Abstract
Objectives:
Aiming to uncover the genetic basis of abdominal obesity, we performed a genome-wide association study (GWAS) meta-analysis of trunk fat mass adjusted by trunk lean mass (TFMadj) and followed by a series of functional investigations.
Subjects:
A total of 11,569 subjects from 6 samples were included into the GWAS meta-analysis.
Methods:
Meta-analysis was performed by a weighted fixed-effects model. In silico replication analysis was performed in the UK-Biobank (UKB) sample (N=331,093) and in the GIANT study (N up to 110,204). Cis-expression QTL (cis-eQTL) analysis, dual-luciferase reporter assay and electrophoresis mobility shift assay (EMSA) were conducted to examine the functional relevance of the identified SNPs. At last, differential gene expression analysis (DGEA) was performed.
Results:
We identified an independent SNP rs12409479 at 1p21 (MAF=0.07, p=7.26×10−10), whose association was replicated by the analysis of TFM in the UKB sample (one sided p=3.39×10−3), and was cross-validated by the analyses of BMI (one-sided p=0.03) and WHRadj (one-sided p=0.04) in the GIANT study. Cis-eQTL analysis demonstrated that allele A at rs12409479 was positively associated with PTBP2 expression level in subcutaneous adipose tissue (N=385, p=4.15×10−3). Dual-luciferase reporter assay showed that the region repressed PTBP2 gene expression by downregulating PTBP2 promoter activity (p<0.001), and allele A at rs12409479 induced higher luciferase activity than allele G did (p=4.15×10−3). EMSA experiment implied that allele A was more capable of binding to unknown transcription factors than allele G. Lastly, DGEA showed that the level of PTBP2 expression was higher in individuals with obesity than in individuals without obesity (N=20 and 11, p=0.04 and 9.22×10−3), suggesting a regulatory role in obesity development.
Conclusions:
Taken together, we hypothesize a regulating path from rs12409479 to trunk fat mass development through its allelic specific regulation of PTBP2 gene expression, thus providing some novel insight into the genetic basis of abdominal obesity.
Keywords: abdominal obesity, trunk fat mass, 1p21, rs12409479, PTBP2
Introduction
Obesity is a chronic metabolic disease characterized by an excessive accumulation of body fat. It is one of the most severe public health problems, associated with a series of disorders including diabetes, cancer, cardiovascular disease, osteoarthritis and obstructive sleep disease 1. Approximately 2.1 billion people globally are overweight or affected by obesity, and the prevalence has been increasing substantially 2. In the United States alone, the total economic cost related to obesity was more than $215 billion annually as of 2010 3. Obesity has been a global leading cause of morbidity and mortality that imposes an enormous burden on public health.
Though obesity can be attributable to lifestyle and cultural factors, such as lack of exercise and excessive calorie intake, the presence of an underlying genetic effect is indisputable 4. The heritability of obesity is up to 70% 5, which is significantly higher than that of many other complex traits such as hypertension (29%) 6. Family, twin, and adoption studies have also indicated a strong genetic determination of human obesity 7, 8. In developed countries, 60%–70% of the variation in obesity-related phenotypes appears to be heritable 9, 10.
Body mass index (BMI), defined as the body weight in kg divided by squared height in meters (kg/m2), is currently the standard measure of obesity due to its simplicity. To date, a variety of genome-wide association studies (GWASs) and meta-analyses have been conducted and hundreds of genetic variants have been identified for BMI 11, 12, accounting for 5% of total heritability.
Despite its popularity and simplicity, BMI is never the ideal phenotype to measure obesity. Human body mass is a complex composite of fat mass, lean mass, bone mass and other soft tissues. Among them, it is only fat mass that induces obesity and causes a series of adverse clinical manifestations 13. Therefore, fat mass is the only accurate and ideal phenotype to measure obesity. Nonetheless, research using fat mass as a measure for obesity is sparse, partly due to the difficulty of accurately measuring and collecting the phenotype.
Among various types of fat-induced obesity, abdominal obesity is perhaps the most severe. Fat stored in the abdomen is more harmful than fat stored at other body regions. For example, truncal adiposity confers a 3-fold increased risk for heart disease in women compared with accumulation of body fat in the gluteal femoral region 14. Moreover, fat mass stored more centrally leads people to be more susceptible to cardiovascular diseases and endocrine disorders 15. Truncal adiposity is also related to a reduction in endogenous growth hormone in women without generalized obesity 16, and with other health conditions, such as skeletal damage 17, 18, pain 19 and liver disease 20. The correlation between an accumulation of abdominal fat with an increased risk of metabolic disturbances is also found in children 21.
In the present study, we aim to uncover the genetic basis of abdominal obesity by conducting a genome-wide association meta-analysis of trunk fat mass in 11,569 subjects. We then perform a series of functional experiments and analyses to fine-map the causal variants and genes. We conduct eQTL analysis, in vitro dual-luciferase assay and EMSA assay to investigate the functional relevance of the identified variants. We also perform differential gene expression analysis in humans to examine the association of relevant gene expression with obesity susceptibility.
Methods
Ethics statement
Samples used in this study are from multiple research and/or clinical centers. All samples were approved by the respective institutional ethics review boards, and all participants signed informed consent documents before being enrolled into the study.
Study populations
Six GWAS samples were incorporated in our study, of which three were from in-house studies and the other three were accessed through the NCBI Database of Genotypes and Phenotypes (dbGAP). The in-house samples include two of European population, with 1,000 and 2,286 unrelated individuals, and one of Chinese Han population with 1,627 unrelated individuals 22. The fourth sample was derived from the Framingham Heart Study (FHS), a longitudinal and prospective cohort comprising over 16,000 participants of European ancestry spanning three generations 23. A total of 6,004 phenotyped individuals were included. Both the fifth and sixth samples were derived from the Women’s Health Initiative (WHI) observational study, a partial factorial randomized and longitudinal cohort with over 12,000 genotyped women, aged 50-79 years, of African-American or Hispanic ancestry 24. The fifth sample comprises 847 phenotyped individuals of African-American ancestry, and the sixth sample comprises 445 phenotyped individuals of Hispanic ancestry.
Phenotype measurements and modeling
Trunk fat mass was measured by dual energy x-ray absorptiometry (DXA) machines (either Lunar Corp., Madison, WI, USA; or Hologic Inc, Bedford, MA, USA). In all the samples, covariates, including gender, age, age^2, height, height^2 and the first ten principal components derived from genome-wide genotype data were screened for significance with the stepwise linear regression model implemented in the stepAIC function of R package MASS. To correct for the effect of lean mass, trunk lean mass was taken as covariate as well. Regression residuals were transformed into a standard normal distribution by the inverse quantiles of standard normal distribution. Normalized residuals were used for subsequent association analysis.
Genotyping and quality control
All GWAS samples were genotyped by high-throughput SNP genotyping arrays (Affymetrix Inc., Santa Clara, CA, USA; or Illumina Inc., San Diego, CA, USA within individual samples) according to the manufacturers’ protocols. Quality control (QC) within each sample was implemented at both the individual level and SNP level. At the individual level, sex compatibility was checked by imputing sex from X-chromosome genotype data with Plink 25. Individuals of ambiguous imputed sex or with inconsistent imputed sex with the reported one were removed. At the SNP level, SNPs violating the Hardy-Weinberg equilibrium (HWE) rule (p-value<1.0×10−5) were removed. In the familial sample FHS, genotypes having the Mendel error were set to missing.
Genotype imputation
GWAS samples were imputed to the 1000 Genomes Project phase 3 sequence variants (as of May 2013) 26. Genotype imputation reference panels of 503 individuals of European ancestry, 504 individuals of East Asian ancestry, 661 of African ancestry, and 347 of admixed American ancestry were downloaded from the project website. Each GWAS sample was imputed by respective reference panel with the closest ancestry. Haplotypes of bi-allelic variants, including SNPs and bi-allelic insertions/deletions (indels), were extracted to form reference panels for imputation. As a QC procedure, variants with zero or one copy of minor alleles were removed.
Prior to imputation, a consistency test of allele frequency between the GWAS sample and the reference sample was examined with the chi-square test. To correct for potential mis-strandedness, GWAS SNPs that failed the consistency test (p<1.0×10−6) were transformed into the reverse strand. SNPs that again failed the consistency test were removed from the GWAS sample.
Imputation was performed with FISH, an algorithm that we developed to impute the diploid genome without the need to phase genotype into haplotype27. The method calculates an exact form of transition matrix in the segmental hidden Markov model, and is therefore accurate, fast and computer memory efficient. The software parameters were set by default.
Individual study association testing
Each GWAS sample was tested for association between normalized phenotype residual and genotyped and imputed genotypes under an additive mode of inheritance. For unrelated samples, association was examined by the linear regression model with MACH2QTL 28, in which allele dosage was taken as predictor for phenotype. For the FHS sample, a mixed linear model was used to account for genetic relatedness within each pedigree 29. Genomic control inflation 30 was estimated for each individual GWAS.
Meta-analysis
Summary association signals from individual samples were combined for meta-analysis. Meta-analysis was performed by a weighted fixed-effects model with METAL 31, where weight was proportional to the inverse variance of regression coefficient. As a QC procedure, only common or less common (MAF≥0.01) and well imputed (imputation r2≥0.3 in at least two samples) SNPs were included for analysis. Between-study heterogeneity effect was measured by I2 and Cochran’s Q p-value 32. Genomic control inflation factor was estimated in meta-analysis.
Sensitivity analysis was performed by removing each study in turn and performing the same meta-analysis in the remaining samples.
In silico replication analysis
To verify the association signals of our identified SNP, we performed in silico replication analyses in three large-scale studies. The first was the GWAS scan of trunk fat mass in up to 331,093 UK-Biobank participants, in which trunk fat mass was measured by impedance approach. The analysis was performed by Dr. Neale’s lab and the results were kindly released to be publicly available before publication. It should be noted that the phenotype being analyzed is trunk fat mass without adjusting for trunk lean mass. Summary association results were downloaded from the following link (https://www.dropbox.com/s/zp05s9ucxjodf6b/23128.assoc.tsv.gz?dl=0).
The second and third studies were two obesity-related traits analyzed by the Genetic Investigation of Anthropometric Traits (GIANT) consortium: BMI and waist-hip ratio adjusted for BMI (WHRadj) 33, 34. Cross-validation in these two related traits strengthens the confidence towards true association in the present study. Summary association results were downloaded from the GIANT consortium website (http://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_file).
Cis-expression quantitative trait loci (cis-eQTL) analysis
To investigate the association between the identified SNP polymorphisms and the nearby gene expressions, we performed cis-eQTL analysis. The datasets we used were derived from the GTEx project 35. The GTEx project collected RNA-sequenced multiple human tissues (up to 11,614) from donors who were also densely genotyped, and analyzed for associations between SNPs and global RNA expression within individual tissues. Three obesity-related tissues, including subcutaneous adipose (N=385), visceral adipose (N=313) and whole blood (N=369), were analyzed. Single tissue eQTL association results were downloaded from the GTEx website (http://www.gtexportal.org/home/).
Dual-luciferase reporter assay
We conducted a dual-luciferase reporter assay to determine whether the regulation of PTBP2 expression by rs12409479 was through regulating its promoter activity. Human embryonic kidney cell line 293T (HEK293T cells) were cultured in DEME high glucose (Hyclone, Thermo Fisher Scientific Inc.) supplemented with 10% fetal bovine serum (FBS, Tianhang Inc.), 100 U mL−1 penicillin, and 100μg mL−1 streptomycin. Cells were maintained at 37°C in a water-saturated atmosphere under 5% CO2 in air, and were cultured to 60%−80% in 12-well or 24-well culture plate before transfection.
Luciferase reporter plasmid containing PTBP2 promoter was constructed by inserting a 2095 bp sequence containing PTBP2 promoter (ENST00000370197.5; PTBP2-201) into the KpnI and HindIII sites of the pGL3-basic vector (Promega Corporation, Madison, WI, USA), which expresses the Firefly luciferase gene. This pGL3-PTBP2-Promoter construct served as a negative control. For the experimental group, a 225 bp DNA sequence centered at rs12409479 (chr1:968,006,63-968,008,87) was cloned into the 5′-end of the above pGL3-PTBP2-Promoter vector, forming the pGL3-rs12409479-PTBP2-Promoter construct. We cloned two types of sequence, each containing either allele A or allele G of rs12409479. The success of plasmid construction was confirmed by DNA Sanger sequencing. The construct was then transfected into the 293T cells using the jetPRIME transfection reagent (PolyPlus-transfection, France). As an internal control for transfection efficiency, the pRL-TK vector (Promega Corporation, Madison, WI, USA) expressing the Renilla luciferase gene was co-transfected at the same time. The cells were collected for luciferase activity measurement after 24h of transfection. We conducted 8 replicates under each experimental condition, and 4 replicates under the null condition.
Luciferase activity was measured using the dual-luciferase reporter system (Promega Corporation, Madison, WI, USA) following the manufacturer’s instructions. The ratio of Firefly to Renilla luciferase activity was compared between the pGL3-rs12409479-PTBP2-Promoter construct and the pGL3-PTBP2-Promoter control construct. We also compared the control construct with the null pGL3-basic vector containing no PTBP2 promoter to examine the transcription efficacy of the promoter, and compared between two allelic experiment groups. Statistical significance was tested using an unpaired Student’s t-test.
Electrophoretic mobility shift assay (EMSA)
We further conducted EMSA to explore whether the oligonucleotide sequence containing the identified SNP binds to transcription factors, and whether the binding activity is allelic specific. Oligonucleotides including biotin-labeled probes and competitor probes were synthetized by Sangon Biotech (Shanghai, China), with HPLC purifying approach. 5×binding buffer was purchased from Beyotime Biotech (Shanghai, China). Total protein was extracted from the 293T cells and stored in aliquots at −80°C until use.
For each sequence containing either a major or minor allele, the following three conditions were conducted in a total volume of 10μl: 1) Negative control: 1μl of biotin-labeled probe (0.07 μM) was incubated with 2ul of 5×binding buffer containing poly (dI-dC), a non-specific competitor of DNA. The mixture was incubated at 25°C for 20 min. 2) Binding reaction: Before adding biotin-labeled probes for 20min at room temperature, 5×binding buffer was pre-incubated with total protein at 25°C for 10 min. 3) Competition reaction: 100-fold excess of unlabeled probe was added to the reaction mixture (5×binding buffer and total protein) for 20min prior to the addition of the labeled probe. The binding reaction products were then separated via electrophoresis at 80V for 2 hours on 6% nondenaturing polyacrylamide gel with 0.5×Tris-borate-EDTA running buffer. Finally, the DNA-protein complexes were detected using the Chemiluminescent EMSA kit (Beyotime, China) according to the manufacturer’s instructions.
Differential gene expression analysis
To establish a relationship between the relevant genes and obesity development, we examined differential gene expression levels in datasets accessed through the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo/).
We restricted the datasets to fat or adipose tissue-related human subjects/tissues. After searching the GEO web portal using the terms “adipose tissue,” “adiposity,” and “adipose stem cell,” we identified 30 datasets. After careful examination of the retrieved items, 7 datasets were available for analysis, as described in a previous study 36. In each dataset, we grouped subjects into case (obesity) group and control (non-obesity) group. Genome-wide gene expressions were logarithm-transformed and normalized. Normalized expression levels were compared between the two groups using the GEO online tool GEO2R (ncbi.nlm.nih.gov/geo/geo2r/). The expression difference was characterized by logarithm transformed fold change (logFC) and p-value.
Results
Meta-analysis
Basic characteristics of the GWAS meta-analysis samples are listed in Supplemental Table 1. Principal component analysis (PCA) was applied to each individual sample, and no population outlier was observed. A total of 28,855,047 bi-allelic variants were imputed into the 1000 Genomes Project reference panel. After filtering out rare variants and variants of poor imputation certainty, 12,061,510 variants were retained for association analysis. After adjusting the phenotype by PCA in each individual study, the genomic control inflation factor was small (λ=1.04), indicating the limited effect of population stratification. A logarithmic quantile-quantile (QQ) plot of meta-analysis test statistics shows a marked deviation in the tail of the distribution, suggesting the possible existence of a true association (Supplemental Figure 1).
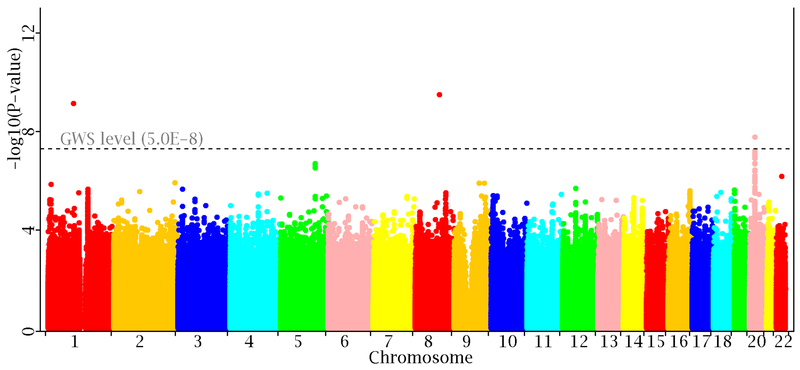
A total of 3 SNPs at 3 genomic loci were identified at the genome-wide significance level (GWS, 5.0×10−8): 1p21 (rs12409479, p=7.26×10−10), 8q21.3 (rs2091921, p=3.21×10−10), and 20p11 (rs4813371, p=1.71×10−8). At a less stringent significance level 1.0×10−7, 5 additional SNPs were identified. Of those, 4 are located in the same locus 20p11 as rs4813371, and are in strong LD with the latter. The fifth SNP rs6011111 (p=9.85×10−8) presented a significant heterogeneity effect (I2=56.8%, Q p=0.04). Manhattan plot of the GWAS meta-analysis is shown in Figure 1.
Figure 1. Manhattan plot of the meta-analysis.
The results from the meta-analysis are plotted against the position on each of the 22 chromosomes. The black dashed line indicates the threshold for genome wide significance level (α=5.0×10−8).
Among the 3 identified loci, 20p11 was previously reported for body fat mass (BFM), with the lead SNP being rs2069126 36. rs2069126 is associated with TFMadj in the present study as well (p=1.21×10−6), though the signal does not reach the GWS level. rs2069126 and rs4813371 are 10.0 kb apart, and the two SNPs have modest LD (r2=0.33), implying that their signals arise from same association source.
Both rs2091921 and rs12409479 at the remaining two identified loci are polymorphic in the three samples (OOS, KCOS and FHS) of European population and in the WHI-HIS Hispanic population sample, but are monomorphic in the samples of Chinese population (COS) and of African population (WHI-AFR). Sensitivity analysis shows that the association at rs2091921 is entirely driven by the KCOS sample (p=5.49×10−10), but is not significant in any of the other samples. Meta-analysis of the remaining samples excluding the KCOS sample reveals that the association becomes non-significant (p=0.12). At rs12409479, the association is mainly driven by the KCOS sample as well (p=6.22×10−8), but is also significant in the larger FHS sample (p=0.01). For meta-analysis of the remaining samples excluding the KCOS sample, the association remains significant (p=2.12×10−3). Given the above sensitivity analysis results, we disregarded rs2091921 and focused on rs12409479 at 1p21 only.
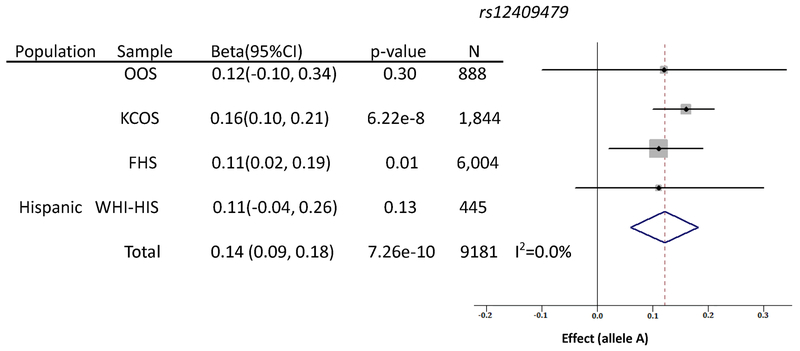
rs12409479 is a common SNP (MAF=0.07) in the European population. Allele A at this SNP tends to increase normalized trunk fat mass by 0.14 standard deviations per copy. It is located 386 kb 5′-upstream of PTBP2 gene. The forest plot is displayed in Figure 2, and the regional plot drawn by LocusZoom 37 is shown in Supplemental Figure 2.
Figure 2. Forest plot of rs12409479 meta-analysis.
Regression coefficient (beta) and its 95% confidence interval (CI) are presented in un-transformed estimates from individual studies. “Total” refers to the combined meta-analysis.
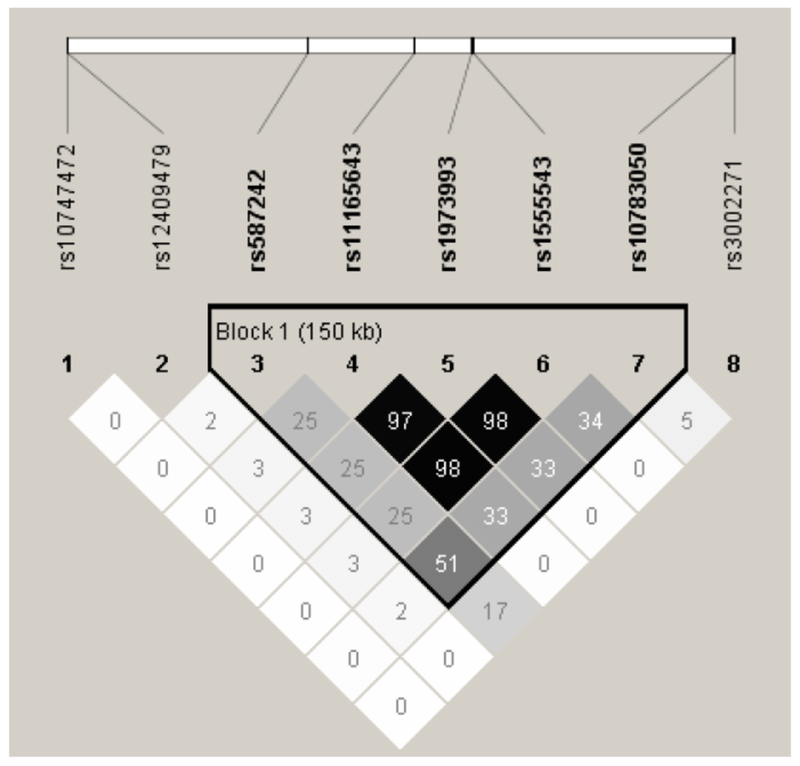
The locus 1p21 encompassing rs12409479 was previously identified for BMI and weight, tagged by multiple SNPs 12, 34, 38–40. The LD patterns between rs12409479 and these SNPs are plotted in Figure 3, and the association signals of these SNPs in the present study are listed in Supplemental Table 2. It is clear that rs12409479 is independent of any of the other reported SNPs (r2=0.00-0.03), implying that it represents an independent signal.
Figure 3. Linkage disequilibrium pattern between rs12409479 and previously identified SNPs.
A total of 7 SNPs were reported by previously GWAS meta-analyses of BMI and body weight (rs10783050, rs11165643, rs10747472, rs3002271, rs587242, rs1555543, rs1973993). Linkage disequilibrium (LD) pattern between each pair of these SNPs including rs12409479 and the LD r2 value are displayed with Haploview 53. Five SNPs are clustered into a single haplotype block spanning 150 kb. rs12409479 is in very low LD (r2=0.00-0.03) with any of the other SNPs, indicating that it is independent of all previous SNPs.
In silico replication analysis
We tested whether rs12409479 was significant in previous large GWAS meta-analyses of obesity-related traits. The first trait is trunk fat mass, analyzed in the UK-Biobank participants (N=331,093). The effect direction is consistent and the p-value is significant (one-sided p=3.39×10−3) in the UK-Biobank analysis, increasing the likelihood of a true association signal. We then surveyed the summary results released by the GIANT consortium for BMI and WHRadj 33, 34. The effect directions in both traits are consistent, in that allele A corresponds to higher BMI and WHRadj levels. For BMI, a weak association is observed in the sub-group of men of 50 years or older (N=93,201, p=0.06). The association is also weak for WHRadj, (N=110,204, p=0.07). Considering the one-sided nature of the replication effort, they are indeed nominally significant (one-sided p=0.03 and 0.04). The main results are listed in Table 1.
Table 1.
Main association results of rs12409479 in studied samples
| Trait | Sample | Beta | SE | P | N | I2(%) |
|---|---|---|---|---|---|---|
| GWAS meta-analysis | ||||||
| TFMadj | OOS | 0.12 | 0.11 | 0.3 | 888 | - |
| KCOS | 0.16 | 0.03 | 6.22×10−8 | 1844 | - | |
| FHS | 0.11 | 0.04 | 0.01 | 6004 | - | |
| WHI-HIS | 0.11 | 0.08 | 0.13 | 445 | - | |
| Meta | 0.14 | 0.02 | 7.26×10−10 | 9181 | 0.0 | |
| Cross-replication | ||||||
| TFM | UKB | 0.01 | 5.12×10−3 | 3.39×10−3 | 331,093 | - |
| BMI | GIANT | 0.03 | 0.02 | 0.03 | 61,506 | - |
| WHRadj | GIANT | 0.02 | 0.01 | 0.04 | 110,204 | - |
Notes: Beta is the regression coefficient of normalized phenotypic residual on the effect allele A. OOS, Omaha osteoporosis study; KCOS, Kansas City osteoporosis study; FHS, Framingham Heart Study; WHI-HIS was the Women’s Health Initiative (WHI) cohort Hispanic subsample. TFM, trunk fat mass; TFMadj, trunk fat mass adjusted by trunk lean mass; BMI, body mass index; WHRadj, waist-hip ratio adjusted by BMI; UKB, UK-Biobank. Nominally significant P values are marked in bold.
Cis-eQTL analysis
We performed cis-eQTL analysis in subcutaneous adipose tissue, visceral adipose tissue and whole blood of the GTEx project datasets. The results show that rs12409479 polymorphisms are significantly associated with PTBP2 gene expression in subcutaneous adipose tissue. Specifically, allele A is associated with a higher level of gene expression (p=4.15×10−3, N=385). The association is nonetheless not observed in either visceral adipose tissue (p=0.17, N=313) or whole blood (p=0.56, N=369), implying that the regulation pattern is specific to the subcutaneous adipose tissue.
Dual-luciferase reporter assay
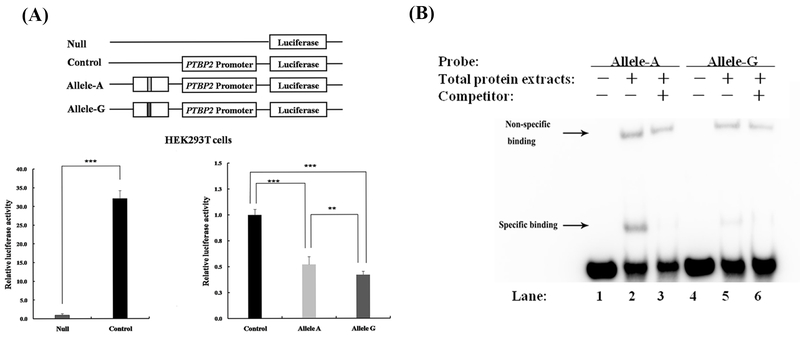
To determine whether the region encompassing rs12409479 regulates PTBP2 promoter activity, we conducted dual-luciferase reporter assay in HEK293T cells. Specifically, a 225 bp rs12409479-centered DNA fragment was cloned into the luciferase reporter vector containing PTBP2 promoter. These constructs were transfected into HEK293T cells along with pRL-TK vector.
The luciferase reporter assay results are displayed in Figure 4A. Compared to the null pGL3-basic vector, the control vector containing the PTBP2 promoter significantly increases luciferase activity by over 30 fold (p=3.65×10−7), proving the successful construction of the PTBP2 promoter in promoting luciferase gene expression. Compared to the pGL3-PTBP2-Promoter vector, the pGL3-rs12409479-PTBP2-Promoter vector containing either allele A or allele G significantly reduces PTBP2 expression (p=8.52×10−10 or 1.82×10−11), with the reduction magnitude being up to 50%. This significant reduction suggests that the sequence containing rs12409479 downregulates PTBP2 gene expression by repressing PTBP2 promoter activity. Between the two allele constructs, the expression of allele A is significantly higher than that of allele G (p=4.15×10−3), implying that the regulation is allelic specific.
Figure 4. Dual-luciferase reporter assay and EMSA results.
(A) Dual-luciferase reporter assay result. The pGL3-PTBP2-Promoter vector was set as a control. Sequence construct containing either allele A or allele G was inserted upstream of the gene promoter. Eight replicates were conducted under each of the three conditions. Compared to the null pGL3-basic vector, the vector containing the PTBP2 promoter increases luciferase expression (p=3.65×10−7). Both allele A construct and allele G construct significantly reduce luciferase activity (both p<0.01), suggesting that the sequence containing rs12409479 represses PTBP2 promoter activity. Between the two allele constructs, the expression of allele A is significantly higher than that of allele G (p=4.15×10−3), implying that the regulation is allelic specific. (B) EMSA result. EMSA was conducted using a biotin labeled A-allele probe or G-allele probe with total protein extracted from HEK293T cells, with or without unlabeled A-allele probe or G-allele probe competitor. The shifted bands at the same migration position are observed in labeled probes for both allele A (lane 2) and allele G (lane 5) when compared to the controls (lane 1 and lane 4), implying their binding affinity to the same transcription factor. The two shifted bands are completely eliminated by the addition of 100-fold excess unlabeled A-allele probe (lane 3) and G-allele probe (lane 6), demonstrating that the binding is specific to the sequence motif containing rs12409479. Between the two alleles, the signal intensity for allele A (lane 2) is stronger than that for allele G (lane 5, too weak to be observed without zooming in the figure), suggesting that the former is more capable of binding to transcription factor than the latter. *: p<0.05; **: p<0.01; ***: p<0.001.
EMSA
We next performed EMSA to explore the possibility that rs12409479 binds to unknown transcription factors in an allelic specific manner. The binding reactions were performed with labeled and competitor probes containing allele A or allele G of rs12409479. The labeled probe sequences are as follows:
forward primer
5′-biotin-TATAAATGGTGGGAAAAAGAAGAATATTTTCT[A/G]GCACATTAAAACTATATGAAGTTCAAATTTCA-3′;
reverse primer
5′-biotin-TGAAATTTGAACTTCATATAGTTTTAATGTGC[T/C]AGAAAATATTCTTCTTTTTCCCACCATTTATA-3′.
The EMSA results are shown in Figure 4B. The shifted bands at the same migration position are observed in labeled probes for both allele A (lane 2) and allele G (lane 5) when compared to negative controls (lane 1 and lane 4), demonstrating their binding affinity to the same transcription factor. The two shifted bands are completely eliminated by the addition of 100-fold excess unlabeled A-allele probe (lane 3) and G-allele probe (lane 6), indicating that the binding is specific to the sequence motif containing rs12409479. Between the two alleles, the signal intensity for allele A (lane 2) is stronger than that for allele G (lane 5), suggesting that the former is more capable of binding to the transcription factor than the latter. Collectively, the EMSA results reveal differential binding activities of the two alleles to the same transcription factor.
Differential gene expression analysis (DGEA)
To provide biological insight into the role of PTBP2 in fat development, we explored its differential expression in datasets accessed through the GEO repository. Of the seven datasets accessed through the GEO repository, the expression of PTBP2 is significantly different in two datasets (GSE29718, N=20, p=0.04; GSE9624, N=11, p=9.22×10−3). The effect directions in both datasets are consistent in that obese subjects tend to have increased gene expression compared to non-obese subjects.
Discussion
In the present study, we have performed a genome-wide association meta-analysis of TFMadj in 11,569 participants from 6 GWAS samples, and have identified a functional variant rs12409479 at 1p21, which is independent of any previously reported variants for obesity-related traits. Moreover, through a series of functional investigations, we have hypothesized a regulating path from rs12409479 to trunk fat mass development through its regulation of PTBP2 gene expression.
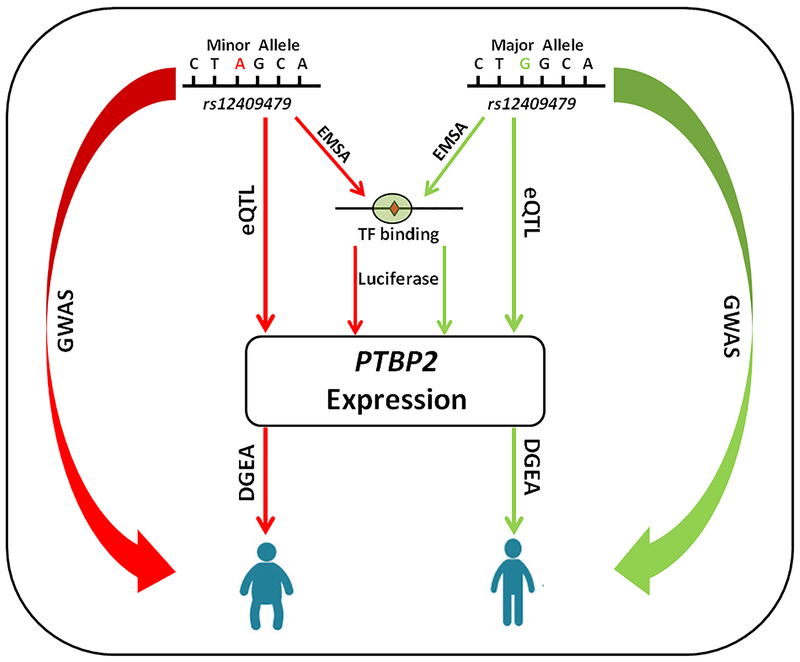
Abdominal fat is located in two major compartments: subcutis and viscera. Truncal subcutaneous fat may exert a greater effect on risk factors than visceral fat does because of its larger mass 41–43. Combining the genetic association analyses with the bioinformatical and functional investigations, we have hypothesized a regulating path from rs12409479 to fat mass development through its allele specific regulation of PTBP2 gene expression, as displayed in Figure 5. GWAS, dual luciferase, EMSA and cis-eQTL experiments consistently supported the upregulated role of allele A in regulating PTBP2 expression, which in turn upregulated the accumulation of trunk fat mass.
Figure 5. Hypothesized regulating path from rs12409479 to trunk fat development.
Based on our GWAS and functional investigation results, we hypothesized a regulating path from rs12409479 to trunk fat through PTBP2 gene. In this diagram, rs12409479 binds to an unknown transcription factor to form a complex that regulates PTBP2 gene expression. Specifically, allele A is more capable of binding to the transcription factor (EMSA) and upregulates PTBP2 (eQTL). PTBP2 in turn regulates trunk fat development in an upward direction (differential gene expression analysis). Consequently, allele A of rs12409479 upregulates trunk fat mass, as evidenced by the association study. The red lines represent upregulation and the green lines represent downregulation.
In this study, we used fat mass instead of BMI as the phenotype to measure obesity. BMI is calculated with total body mass, which is composed of not only fat mass but also lean mass, bone and other soft tissues. Therefore, BMI is unable to discriminate fat mass from other compartments. On the other hand, the majority of human body weight comes from lean mass, while obesity is mainly induced by the excessive storage of fat mass. Unlike fat mass, lean mass has a positive effect on physical fitness, higher caloric expenditure and exercise capacity 44. Therefore, using BMI to classify obesity may provide misleading information about body fat content and risk assessment.
PTBP2 encodes an RNA binding protein that is involved in several post-transcriptional regulatory events of gene expression, including exon splicing. High levels of PTBP2 expression are observed in adult brain and muscles, suggesting its role in the physiology of cardiometabolic phenotypes 45. It is notable that the central nervous system (CNS) plays an important role in obesity etiology. In addition, overexpression of the PTBP2 protein exhibited multiple functions related to the stability and splicing profile of the Nova1 gene, which abolishes brown adipogenesis 46. Several variants previously established for obesity-associated traits, such as BMI, weight and waist circumference, are close to PTBP2 12, 34, 38–40, 47, implying that it might play an important role in human fat storage and obesity development.
Regulatory variants in noncoding regions are thought to explain much more heritability than coding regions for certain diseases 48. Most regulatory elements are located in introns and intergenic regions 49. In addition, recent studies have demonstrated that regulatory risk variants could bind to transcription factors and induce allele-specific effects on the expression of distal gene by long-range interactions 49–51. In this study, the identified SNP rs12409479 is located 386 kb upstream from the PTBP2 gene. Its allele-specific effects on PTBP2 gene expression have been verified by eQTL analysis and functional experiments. Dual-luciferase reporter assay results imply that the region acts as a repressor to PTBP2 expression. Extensive functional investigations are warranted to verify the biological mechanism.
Though ~12 million variants were analyzed, the genome-wide significance level was set at 5.0×10−8. This is because most of the common variants are locally abundant due to the linkage disequilibrium pattern. It has been estimated that the effective number of independent common variants is approximately one million throughout the genome, resulting in a genome-wide significance level of 5.0×10−8 52. Even at a more stringent significance level, the identified novel SNP rs12409479 remains significant at the discovery stage (p=7.26×10−10).
Certain limitations exist in the present study. First, the discovery samples are from diverse populations, which may complicate the interpretation of the results in the presence of population specificity for associated loci. However, the identified SNP rs12409479 is polymorphic only in European and Hispanic populations, and is monomorphic in East Asian and African populations. The relatively limited sample size of Hispanic population (WHI-HIS N=445, 5%) has a minor effect on the association results. Therefore, we do not expect that trans-ancestral samples would pose a problem for the interpretation of our results, particularly at rs12409479. Second, the EMSA assay could only qualitatively visualize the binding affinity to an unknown transcription factor, but could not identify the specific transcription factor. Experiments with finer resolution, such as DNA pull-down assay followed by protein mass spectrum or super-sift assay with a hypothesized protein antibody, may identify the specific transcription factor, which is out of the scope of the present study.
In conclusion, by performing a genome-wide association meta-analysis of TFMadj in 11,569 subjects from 6 GWAS samples, we have identified an independent functional variant rs12409479 at 1p21, and have hypothesized a regulating path from rs12409479 to trunk fat mass development through its allelic specific regulation of PTBP2 gene expression. Our findings may provide useful insights that further improve our understanding of abdominal obesity.
Supplementary Material
Acknowledgments
We are grateful to Loula M Burton at the Tulane university for editing the manuscript. We appreciate all the volunteers who participated into this study. We are grateful to the GIANT consortium, the UK-Biobank, and Dr. Neale’s lab researchers for releasing large-scale summary association results for replication.
This study was partially supported by the national natural science foundation of China (31571291 to LZ, 31771417 and 31501026 to YFP, 81460223 to RH), the natural science foundation of Jiangsu province of China (BK20150323 to YFP), the natural science foundation of Inner Mongolia of China (2014MS08130 to RH), the scientific research foundation for the returned overseas Chinese scholars, ministry of education (to YFP), the NIH (R01 AR069055, U19 AG055373, R01 MH104680, R01 AR059781 and P20 GM109036 to HWD), the Edward G. Schlieder Endowment (to HWD), the startup funding project of Soochow university (Q413900214 to LZ and Q413900114 to YFP) and a project of the priority academic program development of Jiangsu higher education institutions.
The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Whole Body and Regional Dual X-ray Absorptiometry (DXA) dataset was provided by NIH grants R01 AR/AG 41398. The datasets used for the analyses described in this manuscript were obtained from dbGaP (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) through dbGaP accession phs000342.v14.p10.
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, and the US Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2,32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26,42129-32, and 44221. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by the Women’s Health Initiative (WHI), and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. Funding for WHI SHARe genotyping was provided by NHLBI contractN02-HL-64278.The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000200.v10.p3.
Footnotes
Complicance with ethical standards
Conflict of interest: The authors declare that they have no conflict interest.
Supplementary information is available at IJO’s website.
References
- 1.Haslam DW, James WP. Obesity. Lancet 2005; 366(9492): 1197–209. [DOI] [PubMed] [Google Scholar]
- 2.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014; 384(9945): 766–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammond RA, Levine R. The economic impact of obesity in the United States. Diabetes, metabolic syndrome and obesity : targets and therapy 2010; 3: 285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleich S, Cutler D, Murray C, Adams A. Why is the developed world obese? Annual review of public health 2008; 29: 273–95. [DOI] [PubMed] [Google Scholar]
- 5.Walley AJ, Blakemore AI, Froguel P. Genetics of obesity and the prediction of risk for health. Human molecular genetics 2006; 15 Spec No 2: R124–30. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal A, Williams GH, Fisher ND. Genetics of human hypertension. Trends in endocrinology and metabolism: TEM 2005; 16(3): 127–33. [DOI] [PubMed] [Google Scholar]
- 7.Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Molecular and cellular endocrinology 2014; 382(1): 740–57. [DOI] [PubMed] [Google Scholar]
- 8.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behavior genetics 1997; 27(4): 325–51. [DOI] [PubMed] [Google Scholar]
- 9.Comuzzie AG, Allison DB. The search for human obesity genes. Science 1998; 280(5368): 1374–7. [DOI] [PubMed] [Google Scholar]
- 10.Segal NL, Allison DB. Twins and virtual twins: bases of relative body weight revisited. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity 2002; 26(4): 437–41. [DOI] [PubMed] [Google Scholar]
- 11.Pei YF, Zhang L, Liu Y, Li J, Shen H, Liu YZ et al. Meta-analysis of genome-wide association data identifies novel susceptibility loci for obesity. Hum Mol Genet 2014; 23(3): 820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015; 518(7538): 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srikanthan P, Horwich TB, Tseng CH. Relation of Muscle Mass and Fat Mass to Cardiovascular Disease Mortality. The American journal of cardiology 2016; 117(8): 1355–60. [DOI] [PubMed] [Google Scholar]
- 14.Rexrode KM, Carey VJ, Hennekens CH, Walters EE, Colditz GA, Stampfer MJ et al. Abdominal adiposity and coronary heart disease in women. Jama 1998; 280(21): 1843–8. [DOI] [PubMed] [Google Scholar]
- 15.Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, Overvad K et al. General and abdominal adiposity and risk of death in Europe. The New England journal of medicine 2008; 359(20): 2105–20. [DOI] [PubMed] [Google Scholar]
- 16.Miller KK, Biller BM, Lipman JG, Bradwin G, Rifai N, Klibanski A. Truncal adiposity, relative growth hormone deficiency, and cardiovascular risk. The Journal of clinical endocrinology and metabolism 2005; 90(2): 768–74. [DOI] [PubMed] [Google Scholar]
- 17.Russell M, Mendes N, Miller KK, Rosen CJ, Lee H, Klibanski A et al. Visceral fat is a negative predictor of bone density measures in obese adolescent girls. The Journal of clinical endocrinology and metabolism 2010; 95(3): 1247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greco EA, Francomano D, Fornari R, Marocco C, Lubrano C, Papa V et al. Negative association between trunk fat, insulin resistance and skeleton in obese women. World journal of diabetes 2013; 4(2): 31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segura-Jimenez V, Castro-Pinero J, Soriano-Maldonado A, Alvarez-Gallardo IC, Estevez-Lopez F, Delgado-Fernandez M et al. The association of total and central body fat with pain, fatigue and the impact of fibromyalgia in women; role of physical fitness. Eur J Pain 2016; 20(5): 811–21. [DOI] [PubMed] [Google Scholar]
- 20.Canale MP, Manca di Villahermosa S, Martino G, Rovella V, Noce A, De Lorenzo A et al. Obesity-related metabolic syndrome: mechanisms of sympathetic overactivity. International journal of endocrinology 2013; 2013: 865965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor RW, Jones IE, Williams SM, Goulding A. Evaluation of waist circumference, waist-to-hip ratio, and the conicity index as screening tools for high trunk fat mass, as measured by dual-energy X-ray absorptiometry, in children aged 3–19 y. The American journal of clinical nutrition 2000; 72(2): 490–5. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Choi HJ, Estrada K, Leo PJ, Li J, Pei YF et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Human molecular genetics 2014; 23(7): 1923–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nature genetics 2009; 41(11): 1199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Controlled clinical trials 1998; 19(1): 61–109. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81(3): 559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA et al. A map of human genome variation from population-scale sequencing. Nature 2010; 467(7319): 1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Pei YF, Fu X, Lin Y, Wang YP, Deng HW. FISH: fast and accurate diploid genotype imputation via segmental hidden Markov model. Bioinformatics 2014; 30(13): 1876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34(8): 816–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Li J, Pei YF, Liu Y, Deng HW. Tests of association for quantitative traits in nuclear families using principal components to correct for population stratification. Annals of human genetics 2009; 73(Pt 6): 601–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devlin B, Roeder K. Genomic control for association studies. Biometrics 1999; 55(4): 997–1004. [DOI] [PubMed] [Google Scholar]
- 31.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26(17): 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003; 327(7414): 557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS genetics 2015; 11(10): e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Justice AE, Winkler TW, Feitosa MF, Graff M, Fisher VA, Young K et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nature communications 2017; 8: 14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45(6): 580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei YF, Ren HG, Liu L, Li X, Fang C, Huang Y et al. Genomic variants at 20p11 associated with body fat mass in the European population. Obesity (Silver Spring) 2017; 25(4): 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26(18): 2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nature genetics 2009; 41(1): 18–24. [DOI] [PubMed] [Google Scholar]
- 39.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L et al. Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults. PLoS genetics 2017; 13(4): e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature genetics 2010; 42(11): 937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abate N, Garg A, Peshock RM, Stray-Gundersen J, Grundy SM. Relationships of generalized and regional adiposity to insulin sensitivity in men. The Journal of clinical investigation 1995; 96(1): 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abate N, Garg A, Peshock RM, Stray-Gundersen J, Adams-Huet B, Grundy SM. Relationship of generalized and regional adiposity to insulin sensitivity in men with NIDDM. Diabetes 1996; 45(12): 1684–93. [DOI] [PubMed] [Google Scholar]
- 43.Grundy SM. Obesity, metabolic syndrome, and cardiovascular disease. The Journal of clinical endocrinology and metabolism 2004; 89(6): 2595–600. [DOI] [PubMed] [Google Scholar]
- 44.Romero-Corral A, Somers VK, Sierra-Johnson J, Thomas RJ, Collazo-Clavell ML, Korinek J et al. Accuracy of body mass index in diagnosing obesity in the adult general population. Int J Obes (Lond) 2008; 32(6): 959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romanelli MG, Diani E, Lievens PM. New insights into functional roles of the polypyrimidine tract-binding protein. International journal of molecular sciences 2013; 14(11): 22906–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin JC, Lu YH, Liu YR, Lin YJ. RBM4a-regulated splicing cascade modulates the differentiation and metabolic activities of brown adipocytes. Scientific reports 2016; 6: 20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015; 518(7538): 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjalmsson BJ, Xu H et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. American journal of human genetics 2014; 95(5): 535–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corradin O, Scacheri PC. Enhancer variants: evaluating functions in common disease. Genome medicine 2014; 6(10): 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu NQ, Ter Huurne M, Nguyen LN, Peng T, Wang SY, Studd JB et al. The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nature communications 2017; 8: 14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng HF, Forgetta V, Hsu YH, Estrada K, Rosello-Diez A, Leo PJ et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 2015; 526(7571): 112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genetic epidemiology 2008; 32(4): 381–5. [DOI] [PubMed] [Google Scholar]
- 53.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21(2): 263–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.