Abstract
The nucleus houses, organizes, and protects chromatin to ensure genome integrity and proper gene expression, but how the nucleus adapts mechanically to changes in the extracellular environment is poorly understood. Recent studies have revealed that extracellular physical stresses induce chromatin compaction via mechanotransductive processes. We report that increased extracellular multivalent cations lead to increased heterochromatin levels through activation of mechanosensitive ion channels (MSCs), without large-scale cell stretching. In cells with perturbed chromatin or lamins, this increase in heterochromatin suppresses nuclear blebbing associated with nuclear rupture and DNA damage. Through micromanipulation force measurements, we show that this increase in heterochromatin increases chromatin-based nuclear rigidity, which protects nuclear morphology and function. In addition, transduction of elevated extracellular cations rescues nuclear morphology in model and patient cells of human diseases, including progeria and the breast cancer model cell line MDA-MB-231. We conclude that nuclear mechanics, morphology, and function can be modulated by cell sensing of the extracellular environment through MSCs and consequent changes to histone modification state and chromatin-based nuclear rigidity.
INTRODUCTION
The nucleus is the organelle within the cell that contains, organizes, and mechanically protects the genome. Nuclear mechanics dictate proper genome organization and deformations, both of which can alter gene expression (Cremer and Cremer, 2001; Tajik et al., 2016; Cho et al., 2017; Kirby and Lammerding, 2018; Stephens et al., 2019). Consequently, abnormal nuclear morphology and mechanics occur across a spectrum of human diseases and conditions, including heart disease, muscular dystrophy, aging, and many cancers (Butin-Israeli et al., 2012). These conditions are linked to perturbations of the major mechanical components of the nucleus, chromatin, and lamins (Stephens et al., 2018a), and they result in nuclear rupture and abnormal deformations termed “blebs” (Goldman et al., 2004; Vargas et al., 2012; Furusawa et al., 2015; Robijns et al., 2016; Stephens et al., 2018b). Cellular and extracellular compressive forces, such as those due to actin and migration through tight spaces, further exacerbate nuclear morphological instability (Khatau et al., 2009; Tamiello et al., 2013; Denais et al., 2016; Hatch and Hetzer, 2016; Raab et al., 2016; Tocco et al., 2017). Subsequently, abnormal nuclear shape and rupture lead to mislocalization of nuclear proteins, DNA damage, and altered transcription, all of which are thought to cause cellular dysfunction in human diseases (Shimi et al., 2008; Isermann and Lammerding, 2013; Pfeifer et al., 2018; Xia et al., 2018). It is unclear how the cell natively regulates nuclear mechanics and morphology to prevent these outcomes.
Treatment of cells with a broad histone demethylase inhibitor to increase heterochromatin and nuclear rigidity has been shown to rescue nuclear shape in cases of abnormal morphology (Stephens et al., 2018b). Alterations to chromatin compaction on both short and long time scales can be achieved through changes in extracellular media or forces applied to the cell, as shown in studies that varied osmolality (Albiez et al., 2006; Irianto et al., 2013), cell substrate stretching (Heo et al., 2015, 2016; Le et al., 2016; Gilbert et al., 2018), substrate patterning (Versaevel et al., 2012; Jain et al., 2013), compression (Damodaran et al., 2018), and cell migration (Gerlitz and Bustin, 2010; Jacobson et al., 2018; Segal et al., 2018). Direct loading of the cell results in activation of mechanosensitive ion channels (MSCs) in the plasma membrane, resulting in transient calcium influx. This has been shown by stretching the underlying cell substrate (Kim et al., 2014; Heo et al., 2015), pulling a bead attached to the cell (Kim et al., 2015), or pressurized application of fluid to the cell surface (Sullivan et al., 1997). This mechanotransductive response induces chromatin compaction in mesenchymal stem cells. Compaction depends on the heterochromatin methyltransferase EZH2 (Heo et al., 2015) and increases in the facultative heterochromatin marker H3K27me3 (Heo et al., 2016; Le et al., 2016). This pathway can be blocked by inhibition of MSCs in the plasma membrane via GdCl3 or tarantula venom GsMTx4 (Suchyna et al., 2000; Ermakov et al., 2010; Bae et al., 2011; Gnanasambandam et al., 2017). However, activation of MSCs and subsequent increases in heterochromatin have not been observed in differentiated cells. Nonetheless, this heterochromatin-forming mechanotransduction pathway is a candidate for native regulation of nuclear mechanics and morphology, which facilitate proper nuclear function.
Prior studies have focused on mechanically stressing the entire cell to increase heterochromatin, but the effects of stress on the physical properties of the cell nucleus have not been fully investigated. We sought to determine whether activation of MSCs, without whole-cell stretching, could trigger increases in heterochromatin that could regulate nuclear morphology via increased nuclear rigidity. Previous work showed that extracellular multivalent cations interact with and distort negatively charged phospholipids, increasing membrane tension and bending (Gleisner et al., 2016; Ali Doosti et al., 2017). Following this result, we observed that adding divalent cations (via MgCl2 or CaCl2) or cationic polyamines (spermidine) to normal cell culture media increased the occurrence of transient calcium influx via MSCs, while internal cell ion contents remained unchanged. MSC activation was followed by an increase in heterochromatin levels across a variety of cell types.
Remarkably, the cation treatments rescued nuclear shape in cells displaying aberrant morphology (such as nuclear blebs) that had been induced by direct perturbation of either chromatin or lamin B. This effect is due to the cation-induced increases in heterochromatin, which we find to be dependent on histone methyltransferases. The heterochromatin increases were also associated with an increase in nuclear rigidity, providing a mechanism to explain the morphology rescue. Furthermore, supplementing media with increased cations similarly increased heterochromatin and rescued nuclear morphology in models of the accelerated aging disease Hutchison–Gilford progeria syndrome (HGPS) and breast cancer, as well as in patient HGPS cells. Our results show that the cell can modulate nuclear mechanics and morphology in response to the extracellular environment through MSCs that cue changes in histone modification state, driving heterochromatin formation and increased chromatin-based nuclear rigidity. Our report provides the mechanistic basis for a pathway that could be used to treat nuclear blebs, a hallmark of human disease and cellular dysfunction.
RESULTS AND DISCUSSION
Increased extracellular divalent cations increase heterochromatin levels through activation of MSCs
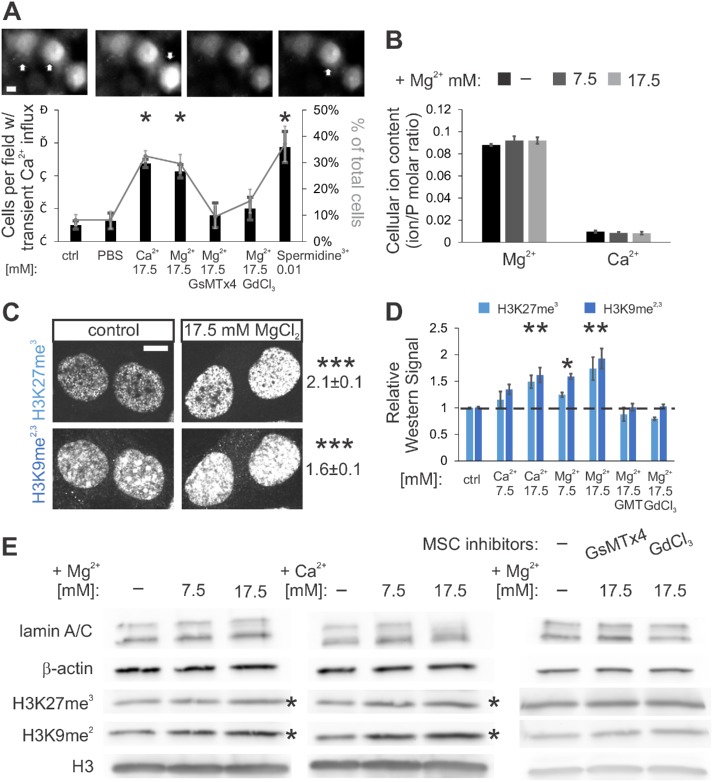
Previous work has revealed that cell stretching activates MSCs, leading to transient calcium influxes (Kim et al., 2015) and increased chromatin compaction/ heterochromatin in mesenchymal stem cells (Heo et al., 2015, 2016; Le et al., 2016). To activate MSCs without physically stretching the cell, we increased extracellular divalent ion concentrations; this is known to increase membrane tension (Gleisner et al., 2016; Ali Doosti et al., 2017). Incubation of mouse embryonic fibroblast (MEF) cells in increased extracellular levels of divalent cations (7.5 or 17.5 mM MgCl2 or CaCl2) or polyamines (0.01 mM spermidine3+) resulted in the hypothesized transient calcium influxes associated with activation of mechanosensitive channels, as measured by Oregon Green BAPTA-1 AM imaging (Figure 1A, 8% in control vs. 30% of cells with increased cations). Increased extracellular divalent cation levels also increased facultative (H3K27me3) and constitutive (H3K9me2,3) heterochromatin, as measured by immunofluorescence (IF) and Western blots (Figure 1, C–E, 24-h treatment, 40–110% increase). Treatment with MSC inhibitors blocked both transient calcium influxes and increases in heterochromatin (GsMTx4 and GdCl3, Figure 1, A, D, and E, and Supplemental Figure S1E), similar to its effect in cell stretching experiments (Heo et al., 2015, 2016).
FIGURE 1:
Increased extracellular divalent cations activate MSCs, which increased transient calcium influxes and heterochromatin formation. (A) Representative images and plot of calcium influx frequency from Oregon Green BAPTA-1 AM. White arrows denote increased intensity due to calcium influx and binding to the reporter. Data shown for MEF cells treated with increased extracellular divalent cations and polyamines and controls (from left to right, n = 13, 4, 4, 11, 5, 9, 4 fields of view, each with ≥9 cells). GsMTx4 (GMT) and GdCl3 are MSC inhibitors. (B) ICP-MS analysis of cellular Mg2+ and Ca2+ ion contents relative to phosphorous (P) content for cells treated with 0, 7.5, or 17.5 mM MgCl2 (n = 8, 8, and 5, respectively). (C) Representative images of MEF nuclei and average fold change in IF signals relative to control (1.0) for heterochromatin (H3K27me3 and H3K9me2,3) in cells incubated with or without additional MgCl2 for 24 h (n = 3 sets, each set >50 cells, p < 0.001). (D, E) Quantification of Western blots and representative blots from cells in standard medium or supplemented for 24 h with MgCl2, CaCl2, or MgCl2 and MSC inhibitors (GsMTx4 and GdCl3) for 24 h (n ≥ 3). Asterisks in E mark changing blots. Supplemental Figure S1E shows MgCl2 with and without MSC inhibitors in the same blot. Scale bar = 10 µm. Error bars represent standard error. Asterisks denote statistically significant differences from control.
The addition of divalent cations to the medium did not significantly alter cell viability, growth, nuclear size, osmolality, or actin content (Supplemental Figures S1, S2K, and S3B). Inductively coupled plasma mass spectrometry (ICP-MS) analysis, which quantifies levels of intracellular ions, measured no stable change in intracellular Mg2+ and Ca2+ relative to phosphorous (Figure 1B) in cells treated with extracellular MgCl2 for 24 h. This confirms that calcium influx is transient and that chromatin condensation is not due to physicochemical ion-chromatin interactions (e.g., Poirier et al., 2002) or intracellular changes in ion levels (as in Guilak et al., 2002, and Finan et al., 2009). The lack of physicochemical effects is unsurprising because the addition of 17.5 mM MgCl2 causes insignificant changes (50 mOsm, ∼15%) in the osmolality of the media, in contrast to other studies, which altered osmolality 50–100% (150–200 mOsm; Guilak et al., 2002; Finan et al., 2009). Thus, MSCs activated by extracellular divalent cations can induce transient calcium influxes and higher levels of heterochromatin independently of cell stretching.
Extracellular multivalent cation transduction suppresses nuclear blebbing
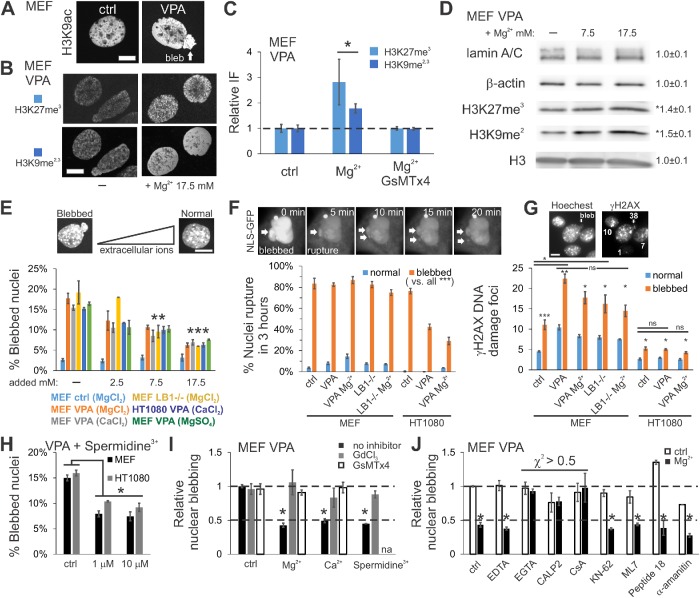
To determine the impact of mechanotransduction-based heterochromatin formation on nuclear morphology, we investigated the effects of added extracellular MgCl2 on cells with disrupted nuclear morphology. We have previously shown that treatment with the histone deacetylase inhibitor valproic acid (VPA) increased decompacted euchromatin and induced abnormal nuclear ruptures and bleb deformations (Figure 2A; Stephens et al., 2018b). On the contrary, we showed that aberrant nuclear morphology could be rescued by increases in heterochromatin by the histone demethylase inhibitor methylstat (Stephens et al., 2018b). Here, we instead increased levels of extracellular MgCl2 in VPA-treated cells with the goal of rescuing aberrant nuclear blebbing via mechanotransduction. Similar to its effects on untreated wild-type (WT) cells, adding extracellular MgCl2 to VPA-treated cells resulted in an increase in heterochromatin, as measured by IF and Western blots (Figure 2, B–D). There was also a corresponding decrease in nuclear blebbing that depended on MSCs (Figure 2, E, H, and I). VPA-treated cells displayed nuclear blebs in 17% of cells, while adding 2.5 mM extracellular MgCl2 decreased nuclear blebbing to ∼12%, 7.5 mM to ∼10%, and 17.5 mM to ∼6.5%, showing a trend toward the 3–4% blebbing observed in control cells (Figure 2E). Time-course analysis revealed that VPA-based nuclear blebbing is rescued coincident with an increase in heterochromatin after 8 h of incubation in increased extracellular MgCl2, but not after 2 h, while euchromatin remains constant (Supplemental Figure S2, B–D). Thus, mechanotransduction via MSCs provides a cell-native way to increase heterochromatin and rescue nuclear shape.
FIGURE 2:
Increased extracellular divalent cations or cationic polyamines suppress nuclear blebbing. (A) Representative IF images showing that VPA increases euchromatin (H3K9ac) and nuclear blebbing relative to control (ctrl). (B) Representative images and (C) average heterochromatin (H3K27me3 and H3K9me2,3) IF signals in VPA in media with normal and additional MgCl2 and with or without MSC inhibitor GsMTx4 (n = 3 sets, 20–53 cells, WT scaled to 1). (D) Western blot from VPA-treated MEF cells in standard or Mg/CaCl2 supplemented medium for 24 h (n = 4 each). (E) Percentages of nuclei displaying a nuclear bleb on treatment with additional extracellular divalent cations in MEF and HT1080 cells. (F) Images depict nuclear rupture and loss of nuclear contents and aggregates. Percentages of nuclei undergoing rupture, observed via NLS-GFP and (G) number of H2AX DNA damage foci for nuclei with normal shape (blue) or blebs (orange; n = 3 experiments, 50–200 cells each). (H) Percentages of nuclei displaying a nuclear bleb on treatment with spermidine3+ in MEF and HT1080 cells (for IF, see Supplemental Figure S2G) (I) Graph of relative nuclear blebbing for VPA-treated cells with increased extracellular divalent cations or polyamines and MSC inhibitors GsMTx4 and GdCl3 (VPA without inhibitor [ctrl] scaled to 1). (J) Relative nuclear blebbing frequency for VPA-treated cells without (white) or with 17.5 mM MgCl2 added (black) and addition of various inhibitors: 0.5 mM EDTA, Mg2+ chelator; 0.5 mM EGTA, Ca2+ chelator; 20 µM CALP2, calmodulin inhibitor; 5 µM CsA, calcineurin inhibitor; 8 µM KN-62, CAM KII inhibitor; 10 µM ML7 and 10 µM peptide 18, MLCK inhibitors; 10 µM α-amanitin, Pol II inhibitor/transcription. Nuclear blebbing experiments, n = 3–5 experiments of 90–200 cells each. Scale bar = 10 µm. Error bars represent standard error. Asterisks denote statistically significant differences (p or χ2 < 0.05).
This native mechanotransduction response is general and physiologically relevant. An alternative cell model of nuclear rupture and blebbing is the lamin B1 null (LB1-/-) cell line (Vargas et al., 2012; Shimi et al., 2015), which also has decreased heterochromatin (Supplemental Figure S2E; Camps et al., 2014; Stephens et al., 2018b). Similar to VPA-treated cells, LB1-/- cells exhibited increases in heterochromatin and decreased nuclear blebbing in the presence of additional extracellular divalent cations (Figure 2E, yellow bar, Supplemental Figure S2E). Suppression of nuclear blebbing and the corresponding increase in heterochromatin in perturbed cells was similar when using a different divalent cation (CaCl2, gray), counterion (MgSO4, green), or cell type (HT1080, dark blue) (Figure 2E; Supplemental Figure S2, A, CaCl2, and F, HT1080 MgCl2). VPA-treated cells exposed to the physiologically relevant cationic polyamine spermidine3+ at physiological concentrations (µM) revealed a similar decrease in nuclear blebbing. Again, this effect was dependent on MSCs and coincided with increased heterochromatin (Figure 2, H and I, and Supplemental Figure S2G). The low concentration used further rules out an osmotic origin for the observed effects. This finding reveals that mechanotransduction-based heterochromatin formation and nuclear morphology rescue is general and can be activated by physiologically relevant molecules. Thus, the extracellular environment sensed by mechanosensitive channels in the cell membrane can influence nuclear morphology by increasing heterochromatin over hours, which in turn suppresses the biophysical effects of direct chromatin or lamin perturbations that cause abnormal morphology.
Physiological impact of nuclear blebs suppressed by mechanotransduction
Nuclear blebs are believed to be linked with nuclear dysfunction (Stephens et al., 2019), but existing data come from observations of cell migration through constricting pores (Denais et al., 2016; Raab et al., 2016; Pfeifer et al., 2018; Xia et al., 2018). To further study the link between blebs and dysfunction, we assayed nuclear rupture and DNA damage in nonmigrating cells. We quantified nuclear shapes of blebbed nuclei compared with normal (elliptically shaped) nuclei to determine whether suppression of blebs corresponds to suppression of nuclear dysfunction. Live-cell imaging of NLS-GFP in MEF cells for 3 h revealed that normally shaped nuclei had a low probability of rupture (∼7%), while blebbed nuclei were >10-fold more likely to rupture (>80%; Figure 2F). Thus, on rescuing nuclear shape via increased extracellular MgCl2, nuclear ruptures decrease significantly (Supplemental Figure S2J). Furthermore, blebbed nuclei displayed twofold more DNA damage as measured by γH2AX IF (Figure 2G). These differences between normal and blebbed nuclei were consistent across conditions (WT, VPA, LB1-/-, and with or without additional extracellular MgCl2 for MEF and HT1080 cells). Thus, the inhibition of nuclear blebs by the addition of extracellular cations suppresses deleterious functional effects, particularly, nuclear rupture and DNA damage.
Protection of nuclear and genomic stability is dependent on the mechanotransduction pathway described earlier, as inhibition of MSCs via GsMTx4 or GdCl3 inhibited the ability of increased extracellular divalent cations or polyamines to suppress chromatin-decompaction-induced nuclear blebs (Figure 2I). Thus, we hypothesize that effects on the nucleus are dependent on calcium influx and signaling within the cell. Further experiments reveal that chelating extracellular calcium with 0.5 mM egtazic acid (EGTA) blocks nuclear morphology rescue, while EDTA-based magnesium chelation does not (Figure 2J). Calcium secondary messengers calmodulin and calcineurin were also essential to extracellular-cation–based bleb suppression (Figure 2J, inhibitors CALP2 and cyclosporine A [CsA], respectively). However, two major mechanotransduction signaling kinases, Ca2+/calmodulin-dependent kinase II and myosin light chain kinase (MLCK), were dispensable (Figure 2J, KN-62, ML7, and peptide 18, respectively). Inhibition of global transcription also did not suppress morphology rescue (Figure 2J, α-amanitin). Thus, cellular mechanotransduction of extracellular cues by MSCs, calcium, and calcium secondary messengers constitutes a novel pathway that can increase heterochromatin formation and rescue nuclear morphology and function.
Our finding that nuclear shape changes correlate with altered nuclear function is consistent with previous research. Several studies have found that the ability of the nucleus to maintain its shape directly impacts genome stability and transcription (reviewed in Stephens et al., 2019). In particular, nuclear blebbing coincides with nuclear rupture (Vargas et al., 2012; Tamiello et al., 2013; Hatch and Hetzer, 2016; Robijns et al., 2016; Stephens et al., 2018b), which in turn has been tied to DNA damage (Denais et al., 2016; Raab et al., 2016; Pfeifer et al., 2018; Xia et al., 2018). Other experiments have found that nuclear blebs can disrupt the cell cycle (Cho et al., 2019) and proper transcription, as chromatin within blebs exhibits decreased transcription (Shimi et al., 2008; De Vos et al., 2011; Helfand et al., 2012; Bercht Pfleghaar et al., 2015). Here, we have additionally shown that the rescue of nuclear shape by mechanotransduction corresponds with a recovery of nearly normal low levels of nuclear rupture and DNA damage. Therefore, activation of the MSCs studied here serves as a native and tunable mechanism to regulate nuclear stability and function via shape maintenance.
Transduction of extracellular cues to rescue nuclear morphology is dependent on methyltransferases to increase heterochromatin levels
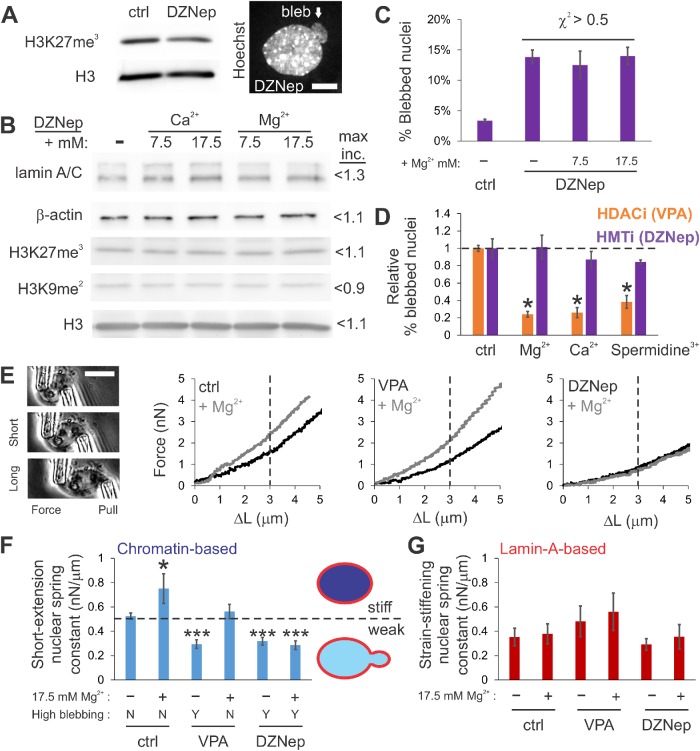
To determine whether rescue of nuclear morphology by extracellular divalent cations was dependent on the corresponding increases in heterochromatin, we cotreated cells with additional MgCl2 and the broad histone methyltransferase inhibitor (HMTi ) 3-deazaneplanocin-A (DZNep) (Miranda et al., 2009). DZNep-treated cells have decreased heterochromatin and display increased levels of nuclear blebbing, similar to VPA (Figure 3A; Stephens et al., 2018b). In stark contrast to control and VPA-treated cells, cells cotreated with DZNep and additional extracellular cations displayed no increase in heterochromatin markers, as measured by Western blots and IF, and nuclear blebbing was not suppressed (Figure 3, B–D; Supplemental Figure S2I, IF). This is a key result because both histone deacetylase and methyltransferase inhibitors result in net chromatin decompaction, weakened chromatin-based nuclear rigidity, and increased nuclear blebbing. However, only the HMTi prevents transduction of these extracellular environmental cues into nuclear morphological rescue, indicating that heterochromatin formation is essential to the process.
FIGURE 3:
Increased extracellular divalent cations suppress nuclear blebbing via methyltransferase activity to increase heterochromatin and chromatin-based nuclear rigidity. (A) Representative Western blot and an example of a blebbed nucleus after DZNep treatment, which decompacts chromatin via heterochromatin loss. (B) Western blots of DZNep-treated MEF cells in standard media or supplemented with divalent cations for 24 h (n = 4, p > 0.05). Maximum signal increase is listed. IF confirms this result (Supplemental Figure S2I). (C) Graph of percentages of DZNep-treated nuclei displaying a nuclear bleb and (D) relative nuclear blebbing for MEF VPA (orange) or MEF DZNep (purple) on addition of extracellular cations (control scaled to 1). For all nuclear blebbing experiments, n = 3–5 experiments of 90–400 cells each. (E) Representative micromanipulation nuclear force measurement images and force vs. extension plots for MEF V-/- control (ctrl), VPA, and DZNep preincubated cells in normal (black) or media supplemented with 17.5 mM MgCl2 (gray). Vertical line separates mechanical regimes where chromatin dominates short extension and lamin A dominates strain stiffening at longer extension force response. (F, G) Graph of average nuclear spring constants measured by micromanipulation for (F) chromatin-based short extension (<3 µm) and (G) lamin-A–based strain stiffening (n = 7–14, calculated as spring constant >3 µm – spring constant <3 µm). Bottom panel in F denotes whether high nuclear blebbing is present as yes (Y) or no (N). Statistical significance relative to WT (ctrl) untreated is reported for F and G. Graphic depicts weaker short-extension nuclear spring constant due to decompacted chromatin that results in increased nuclear blebbing, while stronger nuclei do not display increased nuclear blebbing. Scale bar = 10 µm. Error bars represent standard error. Asterisks denote statistically significant differences (p or χ2 < 0.05).
Heterochromatin formation rescues nuclear morphology by increasing the nuclear spring constant
Micromanipulation force measurements of isolated nuclei provide a way to measure the separate mechanical contributions of chromatin to short-extension stiffness and lamin A/C to strain stiffening at long extensions (Banigan et al., 2017; Stephens et al., 2017, 2018b). Nuclear spring constants were measured in MEF vimentin null (V-/-) cells for ease of nucleus isolation. MEF V-/- nuclei exhibit similar physical properties to WT (Stephens et al., 2017a, 2018), including nuclear bleb suppression via MgCl2 (Supplemental Figure S2H). Micromanipulation force measurements of isolated nuclei revealed that both VPA- and DZNep-treated nuclei, which display increased nuclear blebbing, were ∼40% softer than control in the chromatin-dominated regime, but were similar to control in the lamin A/C-dominated regime (Figure 3, E–G). This is consistent with previous reports that chromatin-based rigidity dictates nuclear morphology, independent of lamins (Stephens et al., 2018b).
Because extracellular divalent cations increased heterochromatin levels, we predicted that cation treatment would increase chromatin-based nuclear spring constants. In all conditions, before performing nucleus isolation and force measurements, the medium was exchanged for fresh standard medium to avoid physicochemical chromatin condensation. Control MEF cells incubated for 24 h in increased extracellular MgCl2 (17.5 mM) displayed a significant increase in the chromatin-based short-extension spring constant and no change in strain stiffening at long extensions (Figure 3, E–G). Similar to control, nuclei from cells cotreated with VPA and additional extracellular MgCl2 exhibited a significant increase in the short-extension nuclear spring constant. The treatment rescued chromatin-based nuclear stiffness to a level comparable to control nuclei (Figure 3, E and F) and decreased nuclear blebbing to near control levels (Figure 3D). Both lamin A/C mechanics, as measured by the increase in the nuclear spring constant at long extensions, and lamin A/C content were unchanged (Figure 3G; Western blots Figures 1E, 2D, and 3B). This result supports our hypothesis that MSCs can regulate histone modification state, which dictates chromatin-based nuclear rigidity and in turn, the nucleus’ ability to maintain normal shape and function.
We further tested this hypothesis by using DZNep as a downstream blockade to heterochromatin formation. Cells cotreated with DZNep and increased extracellular MgCl2 (17.5 mM) did not exhibit a rescued, stiff nuclear spring constant, but instead remained weak (gray line, Figure 3E, DZNep). This demonstrates that the ability to increase the short-extension nuclear force response depends on heterochromatin formation via histone methyltransferase activity (Figure 3F, ctrl and VPA vs. DZNep). Consequently, the inability of DZNep-treated cells to form heterochromatin, even in response to extracellular ions, results in weaker nuclei that remain susceptible to nuclear blebbing, which coincides with nuclear dysfunction. This is likely due to heterochromatin’s mechanical contribution since global transcription inhibition does not disrupt divalent-cation-induced bleb suppression (Figure 2J, α-amanitin). Thus, in these experiments, MSCs sensing extracellular ionic conditions cue histone methyltransferases to increase heterochromatin formation, which stiffens chromatin-based nuclear rigidity and maintains or rescues normal nuclear shape.
Increased extracellular cation mechanotransduction rescues nuclear shape in model and patient cells of human diseases
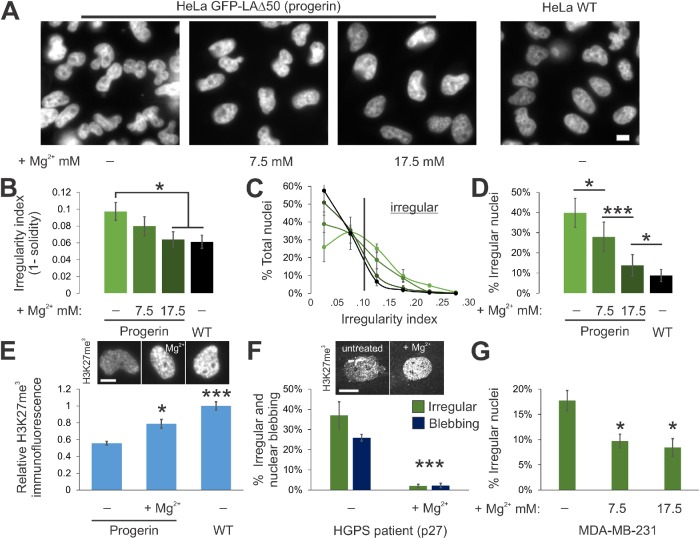
We asked whether this mechanotransductive process can rescue nuclear morphology in diseased cells. A cellular model of a disease with abnormal nuclear morphology is an ectopic expression of the mutant lamin A progerin, which is associated with the accelerated aging disease HGPS (Goldman et al., 2004; Butin-Israeli et al., 2012) and normal human aging (Rodriguez et al., 2009). As with many other known cases of abnormal nuclear morphology, GFP-progerin HeLa nuclei exhibited decreased heterochromatin levels, as measured by IF and Western blots of H3K27me3 (Figure 4E; Supplemental Figure S3A; Shumaker et al., 2006; McCord et al., 2013). GFP-progerin–expressing HeLa cells do not display discrete nuclear blebs, but instead display abnormal overall nuclear shape, which can be quantified by a nuclear irregularity index (Figure 4, A and B; see Material and Methods [Stephens et al., 2018b]). Similar to observations in MEF and HT1080 cells, extracellular MgCl2 restored both nuclear elliptical shape and heterochromatin levels to near HeLa WT levels (Figure 4, A–E; Supplemental Figure S3). The nuclear area remained similar with the addition of extracellular MgCl2 (Supplemental Figure S3B). Rescue of nuclear shape was inhibited when HeLa GFP-progerin cells were cotreated with the MSC inhibitor GsMTx4 and extracellular MgCl2, confirming the dependence of rescue on MSCs (Supplemental Figure S3C).
FIGURE 4:
Increased extracellular divalent cations rescue nuclear shape in models and patient cells of human diseases. (A) Representative images of GFP-progerin–expressing and WT HeLa nuclei labeled with Hoechst. (B–D) Graphs of nuclear irregularity index showing (B) averages, (C) histograms, and (D) percentages of irregular nuclei (irregularity index >0.10) for HeLa expressing GFP-progerin with various amounts of extracellular MgCl2 and WT without additional MgCl2 (n = 3 experiments each with 70–280 cells). (E) Representative images and graph of relative IF signal for facultative heterochromatin marker H3K27me3 for HeLa GFP-progerin in normal or MgCl2 supplemented media compared with HeLa WT (n = 3 experiments, 7.5 or 17.5 mM MgCl2 compared with HeLa progerin-GFP untreated, all sets >70 cells). (F) Graph of percentages of nuclei that are irregularly shaped (green, irregularity index >0.05) and display nuclear blebs (dark blue) for HGPS human fibroblast patient cells in normal or MgCl2 supplemented media (n ≥ 3 experiments, 30–78 cells per set). Representative images of facultative heterochromatin IF are shown above the graph. (G) Graph of percentages of irregular nuclei (irregularity index >0.10) in breast cancer model cell line MDA-MB-231 in normal or MgCl2 supplemented media (n = 4 experiments, each >120 cells). Scale bar = 10 µm. Error bars represent standard error. Asterisks denote statistically significant differences (p or χ2 < 0.05).
We next tested whether these observations hold for progeria (HGPS) patient cells and a breast cancer model cell line (MDA-MB-231). Progeria patient cells display abnormal nuclear shape and nuclear blebbing, but overall, patient cell nuclei are less irregular than HeLa cell nuclei. In progeria patient cells, increasing extracellular divalent cation concentration was sufficient to significantly decrease the percentage of irregular nuclei (here, given by index >0.05) from 37% to 2% and nuclear blebbing from 26% to 2% (Figure 4F), with no change in nuclear size (Supplemental Figure S3B). Similarly, breast cancer model MDA-MB-231 cells treated with extracellular cations exhibited decreased mean nuclear irregularity index (from 0.07 without treatment to 0.06 with ions) and a decrease from 17% irregular nuclei without treatment to 8% irregular nuclei with additional cations (Figure 4G, index >0.10). Together, these data provide evidence that extracellular cues and chromatin contribute to nuclear organization and shape in a range of physiological conditions, including in a human aging syndrome and a model of cancer.
Cells mechanotransduce extracellular signals to regulate chromatin state, nuclear rigidity, and nuclear morphology
We previously showed that changes to chromatin histone modification state modulate cell nuclear mechanics and shape (Stephens et al., 2017, 2018b). These changes in mechanics and shape impact nuclear stability and function (Figure 2, F and G). Here, we have shown that native cellular mechanotransduction via MSCs can protect the nucleus against aberrant nuclear morphology. Alterations in the composition of the extracellular medium can induce heterochromatin formation by histone methyltransferases to modulate nuclear mechanics and morphology, without direct force application to cells. Our findings are supported by the use of multiple biochemical interventions that affect several steps in the cellular response to the extracellular ionic composition.
These experiments provide evidence that chromatin-based mechanics are tunable, physiologically relevant, and can be modulated via mechanotransduction across a range of cells. These effects are triggered by MSCs embedded in the plasma membrane. Thus, direct stretching of cells (Heo et al., 2016; Le et al., 2016), cell–cell interactions, compression (Versaevel et al., 2012; Jain et al., 2013; Damodaran et al., 2018), ion composition (Figures 1 and 2), or charged proteins could all elicit this effect. Possible channels governing these effects are TRPM7, Piezo1/2, and other TRP channels (Liu et al., 2015; Ranade et al., 2015; Weng et al., 2018). Identifying which particular channel or channels are responsible could provide a therapeutic target for treating nuclear dysfunction in human diseases.
More generally, our results suggest that changes in both the mechanical environment and the extracellular physicochemical composition during both differentiation and disease could impact nuclear function through alterations to nuclear mechanics and shape. The fact that our experimental perturbation involves only the charge composition of the extracellular environment highlights that this response is not dependent on distortion of the cell as a whole, but rather only on physicochemical signals received at and transduced through the cell membrane. Thus, these channels, which are prevalent in diverse tissues including skin, joint, bone, muscle, neural, and heart, could be involved in a number of different physiological processes (Ranade et al., 2015). Altogether, our results illuminate a general mechanism by which cells can mechanically adapt to different extracellular environments and maintain robust and normal nuclear morphology and function.
MATERIALS AND METHODS
Cell growth
MEF, HT1080, and MDA-MB-231 cells were cultured in DMEM (Corning) complete with 1% Pen Strep (Fisher) and 10% fetal bovine serum (FBS; HyClone) at 37°C and 5% CO2. HGPS patient cells were cultured in MEM complete medium with 1% Pen Strep and 15% FBS (Goldman et al., 2004). HeLa progerin (GFP-LAΔ50) cells were grown in DMEM containing 1 mg/ml G418 also at 37°C and 5% CO2. As outlined in Taimen et al. (2009), progerin expression was induced via 2 μg/ml deoxycycline treatment for 24 h. On reaching confluency, cells were trypsinized, replated, and diluted into fresh media.
Drug treatment
Cells were treated with histone deacetylase inhibitor VPA at 2 mM for 16–24 h to accumulate decompacted euchromatin, as previously reported in Stephens et al. (2017). To deplete compact heterochromatin, cells were treated with HMTi DZNep at 0.5 μM for 16–24 h (Miranda et al., 2009). To inhibit mechanosensitive ions channels, cells were treated with either GsMTx4 (2.5 μM) or GdCl3 (10 μM) for 16–24 h.
IF
IF experiments were conducted as described previously in Stephens et al. (2017). Cells were seeded on cover glasses in six-well plates and incubated at 37°C and 5% CO2. Once at 80–90% confluence, cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (PBS) for 15 min at room temperature. Cells were then washed three times for 10 min each with PBS, permeabilized with 0.1% Triton X-100 (USBiological) in PBS for 15 min, and washed with 0.06% Tween 20 (USBiological) in PBS for 5 min followed by two more washes in PBS for 5 min each at room temperature. Cells were then blocked for 1 h at room temperature using a blocking solution consisting of 10% goat serum (Sigma Aldrich) in PBS. Primary antibodies were diluted in the blocking solution at the following concentrations: H3K27me3 1:1600 (C36B11, Cell Signaling Technology), H3K9me2-3 1:100 (6F12, Cell Signaling Technology), γH2AX conjugated to Alexa 488 1:300 (20E3, Cell Signaling Technology), lamin A/C 1:10,000 (Active Motif), and lamin B1 1:500 (ab16048, Abcam). After being incubated with the primary antibodies overnight at 4°C in the dark, cells were washed with PBS three times for 5 min each. Next, cells were incubated with anti-mouse or anti-rabbit Alexa 488 or 594 (Life Technologies, 2 mg/ml) fluorescent secondary antibodies diluted at 1:500 in blocking solution for 1 h at room temperature in the dark. Cells were washed with 1 μg/ml Hoechst 33342 (Life Technologies) in PBS for 5 min and then washed three more times with PBS. Finally, cover slides were mounted onto microscope slides using ProLong Gold antifade reagent (Life Technologies) and allowed to dry overnight at room temperature.
Western blots
Western blots were carried out as described previously (Stephens et al., 2017, 2018b). Protein was extracted via whole cell lysates (Sigma) or histone extraction kit (Abcam). Protein was loaded and run in 4–12% gradient SDS–polyacrylamide protein gels (LI-COR) for an hour at 100 V. Gels were then transferred to nitrocellulose blotting membrane with 0.2-μm pores (GE Healthcare) via wet transfer for 2 h at 100 V. The membrane was then washed three times in TBST for 5 min each before blocking in either 5% nonfat milk or bovine serum albumin (BSA) in TBST for an hour at room temperature. Primary antibody was diluted into 5% milk or BSA, depending on company specifications, added to blotting membrane, and allowed to shake and incubate overnight at 4°C. Primary antibodies were diluted in the blocking solution at the following concentrations: heterochromatin markers H3K27me3 1:500 (Millipore) and H3K9me2 1:500 (Millipore), euchromatin marker H3K9ac 1:2000 (Cell Signaling Technology), total histone control H3 1:2,000 (Cell Signaling Technology), lamin A/C 1:2,000 (Active Motif), and loading control β-actin 1:6000 (LI-COR). The next day the membranes were washed four times with TBST before incubation in secondary antibodies conjugated to HRP (Millipore, 12-348 and 12-349) for 1 h at room temperature. The membranes were again washed with TBST three times before chemiluminescence (PerkinElmer, NEL104001EA), and visualization via UVP imager. Quantification of Western blots was done in ImageJ.
Imaging and analysis
IF images were acquired with an IX-70 Olympus widefield microscope using a 60× oil 1.4 NA Olympus objective with an Andor iXon3 EMCCD camera using Metamorph. Image stacks with a step size of 0.4 μm were also acquired using a Yokogawa CSU-X1 spinning disk Leica confocal microscope with a 63× oil 1.4 NA objective and a Photometrics Evolve 512 Delta camera using Metamorph. Exposure times for DAPI, Rhodamine, and FITC were between 50 and 600 ms. Images were saved with Metamorph and transferred to ImageJ for analysis. Nuclei were selected by ImageJ threshold detection in the brightest plane or drawn by hand around Hoechst fluorescence if nuclei were too close together. Background fluorescence was determined by quantifying a 30 × 30 pixel area with no cells. Average intensity values of the nuclei were acquired and the average background signal was subtracted using Excel (Microsoft). For comparisons between cell types or treatment conditions, relative intensities were reported as fold intensity relative to WT or untreated nuclei. Each nucleus was scored as blebbed if a protrusion larger than 1 µm in diameter was present, as detailed previously (Stephens et al., 2018b). Solidity measurements were acquired in ImageJ following threshold detection. Statistical significance was determined for nuclear spring constants, IF, and nuclear irregularity index measurements via the t test. The chi-squared test was used to determine the statistical significance of changes in nuclear blebbing percentages.
DNA damage γH2AX foci counting was accomplished in ImageJ. Line scans of 10 focus spots were Gaussian curve fit to determine the median full width at half maximum sigma of 3.2 pixels. The FeatureJ Laplacian function with this sigma was used to process the image. This resulting image was then inverted. Next, we used the ImageJ find maxima function, with an empirically determined tolerance of 40 for MEF and 25 for HT1080, to select foci. These data were logged in Excel along with determination of whether individual nuclei were normal shaped or blebbed.
Live-cell imaging
Cells were grown to the desired confluence in cell culture dishes containing glass coverslip bottoms (In Vitro Scientific). The dishes were treated with 1 μg/ml Hoechst 33342 (Life Technologies) for 10 min and then imaged on a widefield microscope as described above.
Live-cell time lapse imaging and analysis of transient calcium influx
MEF cells were grown in cell culture dishes containing glass coverslip bottoms (In Vitro Scientific). Cells were loaded with 6.25 µg of Oregon Green 488 BAPATA1–AM for 30 min. Media was removed, cells were washed once with PBS, and then fresh media was added. Cells were imaged with a 60× objective widefield microscope every 3 s for 7–10 min. Data were analyzed in ImageJ where 50 × 50 pixel squares were placed within each nucleus and intensity measurements taken for every frame. Peaks in fluorescence of >10% intensity and longer than three time points were counted as calcium transient influx.
Live-cell time lapse imaging and analysis of nuclear rupture
MEF WT, MEF LB1-/-, or HT1080 WT cells were grown in cell culture dishes containing glass coverslip bottoms (In Vitro Scientific). Cells were treated 1 or 2 d prior to imaging with Cell Light Nucleus-RFP (NLS-RFP Fisher Scientific). Cells were untreated or treated with VPA and/or 17.5 mM MgCl2 1 d prior to imaging. Cells were imaged with a 40× objective on an environmental incubation (37°C and 5% CO2) Delta Vision microscope at 5-min intervals for 3 h. Cells were scored for nuclear blebs by assaying shape and nuclear ruptures by observing RFP spilling out of the nucleus and into the cytoplasm as outlined in Robijns et al. (2016).
Micromanipulation force measurement of an isolated nucleus
Micromanipulation force measurements were conducted as described previously in Stephens et al. (2017). MEF V-/- cells were used for their ease of nucleus isolation from a living cell and have similar nuclear force response to WT nuclei (Stephens et al., 2017, 2018b). The nucleus was isolated by using small amounts of detergent (0.05% Triton X-100 in PBS) locally sprayed onto a living cell via a micropipette. This gentle lysis allows for a second micropipette to retrieve the nucleus from the cell via slight aspiration and nonspecific adherence to the inside of the micropipette. Another micropipette was attached to the opposite end of the nucleus in a similar manner. This “force” micropipette was precalibrated for its deflection spring constant, which is on the order of nN/µm. A custom computer program written in LabView was then run to move the “pull” micropipette and track the position of both the pull and force pipettes. The pull pipette was instructed to move 5 µm at 45 nm/s. The program then tracked the distance between the pipettes to provide a measure of nucleus extension ∼3 µm. Tracking the distance that the force pipette moved/deflected multiplied by the premeasured spring constant provides a calculation of force exerted. Calculations were done in Excel to produce a force-extension plot from which the best-fit slope of the line provided a spring constant for the nucleus (nN/µm). Each nucleus was stretched two to four times to provide an accurate and reproducible measurement of the nuclear spring constant. Short-extension chromatin-based spring constants were measured from 0 to 3 µm and strain-stiffening lamin-A–based spring constants were measured as the difference between the short (0–3 µm) and long (3–6 µm) spring constants.
ICP-MS analysis of cellular ion contents
MEF cells were grown to 50–60% confluency in 225-cm2 flasks and then changed into 25 ml of fresh growth medium only, growth medium containing 7.5 mM MgCl2, and growth medium containing 17.5 mM MgCl2, respectively. These cells were further incubated for 24 h at 37°C and 5% CO2 and then harvested through trypsinization. Cell suspensions were centrifuged and aspirated to remove supernatant, and then 10 ml of fresh growth medium containing 100 µM Gd-DOTA (gadolinium-tetraazacyclododecane tetraacetic acid) was added. Cell mixtures were mixed for 5 min by gentle finger tapping and then centrifuged and aspirated to carefully remove supernatant. Cell pellet samples and drops of background 100 µM Gd-DOTA media were dried and then digested in 150 µl 67% nitric acid (Sigma, Trace grade). After digestion, 3000 µl MQ-H2O (the final nitric acid concentration will be 3% [wt]) was added to the samples. The 10- and 100×-diluted samples were further prepared by adding 3% nitric acid in MQ-H2O. All undiluted and diluted samples were then ready for ICP-MS analysis. Thermo iCAP Q (Quantitative Bio-element Imaging Center [QBIC], Northwestern University) was used to determine total metal contents using one set of trace element calibration standards (200, 100, 40, 20, 10, 5, 1, and 0.1 ppb). Sample element concentrations (ppb) were determined in kinetic energy discrimination mode with 10% H2/He gas to minimize polyatomic interferences. ICP-MS data were analyzed via Microsoft Excel 2012. Extracellular element content was removed by using Gd-DOTA as tracer to subtract extracellular medium background since Gd-DOTA species remain outside of cells (Aime et al., 2002). Thus, cellular Mg, Ca, and P element contents were calculated by subtracting the extracellular element content from the total (cellular + extracellular) element content. The molar ratios of cellular Mg and Ca content, relative to cellular phosphorus content (Mg/P and Ca/P), were used to normalize cellular metal content for different experiments, because this normalization avoids inherent deviations from cell number and volume measurements for each experiment.
Supplementary Material
Acknowledgments
We thank Yixian Zheng for providing us with MEF LB1-/- cells (Shimi et al., 2015). We thank Aykut Erbas¸, Sumitabha Brahmachari, and Ronald Biggs for helpful discussions. We thank Reza Vafabakhsh and Michael Schamber for advising us regarding the transient calcium influx experiments. We thank QBIC at Northwestern University for ICP-MS analysis. We thank the Northwestern Biological Imaging Facility and Director Jessica Hornick for use of the Delta Vision microscope for time lapse imaging. A.D.S. is supported by Pathway to Independence Award NIHGMS K99GM123195. A.D.S., P.Z.L, E.J.B., and J.F.M. are supported by National Science Foundation (NSF) Grants DMR-1206868 and MCB-1022117 and by National Institutes of Health (NIH) grants GM-105847, CA-193419, and, via subcontract, DK-107980. S.A.A. and R.D.G. are supported by NIH GM-106023, CA 193419, and Progeria Research Foundation PRF 2013-51. H.C. and T.V.O. are supported by Physical Science–Oncology Center, National Cancer Institute, Grant U54CA193419. L.M.A. and V.B. are supported by NIH grants R01CA200064 and R01CA155284, NSF grant CBET-1240416, and the Lungevity Foundation. This work was funded by the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust through a Postdoctoral Fellowship to A.D.S.
Abbreviations used:
- CsA
cyclosporine A
- DZNep
3-deazaneplanocin-A
- FBS
fetal bovine serum
- HGPS
Hutchinson–Gilford progeria syndrome
- HMTi
histone methyltransferase inhibitor
- ICP-MS
inductively coupled plasma mass spectrometry
- IF
immunofluorescence
- LB1-/-
lamin B1 null
- MEF
mouse embryonic fibroblast
- MLCK
myosin light chain kinase
- MSC
mechanosensitive ion channel
- PBS
phosphate-buffered saline
- V-/-
vimentin null
- VPA
valproic acid
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-05-0286) on June 19, 2019.
REFERENCES
- Aime S, Cabella C, Colombatto S, Geninatti Crich S, Gianolio E, Maggioni F. (2002). Insights into the use of paramagnetic Gd(III) complexes in MR-molecular imaging investigations. J Magn Reson Imaging , 394–406. [DOI] [PubMed] [Google Scholar]
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, et al. (2006). Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res , 707–733. [DOI] [PubMed] [Google Scholar]
- Ali Doosti B, Pezeshkian W, Bruhn DS, Ipsen JH, Khandelia H, Jeffries GDM, Lobovkina T. (2017). Membrane tubulation in lipid vesicles triggered by the local application of calcium ions. Langmuir , 11010–11017. [DOI] [PubMed] [Google Scholar]
- Bae C, Sachs F, Gottlieb PA. (2011). The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry , 6295–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banigan EJ, Stephens AD, Marko JF. (2017). Mechanics and buckling of biopolymeric shells and cell nuclei. Biophys J , 1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercht Pfleghaar K, Taimen P, Butin-Israeli V, Shimi T, Langer-Freitag S, Markaki Y, Goldman AE, Wehnert M, Goldman RD. (2015). Gene-rich chromosomal regions are preferentially localized in the lamin B deficient nuclear blebs of atypical progeria cells. Nucleus , 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. (2012). Nuclear lamin functions and disease. Trends Genet , 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Wangsa D, Falke M, Brown M, Case CM, Erdos MR, Ried T. (2014). Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB J , 3423–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Irianto J, Discher DE. (2017). Mechanosensing by the nucleus: from pathways to scaling relationships. J Cell Biol , 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Vashisth M, Abbas A, Majkut S, Vogel K, Xia Y, Ivanovska IL, Irianto J, Tewari M, Zhu K, et al. (2019). Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Dev Cell , 920–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. (2001). Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet , 292–301. [DOI] [PubMed] [Google Scholar]
- Damodaran K, Venkatachalapathy S, Alisafaei F, Radhakrishnan AV, Sharma Jokhun D, Shenoy VB, Shivashankar GV. (2018). Compressive force induces reversible chromatin condensation and cell geometry dependent transcriptional response. Mol Biol Cell , 3039–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, et al. (2011). Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet , 4175–4186. [DOI] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. (2016). Nuclear envelope rupture and repair during cancer cell migration. Science , 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakov YA, Kamaraju K, Sengupta K, Sukharev S. (2010). Gadolinium ions block mechanosensitive channels by altering the packing and lateral pressure of anionic lipids. Biophys J , 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan JD, Chalut KJ, Wax A, Guilak F. (2009). Nonlinear osmotic properties of the cell nucleus. Ann Biomed Eng , 477–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M. (2015). Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun , 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G, Bustin M. (2010). Efficient cell migration requires global chromatin condensation. J Cell Sci , 2207–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert H, Mallikarjun V, Dobre O, Jackson MR, Pedley R, Gilmore AP, Richardson SM, Swift J. (2018). Nuclear decoupling is part of a rapid protein-level cellular response to high-intensity mechanical loading. Biorxiv. 10.1101/317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleisner M, Kroppen B, Fricke C, Teske N, Kliesch TT, Janshoff A, Meinecke M, Steinem C. (2016). Epsin N-terminal homology domain (ENTH) activity as a function of membrane tension. J Biol Chem , 19953–19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasambandam R, Ghatak C, Yasmann A, Nishizawa K, Sachs F, Ladokhin AS, Sukharev SI, Suchyna TM. (2017). GsMTx4: mechanism of inhibiting mechanosensitive ion channels. Biophys J , 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS. (2004). Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA , 8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak F, Erickson GR, Ting-Beall HP. (2002). The effects of osmotic stress on the viscoelastic and physical properties of articular chondrocytes. Biophys J , 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW. (2016). Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol , 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Wang Y, Pfleghaar K, Shimi T, Taimen P, Shumaker DK. (2012). Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J Pathol , 735–745. [DOI] [PubMed] [Google Scholar]
- Heo SJ, Driscoll TP, Thorpe SD, Nerurkar NL, Baker BM, Yang MT, Chen CS, Lee DA, Mauck RL. (2016). Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. Elife , e18207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SJ, Thorpe SD, Driscoll TP, Duncan RL, Lee DA, Mauck RL. (2015). Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Sci Rep , 16895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Swift J, Martins RP, McPhail GD, Knight MM, Discher DE, Lee DA. (2013). Osmotic challenge drives rapid and reversible chromatin condensation in chondrocytes. Biophys J , 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann P, Lammerding J. (2013). Nuclear mechanics and mechanotransduction in health and disease. Curr Biol , R1113–R1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson EC, Perry JK, Long DS, Olins AL, Olins DE, Wright BE, Vickers MH, O’Sullivan JM. (2018). Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells. BMC Biol , 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Iyer KV, Kumar A, Shivashankar GV. (2013). Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci USA , 11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. (2009). A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA , 19017–19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TJ, Joo C, Seong J, Vafabakhsh R, Botvinick EL, Berns MW, Palmer AE, Wang N, Ha T, Jakobsson E, et al. (2015). Distinct mechanisms regulating mechanical force-induced Ca(2)(+) signals at the plasma membrane and the ER in human MSCs. Elife , e04876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TJ, Sun J, Lu S, Qi YX, Wang Y. (2014). Prolonged mechanical stretch initiates intracellular calcium oscillations in human mesenchymal stem cells. PLoS One , e109378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby TJ, Lammerding J. (2018). Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol , 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HQ, Ghatak S, Yeung CY, Tellkamp F, Gunschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, Wickstrom SA. (2016). Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol , 864–875. [DOI] [PubMed] [Google Scholar]
- Liu Y-S, Liu Y-A, Huang C-J, Yen M-H, Tseng C-T, Chien S, Lee OK. (2015). Mechanosensitive TRPM7 mediates shear stress and modulates osteogenic differentiation of mesenchymal stromal cells through Osterix pathway. Sci Rep , 16522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. (2013). Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res , 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. (2009). DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther , 1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer CR, Xia Y, Zhu K, Liu D, Irianto J, Garcia VMM, Millan LMS, Niese B, Harding S, Deviri D, et al. (2018). Constricted migration increases DNA damage and independently represses cell cycle. Mol Biol Cell , 1948–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MG, Monhait T, Marko JF. (2002). Reversible hypercondensation and decondensation of mitotic chromosomes studied using combined chemical-micromechanical techniques. J Cell Biochem , 422–434. [DOI] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam HR, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Dumenil AM, Manel N, Piel M. (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science , 359–362. [DOI] [PubMed] [Google Scholar]
- Ranade SS, Syeda R, Patapoutian A. (2015). Mechanically activated ion channels. Neuron , 1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robijns J, Molenberghs F, Sieprath T, Corne TD, Verschuuren M, De Vos WH. (2016). In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci Rep , 30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S, Coppede F, Sagelius H, Eriksson M. (2009). Increased expression of the Hutchinson-Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur J Hum Genet , 928–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal T, Salmon-Divon M, Gerlitz G. (2018). The heterochromatin landscape in migrating cells and the importance of H3K27me3 for associated transcriptome alterations. Cells , 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. (2015). Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell , 4075–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD. (2008). The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev , 3409–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. (2006). Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA , 8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell , 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF. (2018a). Separate roles for chromatin and lamins in nuclear mechanics. Nucleus , 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF. (2019). Chromatin’s physical properties shape the nucleus and its functions. Curr Opin Cell Biol , 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, Adam SA, Goldman RD, Marko JF. (2018b). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell , 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchyna TM, Johnson JH, Hamer K, Leykam JF, Gage DA, Clemo HF, Baumgarten CM, Sachs F. (2000). Identification of a peptide toxin from Grammostola spatulata spider venom that blocks cation-selective stretch-activated channels. J Gen Physiol , 583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MJ, Sharma RV, Wachtel RE, Chapleau MW, Waite LJ, Bhalla RC, Abboud FM. (1997). Non-voltage-gated Ca2+ influx through mechanosensitive ion channels in aortic baroreceptor neurons. Circ Res , 861–867. [DOI] [PubMed] [Google Scholar]
- Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, et al. (2009). A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci USA , 20788–20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajik A, Zhang Y, Wei F, Sun J, Jia Q, Zhou W, Singh R, Khanna N, Belmont AS, Wang N. (2016). Transcription upregulation via force-induced direct stretching of chromatin. Nat Mater , 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiello C, Kamps MA, van den Wijngaard A, Verstraeten VL, Baaijens FP, Broers JL, Bouten CC. (2013). Soft substrates normalize nuclear morphology and prevent nuclear rupture in fibroblasts from a laminopathy patient with compound heterozygous LMNA mutations. Nucleus , 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocco VJ, Li Y, Christopher KG, Matthews JH, Aggarwal V, Paschall L, Luesch H, Licht JD, Dickinson RB, Lele TP. (2017). The nucleus is irreversibly shaped by motion of cell boundaries in cancer and non-cancer cells. J Cell Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. (2012). Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus , 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaevel M, Grevesse T, Gabriele S. (2012). Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun , 671. [DOI] [PubMed] [Google Scholar]
- Weng Y, Yan F, Chen R, Qian M, Ou Y, Xie S, Zheng H, Li J. (2018). PIEZO channel protein naturally expressed in human breast cancer cell MDA-MB-231 as probed by atomic force microscopy. AIP Advances , 055101. [Google Scholar]
- Xia Y, Ivanovska IL, Zhu K, Smith L, Irianto J, Pfeifer CR, Alvey CM, Ji J, Liu D, Cho S, et al. (2018). Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J Cell Biol , 3796–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.