Abstract
Hepatic encephalopathy is a frequent and debilitating complication of liver disorders. Lactulose is an established and reasonably effective treatment, yet with incompletely understood mechanisms of action. The aims of this study were to examine how the faecal microbiota composition changed before, during and after lactulose treatment in a large animal model. Healthy, privately owned dogs (n = 18) completed a prospective cohort study. Faecal samples were collected weekly, while the subjects were either on their usual diet (week 1), or a standardised diet (weeks 2–9), with added oral lactulose in weeks 6–7. DNA extraction and 16S rRNA gene sequencing were undertaken. Faecal samples from week 7 had a significantly lower microbiota richness/diversity, based on observed operational taxonomic units, Shannon/Chao1 indexes and Pielou’s Evenness. Beta diversity based on UniFrac distances was significantly different in week 7 compared to weeks 1, 5 and 9. At the phylum level, week 7 was associated with a significant increase of Firmicutes and Actinobacteria, and a decrease of Bacteroidetes and Fusobacteria, when compared to weeks 5 and 9. In summary, we have shown that lactulose induces a reversible qualitative and quantitative change of the faecal microbiota, which may explain its clinical efficacy in the management of hepatic encephalopathy.
Subject terms: Microbial ecology, Liver cirrhosis
Introduction
Hepatic encephalopathy (HE) is a frequent and debilitating neurological complication in patients with liver disease. Severe (grades 3–4) HE is associated with higher in-hospital and 30-day mortality, independently of extrahepatic organ failures1, and higher liver transplantation 90-day wait list mortality2. Covert (minimal and grade 1) HE directly results in human morbidity, being an independent predictor of reduced health-related quality of life and poor sleep quality3. Furthermore, HE contributes to a substantial economic burden. In the USA alone, total HE-related hospitalisation charges amounted to $7.245 billion in 20094, and up to $58,625 per patient in 20125.
The pathogenesis of HE is not fully understood6. Several neurotoxins and precipitating factors have been implicated, with ammonia being the most well characterised one. In advanced liver disease, this gut-derived neurotoxin may accumulate in the blood and in the brain, due to lack of hepatic conversion into urea and subsequent urinary excretion7. Clinically, in human cirrhosis, plasma ammonia has been correlated with both the severity of HE and the frequency of other organ failures, and was identified as an independent predictor of 28-day mortality8.
Lactulose, a synthetic non-absorbable disaccharide, is a commonly used medication, with or without the addition of the antibiotic rifaximin, for both the treatment and prevention of HE, with a reasonable evidence of efficacy and added benefits in reducing morbidity and mortality9,10. The postulated benefits of lactulose include: (1) decreased colonic transit time and pH, leading to decreased ammonia production and absorption; (2) increased bacterial assimilation of ammonia; (3) decreased bacterial generation of ammonia; (4) production shift from toxic to non-toxic short-chain fatty acids (SCFA); and (5) reduced bacterial DNA translocation11,12. Yet, its mechanisms of action remain incompletely elucidated.
Faecal dysbiosis is known to occur in covert and overt HE. Cirrhotic patients with minimal HE (MHE) harbour a higher proportion of urease-producing Streptococcus salivarius in stool samples, positively correlating with serum ammonia accumulation13. Additionally, the cirrhosis dysbiosis ratio (CDR), a previously validated ratio of autochthonous to non-autochthonous taxa in stool samples of cirrhotic patients, is lowered after development of severe HE, indicating worsened dysbiosis, and associated with 30-day mortality and organ failure14. Moreover, the presence of specific bacterial families (Alcaligenaceae, Porphyromonadaceae, Enterobacteriaceae) is strongly associated with poor cognition and inflammation in HE patients15. Considering the ongoing evidence regarding microbiome disruption in cirrhosis and HE16, it is likely that manipulation of the microbiota may contribute to improved outcomes. Interventions with proposed positive impact have so far included probiotics17, diet18 and faecal microbiota transplantation19.
The impact of lactulose in ameliorating cirrhosis and HE-associated faecal dysbiosis is controversial. A direct impact has been supported by studies based on culture-dependent methodologies, namely: increased Bifidobacterium, Lactobacillus and Bacteroidaceae colonies, and reduced Enterobacteriaceae, Enterococcus and yeasts in patients with MHE, alongside with improved blood ammonia, psychometric tests and reduced risk of developing overt HE20; and increased total aerobic and anaerobic bacterial counts, and lactobacilli in cirrhotic patients without clinical HE, alongside with decreased faecal pH21. Conversely, studies based on culture-independent techniques have not substantiated an effect of lactulose in the microbiome of cirrhotic patients without HE22 and have reported only a minimal change in cirrhotic patients with HE14, including after lactulose withdrawal23.
However, no study using next generation sequencing techniques has assessed quantitative and qualitative changes of the intestinal or faecal microbiome, i.e. changes based on both the abundance and the presence or absence of microbial communities, after lactulose treatment in patients entirely naïve to lactulose. The effects of oral lactulose on the faecal microbiome of healthy humans have only been evaluated through either culture-based24–26 or culture-independent methods targeting predominant bacterial groups27–30. As diet was only standardised in two of those studies24,30, it seems likely that a variable diet could have impacted results31.
The human faecal microbiome is closer to the canine faecal microbiome when compared to pigs or mice32. Dogs evolved to cohabit with people, and hence adapted to a similar diet33. As they are frequently kept as companion animals, they are also exposed to the same environment. In addition, dogs can equally suffer from HE and lactulose is frequently used as supportive treatment in this condition34. Consequently, companion dogs can represent a useful comparative model to explore the faecal microbiome in HE as well as the effects of certain interventions on its composition, richness and function.
Therefore, the aims of this study were to investigate the magnitude and duration of quantitative and qualitative changes of the faecal microbiota by lactulose in healthy privately-owned dogs fed a standardised commercial diet. It was hypothesised that oral lactulose administration would significantly and transiently change the faecal microbiota in healthy dogs.
Methods
Prospective cohort study design
Dogs owned by members of staff at the Hospital for Small Animals, the Royal (Dick) School of Veterinary Studies, University of Edinburgh, were recruited with the following inclusion criteria: no current history of any disease; up to date vaccination and deworming records; and no current or recent administration of medications. A faecal sample was requested to be collected from each subject weekly, pertaining to the interventions schematised in Fig. 1.
Figure 1.
Longitudinal interventions in cohort of healthy owned dogs, designed to assess the faecal microbiota.
The standardised diet was a commercial maintenance diet for adult dogs (Hill’s™ Science Plan™ Canine Adult Advanced Fitness™ Large Breed with Chicken, Hill’s Pet Nutrition Ltd., Guildford, UK) and the lactulose was a 3.5 g/5 ml oral solution (Sandoz Ltd, Hampshire, UK). The dose of lactulose was calculated at 0.5 ml/kg and given every 12 hours, unless excessively soft or unformed faeces were noticed, at which point the subjects’ owners would notify one of the authors (MFF) and subsequently decrease the dose by 25% each time, aiming to achieve a soft faecal consistency that would be still amenable for manual collection.
Faecal samples were collected into plain bijoux tubes, kept frozen at −20 °C for a maximum of 24 hours and transferred afterwards to a −80 °C archiving freezer.
Informed consent was obtained from each subject’s owner. The study was approved by the University of Edinburgh’s Veterinary Ethical Review Committee (reference number 112–14) and carried out in accordance with the institution’s relevant guidelines and regulations.
Faecal DNA extraction, amplification and sequencing
Each sample was defrosted, manually homogenised and DNA extraction performed with a commercial kit (PowerSoil® DNA Isolation Kit, MO BIO Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions35. Amplification of DNA was undertaken with PCR of the hypervariable V4 region of the 16S ribosomal RNA (rRNA) gene, using dual-indexing primers (515F/806R), followed by amplicon quantification (Quant-iT™ PicoGreen®, Invitrogen, Ltd., Paisley, UK) and pooling, as previously described36,37. Standard 16S rRNA library preparation and sequencing with the Illumina® MiSeq® (v2 150PE) platform were performed by Edinburgh Genomics (The University of Edinburgh, UK).
Data analysis
Software packages for data analysis included the Quantitative Insights Into Microbial Ecology (QIIME2™, https://qiime2.org/) pipeline, RStudio® (version 1.1.453, © 2009–2018 RStudio, Inc., Boston, MA, US) with the package qiime2R (v0.12), as well as GraphPad Prism® (version 7, GraphPad Software Inc., La Jolla, CA, US).
The paired-end raw reads (Supplementary Data Mendeley Data) were analysed using QIIME2™. Briefly, sequence data with sequence quality information was imported through the “fastq manifest” format PairedEndFastqManifestPhred33. Demultiplexed sequence counts were summarised showing a median of 58,478 sequences per sample, with a minimum of 27,297 and maximum of 134,220. Sequence quality trimming, chimera filtering and feature table construction was performed through the DADA2 pipeline38. Mapping of feature identifiers to sequences was created using links for the Basic Local Alignment Search Tool (BLAST)39, and multiple sequence alignment of the sequences was undertaken by the Fast Fourier Transform (MAFFT) program40. This was followed by filtering of the alignment with the mask plugin41, generation of a phylogenetic tree with the FastTree program42 and application of midpoint rooting. The QIIME2™ q2-diversity plugin was used for rarefaction analysis and computation of alpha diversity metrics (observed operational taxonomic units [OTUs], Shannon diversity index, Chao1 and Pielou’s Evenness), as well as beta diversity metrics (weighted and unweighted UniFrac distances). Finally, assigning taxonomy to sequences was performed with a pre-trained Naïve Bayes classifier (Greengenes 13_8 99% OTUs) through the QIIME2™ q2-feature-classifier plugin43.
A human-extrapolated CDR: ratio of commensal autochthonous taxa (Lachnospiraceae, Ruminococcaceae, Veillonellaceae, and Clostridiales Insertae Sedis XIV) to potentially pathogenic non-autochthonous taxa (Enterobacteriaceae and Bacteroidaceae)44, was calculated without the inclusion of Clostridiales Insertae Sedis XIV, as this taxon was not detected in the studied population.
Statistical analyses used included descriptive statistics, as well as the following inferential statistical tests: Kruskal-Wallis test to compare differences between subjects regarding alpha diversity metrics; Wilcoxon matched-pairs signed rank test to compare differences between weeks regarding alpha diversity metrics, taxonomy frequencies and the CDR; paired t-test to compare differences between weeks regarding the presence of Lachnospiraceae; and Permutational Multivariate Analysis of Variance (PERMANOVA) test to compare differences between subjects and weeks regarding beta diversity metrics45. Normality was assessed with the Shapiro-Wilk test. Statistical significance level was set at p < 0.05.
Results
Population’s baseline characteristics
A total of 21 dogs were enrolled, with a median age of 5 years (range of 2–10 years). Sex distribution included 12 females (11 neutered) and 9 males (7 neutered). Just over half (n = 11) of the cohort was represented by crossbred dogs, with the remaining 10 dogs distributed as follows: two each of English Cocker Spaniel, English Springer Spaniel, Border Collie and Podenco; and one each of Boston Terrier and Labrador Retriever.
Adherence to study protocol and side effects related to lactulose administration
Three dogs did not complete the study for the following reasons: acute diarrhoea following dietary indiscretion (n = 1); requirement of a NSAID (meloxicam) for pain management secondary to presumptive degenerative joint disease (n = 1); and lip fold dermatitis after being licked by another dog in the household once starting lactulose (n = 1). In addition, two faecal samples (one each from weeks 4 and 8) were accidentally not collected. In total, 172 faecal samples were analysed, one of which failed sequencing (week 6). Side effects associated with lactulose administration included excessively soft faeces (n = 7) and unformed faeces (n = 1), all resolving after a dose reduction of 25% and 50%, respectively, therefore not requiring a drop out from the study.
Alpha diversity
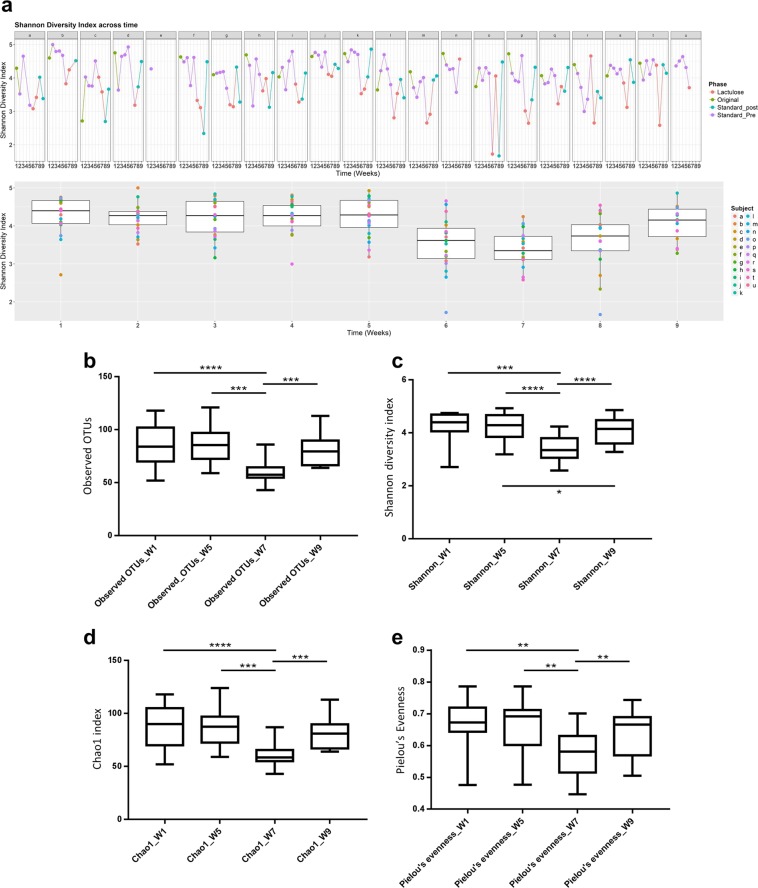
Community richness and diversity values were different between subjects regarding the following metrics: observed OTUs (p < 0.0001), Shannon diversity index (p = 0.0025), Chao1 index (p < 0.0001) and Pielou’s Evenness (p < 0.0001). Assessment of community richness and diversity across time is shown in Fig. 2. There was a reduction of all the above metrics at week 7 (standardised diet and oral lactulose), when compared to weeks 1 (usual diet), 5 (standardised diet) and 9 (standardised diet after having stopped lactulose). Conversely, values from weeks 1, 5 and 9 didn’t differ from each other apart from Shannon diversity index values between week 5 and 9 (Table 1).
Figure 2.
Variation of the canine faecal microbiota, assessed by alpha diversity metrics. (a) Shannon diversity index across a cohort of healthy owned dogs (letters a-u) while either on their usual diet (Original, week 1), a standard diet (Standard_Pre, weeks 2–5), a standard diet and oral lactulose (Lactulose, weeks 6–7), or a standard diet after having stopped lactulose (Standard_Post, weeks 8–9). (b–e) Box and whiskers plots (median, 25th and 75th quartiles), depicting different alpha diversity metrics at selected weeks: observed operating taxonomic units (OTUs) (b); Shannon diversity index (c); Chao1 index (d); and Pielou’s Evenness. (e) *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 (Wilcoxon matched-pairs signed rank test).
Table 1.
Statistical significances (p) of alpha diversity metrics for the canine faecal microbiota when compared across time.
| Metric | Weeks 1 vs 5 | Weeks 1 vs 7 | Weeks 1 vs 9 | Weeks 5 vs 7 | Weeks 5 vs 9 | Weeks 7 vs 9 |
|---|---|---|---|---|---|---|
| Observed OTUsa | 0.8056 | <0.0001 | 0.7904 | 0.0002 | 0.3516 | 0.0003 |
| Shannon diversity index | 0.9843 | 0.0003 | 0.0814 | <0.0001 | 0.0268 | <0.0001 |
| Chao1 index | 0.7453 | <0.0001 | 0.7379 | 0.0002 | 0.2690 | 0.0003 |
| Pielou’s Evenness | 0.7680 | 0.004 | 0.0898 | 0.0010 | 0.0898 | 0.009 |
Cohort of healthy owned dogs while either on their usual diet (week 1), a standard diet (week 5), a standard diet and oral lactulose (week 7), or a standard diet after having stopped lactulose (week 9). Statistical test used: Wilcoxon matched-pairs signed rank test.
aOperational Taxonomic Units.
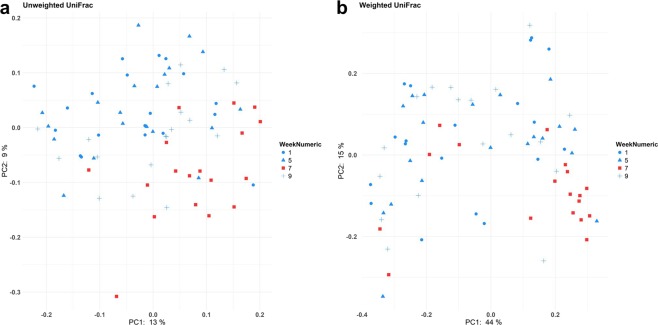
Beta diversity
Bacterial community presence/absence (qualitative) and abundance (quantitative) assessments with unweighted and weighted UniFrac distances, respectively, are illustrated in Fig. 3. These were overall different between subjects (p < 0.001). When analysed across time, the distances at week 7 were different from weeks 1, 5 and 9. However, values at weeks 1, 5 and 9 didn’t differ from each other (Table 2).
Figure 3.
Variation of the canine faecal microbiota, assessed by beta diversity metrics. (a,b) Principal coordinate analysis based on unweighted (a) and weighted (b) UniFrac distances. Cohort of healthy owned dogs while either on their usual diet (week 1), a standard diet (week 5), a standard diet and oral lactulose (week 7), or a standard diet after having stopped lactulose (week 9). Samples from week 7 were significantly different (p < 0.01, PERMANOVA test) from weeks 1, 5 and 9, while none of the latter differed from each other.
Table 2.
Statistical significances (p) of beta diversity metrics for the canine faecal microbiota when compared across time.
| Metric | Weeks 1 vs 5 | Weeks 1 vs 7 | Weeks 1 vs 9 | Weeks 5 vs 7 | Weeks 5 vs 9 | Weeks 7 vs 9 |
|---|---|---|---|---|---|---|
| Unweighted UniFrac | 0.534 | 0.001 | 0.136 | 0.001 | 0.272 | 0.012 |
| Weighted UniFrac | 0.474 | 0.001 | 0.978 | 0.002 | 0.645 | 0.001 |
Cohort of healthy owned dogs while either on their usual diet (week 1), a standard diet (week 5), a standard diet and oral lactulose (week 7), or a standard diet after having stopped lactulose (week 9). Statistical test used: PERMANOVA.
Taxonomy
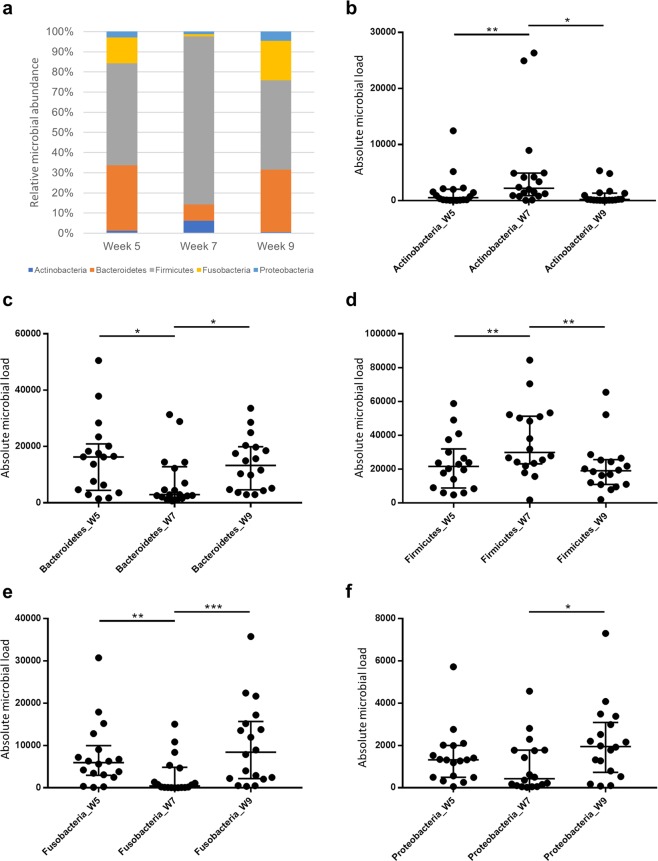
The relative and absolute distribution of phyla abundance across time is depicted in Fig. 4. Irrespective of week number (5, 7 or 9), the most abundant phylum was Firmicutes, followed by Bacteroidetes. A shift of phyla was observed at week 7, with the third most abundant phylum being Actinobacteria, followed by Fusobacteria and Proteobacteria. In contrast, for both weeks 5 and 9, Fusobacteria was the third most abundant phylum, followed by Proteobacteria and Actinobacteria. Moreover, when compared to weeks 5 and 9, week 7 was associated with a higher abundance of both Firmicutes (p = 0.0056 and 0.0047, respectively) and Actinobacteria (p = 0.0090 and 0.0104, respectively), and lower abundance of both Bacteroidetes (p = 0.0304 and 0.0120, respectively) and Fusobacteria (p = 0.0040 and 0.0008, respectively). The abundance of these phyla was similar between weeks 5 and 9 (Firmicutes, p = 0.6397; Actinobacteria, p = 0.6095; Bacteroidetes, p = 0.8650; and Fusobacteria, p = 0.1964).
Figure 4.
Abundance of phyla in the canine faecal microbiota. Cohort of healthy owned dogs while either on a standard diet (week 5), a standard diet and oral lactulose (week 7), or a standard diet after having stopped lactulose (week 9). (a) Median relative abundance of different phyla. (b–f) Dot plots (including bars for median, 25th and 75th quartiles) depicting absolute abundance of: Actinobacteria (b); Bacteroidetes (c); Firmicutes (d); Fusobacteria (e); and Proteobacteria (f). *p < 0.05; **p < 0.01; ***p < 0.001 (Wilcoxon matched-pairs signed rank test).
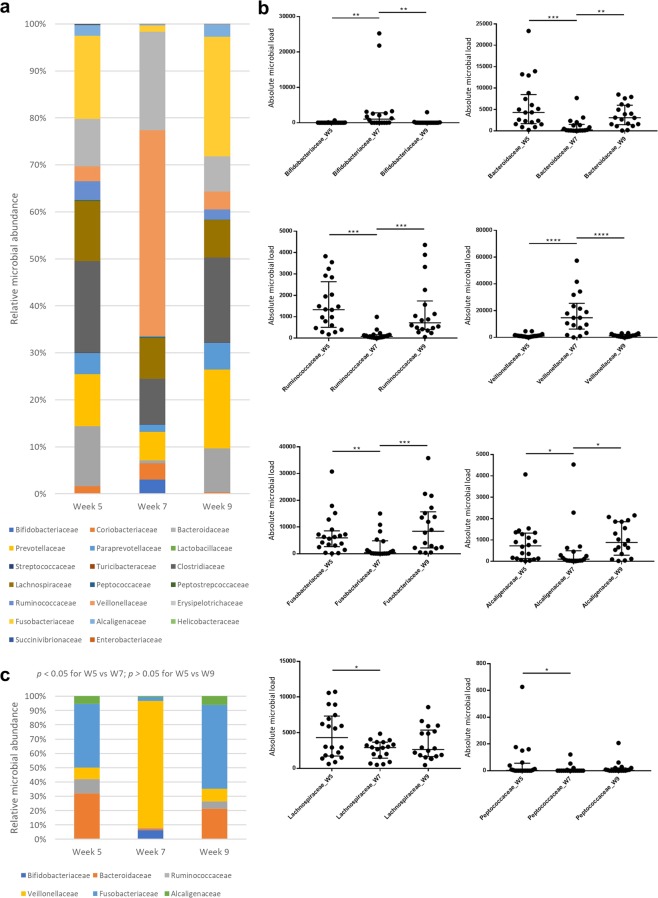
A total of 20 families were found to be present in the microbiota of at least half of the subjects at one or more time points (Fig. 5a). Different abundances across time were found in eight families (Fig. 5b), six of which showed a different abundance in week 7 when compared to weeks 5 and 9 (Fig. 5c), while both these time points had similar abundances. Significant changes at week 7 included an increased representation of Veillonellaceae and Bifidobacteriaceae, and a decreased abundance of Fusobacteriaceae, Bacteroidaceae, Ruminococcaceae, Alcaligenaceae, Lachnospiraceae and Peptococcaceae.
Figure 5.
Abundance of families in the canine faecal microbiota. Cohort of healthy owned dogs while either on a standard diet (week 5), a standard diet and oral lactulose (week 7), or a standard diet after having stopped lactulose (week 9). (a) Median relative abundance of families with counts present in at least 50% of dogs in at least one week group. (b) Dot plots (including bars for median, 25th and 75th quartiles) depicting absolute abundance of statistically significant families (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; Wilcoxon matched-pairs signed rank test). (c) Median relative abundance of statistically significant families (Wilcoxon matched-pairs signed rank test).
Cirrhosis dysbiosis ratio
When analysing the CDR across time (Fig. 6), the highest values were obtained at week 7 with a median of 25.07 (range of 0.08–460.04). These were different from week 1 (median 3.19, range 0.18–30.98, p = 0.0079), week 5 (median 2.54, range 0.18–29.78, p = 0.0003) and week 9 (median 2.10, range 0.24–15.75, p < 0.0001). Conversely, the CDRs calculated for weeks 1, 5 and 9 were similar to each other (p = 0.5678 for week 1 vs 5; p = 0.2288 for week 1 vs 9; and p = 0.5509 for week 5 vs 9).
Figure 6.
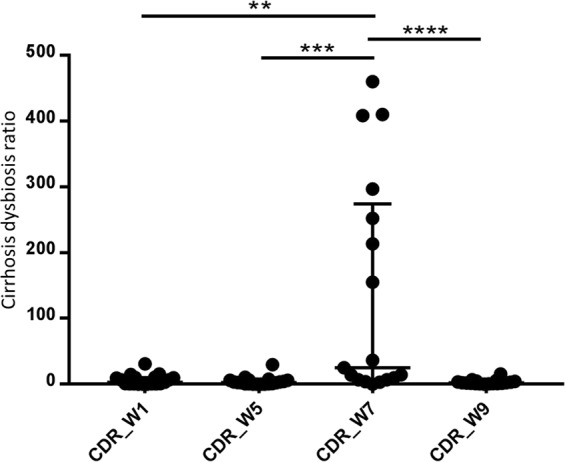
Cirrhosis dysbiosis ratio (CDR), calculated from the canine faecal microbiota, extrapolated from humans. Cohort of healthy owned dogs while either on their usual diet (week 1), a standard diet (week 5), a standard diet and oral lactulose (week 7), or a standard diet after having stopped lactulose (week 9). **p < 0.01; ***p < 0.001; ****p < 0.0001 (Wilcoxon matched-pairs signed rank test).
Discussion
To date, there are no publications employing untargeted or global culture-independent methodologies to assess the faecal microbiome in healthy people receiving lactulose. In people with HE, further limitations often apply, given the common use of standard of care treatment by the time of study enrolment (lactulose, rifaximin, antacids), precluding an evaluation of the microbiome in non-treated HE14,18.
To the authors’ knowledge, this study is the first to investigate the effect of lactulose on the canine microbiota. Collectively, lactulose induced a reversible reduction and qualitative modulation of the faecal microbiota diversity in this population of healthy dogs, while on a commercial standardised diet. Both alpha and beta diversity metrics were affected and specific taxa were implicated in this change.
The impact of lactulose has been studied in healthy mice and pigs through culture-independent methods, most showing an increased alpha diversity, which is in contrast to the present study46–49. However, experimental animals are kept in laboratory-controlled conditions and both species are known to have a more distant microbiome from humans relative to dogs32. Assessing companion dogs, alongside maintaining the advantage of dietary control, may be therefore valuable given their natural shared environment with humans31.
No significant changes of the microbiota were observed in this study due to diet change alone. This is likely explained by the relative similarity of composition of commercial diets (most of the original diets consisted of various brands of dry dog food), in comparison to homemade or raw food50,51. There were marked inter-individual differences concerning overall alpha and beta diversity, which was not surprising, given the non-relatedness of the subjects and the diversity of breeds evaluated52,53.
Significant changes in the abundance of main phyla were observed with the introduction of lactulose and matched by changes noted at family taxa. Within the Firmicutes and Actinobacteria, Veillonellaceae and Bifidobacteriaceae increased; and decreases in Bacteroidaceae and Fusobacteriaceae likely reflect lower abundances of Bacteroidetes and Fusobacteria.
Veillonellaceae was the most abundant family after the administration of lactulose. Its significance in liver disease has been controversial, as previous studies have reported both increases and decreases in human cirrhosis with and without HE13,14. Bacteria of this family convert lactate to acetate and butyrate54. Previously, the latter has been positively correlated with the presence of Veillonellaceae in canine faeces55. Increases in microorganisms producing these SCFA could be beneficial, as in people, acetate was negatively correlated with pro-inflammatory cytokines in cirrhosis, and butyrate was protective against the development of HE16. The administration of lactulose has been associated with increased acetate and butyrate production, together with decreased branch-chained fatty acids isobutyrate and isovalerate46,47,56. Similarly to the present study, several others have reported increased numbers of faecal Bifidobacteriaceae24–30, which produce lactate and acetate57, as well as contribute to metabolic cross-feeding, stimulating other butyrate-producing bacteria58.
Reductions of both Fusobacteriaceae and Alcaligenaceae induced by oral lactulose have not been previously reported. This finding is potentially significant, as their presence in the stools of patients with cirrhosis and HE has been associated with worsened inflammation/endotoxaemia and poor performance on cognitive tests15. On the contrary, decreases in Bacteroidaceae24,26 and Ruminococcaceae30 have been previously reported with lactulose administration. Bacteroides spp., are known to produce isovalerate59, as well as pro-inflammatory and barrier-disruptive neurotoxins/enterotoxins60, and β-glucuronidase61, a potential carcinogenic. Reduced faecal activity levels of this enzyme have been demonstrated previously with lactulose administration26,62. Besides producing butyrate63, Ruminococcaceae and Lachnospiraceae are also known to produce β-glucuronidase64. The lack of a significant increase in Lactobacillaceae with lactulose administration has been demonstrated before; hence, this study confirms that it is not a major hallmark of lactulose use24,25,29,30.
CDR is a measure of dysbiosis in humans: healthy people are reported to have a higher ratio than patients with cirrhosis, and the presence of HE was associated with an even lower CDR14. Although extrapolated from studies in people, the increase of CDR during lactulose administration could represent improvement of dysbiosis, despite an overall lower microbiota diversity and richness.
Limitations of this study include the small number of dogs, the collection of voided faecal samples rather than mucosal or luminal colonic samples, the lack of storage buffer/cryoprotectant and the fact that 16S rRNA gene sequencing allows assessment of taxonomy and abundance, but no species level resolution, nor extrapolation of functional data from the microbiome. However, given the longitudinal study design, each subject could serve as their own control, allowing observation of general trends of microbiota changes. For ethical reasons, invasive collection methods were not employed in these privately owned animals47,65. Short-term storage of faecal samples without cryopreservative or buffer is likely to not impact on major phyla distribution66,67. To investigate functional changes of the microbiome, high-throughput techniques (whole metagenome shotgun sequencing), metabolomics or metatranscriptomics could have been performed, but this was not within the scope of the current study.
In conclusion, lactulose can drive a reversible quantitative and qualitative modulation of the faecal microbiota in this dog model. Future research is warranted to focus on the investigation of microbiome dynamics in canine models of spontaneous naturally occurring HE (e.g. congenital portosystemic shunts)68. This could include similar longitudinal studies to assess effects before and after lactulose treatment or the correlation or comparison with other management strategies. Allowing for more targeted treatment endpoints could not only advance knowledge but also improve outcomes in HE.
Acknowledgements
The authors would like to thank Edinburgh Genomics, The University of Edinburgh, for performing the DNA sequencing and generating the raw data. Edinburgh Genomics is partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1). The authors would also like to thank the owners who consented their dogs to participate in this study. This study was supported by a Royal (Dick) School of Veterinary Studies Clinical Research Grant. The funding source had no involvement in: the study design; the collection, analysis and interpretation of data; the writing of the manuscript; or the decision to submit the manuscript for publication. J.J.S. is a University of Edinburgh Chancellor’s Fellow based at the Roslin Institute. He is supported by strategic funding from the Biotechnology and Biosciences Research Council (BB/P013759/1 and BB/P013732/1). R.J.M. and M.S. were supported by BBSRC through the Institute Strategic Programme funding (BB/J004235/1 and BB/P013740/1).
Author Contributions
J.J.S., R.J.M. and A.G.G. developed study concept and design. M.F.F., J.J.S., D.N.C., S.M.C. and D.E.G. acquired data. M.F.F., S.S.S. and M.S. analysed and interpreted data. M.F.F. and M.S. performed statistical analysis. M.F.F. drafted the manuscript. All authors reviewed the manuscript. J.J.S., D.N.C., S.M.C., D.E.G., R.J.M. and A.G.G. provided administrative, technical and/or material support. S.S.S., J.J.S., R.J.M., A.G.G. and M.S. undertook study supervision.
Data Availability
The datasets generated and/or analysed during the current study are available in the Mendeley Data repository, 10.17632/8ctyv86ccp.1.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Adam George Gow and Mazdak Salavati jointly supervised this work.
References
- 1.Bajaj JS, et al. Hepatic Encephalopathy Is Associated With Mortality in Patients With Cirrhosis Independent of Other Extrahepatic Organ Failures. Clin Gastroenterol Hepatol. 2017;15:565–574 e564. doi: 10.1016/j.cgh.2016.09.157. [DOI] [PubMed] [Google Scholar]
- 2.Gadiparthi C, et al. Waitlist Outcomes in Liver Transplant Candidates with High MELD and Severe Hepatic Encephalopathy. Dig Dis Sci. 2018;63:1647–1653. doi: 10.1007/s10620-018-5032-5. [DOI] [PubMed] [Google Scholar]
- 3.Labenz C, et al. Prospective evaluation of the impact of covert hepatic encephalopathy on quality of life and sleep in cirrhotic patients. Aliment Pharmacol Ther. 2018;48:313–321. doi: 10.1111/apt.14824. [DOI] [PubMed] [Google Scholar]
- 4.Stepanova M, Mishra A, Venkatesan C, Younossi ZM. In-hospital mortality and economic burden associated with hepatic encephalopathy in the United States from 2005 to 2009. Clin Gastroenterol Hepatol. 2012;10:1034–1041 e1031. doi: 10.1016/j.cgh.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Neff G, Zachry W., 3rd Systematic Review of the Economic Burden of Overt Hepatic Encephalopathy and Pharmacoeconomic Impact of Rifaximin. Pharmacoeconomics. 2018;36:809–822. doi: 10.1007/s40273-018-0641-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ochoa-Sanchez R, Rose CF. Pathogenesis of Hepatic Encephalopathy in Chronic Liver Disease. J Clin Exp Hepatol. 2018;8:262–271. doi: 10.1016/j.jceh.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parekh PJ, Balart LA. Ammonia and Its Role in the Pathogenesis of Hepatic Encephalopathy. Clin Liver Dis. 2015;19:529–537. doi: 10.1016/j.cld.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Shalimar, Sheikh Mohammed Faisal, Mookerjee Rajeshwar P., Agarwal Banwari, Acharya Subrat Kumar, Jalan Rajiv. Prognostic Role of Ammonia in Patients With Cirrhosis. Hepatology. 2019;70(3):982–994. doi: 10.1002/hep.30534. [DOI] [PubMed] [Google Scholar]
- 9.Gluud, L. L., Vilstrup, H. & Morgan, M. Y. Non-absorbable disaccharides versus placebo/no intervention and lactulose versus lactitol for the prevention and treatment of hepatic encephalopathy in people with cirrhosis. Cochrane Database Syst Rev. CD003044, 10.1002/14651858.CD003044.pub4 (2016). [DOI] [PMC free article] [PubMed]
- 10.American Association for the Study of Liver, D. & European Association for the Study of the, L Hepatic encephalopathy in chronic liver disease: 2014 practice guideline by the European Association for the Study of the Liver and the American Association for the Study of Liver Diseases. J Hepatol. 2014;61:642–659. doi: 10.1016/j.jhep.2014.05.042. [DOI] [PubMed] [Google Scholar]
- 11.Clausen MR, Mortensen PB. Lactulose, disaccharides and colonic flora. Clinical consequences. Drugs. 1997;53:930–942. doi: 10.2165/00003495-199753060-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moratalla A, et al. Lactulose reduces bacterial DNA translocation, which worsens neurocognitive shape in cirrhotic patients with minimal hepatic encephalopathy. Liver Int. 2017;37:212–223. doi: 10.1111/liv.13200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, et al. Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. Am J Gastroenterol. 2013;108:1601–1611. doi: 10.1038/ajg.2013.221. [DOI] [PubMed] [Google Scholar]
- 14.Bajaj JS, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940–947. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajaj JS, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168–175. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iebba V, et al. Combining amplicon sequencing and metabolomics in cirrhotic patients highlights distinctive microbiota features involved in bacterial translocation, systemic inflammation and hepatic encephalopathy. Sci Rep. 2018;8:8210. doi: 10.1038/s41598-018-26509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajaj JS, et al. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment Pharmacol Ther. 2014;39:1113–1125. doi: 10.1111/apt.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajaj JS, et al. Diet affects gut microbiota and modulates hospitalization risk differentially in an international cirrhosis cohort. Hepatology. 2018;68:234–247. doi: 10.1002/hep.29791. [DOI] [PubMed] [Google Scholar]
- 19.Bajaj JS, et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology. 2017;66:1727–1738. doi: 10.1002/hep.29306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziada DH, Soliman HH, El Yamany SA, Hamisa MF, Hasan AM. Can Lactobacillus acidophilus improve minimal hepatic encephalopathy? A neurometabolite study using magnetic resonance spectroscopy. Arab J Gastroenterol. 2013;14:116–122. doi: 10.1016/j.ajg.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Riggio O, et al. Effect of lactitol and lactulose administration on the fecal flora in cirrhotic patients. J Clin Gastroenterol. 1990;12:433–436. doi: 10.1097/00004836-199008000-00016. [DOI] [PubMed] [Google Scholar]
- 22.Sarangi AN, Goel A, Singh A, Sasi A, Aggarwal R. Faecal bacterial microbiota in patients with cirrhosis and the effect of lactulose administration. BMC Gastroenterol. 2017;17:125. doi: 10.1186/s12876-017-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajaj JS, et al. A longitudinal systems biology analysis of lactulose withdrawal in hepatic encephalopathy. Metab Brain Dis. 2012;27:205–215. doi: 10.1007/s11011-012-9303-0. [DOI] [PubMed] [Google Scholar]
- 24.Terada A, Hara H, Kataoka M, Mitsuoka T. Effect of Lactulose on the Composition and Metabolic Activity of the Human Faecal Flora. Microbial Ecology in Health and Disease. 1992;5:43–50. doi: 10.3109/08910609209141303. [DOI] [Google Scholar]
- 25.Bouhnik Y, et al. Lactulose ingestion increases faecal bifidobacterial counts: a randomised double-blind study in healthy humans. Eur J Clin Nutr. 2004;58:462–466. doi: 10.1038/sj.ejcn.1601829. [DOI] [PubMed] [Google Scholar]
- 26.Ballongue J, Schumann C, Quignon P. Effects of lactulose and lactitol on colonic microflora and enzymatic activity. Scand J Gastroenterol Suppl. 1997;222:41–44. doi: 10.1080/00365521.1997.11720716. [DOI] [PubMed] [Google Scholar]
- 27.Tuohy KM, et al. A Human Volunteer Study to Determine the Prebiotic Effects of Lactulose Powder on Human Colonic Microbiota. Microbial Ecology in Health and Disease. 2002;14:165–173. doi: 10.1080/089106002320644357. [DOI] [Google Scholar]
- 28.De Preter V, et al. Effect of lactulose and Saccharomyces boulardii administration on the colonic urea-nitrogen metabolism and the bifidobacteria concentration in healthy human subjects. Aliment Pharmacol Ther. 2006;23:963–974. doi: 10.1111/j.1365-2036.2006.02834.x. [DOI] [PubMed] [Google Scholar]
- 29.Vanhoutte T, et al. Molecular monitoring of the fecal microbiota of healthy human subjects during administration of lactulose and Saccharomyces boulardii. Appl Environ Microbiol. 2006;72:5990–5997. doi: 10.1128/AEM.00233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venema K, van Nuenen MHMC, van den Heuvel EG, Pool W, van der Vossen JMBM. The Effect of Lactulose on the Composition of the Intestinal Microbiota and Short-chain Fatty Acid Production in Human Volunteers and a Computer-controlled Model of the Proximal Large Intestine. Microbial Ecology in Health and Disease. 2003;15:94–105. doi: 10.1080/08910600310019895. [DOI] [Google Scholar]
- 31.Dong Tien S., Gupta Arpana. Influence of Early Life, Diet, and the Environment on the Microbiome. Clinical Gastroenterology and Hepatology. 2019;17(2):231–242. doi: 10.1016/j.cgh.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coelho LP, et al. Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome. 2018;6:72. doi: 10.1186/s40168-018-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 34.Lidbury JA, Cook AK, Steiner JM. Hepatic encephalopathy in dogs and cats. J Vet Emerg Crit Care (San Antonio). 2016;26:471–487. doi: 10.1111/vec.12473. [DOI] [PubMed] [Google Scholar]
- 35.PowerSoil® DNA Isolation Kit Instruction Manual, MO BIO Laboratories, Inc, https://www.qiagen.com/us/resources/download.aspx?id=5c00f8e4-c9f5-4544-94fa-653a5b2a6373&lang=en (2014).
- 36.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lane, D. In Nucleic Acid Techniques in Bacterial Systematics (eds Stackebrandt, E. & Goodfellow, M.) 115–175 (John Wiley and Sons, 1991).
- 42.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bokulich NA, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bajaj Jasmohan S., Vargas Hugo E., Reddy K. Rajender, Lai Jennifer C., O’Leary Jacqueline G., Tandon Puneeta, Wong Florence, Mitrani Robert, White Melanie B., Kelly Megan, Fagan Andrew, Patil Rohan, Sait Shaimaa, Sikaroodi Masoumeh, Thacker Leroy R., Gillevet Patrick M. Association Between Intestinal Microbiota Collected at Hospital Admission and Outcomes of Patients With Cirrhosis. Clinical Gastroenterology and Hepatology. 2019;17(4):756-765.e3. doi: 10.1016/j.cgh.2018.07.022. [DOI] [PubMed] [Google Scholar]
- 45.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 46.Zhai Shixiang, Zhu Limeng, Qin Song, Li Lili. Effect of lactulose intervention on gut microbiota and short chain fatty acid composition of C57 BL /6J mice. MicrobiologyOpen. 2018;7(6):e00612. doi: 10.1002/mbo3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao B, et al. Lactulose Differently Modulates the Composition of Luminal and Mucosal Microbiota in C57BL/6J Mice. J Agric Food Chem. 2016;64:6240–6247. doi: 10.1021/acs.jafc.6b02305. [DOI] [PubMed] [Google Scholar]
- 48.Chae JP, Pajarillo EAB, Park CS, Kang DK. Lactulose increases bacterial diversity and modulates the swine faecal microbiome as revealed by 454-pyrosequencing. Anim Feed Sci Tech. 2015;209:157–166. doi: 10.1016/j.anifeedsci.2015.07.018. [DOI] [Google Scholar]
- 49.Chae JP, Pajarillo EA, Oh JK, Kim H, Kang DK. Revealing the combined effects of lactulose and probiotic enterococci on the swine faecal microbiota using 454 pyrosequencing. Microb Biotechnol. 2016;9:486–495. doi: 10.1111/1751-7915.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herstad KMV, et al. A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client-owned dogs. BMC Vet Res. 2017;13:147. doi: 10.1186/s12917-017-1073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, An JU, Kim W, Lee S, Cho S. Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog. 2017;9:68. doi: 10.1186/s13099-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Middleton RP, et al. Metabolic Differences between Dogs of Different Body Sizes. J Nutr Metab. 2017;2017:4535710. doi: 10.1155/2017/4535710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hand D, Wallis C, Colyer A, Penn CW. Pyrosequencing the canine faecal microbiota: breadth and depth of biodiversity. PLoS One. 2013;8:e53115. doi: 10.1371/journal.pone.0053115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolenbrander Paul. The Prokaryotes. New York, NY: Springer US; 2006. The Genus Veillonella; pp. 1022–1040. [Google Scholar]
- 55.Sandri, M., Dal Monego, S., Conte, G., Sgorlon, S. & Stefanon, B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. Bmc Veterinary Research. 13, 10.1186/s12917-017-0981-z (2017). [DOI] [PMC free article] [PubMed]
- 56.Bothe Melanie, Maathuis Annet, Bellmann Susann, van der Vossen Jos, Berressem Dirk, Koehler Annalena, Schwejda-Guettes Susann, Gaigg Barbara, Kuchinka-Koch Angelika, Stover John. Dose-Dependent Prebiotic Effect of Lactulose in a Computer-Controlled In Vitro Model of the Human Large Intestine. Nutrients. 2017;9(7):767. doi: 10.3390/nu9070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito Y, et al. Effect of lactulose on short-chain fatty acids and lactate production and on the growth of faecal flora, with special reference to Clostridium difficile. J Med Microbiol. 1997;46:80–84. doi: 10.1099/00222615-46-1-80. [DOI] [PubMed] [Google Scholar]
- 60.Lukiw WJ. The microbiome, microbial-generated proinflammatory neurotoxins, and Alzheimer’s disease. J Sport Health Sci. 2016;5:393–396. doi: 10.1016/j.jshs.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallace BD, et al. Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem Biol. 2015;22:1238–1249. doi: 10.1016/j.chembiol.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Preter V, et al. Effect of dietary intervention with different pre- and probiotics on intestinal bacterial enzyme activities. Eur J Clin Nutr. 2008;62:225–231. doi: 10.1038/sj.ejcn.1602706. [DOI] [PubMed] [Google Scholar]
- 63.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 64.Gloux K, et al. A metagenomic beta-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4539–4546. doi: 10.1073/pnas.1000066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bajaj JS, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horng KR, Ganz HH, Eisen JA, Marks SL. Effects of preservation method on canine (Canis lupus familiaris) fecal microbiota. PeerJ. 2018;6:e4827. doi: 10.7717/peerj.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song, S. J. et al. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitability for Field Studies. mSystems. 1, 10.1128/mSystems.00021-16 (2016). [DOI] [PMC free article] [PubMed]
- 68.Tivers MS, et al. Hyperammonemia and systemic inflammatory response syndrome predicts presence of hepatic encephalopathy in dogs with congenital portosystemic shunts. PLoS One. 2014;9:e82303. doi: 10.1371/journal.pone.0082303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Mendeley Data repository, 10.17632/8ctyv86ccp.1.