Abstract
Cadmium (Cd) is one of the most toxic heavy metals and a non‐essential element to all organisms, including plants; however, the genes involved in Cd resistance in plants remain poorly characterised.
To identify Cd resistance genes in rice, we screened a rice cDNA expression library treated with CdCl2 using a yeast (Saccharomyces cerevisiae) mutant ycf1 strain (DTY167) and isolated two rice phytochelatin synthases (OsPCS5 and OsPCS15).
The genes were strongly induced by Cd treatment and conferred increased resistance to Cd when expressed in the ycf1 mutant strain. In addition, the Cd concentration was twofold higher in yeast expressing OsPCS5 and OsPCS15 than in vector‐transformed yeast, and OsPCS5 and OsPCS15 localised in the cytoplasm. Arabidopsis thaliana plants overexpressing OsPCS5/‐15 paradoxically exhibited increased sensitivity to Cd, suggesting that overexpression of OsPCS5/‐15 resulted in toxicity due to excess phytochelatin production in A. thaliana.
These data indicate that OsPCS5 and OsPCS15 are involved in Cd tolerance, which may be related to the relative abundances of phytochelatins synthesised by these phytochelatin synthases.
Keywords: Cadmium, heavy metal tolerance, Oryza sativa, phytochelatin synthase, YCF1
Introduction
Contamination by toxic heavy metals is correlated with the degree of industrialisation through mining and combustion of fossil fuels and damages the environment and human health (Nriagu & Pacyna 1988). Among heavy metals, copper (Cu) and zinc (Zn) function as cofactors of enzymes involved in redox reactions and electron transfer reactions in all living organisms (Mengel & Kirkby 2001). Non‐essential heavy metals, including arsenic (As), cadmium (Cd), mercury (Hg) and lead (Pb), have potentially toxic effects, such as inactivation of enzymes and promotion of oxidative stress (Van Assche & Clijsters 1990; Mengel & Kirkby 2001). Phytoremediation is a technology that uses plants to remove heavy metals from polluted areas and is considered a cost‐effective and environmentally friendly technology (Raskin et al. 1997; Salt et al. 1998; Krämer 2005). Because natural hyperaccumulating plants produce low biomass, grow slowly and only accumulate a specific element (Cunningham et al. 1995), genetic and molecular studies are required for the optimisation and improvement of phytoremediation to enhance metal tolerance and accumulation.
Cadmium (Cd) is a hazardous environmental contaminant that is responsible for severe human health problems. It causes intracellular damage via inactivation or denaturation of proteins by binding to free sulphydryl residues, by displacing cofactors from a variety of proteins containing transcription factors and by causing oxidative stress, which induces membrane breakage (Liao et al. 2002; Kim et al. 2008). Animals are exposed to Cd through many different channels, but mainly via food intake. Farm products become contaminated with Cd when grown in Cd‐contaminated soils (Kim et al. 2008). Accumulation of Cd in plants induces chlorosis, wilting, growth reduction and cell death (Herbette et al. 2006). Cd detoxification mechanisms in plants include production of cysteine‐rich peptides such as glutathione (GSH), metallothioneins (MT) and phytochelatins (PC) (Rauser 1999; Cobbett 2000a,b).
Phytochelatins are heavy metal‐binding peptides with a general structure of (γ‐Glu‐Cys)n‐Gly, where n ranges from 2 to 11, and are synthesised enzymatically following exposure to various heavy metals (Rauser 1990; Cobbett 2000b). These peptides are synthesised from reduced GSH by phytochelatin synthase (PCS), which is induced by heavy metals such as Cd (Cobbett 2000b; Vatamaniuk et al. 2000). In addition, PC are required for Cd tolerance in plants, and mutants lacking the ability to synthesise PC are hypersensitive to Cd (Howden et al. 1995). In Arabidopsis thaliana, cad1 and cad2 mutants are deficient in PC synthesis because of mutations in γ‐glutamylcysteine synthetase (cad2) and PCS (cad1), respectively (Howden & Cobbette 1992; Howden et al. 1995; Cobbette et al. 1998). PC bind heavy metals such as Cd, Cu and As with high affinity, and the complexes formed in the cytosol are subsequently localised to vacuoles for detoxification (Grill et al. 1985; Salt 1995; Maitani et al. 1996; Zenk 1996; Schmöger et al. 2000).
Phytochelatin synthases are essential proteins that play important roles in heavy metal detoxification. PCS genes were first isolated from A. thaliana, Schizosaccharomyces pombe and Triticum aestivum (Clemens et al. 1999; Ha et al. 1999; Vatamaniuk et al. 1999), although many PCS genes have since been cloned in several plant species (Shen et al. 2010). In addition, overexpression of the AtPCS1 gene in Escherichia coli, Saccaromyces cerevisiae, Nicotiana tabacum and Brassica juncea enhances tolerance to and accumulation of Cd (Vatamaniuk et al. 1999; Merle et al. 2003; Pomponi et al. 2006; Gasic & Korban 2007). Expression of TaPCS1 in Nicotiana glauca also enhances tolerance to and accumulation of Cd and Pb (Martinez et al. 2006). Overexpression of AtPCS1 enhances Cd sensitivity with a high level of PC in A. thaliana and leads to Cd hypersensitivity without increasing the level of PC in tobacco (Lee et al. 2003a,b; Li et al. 2004; Wojas et al. 2008). Although there are many reports on the functions of PCS in heavy metal detoxification in plants, the effects on plant phenotypes have rarely been explained. Overexpression of PCS genes from different species using a model plant such as A. thaliana could reveal their properties and functions and address their application in the improvement of phytoremediation.
Yeast cadmium factor (YCF1) is a member of the ATP‐binding cassette (ABC) transporter family and confers Cd resistance in S. cerevisiae. It functions as a transporter to sequester GSH‐conjugated Cd in vacuoles (Li et al. 1997). To extend our knowledge of the Cd tolerance mechanism in rice, we constructed a rice cDNA library following treatment with CdCl2, and performed screening using the S. cerevisiae ycf1 mutant strain DTY167. We isolated two PCS, OsPCS5 and OsPCS15, via this screening. OsPCS15 is a novel PCS in rice. Both genes responded to Cd and the proteins were localised in the cytoplasm in yeast. In addition, A. thaliana plants overexpressing OsPCS5/‐15 exhibited hypersensitivity to Cd.
Material and Methods
Yeast, bacterial strains and plant material
The YCF1 null mutant, DTY167 (MATα ura3‐52 leu2‐3,‐112 his‐200 trp1‐901 lys2‐801 suc2‐9 ycf1::hisG), and wild type (WT), DTY165 (MATα ura3‐52 his6 leu2‐3,‐112 his3‐ Delta 200 trp1‐901 lys2‐801 suc2‐ Delta), S. cerevisiae strains were used for rice cDNA library screening and control experiments, respectively (Li et al. 1996). The E. coli strain XL‐1 Blue MRF′ (Stratagene, La Jolla, CA, USA) was used for cloning. All A. thaliana plants used in this study were the Columbia (Col‐0) ecotype. Plants were germinated on MS (Murashige & Skoog 1962) nutrient medium (pH 5.8) containing 3% sucrose and 0.25% phytagel and maintained in a growth chamber at 22 °C with a 16/8 h light/dark cycle.
Screening of the rice (Oriza sativa cv. Milyang 117) cDNA expression library
A rice cDNA expression library was inserted into the pAD‐GAL4‐2.1 vector from rice callus treated with CdCl2. S. cerevisiae DTY167 cells were transformed with the rice cDNA library using the lithium acetate transformation method (Gietz & Schiestl 1995). The transformants were selected for leucine prototrophy on SD‐leu medium, transferred to liquid SD‐leu medium, grown overnight at 30 °C and streaked on SD‐leu solid medium containing 50 μm CdCl2. Strains exhibiting high tolerance to Cd were transferred to YPD liquid medium and grown overnight at 30 °C. DNA was extracted from the yeast cells and transformed into E. coli. The colonies were analysed by restriction digestion and sequencing.
Cloning of OsPCS genes
A fragment of OsPCS5 cDNA was amplified by PCR using the gene‐specific primers 5′‐CTCGAGATGGCAGCGATGGCATCCCTG‐3′ and 5′‐TACTAGTCCACCTCCATGGGATTGTGGCACAGGATC‐3′, which contained XhoI and SpeI sites, respectively. In addition, OsPCS15 cDNA was amplified using the gene‐specific primers 5′‐CTCGAGATGGCGTCTAAACCAAGCAGCCGAGCGGAA‐3′ and 5′‐TACTAGTCCACCTCCGCATTGTTCCCAAGGTTGTGG‐3′, which contained XhoI and SpeI sites, respectively. The genes were then cloned into the pGEM‐T easy vector (Promega, Madison, WI, USA) and used for various plasmid constructs.
Metal treatments and RNA gel blot analysis
For heavy metal treatments, suspension culture cells induced from rice (Oryza sativa L. Milyang 117) embryos were used. The suspension cells were maintained at 25 °C with shaking at 90–100 rpm in 100‐ml flasks containing liquid R2 medium supplemented with 2 mg·l−1 2, 4‐D and 3% sucrose, and subcultured every week. Rice suspension cells were treated in the dark with 50 μm CdCl2 or 500 μm Na2HAsO4 in the original flasks. Each sample was harvested on filter papers by vacuum filtration. Harvested cells were immediately frozen in liquid nitrogen and stored at −70 °C.
Rice suspension cells were ground to a fine powder with a mortar and pestle. RNA gel blot analysis was carried out as previously described (Park et al. 2002; Xu et al. 2007). A total of 20 μg of total RNA were separated on a denatured 1.5% formaldehyde/agarose gel, transferred onto a nylon membrane (GeneScreenPlus, NEN Life Science Products, Boston, MA, USA), and hybridised with OsPCS5/‐15 gene‐specific probes. To verify equal loading, rRNA was visualised by staining with ethidium bromide. Hybridisation and washing were carried out under high stringency conditions. The blots were air‐dried and exposed to X‐ray film at −70 °C.
Yeast complementation assay for Cd tolerance
The WT S. cerevisiae (DTY165) and ycf1 mutant (DTY167) cells were transformed with empty vectors (WT/vector and ycf1/vector, respectively). ycf1 mutant (DTY167) cells were also transformed with pXY‐OsPCS5 and pXY‐OsPCS15, respectively. The transformed cells were grown overnight to an optical density at 600 nm (OD600) of 1.7. Aliquots of the cell suspensions were then serially diluted and spotted on solid YPD medium containing or lacking 70 μm CdCl2. Colonies were visualised after incubating the plates for 2–4 days at 30 °C. In addition, the strains were grown overnight at 30 °C in liquid minimal selective medium containing 2% glucose. The cultures were diluted in minimal medium to an OD600 of 0.1 in the presence of various concentrations of CdCl2 and incubated for an additional 24 h, after which growth was determined by measuring OD600 (Ghosh et al. 1999).
Cadmium content and subcellular localisations of OsPCS5 and OsPCS15
Transformed yeast cells were grown in liquid YPD medium containing 20 μm Cd2+, incubated at 30 °C for 24 h, washed in sterilised deionised water, and harvested by centrifugation. Finally, the yeast samples were acidified with HNO3 and incubated at 65 °C for 30 min. The samples were centrifuged at 5000 g for 10 min to remove debris, and the Cd2+ content of the supernatant was determined using inductively coupled plasma optical emission spectroscopy (ICP‐OES; Optima 4300DV/5300DV; Perkin‐Elmer, Waltham, MA, USA).
To observe the subcellular localisations of OsPCS5 and OsPCS15 proteins in yeast, OsPCS5::GFP and OsPCS15::GFP fusion constructs were prepared, respectively. Each cDNA was fused to the coding region of GFP under control of the cauliflower mosaic virus (CaMV) 35S promoter. Transformed yeast cells were grown overnight at 30 °C, subcultured at a 1:1000 dilution in the medium and grown at 30 °C to an OD600 of ~0.7. Cells in log phase were examined at 100× magnification on slides using an Olympus FV1000 confocal microscope (Olympus America, Center Valley, PA, USA) with an excitation wavelength of 488 nm.
Generation of transgenic A. thaliana plants and Cd resistance assays
To construct transgenic A. thaliana plants, OsPCS5 and OsPCS15 cDNA were cloned into the plant GFP expression vector (pCAMBIA1302). In this vector, OsPCS5 and OsPCS15 cDNA were expressed under the control of the CaMV 35S promoter. The recombinant plasmids were transformed into Agrobacterium tumefaciens GV3101, and transgenic A. thaliana plants were generated using the floral dip method (Clough & Bent 1998). Homozygous T3 progeny of transgenic plants were selected and used for further experiments following growth under a 16 h/8 h light/dark cycle at 22 °C. The protein levels of OsPCS5/‐15 transgenic plants grown for 3 weeks on plates were confirmed by Western blot analysis with an anti‐GFP antibody.
To examine the effect of Cd on OsPCS5/‐15 transgenic A. thaliana plants, WT (ecotype Columbia) and transgenic seeds were germinated on half‐strength MS medium solidified with phytagel (2.5 g·l−1; Murashige & Skoog 1962) containing 0, 50 or 70 μm CdCl2. After germination, plates were positioned vertically to check root growth. Root lengths of WT and OsPCS5/‐15 transgenic A. thaliana plants were measured after 10 days.
Results
Isolation of rice PCS and sequence comparison with other plant PCS
To isolate Cd resistance genes in rice, an O. sativa cDNA library was introduced into the ycf1 yeast mutant strain DTY167 (Δycf1). Approximately 4 × 104 independent transformants were plated on agar medium containing Cd and grown for 4 days. A total of 120 colonies were selected and DNA was extracted and amplified using E. coli. The inserts were sequenced and compared with sequences in the GenBank® database. Among them, we isolated two PCS that conferred strong Cd tolerance to Δycf1 mutant yeast. These PCS encode polypeptides with predicted masses of 55.7 and 57.4 kDa. Based on the nomenclature of PCS in rice (Shen et al. 2010), one had already been identified as OsPCS5 and the other was a novel PCS gene in rice. Therefore, the latter was named OsPCS15 because 14 PCS or PCS‐like genes were previously identified in the rice genome using bioinformatics methods (Shen et al. 2010).
The deduced amino acid sequences were aligned with those of well‐characterised PCS such as OsPCS1 (O. sativa), AtPCS1 (A. thaliana) and TaPCS1 (T. aestivum), and the PC domains were well conserved in the two OsPCS (Fig. 1). The amino acid sequence of OsPCS5 displayed 76.3% identity and 83.5% similarity with the TaPCS1 sequence, and the PC domain was more conserved, with 91.7% identity to the corresponding PC domain of TaPCS1. In addition, OsPCS15 showed 67.5% identity and 73.8% similarity with the TaPCS1 sequence. The PC domain also exhibited 78.8% identity to that of the TaPCS1 protein. Although OsPCS15 showed approximately 10% lower similarity to TaPCS1 than OsPCS5, amino acid sequences in the PC domain of OsPCS15 were well conserved. These results imply that the OsPCS15 protein is a novel PCS family protein and may function as a PCS in rice.
Figure 1.

Alignment of the deduced amino acid sequence of plant PCS genes. The amino acid sequences of Oryza sativa (OsPCS1, OsPCS5 and OsPCS15), Arabidopsis thaliana (AtPCS1, AAD41794) and Triticum aestivum (TaPCS1, AAD50592) PCS were aligned using the CLUSTAL W alignment program (Thompson et al. 1994). Identical amino acid residues are shown in black boxes and similar amino acid residues are shown in grey boxes. The putative PC domain predicted by the domain prediction program (NCBI web server, http://www.ncbi.nih.gov/BLAST/) is underlined.
Expression patterns of OsPCS5 and OsPCS15 genes following exposure to heavy metals
To investigate whether the PCS5/‐15 genes are expressed in response to heavy metal stress, we performed northern blot analysis with total RNA isolated from rice suspension cells treated with 50 μm Cd2+or 500 μm As2+ . As shown in Fig. 2A, expression of the OsPCS5 transcript was greatly increased at 12 h and remained elevated for 24 h after treatment with Cd or As. OsPCS15 transcript levels showed similar expression patterns (Fig. 2B). Exposure to these toxic metals increases GSH synthesis and they are detoxified by PC (Rauser 1990; Verbruggen et al. 2009). Expression of the OsPCS5/‐15 genes in response to Cd and As supports a role for OsPCS5/‐15 proteins in PC synthesis.
Figure 2.
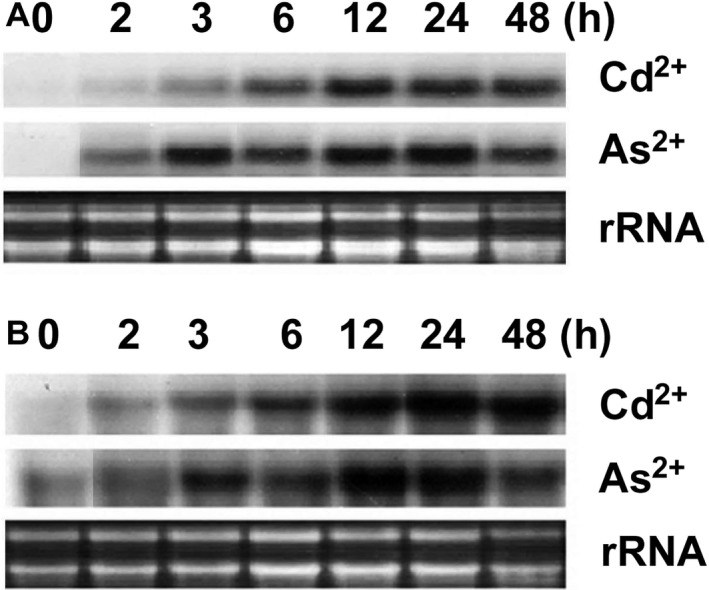
Expression patterns of OsPCS5 and OsPCS15 genes in rice. Northern blot analyses of OsPCS5 (A) and OsPCS15 (B) in response to treatment with 50 μm Cd2+ and 500 μm As5+ in rice cell cultures. Each lane was loaded with 20 μg total RNA prepared from rice suspension‐cultured cells at the indicated time points and separated on a 1.5% formaldehyde/agarose gel. The gel was transferred onto a nylon membrane and hybridised with 32P‐labelled OsPCS5 or OsPCS15 cDNA. To verify equal loading of total RNA, rRNA was visualised by staining with ethidium bromide.
Complementation of OsPCS5/‐15 for Cd tolerance in the yeast ycf1 mutant
Sequestration of Cd and other toxic metals in vacuoles is a well‐characterised mechanism of detoxification (Rea et al. 1998). YCF1 in S. cerevisiae is a crucial factor for the transport of toxic metals into vacuoles and is involved in toxic metal tolerance (Li et al. 1997). To characterise the role of OsPCS5/‐15 in Cd tolerance, S. cerevisiae ycf1 mutant (Δycf1) cells (DTY167) were transformed with constructs containing an empty vector, OsPCS5 or OsPCS15, and WT cells (DTY165) were transformed with an empty vector as a control. The yeast cells were serially diluted and spotted on to YPD agar medium lacking or containing 70 μm CdCl2 (Fig. 3A). Δycf1 cells expressing OsPCS5 or OsPCS15 grew much better than those transformed with an empty vector. They also grew better than WT yeast transformed with an empty vector.
Figure 3.
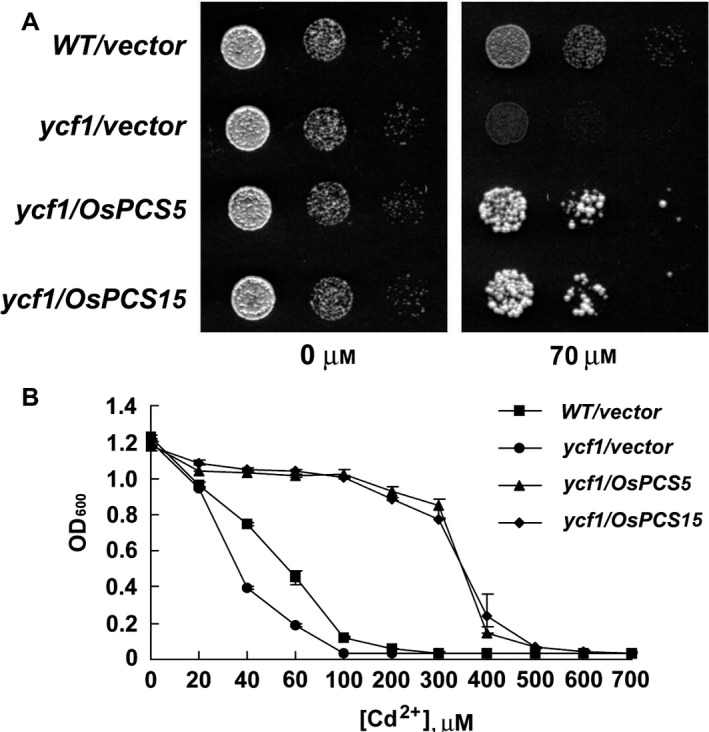
Expression of OsPCS5 and OsPCS15 confers enhanced Cd2+ tolerance in a Δycf yeast mutant. A: Enhanced growth of OsPCS5‐ and OsPCS15‐transformed yeasts on YPD agar plates supplemented with 70 μm CdCl2. Yeast Δycf mutant DTY167 and WT DTY165 cells were transformed with a construct containing the empty vector, OsPCS5 or OsPCS15. Ten‐fold serial dilutions of Δycf mutant and WT yeast cells expressing the indicated plasmid were spotted (2 μl) onto plates containing or lacking 70 μm CdCl2, and incubated at 30 °C for 2–4 days. B: Suppression of Cd2+ hypersensitivity in the yeast Δycf mutant DTY167 by plasmid‐borne OsPCS5 and OsPCS15. Yeast cells expressing OsPCS5 or OsPCS15 were normalised to OD600 and grown in liquid SD media containing different concentrations of Cd2+. After incubation at 30 °C for 24 h, cell growth was determined by measuring OD600. Squares (■), circles (●), triangles (▲) and diamonds (◆) indicate the empty vector (WT), empty vector (ycf1), OsPCS5 (ycf1) and OsPCS15 (ycf1), respectively.
To confirm the Cd tolerance phenotype of OsPCS5‐ and OsPCS15‐expressing yeasts, the cells were grown in liquid SD medium containing various concentrations of CdCl2 and absorbance of cells at 600 nm was measured spectrophotometrically. In medium lacking Cd, growth of OsPCS5‐ and OsPCS15‐transformed Δycf1 cells was similar to that of empty vector‐transformed WT and Δycf1 cells. In the presence of 20, 40 or 60 μm CdCl2, the density of empty vector‐transformed WT and Δycf1 cells decreased in a concentration‐dependent manner. However, the densities of OsPCS5‐ and OsPCS15‐transformed Δycf1 cells were maintained at 1.0 at OD600 in the presence of 300 μm CdCl2 (Fig. 3B). These results indicate that overexpression of OsPCS5 or OsPCS15 in Δycf1 yeast cells confers strong tolerance to CdCl2, suggesting that the OsPCS5 or OsPCS15 protein function in Cd tolerance in yeast.
Cadmium accumulation and subcellular localisation of OsPCS5/‐15 proteins
Phytochelatins are synthesised by cytosolic PC synthase and rapidly induced by Cd. Thereafter, PC chelate Cd and form several complexes to inactivate Cd from free Cd ions in plant cells (DalCorso et al. 2008). To determine Cd accumulation in OsPCS5‐ and OsPCS15‐transformed Δycf1 yeast cells, we analysed the Cd content of empty vector‐, OsPCS5‐ and OsPCS15‐transformed Δycf1 yeast cells grown in liquid YPD medium containing 20 μm CdCl (Fig. 4). The Cd content of ycf1‐OsPCS5/‐15 cells was 2.5‐fold higher than that of the ycf1‐vector control cells. These results imply that OsPCS5/‐15 can contribute to Cd resistance via sequestration of Cd in vacuoles through the formation of PC–Cd complexes.
Figure 4.
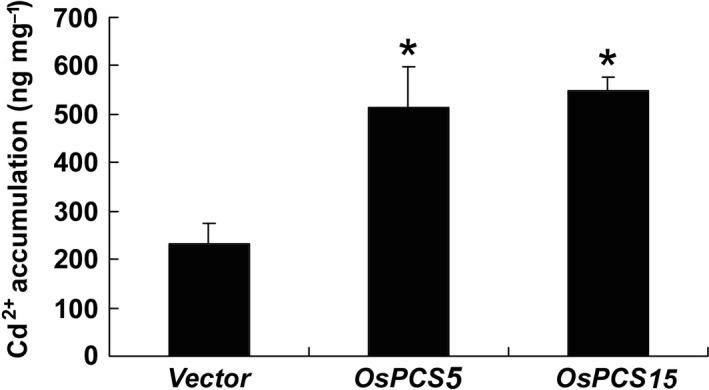
Yeast cells expressing OsPCS5 or OsPCS15 accumulate more Cd2+ than control cells of the Δycf mutant DTY167. Transformed cells were grown in liquid YPD medium containing 20 μm Cd2+ at 30 °C for 24 h. The Cd2+ content of the samples was measured by ICP‐OES. Results are averages (±SE) from three independent experiments performed using three different colonies. Asterisks indicate statistical significance (P < 0.05, Student's t‐test) of differences between yeast cells expressing OsPCS5/‐15 and vector control cells.
Arabidopsis thaliana PCS1 localises in the cytosol and generates metal‐binding PC from GSH (Blum et al. 2010). The ability of OsPCS5/‐15 to complement the Δycf1 yeast mutant may result from the localisation of heterologous proteins in the cytosol of yeast cells. To investigate the localisations of OsPCS5/‐15 proteins, OsPCS5/‐15 proteins tagged with GFP were expressed in yeast cells. GFP signals were visualised using confocal microscopy. Free GFP protein was used as a cytosolic marker protein. Fluorescence of the OsPCS5/‐15::GFP proteins was observed in the cytosol, as was fluorescence of free GFP protein (Fig. 5). These results strongly indicate that OsPCS5/‐15 proteins localise in the cytoplasm in yeast.
Figure 5.
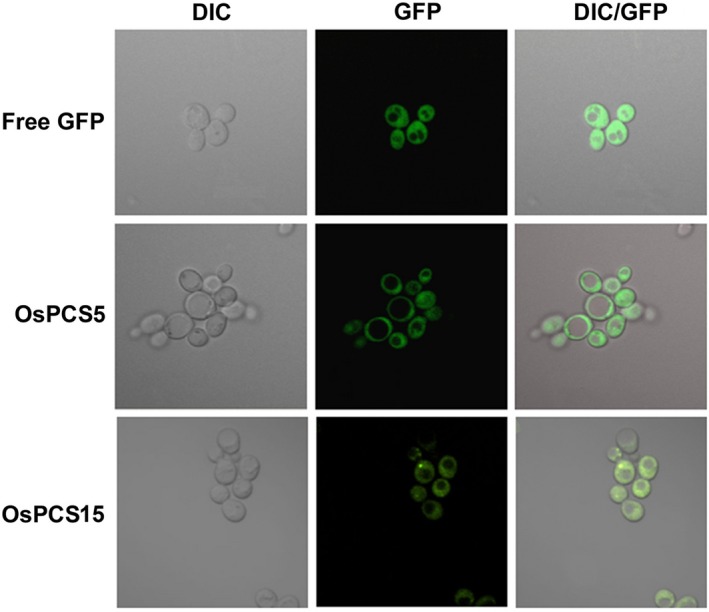
Subcellular localisations of OsPCS5 and OsPCS15 proteins tagged with GFP in yeast cells. Saccharomyces cerevisiae DTY167 cells were transformed with free GFP, OsPCS5::GFP or OsPCS15::GFP, and observed via confocal fluorescence microscopy. DIC and GFP merged images were generated using Olympus software. DIC, differential interference contrast; GFP, green fluorescent protein; DIC/GFP, merged images of DIC and GFP.
Overexpression of OsPCS5 and OsPCS15 and Cd hypersensitivity in A. thaliana
To determine the effects of OsPCS5/‐15 overexpression on Cd tolerance in A. thaliana, we generated OsPCS5/‐15 transgenic A. thaliana plants harbouring plant expression vectors under the control of the CaMV 35S promoter. Individual transgenic plants were selected via hygromycin resistance, followed by Western blot analysis with an anti‐GFP antibody. Three homozygous lines showing the highest expression levels were selected and analysed for Cd tolerance at increasing Cd concentrations (Fig. 6). There were no significant phenotypic differences between OsPCS5/‐15 transgenic plants grown in the absence of Cd. Intriguingly, when the transgenic plant lines were treated with 50 and 70 μm CdCl2, root growth was significantly inhibited. As shown in Fig. 6A, root growth in OsPCS5 transgenic plants was reduced by approximately 33% following treatment with 50 and 70 μm CdCl2. In the OsPCS15 transgenic plant lines, root growth was reduced by 26% following exposure to the same Cd treatments (Fig. 6B). Paradoxically, ectopic expression of PCS by OsPCS5/‐15 increased sensitivity to Cd in A. thaliana.
Figure 6.
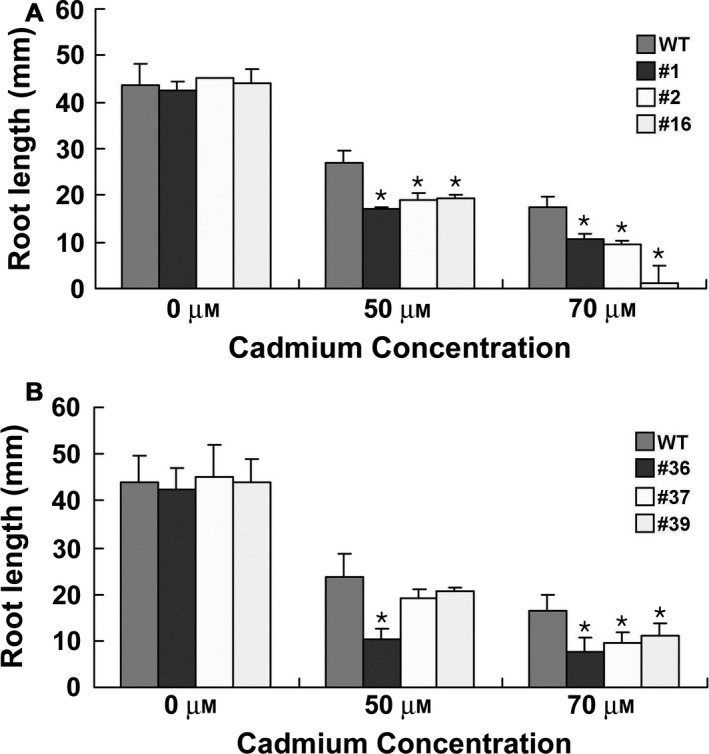
Effects of OsPCS5 and OsPCS15 overexpression on root elongation of plants grown under Cd stress. Seeds were germinated on MS medium containing 0, 50 or 70 μm CdCl2, and Petri dishes were placed in a vertical orientation upon onset of growth. Root lengths of each overexpression line and the WT were measured after 10 days of growth. Results are averages (±SE) of 30 plants. Asterisks indicate statistical significance (P < 0.05, Student's t‐test) of differences between OsPCS5/‐15 overexpressing transgenic plants and WT plants.
Discussion
Heavy metals are a major hazard for ecosystems and human health. Among hazardous toxic metals, Cd is the most toxic to living organisms (He et al. 2015; Song et al. 2015). In particular, Cd accumulation causes a range of adverse effects in higher plants. Cd disturbs plant physiological processes, such as respiration, transpiration and photosynthesis (Toppi & Gabbrielli 1999). Understanding the mechanisms responsible for tolerance to and accumulation of Cd is important for developing efficient strategies to tackle Cd stress in plants.
We generated a rice cDNA library with the ycf1 yeast mutant strain DTY167 (Δycf1) and screened approximately 4 × 104 independent transformants. Two PCS (OsPCS5 and OsPCS15) were isolated as candidates that confer strong Cd tolerance in the Δycf1 mutant yeast (Fig. 1). Examination of the genome of the O. sativa subsp. japonica variety Zhonghua 11 revealed that OsPCS15 encodes a novel PCS (Shen et al. 2010), suggesting a divergence of genes among varieties within the species. Based on previous reports, Cd is the best activator of PCS (Grill et al. 1989; Hayashi et al. 1991). In addition, the A. thaliana PCS (AtPCS1) was identified as a cDNA that suppresses the Cd‐sensitive phenotype of Brewer's yeast ycf1 mutant (Vatamaniuk et al. 1999). Therefore, our screen effectively isolated candidates that confer Cd tolerance in the Δycf1 yeast mutant.
Multiple PCS genes have been identified in A. thaliana (Ha et al. 1999; Vatamaniuk et al. 1999), wheat (Clemens et al. 1999), soybean (Oven et al. 2002) and rice (Shen et al. 2010). Transgenic plants overexpressing PCS genes exhibit increased synthesis of PC and elevated Cd content (Gisbert et al. 2003; Li et al. 2006; Pomponi et al. 2006), and silencing of a PCS gene via RNAi reduces Cd accumulation in rice seeds (Li et al. 2007). On the other hand, overexpression of AtPCS1 in A. thaliana and tobacco increases hypersensitivity to Cd (Lee et al. 2003a; Wojas et al. 2008). These results suggest that none of these PCS genes would be suitable for transforming plants to tolerate heavy metal accumulation. Further studies on the isolation and function of PCS genes in various plant species are required.
Phytochelatin synthases catalyse PC synthesis in plants and PC play an important role in heavy metal homeostasis and detoxification by chelating and sequestering heavy metals (Clemens 2006). Therefore, it is crucial to check PCS expression patterns following treatment with heavy metals. Indeed, PCS genes are expressed following exposure to heavy metals such as Cd, Pb and Zn in rice (Shen et al. 2010). In this study, OsPCS5/‐15 were expressed after treatment with Cd and As, indicating that OsPCS5/‐15 are specific to Cd and As (Fig. 2). OsPCS7 expression is only induced by Hg and Pb, while OsPCS9 expression is induced by Cd and Zn (Shen et al. 2010). These results and our data imply that many PCS genes exist in the rice genome to respond specifically to different heavy metals.
In addition to the significant expression of OsPCS5/‐15 genes following treatment with Cd and As, OsPCS5/‐15 expression in yeast resulted in the tolerance phenotype upon Cd2+ exposure, supporting the suggestion that expression of these genes was responsible for Cd2+ resistance (Fig. 3). OsPCS5/‐15 expression in the yeast mutant Δycf1 enabled cells to grow in a ten‐fold higher Cd2+ concentration than control yeast cells (Fig. 3B). These results concerning the association of metal tolerance with OsPCS5/‐15 expression in yeast provide molecular evidence that PC play a general role in metal homeostasis. PC function as cytosolic chelators and the PC–Cd complexes formed are then sequestered in vacuoles by transporters such as the ABC‐type transporter HMT1 (Ortiz et al. 1992).
In contrast with overexpression of OsPCS5/‐15 genes in yeast, we generated OsPCS5/‐15 transgenic A. thaliana plants using the CaMV 35S promoter. Transgenic A. thaliana plants expressing OsPCS5/‐15 were hypersensitive to Cd (Fig. 6). Similarly, overexpression of PCS genes in A. thaliana, tobacco and rice plants decreases Cd tolerance (Lee et al. 2003a; Li et al. 2004; Wojas et al. 2008; Wang et al. 2012). These results might indicate the activity of PCS from species‐dependent differences generated by the transgenes. The various effects of overexpression may also result from functional differences between PCS from diverse plants. Therefore, not all PCS genes will be suitable for transforming plant species for phytoremediation.
In addition, a study has indicated that Cd accumulation in shoots is higher in Cd‐tolerant transgenic Arabidopsis plants expressing the AtPCS1 gene than in WT and Cd‐intolerant transgenic Arabidopsis plants (Lee et al. 2003b). This is consistent with previous data that Cd‐tolerant transgenic plants overexpressing genes related to synthesis of GSH and PC show increased Cd accumulation in their shoots (Zhu et al. 1999a,b; Domínguez‐Solís et al. 2001). It was also reported that PCS protein is constitutively expressed and activated in the presence of heavy metals (Steffens 1990; Zenk 1996). Therefore, Cd accumulation is correlated with Cd tolerance. Here we speculate that Cd concentration in transgenic A. thaliana plants expressing OsPCS5/‐15 will be similar to that of WT plants. Further progress on the biochemical and functional characterisation of OsPCS5/‐15 genes should focus on PC synthesis and Cd accumulation in transgenic A. thaliana expressing the OsPCS5/‐15 genes.
Author contributions
H.C.P. and W.S.C. designed the study and wrote the manuscript. H.C.P., J.E.H., Y.J., Y.J.K. and X.C.N. performed the experiment and analysed the data. C.Y.K. prepared experimental material and analysed the data. All authors have read and approved the manuscript for publication.
Conflict of Interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Institute of Ecology (NIE‐C‐2019‐15), the Next‐Generation BioGreen 21 Program (#PJ01325401) funded by RDA, and partly by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106.02‐2017.09.
Contributor Information
H. C. Park, Email: hcpark@nie.re.kr.
W. S. Chung, Email: chungws@gnu.ac.kr.
References
- Blum R., Meyer K.C., Wünschmann J., Lendzian K.J., Grill E. (2010) Cytosolic action of phytochelatin synthase. Plant Physiology, 153, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S. (2006) Evolution and function of phytochelatin synthases. Journal of Plant Physiology, 163, 319–332. [DOI] [PubMed] [Google Scholar]
- Clemens S., Kim E.J., Neumann D., Schroeder J.I. (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO Journal, 18, 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . The Plant Journal, 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cobbett C.S. (2000a) Phytochelatins and their roles in heavy metal detoxification. Plant Physiology, 123, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett C.S. (2000b) Phytochelatin biosynthesis and function in heavy‐metal detoxification. Current Opinion in Plant Biology, 3, 211–216. [PubMed] [Google Scholar]
- Cobbette C.S., May M.J., Howden R., Rolls B. (1998) The glutathione‐deficient, cadmium‐sensitive mutant, cad2‐1, of Arabidopsis thaliana is deficient in γ‐glutamylcysteine synthetase. The Plant Journal, 16, 73–78. [DOI] [PubMed] [Google Scholar]
- Cunningham S.D., Betri W.R., Huang J.W. (1995) Phytoremediation of contaminated soils. Trends in Biotechnology, 13, 393–397. [Google Scholar]
- DalCorso G., Farinati S., Maistri S., Furini A. (2008) How plants cope with cadmium: staking all on metabolism and gene expression. Journal of Integrative Plant Biology, 50, 1268–1280. [DOI] [PubMed] [Google Scholar]
- Domínguez‐Solís J.R., Gutiérrez‐Alcalá G., Romero L.C., Gotor C. (2001) The cytosolic O‐acetylserine(thiol)lyase gene is regulated by heavy metals and can function in cadium tolerance. Journal of Biological Chemistry, 276, 9297–9302. [DOI] [PubMed] [Google Scholar]
- Gasic K., Korban S.S. (2007) Expression of Arabidopsis phytochelatin synthase in Indian mustard (Brassica juncea) plants enhances tolerance for Cd and Zn. Planta, 225, 1277–1285. [DOI] [PubMed] [Google Scholar]
- Ghosh M., Shen J., Rosen B.P. (1999) Pathways of As (III) detoxification in Saccharomyces cerevisiae . Proceedings of the National Academy of Sciences of the United States of America, 96, 5001–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. (1995) Transforming yeast with DNA. Methods in Molecular Biology, 5, 255–269. [Google Scholar]
- Gisbert C., Ros R., De Haro A., Walker D.J., Bernal M.P., Serrano R., Navarro‐Aviñó J. (2003) A plant genetically modified that accumulates Pb is especially promising for phytoremediation. Biochemical and Biophysical Research Communications, 303, 440–445. [DOI] [PubMed] [Google Scholar]
- Grill E., Winnacker E.L., Zenk M.H. (1985) Phytochelatin: the principal heavy‐metal complexing peptides of higher plants. Science, 230, 674–676. [DOI] [PubMed] [Google Scholar]
- Grill E., Loeffer S., Winnacker E.L., Zenk M.H. (1989) Phytochelatins, the heavy‐metal‐binding peptides of plants, are synthesized from glutathione by a specific γ‐glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proceedings of the National Academy of Sciences of the United States of America, 86, 6838–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S.B., Smith A.P., Howden R., Dietrich W.M., Bugg S., O'Connel M.J., Goldsbrough P.B., Cobbett C.S. (1999) Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe . The Plant Cell, 11, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Nakagawa C.W., Mutoh N. (1991) Two pathways in the biosynthesis of cadystins (γ‐EC)n‐G in the cell‐free system of the fission yeast. Biochemistry and Cell Biology, 69, 115–121. [DOI] [PubMed] [Google Scholar]
- He S.Y., He Z.L., Yang X.E., Stoffella P.J., Baligar V.C. (2015) Soil biogeochemistry, plant physiology, and phytoremediation of cadmium‐contaminated soils. Advances in Agronomy, 134, 135–225. [Google Scholar]
- Herbette S., Taconnat L., Hugouvieux V., Piette L., Magniette M.‐L.M., Cuine S., Auroy P., Richaud P., Forestier C., Bourguignon J., Renou J.‐P., Vavasseur A., Leonhardt N. (2006) Genome‐wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie, 88, 1751–1765. [DOI] [PubMed] [Google Scholar]
- Howden R., Cobbette C.S. (1992) Cadmium sensitive mutants of Arabidopsis thaliana . Plant Physiology, 99, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Goldsbrough P.B., Andersen C.R., Cobbette C.S. (1995) Cadmium‐sensitive, cad1 mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiology, 107, 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.‐Y., Kim D.‐Y., Shim D., Song W.‐Y., Lee J., Schroeder J.I., Kim S., Moran N., Lee Y. (2008) Expression of the novel wheat gene TM20 confers enhanced cadmium tolerance to baker's yeast. Journal of Biological Chemistry, 283, 15893–15902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer U. (2005) Phytoremediation: novel approaches to cleaning up polluted soils. Current Opinion in Biotechnology, 16, 133–141. [DOI] [PubMed] [Google Scholar]
- Lee S., Moon J.S., Ko T.S., Petros D., Goldsbrough P.B., Korban S.S. (2003a) Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiology, 131, 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Petros D., Moon J.S., Ko T.S., Goldsbrough P.B., Korban S.S. (2003b) Higher levels of ectopic expression of Arabidopsis phytochelatin synthase do not lead to increased cadmium tolerance and accumulation. Plant Physiology and Biochemistry, 41, 903–910. [Google Scholar]
- Li Z.‐S., Szczypka M., Lu Y.‐P., Thiele D.J., Rea P.A. (1996) The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S‐conjugate pump. Journal of Biological Chemistry, 271, 6509–6517. [DOI] [PubMed] [Google Scholar]
- Li Z.‐S., Lu Y.‐P., Zhen R.‐G., Szczypka M., Thiele D.J., Rea P.A. (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1‐catalyzed transport of bis(glutathionato)cadmium. Proceedings of the National Academy of Sciences of the United States of America, 94, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Dhankher O.P., Carreira L., Lee D., Chen A., Schroeder J.I., Balish R.S., Meagher R.B. (2004) Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant and Cell Physiology, 45, 1787–1797. [DOI] [PubMed] [Google Scholar]
- Li J.C., Guo J.B., Xu W.Z., Ma M. (2006) Enhanced cadmium accumulation in transgenic tobacco expressing the phytochelatin synthase gene of Cynodon dactylon . Journal of Integrative Plant Biology, 48, 928–937. [Google Scholar]
- Li J.C., Guo J.B., Xu W.Z., Ma M. (2007) RNA interference‐mediated silencing of phytochelatin synthase gene reduce cadmium accumulation in rice seeds. Journal of Integrative Plant Biology, 49, 1032–1037. [Google Scholar]
- Liao V.H.‐C., Dong J., Freedman J.H. (2002) Molecular characterization of a novel, cadmium‐inducible gene from the nematode Caenorhabditis elegans . Journal of Biological Chemistry, 277, 42049–42059. [DOI] [PubMed] [Google Scholar]
- Maitani T., Kubota H., Sato K., Yamada T. (1996) The composition of metals bound to class III metallothionein (phytochelatin and its desglycyl peptide) induced by various metals in root cultures of Rubia tinctorum . Plant Physiology, 110, 1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Bernal P., Almela C., Vélez D., Agustin G.P., Serrano R., Avinó N.J. (2006) An engineered plant that accumulates higher levels of heavy metals than Thlaspi caerulescens, with yields of 100 times more biomass in mine soils. Chemosphere, 64, 478–485. [DOI] [PubMed] [Google Scholar]
- Mengel K., Kirkby E.A. (2001) Principles of plant nutrition (5thedition). Kluwer Academic, Dordrecht, the Netherlands. [Google Scholar]
- Merle S.S., Cuiné S., Carrier P., Pradines L.C., Luu D.T., Peltier G. (2003) Enhanced toxic metal accumulation in engineered bacterial cells expressing Arabidopsis thaliana phytochelatin synthase. Applied and Environmental Microbiology, 69, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiologia Plantarum, 15, 473–497. [Google Scholar]
- Nriagu J.O., Pacyna J.M. (1988) Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature, 333, 134–139. [DOI] [PubMed] [Google Scholar]
- Ortiz D.F., Kreppel L., Speiser D.M., Dcheel G., McDonald G., Ow D.W. (1992) Heavy metal tolerance in the fission yeast requires an ATP‐binding cassette‐type vacuolar membrane transporter. EMBO Journal, 11, 3491–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oven M., Page J.E., Zenk M.H., Kutchan T.M. (2002) Molecular characterization of the homo‐phytochelatin synthase of soybean Glycine max: relation to phytochelatin synthase. Journal of Biological Chemistry, 277, 4747–4754. [DOI] [PubMed] [Google Scholar]
- Park H.C., Kang Y.H., Chun H.J., Koo J.C., Cheong Y.H., Kim C.Y., Kim M.C., Chung W.S., Kim J.C., Yoo J.H., Koo Y.D., Koo S.C., Lim C.O., Lee S.Y., Cho M.J. (2002) Characterization of a stamen‐specific cDNA encoding a novel plant defensin in Chinese cabbage. Plant Molecular Biology, 50, 59–69. [DOI] [PubMed] [Google Scholar]
- Pomponi M., Censi V., Girolamo D.V., Paolis A.D., Toppi L.D., Aromolo R., Costantino P., Cardarelli M. (2006) Overexpression of Arabidopsis phytochelatin synthase in tobacco plants enhances Cd2+ tolerance and accumulation but not translocation to the shoot. Planta, 223, 180–190. [DOI] [PubMed] [Google Scholar]
- Raskin I., Smith R.D., Salt D.E. (1997) Phytoremediation of metals: using plants to remove pollutants from the environment. Current Opinion in Biotechnology, 8, 221–226. [DOI] [PubMed] [Google Scholar]
- Rauser W.E. (1990) Phytochelatins. Annual Review of Biochemistry, 59, 61–86. [DOI] [PubMed] [Google Scholar]
- Rauser W.E. (1999) Structure and function of metal chelators produced by plants: the case for organic acids, amino acids, phytin and metallothioneins. Cell Biochemistry and Biophysics, 32, 19–48. [DOI] [PubMed] [Google Scholar]
- Rea P.A., Li Z.S., Lu Y.P., Drozdowicz Y.M., Martinoia E. (1998) From vacuolar GS‐X pumps to multispecific ABC transporters. Annual Review of Plant Physiology and Plant Molecular Biology, 49, 727–760. [DOI] [PubMed] [Google Scholar]
- Salt D.E. (1995) MgATP‐dependent transport of phytochelatins across the tonoplast of oat roots. Plant Physiology, 107, 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt D.E., Smith R.D., Raskin I. (1998) Phytoremediation. Annual Review of Plant Physiology and Plant Molecular Biology, 49, 643–668. [DOI] [PubMed] [Google Scholar]
- Schmöger M.E., Oven M., Grill E. (2000) Detoxification of arsenic by phytochelatins in plants. Plant Physiology, 122, 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G.‐M., Zhu C., Du Q.‐Z. (2010) Genome‐wide identification of PHYTOCHELATIN and PHYTOCH_SYNTH domain‐containing phytochelatin family from rice. Electronic Journal of Biology, 6, 73–79. [Google Scholar]
- Song W.E., Chen S.B., Liu J.F., Chen L., Song N.N., Li N., Liu B. (2015) Variation of Cd concentration in various rice cultivars and derivation of cadmium toxicity thresholds for paddy soil by species‐sensitivity distribution. Journal of Integrative Agriculture, 14, 1845–1854. [Google Scholar]
- Steffens J.C. (1990) The heavy metal‐binding peptides of plants. Annual Review of Plant Physiology and Plant Molecular Biology, 41, 553–575. [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toppi L.S.D., Gabbrielli R. (1999) Response to cadmium in higher plants. Environmental and Experimental Botany, 41, 105–130. [Google Scholar]
- Van Assche F., Clijsters H. (1990) Effects of metals on enzyme activity in plants. Plant, Cell and Environment, 13, 195–206. [Google Scholar]
- Vatamaniuk O.K., Mari S., Lu Y.P., Rea P.A. (1999) AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proceedings of the National Academy of Sciences of the United States of America, 96, 7110–7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk O.K., Mari S., Lu Y.P., Rea P.A. (2000) Mechanism of heavy metal ion activation of phytochelatin (PC) synthase: blocked thiols are sufficient for PC synthase‐catalyzed transpeptidation of glutathione and related thiol peptides. Journal of Biological Chemistry, 275, 31451–31459. [DOI] [PubMed] [Google Scholar]
- Verbruggen N., Hermans C., Schat H. (2009) Mechanisms to cope with arsenic or cadmium excess in plants. Current Opinion in Plant Biology, 12, 364–372. [DOI] [PubMed] [Google Scholar]
- Wang F., Wang Z., Zhu C. (2012) Heteroexpression of the wheat phytochelatin synthase gene (TaPCS1) in rice enhances cadmium sensitivity. Acta Biochimica et Biophysica Sinica, 44, 886–893. [DOI] [PubMed] [Google Scholar]
- Wojas S., Clemens S., Hennig J., Sklodowska A., Kopera E., Schat H., Bal W., Antosiewicz D.M. (2008) Overexpression of phytochelatin synthase in tobacco: distinctive effects of AtPCS1 and CePCS genes on plant response to cadmium. Journal of Experimental Botany, 59, 2205–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wang X., Chen J. (2007) Zinc finger protein 1 (ThZF1) from salt cress (Thellungiella halophila) is a Cys‐2/His‐2‐type transcription factor involved in drought and salt stress. Plant Cell Reports, 26, 497–506. [DOI] [PubMed] [Google Scholar]
- Zenk M.H. (1996) Heavy metal detoxification in higher plants – a review. Gene, 179, 21–30. [DOI] [PubMed] [Google Scholar]
- Zhu Y.L., Pilon‐Smits E.A.H., Jouanin L., Terry N. (1999a) Overexpression of glutathione synthetase in Indian mustard enhances cadmium accumulation and tolerance. Plant Physiology, 119, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.L., Pilon‐Smits E.A.H., Tarun A.S., Weber S.U., Jouanin L., Terry N. (1999b) Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing γ‐glutamylcysteine synthetase. Plant Physiology, 121, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]


