Abstract
Synaptobrevin/vesicle-associated membrane protein 2 (VAMP2) is an essential soluble N-ethyl maleimide–sensitive factor attachment protein receptor (SNARE) protein that has been extensively studied in its role in synaptic vesicle fusion. However, sorting and trafficking of VAMP2 within the endosomal system is not well understood. Here, we use the yeast VAMP2 homologue Snc1 to investigate the pathways and signals required for endocytic trafficking. We identify two genetically distinct retrieval pathways from the endosomal system: a plasma membrane recycling pathway that requires the Rcy1 F-box protein and a retrograde pathway originating from the multivesicular/prevacuole endosome dependent on the Snx4-Atg20 sorting nexin complex. Lysine residues within the transmembrane domain of Snc1 are necessary for presentation of a Snx4-Atg20–dependent sorting signal located within its juxtamembrane region. Mutations of the transmembrane lysine residues ablate retrograde sorting and subject Snc1 to quality control via sorting into the degradative multivesicular endosome pathway. Degradative sorting requires lysine residues in the juxtamembrane region of Snc1 and is mediated by the Rsp5 ubiquitin ligase and its transmembrane adapters, Ear1 and Ssh4, which localize to endosome and vacuole membranes. This study shows that Snc1 is trafficked between the endosomal system and the Golgi apparatus via multiple pathways and provides evidence for protein quality control surveillance of a SNARE protein in the endo-vacuolar system.
INTRODUCTION
Soluble N-ethyl maleimide–sensitive factor attachment protein receptor (SNARE) proteins play a central role in membrane fusion, driving important biological processes, including intracellular transport, cell fertilization, viral infection, and neurotransmitter release (Han et al., 2017). The best studied SNARE proteins are those involved in the fusion of synaptic vesicles with the presynaptic membrane in neurons during neurotransmitter release, which include synaptobrevin/vesicle-associated membrane protein 2 (VAMP2), syntaxin, and SNAP-25. In particular, VAMP2 has been extensively studied, in part because its transmembrane domain and juxtamembrane region play important regulatory roles in controlling SNARE complex formation and membrane fusion (Quetglas et al., 2000, 2002; Kweon et al., 2003; Bowen and Brunger, 2006; Han et al., 2016), where basic and aromatic residues in the juxtamembrane region and transmembrane domain have been found to play critical roles in calcium-dependent regulation of neurotransmitter release (Quetglas et al., 2000, 2002). Furthermore, the VAMP2 transmembrane domain contains unique structural features that promote structural flexibility of the juxtamembrane region, which has been proposed to help facilitate the final step of hemifusion to fusion transition in membrane fusion (Han et al., 2016).
Little is known about how VAMP2, and v-SNAREs (also termed “R-SNAREs”) in general are sorted into transport carriers that mediate trafficking between organelles in a monomeric state so that they may participate in trans-SNARE complex formation that catalyzes vesicle fusion with the target membrane. When expressed in cultured cells, VAMP2 has been shown to localize to secretory vesicles, including dense core granules and synaptic-like microvesicles and endosomal compartments (Papini et al., 1995; Quetglas et al., 2002; Kubo et al., 2015). The best described sorting signals for R-SNARE proteins, including VAMP2, VAMP3, and VAMP8, are sequence-specific motifs that bind the CALM clathrin adaptor required for endocytosis (Miller et al., 2011). Interestingly, the crystal structure of the VAMP8-CALM complex suggests that binding of CALM can occur only when VAMP8 is in a monomeric state (Miller et al., 2011). Therefore, understanding the intracellular trafficking of SNARE proteins can give us insight into how this may act as another layer of regulation of its activity. Here, we investigate the intracellular trafficking pathways of the yeast VAMP2 homologue, Snc1, which has often been used as a reporter for studying endocytic retrieval pathways, but its own trafficking itinerary and the factors that regulate it have not been fully elucidated.
Snc1 is a plasma membrane-directed v-SNARE required for fusion of secretory vesicles with the plasma membrane. It is subsequently retrieved from the plasma membrane by endocytosis and returned to the Golgi apparatus via a plasma membrane recycling pathway (Lewis et al., 2000; Hettema et al., 2003; Ma et al., 2017). Interestingly, the regions in Snc1 that are analogous to the VAMP2 juxtamembrane region and transmembrane domain have been shown to play important roles in sorting of Snc1 between organelles of the secretory and endocytic pathways (Lewis et al., 2000). However, previous intracellular trafficking studies targeting the Snc1 transmembrane domain have used an incomplete sequence (Lewis et al., 2000; Reggiori et al., 2000), which was revealed by structural studies (Bowen and Brunger, 2006; Han et al., 2016). Hence, the role of the Snc1 transmembrane domain in intracellular sorting and trafficking is incompletely understood.
Numerous membrane trafficking factors have been identified that are required for proper Snc1 trafficking and localization within the Golgi and endosomal system, including Snx4-Atg20, Rcy1, Drs2/Cdc50, coatomer I, Gcs1 Arf-GAP, numerous endocytosis proteins, and cytoskeleton components (Galan et al., 2001; Hettema et al., 2003; Robinson et al., 2006; Furuta et al., 2007; Burston et al., 2009; Xu et al., 2013, 2017; Boscheron et al., 2016; Ma et al., 2017; MacDonald and Piper, 2017), but the specific roles of these factors and the postendocytic trafficking itinerary of Snc1 are poorly understood.
The Snx4-Atg20 sorting nexin containing a Bin-Amphiphysin-Rvs161 homology (BAR) domain (SNX-BAR) heterodimer is proposed to mediate plasma membrane recycling of Snc1 by functioning as a coat protein for carriers that mediate retrieval of Snc1 to the Golgi apparatus (Hettema et al., 2003; Ma et al., 2017). However, in snx4Δ cells, there is still a substantial proportion of GFP-Snc1 that resides in the plasma membrane, raising the possibility that multiple pathways may mediate retrieval of internalized Snc1 to the Golgi. Moreover, recent evidence indicates that Snx4-Atg20 decorates late/prevacuolar endosomes (PVE) (Arlt et al., 2015; Ma et al., 2017), and Snc1 can mediate membrane fusion with the PVE (Paumet et al., 2004). Recently published data from Glick and colleagues (Day et al., 2018) and from MacDonald and Piper (2017) have challenged the long-held view that PVE-to-Golgi trafficking constitutes an essential segment of plasma membrane recycling pathways. Here, we address the role of Snx4-Atg20 and Rcy1, as they comprise core components of Snc1 retrograde and recycling trafficking pathways. Rcy1 is a F box protein that forms a complex with the GTP-bound form of Ypt31/32 GTPases (Chen et al., 2005). In addition, Rcy1 associates with an aminophospholipid flippase, Drs2, and this interaction is required for plasma membrane recycling of Snc1 (Furuta et al., 2007; Hanamatsu et al., 2014). At present, it is unclear whether Snx4-Atg20 and Rcy1 function on the same, or distinct, Snc1 retrograde trafficking pathways.
The terminal fate for cargo of the endocytic pathway is degradation within the vacuole, and many different routes of retrieval have been described that rescue endocytosed cargoes from this fate (Feyder et al., 2015). However, endocytic cargoes can also be targeted for degradation in the vacuole lumen via the multivesicular body (MVB) pathway, where they are ubiquitylated by the Rsp5 ubiquitin ligase and sorted into intraluminal vesicles via sequential action of endosomal sorting complexes required for transport (ESCRT) proteins (Piper and Katzmann, 2007). This route is also proposed to be utilized by protein quality control pathways that target integral membrane proteins for degradation; however, to date, little evidence exists documenting surveillance of nonnative protein conformations. Rather, most evidence documents sorting of proteins, such as nutrient transport proteins, when in a particular native conformation, or native proteins that are subject to relatively rapid degradation.
Here we elucidate the trafficking itinerary of Snc1 and demonstrate that it traffics via distinct Rcy1-dependent and Snx4-dependent pathways. We also investigate the mechanism by which Snc1 is incorporated into Snx4-retrograde carriers, giving us insight into how SNAREs, in general, can be packaged into transport carriers. We also investigate protein quality control of Snc1 and identify cis- and trans-acting factors that mediate quality control surveillance of Snc1.
RESULTS
Snc1 is trafficked via multiple endocytic retrograde/recycling pathways
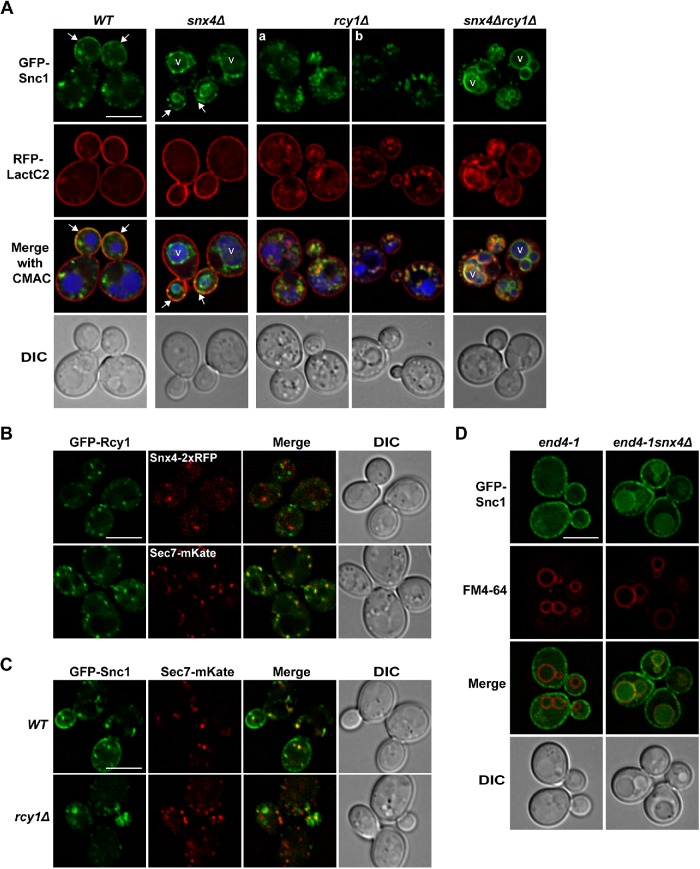
Snc1 was first established as a cargo of an endosome-to-Golgi retrograde pathway defined by the genetic requirement for SNX4, a gene encoding a SNX-BAR protein that dimerizes with the related SNX-BAR proteins, Atg20 and Snx41 (Hettema et al., 2003). A second gene required for retrograde trafficking of GFP-Snc1 is RCY1, which encodes a protein proposed to regulate trafficking through the Golgi and/or from the early endosome to the Golgi (Wiederkehr et al., 2000; Galan et al., 2001; Chen et al., 2011; Sun and Drubin, 2012). However, it is unclear whether Rcy1 and Snx4 are components of the same or different trafficking pathways. The distinct localization patterns of GFP-Snc1 in rcy1Δ and snx4Δ (and atg20Δ) cells is consistent with Snc1 trafficking via multiple retrograde pathways (Hettema et al., 2003; Sun and Drubin, 2012; Ma et al., 2017; Xu et al., 2017). To directly test whether the Snx4 and Rcy1 pathways traffic Snc1 independently of each other, we expressed GFP-Snc1 in snx4Δ, rcy1Δ, and snx4Δrcy1Δ double mutant cells (Figure 1A) (BY4742 strain background). GFP-Snc1 exhibited distinct localizations in each of these strains. In wild-type cells, GFP-Snc1 localizes to endosome and Golgi compartments, which appear as multiple puncta, and to the plasma membrane. No GFP-Snc1 is observed on the vacuole membrane, and we speculate that this is due to efficient export from the PVE prior to its fusion with the vacuole membrane (Figure 1A). In snx4Δ cells, GFP-Snc1 accumulated on the vacuole membrane but still retained partial polarized plasma membrane distribution, as we and others have previously reported (Hettema et al., 2003; Ma et al., 2017). In rcy1Δ cells, GFP-Snc1 accumulated on internal compartments, including tubulo-vesicular clusters near the bud neck of budding cells, as reported previously (Chen et al., 2005; Robinson et al., 2006; Furuta et al., 2007; Sun and Drubin, 2012; Xu et al., 2017). On examination of the volume of rcy1Δ cells, we note that in some cells GFP-Snc1 appears to reside in tubules that appear to be invaginations of the plasma membrane, which we identified by RFP-LactC2, an engineered probe that reports phosphatidylserine (PS), which is enriched on the inner leaflet of the plasma membrane (Figure 1A; an extreme example of these membrane invaginations is presented in Figure 1Ab) (Yeung et al., 2008). Previously reported electron microscopy characterization of rcy1Δ cells also describe morphologically similar aberrant elongated membrane structures that are present in rcy1Δ cells, and this expanded membrane network was reported to contain Tlg1, a t-SNARE that resides on trans-Golgi network (TGN)/endosome compartments, by immunoelectron microscopy analysis (Wiederkehr et al., 2000). In snx4Δrcy1Δ cells, GFP-Snc1 exhibited a distinct localization phenotype compared with either snx4Δ cells or rcy1Δ cells, where GFP-Snc1 accumulated on internal compartments and the vacuole membrane, but little was detected on the plasma membrane (Figure 1A). The distinct distributions of GFP-Snc1 in snx4Δ, rcy1Δ, and snx4Δrcy1Δ double mutant cells suggests that Snx4 and Rcy1 operate on different Snc1 trafficking pathways.
FIGURE 1:
Epistasis analysis of GFP-Snc1 trafficking pathways. (A) Micrographs of GFP-Snc1 and RFP-LactC2 coexpressed in wild-type and the indicated mutant cells. Arrows point to the plasma membrane of budding cells. Panel a shows a representative localization of GFP-Snc1 and RFP-LactC2 that accumulate near in or near the budding cell. Panel b shows an example where GFP-Snc1 compartments appear to be connected to PM invaginations, marked by RFP-LactC2. Vacuoles are visualized using CMAC. (B) Micrographs of rcy1Δ cells showing GFP-Rcy1 colocalization with Snx4-2xRFPor Sec7-mKate, Pearson’s correlations are Rave = 0.22 (n = 49), Rave = 0.64 (n = 43), respectively. (C) Micrographs of wild-type and rcy1Δ cells expressing GFP-Snc1 and Sec7-mKate are shown. (D) Micrographs of end4-1 and end4-1snx4Δ cells expressing GFP-Snc1 are shown. Vacuole membranes were visualized using FM4-64 dye. An approximate medial single Z section is shown unless otherwise indicated. Scale bars indicate 5 μm.
Both Rcy1 and Snx4-Atg20 have been shown to play a role in trafficking of PS-containing membrane within the endocytic system (Sun and Drubin, 2012; Ma et al., 2018). We have previously shown that Snx4-Atg20 binds preferentially to PS-rich membrane in vitro, and in vivo contributes to removing PS from the endosome prior to its fusion with the vacuole membrane (Ma et al., 2018). To determine the relative contributions of Rcy1 and Snx4-Atg20 to plasma membrane enrichment of PS, we examined RFP-LactC2 localization in snx4Δrcy1Δ cells (Figure 1A). In wild-type cells, RFP-LactC2 localized almost exclusively to the plasma membrane (Figure 1A). In rcy1Δ cells, we confirm that RFP-LactC2 accumulated inside the cell, colocalizing with GFP-Snc1, suggesting that Snc1 and PS are cotransported in the Rcy1-dependent pathway (Sun and Drubin, 2012). In snx4Δ cells, RFP-LactC2 localization was indistinguishable from that of wild-type cells, indicating that the Snx4 pathway does not play a significant role in restricting localization of PS to the plasma membrane (Figure 1A). This is in agreement with the proposal that Rcy1 functions on an endosome-to-plasma membrane recycling pathway (MacDonald and Piper, 2017). Interestingly, in snx4Δrcy1Δ double mutant cells, RFP-LactC2 and GFP-Snc1 accumulated on the vacuole membrane, with very little probe on other compartments. These data indicate that the PS that accumulates internally in rcy1Δ cells accesses the PVE, where Snx4-Atg20 retrograde trafficking prevents its accumulation on the vacuole membrane (Figure 1A). These results further support the conclusion that Rcy1 and Snx4 function in two distinct trafficking pathways.
To further test the hypothesis that Rcy1 and Snx4 function on distinct Snc1 trafficking pathways, we used fluorescence microscopy to compare the subcellular localizations of Rcy1 and Snx4. Snx4-2xRFP was expressed from the native SNX4 locus (as previously described [Ma et al., 2017]) and GFP-Rcy1 was expressed in rcy1Δ cells from a plasmid confirmed to complement rcy1Δ in vivo. We have previously shown that Snx4-2xGFP localizes to endosomes that are decorated with other endosomal sorting factors, such as retromer (Ma et al., 2017). When coexpressed, it is clear there is little-to-no colocalization of Snx4 and Rcy1 (Pearson’s correlations, Rave = 0.22 [n = 49]), strongly suggesting that they reside on distinct compartments (Figure 1B). Furthermore, we confirmed that Rcy1 localizes to organelles decorated with Sec7, a peripheral resident of late Golgi compartments (Pearson’s correlations, Rave = 0.64 [n = 43]) (Chen et al., 2005; Day et al., 2018), and concentrates at sites of polarized growth (Galan et al., 2001) (Figure 1B). On the basis of the Rcy1 localization, we wondered whether Rcy1 plays a role in transport of GFP-Snc1 out of the Sec7-positive TGN compartments. However, whereas GFP-Snc1 and Sec7-mKate do partially colocalize in wild-type cells, they do not colocalize in rcy1Δ cells (Figure 1C), suggesting that Rcy1 is required for GFP-Snc1 to access Sec7-decorated compartments. Taken together, these data firmly establish that Rcy1 and Snx4 are components of two distinct Snc1 trafficking pathways.
To determine whether Rcy1 is required for GFP-Snc1 to access Snx4-dependent pathways, we expressed GFP-Snc1 in a clathrin-dependent endocytosis-deficient strain, end4-1 (Wesp et al., 1997), which accumulates endocytic cargo at the plasma membrane, and in end4-1snx4Δ double mutant cells, which are deficient in both endocytosis and Snx4-dependent retrograde trafficking. Interestingly in end4-1snx4Δ cells, GFP-Snc1 accumulated on both the vacuole membrane, marked by FM4-64 dye (Vida and Emr, 1995), and the plasma membrane, indicating that GFP-Snc1 is trafficked to endo-vacuolar organelles independently of endocytosis (Figure 1D).
Snc1 retrograde sorting signal is in its juxtamembrane region
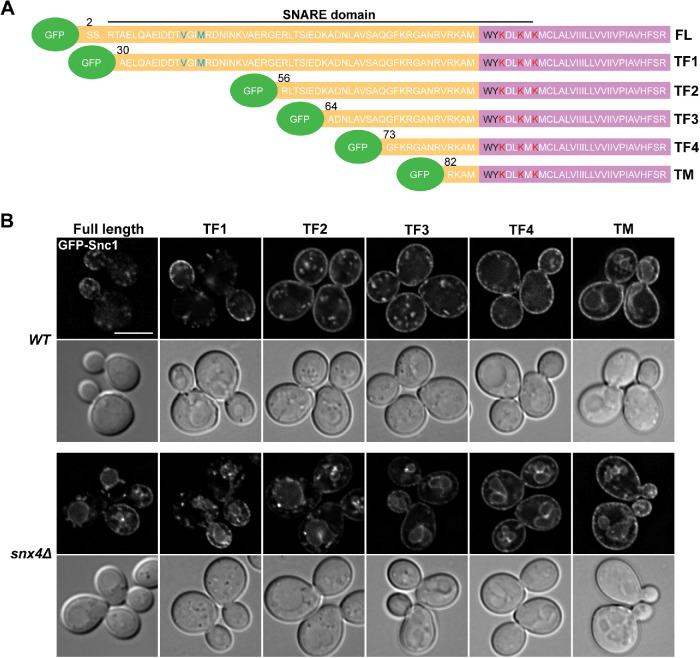
To understand how Snx4-dependent sorting of Snc1 is conferred, we conducted a truncation analysis in wild-type versus snx4Δ cells to identify the minimum sequence requirement for proper GFP-Snc1 localization. We systematically truncated Snc1 at the N-terminus and visualized its localization with N-terminally fused GFP, expressed in wild-type cells (Figure 2A). Full-length GFP-Snc1 localizes to the plasma membrane, concentrating at sites of polarized growth, and to intracellular compartments (Lewis et al., 2000). Similarly, GFP-Snc1 with the SNARE motif intact (Strop et al., 2008) but lacking the N-terminal segment (here referred to as “GFP-Snc1-TF1”) also exhibited this localization (Figure 2B). We note that when the previously identified endocytic signal, requiring V40 and M43, was deleted, GFP-Snc1 is homogenously distributed on the plasma membrane rather than concentrated at the daughter cell (Figure 2B), confirming previous reports that endocytic recycling is required for the polarized distribution of GFP-Snc1 (Lewis et al., 2000; Valdez-Taubas and Pelham, 2003). Additional truncations (TF2, TF3, and TF4 in Figure 2) revealed that most of the cytosolic region, including the SNARE motif (Strop et al., 2008), is dispensable for Snc1 trafficking via the Snx4 pathway. However, there is a distinct requirement for the transmembrane domain and the adjacent 13 amino acids of the cytosolic domain of Snc1 for Snx4-dependent trafficking. This is evident in the localization of Snc1-TM in wild-type cells, which is indistinguishable from its localization in snx4Δ cells. Strikingly, this vacuolar localization of GFP-Snc1-TM is reminiscent of full-length GFP-Snc1 in snx4Δ or snx4Δrcy1Δ cells (Figures 1A and 2B), suggesting that the juxtamembrane region present in GFP-Snc1-TF4 but absent in GFP-Snc1-TM (amino acids 73–81) contains a sorting signal that confers trafficking via the Snx4-dependent retrograde pathway. Importantly, we also found that Snc1 truncation mutants that lack K63, which has been proposed to be ubiquitylated in Rcy1-dependent trafficking of Snc1 (Chen et al., 2011), still retain Snx4-dependent localization. This further establishes that the Snx4-dependent Snc1 trafficking pathway is distinct from the Rcy1 pathway, and importantly, that trafficking via these different pathways requires different sorting signals that are encoded within the Snc1 protein.
FIGURE 2:
Truncation analysis of Snc1 identifies juxtamembrane region to be important for Snx4-dependent trafficking. (A) Schematic of Snc1 truncation mutants. The cytosolic domain of Snc1 is orange and the transmembrane domain is purple. The V40 and M43 residues shown previously to be required for endocytosis are shown in blue. The conserved aromatic residues W86 and Y87 are shown in black and “snorkeling” lysine residues are shown in red. The first residue of Snc1 in each truncation is indicated. (B) Micrographs showing wild-type and snx4Δ cells expressing full-length GFP-Snc1 or indicated truncation mutants. An approximate medial single Z section is shown unless otherwise indicated. Scale bar indicates 5 μm.
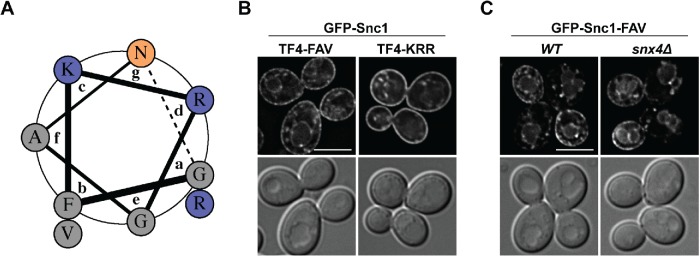
The truncation analysis indicates that a nine–amino acid sequence (amino acids 73–81) in the juxtamembrane region of Snc1 is required for Snx4-dependent retrograde trafficking of GFP-Snc1 (Figure 2). This sequence, as well as the sequence of the transmembrane domain, is highly conserved in synaptobrevins across all species and has been extensively studied in the mammalian homologue VAMP2 (Quetglas et al., 2000, 2002; Kweon et al., 2003; Bowen and Brunger, 2006; Han et al., 2016). Structural analyses of the juxtamembrane region of human monomeric VAMP2 indicate that this region is composed of an alpha helix (VAMP2 amino acids 75–84; Snc1 amino acids 72–81) termed the juxtamembrane helix (JMH) and an unstructured flexible linker region (VAMP2 amino acids 85–89; Snc1 amino acids 82–86) leading to the helical transmembrane domain (Han et al., 2016). Structures of VAMP2 and Snc1 in their plasma membrane cis SNARE complexes show that the entire juxtamembrane region (JMH and flexible linker) forms an alpha helix that extends into the helical transmembrane domain (Strop et al., 2008; Stein et al., 2009). Remarkably, the nine-amino acid region we identified to confer Snx4-dependent retrograde trafficking of Snc1 corresponds exactly to the JMH proposed to form in unbound VAMP2. To further analyze this region, we used helical wheel analysis to assess helical characteristics of the nine-amino acid sequence and found that the predicted helix contains amphipathic features, where one face is composed of basic residues (blue) and the opposite face is composed of uncharged and aromatic residues (gray) (Figure 3A). To test whether these features are important for retrograde trafficking of Snc1, we created point mutations in this region in GFP-Snc1-TF4 and found that substitutions of the basic residues (K75, R76, R80) with alanines had little effect on GFP-Snc1-TF4 localization, but substitutions of the aromatic and uncharged residues (F74A, A78S, V81A) resulted in vacuole membrane localization, reminiscent of GFP-Snc1-TM (Figure 3B). Importantly, the same mutations (FAV→ASA) in full-length GFP-Snc1 also result in vacuole membrane localization of GFP-Snc1 in wild-type cells that is indistinguishable from its localization in snx4Δ cells (Figure 3C).
FIGURE 3:
Mutations in the predicted Snc1 JMH result in localization to the vacuole membrane. (A) Helical wheel representation of the predicted structural features of the nine-amino acid region required for retrograde trafficking of Snc1. Basic residues are colored blue and hydrophobic residues are colored gray. Note the amphipathic nature of this segment, with charged residues on one face. (B) Micrographs of wild-type cells expressing GFP-Snc1-TF4 truncation mutant containing indicated point mutations in the JMH. (C) Micrographs of wild-type and snx4Δ cells expressing full-length GFP-Snc1 containing the indicated point mutations in the predicted JMH. An approximate medial single Z section is shown unless otherwise indicated. Scale bars indicate 5 μm.
Positively charged residues in the Snc1 transmembrane domain are required for Snx4-dependent retrograde trafficking
An unusual feature of the VAMP2 and Snc1 transmembrane domains is the presence of lysine residues within the membrane-spanning segment (see Figure 7B later in the article) that lie C-terminal to two aromatic residues at the cytosol-membrane interface that marks the start of the transmembrane domain. These hydrophobic residues (W89, W90 in VAMP2 and W86, Y87 in Snc1) anchor into the membrane bilayer (Kweon et al., 2003), forcing the unusually long transmembrane domain of VAMP2 to tilt 35° relative to the bilayer normal (Bowen and Brunger, 2006), allowing the lysine side chains to “snorkel” to the membrane–cytosol interface (Bowen and Brunger, 2006; Borisovska et al., 2012). Curiously, mutation of one of the homologous aromatic residues in yeast Snc1 (W86; W89 in VAMP2) results in localization of this mutant protein to the vacuole; that is, it results in the loss of retrograde trafficking via the Snx4-Atg20–dependent pathway (Lewis et al., 2000). A compelling hypothesis is that the orientation of the transmembrane domain within the membrane, conferred by the hydrophobic and lysine residues, orients the juxtamembrane segment containing the Snx4-dependent sorting signal. Consistent with this, molecular dynamics modeling of mammalian VAMP2 demonstrates that the sequence and orientation of the VAMP2 transmembrane domain directly affects the helicity of the JMH (Han et al., 2016).
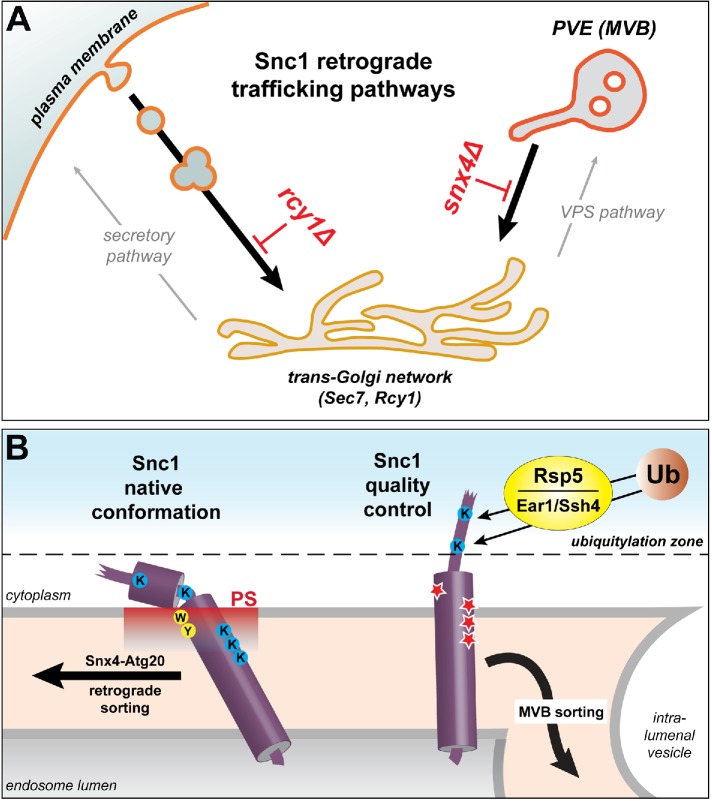
FIGURE 7:
Model of Snc1 trafficking pathways and structural features of Snc1 required for Snx4-dependent trafficking. (A) Snc1 retrograde trafficking pathways in the endo-vacuolar system. Anterograde Snc1 trafficking bifurcates at the TGN, where it can be directed to the plasma membrane via the secretory pathway or the endo-vacuolar system via the VPS pathway. On endocytosis of Snc1 from the plasma membrane, Rcy1 is required for its entry into the TGN, a compartment in which both Sec7 and Rcy1 reside. Retrieval from the PVE to the TGN is mediated by Snx4-Atg20. (B) Structural features of Snc1 that play vital roles in Snx4-dependent trafficking of native Snc1 and sorting of mutant Snc1 proteins into the degradative MVB pathway. In wild-type Snc1, aromatic residues (yellow) anchor into the membrane bilayer, forcing the transmembrane domain to tilt (Kweon et al., 2003; Bowen and Brunger, 2006), allowing downstream basic lysine residues (blue) to snorkel and interact with the membrane surface. This orientation of the transmembrane domain allows proper display of the Snx4-Atg20–dependent sorting signal located in the JMH. We propose that lysine residues within this region and those in the transmembrane segment that snorkel to the membrane surface promote Snc1 residence in PS-rich regions (red) of the endosome membrane, which facilitates sorting into the Snx4-Atg20 retrograde pathway (Ma et al., 2018). Mutant Snc1-DDD and Snc1-W86R (Lewis et al., 2000) (indicated in red stars) are subjected to protein quality control by sorting into the degradative MVB pathway, requiring ubiquitylation of Rsp5, working with the Ear1 and/or Ssh4 adapters. We propose that these mutations cause unfolding of the juxtamembrane and the ‟top” of the transmembrane helices, allowing access of Lys75 and Lys83 to the “ubiquitylation” zone defined by Sardana et al. (2019).
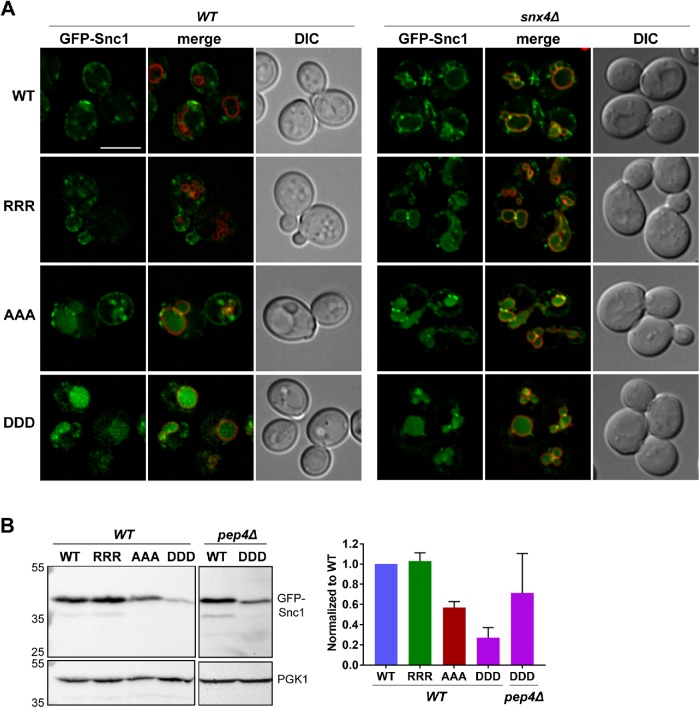
A previous study showed that single mutations of individual lysine residues within the TMD (K88, K91, K93) to alanine do not affect Snc1 trafficking (Lewis et al., 2000), so we determined the localization of a triple mutant in which different amino acid substitutions at positions K88, K91, and K93 in the full-length GFP-Snc1 protein were assayed in wild-type cells (Figure 4A). The three mutant constructs contain lysine to arginine substitutions, which preserves the charge of these residues, lysine to alanine substitutions, which neutralizes the charge but otherwise is not expected to change the helical structure of the transmembrane domain, or lysine to aspartate mutations, which reverses the charge of these residues. To eliminate expression variability due to expression from plasmids, we integrated each of the four GFP-Snc1 constructs into the URA3 locus in wild-type cells. When K88, K91, and K93 were mutated to RRR, GFP-Snc1 localization remained unchanged (Figure 4A). When KKK was mutated to AAA, GFP-Snc1 accumulates in the vacuole (identified by FM4-64), suggesting that export of GFP-Snc1 from the endosome is defective and that it is targeted for degradation within the vacuole; however, GFP-Snc1-AAA still localized to internal compartments and the plasma membrane of the budding cell, suggesting that the recycling defect was only partial (Figure 4A). When KKK was mutated to DDD, almost all of the GFP-Snc1 accumulated in the vacuole (Figure 4A), with a concomitant loss of plasma membrane localization, suggesting that the aspartate substitutions at these positions resulted in a more severe recycling defect compared with the alanine substitutions, and subsequent targeting for degradation in the vacuole. These results suggest that the charge at these positions, rather than stereospecific features of the lysine side chains, is important for Snc1 trafficking. We further tested the localization of these Snc1 mutants in snx4Δ cells and found that GFP-Snc1-DDD localized to the vacuole lumen in snx4Δ cells as it does in wild-type cells, while GFP-Snc1-AAA localization presented an additive effect where it accumulated both on the vacuole membrane and in the vacuole lumen. This indicates that while K88, K91, and K93 are required for Snx4-dependent trafficking, they also play a role in targeting Snc1 into the degradative pathway.
FIGURE 4:
Positively charged residues within the Snc1 transmembrane domain prevent targeting to the vacuole lumen. (A) Micrographs of wild-type or snx4Δ cells expressing wild-type or indicated GFP-Snc1 mutants. GFP-SNC1 genes were integrated into the URA3 locus to prevent plasmid expression variability. Vacuoles are visualized using FM4-64 dye. Scale bar indicates 5 μm. (Β) Cell lysates of wild-type or pep4Δ cells expressing the indicated GFP-Snc1 variants were probed with anti-GFP antibody and the amount of GFP-Snc1 was determined by semiquantitative Western blot analysis. Anti-PGK1 immunoblot was used as protein loading control. The positions of molecular mass (kDa) markers are indicated. The results from three experiments were averaged and standard error of the mean indicated.
We further characterized these mutants by quantifying the amount of wild-type and mutant GFP-Snc1 proteins in wild-type cells using immunoblot analysis. Protein levels are maintained in the GFP-Snc1-RRR mutant compared with wild-type GFP-Snc1 but decreased by 42% ± 6% when expressed as GFP-Snc1-AAA and 72% ± 3% when expressed as GFP-Snc1-DDD, consistent with the vacuolar localization of GFP-Snc1-AAA and GFP-Snc1-DDD (Figure 4B). We note that in GFP-Snc1-AAA and GFP-Snc1-DDD samples, we do not see a corresponding increase in free GFP. However, when we immunoblotted for GFP-Snc1-DDD in a strain lacking PEP4, the major vacuolar protease, we see an increase in the level of full-length GFP-Snc1-DDD, indicating that the decrease in GFP-Snc1-DDD levels in wild-type cells is in fact due to vacuolar degradation (Figure 4B).
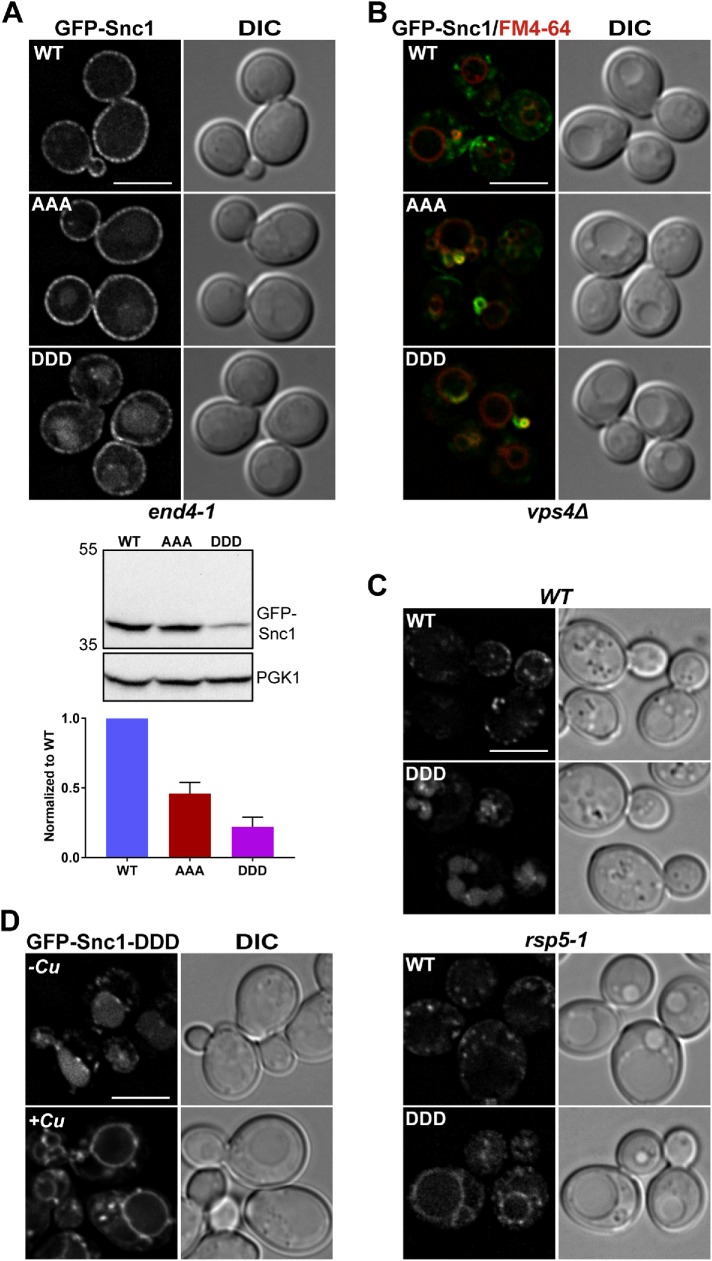
To test whether GFP-Snc1-AAA and GFP-Snc1-DDD are bona fide retrograde trafficking mutants, we first asked whether these mutants can still traffic to the plasma membrane. To do this, we first tested whether GFP-Snc1-AAA and GFP-Snc1-DDD accumulate on the plasma membrane of end4-1 cells. Wild-type GFP-Snc1 accumulated exclusively on the plasma membrane (Figure 5A). Similarly, both GFP-Snc1-AAA and GFP-Snc1-DDD also accumulated on the plasma membrane, indicating that these mutant proteins are trafficked via the secretory pathway to the plasma membrane (Figure 5A). Notably, both GFP-Snc1-AAA and GFP-Snc1-DDD also exhibited vacuolar localization, with brighter GFP-Snc1-DDD signal in the vacuole lumen. This vacuolar localization also corresponds to a decrease in the protein level of Snc1, similar to GFP-Snc1-AAA and GFP-Snc1-DDD expression levels in wild-type cells (Figure 5A). This is particularly interesting because it suggests that these forms of Snc1 follow an endocytosis-independent trafficking route to the vacuole, reminiscent of the trafficking itinerary we have established for wild-type GFP-Snc1 when SNX4 is ablated (Figure 1). Therefore, we conclude that the conserved Lys residues in the transmembrane domain (K88, K91, and K93) are required for Snx4-mediated retrograde trafficking of Snc1 from the endosome to the Golgi.
FIGURE 5:
Snc1 transmembrane domain mutants are defective for trafficking via the Snx4 retrograde pathway. (A) Micrographs of end4-1 cells expressing the indicated GFP-Snc1 constructs. Note that all GFP-Snc1 mutants tested localize to both the plasma membrane and the vacuole labeled by FM4-64. Cell lysates of end4-1 cells expressing the indicated GFP-Snc1 point mutants were probed with anti-GFP and the amount of GFP-Snc1 was determined by semiquantitative Western blot analysis. Anti-PGK1 immunoblot was used as protein loading control. The positions of molecular mass (kDa) markers are indicated. The results from three experiments were averaged and standard error of the mean indicated. (B) Micrographs of vps4Δ cells expressing indicated GFP-Snc1 constructs. Note that all GFP-Snc1 mutants tested localize to the “class E compartment” and were absent from the vacuole. Class E compartments and vacuoles were visualized using FM4-64 dye. (C) Micrographs of rsp5-1 cells and isogenic wild-type cells expressing GFP-Snc1 or GFP-Snc1-DDD. Note that GFP-Snc1-DDD localizes to the vacuole membrane instead of the vacuole lumen in rsp5-1 cells. (D) Micrographs of wild-type cells coexpressing Rsp5-DUb and GFP-Snc1-DDD. Rsp5-DUb is driven by the CUP1 promoter and to induce expression, cells were incubated with 50 μM CuCl2 for 4 h at 30°C. An approximate medial single Z section is shown unless otherwise indicated. Scale bars indicate 5 μm.
The appearance of mutant GFP-Snc1-DDD in the vacuole lumen, and the corresponding loss of its recycling, suggests that it is targeted for degradation within the vacuole. Membrane proteins in the endosome can be targeted for degradation via the MVB pathway, in which they are sorted into intraluminal vesicles in a process mediated by ESCRTs and on endosome/MVB fusion with the vacuole, are degraded by vacuolar proteases (Piper and Katzmann, 2007). The presence of acidic residues in the transmembrane domain of MVB-targeted proteins has been described to trigger sorting into the degradative MVB pathway as a way of differentiating sorting of proteins to the vacuole membrane versus the vacuole lumen (Reggiori et al., 2000). To determine whether the vacuole lumen accumulation of mutant GFP-Snc1 is due to sorting into the MVB pathway, we expressed GFP-Snc1-AAA and GFP-Snc1-DDD in a vps4Δ background, which accumulates MVB-targeted cargo in an enlarged late endosome-like structure, termed the “class E compartment,” and prevents it from reaching the vacuole (Babst et al., 1997). Strikingly, both GFP-Snc1-AAA and GFP-Snc1-DDD are trapped in the class E compartment and are completely absent from the vacuole, marked by FM4-64 dye, suggesting that GFP-Snc1-AAA and GFP-Snc1-DDD reach the vacuole via the degradative MVB pathway (Figure 5B). However, it is not clear from these results whether sorting into the Snx4 pathway is defective or whether MVB sorting occurs upstream of and/or overrides Snx4-Atg20–dependent sorting. To test this, we reasoned that if GFP-Snc1-DDD was prevented from entering into the MVB pathway, we could assess the effect of the Asp mutations to retrograde trafficking. Targeting of proteins into the MVB pathway requires ubiquitylation of the proteins, often by the cytosolic Rsp5 ubiquitin ligase (Belgareh-Touzé et al., 2008). Rsp5 is a member of the Nedd4 (neural-precursor-cell-expressed, developmentally down-regulated) family of E3 ubiquitin ligases and has a wide variety of functions in the cell, including conferring MVB sorting in the endosomal system (Belgareh-Touzé et al., 2008). Because RSP5 is an essential gene, we utilized two different mutant Rsp5 reagents to test whether GFP-Snc1-DDD regained wild-type localization when Rsp5 function was compromised. First, we expressed GFP-Snc1 and GFP-Snc1-DDD in rsp5-1 cells, which contain a hypomorphic RSP5 allele (Wang et al., 1999), and observed that GFP-Snc1-DDD accumulated on the vacuole membrane rather than the vacuole lumen, as in wild-type cells (Figure 5C). Next, we utilized an engineered fusion protein, Rsp5-DUb driven by the CUP1 promoter, which contains the catalytic domain of a deubiquitinating enzyme (Ubp7) fused to Rsp5 that acts in a dominant manner to deubiquitinate Rsp5 substrates (Stringer and Piper, 2011). When coexpressed with Rsp5-DUb (in wild-type cells), GFP-Snc1-DDD accumulated on the vacuole membrane, similar to rsp5-1 cells (Figure 5D). We conclude from these results that GFP-Snc1-DDD fails to be sorted into the Snx4 retrograde pathway and furthermore that Rsp5 plays a role in MVB targeting of mutant GFP-Snc1.
Mutations in the Snc1 transmembrane domain are recognized by the endosomal quality control system
How is GFP-Snc1-DDD recognized for targeting into the degradative MVB pathway? When we compared the subcellular localization of GFP-Snc1 and GFP-Snc1-DDD, we noticed that GFP-Snc1-DDD accumulates in the vacuole lumen (in wild-type cells and snx4Δ cells) while wild-type Snc1 accumulates predominantly on the vacuole membrane (in snx4Δ cells) (Figures 1 and 4). Thus, the sole absence of Snx4-Atg20–dependent recycling is not sufficient to target Snc1 into the MVB pathway. However, severe mutations in the transmembrane domain of Snc1 (e.g., Snc1-AAA, Snc1-DDD) cause both loss of Snx4-dependent retrograde trafficking and gain of Rsp5-dependent sorting into the MVB pathway (Figure 5). This suggests that the transmembrane domain of Snc1 also plays a vital role in “protein quality control” of Snc1.
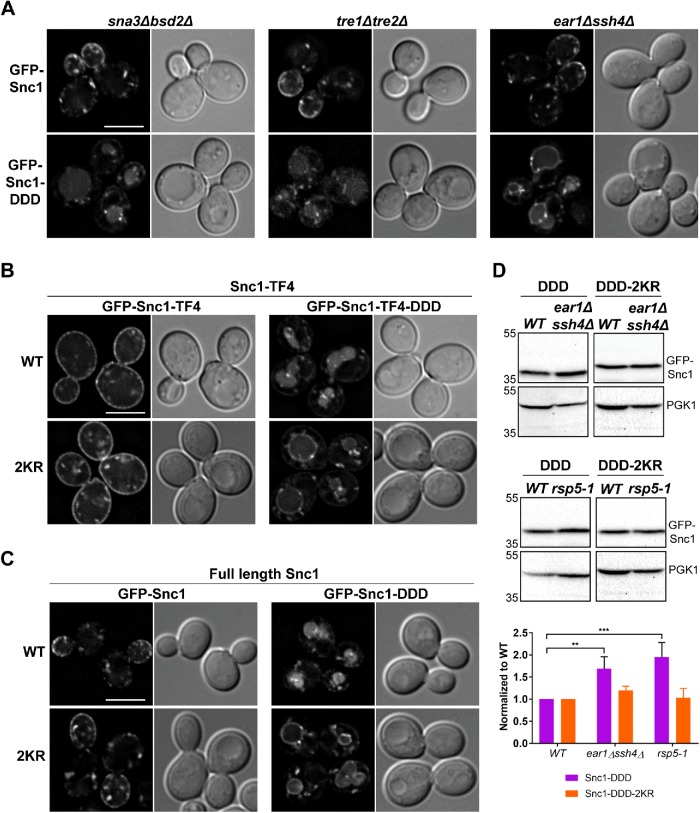
Rsp5 functions at many locations in the cell to ubiquitylate structurally and functionally diverse substrates. Some of these substrates can be directly recognized by Rsp5, while others are recognized through adaptor proteins that then recruit Rsp5 (Belgareh-Touzé et al., 2008). Since the mutations in Snc1-DDD that promote Rsp5-dependent degradation reside inside the membrane bilayer and Rsp5 is a cytosolic protein, we reasoned that MVB targeting of Snc1-DDD requires a transmembrane protein adaptor. To identify such a Rsp5 adaptor, we took a candidate approach using two criteria: 1) the adaptor must be a transmembrane protein and 2) the adaptor must reside, at least partly, in the endosome and/or vacuole. Six Rsp5 adaptors fit these criteria (Figure 6A), some of which have been shown to function redundantly, so we tested GFP-Snc1-DDD localization in all single mutants as well as in double mutants for functionally redundant adaptors. Of these, we identify Ear1 and Ssh4, which are paralogues and function redundantly for several proteins targeted into the vacuole via the canonical vacuolar protein sorting (VPS) pathway (Léon et al., 2008; Sardana et al., 2019), to be important for GFP-Snc1-DDD degradation. When expressed in ear1Δssh4Δ cells, GFP-Snc1-DDD accumulates on the vacuole membrane instead of the solely in the vacuole lumen (Figure 6A). This is consistent with a recent study that proposed Ear1 and Ssh4 to function redundantly in endo-vacuolar organelle quality control (Sardana et al., 2019). Importantly, we note that wild-type GFP-Snc1 localization was normal in all strains tested, suggesting that Rsp5 and its adaptors do not play a role in normal trafficking of Snc1 and that this dependence of GFP-Snc1-DDD trafficking on Rsp5 is indicative of a bona fide protein quality control surveillance (Figure 6A).
FIGURE 6:
GFP-Snc1-DDD is targeted into the MVB pathway by an endosomal quality control system. (A) Micrographs showing GFP-Snc1 and GFP-Snc1-DDD expressed in cells that are null for the indicated Rsp5 adaptors. Note that GFP-Snc1-DDD accumulates at the vacuole membrane in ear1Δssh4Δ cells. (B) Micrographs showing GFP-Snc1-TF4 constructs with indicated mutations expressed in wild-type cells. (C) Micrographs showing full-length GFP-Snc1 constructs with indicated mutations expressed in wild-type cells. An approximate medial single Z section is shown unless otherwise indicated. Scale bars indicate 5 μm. (D) Cell lysates of indicated strains expressing GFP-Snc1-DDD or GFP-Snc1-DDD-2KR were probed with anti-GFP and the amount of GFP-Snc1 was determined by semiquantitative Western blot analysis. Anti-PGK1 immunoblot was used as protein loading control. The positions of molecular mass (kDa) markers are indicated. The results from at least three experiments were averaged and the standard error of the mean was indicated. Two-way analysis of variance was used to determine statistical significance (GraphPad Prism 7.01). **p ≤ 0.01; ***p ≤ 0.001. Note that the differences seen in the levels of GFP-Snc1-DDD-2KR across all strains are not significant.
Next, we tested whether lysine residues in the cytosolic region of Snc1-DDD are required for targeting into the MVB pathway. First, we constructed GFP-Snc1-TF4-DDD to test whether the lysine residues present in the smallest truncation mutant that retain Snx4-dependent trafficking are sufficient for targeting into the MVB pathway. Similar to the localization of full-length GFP-Snc1-DDD, GFP-Snc1-TF4-DDD localized almost exclusively to the vacuole lumen (Figure 6B). Since GFP-Snc1-TF4-DDD traffics to the vacuole lumen, we can assume that the lysine residues not included in this truncation mutant are not required for MVB targeting of Snc1. Therefore, we tested the two cytosolic lysine residues present in Snc1-TF4 (K75 and K83) and mutated both to arginine (GFP-Snc1-TF4-DDD-2KR). When expressed in wild-type cells, GFP-Snc1-TF4-DDD-2KR localized to the vacuole membrane while GFP-Snc1-TF4-2KR maintained wild-type localization (Figure 6B), indicating that these two lysine residues are necessary for MVB targeting of mutant Snc1 but do not play a role in normal trafficking of Snc1. We also tested the contribution of these two lysine residues in MVB targeting of full-length Snc1-DDD and found that GFP-Snc1-DDD-2KR also localized to the vacuole membrane while GFP-Snc1-2KR maintained wild-type localization, indicating that these two lysine residues are critical for MVB targeting of Snc1-DDD (Figure 6C). To corroborate these results, we conducted immunoblot analysis of GFP-Snc1-DDD and GFP-Snc1-DDD-2KR expressed in wild-type or ear1Δssh4Δ cells and observed that the amount of GFP-Snc1-DDD is increased in ear1Δssh4Δ cells compared with wild-type cells, and the amounts of GFP-Snc1-DDD-2KR are equivalent in wild-type and ear1Δssh4Δ cells (Figure 6D). We also analyzed the protein levels of GFP-Snc1-DDD and GFP-Snc1-DDD-2KR in rsp5-1 cells and similarly found that protein levels of GFP-Snc1-DDD, but not of GFP-Snc1-DDD-2KR, increased in rsp5-1 cells. These results support our conclusion that Rsp5 and its adaptors Ear1 and Ssh4 are required for degradation of Snc1-DDD and furthermore, that this degradation is dependent on K75 and K83 in the juxtamembrane region of Snc1.
DISCUSSION
Results presented here demonstrate that GFP-Snc1 is retrieved from the endosomal network via two distinct pathways that are defined by requirements for Snx4 and Rcy1 (Figure 7A). Previous studies firmly established that GFP-Snc1 is broadly distributed between secretory and endosomal organelles of wild-type cells, and in snx4Δ and atg20Δ cells it accumulates on the vacuole, indicating that Snx4-Atg20 mediates retrieval from the endosome, likely back to the Golgi (Lewis et al., 2000; Hettema et al., 2003; Ma et al., 2017). The Rcy1-dependent pathway functions downstream of endocytosis, as endocytosis-deficient GFP-Snc1 (GFP-Snc1 containing V40A and M43A) localizes predominantly to the plasma membrane in rcy1Δ cells, as it does in wild-type cells (Galan et al., 2001; Chen et al., 2005). We show here that in snx4Δend4-1 cells, GFP-Snc1 localizes to the plasma membrane and the vacuole membrane, indicating that delivery to the endosome occurs independently of endocytosis (Figure 1D). Rcy1 is proposed to be a component of an endosome-to-plasma membrane recycling circuit that was initially described by Wiederkehr et al. (2000) and elaborated by MacDonald and Piper (2017). The polarized distribution pattern of the rcy1Δ compartment that appears in rcy1Δ cells (Wiederkehr et al., 2000; Galan et al., 2001; Chen et al., 2005, 2011; Furuta et al., 2007; Sun and Drubin, 2012; MacDonald and Piper, 2017) (Figure 1) suggests that this abnormal compartment is of endocytic origin. Consistent with this, the membrane of the rcy1Δ compartment contains PS (Figure 1) and phosphatidylinositol 4,5-bisphospate (PI[4,5]P2), two lipids that are enriched in the cytoplasmic leaflet of the plasma membrane, and decorated with endocytosis-associated actin patches (Sun and Drubin, 2012). Furthermore, initial characterization of rcy1Δ cells describe the rcy1Δ compartment to contain Tlg1, a marker of the TGN/early endosome, further indicating that this compartment is of endocytic origin (Wiederkehr et al., 2000). These observations can be explained by positing a role for Rcy1 in the delivery of endocytosed plasma membrane to a compartment decorated with Sec7 and Rcy1, considered to be the TGN (Figure 1) (Chen et al., 2005; Day et al., 2018). Interestingly, our results (Figure 1) show that GFP-Snc1 prominently decorates the vacuole membrane of snx4Δrcy1Δ cells, indicating that GFP-Snc1 can access the PVE independently of the Rcy1-dependent pathway. This may be via a trafficking pathway from the aberrant rcy1Δ compartment, or more likely, due to trafficking from the Golgi to the PVE (and deficient retrieval from the endosome). On the basis of the fusion-competent SNARE complexes containing Snc1 (discussed in the next paragraph), we propose that the TGN is also the destination of the Snx4-dependent retrieval pathway.
We propose a model in which GFP-Snc1 is exported from the TGN via two pathways: the secretory pathway, which mediates delivery of newly synthesized and endocytosed material to the cell surface, and the VPS pathway, which transports material from the Golgi to the PVE (Figure 7A). Whereas delivery of plasma membrane-derived, endocytosed GFP-Snc1 to the TGN requires Rcy1, GFP-Snc1 retrieval from the PVE is mediated by Snx4-Atg20 (this study and Ma et al., 2017). This model is supported by published observations that Snc1 (and its close homologue, Snc2) forms fusion-competent SNARE complexes with multiple Q-SNARE complexes that reside in the Golgi/early endosome, the plasma membrane, and the PVE (Kienle et al., 2009). Snc1 (and Snc2) is the R-SNARE of Golgi-derived vesicles that mediates fusion via the plasma membrane-localized Sso1/Sec9 Q-SNARE (McNew et al., 2000), and we suggest that Snc1 (and Snc2) also functions as an R-SNARE on Golgi-derived vesicles of the VPS pathway that fuse with the PVE via the Pep12/Syn8/Vti1 Q-SNARE complex (Lewis and Pelham, 2002). On this basis, we further suggest that Snx4-dependent retrieval of Snc1 requires that Snc1 is dissociated from the postfusion SNARE complex. An important consequence of this is that Snc1 will be competent to participate in fusion with its cognate Golgi-localized Q-SNARE (Tlg1/Tlg2/Vti1) to complete retrograde trafficking (Paumet et al., 2001).
Consistent with the existence of two distinct Snc1 retrieval pathways, two distinct retrieval signals have now been identified in Snc1. Ubiquitylation of Snc1 has been shown to confer plasma membrane recycling in a manner dependent on recognition by coatomer I (Chen et al., 2011; Xu et al., 2017). Here, we identified structural features of the Snc1 juxtamembrane region and transmembrane domain that confer retrograde sorting of Snc1 via the Snx4-dependent pathway (Figure 7B). We identified three amino acid substitutions in the juxtamembrane region (F74, A78, V81) and three substitutions in the transmembrane domain (K88, K91, K93) that ablate Snx4-dependent retrieval. Molecular dynamics modeling of VAMP2 suggests that F74, A78, and V81 resides in the JMH, and that this conformation is directly coupled to the orientation of the transmembrane domain, indicating that mutations in the transmembrane domain (including K88, K91, and K93) can result in conformational changes to the juxtamembrane region that ablate the Snx4 signal (Han et al., 2016). As Snx4-Atg20 is a peripheral membrane protein complex, and these residues of Snc1 lie within the cytoplasm or near the cytosol-membrane interface, they may be recognized by Snx4-Atg20 directly. That the charge, rather than stereospecific features, at the positions of the lysine residues (K88, K91, K93) in the transmembrane domain, is essential for retrieval (Figure 4) argues that these residues are not directly recognized by a trans-acting sorting factor. In this regard, we note that our previous study of lipid-binding preferences of yeast SNX-BAR proteins established that Snx4-Atg20 preferentially binds to and tubulates liposomes containing PS (Ma et al., 2018). Topologically, the critical K88, K91, and K93 residues in the Snc1 transmembrane domain reside in the cytosolic leaflet of the endosome membrane and snorkel to the membrane–cytosol interface. As PS is an anionic lipid, local enrichment of PS about the Snc1 transmembrane domain may promote incorporation of Snc1 into PS-enriched endosome-derived transport carriers coated by Snx4-Atg20. In addition, capture may be promoted by residues (F74, A78, V81) in the juxtamembrane region of Snc1 that may be recognized by Snx4-Atg20. We note that in cells that lack PS (i.e., cho1Δ), Snc1 accumulates on intracellular compartments but does not exhibit vacuolar localization (Takeda et al., 2014; Hankins et al., 2015), suggesting that Snx4-dependent retrograde trafficking of Snc1 may be kinetically delayed but not ablated. Interestingly, in cho1Δ cells, the amount of phosphatidylinositol, the other major anionic lipid species of the yeast cell, is substantially increased (by 8%) (Fairn et al., 2011), and this may partially compensate for the loss of PS in the endosome membrane. Evidence for this comes from the addition of exogenous lysoPS, which can be taken up by the cell and converted to PS, and concomitantly results in the decease of PI in the cell (Fairn et al., 2011), suggesting that a balance of PS and PI is maintained in the cell and the presence of one may partially compensate for the loss of the other.
A protein–lipid electrostatic interaction underlies the clustering of syntaxin-1A, which participates in the neuronal exocytic SNARE complex, and PI(4,5)P2 in the plasma membrane (van den Bogaart et al., 2011) and molecular dynamics simulations suggest that the presence of PS also contributes to the formation of these nanodomains (van den Bogaart et al., 2011). We propose that Snc1 may utilize a similar mechanism, where the snorkeling basic residues in the Snc1 transmembrane domain interact with PS and/or PI3P in the endosome membrane to sequester Snc1, potentiating Snx4-Atg20-mediated sorting and export.
In snx4Δ cells, wild-type GFP-Snc1 fails to be retrieved from the endosome and it therefore accumulates in the vacuole membrane. Our analysis of the GFP-Snc1-DDD retrieval-defective mutant protein, however, led to the fortuitous discovery that it is sorted into the vacuolar degradative pathway via the Rsp5 ubiquitin ligase and the redundant Ear1 and Ssh4 Rsp5 adaptor proteins. Structural and molecular dynamics simulations of VAMP2 indicate that the Lys-to-Asp mutations that we introduced alter the native conformation of this region, which is conserved with Snc1 (Han et al., 2016). That is, these mutations compromise the folding, hence the “quality,” of the Snc1 TMD and surrounding residues, resulting in its targeting for degradation via the MVB pathway. A recent study by Emr and colleagues defined the structural features required for Ssh4-directed ubiquitylation of an engineered membrane protein by Rsp5 on mistargeting to the vacuole membrane. For this protein, they found that cytoplasmic lysine residues within 10–38 residues from its single TMD could be ubiquitylated and proposed a “ubiquitination zone” that ensures organelle quality control by promiscuous ubiquitylation-dependent turnover of integral membrane proteins (Sardana et al., 2019). In contrast, turnover of GFP-Snc1-DDD provides an example of protein quality control in the endo-vacuolar system that is triggered by a nonnative protein conformation. Interestingly, in Snc1-DDD, the two lysine residues that are required for Ear1/Ssh4/Rsp5-dependent vacuole targeting are three and 11 residues away from the membrane (Figure 6). The requirement for these residues (K75 and K83) in MVB sorting of Snc1-DDD could be explained if the Lys-to-Asp mutations in the Snc1-DDD mutant cause this portion of the TMD to extend into the cytoplasm, thereby poising these lysine residues to be ubiquitylated. Interestingly, Sardana et al. (2019) found that wild-type Snc1 also undergoes Ssh4-dependent degradation in snx4Δ cells; however, in the strain used for our studies (BY4742), integral membrane proteins accumulate on the vacuole membrane (Katzmann et al., 2004). We note that in native Snc1 there are three lysine residues within 10–38 residues of the TMD (i.e., within the ubiquitination zone), but native Snc1 is not significantly targeted to the vacuole in wild-type or even snx4Δ cells, underscoring the role of the TMD and JMH in governing access of K75 and K83 to the ubiquitylation machinery. We propose that K75 and K83 are protected from ubiquitylation due to interaction of the JMH region with the membrane (Figure 7B). Our results provide evidence for bona fide protein quality control surveillance of a SNARE protein.
MATERIALS AND METHODS
Yeast strains and culture conditions
Yeast strains were constructed in BY4742 (MATα his3-1, leu2-0, met15-0, and ura3-0) by homologous recombination of gene-targeted, PCR-generated DNAs using the method of Longtine et al. (1998). Mutant strains used were either derived from the EUROSCARF KANMX deletion collection (Open Biosystems/Thermo Scientific, Waltham, MA) or produced by replacement of the complete reading frame with the HIS3MX6 or URA3 cassette. Gene deletions were confirmed by PCR amplification of the deleted locus. Cells were grown in standard synthetic complete medium lacking nutrients required to maintain selection for auxotrophic markers and/or plasmids (Sherman et al., 1979).
For copper induction of Rsp5-DUb expression, cells were grown to early log phase in standard synthetic complete medium and incubated with 50 μM CuCl2 for 4 h at 30°C.
Live cell light microscopy and image analysis
Yeast cells from cultures grown to OD600 ≈ 0.5 were mounted in growth medium, and three-dimensional image stacks were collected at 0.3-µm z increments on a DeltaVision workstation (Applied Precision) based on an inverted microscope (IX-70; Olympus) using a 100 × 1.4 NA oil immersion lens. Images were captured at 24°C with a front-illuminated sCMOS, 2560 × 2160 pixels camera and deconvolved using the iterative-constrained algorithm and the measured point spread function. To visualize vacuole morphology, yeast cells were labeled with 7-aminochloromethylcoumarin (CMAC) (Life Technologies) at a concentration of 100 μM for 30 min in synthetic medium at room temperature. To visualize vacuole membrane, yeast cell cultures were incubated with FM4-64 lipophilic dye (Life Technologies) at 32 nM at 30°C for 20 min, washed, and resuspended in fresh medium and grown for 90 min to allow FM4-64 to transit to the vacuole membrane (adapted from Vida and Emr, 1995). Image analysis and preparation was done using Softworx 6.1 (Applied Precision Instruments) and ImageJ v1.50d (Rasband).
GFP-Snc1 immunoblotting
For quantitative Western blot analysis of GFP-Snc1, cells were grown under standard vegetative, as described above. Typically 3.0 × 107 cells were harvested per yeast strain by centrifugation and lysed by glass bead agitation in SDS–Page sample buffer. Polyacrylamide gels (10%) were loaded with 0.5 × 107 cell equivalents and transferred onto standard 0.45 μm nitrocellulose. Anti-GFP primary mouse monoclonal antibody (1814460, Roche) was diluted 1:2500 and Santa Cruz (sc-2055) goat anti-mouse HRP–conjugated antibody was used at 1:5000. Anti-phosphoglycerate kinase (Pgk)1 at 1:5000 (Life Technologies) was used as loading controls. All enhanced chemiluminescence (ECL) blots were development on a Chemidoc-XRS+(Bio-Rad) and band intensities were quantified using Quanity One 1D analysis software (Bio-Rad).
Plasmid construction
Each full-length GFP-Snc1 yeast expression construct was made by amplification of gBlocks Gene Fragments (IDT) of Snc1 cDNA sequence containing the necessary point mutations described. Each gBlocks Gene Fragment contains BamHI and HindIII restriction sites to allow cloning into the pGOGFP backbone, a permutation of pRS416 (Cowles et al., 1997), which contains BglII and HindIII sites. GFP-Snc1 truncation mutants were constructed using primers designed to amplify the described truncations and cloned into pGOGFP using BamHI/BglII and HindIII. GFP-Rcy1 was constructed by amplification of Rcy1 locus from the yeast genome and cloned into pGOGFP using BamHI/BglII and HindIII. Genomic integration of GFP-Snc1 was done by amplification of GFP-Snc1-URA3 from pRS416-GFP plasmids using primers containing homology to flanking regions of ura3Δ0 and transformation into wild-type BY4742 cells.
Acknowledgments
We are grateful to colleagues for discussions and critical reading of the manuscript and especially thank Chris MacDonald and Rob Piper for sharing the Rsp5-DUb construct and yeast strains. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number GM-060221 and in part by the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM-007223.
Abbreviations used:
- BAR
Bin-Amphiphysin-Rvs161
- CMAC
7-Aminochloromethylcoumarin
- ESCRT
endosomal sorting complexes required for transport
- JMH
juxtamembrane helix
- MVB
multivesicular body
- PGK
phosphoglycerate kinase
- PS
phosphatidylserine
- PVE
prevacuolar endosome
- SNARE
soluble N-ethyl maleimide–sensitive factor attachment protein receptor
- SNX-BAR
sorting nexin containing a BAR domain
- TGN
trans-Golgi network
- TMD
transmembrane domain
- VAMP2
vesicle-associated membrane protein 2
- VPS
vacuolar protein sorting.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-02-0117) on May 8, 2019.
REFERENCES
- Arlt H, Auffarth K, Kurre R, Lisse D, Piehler J, Ungermann C. (2015). Spatiotemporal dynamics of membrane remodeling and fusion proteins during endocytic transport. Mol Biol Cell , 1357–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. (1997). Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J , 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh-Touzé N, Léon S, Erpapazoglou Z, Stawiecka-Mirota M, Urban-Grimal D, Haguenauer-Tsapis R. (2008). Versatile Role of the Yeast Ubiquitin Ligase Rsp5p in Intracellular Trafficking, London, UK: Portland Press Ltd. [DOI] [PubMed] [Google Scholar]
- Borisovska M, Schwarz YN, Dhara M, Yarzagaray A, Hugo S, Narzi D, Siu SW, Kesavan J, Mohrmann R, Böckmann RA. (2012). Membrane-proximal tryptophans of synaptobrevin II stabilize priming of secretory vesicles. J Neurosci , 15983–15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheron C, Caudron F, Loeillet S, Peloso C, Mugnier M, Kurzawa L, Nicolas A, Denarier E, Aubry L, Andrieux A. (2016). A role for the microtubule+ end protein Bik1 (CLIP170) and the Rho1 GTPase in Snc1 trafficking. J Cell Sci, jcs.190330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen M, Brunger AT. (2006). Conformation of the synaptobrevin transmembrane domain. Proc Natl Acad Sci USA , 8378–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston HE, Maldonado-Baez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. (2009). Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol , 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Chen S, Tokarev AA, Liu F, Jedd G, Segev N. (2005). Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol Biol Cell , 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Shah AH, Segev N. (2011). Ypt31/32 GTPases and their F-Box effector Rcy1 regulate ubiquitination of recycling proteins. Cell Logist , 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. (1997). The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell , 109–118. [DOI] [PubMed] [Google Scholar]
- Day KJ, Casler JC, Glick BS. (2018). Budding yeast has a minimal endomembrane system. Dev Cell , 56–72.e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Hermansson M, Somerharju P, Grinstein S. (2011). Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat Cell Biol , 1424–1430. [DOI] [PubMed] [Google Scholar]
- Feyder S, De Craene J-O, Bär S, Bertazzi D, Friant S. (2015). Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int J Mol Sci , 1509–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. (2007). Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p–Rcy1p pathway. Mol Biol Cell , 295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J-M, Wiederkehr A, Seol JH, Haguenauer-Tsapis R, Deshaies RJ, Riezman H, Peter M. (2001). Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol Cell Biol , 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pluhackova K, Böckmann RA. (2017). The multifaceted role of SNARE proteins in membrane fusion. Front Physiol , 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pluhackova K, Bruns D, Böckmann RA. (2016). Synaptobrevin transmembrane domain determines the structure and dynamics of the SNARE motif and the linker region. Biochim Biophys Acta , 855–865. [DOI] [PubMed] [Google Scholar]
- Hanamatsu H, Fujimura-Kamada K, Yamamoto T, Furuta N, Tanaka K. (2014). Interaction of the phospholipid flippase Drs2p with the F-box protein Rcy1p plays an important role in early endosome to trans-Golgi network vesicle transport in yeast. J Biochem , 51–62. [DOI] [PubMed] [Google Scholar]
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR. (2015). Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell , 4674–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HRB. (2003). Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J , 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. (2004). Multivesicular Body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol Biol Cell , 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienle N, Kloepper TH, Fasshauer D. (2009). Phylogeny of the SNARE vesicle fusion machinery yields insights into the conservation of the secretory pathway in fungi. BMC Evol Biol , 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Kobayashi M, Nozaki S, Yagi C, Hatsuzawa K, Katoh Y, Shin H-W, Takahashi S, Nakayama K. (2015). SNAP23/25 and VAMP2 mediate exocytic event of transferrin receptor-containing recycling vesicles. Biol Open , 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon D-H, Kim CS, Shin Y-K. (2003). Insertion of the membrane proximal region of the neuronal SNARE coiled coil into the membrane. J Biol Chem , 12367–12373. [DOI] [PubMed] [Google Scholar]
- Léon S, Erpapazoglou Z, Haguenauer-Tsapis R. (2008). Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol Biol Cell , 2379–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HRB. (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell , 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. (2002). A new yeast endosomal SNARE related to mammalian syntaxin 8. Traffic , 922–929. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Ma M, Burd CG, Chi RJ. (2017). Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic , 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Kumar S, Purushothaman L, Babst M, Ungermann C, Chi RJ, Burd CG. (2018). Lipid trafficking by yeast Snx4 family SNX-BAR proteins promotes autophagy and vacuole membrane fusion. Mol Biol Cell , 2190–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Piper RC. (2017). Genetic dissection of early endosomal recycling highlights a TORC1-independent role for Rag GTPases. J Cell Biol , 3275–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Söllner TH, Rothman JE. (2000). Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature , 153. [DOI] [PubMed] [Google Scholar]
- Miller SE, Sahlender DA, Graham SC, Höning S, Robinson MS, Peden AA, Owen DJ. (2011). The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell , 1118–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papini E, Rossetto O, Cutler DF. (1995). Vesicle-associated membrane protein (VAMP)/synaptobrevin-2 is associated with dense core secretory granules in PC12 neuroendocrine cells. J Biol Chem , 1332–1336. [DOI] [PubMed] [Google Scholar]
- Paumet F, Brügger B, Parlati F, McNew JA, Söllner TH, Rothman JE. (2001). A t-SNARE of the endocytic pathway must be activated for fusion. J Cell Biol , 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumet F, Rahimian V, Rothman JE. (2004). The specificity of SNARE-dependent fusion is encoded in the SNARE motif. Proc Natl Acad Sci USA , 3376–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ. (2007). Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol , 519–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quetglas S, Iborra C, Sasakawa N, De Haro L, Kumakura K, Sato K, Leveque C, Seagar M. (2002). Calmodulin and lipid binding to synaptobrevin regulates calcium-dependent exocytosis. EMBO J , 3970–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quetglas S, Leveque C, Miquelis R, Sato K, Seagar M. (2000). Ca2+-dependent regulation of synaptic SNARE complex assembly via a calmodulin-and phospholipid-binding domain of synaptobrevin. Proc Natl Acad Sci USA , 9695–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Black MW, Pelham HR. (2000). Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol Biol Cell , 3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, Poon PP, Schindler C, Murray LE, Kama R, Gabriely G, Singer RA, Spang A, Johnston GC, Gerst JE. (2006). The Gcs1 Arf-GAP mediates Snc1, 2 v-SNARE retrieval to the Golgi in yeast. Mol Biol Cell , 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Zhu L, Emr SD. (2019). Rsp5 Ubiquitin ligase–mediated quality control system clears membrane proteins mistargeted to the vacuole membrane. J Cell Biol , 234–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence LW. (1979). Methods in Yeast Genetics: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1–98. [Google Scholar]
- Stein A, Weber G, Wahl MC, Jahn R. (2009). Helical extension of the neuronal SNARE complex into the membrane. Nature , 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer DK, Piper RC. (2011). A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol , 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strop P, Kaiser SE, Vrljic M, Brunger AT. (2008). The structure of the yeast plasma membrane SNARE complex reveals destabilizing water-filled cavities. J Biol Chem , 1113–1119. [DOI] [PubMed] [Google Scholar]
- Sun Y, Drubin DG. (2012). The functions of anionic phospholipids during clathrin-mediated endocytosis site initiation and vesicle formation. J Cell Sci , 6157–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Yamagami K, Tanaka K. (2014). Role of phosphatidylserine in phospholipid flippase-mediated vesicle transport in Saccharomyces cerevisiae. Eukaryot Cell , 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas J, Pelham HR. (2003). Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol , 1636–1640. [DOI] [PubMed] [Google Scholar]
- van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, Dier M, Hell SW, Grubmüller H, Diederichsen U, Jahn R. (2011). Membrane protein sequestering by ionic protein–lipid interactions. Nature , 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol , 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Yang J, Huibregtse JM. (1999). Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol , 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn A, Riezman H. (1997). End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell , 2291–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A, Avaro S, Prescianotto-Baschong C, Haguenauer-Tsapis R, Riezman H. (2000). The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J Cell Biol , 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR. (2013). Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol , 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Hankins HM, MacDonald C, Erlinger SJ, Frazier MN, Diab NS, Piper RC, Jackson LP, MacGurn JA, Graham TR. (2017). COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. eLife , e28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. (2008). Membrane phosphatidylserine regulates surface charge and protein localization. Science , 210–213. [DOI] [PubMed] [Google Scholar]