Abstract
Background & Aims:
Little is known about the factors that affect outcomes of patients with biliary atresia and there are no medical therapies that increase biliary drainage.
Methods:
Liver biopsies and clinical data were obtained from infants with cholestasis and from children without liver disease (controls); mRNA was isolated, randomly assigned to discovery (n=121) and validation sets (n=50), and analyzed by RNAseq. Using the Superpc R package followed by Cox regression analysis, we sought to identify gene expression profiles that correlated with survival without liver transplantation at 24 months of age. We also searched for combinations of gene expression patterns, clinical factors, and laboratory results obtained at diagnosis and at 1 and 3 months after surgery that associated with transplant-free survival for 24 months of age. We induced biliary atresia in BALB/c mice by intraperitoneal administration of Rhesus rotavirus type A. Mice were given injections of the antioxidants N-acetyl-cysteine (NAC) or manganese (III) tetrakis-(4-benzoic acid)porphyrin. Blood and liver tissues were collected and analyzed by histology and immunohistochemistry.
Results:
We identified a gene expression pattern of 14 mRNAs associated with shorter vs longer survival times in the discovery and validation sets (P<.001). This gene expression signature, combined with level of bilirubin 3 months after hepatoportoenterostomy, identified children that survived for 24 months with an area under the curve value of 0.948 in the discovery set and 0.813 in the validation set (P<.001). Computer models correlated a cirrhosis-associated transcriptome with decreased times of transplant-free survival; this transcriptome included activation of genes that regulate the extracellular matrix and numbers of activated stellate cells and portal fibroblasts. Many mRNAs expressed at high levels in liver tissues from patients with 2-year transplant-free survival had enriched scores for glutathione metabolism. Among mice with biliary atresia given injections of anti-oxidants, only NAC reduced histologic features of liver damage and serum levels of aminotransferase, gamma-glutamyl transferase, and bilirubin. NAC also reduced bile duct obstruction and liver fibrosis and increased survival times.
Conclusions:
In studies of liver tissues from infants with cholestasis, we identified a 14-gene expression pattern that associated with transplant-free survival for 2 years. mRNAs encoding proteins that regulate fibrosis genes were increased in liver tissues from infants that did not survive for 2 years whereas mRNAs that encoded proteins that regulate glutathione metabolism were increased in infants that survived for 2 years. NAC reduced liver injury and fibrosis in mice with biliary atresia, and increased survival times. Agents such as NAC that promote glutathione metabolism might be developed for treatment of biliary atresia.
Keywords: chronic liver disease, cholestasis, biomarker, risk factor
Graphical Abstract
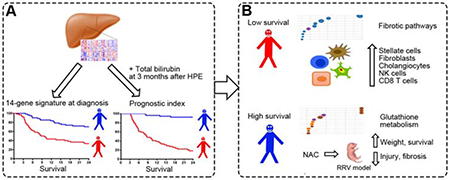
Lay Summary
The liver expression profile of 14 genes at diagnosis predicts the 2-year survival in children with biliary atresia and identifies glutathione metabolism as putative drug target.
Introduction
Biliary atresia (BA), the most common neonatal cholangiopathy, results from a fibro-inflammatory obstruction of the extrahepatic bile duct and induces a rapidly progressive hepatic fibrosis.1 Despite advances in understanding key factors relevant to etiology and pathogenesis of disease,2 the only treatment is the hepatoportoenterostomy (HPE), which may restore bile drainage in some patients, but does not stop the progressive fibrosis that is typical of biliary atresia.3 In studies designed to identify triggers of biliary injury using mouse and zebrafish models, viruses and environmental toxins have been shown to injure cholangiocytes and activate the innate and adaptive immune systems.1, 4 For example, dendritic cells and NK cells populate the livers of infants at the time of diagnosis and are directly implicated in the epithelial breach and the promotion of an adaptive inflammatory response that amplifies the cholangiocyte injury and obstructs the duct lumen.5–8 However, the use of corticosteroids to suppress the inflammatory response after HPE did not improve biliary drainage in a double-blind, placebo-controlled study,9 but appeared to reduce the serum total bilirubin if given to younger infants suggesting that stages of disease may be an important factor in treatment response.10
Despite the association of young age with treatment response, age alone does not predict clinical outcome nor correlates reliably with liver histological scoring or transcriptional profiles.11–13 With substantial variability in clinical course, the discovery of predictive biomarkers at the time of diagnosis would be invaluable to the field by enabling the customization of treatment protocols and the stratification of patients into clinical trials. In search of predictive biomarkers, we analyzed a comprehensive platform containing clinical and laboratory data and the hepatic transcriptional profiles at diagnosis. Our analyses identified a 14-gene signature prior to surgical intervention that best predicted low survival with the native liver at 2 years of age in a discovery cohort, which was reproduced in a validation cohort. This signature was linked to an abundance of activated hepatic stellate cells and portal fibroblasts, and to a transcriptional evidence of oxidative stress. Testing the relevance of this molecular signature in neonatal mouse models of biliary atresia and fibrosis, we found that the use of the anti-oxidant N-Acetyl-Cysteine improved the clinical outcome and suppressed biliary injury and liver fibrosis.
Materials and Methods
Patients
Liver biopsies and clinical data were obtained from infants with cholestasis enrolled into a prospective study (ClinicalTrials.gov Identifier: ) of the NIDDK-funded Childhood Liver Disease Research Network (www.childrennetwork.org) and from infants evaluated at Cincinnati Children’s Hospital Medical Center. The study protocols were approved by the human research review boards of all participating institutions. The diagnosis of BA was defined by an abnormal intraoperative cholangiogram and histological demonstration of obstruction of extrahepatic bile ducts. Liver biopsies were obtained at the time of HPE/diagnosis. Follow-up visits for clinical data collection occurred at 1 and 3 months after HPE and at age of 24 months. Normal controls (NC) consisted of liver biopsy samples obtained from 7 deceased-donor children aged 22-42 months as described previously.14
RNA sequencing and supervised principal components for Cox regression
Details of RNAseq analyses are described in Supplementary Methods. All RNAseq data have been deposited to GEO database under accession number GSE122340. The correlation between patient short-term survival data (survival with native liver at 24 months of age) and the normalized gene expression data were subjected to supervised principal component analysis using the Superpc R package, followed by Cox regression analysis (Superpc Cox) as reported previously.15 Steps for Superpc Cox are described in Supplementary Methods. We first trained the Superpc Cox regression model on the discovery cohort (n=121) and tested the reproducibility of the predictor of survival in the validation cohort (n=50). By ranking the patients into low survival and high survival groups based on the predicted score in a descending manner, study subjects were dichotomized into 2 groups, with those with relatively higher predicted scores being assigned to the low survival group. To examine if the transition to non-survival delineates a transcriptional trajectory, we performed principal component analysis to cluster all 171 BA patients and 7 normal controls as described previously.16
Prognostic index
We developed a prognostic index by the linear combination of 2 parameters (the 14-gene prognostic signature and total bilirubin at 3 months after HPE) weighted with regression coefficients from the multivariate Cox regression model in the discovery cohort using the following formula to generate an index:
Applying this formula to the data in the discovery cohort, subjects were ranked based on the index in a descending manner and dichotomized into a high index (low survival) and a low index (high survival) groups, which again dichotomized study subjects into 2 survival groups. This prognostic index was applied to the validation cohort following the same approach.
Transcriptional overlap with hepatopathies, pathway enrichment analysis, and estimation of cell abundance
Analytical methods are described in Supplementary Methods. The detailed information of studies for liver diseases and their animal models are summarized in Supplementary Table 1. Details are described in Supplementary Methods.
Standard and modified neonatal mouse model of biliary atresia and anti-oxidant treatment
To directly test the functional relationship between liver fibrosis, antioxidant-related genes, and survival, we performed experiments in standard and modified mouse models of BA. For standard mouse model, biliary atresia is induced by the intraperitoneal (I.P.) administration of 1.5×106 fluorescence forming units (ffu) Rhesus rotavirus type A (RRV) to BALB/c mice in day 1 of life. The modification consisted of a delay in I.P. inoculation of 1.875×106 ffu of RRV until day 3 of life to produce an ongoing hepatic injury, hyperbilirubinemia, and fibrosis. Detailed testing the effect of anti-oxidants in this model and phenotyping are described in Supplementary Methods.
Histopathology, immunostaining, colorimetric assays, and real-time PCR
Details are described in Supplementary Methods. Primer sequences are listed in Supplementary Table 2.
Statistical analyses
Detailed statistical analyses are described in Supplementary Methods.
Results
The hepatic transcriptome contains a 14-gene signature identifying patients with high and low survival
To explore the relationship between gene expression profiling and patient survival with the native liver at 2 years of age, we developed an analytic pipeline that started with the random assignment of 171 human liver biopsies collected at the time of diagnosis into discovery (N=121) and validation (N=50) cohorts (Figure 1A). Analyses of clinical variables of both cohorts showed no differences in sex, age, biochemical indicators of liver injury or cholestasis (serum levels of TB, AST, ALT, GGT), platelet count, presence of syndromic BA (BASM), histological inflammation, fibrosis, or survival (Supplementary Table 3 and Supplementary Figure 1).
Figure 1. A 14-gene signature that predicts patient’s survival at 2 years of age.
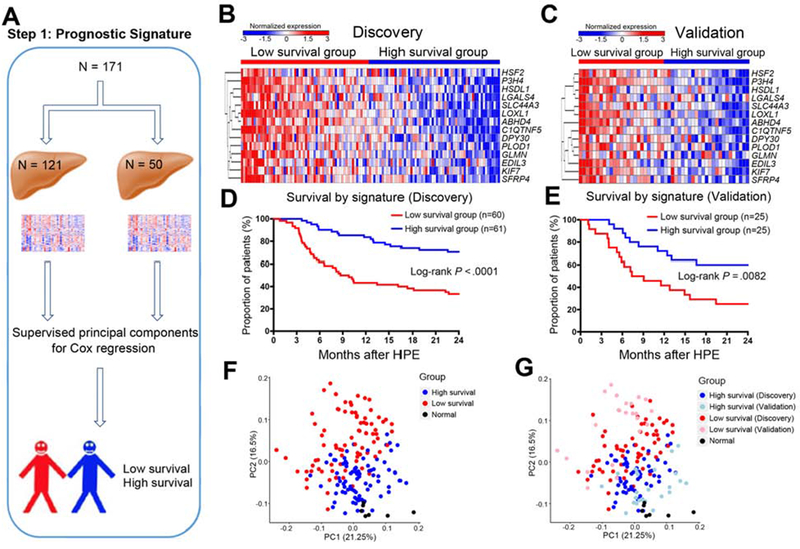
Panel A shows the analytic pipeline that started with the random assignment of 171 human liver biopsies collected at the time of diagnosis into discovery (N=121) and validation (N=50) cohorts, then Supervised principal components for Cox regression was applied to dichotomize patients into low and high survival groups. Heatmaps in B and C show the expression of 14 genes in discovery and validation cohorts, respectively (levels of gene expression are shown as color variation from red [high] to blue [low]). Euclidean distance was used in hierarchical clustering of genes. Graphs in D and E show Kaplan-Meier plots dichotomizing individual cohorts into 2 groups based on high or low survival based on predicted scores generated by Superpc Cox regression. Log-rank test was used to compare the survival distributions of two groups. Graphs in F and G show PCA clustering of all 171 biliary atresia patients and 7 normal controls.
To identify prognostic gene signatures, we performed Superpc and survival analyses using liver RNAseq data from the discovery cohort. We identified 14 genes associated with survival in the discovery cohort, which was reproducible when we applied the same analytical approach to RNAseq from the validation cohort (Figure 1B and C and Supplementary Table 4). Using the 14-gene signature, the patients were dichotomized into 2 groups based on the predicted scores, with significant survival differences between two groups both in the discovery (Log-rank P < 0.0001; Figure 1D) and validation cohorts (Log-rank P = 0.0082; Figure 1E). The median length of follow up is 9 and 8 months for low survival groups in discovery and validation cohorts, respectively, and 23 months for high survival groups in both cohorts. In the low survival groups, only 33% (discovery cohort) and 24% (validation cohort) were alive with the native liver, in contrast with 70% (discovery cohort) and 60% (validation cohort) in the high survival groups (Figure 1D and E). Applying principal component analysis clustering to these groups and a group of normal controls, we observed that normal subjects were positioned closer to those with high survival, with a similar separation pattern reproduced in the validation cohorts (Figure 1F–G). Analyses of the clinical parameters between these groups showed differences in age and histological fibrosis, with younger subjects in the high survival group in the discovery (median [25-75%] days: 52 [40-73] vs 68 [55-78], P=0.0001) and validation cohorts (50 [37-71] vs 59 [55-82], P = 0.034), and relatively higher degree of fibrosis in the low survival group in the discovery cohort (P = 0.0039) and validation cohorts (P = 0.016; Supplementary Table 5). These data uncovered a 14-gene signature at the time of diagnosis that successfully predicted the 2-year survival of patients with biliary atresia, with an initial association among survival, age and fibrosis at diagnosis.
The 14-gene signature and total bilirubin after HPE form an index with high predictive characteristics
To directly examine how the 14-gene signature performs as a predictor of outcome, we applied uni- and multivariate Cox regression analyses to a platform of data elements containing the signature, age at diagnosis, and other clinical parameters obtained at diagnosis and at 1 and 3 months after HPE. In a univariate analysis, the 14-gene signature at diagnosis, high serum TB, AST, and ALT at 1 and/or 3 months after HPE were associated with decreased survival (Supplementary Table 6 and Supplementary Table 7). However, applying multivariate analyses to the discovery cohort, only the 14-gene signature (hazard ratio [HR] = 2.2 [95% confidence interval = 1.4-3.6], P = 0.002) and the persistent elevation of TB at 3 months after HPE (HR = 1.2 [1.12-1.29], P < 0.0001) were relative risk factors for low survival (Table 1), with similar findings in the validation cohort (Supplementary Table 7).
We next developed a prognostic index by linearly combining the 14-gene signature and TB at 3 months weighed with regression coefficients in the discovery cohort (Figure 2A). In this approach, a higher index indicates lower survival. When generated for the discovery cohort, the prognostic index dichotomized the patients into 2 groups, with a low survival of 20% in the group with a high index and 90% with a low index (Log-rank P < 0.0001, Figure 2B), which was reproduced in the validation cohort (survival of 16% and 70%, respectively; Log-rank P = 0.0004; Figure 2C). Testing the performance of the prognostic index to predict a 2 year-survival generated an AUC of 0.948 in the discovery and 0.813 in the validation cohorts (Figures 2D and E), with corresponding model c-statistic of 0.862 and 0.763, sensitivity of 0.886 and 0.825, specificity of 0.867 and 0.821, PPV of 0.822 and 0.801, NPV of 0.907 and 0.811, respectively. A comparative analysis of against the 14-gene signature and TB showed that TB (total bilirubin) 3 months after HPE performs better than the 14-gene signature and that the prognostic index outperforms the 14-gene signature and TB independently in all parameters analyzed (Supplementary Table 8). These data suggested that a prognostic index that combines the signature at diagnosis and the TB as a surrogate of surgical response has a better accuracy in predicting survival.
Figure 2. Prognostic index of 2-year survival with the native liver.
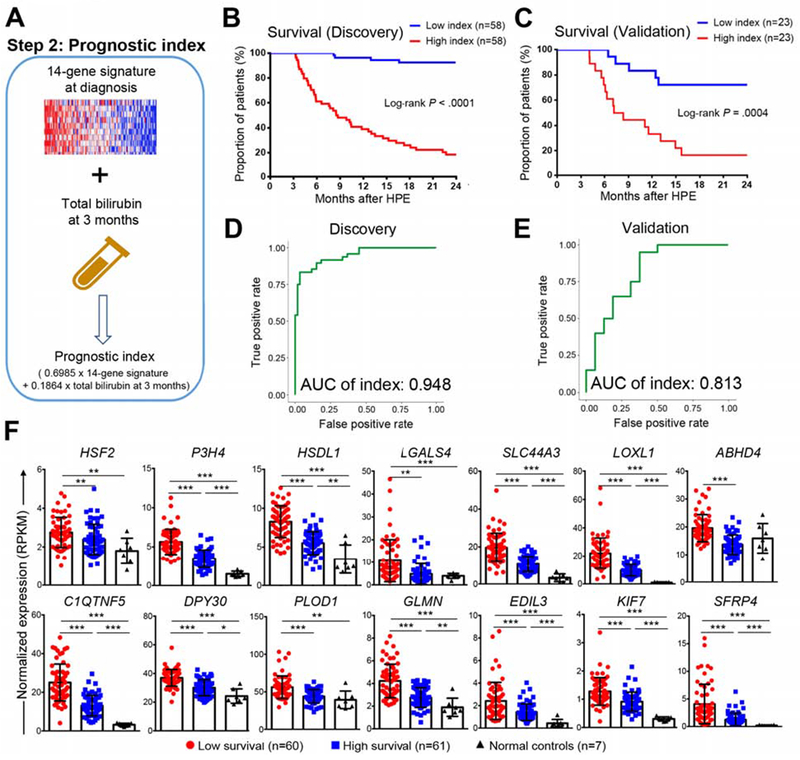
Panel A shows a strategy that combines the 14-gene signature and serum total bilirubin at 3 months after hepatoportoenterostomy to generate a prognostic index. Kaplan-Meier plots in B and C dichotomizes the cohort into groups of high or low survival based on prognostic index for individual patients. Log-rank test was used to compare the survival distributions of two groups. In D and E, ROC curves generated by the prognostic index at 2 years of age for D discovery and E validation cohorts. AUC of index is shown. In panel F, boxplots of expression levels of 14 genes in the low and high survival groups in discovery cohort and normal controls group. Expression levels are represented as mean ± SD of normalized expression (RPKM value). P value was calculated by Wilcoxon rank sum test. *P < 0.05, **P < 0.01, ***P < 0.001.
The hepatic expression signature overlaps with human and experimental fibrosis
Searching for prominent biological processes linked to the 2-year outcome, we first compared the expression levels of individual genes in the high survival group with those in the low survival group and a group of normal controls. There was a gradation of expression levels, with the expression for all 14 genes peaking in the low survival group, followed by lower levels in the high survival group, and the lowest level in the control group (Figure 2F). This suggested that the induction of gene expression in the liver occurs in all patients with biliary atresia, but it has the highest degree of expression in the low survival group that more directly relates to the poor outcome.
Next, to obtain insight into predominant biological processes related to outcome prediction, we mined the entire transcriptome platform to identify genes that were differentially expressed between the low and high survival groups (Figure 3A). Setting a threshold of 1.5 fold change and a Benjamini-Hochberg adjusted P <0.05, we identified 820 upregulated and 123 downregulated genes in the discovery cohort and 1,218 upregulated and 46 downregulated genes in the validation cohort (Figure 3B and C).
Figure 3. Low survival group exhibits a prominent gene signatures of cirrhosis.
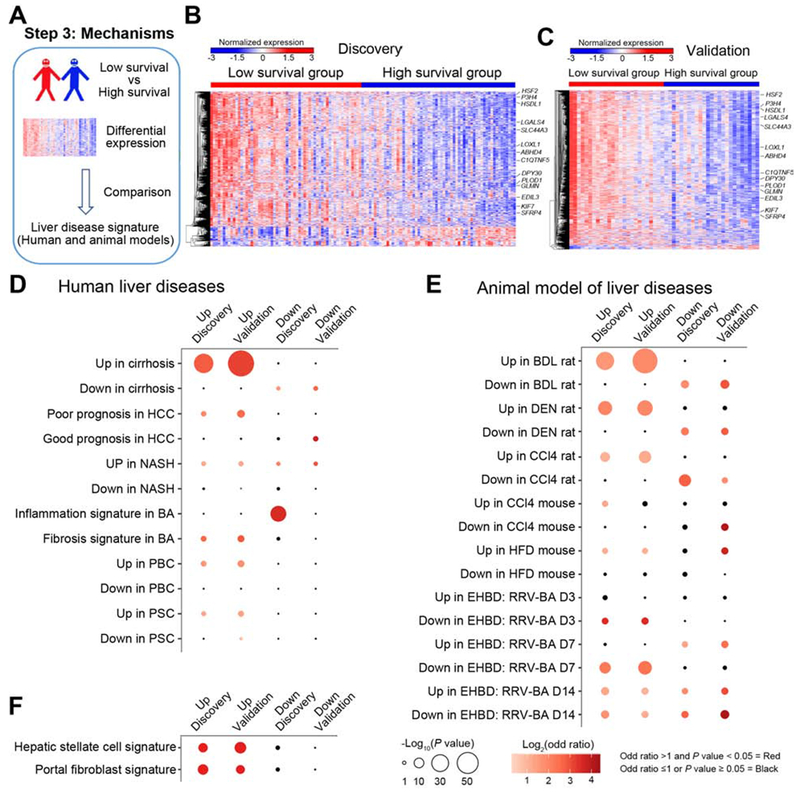
Panel A shows the use of differentially expressed genes between high and low survival groups compared with signatures of other liver diseases and animal models. The expression heatmaps of differentially expressed genes are shown for the discovery (B) and validation (C) cohorts (levels of gene expression are shown as color variation from red [high] to blue [low], with Euclidean distance used in hierarchical clustering). 14 genes are identified in the heatmaps. Pairwise overlapping comparisons were performed between upregulated or downregulated genes with gene signatures of human liver diseases (shown in D), animal models of human liver diseases (E), and hepatic stellate cells and portal fibroblasts (F). Overlaps were examined by Fisher’s exact test and shown as dots whose size is proportional to −log10(P value) and gradation of red is proportional to log2(odd ratio). Overlaps with −log10(P value) >1.3 and log2(odd ratio) >0 were defined as significant overlaps and are shown in red; the color black depicts overlaps with odds ratio ≤1 or P value > 0.05.
Investigating whether this comprehensive gene panel may be specific for biliary atresia or contain expression signatures of pathogenic mechanisms shared by other diseases, we performed pairwise overlapping analyses between differentially expressed genes and a variety of gene signatures published for diseased human livers and livers and extrahepatic bile ducts for animal models of liver disease (Supplementary Table 1). These gene signatures were derived by comparing disease livers and liver/extrahepatic bile ducts with normal controls. The most consistent finding was a significant overlap of up-regulated genes in the low survival group with those in human cirrhotic livers for the discovery and validation cohorts, followed by hepatocellular carcinoma (HCC), nonalcoholic steatohepatitis (NASH), a fibrosis signature previously published for livers of infants with biliary atresia, primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) (Figure 3D).11 Among the experimental models, the most significant overlap was with gene signatures of chronic liver injury induced by bile duct ligation, diethyl-nitrosamine, and carbon tetrachloride (Figure 3E). In the analysis of extrahepatic bile ducts from the murine model of rotavirus-induced experimental biliary atresia,17 there was an inverse relationship with the time of inflammatory obstruction (7 days after virus infection). Based on these findings, we applied the same strategy to cells known to be the main producers of extracellular matrix and found significant transcriptional overlaps with activated hepatic stellate cells and portal fibroblasts (Figure 3F).18, 19 Combined, these data pointed to a close relationship between upregulated genes in the low survival group and hepatic fibrosis. They also provided an initial link to the activation of cells known to produce extracellular matrix.
Transcriptional modeling identifies the expression and cellular source of extracellular matrix
To investigate the molecular basis for the relationship between the low-survival signature and fibrosis in other diseases and experimental models, we applied pathway enrichment analyses to the up-regulated genes in the low survival group (Figure 4A). Nine of the top 10 most statistically significant pathways related to the expression of extracellular matrix, including glycoproteins, collagen, and proteoglycans (Figure 4B, Supplementary Figure 2A). Based on the findings of a transcriptional overlap with aHSCs and aPFs discussed above and on the major roles of these cells as effectors of liver fibrosis,20, 21 we estimated their relative abundance by ssGSEA using gene expression enriched for individual cell types as described previously.22 Transcriptional estimation of aHSCs, aPFs, and cholangiocytes showed a gradation of cellular abundance, with a peak in the low survival group, a decrease in the high survival group, and the lowest in the normal group (Figure 4C–E). These transcriptional estimates were validated in a set of liver biopsy samples immunostained to detect α-SMA and cytokeratin 7 (Supplementary Figure 3A–G) and by the expression of the gene markers ACTA2 (for α-SMA) and KRT7 and KRT19 (for cytokeratins 7 and 19; Supplementary Figure 3H and I). These data suggested that these cells may contribute to the prominent fibrosis and lower survival in biliary atresia.23
Figure 4. Extracellular matrix formation related gene signature and cellular source, CD8T and NK cells are enriched in the low survival group.
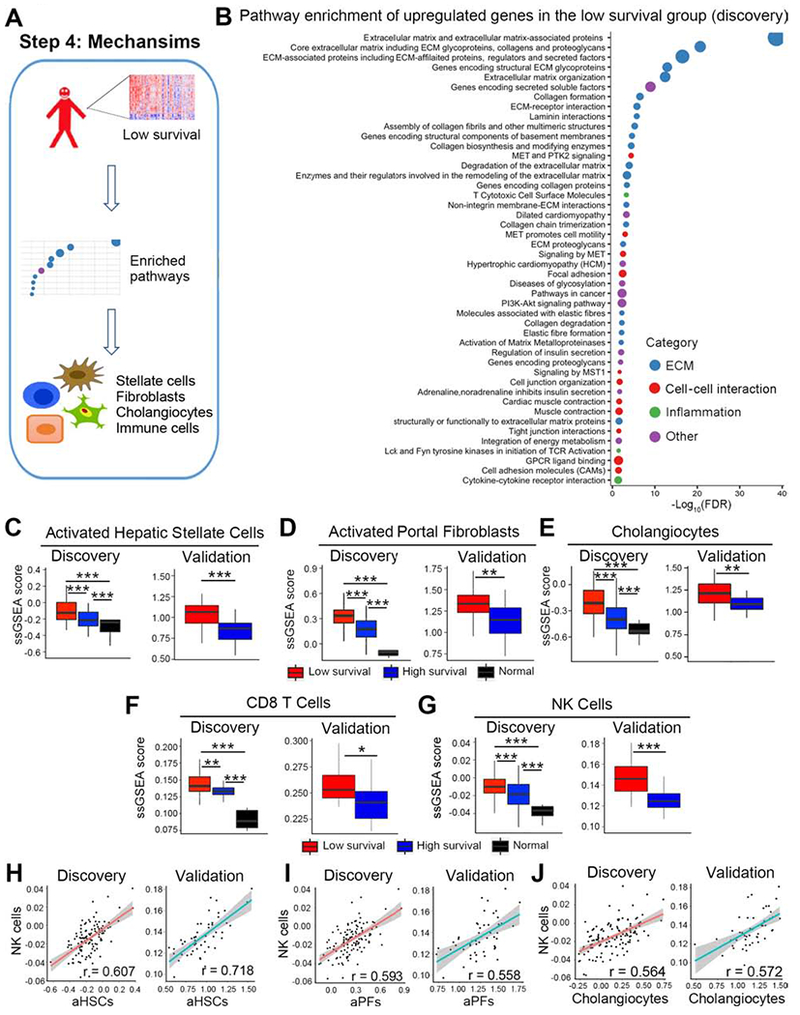
Panel A shows an analytic pipeline identifying enriched pathways and cell types in the low survival group and the quantification of the relative abundance of individual cell types. Dotplot shown in B depicts pathways that are enriched in upregulated genes of the low survival group in the discovery cohort. Pathway terms were ranked according to their −log10(FDR) values and categorized into extracellular matrix (ECM), cell-cell interaction, inflammation and other. The dot sizes are proportional to number of genes. For panels C-G, boxplots show enrichment of activated hepatic stellate cells (aHSC, C), activated portal fibroblasts (aPFs, D), cholangiocytes (E), CD8 T cells (F), and NK cells (G). Enrichments are represented as mean ± SD of ssGSEA scores. P value was calculated by Wilcoxon rank sum test. Scatterplots shown in H-J represent linear correlation between NK cells and aHSCs, aPFs, and cholangiocytes. Pearson correlation coefficient r is shown, with the grey area representing 95% confidence limits. *P < 0.05, **P < 0.01, ***P < 0.001.
Based on the increasing recognition that hepatic immune cells play a role as inducers of fibrogenesis and in the pathogenesis of biliary atresia,1, 24, 25 we applied ssGSEA to quantify the abundance of innate and adaptive immune cell types using gene signatures from two independent studies (Bindea et al26 and Charoentong et al22). The abundance of liver CD8-T and NK cells was greater in the low survival group in the discovery and validation cohorts (Figure 4F and G using gene signatures reported by Bindea et al 26; data using signatures from Charoentong et al 22 are shown in Supplementary Figure 4A and B). The same strategy identified differences in dendritic cells, eosinophils, neutrophils, and subtypes of T cells, but they could not be reproduced in the validation cohort or when we compared the signatures using the two published methods (Supplementary Figure 5). Using Pearson correlation to explore the potential functional relationship between CD8-T and NK cells with aHSCs, aPFs, and cholangiocytes, we found the best correlation with NK cells (Figure 4H–J and Supplementary Figure 4C–E), while the correlation with CD8-T cells was low or not reproducible (Supplementary Figure 4F–K). Altogether, these data provide evidence that the low survival signature is linked to an increased production of extracellular matrix in the liver, with a relative abundance of aHSCs and aPFs, and a potential functional relationship between these cells and NK cells.
Gene signature in the high survival group is enriched for glutathione metabolism
Having demonstrated that biological pathways and cells converge to a predominant fibrosis signature in subjects with poor outcome, we reasoned that analysis of the transcriptional signature of those with improved survival may give insight into mechanisms of hepatocellular protection and identify therapeutic targets. To this end, we applied pathway enrichment analyses to genes upregulated in the high survival group (Figure 5A). Among the pathways with the highest statistical significance, 6 related to oxidation and at least 4 were enriched for glutathione metabolism, implying a transcriptional response to oxidative radicals (Figure 5B and Supplementary Figure 2B). Some secondary pathways related to hepatocytes (ex: bile acid metabolism) are also featured in Figure 5B, but the lower strength of significance implies a similar functional representation of these cells among the low- and high-survival groups. Further analyzing the glutathione gene signature, we found the glutathione metabolism-related genes increased in the high survival group (Figure 5C). When expressed as a ratio to ssGSEA scores for aHSCs, aPFs, and cholangiocytes, glutathione metabolism-related genes increased in all three cell types in the high survival group in the discovery and validation cohorts (Figure 5D–F). These findings raised the possibility that an effective anti-oxidative response may suppress hepatic fibrosis and promote improved survival in biliary atresia.
Figure 5. Gene signature in the high survival group is enriched by glutathione metabolism.
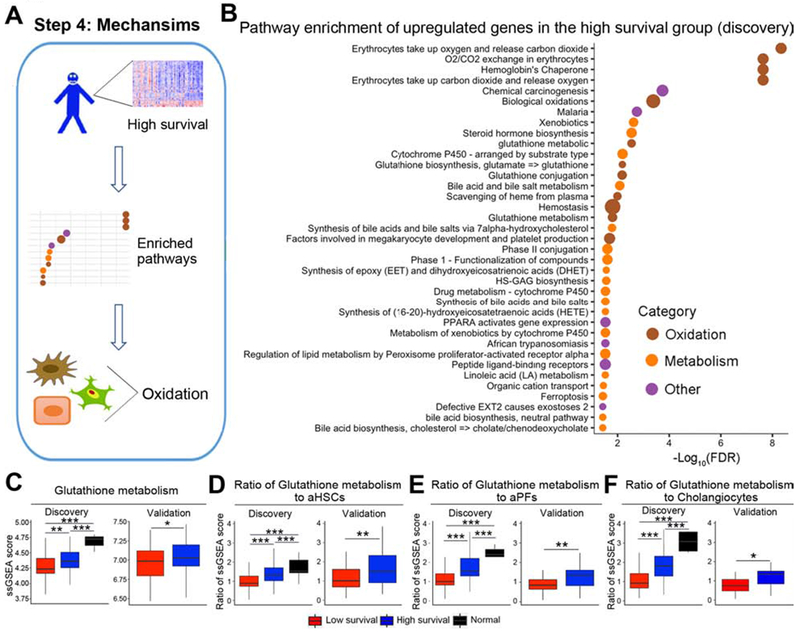
Panel A shows an analytic pipeline identifying enriched pathways and cell types in the high survival group and the quantification of the oxidation signatures relative to the abundance of individual cell types. The dotplot shown in B depicts pathways that are enriched in upregulated genes of the high survival group in the discovery cohort. Pathway terms were ranked according to their −log10(FDR) values and categorized into oxidation, metabolism and other. The dot sizes are proportional to number of genes. For panels C-F, boxplots show ssGSEA scores for glutathione metabolism (C) and ratios of ssGSEA scores for glutathione metabolism and aHSCs (D), aPFs (E), and cholangiocytes (F). *P < 0.05, **P < 0.01, ***P < 0.001.
Treatment with a glutathione-related antioxidant suppresses injury and fibrosis in both standard and modified RRV models
To examine the potential impact of antioxidants on injury and hepatic fibrosis of neonates, we used the standard RRV mouse model and a modified model by delaying its injection to 3 days of life. The development of this modified model was necessary because the standard model with RRV infection in the first 24 hours of birth allows for studies of biliary injury (early phases of pathogenesis of biliary atresia) but not of fibrosis due to early death of neonatal mice between 10-14 days.27 Although some studies show liver fibrosis at day 14 after RRV infection in the standard model, the degree of fibrosis is mild.28, 29 In initial experiments to validate the modified model of delayed RRV infection, we examined EHBDs after RRV administration throughout the suckling period and found focal epithelial injury and inflammatory obstruction 7 days later, followed by improvement of the epithelial injury, decreased inflammation, and restoration of the duct lumen by days 14 (Supplementary Figure 6A). In the liver, portal tracts had severe inflammation and increased number of duct profiles, with Sirius red staining showing fibrosis that intensified from days 7 to 19 (Supplementary Figure 6B and C).
Next, directly testing the hypothesis that antioxidants can suppress injury, liver fibrosis and improve survival, we injected NAC (150 mg/kg per day) or MnTBAP (5 mg/kg per day) i.p. beginning 12 hours after RRV inoculation in standard model (Figure 6A) and in modified model (Figure 7A). In control non-infected mice, the administration of similar doses of NAC or MnTBAP had no effect on growth or survival through 15 days of life, and mice displayed no unusual behavior suggestive of adverse effects of the drugs (data not shown). In both the standard and modified models, mice treated with NAC had better weight gain and survival (80% survival in at day 14 in standard model, 100% survival beyond 19 days after infection or 22 days of age in modified model) with lower levels of serum total bilirubin, ALT, and GGT when compared to RRV+PBS, while the administration of MnTBAP was similar to RRV+PBS with lower weight and survival (Figure 6B–F and Figure 7B–F). Treatment with NAC substantially decreased epithelial injury and inflammation and prevented EHBD obstruction (Figure 6G and Figure 7G). In liver, NAC significantly reduced portal inflammation, the extent of Sirius red staining and the presence of αSMA+ cells (Figure 6H–J and Figure 7I–J). In contrast, H&E, Sirius red and αSMA staining following MnTBAP was similar to RRV+PBS(not shown). Using real-time PCR, we did not observe significant changes of fibrosis associated genes in the standard model probably due to mild degree of fibrosis (Supplementary Figure 7A). Strikingly, treatment with NAC decreased the expression of the fibrosis-associated genes Acta2, Col1a1, Timp1 and Tgfb1 in the modified model (Supplementary Figure 7B). These data pointed to a beneficial effect of the glutathione-related antioxidant NAC in the suppression of neonatal injury and fibrosis.
Figure 6. N-acetyl-cysteine (NAC) suppresses liver injury in the standard mouse model of biliary atresia.
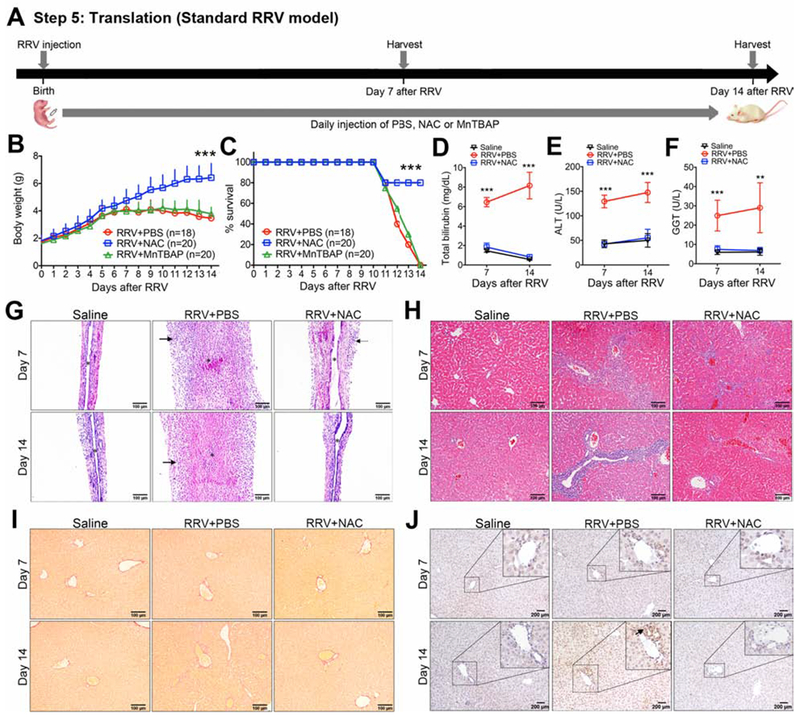
Panel A shows the experimental design of an RRV induced BA model, with 1.5×106 ffu of RRV injected intraperitoneally (i.p.) into newborn BALB/C mice, followed by the daily i.p. injection of PBS, NAC, or MnTBAP until day 14. Tissues and serum samples were harvest at days 7 and day 14 after RRV injection. Daily body weights (mean+ SD; N=18-26 mice per group from three independent experiments; shown in B), percent survival (C), and serum total bilirubin, ALT and GGT (D-F; mean ± SD; N=6 mice per group). **P < 0.01, ***P < 0.001. Representative EHBD (G) and liver (H) sections with H&E staining at Day 7 or Day 14 after RRV infection or wild type saline controls without RRV infection. In panel G, Arrows denote areas of periductal inflammation; asterisk denotes duct lumen. Representative liver sections with Sirius red (I) and anti-αSMA (J) staining at Day 7 or Day 14 after RRV infection or wild type saline controls without RRV infection.
Figure 7. N-acetyl-cysteine (NAC) suppresses liver injury and fibrosis in a modified mouse model of biliary atresia.
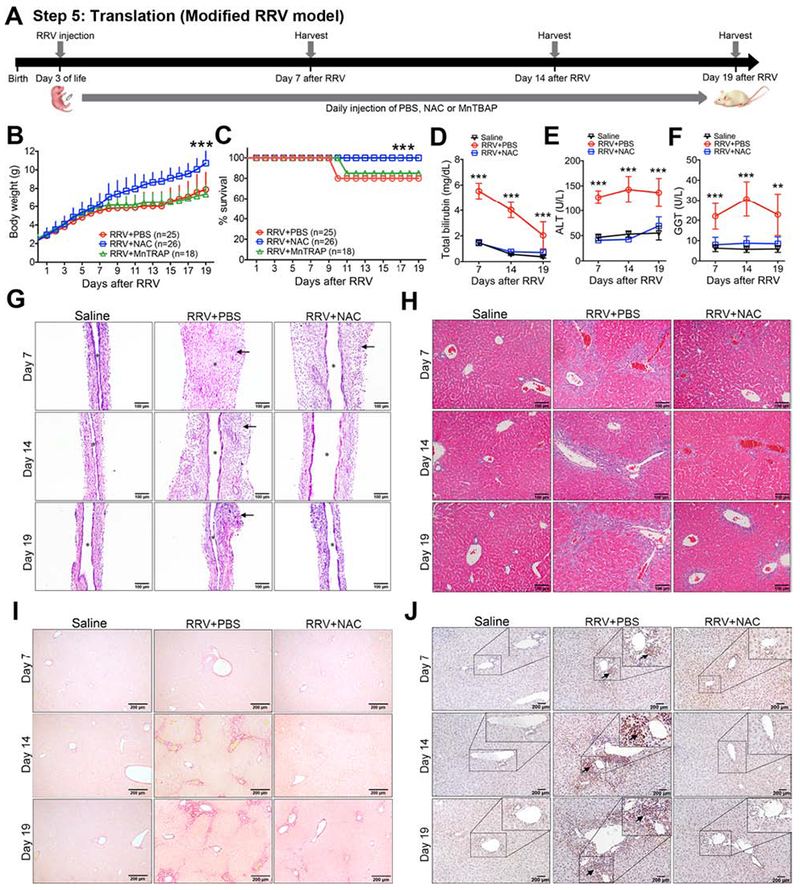
Panel A shows the experimental design of an RRV induced injury and liver fibrosis model, with 1.875×106 ffu of RRV injected intraperitoneally (i.p.) into newborn BALB/C mice at day 3 of life, followed by the daily i.p. injection of PBS, NAC, or MnTBAP until day 22 of life (19 doses), at which time tissues and serum samples were harvest. Daily body weights (mean+ SD; N=18-26 mice per group from three independent experiments; shown in B), percent survival (C), and serum total bilirubin, ALT and GGT (D-F; mean ± SD; N=6-12 mice per group). **P < 0.01, ***P < 0.001. Representative EHBD (G) and liver (H) sections with H&E staining at Day 7 or Day 14 after RRV infection or wild type saline controls without RRV infection. In panel G, arrows denote areas of periductal inflammation; asterisk denotes duct lumen. Representative liver sections with Sirius red (I) and anti-αSMA (J) staining at Day 7 or Day 14 after RRV infection or wild type saline controls without RRV infection. In panel J, arrows denote α-SMA positive cells.
Discussion
We found that the liver of infants with biliary atresia have a 14-gene signature at diagnosis that predicts survival with the native liver at 2 years of age. This predictive feature is not shared by other clinical or biochemical parameters at diagnosis; however, the combination of the molecular signature with the serum total bilirubin level 3 months after HPE further improved the predictive properties as an index of survival. The superiority of the prognostic index over the 14-gene signature and total bilirubin 3 months after HPE as individual parameters does not minimize the unique value of the 14-gene signature due to its relative predictive properties for long-term outcome at the time of diagnosis (prior to surgical intervention). Functional analysis of the gene signature in the group with low survival revealed a prominent footprint for extracellular matrix production, which was further supported by digital modeling pointing to a high abundance of activated stellate cells and portal fibroblasts. In the high survival group, pathway enrichment analyses identified an increased expression of genes related to glutathione metabolism. Consistent with this signature, the use of NAC in both standard RRV model and a neonatal mouse model of chronic injury decreased serum ALT, GGT and bilirubin, suppressed fibrosis, and fostered long-term survival.
The ability of a 14-gene signature to identify groups of infants with high survival supports the existence of key biological processes that may play a role in the likelihood of response to surgical intervention and in the activation of cellular processes to promote tissue repair. Looking at the potential relationship between the 14-gene signature and a young age at HPE being linked to better outcome, the ages differed between the two groups, but a multivariate analysis did not identify age as an important outcome factor. A similar relationship was reported previously, but the substantial overlap of ages in the high and low survival groups in that report limited the use of age as a predictor of outcome.11 The 14-gene expression signature does not contain the inflammation profile associated with an improved outcome reported in this previous study, thus raising the possibility that the strength of the association in that study was falsely high due to a small sample size, as supported by the lack of association when we included the larger sizes for the discovery and validation cohorts reported here. The 14-gene signature does reproduce the previous report that a fibrosis expression signature is associated with low survival. A discrepancy to note is the lack of significant correlation between histological fibrosis and outcome,11 which was reported previously and reproduced here. A potential explanation being the sampling artifact derived from a heterogeneous tissue injury in which the examination of a 5-6μ thin section by histology may not include the neighboring fibrosis. In contrast, the RNA is isolated from a relatively larger tissue fragment likely to contain a greater representation of tissue. Another possibility is the influence of other biological factors influencing outcome, such as the oxidative stress which we explore mechanistically in animal models. These factors notwithstanding, our study and the previous report have similar findings that a prominent expression of matrix genes adversely affects clinical outcome.11 The combination of these data with the prominent signature of activated stellate cells and portal fibroblasts underscores the existence of a hepatic environment that is functionally primed to induce ongoing fibrosis.
Predictive algorithms for gene function point to the activation of glutathione pathways in infants with improved survival. Conversely, those with poor survival had low levels of expression of genes related to glutathione metabolism when analyzed as a group as well as when normalized by the levels in stellate cells, portal fibroblasts, and cholangiocytes implying a substantial level of oxidative stress. These findings are particularly relevant in view of the increasing experimental evidence that oxidative stress is an important mechanism of biliary injury, obstructive phenotype, and expression of fibrosis genes in a toxin model of biliary atresia in zebrafish.30–32 In such a model, biliatresone, a naturally occurring biliary toxin, injures cholangiocytes by transiently depleting glutathione, which leads to a loss of cholangiocyte polarity, disruption of epithelial integrity, and activation of fibrosis genes.31 Using pharmacologic and genetic means to manipulate the glutathione-redox homeostasis, investigators demonstrated the impact of NAC in suppressing cholangiocyte injury.30 Here, we tested the effect of NAC on injury and fibrosis in both standard RRV mouse model and a modified mouse model of transient injury and obstruction of extrahepatic bile ducts that allows for prolonged survival and the development of hepatic fibrosis. In both models, the prophylactic administration of NAC protected against cell injury (shown by lower ALT and GGT) and improved biliary flow (shown by lower serum bilirubin); it also suppressed fibrosis in the chronic injury model. The specificity of this metabolic pathway is supported by the lack of improvement in mice treated with the antioxidant MnTBAP. These data suggest that the availability of glutathione may be an important determinant of improved survival in infants with biliary atresia and identify glutathione metabolism as a potential therapeutic target in biliary atresia.
The hepatic population of inflammatory cells in infants with biliary atresia has been linked to pathogenesis of disease, with their abundance at diagnosis suggesting an earlier stage of tissue injury.33 Their relationship to fibrogenesis, however, is less clear. In mouse models of fibrosis, CD8 T cells may promote fibrosis by activation of stellate cells, whereas NK cells attenuate liver fibrosis by killing these cells.34, 35 In patients with hepatitis C virus infection, the level of intrahepatic NK cells was inversely correlated with fibrosis stage. Without access to fresh liver tissue from infants with BA to directly quantify cells and correlate with fibrosis, we used validated gene expression-based computational models to quantify the relative abundance of immune cells between the high and low survival groups. Among all cell types, only NK cells reproducibly correlated with stellate cells and portal fibroblasts (as well as with the genes encoding the activation marker ACTA2/α-SMA) in a positive fashion regardless of the predictive model. We recognize the intrinsic limitation of analyses of whole liver RNA expression as they do not allow for the detection of important biological changes that may be present in less abundant non-hepatocyte cells. Our approach estimated these cells in relative values across the cellular ecosystem, but the expression of RNAs as an average for each cell type cannot make visible the functional changes in subgroups of cells that may be important to mechanisms of disease (such as transdifferentiation) and tissue pathology. For such studies, the use of single cell RNA sequencing will give a more detailed and accurate map of the cellular landscape of biliary atresia.
In closing, the relationship between a 14-gene signature at diagnosis and the 2-year survival with the native liver provides insight into staging of liver disease and the development of new therapies. In relation to staging of disease, the validation of a prominent fibrosis-related signature in a separate cohort and its relationship to poor outcome provide evidence that fibrosis at diagnosis is a hallmark of poor survival with the native liver, a concept that has been suggested by previous reports.11–13 In these patients, fibrosis may represent a later stage of liver disease that may not be uniformly correlated with the age at diagnosis, or it may represent a rapid fibrosis program that is poorly understood and limited to a subgroup of patients. A potential application of the 14-gene signature is to guide clinical trials or the development of new care protocols that take into account the most prominent biological processes at the time of diagnosis. For example, infants with low expression of the 14 genes may be assigned to new trials evaluating the effect of new adjuvant therapies to enhance bile drainage after HPE; conversely, those with a uniformly high gene expression may be prioritized for anti-fibrotic therapies, or perhaps not undergo HPE and be considered for close clinical follow-up with timely evaluation for liver transplantation (especially if the expression is present in infants >75 days). Before these uses, the predictive power of the gene signature requires a prospective validation in which the assignment of outcome in individual patients is made at diagnosis, with long-term follow-up of clinical outcome to validate or negate the prediction. A particularly appealing possibility is the design of a clinical trial designed to infants with high expression of glutathione aimed at testing the ability of NAC to suppress In relation to new therapies, the enrichment of glutathione-related genes points to their potential role in promoting survival as well as in the suppression of fibrosis, both supported by our data in neonatal mice treated with NAC, a compound that has been shown to stimulate bile acid-independent bile flow.36 The known safety profile of NAC in acute liver failure, its use in other pediatric diseases,37 and the data presented here form the foundation for a potential trial aimed at decreasing cellular injury and suppressing fibrosis, perhaps targeting infants that have lower fibrosis by the 14-gene signature or those with good response after HPE, in whom fibrosis progresses despite ongoing bile flow.
Supplementary Material
What You Need to Know.
BACKGROUND AND CONTEXT
Biliary atresia is a severe obstructive cholangiopathy of neonates, with variable response to surgical treatment. Without effective medical treatment, the liver disease rapidly progresses to cirrhosis.
NEW FINDINGS
We identified a hepatic expression signature of 14 genes at diagnosis with a high predictive performance for survival with the native liver at 2 years of age. The signature was enriched by genes related to glutathione metabolism. Experimentally, the activation of this pathway with n-acetyl-cysteine decreased tissue injury and cholestasis and improved survival of young mice.
LIMITATIONS
The performance of the 14-gene signature in predicting the 2 year survival requires a prospective validation in new cohorts.
IMPACT
The 14-gene signature at diagnosis may be a valuable tool to guide enrollment of subjects into clinical trials and forms a rationale for an interventional study to assess the efficacy of n-acetyl-cysteine in improving clinical outcome.
Acknowledgments
Grant support
Supported by the NIH grants DK-64008 and DK-83781 to JAB and by the Gene Analysis Cores of the Digestive Health Center (DK-78392). The work was also supported by U01 and UL1 grants from the National Institute of Diabetes, Digestive and Kidney Diseases: DK 62497 and UL1TR000077 (Cincinnati Children’s Hospital Medical Center), DK 62470 and UL1TR002378 (Children’s Healthcare of Atlanta), DK 62481 and UL1TR001878 (The Children’s Hospital of Philadelphia), DK 62456 (The University of Michigan), DK 84536 and UL1TR001108 (Riley Hospital for Children), DK 84575 and UL1TR002319 (Seattle Children’s Hospital), DK 62500 and UL1TR000004 (UCSF Children’s Hospital), DK 62503 (Johns Hopkins School of Medicine), DK 62445 (Mount Sinai School of Medicine), DK 62466 and UL1TR001857 (Children’s Hospital of Pittsburgh of UPMC), DK 62453 and UL1TR002535 (Children’s Hospital Colorado), DK 62452 (Washington University School of Medicine), DK 84538 and UL1TR000130 (Children’s Hospital Los Angeles), DK 62436 and UL1TR000150 (Ann & Robert H Lurie Children’s Hospital of Chicago), DK103149 (Texas Children’s Hospital), DK103135 (The Hospital for Sick Children), DK103140 (University of Utah).
Abbreviations
- APRI
AST to platelet ratio index
- AUC
area under the curve
- BA
biliary atresia
- BDL
bile duct ligated
- ECM
extracellular matrix
- EHBD
extra-hepatic bile duct
- FPKM
fragments per kilobase of exon per million mapped reads
- GGT
gamma-glutamyl transferase
- HPE
hepatoportoenterostomy
- aHSCs
activated hepatic stellate cells
- HR
hazard ratio
- LT
liver transplantation
- MnTBAP
Manganese (III) tetrakis-(4-benzoic acid)porphyrin
- NAC
N-acetyl-cysteine
- NK cells
natural killer cells
- NC
normal controls
- NASH
nonalcoholic steatohepatitis
- aPFs
activated portal fibroblasts
- RPKM
reads per kilobase of exon per million mapped reads
- ROC
receiver operating characteristic
- ROS
reactive oxygen species
- RRV
rhesus rotavirus
- Superpc
supervised principal components for Cox regression
- ssGSEA
single sample gene set enrichment analysis
- TB
total bilirubin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Nothing to disclose
Transcript Profiling: GSE122340
Writing Assistance: None
References
Author names in bold designate shared co-first authorship
- 1.Asai A, Miethke A, Bezerra JA. Pathogenesis of biliary atresia: defining biology to understand clinical phenotypes. Nat Rev Gastroenterol Hepatol 2015; 12:342–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilgore A, Mack CL. Update on investigations pertaining to the pathogenesis of biliary atresia. Pediatr Surg Int 2017;33:1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam PKH, Chung PHY, St Peter SD, et al. Advances in paediatric gastroenterology. Lancet 2017;390:1072–1082. [DOI] [PubMed] [Google Scholar]
- 4.Luo Z, Jegga AG, Bezerra JA. Gene-disease associations identify a connectome with shared molecular pathways in human cholangiopathies. Hepatology 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shivakumar P, Sabla GE, Whitington P, et al. Neonatal NK cells target the mouse duct epithelium via Nkg2d and drive tissue-specific injury in experimental biliary atresia. J Clin Invest 2009;119:2281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezerra JA, Tiao G, Ryckman FC, et al. Genetic induction of proinflammatory immunity in children with biliary atresia. Lancet 2002;360:1653–9. [DOI] [PubMed] [Google Scholar]
- 7.Mack CL, Falta MT, Sullivan AK, et al. Oligoclonal expansions of CD4+ and CD8+ T-cells in the target organ of patients with biliary atresia. Gastroenterology 2007; 133:278–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxena V, Shivakumar P, Sabla G, et al. Dendritic cells regulate natural killer cell activation and epithelial injury in experimental biliary atresia. Sci Transl Med 2011. ;3:102ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bezerra JA, Spino C, Magee JC, et al. Use of corticosteroids after hepatoportoenterostomy for bile drainage in infants with biliary atresia: the START randomized clinical trial. JAMA 2014;311:1750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davenport M, Stringer MD, Tizzard SA, et al. Randomized, double-blind, placebo-controlled trial of corticosteroids after Kasai portoenterostomy for biliary atresia. Hepatology 2007;46:1821–7. [DOI] [PubMed] [Google Scholar]
- 11.Moyer K, Kaimal V, Pacheco C, et al. Staging of biliary atresia at diagnosis by molecular profiling of the liver. Genome Med 2010;2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weerasooriya VS, White FV, Shepherd RW. Hepatic fibrosis and survival in biliary atresia. J Pediatr 2004;144:123–5. [DOI] [PubMed] [Google Scholar]
- 13.Pape L, Olsson K, Petersen C, et al. Prognostic value of computerized quantification of liver fibrosis in children with biliary atresia. Liver Transpl 2009;15:876–82. [DOI] [PubMed] [Google Scholar]
- 14.Bessho K, Mourya R, Shivakumar P, et al. Gene expression signature for biliary atresia and a role for interleukin-8 in pathogenesis of experimental disease. Hepatology 2014;60:211–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol 2004;2:E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Yella J, Chen J, et al. Unsupervised gene expression analyses identify IPF-severity correlated signatures, associated genes and biomarkers. BMC Pulm Med 2017;17:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessho K, Shanmukhappa K, Sheridan R, et al. Integrative genomics identifies candidate microRNAs for pathogenesis of experimental biliary atresia. BMC Syst Biol 2013;7:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang DY, Goossens N, Guo J, et al. A hepatic stellate cell gene expression signature associated with outcomes in hepatitis C cirrhosis and hepatocellular carcinoma after curative resection. Gut 2016;65:1754–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwaisako K, Jiang C, Zhang M, et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A 2014;111:E3297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells RG. The portal fibroblast: not just a poor man’s stellate cell. Gastroenterology 2014;147:41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells RG, Schwabe RF. Origin and function of myofibroblasts in the liver. Semin Liver Dis 2015;35:e1. [DOI] [PubMed] [Google Scholar]
- 22.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248–262. [DOI] [PubMed] [Google Scholar]
- 23.Roskams T, Desmet V. Ductular reaction and its diagnostic significance. Semin Diagn Pathol 1998;15:259–69. [PubMed] [Google Scholar]
- 24.Lakshminarayanan B, Davenport M. Biliary atresia: A comprehensive review. J Autoimmun 2016;73:1–9. [DOI] [PubMed] [Google Scholar]
- 25.Zagory JA, Nguyen MV, Wang KS. Recent advances in the pathogenesis and management of biliary atresia. Curr Opin Pediatr 2015;27:389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- 27.Shivakumar P, Campbell KM, Sabla GE, et al. Obstruction of extrahepatic bile ducts by lymphocytes is regulated by IFN-gamma in experimental biliary atresia. J Clin Invest 2004;114:322–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keyzer-Dekker CM, Lind RC, Kuebler JF, et al. Liver fibrosis during the development of biliary atresia: Proof of principle in the murine model. J Pediatr Surg 2015;50:1304–9. [DOI] [PubMed] [Google Scholar]
- 29.Zagory JA, Fenlon M, Dietz W, et al. PROMININ-1 promotes biliary fibrosis associated with biliary atresia. Hepatology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Lorent K, Wilkins BJ, et al. Glutathione antioxidant pathway activity and reserve determine toxicity and specificity of the biliary toxin biliatresone in zebrafish. Hepatology 2016;64:894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waisbourd-Zinman O, Koh H, Tsai S, et al. The toxin biliatresone causes mouse extrahepatic cholangiocyte damage and fibrosis through decreased glutathione and SOX17. Hepatology 2016;64:880–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koo KA, Waisbourd-Zinman O, Wells RG, et al. Reactivity of Biliatresone, a Natural Biliary Toxin, with Glutathione, Histamine, and Amino Acids. Chem Res Toxicol 2016;29:142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bessho K, Bezerra JA. Biliary atresia: will blocking inflammation tame the disease? Annu Rev Med 2011. ;62:171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gur C, Doron S, Kfir-Erenfeld S, et al. NKp46-mediated killing of human and mouse hepatic stellate cells attenuates liver fibrosis. Gut 2012;61:885–93. [DOI] [PubMed] [Google Scholar]
- 35.Muhanna N, Abu Tair L, Doron S, et al. Amelioration of hepatic fibrosis by NK cell activation. Gut 2011. ;60:90–8. [DOI] [PubMed] [Google Scholar]
- 36.Ballatori N, Truong AT. Relation between biliary glutathione excretion and bile acid-independent bile flow. Am J Physiol 1989;256:G22–30. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins DD, Wiest DB, Mulvihill DM, et al. Fetal and Neonatal Effects of N-Acetylcysteine When Used for Neuroprotection in Maternal Chorioamnionitis. J Pediatr 2016;168:67–76 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


