Abstract
Nonalcoholic fatty liver disease (NAFLD) represents a spectrum of disease ranging from simple steatosis to more severe forms of liver injury including nonalcoholic steatohepatitis (NASH), fibrosis, and hepatocellular carcinoma (HCC). In humans, only 20–40% of patients with fatty liver progress to NASH, and mice fed a high-fed diet (HFD) develop fatty liver but are resistant to NASH development. To understand how simple steatosis progresses to NASH, we examined hepatic expression of anti-inflammatory microRNA-223 (miR-223) and found that this miRNA was highly elevated in hepatocytes in HFD-fed mice and in human NASH samples. Genetic deletion of the miR-223 induced a full spectrum of NAFLD in long-term HFD-fed mice including steatosis, inflammation, fibrosis and HCC. Furthermore, microarray analyses revealed that compared to wild-type mice, HFD-fed miR-223 knockout (miR-223KO) mice had greater hepatic expression of many inflammatory genes and cancer-related genes, including (C-X-C motif) chemokine 10 (Cxcl10) and transcriptional co-activator with PDZ-binding motif (Taz), two well-known factors to promote NASH development. In vitro experiments demonstrated that Cxcl10 and Taz are two downstream targets of miR-223 and overexpression of miR-223 reduced their expression in cultured hepatocytes. Hepatic levels of miR-223, CXCL10 and TAZ mRNA were elevated in human NASH samples, which positively correlated with hepatic levels of several miR-223 targeted genes as well as several pro-inflammatory, cancer-related and fibrogenic genes. Conclusion, HFD-fed miR-223KO mice develop a full spectrum of NAFLD, representing a clinically relevant mouse NAFLD model. MiR-223 plays a key role in controlling steatosis-to-NASH progression by inhibiting hepatic Cxcl10 and Taz expression. MiR-223 may be a novel therapeutic target for the treatment of NASH.
Keywords: hepatocellular carcinoma, fatty liver, inflammation, CXCL10, TAZ
Introduction
Nonalcoholic fatty liver disease (NAFLD) represents a spectrum of disorder ranging from hepatocellular steatosis to nonalcoholic steatohepatitis (NASH), fibrosis, cirrhosis and hepatocellular carcinoma (HCC).(1–3) Due to its approximately 25% prevalence worldwide, NAFLD is the major cause of chronic liver diseases, emerging as a significant public health problem.(4–7) NASH is characterized microscopically by steatosis, ballooned hepatocytes with Mallory-Denk hyaline bodies, inflammatory cell infiltration with predominant neutrophil infiltration, and pericellular fibrosis.(1–3) Individuals with simple steatosis are believed to be a low risk of adverse outcomes, whereas patients with NASH without treatment likely progress to cirrhosis and HCC.(1–3) A large number of molecular pathways have been identified to promote the steatosis to NASH and liver cancer; (1–3) however, the exact mechanisms by which inflammation is induced and regulated in NASH remain largely unknown.
Hepatic neutrophil infiltration is a hallmark of NASH and plays an important role in promoting hepatocyte damage in the progression of NASH via the generation of reactive oxygen species, the production of proinflammatory mediators, and the formation of neutrophil extracellular traps.(1–3, 8, 9) In addition, neutrophil-hepatic stellate cell interactions may also drive the progression of liver fibrosis in experimental steatohepatitis.(10) Emerging evidence suggests that neutrophil activation and functions are regulated by many proinflammatory mediators, cytokines, growth factors, transcription factors, and epigenetic factors (e.g. microRNAs [miRNAs]).(11) Among miRNAs, miR-223 is expressed at the highest levels in neutrophils, playing a critical role in attenuating neutrophil maturation and activation.(12–14) We have previously demonstrated that miR-223 plays an important role in attenuating alcoholic liver injury and acetaminophen-induced liver injury by targeting Il6 and Ikka genes in neutrophils.(15, 16) However, a recent study suggests that miR-223 does not play a role in controlling acute liver injury in several mouse models and in controlling liver fibrosis induced by chronic CCl4 treatment.(17) The part of reasons for the discrepancy between this study and ours may be due to the different wild-type control mice used (wild-type littermate control mice were used in our study(15, 16) but not in other study(17)). Interestingly, several recent studies reported that levels of miR-223 were upregulated in the liver in a mouse model of NASH (18) and were elevated in the serum in NASH patients.(19) However, the function of miR-223 in the pathogenesis of NASH and whether miR-223 regulates steatosis-NASH conversion remain unclear.
In the current study, we found that miR-223 was upregulated in hepatocytes in HFD-fed mice and in NASH patients. More interestingly, genetic deletion of the miR-223 promoted the progression of high-fat diet (HFD)- or methionine-choline deficient diet (MCD)-induced NASH as well as long-term HFD-induced liver cancer in mice. These data suggest that miR-223 protects against NASH development, which is likely due to the anti-inflammatory functions of miR-223 in neutrophils.(13–16) Here we identified a novel mechanism by which miR-223 prevents the progression of NASH and liver cancer by directly targeting several inflammatory genes and oncogenes in hepatocytes. Among these targeted genes, both (C-X-C motif) chemokine 10 (Cxcl10) and transcriptional coactivator with PDZ-binding motif (Taz) (also known as WW domain-containing transcription regulator 1 [Wwtr1]), have been shown to play an important role in promoting the progression of NASH as deletion of either of them markedly ameliorated NASH development.(20–22) CXCL10 (also known as interferon gamma-induced protein 10) not only activates and promotes inflammatory cells (e.g. macrophages)(20) but also directly induces hepatocyte damage by targeting CXCR3,(21, 23) resulting in liver injury and inflammation. TAZ, together with another transcriptional co-activator yes-associated protein (YAP), is the major downstream effector of the Hippo signaling pathway, playing an important role in promoting liver inflammation, regeneration, and carcinogenesis.(22, 24, 25) Finally, in NASH patients, hepatic expression of Cxcl10 and Taz correlated well with expression of several potential miR-223 targeted genes, and inflammatory genes, suggesting that miR-223 also plays an important role in ameliorating NASH by regulating Cxcl10, Taz, and other targeted genes in NASH patients.
Materials and Methods
Mice, HFD feeding model, and MCD feeding model
MiR-223−/− (miR-223 KO) mice (The Jackson Laboratory, Bar Harbor, ME) were crossed with C57BL/6J mice to obtain female miR-223+/− mice. Female miR-223+/− and male miR-223−/y mice (the miR-223 locus is on the X chromosome) were bred at the NIAAA FLAC facility to generate male miR-223−/y (male miR-223KO) and wild-type (WT) miR-223+/y littermate controls. All male miR-223+/y littermates were used as WT controls for male miR-223KO (miR-223−/y) mice in our study. Male mice were fed an HFD (60 kcal% fat; catalog no. D12492, Research Diets, Inc., New Brunswick, NJ) or a chow diet (10 kcal% fat) as a control diet (CD) for three months or one year. Male mice were also fed an MCD diet (MP Biomedicals, catalog # 0296043920) or the methionine/choline control diet (MP Biomedicals, catalog # 0296044110) for 4 weeks. The study was approved by the National Institute on Alcohol Abuse and Alcoholism Animal Care and Use Committee.
Liver tissues from NASH patients
Normal (n=10) and NASH human liver samples (n=14) were obtained from the National Institutes of Health-funded Liver Tissue Cell Distribution System at the University of Minnesota, which was funded by NIH contract # HHSN276201200017C. The general characteristics of normal and NASH human liver samples are listed in Supporting Table S1.
Microarray analyses of mouse liver samples
Total RNAs were isolated from liver tissues of 3-month HFD-fed WT and miR-223KO mice and subjected to microarray analysis as described in Supporting materials. Full microarray data have been uploaded to NCBI’s Gene Expression Omnibus (The GEO accession number is GSE129080).
MiR-223 in situ hybridization
MiRNA in situ hybridization was performed as described previously.(26, 27) In brief, 10 μm thick cryosections of mouse liver tissues were fixed with fresh cold 4% paraformaldehyde (PFA) for overnight at 4°C. After predigestion, the sections were hybridized with FAM-labeled U6 or FAM-labeled miR-223 probe (Qiagen, Valencia, CA) in hybridization solution at 55°C for 1 hour. After washing in 0.1×SSC buffer, the slides were applied with sheep anti-FAM-POD (Abcam, Cambridge, MA) diluted 1:250 in blocking solution and incubated for 60 min at room temperature. Tyramine signal amplification (TSA) kit (Perkin Elmer) was performed to amplify the signal. Meanwhile, the cell marker protein was done by incubating primary antibody (HNF-4α [Abcam] for 1 hour at room temperature, and then with the Alexa Fluor–conjugated secondary antibody Alexa Fluor 549 goat anti-rabbit IgG (H+L) (Vector Laboratories Technology) or Alexa Fluor 549 goat anti-mouse IgG (H+L) (Cell Signaling Technology) for 1 hour at room temperature. Slides were incubated with 1 mg/mL 4’, 6’-diamino-2-phenylindole (DAPI) for 5 min for nuclear staining. The images were obtained by using LSM 710 confocal microscope (Zeiss, Thornwood, NY) with filters allowing detection of FITC and Alexa 549.
Similar procedures for miR-223 in situ hybridization were done in formalin-fixed human NASH samples and albumin was used for hepatocyte marker using an anti-Albumin antibody from Millipore (Millipore, Billerica, MA).
Statistical analysis
Data are expressed as the means ± SEM and were analyzed using GraphPad Prism software (v. 5.0a; GraphPad Software, La Jolla, CA). To compare values obtained from two groups, the Student t test was performed. Data from multiple groups were compared with one-way ANOVA followed by Tukey’s post hoc test. P values of <0.05 were considered significant.
Many other methods are included in supporting materials including real-time quantitative PCR primer sequences in supporting Table S2.
Results
Upregulation of miR-223 in hepatocytes from HFD-fed mice and NASH patients
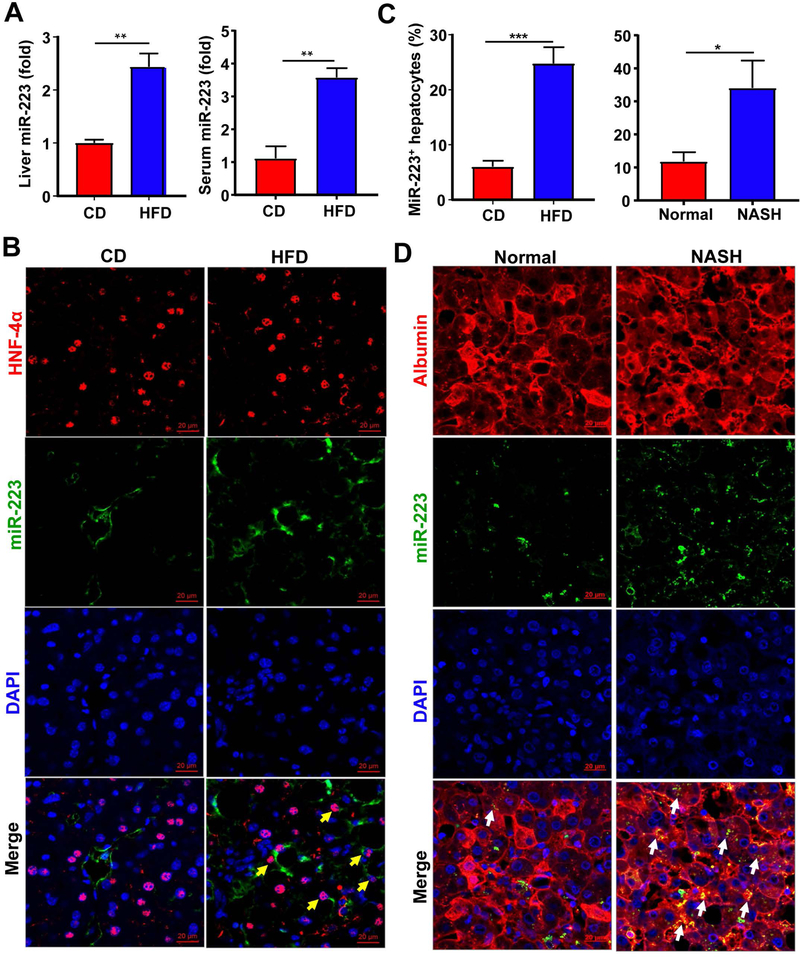
To investigate the role of miR-223 in the development of NASH, we firstly examined serum and hepatic miR-223 levels in HFD-fed mice and NASH patients. As shown in Fig. 1A, hepatic and serum levels of miR-223 were significantly upregulated after 3-month HFD feeding. To determine which cell types in the liver express miR-223, we performed in situ hybridization analyses and immunofluorescent staining of the hepatocyte marker HNF-4α or the neutrophil marker MPO. As expected, miR-223 was detected in many neutrophils in the liver samples from HFD-fed mice (double staining of miR-223 and MPO in Supporting Fig. S1A). Interestingly, miR-223 was also detected in many HNF-4α+ hepatocytes with significantly higher levels in HFD-fed mice compared to CD-fed mice. (Fig. 1B,C). Although the relative miR-223 expression levels in the liver (hepatocytes) were significantly increased after HFD feeding, these levels in the liver (hepatocytes) were much lower than those in neutrophils as evidenced by the CT value from RT-qPCR analysis (data not shown).
Figure 1. miR-223 is highly elevated in hepatocytes in HFD-fed mice and NASH patients.
(A-C) C57BL/6J mice were fed an HFD or control diet (CD) for three months. Liver and serum samples were collected and subjected to the measurement of miR-223 by RT-qPCR (panel A). Frozen liver tissue sections were also subjected to miR-223 in situ hybridization along with immunofluorescence staining of hepatocyte marker HNF-4α. Representative images of miR-223 expression (green), HNF-4α (red) and nuclei (blue) are shown (panel B). MiR-223+ hepatocytes were counted (panel C). (C-D) Formalin-fixed human liver tissue sections were subjected to miR-223 in situ hybridization and albumin staining for hepatocytes. Albumin+MiR-223+ hepatocytes were counted (panel C). Representative images of miR-223 (green) and nuclei (blue) are shown (Panel D). Arrows indicate miR-223+ hepatocytes. Values represent means ± SEM (n=8–12). *P< 0.05, **P< 0.01, ***P<0.001.
Similarly, in situ hybridization analyses of formalin-fixed human liver samples revealed that many MPO+ neutrophils express miR-223 (Supporting Fig. S1B). To detect the co-localization of miR-223 and hepatocytes, we performed albumin and miR-223 double staining (HNF staining did not work well in formalin-fixed tissues). Few albumin+ hepatocytes expressed miR-223 in healthy liver samples, whereas miR-223 was strongly detected in albumin+ hepatocytes from NASH patients (Fig. 1C,D).
MiR-223KO mice develop NASH after 3-month HFD feeding or MCD feeding
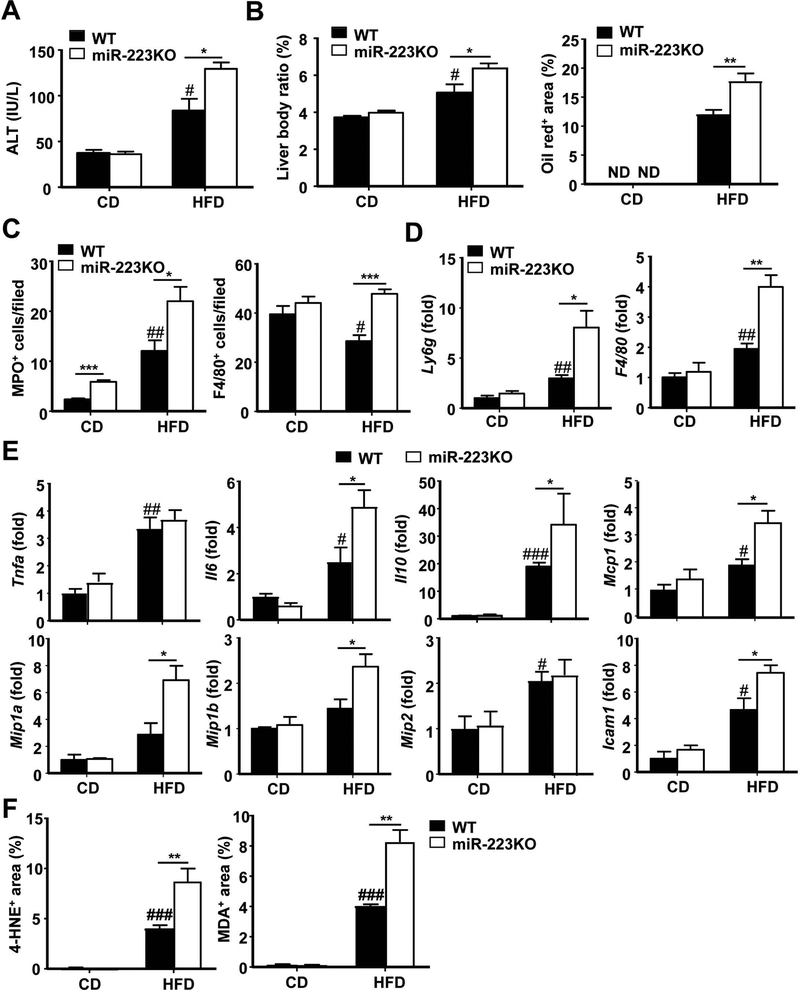
To further understand the function of miR-223 in NASH, male miR-223KO and their WT littermate control mice were fed an HFD and CD. Both groups of mice had similar body weight gain after HFD feeding (Supporting Fig. S2A). Interestingly, compared to WT mice, miRNA-223KO mice developed more severe phenotypes of NASH, such as greater steatosis, higher levels of serum ALT (Fig. 2A,B and Supporting Fig. S2B–D), and greater degree of hepatic neutrophil and macrophage infiltration as demonstrated by MPO (a marker for neutrophils) and F4/80 (a marker for macrophages) immunostaining (Fig. 2C and Supporting Fig. S3A,B) and RT-qPCR analysis of hepatic expression of Ly6g (a marker for neutrophils) and F4/80 (Fig. 2D). Interestingly, both the ratio and the level of circulating neutrophils were lower in HFD-fed miR-223KO mice than that in WT mice (Supporting Fig. S3C). Next, we wanted to know the change of other immune cells in HFD-fed WT and miR-223KO mice by performing flow cytometric analyses. As shown in Supporting Fig. S3D, the numbers of CD3+ T, CD19+ B, and NK1.1+ cells were comparable between HFD-fed WT and miR-223KO mice. Furthermore, hepatic mRNA expression of inflammatory cytokines (Interleukin [Il]6, Il10), chemokines (Monocyte chemotactic protein 1 [Mcp1], Macrophages inflammatory protein [Mip] 1a, and Mip1b) and Intercellular adhesion molecule 1(Icam1) in miR-223KO mice was much higher than those in WT mice after HFD feeding for 3 months (Fig. 2E). Additionally, compared to HFD-fed WT mice, HFD-fed miR-223KO mice had greater hepatic levels of several steatogenic genes (Sterol regulatory element-binding protein 1 [Srebp1c], Stearoyl-CoA desaturase-1 [Scd1], Acetyl-coenzyme A carboxylase [Acc1], Cell death-inducing DNA fragmentation factor alpha-like effector A [Cidea]) (Supporting Fig. S3E) and greater hepatic levels of lipid peroxide including malonaldehyde (MDA) and 4-hydroxynonenal (4-HNE, oxidative stress marker) (Fig. 2F and Supporting Fig. S3F).
Figure 2. miR-223KO mice are more susceptible to 3m-HFD-induced liver injury, steatosis and inflammation.
WT and miR-223KO mice were fed an HFD or CD for three months. Serum and liver tissue samples were collected. (A) Serum ALT levels were measured. (B) Liver body ratio was measured. Lipids was stained with Oil Red and the quantitation of Oil Red+ area per field was determined. Representative images are shown in supporting Fig. S2D. (C) Liver tissues were also subjected to MPO and F4/80 immunostaining and the quantitation of MPO+ and F4/80+ cells per field was determined. Representative images are shown in Supporting Fig. S3A–B. (D, E) RT-qPCR analyses of liver Ly6g and F4/80 mRNA, and other inflammatory mediators. (F) Quantification of 4-hydroxynonenal (HNE)+ and malonaldehyde (MDA)+ area per field. Values represent means ± SEM (n=8–12). *P< 0.05, **P< 0.01, ***P<0.001 in comparison with corresponding WT groups; #P<0.05, ##P<0.01, ###P<0.001 in comparison with WT CD group.
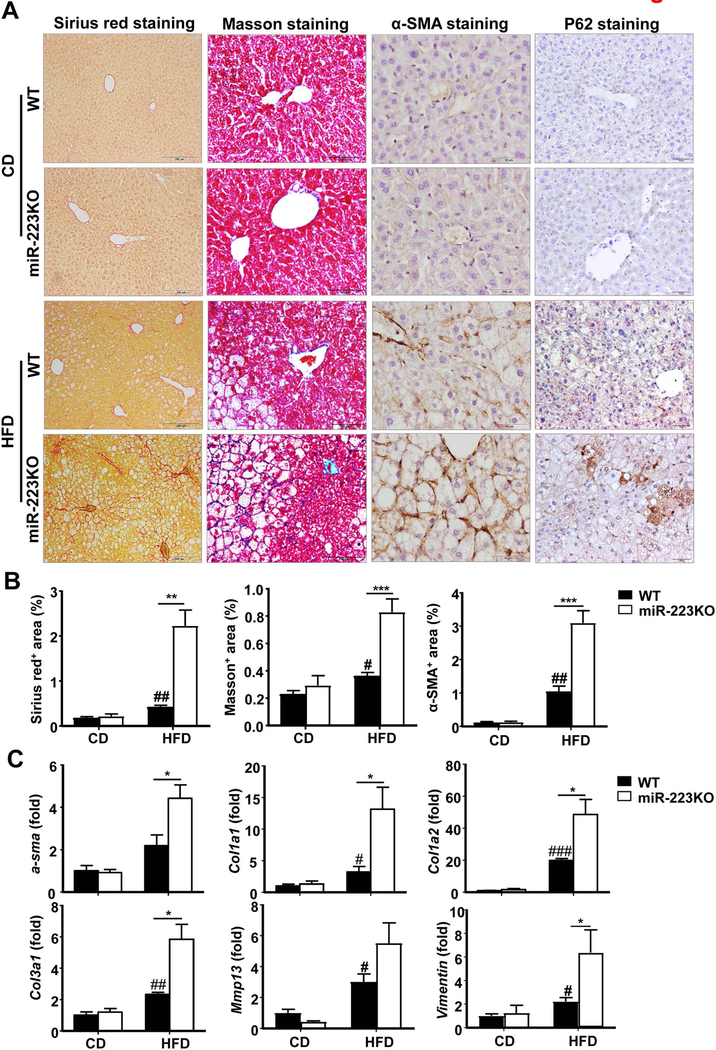
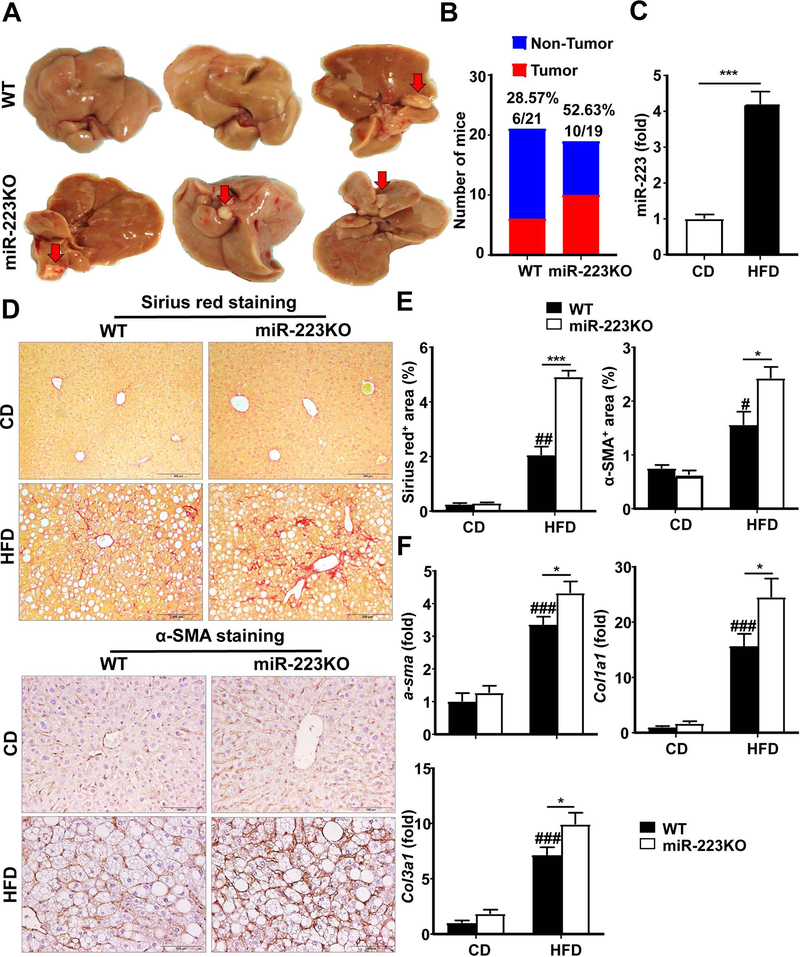
Fibrosis, a typical feature of human NASH, was usually not robust in HFD-fed mice,(28) whereas miR-223KO mice developed greater liver fibrosis than WT mice after HFD feeding as determined by Sirius red staining, Masson staining and α-smooth muscle actin [SMA] staining) (Fig. 3A,B), and RT-qPCR analysis of hepatic α-sma, collagen (Col1a1, Col1a2, and Col3a1) and Vimentin mRNAs (Fig. 3C), and western blotting analyses of fibrotic markers (Supporting Fig. S4A). To determine whether miR-223 can directly affect hepatic stellate cell (HSC) activation, we isolated primary HSCs from WT and miR-223KO mice and cultured them for 1 day or 5 days in vitro. As shown in Supporting Fig. S4B, HSCs from miR-223KO mice expressed higher levels of α-sma, collagens (Col1a1, Col3a1, and Col4a1) and Tgf-β1 than HSCs from WT mice, suggesting miR-223 can directly attenuate HSC activation.
Figure 3. miR-223KO mice are more susceptible to HFD-induced fibrosis.
WT and miR-223KO mice were fed an HFD or CD for three months. Liver tissue samples were collected. (A) Representative images of Sirius red staining, Masson staining, α-SMA staining and P62 staining of liver tissue sections are shown. (B) Quantification of fibrotic area per field. (C) RT-qPCR analyses of hepatic fibrogenesis genes. Values represent means ± SEM (n=8–12). *P< 0.05, **P< 0.01, ***P< 0.001 in comparison with WT HFD groups; #P<0.05, ##P<0.01, ###P<0.001 in comparison with WT CD groups.
Mallory-Denk bodies together with ballooning hepatocytes, which are characteristic hallmarks of NASH, were also examined. As illustrated in Supporting Fig. S4C, keratin 8 and keratin 18, two major components of Mallory-Denk bodies,(29) were significantly upregulated in miR-223KO mice compared with WT mice after HFD feeding. Furthermore, stress protein p62, which is another marker for Mallory-Denk bodies,(29) was markedly expressed in hepatocytes in miR-223KO mice, but not in WT mice after HFD feeding as demonstrated by immunohistochemistry analyses (Fig. 3A).
The role of miR-223 was also tested in another model of NASH induced by MCD feeding. As illustrated in Supporting Fig. S5A, hepatic expression of miR-223 was highly elevated in MCD-fed mice compared to CD-fed mice. Compared to MCD-fed WT mice, MCD-fed miR-223 KO mice had higher levels of serum ALT (Supporting Fig. S5B) and greater mRNA expression of various proinflammatory mediators and fibrogenic markers in the liver (Supporting Fig. S5C), suggesting that miR-223 protects against MCD-induced NASH.
Genes involved in cancer and hyperplasia/hyperproliferation are abnormally expressed in the livers of miR-223KO mice after HFD feeding
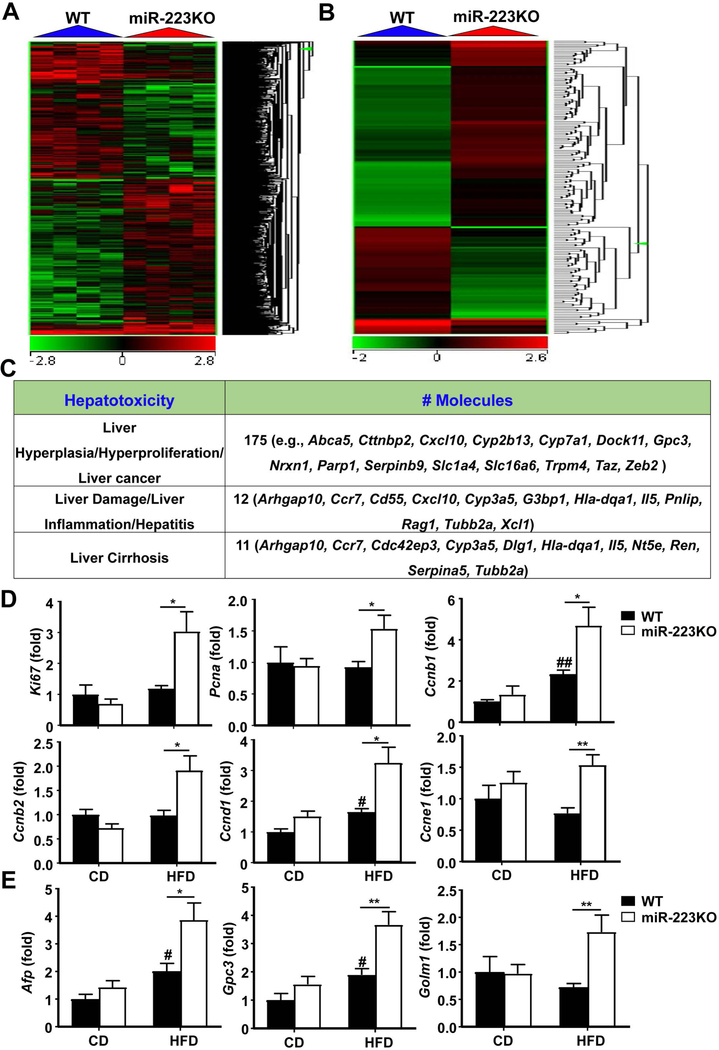
To better explore the mechanisms underlying the abnormalities observed in HFD-fed miR-223KO mice, we examined hepatic gene expression in 3-month-HFD-fed WT and miR-223KO mice by microarray analysis. Ingenuity Pathway Analysis of molecular and cellular functions of a stringent set (p<0.05) of hepatic genes identified 357 upregulated and 327 downregulated genes in HFD-fed miR-223KO mice compared with HFD-fed WT mice. The top diseases associated with these dysregulated genes are cancer, organismal injury and abnormalities, and inflammatory response (Supporting Fig. S6A, and Supporting Table S3). The heat map analysis showed great differences (>2 fold) in gene expression profile in WT and miR-223KO mice (Fig. 4A). The differential gene expression related to liver cancer, inflammation, and cirrhosis was further analyzed, which specifically highlighted the significant abnormal expression of 175 genes (such as Abca5, Cttnbp2, Cxcl10, Cyp2b13, Cyp7a1, Dock11, Gpc3, Nrxn1, Parp1, Serpinb9, Slc1a4, Slc6a6, Trpm4, Taz, and Zeb2) that are involved in liver cancer, 12 genes (such as Arhgap10, Ccr7, Cd55, Cxcl10, Cyp3a5, G3bp1, Hla-dqa1, Il5, Pnlip, Rag1, Tubb2a, and Xcl1) that are involved in liver damage and inflammation (Fig. 4B,C).
Figure 4. Upregulation of hepatocarcinogenesis-related genes in miR-223KO mice versus WT mice after 3m-HFD feeding.
WT and miR-223KO mice were fed an HFD or CD for three months. Liver tissue samples were collected, and then subjected to microarray analysis. (A) Heat map of gene expression (>2-fold changes) from microarray analysis is shown. (B) Heat map of 172 liver cancer-related genes expression from microarray analysis is shown. (C) Ingenuity Pathway Analysis of liver cancer, inflammation, and cirrhosis in WT and miR-223KO mice. (D) RT-qPCR analyses of hepatic proliferation markers. (E) RT-qPCR analyses of HCC markers. Values represent means ± SEM (n=4 in panels A-C; n=8 in panels D-E). *P< 0.05, **P< 0.01 in comparison with WT HFD groups; #P<0.05, ##P<0.01 in comparison with WT CD groups.
Microarray analysis revealed that the proliferation markers (Ki67, Pcna, Ccnb1, Ccnb2, Ccnd1, and Ccne1) and HCC markers (Gpc3 and Golm1) were significantly elevated in miR-223KO mice compared with WT mice after 3m-HFD feeding; whereas hepatic expression of the NF-κB pathway-related genes such as Chuk, Nfkbia, and Nfkb1 was comparable between these two groups (Supporting Fig. S6B, C). The upregulation of several genes from the microarray data was confirmed by RT-qPCR analysis (Fig. 4D,E). In addition, microarray data revealed comparable levels of hepatic Afp mRNA between HFD-fed WT and miR-223KO mice (Supporting Fig. S6B), but RT-qPCR detected significantly higher levels of hepatic Afp mRNA in HFD-fed miR-223KO mice than in HFD-fed WT mice (Fig. 4E). Moreover, some proliferation marker protein levels (PCNA, CCND1 and CCNE1) and HCC marker protein levels (GPC3 and GOLM1) were upregulated in the livers from 3m-HFD-fed miR-223KO mice compared with WT mice; whereas p-mTOR and total mTOR, and p53 protein levels in 3m-HFD-fed WT and miR-223KO were comparable (Supporting Fig. S6D).
MiR-223KO mice are more susceptible to long-term HFD-induced NASH and liver cancer development
To address whether miR-223KO mice are more susceptible to hepatocarcinogenesis, WT and miR-223KO mice were fed with a HFD for one year. As demonstrated in Fig. 5A,B, ten of 19 miR-223KO mice (52.63%) developed liver tumors (1–3 macroscopic tumors observed per mouse), whereas 6 of 21 WT mice (28.57%) developed HCC after long-term HFD feeding. Hepatic miR-223 levels were upregulated by approximately 4-fold after HFD feeding for one year (Fig. 5C). Interestingly, although serum ALT and AST levels, liver weight, liver body ratio, and steatosis were comparable in WT and miR-223KO mice after one-year HFD feeding, miR-223KO mice had greater liver fibrosis than WT mice, as demonstrated by H&E staining, Sirius red staining and α-SMA staining (Fig. 5D,E and Supporting Fig. S7A–C) and by RT-qPCR analyses of fibrogenic markers (α-sma, Col1α1 and Col3α1) (Fig. 5F).
Figure 5. miR-223KO mice show higher tumor incidence and greater degree of liver fibrosis compared with WT mice after HFD feeding for one year.
WT (n=21) and miR-223KO mice (n=19) were fed an HFD or CD for one year. Liver tissue samples were collected. (A) Representative photographs of liver tumors that developed in WT and miR-223KO mice are shown. Arrows indicate liver tumor. (B) Liver tumor incidence in WT and miR-223KO mice was analyzed. (C) RT-qPCR analysis of hepatic miR-223 levels after HFD feeding for one year. (D) Representative Sirius red staining and α-SMA staining of liver tissue sections are shown. (E) Quantification of fibrotic area per field. (F) RT-qPCR analyses of hepatic fibrogenesis genes. Values represent means ± SEM (n=6–21). *P< 0.05, ***P< 0.001 in comparison with WT HFD groups; #P<0.05, ##P<0.01, ###P<0.001 in comparison with WT CD groups.
Next, we further characterized the liver inflammatory response in WT and miR-223KO mice after long-term HFD feeding. As illustrated in Supporting Fig. S8A–D, compared to WT mice, miR-223KO mice had a greater number of neutrophils and macrophages as demonstrated by immunostaining and RT-qPCR analyses of neutrophil and macrophage markers (Ly6g and F4/80). However, both the percentage and the level of circulating neutrophils were significantly lower in miR-223KO mice compared with WT mice (Supporting Fig. S8E). Furthermore, expression of chemokine Mip2, and chemokine receptor Ccr2, but not other cytokines and chemokines, were significantly elevated in miR-223KO mice compared with WT mice (Supporting Fig. S9).
MiR-223KO mice are associated with greater elevation of hepatic Cxcl10 and Taz than WT mice after 3m- and 1y-HFD feeding, and MCD feeding
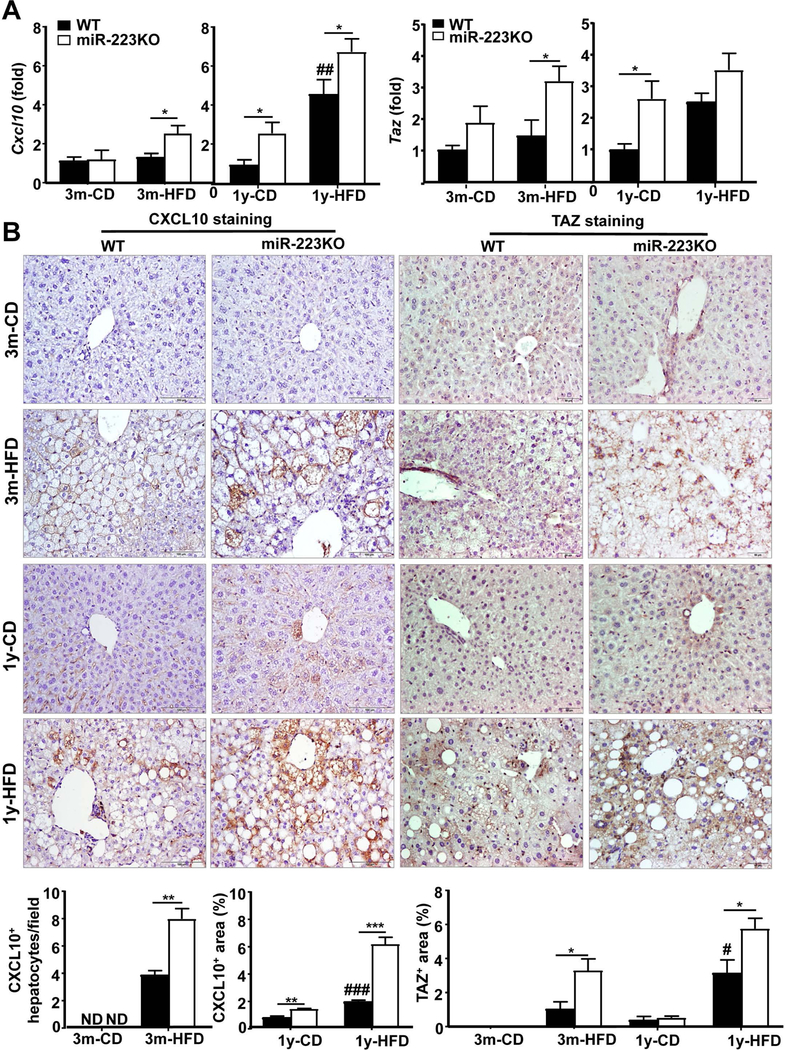
To search for the potential target of miR-223, we used a bioinformatics approach (microRNA.org database: http://www.microrna.org). This database predicts microRNA targets and target downregulation scores. By using this database, we went through the most upregulated genes in miR-223KO mice versus WT mice in our microarray data and identified 7 potential target genes of miR-223 (Cxcl10, Taz, Serpinb9, Nrxn1, Slc1a4, Slc16a6, and Dock11). The levels of these genes were higher in miR-223KO mice compared with WT mice (microarray data, Supporting Fig. S10A). We paid more attention to Cxcl10 and Taz because both CXCL10 and TAZ have been shown to play a pivotal role in the pathogenesis of experimental NASH.(20–22) As illustrated in Fig. 6A, RT-qPCR analyses demonstrated that hepatic expression of both Cxcl10 and Taz was higher in 3m- and 1y-HFD-fed miR-223KO than that in WT mice. RT-qPCR analyses also confirmed that compared to HFD-fed WT mice, HFD-fed miR-223KO mice were associated with higher hepatic expression of several other miR-223 targeted genes including Serpinb9, Slc1a4, Slc16a6, and Dock11 (Supporting Fig. S10B). Interestingly, although hepatic expression of Cxcl10 was higher in HFD-fed miR-223KO mice than in WT mice, hepatic expression of other CXCR3 ligands (Cxcl9 and Cxcl11) and Cxcr3 was comparable between these two groups (Supporting Fig. S10C). Finally, immunohistochemistry staining analyses demonstrated that hepatocytes from miR-223KO mice expressed much higher levels of CXCL10 and TAZ protein than those from WT mice after 3m- or 1y-HFD feeding (Fig. 6B). Serum levels of CXCL10 protein were also higher in HFD-fed miR-223KO verse WT mice (Supporting Fig. S11A). Greater levels of potential target genes including Cxcl10, Taz, Nrxn1, and HCC marker Golm1 in miR-223 KO mice versus WT mice were also observed in the MCD-induced NASH model (Supporting Fig. S11B), but other potential target gene levels and HCC marker Gpc3 were not significantly increased in MCD-fed miR-223KO mice compared with WT mice.
Figure 6. A greater number of hepatocytes strongly expressed CXCL10 and TAZ in HFD-fed miR-223KO mice versus WT mice.
WT and miR-223KO mice were fed an HFD or CD for three months or one year. (A) Liver tissues were collected for RT-qPCR analyses of Cxcl10 and Taz. (B) Liver tissues were also subjected to immunohistochemistry analyses. Representative CXCL10 staining and TAZ staining of liver tissue sections. Quantification of CXCL10+ hepatocytes or CXCL10+ area and TAZ+ area per field was quantified. Values represent means ± SEM (n=8–12). *P< 0.05, **P<0.01, ***P< 0.001 in comparison with corresponding WT groups; #P<0.05, ##P<0.01, ###P<0.001 in comparison with WT CD groups.
Cxcl10 and Taz are miR-223 targeted genes in hepatocytes
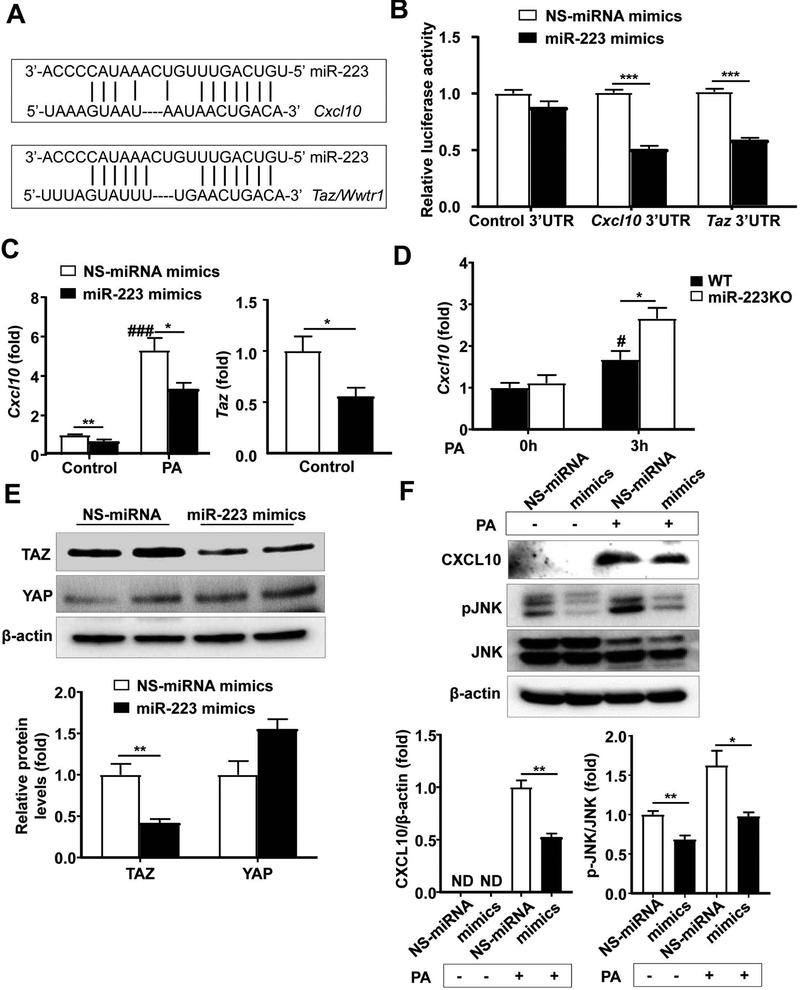
The bioinformatics approach analysis of the target prediction of miR-223 revealed that both Cxcl10 and Taz are the miR-223 potential targeted genes (Fig. 7A). To further confirm this notion, we performed Dual-Luciferase assay by co-transfecting luciferase vector containing the 3’ untranslated region (3’UTR) of Cxcl10 or Taz and miR-223 mimics or non-specific (NS)-miRNA mimics. As shown in Fig. 7B, overexpression of miR-223 in mouse hepatocyte cell line AML12 cells significantly reduced luciferase activity from Cxcl10 3’UTR or Taz 3’UTR vector, but not from control 3’UTR vector. Next, we wondered whether overexpression of miR-223 attenuated expression of both genes. As illustrated in Fig. 7C, overexpression of miR-223 via the transfection of miR-223 mimics markedly suppressed expression of both Cxcl10 and Taz in mouse hepatocytes. Previous studies reported that free fatty acid (such PA) upregulated CXCL10 in hepatocytes.(30, 31) Here we demonstrated that PA treatment upregulated Cxcl10 mRNA expression in hepatocytes, and such upregulation was markedly inhibited by overexpression of miR-223 mimics (Fig. 7C). In addition, PA treatment induced much higher levels of Cxcl10 mRNA expression in miR-223KO hepatocytes than those in WT hepatocytes (Fig. 7D). Furthermore, western blotting analyses revealed that overexpression of miR-223 reduced TAZ but not YAP protein expression in hepatocytes (Fig. 7E). In addition, overexpression of miR-223 suppressed PA-induced CXCL10 protein expression and PA-induced JNK activation (Fig. 7F), which is known to be activated by CXCL10.(32)
Figure 7. miR-223 regulates Cxcl10 and Taz expression in hepatocytes.
(A) Bioinformatics approach analyses of the target prediction of miR-223. (B) Dual-Luciferase activity assay was performed to verify binding between miR-223 and CXCL10 or TAZ. AML12 cells were co-transfected with control luciferase vector, Cxcl10 3’UTR vector or Taz 3’UTR vector and miR-223 mimics or non-specific (NS)-miRNA mimics for 48h. Relative luciferase activity was determined. (C) AML12 cells were transfected with NS-miRNA mimics and miR-223 mimics for 24h, and then challenged with 0.1 mM palmitic acid (PA) for 3h. The Cxcl10 and Taz mRNA levels were measured by RT-qPCR. (D) Primary hepatocytes from WT and miR-223KO mice were treated with PA for 3h. The Cxcl10 mRNA expression was measured by RT-qPCR. (E) AML12 cells were transfected with NS-miRNA mimics and miR-223 mimics for 24h. The YAP and TAZ proteins were measured by western blotting. (F) AML12 cells were transfected with NS-miRNA mimics and miR-223 mimics for 24h, and then treated with 0.1 mM PA for 24h. The protein levels in whole hepatocyte lysates were determined by Western blot analyses. Values represent means ± SEM from three independent experiments. *P< 0.05 as indicated; #P<0.05, ###P<0.001 in comparison with control NS-miRNA mimic groups in panel C or 0h WT group in panel D.
Upregulation and correlation of CXCL10 and TAZ and several other proven targets of miR-223 in NASH patients,
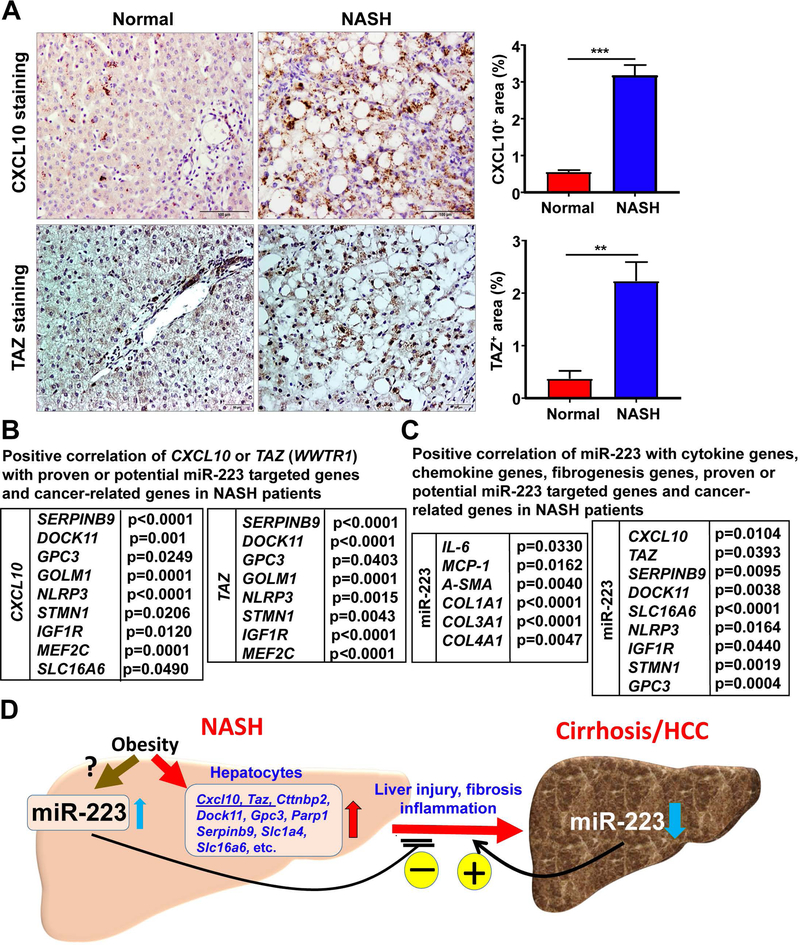
The data from published microarray analysis of NASH patient samples (33) revealed that hepatic expressions of CXCL10, TAZ, and many other potential and proven miR-223 targeted genes were significantly upregulated in NASH patients, but not in patients with fatty liver compared with healthy samples (Supporting Fig. S12). These targeted genes include: CXCL10, TAZ, SERPINB9, DOCK11, GPC3, GOLM1, NLRP3,(34) STMN1,(35) IGF-1R,(36) and MEF2C(12). Upregulation of hepatic CXCL10 and TAZ protein in NASH patients was further confirmed by immunohistochemistry staining analysis (Fig. 8A).
Figure 8. Several potential targets of miR-223 positively correlate with CXCL10 and TAZ expression in NASH patients.
(A) Normal liver and NASH samples were subjected to immunohistochemistry analysis of CXCL10 and TAZ. Representative images are shown, and quantification of CXCL10+ and TAZ+ area per field was performed. Values represent means ± SEM. **P< 0.01, ***P< 0.001 as indicated. (B) The gene expression profiles in healthy control liver, fatty liver, and NASH patients were obtained from published microarray data (the accession number E-MEXP-3291 [http://www.webcitation.org/5zyojNu7T]). Positive correlation of CXCL10 or TAZ with several proven or potential miR-223 targeted genes and cancer-related genes in NASH patient liver samples. P value is indicated. (C) Gene expression profiles in the livers of normal (n=10) and NASH (n=14) patients were analyzed by RT-qPCR. Positive correlation of miR-223 with cytokine, chemokine, and fibrogenic genes, and proven or potential miR-223 targeted genes and cancer-related gene in NASH patients. P value is indicated. (D) A model depicting the critical role of miR-223 in controlling the progression of NASH. Obesity associated fatty liver upregulates miR-223, which subsequently attenuates many downstream target genes including pro-inflammatory and oncogenic genes, thereby ameliorating NASH progression. In contrast, miR-223 is markedly downregulated in HCC. Such downregulation of miR-223 accelerates HCC progression. Thus, miR-223 is a critical regulator of NASH progression and could be a novel therapeutic target for the treatment of NASH and liver cancer.
These findings prompted us to explore whether there is a correlation between miR-223 and CXCL10 or TAZ in NASH patients. Because miRNA was not included in microarray data analysis, we were unable to analyze the correlation between miR-223 and CXCL10 or TAZ. Instead, we analyzed the correlation of CXCL10 or TAZ with several other miR-223 potential and proven targeted genes in NASH patients. As shown in Fig. 8B and Supporting Fig. S13, hepatic expression of CXCL10 or TAZ positively correlated with several other miR-223 potential or proven targeted genes and cancer-related genes in NASH patients. These genes include SERPINB9, DOCK11, SLC16A6, GPC3, GOLM1, NLRP3, STMN1, IGF1R, and MEF2C.
Remarkably, several cytokine and chemokine genes (IL6, MCP1, MIP1B, and CXCL1) and fibrogenesis genes (α-SMA, COL1A1, COL1A2, and COL4A1) were also markedly increased in NASH, which are positively correlated with CXCL10 and TAZ levels (Supporting Fig. S14, S15), suggesting that CXCL10 and TAZ expression are positively correlated with inflammation and fibrosis in NASH patients.
To further confirm whether miR-223 positively correlates with cancer-related genes, cytokine and chemokine genes and fibrogenesis genes, human liver tissues from normal (n=10) and NASH (n=14) patients were obtained and subjected to RT-qPCR analysis. As shown in Supporting Fig. S16A, cytokine and chemokine genes (IL-6, MCP1, and MIP1A), potential and proven miR-223 targeted genes (CXCL10, TAZ, SERPINB9, MEF2C, IGF-1R, and STMN1), and cancer-related gene (GPC3) were significantly increased in NASH samples compared with normal samples. Importantly, the levels of miR-223 positively correlated with several cytokine, chemokine, and fibrogenesis genes, and potential or proven miR-223 targeted genes and cancer-related genes in NASH patients (Fig. 8C and Supporting Fig. 16B). These genes include IL-6, MCP1, A-SMA, COL1A1, COL3A1, COL4A1, CXCL10, TAZ, SERPINB9, DOCK11, SLC16A6, NLRP3, IGF1R, STMN1, and GPC3.
Discussion
NAFLD represents a spectrum of disorders from simple steatosis to severe forms of liver injury including NASH and liver cancer; however, how the progression from steatosis to NASH is induced and regulated is not fully understood.(1–3) In the current study, we provided several novel findings demonstrating miR-223 is a critical regulator of NASH progression by targeting several inflammatory genes and oncogenes including Cxcl10 and Taz in hepatocytes. Firstly, we demonstrated that miR-223 is highly elevated in hepatocytes from mouse fatty liver, acting as a negative feedback to prevent the progression from simple steatosis to NASH and liver cancer in mouse models. Secondly, we identified several downstream targets of miR-223 in hepatocytes that may explain how miR-223 ameliorates NASH and liver cancer; Finally, miR-223 is also highly elevated in hepatocytes from NASH patients, and several miR-223 target genes are elevated and positively correlated each other in these NASH patients. We have integrated all of these findings in a summarized figure (Fig. 8D) depicting how miR-223 restrains NASH and liver cancer.
MiR-223KO mice develop NASH with liver fibrosis and cancer after HFD feeding: a mouse model of NASH.
Normal C57BL/6 mice develop severe steatosis but are usually resistant to NASH and cancer after long-term HFD feeding.(28) In the current study, we demonstrated that compared to WT littermates, miR-223KO mice develop much more severe features of NASH with fibrosis, as demonstrated by greater liver injury, inflammation, oxidative stress, fibrosis in miR-223KO mice versus WT mice after 3-month HFD feeding. In addition, we showed that p62, the marker for Mallory-Denk body (29) that is the major feature of human NASH,(37) was strongly expressed in hepatocytes from miR-223KO mice, but not in WT mice after HFD feeding. Furthermore, miR-223KO mice was reported to exhibit an increased severity of systemic insulin resistance compared with WT mice after HFD feeding.(38) Finally, miR-223KO mice had a greater susceptibility to hepatocarcinogenesis with stronger liver fibrosis and inflammatory response after long-term HFD feeding compared to WT littermates. Collectively, HFD-fed miR-223KO mice display all the physiological, metabolic, and histological endpoints of human NASH. (1–3) Thus, HFD-fed miRNA-223 KO mice can be used as a clinically relevant mouse model (28) to mimic the pathogenesis progression for the study of human NASH and its associated liver cancer.
The mechanisms by which miRNA-223 restrains NASH.
MiRNA-223 is considered a neutrophil-specific miRNA, playing an important role in suppressing liver inflammation and injury in several liver injury models by reducing neutrophil activation,(14–16) which likely also applies to the protective effect of miR-223 in the HFD-induced NASH development. In the current paper, we identified a novel mechanism by which miR-223 in hepatocytes acts as anti-inflammatory molecule in NASH by directly targeting several inflammatory genes. Firstly, by performing miR-223 in situ hybridization, we clearly detected miR-223 in hepatocytes with marked elevation after HFD feeding. This elevation was likely due to exosomal miR-223 transfer because hepatic pre-miRNA-223 expression was not upregulated in hepatocytes (data not shown) and it is known that miR-223 can be transferred from cells to cells via the exosome.(39) Further studies are required to confirm this notion. Secondly, microarray analyses demonstrated that hepatic Cxcl10 mRNA was highly elevated in HFD-fed miR-223KO mice compared to WT mice. Furthermore, by using bioinformatics approach and in vitro transfection experiments, we demonstrated that Cxcl10 is a direct target of and inhibited by miR-223 in hepatocytes. Several previous studies demonstrated that hepatocyte-derived CXCL10 plays a key role in the pathogenesis and development of NASH,(20, 21, 30, 40) thus miR-223 inhibition of CXCL10 in hepatocytes likely contributes, at least in part, to the anti-NASH effect of miR-223. Thirdly, in addition to CXCL10, of course, several other miR-223 targets in hepatocytes may be also involved in the progression of steatosis to NASH. In this paper, we focused on TAZ because this protein, a downstream molecule of the Hippo signaling pathway, is known to play an important role in promoting liver inflammation and NASH progression.(22) We demonstrated that the level of Taz was significantly increased in the livers of miR-223KO mice compared with WT mice and that Taz is a downstream target of and inhibited by miR-223 in hepatocytes. Thus, miR-223-mediated inhibition of the anti-inflammatory molecule TAZ likely also contributes to the anti-NASH function of miR-223. Finally, in situ hybridization and immunochemical staining demonstrated that miR-223 and its downstream targets (CXCL10 and TAZ) were also markedly increased in hepatocytes in human NASH samples. CXCL10 and TAZ expression levels positively correlated with several cytokine and chemokine genes (IL6, MCP1, MIP1B, and CXCL1) and fibrogenesis genes (α-SMA, COL1A1, COL1A2, and COL4A1) in NASH patients. Most importantly, from the data of RT-qPCR analyses of gene profiles in normal and human NASH samples, we demonstrated that the levels of miR-223 positively correlated with several cytokine and chemokine genes (IL-6, MCP1), fibrogenesis genes (A-SMA, COL1A1, COL3A1, and COL4A1), and potential or proven miR-223 targeted genes (CXCL10, TAZ, SERPINB9, DOCK11, SLC16A6, NLRP3, IGF1R, and STMN1), and cancer-related gene (GPC3) in NASH patients. Collectively, our data suggest that hepatic miR-223 restrains liver inflammation to prevent the progression from fatty liver to NASH, at least by targeting various pro-inflammatory mediators including CXCL10 and TAZ expression in hepatocytes (see summarized Fig. 8D).
The mechanisms by which miRNA-223 restrains NASH-associated liver cancer.
Long-term HFD feeding generated a greater number and size of liver cancer in miR-223KO mice than in WT mice, suggesting that miR-223 restrains NASH-associated HCC. The obvious underlying mechanism for this is that miR-223 attenuates NASH progression as discussed above. Interestingly, the top list of group genes from Ingenuity Pathway Analysis and heatmap diagram of microarray data is associated with liver cancer, including abnormal expression of 172 genes (such as Abca5, Cttnbp2, Cxcl10, Cyp2b13, Cyp7a1, Dock11, Gpc3, Nrxn1, Parp1, Serpinb9, Slc1a4, Slc6a6, Trpm4, Wwtr1, and Zeb2), suggesting miR-223 attenuates HCC by regulating these cancer-associated genes. Moreover, by using the microRNA.org database (41) to analyze miR-223 targeted genes among these upregulated genes, we identified seven potential targets of miR-223 that are related to liver carcinogenesis, including Cxcl10, Taz, Serpinb9, Nrxn1, Slc1a4, Slc16a6, and Dock11. Among these genes, Taz is a well-documented oncogenic gene that encodes a protein to promote liver cancer development.(24, 42, 43) Therefore, miR-223 can directly target multiple oncogenic genes including Taz, thereby attenuating NASH-associated HCC. Interestingly, previous studies have well demonstrated that miR-223 is markedly downregulated in human HCC and liver cirrhosis compared to normal healthy livers, (35, 44) such miR-223 reduction likely leads the upregulation of miR-223-targeted inflammatory and oncogenic genes, thereby accelerating HCC progression.
In conclusion, in addition to the well documented anti-inflammatory functions of miR-223 in neutrophils, miR-223 can also target several inflammatory and oncogenic genes (e.g. Cxcl10 and Taz) in hepatocytes, thereby preventing NASH progression. Overexpression of miR-223 could be a novel therapeutic strategy for the treatment of NASH.
Supplementary Material
Financial support:
This work was supported by the intramural program of NIAAA, NIH (B.G.)
Abbreviations:
- Acc1
acetyl-coenzyme A carboxylase
- Afp
alpha-Fetoprotein
- ALT
alanine aminotransferase
- Ccnb1
Cyclin B1
- Ccnb2
Cyclin B2
- Ccnd1
Cyclin D1
- Ccne1
Cyclin E1
- CD
control diet
- Cidea
Cell death-induced DNA fragmentation factor alpha-like effector A
- Cideb
Cell-death-inducing DFF45-like effector b
- Col1α1
Collagen type 1 alpha 1
- Col1α2
Collagen type 1 alpha 2
- Col3α1
Collagen type 3 alpha 1
- CPT-1
carnitine palmitoyltransferase 1
- CXCL
(C-X-C motif) chemokine
- CXCR3
(C-X-C motif) chemokine receptor 3
- Dock11
Dedicator of cytokinesis 11
- Fas
fatty acid synthase
- FFA
free fatty acid
- Golm1
Golgi membrane protein 1
- Gpc3
Glypican-3
- H&E
hematoxylin and eosin
- HFD
high-fat diet
- 4-HNE
4-Hydroxynonenal
- Icam-1
intercellular adhesion molecule 1
- Igf1r
Insulin-like growth factor I receptor
- MCP-1
monocyte chemotactic protein 1
- MDA
malonaldehyde
- Mef2c
myocyte enhancer factor 2C
- Mip
macrophage inflammatory protein
- miR-223
microRNA-223
- miR-223KO
miR-223 knockout
- Mmp-13
Matrix Metallopeptidase 13
- MPO
myeloperoxidase
- mRNA
messenger RNA
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- Nlrp3
NLR family, pyrin domain containing 3
- Nrxn1
Neurexin I
- PPAR
Peroxisome proliferator-activated receptor
- SCD-1
Stearoyl-CoA desaturase-1
- Serpinb9
Serpin family B member 9
- Slc1a4
Solute Carrier Family 1 member 4
- Slc16a6
Solute Carrier Family 16 member 6
- α-SMA
α-smooth muscle actin
- Srebp-1c
sterol regulatory element-binding protein 1
- TAZ
transcriptional coactivator with PDZ-binding motif
- WT
wild-type
- Wwtr1
WW domain-containing transcription regulator protein 1
Footnotes
Potential conflict of interest: Nothing to report
REFERENCES:
- 1.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018;24:908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibrahim SH, Hirsova P, Gores GJ. Non-alcoholic steatohepatitis pathogenesis: sublethal hepatocyte injury as a driver of liver inflammation. Gut 2018;67:963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machado MV, Diehl AM. Pathogenesis of Nonalcoholic Steatohepatitis. Gastroenterology 2016;150:1769–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Younossi Z, Tacke F, Arrese M, Sharma BC, Mostafa I, Bugianesi E, et al. Global Perspectives on Non-alcoholic Fatty Liver Disease and Non-alcoholic Steatohepatitis. Hepatology 2018. [DOI] [PubMed] [Google Scholar]
- 5.Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018;67:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology 2017;65:1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter TG, Charlton M. The Triumph of Bacchus: The Emergence of Nonalcoholic Steatohepatitis and Alcoholic Liver Disease as the Leading Causes of Mortality from Cirrhosis. Hepatology 2018. [DOI] [PubMed] [Google Scholar]
- 8.Gao B, Tsukamoto H. Inflammation in Alcoholic and Nonalcoholic Fatty Liver Disease: Friend or Foe? Gastroenterology 2016;150:1704–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Windt DJ, Sud V, Zhang H, Varley PR, Goswami J, Yazdani HO, et al. Neutrophil extracellular traps promote inflammation and development of hepatocellular carcinoma in nonalcoholic steatohepatitis. Hepatology 2018;68:1347–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, Xu MJ, Cai Y, Wang W, Jiang JX, Varga ZV, et al. Neutrophil-Hepatic Stellate Cell Interactions Promote Fibrosis in Experimental Steatohepatitis. Cell Mol Gastroenterol Hepatol 2018;5:399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogt KL, Summers C, Chilvers ER, Condliffe AM. Priming and de-priming of neutrophil responses in vitro and in vivo. Eur J Clin Invest 2018;48 Suppl 2:e12967. [DOI] [PubMed] [Google Scholar]
- 12.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008;451:1125–1129. [DOI] [PubMed] [Google Scholar]
- 13.Yuan X, Berg N, Lee JW, Le TT, Neudecker V, Jing N, Eltzschig H. MicroRNA miR-223 as regulator of innate immunity. J Leukoc Biol 2018;104:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye D, Zhang T, Lou G, Liu Y. Role of miR-223 in the pathophysiology of liver diseases. Exp Mol Med 2018;50:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Feng D, Li M, Gao Y, Ramirez T, Cao H, et al. Hepatic mitochondrial DNA/Toll-like receptor 9/MicroRNA-223 forms a negative feedback loop to limit neutrophil overactivation and acetaminophen hepatotoxicity in mice. Hepatology 2017;66:220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, He Y, Zhou Z, Ramirez T, Gao Y, Gao Y, et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47(phox)-oxidative stress pathway in neutrophils. Gut 2017;66:705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schueller F, Roy S, Loosen SH, Alder J, Koppe C, Schneider AT, et al. miR-223 represents a biomarker in acute and chronic liver injury. Clin Sci (Lond) 2017;131:1971–1987. [DOI] [PubMed] [Google Scholar]
- 18.Katsura A, Morishita A, Iwama H, Tani J, Sakamoto T, Tatsuta M, et al. MicroRNA profiles following metformin treatment in a mouse model of non-alcoholic steatohepatitis. Int J Mol Med 2015;35:877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015;64:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomita K, Freeman BL, Bronk SF, LeBrasseur NK, White TA, Hirsova P, Ibrahim SH. CXCL10-Mediates Macrophage, but not Other Innate Immune Cells-Associated Inflammation in Murine Nonalcoholic Steatohepatitis. Sci Rep 2016;6:28786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Shen J, Man K, Chu ES, Yau TO, Sung JC, et al. CXCL10 plays a key role as an inflammatory mediator and a non-invasive biomarker of non-alcoholic steatohepatitis. J Hepatol 2014;61:1365–1375. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Zheng Z, Caviglia JM, Corey KE, Herfel TM, Cai B, et al. Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell Metab 2016;24:848–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Wu WK, Xu W, Man K, Wang X, Han J, et al. C-X-C Motif Chemokine 10 Impairs Autophagy and Autolysosome Formation in Non-alcoholic Steatohepatitis. Theranostics 2017;7:2822–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W, Khan SK, Liu Y, Xu R, Park O, He Y, et al. Hepatic Hippo signaling inhibits protumoural microenvironment to suppress hepatocellular carcinoma. Gut 2018;67:1692–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y, Feng D, Ma X, Fan S, Gao Y, Fu K, et al. Pregnane × Receptor Regulates Liver Size and Liver Cell Fate by Yes-Associated Protein Activation in Mice. Hepatology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen BS, Moller T, Holmstrom K. Chromogen detection of microRNA in frozen clinical tissue samples using LNA probe technology. Methods Mol Biol 2014;1211:77–84. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen BS, Holmstrom K. Combined microRNA in situ hybridization and immunohistochemical detection of protein markers. Methods Mol Biol 2013;986:353–365. [DOI] [PubMed] [Google Scholar]
- 28.Farrell G, Schattenberg JM, Leclercq I, Yeh MM, Goldin R, Teoh N, Schuppan D. Mouse models of nonalcoholic steatohepatitis Towards optimization of their relevance to human NASH. Hepatology 2018. [DOI] [PubMed] [Google Scholar]
- 29.Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, et al. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res 2007;313:2033–2049. [DOI] [PubMed] [Google Scholar]
- 30.Ibrahim SH, Hirsova P, Tomita K, Bronk SF, Werneburg NW, Harrison SA, et al. Mixed lineage kinase 3 mediates release of C-X-C motif ligand 10-bearing chemotactic extracellular vesicles from lipotoxic hepatocytes. Hepatology 2016;63:731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahan RH, Porsche CE, Edwards MG, Rosen HR. Free Fatty Acids Differentially Downregulate Chemokines in Liver Sinusoidal Endothelial Cells: Insights into Non-Alcoholic Fatty Liver Disease. PLoS One 2016;11:e0159217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahin H, Borkham-Kamphorst E, do ON, Berres ML, Kaldenbach M, Schmitz P, et al. Proapoptotic effects of the chemokine, CXCL 10 are mediated by the noncognate receptor TLR4 in hepatocytes. Hepatology 2013;57:797–805. [DOI] [PubMed] [Google Scholar]
- 33.Lake AD, Novak P, Fisher CD, Jackson JP, Hardwick RN, Billheimer DD, et al. Analysis of global and absorption, distribution, metabolism, and elimination gene expression in the progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos 2011;39:1954–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neudecker V, Haneklaus M, Jensen O, Khailova L, Masterson JC, Tye H, et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med 2017;214:1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 2008;135:257–269. [DOI] [PubMed] [Google Scholar]
- 36.Jia CY, Li HH, Zhu XC, Dong YW, Fu D, Zhao QL, et al. MiR-223 suppresses cell proliferation by targeting IGF-1R. PLoS One 2011;6:e27008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunt EM, Kleiner DE, Wilson LA, Sanyal AJ, Neuschwander-Tetri BA, Nash CRN. Improvements in Histologic Features and Diagnosis associated with Improvement in Fibrosis in NASH: Results from the NASH Clinical Research Network Treatment Trials. Hepatology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 2012;125:2892–2903. [DOI] [PubMed] [Google Scholar]
- 39.Neudecker V, Brodsky KS, Clambey ET, Schmidt EP, Packard TA, Davenport B, et al. Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomita K, Kabashima A, Freeman BL, Bronk SF, Hirsova P, Ibrahim SH. Mixed Lineage Kinase 3 Mediates the Induction of CXCL10 by a STAT1-Dependent Mechanism During Hepatocyte Lipotoxicity. J Cell Biochem 2017;118:3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008;36:D149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel SH, Camargo FD, Yimlamai D. Hippo Signaling in the Liver Regulates Organ Size, Cell Fate, and Carcinogenesis. Gastroenterology 2017;152:533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagenbeek TJ, Webster JD, Kljavin NM, Chang MT, Pham T, Lee HJ, et al. The Hippo pathway effector TAZ induces TEAD-dependent liver inflammation and tumors. Sci Signal 2018;11. [DOI] [PubMed] [Google Scholar]
- 44.Yu G, Chen X, Chen S, Ye W, Hou K, Liang M. MiR-19a, miR-122 and miR-223 are differentially regulated by hepatitis B virus × protein and involve in cell proliferation in hepatoma cells. J Transl Med 2016;14:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.