Abstract
Background
Neuroanatomical pattern classification using support vector machines (SVMs) has shown promising results in classifying Multiple Sclerosis (MS) patients based on individual structural magnetic resonance images (MRI).
Objectives
To determine whether pattern classification using SVMs facilitates predicting conversion to clinically definite multiple sclerosis (CDMS) from clinically isolated syndrome (CIS).
Methods
We used baseline MRI data from 364 patients with CIS, randomised to interferon beta-1b or placebo. Non-linear SVMs and 10-fold cross-validation were applied to predict converters/non-converters (175/189) at two years follow-up based on clinical and demographic data, lesion-specific quantitative geometric features and grey-matter-to-whole-brain volume ratios. We applied linear SVM analysis and leave-one-out cross-validation to subgroups of converters (n=25) and non-converters (n=44) based on cortical grey matter segmentations.
Results
Highest prediction accuracies of 70.4% (p=8e-5) were reached with a combination of lesion-specific geometric (image-based) and demographic/clinical features. Cortical grey matter was informative for the placebo group (acc.: 64.6%, p=0.002) but not for the interferon group. Classification based on demographic/clinical covariates only resulted in an accuracy of 56% (p=0.05). Overall, lesion geometry was more informative in the interferon group, EDSS and sex were more important for the placebo cohort.
Conclusions
Alongside standard demographic and clinical measures, both lesion geometry and grey matter based information can aid prediction of conversion to CDMS.
Introduction
The advance of non-invasive imaging techniques, such as magnetic resonance imaging (MRI), has made a large impact on the study of Multiple Sclerosis (MS). Measures of abnormalities derived from structural MRI are useful in the context of early diagnosis, treatment planning and monitoring of disease progression. To date, however, MRI data is mainly used in a qualitative way to assess the dissemination of MS lesions in space and time.
The most common quantitative measure is lesion load, i.e. the total lesion volume. Previous work on using lesion load for classification has been inconclusive (Aban et al. 2008), (Zivadinov et al. 2005; MacKay Altman et al. 2012). Other studies have shown that conventional MRI measures have rather low predictive value and are therefore poor indicators for determining the clinical outcomes in MS (Lovblad et al. 2010). Existing quantitative methods that are used for the analysis of MS lesions are to a large extent inapt to fully reflect the given data (Filli et al. 2012; Locatelli et al. 2004) (Bates et al. 2003) (Wei et al. 2004) (Ge et al. 2014).
Clinically isolated syndromes (CIS), such as optic neuritis, brainstem or spinal cord syndromes are the first clinical presentations of MS in about 85% of cases (Scalfari et al. 2010; Confavreux and Vukusic 2006). Over time, most patients with a CIS, with additional clinically silent brain lesions on MRI that suggest disseminated CNS disease, develop relapsing-remitting MS (RRMS) and have a substantial risk for later progression of disability(Brex et al. 2002a; M. Tintore et al. 2006; Fisniku et al. 2008b).
The identification of patients with CIS who will develop clinically definite MS (CDMS) in the short-term is particularly relevant from a clinical, therapeutic(Calabrese et al. 2011), and economical point of view (Beer and Kesselring 1994).
A number of clinical features including laboratory investigations and MRI abnormalities have been associated with an increased risk for conversion to CDMS. Among these, MRI is the most informative marker (M. Tintore et al. 2005; M. Tintore et al. 2001; Morrissey et al. 1993; O'Riordan et al. 1998; Brex et al. 2002b) to monitor treatment efficacy. Several studies have confirmed the predictive value of baseline and follow-up MR imaging for conversion to CDMS, depending on the population, follow-up duration, and treatment intervention. To date, however, no reliable method exists to predict who will, and will not, develop MS amongst those with CIS.
One of the reasons for the limited impact of the findings on clinical practice is that neuroimaging studies have typically reported population differences between groups. For neuroimaging to be useful in a clinical setting, however, inferences have to be made at the level of the individual rather than the group. One analytical method that allows such inference is multivariate pattern classification based on support vector machines (SVMs) which are sensitive to spatially distributed and subtle effects in the brain that would be otherwise undetectable using traditional methods which focus on gross differences at group level (for a review on SVMs applied to neurological conditions see (Orru et al. 2012)).
Our present study aims to determine whether multivariate neuroanatomical pattern classification facilitates predicting conversion to CDMS in individuals with a CIS based on structural MRI features and clinical characteristics in a two-year follow-up. A combination of different MRI-based features has been shown to improve results as compared to using only one measurement (Wottschel et al. 2015). The study showed that a linear SVM correctly predicted CDMS in 71% of patients at 1 year, and in 68% at 3 years of follow-up based on different combinations of lesional and clinical features.
A recent study on lesional geometry in MS used 3D-printing of lesions to qualitatively rate their shape and surface characteristics (Newton et al. 2017). Earlier work by Goldberg-Zimring et al. (Goldberg-Zimring et al. 2003) studied longitudinal changes in lesion geometry as captured by spherical harmonics. The study also found that lesional shape was more variable over time than size or volume. Although providing valuable insights, the methods presented in these two studies are not easily applicable to studies with more than a handful of patients.
On a larger scale, Gourraud et al (2013) (Gourraud et al. 2013) used the topological arrangement of lesional clusters (e.g. many small clusters vs. few big clusters) as an additional trait in a GWAS analysis; however, the study did not consider measures derived from individual lesions.
The present study demonstrates that a rich feature set incorporating multiple types of data and in combination with a classifier that is well suited for high-dimensional, multi-modal settings, SVMs, provide an alternative approach. Unlike previous work(Wottschel et al. 2015), we employed kernel-based SVMs in a non-linear classification scheme using a large number of quantitative features in a large population of 175 converters and 189 non-converters. By solving the classification problem in a higher dimensional auxiliary space, kernel-based SVMs can more easily adapt to the structure of the data (Schölkopf and Smola, 2001). Most importantly, in addition to traditional demographic, clinical and lesional measures, we included aspects of lesion geometry derived from Minkowski functionals (Legland et al., 2007). These functionals provide quantitative measures that are closely related to standard geometric quantities such as volume, surface area and width of individual lesions.
Furthermore, there is increasing evidence that grey matter (GM) atrophy in CIS predicts conversion to CDMS (Calabrese et al. 2011; Raz et al. 2010; Jenkins et al. 2011). Therefore, we also include measures of GM volume and demonstrate how predictive performance of non-linear SVM can be
Finally, in a separate analysis, we used linear SVM to investigate whether SVMs can differentiate CIS from CDMS based on cortical grey matter characteristics.
Methods
Data and the BENEFIT study
This is a post-hoc analysis of MRI and clinical data from the BENEFIT study(Kappos et al. 2006; Frederik Barkhof et al. 2007). Briefly, BENEFIT was a double-blind, placebo-controlled, randomized, parallel-group, multicenter (total of 98 centers), phase 3 study that evaluated the safety, tolerability and efficacy of interferon beta-1b (Betaferon/Betaseron; Bayer HealthCare Pharmaceuticals Inc) in patients with a monofocal or multifocal presentation of the disease, a first demyelinating event suggestive of MS and at least two clinically silent lesions on a T2-weighted brain magnetic resonance (MRI) scan. Within 60 days of the onset of the first clinical event, and after providing written informed consent, patients (n=468, aged 18-45 years) were randomly assigned, in a 5:3 ratio, to interferon beta-1b 250 μg (n=292) or placebo (n=176) subcutaneously every other day for 2 years or until CDMS was diagnosed by use of the modified Poser criteria (Bakshi et al. 2005) and confirmed by a central committee. Patients were then eligible to enter the follow-up phase with open-label interferon beta-1b (Kappos et al. 2007a). During this double-blind phase, visits were scheduled for collection of Expanded Disability Status Scale (EDSS), MRI, and other efficacy data and for safety data at months 3, 6, 9, 12, 18, and 24.
MRI acquisition and pre-processing
Baseline MR images were obtained using an oblique axial acquisition plane and gadolinium diethylenetriamine pentaacetic acid (0.1 mmol/kg of body weight) as contrast agent (T1c). The MR imaging protocol included a T1-weighted (T1w) spin echo after contrast administration with repetition times of 400 to 700 milliseconds and echo times of 5 to 25 milliseconds. A dual echo T2-weighted (T2w) spin echo with repetition times of 2000 to 3000 milliseconds and echo times of 20 to 40 milliseconds (first echo) and 60 to 100 milliseconds (second echo) was acquired as well. The field of view for all examinations was 25 cm with a 256x256–pixel matrix, resulting in a pixel size of roughly 1x1 mm2. Images were acquired in 2 interleaved sets with a 3-mm gap, resulting in whole-brain coverage with contiguous 3-mm-thick sections.
All MRI scans were performed with 0.1 mmol/kg gadolinium (Gd)-DTPA. The numbers and volumes of hyperintense lesions on T2-weighted images and Gd-enhancing lesions on T1-weighted images were centrally evaluated by the MRI Analysis Centre in Amsterdam, which was kept blinded to treatment allocation.
All of an individual patient's MRI was performed using the same machine. Assessment of the MRIs was performed centrally at the Image Analysis Centre at the Vrije Universiteit Medical Center. The quality assessment included a trial-specific standard operating procedure, training specifications (including a dummy-run procedure for site selection), and internal quality control procedures. During the trial each MRI was subjected to a routine quality control procedure, and additional MRIs were obtained when quality standards were not met.
The numbers and volumes of hyperintense lesions on T2-weighted images (T2 lesions) and Gd-enhancing (Gd lesions) and hypointense lesions on T1-weighted images (T1-hypointense lesions) were determined by evaluators who were blinded to treatment allocation but not to MRI order. Because all the MRIs were performed after Gd administration, T1-hypointense lesions were defined as the subset of new or enlarging lesions on T2-weighted images that were isointense to hypointense relative to gray matter on T1-weighted images. Lesions were identified by trained radiologists who marked the MS lesions on hard copy; their volumes were subsequently determined electronically by trained technicians using in-house–developed software (Show_Images version 3.6.3) based on local thresholding (Barkhof et al. 2007).
All MRI scans used in the present study were performed at 1.5T and included transaxial contiguous 3-mm dual-echo images. T1-weighted images with sufficient image quality together with their corresponding lesion masks, as well as clinical data at two year follow-up, had to be comparable across scanners. We employed subsets of placebo patients and interferon beta-1b patients who either converted to CDMS (conv+) or did not convert (conv-) (see Table 1). Definition of the outcome measure was based on the double blind phase.
Table 1.
Baseline clinical and MRI statistics of subjects used for a) non-linear SVM analysis (T1-Gd data set), and b) linear SVM analysis (cortical grey matter maps). */+/# indicate significant differences between groups (p < 0.05).
Abbreviations: T treated, NT not treated, SD standard deviation, IQR interquartile range, EDSS expanded disability status scale, bs baseline, y1 year1, Total non-linear SVM) or cortical (linear SVM) GM volume - as obtained from the SPM/VBM8 pipeline, PGMVC percentage gray matter volume change, TCDMS time to CDMS (days)
| Non-linear SVM | Linear SVM | |||||||
|---|---|---|---|---|---|---|---|---|
| placebo | IFN-b | placebo | IFN-b | |||||
| conv- | conv+ | conv- | conv+ | conv- | conv+ | conv- | conv+ | |
| N | 22 | 39 | 49 | 50 | 25 | 44 | 49 | 49 |
| Mean age, years (SD) | 30.6 (7.5) | 29.2 (8.0) | 31.4 (8.1) * | 27.9 (6.8) * | 34.1 (6.3) + | 29.0 (6.7) + | 31.1 (7.8) | 29.8 (7.6) |
| m/f (ratio) | 6/16 (1:2.7) | 12/27 (1:2.2) | 21/28 (1:1.3) | 13/37 (1:2.8) | 8/17 (1:2.1) | 12/32 (1:2.7) | 18/31 (1:1.72) | 12/37 (1:3.08) |
| TCDMS, median (IQR) | 1780 (1805-857) # | 249 (627-74) | 1807 (1820-1794) # | 432 (824-111) | 1798 (1808-1497) | 303 (709-138) | 1808 (1822-1791) | 423 (1027-204) |
| EDSS, median (IQR) | 1.0 (2.0-0.0) | 1.5 (2.0-1.0) | 2.0 (2.0-1.0) | 2.0 (2.5-1.0) | 1.5 (2.0–1.0) | 1.5 (2.0–1.0) | 1.5 (2.0–1.0) | 1.5 (2.0–1.0) |
| N relapse, median (IQR) | 0 (0-0) # | 1.0 (1.0-0.75) # | 0 (0-0) + | 1.0 (1.0-0) + | 0 (0-0) * | 1.0 (1.0-0.75) * | 0 (0-0) + | 1.0 (1.0-0) + |
| N T2 lesions at m24, median (IQR) | 0 (1.25-0) | 0.5 (3.0-0) | 0 (0.75-0) | 0 (0.75-0) | 0 (1.25-0) | 0.5 (3.0-0) | 0 (0.75-0) | 0 (0.75-0) |
| N Gd-lesions at m24, median (IQR) | 0 (0-0) | 0 (1.0-0) | 0 (1-0) | 0 (0.0-0) | 0 (0-0) | 0 (1.0-0) | 0 (1-0) | 0 (0-0) |
| Total (cortical^) GM volume [ml], mean (SD) | 554.7 (177.3) | 575.6 (234.2) | 651.0 (217.1) | 587.2 (265.4) | 251.52 (102.37)^ | 284.0 (56.96) ^ | 303.24 (27.88) ^ | 293.64 (12.75) ^ |
Statistical analysis of demographic data
Statistical analysis was performed using IBM SPSS Version 19.0. Group comparisons were performed using the Student's t-test, chi2-test or Wilcoxon rank sum test when appropriate. Values are reportet as mean (SD) or median (IQR). A statistical level of p < 0.05 was considered significant.
Classification based on lesion-specific geometric measures and feature combinations using non-linear SVMs
For this approach, we considered a classification and prediction model using kernel-based (radial basis functions) support vector machines. As input to the classifier we used detailed geometric features of individual lesions derived from the available MRI data (T2-, T1-, Gd-lesions).
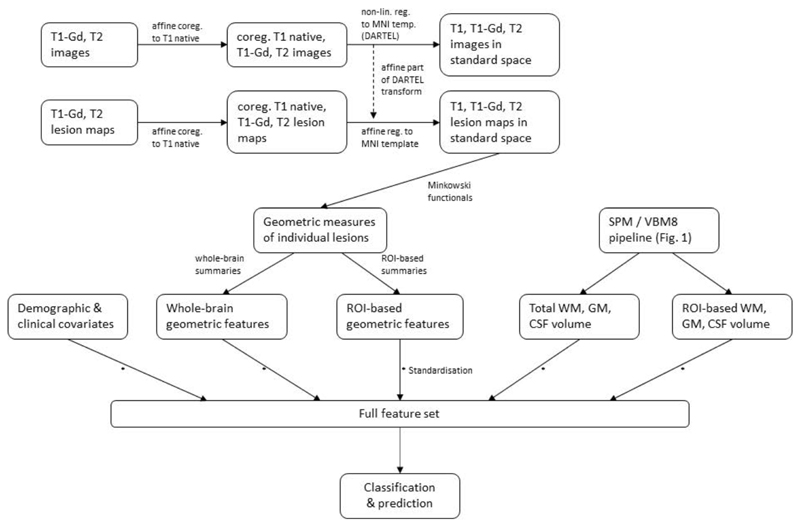
First, the binary lesion masks were used to identify individual lesions. Minkowski functionals(Legland et al. 2011) were then used to extract and quantify the geometry of each lesion. In standard 3D Euclidean space, Minkowski functionals are directly related to the geometric quantities volume, surface area, mean breadth and the Euler-Poincare (EP) characteristic(Arns et al. 2001; Lang et al. 2001). More details on the computation of geometric features as well as feature reduction and additional preprocessing steps (see also Figure 2) can be found in the supplementary materials.
Figure 2.
Preprocessing pipeline for geometry-based lesion measures and subsequent classification with nonlinear SVMs.
We also considered a parcellation of the whole brain into 13 regions of interest (ROIs) in order to add some amount of spatial information about lesion location to the feature set. (See supplementary materials for a description of the 13 ROIs.) Lesion dissemination in space is a standard requirement in the criteria for the diagnosis of CDMS (McDonald et al. 2001; C. H. Polman et al. 2005). Ideally, including spatial information will aide classification and prediction accuracy.
Classification based on linear SVMs using cortical grey matter segmentations
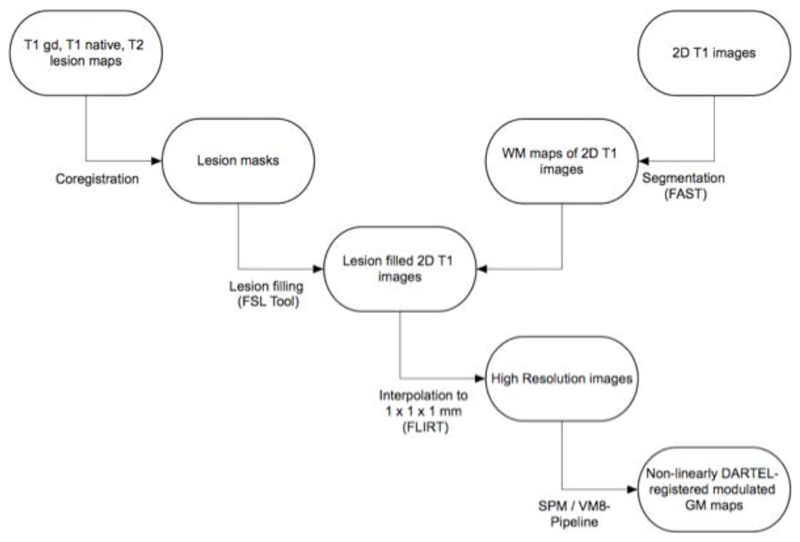
For linear pattern classification analysis, we used LIBSVM, a library for SVMs (http://www.csie.ntu.edu.tw/-cjlin/libsvm), running under Matlab 7.1 (MathWorks, USA). Two-dimensional T1-weighted images (Placebo: n=44 converters, 25 non-converters; IFN-b: n=49 converters, 49 non-converters) with sufficient image quality aquired on three different 1.5T scanners were interpolated to an isotropic resolution of 1 mm3,(Richert et al. 2006). Images were processed using FMRIBs software libray FSL for coregistration, segmentation (FAST), lesion filling, and interpolation (FLIRT). Interpolated images were visually checked, and then normalized, segmented and modulated using Statistical Parametric Mapping software (SPM8; Wellcome Department of Imaging Neurosciences, University College London) and Voxel-Based Morphometry (VBM8) toolbox (http://dbm.neuro.uni-jena.de/vbm8/) and DARTEL.
For the grey matter classification task, the total number of dimensions was determined by the number of voxels within the cortical GM mask. To implement a linear SVM, a kernel matrix was created from the images based on correlation, i.e. the similarity between each pair of subjects. Details on the rationale for using linear SVMs in this setting, as well as advantages and limitations of SVMs in general can be found in the supplementary materials.
Predictive performance was evaluated using leave-one-out cross-validation; details are given in the supplementary materials.
Results
The demographic and clinical characteristics of patients are summarised in Table 1.
Classification based on feature combinations
A summary of balanced prediction accuracies of kernel SVMs trained on different combinations of input features is given in Table 2 and visualised in Figure 3. All values are based on nonlinear SVMs using an RBF kernel. Parameters were optimised through grid search. P-values are based on permutation tests of the class labels (1000 permutations per model). One way of estimating the importance of different features is to visualise the corresponding SVM weights. Since it is not possible to obtain an analytic expression for the weights and thus separate them in the nonlinear case, any illustration has to rely on weights based on linear SVMs. Even in the linear case the plots are meant purely as a qualitative way of visualising the importance of input variables relative to one another. In addition, the weights do not indicate whether any interaction between features are relevant during classification. No quantitative assessments should be drawn directly from the magnitude of individual weights.
Table 2.
Prediction accuracy of optimised kernel SVM based on different combinations of input features; including 95% confidence intervals and corresponding p-values.
* Summaries include total, mean, median, minimum, maximum and standard deviation (SD) of lesion volume, surface area and mean breadth, respectively. Measures are based on T1-Gd MRI data. ** Total, mean and SD of volume, surface area and mean breadth (excluding median, minimum, maximum measures), and EP.
Abbreviations: DC -demographic and clinical covariates, EDSS - expanded disability status scale, EP - Euler-Poincare characteristic, GEO-brain - whole-brain summaries of geometric measures, GM - grey matter, ROI - region of interest.
| model | input features [resulting number of features] |
placebo group (N=61, conv+: 39, conv-: 22) |
IFN-b group (N=99, conv+: 50, conv-: 49) |
|---|---|---|---|
| M1 | demographic and clinical
covariates (DC): age, sex, EDSS [3 features] |
56.7% [48.5–64.9%] (p=0.053) [77:63] * |
55.5% [49.0-62.0%] (p=0.051) [98:126] * |
| M2 | DC, lesion count, lesion load (i.e.
total lesion volume) [5 f.] |
50.1% [37.6-62.6%] (p=0.537) |
57.6% [47.9-67.3%] (p=0.072) |
| M3 | DC, GM volume ratio (GM) [4 f.] | 54.3% [41.8-66.8%] (p=0.261) |
58.6% [48.9-68.3%] (p=0.048) |
| M4 | DC, GM, lesion count, lesion load [6 f.] | 60.9% [48.7-73.1%] (p=0.046) |
60.6 [51.0-70.2%] (p=0.020) |
| M5 | DC, GM, count, load,
Euler-Poincare characteristic (EP) [7 f.] |
56.6% [44.2-69.0%] (p=0.145) |
54.5% [44.7-64.3%] (p=0.193) |
| M6 | whole-brain summaries* of
geometric measures (GEO-brain) [21 f.] |
57.9% [45.5-70.3%] (p=0.126) |
58.5% [48.8-68.2%] (p=0.046) |
| M7 | DC, GM, GEO-brain [25 f.] |
67.6% [55.9-79.3%] (p=0.0022) |
64.6% [55.2-74.0%] (p=0.0017) |
| M8 | DC, GM by ROI, geometric measures
by ROI (GEO-ROI) [276 f.] |
51.2% [38.7-63.7%] (p=0.468) |
59.6% [49.9-69.3%] (p=0.035) |
| M9 | DC, GM, lesion count, subset** of
GEO- brain [15 f.] |
57.8% [45.4-70.2%] (p=0.133) |
70.4% [61.4-79.4%] (p=8e-5) |
| M10 | DC, GM by ROI, lesion count,
subset** of GEO-ROI [159 f.] |
50.5% [38.0-63.0%] (p=0.491) |
66.6% [57.3-75.9%] (p=0.0004) |
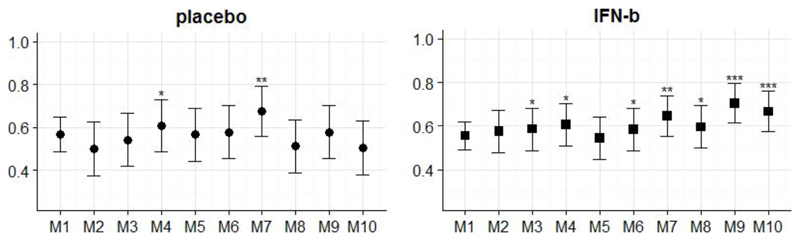
Figure 3.
Prediction accuracies of models as described in Table 2. Bars indicate 95% confidence interval. Significance levels based on permutation testing: * (p < 0.05), ** (p < 0.005), *** (p < 0.0005).
In general, we found that our trained classifiers were more likely to distinguish correctly between converters and non-converters in the INF-b treated cohort compared to the placebo treated group.
The simplest model (M1) utilised demographic (sex, age) and clinical (EDSS score) covariates only (accuracy: 56%, p=0.05). Including grey matter-to-whole-brain volume ratios (M3) increased prediction accuracy by a few per cent in the INF-b but not in the placebo group.
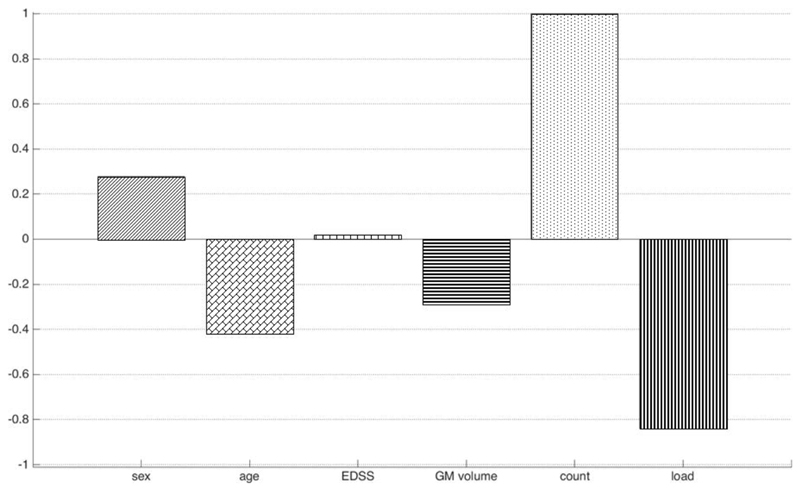
Model M2 included “traditional” measures such as lesion count and total lesion load (i.e. total lesion volume). These were combined with the grey matter volume feature in M4. Figure 4 shows the relative magnitude of SVM weights (again, resulting from training a linear SVM) for each input feature with classification accuracy equal to 57.6% (p=0.07).
Figure 4.
M4. Individual weights from a linear SVM trained on features including demographic and clinical covariates, grey matter volume, lesion count and total lesion load; based on T1-Gd MRI data of IFN-b treated patients (converters: 50, non-converters: 49). Positive (negative) weights indicate that a larger feature value will drive prediction towards conversion (non-conversion). For sex, being female (male) corresponds to the positive (negative) axis. Weights are scaled relatively to the largest individual weight, which is set equal to one. Note that weights should only be interpreted qualitatively, not quantitatively.
M6 was solely based on whole-brain summary measures of lesion-based geometry and performed on a similar level as using demographic/clinical features only. Combining the two kinds of input information and adding grey matter volume (M7) lead to a significant improvement of classification accuracy, reaching 64.6% (p=0.0017) for the INF-b group.
Model M8 represented the largest possible feature set by splitting the summary measures of lesion geometry into 13 ROIs, plus including all other available covariates. Classifier performance was markedly reduced, most likely due to large amounts of redundant or irrelevant (i.e. equal to zero) input features. Dimensionality reduction via PCA space was only partly able to ameliorate this issue.
In M9 half of the summary statistics were excluded in order to lower the level of redundancy in the data. Only total, mean and standard deviation measures were retained in the feature set which resulted in a prediction accuracy of 70.4% (p=8e-5). Note that splitting the same features by ROIs again reduced overall performance (cf. M10).
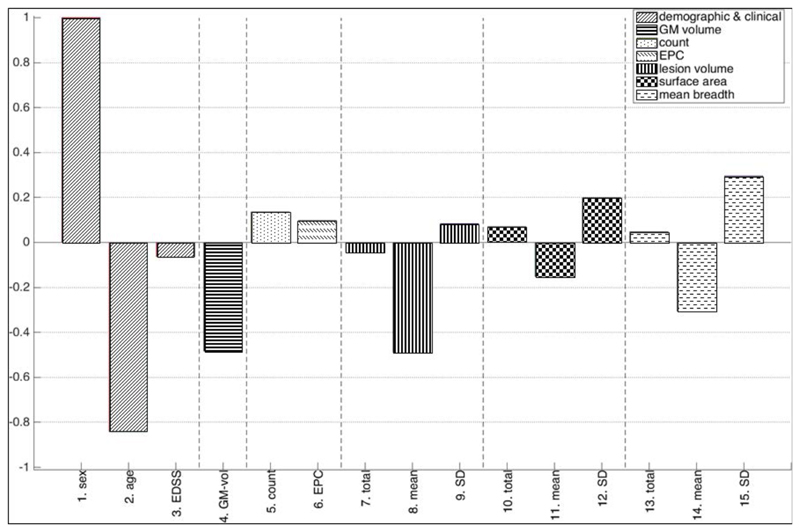
Figure 5 displays SVM weights for M9 of the corresponding linear classifier. The root mean square values (RMS, or quadratic means) of SVM weights shown in Figure 6 (accuracy: 60.5%, p=0.019) allow for a comparison between different kinds of features and their variability.
Figure 5.
M9. Individual weights from a linear SVM trained on demographic, clinical, grey matter volume and geometry based features derived from T1-Gd MRI data of IFN-b treated patients (converters: 50, non-converters: 49). Positive (negative) weights indicate that a larger feature value will drive prediction towards conversion (non-conversion). For sex, being female (male) corresponds to the positive (negative) axis. Weights are scaled relatively to the largest individual weight, which is set equal to one. Note that weights should only be interpreted qualitatively, not quantitatively.
Abbreviations: EDSS - expanded disability status scale, EPC - Euler-Poincare characteristic, GM - grey matter, SD – standard deviation.
Figure 6.
M9. Root mean squares summaries of the weights in Fig. 4 indicating the relative importance of different kinds of input features during classification. Weights are scaled relatively to the largest individual weight, which is set equal to one. Note that weights should only be interpreted qualitatively, not quantitatively.
For reference, training a kernel SVM with input features as specified in model M9 on T2 weighted lesion data resulted in a prediction accuracy of about 60% (p<0.002); and similarly for T1 weighted black-hole lesion data (accuracy 64%, p<0.0008). We also combined feature sets based on different imaging modalities, at the cost of increasing redundancy, which gave an accuracy of about 63% (p<0.01). Note that for the combined-modality model the size of available data was reduced by roughly 20% as not all imaging modalities were available for all subjects.
Classification based on cortical grey matter segmentations
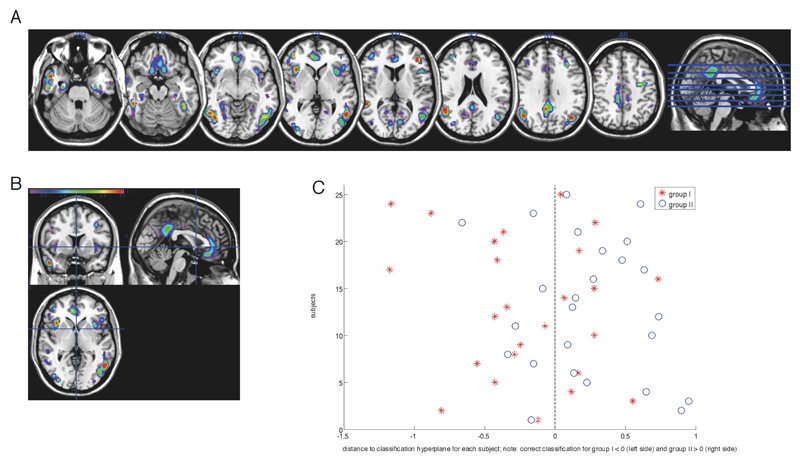
The average accuracy from the 500 bootstraps was 71.2% (95% confidence interval: 70.7-71.6%), which means that, on average, SVMs correctly predicted CDMS in 71.2% of placebo-treated individuals, with a sensitivity of 64% (converters correctly identified) and specificity of 78.3% (non-converters correctly identified). Middle and medial frontal, superior and middle temporal, anterior and posterior cingulate, middle temporal, fusiform, middle occipital, and insular regions contributed most to the classification accuracy as shown here for the matched placebo groups (Figure 7).
Figure 7.
Weight vector maps showing the most discriminating brain regions between placebo groups for the matched placebo groups (top 10 %; accuracy 66%). (A, B) Regions that contributed most to classification accuracy in the matched placebo groups are shown in red, in axial, coronal and sagittal views (z = [-28, -18, -8, 2, 10, 22, 31]). (C) Projection of each subject onto the weight vector; with positive patterns (blue circles) discriminating for conv+, and negative patterns (red crosses) discriminating for conv-CDMS could not be predicted based on cortical segmentations of the treated patients (total accuracy 48%; sens.: 42.9%, spec.: 53.1; p=0.6)
Discussion
Our study aims to determine whether multivariate neuroanatomical pattern classification facilitates predicting conversion to CDMS in individuals with a CIS based on structural MRI features and clinical characteristics in a two-year follow-up. We used geometric measures of individual lesions computed from MRI data in combination with clinical information from patients with CIS to predict conversion to CDMS. We included a large set of quantitative measures on individual lesions to perform classification into one of two groups, converters and non-converters, based on SVMs. In addition to providing an accurate classifier, we aimed to determine which types of features are most important for successful classification.
In the IFN-b group the best performance was achieved with non-linear classification and model M9 utilizing demographic and clinical covariates, global grey matter volume, lesion count as well as total, mean and standard deviation measures of lesion geometrics. In the placebo group linear classification using cortical grey matter segmentations yielded the highest accuracy. It should be noted that using prediction accuracies directly for model comparison is not suitable, since differences could arise by chance. The focus should therefore lie on models that reached statistically significant accuracies. The classification accuracies were only moderate (not exceeding 70%) which limits the applicability of the model for predicting the onset of disease in the brain scans of CIS patients in clinical praxis. In general, this application is challenging for classification, as the differences observed in CIS are likely to be much more subtle then those observed in established MS. Not all CIS patients develop MS, and in those who do, disability is highly variable(Fisniku et al. 2008b).
Furthermore, it cannot be excluded that some of the patients in the non-converter group may still develop MS in the long-term. Our results, however, are consistent with other studies on prediction of disease onset in MS (Wottschel et al. 2015) and Alzheimer’s disease(Young et al. 2013) which reported similar classification accuracies.
Classification based on feature combinations
Consistent with the results from earlier studies (Wottschel et al. 2015),55, the demographic attributes age and sex as well as the EDSS score carry a considerable amount of information about the disease and its progression (see Fig 6). When considering the full set of all geometric measures, each summarised in a ROI specific value (M10 in Table 2), a large number of these features will contain similar or even redundant information. However, it should be noted that imaging data of CIS patients may not necessarily be well suited for splitting into ROIs because most CIS patients have relatively few or even only one lesion.a This is due to the fact that the gain in spatial information about lesion locations may be offset by the inclusion of a large number of uninformative, zero-valued features for regions where no lesion is present.
Thus, feature selection or feature reduction becomes a necessity when optimising the classification procedure. A PCA transformation of input features reduces some of this redundancy and, in principle, SVMs can cope with a certain level of inter-dependency between features.
Nevertheless, classification outcomes are still likely to be affected by a lack of distinct information across the feature set. Therefore in this study, a partition of geometric features according to different brain regions turned out to be useful only to some extent.
Ideally, as classification outcome varies depending on the combination of features used as input, one would like to find the most informative subset among all available features. This is hindered by the fact that an exhaustive combinatorial search across all features is computationally infeasible; and it would lead to overfitting by selecting a feature set that is specifically suited for the available data set but is unlikely to generalise well to new data. As a way out we considered pre-specified subsets of possible feature combinations as presented in Table 2.
Principally, the examination of weights of individual features for the support vectors across different models can help inform which kinds of features are driving the classification procedure. A comparison of the magnitude of SVM weights provides a qualitative assessment of relative importance of different input features. With regard to our set of geometry-based features, a comparison across different (linear) classifier weights indicated that the EP characteristic is often more significant than a simple lesion count. Among clinical and demographic covariates, age seemed to be the single best predictor with younger patients showing a higher probability to convert to CDMS (cf. negative weight on age in Figures 4 and 5).
When comparing placebo and IFN-b groups, lesion geometry played a bigger role in the classifier for the IFN-b group. EDSS and sex were more important for the placebo cohort, less so for the IFN-b group. Both features carried predominantly positive weights in all SVM models, indicating that higher EDSS at baseline as well as being female are predictors for conversion. Additionally, we found that the grey matter-to-whole-brain volume ratio in most cases was more informative than either EDSS or sex.
Active plaques typically are associated with Gd- enhancement on MRI and most likely represent the pathological substrate of the attacks in MS (Filippi et al. 2012; Popescu et al. 2013). Therefore, we concentrated here on T1-Gd imaging data. However, a comparison of weights from SVMs trained on T2 or T1 black hole MRI data showed very small variation, especially with respect to the sign of individual weights, across different modalities within each cohort (placebo or IFN-b), and much larger differences between the two cohorts.
It should be noted that a quantitative comparison of prediction accuracies and/or p-values across models is to be avoided. A statistically rigorous method for model comparison would need to be based on, for example, an appropriate permutation method, in order to assess the probability of finding spurious differences between any two given models. We are not aware of the existence of such a method in the context of a cross-validation framework, like the one we use in this work. Although beyond the scope of the current study, the need for the development of an appropriate permutation method for this setting could provide a basis for future work.
Classification based on cortical grey matter segmentations
Using cortical grey matter segmentations, we show that linear SVMs moderately predicted individual conversion/non-conversion to CDMS in 71.2% of placebo-treated CIS patients. Although not exceptionally high, it is worth noting that the accuracy represents the average of 78% specificity and 64% sensitivity. The latter of which might be explained by the possibility that some of the CDMS patients categorized as converters do not show cortical pathology. Notably, patients experiencing a rapid transformation within 1 year show cortical atrophy in contrast to patients with slower disease progression (Perez-Miralles et al. 2013). We cannot exclude that those converters who were wrongly classified as non-converters are patients with a slower disease progression. In a 2-year follow-up the non-converters are likely to still develop CDMS, some of them maybe even shortly after the last follow-up. Structural grey matter changes, i.e. both, cortical lesions and grey matter atrophy are considered as promising characteristics to track the conversion from CIS to CDMS 20, (Calabrese et al. 2010), (M Tintore et al. 2008), (Kelly et al. 1993).
Some studies, however, failed to find a significant cortical atrophy in CIS compared to NC(Dalton et al. 2004; Ceccarelli et al. 2008; Ramasamy et al. 2009) while others have observed a significant cortical atrophy only in CIS(Bergsland et al. 2012) having a dissemination in space (DIS) of the lesions or only in selected brain regions, such as the hippocampus and the deep grey matter(Sastre-Garriga et al. 2005). Multivariate analysis of cortical thickness in patients with CIS who convert early to MS identified atrophy of superior frontal gyrus, thalamus, putamen and cerebellum as independent predictors of conversion to MS20. CIS with atrophy of such areas had a double risk of conversion.
These heterogeneous and partly contradictory findings may be explained e.g. by the clinical and paraclinical heterogeneity of patients with CIS, who may have quite different WM lesion load, and, in some cases, will never develop definite MS (Calabrese et al. 2011).
Previously, it was shown that regional GM atrophy is relevant in patients with CIS who convert early to MS(Calabrese et al. 2011; Raz et al. 2010), but about 20 % of CIS patients do not convert to MS after two decades (Fisniku et al. 2008b). An accuracy of 70% in our linear SVM analysis reveals that cortical GM patterns played a role for discrimination of converters and non-converters in the placebo-treated patients. In contrast, converters and non-converters could not be discriminated in the IFN-b groups. In our study, group labeling was based on conversion/non-conversion to CDMS at two year follow-up. However, at follow-up, parts of those patients which would have been expected to convert under placebo changed to non-converters or better “responders” under therapy. The latter might explain why we could not discriminate the treated group accurately using linear SVM.
Limitations
Our study further supports the idea that structural neuroimaging can inform prediction of CDMS. There are some limitations, however, that should be taken into account.
A crucial prerequisite for reliable image classification at the single-case level is the identification of morphometric criteria for distinguishing individuals. Beyond the impact of key demographic, clinical and lesion-associated MRI parameters on the 'natural risk' of CDMS (C. Polman et al. 2008), subclinical structural damage in the brain (i.e. changes in the normal-appearing brain tissue) might also occur early in the disease. However, its functional consequences in CIS patients might be negligible (Kappos et al. 2007a). Recently, mapping brain regions with diagnostic information using a “searchlight approach” and leave-one-out cross-validation to identify neuroanatomical patterns relevant for individual classification of patients and controls has been performed successfully in relapsing-remitting MS (Weygandt et al. 2011). Hotspots of MS associated tissue alterations which are highly informative about the clinical status have been identified in different normal-appearing areas of the brain. We infer from these results, that the approach could be promising also for classification and prognosis of patients with CIS.
The smaller sample size of the non-converters in the linear SVM analysis using GM segmentations may have limited the converters vs non-converters classification accuracy, and thus the findings may have been influenced by the heterogeneity of the non-converters subgroup. To account for this, we controlled for the potential effects of covariates, such as age, gender, and scanner.
Generally, it is unclear whether supervised MRI-based pattern recognition can achieve the level of sensitivity and specificity needed in order to be integrated into clinical applications. In the future, feature selection methods and the use of larger, independent test data may further increase classification accuracy. Furthermore, the shift from single predictive models to ensembles of classifiers may produce more generalisable diagnostic biomarkers by averaging the diagnostic decisions of numerous predictive models (Koutsouleris et al. 2010). Additionally, it will be interesting to investigate whether other para-clinical markers e.g. synthesis of oligoclonal bands (M Tintore et al. 2008) and genetic factors (Kelly et al. 1993) can improve SVM-based classification accuracy.
The IFN and the placebo group showed different features to have a role in determining conversion from CIS to CDMS. Early initiation of treatment at the stage of CIS has been demonstrated to delay conversion to CDMS, and to positively affect the evolution of clinical and MRI disease aspect (Kappos et al. 2007b). Assessment of the balance between burden of treatment and effects on outcome in patients with CIS is clinically important because a considerable proportion of patients will develop CDMS and eventually progress to substantial disability in the following years (Kappos et al. 2009). However, about 15–20% might not develop CDMS, not even after 20 years (Fisniku et al. 2008b, 2008a). In the context of early intervention, multivariate pattern classification may constitute the framework for MRI based discrimination of those individuals who will benefit from treatment from those who will do well without intervention.
Conclusions
The main potential of SVM-based classification is that it might be useful for predicting the clinical transition to MS at the individual level. Automatic pattern classification methods have been considered to promote a potentially accessible and objective way to improve clinical decision making, and may present a measure of the risk of developing MS in individuals with CIS, if sufficiently accurate.
We demonstrated that MRI data of MS lesions contain more information about the disease than is currently utilized in clinical assessments. Both lesion geometry and grey matter based information can aid prediction of conversion to CDMS. Inclusion of other lesional or degenerative MRI features may in the future further improve classification accuracy.
Supplementary Material
Figure 1.
Preprocessing pipeline.
Footnotes
With other data sets that comprise of larger numbers of lesions per patients, we found that the segmentation into white matter track regions can substantially increase prediction accuracies compared to those feature sets that rely on whole brain summaries only.
Compliance with Ethical Standards:
Conflict of Interest:
Kerstin Bendfeldt declares that she has no conflict of interest.
Bernd Taschler declares that he has no conflict of interest
Laura Gaetano declares that she has no conflict of interest
Philip Madoerin declares that he has no conflict of interest.
Pascal Kuster declares that he has no conflict of interest
Nicole Mueller-Lenke declares that she has no conflict of interest
Michael Amann declares that he has no conflict of interest.
Hugo Vrenken declares that he has no conflict of interest
Viktor Wottschel declares that he has no conflict of interest
Frederik Barkhof declares that he has no conflict of interest.
Stefan Borgwardt declares that he has no conflict of interest
Stefan Klöppel declares that he has no conflict of interest
Eva-Maria Wicklein declares that she has no conflict of interest.
Ludwig Kappos declares that he has no conflict of interest
Gilles Edan declares that he has no conflict of interest
Mark S. Freedman declares that he has no conflict of interest.
Xavier Montalbán declares that he has no conflict of interest.
Hans-Peter Hartung declares that he has no conflict of interest.
Christoph Pohl † - no conflict of interest.
Rupert Sandbrink declares that he has no conflict of interest
Rupert Sandbrink declares that he has no conflict of interest.
Bernd Taschler declares that he has no conflict of interest
Till Sprenger has received research grants from the Swiss MS Society, Swiss National Research Foundation, EFIC-Grünenthal and Novartis Pharmaceuticals Switzerland. Till Sprengers current and/or previous employers have received compensation for consultation and speaking activities from Mitsubishi Pharma, Eli Lilly, Sanofi Genzyme, Novartis, ATI, Actelion, Electrocore, Biogen Idec, Teva and Allergan.
Ernst-Wilhelm Radue declares that he has no conflict of interest
Jens Wuerfel declares that he has no conflict of interest.
Thomas E. Nichols declares that he has no conflict of interest.
Ethical approval:
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent:
Informed consent was obtained from all individual participants included in the study.
References
- Aban IB, Cutter GR, Mavinga N. Inferences and Power Analysis Concerning Two Negative Binomial Distributions with An Application to MRI Lesion Counts Data. Computational statistics & data analysis. 2008;53(3):820–833. doi: 10.1016/j.csda.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arns CH, Knackstedt MA, Pinczewski WV, Mecke KR. Euler-Poincaré characteristics of classes of disordered media. Physical Review E. 2001;63(3) doi: 10.1103/PhysRevE.63.031112. 031112. [DOI] [PubMed] [Google Scholar]
- Bakshi R, Dandamudi VS, Neema M, De C, Bermel RA. Measurement of brain and spinal cord atrophy by magnetic resonance imaging as a tool to monitor multiple sclerosis. J Neuroimaging. 2005;15(4 Suppl):30S–45S. doi: 10.1177/1051228405283901. [Research Support, Non-U.S. Gov't Review] [DOI] [PubMed] [Google Scholar]
- Barkhof F, Polman CH, Radue E-W, Kappos L, Freedman MS, Edan G, et al. Magnetic Resonance Imaging Effects of Interferon Beta-1b in the BENEFIT Study: Integrated 2-Year Results. Arch Neurol. 2007;64(9):1292–1298. doi: 10.1001/archneur.64.9.1292. [DOI] [PubMed] [Google Scholar]
- Barkhof F, Polman CH, Radue EW, Kappos L, Freedman MS, Edan G, et al. Magnetic resonance Imaging effects of interferon beta-1b in the BENEFIT study - Integrated 2-year results. Archives of Neurology. 2007;64(9):1292–1298. doi: 10.1001/archneur.64.9.1292. [DOI] [PubMed] [Google Scholar]
- Bates E, Wilson SM, Saygin AP, Dick F, Sereno MI, Knight RT, et al. Voxel-based lesion-symptom mapping. Nat Neurosci. 2003;6(5):448–450. doi: 10.1038/nn1050. [DOI] [PubMed] [Google Scholar]
- Beer S, Kesselring J. High prevalence of multiple sclerosis in Switzerland. Neuroepidemiology. 1994;13(1–2):14–18. doi: 10.1159/000110353. [DOI] [PubMed] [Google Scholar]
- Bergsland N, Horakova D, Dwyer MG, Dolezal O, Seidl ZK, Vaneckova M, et al. Subcortical and Cortical Gray Matter Atrophy in a Large Sample of Patients with Clinically Isolated Syndrome and Early Relapsing-Remitting Multiple Sclerosis. American Journal of Neuroradiology. 2012 doi: 10.3174/ajnr.A3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brex PA, Ciccarelli O, O'Riordan JI, Sailer M, Thompson AJ, Miller DH. A longitudinal study of abnormalities on MRI and disability from multiple sclerosis. N Engl J Med. 2002a;346(3):158–164. doi: 10.1056/NEJMoa011341. [Research Support, Non-U.S. Gov't] [DOI] [PubMed] [Google Scholar]
- Brex PA, Ciccarelli O, O'Riordan JI, Sailer M, Thompson AJ, Miller DH. A longitudinal study of abnormalities on MRI and disability from multiple sclerosis. New England Journal of Medicine. 2002b;346(3):158–164. doi: 10.1056/NEJMoa011341. [DOI] [PubMed] [Google Scholar]
- Calabrese M, Filippi M, Gallo P. Cortical lesions in multiple sclerosis. Nature reviews. Neurology. 2010;6(8):438–444. doi: 10.1038/nrneurol.2010.93. [Review] [DOI] [PubMed] [Google Scholar]
- Calabrese M, Rinaldi F, Mattisi I, Bernardi V, Favaretto A, Perini P, et al. The predictive value of gray matter atrophy in clinically isolated syndromes. Neurology. 2011;77(3):257–263. doi: 10.1212/WNL.0b013e318220abd4. [DOI] [PubMed] [Google Scholar]
- Ceccarelli A, Rocca MA, Pagani E, Colombo B, Martinelli V, Comi G, et al. A voxel-based morphometry study of grey matter loss in MS patients with different clinical phenotypes. NeuroImage. 2008;42(1):315–322. doi: 10.1016/j.neuroimage.2008.04.173. [DOI] [PubMed] [Google Scholar]
- Confavreux C, Vukusic S. Natural history of multiple sclerosis: a unifying concept. Brain. 2006;129(3):606–616. doi: 10.1093/brain/awl007. [DOI] [PubMed] [Google Scholar]
- Dalton CM, Chard DT, Davies GR, Miszkiel KA, Altmann DR, Fernando K, et al. Early development of multiple sclerosis is associated with progressive grey matter atrophy in patients presenting with clinically isolated syndromes. Brain. 2004;127(5):1101–1107. doi: 10.1093/brain/awh126. [DOI] [PubMed] [Google Scholar]
- Filippi M, Rocca MA, Barkhof F, Bruck W, Chen JT, Comi G, et al. Association between pathological and MRI findings in multiple sclerosis. Lancet neurology. 2012;11(4):349–360. doi: 10.1016/S1474-4422(12)70003-0. [Comparative Study Review] [DOI] [PubMed] [Google Scholar]
- Filli L, Hofstetter L, Kuster P, Traud S, Mueller-Lenke N, Naegelin Y, et al. Spatiotemporal distribution of white matter lesions in relapsing-remitting and secondary progressive multiple sclerosis. Multiple Sclerosis. 2012;18(11):1577–1584. doi: 10.1177/1352458512442756. [DOI] [PubMed] [Google Scholar]
- Fisniku LK, Brex PA, Altmann DR, Miszkiel KA, Benton CE, Lanyon R, et al. Disability and T2 MRI lesions: a 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain. 2008a;131(Pt 3):808–817. doi: 10.1093/brain/awm329. [DOI] [PubMed] [Google Scholar]
- Fisniku LK, Brex PA, Altmann DR, Miszkiel KA, Benton CE, Lanyon R, et al. Disability and T2 MRI lesions: a 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain. 2008b;131(3):808–817. doi: 10.1093/brain/awm329. [DOI] [PubMed] [Google Scholar]
- Ge T, Muller-Lenke N, Bendfeldt K, Nichols TE, Johnson TD. Analysis of Multiple Sclerosis Lesions Via Spatially Varying Coefficients. Ann Appl Stat. 2014;8(2):1095–1118. doi: 10.1214/14-aoas718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg-Zimring D, Achiron A, Guttmann CRG, Azhari H. Three-dimensional analysis of the geometry of individual multiple sclerosis lesions: Detection of shape changes over time using spherical harmonics. Journal of Magnetic Resonance Imaging. 2003;18(3):291–301. doi: 10.1002/jmri.10365. [DOI] [PubMed] [Google Scholar]
- Gourraud PA, Sdika M, Khankhanian P, Henry RG, Beheshtian A, Matthews PM, et al. A genome-wide association study of brain lesion distribution in multiple sclerosis. Brain. 2013;136:1012–1024. doi: 10.1093/brain/aws363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Ciccarelli O, Atzori M, Wheeler-Kingshott CAM, Miller DH, Thompson AJ, et al. Early pericalcarine atrophy in acute optic neuritis is associated with conversion to multiple sclerosis. Journal of Neurology Neurosurgery and Psychiatry. 2011;82(9):1017–1021. doi: 10.1136/jnnp.2010.239715. [DOI] [PubMed] [Google Scholar]
- Kappos L, Freedman MS, Polman CH, Edan G, Hartung HP, Miller DH, et al. Effect of early versus delayed interferon beta-1b treatment on disability after a first clinical event suggestive of multiple sclerosis: a 3-year follow-up analysis of the BENEFIT study. Lancet. 2007a;370(9585):389–397. doi: 10.1016/S0140-6736(07)61194-5. [DOI] [PubMed] [Google Scholar]
- Kappos L, Freedman MS, Polman CH, Edan G, Hartung HP, Miller DH, et al. Effect of early versus delayed interferon beta-1b treatment on disability after a first clinical event suggestive of multiple sclerosis: a 3-year follow-up analysis of the BENEFIT study. Lancet. 2007b;370(9585):389–397. doi: 10.1016/S0140-6736(07)61194-5. [DOI] [PubMed] [Google Scholar]
- Kappos L, Freedman MS, Polman CH, Edan G, Hartung HP, Miller DH, et al. Long-term effect of early treatment with interferon beta-1b after a first clinical event suggestive of multiple sclerosis: 5-year active treatment extension of the phase 3 BENEFIT trial. Lancet neurology. 2009;8(11):987–997. doi: 10.1016/S1474-4422(09)70237-6. [DOI] [PubMed] [Google Scholar]
- Kappos L, Polman CH, Freedman MS, Edan G, Hartung HP, Miller DH, et al. Treatment with interferon beta-1b delays conversion to clinically definite and McDonald MS in patients with clinically isolated syndromes. Neurology. 2006;67(7):1242–1249. doi: 10.1212/01.wnl.0000237641.33768.8d. [DOI] [PubMed] [Google Scholar]
- Kelly MA, Cavan DA, Penny MA, Mijovic CH, Jenkins D, Morrissey S, et al. The influence of HLA-DR and-DQ alleles on progression to multiple sclerosis following a clinically isolated syndrome. Human immunology. 1993;37(3):185–191. doi: 10.1016/0198-8859(93)90184-3. [DOI] [PubMed] [Google Scholar]
- Koutsouleris N, Patschurek-Kliche K, Scheuerecker J, Decker P, Bottlender R, Schmitt G, et al. Neuroanatomical correlates of executive dysfunction in the at-risk mental state for psychosis. Schizophrenia research. 2010;123(2-3):160–174. doi: 10.1016/j.schres.2010.08.026. [Research Support, Non-U.S. Gov't] [DOI] [PubMed] [Google Scholar]
- Lang C, Ohser J, Hilfer R. On the analysis of spatial binary images. Journal of microscopy. 2001;203(3):303–313. doi: 10.1046/j.1365-2818.2001.00899.x. [DOI] [PubMed] [Google Scholar]
- Legland D, Kiêu K, Devaux M-F. Computation of Minkowski measures on 2D and 3D binary images. Image Analysis & Stereology. 2007;26(2):83–92. [Google Scholar]
- Locatelli L, Zivadinov R, Grop A, Zorzon M. Frontal parenchymal atrophy measures in multiple sclerosis. Mult Scler. 2004;10(5):562–568. doi: 10.1191/1352458504ms1093oa. [DOI] [PubMed] [Google Scholar]
- Lovblad KO, Anzalone N, Dorfler A, Essig M, Hurwitz B, Kappos L, et al. MR imaging in multiple sclerosis: review and recommendations for current practice. AJNR American journal of neuroradiology. 2010;31(6):983–989. doi: 10.3174/ajnr.A1906. [Research Support, Non-U.S. Gov't Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay Altman R, Petkau AJ, Vrecko D, Smith A. A longitudinal model for magnetic resonance imaging lesion count data in multiple sclerosis patients. Statistics in medicine. 2012;31(5):449–469. doi: 10.1002/sim.4394. [Research Support, Non-U.S. Gov't] [DOI] [PubMed] [Google Scholar]
- McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50(1):121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- Morrissey SP, Miller DH, Kendall BE, Kingsley DPE, Kelly MA, Francis DA, et al. The Significance of Brain Magnetic-Resonance-Imaging Abnormalities at Presentation with Clinically Isolated Syndromes Suggestive of Multiple-Sclerosis - a 5-Year Follow-up-Study. Brain. 1993;116:135–146. doi: 10.1093/brain/116.1.135. [DOI] [PubMed] [Google Scholar]
- Newton BD, Wright K, Winkler MD, Bovis F, Takahashi M, Dimitrov IE, et al. Three-Dimensional Shape and Surface Features Distinguish Multiple Sclerosis Lesions from Nonspecific White Matter Disease. Journal of Neuroimaging. 2017;27(6):613–619. doi: 10.1111/jon.12449. [DOI] [PubMed] [Google Scholar]
- O'Riordan JI, Thompson AJ, Kingsley DPE, MacManus DG, Kendall BE, Rudge P, et al. The prognostic value of brain MRI in clinically isolated syndromes of the CNS - A 10-year follow-up. Brain. 1998;121:495–503. doi: 10.1093/brain/121.3.495. [DOI] [PubMed] [Google Scholar]
- Orru G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using Support Vector Machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neuroscience and biobehavioral reviews. 2012;36(4):1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. [Research Support, Non-U.S. Gov't Review] [DOI] [PubMed] [Google Scholar]
- Perez-Miralles F, Sastre-Garriga J, Tintore M, Arrambide G, Nos C, Perkal H, et al. Clinical impact of early brain atrophy in clinically isolated syndromes. Mult Scler. 2013;19(14):1878–1886. doi: 10.1177/1352458513488231. [DOI] [PubMed] [Google Scholar]
- Polman C, Kappos L, Freedman M, Edan G, Hartung HP, Miller D, et al. Subgroups of the BENEFIT study: Risk of developing MS and treatment effect of interferon beta-1b. Journal of Neurology. 2008;255(4):480–487. doi: 10.1007/s00415-007-0733-2. [DOI] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Edan G, Filippi M, Hartung H-P, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the ldquoMcDonald Criteriardquo. Annals of Neurology. 2005;58(6):840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- Popescu BFG, Pirko I, Lucchinetti CF. Pathology of multiple sclerosis: where do we stand? CONTINUUM: Lifelong Learning in Neurology. 2013;19(4, Multiple Sclerosis):901–921. doi: 10.1212/01.CON.0000433291.23091.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy DP, Benedict RHB, Cox JL, Fritz D, Abdelrahman N, Hussein S, et al. Extent of cerebellum, subcortical and cortical atrophy in patients with MS: A case-control study. Journal of the Neurological Sciences. 2009;282(1–2):47–54. doi: 10.1016/j.jns.2008.12.034. [DOI] [PubMed] [Google Scholar]
- Raz E, Cercignani M, Sbardella E, Totaro P, Pozzilli C, Bozzali M, et al. Gray- and White-Matter Changes 1 Year after First Clinical Episode of Multiple Sclerosis: MR Imaging. Radiology. 2010;257(2):448–454. doi: 10.1148/radiol.10100626. [DOI] [PubMed] [Google Scholar]
- Richert ND, Howard T, Frank JA, Stone R, Ostuni J, Ohayon J, et al. Relationship between inflammatory lesions and cerebral atrophy in multiple sclerosis. Neurology. 2006;66(4):551–556. doi: 10.1212/01.wnl.0000197982.78063.06. [Research Support, N.I.H., Intramural] [DOI] [PubMed] [Google Scholar]
- Sastre-Garriga J, Ingle GT, Chard DT, Cercignani M, Ramio-Torrenta L, Miller DH, et al. Grey and white matter volume changes in early primary progressive multiple sclerosis: a longitudinal study. Brain. 2005;128(6):1454–1460. doi: 10.1093/brain/awh498. [DOI] [PubMed] [Google Scholar]
- Scalfari A, Neuhaus A, Degenhardt A, Rice GP, Muraro PA, Daumer M, et al. The natural history of multiple sclerosis, a geographically based study 10: relapses and long-term disability. Brain. 2010;133(7):1914–1929. doi: 10.1093/brain/awq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölkopf B, Smola AJ. Learning with Kernels. MIT Press; 2001. [Google Scholar]
- Tintore M, Rovira A, Brieva L, Grive E, Jardi R, Borras C, et al. Isolated demyelinating syndromes: comparison of CSF oligoclonal bands and different MR imaging criteria to predict conversion to CDMS. Mult Scler. 2001;7(6):359–363. doi: 10.1177/135245850100700603. [DOI] [PubMed] [Google Scholar]
- Tintore M, Rovira A, Rio J, Nos C, Grive E, Tellez N, et al. Is optic neuritis more benign than other first attacks in multiple sclerosis? Ann Neurol. 2005;57(2):210–215. doi: 10.1002/ana.20363. [DOI] [PubMed] [Google Scholar]
- Tintore M, Rovira A, Rio J, Nos C, Grive E, Tellez N, et al. Baseline MRI predicts future attacks and disability in clinically isolated syndromes. Neurology. 2006;67(6):968–972. doi: 10.1212/01.wnl.0000237354.10144.ec. [Comparative Study Research Support, Non-U.S. Gov't] [DOI] [PubMed] [Google Scholar]
- Tintore M, Rovira A, Rio J, Tur C, Pelayo R, Nos C, et al. Do oligoclonal bands add information to MRI in first attacks of multiple sclerosis? Neurology. 2008;70(13 Part 2):1079–1083. doi: 10.1212/01.wnl.0000280576.73609.c6. [DOI] [PubMed] [Google Scholar]
- Wei X, Guttmann CR, Warfield SK, Eliasziw M, Mitchell JR. Has your patient's multiple sclerosis lesion burden or brain atrophy actually changed? Mult Scler. 2004;10(4):402–406. doi: 10.1191/1352458504ms1061oa. [Research Support, Non-U.S. Gov't Research Support, U.S. Gov't, P.H.S.] [DOI] [PubMed] [Google Scholar]
- Weygandt M, Hackmack K, Pfüller C, Bellmann–Strobl J, Paul F, Zipp F, et al. MRI Pattern Recognition in Multiple Sclerosis Normal-Appearing Brain Areas. PloS one. 2011;6(6):e21138. doi: 10.1371/journal.pone.0021138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wottschel V, Alexander DC, Kwok PP, Chard DT, Stromillo ML, De Stefano N, et al. Predicting outcome in clinically isolated syndrome using machine learning. Neuroimage Clin. 2015;7:281–287. doi: 10.1016/j.nicl.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S, et al. Accurate multimodal probabilistic prediction of conversion to Alzheimer's disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivadinov R, Grop A, Sharma J, Bratina A, Tjoa CW, Dwyer M, et al. Reproducibility and accuracy of quantitative magnetic resonance imaging techniques of whole-brain atrophy measurement in multiple sclerosis. J Neuroimaging. 2005;15(1):27–36. doi: 10.1177/1051228404271010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.