SUMMARY
The catalytic activity of mitogen-activated protein kinases (MAPKs) is dynamically modified in plants. Since MAPKs have been shown to play important roles in a wide range of signaling pathways, the ability to monitor MAPK activity in living plant cells would be valuable. Here, we report the development of a genetically encoded MAPK activity sensor for use in Arabidopsis thaliana. The sensor is composed of yellow and blue fluorescent proteins, a phosphopeptide binding domain, a MAPK substrate domain and a flexible linker. Using in vitro testing, we demonstrated that phosphorylation causes an increase in the Förster resonance energy transfer (FRET) efficiency of the sensor. The FRET efficiency can therefore serve as a readout of kinase activity. We also produced transgenic Arabidopsis lines expressing this sensor of MAPK activity (SOMA) and performed live-cell imaging experiments using detached cotyledons. Treatment with NaCl, the synthetic flagellin peptide flg22 and chitin all led to rapid gains in FRET efficiency. Control lines expressing a version of SOMA in which the phosphosite was mutated to an alanine did not show any substantial changes in FRET. We also expressed the sensor in a conditional loss-of-function double-mutant line for the Arabidopsis MAPK genes MPK3 and MPK6. These experiments demonstrated that MPK3/6 are necessary for the NaCl-induced FRET gain of the sensor, while other MAPKs are probably contributing to the chitin and flg22-induced increases in FRET. Taken together, our results suggest that SOMA is able to dynamically report MAPK activity in living plant cells.
Keywords: Arabidopsis thaliana, MAP kinase, FRET sensor, technical advance, live-cell imaging
INTRODUCTION
Live cell imaging is a powerful method for studying a variety of cellular processes in plants (Grossmann et al., 2018). Genetically encoded biosensors are critical tools for performing live-cell imaging because they allow the researcher to specifically monitor a particular biomolecule within the cell. A wide range of fluorescent biosensors have been used in plants, including those that report on the status of calcium, glucose, inorganic phosphate, zinc, ATP, hydrogen peroxide, pH, auxin, gibberellic acid and abscisic acid (Nagai et al., 2004; Deuschle et al., 2006; Zhao et al., 2011; Brunoud et al., 2012; Gjetting et al., 2012; Jones et al., 2014; Lanquar et al., 2014; Waadt et al., 2014; Hernandez-Barrera et al., 2015; Mukherjee et al., 2015; De Col et al., 2017; Rizza et al., 2017). Biosensors for measuring the activity of protein kinases have been described for use in animal cells, but to date none have been reported for plants.
The most widely used sensors of kinase activity in animal cells are based on the process of Förster resonance energy transfer (FRET) (Aoki et al., 2012; Sample et al., 2014). These sensors carry a substrate domain specific for a kinase of interest and a phosphopeptide-binding domain. When the kinase of interest is active within a cell it will phosphorylate the sensor. The phosphopeptide-binding domain then drives a conformational change that leads to a change in the ratio of fluorescence from the two fluorophores, which can be monitored in living samples using confocal microscopy. Kinase activity sensors of this type have been used to measure mitogen-activated protein kinase (MAPK) activation in animal cell cultures, Caenorhabditis elegans, zebrafish embryos and mice (Kamioka et al., 2012; Tomida et al., 2012; Aoki et al., 2013; Ryu et al., 2015; Sari et al., 2018).
We were interested in developing a biosensor for MAPK activity in Arabidopsis because of the central role that these kinases play in a wide range of signaling pathways. Our current understanding of MAPK function in Arabidopsis is based on extensive genetic and biochemical studies which have demonstrated roles for MAPK signaling in both stress responses and developmental regulation (Rodriguez et al., 2010; Moustafa et al., 2014; Xu and Zhang, 2015; Liu and He, 2017; Devendrakumar et al., 2018; Komis et al., 2018; Zhang et al., 2018). Of the 20 MAPK genes present in the Arabidopsis genome, mutant phenotypes have been associated with mpk1, mpk3, mpk4, mpk6, mpk8, mpk9, mpk10, mpk12, mpk17 and mpk18 (Petersen et al., 2000; Bush and Krysan, 2007; Wang et al., 2007, 2008; Hord et al., 2008; Ren et al., 2008; Jammes et al., 2009; Walia et al., 2009; Galletti et al., 2011; Takahashi et al., 2011; Zeng et al., 2011; Stanko et al., 2014; Enders et al., 2017; Zhang et al., 2017; Frick and Strader, 2018). The biological processes regulated by these MAPKs include biotic and abiotic stress responses, cell division and developmental patterning.
Genetic studies typically allow one to identify a pathway or process in which a particular gene product functions, but understanding the precise molecular mechanism by which it functions requires biochemical analysis. In the case of MAPK signaling, understanding the activation status of the kinase in different biological contexts has proved to be a subject of great interest to the field. The catalytic activity of a MAPK is switched on by dual phosphorylation of its conserved activation loop by an upstream MAPK kinase (MAPKK) and switched off by dephosphorylation of those residues by protein phosphatases (Rodriguez et al., 2010). In its activated state, MAPK is able to phosphorylate a wide range of substrate molecules in a cell. Current methods for interrogating the activation status of MAPKs in Arabidopsis involve homogenizing samples and extracting protein for in vitro analysis (Su et al., 2017; Sun et al., 2018). Although these methods have been very successful in documenting patterns of MAPK activation in response to a number of different biotic and abiotic stress treatments, one limitation of these studies is that they provide an average of the kinase activation across all the cells in the sample. Furthermore, it has been well documented that MAPK activity does not occur uniformly in all cell types in different mammalian signaling networks (Lahav et al., 2004; Spencer et al., 2009; Albeck et al., 2013), suggesting that Arabidopsis MAPKs may also display heterogeneity in response to a stimulus. Developing tools that allow for the observation of MAPK activation with single-cell resolution in living samples therefore has the potential to improve our understanding of how signaling is integrated across scales from cell to tissue to organism. Here, we describe the development of a genetically encoded biosensor designed to report the activity of MAPKs in Arabidopsis thaliana.
RESULTS
Construction of the sensor of MAP kinase activity (SOMA)
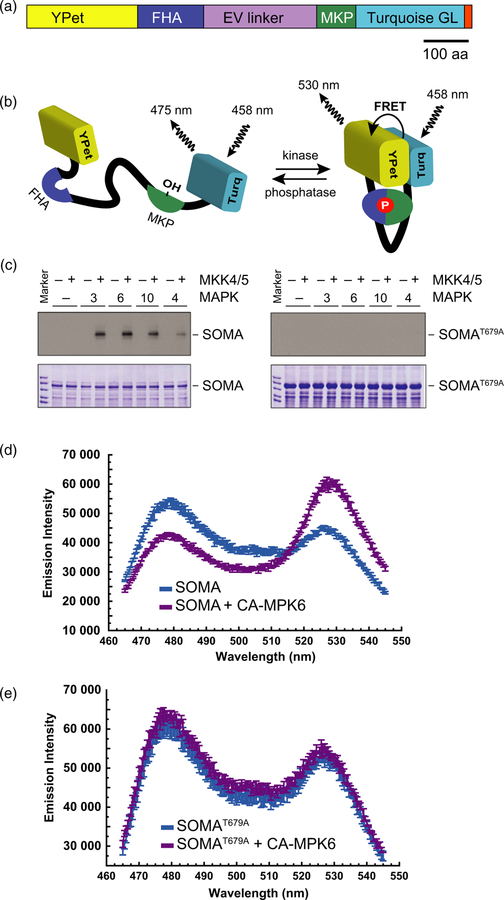
In order to produce a genetically encoded biosensor that would report MAPK activity in Arabidopsis, we made use of a previously established MAPK activity sensor developed for use in mammalian cells (Komatsu et al., 2011). In that study, the authors evaluated a number of different fluorophore combinations and flexible linkers and identified that the combination of YPet, Turquoise GL and the 244-amino-acid version of the EV linker allowed for one of the strongest FRET gains. We took that mammalian sensor and replaced the mammalian substrate domain with DNA encoding an 80-amino-acid region surrounding threonine 64 of the Arabidopsis MAP kinase phosphatase (MKP1) protein (Ulm et al., 2001). Threonine 64 of MKP1 has been previously shown to be phosphorylated by MAP kinase 6 (MPK6) (Park et al., 2011). The functional domains of SOMA are the YPet yellow fluorescent protein, the FHA1 phosphopeptide-binding domain of yeast RAD53, a 244-amino-acid version of the EV linker, 80 amino acids of Arabidopsis MPK1 and the Turquoise GL blue fluorescent protein (Figure 1a) (Sun et al., 1998; Nguyen and Daugherty, 2005; Goedhart et al., 2010; Komatsu et al., 2011). We also introduced a serine to aspartic acid substitution at the +3 position with respect to the phosphothreonine site in the MKP1 substrate domain of SOMA since previous work had shown that an aspartic acid at this position enhances FHA1 affinity for the phosphorylated form of the substrate (Yongkiettrakul et al., 2004; Komatsu et al., 2011).
Figure 1.
Structure and in vitro testing of the sensor of mitogen-activated protein kinase (MAPK) activity (SOMA).
(a) Domain structure of the SOMA protein. YPet is a yellow fluorescent protein, FHA is the FHA1 phosphopeptide-binding domain of yeast RAD53, EV linker is a flexible linker domain, MPK is an 80-amino-acid segment of the Arabidopsis MKP1 protein containing a MPK6 phosphorylation site, and Turquoise GL is a blue fluorescent protein. The small segment at the extreme C-terminus of the diagram indicates the location of the nuclear localization or nuclear exclusion signal. aa, amino acid.
(b) Phosphorylation of SOMA within the MKP1 domain is expected to produce a conformation shift that increases Förster resonance energy transfer (FRET) efficiency due to the enhanced affinity of the FHA1 domain for the phosphorylated form of the substrate domain. Removal of that phosphate by a phosphatase is expected to cause SOMA to revert to a conformation with lower FRET efficiency.
(c) In vitro kinase assays performed using proteins extracted from Escherichia coli. SOMA and SOMAT679A as substrates were incubated in the presence of the indicated Arabidopsis MAPK protein with (+) and without (−) constitutively active Arabidopsis MKK4 and MKK5. Reaction products were separated on gels and the incorporation of 32P into the substrates was evaluated via autoradiography (upper panels). Lower panels show the loading controls stained with Coomassie brilliant blue.
(d), (e) In vitro FRET assays using E. coli expressed SOMA and SOMAT679A performed in the presence or absence of E. coli expressed constitutively active MPK6 (CA-MPK6). The emission spectra of SOMA and SOMAT679A produced by excitation of the Turquoise GL domain with 435 nm light is shown.
The rationale for the design of this sensor is outlined in Figure 1(b). When the sensor is in an unphosphorylated state, the FHA1 domain has low affinity for the substrate domain and the ratio of YPet to Turquoise GL fluorescent emission is consequently low in response to Turquoise GL excitation. When MAPK phosphorylates the sensor’s substrate domain, the affinity of FHA1 for the substrate domain should increase substantially. Binding of FHA1 to the substrate domain then causes an increase in the ratio of YPet to Turquoise GL fluorescent emission in response to Turquoise GL excitation (Komatsu et al., 2011). Removal of the phosphate group from the sensor by a phosphatase will cause the sensor to return to a state where the ratio of YPet to Turquoise GL emission is low upon Turquoise GL excitation. The sensor therefore provides a readout of the balance between the competing activities of the kinases and phosphatases that recognize the sensor’s substrate domain. In the case of SOMA, we chose a substrate domain that has previously been shown to be phosphorylated by MPK6 in Arabidopsis (Park et al., 2011).
In order to determine if the 80-amino-acid portion of MKP1 included in SOMA could be phosphorylated by Arabidopsis MAPKs in the context of the full-length SOMA protein, we performed in vitro kinase assays using recombinant proteins purified from Escherichia coli. For these experiments, we also constructed a mutant form of the sensor, SOMAT679A, in which the threonine that was expected to be phosphorylated was mutated to an alanine. We incubated these sensor constructs with Arabidopsis MPK3, MPK4, MPK6 and MPK10 that had been activated by co-incubation with constitutively active versions of Arabidopsis MAP kinase kinase 4 (MKK4) and MAP kinase kinase 5 (MKK5) (Ren et al., 2002). Phosphorylation of SOMA was detected for all of these kinases, although the intensity of the signal produced by MPK4 was weaker than that of the other kinases (Figure 1c). No phosphorylation was detected in any of the SOMAT679A reactions. These results indicate that the previously characterized MKP1 phosphorylation can be recognized by MAPKs in the context of SOMA and that there are no additional MAPK phosphorylation sites in the sensor.
To determine if the phosphorylation status of SOMA affected the ratio of YPet to Turquoise GL emission, we performed in vitro experiments using proteins expressed in E. coli. We incubated SOMA and SOMAT679A in the presence or absence of a constitutively active version of MPK6 (CA-MPK6), and then the fluorescence emission profile was determined with a scanning spectrofluorimeter (Berriri et al., 2012). Using an excitation wavelength targeting the Turquoise GL fluorophore, we observed emission peaks corresponding to both Turquoise GL and YPet (Figure 1d, e). In the case of SOMA, co-incubation with CA-MPK6 caused a reduction in Turquoise GL emission and an increase in YPet emission relative to SOMA alone. This result suggested that phosphorylation of the sensor caused an increase in FRET efficiency, although additional experimentation would be needed to verify that the mechanism responsible for the change in YPet to Turquoise emission is differential FRET efficiency. Supporting this conclusion, we observed that CA-MPK6 did not change the ratio of YPet to Turquoise GL emission of SOMAT679A upon Turquoise GL excitation. To further explore if phosphorylation of SOMA is responsible for the observed change in the ratio of YPet to Turquoise GL emission upon Turquoise GL excitation shown in Figure 1(d), we performed an experiment using lambda protein phosphatase and observed that phosphatase strongly reduced the ratio of YPet to Turquoise GL emission upon Turquoise GL excitation displayed by the SOMA + CA-MPK6 reactions (Figure S1 in the online Supporting Information). Taken together, these results indicate that phosphorylation of threonine 679 of SOMA causes an increase in the ratio of YPet to Turquoise GL emission upon excitation of Turquoise GL in vitro.
In vivo testing of SOMA
In order to determine if SOMA was effective in vivo, we generated transgenic lines expressing four different versions of SOMA as outlined in Table 1. These lines expressed SOMA or SOMAT679A tagged with either the SV40 nuclear localization signal (NLS) or a human immunodeficiency virus 1 (HIV-1) nuclear export signal (NES) (Kalderon et al., 1984; Wen et al., 1995). Expression was driven by the cauliflower mosaic virus 35S promoter in the case of the –NES-tagged sensors and the Arabidopsis Ubiquitin 10 (UBQ10) promoter in the case of the –NLS-tagged sensors (Odell et al., 1985; Grefen et al., 2010). Two independent transgenic lines were characterized for each of the four sensor constructs (Table 1). The transgenic lines expressing the SOMA constructs did not display any obvious phenotypic differences when compared with wild-type control plants (Figures S2 and S3.)
Table 1.
Transgenic lines produced for this study
| Line name | ABRC stock no. | Plasmid | Promoter | Localization | Background |
|---|---|---|---|---|---|
| SOMA-NLS-1 | CS71683 | pSOMA-NLS | UBQ10 | Nucleus | Col-0 |
| SOMA-NLS-2 | CS71684 | pSOMA-NLS | UBQ10 | Nucleus | Col-0 |
| SOMA-NES-1 | CS71685 | pSOMA-NES | CaMV 35S | Cytoplasm | Col-0 |
| SOMA-NES-2 | CS71686 | pSOMA-NES | CaMV 35S | Cytoplasm | Col-0 |
| SOMAT679A-NLS-1 | CS71687 | pSOMAT679A-NLS | UBQ10 | Nucleus | Col-0 |
| SOMAT679A-NLS-2 | CS71688 | pSOMAT679A-NLS | UBQ10 | Nucleus | Col-0 |
| SOMAT679A-NES-1 | CS71689 | pSOMAT679A-NES | CaMV 35S | Cytoplasm | Col-0 |
| SOMAT679A-NES-2 | CS71690 | pSOMAT679A-NES | CaMV 35S | Cytoplasm | Col-0 |
| MPK6YG SOMA-NES | CS71691 | pSOMA-NES | CaMV 35S | Cytoplasm |
mpk3 mpk6 Pmpk6: MPK6YG |
Localization indicates the subcellular localization of the sensor protein. ABRC stock no. indicates the stock number for ordering seed from the Arabidopsis Biological Resource Center.
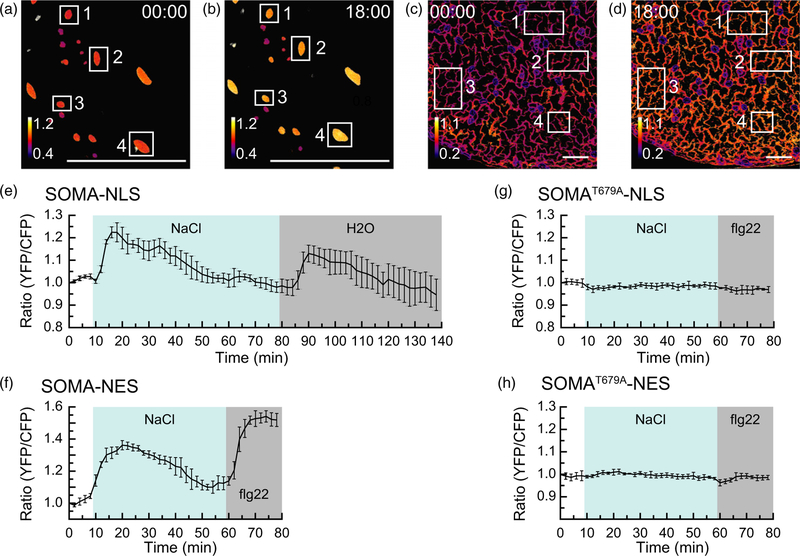
We next wanted to determine if treatments known to activate MAPK activity in Arabidopsis caused a change in the ratio of YPet to Turquoise GL emission of SOMA in planta. To do this we mounted 5-day-old detached cotyledons in water in a microfluidic device suitable for live-cell imaging and used confocal microscopy to measure the ratio of YPet to Turquoise GL emission in response to Turquoise GL excitation (Vang et al., 2018). This imaging chamber holds a cotyledon in a volume of about 30 µl of liquid that can be exchanged with a fresh solution in less than 1 min via microfluidic passive pumping. As a result, one can observe a sample before, during and after the application of a desired chemical treatment. Using this imaging system, a rapid gain in the ratio of YPet to Turquoise GL emission was observed with both SOMA-NLS and SOMA-NES within 2–4 min of treatment with 150 mM NaCl (Figure 2a–e,g). NaCl stress has previously been shown to activate MPK3/6 in Arabidopsis (Droillard et al., 2004; Yu et al., 2010). The ratio of YPet to Turquoise GL emission returned to pre-treatment levels 35–45 min after the initial peak. A follow-up treatment with water to induce hypo-osmotic shock or the synthetic flagellin peptide flg22 to double-stress the samples with hypo-osmotic shock plus an elicitor stimulus caused a second gain in the ratio of YPet to Turquoise GL emission. These results demonstrate that the ratio of YPet to Turquoise GL emission of SOMA rapidly changes in response to treatments known to activate MAPK activity in Arabidopsis. The observation that the ratio of YPet to Turquoise GL emission drops back to pre-treatment levels after about 35–45 min and can then be re-stimulated by a final hypo-osmotic shock indicates that the sensor remains viable for the duration of the experiment and that its changes are reversible.
Figure 2.
The in vivo response of sensor of mitogen-activated protein kinase (MAPK) activity (SOMA) to NaCl stress.
(a)–(d) Processed confocal images of the cotyledon epidermis from the SOMA-NLS-1 (a,b) and SOMA-NES-1 (c,d) transgenic lines depicting the ratio of YPet to Turquoise GL emission produced by exciting Turquoise GL. The calibration bar in the lower left indicates the numerical scale corresponding to the heat map. The scale bar on the bottom indicates 100 µm. Time stamps indicate when the image was collected in minutes:seconds. Images at time 00:00 were collected before treatment, while those at time point 18:00 were collected 12 min after treatment with 150 mM NaCl. Numbered rectangles indicate the regions of interest (ROIs) used to measure YPet and Turquoise GL emission. (e), (f) The average ratio of YPet to Turquoise GL emission produced by exciting Turquoise GL was determined using the ROIs shown in (a–d), normalized to the starting value for each ROI, and plotted versus time. The shaded background on each graph indicates when the sample was exposed to a given treatment. During the first 10 min of each experiment the samples were incubated in pure water. ‘NaCl’ indicates 150 mM NaCl, ‘H2O’ indicates pure water, and ‘flg22’ indicates that 1 µm flg22 was present in the imaging chamber during the shaded time period. Error bars indicate standard deviation. ‘Ratio (YFP/CFP)’ indicates the normalized value of YPet emission divided by Turquoise GL emission after excitation of Turquoise GL with 458 nm light. (g), (h) Transgenic lines SOMAT679A-NLS-1 and SOMAT679A-NES-1 were analyzed as above using the ROIs indicated in Figure S17. Videos are available as Movies S1–S4. These experiments were repeated at least five times with similar results.
No substantial changes in the ratio of YPet to Turquoise GL emission were observed in experiments using lines expressing SOMAT679A-NLS and SOMAT679A-NES (Figure 2f,h), indicating that the known phosphorylation site of SOMA is necessary for stress-induced changes in the ratio of YPet to Turquoise GL emission in vivo. In addition, treatment of SOMA-NLS and SOMA-NES lines with water as a control did not result in any substantial changes in the ratio of YPet to Turquoise GL emission (Figure S4), demonstrating that the imaging protocol did not affect the sensors. Similar results were obtained using independent transgenic lines for each of the four sensor constructs (Figure S5). We also observed that the guard cells of plants expressing SOMA-NLS did not display any change in the ratio of YPet to Turquoise GL emission when treated with NaCl, in contrast to neighboring pavement cells (Figure S6).
The NaCl-induced change in the ratio of YPet to Turquoise GL emission of SOMA is dependent on MPK3/6
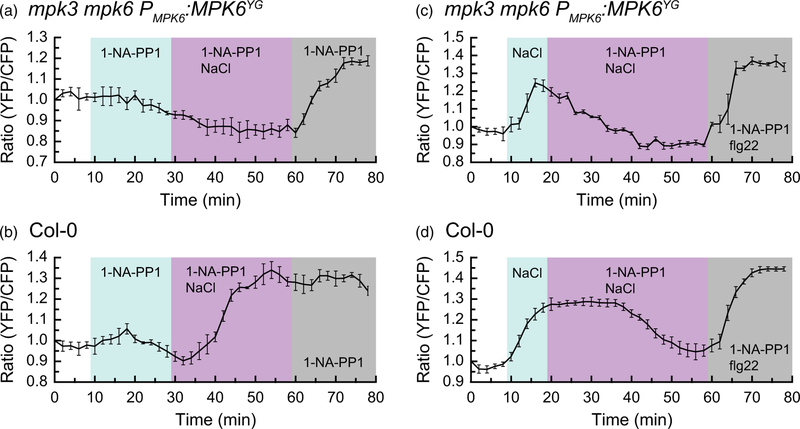
We next wanted to determine if the change in the ratio of YPet to Turquoise GL emission triggered by NaCl stress in cotyledons was dependent on MAPK activity. Because MPK3 and MPK6 have been shown to be highly redundant kinases, we wanted to test SOMA in an mpk3 mpk6 double-mutant background (Wang et al., 2007). It is also known, however, that mpk3 mpk6 double mutants are embryo lethal (Wang et al., 2007). We therefore made use of mpk3 mpk6 Pmpk6:MPK6YG, a conditional loss-of-function mpk3 mpk6 double-mutant line (Xu et al., 2014). This line is homozygous for T-DNA knockout mutations of MPK3 and MPK6. It also expresses MPK6YG, which is a version of MPK6 that is sensitive to 1-NA-PP1, a bulky ATP analog that does not bind efficiently to the ATP-binding pockets of wild-type kinases but acts as a potent inhibitor of MPK6YG (Bishop et al., 2000; Xu et al., 2014). Working with the mpk3 mpk6 Pmpk6:MPK6YG line therefore allowed us to use a chemical–genetic approach to switch off MPK3/ 6 activity during live-cell imaging.
We introduced SOMA-NES into the mpk3 mpk6 Pmpk6: MPK6YG line via genetic crossing and tested the effect of the 1-NA-PP1 inhibitor on NaCl-induced changes in the ratio of YPet to Turquoise GL emission. A detached cotyledon from the SOMA-NES mpk3 mpk6 Pmpk6:MPK6YG line was mounted in water in a HybriWell and observed using confocal microscopy. Addition of 1-NA-PP1 to the sample did not cause a substantial change in FRET efficiency. After 20 min of incubation in 1-NA-PP1, the solution in the imaging chamber was replaced with 150 mM NaCl + 10 µm 1-NA-PP1. No change in the ratio of YPet to Turquoise GL emission was observed, suggesting that MPK3/6 activity is required for the previously described NaCl-induced change in the ratio of YPet to Turquoise GL emission seen with SOMA. At the 60 min time-point, the solution in the imaging chamber was replaced with 10 µm 1-NA-PP1. The effect of this final solution change was to produce a hypo-osmotic shock to the sample while maintaining the presence of 10 µm 1-NA-PP1. With this hypo-osmotic shock, we observed a rapid gain in the ratio of YPet to Turquoise GL emission. This result suggests that a MAPK other than MPK3/6 may be responsible for this hypo-osmotic-induced change in the ratio of YPet to Turquoise GL emission. It should be noted that NaCl stress has been shown to induce the kinase activities of MP3/6 but not MPK4 (Droillard et al., 2004). By contrast, hypo-osmotic shock induces MPK3/6 and MPK4 (Droillard et al., 2004). Therefore MPK4 is a good candidate for the kinase driving the change in the ratio of YPet to Turquoise GL emission of SOMA caused by hypo-osmotic shock.
As a control, we performed the same experiment as described above using SOMA-NES in the wild-type Col-0 genetic background. In that case, we observed a strong increase in the ratio of YPet to Turquoise GL emission upon NaCl treatment, even in the presence of 1-NA-PP1 (Figure 3b), suggesting that the inhibitor does not affect wild-type MAPKs. When the final hypo-osmotic shock was applied to this sample, the initial NaCl-induced increase in the ratio of YPet to Turquoise GL emission had not diminished, so no additional gain was observed.
Figure 3.
MPK6 activity is needed for the NaCl-induced increase in the ratio of YPet to Turquoise GL emission of sensor of mitogen-activated protein kinase (MAPK) activity (SOMA).
The SOMA-NES in the cotyledon epidermis was analyzed in wild-type Col-0 and mpk3 mpk6 Pmpk6:MPK6YG genetic backgrounds. Graphs depict the average ratio of YPet to Turquoise GL emission produced by exciting Turquoise GL using the regions of interest shown in Figure S18. The shaded background on each graph indicates when the sample was exposed to a given treatment. During the first 10 min of each experiment the samples were incubated in pure water. ‘1-NA-PP1’ indicates 10 µm 1-NA-PP1, ‘1-NA-PP1 NaCl’ indicates 10 µm 1-NA-PP1 + 150 mM NaCl, and ‘1-NA-PP1 flg22’ indicates 10 µm 1-NA-PP1 + 1 µm flg22 was present in the imaging chamber during the shaded time period. Error bars indicate standard deviation. ‘Ratio (YFP/CFP)’ indicates the normalized value of YPet emission divided by Turquoise GL emission after excitation of Turquoise GL with 458 nm light. Videos are available as Movies S5–S8. 1-NA-PP1 is an ATP analog. These experiments were repeated at least four times with similar results.
Next we wanted to determine if 1-NA-PP1 could reverse the NaCl-induced gain in the ratio of YPet to Turquoise GL emission of the SOMA-NES mpk3 mpk6 Pmpk6:MPK6YG line. For this experiment, an initial 150 mM NaCl treatment caused the expected increase in the ratio of YPet to Turquoise GL emission. Ten minutes after that NaCl treatment, the sample was exposed to 150 mM NaCl + 10 µm 1-NA-PP1. Addition of 1-NA-PP1 caused the ratio of YPet to Turquoise GL emission to drop back to the pre-treatment level within 20 min (Figure 3c). By contrast, 1-NA-PP1 did not affect the duration of the NaCl-induced gain in the ratio of YPet to Turquoise GL emission with SOMA-NES in the wild-type Col-0 background (Figure 3d), which took about 35 min to return pre-treatment levels. Therefore, 1-NA-PP1 was able to reverse the NaCl-induced gain in the ratio of YPet to Turquoise GL emission of SOMA in the mpk3 mpk6 Pmpk6:MPK6YG background, suggesting that MPK3/6 activity is necessary for the sustained, elevated ratio of YPet to Turquoise GL emission triggered by NaCl treatment. As before, a final hypo-osmotic/flg22 treatment produced a gain in the ratio of YPet to Turquoise GL emission at the end of the imaging period for both samples. Additional control experiments using DMSO in place of 1-NA-PP1 confirmed that the effects we observed were due to 1-NA-PP1 rather than the DMSO solvent (Figure S7).
The response to flg22 is heterogeneous
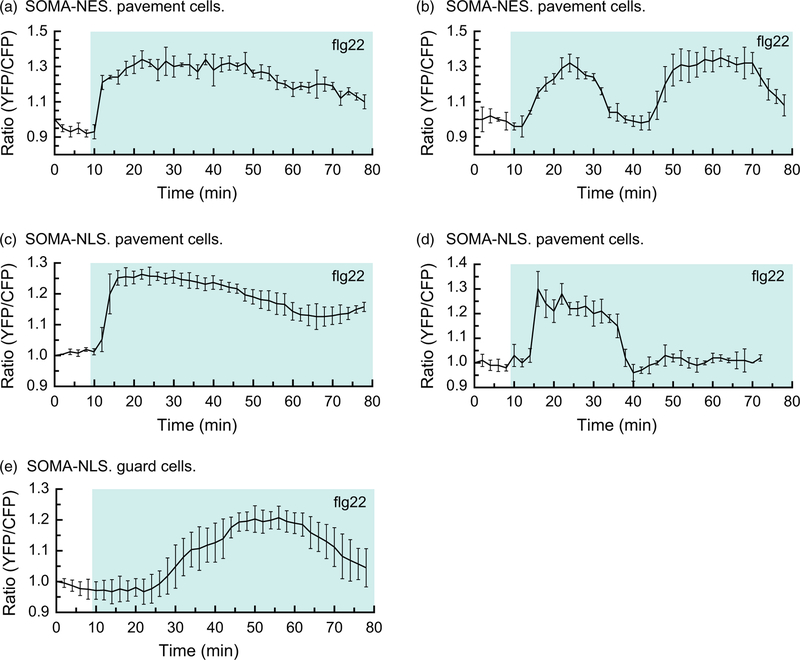
flg22 is a synthetic peptide whose sequence is derived from the bacterial elicitor flagellin (Gomez-Gomez et al., 1999). flg22 is known to rapidly induce the activity of MPK3/4/6 via the plasma membrane-bound receptor FLS2 (Gomez-Gomez and Boller, 2000; Asai et al., 2002). We were therefore interested in characterizing the response to flg22 treatment of plants expressing SOMA. For this experiment, 5-day-old detached cotyledons were treated with 1 µm flg22 during the imaging process. In the case of SOMA-NES, we observed a heterogeneous pattern of changes in the ratio of YPet to Turquoise GL emission (Figure 4a,b). In all the pavement cells in the sample, we observed a rapid gain in the ratio of YPet to Turquoise GL emission within a few minutes of flg22 treatment. In a subset of the pavement cells, the ratio of YPet to Turquoise GL emission rapidly dropped back to the pre-treatment levels within 30 min, followed by a second rapid gain in the ratio of YPet to Turquoise GL emission to produce the biphasic pattern seen in Figure 4(a). In neighboring regions of the same cotyledon that is shown in Figure 4(a), the initial gain in the ratio of YPet to Turquoise GL emission did not drop down to pre-treatment levels until 60 min after exposure to flg22 (Figure 4b). These results demonstrated that the ratio of YPet to Turquoise GL emission of SOMA can rapidly increase and decrease in vivo, and that the pattern of changes induced by flg22 treatment can vary within a single cotyledon.
Figure 4.
The response of sensor of mitogen-activated protein kinase (MAPK) activity (SOMA) to flg22 is heterogeneous.
The response of the SOMA-NES-1 and SOMA-NLS-1 transgenic lines to flg22 treatment was evaluated by analyzing cotyledon epidermal cells. Graphs depict the average ratio of YPet to Turquoise GL emission produced by exciting Turquoise GL using the regions of interest shown in Figure S19. The shaded background on each graph indicates when the sample was exposed to a given treatment. During the first 10 min of each experiment the samples were incubated in pure water. ‘flg22’ indicates that 1 µm flg22 was present in the imaging chamber during the shaded time period. Error bars indicate standard deviation. ‘Ratio (YFP/CFP)’ indicates the normalized value of YPet emission divided by Turquoise GL emission after excitation of Turquoise GL with 458 nm light. Videos are available as Movies S9–S11. These experiments were repeated at least five times with similar results.
In the case of SOMA-NLS, we also observed distinct patterns of the ratio of YPet to Turquoise GL emission in response to flg22 treatment. In pavement cells we saw a rapid increase in the ratio of YPet to Turquoise GL emission shortly after flg22 treatment in all samples tested. In some cases, the gain returned to pre-treatment levels within 30 min of exposure to flg22 (Figure 4c) while in other examples the gain was sustained for over 60 min (Figure 4d). In addition, we consistently observed that gain in the ratio of YPet to Turquoise GL emission displayed by SOMA-NLS in guard cells was substantially delayed when compared with pavement cells in the same sample (Figure 4d,e). In pavement cells, the flg22-induced gain begins within 4 min of flg22 exposure, while in guard cells it begins about 15 min after flg22 has been added to the sample. We did not observe any change in the ratio of YPet to Turquoise GL emission with SOMA-NES in guard cells, but this may be due to technical challenges associated with imaging the cytoplasm of guard cells with our imaging system.
Repetition of the flg22 experiments using independent transgenic lines for each sensor construct produced similar results (Figure S8). In addition, experiments performed using the SOMA-NES mpk3 mpk6 Pmpk6:MPK6YG line indicated that 1-NA-PP1 did not affect flg22-and chitin-induced changes in the ratio of YPet to Turquoise GL emission (Figure S9). These results suggest that MAPKs in addition to MPK3/6 are able to drive the flg22-and chitin-induced changes in the ratio of YPet to Turquoise GL emission of SOMA. Since MPK4 is known to be activated by flg22 and chitin, it is a likely candidate (Ichimura et al., 2006).
Chitin induces a rapid FRET gain in guard cells
Like flg22, chitin is a pathogen-associated elicitor known to trigger rapid activation of MPK3/4/6 (Yamada et al., 2016). Using the same experimental system as described above, 5-day-old cotyledons were exposed to 40 mg ml−1 chitin during imaging. As with flg22, we observed a rapid gain in the ratio of YPet to Turquoise GL emission in pavement cells with SOMA-NES and SOMA-NLS (Figure 5a–b). The most pronounced difference between the response of cotyledons to chitin versus flg22 was seen in guard cells where chitin induced a gain in the ratio of YPet to Turquoise GL emission shortly after treatment, in contrast to the substantially delayed gain caused by flg22 exposure in guard cells. In addition, SOMA-NLS in guard cells often produced a strong second peak of YPet versus Turquoise GL emission intensity about 45 min after the initial treatment. Similar results were obtained with experiments performed using independent transgenic lines for each sensor construct (Figure S10).
Figure 5.
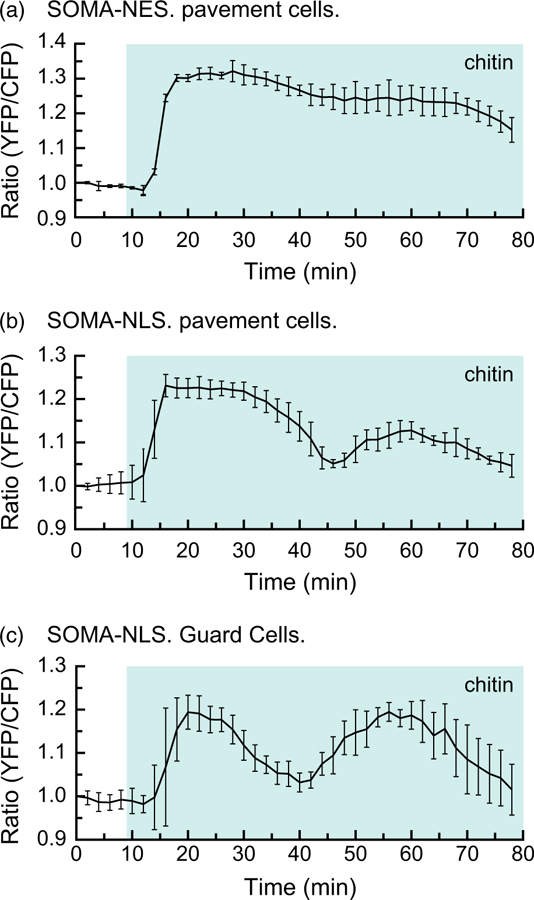
Chitin induces a rapid increase in the ratio of YPet to Turquoise GL emission in guard cells.
The response of the SOMA-NES-1 and SOMA-NLS-1 transgenic lines to chitin treatment was evaluated by analyzing cotyledon epidermal cells. Graphs depict the average ratio of YPet to Turquoise GL emission produced by exciting Turquoise GL using the regions of interest shown in Figure S20. The shaded background on each graph indicates when the sample was exposed to a given treatment. During the first 10 min of each experiment the samples were incubated in pure water. ‘chitin’ indicates that 40 mg ml−1 chitin was present in the imaging chamber during the shaded time period. Error bars indicate standard deviation. ‘Ratio (YFP/CFP)’ indicates the normalized value of YPet emission divided by Turquoise GL emission after excitation of Turquoise GL with 458 nm light. Videos are available as Movies S12 and S13. These experiments were repeated at least five times with similar results.
DISCUSSION
Reversible protein phosphorylation is a fundamental mechanism used to regulate a myriad of signal transduction pathways. Modulation of protein kinase activity therefore plays an essential role in cellular signaling. For this reason, there is considerable scientific interest in measuring the activation status of protein kinases in living systems. Established methods for monitoring the activation status of protein kinases involve first homogenizing tissues or organisms and then analyzing the kinase of interest in vitro. There are two major drawbacks to this approach: (i) potential heterogeneity of kinase activity within the tissue is lost due to the homogenization process and (ii) the living sample must be destroyed in order to analyze it, which means that time-course experiments on a single sample cannot be performed. Genetically encoded biosensors provide an alternative to these destructive methods for monitoring kinase activity in living cells (Aoki et al., 2012; Sample et al., 2014).
Genetically encoded biosensors for measuring MAPK activity have been developed for use in animal cells (Kamioka et al., 2012; Tomida et al., 2012; Aoki et al., 2013; Ryu et al., 2015; Sari et al., 2018). We were therefore interested in determining if one of these sensors could be adapted for use in Arabidopsis. To accomplish this, we replaced the substrate domain of an animal MAPK sensor with an 80-amino-acid portion of Arabidopsis MKP1 (Ulm et al., 2001). This substrate domain had been previously shown to contain a threonine residue (T64) that can be phosphorylated by MPK6 (Park et al., 2011). Our in vitro testing confirmed that this threonine can be efficiently phosphorylated by Arabidopsis MAPKs in the context of our biosensor and that this phosphorylation caused a gain of about 50% in the ratio of YPet to Turquoise GL emission, which is similar to the gain reported for kinase activity sensors used in animal cells (Komatsu et al., 2011). These experiments indicated that our sensor of MAPK activity (SOMA) is able to report the presence of activated MAPKs through an increase in the ratio of YPet to Turquoise GL emission in response to Turquoise GL excitation in vitro.
We were next interested in evaluating the performance of SOMA in vivo. The following evidence supports the idea that SOMA is able to act as a sensor of MAPK activity in living plants. To begin with, we have demonstrated that three stress treatments known to activate MAPK activity in Arabidopsis all cause rapid gains in the ratio of YPet to Turquoise GL emission of SOMA: 150 mM NaCl, 1 µm flg22 and 40 mg ml−1 chitin. Next, using lines expressing the phosphosite mutant SOMAT679A, we demonstrated that these stress-induced gains in the ratio of YPet to Turquoise GL emission depend on the known MAPK phosphorylation site present in the sensor. Finally, we tested SOMA in the mpk3 mpk6 Pmpk6:MPK6YG genetic background, which allowed us to use a chemical–genetic approach to selectively switch off MPK3/6 activity using the 1-NA-PP1 inhibitor (Xu et al., 2014). These experiments demonstrated that MPK3/6 activity is necessary to drive the gain in the ratio of YPet to Turquoise GL emission of SOMA induced by NaCl stress. Taken together, these results show that the NaCl-induced increases in the ratio of YPet to Turquoise GL emission that we observe with SOMA require the known MPK3/6 phosphorylation site present in SOMA and the activity of MPK3/ 6. These results are consistent with a model in which SOMA acts as a reporter of MAPK activity in vivo.
The MAPK gene family in Arabidopsis is composed of 20 members, and it has been shown that many stress treatments activate MPK3, MPK6, MPK4 and others (Rodriguez et al., 2010). We were therefore interested in understanding the extent to which SOMA may be reporting the activities of MAPKs in addition to MPK3/6. As discussed above, the gain in the ratio of YPet to Turquoise GL emission of SOMA caused by NaCl stress appears to be due to MPK3/6 activity. The gains in the ratio of YPet to Turquoise GL emission of SOMA caused by hypo-osmotic stress and flg22 treatment, however, are not diminished by 1-NA-PP1 when tested in the mpk3 mpk6 Pmpk6:MPK6YG background. These results suggest that MAPKs other than MPK3/6 are able to drive changes in the ratio of YPet to Turquoise GL emission of SOMA. It is interesting to note that both hypo-osmotic shock and flg22 are known to activate MPK4 in addition to MPK3/6, and flg22 treatment also activates MPK1, MPK11 and MPK13 (Bethke et al., 2012; Nitta et al., 2014). By contrast, stress due to elevated NaCl appears to activate MPK3/ 6 but not MPK4 (Droillard et al., 2004). These results are consistent with a model in which SOMA acts as a MAPK activity reporter for MPK4 as well as MPK3/6. Further genetic testing with additional genetic backgrounds will be needed to determine the full extent to which SOMA is able to report the activities of different MAPK isoforms. These experiments are complicated by the embryo lethality of mpk3 mpk6 double mutants and the severe dwarf phenotype of mpk4 single mutants (Ichimura et al., 2006; Suarez-Rodriguez et al., 2007; Wang et al., 2007).
The SOMA sensor, and similar MAPK sensors developed for use in animal cells, function by reporting the phosphorylation status of a substrate domain known to be phosphorylated by the kinase of interest. Because of this situation, it might be more accurate to think of these sensors as ‘phosphorylation status reporters’ since they integrate the net kinase and phosphatase activities acting on that specific phosphosite. In the case of SOMA, we have presented experimental evidence that MAPKs are able to target the sensor. It remains a possibility that other kinases may also be able to phosphorylate SOMA. Future genetic and bio-chemical studies will be needed to resolve that question.
The gain in the ratio of YPet to Turquoise GL emission displayed by kinase activity reporters such as SOMA is believed to occur due to binding of the forkhead (FHA1) domain to the phosphorylated substrate domain of the sensor (Komatsu et al., 2011; Sample et al., 2014). In order for the reporter to provide an up-to-date readout of kinase activity, it is important that phosphatases are able to efficiently dephosphorylate the sensor. Work with similar kinase sensors in animal cells has indicated that the FHA1 domain used in SOMA does not bind so tightly to phosphorylated substrate domains that it precludes efficient access by phosphatases (Komatsu et al., 2011). Our observation that the ratio of YPet to Turquoise GL emission of SOMA can rapidly drop in vivo after an initial stress-induced gain suggests that, like its animal cell precursor, SOMA is able to be efficiently de-phosphorylated (Figure 4a,c).
Previous work using in vitro methods to assay MAPK activation status has indicated that stress treatments such as NaCl, flg22 and chitin typically cause MPK3/4/6 to be transiently activated for a time period ranging from about 30 to 60 min (Droillard et al., 2004; Wan et al., 2004; Ichimura et al., 2006; Meszaros et al., 2006; Doczi et al., 2007; Suarez-Rodriguez et al., 2007; Yu et al., 2010; Yamada et al., 2016). The patterns of changes in the ratio of YPet to Turquoise GL emission that we observed in vivo with SOMA are consistent with this type of transient MAPK activation. Because SOMA allows us to observe living cells in the context of intact tissue, however, we were also able to document heterogeneity at the cellular level. A common type of heterogeneity was the observation that some cells produced relatively transient peaks of the ratio of YPet to Turquoise GL emission lasting about 25 min, while other cells in the same sample maintained their elevated levels for over 60 min. In addition, we also observed that guard cells display distinct patterns of the ratio of YPet to Turquoise GL emission when compared with pavement cells in the same sample. It should be noted that one potential source of heterogeneity during this type of imaging experiment would be non-uniform exposure of the sample to the chemical treatment, such as flg22. We have previously characterized the imaging chambers used for this study, and based on that analysis we expect that the regions of the cotyledons analyzed here should have been uniformly exposed to the chemical treatments (Vang et al., 2018). Nevertheless, it is important to consider both technical and biological explanations for heterogeneous responses. If the heterogeneity is biological in origin, its biological significance is unknown at this time, but our ability to observe it highlights the potential for biosensors such as SOMA to reveal potentially new dimensions to signaling pathways that could not be observed using established in vitro methods.
In the present study, our objective has been to document the performance of a genetically encoded biosensor designed to report the activity of Arabidopsis MAPKs. To accomplish this we performed in vitro testing with E. coli expressed proteins and in vivo testing using the cotyledon epidermis as a model system. We chose the cotyledon epidermis because it provides a convenient and simple subject for confocal imaging. Because MAPK signaling has been implicated in a wide range of signaling pathways in Arabidopsis, it will be of interest to determine the extent to which SOMA can be used to study additional signaling pathways in different organ and tissue systems. While beyond the scope of the present study, such future work has the potential to reveal new aspects of MAPK signaling that can only be observed using live-cell imaging methods.
EXPERIMENTAL PROCEDURES
Plasmid construction
The plasmid backbones used to construct our SOMA plasmids were a kind gift of Michiyuki Matsuda and Kazuhiro Aoki. Specifically, we used the previously described AKAR3 construct (Komatsu et al., 2011) and swapped in the following functional domains using conventional restriction enzyme cloning: ECFP was replaced with Turquoise-GL, the shorter linker was replaced with the 244-amino-acid EV linker and the substrate domain of AKAR3 was replaced with DNA encoding for amino acids 15–94 of the Arabidopsis MKP1 protein. We also introduced a serine to aspartic acid substitution within the MKP1 coding sequence, which corresponds to the +3 position with respect to the phosphothreonine site in the MKP1 substrate domain, as previous work has shown that an aspartic acid at this position enhances the affinity of FHA1 for the phosphorylated form of the substrate (Yongkiettrakul et al., 2004; Komatsu et al., 2011). To make the phosphosite mutant form of SOMA, site-directed mutagenesis was then performed to change the coding sequence at the threonine 679 position of the sensor protein to alanine. The resulting SOMA and SOMAT679A protein-coding sequences were then cloned into a derivative of pET-32(a)+ (Millipore Sigma, http://www.merckmillipore.com/) for expression of recombinant proteins in E. coli. Plasmid maps for pET-SOMA and pET-SOMA-T679A are provided in Figures S11 and S12, and the full DNA sequence for each is provided in Data S1 and S2. For targeted expression, our sensors in the nuclei of transgenic plants, the SOMA and SOMAT679A protein-coding sequences, were moved into the binary vector pCAMBIA-1300 to produce pSOMA-NLS and pSOMAT679A-NLS. For targeted expression in the cytoplasm of transgenic plants, the SOMA and SOMAT679A protein-coding sequences were moved into a derivative of the binary vector pRI 201-AN (Takara Bio, http://www.takarabio.com/) to produce pSOMA-NES and pSOMAT679A-NES. Plasmid maps for these constructs are provided in Figures S13–S16, and the full DNA sequence for each is provided in Data S3–S6. Plasmid DNA for pET-SOMA (Addgene no. 118935), pET-SOMA-T679A (Addgene no. 118936), pSOMA-NLS (Addgene no. 118937), pSOMAT679A-NLS (Addgene no. 118938), pSOMA-NES (Addgene no. 118939) and pSOMAT679A-NES (Addgene no. 118940) is available through Addgene (http://www.addgene.org/).
In vitro FRET assay
Recombinant SOMA, SOMAT679A and MPK6-CA were expressed in the Rosetta2 strain of E. coli (Millipore Sigma). MPK6-CA is a constitutively active form of Arabidopsis MPK6 (Berriri et al., 2012). A plasmid expressing MPK6-CA was a kind gift from Jean Colcombet. Escherichia coli cells expressing the recombinant proteins were treated with Bug Buster (Millipore Sigma). Then 7 µl of the SOMA cleared lysate was added to a reaction containing 25 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol (TRIS)-HCl, 0.5 mM DTT, 5 mM MgCl2, 0.1 mM ATP and 100 mM NaCl and water to a total volume of 200 µl. For reactions including MPK6-CA, 4 µl of the MPK6-CA cleared lysate was also added. The reactions were then incubated at 25°C for 30–60 min and the fluorescent emission spectrum was measured using a QuantaMaster 40 Spectrofluorometer. Excitation was performed at 420 nm and the emission range analyzed was 465–545 nm. The slit size for both excitation and emission was 5 nm with a step size of 0.5 nm. For the lambda protein phosphatase (LPP) experiments, 2 µl of LPP, 20 µl of 10 mM MgCl2 and 20 µl of 10× NEBuffer Pack for Protein MetalloPhosphatases (PMP) (New England Biolabs, http://www.neb.com/) were added to the reactions after 60 min of incubation, and the reaction was further incubated at 30°C for another 15 min.
In vitro kinase assay
Recombinant His-tagged MPK3, MPK4, MPK6 and MPK10 (7.5 µg each) were activated by incubation with a mixture of 0.125 µg recombinant MKK4DD and 0.125 µg MKK5DD as previously described (Liu and Zhang, 2004). Activated MAPKs were then used to phosphorylate recombinant SOMA and SOMA-T679A proteins (1:20 enzyme:substrate weight ratio) in the kinase reaction buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2 and 1 mM DTT) with 25 µm ATP and [γ-32P] ATP (0.1 µCi per reaction). The reactions were stopped by the addition of SDS-loading buffer after 30 min. After resolution in a 10% SDS-polyacrylamide gel, all the proteins were visualized by Coomassie blue staining, and the phosphorylated SOMA was visualized by autoradiography.
Transgenic lines
Transgenic lines expressing SOMA-NLS, SOMAT679A-NLS, SOMA-NES and SOMAT679A-NES were produced via the floral dip method using the GV3101 strain of Agrobacterium tumefaciens (Clough and Bent, 1998). Primary transformants expressing the sensor constructs were identified by screening with an epifluorescence microscope 4-day-old seedlings germinated on 1% agar (w/v) plates containing 0.5× Murashige and Skoog basal salt mixture. Seedlings showing strong fluorescence were transferred to soil and seed was collected. Two independent transgenic lines for each of the four sensor constructs were produced as shown in Table 1. Seed for the transgenic lines is available via the Arabidopsis Biological Resource Center (ABRC) (http://abrc.osu.edu/), and the ABRC stock number for each line is shown in Table 1.
Plant material and growth conditions
To prepare seedlings for confocal imaging, seeds were surface sterilized using 95% ethanol and then plated onto growth media composed of 1% agar (w/v) containing 0.5× Murashige and Skoog basal salt mixture in 130 mm square Petri dishes. The samples were incubated in the dark at 4°C for 1–3 days to stratify the seeds and then placed under constant light at 20–25°C with the plates in a vertical orientation. Cotyledons were harvested for imaging after the plates had been under light for 4–5 days.
Confocal microscopy
Detached cotyledons were prepared for imaging on the confocal microscope using the HybriWellTM method as previously described (Vang et al., 2018). Briefly, a 5-µl drop of ultrapure water was placed on a 45 mm 9× 55 mm microscope cover glass. Then, a cotyledon from a 4–5-day-old seedling was placed abaxial side down on top of the droplet. Cotyledons were gently submerged by dripping water around them with a pipette. Excess water was then removed, and a HybriWellTM (Grace Bio-Labs, http://gracebio.com/, cat. no. 611102) was gently placed on the coverslide with the cotyledon in the center to form a 150-µm deep imaging chamber with a volume of 30 µl. The dimensions of the chamber are such that the cotyledon is gently held in place by pressure from the top of the chamber, but liquid can still pass between the abaxial side of the cotyledon and the coverglass (Vang et al., 2018). Then 400 µl of ultrapure water was injected through one of the HybriWellTM ports using a pipettor to fill the 30-µl chamber with water and expel any air bubbles. A 200-µl droplet of ultrapure water was then placed on one of the ports to prevent the chamber from drying out. The HybriWells containing mounted seedlings were then placed in covered Petri dishes and equilibrated by incubating at 20–23°C under constant light for 6–12 h prior to imaging.
Confocal microscopy was performed using either a Zeiss LSM 510 Meta or a Zeiss LSM 780 (http://www.zeiss.com/) with a 20× objective. Samples were excited at 458 nm with 1% power for the LSM 510 and 5% power for the LSM 780. Emission was measured between 463 and 517 nm for Turquoise GL and between 534 and 570 nm for YPet. Z-stacks were collected every 2 min with an optical slice thickness of 2.92 µm for the LSM 510 and 2 µm for the LSM 780.
Chemical treatments were added to the samples during imaging by pipetting 300 µl of solution containing the treatment onto one port of the HybriWell. For flg22 treatment, we used a 1 µm solution of synthetic flg22 peptide (QRLSTGSRINSAKDDAAGLQIA) (PhytoTechnology Laboratories, http://phytotechlab.com/, cat. no. P6622) in water. For chitin treatment, 200 mg of chitin from shrimp shells (Sigma, http://www.sigmaaldrich.com/, cat. no. C7170–100G) was added to a 1.5 ml Eppendorf tube containing two steel pellets and ground at 25 Hz for 10 min using a Retsch MM 200 mixer mill (Retsch, http://www.retsch.com/). The pulverized chitin was then added to 5 ml of water and vortexed briefly prior to use. The ATP analog 1-NA-PP1 (MedChemExpress, http://www.medchemexpress.com/, cat. no. HY-13941) was obtained from the manufacturer as a 10 mM stock solution dissolved in dimethyl sulfoxide (DMSO).
Image analysis
Post-processing of the raw image data was performed using Fiji (Schindelin et al., 2012). First, the ‘Z-projection’ function was performed on an image stack using the ‘Max Intensity’ setting. The resulting projection was then separated into two separate images, one for the Turquoise GL emission channel and one for the YPet emission channel. The ‘Subtract Background’ function was performed on both images, with the ‘rolling ball radius’ set as the default 50 pixels. A mask was then created from the YPet channel using the ‘Convert to Mask’ function. The background subtracted YPet and Turquoise GL images were then converted into 32-bit images. These 32-bit images were then multiplied by the Mask file. The resulting YPet image was divided by the resulting Turquoise GL image using the ‘Image Calculator’ function to create to a ratio image representing the ratio of YPet to Turquoise emission. Finally, the ‘Threshold’ function was performed using the default values, with the ‘NaN background’ option enabled. The ‘Fire’ lookup table was then applied to the final ratio image.
To measure the ratio of YPet to Turquoise GL emission, a region of interest (ROI) was selected within the ratio image using Fiji and the average ratio value within that ROI was then measured. For time-course experiments, the change in the YPet to Turquoise GL emission ratio was normalized to the initial value of the YPet to Turquoise GL emission ratio and plotted versus time. The initial pixel intensity values for the raw image data in the YPet and Turquoise GL channels are presented for each experiment in Table S1. The ROIs used for analyzing the data presented in Figures 1–5 are shown in Figures S17–S20. Movies S1–S13 present time-course videos of the ratio image data for selected experiments.
Supplementary Material
Data S1. Annotated DNA sequence of plasmid pET-SOMA.
Movie S12. Figure 5(a): SOMA-NES chitin.
Movie S2. Figure 2(f): SOMA (sensor of MAPK activity)-NES NaCl.
Movie S3. Figure 2(g): SOMAT679A-NLS NaCl.
Movie S4. Figure 2(h): SOMAT679A-NES NaCl.
Movie S5. Figure 3(a): SOMA-NES mpk3 mpk6 Pmpk6:MPK6YG 1-NA-PP1 NaCl.
Movie S6. Figure 3(b): SOMA-NES Col-0 1-NA-PP1 NaCl.
Movie S7. Figure 3(c): SOMA-NES mpk3 mpk6 Pmpk6:MPK6YG 1-NA-PP1 NaCl.
Movie S8. Figure 3(d): SOMA-NES Col-0 1-NA-PP1 NaCl.
Data S2. Annotated DNA sequence of plasmid pET-SOMA-T679A.
Figure S1. In vitro Förster resonance energy transfer assay with lambda protein phosphatase.
Figure S10. Chitin treatment of SOMA (sensor of MAPK activity)-NLS-2, SOMA-NES-2, SOMAT679A-NLS-2, and SOMAT679A-NES-2.
Figure S11. Map of plasmid pET-SOMA.
Figure S12. Map of plasmid pET-SOMA-T679A.
Figure S13. Map of plasmid pSOMA-NLS.
Figure S14. Map of plasmid pSOMA-T679A-NLS.
Figure S15. Map of plasmid pSOMA-NES.
Figure S16. Map of plasmid pSOMA-T679A-NES.
Figure S17. Regions of interest for Figure 2.
Figure S18. Regions of interest for Figure 3.
Data S3. Annotated DNA sequence of plasmid pSOMA-NLS.
Figure S19. Regions of interest for Figure 4
Figure S2. Seedling phenotypes of SOMA (sensor of MAPK activity) transgenic lines.
Figure S20. Regions of interest for Figure 5.
Figure S3. Rosette phenotypes of SOMA (sensor of MAPK activity) transgenic lines.
Figure S4. Control treatment with water only for SOMA (sensor of MAPK activity)-NLS and SOMA-NES.
Figure S5. SOMA (sensor of MAPK activity)-NLS-2, SOMA-NES-2, SOMAT679A-NLS-2 and SOMAT679A-NES-2 NaCl treatment.
Figure S6. The in vivo response of guard cells expressing SOMA (sensor of MAPK activity)-NLS-1 to NaCl stress.
Figure S7. The DMSO controls for SOMA (sensor of MAPK activity)-NES in mpk3 mpk6 Pmpk6:MPK6YG.
Figure S8. flg22 treatment of SOMA (sensor of MAPK activity)-NLS-2, SOMA-NES-2, SOMAT679A-NLS-2 and SOMAT679A-NES-2.
Figure S9. flg22 and chitin treatment of SOMA (sensor of MAPK activity)-NES in mpk3 mpk6 Pmpk6:MPK6YG.
Data S4. Annotated DNA sequence of plasmid pSOMA-T679A-NLS.
Table S1. Initial pixel intensity values from raw image data.
Data S5. Annotated DNA sequence of plasmid pSOMA-NES.
Data S6. Annotated DNA sequence of plasmid pSOMA-T679A-NES.
Movie S1. Figure 2(e): SOMA (sensor of MAPK activity)-NLS NaCl.
Movie S11. Figure 4(d): SOMA-NLS flg22.
ACKNOWLEDGEMENTS
The authors wish to thank Michiyuki Matsuda and Kazuhiro Aoki for generously sharing their FRET sensor constructs. This work was supported by the National Science Foundation (NSF MCB-1137950). The authors wish to thank John Seagrist for technical assistance with this project.
Footnotes
CONFLICT OF INTERESTS
The authors declare no competing financial interests.
REFERENCES
- Albeck JG, Mills GB and Brugge JS (2013) Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49, 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Komatsu N, Hirata E, Kamioka Y and Matsuda M (2012) Stable expression of FRET biosensors: a new light in cancer research. Cancer Sci 103, 614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Kumagai Y, Sakurai A, Komatsu N, Fujita Y, Shionyu C and Matsuda M (2013) Stochastic ERK activation induced by noise and cell-to-cell propagation regulates cell density-dependent proliferation. Mol. Cell, 52, 529–540. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM and Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Berriri S, Garcia AV, Frei dit Frey N, Rozhon W, Pateyron S, Leon-hardt N, Montillet JL, Leung J, Hirt H and Colcombet J (2012) Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell, 24, 4281–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Pecher P, Eschen-Lippold L, Tsuda K, Katagiri F, Glaze-brook J, Scheel D and Lee J (2012) Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact 25, 471–480. [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT et al. (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature, 407, 395–401. [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature, 482, 103–106. [DOI] [PubMed] [Google Scholar]
- Bush SM and Krysan PJ (2007) Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot 58, 2181–2191. [DOI] [PubMed] [Google Scholar]
- Clough SJ and Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16, 735–743. [DOI] [PubMed] [Google Scholar]
- De Col V, Fuchs P, Nietzel T et al. (2017) ATP sensing in living plant cells reveals tissue gradients and stress dynamics of energy physiology. Elife, 6, Elife 10.7554/eLife.26770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S and Frommer WB (2006) Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell, 18, 2314–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devendrakumar KT, Li X and Zhang Y (2018) MAP kinase signalling: interplays between plant PAMP-and effector-triggered immunity. Cell. Mol. Life Sci 75, 2981–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doczi R, Brader G, Pettko-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M and Hirt H (2007) The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell, 19, 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droillard MJ, Boudsocq M, Barbier-Brygoo H and Lauriere C (2004) Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett 574, 42–48. [DOI] [PubMed] [Google Scholar]
- Enders TA, Frick EM and Strader LC (2017) An Arabidopsis kinase cascade influences auxin-responsive cell expansion. Plant J 92, 68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick EM and Strader LC (2018) Kinase MPK17 and the peroxisome division factor PMD1 influence salt-induced peroxisome proliferation. Plant Physiol 176, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R, Ferrari S and De Lorenzo G (2011) Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide-or flagellin-induced resistance against Botrytis cinerea. Plant Physiol 157, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjetting KS, Ytting CK, Schulz A and Fuglsang AT (2012) Live imaging of intra-and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot 63, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Hink MA, Vischer NO, Jalink K and Gadella TW Jr (2010) Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods, 7, 137–139. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L and Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Felix G and Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18, 277–284. [DOI] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K and Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64, 355–365. [DOI] [PubMed] [Google Scholar]
- Grossmann G, Krebs M, Maizel A, Stahl Y, Vermeer JEM and Ott T (2018) Green light for quantitative live-cell imaging in plants. J. Cell Sci 131, 10.1242/jcs.209270 [DOI] [PubMed] [Google Scholar]
- Hernandez-Barrera A, Velarde-Buendia A, Zepeda I, Sanchez F, Quinto C, Sanchez-Lopez R, Cheung AY, Wu HM and Cardenas L (2015) Hyper, a hydrogen peroxide sensor, indicates the sensitivity of the Arabidopsis root elongation zone to aluminum treatment. Sensors (Basel), 15, 855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CL, Sun YJ, Pillitteri LJ, Torii KU, Wang H, Zhang S and Ma H (2008) Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant, 1, 645–658. [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K and Shirasu K (2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem 281, 36969–36976. [DOI] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl Acad. Sci. USA 106, 20520–20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Danielson JA, Manojkumar SN, Lanquar V, Grossmann G and Frommer WB (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife, 3, e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF and Smith AE (1984) Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature, 311, 33–38. [DOI] [PubMed] [Google Scholar]
- Kamioka Y, Sumiyama K, Mizuno R, Sakai Y, Hirata E, Kiyokawa E and Matsuda M (2012) Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct. Funct 37, 65–73. [DOI] [PubMed] [Google Scholar]
- Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y and Matsuda M (2011) Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell, 22, 4647–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G, Samajova O, Ovecka M and Samaj J (2018) Cell and developmental biology of plant mitogen-activated protein kinases. Annu. Rev. Plant Biol 69, 237–265. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB and Alon U (2004) Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet 36, 147–150. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Grossmann G, Vinkenborg JL, Merkx M, Thomine S and Frommer WB (2014) Dynamic imaging of cytosolic zinc in Arabidopsis roots combining FRET sensors and RootChip technology. New Phytol 202, 198–208. [DOI] [PubMed] [Google Scholar]
- Liu Y and He C (2017) A review of redox signaling and the control of MAP kinase pathway in plants. Redox. Biol 11, 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y and Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell, 16, 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros T, Helfer A, Hatzimasoura E et al. (2006) The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J 48, 485–498. [DOI] [PubMed] [Google Scholar]
- Moustafa K, AbuQamar S, Jarrar M, Al-Rajab AJ and Tremouillaux-Guiller J (2014) MAPK cascades and major abiotic stresses. Plant Cell Rep 33, 1217–1225. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Banerjee S, Wheeler A, Ratliff LA, Irigoyen S, Garcia LR, Lockless SW and Versaw WK (2015) Live imaging of inorganic phosphate in plants with cellular and subcellular resolution. Plant Physiol 167, 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Yamada S, Tominaga T, Ichikawa M and Miyawaki A (2004) Expanded dynamic range of fluorescent indicators for Ca(2 + ) by circularly permuted yellow fluorescent proteins. Proc. Natl Acad. Sci. USA 101, 10554–10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AW and Daugherty PS (2005) Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol 23, 355–360. [DOI] [PubMed] [Google Scholar]
- Nitta Y, Ding P and Zhang Y (2014) Identification of additional MAP kinases activated upon PAMP treatment. Plant Signal Behav 9, e976155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F and Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature, 313, 810–812. [DOI] [PubMed] [Google Scholar]
- Park HC, Song EH, Nguyen XC et al. (2011) Arabidopsis MAP kinase phosphatase 1 is phosphorylated and activated by its substrate AtMPK6. Plant Cell Rep 30, 1523–1531. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H et al. (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell, 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Ren D, Yang H and Zhang S (2002) Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem 277, 559–565. [DOI] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J and Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl Acad. Sci. USA 105, 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza A, Walia A, Lanquar V, Frommer WB and Jones AM (2017) In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat. Plants, 3, 803–813. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M and Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol 61, 621–649. [DOI] [PubMed] [Google Scholar]
- Ryu H, Chung M, Dobrzynski M, Fey D, Blum Y, Lee SS, Peter M, Kholodenko BN, Jeon NL and Pertz O (2015) Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol 11, 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample V, Mehta S and Zhang J (2014) Genetically encoded molecular probes to visualize and perturb signaling dynamics in living biological systems. J. Cell Sci 127, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sari DWK, Akiyama R, Naoki H, Ishijima H, Bessho Y and Matsui T (2018) Time-lapse observation of stepwise regression of Erk activity in zebrafish presomitic mesoderm. Sci. Rep 8, 4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM and Sorger PK (2009) Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature, 459, 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanko V, Giuliani C, Retzer K, Djamei A, Wahl V, Wurzinger B, Wilson C, Heberle-Bors E, Teige M and Kragler F (2014) Timing is everything: highly specific and transient expression of a MAP kinase determines auxin-induced leaf venation patterns in Arabidopsis. Mol. Plant, 7, 1637–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Zhang M, Zhang L, Sun T, Liu Y, Lukowitz W, Xu J and Zhang S (2017) Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell, 29, 526–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF and Krysan PJ (2007) MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol 143, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS and Stern DF (1998) Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science, 281, 272–274. [DOI] [PubMed] [Google Scholar]
- Sun T, Nitta Y, Zhang Q, Wu D, Tian H, Lee JS and Zhang Y (2018) Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep 19, e45324 10.15252/embr.201745324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Mizoguchi T, Yoshida R, Ichimura K and Shinozaki K (2011) Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell, 41, 649–660. [DOI] [PubMed] [Google Scholar]
- Tomida T, Oda S, Takekawa M, Iino Y and Saito H (2012) The temporal pattern of stimulation determines the extent and duration of MAPK activation in a Caenorhabditis elegans sensory neuron. Sci. Signal 5, ra76. [DOI] [PubMed] [Google Scholar]
- Ulm R, Revenkova E, di Sansebastiano GP, Bechtold N and Paszkowski J (2001) Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang S, Seitz K and Krysan PJ (2018) A simple microfluidic device for live cell imaging of Arabidopsis cotyledons, leaves, and seedlings. Biotechniques, 64, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED and Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife, 3, e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia A, Lee JS, Wasteneys G and Ellis B (2009) Arabidopsis mitogen-activated protein kinase MPK18 mediates cortical microtubule functions in plant cells. Plant J 59, 565–575. [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang S and Stacey G (2004) Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol 5, 125–135. [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC and Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell, 19, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bruffett K, Lee J, Hause G, Walker JC and Zhang S (2008) Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell, 20, 602–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY and Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell, 82, 463–473. [DOI] [PubMed] [Google Scholar]
- Xu J and Zhang S (2015) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20, 56–64. [DOI] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D and Zhang S (2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77, 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Shirakawa T et al. (2016) The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J 35, 2468–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongkiettrakul S, Byeon IJ and Tsai MD (2004) The ligand specificity of yeast Rad53 FHA domains at the +3 position is determined by nonconserved residues. Biochemistry, 43, 3862–3869. [DOI] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y and Zhang W (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188, 762–773. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Chen JG and Ellis BE (2011) AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J 67, 895–906. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wu H, Su J, Wang H, Zhu Q, Liu Y, Xu J, Lukowitz W and Zhang S (2017) Maternal control of embryogenesis by MPK6 and its upstream MKK4/MKK5 in Arabidopsis. Plant J 92, 1005–1019. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su J, Zhang Y, Xu J and Zhang S (2018) Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol 45, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J et al. (2011) An expanded palette of genetically encoded Ca(2)(+) indicators. Science, 333, 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Annotated DNA sequence of plasmid pET-SOMA.
Movie S12. Figure 5(a): SOMA-NES chitin.
Movie S2. Figure 2(f): SOMA (sensor of MAPK activity)-NES NaCl.
Movie S3. Figure 2(g): SOMAT679A-NLS NaCl.
Movie S4. Figure 2(h): SOMAT679A-NES NaCl.
Movie S5. Figure 3(a): SOMA-NES mpk3 mpk6 Pmpk6:MPK6YG 1-NA-PP1 NaCl.
Movie S6. Figure 3(b): SOMA-NES Col-0 1-NA-PP1 NaCl.
Movie S7. Figure 3(c): SOMA-NES mpk3 mpk6 Pmpk6:MPK6YG 1-NA-PP1 NaCl.
Movie S8. Figure 3(d): SOMA-NES Col-0 1-NA-PP1 NaCl.
Data S2. Annotated DNA sequence of plasmid pET-SOMA-T679A.
Figure S1. In vitro Förster resonance energy transfer assay with lambda protein phosphatase.
Figure S10. Chitin treatment of SOMA (sensor of MAPK activity)-NLS-2, SOMA-NES-2, SOMAT679A-NLS-2, and SOMAT679A-NES-2.
Figure S11. Map of plasmid pET-SOMA.
Figure S12. Map of plasmid pET-SOMA-T679A.
Figure S13. Map of plasmid pSOMA-NLS.
Figure S14. Map of plasmid pSOMA-T679A-NLS.
Figure S15. Map of plasmid pSOMA-NES.
Figure S16. Map of plasmid pSOMA-T679A-NES.
Figure S17. Regions of interest for Figure 2.
Figure S18. Regions of interest for Figure 3.
Data S3. Annotated DNA sequence of plasmid pSOMA-NLS.
Figure S19. Regions of interest for Figure 4
Figure S2. Seedling phenotypes of SOMA (sensor of MAPK activity) transgenic lines.
Figure S20. Regions of interest for Figure 5.
Figure S3. Rosette phenotypes of SOMA (sensor of MAPK activity) transgenic lines.
Figure S4. Control treatment with water only for SOMA (sensor of MAPK activity)-NLS and SOMA-NES.
Figure S5. SOMA (sensor of MAPK activity)-NLS-2, SOMA-NES-2, SOMAT679A-NLS-2 and SOMAT679A-NES-2 NaCl treatment.
Figure S6. The in vivo response of guard cells expressing SOMA (sensor of MAPK activity)-NLS-1 to NaCl stress.
Figure S7. The DMSO controls for SOMA (sensor of MAPK activity)-NES in mpk3 mpk6 Pmpk6:MPK6YG.
Figure S8. flg22 treatment of SOMA (sensor of MAPK activity)-NLS-2, SOMA-NES-2, SOMAT679A-NLS-2 and SOMAT679A-NES-2.
Figure S9. flg22 and chitin treatment of SOMA (sensor of MAPK activity)-NES in mpk3 mpk6 Pmpk6:MPK6YG.
Data S4. Annotated DNA sequence of plasmid pSOMA-T679A-NLS.
Table S1. Initial pixel intensity values from raw image data.
Data S5. Annotated DNA sequence of plasmid pSOMA-NES.
Data S6. Annotated DNA sequence of plasmid pSOMA-T679A-NES.
Movie S1. Figure 2(e): SOMA (sensor of MAPK activity)-NLS NaCl.
Movie S11. Figure 4(d): SOMA-NLS flg22.