Abstract
The human disease lymphatic filariasis causes the debilitating effects of elephantiasis and hydrocele. Lymphatic filariasis currently affects the lives of 90 million people in 52 countries. There are three nematodes that cause lymphatic filariasis, Brugia malayi, Brugia timori, and Wuchereria bancrofti, but 90% of all cases of lymphatic filariasis are caused solely by W. bancrofti (Wb). Here we use population genomics to reconstruct the probable route and timing of migration of Wb strains that currently infect Africa, Haiti, and Papua New Guinea (PNG). We used selective whole genome amplification to sequence 42 whole genomes of single Wb worms from populations in Haiti, Mali, Kenya, and PNG. Our results are consistent with a hypothesis of an Island Southeast Asia or East Asian origin of Wb. Our demographic models support divergence times that correlate with the migration of human populations. We hypothesize that PNG was infected at two separate times, first by the Melanesians and later by the migrating Austronesians. The migrating Austronesians also likely introduced Wb to Madagascar where later migrations spread it to continental Africa. From Africa, Wb spread to the New World during the transatlantic slave trade. Genome scans identified 17 genes that were highly differentiated among Wb populations. Among these are genes associated with human immune suppression, insecticide sensitivity, and proposed drug targets. Identifying the distribution of genetic diversity in Wb populations and selection forces acting on the genome will build a foundation to test future hypotheses and help predict response to current eradication efforts.
Keywords: parasite, genomics, population genetics
Introduction
Infectious diseases have shaped the history of the human populations and are today one of the main burdens on human health. Human health is disproportionately affected in developing regions where access to medical care or sanitation are lacking. In addition to clinical studies and laboratory investigations, knowledge of the evolutionary history of a pathogen can improve our understanding of the current threat of infection and possible treatment or containment strategies.
Lymphatic filariasis (LF) is a human disease currently infecting over 90 million people across 52 countries and is the second leading cause of permanent and long-term disability worldwide (Ottesen et al. 2008; World Health Organization 2010, 2012). Disability is associated with recurrent adenolymphangitis, hydrocele, and elephantiasis that result in a loss of 5.9 million disability-adjusted-life-years (Gyapong et al. 2005; World Health Organization 2015, 2016). A majority of cases of LF, >90%, are caused by the nematode Wuchereria bancrofti (Wb) with remaining cases caused by Brugia malayi and Brugia timori (World Health Organization 2015). Wb is distributed throughout the tropics, whereas B. malayi and B. timori are endemic to South East Asia and Indonesia (World Health Organization 2015). Current treatments involve the drug Diethylcarbamazine (DEC), although DEC can cause serious complications (including encephalopathy and death) in patients who may also have onchocerciasis (caused by infection with Onchocerca volvulus) or loiasis (caused by Loa loa) as these species have overlapping distributions. Alternative use drugs such as ivermectin may kill only the larval blood stage (microfilariae, MF) and not the adult worms, thus having limited long-term effects on the worm populations (Centers for Disease Control and Prevention 2018).
Wb is highly specialized for human hosts and efforts to culture it in vivo (silver-leaf monkeys, Trachypithecus cristatus; Palmieri et al. 1983) or in vitro have failed to produce viable adult worms (Zaraspe and Cross 1986; Franke et al. 1987, 1990). Thus, much of what we know about Wb functional biology is derived from studies of B. malayi in nonprimates. However, the vast difference in distribution, host preference, and disease incidence between filarial nematodes means we must make a renewed effort to understand Wb biology.
Due to experimental limitations, genomics (and other omics) could offer an alternative approach to understanding Wb biology. However, here there are two main issues: (1) obtaining sufficient DNA quantity for genomics and (2) a need for single worms. Adult worm life-stages have sufficient DNA quantity but are inaccessible without invasive surgical methods. Obtaining the blood stage of Wb, MF, is less invasive but since multiple adult worms reproduce within a human host the sample will contain DNA from multiple genomes. Whole genome amplification (WGA) has been utilized to generate sufficient template for sequencing in other parasites but the presence of extraneous host DNA in a blood sample limits the use of commercially available techniques for Wb MF. In a previous study of Wb, Small et al. (2016) used WGA on individual Wb worms dissected from experimentally infected mosquitoes (Erickson et al. 2013). However, this process is not scalable to a field setting due to the low infectivity rate of Wb in mosquitoes (∼2%; Paily et al. 2009).
Here we utilize selective whole genome amplification (sWGA) to specifically enrich Wb DNA from single MF while avoiding amplification of Human DNA present in a host blood sample (Leichty and Brisson 2014; Clarke et al. 2017). This allows us to effectively utilize readily available field sample as well as sequence single worms. We used sWGA to generate 42 new genome sequences from Wb worms collected prior to mass drug administration from Haiti, Africa, and Papua New Guinea (PNG). We also use long-read sequencing to improve upon the current Wb reference genome (Small et al. 2016).
Our specific goals were to (1) identify the geographic origins of Wb infections, (2) test hypotheses of Wb migration by estimating the timing and migration, and (3) identify a list of candidate genes presented as highly differentiated between populations. We place our results into a context of LF elimination and detail how our methods can be applied to other parasitic nematodes.
Results
Improved W. bancrofti Genome Sequence and Gene Prediction
We used long-reads generated with PacBio sequencing to scaffold the current Wb genome assembly (PRJNA275548). PacBio sequencing produced 1.2 million subreads with an average of 5.0 kb in length. Scaffolding resulted in 856 scaffolds, with an N50 of 537 kb and with 198,000 introduced Ns. Scaffolds were assigned to chromosomes using ragout v1.7 (Kolmogorov et al. 2014) which resulted in 17 scaffolds longer than 100 kb, an N50 of 12.37 and 1.4 Mb of introduced Ns. The final assembly size was 88.46 Mb with the longest scaffold of 24.2 Mb corresponding to the X chromosome (supplementary table S1, Supplementary Material online). Benchmarking analysis using BUSCO (Simão et al. 2015) identified 97.5% of tested orthologs as complete. Visualization of the scaffolding and comparisons to B. malayi for the 17 largest scaffolds (length >100 kb) are presented in supplementary figure S1 (Supplementary Material online). We sequenced an RNA library to aid in gene predictions. Sequencing generated 600 million reads of which 1.7% mapped to the Wb genome and 98.3% mapped to human genome (Hg19). Analysis with Maker3 (Cantarel et al. 2008) resulted in 9,651 predicted genes, including 9,517 assigned to the 17 longest scaffolds.
Genome Sequencing for 42 Individual Worms
We used the program sWGA (Clarke et al. 2017) to design 9 primers that amplify Wb DNA while minimizing human DNA (supplementary table S5, Supplementary Material online). We used sWGA to amplify 10 MF from each of 26 infected human blood samples collected from Mali (N = 5), PNG (N = 11), Kenya (N = 5), and Haiti (N = 5) (Supplementary Material online). After amplification, we assessed the ratio of Wb to human DNA using qPCR and selected 42 individual worms with the highest ratio of Wb: Human DNA for sequencing. After quality control and adapter trimming, we retained, on average, 40 million reads per individual worm with ∼90% of reads mapping to Wb.
We compared these 42 newly sequenced genomes with 13 previously sequenced genomes from PNG (Small et al. 2016) and a single genome from Mali (Desjardins et al. 2013). We removed highly consanguineous worms, defined as first and second order relationships, identified using KING (Manichaikul et al. 2010). We also removed individual worms with high heterozygosity across the mitochondrial genome since this suggested that the sequences derive from multiple individuals. After further quality filtering, we were left with a total of 47 individual worms (supplementary table S2, Supplementary Material online). Final samples sizes of diploid individuals were: Haiti = 7, Mali = 11, Kenya = 9, PNG = 20.
We identified 403,487 single nucleotide polymorphisms (SNPs) among the 47 Wb individuals, with an average Ts/Tv ratio of 3.58 and average coverage per called genotype of 141×. More SNPs were discovered in intergenic regions than expected, observed: expected ratio 1.16, with fewer SNPs than expected in introns (0.53) and protein-coding regions (0.60) (supplementary table S3, Supplementary Material online).
Distribution of Genetic Diversity among W. bancrofti Populations
The median value of genetic diversity was not statistically different among populations (Haiti = 0.00084, Mali = 0.00072, Kenya = 0.00076, and PNG = 0.00074, supplementary fig. S2A, Supplementary Material online) with highest genetic variance in Haiti population. The median value of Tajima’s D statistic (Tajima 1989) was greater than 0 among African and Haitian populations (Mali = 0.4202, Kenya = 0.2446, Haiti = 0.3159). By contrast, the median value of Tajima’s D statistic in PNG was −0.2151; supplementary fig. S2B, Supplementary Material online). The scaled site-frequency spectrum is consistent with that expected given the median Tajima’s D statistic values, with an excess of intermediate variants in the Haiti and Africa populations but not in the PNG population (supplementary fig. S2C, Supplementary Material online). The correlation among genotypes followed the same expectation, with higher correlations in the African and Haitian populations compared with the PNG population (supplementary fig. S2D, Supplementary Material online). These trends indicate that African and Haitian populations have likely undergone recent population contractions, whereas the PNG population size has likely remained constant.
Population Structure and Differentiation among Populations
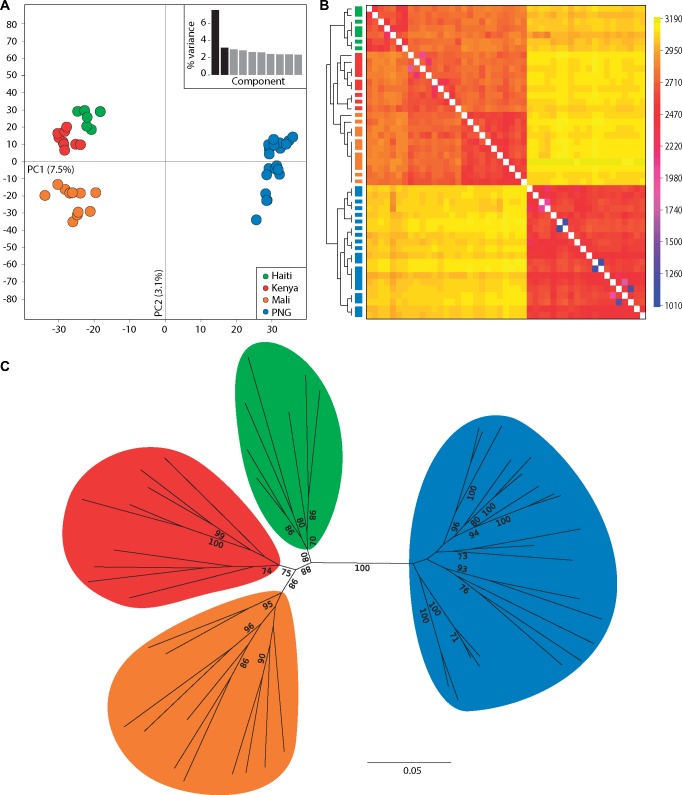
Principal component analysis (PCA) separated individuals according to their geographical origins (fig. 1A; PCA for each chromosome in supplementary fig. S4, Supplementary Material online). Principal component (PC)1, explained 7.5% of the variance, and separated PNG from all other populations. PC2, explained 3.1% of the variance and divided African and Haiti populations. FST values also support higher drift among PNG and African populations (supplementary table S6, Supplementary Material online). Phylogenetic reconstruction, using genome-wide SNPs, supports the PCA where worms are reciprocally monophyletic in regards to geographic location (fig. 1C). FineSTRUCTURE (Lawson et al. 2012) analysis identifies multiple individual worms with a high coancestry (fig. 1B). There is a clear distinction between worms sampled from PNG versus those of Africa and Haiti, with some additional within population coancestry. Worms with high coancestry also sampled from the same host infection are shown as merged blocks in figure 2B (dendrogram with sample labels are presented in supplementary fig. S3, Supplementary Material online).
Fig. 1.
Genetic structure and ancestry among Wuchereria bancrofti populations. (A) PCA of 10,000 SNPs across the genome after removing sites in high LD. Principal component (PC) 1 separates Haiti (green), Mali (orange), Kenya (red) from the population of Papua New Guinea (blue). PC 2 separates Kenya, Mali, and Haiti. Percent variation explained by each component is summarized in the bar plot (inset). (B) Coancestry matrix (a summary of nearest neighbor haplotype relationships in the data set) of Wb populations using fineSTRUCTURE. Dendrograms relating individuals to the coancestry matrix are along the vertical axis with populations denoted by colors corresponding to PCA. Cooler colors represent higher coancestry. Fused boxes of similar color denote worms sampled from the same host infection. A plot with individual sample labels on the vertical axis is available in supplementary figure S3 (Supplementary Material online). (C) Whole genome SNP phylogeny using SNPhylo highlights that individuals from the same population are monophyletic. Support is shown for bootstrap values greater than 70.
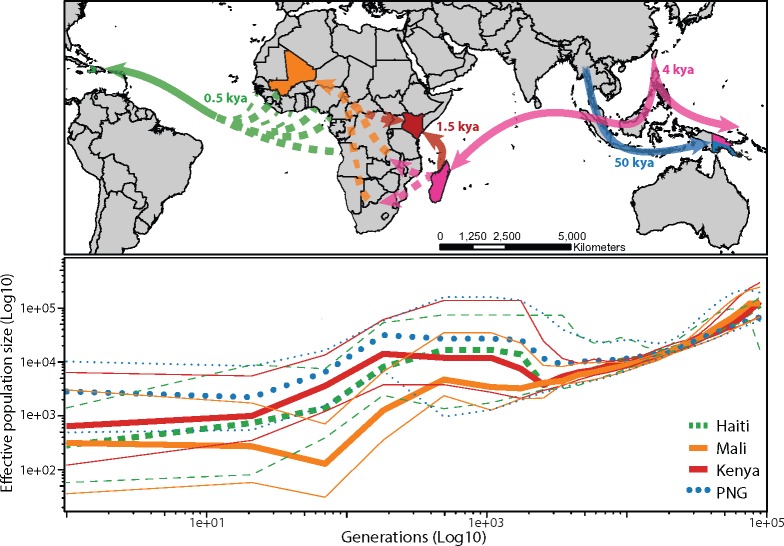
Fig. 2.
Demographic history of Wuchereria bancrofti. (A) Map representing potential routes of Wb dispersal highlighting sampled populations and shared ancestry: Haiti (green), Mali (orange), Kenya (red), Papua New Guinea (blue). Pink colors represent unsampled and thus inferred population ancestry connecting Wb populations (color figure available online). (B) Reconstruction of change in effective population size for each Wb population using MSMC2 and PopSizeABC. Thinner shaped lines of same shape represent 95% confidence intervals for the effective population sizes.
Cross-validation of ADMIXTURE (Alexander et al. 2009) runs was lowest for two ancestral clusters (supplementary fig. S9, Supplementary Material online). Two ancestral clusters separate the individual worms into a population containing all PNG worms and a second population with worms from Africa and Haiti (supplementary fig. S5, Supplementary Material online). Additional K values, K = {2, 5}, further subdivides populations by geography. At K = 3, Mali individuals are assigned into a new ancestral population. At K = 4, the PNG individuals are partially assigned to two ancestral populations. At K = 5, each geographic population has a distinct ancestral source population.
Genetic Relatedness and Inbreeding in W. bancrofti Populations
For each population, we compared the mean lengths of identity by descent (IBD) tracts among worms residing within the same host infection (IBDWITHIN) and in different host infections (IBDBETWEEN) (supplementary fig. S8A and table S9, Supplementary Material online). In Kenya and PNG populations, IBDWITHIN was significantly greater than IBDBETWEEN (P-value < 0.01). In Haiti and Mali populations IBDWITHIN and IBDBETWEEN values were not significantly different, although Haiti follows a similar trend to PNG and Kenya (supplementary table S9, Supplementary Material online). Inbreeding coefficients were highest in Mali (FROH = 0.192) and lowest in PNG (FROH = 0.102). Values from Kenya and Haiti were similar and intermediate: Kenya FROH = 0.136, Haiti FROH = 0.139 (supplementary fig. S8B, Supplementary Material online).
Demographic History of W. bancrofti Populations
We tested the fit of alternative models of divergence with and without postdivergence migration. We used ∂a∂i (Gutenkunst et al. 2009) to model the shared demographic history between each pair of Wb populations. The best fit model to the data was a strict isolation model (no migration) between populations after divergence (Likelihood Ratio Test of rejecting the null model of no migration, P-value = 0.30). Demographic scenarios evaluated in ABLE (Beeravolu et al. 2018) also supported a model with no postdivergence gene flow (supplementary table S4, Supplementary Material online).
Estimates of contemporary effective population size and divergence times from the best fit model are presented in table 1 (panel C in supplementary fig. S7 and Model2-1 in supplementary table S4, Supplementary Material online). The four Wb populations last shared a common ancestor ∼50 kya (kya = thousand years ago; assuming one generation per year) with an ancestral effective population size of ∼98,000–125,000. The most recent common ancestor between African Wb populations is 1.4–2.0 kya with an ancestral size of ∼697–987. The divergence time between the Mali and Haitian populations of Wb is 0.3–0.5 kya with an ancestral size of ∼8,000–32,000 (table 1).
Table 1.
Demographic Parameter Estimates.
| Population | Effective-Size | Divergence (years) |
|---|---|---|
| Haiti | 50–1,100 | — |
| Mali | 30–1,200 | — |
| Haiti–Mali | 8,318–32,000 | 394–502 |
| Kenya | 100–6,000 | — |
| Haiti–Mali–Kenya | 697–2,500 | 1,450–1,898 |
| Papua New Guinea | 500–10,000 | — |
| Haiti–Mali–Kenya–PNG | 98,000–125,000 | 37,000–57,000 |
The historical population trajectories of each Wb populations, estimated using both an HMM coalescent approach (Malaspinas et al. 2016) and the genome-wide site-frequency spectrum (Boitard et al. 2016) support a continued decline in Wb population sizes since ∼0.5 kya to present (fig. 2B). The trajectories of Mali, Kenya, and Haiti diverge between 1.5 and 3.0 kya, with the Mali population declining from an effective population size of ∼2,000 down to ∼300. Kenya and Haiti populations follow similar but not identical trajectories with an initial increase in size to 10,000 followed by a decrease to an effective-size of 700 and 300, respectively (fig. 2B and table 1). The PNG population trajectory diverged ∼10kya with an initial ancestral size of ∼30,000 and then declining to 3,000.
Local Adaptation in Populations of W. bancrofti
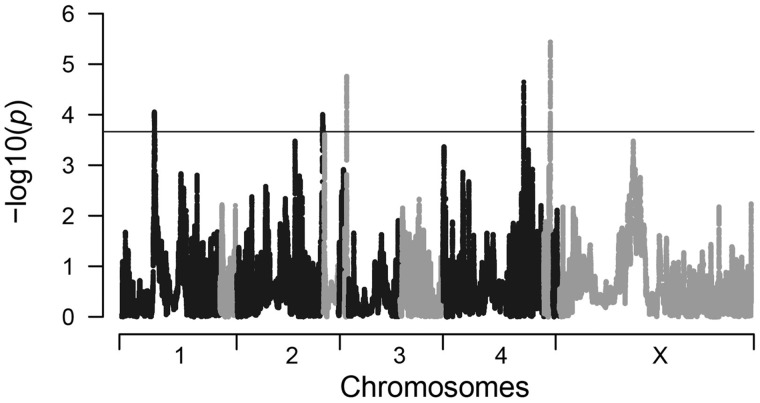
We used the haplotype-based approach in hapFLK (Fariello et al. 2013) with a goal to provide a candidate list of genes that are possible targets of local adaptation. We identified 9 regions, containing 18 genes, as outliers following an False Discovery Rate (FDR) correction (fig. 3). Among the outlier regions we determined the focal population using eigen analysis as described in Fariello et al. (2013). We determined that five genes in Haiti, five genes in Mali, two genes in Kenya, and six genes shared between Mali and Kenya were candidates of local adaptation (table 2). Functional annotations are listed in table 2 (full database ID in supplementary table S7, Supplementary Material online). A test for functional enrichment on candidate genes was not significant after a correction for multiple comparisons (supplementary table S8, Supplementary Material online).
Fig. 3.
Manhattan plot of local adaptation in Wuchereria bancrofti. Results of selection scan using hapFLK. Scaffolds are listed along the horizontal axis alternating between black and gray for contrast. The horizontal line represents an FDR of 10%. Outlier regions above the line were annotated and examined for gene function and presented in table 2.
Table 2.
Genes Present in Locally Adapted Haplotypes.
| Population | Chr | Location | Candidate Gene | Description | WBGene ID |
|---|---|---|---|---|---|
| Haiti | Chr1_0 | 7,448,651–7,451,875 | Madf-4 | Alcohol dehydrogenase transcription factor Myb/SANT-like | WBGene00011575 |
| 7,454,768–7,459,018 | CELE_F35G12.12 | 26S proteasome non-ATPase regulatory subunit 5 | WBGene00009445 | ||
| Chr4_0 | 10,151,016–10,153,158 | SMIM-1 | Small integral membrane protein 12 | WBGene00102523 | |
| Chr4_1 | 827,644–828,549 | Sre-1 | Serpentine receptor class epsilon-21 | WBGene00008126 | |
| Chr4_2 | 358,437–361,758 | Tag-196 | Cathepsin F; cysteine protease | WBGene00007055 | |
| Mali | Chr2_1 | 127,622–129,024 | Nurf-1 | Nucleosome-remodeling factor | WBGene00009180 |
| 130,750–132,216 | Hypothetical | NA | NA | ||
| 133,580–134,715 | Rsr-2 | Serine/arginine repetitive matrix 2 | WBGene00013260 | ||
| 140,713–142,929 | Nurf-1 | Nucleosome-remodeling factor | WBGene00009180 | ||
| 149,606–154,128 | Nurf-1 | Nucleosome-remodeling factor | WBGene00009180 | ||
| 171,145–173,380 | Snf-3 | Na/Cl betaine and GABA transporter | WBGene00004902 | ||
| Chr4_0 | 147,980–150,998 | Ctps-1 | CTP synthase | WBGene00012316 | |
| Mali–Kenya | ChrX_0 | 8,583,783–8,585,683 | Dif-1 | Congested-like trachea protein-related | WBGene00000996 |
| 8,589,182–8,592,937 | Cdgs-1 | Phosphatidate cytidylyltransferase | WBGene00016384 | ||
| 8,595,957–8,597,834 | Ric-3 | Resistance to inhibitors of cholinesterase protein 3 | WBGene00004363 | ||
| 8,599,957–8,602,020 | Dmd-7 | DM (doublesex/MAB-3) domain family | WBGene00019521 | ||
| 8,603,640–8,606,644 | Cav-1 | Caveolin-1 | WBGene00000301 | ||
| 8,618,382–8,623,441 | PAN-3 | NA | NA | ||
| Kenya | Chr2_0 | 7,311,511–7,317,467 | Hypothetical | NA | NA |
| ChrX_0 | 9,145,086–9,145,648 | Slo-1 | Large-conductance calcium-activated potassium channel | WBGene00004830 |
Note.— WBGene ID is WormBase gene ID (www.wormbase.org). Chr Scaffolds. NA, not applicable.
Potential targets for antiparasite drugs.
Discussion
The goals of our study were to (1) identify the geographic origins of Wb infections, (2) determine how Wb spread around the tropics, and (3) identify any region-specific factors that have affected the distribution of genetic diversity among the Wb populations. Our results correlate Wb spread to patterns of human migration and build a foundation of resources for further study of LF.
Geographic Origins of W. bancrofti Infections and Human Migrations
In examining the origins of Wb populations, we need to compare with studies on historical human migration. We recognize that this literature is vast and thus do not attempt to reference all possible source materials. We instead direct the reader to recent reviews which contain the proper depth of citations.
Africa: Mali and Kenya
The Austronesians (Austronesian-speaking peoples) migrated from Southern Borneo across the Indian Ocean to Madagascar ∼1.5–1.8 kya (Bellwood et al. 2006; Gray et al. 2009; Brucato et al. 2016, 2019). We hypothesize that Wb was spread from Island Southeast Asia (ISEA) to Madagascar during the Austronesian migration with later migrations spreading Wb to continental Africa. Although recent research proposes that the Austronesians may have also colonized the Comoros and parts of Eastern Africa (Brucato et al. 2019). If not directly by the Austronesians, then Wb may have spread to continental Africa during admixture among the Malagasy and Bantu speaking people of Africa (∼1.8 kya) (Pierron et al. 2014, 2017). Other have proposed that the Austronesian migration spread Wb based solely on historical medical records and art depicting elephantiasis (Laurence 1968, 1989; Hoeppli 1969; Fagg 1977). Although other diseases may produce similar symptoms (e.g., leprosy).
We estimated the ancestral effective population size of the African populations of Wb at ∼697–987. We would expect a Wb population infecting a small migrating host population to have a lower effective population size. However, the size of the ancestral Wb population in Southern Borneo is unclear. If we compare to the ancestral effective population size of Wb populations, ∼98,000–125,000, then the migration resulted in a 100-fold reduction. Though, it is unclear if this is representation of the ancestral Wb population in Southern Borneo. This can easily be resolved by future studies adding Wb populations Madagascar, where bancroftian LF is still prevalent (Garchitorena et al. 2018), and ISEA.
New World: Haiti
Wb would have multiple routes into the New World with migrating human populations (reviewed by Nielsen et al. 2017). We do not possess samples from other countries in South America, so can only address the origins of Wb infections in Haiti. The transatlantic slave trades transported large numbers of Western and Central Africans to the New World (estimates of 4,000 to Jamaica in 1518 CE, Wynter 1984). Since the divergence time between the Mali and Haiti populations of Wb was ∼0.3–0.5 kya (fig. 2 and table 1), it is likely that Wb was also transported to Haiti during the slave trade.
There are many examples of parasites spread by the transatlantic slave trade (Scott 1943; Tatem et al. 2006; Crellen et al. 2016). The estimated ancestral effective population size between Haiti and Mali is 8,000–32,000. One hypothesis is that Haiti may represent a mixture of multiple Wb populations from Africa consistent with slaves captured from multiple African countries. However, when we tested models including multiple introductions into Haiti, they were not more likely than a single introduction event (Supplementary Material online). Another possible explanation is that Wb populations increased in diversity as it spread across Africa. Additional samples from Central and Northern African countries would be required to determine how Wb diversity is liked to geographic expansion in Africa.
Papua New Guinea
The first humans reached Oceania around 40–50 kya (Nielsen et al. 2017). We estimate an Most Recent Common Ancestor between PNG and Haiti/African populations of ∼50 kya, with a more recent admixture event ∼5 kya (table 1). We hypothesize that the Melanesians spread Wb to PNG ∼50 kya and then ∼5 kya the Austronesian migration transported another Wb populations to PNG. The pattern of admixture in the PNG population is evident from the ADMIXTURE analysis (K = 4, supplementary fig. S5, Supplementary Material online) and recreated in the admixture simulations based on our model (supplementary fig. S6, Supplementary Material online). Human admixture between the Austronesians and Papuans is supported by language and genetic data, although estimates of proportion and timing vary (Bellwood et al. 2006; Gray et al. 2009; Xu et al. 2012; Skoglund et al. 2016).
Given the common ancestry it is likely that the Wb population that spread to Africa was founded from the same ancestral population that later admixed into PNG. A potential future test to elucidate Wb admixture would be to utilize Wb populations present on the islands of the South Pacific. The islands of the South Pacific were colonized by the expansion of the Austronesians into Oceania ∼3 kya and then later by Papuans (Skoglund et al. 2016).
Locally Adapted Haplotypes
As Wb spread to different regions it would have come into contact with novel environments, including new mosquito vectors, cotransmitted pathogens, and variations in human immune responses. Uncovering locally adapted genes may provide clues as to why Wb has been successful at colonizing and infecting human populations (e.g., in contrast to other filarial nematodes, B. malayi and B. timori). Functional annotation revealed that the genes in these regions are associated with a variety of functions including human host interaction, reproduction, and response to chemical toxicity (table 2).
Acetylcholinesterase inhibitors are a primary action of carbamates and organophosphates pesticides against arthropods (Casida and Durkin 2013; Verma et al. 2017). Ric-3, identified as conferring resistant to Acetylcholinesterase inhibitors, was first identified in Caenorhabditis elegans (Nguyen et al. 1995). Resistance to Acetylcholinesterase inhibitors may protect Wb from pesticides used to kill mosquito vectors. Future use of Acetylcholinesterase inhibitors as an antihelminthic, while promising in Trichuris muris (Sundaraneedi et al. 2018), may have limited efficacy on Wb populations in Mali and Kenya. Tracking changes in allele frequency at Ric-3 during pesticide application may provide further insights as to the role of these genes in Wb and would present a novel mechanism for insecticide as a nematicide.
Four other outlier genes have been identified as potential targets for new antiparasite drugs: Na/Cl Betaine and GABA transporter (Snf-3) (Jiang et al. 2005; Mullen et al. 2006; Casida and Durkin 2013; O’Neill et al. 2016), PAN domain protein (Kumar et al. 2007; O’Neill et al. 2016), Calcium-activated potassium channel (Slo-1) (Welz et al. 2011), and Phosphatidate Cytidylyltransferase (Cdgs-1) (Narayan et al. 1989; Crowther et al. 2010) (labeled with superscript [a] in table 2). All of our samples were collected prior to mass drug administration; thus, the signal of local adaptation may be in response to other environmental factors rather than drug resistance. However, studies creating lists of candidate drug targets should examine diversity in Wb populations prior to development as drug resistance could exist in a Wb population before drug treatment has even begun.
Only one of the genes in our candidate list had a nonsynonymous: synonymous substitution ratio >1, but its function was unknown (Kenya: Chr2_0). It is possible that the high differentiation in haplotype frequencies detected among populations is caused by demographic factors, including founder effects and small effective population sizes. In a study of O. volvulus the authors concluded that although sensitivity to Ivermectin does vary among test populations that the distribution of genetic diversity is most strongly influenced by population history (Doyle et al. 2017). Thus, we cannot determine without further testing whether these haplotypes are responding to selective forces or demography.
Applications to Ongoing Elimination
The World Health Organization classifies LF as a vulnerable infectious disease meaning that it could potentially be completely eradicated. Wb is the main causative agent of LF (>90% of cases); so, to be successful in eliminating LF we must drive Wb extinct. Our study provides novel tools and data that could help with the agenda. First, our estimates of diversity and relatedness in four endemic Wb populations prior to mass drug administration provide a baseline to evaluate the effectiveness of treatment in each region as well as to identify regions that might need specific renewed efforts. Second, our characterization of genetic polymorphisms provides a list of informative genetic markers that can now be used to efficiently monitor changes in genetic diversity by sequencing amplicons or genotyping (see similar approaches in Plasmodium [Friedrich et al. 2016; Redmond et al. 2018]). Finally, although our data emphasize the limited gene flow between continental populations, they also showed the ability of Wb to successfully migrate between continents and adapt to new environments and vectors which serves as a cautionary note until all populations are successfully eliminated.
Materials and Methods
Sample Collection
Samples used in this study from PNG were collected under IRB approved by Case Western Reserve University and PNG—Institute for Medical Research. Haitian samples were collected under IRB protocols reviewed and approved by the CDC IRB and the ethics committee of Hopital Ste. Croix in Leogane, Haiti. The samples from Mali were obtained as part of an NIAID and the University of Bamko-approved protocol (#02 -I-N200). The samples from Kenya were obtained from the South Kenyan Coast (Msambweni District) as part of a protocol approved by the Kenya Medical Research Institute National Ethical Review Committee and the Institutional Review Board for Human Studies at Case Western Reserve University. Information pertaining to samples and worms are available in supplementary table S2 (Supplementary Material online).
Improvement of W. bancrofti Genome Assembly and Annotations
The continuity of the current Wb genome assembly was improved through the addition of PacBio reads. PacBio sequences were error corrected using proovread v2.06 (Hackl et al. 2014) before being used to scaffold the current Wb genome using SSPACE-LongRead (Boetzer and Pirovano 2014). The final assembly was gap-closed using PBJelly (English et al. 2012). The improved Wb genome was further scaffolded using Ragout v1.2 (Kolmogorov et al. 2014) with alignments to B. malayi (PRJNA10729) and L. loa (PRJNA60051). Sequenced RNA was assembled into transcripts and Maker3 was used to finish annotations. VCFs were annotated using SnpEff v3.4 (Cingolani et al. 2012). See Supplementary Material online for additional details on template preparation, sequencing, and assembly.
sWGA and Population Sequencing
The program sWGA v0.3.0 (Clarke et al. 2017) was used to design primers for WGA of Wb while avoiding amplification of both Human sequences (Hg19) and Wb mitochondrial DNA (Ramesh et al. 2012). Ten MF were isolated from each individual blood sample for a total of 200 MF and DNA was isolated following methods in Small et al. (2016). After amplification, 42 samples were selected for library preparation and sequencing to include a minimum of 10 samples per geographic location. Primers are available in supplementary table S5 (Supplementary Material online), further details are available in Supplementary Material online.
Variant Calling
Reads passing quality controls were mapped to the improved Wb genome sequence (SAMN10411877) using BWA v0.7.13 (Li and Durbin 2009). Variants were called for each individual sample separately using GATK v3.1 HaplotypeCaller v3.5 (McKenna et al. 2010). The final set of SNPs containing all individuals was filtered to remove putatively repetitive and paralogous sequences (further details are available in Supplementary Material online).
Population Structure
Population structure was analyzed using parametric and nonparametric methods: PCA (as implemented in scikit-allel), ADMIXTURE v1.3.0 (Alexander et al. 2009), and phylogeny of genome-wide SNPs (Lee et al. 2014). The VCF file for structure analysis was pruned to remove SNPs in linkage disequilibrium using modules in the scikit-allel package and an r2 threshold of 0.05. Further filters were used to remove singletons, multiallelic sites, and sites with greater than 20% missing data resulting in 10340 positions distributed throughout all the autosomal scaffolds.
ADMIXTURE was run for K (number of ancestral populations) from 2 to 5 with 5-fold cross-validation. Each ADMIXTURE analysis was repeated 30 times with different seeds. In order to better understand the different solutions reported by ADMIXTURE, each value of K was input to the online version of CLUMPAK (Kopelman et al. 2015). ADMIXTURE was run a second time on the PNG population alone as well as the African and Haitian populations to examine population structure within each cluster. Simulations for ADMIXTURE were done using msmove (github.com/geneva/msmove) and the best fit demographic model. Results were analyzed in the same method as noted above.
The program fineSTRUCTURE v2.1.1 (Lawson et al. 2012) was used to explore genetic relatedness among Wb worms for all populations. A VCF file was prepared by phasing SNPs using read-informative phasing available in SHAPEIT2 v2.r837 (Delaneau et al. 2013). SHAPEIT2 was run in “assemble” mode with the following options: “–states 200 –window 0.5 –rho 0.000075 –effective-size 14000.” Results were plotted using programs available in the fineSTRUCTURE software package.
Diversity
Nucleotide diversity and Tajima’s D statistic (Tajima 1989) were calculated in 10,000 bp nonoverlapping windows and plotted for each population using the Python package scikit-allel v1.1.0 and Matplotlib v2.0.2. The scaled site-frequency spectrum (multiplied by the scaling factor k * (n − k)/n, where k is the minor allele count and n is the number of chromosomes) was calculated for each population on the Linkage Disequilibrium thinned and folded variant set in scikit-allel. The decay of the correlation between genotypes was plotted as the average correlation between genotypes in bins of 100 bp windows using scikit-allel and custom python scripts.
IBD tracts were calculated using fastIBD (Browning and Browning 2011) on phased data. A VCF file was prepared by phasing SNPs using read-informative phasing available in SHAPEIT2 v2.r837 (Delaneau et al. 2013). SHAPEIT2 was run in “assemble” mode with the following options: “–states 200 –window 0.5 –rho 0.000075 –effective-size 14000.” Inbreeding coefficient was calculated using Plink v1.9 (Purcell et al. 2007) on runs of homozygosity.
Demography
Estimates of effective population size were calculated using the MSMC2 algorithm (Malaspinas et al. 2016) on individual genomes with ten bootstrap replicates. MSMC2 input files were constructed using scripts available at msmc-tools (https://github.com/stschiff/msmc-tools), a positive mappability mask (http://lh3lh3.users.sourceforge.net/snpable.shtml) and a negative mask removing sites covered by less than ten reads in each individual genome. A summary for each population was plotted using the R package ggplot2 after linear extrapolation to synchronize time epochs (stsmall/Wb_sWGA/msmc2_interpolate.py).
A modified version of PopSizeABC (Boitard et al. 2016) (github.com/stsmall/popsizeabc) was used to estimate the piece-wise changes in effective population size for each Wb population. Five lakhs simulation data sets were generated using msprime v0.4.0 (Kelleher et al. 2016) with accompanied summary statistics calculated using scripts in github.com/stsmall/popsizeabc. Residuals and data informativeness were evaluated using the “plot” function in the R package “abc” package v2.1 (Csilléry et al. 2012); neural-net option and tolerance of 0.001. Results from PopSizeABC and MSMC2 were combined into a single plot by overlapping coestimated times.
We used ∂a∂i v1.6.3 (Gutenkunst et al. 2009) to model the shared demographic history between pairs of populations. The folded-site-frequency spectrum was used to compute the likelihood of the data under three different models: (1) an isolation-with-migration model with symmetrical migration, (2) an isolation-migration model with asymmetrical migration, and (3) a pure isolation model (migration rates set to 0). Each model allowed for the estimate of the ancestral population size as well as the daughter population sizes (modeled as exponential population size change after divergence). Our optimization procedure utilized methods and scripts available from https://github.com/kern-lab/miscDadiScripts.
The block-site-frequency-spectrum was used to compute the likelihoods of multiple-population models using the program ABLE v0.1.2 (Beeravolu et al. 2018). Models assumed stepwise changes in population size under the following scenarios: (1) populations diverge without migration, (2) populations diverge followed by asymmetrical migration, (3) populations diverge followed by admixture in the PNG population (supplementary fig. S7, Supplementary Material online). Each model was run for using the global search Controlled Random Search with 50,000 local trees searches. Confidence intervals of parameter estimates were inferred using the methods outlined in Beeravolu et al. (2018). Estimates were transformed into year using a mutation rate of 2.9E-9 per generation (Denver et al. 2009) and assuming one generation per year (estimated 8–14 months in Paily et al. [2009] and Farrar et al. [2013]).
Natural Selection and Local Adaptation
HapFLK was run on each chromosome in the African and Haitian populations of Wb using PNG as outgroup. HapFLK allows for the detection of positive selection from multiple-population samples using haplotype information. The number of clusters was calculated using fastPHASE cross-validation on each scaffold. P-values for hapFLK results were calculated using scripts available at https://forge-dga.jouy.inra.fr/projects/hapflk and following methods in Fariello et al. (2013). Candidate regions were identified using an FDR of 10% in the R package q-value v2.12.0. Population with the fixed or near fixed allele were identified using local population trees and eigenvalue analysis as described in Fariello et al. (2013).
Outlier regions were queried against the nr_db database downloaded from NCBI (date of download January 31, 2018). Protein sequences were obtained from improved annotations (see above section Reference Genome Improvement). The top five hits, sorted by e-value, were retained and gene ontology inferred by comparison to B. malayi (Ghedin et al. 2007) and C. elegans genomes (C. elegans Sequencing Consortium 1998). Genome enrichment tests were performed using the Fisher’s exact-test option in Panther (Tang et al. 2017) with comparison to annotations of C. elegans.
Data Availability
Wuchereria bancrofti improved genome assembly: SSBO00000000. All BAM files associated with the sWGA experiments aligned to SAMN10411877: SAMN10423675-SAMN10423716.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Patrick J. Lammie for contributing samples from Haiti as well as providing comments on the manuscript. This work was supported by grants from the National Institutes of Health (AI103263 to P.A.Z.) and the Clinical and Translational Science Collaborative of Cleveland (UL1TR000439 to P.A.Z.).
References
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeravolu CR, Hickerson MJ, Frantz LAF, Lohse K.. 2018. ABLE: blockwise site frequency spectra for inferring complex population histories and recombination. Genome Biol. 19(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood P, Fox JJ, Tryon D.. 2006. The Austronesian: historical and comparative perspectives. Canberra (Australia: ): ANU E Press. [Google Scholar]
- Boetzer M, Pirovano W.. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15:211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitard S, Rodríguez W, Jay F, Mona S, Austerlitz F.. 2016. Inferring population size history from large samples of genome-wide molecular data—an approximate Bayesian computation approach. PLoS Genet. 123:e1005877.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR.. 2011. A fast, powerful method for detecting identity by descent. Am J Hum Genet. 882:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucato N, Fernandes V, Cox MP, Boland A, Deleuze J-F, Ricaut F-X, Rito T, Pereira L, Besse C, Sudoyo H, et al. 2019. Evidence of Austronesian genetic lineages in East Africa and South Arabia: complex dispersal from Madagascar and Southeast Asia. Genome Biol Evol. 113:748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucato N, Kusuma P, Cox MP, Pierron D, Purnomo GA, Adelaar A, Kivisild T, Letellier T, Sudoyo H, Ricaut FX.. 2016. Malagasy genetic ancestry comes from an historical Malay trading post in Southeast Borneo. Mol Biol Evol. 339:2396–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282(5396):2012–2018. [DOI] [PubMed]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Alvarado AS, Yandell M.. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 181:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casida JE, Durkin KA.. 2013. Neuroactive insecticides: targets, selectivity, resistance, and secondary effects. Annu Rev Entomol. 58:99–117. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. 2018. Lymphatic filariasis treatment. Centers for Disease Control and Prevention [Internet]. Available from: https://www.cdc.gov/parasites/lymphaticfilariasis/treatment.html
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 62:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke EL, Sundararaman SA, Seifert SN, Bushman FD, Hahn BH, Brisson D.. 2017. swga: a primer design toolkit for selective whole genome amplification. Bioinformatics 33:2071–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crellen T, Allan F, David S, Durrant C, Huckvale T, Holroyd N, Emery AM, Rollinson D, Aanensen DM, Berriman M, et al. 2016. Whole genome resequencing of the human parasite Schistosoma mansoni reveals population history and effects of selection. Sci Rep. 6:20954.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther GJ, Shanmugam D, Carmona SJ, Doyle MA, Hertz-Fowler C, Berriman M, Nwaka S, Ralph SA, Roos DS, van Voorhis WC, et al. 2010. Identification of attractive drug targets in neglected- disease pathogens using an in silico approach. PLoS Negl Trop Dis. 4:e804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csilléry K, François O, Blum M.. 2012. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol Evol. 33:475–479. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J.. 2013. Haplotype estimation using sequencing reads. Am J Hum Genet. 934:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledo JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M, et al. 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci U S A. 10638:16310–16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CAC, Cerqueira GGC, Goldberg JM, Dunning Hotopp JC, Haas BJ, Zucker J, Ribeiro JMC, Saif S, Levin JZ, Fan L, et al. 2013. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nat Genet. 455:495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SR, Bourguinat C, Nana-Djeunga HC, Kengne-Ouafo JA, Pion SDS, Bopda J, Kamgno J, Wanji S, Che H, Kuesel AC, et al. 2017. Genome-wide analysis of ivermectin response by Onchocerca volvulus reveals that genetic drift and soft selective sweeps contribute to loss of drug sensitivity. PLoS Negl Trop Dis. 117:e0005816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 711:e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson SM, Thomsen EK, Keven JB, Vincent N, Koimbu G, Siba PM, Christensen BM, Reimer LJ.. 2013. Mosquito-parasite interactions can shape filariasis transmission dynamics and impact elimination programs. PLoS Negl Trop Dis. 7:e2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagg B. 1977. Nok terracottas. 1st ed. London and Lagos: Ethnographica for the National Museum. [Google Scholar]
- Fariello MI, Boitard S, Naya H, SanCristobal M, Servin B.. 2013. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics 1933:929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar J, Hotez PJ, Junghanss T, Kang G, Lalloo D, White NJ.. 2013. Manson’s tropical diseases. 23rd ed Elsevier, USA:Saunders Ltd. [Google Scholar]
- Franke ED, Riberu W, Wiady I.. 1987. In vitro cultivation of third stage larvae of Wuchereria bancrofti to the fourth stage. Am J Trop Med Hyg. 372:370–375. [DOI] [PubMed] [Google Scholar]
- Franke ED, Riberu W, Wiady I.. 1990. Evaluation of medium supplements for in vitro cultivation of Wuchereria bancrofti. J Parasitol. 762:262–265. [PubMed] [Google Scholar]
- Friedrich LR, Popovici J, Kim S, Dysoley L, Zimmerman PA, Menard D, Serre D.. 2016. Complexity of infection and genetic diversity in Cambodian Plasmodium vivax. PLoS Negl Trop Dis. 103:e0004526.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garchitorena A, Raza-Fanomezanjanahary EM, Mioramalala SA, Chesnais CB, Ratsimbasoa CA, Ramarosata H, Bonds MH, Rabenantoandro H.. 2018. Towards elimination of lymphatic filariasis in southeastern Madagascar: successes and challenges for interrupting transmission PLoS Negl Trop Dis. 12:e0006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, et al. 2007. Draft genome of the filarial nematode parasite Brugia malayi. Science 3175845:1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RD, Drummond AJ, Greenhill SJ.. 2009. Language phylogenies reveal expansion pulses and pauses in pacific settlement. Science 3235913:479–483. [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5:e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyapong JO, Kumaraswami V, Biswas G, Ottesen EA, Biswasm G, Ottesen EA.. 2005. Treatment strategies underpinning the global programme to eliminate lymphatic filariasis. Expert Opin Pharmacother. 62:179–200. [DOI] [PubMed] [Google Scholar]
- Hackl T, Hedrich R, Schultz J, Förster F.. 2014. Proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 3021:3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppli R. 1969. Parasitic diseases in Africa & the Western Hemisphere. Acta Trop. Suppl 10:1-240. [PubMed] [Google Scholar]
- Jiang G, Zhuang L, Miyauchi S, Miyake K, Fei YJ, Ganapathy V.. 2005. A Na+/Cr-coupled GABA transporter, GAT-1, from Caenorhabditis elegans: structural and functional features, specific expression in GABA-ergic neurons, and involvement in muscle function. J Biol Chem. 2803:2065–2077. [DOI] [PubMed] [Google Scholar]
- Kelleher J, Etheridge AM, McVean G.. 2016. Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS Comput Biol. 125:e1004842.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Raney B, Paten B, Pham S.. 2014. Ragout—a reference-assisted assembly tool for bacterial genomes. Bioinformatics 3012:i302–i309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I.. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 155:1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Chaudhary K, Foster JM, Novelli JF, Zhang Y, Wang S, Spiro D, Ghedin E, Carlow C.. 2007. Mining predicted essential genes of Brugia malayi for nematode drug targets. PLoS One 211:e1189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence BR. 1968. Elephantiasis and Polynesian origins. Nature 2195154:561–563. [DOI] [PubMed] [Google Scholar]
- Laurence BR. 1989. The global dispersal of bancroftian filariasis. Parasitology Today 58:260–264. [DOI] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D.. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8:e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Guo H, Wang X, Kim C, Paterson AH.. 2014. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15:162.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichty AR, Brisson D.. 2014. Selective whole genome amplification for resequencing target microbial species from complex natural samples. Genetics 1982:473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2514:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas AS, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergström A, Athanasiadis G, Cheng JY, Crawford JE, et al. 2016. A genomic history of Aboriginal Australia. Nature 5387624:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM.. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 2622:2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 209:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen GP, Mathews EA, Saxena P, Fields SD, McManus JR, Moulder G, Barstead RJ, Quick MW, Rand JB.. 2006. The Caenorhabditis elegans snf-11 gene encodes a sodium-dependent GABA transporter required for clearance of synaptic GABA. Mol Biol Cell 177:3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S, Dani HM, Misra UK.. 1989. Induction of phosphatidylcholine biosynthesis via CDPcholine pathway in lung and liver of rats following intratracheal administration of DDT and endosulfan. J Biochem Toxicol. 44:205–210. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Alfonso A, Johnson CD, Rand JB.. 1995. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics 1402:527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E.. 2017. Tracing the peopling of the world through genomics. Nature 5417637:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill M, Ballesteros C, Tritten L, Burkman E, Zaky WI, Xia J, Moorhead A, Williams SA, Geary TG.. 2016. Profiling the macrofilaricidal effects of flubendazole on adult female Brugia malayi using RNAseq. Int J Parasitol Drugs Drug Resist. 6:288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EA, Hooper PJ, Bradley M, Biswas G.. 2008. The global programme to eliminate lymphatic filariasis: health impact after 8 years. PLoS Negl Trop Dis. 210:e317.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paily KP, Hoti SL, Das PK.. 2009. A review of the complexity of biology of lymphatic filarial parasites. J Parasit Dis. 33(1–2):3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri JR, Connor DH, Purnomo, Marwoto HA.. 1983. Bancroftian filariasis. Wuchereria bancrofti infection in the silvered leaf monkey (Presbytis cristatus). Am J Pathol. 112:383–386. [PMC free article] [PubMed] [Google Scholar]
- Pierron D, Heiske M, Razafindrazaka H, Rakoto I, Rabetokotany N, Ravololomanga B, Rakotozafy LM-A, Rakotomalala MM, Razafiarivony M, Rasoarifetra B, et al. 2017. Genomic landscape of human diversity across Madagascar. Proc Natl Acad Sci U S A. 114:E6498–E6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron D, Razafindrazaka H, Pagani L, Ricaut F-X, Antao T, Capredon M, Sambo C, Radimilahy C, Rakotoarisoa J-A, Blench RM, et al. 2014. Genome-wide evidence of Austronesian–Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc Natl Acad Sci U S A. 1113:936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 813:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A, Small ST, Kloos ZA, Kazura JW, Nutman TB, Serre D, Zimmerman PA.. 2012. The complete mitochondrial genome sequence of the filarial nematode Wuchereria bancrofti from three geographic isolates provides evidence of complex demographic history. Mol Biochem Parasitol. 1831:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond SN, MacInnis BM, Bopp S, Bei AK, Ndiaye D, Hartl DL, Wirth DF, Volkman SK, Neafsey DE.. 2018. De novo mutations resolve disease transmission pathways in clonal malaria. Mol Biol Evol. 357:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott HH. 1943. The influence of the slave-trade in the spread of tropical disease. Trans R Soc Trop Med Hyg. 37:169–188. [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 3119:3210–3212. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Posth C, Sirak K, Spriggs M, Valentin F, Bedford S, Clark GR, Reepmeyer C, Petchey F, Fernandes D, et al. 2016. Genomic insights into the peopling of the Southwest Pacific. Nature 5387626:510–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ST, Reimer LJ, Tisch DJ, King CL, Christensen BM, Siba PM, Kazura JW, Serre D, Zimmerman PA.. 2016. Population genomics of the filarial nematode parasite Wuchereria bancrofti from mosquitoes. Mol Ecol. 257:1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaraneedi M, Eichenberger RM, Al-Hallaf R, Yang D, Sotillo J, Rajan S, Wangchuk P, Giacomin PR, Keene FR, Loukas A, et al. 2018. Polypyridylruthenium(II) complexes exert in vitro and in vivo nematocidal activity and show significant inhibition of parasite acetylcholinesterases. Int J Parasitol Drugs Drug Resist. 81:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1233:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Thomas PD, Kang D, Mills C, Muruganujan A, Huang X, Mi H.. 2017. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatem AJ, Rogers DJ, Hay SI.. 2006. Global transport networks and infectious disease spread. Adv Parasitol. 62:293–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Kashyap SS, Robertson AP, Martin RJ.. 2017. Functional genomics in Brugia malayi reveal diverse muscle nAChRs and differences between cholinergic anthelmintics. Proc Natl Acad Sci U S A. 11421:5539–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz C, Krüger N, Schniederjans M, Miltsch SM, Krücken J, Guest M, Holden-Dye L, Harder A, von Samson-Himmelstjerna G.. 2011. SLO-1-channels of parasitic nematodes reconstitute locomotor behaviour and emodepside sensitivity in Caenorhabditis elegans slo-1 loss of function mutants. PLoS Pathog. 74:e1001330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2010. Global programme to eliminate lymphatic filariasis.
- World Health Organization. 2012. Global Programme to eliminate lymphatic filariasis: progress report on mass drug administration. Wkly Epidemiol Rec. 37:345–356. [Google Scholar]
- World Health Organization. 2015. Lymphatic filariasis factsheet.
- World Health Organization. 2016. Global programme to eliminate lymphatic filariasis: progress report, 2015.
- Wynter S. 1984. New Seville and the conversion of Bartolome de Las Casas. Jam J. 17:25–32. [Google Scholar]
- Xu S, Paguach I, Stoneking M, Kayser M, Jin L, The HUGO Pan-Asian SNP Consortium. 2012. Genetic dating indicates the the Asian-Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion. Proc natl Acad Sci U.S.A. 109(12):4574–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaraspe G, Cross JH.. 1986. Attempt to culture Wuchereria bancrofti in vitro. Southeast Asian J Trop Med Public Health 174:579–581. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Wuchereria bancrofti improved genome assembly: SSBO00000000. All BAM files associated with the sWGA experiments aligned to SAMN10411877: SAMN10423675-SAMN10423716.