Abstract
Understanding how cellular activities impact genome stability is critical to multiple biological processes including tumorigenesis and reproductive biology. The fungal pathogen Candida albicans displays striking genome dynamics during its parasexual cycle as tetraploid cells, but not diploid cells, exhibit genome instability and reduce their ploidy when grown on a glucose‐rich “pre‐sporulation” medium. Here, we reveal that C. albicans tetraploid cells are metabolically hyperactive on this medium with higher rates of fermentation and oxidative respiration relative to diploid cells. This heightened metabolism results in elevated levels of reactive oxygen species (ROS), activation of the ROS‐responsive transcription factor Cap1, and the formation of DNA double‐strand breaks. Genetic or chemical suppression of ROS levels suppresses each of these phenotypes and also protects against genome instability. These studies reveal how endogenous metabolic processes can generate sufficient ROS to trigger genome instability in polyploid C. albicans cells. We also discuss potential parallels with metabolism‐induced instability in cancer cells and speculate that ROS‐induced DNA damage could have facilitated ploidy cycling prior to a conventional meiosis in eukaryotes.
Keywords: Candida albicans, DNA double‐strand breaks, parasexual cycle, reactive oxygen species, tetraploid
Subject Categories: Cell Cycle; DNA Replication, Repair & Recombination; Microbiology, Virology & Host Pathogen Interaction
Introduction
Polyploidy, a state where cells contain more than two copies of the genome, is ubiquitous among eukaryotic organisms. In Arabidopsis, polyploid cells can arise via gamete fusion or whole‐genome duplication (Bomblies & Madlung, 2014) and provide advantages such as heterosis, gene redundancy, and enhanced nutrient uptake due to polyploid cells being larger than diploid cells (Comai, 2005; Chao et al, 2013). In humans, polyploidization is known to occur in heart muscle, liver, brain, and blood cells and is associated with both normal development and pathological conditions (Gentric & Desdouets, 2014). Polyploid cells form in uterine muscle tissue as a normal part of pregnancy and are thought to aid in embryonic implantation (Mori et al, 2011; Sroga et al, 2012). Cancer progression is also associated with polyploidization; aneuploid tumors often arise through formation of a tetraploid intermediate and promote malignancy by disrupting regulators of cell growth (Storchova & Pellman, 2004; Fujiwara et al, 2005; Zack et al, 2013).
Polyploid forms have also been characterized in diverse fungal lineages, including the budding yeast Saccharomyces cerevisiae which can exist in ploidy states from haploid (1n) to tetraploid (4n) (Zhu et al, 2016). Higher ploidy states can arise via gamete fusion during a conventional sexual cycle or via asexual processes such as mitotic collapse. A characteristic feature of polyploid cells is increased genome instability, with tetraploid cells exhibiting rates of chromosome loss that are orders of magnitude greater than those of diploid cells (Mayer & Aguilera, 1990). Polyploidy and genome instability can facilitate evolution, with higher ploidy states undergoing adaptation faster than diploid or haploid cells due to higher mutation rates with greater fitness effects (Selmecki et al, 2015). Ploidy shifts in S. cerevisiae cells are induced by a variety of environmental stresses, including culture in the presence of high salt or ethanol (Gerstein et al, 2006; Voordeckers et al, 2015).
Ploidy variation has also been described in the opportunistic fungal pathogen Candida albicans, a commensal species that can seed both debilitating mucosal and life‐threatening systemic infections. Candidiasis is a major problem in the hospital setting and is exacerbated by the prevalence of drug‐resistant C. albicans strains (Ksiezopolska & Gabaldon, 2018). Natural C. albicans isolates are diploid although rare haploid forms have been isolated, albeit with reduced fitness (Hickman et al, 2013). As in S. cerevisiae, ploidy shifts can enable adaptation; diploid C. albicans cells exposed to the antifungal drug fluconazole form a transient tetraploid state that generates drug‐resistant aneuploid progeny via aberrant divisions (Harrison et al, 2014).
Tetraploid C. albicans cells are also formed by fusion of diploid cells during mating (Hull et al, 2000; Magee & Magee, 2000; Bennett & Johnson, 2003). Sexual reproduction in C. albicans is unusual in that cells must undergo an epigenetic switch from the sterile “white” state to the mating‐competent “opaque” state (Lockhart et al, 2002; Miller & Johnson, 2002; Bennett, 2015). In addition, rather than undergoing a conventional meiosis, tetraploid cells return to a diploid (or aneuploid) state via aberrant mitotic divisions, a process termed “concerted chromosome loss” (CCL; Bennett & Johnson, 2003; Forche et al, 2008; Hickman et al, 2015). Growth on S. cerevisiae “pre‐sporulation” (PRE‐SPO) medium at 37°C induces CCL in tetraploid cells, whereas diploid cells are stable under these culture conditions. In addition to genome instability, many tetraploid cells undergo cell death during growth on PRE‐SPO medium whereas most diploid cells remain viable. However, the mechanism whereby PRE‐SPO medium induces polyploid‐specific responses in C. albicans has yet to be addressed.
In this work, we propose that a metabolic shift triggers genome instability in polyploid C. albicans cells. Transcriptional and metabolic profiling reveals that C. albicans cells grown on glucose‐rich PRE‐SPO medium are metabolically hyperactive, with increased rates of both fermentation and oxidative respiration. Critically, cellular metabolism is further upregulated in polyploid cells relative to diploid cells. We demonstrate that elevated metabolic flux results in the production of reactive oxygen species (ROS) and DNA damage, and that the latter is a driver of genome instability in polyploid cells. These studies therefore establish links between metabolic flux, ROS, oxidative stress, and DNA damage in C. albicans, and that this mechanism selectively impacts genome integrity in polyploid cells. We discuss parallels with tumorigenesis in mammalian cells and propose that metabolism‐induced genome instability may have promoted primitive ploidy cycles prior to a conventional meiosis.
Results
Polyploid Candida albicans cells undergo CCL on PRE‐SPO medium
A conventional meiosis has not been observed in C. albicans despite its genome containing numerous conserved meiosis genes (Tzung et al, 2001; Butler et al, 2009). Instead, tetraploid mating products reduce their ploidy via a parasexual program of CCL, during which tetraploid cells return to a diploid or aneuploid state (Bennett & Johnson, 2003; Forche et al, 2008). Concerted chromosome loss can be efficiently induced by incubation of tetraploid C. albicans cells on S. cerevisiae PRE‐SPO medium at 37°C (Bennett & Johnson, 2003). Tetraploid cells experience extensive cell death during the first 3 days of growth on PRE‐SPO medium followed by a rebound in viability, whereas diploid cells largely remain viable under these conditions (Fig 1A), as previously reported (Bennett & Johnson, 2003). We find that tetraploid cells grown on PRE‐SPO medium also exhibit erratic microtubule and nuclear dynamics, indicative of aberrant nuclear divisions on this medium (Fig 1B).
Figure 1. Tetraploid C. albicans cells undergo genome instability and cell death on PRE‐SPO medium.
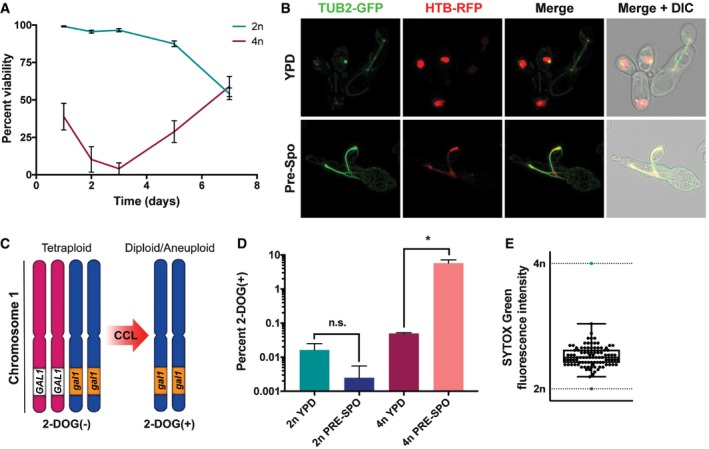
- Viability of diploid (SC5314) and tetraploid (RBY18) cells grown on PRE‐SPO medium at 37°C (n = 5, error bars represent SEM).
- A tetraploid β‐tubulin (TUB2)‐GFP, histone H2B (HTB)‐RFP strain was cultured on YPD or PRE‐SPO medium at 30 or 37°C, respectively, for 24 h. Cell images indicate maximum intensity Z projections for GFP and RFP channels, and merged images of the GFP/RFP and GFP/RFP/DIC channels.
- Assay to monitor for loss of GAL1 in tetraploid strain RBY18. Strain RBY18 is heterozygous for the GAL1 gene, and only cells that have lost GAL1 function can grow on 2‐DOG medium.
- Diploid strain RBY1 and tetraploid strain RBY18 were cultured on YPD or PRE‐SPO medium at 30 or 37°C, respectively, for 7 days and plated onto 2‐DOG medium to monitor for GAL1 loss. (*) denotes a statistically significant difference between the indicated sample groups (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
- 2‐DOGR progeny arising from RBY18 on PRE‐SPO medium were DNA‐stained and their ploidy determined via flow cytometry. Box extends from the 25th to 75th percentiles, center line represents the median value, and upper and lower whiskers represent the maximum and minimum values from the analyzed progeny (n = 94).
To monitor genome stability, we used a tetraploid strain, RBY18, that is heterozygous for GAL1 on chromosome 1 (GAL1/GAL1/gal1/gal1) (Fig 1C). Only strains that have lost GAL1 function are viable on 2‐deoxygalactose (2‐DOG) medium (Gorman et al, 1992), providing a selection for tetraploid cells that have lost the two GAL1 alleles and undergone a reduction in ploidy (Bennett & Johnson, 2003). Genome stability in diploid cells was monitored using strain RBY1, which is also heterozygous for GAL1 on chromosome 1 (GAL1/gal1). When RBY18 was cultured on PRE‐SPO medium at 37°C for 7 days, 5.8% of cells lost GAL1 and became 2‐DOGR, an increase of 116‐fold over cells grown on YPD medium (0.05% 2‐DOGR cells; Fig 1D). In contrast, there was no significant difference in GAL1 loss in diploid RBY1 cells cultured on PRE‐SPO v. YPD medium (Fig 1D). Flow cytometric analysis of DNA content in 2‐DOGR colonies recovered from PRE‐SPO medium indicated that tetraploid (4n) cells had reduced their ploidy and now exhibited a DNA content between diploid (2n) and triploid (3n) (Fig 1E).
Transcriptional profiling of diploid and tetraploid cells on YPD and PRE‐SPO media
RNA sequencing (RNA‐seq) was used to examine gene expression of C. albicans cells grown on standard YPD medium (at 30°C) or on PRE‐SPO medium (at 37°C). YPD cultures were performed at the lower temperature as extensive filamentation occurs during growth on YPD, but not PRE‐SPO, medium at 37°C. Both diploid and tetraploid cells were analyzed on these media at 3, 6, 12, and 24 h time points.
RNA‐seq analysis revealed that diploid and tetraploid cells showed similar gene expression profiles on both YPD and PRE‐SPO media, despite clear differences in cell survival and genome stability on the two media (Fig 2A and Table EV1). Notably, gene ontology (GO) term analysis revealed that processes related to DNA double‐strand break (DSB) formation, DNA repair, DNA recombination, and meiosis were significantly enriched among differentially expressed genes between cells grown on the two media (24 h time point, Fig 2B), with an upregulation of these processes on PRE‐SPO relative to YPD medium (Fig 2C). However, no obvious ploidy‐specific expression patterns of meiosis‐specific genes were observed (Fig EV1). We also conducted GO term analysis on differentially expressed genes between diploid and tetraploid cells grown on PRE‐SPO medium. At both 3 h and 6 h, nicotinamide binding and acetate metabolism genes were enriched among differentially expressed genes (Appendix Tables S1 and S2), while at 12 h and 24 h DNA replication, regulation of helicase activity and DNA binding genes were identified (Appendix Tables S3 and S4). These results implicated metabolism and DNA damage as potentially relevant to the distinct behavior of tetraploid cells grown on PRE‐SPO v. YPD media.
Figure 2. Transcriptional profiling of Candida albicans diploid and tetraploid cells on YPD and PRE‐SPO medium.
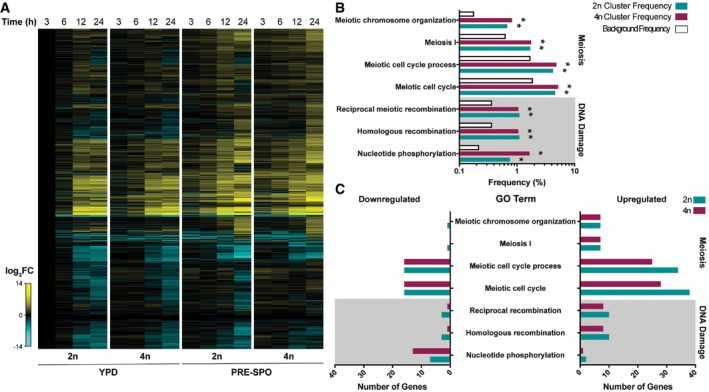
- Heatmap of RNA‐seq data showing global gene expression changes in diploid (SC5314, 2n) and tetraploid (RBY18, 4n) cells on YPD and PRE‐SPO medium over 24 h.
- Results of gene ontology (GO) term enrichment analysis of differentially expressed genes between YPD and PRE‐SPO media at 24 h in diploid (2n) and tetraploid (4n) cells. Background frequency represents the total number of genes annotated to each GO term in the C. albicans genome divided by the total number of genes in the genome. (*) denotes a statistically significant enrichment of genes annotated to the indicated GO term (P < 0.05, Fisher's exact test).
- Number of genes in each category that are upregulated or downregulated on PRE‐SPO medium v. YPD medium in diploid (2n) and tetraploid (4n) cells for the indicated GO terms.
Figure EV1. Transcriptional profiling of meiosis genes on YPD and PRE‐SPO media.
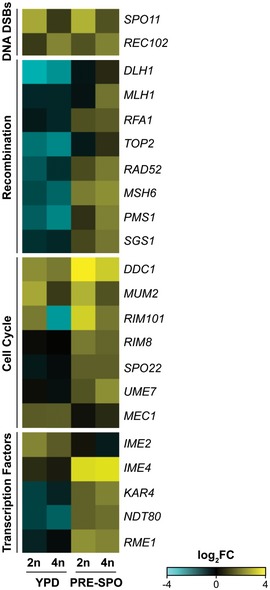
Heatmap of RNA‐seq data for C. albicans homologs to “meiotic” genes in diploid and tetraploid cells on YPD and PRE‐SPO media at 24 h, including programmed formation of DNA double‐strand breaks (DSBs), recombination, coordination of the meiotic cell cycle, and transcription factors.
Inspection of metabolic gene expression also revealed that glycolytic genes were induced earlier on PRE‐SPO than on YPD medium in both diploid and tetraploid cells (compare 6 h time points, Fig 3A). In addition, oxidative stress‐associated genes, including those encoding the Sod3 superoxide dismutase and the Ddr48 DNA damage response factor, were activated at earlier time points on PRE‐SPO medium. These results indicate that growth on PRE‐SPO medium potentially results in enhanced metabolic activity and increased activation of oxidative stress responses than that on YPD medium.
Figure 3. Analysis of metabolic and oxidative stress responses in diploid and tetraploid cells grown on YPD and PRE‐SPO medium.
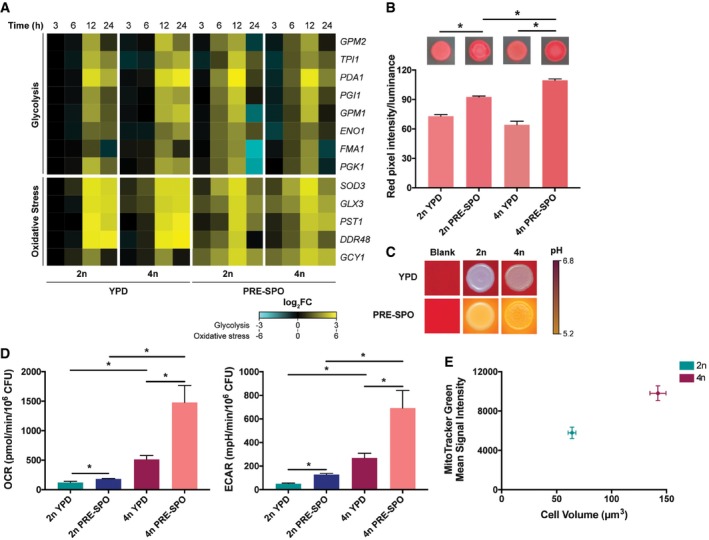
- Heatmap of RNA‐seq data for glycolysis and oxidative stress genes in diploid (2n) and tetraploid (4n) cells grown on YPD or PRE‐SPO medium over 24 h.
- TTC assay results in diploid and tetraploid cells grown on YPD or PRE‐SPO medium at 30 or 37°C, respectively. Cells were grown for 24 h at which point a TTC overlay solution was applied and color allowed to develop for 15 min. Calculated values represent the total amount of reddening. (*) denotes a significant difference between the indicated groups (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
- Diploid and tetraploid strains were spot‐inoculated onto YPD or PRE‐SPO medium supplemented with the pH indicator bromocresol purple and cultured for 24 h at 30 or 37°C, respectively.
- Analysis of metabolic flux in diploid and tetraploid cells following growth for 24 h on YPD or PRE‐SPO medium (30 or 37°C, respectively), using a Seahorse instrument. OCR values represent rates of aerobic respiration, while ECAR values represent rates of fermentation. (*) denotes a significant difference between the indicated experimental groups (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
- Cell size and mean MitoTracker Green signal intensity in diploid and tetraploid cells grown on YPD medium for 24 h at 30°C (n = 100 and n = 5 for cell volume and MitoTracker Green measurements, respectively, error bars represent SEM).
Candida albicans tetraploid cells are metabolically hyperactive on PRE‐SPO medium
Given that C. albicans cells show earlier activation of glycolytic and oxidative stress genes on PRE‐SPO versus YPD medium, we compared the metabolic activity of diploid and tetraploid cells on these media using several complementary approaches. First, a 2,3,5‐triphenyltetrazolium chloride (TTC) overlay technique was performed as this method can assess activity of the electron transport chain in microbial populations (Ogur et al, 1957; Rich et al, 2001; Morales et al, 2013). TTC is white and can passively diffuse through the plasma membrane to interact with mitochondria, where it is converted to red 1,3,5‐triphenylformazan (TPF). We grew equivalent numbers of diploid and tetraploid C. albicans cells on YPD and PRE‐SPO media at 30 and 37°C, respectively, and after 24 h, TTC was applied to each plate. We found that diploid and tetraploid colonies generated greater reddening on PRE‐SPO medium than on YPD medium, with tetraploid cells also producing more color than diploid cells suggestive of heightened respiratory activity (Fig 3B).
Next, we examined media acidification which is indicative of the formation of acidic fermentation end products such as acetate (Morales et al, 2013). YPD and PRE‐SPO media were supplemented with the pH indicator bromocresol purple, which is yellow at pH values below 5.2 and purple at pH values above 6.8. Equal numbers of diploid and tetraploid cells were inoculated onto YPD and PRE‐SPO media and grown for 24 h at 30 and 37°C, respectively. Diploid and tetraploid cells both acidified PRE‐SPO medium whereas the pH was largely unchanged with cells grown on YPD medium (Fig 3C), suggesting increased production of acidic fermentation end products during growth on PRE‐SPO medium.
To further characterize the metabolic activities of diploid and tetraploid cells, aerobic respiratory and fermentative rates were assayed by measuring the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), respectively. C. albicans cells were grown on YPD and PRE‐SPO media for 24 h at 30 and 37°C, respectively, and transferred to a Seahorse instrument to measure metabolic rates. Interestingly, we found that metabolic activities were considerably higher in polyploid cells than in diploid cells; both OCR and ECAR were ~4‐fold higher (YPD medium) or ~8‐fold higher (PRE‐SPO medium) in tetraploid cells compared to diploid cells (Fig 3D). We also found that cells grown on PRE‐SPO medium exhibited significantly higher OCR and ECAR values relative to the same cells grown on YPD medium. Thus, tetraploid cells showed a 2.9‐fold increase in OCR and a 2.1‐fold increase in ECAR when grown on PRE‐SPO relative to YPD, while diploid cells exhibited a 1.5‐fold increase in OCR and 1.9‐fold increase in ECAR between the two media (Fig 3D). The OCRs that we observed were due to mitochondrial respiration and not other oxygen‐consuming processes as these were blocked by addition of the electron transport chain inhibitors antimycin A and oligomycin together with the alternative oxidase inhibitor salicylhydroxamic acid (SHAM) (Fig EV2A). ECARs were not inhibited by this treatment (Fig EV2B). Together, these results establish that both cell ploidy and culture media impact C. albicans metabolism; growth on PRE‐SPO medium enhances metabolic activity relative to that on YPD medium, and tetraploid cells are also considerably more active than diploid cells.
Figure EV2. Oxygen consumption on PRE‐SPO medium is inhibited by restricting mitochondrial respiration.
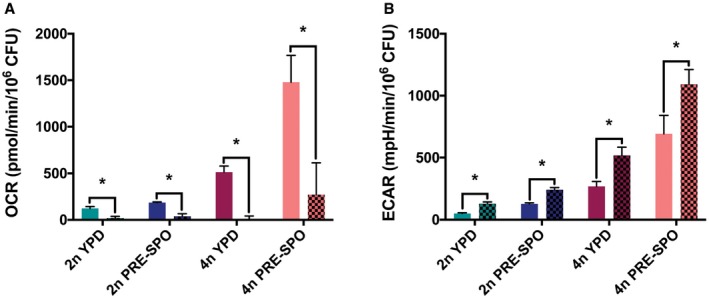
- Oxygen consumption rates (OCR) in diploid and tetraploid cells following growth for 24 h on YPD and PRE‐SPO media at 30 and 37°C, respectively. OCR was compared before (bars with no pattern) and after (bars with checkered pattern) treatment with the electron transport chain (ETC) inhibitors antimycin A (5 μM), rotenone (5 μM), and the alternative oxidase inhibitor SHAM (500 μM). (*) denotes a significant difference between indicated experimental groups (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
- Extracellular acidification rates (ECAR) in diploid and tetraploid cells grown for 24 h on YPD and PRE‐SPO media at 30 and 37°C, respectively, before (bars with no pattern) and after (bars with checkered pattern) treatment with the same ETC inhibitors as in (A). (*) denotes a significant difference between the indicated groups (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
Given that tetraploid cells exhibited a higher OCR than diploid cells, we sought to probe the relationship between ploidy and mitochondrial content. Diploid and tetraploid cells were grown on YPD medium for 24 h at 30°C and stained with the mitochondrial marker MitoTracker Green. Cells were analyzed by flow cytometry to determine mitochondrial content and by microscopy to examine cell size. Tetraploid cells had a cell volume ~2.2‐fold greater than diploid cells, and this correlated with an increased MitoTracker Green signal indicating an increased mitochondrial content (Fig 3E). These results indicate that C. albicans tetraploid cells are more metabolically active than diploid cells due, at least in part, to their larger size and increased mitochondrial content.
Candida albicans cells experience elevated oxidative stress on PRE‐SPO medium
Transcriptional profiling showed that oxidative stress genes were upregulated more rapidly in C. albicans cells grown on PRE‐SPO than on YPD medium (Fig 3A), and we therefore directly evaluated oxidative stress responses on the two media. To determine whether reactive oxygen species (ROS) were present, diploid and tetraploid cells were grown for 24 h on YPD and PRE‐SPO media at 30 and 37°C, respectively, and stained with CellROX Green. This is a membrane permeant dye that fluoresces upon oxidation by ROS and DNA binding. No observable fluorescent signal was present in cells grown on YPD medium when examined via microscopy, whereas both diploid and tetraploid cells grown on PRE‐SPO medium stained positively indicating that ROS are generated on this medium (Fig 4A). Flow cytometric analysis of CellROX green‐stained cells corroborated cell microscopic images, with an increased fluorescence signal in diploid/tetraploid cells on PRE‐SPO relative to YPD medium (Fig 4B). Notably, tetraploid cells grown on PRE‐SPO medium exhibited the strongest signal, with 2.0‐fold higher fluorescence levels than diploid cells on this medium (Fig 4B).
Figure 4. Candida albicans cells accumulate ROS and activate oxidative stress responses when cultured on PRE‐SPO medium.
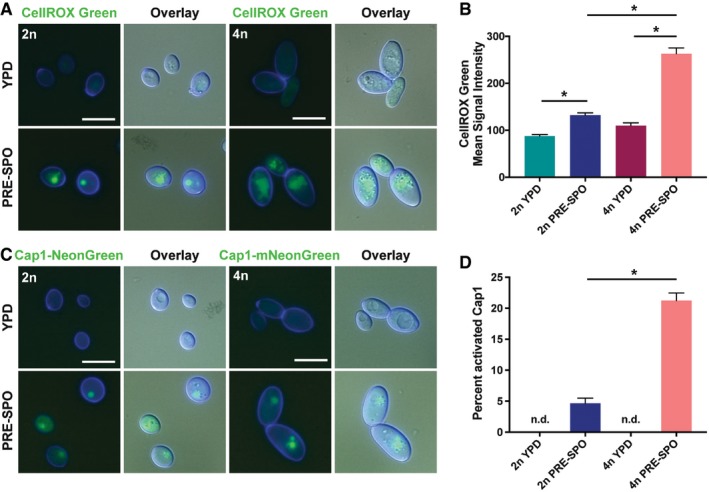
- Diploid and tetraploid cells were grown on YPD or PRE‐SPO medium at 30 or 37°C, respectively, for 24 h and stained with CellROX Green. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
- Diploid and tetraploid cells were grown on YPD or PRE‐SPO medium at 30 or 37°C, respectively, for 24 h, stained with CellROX Green, and signal analyzed via flow cytometry. (*) denotes a significant difference between the indicated groups (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
- Diploid and tetraploid cells containing a fluorescently labeled version of the transcription factor Cap1 (Cap1‐mNeonGreen) were grown on YPD or PRE‐SPO medium at 30 or 37°C, respectively, for 24 h. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
- Quantitation of the percentage of diploid and tetraploid cells with activated Cap1 on YPD or PRE‐SPO medium. “n.d.” indicates that no cells with activated Cap1 were detected. (*) denotes a significant difference between the indicated groups (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
Previous work showed that C. albicans also produces ROS during hyphal morphogenesis (Rossi et al, 2017). Consistent with this finding, cells grown on YPD medium at 37°C, a growth condition which induces filamentation, stained positively with CellROX Green (Fig EV3A). However, a fluorescence signal was absent in cells grown on SCD medium at 37°C (Fig EV4A), a growth condition where filamentation does not occur, indicating that growth at 37°C is not sufficient for ROS production by C. albicans cells.
Figure EV3. Tetraploid cell response to growth on YPD medium at 37°C.
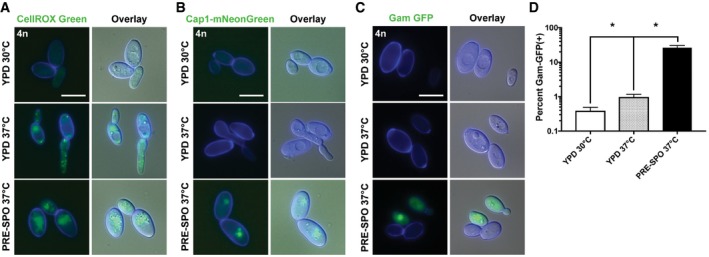
- Tetraploid cells were grown on YPD medium at 30 or 37°C, or PRE‐SPO medium at 37°C, for 24 h and stained with CellROX Green. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
- Tetraploid cells containing a fluorescently labeled version of the Cap1 transcription factor (Cap1‐mNeonGreen) were grown on YPD medium at 30 or 37°C, or PRE‐SPO medium at 37°C, for 24 h. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
- A diploid strain expressing a Tet‐ON Gam‐GFP reporter was cultured on YPD medium at 30 or 37°C, or PRE‐SPO medium at 37°C, in the presence (Gam‐GFP ON) of doxycycline for 24 h. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
- Flow cytometric analysis of the tetraploid Gam‐GFP reporter strain grown on YPD medium at 30 or 37°C, or PRE‐SPO medium at 37°C in the presence of doxycycline for 24 h. (*) denotes a significant difference between the indicated groups (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
Figure EV4. Quantification of Gam‐GFP signal as a readout of DNA damage.
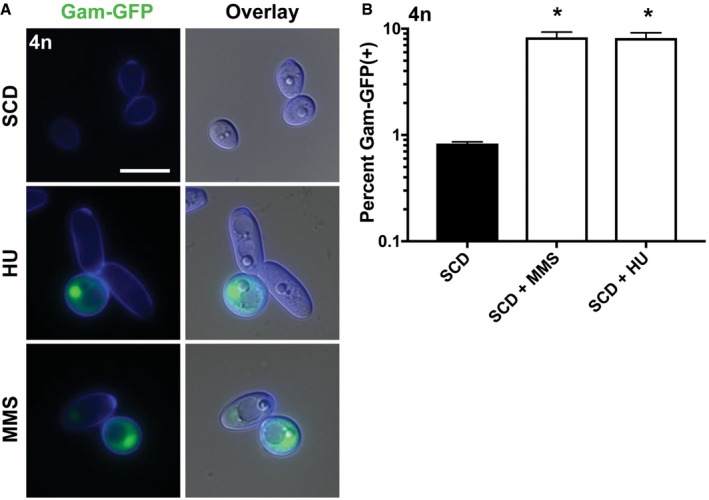
-
ATetraploid cells expressing a Gam‐GFP reporter were cultured on SCD medium (37°C for 24 h) with or without the DNA‐damaging agents HU (20 mM) or MMS (0.01%) and analyzed via fluorescence microscopy. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
-
BFlow cytometric analysis of the tetraploid Gam‐GFP reporter strain grown on SCD medium (37°C for 24 h) with or without HU (20 mM) or MMS (0.01%). (*) denotes a significant difference between untreated SCD medium and that supplemented with the indicated agent (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
We next examined oxidative stress responses using a fluorescently tagged Cap1 protein, a transcription factor that regulates multiple oxidative stress response pathways in C. albicans including antioxidant defense, protein degradation, and drug resistance (Wang et al, 2006). Cap1 is distributed homogenously between the nucleus and the cytoplasm under non‐stress conditions, but accumulates in the nucleus in response to oxidative stress to activate antioxidant gene expression (Patterson et al, 2013). To assay for Cap1 activation, diploid and tetraploid strains were generated expressing fluorescently labeled Cap1‐mNeonGreen. These reporter strains were grown for 24 h on YPD and PRE‐SPO media at 30 and 37°C, respectively, and imaged to examine Cap1 localization. Diploid and tetraploid cells grown on PRE‐SPO medium exhibited a clear nuclear Cap1 signal, whereas cells grown on YPD exhibited only a diffuse signal (Fig 4C). Growth at 37°C was not solely responsible for the observed phenotypes on PRE‐SPO medium, as tetraploid cells grown on YPD medium at 37°C also did not exhibit a nuclear Cap1 signal (Fig EV3B). Quantitation of viable cells revealed that more tetraploid cells than diploid cells activated Cap1 during PRE‐SPO culture (Fig 4D). These results establish that tetraploid cells experience higher levels of oxidative stress than diploid cells when cultured on this medium.
Suppression of ROS protects cells from oxidative stress and instability
To determine whether oxidative stress contributes to decreased tetraploid cell viability, PRE‐SPO medium was supplemented with varying levels of the antioxidants ascorbic acid, dithiothreitol (DTT), or glutathione. Each of these antioxidants significantly increased viability on PRE‐SPO medium in a dose‐dependent matter, with higher concentrations providing greater protection (Fig 5A). Antioxidants were also tested for their ability to mitigate genome instability in tetraploid cells grown on PRE‐SPO medium. All of the tested antioxidants significantly reduced genome instability, with higher concentrations again providing a greater protective effect (Fig 5B). Finally, antioxidants were examined at the concentrations that provided the greatest protective effect against cell death and genome instability to determine whether oxidative stress responses were reduced. At these optimized concentrations (glutathione, 1 mM; DTT, 1 mM; ascorbic acid, 20 mM), each antioxidant significantly reduced Cap1 activation in cells grown on PRE‐SPO medium (Fig 5C).
Figure 5. Antioxidants protect against cell death and genome instability in tetraploid cells grown on PRE‐SPO medium.
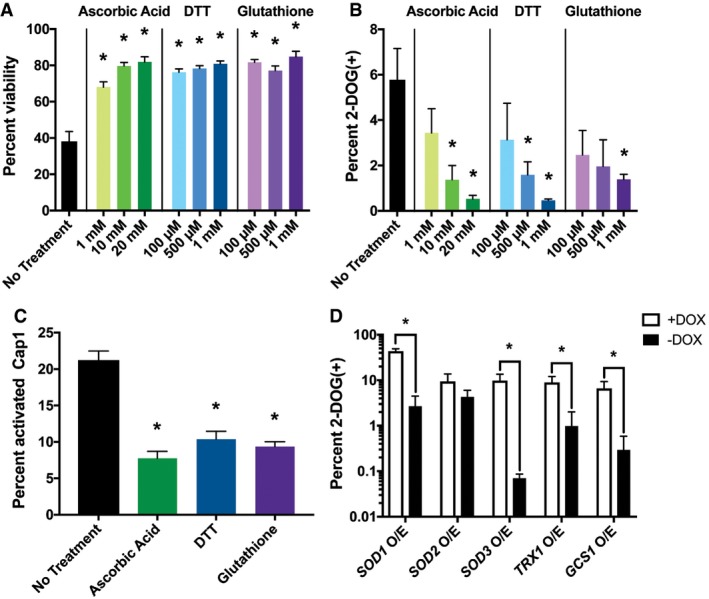
- Analysis of cell viability in tetraploid cells after culture on PRE‐SPO medium (37°C for 24 h) when supplemented with ascorbic acid, DTT, or glutathione at the indicated concentrations. (*) denotes a significant difference in viability between cells with and without antioxidant (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
- Analysis of GAL1 loss (2‐DOG resistant colonies) in tetraploid strain RBY18 after growth on PRE‐SPO medium with or without antioxidants for 7 days at 37°C. (*) denotes a significant difference between the percentage of 2‐DOG(+) colonies with and without antioxidant (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
- Tetraploid cells expressing Cap1‐mNeonGreen were grown on PRE‐SPO medium supplemented with 20 mM ascorbic acid, 1 mM DTT, or 1 mM glutathione for 24 h at 37°C. Cell images were analyzed to determine the percentage of viable cells with activated Cap1. (*) denotes a significant difference in Cap1 activation between cells with and without antioxidant (P < 0.05, Student's t‐test, n = 5, error bars represent SEM).
- Assay to monitor for loss of GAL1 from tetraploid RBY18 overexpressing the indicated genes. Cells were grown for 7 days at 37°C on PRE‐SPO medium in the presence (gene OFF) or absence (gene ON) of doxycycline (DOX). (*) denotes a significant difference between + and − DOX conditions (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
We also assessed the ability of endogenous enzymatic activities to protect against genome instability on PRE‐SPO medium. Tetraploid strains were constructed containing a doxycycline repressible “Tet‐OFF” cassette to regulate overexpression (O/E) of antioxidant genes including SOD1 and SOD3, encoding two cytosolic superoxide dismutases (Lamarre et al, 2001; Hwang et al, 2002), SOD2, a predicted mitochondrial superoxide dismutase (Rhie et al, 1999), TRX1, encoding the major thioredoxin (da Silva Dantas et al, 2010), and GCS1, encoding a key enzyme in glutathione biosynthesis (Baek et al, 2004). While each of these genes is linked to antioxidant responses, we note that SOD1 and SOD3 have also been recently shown to minimize glucose uptake in C. albicans (Broxton et al, 2018). Ectopic expression of each of the tested enzymes except SOD2 rescued genome instability of tetraploid cells (Fig 5D). Together, these results indicate that oxidative stress contributes to both cell death and chromosome instability of tetraploid C. albicans cells grown on PRE‐SPO medium.
DNA double‐strand breaks are generated by growth on PRE‐SPO medium
A key mechanism by which ROS induces cellular toxicity is through structural changes to DNA including base modifications, DNA cleavage events, and DNA–protein cross‐links (Jena, 2012). Given that detectable levels of ROS accrue in cells during growth on PRE‐SPO medium and that these cells exhibit activation of oxidative stress responses, we examined whether DNA double‐strand breaks (DSBs) are formed.
To track formation of DSBs in C. albicans, we generated a reporter using a viral Gam protein that binds irreversibly to sites of DSBs. Fluorescent labeling of Gam has been shown to enable the visualization of DSBs in live mammalian and bacterial cells (Shee et al, 2013; Belenky et al, 2015). As a control, a tetraploid C. albicans strain containing a doxycycline‐inducible (Tet‐ON) Gam‐GFP construct was treated with methyl methanesulfonate (MMS) or hydroxyurea (HU), and both DNA‐damaging agents resulted in a strong nuclear signal that was also detected by flow cytometry (Fig EV4A and B).
To assay for DSB formation on different media, diploid and tetraploid Gam‐GFP reporter strains were grown for 24 h on YPD and PRE‐SPO media at 30 and 37°C, respectively, and examined by fluorescence microscopy and flow cytometry. An intense nuclear signal was detected in both diploid and tetraploid cells cultured on PRE‐SPO medium, whereas no signal was observed on YPD medium (Fig 6A). This finding was corroborated by flow cytometry where 57% of tetraploid cells became Gam‐GFP positive after 24 h on PRE‐SPO medium versus 0.2% on YPD medium (see +DOX cells, Fig 6B). Growth at 37°C was not solely responsible for DSBs on PRE‐SPO medium as tetraploid cells cultured on YPD medium at 37°C did not exhibit a nuclear Gam‐GFP signal and the percentage of Gam‐GFP‐positive cells increased minimally, albeit significantly, relative to growth on YPD medium at 30°C (Fig EV3C and D).
Figure 6. Oxidative stress contributes to DNA double‐strand break formation during Candida albicans growth on PRE‐SPO medium.
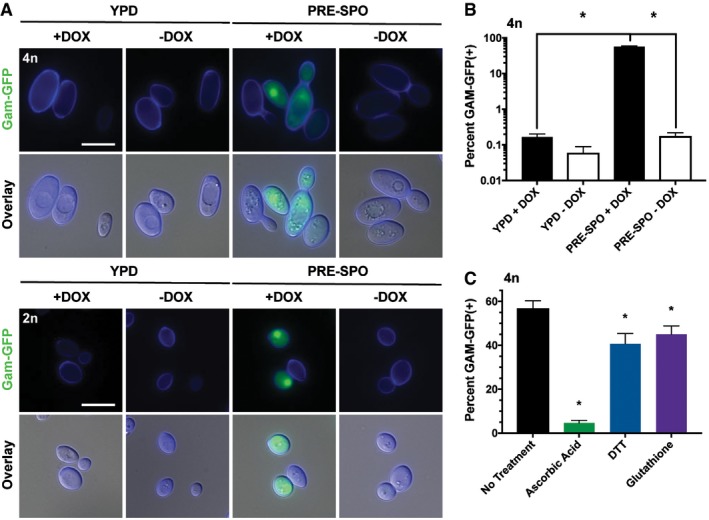
- Diploid (2n) and tetraploid (4n) strains expressing a Gam‐GFP reporter were cultured on YPD or PRE‐SPO medium at 30 or 37°C, respectively, in the presence (Gam‐GFP ON) or absence (Gam‐GFP OFF) of doxycycline (DOX) for 24 h. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
- Flow cytometric analysis of the tetraploid Gam‐GFP reporter strain grown on YPD or PRE‐SPO medium at 30 or 37°C, respectively, in the presence or absence of doxycycline for 24 h. (*) denotes a significant difference in the percentage of Gam‐GFP(+) cells between experimental groups (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
- Flow cytometric analysis of the tetraploid Gam‐GFP reporter strain grown on PRE‐SPO medium with or without 20 mM ascorbic acid, 1 mM DTT, or 1 mM glutathione at 37°C for 24 h. (*) denotes a significant difference between Gam‐GFP cells with and without antioxidant (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
We next examined if protection from oxidative stress would reduce DSB formation in tetraploid cells grown on PRE‐SPO medium. The tetraploid Gam‐GFP reporter strain was cultured with and without antioxidants on PRE‐SPO medium for 24 h and cells examined using fluorescence microscopy and flow cytometry. Interestingly, all antioxidants significantly reduced the formation of DSBs on PRE‐SPO medium, with ascorbic acid blocking most DSB formation (Fig 6C). These results demonstrate that C. albicans tetraploid cells experience extensive DSB formation when cultured on PRE‐SPO medium and that chemical agents that diminish ROS levels can reduce these high levels of DNA damage.
Tetraploid cells exposed to DNA‐damaging agents undergo ploidy reduction
DNA damage (including DSBs) can impact genome integrity (Jackson & Bartek, 2009; Mehta & Haber, 2014), and we therefore tested the effect of hydroxyurea (HU) and methyl methanesulfonate (MMS) on C. albicans cell viability and genome stability. As shown above, both DNA‐damaging agents promote DSB formation in tetraploid C. albicans cells using the Gam‐GFP reporter (Fig EV3A and B). When assayed for sensitivity to MMS and HU at 30 and 37°C, tetraploid cells were more sensitive to both compounds compared to diploid cells, and this was particularly evident at 37°C (Fig 7A). To examine whether DNA damage promotes chromosome instability, the tetraploid RBY18 strain was exposed to HU, MMS, or ultraviolet (UV) radiation and cells assayed for those that had lost GAL1 and become 2‐DOGR. Exposure of cells to each of these genotypic stresses caused significantly elevated frequencies of 2‐DOGR colonies (Fig 7B), indicating that DNA damage led to increased chromosome instability in tetraploid cells.
Figure 7. DNA damage induces chromosome instability in tetraploid Candida albicans cells.
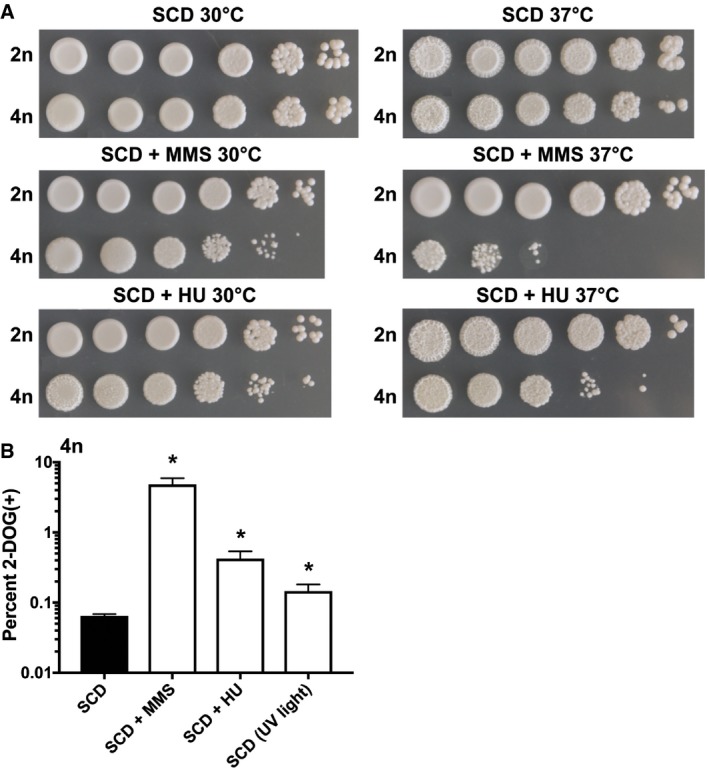
- 10‐fold dilutions of diploid and tetraploid cells were spot‐inoculated onto SCD medium with or without the DNA‐damaging agent hydroxyurea (HU, 20 mM) or methyl methanesulfonate (MMS, 0.01%). Cells were grown for 48 h at 30 or 37°C and the plates imaged.
- Assay to monitor for loss of GAL1 (2‐DOG resistant cells) arising from tetraploid strain RBY18 after culture at 37°C on SCD medium alone, SCD supplemented with HU (20 mM) or MMS (0.01%), or SCD medium with exposure to UV light for 5 s a day for 7 days. (*) denotes a significant difference between control SCD medium and the indicated treatment (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
Evidence that oxidative stress induces genome instability in tetraploid cells
The experiments described above indicate that endogenous ROS are associated with DNA damage and that this can induce genome instability in tetraploid C. albicans cells. We next sought to characterize if exogenous oxidative stress could recapitulate the phenotypes seen on PRE‐SPO medium and cause chromosome instability. Tetraploid C. albicans cells were exposed to hydrogen peroxide (H2O2), as well as the oxidative stress‐inducing agents paraquat (PQT) and piperlongumine (PL). PQT generates superoxide anions within the mitochondrial respiratory chain (Cocheme & Murphy, 2008), whereas PL is a potential anticancer compound that increases ROS and selectively kills cancer cells (Karki et al, 2017). When exposed to H2O2, PQT, or PL for 24 h at 37°C, tetraploid cells stained positively for ROS and Cap1 protein translocated into the nucleus, consistent with cells experiencing increased oxidative stress (Fig 8A and B). A nuclear Gam‐GFP signal was also observed in tetraploid cells exposed to each of these agents, indicative of DSB formation in the presence of these stressors (Fig 8C and D). Moreover, analysis of GAL1 marker stability revealed that H2O2, PQT, and PL induced significantly elevated levels of genome instability in tetraploid cells (Fig 8E). Together, these results establish that exposure to oxidative stress can induce DSB formation and chromosome instability in tetraploid cells in a manner that parallels that observed during growth on PRE‐SPO medium.
Figure 8. Tetraploid cells exposed to exogenous oxidative stress experience increased DSB formation and chromosome loss.
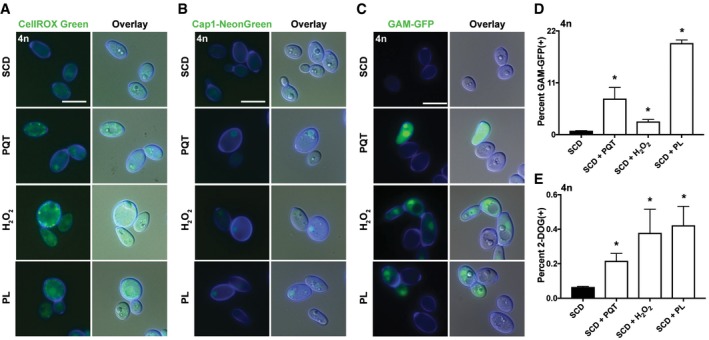
- Tetraploid cells were grown on SCD medium with or without the oxidative stress‐inducing agents paraquat (PQT; 600 μg/ml), hydrogen peroxide (H2O2; 2 mM), or piperlongumine (PL; 100 μg/ml) for 24 h at 37°C and stained with CellROX Green. Cell images indicate calcofluor white staining (cell wall; blue), GFP, and a merged image of GFP/DAPI/DIC channels. Scale bar = 10 μm.
- Tetraploid cells expressing Cap1‐mNeonGreen were grown on SCD medium with or without PQT (600 μg/ml), H2O2 (2 mM), or PL (100 μg/ml) for 24 h at 37°C. Cell images as in (A). Scale bar = 10 μm.
- Tetraploid cells expressing a Gam‐GFP reporter were cultured on SCD medium with or without PQT (600 μg/ml), H2O2 (2 mM), or PL (100 μg/ml) for 24 h at 37°C and analyzed via fluorescence microscopy. Cell images as in (A). Scale bar = 10 μm.
- Flow cytometric analysis of the tetraploid Gam‐GFP reporter strain grown on SCD with or without PQT (600 μg/ml), H2O2 (2 mM), or PL (100 μg/ml) for 24 h at 37°C. (*) denotes a significant difference in Gam‐GFP values between control and test agent (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
- 2‐DOG assay to monitor for loss of GAL1 function from tetraploid strain RBY18 after culture on SCD medium with or without PQT (600 μg/ml), H2O2 (2 mM), or PL (100 μg/ml) at 37°C for 7 days. (*) denotes a significant difference between 2‐DOG resistant colonies between control and the indicated agent (P < 0.05, Student's t‐test, n = 3, error bars represent SEM).
Discussion
Ploidy change is a common occurrence in eukaryotic cells being integral to sexual reproduction, tumorigenesis, and the development of certain somatic tissues (Storchova & Pellman, 2004; Bennett et al, 2014; Todd et al, 2017). A striking example of ploidy change occurs in C. albicans where diploid cells mate to form tetraploid cells that then reduce their ploidy via aberrant mitotic divisions (Bennett & Johnson, 2003; Forche et al, 2008; Bennett, 2015). Ploidy reduction is promoted by growth of tetraploid cells on PRE‐SPO medium whereas diploid cells are stable under these conditions. In this work, we reveal that nutritional cues trigger both genomic instability and cell death in tetraploid cells and identify causal links to metabolism, ROS production, and DNA double‐strand break (DSB) formation.
The elevated metabolism of polyploid cells enhances ROS levels
Candida albicans tetraploid cells are relatively stable during standard culture although long‐term passaging, as in S. cerevisiae, causes cells to reduce their ploidy toward the diploid state (Gerstein et al, 2006; Hickman et al, 2015; Selmecki et al, 2015). Growth on PRE‐SPO medium influences genome instability in a ploidy‐specific manner; tetraploid cells, but not diploid cells, show reduced viability and increased chromosomal instability when grown at 37°C on this medium. Expression profiling revealed limited differences between diploid and tetraploid C. albicans cells, consistent with studies in S. cerevisiae where ploidy and cell size have only a modest effect on the transcriptome (Galitski et al, 1999; Wu et al, 2010). The accelerated induction of glycolytic and oxidative stress response genes, however, suggested that increased metabolic activity could contribute to the phenotypes observed on PRE‐SPO medium.
A direct assessment of metabolism revealed that PRE‐SPO‐cultivated C. albicans cells showed increased fermentative and aerobic respiratory rates compared to those on standard YPD medium. We also found that tetraploid cells exhibited metabolic activities that were fourfold to eightfold higher than diploid cells, while cell size and mitochondrial content were doubled in tetraploid cells relative to diploid cells. In line with increased metabolic flux and induction of oxidative stress, cells grown on PRE‐SPO showed higher ROS levels and activation of the ROS‐responsive transcription factor Cap1 compared to cells grown on YPD. Importantly, supplementation of PRE‐SPO medium with antioxidants or overexpression of endogenous antioxidant enzymes suppressed both ROS levels and the oxidative stress response. The addition of antioxidants may protect against oxidative stress, in part, by lowering the overall redox potential as shown for S. cerevisiae cells (Bednarska et al, 2008).
Endogenous ROS induces DNA damage and chromosome instability in tetraploid cells
Reactive oxygen species production is known to promote DNA damage in multiple cell types (Storr et al, 2013; Tafani et al, 2016). In line with this, we observed increased ROS‐induced DSB formation in C. albicans cells grown on PRE‐SPO medium. Supplementation of this medium with antioxidants mitigated both DSB formation and genetic instability in tetraploid cells. This suggests a model whereby a hyperactive metabolism on PRE‐SPO medium generates sufficient ROS to cause DSB formation, which in turn results in genome instability and cell death in polyploid C. albicans cells.
We also note that C. albicans tetraploid cells were more susceptible to treatment with DNA‐damaging agents than diploid cells. Thus, tetraploid cells showed increased cell death and chromosomal instability when treated with MMS or HU than diploid cells. This result mirrors studies in S. cerevisiae where tetraploid cells were more sensitive to DSB‐inducing agents than diploid cells and tetraploid cells were also more reliant on homologous recombination for DNA repair, particularly at elevated temperatures (Storchova et al, 2006; Storchova, 2014). Cells of higher ploidy may be more susceptible to DNA damage and chromosome mis‐segregation due to the altered geometry of the mitotic spindle as cell size increases (Storchova et al, 2006; Storchova, 2014). Treatment of C. albicans cells with exogenous sources of oxidative stress also recapitulated the phenotypes on PRE‐SPO medium. Treatment of tetraploid cells with these agents therefore resulted in elevated levels of ROS, DSB formation, and chromosome instability. This reveals similarities to studies in S. cerevisiae where deletion of DNA repair genes was found to promote chromosome rearrangements due to increased oxidative DNA damage (Degtyareva et al, 2008).
Taken together, these results suggest that polyploid C. albicans cells, but not diploid cells, experience genome instability and cell death on PRE‐SPO medium due to two main factors. First, tetraploid cells are more metabolically active on this medium than diploid cells, thereby generating higher levels of ROS and DNA damage. Second, tetraploid cells are also markedly more sensitive to DNA damage than diploid cells, so that these cells are more prone to damage‐induced instability and cell killing. Culture on PRE‐SPO medium therefore produces sufficient ROS/DNA damage to cause extensive genome instability in tetraploid cells, whereas ROS levels do not reach the threshold necessary for instability in diploid cells.
Polyploidy, cancer, and ROS in mammalian cells
Cancer progression is a complex process and yet several parallels exist between the processes supporting tumorigenesis and those observed in polyploid fungal cells. Strikingly, ~90% of all solid tumors are aneuploid and many of these forms arise due to the inherent genome instability of polyploid cells (Coward & Harding, 2014). Both polyploid and aneuploid forms facilitate karyotypic variation, tumor progression, and the emergence of drug resistance. Polyploid C. albicans cells similarly exhibit increased genome instability, and the resulting karyotypic diversity can promote adaptation and resistance to antifungal drugs (Bennett & Johnson, 2003; Harrison et al, 2014; Hirakawa et al, 2017).
While links between ploidy and genome instability are well recognized both in yeast and in mammalian cells (Storchova & Kuffer, 2008), those between ploidy and metabolism are considerably more complex. Cancer polyploid cells are larger than diploid cells and can display a higher metabolic rate commensurate with their size (Coward & Harding, 2014; Donovan et al, 2014). It also appears that this increase in metabolism promotes genome instability in certain cancer cells. For example, polyploid prostate and mammary epithelial cells display higher ROS levels than diploid cells due to their increased mitochondrial content (Roh et al, 2012). Increased ROS levels can, in turn, induce DNA damage and genome instability (Samper et al, 2003; Radisky et al, 2005), while expression of a mitochondrial superoxide dismutase suppressed genome instability and tumor formation caused by ROS (van de Wetering et al, 2008). These results suggest that, as in C. albicans, connections exist between endogenous ROS and genome instability in certain cancer cells.
It should be noted, however, that eukaryotic cells do not exhibit a universal correlation between cell size and metabolic activity. For example, varying the size of hepatocytes led to a nonlinear relationship with mitochondrial respiration, with optimal rates of oxidative phosphorylation observed at intermediate cell sizes (Miettinen et al, 2014). In the case of hepatocytes, this may be a functional adaptation that limits cellular ROS accumulation in the toxic liver environment (Schoenfelder & Fox, 2015).
It is therefore apparent that metabolic cues are implicated in ROS production, DNA damage, and chromosomal instability in both fungal and animal cells. Studies of unicellular eukaryotes could therefore provide additional mechanistic insights into links between metabolism, ROS, and tumorigenesis.
A model for metabolism‐induced ROS in the evolution of eukaryotic ploidy cycles?
A central unresolved question in biology is how did sexual reproduction evolve in eukaryotes? The emergence of sex would have enhanced DNA repair yet came with significant costs including the twofold cost of sex (de Visser & Elena, 2007; Bernstein et al, 2011; Goddard, 2016; McDonald et al, 2016). Intriguingly, sex may have been especially advantageous in dealing with oxidative stress from the endosymbiont that formed the pre‐mitochondrion, as this aerobic bacterium would have generated high levels of ROS (Blackstone, 1995; Bernstein et al, 2011; Horandl & Speijer, 2018). Sex would have been particularly effective in repairing subsequent ROS‐induced DNA damage via homologous recombination. In support of a connection between ROS and sex, oxidative stress still promotes sexual reproduction in multiple extant eukaryotes (Reeves & Jackson, 1974; Bernstein & Johns, 1989; Nedelcu et al, 2004).
The current work suggests that ROS could have facilitated ploidy cycling as a forerunner to meiosis in the pre‐eukaryote. High levels of ROS would have been particularly detrimental to cells of higher ploidy and, thus, if haploid cells generated diploid or polyploid cells (e.g., by cell fusion or endoreduplication), then the resulting cells may have been forced to revert to a lower ploidy state. In this model, ploidy reduction would have been a natural consequence of the high levels of ROS produced by early eukaryotes (Horandl & Speijer, 2018), and a primitive ploidy cycle could have occurred without meiosis.
This model, while speculative, aligns with other aspects of meiosis in extant eukaryotes. For example, DSBs are essential for meiotic recombination with their production regulated by the Spo11 endonuclease (Lam & Keeney, 2014). Spo11 is dispensable for meiosis, however, if DSBs are generated by an alternative mechanism such as γ radiation (Dernburg et al, 1998). It is therefore possible that ROS‐induced DSBs were the causal agents of ploidy reduction in pre‐eukaryotes, and that this step subsequently came under the control of Spo11 to allow for tighter regulation of potentially lethal DNA breaks.
Conclusions
In this study, we establish connections between metabolism, endogenous ROS, DNA damage, and genome instability, and establish how polyploid cells can be selectively induced to reduce their ploidy even in the absence of a conventional meiosis. These studies parallel those in certain cancer cells where polyploidy, ROS, and chromosome instability are intertwined, indicating that similar mechanisms may impact genome stability in diverse eukaryotes. Our results also provide a speculative model for how metabolism‐induced processes could have enabled a rudimentary ploidy cycle in evolution. Further studies are necessary to more precisely define the relationship between metabolism, ROS, and genome integrity, as well as their impact in multiple eukaryotic species.
Materials and Methods
Media and chemicals
Yeast extract peptone dextrose (YPD) and synthetic complete dextrose (SCD) media were prepared as described (Guthrie & Fink, 1991). Saccharomyces cerevisiae pre‐sporulation (“PRE‐SPO”) medium contained 0.8% yeast extract, 0.3% peptone, 10% glucose (added prior to autoclaving), and 2% agar. Quercetin hydrate (Acros Organics, #AC174070100), curcumin (Acros Organics, #AC218580100), L‐ascorbic acid (Acros Organics, #AC352680050), L‐glutathione (Sigma‐Aldrich, Cat. No. G4251‐10G), dithiothreitol (Fisher Scientific, #BP172‐25), paraquat dichloride hydrate (Sigma‐Aldrich, #36541‐100MG), hydrogen peroxide (Sigma‐Aldrich, #H1009‐100ML), piperlongumine (Cayman Chemical, #20069‐09‐4), methyl methanesulfonate (Acros Organics, #AC156890050), and hydroxyurea (MP Biomedicals, #0210202310‐10 g) were added to the indicated media once the media had cooled to approximately 40°C, at which point the media was poured and plates were used for assays within 24 h.
Strain construction
A CAP1‐mNeonGreen cassette was amplified from pRB895 (pSFS2A‐mNeonGreen) (Frazer et al, 2019) using oligonucleotides 4564 and 4565 (see Appendix Table S5), which contain homology to the CAP1 gene and the mNeonGreen plasmid. This cassette was transformed into the diploid strain SC5314 and the tetraploid strain RBY18 to create strains CAY9331 (diploid) and CAY9332 (tetraploid).
The plasmid pRS11 was generated by first synthesizing a C. albicans codon‐optimized Gam gene flanked by two SalI restriction sites, which was then digested with SalI and cloned into the pNIM1 vector (Park & Morschhauser, 2005). pRS11 was linearized with restriction enzymes SacII and ApaI and transformed into the diploid strain SC5314 and the tetraploid strain RBY18 to generate strains CAY7504 (diploid) and CAY7571 (tetraploid).
The plasmid pLC605 contains a Tet‐OFF promoter sequence which can be fused to genes of interest to generate overexpression (O/E) cassettes (Veri et al, 2018). Tet‐OFF O/E cassettes were created for the antioxidant enzymes encoded by SOD1, SOD2, SOD3, TRX1, and GCS1 by PCR amplification of pLC605 using oligos 5547/5548, 4823/4824, 4702/4703, 4813/4814, and 4817/4818, respectively (see Appendix Table S5). These cassettes were transformed into RBY18 to generate strains CAY10665 (SOD1 O/E), CAY9516 (SOD2 O/E), CAY9388 (SOD3 O/E), CAY9501 (TRX1 O/E), and CAY9507 (GCS1 O/E).
Quantification of chromosome loss
Tetraploid stain RBY18 was cultured in quadrants on media of interest and incubated for 7 days at 37°C. Cells were then scraped off each plate and suspended in sterile PBS. Two successive 1:100 dilutions were made in sterile PBS. 100 μl of the undiluted and 1:100 dilution cell suspensions was plated onto SC 2‐DOG medium, and 100 μl of the 1:100 dilution and 1:10,000 dilution cell suspensions was plated onto YPD medium. These plates were incubated at 30°C for 48 h, at which point the plates were collected and the total number of colonies were counted on each plate. Each experiment was performed in biological triplicate.
Viability assays
Strains SC5314 and RBY18 were collected from YPD medium, and approximately 6 × 107 cells from each strain were top‐spread onto YPD and PRE‐SPO media and incubated at 30 and 37°C, respectively. At 1 and 5 days, cells were collected, stained with 0.5 mg/ml propidium iodide, and imaged using a fluorescence microscope (Zeiss Z1 Axio Observer, Zeiss filter set 45, 250 ms exposures). Images were then viewed with Fiji image processing software, and ~300 cells were scored for viability, with PI‐positive cells representing dead yeast cells (Dudgeon et al, 2008). Each experiment was performed in biological quintuplicate.
RNA sequencing
Strains SC5314 and RBY18 were collected from YPD medium; approximately 6 × 107 cells from each strain were top‐spread onto YPD and PRE‐SPO media and incubated at 30 and 37°C, respectively. Cells were collected at 3, 6, 12, and 24 h, flash‐frozen in liquid nitrogen, and stored at −80°C. Each experiment was performed in biological duplicate. RNA was purified using the Ambion RiboPure Yeast RNA Purification Kit (ThermoFisher, #AM1926) and RNA integrity assessed using an Agilent Bioanalyzer 2100. mRNA libraries were prepared using the Illumina TruSeq Stranded mRNA Library Preparation Kit (Illumina, #20020595). Libraries were then pooled and sequenced on an Illumina HiSeq 4000 platform generating 100‐bp paired‐end reads (at The Ohio State University Comprehensive Cancer Center, Columbus, OH). Reads were aligned to the C. albicans SC5314 reference genome (Assembly 21) using the Spliced Transcripts Alignment to a Reference (STAR) aligner (Dobin et al, 2013). Differential expression analysis was conducted using edgeR. Gene ontology enrichment analysis was conducted using the Gene Ontology Consortium enrichment analysis tool (Mi et al, 2017).
2,3,5‐Triphenyltetrazolium chloride (TTC) reduction assay
To assess metabolic activity of C. albicans cells on YPD and PRE‐SPO media, a TTC overlay technique was used (Ogur et al, 1957). Approximately 2 × 107 cells of SC5314 and RBY18 strains were spotted onto YPD and PRE‐SPO media and incubated for 24 h at 30 and 37°C, respectively. Prior to applying the TTC overlay solution, each plate was imaged for a 0 min reference. A TTC overlay agar solution (0.067 M potassium phosphate buffer pH 7.0, 1.5% agar, 0.1% TTC) was warmed to 55°C and 10 ml added onto the plates containing the spot‐inoculated colonies. The flooded plates were imaged 15 min after the addition of the overlay solution. Each experiment was performed in biological quintuplicate.
For image quantitation, collected images were viewed with Fiji image processing software. Spot colonies were selected, and mean RGB breakdowns of pixel values for each colony were determined using the color histogram tool. The relative luminance (Y) of each colony was calculated by plugging the RGB values into the formula Y = 0.2126R + 0.7152G + 0.0722B, and the percent of red light contributing to the overall luminance was calculated using the formula (0.2126R/Y) × 100. The percent red light of overall luminance was calculated for both the 0 min and 15 min images, and the 15 min value was normalized to the 0 min value for a final red pixel intensity value.
Aerobic respiratory and fermentative rate assays
Strains SC5314 and RBY18 were collected from YPD medium, and approximately 6 × 107 cells from each strain were top‐spread onto YPD and PRE‐SPO media and incubated at 37°C for 24 h. An Agilent Seahorse XFe96 sensor cartridge (Agilent Technologies, Cat. No. 102416‐100) was hydrated in a utility plate with each well containing 200 μl of sterile water overnight at 37°C. On the following day, the sensor cartridge was moved into a utility plate containing 200 μl of XF Calibrant (Agilent Technologies, Cat. No. 100840‐000) in each well and incubated for one hour at 37°C. Cells were harvested from each plate and suspended in sterile water to a concentration of approximately 5.0 × 107 CFU/ml. 10 μl of each cell suspension was seeded into wells containing 200 μl of pre‐warmed Seahorse XF assay medium (supplemented with 10 mM glucose, 10 mM sodium pyruvate) in an Agilent Seahorse XF96 cell culture microplate (Agilent Technologies, #102416‐100). Where appropriate, drug injection ports for each well were loaded with 20 μl of 50 μM antimycin A, 50 μM rotenone, and 5 mM salicylhydroxamic acid (SHAM) diluted in Seahorse XF assay medium, representing a final concentration of 5 μM antimycin A, 5 μM rotenone, and 500 μM SHAM after injection. The cell plate was placed into the Seahorse XFe96 instrument and the assay run for fourteen total cycles, with each cycle consisting of one minute of mixing, zero minutes waiting, and three minutes measuring. The first seven measurements were used to collect baseline OCR and ECAR measurements, at which point the antimycin A, rotenone, and SHAM cocktail were injected into each well and seven measurements were recorded to collect treated OCR and ECAR measurements. Upon completion of assay, the exact number of cells in each cell suspension was assessed via hemocytometer counts and oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements were normalized to both the total number of cells seeded into each well and the viability of cells under each growth condition (calculated in the viability assays detailed above). These normalized measurements were then multiplied by 106 for final OCR measurements in the units of pmol/min/106 CFU and ECAR measurements in the units of mpH/min/106 CFU. Each experiment was performed in biological quintuplicate.
Antioxidant enzyme overexpression assays
PRE‐SPO medium plates containing 5 μg/ml doxycycline (DOX) were prepared and used the same day for each assay. Strains CAY10665 (SOD1 O/E), CAY9516 (SOD2 O/E), CAY9388 (SOD3 O/E), CAY9501 (TRX1 O/E), and CAY9507 (GCS1 O/E) were grown on YPD medium and then cultured in quadrants on PRE‐SPO medium with or without DOX and incubated for 7 days at 37°C, at which point cells were harvested and a 2‐DOG assay was conducted as detailed above. Each experiment was performed in biological triplicate.
Stress assays
SC5314 and RBY18 cells were grown overnight in liquid YPD medium at 30°C. The following day, the cultures were diluted 1:50 and grown to mid‐logarithmic phase (approximately 4–5 h). Cell density was determined by hemocytometer count, and cells were diluted to a density of 2 × 108 CFU/ml. Five successive 1:10 serial dilutions were made in sterile phosphate‐buffered saline (PBS) and 5 μl of each cell suspension spotted onto freshly prepared SCD, SCD + 20 mM hydroxyurea, and SCD + 0.01% methyl methanesulfonate media. Plates were incubated for 48 h at 30 and 37°C at which point plates were imaged.
CellROX Green staining
SC5314 and RBY18 were cultured in quadrants on YPD and PRE‐SPO media and incubated for 24 h at 30 and 37°C, respectively. Cells were harvested and washed with sterile PBS. PBS‐washed cells were then suspended in 5 μM CellROX Green Reagent (ThermoFisher Scientific, #C10444) and incubated for 30 min at 37°C. The staining solution was then removed from cells, and cells were washed three times with PBS. CellROX green‐stained cells were then co‐stained with calcofluor white at a final concentration of 0.1 mg/ml to visualize the cell wall. For microscopy, cells were imaged using a fluorescence microscope (Zeiss Z1 Axio Observer; CellROX Green: Zeiss filter set 38, 500 ms exposure; calcofluor white: Zeiss filter set 49, 25 ms exposure), and images viewed and processed with Fiji image processing software. For flow cytometry, a total of 100,000 cells were analyzed on a FACSAria (BD Biosciences), and data were analyzed via FlowJo (TreeStar, Ashland, OR). Each experiment was performed in biological quintuplicate.
MitoTracker Green staining
SC5314 and RBY18 were cultured in quadrants on YPD medium and incubated for 24 h at 30°C. Cells were harvested and washed with sterile PBS. PBS‐washed cells were then suspended in 1 μM MitoTracker Green Reagent (ThermoFisher Scientific, #M7514) and incubated for 30 min at 30°C. The staining solution was then removed, and cells were washed once with PBS. For flow cytometry, a total of 500,000 cells were analyzed on a FACSAria (BD Biosciences) and data were analyzed via FlowJo (TreeStar, Ashland, OR). Each experiment was performed in biological triplicate.
Gam‐GFP assays
A Gam‐GFP reporter strain was used to detect the presence of DSBs in tetraploid C. albicans cells. This strain was grown on the indicated growth medium in the presence of doxycycline (+DOX; 12.5 μg/ml) to induce the Gam‐GFP protein. Plates without doxycycline (−DOX) were included as controls where Gam‐GFP was not induced. CAY7504 (diploid) and CAY7571 (tetraploid) cells containing Gam‐GFP were inoculated into quadrants on the indicated medium and incubated for 24 h at the indicated temperature. Cells were recovered from each plate and suspended in 1 ml of sterile PBS. A total of 500,000 cells from each sample were run on a BD FACSAria (Becton Dickinson Biosciences) and data analyzed using FlowJo software (Tree Star, Ashland, OR). Each experiment was performed in biological triplicate. For microscopy, cells were co‐stained with calcofluor white at a final concentration of 0.1 mg/ml. Cells were imaged using a fluorescence microscope (Zeiss Z1 Axio Observer; Gam‐GFP: Zeiss filter set 38, 250 ms exposure; calcofluor white: Zeiss filter set 49, 25 ms exposures), and images were viewed and processed with Fiji image processing software.
Cap1‐mNeonGreen assays
Strains CAY9331 (diploid) and CAY9332 (tetraploid) expressing Cap1‐mNeonGreen were inoculated into quadrants on the indicated medium. Cells were recovered from each plate and suspended in sterile PBS. Cells were co‐stained with calcofluor white at a final concentration of 0.1 mg/ml. Cells were imaged using a fluorescence microscope (Zeiss Z1 Axio Observer; CAP1‐NeonGreen: Zeiss filter set 38, 250 ms exposure; calcofluor white: Zeiss filter set 49, 25 ms exposure), and images were viewed and processed with Fiji processing software. For quantitation of Cap1 activation, brightness on the GFP channel was adjusted to the maximum observed brightness in the data set and cells that exhibited a robust nuclear signal were counted as Cap1‐activated. Each experiment was performed in biological quintuplicate.
DNA staining for ploidy analysis
Candida albicans cells were grown overnight at 30°C in liquid YPD medium, at which point cells were collected, fixed with 70% ethanol, and incubated for 1 h at 4°C. Cells were then washed twice with 50 mM Tris–Cl, pH 8.0, 5 mM EDTA, resuspended in 2 mg/ml RNase A (Sigma‐Aldrich, #R5503‐500MG) in 50 mM Tris–Cl, pH 8.0, 5 mM EDTA, and incubated for 2 h at 37°C. RNase‐treated cells were pelleted and resuspended in 5 mg/ml pepsin (Sigma‐Aldrich, #P7000‐500G) in 55 mM HCl and incubated for 1 h at 37°C. Cells were then washed twice with 50 mM Tris–Cl, pH 7.5, 5 mM EDTA and suspended in 1 μM SYTOX Green Nucleic Acid Stain (Thermo Fisher Scientific, #S7020) in 50 mM Tris–Cl, pH 7.5, 5 mM EDTA and incubated overnight at 4°C. For ploidy analysis, 100,000 cells were run on an Attune NxT Flow Cytometer (Thermo Fisher Scientific) and data were analyzed via FlowJo (TreeStar, Ashland, OR).
Author contributions
Conceptualization, GJT and RJB; Methodology, GJT, CH, NA, and RJB; Formal Analysis, GJT; Resources, RSS and PB; Writing—Original Draft, GJT and RJB; Supervision, GJT and RJB; Funding Acquisition, RJB.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We would like to thank Matthew Anderson and Joshua Wang for help with RNA‐seq analyses; Marla Tipping and Kate Neville for assistance with Seahorse experiments; Robert Cramer, Iuliana Ene, and Corey Frazer for scientific discussion; Matthew Hurton and Emily Roblee for help with preliminary assays; and Leah Cowen for plasmids. This work was supported by National Institutes of Health grants AI081704/AI122011 to RJB, GM110578/GM094712 to NA and by the NIGMS RI‐INBRE program under grant P20GM103430 to NA, as well as by a Burroughs Wellcome Fund PATH award to RJB.
The EMBO Journal (2019) 38: e101597
Data availability
All RNA‐seq data associated with this project have been uploaded to NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), with accession code PRJNA507025.
References
- Baek YU, Kim YR, Yim HS, Kang SO (2004) Disruption of gamma‐glutamylcysteine synthetase results in absolute glutathione auxotrophy and apoptosis in Candida albicans . FEBS Lett 556: 47–52 [DOI] [PubMed] [Google Scholar]
- Bednarska S, Leroy P, Zagulski M, Bartosz G (2008) Efficacy of antioxidants in the yeast Saccharomyces cerevisiae correlates with their effects on protein thiols. Biochimie 90: 1476–1485 [DOI] [PubMed] [Google Scholar]
- Belenky P, Ye JD, Porter CB, Cohen NR, Lobritz MA, Ferrante T, Jain S, Korry BJ, Schwarz EG, Walker GC et al (2015) Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep 13: 968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD (2003) Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J 22: 2505–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Forche A, Berman J (2014) Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb Perspect Med 4: a019604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ (2015) The parasexual lifestyle of Candida albicans . Curr Opin Microbiol 28: 10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein C, Johns V (1989) Sexual reproduction as a response to H2O2 damage in Schizosaccharomyces pombe . J Bacteriol 171: 1893–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H, Carol Bernstein C, Michod RE (2011) Meiosis as an evolutionary adaptation for DNA repair In DNA Repair, Kruman I. (ed), pp 357–382. Rijeka, Croatia: InTech; [Google Scholar]
- Blackstone NW (1995) Perspective a units‐of‐evolution perspective on the endosymbiont theory of the origin of the mitochondrion. Evolution 49: 785–796 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Madlung A (2014) Polyploidy in the Arabidopsis genus. Chromosome Res 22: 117–134 [DOI] [PubMed] [Google Scholar]
- Broxton CN, He B, Bruno BM, Culotta VC (2018) A role for Candida albicans superoxide dismutase enyzmes in glucose signaling. Biochem Biophys Res Commun 495: 814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL et al (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Dilkes B, Luo H, Douglas A, Yakubova E, Lahner B, Salt DE (2013) Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis . Science 341: 658–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocheme HM, Murphy MP (2008) Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem 283: 1786–1798 [DOI] [PubMed] [Google Scholar]
- Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Coward J, Harding A (2014) Size does matter: why polyploid tumor cells are critical drug targets in the war on cancer. Front Oncol 4: 123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyareva NP, Chen L, Mieczkowski P, Petes TD, Doetsch PW (2008) Chronic oxidative DNA damage due to DNA repair defects causes chromosomal instability in Saccharomyces cerevisiae . Mol Cell Biol 28: 5432–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan P, Cato K, Legaie R, Jayalath R, Olsson G, Hall B, Olson S, Boros S, Reynolds BA, Harding A (2014) Hyperdiploid tumor cells increase phenotypic heterogeneity within Glioblastoma tumors. Mol BioSyst 10: 741–758 [DOI] [PubMed] [Google Scholar]
- Dudgeon DD, Zhang N, Ositelu OO, Kim H, Cunningham KW (2008) Nonapoptotic death of Saccharomyces cerevisiae cells that is stimulated by Hsp90 and inhibited by calcineurin and Cmk2 in response to endoplasmic reticulum stresses. Eukaryot Cell 7: 2037–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ (2008) The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol 6: e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer C, Hernday AD, Bennett RJ (2019) Monitoring phenotypic switching in Candida albicans and the use of next‐gen fluorescence reporters. Curr Protoc Microbiol 53: e76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D (2005) Cytokinesis failure generating tetraploids promotes tumorigenesis in p53‐null cells. Nature 437: 1043–1047 [DOI] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR (1999) Ploidy regulation of gene expression. Science 285: 251–254 [DOI] [PubMed] [Google Scholar]
- Gentric G, Desdouets C (2014) Polyploidization in liver tissue. Am J Pathol 184: 322–331 [DOI] [PubMed] [Google Scholar]
- Gerstein AC, Chun HJ, Grant A, Otto SP (2006) Genomic convergence toward diploidy in Saccharomyces cerevisiae . PLoS Genet 2: e145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard MR (2016) Molecular evolution: sex accelerates adaptation. Nature 531: 176–177 [DOI] [PubMed] [Google Scholar]
- Gorman JA, Gorman JW, Koltin Y (1992) Direct selection of galactokinase‐negative mutants of Candida albicans using 2‐deoxy‐galactose. Curr Genet 21: 203–206 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991) Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; [Google Scholar]
- Harrison BD, Hashemi J, Bibi M, Pulver R, Bavli D, Nahmias Y, Wellington M, Sapiro G, Berman J (2014) A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol 12: e1001815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su CH, Bennett RJ, Wang Y et al (2013) The ‘obligate diploid’ Candida albicans forms mating‐competent haploids. Nature 494: 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MA, Paulson C, Dudley AM, Berman J (2015) Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans . Genetics 200: 781–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa MP, Chyou DE, Huang D, Slan AR, Bennett RJ (2017) Parasex generates phenotypic diversity de novo and impacts drug resistance and virulence in Candida albicans . Genetics 207: 1195–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horandl E, Speijer D (2018) How oxygen gave rise to eukaryotic sex. Proc Biol Sci 285: 20172706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Raisner RM, Johnson AD (2000) Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289: 307–310 [DOI] [PubMed] [Google Scholar]
- Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO (2002) Copper‐ and zinc‐containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148: 3705–3713 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J (2009) The DNA‐damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena NR (2012) DNA damage by reactive species: mechanisms, mutation and repair. J Biosci 37: 503–517 [DOI] [PubMed] [Google Scholar]
- Karki K, Hedrick E, Kasiappan R, Jin UH, Safe S (2017) Piperlongumine induces reactive oxygen species (ROS)‐dependent downregulation of specificity protein transcription factors. Cancer Prev Res (Phila) 10: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiezopolska E, Gabaldon T (2018) Evolutionary emergence of drug resistance in Candida opportunistic pathogens. Genes 9: 461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam I, Keeney S (2014) Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 7: a016634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre C, LeMay JD, Deslauriers N, Bourbonnais Y (2001) Candida albicans expresses an unusual cytoplasmic manganese‐containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J Biol Chem 276: 43784–43791 [DOI] [PubMed] [Google Scholar]
- Lockhart SR, Pujol C, Daniels KJ, Miller MG, Johnson AD, Pfaller MA, Soll DR (2002) In Candida albicans, white‐opaque switchers are homozygous for mating type. Genetics 162: 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Magee PT (2000) Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289: 310–313 [DOI] [PubMed] [Google Scholar]
- Mayer VW, Aguilera A (1990) High levels of chromosome instability in polyploids of Saccharomyces cerevisiae . Mutat Res 231: 177–186 [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rice DP, Desai MM (2016) Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531: 233–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Haber JE (2014) Sources of DNA double‐strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 6: a016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen TP, Pessa HK, Caldez MJ, Fuhrer T, Diril MK, Sauer U, Kaldis P, Bjorklund M (2014) Identification of transcriptional and metabolic programs related to mammalian cell size. Curr Biol 24: 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD (2002) White‐opaque switching in Candida albicans is controlled by mating‐type locus homeodomain proteins and allows efficient mating. Cell 110: 293–302 [DOI] [PubMed] [Google Scholar]
- Morales DK, Grahl N, Okegbe C, Dietrich LE, Jacobs NJ, Hogan DA (2013) Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 4: e00526‐12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Kitazume M, Ose R, Kurokawa J, Koga K, Osuga Y, Arai S, Miyazaki T (2011) Death effector domain‐containing protein (DEDD) is required for uterine decidualization during early pregnancy in mice. J Clin Investig 121: 318–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu AM, Marcu O, Michod RE (2004) Sex as a response to oxidative stress: a twofold increase in cellular reactive oxygen species activates sex genes. Proc Biol Sci 271: 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogur M, St. John R, Nagai S (1957) Tetrazolium overlay technique for population studies of respiration deficiency in yeast. Science 125: 928–929 [DOI] [PubMed] [Google Scholar]
- Park YN, Morschhauser J (2005) Tetracycline‐inducible gene expression and gene deletion in Candida albicans . Eukaryot Cell 4: 1328–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson MJ, McKenzie CG, Smith DA, da Silva Dantas A, Sherston S, Veal EA, Morgan BA, MacCallum DM, Erwig LP, Quinn J (2013) Ybp1 and Gpx3 signaling in Candida albicans govern hydrogen peroxide‐induced oxidation of the Cap1 transcription factor and macrophage escape. Antioxid Redox Signal 19: 2244–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA et al (2005) Rac1b and reactive oxygen species mediate MMP‐3‐induced EMT and genomic instability. Nature 436: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RJ, Jackson RM (1974) Stimulation of sexual reproduction in Phytophthora by damage. J Gen Microbiol 84: 303–310 [Google Scholar]
- Rhie GE, Hwang CS, Brady MJ, Kim ST, Kim YR, Huh WK, Baek YU, Lee BH, Lee JS, Kang SO (1999) Manganese‐containing superoxide dismutase and its gene from Candida albicans . Biochim Biophys Acta 1426: 409–419 [DOI] [PubMed] [Google Scholar]
- Rich PR, Mischis LA, Purton S, Wiskich JT (2001) The sites of interaction of triphenyltetrazolium chloride with mitochondrial respiratory chains. FEMS Microbiol Lett 202: 181–187 [DOI] [PubMed] [Google Scholar]
- Roh M, van der Meer R, Abdulkadir SA (2012) Tumorigenic polyploid cells contain elevated ROS and ARE selectively targeted by antioxidant treatment. J Cell Physiol 227: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DCP, Gleason JE, Sanchez H, Schatzman SS, Culbertson EM, Johnson CJ, McNees CA, Coelho C, Nett JE, Andes DR et al (2017) Candida albicans FRE8 encodes a member of the NADPH oxidase family that produces a burst of ROS during fungal morphogenesis. PLoS Pathog 13: e1006763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E, Nicholls DG, Melov S (2003) Mitochondrial oxidative stress causes chromosomal instability of mouse embryonic fibroblasts. Aging Cell 2: 277–285 [DOI] [PubMed] [Google Scholar]
- Schoenfelder KP, Fox DT (2015) The expanding implications of polyploidy. J Cell Biol 209: 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R et al (2015) Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C, Cox BD, Gu F, Luengas EM, Joshi MC, Chiu LY, Magnan D, Halliday JA, Frisch RL, Gibson JL et al (2013) Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. eLife 2: e01222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Dantas A, Patterson MJ, Smith DA, Maccallum DM, Erwig LP, Morgan BA, Quinn J (2010) Thioredoxin regulates multiple hydrogen peroxide‐induced signaling pathways in Candida albicans . Mol Cell Biol 30: 4550–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroga JM, Ma X, Das SK (2012) Developmental regulation of decidual cell polyploidy at the site of implantation. Front Biosci 4: 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Pellman D (2004) From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 5: 45–54 [DOI] [PubMed] [Google Scholar]
- Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O'Toole E, Pellman D (2006) Genome‐wide genetic analysis of polyploidy in yeast. Nature 443: 541–547 [DOI] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C (2008) The consequences of tetraploidy and aneuploidy. J Cell Sci 121: 3859–3866 [DOI] [PubMed] [Google Scholar]
- Storchova Z (2014) Ploidy changes and genome stability in yeast. Yeast 31: 421–430 [DOI] [PubMed] [Google Scholar]
- Storr SJ, Woolston CM, Zhang Y, Martin SG (2013) Redox environment, free radical, and oxidative DNA damage. Antioxid Redox Signal 18: 2399–2408 [DOI] [PubMed] [Google Scholar]
- Tafani M, Sansone L, Limana F, Arcangeli T, De Santis E, Polese M, Fini M, Russo MA (2016) The interplay of reactive oxygen species, hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxid Med Cell Longev 2016: 3907147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RT, Forche A, Selmecki A (2017) Ploidy variation in fungi: polyploidy, aneuploidy, and genome evolution. Microbiol Spectr 10.1128/microbiolspec.FUNK-0051-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzung KW, Williams RM, Scherer S, Federspiel N, Jones T, Hansen N, Bivolarevic V, Huizar L, Komp C, Surzycki R et al (2001) Genomic evidence for a complete sexual cycle in Candida albicans . Proc Natl Acad Sci USA 98: 3249–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veri AO, Miao Z, Shapiro RS, Tebbji F, O'Meara TR, Kim SH, Colazo J, Tan K, Vyas VK, Whiteway M et al (2018) Tuning Hsf1 levels drives distinct fungal morphogenetic programs with depletion impairing Hsp90 function and overexpression expanding the target space. PLoS Genet 14: e1007270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser JA, Elena SF (2007) The evolution of sex: empirical insights into the roles of epistasis and drift. Nat Rev Genet 8: 139–149 [DOI] [PubMed] [Google Scholar]
- Voordeckers K, Kominek J, Das A, Espinosa‐Cantu A, De Maeyer D, Arslan A, Van Pee M, van der Zande E, Meert W, Yang Y et al (2015) Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet 11: e1005635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, Ying K, Chen WS, Jiang YY (2006) Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans . Free Radic Biol Med 40: 1201–1209 [DOI] [PubMed] [Google Scholar]
- van de Wetering CI, Coleman MC, Spitz DR, Smith BJ, Knudson CM (2008) Manganese superoxide dismutase gene dosage affects chromosomal instability and tumor onset in a mouse model of T cell lymphoma. Free Radic Biol Med 44: 1677–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Rolfe PA, Gifford DK, Fink GR (2010) Control of transcription by cell size. PLoS Biol 8: e1000523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH et al (2013) Pan‐cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YO, Sherlock G, Petrov DA (2016) Whole genome analysis of 132 clinical Saccharomyces cerevisiae strains reveals extensive ploidy variation. G3 (Bethesda) 6: 2421–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Data Availability Statement
All RNA‐seq data associated with this project have been uploaded to NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), with accession code PRJNA507025.


