol silibinin. Cell Cycle Georget Tex. 2013;12:3390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasper M, Schäfer A, Piontek G, Teufel J, Brockhoff G, Ringel F, et al. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro-Oncol. 2010;12:1024–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcato P, Dean CA, Giacomantonio CA, Lee PWK. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle Georget Tex. 2011;10:1378–84. [DOI] [PubMed] [Google Scholar]
- 42.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2010;16:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu S-L, Liu S, Cui W, Shi Y, Liu Q, Duan J-J, et al. Aldehyde dehydrogenase 1A1 circumscribes high invasive glioma cells and predicts poor prognosis. Am J Cancer Res. 2015;5:1471–83. [PMC free article] [PubMed] [Google Scholar]
- 44.Santandreu FM, Brell M, Gene AH, Guevara R, Oliver J, Couce ME, et al. Differences in mitochondrial function and antioxidant systems between regions of human glioma. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 2008;22:757–68. [DOI] [PubMed] [Google Scholar]
- 45.Adam SA, Schnell O, Pöschl J, Eigenbrod S, Kretzschmar HA, Tonn J-C, et al. ALDH1A1 is a marker of astrocytic differentiation during brain development and correlates with better survival in glioblastoma patients. Brain Pathol Zurich Switz. 2012;22:788–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seguin L, Kato S, Franovic A, Camargo MF, Lesperance J, Elliott KC, et al. An integrin β₃-KRAS-RalB complex drives tumour stemness and resistance to EGFR inhibition. Nat Cell Biol. 2014;16:457–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res Off J Am Assoc Cancer Res. 2009;15:4234–41. [DOI] [PubMed] [Google Scholar]
- 48.Schäfer A, Teufel J, Ringel F, Bettstetter M, Hoepner I, Rasper M, et al. Aldehyde dehydrogenase 1A1--a new mediator of resistance to temozolomide in glioblastoma. Neuro-Oncol. 2012;14:1452–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu P, Kumar IS, Brown S, Kannappan V, Tawari PE, Tang JZ, et al. Disulfiram targets cancer stem-like cells and reverses resistance and cross-resistance in acquired paclitaxel-resistant triple-negative breast cancer cells. Br J Cancer. 2013;109:1876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hothi P, Martins TJ, Chen L, Deleyrolle L, Yoon J-G, Reynolds B, et al. High-throughput chemical screens identify disulfiram as an inhibitor of human glioblastoma stem cells. Oncotarget. 2012;3:1124–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.