Abstract
Motivation
Drug repurposing is a potential alternative to the classical drug discovery pipeline. Repurposing involves finding novel indications for already approved drugs. In this work, we present a novel machine learning-based method for drug repurposing. This method explores the anti-similarity between drugs and a disease to uncover new uses for the drugs. More specifically, our proposed method takes into account three sources of information: (i) large-scale gene expression profiles corresponding to human cell lines treated with small molecules, (ii) gene expression profile of a human disease and (iii) the known relationship between Food and Drug Administration (FDA)-approved drugs and diseases. Using these data, our proposed method learns a similarity metric through a supervised machine learning-based algorithm such that a disease and its associated FDA-approved drugs have smaller distance than the other disease-drug pairs.
Results
We validated our framework by showing that the proposed method incorporating distance metric learning technique can retrieve FDA-approved drugs for their approved indications. Once validated, we used our approach to identify a few strong candidates for repurposing.
Availability and implementation
The R scripts are available on demand from the authors.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The traditional drug discovery process is extremely slow and costly. Developing a new drug takes ∼10–17 years and requires anything between $500 million and $2 billion, depending on the drug and pharmaceutical company (Adams and Brantner, 2006; Dickson and Gagnon, 2009; DiMasi et al., 2003). In spite of unprecedented investments in research and development, the number of new Food and Drug Administration (FDA)-approved drugs remains low, reflecting the limitations of the current research and development model (Munos, 2009). In this context, the identification of novel disease indications for approved drugs, known as drug repositioning (or repurposing), is a very effective way to increase the therapeutic arsenal at a very reduced cost (Ashburn and Thor, 2004). In fact, some authors believe that repurposing should be ‘the primary strategy in drug discovery for every broadly focused, research-based pharmaceutical company’ (Tobinick, 2009). Approximately 90% of drug leads fail to move beyond early development and toxicity testing, and many of the few drugs that make it to clinical trials fail because of side-effects or adverse events. Finding new disease indications for existing drugs sidesteps all these issues and can therefore increase the available therapeutic choices at a fraction of the cost of new drug development. Some of the examples of successful repurposed drugs are sildenafil citrate, repurposed from angina to erectile dysfunction; thalidomide, repurposed from morning sickness to multiple myeloma (Novac, 2013) and phenytoin, repurposed from seizures to bipolar disorder (Mariotti et al., 2010).
A number of computational approaches for drug repurposing have been developed (Lotfi Shahreza et al., 2017). Based on available clinical knowledge such as drug chemical or pharmaceutical information, disease biomarkers, target pathways or symptomatology information, these methods can be roughly divided into: (i) drug-based methods, and (ii) disease-based methods (Dudley et al., 2011). In the following, we review some methods from each of the two categories, as well as methods that incorporate knowledge from both categories.
Keiser et al. (2009) proposed a method that predicts several off-target interactions based on the fact that the chemical structure influences the therapeutic effect of a drug. This method calculates a similarity score for each drug–target pair using Similarity Ensemble Approach (Keiser et al., 2007). A new target is proposed for a drug based on this similarity score.
A number of methods have been proposed for drug–target interaction (DTI) prediction based on personal recommendation algorithms were recently reviewed in Alaimo et al. (2016). Cheng et al. (2012) proposed a network-based inference method based on personal recommendation algorithms for DTI prediction. In order to construct the drug–target bipartite network, this method calculates a drug–target similarity score based on the drug–drug structural similarity and target–target genomic sequence similarity. Alaimo et al. (2013) introduced a new model called domain tuned-hybrid for DTI prediction. This method extends the network-based inference algorithm by adding drug–target domain knowledge into the framework. Chen et al. (2015) also proposed two methods called ProbS and HeatS to predict direct drug–disease associations based on the personal recommendation algorithms.
Several methods have been proposed based on the idea that if a drug-exposure gene expression profile inversely correlates with a disease gene expression profile, the drug may have a therapeutic effect on the disease (Lamb et al., 2006; Sirota et al., 2011). These methods support the systematic identification of new indications for already approved drugs (Iorio et al., 2013). The Connectivity Map (CMap) project is one of the first systematic approaches that aims to compare gene expression profiles across experimental conditions (Lamb et al., 2006). CMap has been used to identify cimetidine, an anti-ulcer drug, as potentially efficacious in lung adenocarcinoma, efficacy which was subsequently demonstrated (Sirota et al., 2011). Iorio et al. (2010) proposed a method to find drug similarities. This method constructs a drug–drug network based on consensus response pair-wise similarity. This similarity score is a novel rank-based metric inherited from GSEA (Subramanian et al., 2005). A recent method proposed by Vargas et al. (2018), identified a number of master regulators of Alzheimer disease based on the gene expression data of human hippocampus and transcription regulatory network analysis. Using these master regulators and CMap method (Lamb et al., 2006), six FDA-approved drugs with potential therapeutic effect on the Alzheimer disease were identified. Suthram et al. (2010) introduced a method to find disease similarities by incorporating gene expression microarray data and protein–protein interaction network. This method constructs a disease–disease network based on the functional module activity shared between diseases. The hypothesis is that if two diseases share a common molecular pathology, then the drug used for treating one of them can also be used for treating the other one. Langhauser et al. (2018) proposed a drug repurposing method based on this hypothesis. In particular, they first constructed a disease–disease network in which the diseases are connected to each other based on the number of shared genes, existing physical protein interactions between them, symptom similarity and co-morbidities information. This constructed network revealed number of novel clusters of heterogeneous diseases with common mechanisms. The authors experimentally validated the therapeutic effect of soluble guanylate cyclase, used as a smooth muscle relaxation, on the neurological diseases.
Sun et al. (2017) introduced a new method incorporating the drug, disease and gene information obtained from GenBank (Benson et al., 2002), drugBank (Law et al., 2014) and OMIM (Hamosh et al., 2005) databases. Using these sources of information, they created a network of diseases, drugs and genes. In this network, a drug and a gene are connected if the drug targets one of the gene’s associated proteins. A gene is connected to its associated disease and a disease is connected to its approved drugs. Then, they investigated the novel disease–drug pairs based on this constructed network. Finally, they employed literature mining techniques in order to find scientific articles supporting these novel disease–drug associations. The anti-tubercular effect of the anti-psychotic drug, chlorpromazine, is one of their promising finding.
A novel method proposed by Ghofrani et al. (2006) based on the hypothesis that if a given treatment has a side effect that remedies a given condition, then the drug may be repurposed for the latter condition. As a well-known example, sildenafil citrate was repurposed from angina to erectile dysfunction, when systematic erections were noted in angina patients treated with this drug.
This work describes a machine learning-based method for drug repurposing. This method is based on disease gene expression profiles, large-scale drug-exposure gene expression profiles and the clinical established knowledge between them. Based on these sources of information, our proposed method ranks the drugs that are effective for a given disease by incorporating a distance metric learning (DML) algorithm in which the given disease and its FDA-approved drugs have smaller distances compare to the other disease–drug pairs.
2 Materials and methods
2.1 Disease and drug gene expression data
Disease gene expression microarray data are obtained from NCBI Gene Expression Omnibus (GEO) (Edgar et al., 2002). The pre-processing procedure we use includes transformation and quantile normalization (Irizarry et al., 2003).
The drug data come from two different sources: CMap (Lamb et al., 2006) for breast cancer and rheumatoid arthritis (RA) and National Institutes of Health’s Library of Integrated Network-based Cellular Signatures (http://www.lincsproject.org/) for idiopathic pulmonary fibrosis (IPF). We use two different data sources in order to show the method is reliable and works independently of the source of the drug data. The pre-processing steps for drug gene expression data are included in the Supplementary Materials.
2.2 Framework overview
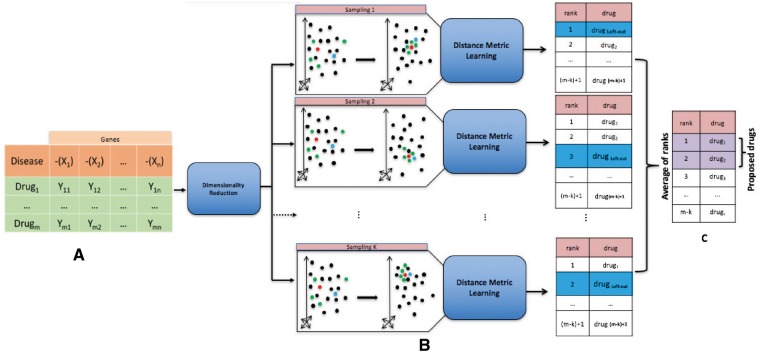
The main idea of this work is to represent a disease gene expression profile and large-scale drug gene expression profiles in a common space where the specific disease and its FDA-approved drugs are mapped closer to each other compared to the other disease–drug pairs. Figure 1 shows the pipeline of the framework. The first step of the framework is to represent drugs and diseases in a meaningful common space.
Fig. 1.
Framework overview. (A) We transform the input matrix into a lower dimensionality matrix by applying LLE (Roweis and Saul, 2000). (B) We incorporate the known relationship between disease and its FDA-approved drugs into this space using DML algorithm. Our hypothesis is that under this new space and the new metric, the clinically relevant drugs get close to the disease. We use the leave-one-out cross validation. In particular, in each round of sampling, we use one of the FDA-approved drugs for the disease (called left-out drug) for validation and we use the rest of them for training the DML algorithm. We then rank the list of drugs from the nearest one to the disease gene expression profile to the farthest one, based on the new learned metric. We repeat this procedure k times (where k is the number of FDA-approved drugs for the disease). (C) For each drug, an average of ranks is calculated over the k rounds of sampling. The proposed drugs are the ones with the lower average rank. In this figure, the red point represents the disease, the blue point represents the left-out drug and the green point represents the FDA-approved drug for the disease used for training the DML algorithm
Let be a collection of gene expression profiles, where m is the number of drugs in this collection. Each is a vector representing a gene expression profile, and n is the number of features of this vector, which represents genes. The input matrix we use subsequently is a combination of the reversed measurements of the genes in the disease profile and the measurements of the same genes in each of the drug profiles. The intuition behind using gene expression representation of drugs and diseases is directly influenced by the rationale behind the CMap method (Lamb et al., 2006). With drug and disease activity represented through their gene expression profiles, it is possible to use a distance measure in this space as a measure of anti-similarity or effectiveness.
The second step of our framework is the incorporation of existing knowledge. This knowledge can be found in the extensive clinical studies about diseases and FDA-approved drugs. These studies have been conducted over several years (costing millions of dollars) and reveal important relationships between certain disease–drug pairs, including drug effectiveness. Our method capitalizes on this kind of information by incorporating it through metric learning algorithms. These algorithms are a class of machine learning-based methods that keep all pairs of ‘similar’ points close, while separating all ‘dissimilar’ pairs, where similarity and dissimilarity can be custom-defined. Here, we use these algorithms to find a suitable distance measure in the gene expression representation space such that the disease–drug pairs that have been clinically proved to be relevant get closer to each other. More specifically, we use the Mahalanobis distance, a distance between two N-dimensional points, that is scaled by the statistical variation in each component of the point (Drăghici, 2011; Mahalanobis, 1936). The Mahalanobis distance between two points and is given by:
| (1) |
where M is a covariance matrix that captures the properties of the multi-dimensional disease–drug space. M is learned such that disease–drug pairs clinically proved to be relevant get close to each other. In order to learn the matrix M, we use the DML method (Xing et al., 2002). This method is based on posing metric learning as a convex optimization problem. This requires reducing the dimension of the gene expression representation because DML algorithms are not applicable in high dimension problems due to high computational complexity (Torresani and Lee, 2006). For this purpose, we use two different methods: Principal Component Analysis (PCA) (Jolliffe, 2002) and Locally Linear Embedding (LLE) (Roweis and Saul, 2000). The details of LLE are included in the Supplementary Materials.
In this step, we transform the input matrix into a lower dimensionality matrix by applying LLE (Roweis and Saul, 2000), and then perform a transformation of the data using DML algorithm. As the final step, the Euclidean distance between the disease gene expression profile and each of the drug-exposure expression profiles is calculated. We then rank the list of the drugs from nearest one to the disease gene expression profile to the farthest one, based on the computed Euclidean distance. We repeat this procedure k times (where k is the number of FDA-approved drugs for the disease), each time using one of the FDA-approved drugs for the disease for the validation and using the rest of them for training the DML algorithm. This is known as the leave-one-out method. For each drug, an average of ranks is calculated over the k rounds of sampling. The proposed drugs are the ones with the lower average rank.
In summary, our proposed framework includes the following steps: (i) we construct the gene expression profiles for each drug and disease; (ii) we incorporate the known relationship between disease and its FDA-approved drugs into this space using DML; under this new space and new metric, the clinically relevant drugs and disease get close to each other; (iii) for each disease, we then retrieve its closest drugs, based on the new learned metric. In particular, let be a collection of FDA-approved drugs for the disease x. In each round of leave-one-out cross validation, we use one of the drugs from this list (called left-out drug) for the validation and we use the rest for training the DML algorithm. We then rank the list of drugs from the nearest one to the disease gene expression profile to the farthest one, based on the new learned metric. We repeat this procedure k times (where k is the number of FDA-approved drugs for the disease x). Finally, for each drug, an average of ranks is calculated over the k rounds of sampling. The proposed drugs are the ones with the lower average rank. The Normalized Discounted Cumulative Gain (NDCG) and the area under the curve (AUC) metrics that we use to assess the performance of a method are now calculated based on the rank of the left-out drug in each round of sampling. The reported NDCG and AUC values for each method are the average value of these metrics over the k rounds of sampling.
2.3 Evaluation
We evaluate the quality of the ranking obtained from each method using two different metrics. First, we use the NDCG metric (Järvelin and Kekäläinen, 2002) to compare the quality of the ranking obtained from each method. This score shows how close a particular ranking is to the ideal ranking. For our purposes, in each round of leave-one-out cross validation, the ideal ranking is the one where the left-out drug is at the top of the ranked list. The reported NDCG value for each method is the average of this metric over all rounds. The details of this metric are included in the Supplementary Materials. We also use the AUC of the receiver-operator characteristic as an alternative assessment of how well the various methods perform in terms of placing the FDA-approved drug for a disease at the top of their ranked list. The reported AUC for each method is the average of this score over all the rounds.
2.4 Assessment
The assessment and comparison of the proposed methods with the existing approaches is based on the statistical power the ability to find FDA-approved drugs for a given disease at the top of the ranked list of drugs, as well as the ability to provide a meaningful ranking of drugs (drugs at the top of the ranked list are most likely to be clinically related to the given disease). We compare the following alternative approaches:
Incorporating gene expression data and calculating drug–disease score (Sirota et al., 2011) for each disease–drug pair.
Incorporating gene expression data and calculating anti-correlation for each disease–drug pair.
Incorporating gene expression data, PCA and calculating Euclidean distance for each disease–drug pair (PCA-ED).
Incorporating gene expression data, LLE and calculating Euclidean distance for each disease–drug pair (LLE-ED).
Incorporating gene expression data and calculating Euclidean distance for each disease–drug pair.
Incorporating gene expression data, clinical studies, PCA, DML and calculating Euclidean distance for each disease–drug pair (PCA-DML).
Incorporating gene expression data, clinical studies, LLE, DML and calculating Euclidean distance for each disease–drug pair (LLE-DML).
The first method is proposed by Sirota et al. (2011). The second method is based on the simple but intuitive idea that a drug would be effective if it counteract the effect of the disease on every single differentially expressed genes (simple anti-correlation of gene and drug expression profiles). The methods 3–7 above are proposed in this manuscript. The novel methods (items 3–7 above) and the anti-correlation method (item 2) are implemented in R. Items 6–7 are implemented using R package dml v1.1.0 (Tang et al., 2015). The method proposed in Sirota et al. (2011) (item 1) is also available in the same programing language by using the R package DrugVsDisease v2.8.0 (Pacini, 2013).
3 Results/discussion
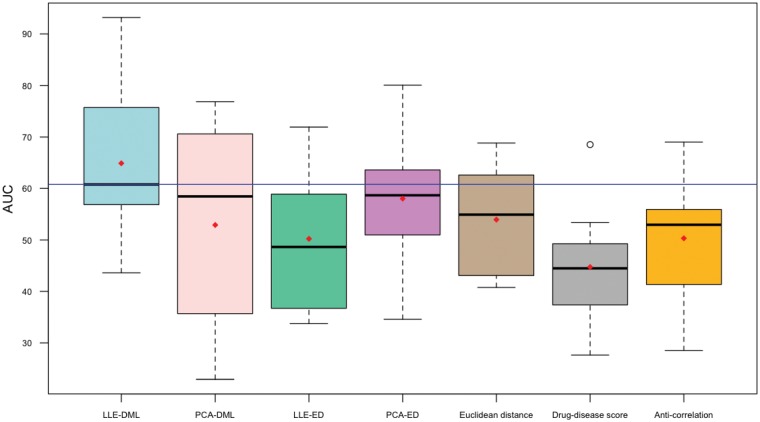
We use three different diseases (breast cancer, IPF and RA) in order to illustrate the capabilities of our proposed methods. As it shown in Table 1 and Figure 2, the proposed LLE-DML method performs better in terms of placing the FDA-approved drugs for their approved indications at the top of their ranked list.
Table 1.
The performances of seven methods compared on the breast cancer, IPF and RA datasets
| LLE-DML |
PCA-DML |
LLE-ED |
PCA-ED |
Euclidean distance |
Drug–disease score Sirota et al. (2011) |
Anti-correlation |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dataset | NDCG | P-value | NDCG | P-value | NDCG | P-value | NDCG | P-value | NDCG | P-value | NDCG | P-value | NDCG | P-value | |
| Breast cancer | GSE26910 | 0.3348 | 0.0140 | 0.1586 | 0.2398 | 0.1317 | 0.5904 | 0.1486 | 0.3207 | 0.1820 | 0.1299 | 0.2533 | 0.0429 | 0.1827 | 0.1239 |
| GSE1299 | 0.3355 | 0.0140 | 0.2695 | 0.0329 | 0.1421 | 0.3956 | 0.1741 | 0.1648 | 0.2095 | 0.0779 | 0.1286 | 0.6783 | 0.2096 | 0.0779 | |
| TCGA-BRCA | 0.2443 | 0.0480 | 0.1465 | 0.3387 | 0.1482 | 0.3237 | 0.1585 | 0.2398 | 0.1538 | 0.2747 | 0.1523 | 0.2847 | 0.1517 | 0.2937 | |
| Idiopathic pulmonary fibrosis | LGRC | 0.2445 | 0.0460 | 0.1609 | 0.3037 | 0.1404 | 0.5894 | 0.1611 | 0.2987 | 0.1652 | 0.2507 | 0.1437 | 0.5325 | 0.1562 | 0.3497 |
| GSE33577 | 0.3639 | 0.0180 | 0.1730 | 0.2088 | 0.1614 | 0.2987 | 0.1609 | 0.3037 | 0.1588 | 0.3247 | 0.1566 | 0.3447 | 0.1560 | 0.3526 | |
| GSE55384 | 0.2468 | 0.0410 | 0.1871 | 0.1528 | 0.1493 | 0.4396 | 0.1671 | 0.2378 | 0.1695 | 0.2238 | 0.1408 | 0.5844 | 0.1654 | 0.2507 | |
| GSE24206 | 0.2579 | 0.0350 | 0.1339 | 0.7572 | 0.1654 | 0.2488 | 0.1934 | 0.1339 | 0.2095 | 0.0899 | 0.2511 | 0.0370 | 0.1719 | 0.2138 | |
| Rheumatoid arthritis | GSE1919 | 0.1955 | 0.1069 | 0.1324 | 0.5554 | 0.1819 | 0.1339 | 0.1337 | 0.5285 | 0.1342 | 0.5155 | 0.1452 | 0.3516 | 0.1377 | 0.4625 |
| GSE15573 | 0.1862 | 0.1209 | 0.1454 | 0.3497 | 0.1474 | 0.3287 | 0.1427 | 0.3846 | 0.1704 | 0.1738 | 0.1358 | 0.4835 | 0.1655 | 0.1928 | |
Note: The NDCG value is determined to be significant based on the bootstrap procedure. Rows with bold text show the highest and the most significant (P-value <0.05) NDCG values. As shown, the LLE-DML method performs better in terms of placing the FDA-approved drugs for their approved indications at the top of their ranked list.
Fig. 2.
The performances of seven methods compared on the breast cancer, IPF and RA datasets based on the AUC of the receiver-operator characteristic. The proposed LLE-DML is superior to all other methods including the one proposed by Sirota et al. (2011)
The detailed results of breast cancer, IPF and RA are shown in separate sections. Among the top ranked drugs, we only chose the FDA-approved ones for further evaluation of their potential therapeutic effect on a given disease. The reason is that off-label use of an FDA-approved drug carry less risk compared to those experimental small molecules that are not yet FDA-approved (Libermann, 2012). We compare the proposed drug repurposing methods with other existing methods by comparing the rankings of FDA-approved drugs, as well as the rankings of other alternative drugs proposed elsewhere in the literature.
3.1 Breast cancer
The first breast cancer dataset is the result of comparing gene expression levels between stroma surrounding invasive breast primary tumors (n = 6) and matched samples of normal stroma (n = 6) using Affymetrix Human Genome U133 Plus2.0 Array. This dataset is available in GEO (ID: GSE26910) (Planche et al., 2011). The second disease dataset compares four breast cancer samples with two healthy samples using Affymetrix Human Genome U133A Array. This dataset is available via GEO (ID: GSE1299) (Mecham et al., 2004). The third dataset is a RNA-Seq data for breast cancer obtained from the Cancer Genome Atlas (TCGA) research network (The Cancer Genome Atlas Research Network, 2012).
The top 10 drugs for breast cancer as identified by each method for the datasets GSE26910, GSE1299 and TCGA-BRCA are included in the Supplementary Materials. Among the top ranked drugs, we chose daunorubicin, ambroxol and ciclopirox that have lower average rank across all the breast cancer datasets for further evaluations. Daunorubicin is one of the drug candidates proposed by our method for treating breast cancer. This drug is an anthracycline used in treatment of leukemia. The anthracyclines are the inhibitors of Human DNA topoisomerase II-alpha. Clinical studies proved that Human DNA topoisomerase II-alpha is a marker of cell proliferation in breast cancer (Lynch et al., 1997). Based on this evidence, Daunorubicin which inhibits Human DNA topoisomerase II-alpha may have a potential therapeutic effect on breast cancer. This hypothesis is under phase I clinical study evaluating the effectiveness of Daunorubicin in treating breast cancer patients (ClinicalTrials.gov identifier: NCT00004207).
The other proposed drug is ambroxol. This drug is an inhibitor of protein Cytochrome P450 3A4 which is used in the treatment of respiratory diseases. The potential therapeutic role of protein Cytochrome P450 3A4 in breast carcinogenesis has been already proved (Modugno et al., 2003). redundant.
Ciclopirox is also one of the top rank drugs proposed by our method for treating breast cancer. This drug is an antifungal medication that is able to inhibit the cell proliferation in breast cancer (Zhou et al., 2010).
3.2 IPF
We analyze four gene expression profiles studying IPF disease. The first such dataset analyzed the whole lung explant from advanced IPF patients using Affymetrix Human Genome U133 Plus 2.0 Array. This dataset is available from GEO (ID: GSE24206) (Meltzer et al., 2011). The second disease dataset is the result of comparing gene expression levels between lung tissue samples from 40 IPF patients and 8 healthy controls using Agilent-014850 Whole Human Genome Microarray 4x44K G4112F (GEO ID: GSE53845) (DePianto et al., 2015). The third IPF dataset used in this study was produced by comparing 93 IPF patients with 30 healthy samples using Agilent-014850 Whole Human Genome Microarray 4x44K G4112F. This dataset is available via GEO (ID: GSE33566) (Yang et al., 2012). The fourth dataset we use is the Lung Genomics Research Consortium dataset. This dataset has gene expression arrays for lung tissues from 61 patients with Interstitial Lung Disease and 17 controls (GEO ID: GSE47460). The top 10 drugs for IPF as identified by each method for these datasets are included in the Supplementary Materials. In this case study, we chose erlotinib, gefitinib and sorafenib that have lower average rank across all the IPF datasets for further evaluation. The first two drugs, erlotinib and gefitinib, are both the inhibitor of the epidermal growth factor receptor (EGFR) which are approved for treatment of advanced or metastatic non-small cell lung cancer. The EGFR inhibitors are able to prevent progression of pulmonary fibrosis (Hardie et al., 2008). The protective effect of gefitinib and erlotinib on lung fibrosis induced by bleomycin is reported in Ishii et al. (2006) and Hardie et al. (2008). These findings suggest that the EGFR inhibitors may have the therapeutic effect on IPF. Sorafenib have been approved for treatment of advanced renal cell carcinoma. The therapeutic effect of Sorafenib on the IPF has been reported in Chen et al. (2013).
3.3 RA
We use two different RA datasets. The first such dataset is the result of comparing gene expression levels between synovial tissues from RA patients (n = 5) and normal donors (n = 5) using Affymetrix Human Genome U95A Array. The details of this study and its biological significance are presented elsewhere (Ungethuem et al., 2010). This dataset is available in GEO (ID: GSE1919). The second RA dataset is produced by comparing 18 RA samples with 15 healthy samples using Illumina human-6 v2.0 expression beadchip microarray platform. This dataset is available via GEO (ID: GSE15573) (Teixeira et al., 2009). The top 10 drugs for RA as identified by each method for these datasets are included in the Supplementary Materials. Sirolimus is an immunosuppressant drug predicted to have a therapeutic effect on RA disease by our proposed method. Rapamycin (sirolimus), a natural product derived from the soil bacteria Streptomyces hygrosopius, was approved for use in organ transplantation (Kahan et al., 2000; Schreiber, 1991; Sehgal et al., 1975; Vezina et al., 1975). Other independent studies have also shown that sirolimus has therapeutic effects on inflammatory arthritis (Cejka et al., 2010). In addition, there is a phase II clinical study aiming to find out if sirolimus could be useful for patients with autoimmune cytopenias such as RA (ClinicalTrials.gov identifier: NCT00392951). Trimipramine and verteporfin are also suggested by our method for treatment of RA disease. Trimipramine is an antidepressant drug. Based on the results published in Grant Macfarlane et al. (1986), this drug is able to reduce the joint pain in RA patients. The effect of laser therapy using verteporfin, an antineovascularization drug, for RA in an animal model has been published in Hendrich et al. (2001).
4 Conclusion
The main goal of computational drug repurposing methods is to find new indications for already approved drugs, in a systematic manner. Using the large number of publicly available datasets for diseases and drugs and by taking advantage of known relations between them, we propose a machine learning-based method for drug repurposing. In particular, our proposed method learns a similarity metric through a supervised machine learning-based algorithm and ranks drugs according to their predicted effectiveness for a disease. We use measurements of more than 20 000 genes in 9 datasets studying 3 different diseases, as well as measurements of around 500 drug instances obtained from CMap and Library of Integrated Network-based Cellular Signatures databases.
The results show that the proposed method provides better drug ranking when compared to the classical approach proposed by Sirota et al. (2011). We also investigate whether the better performance by the LLE-DML method is actually due to the DML algorithm or due to the dimensionality reduction algorithm by retrieving the closest drugs to the disease after transforming the data to lower dimension using the dimensionality reduction algorithm without incorporating DML algorithm (competing methods 3 and 4). The results show that, the LLE-DML method provides better results compared to the methods that are only based on the dimensionality reduction algorithms. This leads us to the conclusion that incorporating a non-linear dimensionality reduction algorithm (LLE) with the DML algorithm is indeed able to transform the data into a space in which the disease–drug pairs that have been clinically proved to be relevant become closer to each other.
The method proposed here was based on transcriptional data alone. However, incorporating transcriptional data with available clinical knowledge such as drug (Guney et al., 2016), chemical or pharmaceutical information (Napolitano et al., 2013), disease biomarkers, target pathways or symptomatology information may yield still better results.
Funding
This study was supported in part by the following grants: [NIH R01 DK089167, NSF 213 DBI-0965741, R42 GM087013]; and by the Robert J. Sokol Endowment in Systems Biology.
Conflict of Interest: none declared.
Supplementary Material
References
- Adams C.P., Brantner V.V. (2006) Estimating the cost of new drug development: is it really $802 million? Health Aff., 25, 420–428. [DOI] [PubMed] [Google Scholar]
- Alaimo S. et al. (2013) Drug–target interaction prediction through domain-tuned network-based inference. Bioinformatics, 29, 2004–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaimo S. et al. (2016) Recommendation techniques for drug–target interaction prediction and drug repositioning. Methods Mol. Biol., 1415, 441–462. [DOI] [PubMed] [Google Scholar]
- Ashburn T.T., Thor K.B. (2004) Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov., 3, 673–683. [DOI] [PubMed] [Google Scholar]
- Benson D.A. et al. (2002) GenBank. Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka D. et al. (2010) Mammalian target of rapamycin signaling is crucial for joint destruction in experimental arthritis and is activated in osteoclasts from patients with rheumatoid arthritis. Arthritis Rheum., 62, 2294–2302. [DOI] [PubMed] [Google Scholar]
- Chen H. et al. (2015) Network-based inference methods for drug repositioning. Comput. Math. Methods Med., 2015, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. et al. (2013) Sorafenib ameliorates bleomycin-induced pulmonary fibrosis: potential roles in the inhibition of epithelial–mesenchymal transition and fibroblast activation. Cell Death Dis., 4, e665.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. et al. (2012) Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol., 8, e1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePianto D.J. et al. (2015) Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax, 70, 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson M., Gagnon J.P. (2009) The cost of new drug discovery and development. Discov. Med., 4, 172–179. [PubMed] [Google Scholar]
- DiMasi J. et al. (2003) The price of innovation: new estimates of drug development costs. J. Health Econ., 22, 151–186. [DOI] [PubMed] [Google Scholar]
- Drăghici S. (2011) Statistics and Data Analysis for Microarrays Using R and Bioconductor. Chapman and Hall/CRC Press, Boca Raton, FL. [Google Scholar]
- Dudley J.T. et al. (2011) Exploiting drug–disease relationships for computational drug repositioning. Brief. Bioinform., 12, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. et al. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghofrani H.A. et al. (2006) Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nat. Rev. Drug Discov., 5, 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant Macfarlane J. et al. (1986) Trimipramine in rheumatoid arthritis: a randomized double-blind trial in relieving pain and joint tenderness. Curr. Med. Res. Opin., 10, 89–93. [DOI] [PubMed] [Google Scholar]
- Guney E. et al. (2016) Network-based in silico drug efficacy screening. Nat. Commun., 7, 10331.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A. et al. (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res., 33, D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie W.D. et al. (2008) EGF receptor tyrosine kinase inhibitors diminish transforming growth factor-α-induced pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol., 294, L1217–L1225. [DOI] [PubMed] [Google Scholar]
- Hendrich C. et al. (2001) Experimental photodynamic laser therapy for rheumatoid arthritis using photosan-3, 5-ALA-induced PPIX and BPD verteporfin in an animal model In: Gerber,B.E. et al. (eds) Lasers in the Musculoskeletal System. Springer, Berlin, Heidelberg, pp. 69–74. [Google Scholar]
- Iorio F. et al. (2010) Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. USA, 107, 14621–14626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F. et al. (2013) Transcriptional data: a new gateway to drug repositioning? Drug Discov. Today, 18, 350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A. et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Ishii Y. et al. (2006) Gefitinib prevents bleomycin-induced lung fibrosis in mice. Am. J. Respir. Crit. Care Med., 174, 550–556. [DOI] [PubMed] [Google Scholar]
- Järvelin K., Kekäläinen J. (2002) Cumulated gain-based evaluation of IR techniques. ACM Trans. Inf. Syst., 20, 422–446. [Google Scholar]
- Jolliffe I. (2002) Principal Component Analysis. Wiley Online Library, Hoboken, NJ. [Google Scholar]
- Kahan B.D. et al. (2000) Efficacy of sirolimus compared with azathioprine for reduction of acute renal allograft rejection: a randomised multicentre study. Lancet, 356, 194–202. [DOI] [PubMed] [Google Scholar]
- Keiser M.J. et al. (2007) Relating protein pharmacology by ligand chemistry. Nat. Biotechnol., 25, 197.. [DOI] [PubMed] [Google Scholar]
- Keiser M.J. et al. (2009) Predicting new molecular targets for known drugs. Nature, 462, 175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. et al. (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science, 313, 1929–1935. [DOI] [PubMed] [Google Scholar]
- Langhauser F. et al. (2018) A diseasome cluster-based drug repurposing of soluble guanylate cyclase activators from smooth muscle relaxation to direct neuroprotection. NPJ Syst. Biol. Appl., 4, 8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V. et al. (2014) Drugbank 4.0: shedding new light on drug metabolism. Nucleic Acids Res., 42, 1091–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libermann T. (2012) Testing new drugs for treatment of melanoma patients applying connectivity map database analysis with melanoma gene signatures. Technical report, DTIC Document. [Google Scholar]
- Lotfi Shahreza M. et al. (2017) A review of network-based approaches to drug repositioning. Brief. Bioinform., 19, 878–892. [DOI] [PubMed] [Google Scholar]
- Lynch B.J. et al. (1997) Human DNA topoisomerase II-alpha: a new marker of cell proliferation in invasive breast cancer. Hum. Pathol., 28, 1180–1188. [DOI] [PubMed] [Google Scholar]
- Mahalanobis P.C. (1936) On the generalized distance in statistics. PNIS, 2, 49–55. [Google Scholar]
- Mariotti V. et al. (2010) Effect of prolonged phenytoin administration on rat brain gene expression assessed by DNA microarrays. Exp. Biol. Med., 235, 300–310. [DOI] [PubMed] [Google Scholar]
- Mecham B.H. et al. (2004) Sequence-matched probes produce increased cross-platform consistency and more reproducible biological results in microarray-based gene expression measurements. Nucleic Acids Res., 32, 74.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer E.B. et al. (2011) Bayesian probit regression model for the diagnosis of pulmonary fibrosis: proof-of-principle. BMC Med. Genomics, 4, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modugno F. et al. (2003) A potential role for the estrogen-metabolizing cytochrome P450 enzymes in human breast carcinogenesis. Breast Cancer Res. Treat., 82, 191–197. [DOI] [PubMed] [Google Scholar]
- Munos B. (2009) Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov., 8, 959–968. [DOI] [PubMed] [Google Scholar]
- Napolitano F. et al. (2013) Drug repositioning: a machine-learning approach through data integration. J. Cheminform., 5, 30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novac N. (2013) Challenges and opportunities of drug repositioning. Trends Pharmacol. Sci., 34, 267–272. [DOI] [PubMed] [Google Scholar]
- Pacini C. (2013) DrugVsDisease: Comparison of Disease and Drug Profiles Using Gene Set Enrichment Analysis. R package version 2.4.0.
- Planche A. et al. (2011) Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS One, 6, 18640.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roweis S.T., Saul L.K. (2000) Nonlinear dimensionality reduction by locally linear embedding. Science, 290, 2323–2326. [DOI] [PubMed] [Google Scholar]
- Schreiber S.L. (1991) Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science, 251, 283–287. [DOI] [PubMed] [Google Scholar]
- Sehgal S. et al. (1975) Rapamycin (AY-22, 989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot., 28, 727–732. [DOI] [PubMed] [Google Scholar]
- Sirota M. et al. (2011) Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med., 3, 96ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P. et al. (2017) Drug repurposing by integrated literature mining and drug–gene–disease triangulation. Drug Discov. Today, 22, 615–619. [DOI] [PubMed] [Google Scholar]
- Suthram S. et al. (2010) Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoS Comput. Biol., 6, e1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. et al. (2015) dml: Distance Metric Learning in R. CRAN. R package version 1.1.0.
- Teixeira V.H. et al. (2009) Transcriptome analysis describing new immunity and defense genes in peripheral blood mononuclear cells of rheumatoid arthritis patients. PLoS One, 4, 6803.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (2012) Comprehensive molecular portraits of human breast tumours. Nature, 490, 61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobinick E. (2009) The value of drug repositioning in the current pharmaceutical market. Drug News and Perspect., 22, 119.. [DOI] [PubMed] [Google Scholar]
- Torresani L., Lee K-c. (2006) Large margin component analysis. In Bernhard,H.S. et al. (eds) Advances in Neural Information Processing Systems, MIT Press, Cambridge, MA, USA, pp. 1385–1392.
- Ungethuem U. et al. (2010) Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis. Physiol. Genomics, 42A, 267–282. [DOI] [PubMed] [Google Scholar]
- Vargas D.M. et al. (2018) Alzheimer’s disease master regulators analysis: search for potential molecular targets and drug repositioning candidates. Alzheimers Res. Ther., 10, 59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina C. et al. (1975) Rapamycin (AY-22, 989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot., 28, 721–726. [DOI] [PubMed] [Google Scholar]
- Xing E.P. et al. (2002) Distance metric learning with application to clustering with side-information. In: Suzanna,B. et al. (eds) Advances in Neural Information Processing Systems, MIT Press, Cambridge, MA, USA, pp. 505–512.
- Yang I.V. et al. (2012) The peripheral blood transcriptome identifies the presence and extent of disease in idiopathic pulmonary fibrosis. PLoS One, 7, 37708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. (2010) The antitumor activity of the fungicide ciclopirox. Int. J. Cancer, 127, 2467–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.