Abstract
An operon is a set of neighboring genes in a genome that is transcribed as a single polycistronic message. Genes that are part of the same operon often have related functional roles or participate in the same metabolic pathways. The majority of all bacterial genes are co-transcribed with one or more other genes as part of a multi-gene operon. Thus, accurate identification of operons is important in understanding co-regulation of genes and their functional relationships. Here, we present a computational system that uses RNA-seq data to determine operons throughout a genome. The system takes the name of a genome and one or more files of RNA-seq data as input. Our method combines primary genomic sequence information with expression data from the RNA-seq files in a unified probabilistic model in order to identify operons. We assess our method’s ability to accurately identify operons in a range of species through comparison to external databases of operons, both experimentally confirmed and computationally predicted, and through focused experiments that confirm new operons identified by our method. Our system is freely available at https://cs.wellesley.edu/~btjaden/Rockhopper/.
Keywords: operon, transcription, polycistronic, bacteria, bioinformatics, RNA-seq
1. Introduction
An operon is a set of consecutive genes on the same strand in a genome that are co-transcribed into a single polycistronic message. Operons were first described by Jacob and Monod [1]. Operons pervade the genomes of bacteria and archaea, and less commonly can be found in eukaryotes such as nematodes [2]. They are a means by which an organism can implement co-expression of related genes. As a result, identification of operons is an important component in elucidating gene regulatory networks.
A variety of experimental approaches have been employed to detect operons, such as RNA polymerase footprinting, primer extension or S1 mapping, northern blotting, RNase protection assays, and RT-PCR. While these experimental methods may be precise and provide strong evidence in support of an operon, they can be relatively costly or time consuming. Thus, a number of computational methods have been developed for systematic identification of operons throughout a genome. These computational methods have the advantage of being fast and efficient, but the operon predictions they generate can have varying degrees of accuracy with respect to their correspondence with experimentally verified operons [3].
Computational methods for predicting operons generally start with directons, which are consecutive genes on the same strand of a genome [4], and use various features indicative of operons to predict whether the genes in the directon are co-transcribed as part of a single operon or not. The features used in computational methods for predicting operons (reviewed in [5, 6]) can be described generally by three categories: (1) primary sequence features, (2) external data source features, and (3) transcript expression features.
In the first category, primary sequence features, the most commonly used feature is intergenic distance [6]. The distribution of intergenic distances for genes known to be part of the same operon is different, and on average shorter in length, than the distribution of intergenic distances for consecutive genes in a genome that are not part of the same operon. Intergenic distance between consecutive genes is straightforward to compute and offers strong predictive power relative to many other features used for operon prediction [6, 7]. Another example of a primary sequence feature that can be used for operon prediction is the presence of transcription signals, such as promoters or terminators - most commonly Rho-independent terminators - at the beginning and end of the candidate operon, respectively, but not between genes within the candidate operon [8]. Codon usage is commonly used for the problem of identifying protein-coding gene sequences in a genome, and an early studied observed different codon usage patterns for genes in the same operon as compared to genes in different operons based on a small sample of genes [9]. Several studies have employed codon usage as a feature in operon prediction under the assumption that codon usage for genes in the same operon is more similar than codon usage for genes not in the same operon [10–12]. However, the results are mixed when evaluating the contribution of codon usage as a feature in operon prediction, with some examples of codon usage showing significant predictive power [10] and other examples showing little or no statistical difference when incorporating this feature into operon prediction [11, 12].
In the second category, external data source features, features for operon prediction have been derived from various data sources beyond an organism’s genomic sequence. One feature in this category is the functional relationship of the protein products from genes in a candidate operon [13]. Functionally related genes as determined from clusters of orthologous groups (COGs) of proteins and participation in similar metabolic pathways can be indicative of genes that are co-transcribed as part of the same operon [14]. Comparative genomics analyses are commonly used to identify these features, as phylogenetic profiles evince neighborhoods of conserved genes [15, 16]. Among operon prediction methods that utilize features from the first two categories but not the third category, transcript expression data, the method based on the STRING database [13] and its corresponding webservers [7, 17] demonstrates some of the highest accuracy of operon predictions [18], and it uses conserved genes profiles as a core feature.
In the third category, transcript expression data, features are determined from whole-genome transcript expression assays. Early studies used microarray data to aid in operon prediction. Co-expression across different conditions of consecutive genes suggests an increased likelihood of the genes belonging to the same operon [19]. Similarly, observed expression from the intergenic region between genes indicates an enhanced likelihood of the genes being co-expressed [20]. In recent years, there has been an explosion in the use of RNA-seq data throughout biological and related sciences. The Sequence Read Archive (SRA) now contains more than 20 peta-bases of sequencing data [21, 22]. Along with this growth in RNA-seq data is its increasing use in characterizing operons. In some cases, methods based on RNA-seq data have been developed and applied to identify operons in specific organisms [23–26]. In other cases, general purpose tools have been developed to predict operons broadly based on RNA-seq data [12, 16, 27].
In order to improve access to the growing number of operon identifications, a number of databases and web servers are available. In some cases, computational predictions of operons are made offline for a large number of genomes and a database allows users to query these pre-computed operon predictions [15, 17, 28]. In other cases, web servers enable real-time prediction of operons based on user supplied data [7, 16]. A number of databases also integrate curated information about experimentally verified operons [29–31]. While the various computational methods and databases for predicting operons are not always in agreement [3, 24], tools for computational identification of operons can be effective, often achieving predictive accuracies of 90% or more [13]. Table 1 provides a summary of fifteen different methods for predicting operons. Four of the methods listed in Table 1 provide a webserver interface for accessing the operon predictions. The final seven columns in the table indicate the features used by each method when making its predictions. The least commonly used feature in this set is codon usage, with only one of the fifteen methods employing this feature, whereas the most commonly used feature is intergenic distance between operon genes, which is used by eleven of the fifteen methods.
Table 1.
Different methods for predicting operons are shown. The first column provides a citation for the method. The second column indicates the year of publication. The third column indicates the number of genomes for which operon predictions were made. The fourth column indicates if a webserver interface is provided for accessing the predictions. The final seven columns indicate features used for predictions: intergenic distance between genes in an operon, conservation of genes as determined through comparative genomics, functional annotation of genes, transcription signals in the form of promoter regions or terminators, codon usage of genes, microarray gene expression data, and RNA-Seq data.
| Reference | Year | # of Genomes | Web-server | IG | Conservation | Function/Annotation | Promoter and/or Terminator | Codon Usage | Micro-array | RNA-Seq |
|---|---|---|---|---|---|---|---|---|---|---|
| [19] | 2002 | 1 | ♦ | ♦ | ||||||
| [20] | 2002 | 1 | ♦ | ♦ | ||||||
| [4] | 2005 | 2 | ♦ | ♦ | ♦ | |||||
| [11] | 2005 | 124 | ♦ | ♦ | ♦ | ♦ | ||||
| [15] | 2009 | >500 | ♦ | ♦ | ♦ | |||||
| [13] | 2010 | 2 | ♦ | ♦ | ♦ | |||||
| [23] | 2010 | 1 | ♦ | ♦ | ||||||
| [8] | 2011 | 3 | ♦ | ♦ | ♦ | |||||
| [17] | 2012 | >1200 | ♦ | ♦ | ♦ | ♦ | ||||
| [16] | 2014 | 2072 | ♦ | ♦ | ♦ | ♦ | ||||
| [12] | 2014 | 4 | ♦ | ♦ | ♦ | |||||
| [26] | 2014 | 1 | ♦ | ♦ | ||||||
| [24] | 2016 | 1 | ♦ | |||||||
| [7] | 2018 | 8 | ♦ | ♦ | ♦ | ♦ | ||||
| [25] | 2018 | 1 | ♦ | ♦ |
One challenge for these computational methods is that, in many cases, they are trained on data from organisms, such as Escherichia coli and Bacillus subtilis, where a large number of operons have been confirmed experimentally, but their applicability to prokaryotes more broadly is difficult to assess without large sets of independently verified operons outside of a few model organisms. Operon prediction is further challenged by the dynamic nature of transcription, as operon structures can change when environmental conditions vary [12]. Sets of consecutive genes may be co-transcribed as part of an operon in one condition but be individually transcribed in another condition, e.g., when alternative promoters occur both before and within operons [32, 33]. Most methods make static predictions of genes forming an operon, rather than dynamic condition-specific predictions.
Rockhopper is a computational system that provides comprehensive analysis of bacterial RNA-seq data [27, 34]. Rockhopper supports a variety of tasks, including aligning sequencing reads to a reference genome, assembling transcripts, identifying transcript boundaries and novel transcripts such as regulatory RNAs, de novo transcript assembly when reference genomes are unavailable, normalizing data from different experiments to enable meaningful inter-experimental integration and comparison, quantifying transcript abundance levels, statistical testing for differential gene expression in different experimental conditions, and visualizing results in a genome browser. Rockhopper also uses RNA-seq data to identify operons throughout genomes. In the remainder of this article, we detail the methodology used by Rockhopper for operon prediction, assess the predictions with more focused wet-lab experiments, compare the results with databases of operon identifications, and evaluate the results provided by other groups who have employed Rockhopper for operon annotation. Though the methods that we describe have applicability to other domains, the remainder of this article focuses on bacterial operon identification, since this is the domain where the vast majority of operons are found and have been studied.
2. Methods
2.1. Input to Rockhopper software system
As input, Rockhopper requires the name of a genome and one or more files of RNA-seq reads. Sequencing read files may be in fastq, qseq, fasta, sam or bam format, and the files optionally may be gzipped. The sequencing reads may correspond to single-end reads or paired-end reads, and they may be strand-specific or strand-ambiguous. Based on the supplied name of a genome, Rockhopper automatically downloads the genome sequence and gene annotations from RefSeq [35]. If a user wishes to analyze data from an organism not found in RefSeq, the user may supply their own genome sequence file in fasta format and their own gene annotation file as input to Rockhopper.
2.2. Mapping sequencing reads
For all RNA-seq data in this study, prior to using Rockhopper to map sequencing reads to a genome, the Trimmomatic [36] tool was used with default parameter settings for adapter trimming and quality filtering. Rockhopper was then employed to align sequencing reads to a genome. Rockhopper maps reads to a genome in a manner similar to that of Bowtie 2 [37], by building an FM-index [38] for the genome based on the Burrows-Wheeler transform [39]. One of the challenges in analyzing RNA-seq data is that reads often map ambiguously to different transcripts, e.g., when a genome contains multiple paralogous genes [40]. The approach used by RNA-seq analysis pipelines for resolving these ambiguities and allocating reads to paralogous genes can have significant effects on downstream analyses such as testing for differential gene expression [41]. Rockhopper uses the straightforward approach of allocating ambiguous reads equally among the genes to which the reads map. In future work, there may be opportunities for evolving Rockhopper’s model, as recent studies have proposed new methods for allocating ambiguous reads that have the potential to result in increased accuracy of downstream analyses [42].
2.3. Computing directon information
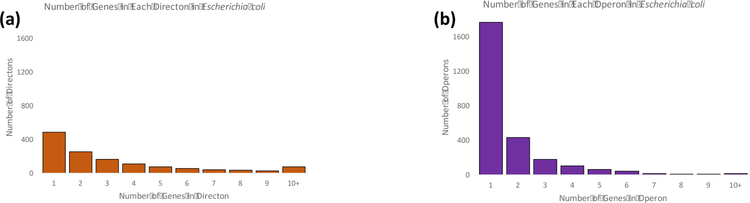
As with many other operon prediction methods [4], Rockhopper begins its identification of operons with directons, which are sets of adjacent genes in a genome on the same strand. Using the input gene annotation files, Rockhopper calculates all directons in a genome before determining which subset of directons are co-transcribed, i.e., are operons. As context, Figure 1a shows the length of directons in the E. coli genome [43]. Here, we use the number of genes in the directon to describe its length rather than, say, the number of nucleotides. As Figure 1a illustrates, there are approximately 500 instances of directons containing a single gene among the 4,386 protein-coding genes in the E. coli genome. Single gene directons occur when a gene is flanked on either side in the genome by a gene on the opposite strand. Similarly, there are approximately 250 instances of length two-gene directons and approximately 150 instances of length three-gene directons. In comparison, Figure 1b shows the length of operons in E. coli as reported by RegulonDB [31]. E. coli is used here because it is arguably the organism where operons have been most extensively studied and because very few other bacterial genomes have large sets of experimentally validated operons. As Figure 1b illustrates, approximately 1,800 genes of the 4,386 genes in the E. coli genome are transcribed individually as single gene operons, and there are approximately 400 instances of pairs of genes being co-transcribed as two-gene operons. While the distributions in Figures 1a and 1b both decrease as the number of genes increases, the rate of decrease is initially sharper for E. coli operons (Figure 1b) than for directons (Figure 1a), suggesting that operons are more likely to be composed of fewer genes than directons.
Figure 1.
(a) The histogram shows how many directons in E. coli are composed of the indicated number of a genes. A directon is defined as a set of consecutive genes on the same strand in a genome. (b) The histogram shows how many operons in E. coli are composed of the indicated number of genes.
2.4. Intergenic distance as a feature of operon prediction
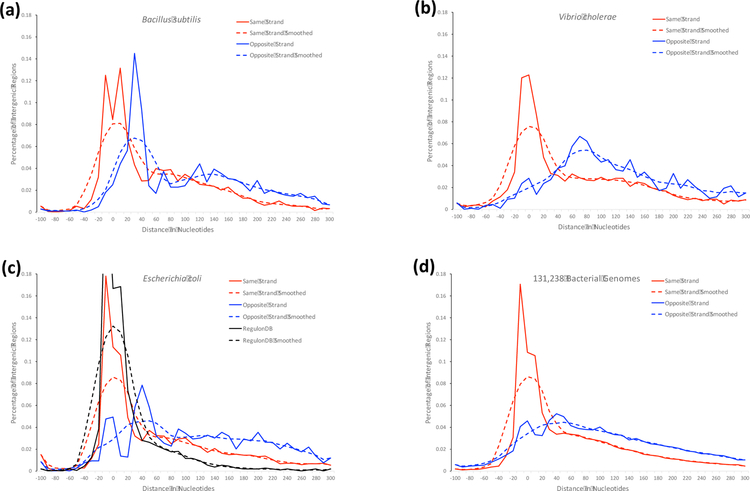
Consistent with most other methods for predicting operons throughout a genome, Rockhopper uses the intergenic distance between genes as a predictive feature in determining operons, as consecutive genes separated by a small intergenic distance are more likely to be co-transcribed than consecutive genes separated by a large intergenic distance [6]. Figures 2a, 2b, and 2c show distributions of the intergenic distances between consecutive genes on the same strand (solid red line) and on opposite strands (solid blue line) for Bacillus subtilis, Vibrio cholerae, and Escherichia coli, respectively. Figure 2c additionally shows the distribution of intergenic distances between consecutive genes that are part of documented operons in RegulonDB (solid black line) in Escherichia coli [31]. As Figure 2c demonstrates, the distribution of intergenic distances for documented operons in RegulonDB (in black) is more similar to the distribution for consecutive genes on the same strand (red) than consecutive genes on the opposite strand (blue) though it has less deviation than the distribution for consecutive genes on the same strand (red). Dashed lines in the figures illustrate versions of the distributions smoothed via an Epanechnikov kernel function [44]. Figures 2a, 2b, and 2c illustrate how different genomes have different propensities for shorter or longer intergenic regions between consecutive genes on the same strand and between consecutive genes on opposite strands. As broader context, Figure 2d shows the same distributions but, rather than for individual genomes, for 131,238 bacterial genomes available from RefSeq [35]. For a particular genome designated by the user, Rockhopper calculates distributions analogous to those shown in Figures 2a, 2b, and 2c. Then, given the intergenic distance in nucleotides between two consecutive genes on the same strand, Rockhopper estimates the probability that the genes correspond to the same operon and the probability that the genes do not correspond to the same operon. The probability that the genes correspond to the same operon is based on the smoothed distribution of intergenic distances between consecutive genes on the same strand (dashed red line in Figure 2) for the relevant genome. The probability that the genes do not correspond to the same operon is based on the smoothed distribution of intergenic distances between consecutive genes on opposite strands (dashed blue line in Figure 2) for the relevant genome.
Figure 2.
The distribution of intergenic distances in nucleotides between consecutive genes on the same strand (solid red line) and between consecutive genes on the opposite strand (solid blue line) is shown for (a) Bacillus subtilis, (b) Vibrio cholerae, (c) Escherichia coli, and (d) 131,238 bacterial genomes. The Escherichia coli figure in (c) additionally shows the distribution of intergenic distances in nucleotides between consecutive genes that are part of documented operons in RegulonDB (solid black line) [31]. Dashed lines in the figures indicate smoothed versions of the distributions as determined with an Epanechnikov kernel.
2.5. Expression patterns as a feature of operon prediction
RNA-seq data evince expression patterns of genes, and these expression patterns contain information about the likelihood that consecutive genes in a genome are part of the same operon [24]. Genes that are co-transcribed as part of the same polycistronic message generally have related expression levels to each other [19, 25]. Like other features for operon identification, expression patterns contain some predictive information but also have limits in their ability to distinguish genes that are co-transcribed from genes that are not co-transcribed. For example, one study observed differential expression of polycistronic genes in 43% of operons in E. coli [26]. Thus, as one feature to aid in its prediction of genes belonging to the same operon, Rockhopper uses the relationship of expression across conditions assayed by RNA-seq experiments. Rockhopper computes two distributions based on the correlation coefficient of two consecutive genes’ expression levels across the assayed conditions [45]. The first distribution corresponds to correlations of consecutive genes on the same strand throughout the genome. The second distribution corresponds to correlations of consecutive genes on opposite strands throughout the genome. These two expression correlation distributions, like the two distributions of intergenic distances described in the previous section, enable Rockhopper to estimate the probability that two consecutive genes are part of the same operon or not, based on the relevant feature.
2.6. Probabilistic model for combining features
In order to combine information from different features suggestive of genes belonging to the same operon into a unified probabilistic model for predicting operons, Rockhopper employs a naïve Bayes classifier. Specifically, let g be consecutive genes on the same strand of a genome. Rockhopper models the probability that g corresponds to an operon, i.e., the genes in g are co-transcribed as part of the same polycistronic message, as
| (1) |
and the probability that g does not correspond to an operon as
| (2) |
The denominator in expressions (1) and (2) above is constant so, assuming independence of the d=2 different predictive features, the probabilities can be expressed as
| (3) |
and
| (4) |
respectively. Note, the products in expressions (3) and (4) above are over the d=2 features corresponding to intergenic distance and expression pattern, as described in the two previous sections above. The prior probability that consecutive genes on the same strand are co-transcribed as part of the same operon is estimated as 1.0 - (# of directons/number of pairs of genes on the same strand) [4]. As points of reference, the prior probability of two consecutive genes on the same strand being part of the same operon is computed by Rockhopper as 69% for genes in V. cholerae, 73% for genes in E. coli, 84% for genes in S. enterica, and 85% for genes in S. pyogenes. Once the probability of g being in the same operon and the probability of g not being in the same operon are computed, g is classified as an operon or not based on whichever probability is greater using the maximum a posteriori (MAP) decision rule
| (5) |
2.7. Evaluation metrics
When evaluating operon predictions, different metrics may be employed to assess the accuracy of the predictions. One possible metric considers whether all genes in an operon are correctly predicted or not. The challenge with such a metric is that some operons contain many genes, as indicated in Figure 1b, and the metric will not distinguish between approaches that correctly predict most but not all genes in an operon versus approaches that do not correctly predict any genes in an operon. Thus, rather than each entire operon as the basic unit of prediction and evaluation, gene pairs are used as the basic unit, where a gene pair is defined here as two consecutive genes on the same strand in a genome. For genome-wide operon predictions, all gene pairs in the genome are identified and a prediction is made for each as to whether the two genes in the pair are co-transcribed as part of the same operon or not. Assuming there exists some external source of information about operons in the genome, the predictions can then be compared to the external source to quantify how many gene pair predictions are correct. A true positive prediction corresponds to a gene pair predicted to be part of the same operon that is part of the same operon in the external source. A false positive prediction corresponds to a gene pair predicted to be part of the same operon that is not part of the same operon in the external source. A true negative prediction corresponds to a gene pair predicted not to be part of the same operon that is not part of the same operon in the external source. And a false negative prediction corresponds to a gene pair predicted not to be part of the same operon that is part of the same operon in the external source. The sensitivity (aka, recall or true positive rate) of the predictions is the percentage of gene pairs belonging to the same operon in the external source that are correctly predicted to be part of the same operon, i.e., the number of true positive predictions divided by the sum of true positive and false negative predictions. The specificity (aka, selectivity or true negative rate) of the predictions is the percentage of gene pairs not belonging to the same operon in the external source that are correctly predicted not to be part of the same operon, i.e., the number of true negative predictions divided by the sum of true negative and false positive predictions.
2.8. RNA-seq data
All RNA-seq data used in this study is publicly available from the Sequence Read Archive [21] with the following accession numbers: ERR2221428, SRR6158171, SRR7268670, SRR7852779 for Bacillus subtilis; SRR7226161, SRR7226168 for Caulobacter vibrioides; SRR031126, SRR031130 for Helicobacter pylori; SRR8142714, SRR8239645, SRR8284752, SRR8327205 for Pseudomonas aeruginosa; SRR3339292, SRR5891555, SRR6170086, SRR6170094 for Shewanella oneidensis; SRR7178745, SRR7298349 for Vibrio cholerae; SRR5456182, SRR5456180 for Streptococcus pyogenes; SRR794912, SRR794915, SRR794917, SRR794919 for Salmonella enterica; SRR794875, SRR794877 for Neisseria gonorrhoeae; SRR794872, SRR794869, SRR794866, SRR794863, SRR794860, SRR794857, SRR794854, SRR794851, SRR794848, SRR794845, SRR794842, SRR794839, SRR794836, SRR794833, SRR794830, SRR794827 for Escherichia coli.
2.9. Rockhopper usage and availability
Rockhopper is available from the website https://cs.wellesley.edu/~btjaden/Rockhopper. In addition to the source code, installation and executables are available for PCs and Macs, and a JAR file is available for any platform. Rockhopper is implemented using the Java programming language and Java version 1.6 or later is required for its execution. Rockhopper can be run either from the command line or via a graphical user interface (GUI). In the past year, Rockhopper was downloaded approximately seven thousand times from four thousand unique IP addresses.
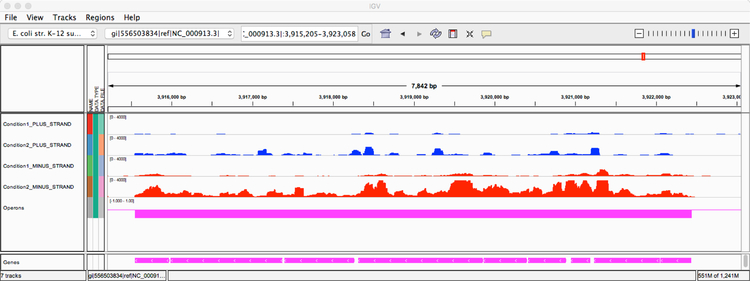
As output for operon predictions, Rockhopper generates a tab-delimited text file. Each line of the file corresponds to an operon prediction. Each operon prediction has five components: the nucleotide coordinate of the start of the coding sequence of the first gene in the operon, the nucleotide coordinate of the end of the coding sequence of the last gene in the operon, the strand of the operon, the number of genes in the operon, and a list containing the names of the genes in the operon. In addition to the text file containing information about all of the operon predictions, Rockhopper enables visualization of the results using the Integrative Genomics Viewer (IGV) genome browser [46]. Figure 3 illustrates the visual output of Rockhopper’s operon predictions along with the location of annotated genes and sequencing reads on both strands of the genome. Figure 3 focuses on a genomic region containing a nine gene operon that has been well studied and is experimentally confirmed [47–50]. In the figure, the first four tracks show that there is the largest expression for this region on the minus strand in the second experimental condition, and there is evidence of expression spanning all nine genes. As the fifth track indicates, Rockhopper predicts that all nine genes in this region are co-transcribed as part of the same operon. While the figure focuses on a specific region, the genome browser included as part of Rockhopper allows interactive exploration of Rockhopper’s operon predictions throughout a genome.
Figure 3.
The visualization created by Rockhopper using the IGV genome browser illustrates a 7,842 basepair region in the E. coli genome, including sequencing reads corresponding to this region from two experimental conditions. The first condition corresponds to minimal medium supplemented with 0.2% D-glucose (SRA SRR794972) and the second condition corresponds to LB medium exposed to 0.5% α-methylglucoside (SRA SRR794863). The region contains nine ATP synthase genes on the minus strand: atpI, atpB, atpE, atpF, atpH, atpA, atpG, atpD, atpC. There are six tracks (rows of information) displayed in the browser. The first track (in blue) indicates sequencing reads observed on the plus strand from the first experimental condition. The second track (in blue) indicates sequencing reads observed on the plus strand from the second experimental condition. The third track (in red) indicates sequencing reads observed on the minus strand from the first experimental condition. The fourth track (in red) indicates sequencing reads observed on the minus strand from the second experimental condition. The fifth track (in magenta) represents an operon prediction from Rockhopper. The sixth track (in magenta) at the very bottom indicates the locations of nine annotated genes on the minus strand in the E. coli genome.
3. Results
3.1. Evaluation of predictions against operon databases
We used Rockhopper to predict operons throughout the genomes of ten bacteria: Neisseria gonorrhoeae FA1090, Salmonella enterica subspecies enterica serovar Typhimurium strain LT2, Streptococcus pyogenes M1 GAS, Escherichia coli strain K-12 substrain MG1655, Caulobacter vibrioides CB15, Helicobacter pylori 26695, Pseudomonas aeruginosa PA01, Bacillus subtilis subsp. subtilis str. 168, Shewanella oneidensis MR-1, and Vibrio cholerae 01 biovar El Tor str. N16961. Table 2 shows the number of operons predicted by Rockhopper in each genome and the number of RNA-seq experiments used to make the predictions. The number of genes in a genome that Rockhopper predicted to be part of a multi-gene operon ranged from 38% for Caulobacter vibrioides to 80% for Vibrio cholerae. Rockhopper’s operon predictions were compared to the operon predictions from the Database of prOkaryotic OpeRons (DOOR2), a leading database of computationally predicted operons in bacteria [16]. The final column in Table 2 indicates the similarity, as measured by the Rand coefficient [51], between Rockhopper’s operon predictions and DOOR’s operon predictions. The overlap between the two sets of predictions ranged between 88% for V. cholerae to 95% for E. coli.
Table 2.
Information on operon predictions from Rockhopper for ten genomes is shown in the table. The first column indicates the genome. The second column indicates the number of RNA-seq experiments from which Rockhopper used data to make its operon predictions. The third column indicates the number of multi-gene operons (operons containing two or more genes) predicted by Rockhopper. The fourth column indicates the total number of genes in all the multi-gene operons predicted by Rockhopper. The fifth column indicates the total number of protein-coding genes in the genome as annotated by RefSeq [35]. The sixth column indicates the similarity of Rockhopper’s operon predictions to DOOR’s operon predictions [16] as measured by the Rand similarity coefficient [51].
| Genome | Number of RNA- Seq Exps |
Multi- Gene Operons |
Number of Genes Part of some Multi- Gene Operon |
Total Genes in Genome |
Similarity to DOOR Predictions |
|---|---|---|---|---|---|
|
Neisseria gonorrhoeae |
2 | 410 | 1114 | 1886 | 91% |
|
Salmonella enterica |
4 | 868 | 2677 | 4549 | 94% |
|
Streptococcus pyogenes |
2 | 412 | 1307 | 1693 | 94% |
|
Escherichia coli |
16 | 838 | 2557 | 4386 | 95% |
|
Caulobacter vibrioides |
2 | 780 | 1428 | 3737 | 92% |
|
Helicobacter pylori |
2 | 301 | 1111 | 1448 | 94% |
|
Pseudomonas aeruginosa |
4 | 1215 | 3755 | 5573 | 92% |
|
Bacillus subtilis |
4 | 880 | 2765 | 4328 | 90% |
|
Shewanella oneidensis |
4 | 724 | 2169 | 4266 | 90% |
|
Vibrio cholerae |
2 | 675 | 2011 | 3510 | 88% |
Operon predictions from both Rockhopper and DOOR were then evaluated against three external operon databases that are based on operons with experimental evidence: RegulonDB contains highly curated information about experimentally verified operons in E. coli [31], Sharma et al. produced an extensive annotation of operons in Helicobacter pylori [23], and DBTBS consists of experimentally supported operons in Bacillus subtilis [29]. Results from comparing Rockhopper’s and DOOR’s predictions to databases of experimentally supported operons are shown in Table 3. The sensitivities in Table 3 suggest that both Rockhopper and DOOR successfully identify most operons in a genome. The specificities in Table 3 suggest a low false positive rate for both approaches, i.e., when consecutive genes are not part of the same operon then the approaches successfully identify most of the genes as not being co-transcribed. Across the three databases, the sensitivity of Rockhopper’s predictions range from 88% to 95%, slightly higher than the sensitivity of DOOR’s predictions, which range from 84% to 93%. The specificity of predictions from Rockhopper and DOOR are comparable, ranging between 81% to 96% and between 80% to 95%, respectively. The slightly higher sensitivity of Rockhopper as compared to DOOR at a comparable specificity suggests that Rockhopper is identifying more operons as it has a higher number of true positive predictions without the cost of increased false positive predictions.
Table 3.
Rockhopper’s operon predictions are compared against external operon databases based on operons with experimental evidence. The first column indicates the genome. The second column indicates the external database that Rockhopper’s predictions are compared against. The third column indicates the sensitivity of DOOR’s operon predictions. The fourth column indicates the specificity of DOOR’s operon predictions. The fifth column indicates the sensitivity of Rockhopper’s operon predictions. The sixth column indicates the specificity of Rockhopper’s operon predictions.
To evaluate how the number of RNA-seq experiments might influence Rockhopper’s performance, we varied the number of E. coli RNA-seq data sets that we used from 2 to 16 when making operon predictions with Rockhopper. We used E. coli here because of the large number of RNA-seq data sets available for this organism and because there is an external database of operons, RegulonDB [31], that is well curated. The sensitivity of Rockhopper’s operon predictions when compared to operons in RegulonDB remained the same at 90% as the number of RNA-seq data sets used by Rockhopper increased from 2 to 16. The specificity of Rockhopper’s operon predictions increased slightly, from 79% to 81%, as the number of RNA-seq data sets used by Rockhopper increased from 2 to 16.
In order to better understand how different features impact the accuracy of Rockhopper’s operon predictions, we calculated the mean values of the features for different sets of predictions - those corresponding to true positive, false positive, true negative, and false negative predictions. Here, we used Rockhopper’s predictions from E. coli as compared to RegulonDB [31], as this is a comprehensive and regularly curated database of experimentally verified operons. For the feature of intergenic distance, the mean distance between pairs of E. coli genes that Rockhopper predicts as belonging to the same operon are 16 nucleotides and 55 nucleotides, for true positive and false positive predictions, respectively. The mean intergenic distance between pairs of E. coli genes that Rockhopper predicts as not belonging to the same operon are 237 nucleotides and 223 nucleotides, for true negative and false negative predictions, respectively. For the feature of expression correlation, the mean correlation of expression between pairs of E. coli genes that Rockhopper predicts as belonging to the same operon are 0.86 and 0.85, for true positive and false positive predictions, respectively. The mean correlation of expression between pairs of E. coli genes that Rockhopper predicts as not belonging to the same operon are 0.39 and 0.62, for true negative and false negative predictions, respectively. These results indicate a significant difference in expression between Rockhopper’s true negative predictions (0.39) and Rockhopper’s false negative predictions (0.62).
3.2. Experimental validation of operon predictions
We hypothesize that some of Rockhopper’s predictions that do not correspond to verified operons in external databases may indeed be genuine operons that have not been identified or validated previously. If this is the case, some of Rockhopper’s predictions that are counted as “false positives” and that contribute to a lower specificity score may indeed be “true positives” and their incorrect classification would be a consequence of comparing the predictions to incomplete operon databases. To test this hypothesis, we performed experiments to evaluate whether some of Rockhopper’s novel operon predictions, i.e., predictions of operons not found in the database, might correspond to genuine operons as opposed to being false predictions. We began by choosing 10 random sets of consecutive genes in E. coli according to the following criteria: each set of genes was predicted by Rockhopper to be co-transcribed as part of a multi-gene operon, some of the genes in the set are identified in RegulonDB as being part of the same operon whereas other genes in the set are not identified in RegulonDB as being part of the same operon, and the genes showed significant expression in the RNA-seq data. The 10 sets of genes ranged in size from 2 genes to 11 genes. For 13 pairs of consecutive genes within these sets that were identified in RegulonDB as being part of the same operon and for 14 pairs of consecutive genes within these sets that were not identified by RegulonDB as being part of the same operon, we performed RT-PCR to test for co-transcription. We found evidence from the RT-PCR experiments supporting co-transcription for 11 of the 13 pairs that RegulonDB reported to be part of the same operon, and we found evidence from the RT-PCR experiments supporting co-transcription for 12 of the 14 pairs that RegulonDB did not report to be part of the same operon [27]. To further understand whether some of Rockhopper’s predictions might correspond to operons that have not previously been identified, we similarly tested some of our novel operon predictions in N. gonorrhoeae using RT-PCR. For six pairs of genes in N. gonorrhoeae that Rockhopper predicted to be co-transcribed as part of the same operon, we tested the pairs for co-transcription using RT-PCR and found evidence of co-transcription for all six [27]. For two pairs of genes in N. gonorrhoeae that Rockhopper predicted not to be co-transcribed, we tested the pairs for co-transcription using RT-PCR and found evidence of co-transcription for one of the two pairs [27]. Thus, in two different genomes, one in which operons have been studied extensively (E. coli) and one in which there has been less examination of operon structures (N. gonorrhoeae), we found that a high percentage of Rockhopper’s novel operon predictions were supported by independent experimental evidence. However, there were also a small number of examples of previously verified operons for which we did not observe evidence of co-transcription in our RT-PCR experiments and one example where Rockhopper did not predict genes to be part of the same operon yet we observed evidence of co-transcription in our RT-PCR experiments. We interpret these findings to support the notion that Rockhopper’s operon predictions are largely accurate and are applicable to different bacterial genomes, but there is still room for improvement both in increasing the sensitivity of the predictions and reducing the false positive rate.
4. Discussion
We evaluated Rockhopper’s operon predictions by comparing them to databases of experimentally determined operons and to a leading computational approach. Rockhopper demonstrates sensitivities between 88% and 95% and specificities between 81% and 96% when its predictions are compared to databases of operons for three bacterial species. Rockhopper shows slightly improved sensitivity and comparable specificity when compared to DOOR 2.0, a leading method for operon prediction [16].
When looking more closely at individual features of Rockhopper’s operon predictions, we observed a large gap in intergenic distance between Rockhopper’s positive predictions (16 nucleotides for true positives and 55 nucleotides for false positives) and Rockhopper’s negative predictions (237 nucleotides for true negatives and 223 nucleotides for false negatives), as expected. Interestingly, for the feature of expression patterns, the gap between positive predictions (0.86 correlation of expression for true positives and 0.85 correlation of expression for false positives) and negative predictions (0.39 correlation of expression for true negatives and 0.62 correlation of expression for false negatives) was less dramatic. These results indicate the importance of combining multiple features for operon prediction, as individual features on their own cannot reliably distinguish co-transcribed genes from non-co-transcribed pairs of genes. Further, these data suggest that there may be opportunities to further improve the predictive accuracy of the system by differentially combining the features. Rockhopper’s Bayesian classification approach makes the simplifying assumption that features are independent of each other. However, further work may benefit from exploring more complex relationships among features.
One of the challenges with evaluating operon prediction methods is that there are few genomes for which large sets of operons have been experimentally confirmed. For the few such genomes where large sets of validated operons exist, it is unclear to what extent the set of confirmed operons is complete. Thus, assessment of predictions as true/false positives/negatives is imperfect. The situation is further complicated as many operons are dynamic in nature, with genes being co-transcribed in one condition but not in another. One study in E. coli found that 36% of operons had internal promoters and terminators that generated multiple transcription units [26]. Rockhopper makes static rather than dynamic operon predictions, though it can be used for dynamic predictions as well. One group built a database called OperomeDB of condition specific operons for more than a hundred bacterial transcriptomes based on Rockhopper’s operon predictions [52]. Another group developed a method for condition specific operon prediction and evaluated their results against Rockhopper’s operon predictions for three genomes, Porphyromonas gingivalis, Escherichia coli, and Salmonella enterica, observing that Rockhopper consistently led to more accurate operon predictions than the condition specific approach [12].
As further evaluation, we assessed Rockhopper’s predictions for two genomes using RT-PCR. For many of Rockhopper’s predictions that did not correspond to known operons and which we had labeled as false positive predictions when comparing them to various databases, we observed evidence that the genes were co-transcribed as part of the same operon and that a number of Rockhopper’s novel operon predictions correspond to previously unknown operons.
Rockhopper’s method for predicting operons throughout a genome has been evaluated by other groups and applied to a broad range of bacteria. In an interesting application, Rockhopper was used to identify operons in E. coli that responded to colorectal cancer [53]. Rockhopper’s operon identifications have been investigated in a wide range of other bacteria and archaea such as Stenotrophomonas maltophilia [54], Methanolobus psychrophilus [55], Listeria monocytogenes [56], Streptomyces avermitilis [57], Clostridium acetobutylicum [58], Oenococcus oeni [59], Lactococcus lactis [60], Streptococcus pneumoniae [61], and Leptospira interrogans [62].
An alternative to Rockhopper’s approach for operon prediction is to use features that are genome-specific or that have variable success in different genomes, such as computationally predicting transcription signals including promoter regions, transcription factor binding sites, and rho-independent terminators. In contrast, Rockhopper was designed so that it can be used for operon prediction in any bacterial genome for which RNA-seq data is available. Our results indicate that the accuracies of its predictions are relatively stable across different genomes.
5. Conclusion
Operons are one mechanism by which organisms can coordinate the expression of proximal genes. Understanding which genes are co-transcribed as part of a single polycistronic message provides information about the functional relationship and regulation of the genes. Computational methods have proven useful as efficient tools for predicting operons throughout a genome. Here, we present one such computational method, Rockhopper. Rockhopper uses RNA-seq data, which is increasingly being generated, to aid in its operon predictions. Rockhopper combines primary sequence information and RNA-seq data in a unified probabilistic model in order to make operon predictions in a genome. Rockhopper demonstrates high accuracy across a range of bacterial genomes. We hope that Rockhopper serves as a useful tool to the community for accurate, efficient, and user-friendly operon identification.
Highlights.
Integrated bioinformatics system for identifying operons in bacterial genomes
Combining computational and experimental data improves accuracy of operon prediction
Provides insight into co-expressed genes, functional relationships, and regulatory networks
Acknowledgments
This work was supported by the National Institutes of Health grant R15 GM102755 (to B.T.). The author would like to thank members of the Rockhopper user community for their valuable feedback on the system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacob F, et al. , The operon: a group of genes whose expression is coordinated by an operator. Comptes Rendus de l’Academie des Sciences, 1960. 250: p. 1727–1729. [PubMed] [Google Scholar]
- 2.Pettitt J, et al. , Operons are a conserved feature of nematode genomes. Genetics, 2014. 197(4): p. 1201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haller R, et al. , The transcriptome of Mycobacterium tuberculosis. Appl Microbiol Biotechnol, 2010. 86(1): p. 1–9. [DOI] [PubMed] [Google Scholar]
- 4.Westover BP, et al. , Operon prediction without a training set. Bioinformatics, 2005. 21(7): p. 880–888. [DOI] [PubMed] [Google Scholar]
- 5.Brouwer RWW, Kuipers OP, and van Hijum SAFT, The relative value of operon predictions. Briefings in Bioinformatics, 2008. 9(5): p. 367–375. [DOI] [PubMed] [Google Scholar]
- 6.Chuang LY, et al. , Features for computational operon prediction in prokaryotes. Brief Funct Genomics, 2012. 11(4): p. 291–9. [DOI] [PubMed] [Google Scholar]
- 7.Taboada B, et al. , Operon-mapper: A Web Server for Precise Operon Identification in Bacterial and Archaeal Genomes. Bioinformatics, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solovyev V and Salamov A, Automatic annotation of microbial genomes and metagenomic sequences, in Metagenomics and its Applications in Agriculture, Li RW, Editor. 2011, Nove Science Publishers, Inc.: Hauppauge; p. 61–78. [Google Scholar]
- 9.Harayama S, Codon usage patterns suggest independent evolution of two catabolic operons on toluene-degradative plasmid TOL pWW0 of Pseudomonas putida. J Mol Evol, 1994. 38(4): p. 328–35. [DOI] [PubMed] [Google Scholar]
- 10.Bockhorst J, et al. , A Bayesian network approach to operon prediction. Bioinformatics, 2003. 19(10): p. 1227–1235. [DOI] [PubMed] [Google Scholar]
- 11.Price MN, et al. , A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res, 2005. 33(3): p. 880–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortino V, et al. , Transcriptome dynamics-based operon prediction in prokaryotes. BMC Bioinformatics, 2014. 15: p. 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taboada B, Verde C, and Merino E, High accuracy operon prediction method based on STRING database scores. Nucleic Acids Res, 2010. 38(12): p. e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galperin MY, et al. , Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res, 2015. 43(Database issue): p. D261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertea M, et al. , OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res, 2009. 37(Database issue): p. D479–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao X, et al. , DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Research, 2014. 42: p. D654–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taboada B, et al. , ProOpDB: Prokaryotic Operon DataBase. Nucleic Acids Res, 2012. 40(Database issue): p. D627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidi SSA and Zhang X, Computational operon prediction in whole-genomes and metagenomes. Brief Funct Genomics, 2017. 16(4): p. 181–193. [DOI] [PubMed] [Google Scholar]
- 19.Sabatti C, et al. , Co-expression pattern from DNA microarray experiments as a tool for operon prediction. Nucleic Acids Research, 2002. 30(13): p. 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tjaden B, et al. , Identifying operons and untranslated regions of transcripts using Escherichia coli RNA expression analysis. Bioinformatics, 2002. 18: p. S337–S344. [DOI] [PubMed] [Google Scholar]
- 21.Kodama Y, Shumway M, and Leinonen R, The sequence read archive: explosive growth of sequencing data. Nucleic Acids Research, 2012. 40(D1): p. D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.SRA Database Growth. Available from: https://www.ncbi.nlm.nih.gov/sra/docs/sragrowth/.
- 23.Sharma CM, et al. , The primary transcriptome of the major human pathogen Helicobacter pylori. Nature, 2010. 464: p. 250–255. [DOI] [PubMed] [Google Scholar]
- 24.Pelly S, et al. , REMap: Operon map of M. tuberculosis based on RNA sequence data. Tuberculosis (Edinb), 2016. 99: p. 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slager J, Aprianto R, and Veening JW, Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Res, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conway T, et al. , Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. MBio, 2014. 5(4): p. e01442–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClure R, et al. , Computational analysis of bacterial RNA-Seq data. Nucleic Acids Research, 2013. 41(14): p. e140–e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vey G and Charles TC, MetaProx: the database of metagenomic proximons. Database, 2014: p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sierro N, et al. , DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res, 2008. 36(Database issue): p. D93–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuda S and Yoshizawa AC, ODB: a database for operon organizations, 2011 update. Nucleic Acids Res, 2011. 39(Database issue): p. D552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos-Zavaleta A, et al. , A unified resource for transcriptional regulation in Escherichia coli K-12 incorporating high-throughput-generated binding data into RegulonDB version 10.0. BMC Biol, 2018. 16(1): p. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guell M, et al. , Transcriptome complexity in a genome-reduced bacterium. Science, 2009. 326: p. 1268–1271. [DOI] [PubMed] [Google Scholar]
- 33.Sorek R and Cossart P, Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nature Reviews Genetics, 2010. 11: p. 9–16. [DOI] [PubMed] [Google Scholar]
- 34.Tjaden B, De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biology, 2015. 16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haft DH, et al. , RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res, 2018. 46(D1): p. D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, and Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014. 30(15): p. 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B and Salzberg S, Fast gapped-read alignment with Bowtie 2. Nature Methods, 2012. 9: p. 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferragina P and Manzini G, Opportunistic data structures with applications. Proceedings of the 41st Annual Symposium on Foundations of Computer Science, 2000: p. 390–398. [Google Scholar]
- 39.Burrows M and Wheeler DJ, A Block Sorting Lossless Data Compression Algorithm 1994, Technical Report 124. Palo Alto, CA: Digital Equipment Corporation. [Google Scholar]
- 40.Hiller D, et al. , Identifiability of isoform deconvolution from junction arrays and RNASeq. Bioinformatics, 2009. 25(23): p. 3056–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, et al. , Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol, 2013. 31(1): p. 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia X, ARSDA: A New Approach for Storing, Transmitting and Analyzing Transcriptomic Data. G3 (Bethesda), 2017. 7(12): p. 3839–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blattner FR, et al. , The complete genome sequence of Escherichia coli K-12. Science, 1997. 277(5331): p. 1453–62. [DOI] [PubMed] [Google Scholar]
- 44.Epanechnikov VA, Non-parametric estimation of a multivariate probability density. Theory of Probability and its Applications, 1969. 14(1): p. 153–158. [Google Scholar]
- 45.Pearson K, Notes on regression and inheritance in the case of two parents. Proceedings of the Royal Society of London, 1895. 58: p. 240–242. [Google Scholar]
- 46.Thorvaldsdottir H, Robinson JT, and Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform, 2013. 14(2): p. 178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones HM, Brajkovich CM, and Gunsalus RP, In vivo 5’ terminus and length of the mRNA for the proton-translocating ATPase (unc) operon of Escherichia coli. J Bacteriol, 1983. 155(3): p. 1279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen J, et al. , The promoters of the atp operon of Escherichia coli K12. Mol Gen Genet, 1984. 193(1): p. 64–71. [DOI] [PubMed] [Google Scholar]
- 49.Walker JE, et al. , DNA sequence around the Escherichia coli unc operon. Completion of the sequence of a 17 kilobase segment containing asnA, oriC, unc, glmS and phoS. Biochem J, 1984. 224(3): p. 799–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasimoglu E, et al. , Transcriptional regulation of the proton-translocating ATPase (atpIBEFHAGDC) operon of Escherichia coli: control by cell growth rate. J Bacteriol, 1996. 178(19): p. 5563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rand WM, Objective criteria for the evaluation of clustering methods. Journal of the American Statistical Association, 1971. 66: p. 846–850. [Google Scholar]
- 52.Chetal K and Janga SC, OperomeDB: A Database of Condition-Specific Transcription Units in Prokaryotic Genomes. Biomed Res Int, 2015. 2015: p. 318217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arthur JC, et al. , Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun, 2014. 5: p. 4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernardini A and Martinez JL, Genome-wide analysis shows that RNase G plays a global role in the stability of mRNAs in Stenotrophomonas maltophilia. Sci Rep, 2017. 7(1): p. 16016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, et al. , Global mapping transcriptional start sites revealed both transcriptional and post-transcriptional regulation of cold adaptation in the methanogenic archaeon Methanolobus psychrophilus. Sci Rep, 2015. 5: p. 9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lobel L and Herskovits AA, Systems Level Analyses Reveal Multiple Regulatory Activities of CodY Controlling Metabolism, Motility and Virulence in Listeria monocytogenes. PLoS Genet, 2016. 12(2): p. e1005870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liot Q and Constant P, Breathing air to save energy--new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis. Microbiologyopen, 2016. 5(1): p. 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venkataramanan KP, et al. , Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum. Biotechnol Biofuels, 2015. 8: p. 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternes PR, et al. , Whole transcriptome RNAseq analysis of Oenococcus oeni reveals distinct intra-specific expression patterns during malolactic fermentation, including genes involved in diacetyl metabolism. Int J Food Microbiol, 2017. 257: p. 216–224. [DOI] [PubMed] [Google Scholar]
- 60.van der Meulen SB, de Jong A, and Kok J, Transcriptome landscape of Lactococcus lactis reveals many novel RNAs including a small regulatory RNA involved in carbon uptake and metabolism. RNA Biol, 2016. 13(3): p. 353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warrier I, et al. , The Transcriptional landscape of Streptococcus pneumoniae TIGR4 reveals a complex operon architecture and abundant riboregulation critical for growth and virulence. bioRxiv, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhukova A, et al. , Genome-Wide Transcriptional Start Site Mapping and sRNA Identification in the Pathogen Leptospira interrogans. Front Cell Infect Microbiol, 2017. 7: p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]