Abstract
Bacteriophages can package part of their host’s genetic material, including antibiotic resistance genes (ARGs), contributing to a rapid dissemination of resistances among bacteria. Phage particles containing ARGs were evaluated in meat, pork, beef and chicken minced meat, and ham and mortadella, purchased in local retailer. Ten ARGs (blaTEM, blaCTX-M-1, blaCTX-M-9, blaOXA-48, blaVIM, qnrA, qnrS, mecA, armA and sul1) were analyzed by qPCR in the phage DNA fraction. The genes were quantified, before and after propagation experiments in Escherichia coli, to evaluate the ability of ARG-carrying phage particles to infect and propagate in a bacterial host. According to microbiological parameters, all samples were acceptable for consumption. ARGs were detected in most of the samples after particle propagation indicating that at least part of the isolated phage particles were infectious, being sul1the most abundant ARG in all the matrices followed by β-lactamase genes. ARGs were also found in the phage DNA fraction of thirty-seven archive chicken cecal samples, confirming chicken fecal microbiota as an important ARG reservoir and the plausible origin of the particles found in meat. Phages are vehicles for gene transmission in meat that should not be underestimated as a risk factor in the global crisis of antibiotic resistance.
Subject terms: Antimicrobial resistance, Bacteriophages
Introduction
Antibiotic resistance is a major threat to public health and food security. A decreasing production of new antimicrobial compounds and a reduced effectiveness of the existing ones caused by the emergence of resistant bacteria is resulting in higher mortality rates1. The uncontrolled use of antibiotics and the ability of bacteria to mutate or acquire external genes is leading to the emergence of resistant strains. Epidemiological studies confirm that antibiotic consumption is directly correlated with the emergence and dissemination of resistance2.
Natural antibiotics have existed for billions of years in bacterial ecosystems as a strategy for adaptation and defense against other bacteria3, resulting in a simultaneous evolution of antibiotic resistance genes (ARGs)4. The resistance originated in environmental reservoirs and transmitted to human pathogens5 has been exacerbated by exposure to antibiotics present in clinical and agricultural settings6,7. Indeed, most of the antibiotics used in medicine, livestock and aquaculture are excreted practically unaltered to the environment, generating a pressure that selects for naturally occurring resistant strains and their accompanying ARG8. Bacteria can be intrinsically resistant to antibiotics or can become resistant by mutation or by acquisition of ARGs by horizontal gene transfer9,10. While conjugation and transformation are well-studied mechanisms, transduction, which is understood as gene transfer mediated by a bacterial virus or bacteriophage/phage, is attracting increasing attention and has perhaps been underestimated11,12.
Three mechanisms of transduction have been described so far; specialized transduction13, mediated by temperate phages that lysogenize the bacterial cell and mobilize the genes adjacent to their insertion site. Generalized transduction14 mediated by virulent phages that package bacterial DNA instead of phage DNA. And the most recent mechanism described, lateral transduction mediated by temperate phages that do not excise from the bacterial chromosome after induction and generate capsids able to package only bacterial DNA located downstream the phage insertion site with great efficiency15. While temperate phages can propagate in a suitable host strain generating phage progeny, transducing particles (containing only bacterial DNA) generated by generalized or lateral transduction, can infect a new host cell but do not possess phage genes and therefore are unable to propagate.
Bacteriophages are globally ubiquitous and extremely abundant, with an estimated 1031 bacteriophages in the biosphere16. There is also a high percentage (from 4 to 68%) of lysogenic bacteria containing inducible prophages in different ecosystems17. Thanks to their structural characteristics, bacteriophages are highly persistent in the environment, the capsid acting as a shield that protects the packaged DNA18,19. Phage particles carrying ARGs have been described in a range of environments, including sludge, soil and sewage20–23. Their origin in sewage is probably human fecal pollution, including healthy humans and hospital wastes, and animal fecal pollution from farms, which is supported by the detection of ARG-containing phage particles in human and animal feces24–26. ARG-containing phage particles have also been found in the human respiratory tract27. Thus, the most plausible origin of these particles, whether in the intestines or respiratory tract, is the microbiota25. Once free ARG-containing phage particles enter the environment, they can end up in food, as shown in a recent study of vegetables classed suitable for human consumption28, and therefore be ingested.
This study focus on the occurrence of ARG-containing phage particles in meat products and, considering that the origin of the particles might be the intestinal microbiota of antibiotic-treated animals, they were also evaluated in chicken cecal samples.
Results
Microbiological parameters of the samples
The morphological types of the colonies grown in TSA were quite homogeneous with few differences. Even if we did not identified the different populations of total aerobic bacteria, we assessed that the majority of the aerobic bacteria were resistant to ampicillin (Table 1) with differences close to 0.5 log10 units between the number of colonies grown in the absence or presence of ampicillin. Dark blue colonies on Chromocult agar were identified as E. coli. These were detected in pork, beef and chicken samples, whereas amp-resistant E. coli (dark blue colonies grown on Chromocult agar containing ampicillin) was detected only in pork and chicken (Table 1). In pork and chicken, the differences were of 0.5 and 0.07 log10 units, respectively between the number of colonies grown with or without ampicillin, showing also that the largest fraction of the E. coli detected was amp-resistant. Somatic coliphages were detected in all samples except mortadella, with an abundance correlating with that of E. coli.
Table 1.
Bacterial and viral indicators.
| Microorganism | Pork | Beef | Chicken | Ham | Mortadella | |
|---|---|---|---|---|---|---|
| n | 10 | 10 | 10 | 5 | 5 | |
| Total aerobic bacteria | % | 80 | 100 | 100 | 100 | 100 |
| Media | 6.2 · 105 | 4.5 · 105 | 2.1 · 106 | 1.0 · 105 | 2.1 · 105 | |
| SD | 3.3 · 102 | 4.3 · 101 | 6.7 · 103 | 1.0 · 103 | 4.2 · 102 | |
| Total aerobic bacteria ampR | % | 40 | 100 | 100 | 100 | 100 |
| Media | 2.4 · 105 | 3.0 · 105 | 6.3 · 105 | 6.7 · 104 | 4.5 · 104 | |
| SD | 1.2 · 101 | 2.0 · 101 | 7.1 · 104 | 8.0 · 104 | 6.6 · 104 | |
| E. coli | % | 30 | 30 | 90 | 0 | 0 |
| Media | 2.3 · 103 | 3.7 · 101 | 3.3 · 103 | <41.6 | <41.6 | |
| SD | 2.7 · 103 | — | 7.3 · 103 | — | — | |
| E. coli ampR | % | 30 | 0 | 90 | 0 | 0 |
| Media | 6.6 · 102 | <41.6 | 2.8 · 103 | <41.6 | <41.6 | |
| SD | 4.7 · 102 | — | 6.7 · 102 | — | — | |
| Somatic coliphages | % | 60 | 50 | 100 | 40 | 0 |
| Media | 3.2 · 102 | 6.7 · 101 | 2.3 · 103 | 1.9 · 101 | <12.5 | |
| SD | 2.5 · 102 | 5.0 · 101 | 2.1 · 103 | 8.8 · 100 | — |
Total aerobic bacteria and E. coli grown in the absence or presence of ampicillin were enumerated as bacterial indicators (CFU/25 g) while somatic coliphages were enumerated as viral indicators (PFU/25 g) in the food samples.
SD Standard deviation.
Minced chicken gave the highest percentage of positive samples for all the indicators and the highest average values of all microorganisms (Table 1), followed by minced pork (not all samples were positive for total aerobic bacteria) and minced beef. E. coli was not detected in ham and mortadella, whereas somatic coliphages were undetectable in mortadella but present in ham, albeit at very low densities.
Detection of ARGs in the DNA from phage particles in meat matrices
The ARGs from the DNA extracted from phage particles in each meat matrix were quantified by qPCR. The samples were considered positive when giving amplification values of the target gene within the limit of quantification (LOQ), which was previously defined with standard curves. Those samples outside the LOQ but within the limit of detection and those that did not show any increase in fluorescence in the qPCR cycles (indeterminate samples) were discounted for the study of gene prevalence.
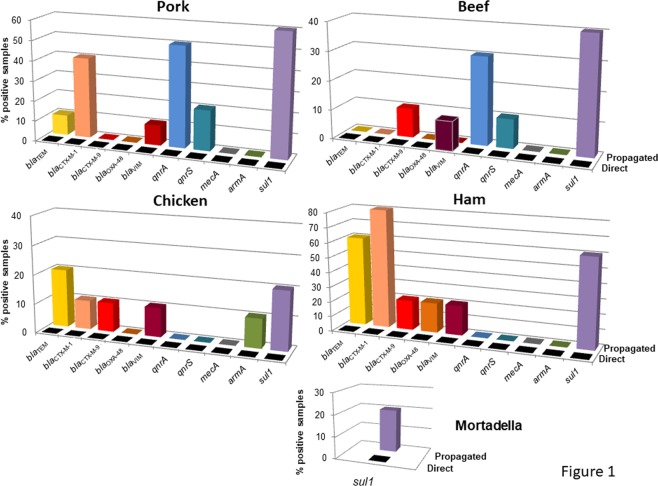
When ARGs were analyzed in the DNA of phage particles extracted directly from each sample, only blaVIM was detected in one beef sample. After propagation, ARGs were identified in DNA from the phage particles in all the matrices, with 10–80% of samples testing positive, depending on the matrix (Fig. 1). The heterogeneous results initially made it difficult to assess which matrix had a higher percentage of positive samples for the different ARGs. In ham, despite only 5 samples being analyzed, 6 ARGs were detected and 4 out of the 5 samples (80%) were positive for blaCTX-M-1. In contrast, in mortadella only sul1 was detected, in only one sample (Fig. 1). Among the minced meats, the highest percentages of samples positive for ARGs were in pork (6 ARGs in up to 60% of samples), followed by chicken and beef (6 ARGs in up to 20% and 5 ARGs in up to 40% of samples, respectively). The most prevalent gene was sul1, followed by blaCTX-M-1 and blaTEM. blaOXA-48 was only detected in one sample of ham, which was the most polluted meat according to the fecal indicators (presence of total aerobic bacteria: 2.24 × 104 CFU/25 g; absence of E. coli but presence of somatic coliphages: 1.25 × 101 PFU/25 g). Accordingly, the same sample of ham also showed the presence of blaTEM, blaCTX-M-1, blaCTX-M-9, blaVIM and sul1.
Figure 1.
Percentage of positive samples for each ARG in each meat matrix before (direct) and after propagation in the E. coli WG5 enrichment culture (propagated). All the values represented are within the limit of quantification.
One chicken sample was positive for armA, a prevalent ARG in poultry. This sample also contained fecal pollution indicators (E. coli and somatic coliphages), although at below average values (Table 1), as well as blaTEM and blaCTX-M-1. The only tested gene undetected in all the samples of this study was mecA.
Abundance of ARGs in the DNA from phage particles
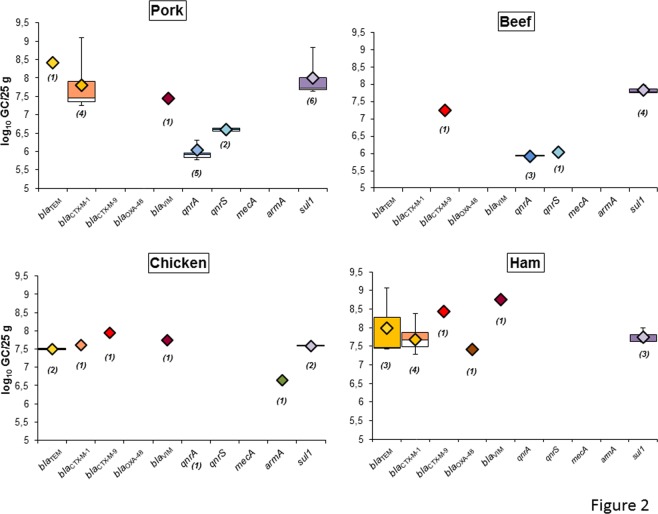
ARG abundance was evaluated in the samples with levels within the LOQ (Fig. 2). This was determined by the last valid Ct for each ARG assay (Table 2) in the standard curve that is consistent in the diverse replicates.
Figure 2.
Abundance of ARGs in the DNA isolated from the phage particles of each meat matrix and ham after propagation in the E. coli WG5 enrichment cultures. Data is represented in boxplots (log10GC/25 g). The diamond represents the average value of the positive samples, the upper squares include samples that present values within the 75th percentile, and in the lower white boxes the values are shown within the 25th percentile. The number of samples appears in brackets.
Table 2.
Oligonucleotides used in this study for PCR and qPCR assays targeting each ARG. For each assay, the amplimer length and the limit of quantification (LOQ) are indicated.
| Target gene | Reaction | Oligonucleotide | Sequence | Amplimer (bp) | LOQ (GC) | Reference |
|---|---|---|---|---|---|---|
| bla TEM | PCR | UP | CTCACCCAGAAACGCTGGTG | 569 | 21 | |
| LP | ATCCGCCTCCATCCAGTCTA | |||||
| qPCR | UP | CACTATTCTCAGAATGACTTGGT | 85 | 7.6 | 65 | |
| LP | TGCATAATTCTCTTACTGTCATG | |||||
| TaqMan TEM | 6FAM-CCAGTCACAGAAAAGCATCTTACGG-MGBNFQ | |||||
| blaCTX-M-1-group | PCR | UP | ACGTTAAACACCGCCATTCC | 356 | 21 | |
| LP | TCGGTGACGATTTTAGCCGC | |||||
| qPCR | UP | ACCAACGATATCGCGGTGAT | 101 | 8.4 | 21 | |
| LP | ACATCGCGACGGCTTTCT | |||||
| TaqMan CTX-M-1 | 6FAM–TCGTGCGCCGCTG-MGBNFQ | |||||
| blaCTX-M-9-group | PCR | UP | ACGCTGAATACCGCCATT | 352 | 26 | |
| LP | CGATGATTCTCGCCGCTG | |||||
| qPCR | UP | ACCAATGATATTGCGGTGAT | 85 | 13 | 26 | |
| LP | CTGCGTTCTGTTGCGGCT | |||||
| TaqMan CTX-M-9 | 6FAM – TCGTGCGCCGCTG- MGBNFQ | |||||
| bla OXA-48 | PCR | UP | CGTTATGCGTGTATTAGCCTTAT | 790 | 25 | |
| LP | TTTTTCCTGTTTGAGCACTTCTTT | |||||
| qPCR | UP | CGGTAGCAAAGGAATGGCAA | 133 | 18.2 | 25 | |
| LP | TGGTTCGCCCGTTTAAGATT | |||||
| TaqMan OXA-48 | 6FAM-CGTAGTTGTGCTCTGGA-MGBNFQ | |||||
| bla VIM | PCR | UP | TCTACATGACCGCGTCTGTC | 748 | 28 | |
| LP | TGTGCTTTGACAACGTTCGC | |||||
| qPCR | UP | AATGGTCTCATTGTCCGTGATG | 61 | 33.9 | 28 | |
| LP | TCGCACCCCACGCTGTA | |||||
| TaqMan VIM | 6FAM –TGATGAGTTGCTTTTGATTG- MGBNFQ | |||||
| sul1 | PCR | UP | TTCATGGGCAAAAGCTTGATG | 965 | 20, 66 | |
| LP | GGCCGGAAGGTGAATGCTA | |||||
| qPCR | UP | CCGTTGGCCTTCCTGTAAAG | 67 | 5.9 | 20 | |
| LP | TTGCCGATCGCGTGAAGT | |||||
| TaqMan sul1 | 6FAM-CGAGCCTTGCGGCGG-MGBNFQ | |||||
| mecA | PCR | UP | GATAGCAGTTATATTTCTA | 434 | 21 | |
| LP | ATACTTAGTTCTTTAGCGAT | |||||
| qPCR | UP | CGCAACGTTCAATTTAATTTTGTTAA | 92 | 10.4 | 67 | |
| LP | TGGTCTTTCTGCATTCCTGGA | |||||
| TaqMan mecA | 6FAM-AATGACGCTATGATCCCAATCTAACTTCCACA-MGBNFQ | |||||
| qnrA | PCR | UP | ACGCCAGGATTTGAGTGAC | 565 | 46 | |
| LP | CCAGGCACAGATCTTGAC | |||||
| qPCR | UP | AGGATTGCAGTTTCATTGAAAGC | 138 | 8.6 | 46 | |
| LP | TGAACTCTATGCCAAAGCAGTTG | |||||
| TaqMan qnrA | 6FAM-TATGCCGATCTGCGCGA-MGBNFQ | |||||
| qnrS | PCR | UP | AAGTGATCTCACCTTCACCGCTTG | 425 | 46 | |
| LP | TTAAGTCTGACTCTTTCAGTGATG | |||||
| qPCR | UP | CGACGTGCTAACTTGCGTGA | 118 | 8.3 | 46 | |
| LP | GGCATTGTTGGAAACTTGCA | |||||
| TaqMan qnrS | 6FAM-AGTTCATTGAACAGGGTGA-MGBNFQ | |||||
| armA | PCR | UP | CAAATGGATAAGAATGATGTT | 774 | 63 | |
| LP | TTATTTCTGAAATCCACT | |||||
| qPCR | UP | GAAAGAGTCGCAACATTAAATGACTT | 94 | 33.4 | 24 | |
| LP | GATTGAAGCCACAACCAAAATCT | |||||
| TaqMan armA | 6FAM-TCAAACATGTCTCATCTATT-MGBNFQ | |||||
| 16SrDNA | qPCR | 338 F | ACTCCTACGGGAGGCAGCAG | 236 | 68 | |
| 518 R | ATTACCGCGGCTGCTGG | |||||
| pGEM | PCR | pGEM7up | TGTAATACGACTCACTAT | Promega |
The abundance of ARGs cannot be considered as an absolute value, because it was ascertained after an enrichment step. Nevertheless, it can be assumed that ARGs with a higher copy number may also have been more abundant in the directly analyzed samples, but at levels below the limit of detection. Also, the detection of ARGs in DNA from phage particles after the enrichment cultures indicates that the phage particles were able to infect and propagate on the host strain and were therefore infectious.
As indicated in the previous section, the only ARG detected in DNA from the phage particles isolated directly from minced beef was blaVIM, at an abundance of 6.2 log10 GC in 25 g of a single sample. This sample showed values of amp-resistant total aerobic bacteria of 2.63 × 104 CFU/25 g, below the average for beef, and an absence of E. coli or amp-resistant E. coli. The inability of the particles harboring blaVIM in this sample to infect E. coli WG5 was demonstrated by the absence of the gene after propagation, suggesting that the particles ceased to be infectious or that the strain was an unsuitable host.
As well as being the most prevalent, sul1 was the most abundant gene on average, reaching densities close to or higher than 108 GC/25 g in all the samples (Fig. 2). In minced pork and ham, blaCTX-M-1 and blaTEM levels were also quite high, reaching 109 GC/25 g (Fig. 2). The genes with the lowest densities were qnrA and qnrS. The other genes gave very heterogeneous signals. In mortadella, only sul1 was detected in a single sample, with values of 107.7 GC/25 g after propagation.
ARGs in DNA in phage particles isolated from chicken feces
We tested 37 archived samples of chicken feces obtained in 2015 from the cecum of broiler chickens at a slaughterhouse covering the area of northeastern Spain. As the chicken meat samples gave the highest values of fecal indicators (Table 1), the ARGs in the phage DNA fraction of chicken fecal samples were evaluated for any correlation with the high levels of ARGs detected in chicken meat.
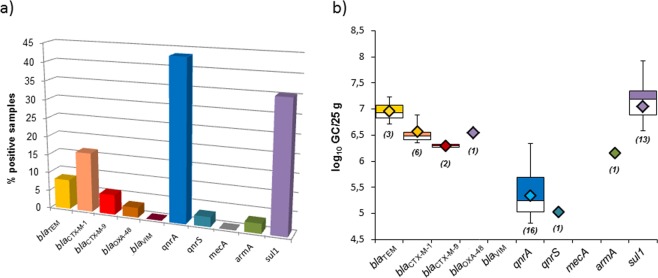
In this set of samples, microbiological parameters were not analyzed and ARGs were only screened in the DNA of phages directly isolated from the feces without any propagation step. As these archive samples had been frozen for three years, it was considered that viable microorganisms and infectious viruses would have been partially or totally inactivated during storage. In addition, the results (Fig. 3) showed that the concentrations of ARGs in the feces were sufficiently high to be detected without further propagation. The most prevalent genes were qnrA and sul1, (Fig. 3a), a pattern more similar to the beef and pork samples than to the chicken meat.
Figure 3.
ARGs in phage particles in chicken feces. (a) Percentage of positive samples for each ARG in the fecal matrix. All the values represented are within the limit of quantification. (b) Abundance of each ARG in chicken feces represented in boxplots (log10GC/25 g). The diamond represents the average value of the positive samples, the colored upper squares include samples that present values within the 75th percentile, and in the lower white boxes the values are shown within the 25th percentile. The number of samples appears in brackets.
The most abundant ARG (Fig. 3b) in chicken feces was again sul1. The abundance of quinolone resistance genes was lower, although qnrA was present in 43% of the samples. Compared to sul1 or qnrA, the β-lactamases blaTEM, blaCTX-M-1 and blaCTX-M-9 were found in a smaller number of samples, but at high densities of 106–107 GC/25 g. The level of blaOXA-48 was similar to that of the other β-lactamases in only one sample, and blaVIM was not detected at all. As in all the other analyses, mecA was not found. Finally, armA was detected in only one sample of chicken feces, as occurred in chicken meat, but with a high abundance, only slightly lower than that of the β-lactamase genes (Fig. 3b).
Discussion
This study focuses on the prevalence of ARGs in bacteriophages, ubiquitous bacterial viruses that have been reported in diverse environments21,22,29–31, in human25 and animal feces26, and more recently in vegetables and agricultural soils28,32. Bacteriophages facilitate ARG transmission between bacteria, as transduction does not require direct contact between “donor” and “recipient” cells, although the transfer mechanism is not completely understood. Phages can transmit ARGs through generalized transduction, as reported for Salmonella phages in chicken meat33 or in veterinary settings34. Particles mobilizing ARGs can also be produced in temperate phages induced from bacterial strains25, resulting in ARG transfer35. Recently a new lateral transduction mechanism has been reported15, by which phages package and spread the bacterial chromosome at high frequencies.
Regardless of the mechanism involved, the injection of the mobilized DNA into the recipient strain depends only on the phage capsid. On the other hand, the ability of the transducing particles to propagate in a host cell requires expression of the phage genes, and particles generated through generalized or lateral transduction only possess bacterial DNA. According with previous studies of our group21,25,28, transducing particles containing only bacterial DNA (including ARG) are abundant. These results are in accordance with other authors that indicate that mostly bacteriophages, understood as phage capsids containing complete phage genomes, rarely carry antibiotic resistance genes36. Instead, transducing particles containing bacterial DNA (including ARG) are not considered when analyzing phage genomes, and might be confounded with bacterial DNA contamination. In the current study however, at least a fraction of the phages were able to propagate, indicating that these were infectious and possessed phage genes in addition to the ARGs. Only one beef sample, in which blaVIM-containing particles were directly isolated, became negative after the enrichment culture. This case suggests the particles were unable to propagate and the ARG may have been lost after injection in the host cell without a subsequent generation of phage progeny. The infectiveness of the particles is an indication of their ability to attach the recipient cell and inject their DNA, the first steps required for transduction. Our attempts to detect transductants that acquired one of the ARGs were unsuccessful, but this limitation could be explained by several causes: the particles in the samples were able to propagate but not able to transduce the ARG; the recipient host did not possess the proper site to allow recombination of the ARG hence the frequency of transduction was too low to be detected; or even that other virulent phages in the samples propagated and killed the transductant cells before they were isolated, considering that the transducing particles might not generate superinfection immunity in the recipient cell37. Probably some of the particles detected in the samples were infectious phages, this is phage capsids with a complete phage genome that incorporated the resistance gene
The food industry has long been subject to a very strict regulation of various microbiological parameters. In minced meat the maximum values of E. coli established by the European Union are between 50 and 500 CFU/g38. Our samples fulfilled the requirements and did not exceed these values, although their microbiota contained a significant population resistant to β-lactam antibiotics. The detected ARGs exhibited considerable heterogeneity in prevalence and/or abundance. Nevertheless, as indicated above, as the abundance is a consequence of the propagation steps, the densities obtained can only be considered as indicative.
All the samples contained sul1 in densities of up to 107–8 GC/25 g. This ubiquity may be explained by the frequent use of sulfonamides in veterinary medicine in the European Union39,40. β-lactamase genes were also highly prevalent, again in accordance with current veterinary use41 and the pervasiveness of extended-spectrum β-lactamase-producing Enterobacteriaceae in animal feces42–44 as well as the presence of ARGs in the phage DNA fraction in animal wastewater. Worthy of note is the detection of blaOXA-48, an emerging carbapenemase that hinders treatment with carbapenems. Besides its clinical significance, it has previously been detected chromosomically in bacteria from food-producing animals45 and in DNA in phage particles isolated from agricultural settings28. Quinolone-resistance genes, particularly qnrA, were prevalent in pork and beef and also present in chicken feces in moderate abundance, in agreement with observations in animal wastewater46. Quinolones are frequently administered in cattle and swine47, although their use in animals is restricted to the treatment of infections48. armA was found in chicken meat and feces in moderate levels. It has previously been reported in Salmonella or E. coli from pigs and poultry, alone or in combination with other ARGs49. Finally, the non-detection of mecA in our samples could be due to its absence, or to its relation to Staphylococcus sp.50; phage particles containing mecA and generated from Staphylococcus might be unable to propagate in E. coli and therefore be undetectable.
Both ham and mortadella undergo thermal processing in their preparation, which largely eliminates microorganisms. Further, it is not ruled out that these products contain preservatives that prevent the growth of microorganisms. In spite of similar levels of culturable microbiota, ham showed more infective ARG-phage particles than mortadella. This could be due to an incorrect manipulation in the shop where it was bought, or perhaps because ham is more commonly consumed than mortadella and therefore suffers more manipulation in a single day. Taking into account that each sample was purchased in a different establishment, cross-contamination may be relatively common since at least one ARG was detected in phage particles in all samples.
The Joint Inter-agency Antimicrobial Consumption and Resistance Analysis Spain47 estimated the antibiotic consumption by the most relevant animal producing species. The report shows that swine group most treated with β-lactam antibiotics and quinolones (58 and 60% respectively), partially explaining the prevalence and ARG abundance in pork and ham in this study, being cattle the second most treated group (27–28) followed byo poultry (6–7%).
The chicken feces were found to contain ARG-packaging phage particles, as previously observed in human feces25, which points towards feces as a source of the phage particles found in chicken meat. Intensive poultry production, one of the fastest growing industries in the world, greatly depends on antimicrobials to prevent and treat diseases. The prophylactic use of antibiotics is difficult to eradicate due to the fear of infections spreading through the chicken farms. However, such an intensive use of antibiotics implies a very high selective pressure and the promotion of resistance transfer events within the chicken microbiota51.
The most prevalent genes in the chicken fecal samples were sul1 and qnrA. Sulfonamides and fluoroquinolones are strongly adsorbed to feces40, persisting for a long time after excretion and elimination. The constant presence of antibiotics may select these resistances in the fecal microbiota, which are subsequently spread in phage particles. Thus, the fecal contamination in the samples, although low, could be the origin of the ARG-containing phage particles detected. The presence of fecal contamination in chicken meat is attributed to breeding conditions and the processing and handling of the meat before selling. When meat is crushed, microorganisms present on the surface can penetrate the whole product, so thorough cooking is vital before consumption52.
Antimicrobials are administered in livestock for preventive and/or therapeutic purposes, but they have also been used as growth promoters to increase the efficiency of animal production, a practice prohibited in Europe since 2006 (EC Regulation No. 1831/2003)53. The selective pressure arising from this massive and indiscriminate use of antibiotics enhances the mobility of antibiotic resistance genes (ARGs) and the subsequent appearance of resistant bacteria54. Moreover, phages persist to certain treatments used for the processing of food or water samples, such as chlorination, thermal treatment or high hydrostatic pressure (HHP)55. Food additives such as EDTA or sodium citrate46 may promote the induction of the lytic cycle of certain prophages that will be spread in the food matrix, in the environment or be ingested and reach the human gut, being able to transduce DNA56.
At the moment there is no legislation to control antibiotic resistant bacteria or the ARGs and genetic elements that mobilize them. The official incorporation of phage detection methods as indicators is only beginning to be envisaged57. Meanwhile, the detection of phages is not yet part of food safety practice, including that of phages potentially capable of transferring ARGs to the natural human microbiota as well as to pathogens.
The expansion of resistances will take the world into a post-antibiotic era, where many common and minor infections will again become life-threatening1 unless urgent methods are taken.
Levels of ARGs in the samples were quite heterogeneous. The most striking result was the prevalence and abundance of sul1 and β-lactamase genes, related with commonly used antibiotics in veterinary medicine, and the non-detection of mecA. The propagation experiments showed that part of the phage particles carrying ARGs were infective. Since infection is the first step in ARG transduction, this finding provides further evidence for the role of phage particles in the horizontal transmission of ARGs between bacteria, or between animals through cross-contamination, or to humans via the food chain.
In the absence of any type of food safety regulations aimed at monitoring bacteriophages, their consumption by the population constitutes a health risk factor. Hence, there is a need for further study of ARG transmission by phages through the food chain and the possible transfer of ARGs between commensal bacteria and pathogens.
Methods
Samples
The minced meat samples used were classified into 3 groups, pork, beef, and chicken, with 10 samples of each; the sliced meat analyzed consisted of 5 samples of ham and 5 of mortadella. All samples were purchased from local retailers in the area of Barcelona (Spain) during 2017–2018. All of them were considered to be fresh samples as they had not undergone any packaging or freezing process and were analyzed within 24 hours after purchase.
Thirty-seven archived samples of chicken feces (cecal contents) of broiler chickens from different flocks and farms from northeastern Spain were collected at the slaughterhouse in 2015 and stored at −20 °C.
Twenty grams of each sample were homogenized in 60 ml of phage buffer, using the Stomacher homogenizer (IUL Instruments GmbH, Königswinter, Germany) for 2 minutes. Stomacher bags with filters (Afora, Barcelona, Spain) were used to improve the separation of solid waste from the liquid fraction containing the microorganisms.
Bacterial and viral indicators
Total aerobic microorganisms and total aerobic microorganisms resistant to ampicillin (amp) in the homogenate were evaluated on Tryptone Soy agar (TSA) or TSA with amp (100 μg/ml) at 37 °C.
Total Escherichia coli and E. coli resistant to amp in the homogenates were determined on Chromocult® Coliform Agar (Merck, Darmstadt, Germany) or Chromocult® Coliform Agar with amp (100 μg/ml) respectively. Incubation was first performed for 2 hours at 37 °C to adapt potentially damaged microorganisms and then overnight at 44 °C. Ten percent of the blue colonies presumed to be E. coli were confirmed with the Indole test and grown in McConkey agar.
Somatic coliphages, proposed as viral indicators of fecal pollution58, were evaluated to determine the presence of fecal viruses in the samples. Ten ml of homogenates obtained as above were centrifuged for 15 minutes at 4000xg and the supernatant was filtered through 0.22 μm low protein binding polyethersulfone membranes (PES) (Millex -GP, Millipore, Bedford, MA). Ten-fold dilutions of the filtrates were analyzed in duplicate for the presence of somatic coliphages following the ISO standard method59 that uses E. coli strain WG5 (ATCC 700078) as the bacterial host. Plates were incubated at 37 °C for 18 h.
Each homogenate was analyzed for all the indicators in duplicate.
Chicken feces
Four g of feces were homogenized in 16 ml of phage buffer using a horizontal agitator at 800 rpm for 10 minutes and then centrifuged at 4000 × g for 25 minutes. The supernatant was collected, and centrifugation was repeated under the same conditions. The final supernatant was filtered through 0.22 μm low protein binding polyethersulfone membranes (PES) (Millex -GP, Millipore, Bedford, MA) and used for the extraction of phage particles and DNA in the particles. In this case, microbial indicators were not analyzed, as these were archival samples kept at −20 °C since their collection in 2015.
Extraction of phage particles
One ml of the homogenate filtered as described above was treated with chloroform (1:10 (v/v)) to remove possible DNA carrier vesicles. Briefly, chloroform was added to the sample, which was vigorously vortexed for 5 minutes and centrifuged at 16000 × g for 5 minutes. The supernatant was recovered and incubated with DNase (100 units/mL of the supernatant) to eliminate any free DNA present in the samples outside the phage particles. DNase was inactivated by heating for 5 minutes at 75 °C and an aliquot was taken as a control. These lysates were used for extraction of DNA in the phage particles or to evaluate the infectivity of the phage particles by propagation.
Controls were performed to confirm the removal of DNA by the DNase treatment and the correct inactivation of DNase by heat treatment. The procedure was done as described previously60.
Extraction of DNA in the phage particles
The protocol for DNA extraction was as previously described61 with some modifications. After the treatment with chloroform and DNase, aliquots of 50 μl were stored at −20 °C as negative controls for the presence of non-encapsidated DNA. The extraction of DNA from the phage capsids was continued with the remaining phage suspension. To break the capsids and release the genetic material, the samples were treated with proteinase K (20 mg/ml) in 250 μl of proteinase K buffer and incubated for 1 h at 55 °C. Encapsidated DNA was extracted by phenol-chloroform (1:1) (v:v) treatment and the aqueous phase was again treated with chloroform (1:1) (v:v) by centrifuging at the same speed and time as in the previous step. The remaining phenol/chloroform was removed by adding the mixture to Phase Lock Gel Tubes (5- Prime, Hucoa Erlöss, Madrid, Spain) and centrifuging following the manufacturer’s instructions. DNA was precipitated using 100% ethanol and 3M sodium acetate, and resuspended in 100 µl of ultrapure water. DNA was quantified using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientifics, Wilmington, DE).
Amplification of ARGs in DNA of phage particles
Quantitative real-time PCR (qPCR) using TaqMan hydrolysis probes was conducted with the StepOne Real Time PCR System (Applied Biosystems) in a 20 μl reaction mixture with the TaqMan® Environmental Master Mix 2.0 (Applied Biosystems). The results were analyzed with the Applied Biosystems StepOne™ Instrument program.
Ten qPCR assays targeting ARGs were performed in 9 μl of the sample DNA or quantified plasmid DNA. The ARGs were selected from groups relevant in clinics, abundant in the environment, or that confer resistance to different antibiotic groups used in veterinary. These included β-lactamase genesthat confer resistance to β-lactam antibiotics (blaTEM, blaCTX-M-1 group, blaCTX-M-9 group, blaOXA-48 and blaVIM), two quinolone resistance genes (qnrA and qnrS), a gene conferring resistance to methicillin (mecA), commonly found in Staphylococcus21,46, sul1, which confers resistance to sulfonamides and is frequently found in environmental and clinical bacterial populations62, and armA, which encodes aminoglycoside resistance and is widely distributed in Enterobacteriaceae63.
For quantification, standards of known concentration containing a fragment of each ARG were prepared. Each ARG was amplified by conventional PCR, using an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Barcelona, Spain) with the primers described in Table 2. The amplicons were purified and cloned into a pGEM-T Easy vector for insertion of PCR products (Promega, Barcelona, Spain). The correct cloning of the fragment in the vector was confirmed by PCR using primer pGEM7up (Table 2) and the construct was used to generate the standard curves as previously described21. The standards were also used as positive controls.
All samples were performed in triplicate (including the standards and negative controls). The number of gene copies (GC) was defined as the mean of the triplicate data obtained. To quantify the ARGs, we considered the GC results obtained within the threshold cycle (Ct) within the limit of quantification (LOQ).
Negative controls to exclude contamination with non-encapsidated DNA
To rule out contamination with DNA outside the phage particles (either bacterial or free DNA present in the samples), the aliquot of each sample taken after DNase treatment and before desencapsidation by proteinase K digestion, and kept at −20 °C, was screened by qPCR for each ARG and for bacterial 16SrDNA (Table 2). These controls were expected to be negative after the removal of all non-encapsidated DNA by the DNase. The ARGs were evaluated by qPCR assays as described above and in Table 2, and the absence of bacterial 16S rDNA was verified by qPCR using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and primers 338F/518R (Table 2).
Propagation cultures
Phage particles in the samples were propagated in enrichment cultures of the strain E. coli WG5 as a host bacterium. This strain, used for the somatic coliphage count, was selected because of its sensitivity to phage infection28 and because it does not harbor any of the ARGs targeted in this study or any prophage64.
To evaluate the infectivity of the phage particles carrying ARGs, the analysis of ARGs by qPCR was carried out both before (direct quantification from the sample) and after phage propagation in the enrichment cultures. As the chicken feces were archive samples and not expected to contain infectious phages, the propagation experiments were only conducted with phages in the meat samples. If phages carrying ARGs were able to infect and propagate on the host strain, the ARG levels would be expected to increase due to the higher concentration of phage particles. If ARG levels decreased, the particles would be considered incapable of propagating.
The propagation culture was prepared with one ml of phage particles after filtration and DNAse treatment and one ml of E. coli WG5 at the exponential phase (OD600 0.3) in 8 ml of Luria-Bertrani broth (LB). This mixture was incubated for 18 h at 37 °C with shaking. After incubation, the phages were purified, and DNA in phage particles was extracted as indicated above.
Acknowledgements
This work was supported by the Spanish Ministerio de Innovación y Ciencia (AGL2016-75536-P), the Agencia Estatal de Investigación (AEI) and the European regional fund (ERF), the Generalitat de Catalunya (2017SGR170) and the Centre de Referència en Biotecnologia (XeRBa). CERCA Programme from the Generalitat de Catalunya is also acknowledged. M.B.-J. has a grant from COLCIENCIAS (Republic of Colombia). P.B.-P. has a grant from the Spanish Ministry of Economy, Industry and Competitiveness (BES-2017-081296). L.R.-R. is supported by the Beatriu de Pinos postdoctoral programme of the Government of Catalonia’s Secretariat for Universities and Research of the Ministry of Economy and Knowledge.
Author Contributions
Conception and design: M.M. Acquisition of data: C.G.-G., P.B.-P., M.C.-C. Laboratory analysis: C.G.-G., M.B.-J., P.Q. Analysis and interpretation of data: M.M., L.R.-R., Drafting Manuscript: C.G.-G., M.M. Review of manuscript: L.R.-R., M.C.-C. and M.M.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. Antibiotic resistance at, https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (2018).
- 2.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T a peer-reviewed J. Formul. Manag. 2015;40:277–83. [PMC free article] [PubMed] [Google Scholar]
- 3.Baquero F, Tedim AP, Coque TM. Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 2013;4:15. doi: 10.3389/fmicb.2013.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perron GG, et al. Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS One. 2015;10:1–19. doi: 10.1371/journal.pone.0069533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen HK, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–9. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 6.Czekalski N, Sigdel R, Birtel J, Matthews B, Bürgmann H. Does human activity impact the natural antibiotic resistance background? Abundance of antibiotic resistance genes in 21 Swiss lakes. Environ. Int. 2015;81:45–55. doi: 10.1016/j.envint.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Wu S, Carvalho PN, Müller JA, Manoj VR, Dong R. Sanitation in constructed wetlands: A review on the removal of human pathogens and fecal indicators. Sci. Total Environ. 2016;541:8–22. doi: 10.1016/j.scitotenv.2015.09.047. [DOI] [PubMed] [Google Scholar]
- 8.Baquero F, Martinez JL, Canton R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008;19:260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Von Wintersdorff, C. J. H. et al. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 7, (2016). [DOI] [PMC free article] [PubMed]
- 10.Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 11.Stanczak-Mrozek KI, Laing KG, Lindsay JA. Resistance gene transfer: Induction of transducing phage by sub-inhibitory concentrations of antimicrobials is not correlated to induction of lytic phage. J. Antimicrob. Chemother. 2017;72:1624–1631. doi: 10.1093/jac/dkx056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spano AJ, et al. In vitro assembly of a prohead-like structure of the Rhodobacter capsulatus gene transfer agent. Virology. 2007;364:95–102. doi: 10.1016/j.virol.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 13.Thierauf A, Perez G, Maloy AS. Generalized transduction. Methods Mol. Biol. 2009;501:267–86. doi: 10.1007/978-1-60327-164-6_23. [DOI] [PubMed] [Google Scholar]
- 14.Thierauf, A., Perez, G. & Maloy, S. In Bacteriophages Methods and Protocols (eds Clokie, M. R. J. & Kropinski, A. M.) 267–286 (Humana Press, 2009).
- 15.Chen J, et al. Genome hypermobility by lateral transduction. Science (80-.). 2018;362:207–212. doi: 10.1126/science.aat5867. [DOI] [PubMed] [Google Scholar]
- 16.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 17.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 2003;6:417–24. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 18.Romanowski G, Lorenz MG, Sayler G, Wackernagel W. Persistence of free plasmid DNA in soil monitored by various methods, including a transformation assay. Appl. Environ. Microbiol. 1992;58:3012–9. doi: 10.1128/aem.58.9.3012-3019.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu B. Degradation of plasmid and plant DNA in water microcosms monitored by natural transformation and real-time polymerase chain reaction (PCR) Water Res. 2006;40:3231–8. doi: 10.1016/j.watres.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 20.Calero-Cáceres William, Melgarejo Ana, Colomer-Lluch Marta, Stoll Claudia, Lucena Francisco, Jofre Juan, Muniesa Maite. Sludge As a Potential Important Source of Antibiotic Resistance Genes in Both the Bacterial and Bacteriophage Fractions. Environmental Science & Technology. 2014;48(13):7602–7611. doi: 10.1021/es501851s. [DOI] [PubMed] [Google Scholar]
- 21.Colomer-Lluch, M., Jofre, J. & Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One6, (2011). [DOI] [PMC free article] [PubMed]
- 22.Lekunberri I, Subirats J, Borrego CM, Balcázar JL. Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 2017;220:981–984. doi: 10.1016/j.envpol.2016.11.059. [DOI] [PubMed] [Google Scholar]
- 23.Balcazar JL. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014;10:e1004219. doi: 10.1371/journal.ppat.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quirós P, et al. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014;58:606–9. doi: 10.1128/AAC.01684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown-Jaque M, et al. Antibiotic resistance genes in phage particles isolated from human feces and induced from clinical bacterial isolates. Int. J. Antimicrob. Agents. 2018;51:434–442. doi: 10.1016/j.ijantimicag.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 26.Colomer-Lluch M, Imamovic L, Jofre J, Muniesa M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 2011;55:4908–4911. doi: 10.1128/AAC.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown-Jaque M, et al. Detection of Bacteriophage Particles Containing Antibiotic Resistance Genes in the Sputum of Cystic Fibrosis Patients. Front. Microbiol. 2018;9:856. doi: 10.3389/fmicb.2018.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larrañaga O, et al. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018;115:133–141. doi: 10.1016/j.envint.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Muniesa M, et al. Bacteriophages and diffusion of beta-lactamase genes. Emerg. Infect. Dis. 2004;10:1134–7. doi: 10.3201/eid1006.030472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunathilaka GU, Tahlan V, Mafiz AI, Polur M, Zhang Y. Phages in urban wastewater have the potential to disseminate antibiotic resistance. Int. J. Antimicrob. Agents. 2017;50:678–683. doi: 10.1016/j.ijantimicag.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Anand T, et al. Abundance of antibiotic resistance genes in environmental bacteriophages. J. Gen. Virol. 2016;97:3458–3466. doi: 10.1099/jgv.0.000639. [DOI] [PubMed] [Google Scholar]
- 32.Ross J, Topp E. Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction. Appl. Environ. Microbiol. 2015;81:7905–13. doi: 10.1128/AEM.02363-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shousha A, et al. Bacteriophages Isolated from Chicken Meat and the Horizontal Transfer of Antimicrobial Resistance Genes. Appl. Environ. Microbiol. 2015;81:4600–6. doi: 10.1128/AEM.00872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilbert M, Csadek I, Auer U, Hilbert F. Antimicrobial resistance-transducing bacteriophages isolated from surfaces of equine surgery clinics – a pilot study. Eur. J. Microbiol. Immunol. 2017;7:296–302. doi: 10.1556/1886.2017.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y-D, Park J-H. Phage Conversion for Beta-Lactam Antibiotic Resistance of Staphylococcus aureus from Foods. J. Microbiol. Biotechnol. 2016;26:263–269. doi: 10.4014/jmb.1508.08042. [DOI] [PubMed] [Google Scholar]
- 36.Enault F, et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2017;11:237–247. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bondy-Denomy J, et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016;10:2854–2866. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rees C.E.D., Doyle L., Taylor C.M. Foodborne Diseases. 2017. Listeria monocytogenes; pp. 253–276. [Google Scholar]
- 39.Garcia-Galan M, Díaz-Cruz M. Combining chemical analysis and ecotoxicity to determine environmental exposure and to assess risk from sulfonamides. TrAC Trends Anal. Chem. 2009;28:804–819. doi: 10.1016/j.trac.2009.04.006. [DOI] [Google Scholar]
- 40.Kemper N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008;8:1–13. doi: 10.1016/j.ecolind.2007.06.002. [DOI] [Google Scholar]
- 41.De Briyne N, Atkinson J, Pokludová L, Borriello SP. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014;175:325. doi: 10.1136/vr.102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesa R. J. Extended-spectrum -lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage) Journal of Antimicrobial Chemotherapy. 2006;58(1):211–215. doi: 10.1093/jac/dkl211. [DOI] [PubMed] [Google Scholar]
- 43.Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008;13:19044. [PubMed] [Google Scholar]
- 44.European Commission. Surveillance of antimicrobial resistance in Europe – Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). 1831/2003/EC (2005).
- 45.Ceccarelli D, et al. Chromosome-encoded blaOXA-48 -like variants in Shewanella spp. from food-producing animals, fish and the aquatic environment. Antimicrob. Agents Chemother. 2016;61:AAC.01013–16. doi: 10.1128/AAC.01013-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colomer-Lluch M, Jofre J, Muniesa M. Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J. Antimicrob. Chemother. 2014;69:1265–1274. doi: 10.1093/jac/dkt528. [DOI] [PubMed] [Google Scholar]
- 47.Alonso, M. et al. Informe JIACRA Spain. Primer análisi integrado del consumo de antibióticos y su relación con la aparición de resistencia. Plan Nacional de Resistencia a Antibióticos, at, http://www.resistenciaantibioticos.es/es/system/files/field/files/informe_jiacra-espana.pdf?file=1&type=node&id=410&force=0 (2018).
- 48.Collignon Peter. Fluoroquinolone Use in Food Animals. Emerging Infectious Diseases. 2005;11(11):1789–1792. doi: 10.3201/eid1111.040630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du X-D, et al. Tn1548-associated armA is co-located with qnrB2, aac(6′)-Ib-cr and blaCTX-M-3 on an IncFII plasmid in a Salmonella enterica subsp. enterica serovar Paratyphi B strain isolated from chickens in China. J. Antimicrob. Chemother. 2012;67:246–248. doi: 10.1093/jac/dkr407. [DOI] [PubMed] [Google Scholar]
- 50.Köck R, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010;15:19688. doi: 10.2807/ese.15.41.19688-en. [DOI] [PubMed] [Google Scholar]
- 51.Xiong W, et al. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. 2018;6:34. doi: 10.1186/s40168-018-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luber P. Cross-contamination versus undercooking of poultry meat or eggs - which risks need to be managed first? Int. J. Food Microbiol. 2009;134:21–8. doi: 10.1016/j.ijfoodmicro.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 53.Hughes, P. & Heritage, J. In Antibiotic growth promoters in food animals (ed. Food and Agriculture Organization of the United Nations (FAO)) 129–152, at, http://www.fao.org/tempref/docrep/fao/007/y5159e/y5159e05.pdf (2004).
- 54.Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14:176–82. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Calero-Cáceres W, Muniesa M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016;95:11–8. doi: 10.1016/j.watres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 56.Tóth I, et al. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 2003;69:7242–7. doi: 10.1128/AEM.69.12.7242-7247.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.U.S. EPA. 2016 Coliphage Experts Workshop: Discussion Topics and Findings. Office of Water. EPA 823-F-16F001 (2016).
- 58.Jofre, J. In Human Viruses in Water (ed. Bosch, A.) 17, 227–249 (Elsevier, 2007).
- 59.Anonymous. ISO 10705-2: Water quality. Detection and enumeration of bacteriophages -part 2: Enumeration of somatic coliphages (2000).
- 60.Colomer-Lluch M, et al. Antibiotic resistance genes in bacterial and bacteriophage fractions of Tunisian and Spanish wastewaters as markers to compare the antibiotic resistance patterns in each population. Environ. Int. 2014;73:167–75. doi: 10.1016/j.envint.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook, J. & Russell, D. Molecular Cloning: A Laboratory Manual. Cold Spring Harb. Lab. Press. Cold Spring Harb. NY 999. at, http://books.google.com/books?id=YTxKwWUiBeUC&printsec=frontcover\npapers2://publication/uuid/BBBF5563-6091-40C6-8B14-06ACC3392EBB (2001).
- 62.Pruden A, Arabi M, Storteboom HN. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012;46:11541–9. doi: 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- 63.Galimand M, Sabtcheva S, Courvalin P, Lambert T. Worldwide Disseminated armA Aminoglycoside Resistance Methylase Gene Is Borne by Composite Transposon Tn1548. Antimicrob. Agents Chemother. 2005;49:2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imamovic, L. et al. Complete genome sequence of Escherichia coli strain WG5. Genome Announc. 6, (2018). [DOI] [PMC free article] [PubMed]
- 65.Lachmayr KL, Kerkhof LJ, Dirienzo AG, Cavanaugh CM, Ford TE. Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 2009;75:203–11. doi: 10.1128/AEM.01254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011;14:236–43. doi: 10.1016/j.mib.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 67.Volkmann H, Schwartz T, Bischoff P, Kirchen S, Obst U. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan) J. Microbiol. Methods. 2004;56:277–86. doi: 10.1016/j.mimet.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 68.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]