Abstract
Objectives:
To demonstrate the utility of a natural language processing (NLP) algorithm for mining kidney stone composition in a large-scale electronic health records (EHR) repository.
Methods:
We developed StoneX, a pattern matching method for extracting kidney stone composition information from clinical notes. We trained the extraction algorithm on manually annotated text mentions of calcium oxalate monohydrate (CaOxM), calcium oxalate dihydrate (CaOxD), hydroxyapatite, brushite, uric acid, and struvite stones. We employed StoneX to identify patients with kidney stone composition data and mine >125 million notes from the our institutional EHR. Analyses performed on the extracted patients included stone type conversions overtime, survival analysis from a second stone surgery, and disease associations by stone composition to validate the phenotyping method against known associations.
Results:
The NLP algorithm identified 45,235 text mentions corresponding to 11,585 patients. Overall, the system achieved PPVs >90% for CaOxM, CaOxD, hydroxyapatite, brushite, and struvite; except for uric acid (PPV=87.5%). Survival analysis from a second stone surgery showed statistically significant differences among stone types (P=0.03). Several phenotype associations were found: uric acid–type 2 diabetes (odds ratio, OR=2.69, 95% confidence intervals, CI=1.91–3.79), struvite–neurogenic bladder (OR=12.27, 95% CI=4.33–34.79), struvite–urinary tract infection (OR=7.36, 95% CI=3.01–17.99), hydroxyapatite–pulmonary collapse (OR=3.67, 95%CI=2.10–6.42), hydroxyapatite–neurogenic bladder (OR=5.23, 95% CI=2.05–13.36), brushite–calcium metabolism disorder (OR=4.59, 95% CI=2.14–9.81), and brushite–hypercalcemia (OR=4.09, 95% CI=1.90–8.80).
Conclusions:
NLP extraction of kidney stone composition from large-scale EHRs is feasible with high precision, enabling high-throughput epidemiological studies of kidney stone disease. These tools will enable high fidelity kidney stone research from the EHR.
Keywords: kidney calculi, natural language processing, precision phenotyping, electronic health records
INTRODUCTION
Kidney stones affect 9% individuals within the United States.1 Over the last few decades, disease prevalence has dramatically risen in all demographic groups.1, 2 Along with the widespread adoption of electronic health records (EHRs) in healthcare, there has been an increasing recognition that EHRs enable the study of variability in disease dynamics and heterogeneity. Phenotypes describe clinical conditions within the EHR, and they are created using a defined set of data elements generated from a computerized query. Specifically for kidney stone disease, methods to phenotype stone formers within large EHR datasets are nonexistent. As in studies utilizing administrative datasets, the identification of kidney stone patients within the EHR is currently limited to using nonspecific administrative coding (e.g., International Classification of Diseases, ICD). While these codes have high validity (PPV >95%),3 no tools are available for more rigorous phenotyping. As a result, detailed data relevant to stone disease are buried within multiple documents and across multiple data points. Large datasets utilized for kidney stone research have to date been unable to advance precision medicine applications for stone disease, in part, because they lack additional phenotyping information, such as stone composition data.
Over the last 2 decades, natural language processing (NLP) methods have been applied in various clinical applications due to the fact that a vast amount of relevant clinical information is stored as free-text form into the EHRs. NLP and machine learning methodologies have been also applied to specific urologic conditions. They include information extraction methods from radical prostatectomy pathology reports,4 identification of patients with prostate biopsies based on information encoded in pathology reports,5 assessment of bladder cancer pathology reports,6 and machine learning algorithms to diagnose kidney stones based on laboratory, vital signs, and demographic information.7
We have developed an NLP method that uses a pattern-matching algorithm for stone composition eXtraction (StoneX) across our institutional EHR. The rationale for a stone composition phenotyping method was as follows. First, as in most institutions, our, stone analyses are typically performed via infrared spectroscopy and are performed at dedicated commercial stone analysis laboratories.8 The test results are not discrete laboratory data within the EHR, but rather contained within a written report that is transcribed by clinicians into unstructured clinical notes. Second, stone composition and urine biochemistries comprise important clinical data that guide preventative and therapeutic treatment based on underlying pathogenesis mechanisms and disease severity.9–11 Finally, our goal is to lay the groundwork for a generalizable NLP algorithm using a clinically accepted delineator of stone disease that can be investigated within other institutional EHRs.
MATERIAL AND METHODS
Study Population
Large-scale data mining is enabled by the Vanderbilt Synthetic Derivative (SD), which is a research-oriented data repository that stores the de-identified version of the Vanderbilt EHR.12 Currently, the SD contains clinical records of 2.9 million patients with >1 billion distinct observations dating back to the 1980s and >125 million clinical notes since 1990s enabling researchers to perform longitudinal studies based on quantitative research methods. The database includes diagnostic and procedure (ICD and CPT) codes, basic demographics (age, gender, race), text from clinical care including discharge summaries, progress notes, problem lists, laboratory values, radiology reports, and medication orders. Out of the total number of patients in the SD, 29,739 have upper urinary tract stone disease diagnoses based on ICD codes. To maintain the de-identified nature of the SD, the SD does not contain outside laboratory scanned records, such as reports of stone composition. Local IRB approval was obtained for this study.
StoneX Design
The StoneX architecture (see Supplementary Figure 1) was designed on top of a supervised machine learning framework, which is described in greater detail below.
Manual Chart Review of Clinical Notes
Manual chart review was performed on 400 randomly sampled notes with mentions of “stone analysis” and “kidney stone”. These notes correspond to 356 distinct patients, with each note having on average 820.3 words. Two clinicians reviewed the notes and manually annotated a total number of 921 text expressions denoting a kidney stone composition (i.e., 2.3 stone composition mentions per note). The chart review was performed using BRAT, a web-based tool for visualization and text annotation.13 Mentions with both % and non-% values of stone compositions were identified for calcium oxalate monohydrate (CaOxM), calcium oxalate dihydrate (CaOxD), hydroxyapatite, brushite, uric acid, and struvite. Supplementary Figure 2 depicts a screenshot of BRAT with two annotations for % hydroxyapatite and % CaOxM. A total of 100 notes were double-reviewed with a high inter-rater agreement (Cohen’s kappa=0.85).
Extraction of Kidney Stone Composition from Clinical Text
We designed our approach using a standard machine learning framework. We randomly selected 70% of the 400 manually reviewed notes for training (training set) and the remaining 30% for evaluation (test set). To automatically check the annotated text mentions of kidney stone composition for the presence of predefined templates, we implemented a text processing method called pattern matching. We represented these patterns with regular expressions, or rules, by analyzing the textual context of kidney stone mentions in the notes.
During the training phase, we performed an optimization process to learn the set of rules that best match the annotated text expressions from the training set (Rule Optimization in Supplementary Figure 1). First, we automatically constructed an exhaustive set of rules such that each annotated mention from the training set is matched by at least one rule from the rule set. Next, we iteratively refined the rules for each stone composition with the goal of maximizing the algorithm performance. The most challenging problem we needed to solve during this step resembles the word-sense disambiguation problem from computational linguistics,14,15 where, instead of disambiguating polysemous words, our goal was to disambiguate rules that match other medical concepts in addition to kidney stone composition concepts. Our solution to this problem was to impose additional constraints on the ambiguous regular expressions after a manual analysis of their matched text expressions. Finally, we used backward elimination to extract the final set of rules that best matched the annotated expressions from the training set. The rules learned during the training phase are listed in Supplementary Table 1.
On the test set, we compared the manually annotated mentions with the ones extracted by the final set of rules and reported the algorithm performance in terms of positive predictive value (PPV), sensitivity, and F1-score, where the F1-score is defined as the harmonic mean between PPV and sensitivity.
Large-Scale Extraction of Kidney Stone Composition
We applied StoneX over all 125 million notes in the EHR to identify all patients with kidney stone composition information. For each stone composition mention identified in a patient note, we recorded the patient identifier, note timestamp, stone composition, and % composition value. Using this phenotyping method, we created a data structure with timestamped kidney stone composition information for each kidney stone patient, in addition to demographic data, ICD, and CPT codes.
Data and Statistical Analyses
We performed several analyses to validate the phenotyping method against known disease patterns and associations. First, we calculated the distribution of kidney stone prevalence by age, race, and sex as well as patient distributions by the number of kidney stone procedures as indicated by the presence of specific CPT codes (Supplementary Table 2). We performed the one-way analysis of variance (ANOVA) to determine whether there were statistically significant differences in the age at first stone composition among the 6 kidney stone types, as well as a post-hoc analysis of multiple comparisons between pairs of stone types based on the pairwise t-test with Bonferroni correction for multiple testing. We employed the Cochran’s Q test to determine whether the patient proportions among the 6 stone types were significantly different followed by a pairwise comparison using the Wilcoxon signed-rank test with Bonferroni correction.
Next, we conducted a survival analysis for an additional kidney stone surgery after a first identified surgery. The outcome of interest was the time from the date of the first kidney stone intervention to the date of the second stone intervention. Patients without a second kidney stone intervention were censored at the date of their last record in the EHR. Patients with multiple CPT codes spanned in <1 month since the date of the first intervention were excluded to account for staged procedures. We employed the Kaplan-Meier method16 to evaluate the survival rate from the first to the second kidney stone surgery and the log-rank test to assess differences between survival curves.
Finally, we performed multiple case-control studies to investigate associations between clinical phenotypes and each of the 6 stone compositions. Patients selected for this analysis had pure or mixed stones >50% composition value that did not change over time; thus, each patient from the analysis mainly presented the clinical characteristics of only one of the 6 stone compositions. The exposed cohort of patients selected for a specific case-control study associated with a stone composition were the patients with the specific stone composition. All the other patients selected for the above-mentioned study constituted the unexposed cohort. We extracted cases and controls associated with the top 50 most prevalent ICD9 codes for each stone composition. The codes pertaining to kidney-stone related symptoms or procedures were excluded by manual review. A final set of 14 ICD9 codes was selected as phenotype variables (Supplementary Table 3). For each phenotype-stone composition pair, we performed multivariate logistic regression adjusting for age at first stone composition mention, race, sex, and thiazide status (Supplementary Table 4), and reported odds ratio (OR) estimates with corresponding 95% confidence intervals (CIs). We included thiazide status into the model was to avoid possible spurious associations with hypertension phenotypes. To account for multiple testing, we Bonferroni-adjusted the significance threshold by dividing α=0.05 to the total number of independent tests (i.e., 5.95 × 10−4 = 0.05 / (14 × 6)). All analyses were performed with R version 3.4.2.
RESULTS
StoneX Evaluation
Table 1 lists the results achieved by StoneX after applying the optimized rules for each stone composition on the test set. The system achieved PPVs >90% for all stone compositions except for % uric acid (PPV=87.5%). Our error analysis revealed that most of the false positives for this stone composition were mainly due to mislabeling of urinary uric acid mentions as uric acid kidney stone events. The majority of false negatives correspond to misspellings and mentions expressed in text in an unusual format (e.g., “55%--{calcium oxalate monohydrate}”) only in the notes from the test set. Additional misclassifications were noticed in enumerations of stone composition mentions where the percent values were placed at the end.
Table 1.
Performance of StoneX on kidney stone composition identification in clinical notes.
| Stone composition | PPV | Sensitivity | F1-score |
|---|---|---|---|
| % CaOxM | 94.9 | 90.2 | 92.5 |
| % CaOxD | 93.8 | 83.3 | 88.2 |
| % Hydroxyapatite | 90.9 | 90.9 | 90.9 |
| % Brushite | 100.0 | 80.0 | 88.9 |
| % Uric acid | 87.5 | 100.0 | 93.3 |
| % Struvite | 100.0 | 100.0 | 100.0 |
CaOxM, calcium oxalate monohydrate; CaOxD, calcium oxalate dihydrate
Demographics and Stone Types
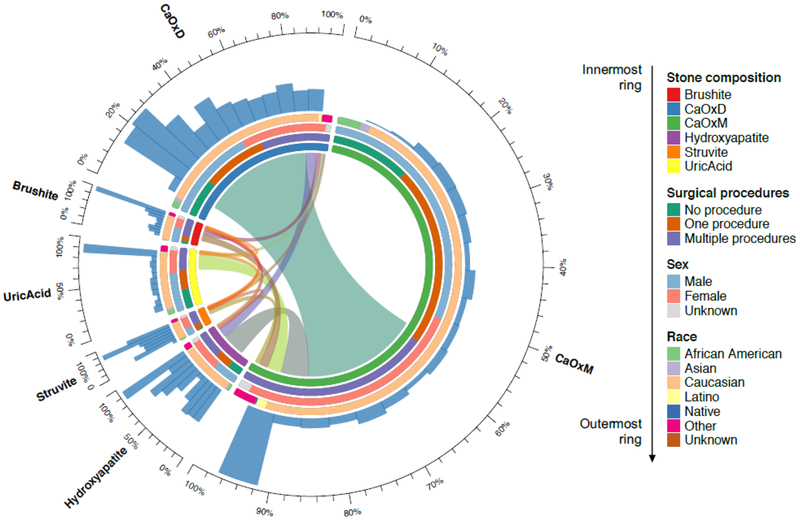
StoneX identified 45,235 text mentions of both % and non-% stone types corresponding to 11,585 patients across >125 million notes. To extract non-% mentions we used the optimized rules without the percent value information. From the extracted 11,585 patients, we performed statistical analysis on 2,417 patients with at least one stone type mention with a percent value. The circos plot from Figure 1 shows baseline statistics for this cohort. Here, patients with mixed stone compositions were included in multiple categories. Most of the patients are male (52.1%) and had CaOxM stones (N=1,965). The histograms on the outermost ring of the circos plot indicate that all stone types except CaOxD have the patient distribution’s peak in the 90–100% composition value range. The chord diagram depicted inside the innermost ring of the circus plot shows that 56% of CaOxM patients have only this stone type while 30% of them have also the CaOxD stone component.
Figure 1.
Circos plot with the analysis of 2,417 kidney stone patients from the entire Vanderbilt HER using StoneX. Patients with missing stone composition subtype percentages were excluded. In the plot, each ring represents a specific analysis and each sector corresponds to one stone type. From innermost to outermost: The center of the plot shows a visual representation of stone type link. Each link width is proportional to the percent of patients with the corresponding stone types. The width of the sector area with no links represents the proportion of patients with only the corresponding stone composition. For example, a large proportion of calcium oxalate monohydrate stones also had calcium oxalate dihydrate, and vice versa. A large proportion of calcium oxalate monohydrate stones were 100% pure. Moving outward, the next 4 colored rings from outer to inner show distributions by stone composition, surgical procedures (none, one, multiple), sex, and race. Further outward shows histograms with proportions of stones with respective percent compositions.
Age at first stone composition was youngest in the brushite and hydroxyapatite groups (see Supplementary Tables 5 and 6). The ANOVA test showed a statistically significant difference in the age at first stone composition among the 6 stone types (P<0.001), while the pairwise t-test revealed a statistically significant difference between CaOxM and each of the other stone types (each P<0.001). Similarly, the Cochran’s Q test indicated significant differences among the stone type proportions (P<0.001) and the Wilcoxon signed-rank test showed significant differences between all pairwise comparisons (each P<0.001). There was a female predominance in the hydroxyapatite and struvite groups (54.5% and 59.2%, respectively). Brushite and struvite stone formers had the highest prevalence of having had multiple surgeries (69.0% and 61.8%, respectively). Supplementary Figure 3 shows the distribution of patients with combinations pure and mixed stone types.
The alluvial diagram in Supplementary Figure 4 visualizes the stone type conversions overtime. This analysis was restricted to pure and mixed stone formers with >50% of a dominant stone type (N=116). The most prominent interconversions in the alluvial diagram are CaOxM↔CaOxD and CaOxM↔uric acid.
Survival Analysis from the Second Stone Surgery
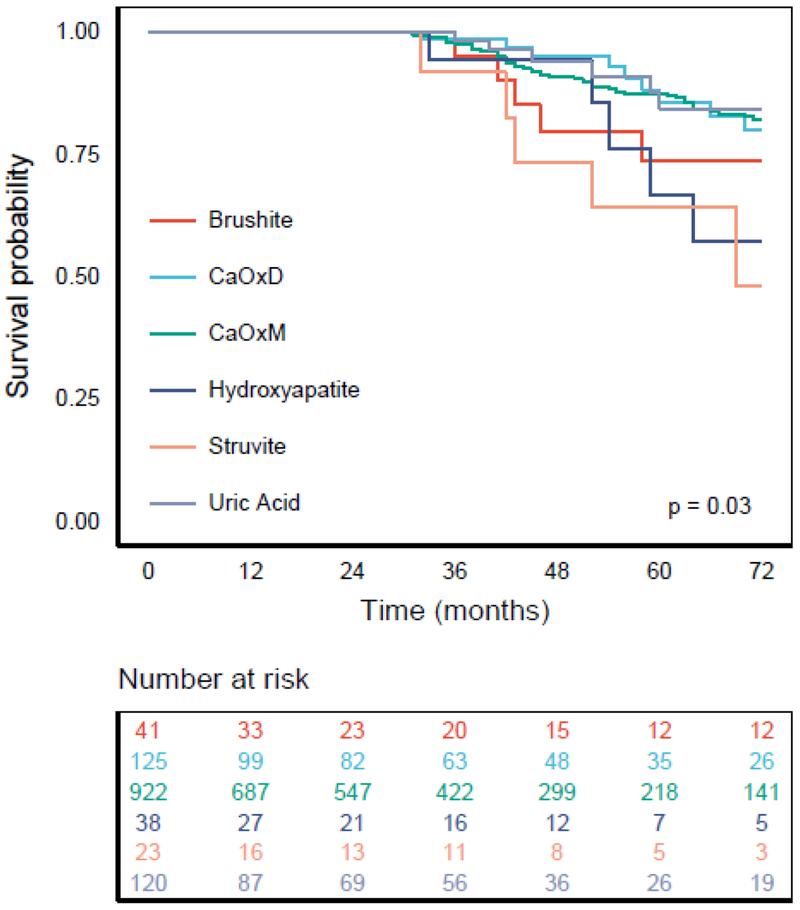
We identified 1,269 patients having only one major stone composition (>50%) and having had at least one stone-related procedure. Figure 2 shows the Kaplan-Meier survival curves by stone type using an observation period of six years since the first kidney stone procedure. We obtained significant survival differences among the stone types (P=0.03). Median survival time for patients with struvite stones was 69 months, and it was not reached for any of the other groups.
Figure 2.
Kaplan-Meier survival estimates from a second kidney stone surgery. Median survival time for patients with struvite stones was 69 months, while this was not reached for other stone types. Log-rank test showed a difference in survival among the groups (P=0.03).
Phenome Association Studies
Phenotype associations were performed on 1,583 patients with pure or mixed stones >50% that did not change over time. Out of the 84 studies performed (14 phenotypes × 6 stone types), 7 associations were found significant after Bonferroni correction (Table 2). The most significant association was between uric acid and type 2 diabetes (OR=2.69, 95% CI=1.91–3.79, P=1.62×10−8), where 40.7% of patients with uric acid stones had type 2 diabetes. Additional significant studies include associations between struvite and neurogenic bladder (OR=12.27, 95% CI=4.33–34.79, P=2.40×10−6) and between struvite and personal history of urinary tract infections (OR=7.36, 95% CI=3.01–17.99, P=1.19×10−5).
Table 2.
Phenotype associations grouped by stone composition.
| Stone composition | ICD9 code | ICD9 code description | Cases | Controls | Odds ratio (95% CI) | P-value |
|---|---|---|---|---|---|---|
| Uric Acid | 250.00 | Diabetes mellitus without mention of complication, type II or unspecified type, not stated as uncontrolled | 319 | 1264 | 2.69 (1.91, 3.79) | 1.62×10−8 |
| Struvite | 596.54 | Neurogenic bladder NOS | 43 | 1540 | 12.27 (4.33, 34.79) | 2.40×10−6 |
| V13.02 | Personal history, urinary (tract) infection | 146 | 1437 | 7.36 (3.01, 17.99) | 1.19×10−5 | |
| Hydroxya patite | 518.0 | Pulmonary collapse | 266 | 1317 | 3.67 (2.10, 6.42) | 5.06×10−6 |
| 596.54 | Neurogenic bladder NOS | 43 | 1540 | 5.23 (2.05, 13.36) | 5.43×10−4 | |
| Brushite | 275.40 | Unspecified disorder of calcium metabolism | 81 | 1502 | 4.59 (2.14, 9.81) | 8.64×10−5 |
| 275.42 | Hypercalcemia | 102 | 1481 | 4.09 (1.90, 8.80) | 3.08×10−4 | |
COMMENT
The NLP-based method proposed in this study accurately extracted kidney stone composition expressions from the notes in our institutional EHR. Specifically, our evaluation showed that stone composition expressions could be identified in clinical text with high precision for the main stone compositions. Furthermore, the large-scale extraction of stone composition from 125 million clinical notes demonstrated that this algorithm has the ability to identify kidney stone patients in big EHR repositories.
Our analyses replicated previous results and explored several applications revealing novel findings. The distribution of stone types identified is similar to other published series with predominance of calcium-based stone types.17,18 Similar to previous studies, we found that calcium oxalate and uric acid stones are predominant in males, struvite and hydroxyapatite stones are more common in females, and uric acid stones occur later in age in both genders.19,20 An interesting finding enabled by the longitudinal health records is the visual analysis for capturing major trends of stone type conversions over time. While stone type conversions have been previously studied in isolation,21,22 the alluvial diagram provides a holistic approach for this type of study. The availability of large longitudinal health records also enabled a specific type of survival analysis, survival from the second stone surgery, which has been previously shown difficult to perform for rare stone types.23 Our data would indicate that struvite stone formers, followed by hydroxyapatite and brushite stone formers are at higher risk for recurrence defined by surgical intervention, compared to calcium oxalate and uric acid stone formers. These data may be helpful towards informing surveillance protocols after surgical intervention. For example, struvite stone formers may benefit from more frequent diagnostic imaging after surgery to determine early stone recurrences, compared to calcium oxalate stone formers.
Furthermore, the clinical utility of our phenotyping method was demonstrated though multiple phenotype associations for each stone composition. For example, the association between uric acid and type 2 diabetes has been well studied, linking uric acid stone disease as an indicator of metabolic syndrome 24,25. The hydroxyapatite–pulmonary collapse association we detected in our experiments has been previously unreported. A possible explanation is that hydroxyapatite stone formers often have more comorbidities including urinary tract infections leading to inpatient hospitalization and therefore more pulmonary related diagnoses. Pulmonary atelectasis is often coded with the same ICD code, and is a common diagnosis after inpatient surgery. Furthermore, previous studies have reported the higher prevalence of struvite and hydroxyapatite stones in patients with neurogenic bladder,26 a recognized contribution of urinary tract infection to struvite formers,27 and a high rate of calcium metabolic abnormality in patients with brushite stones.28 These findings warrant additional investigation into how disease associations inform modifiable risk factors and predict disease based on stone subtype.
Future directions of this work include improving stone composition extraction using a context-based machine learning approach and externally validating our method on EHR systems in other institutions. We also plan to build on our previous work to improve kidney stone composition extraction using assertion classification29 and distributional word embedding models.30 Finally, we plan to expand our algorithm to identify stone events from specific sources of information including surgery, emergency room visits, passed stones, radiographic stone disease, and 24-hour urine data.
Our approach has a number of limitations. First, the extraction of stone composition from notes is sensitive to transcription errors that may occur when stone analysis reports are interpreted. Second, rule-based methods are limited in capturing the surrounding contextual information of specific text expressions. This limitation partially explains the lower performance values achieved for stone composition mentions that occur in multiple contexts. The findings from this work will need external validation. Moreover, despite being commonly used in high-throughput observational studies, we acknowledge the limitation of the phenotyping algorithm using only ICD codes to extract the case/control disease outcomes.
CONCLUSIONS
We demonstrate high performance and clinical utility of an NLP algorithm for large-scale extraction of kidney stone composition from millions of clinical notes. We proposed a visual analysis approach to capture stone type conversions over time and showed a difference in survival from a second stone surgery by stone composition. We conducted a landscape of phenotype associations on kidney stone patients showing the potential of our approach for efficient and cost-effective discovery of novel associations and replication of previous findings by stone composition. Additional applications that could be facilitated by our phenotype algorithm include prognosis and prediction models for kidney stone disease.
Supplementary Material
Acknowledgements:
This study was supported by CTSA award No. UL1 TR002243 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
REFERENCES
- 1.Scales CD Jr., Smith AC, Hanley JM, et al. Prevalence of kidney stones in the United States. Eur Urol. 2012;62:160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tasian GE, Ross ME, Song LH, et al. Annual Incidence of Nephrolithiasis among Children and Adults in South Carolina from 1997 to 2012. Clin J Am Soc Nephro. 2016;11:488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semins MJ, Trock BJ, Matlaga BR. Validity of administrative coding in identifying patients with upper urinary tract calculi. J Urol. 2010;184:190–192. [DOI] [PubMed] [Google Scholar]
- 4.Kim BJ, Merchant M, Zheng C, et al. A natural language processing program effectively extracts key pathologic findings from radical prostatectomy reports. J Endourol. 2014;28:1474–1478. [DOI] [PubMed] [Google Scholar]
- 5.Thomas AA, Zheng C, Jung H, et al. Extracting data from electronic medical records: validation of a natural language processing program to assess prostate biopsy results. World J Urol. 2014;32:99–103. [DOI] [PubMed] [Google Scholar]
- 6.Schroeck FR, Pattison EA, Denhalter DW, et al. Early Stage Bladder Cancer: Do Pathology Reports Tell Us What We Need to Know? Urology. 2016;98:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Bird VY, Ruchi R, et al. Development of a personalized diagnostic model for kidney stone disease tailored to acute care by integrating large clinical, demographics and laboratory data: the diagnostic acute care algorithm - kidney stones (DACA-KS). BMC Med Inform Decis Mak. 2018;18:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krambeck AE, Khan NF, Jackson ME, et al. Inaccurate reporting of mineral composition by commercial stone analysis laboratories: implications for infection and metabolic stones. J Urol. 2010;184:1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearle MS, Goldfarb DS, Assimos DG, et al. Medical management of kidney stones: AUA guideline. J Urol. 2014;192:316–324. [DOI] [PubMed] [Google Scholar]
- 10.Pak CY, Poindexter JR, Adams-Huet B, et al. Predictive value of kidney stone composition in the detection of metabolic abnormalities. Am J Med. 2003;115:26–32. [DOI] [PubMed] [Google Scholar]
- 11.Gambaro G, Croppi E, Coe F, et al. Metabolic diagnosis and medical prevention of calcium nephrolithiasis and its systemic manifestations: a consensus statement. J Nephrol. 2016;29:715–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danciu I, Cowan JD, Basford M, et al. Secondary use of clinical data: the Vanderbilt approach. J Biomed Inform. 2014;52:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenetorp P, Pyysalo S, Topić G, et al. brat: a Web-based Tool for NLP-Assisted Text Annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics 2012. [Google Scholar]
- 14.Firth JR. A synopsis of linguistic theory 1930-1955. Studies in Linguistic Analysis. 1957. [Google Scholar]
- 15.Yarowsky D Unsupervised word sense disambiguation rivaling supervised methods. Proceedings of the 33rd annual meeting on Association for Computational Linguistics 1995. [Google Scholar]
- 16.Kaplan EL, Meier P. Nonparametric-Estimation from Incomplete Observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 17.Knoll T, Schubert AB, Fahlenkamp D, et al. Urolithiasis Through the Ages: Data on More Than 200,000 Urinary Stone Analyses. J Urology. 2011;185:1304–1311. [DOI] [PubMed] [Google Scholar]
- 18.Singh P, Enders FT, Vaughan LE, et al. Stone Composition Among First-Time Symptomatic Kidney Stone Formers in the Community. Mayo Clin Proc. 2015;90:1356–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daudon M, Dore JC, Jungers P, et al. Changes in stone composition according to age and gender of patients: a multivariate epidemiological approach. Urol Res. 2004;32:241–247. [DOI] [PubMed] [Google Scholar]
- 20.Lieske JC, Rule AD, Krambeck AE, et al. Stone Composition as a Function of Age and Sex. Clin J Am Soc Nephro. 2014;9:2141–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandel N, Mandel I, Fryjoff K, et al. Conversion of calcium oxalate to calcium phosphate with recurrent stone episodes. J Urol. 2003;169:2026–2029. [DOI] [PubMed] [Google Scholar]
- 22.Reinstatler L, Stern K, Batter H, et al. Conversion from Cystine to Non-cystine Stones: Incidence and Associated Factors. J Urol. 2018. [DOI] [PubMed] [Google Scholar]
- 23.Rule AD, Lieske JC, Li X, et al. The ROKS nomogram for predicting a second symptomatic stone episode. J Am Soc Nephrol. 2014;25:2878–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meydan N, Barutca S, Caliskan S, et al. Urinary stone disease in diabetes mellitus. Scand J Urol Nephrol. 2003;37:64–70. [DOI] [PubMed] [Google Scholar]
- 25.Daudon M, Traxer O, Conort P, et al. Type 2 diabetes increases the risk for uric acid stones. J Am Soc Nephrol. 2006;17:2026–2033. [DOI] [PubMed] [Google Scholar]
- 26.Matlaga BR, Kim SC, Watkins SL, et al. Changing composition of renal calculi in patients with neurogenic bladder. J Urol. 2006;175:1716–1719; discussion 1719. [DOI] [PubMed] [Google Scholar]
- 27.Schwaderer AL, Wolfe AJ. The association between bacteria and urinary stones. Ann Transl Med. 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krambeck AE, Handa SE, Evan AP, et al. Profile of the brushite stone former. J Urol. 2010;184:1367–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bejan CA, Vanderwende L, Xia F, et al. Assertion modeling and its role in clinical phenotype identification. J Biomed Inform. 2013;46:68–74. [DOI] [PubMed] [Google Scholar]
- 30.Bejan CA, Angiolillo J, Conway D, et al. Mining 100 million notes to find homelessness and adverse childhood experiences: 2 case studies of rare and severe social determinants of health in electronic health records. J Am Med Inform Assoc. 2018;25:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.