Abstract
Clostridioides difficile is a Gram-positive, anaerobic bacterium. It is known that C. difficile is one of the major causes of antibiotic associated diarrhea. The enhanced antibiotic resistance observed in C. difficile is the result of highly resistant spores produced by the bacterium. In Bacillus subtilis, the sin operon is involved in sporulation inhibition. Two proteins coded within this operon, SinR and SinI, have an antagonistic relationship; SinR acts as an inhibitor to sporulation whereas SinI represses the activity of SinR, thus allowing the bacterium to sporulate. In a previous study, we examined the sin locus in C. difficile and named the two genes associated with this operon sinR and sinR’, analogous to sinR and sinI in B. subtilis, respectively. We have shown that SinR and SinR’ have pleiotropic roles in pathogenesis pathways and interact antagonistically with each other. Unlike B. subtilis SinI, SinR’ in C. difficile carries two domains: the HTH domain and the Multimerization Domain (MD). In this study, we first performed a GST Pull-down experiment to determine the domain within SinR’ that interacts with SinR. Second, the effect of these two domains on three phenotypes; sporulation, motility, and toxin production was examined. The findings of this study confirmed the prediction that the Multimerization Domain (MD) of SinR’ is responsible for the interaction between SinR and SinR’. It was also discovered that SinR’ regulates sporulation, toxin production and motility primarily by inhibiting SinR activity through the Multimerization Domain (MD).
Keywords: Clostridium difficile, Clostridioides difficile sin locus, SinR, toxin production, sporulation, motility, gene regulation
INTRODUCTION
Clostridioides (Clostridium) difficile is a Gram-positive, anaerobic bacterium that is the leading cause of antibiotic associated diarrhea and pseudomembranous colitis [1]. The gastrointestinal infections associated with this bacterium, known as C. difficile infection (CDI), mainly affect individuals undergoing antibiotic treatment. CDI is the most common and costly healthcare-associated infection with an estimate of nearly half a million cases and approximately 29,000 deaths occurring annually in the United States [2]. C. difficile produce Toxins A and B, which damage the colonic epithelium, resulting in moderate to severe diarrhea [3]. Due to the strictly anaerobic nature of the vegetative cells, C. difficile survives outside the host in the form of dormant spores, which contribute to the transmission of C. difficile in the health care setting [4].
Once inside the host gastrointestinal tract, C. difficile pathogenesis is associated with spore germination, toxin production and motility of the vegetative cells in certain C. difficile toxigenic strains [5]. C. difficile spores germinate into vegetative cells in the lower gastrointestinal tract since the oxygen concentration at this site is negligible. Certain substances that are present in the intestine, importantly certain bile acids, induce germination of the spore into an actively replicating vegetative cell [6,7]. Following spore germination, the vegetative cells secrete Toxins A and B, that damage the intestinal epithelial cells and cause dissociation of the tight junctions [8]. Although toxins serve as the major virulence factor of C. difficile, motility and adherence were shown to be important for its successful colonization and persistence in the host intestine [9,10].
The genes coding for Toxins A and B, tcdA and tcdB, are localized within a Pathogenicity Locus (PaLoc). Other accessory genes present in the PaLoc include tcdR, tcdC and tcdE. TcdR is an alternative sigma factor that facilitates the transcription of tcdA and tcdB genes, while positively influencing its own transcription [11]. A recent study discovered that SigD, the sigma factor needed for the expression of the flagellar operon, positively regulates tcdR expression. Thus, SigD indirectly influences toxin production [12,13]. Since flagellar expression has been linked to bacterial adherence, linking flagellar production with toxicity ensures C. difficile produces toxins after successfully colonizing the host. Recent studies have also suggested that the regulatory pathways controlling sporulation and toxin production are interconnected in C. difficile. For example, mutations in spo0A, the master regulator of sporulation, affected toxin production in specific C. difficile isolates [14,15]. Mutations in rstA, sigH and codY influenced all three pathways of sporulation, toxin production and motility [16–19].
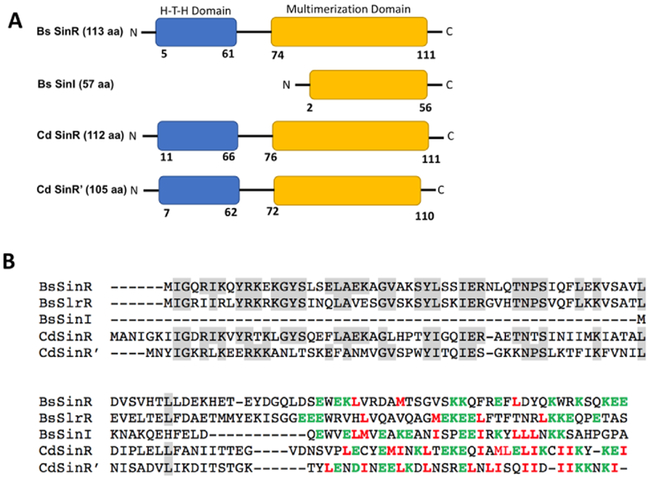
In Bacillus subtilis, the sin (named as such, because of its sporulation inhibition action) operon encodes SinR and SinI. B. subtilis SinR (BsSinR) inhibits sporulation, whereas SinI acts as an inhibitor of BsSinR [20–22]. The C. difficile sin locus also codes for two regulators. However, both of these regulators show homology to BsSinR, hence named as SinR and SinR’ [23]. Previously we have shown that regulators coded in the sin locus positively influence toxin production along with sporulation and motility [23]. A recent study also reported that members of the C. difficile sin locus suppress biofilm formation [24]. Similar to the BsSinI-SinR interaction, the C. difficile SinR’ (CdSinR’) binds to and regulates the activity of SinR [25,26]. An important difference between C. difficile SinR’ and B. subtilis SinI is the presence of a Helix-Turn-Helix DNA binding domain (HTH) in addition to the Multimerization Domain (MD) (Fig. 1A). This suggests that SinR’ could act as a transcriptional activator, independent of SinR. In our previous study, we have shown that in the absence of SinR’ (R20291::sinR’), C. difficile cells produced elevated levels of spores, toxins and were highly motile compared to the R20291 parent strain [23]. This suggested that SinR’ must have a central role in regulating these pathways. It is possible that SinR’ directly regulates key genes in these pathways by acting as a transcriptional activator or repressor. Alternatively, SinR’ could indirectly influence these pathways by regulating SinR, utilizing its Multimerization Domain. To decipher the importance of HTH and MD of CdSinR’ in these pathways, we complemented the R20291:: sinR’ mutant with either the Multimerization or HTH domains of SinR’, observing the resulting phenotypes. Our results indicated that the MD of SinR’ regulates the pleiotropic regulator SinR, indirectly regulating sporulation, toxin production and motility in C. difficile R20291.
Figure 1. Comparison of B. subtilis and C. difficile Sin regulators.
(A) The HTH and MD domains within B. subtilis and C. difficile Sin proteins. (B) Sequence alignment of the C. difficile SinR and SinR’ with Bacillus subtilis SinR (BsSinR), SinI (BsSinI) and SlrR using ClustalW. The hydrophobic and polar residues in C-terminus region that could form intermolecular hydrophobic core are highlighted in green and red, respectively.
MATERIALS AND METHODS
Bacterial strains, Growth Conditions, and Plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. C. difficile strains were grown in an anaerobic chamber maintaining optimum growth environment (10% H2, 10% C02 and 80% N2) at 37°C in TY (Tryptose and Yeast Extract) media supplemented with antibiotics (Erythromycin (Erm; 2.5 μg ml−1), Lincomycin (Linco; 20ug ml−1), Cefoxitin (Cef; 25 μg ml−1), Thiamphenicol (Thio; 15 μg ml−1) when needed. Sporulation assays were performed with C. difficile strains grown in 70:30 media (63 g Bacto-Peptone, 3.5 g Protease Peptone, 11.1 g BHI, 1.5 g Yeast Extract, 1.06 g Tris base, 0.7 g Ammonium Sulfate, and 15 g agar/liter) and they were supplemented with the antibiotics as needed. For conjugation, the Eschericia coli S17–1 cells carrying C. difficile shuttle vectors were grown aerobically at 37°C in Luria-Bertani (LB) broth supplemented with chloramphenicol (25μg ml−1). GeneClean Kit (mpbio) was used for agarose gel DNA purification. C. difficile genomic DNA was isolated using DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer’s instruction. Qiaprep Spin Miniprep Kit was used for plasmid DNA isolation. Standard cloning procedures were used to create the plasmid constructs used in this study.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid |
Relevant features or genotype | Source or reference |
|---|---|---|
| Clostridioides difficile R20291 | Clinical isolate - NAP1/027 ribotype, isolated in 2006 following an outbreak in Stoke Mandeville Hospital, UK | [33] |
| Escherichia coli DH5α | endAI recA1 deoR hsdR17 (rK− mK+) | NEB |
| Escherichia coli S17-1 | Strain with integrated RP4 conjugation transfer function; favors conjugation between E. coli and C. difficile | [34] |
| C. difficileR20291 ::sinR’: | R20291with intron insertion within sinR’ | [23] |
| pRPF185 | E. coli/C. difficile shuttle plasmid | [35] |
| pRG306 | pRPF185 containing sinR’ under inducible tet promoter | This study |
| pBPG004 | pRPF185 containing HTH Domain under inducible tet promoter | This study |
| pBPG003 | pRPF185 containing MD Domain under inducible tet promoter | This study |
| pRG327 | pET16B containing sinR gene with His tag | This study |
| pGST parallel II | GST parallel II vector for GST fusions | [27] |
| pRG331 | GST parallel II containing sinR’ with GST tag | This study |
| pAYC001 | GST parallel II containing HTH with GST tag | This study |
| pAYC002 | GST parallel II containing MD with GST tag | This study |
SinR-6His; SinR-GST pull-down experiment
To express GST tagged SinR’-HTH and MD domains, we cloned their respective coding regions in the pGST-parallel2 expression system [27]. Oligonucleotides used in this study are listed in Table 2. First, the sinR’-HTH and the sinR’-MD regions were PCR amplified with primer ORG619/ORG709 and ORG710/ORG620, respectively, using R20291 chromosomal DNA as a template. The PCR fragments were then cloned in between NcoI and SalI sites of the pGST-parallel2 vector. The resulting plasmid was then transformed into E. coli Rosetta DE3 competent cells to obtain the recombinant strains. To overexpress recombinant proteins, E. coli recombinant strains were grown at 37°C in LB medium containing chloramphenicol (25 μg ml−1). Protein expression was achieved by inducing with 1mM IPTG at 17°C overnight with mild agitation. To perform the pull-down experiment, 200 μgs of whole cell lysate proteins from the E. coli cells expressing various SinR’-GST constructs were mixed with approximately 20 μgs of purified SinR-6His protein and incubated at 4°C for 1 hour. The mixture was then passed through the Ni++ affinity column (Sigma-Aldrich) to trap and elute SinR-6His protein. Whole lysates from E. coli cells expressing GST alone or the full -length SinR’ were also mixed with purified SinR-6His protein and were processed in the same way as the test samples. The elutes from Ni++ columns were then separated using 4-15% gradient SDS-PAGE gels and they were electroblotted onto PVDF membrane. Membranes with immobilized proteins were then probed with either Mouse anti-6His-HRP conjugated (Novagen) antibodies at 1:10,000 dilution or with Rabbit anti-GST (Genescript) antibodies at the dilution of 1:5000 followed by HRP conjugated anti-rabbit (Genescript) secondary antibodies at 1:10,000 dilution. Immunodetection of proteins was performed using Luminata Forte Western HRP Substrate and the Syngene G:BOX imaging system.
Table 2.
Oligos used in this study
| Name | Sequence (5’ → 3’) | Description |
|---|---|---|
| ORG553c | GTTAACAGATCTGAGCTCGTGAGGGAAATAGTAACAATAATGAATTATATAG | sinR’ Forward with SacI-pRPF185 (for pRG306) |
| ORG554 | AAGTTTTATTAAAACTTATAGGATCCTTATATTTTATTCTTTTTTATGATGTCTATAATC | sinR’ Reverse with BamH1 -pRPF185 (for pRG306) |
| ORG700 | AAGTTTTATTAAAACTTATAGGATCCTTAGGAAGTAATATCTTTTATAAGAACATCTGC | sinR’ HTH domain reverse (1-68aa)(For pBPG004) |
| ORG701c | GAGCTCGTGAGGGAAATAATGATAAAAGATATTACTTCCACTGGAAAAACATATTTAGAA | sinR’ MD domain forward (63-105aa)(For pBPG003) |
| ORG619 | CGATACGACCGAAAACCTGTATTTTCAGGGCGCCATGGGGATGAATTATATAGG | sinR’ forward GST with NcoI (pRG331) |
| ORG620 | GCGGCCGCACTAGTTGAGCTCGTCGACTTATATTTTATTCTTTTTTATGATGTC | sinR’ rev GST with Sall (pRG331) |
| ORG710 | GCGCCATGGGTGAGGGAAATAATGATAAAAGATATTACTTCCACTGGAAAAACATATTTA | sinR’ forward (MD) (63-105aa) withNcol (pAYC002) |
| ORG709 | GCGGCCGCACTAGTTGAGCTCGTCGACTTAGGAAGTAATATCTTTTATAAGAACATCTGC | SinR’ reverse domain (HTH) (1-68aa) reverse GST with Sall (pAYC001) |
Expressing sinR’-MD and sinR’-HTH in R20291 ::sinR’
Methods used for the creation of R20291::sinR’ mutant have been described elsewhere [23]. To express sinR’-MD and sinR’-HTH, we cloned them in Clostridial shuttle vector pRPF185 under the tetracycline inducible promoter. The oligonucleotides ORG553 with ORG700 and ORG554 with ORG701 were used to amplify DNA regions in CdSinR’ coding for HTH and MD, respectively. The PCR products and pRPF185 vector were then digested with the restriction enzymes BamHI and SacI before ligation of the vector and PCR products. The resulting plasmid constructs, pBPG003 (with MD) and pBPG004 (with HTH), were then introduced into C. difficile R20291::sinR’ by conjugation following the protocol described previously [23,28]. Transconjugants with the plasmids were grown in the presence of Thiamphenicol (Thio; 15 μg ml−1) and anhydrous tetracycline (ATc; 100 ng ml−1) was used to induce the expression of sinR’ constructs whenever needed. Transconjugants with the vector pRPF185 and the plasmid expressing full length SinR’ were also isolated and used as controls.
Toxin ELISA
Toxin levels of C. difficile test strains were measured using Premier Toxin A&B enzyme linked immunosorbent assay (ELISA) kit from Meridian Diagnostics Inc. (Cincinnati, OH). Purified C. difficile toxins B (Cayman chemical) was used in the ELISA as per the recommended protocol by the manufacturer to construct a standard curve (Supplemental Figure 1). C. difficile cultures were grown in TY broth and harvested by centrifugation at 12 and 24 h of growth. Two hundred microliters of the culture supernatants were used for toxin measurements. The resulting pellets were re-suspended in 200 μl PBS supplemented with PMSF (2mM final concentration) and were sonicated. The cytosolic fraction was collected by centrifugation and 100 μg of cytosolic proteins were used to measure cytosolic toxin content following the manufacturer’s recommended protocol. Toxin ELISA was performed in 3 replicates and independently repeated at least three times.
Motility Assay
Test strains were grown in TY supplemented with Thiamphenicol (Thio; 15 μgml−1) and Lincomycin (Linco; 20 μgml−1) until late exponential phase (12 hours) [23]. Cell density was adjusted to 0.5 OD at 600 nm using fresh TY medium. 5 mL of each culture was stabbed using long pipet tips in 0.3% TY agar tubes or spotted in 0.3% TY agar plates supplemented with Thiamphenicol (Thio; 15 μg ml−1) and anhydrous tetracycline (ATc; 100 ng ml−1). After incubation at 37°C, the motility was quantified by measuring the radius of the cultures at different time points. Motility assay was performed in 4 replicates and independently repeated at least three times.
Sporulation Efficiency
For the sporulation efficiency assay, C. difficile strains were initially grown in TY broth with taurocholate (Tauro; 0.1%) for 10-12 hours. Equal number of cells (normalized by OD600) were then used to inoculate fresh 70:30 medium supplemented with Thiamphenicol (Thio; 15 μgml−1). The cultures were grown until they reached OD600 of 0.6 to 0.8, before inducing with anhydrous tetracycline (ATc; 100 ng ml−1). After 24, 48 and 72 hours of growth, cultures were centrifuged and the pellets were re-suspended in 30 μl of 70:30 medium. 10 μl of culture was added to 990 μl of nuclease free water. Each successive dilution was performed by adding 100 μl of solution to 900 μl of nuclease free water. Serial dilution was performed 5-8 times and 5 μL of each dilution was spotted on 70:30 sporulation agar plates with Thiamphenicol (Thio; 15 μgml−1). Additionally, cells were treated with 95% ethanol for one hour to eliminate vegetative cells and enumerate spores using microscopy [23]. The aforementioned serial dilution was also performed on ethanol treated cells and 5 μl of each dilution was spotted onto 70:30 sporulation agar (63 g Bacto-Peptone, 3.5 g Protease-Peptone, 11.1 g BHI, 1.5 g Yeast-Extract, 1.06 g Tris base, 0.7 g NH4SO4, 15 g agar per liter) supplemented with Thiamphenicol (Thio; 15 μgml−1) [29]. Both ethanol treated and untreated cells were anaerobically incubated for 72 hours at 37°C and sporulation efficiency was calculated from resulting bacterial colony growth as previously described [23,28].
Statistical Analyses
Statistical analyses for sporulation, toxin assay and motility were performed using one way-ANOVA with Dunnett’s multiple comparisons test comparing values to the average of the sinR’ mutant with vector control.
RESULTS AND DISCUSSION
Determining the SinR interacting domain in SinR’
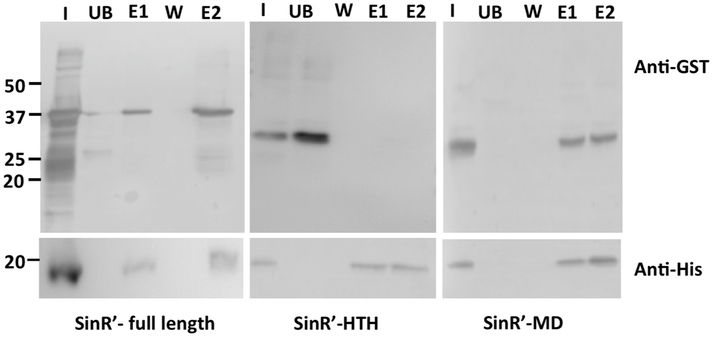
In B. subtilis, the BsSinR protein carries 113 amino acid residues and is composed of a Helix-Turn-Helix (HTH) DNA binding domain and a Multimerization Domain (MD). The HTH domain is made up of residues from 1–69 aa and the MD from 74–111 aa. The BsSinI protein carries just 57 aa residues and contains only the MD [20]. The crystal structure of BsSinI–BsSinR revealed both proteins interacting through their MDs connected by a short linker [30]. Unlike BsSinI, the C. difficile SinR’ carries both the HTH domain and the MD that span from residues 1–63 aa and 76–110 aa, respectively (Fig 1A, 1B). We performed protein-protein interaction studies using the GST-pulldown procedure to determine the SinR’ domain interacting with SinR. We expressed and purified SinR-6His as previously described [23]. The predicted coding regions of HTH and the MD of SinR’ were cloned and expressed in GST-Parallel vector to perform co-purification pull-down experiments. Results showed that full length SinR’ could be pulled ‘ down, forming the 37 kDa SinR-SinR’ complex as expected (Fig. 2). Among the tested ί samples, only the SinR-MD could be co-purified along with SinR-6His, whereas the SinR-HTH was found only in the unbound and wash fractions, showing that it failed to interact with SinR (Fig. 2). These results proved that similar to the B. subtilis SinR-SinI complex, C. difficile SinR’ also interacts with SinR using its Multimerization Domain. In B. subtilis, the interactions of SinR with its partner proteins, SinI and SlrR, are mainly mediated by their C-terminus hydrophobic residues [30]. It is interesting to note that SinR and the MD of SinR’ also possess several hydrophobic residues towards the end of their C-terminus region, which could potentially form an intermolecular hydrophobic core, possibly driving the protein-protein interaction (Fig 1B).
Figure 2. SinR’ interacts with SinR through its Multimerization Domain.
(A) In vitro, protein-protein interactions indicate that SinR’-MD binds tightly to SinR. E. coli whole cell lysates expressing various GST-tagged SinR’ proteins were incubated with SinR-6His proteins and were purified using Ni++ agarose affinity columns. The elutes were probed with Rabbit Anti-GST and with Mouse Anti-His antibodies. Lanes details are as follows:
I. Input 1: Mixture of SinR’-GST expressing E. coli lysate with purified SinR-6His
UB. Unbound from input 1 after passing through Ni++ column
W. First wash
E1. Elute with 50 mM imidazole (GST+SinR-6His)
E2. Elute with 200 mM imidazole (SinR’-GST + SinR-6His)
CdSinR’ regulates toxin production, motility and sporulation by controlling the activity of CdSinR
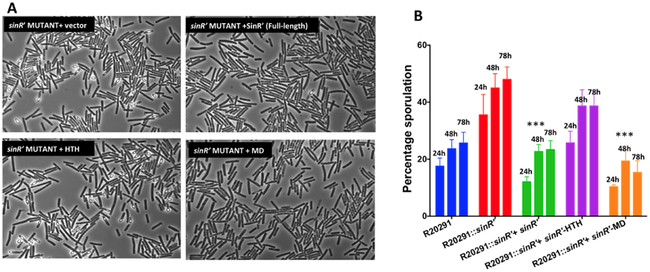
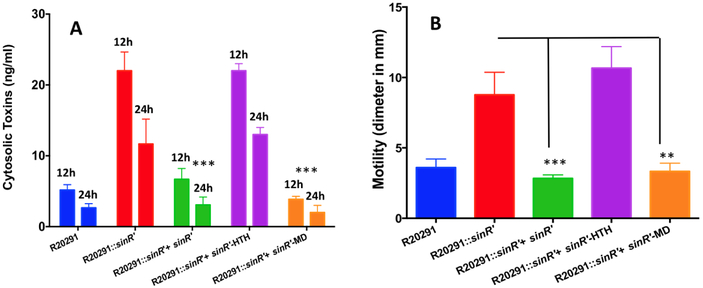
We previously reported that a mutation of sinR’ in C. difficile R20291 strain resulted in hyper sporulation, increased toxin production and elevated motility [23]. Mutants lacking functional SinR’ produced approximately 3.5-fold more spores than the R20291 parent strain (Fig. 3A and 3B). Mutant strains also produced almost 2.5-fold more cytosolic toxin than the parent strain (Fig. 4A). Furthermore, a hyper-motile phenotype was also observed in the sinR’ mutant (Fig. 4B). These results clearly demonstrate that SinR’ negatively regulates these pathways. In our previous study, we determined that SinR positively regulates toxin production, sporulation and motility in C. difficile R20291 [23]. Hence, SinR’ could indirectly regulates these pathways by controlling the activity of SinR. To determine the importance of SinR’ interaction with SinR in regulation of these pathways, we introduced the plasmid constructs that express either full length sinR’ or the truncated sinR’ with either the HTH domain or MD into the R20291::sinR’ mutant. Mutant strain carrying the plasmid alone was used as a control. Phenotypic analyses of the mutant expressing different sinR’ constructs were performed for sporulation, toxin production and motility. As expected, expression of the full length SinR’ complemented the mutant phenotypes and returned sporulation, toxin production and motility to levels similar to those observed in the R20291 parent strain (Fig 3AB, Fig. 4AB). R20291::sinR’ expressing full length SinR’ produced approximately 4.2 fold less spores and approximately 3.5 fold less toxins compared to the mutant R20291::sinR’ (Fig. 3A ,and Fig. 3B). Toxins measured in the culture supernatants of these cultures showed the similar trend as the cytosolic toxins (Supplemental Fig. 2). Comparison of motility between the two strains further strengthened the observed pattern, with the R20291::sinR’ mutant presenting a 2.5 fold increase in diameter of bacterial growth when compared to the complemented strain. Expression of the sinR’-HTH domain showed no complementation of the three phenotypes tested, indicating that it has no role in the relieving of SinR repression from its target genes (Fig. 4B, Supplemental Fig. 2). Complementation of the R20291::sinR’ mutant strain with the MD returns each of the three phenotypes to levels observed in both the R20291 parent strain and the R20291::sinR’ strain expressing the full length SinR’. Strains expressing the MD produced approximately 3.5-fold less toxins and 3.7-fold less spores when compared to R20291::sinR’ mutant strain (Fig. 4B, Supplemental Fig. 2).
Figure 3. Effect of SinR’ HTH and MD on sporulation.
Hyper-sporulation phenotype of sinR’ mutant could be complemented by expressing either full-length SinR’ or the SinR’-MD. (A) Phase contrast microscopy of R20291::sinR’ strain expressing either full-length SinR’ or the truncated SinR’-HTH, SinR’-MD. (B) C. difficile cultures were grown in 70:30 medium under anaerobic conditions and sporulation frequency (CFU/ml of ethanol resistant spores) of R20291::sinR’ and the mutant complemented with different constructs were determined at 24, 48 and 72h of growth. The experiments were repeated at least three times independently. Statistical analysis was performed using one way-ANOVA with Dunnett’s multiple comparisons test comparing values to the average of the sinR’ mutant with vector control (***<0.0005, ** <0.005 p value).
Figure 4. Effect of SinR’ HTH and MD on toxin production and motility.
(A) Toxin production measured by toxins specific ELISA. Cytosolic proteins were collected at 12 and 24 h of growth and the toxins concentrations were measured using ELISA (B) Motility assays of the C. difficile R20291, sinR’ mutant and complemented sinR’ mutant. The experiments were repeated at least three times independently. Statistical analysis was performed using one way-ANOVA with Dunnett’s multiple comparisons test comparing values to the average of the sinR’ mutant with vector control (***<0.0005 p value).
These results suggest that SinR’ regulates sporulation, toxin production and motility, mainly by controlling the repressor activity of SinR in C. difficile. This interaction is very similar to SinI controlling SinR repressor function in B. subtilis to regulate sporulation and biofilm formation [20,30]. This result raises questions regarding the importance of the HTH domain in SinR’ and its role in C. difficile physiology. One possibility is that SinR’ might bind to specific DNA sequences and could regulate specific sets of target genes. Another possibility is that the SinR-SinR’ complex may specifically bind to unique promoters to regulate certain genes. For example, in B. subtilis, the SlrR controls autolysis through interaction with SinR [31,32]. The SinR-SlrR complex binds to and inhibits lytABC and lytF promoters, which direct autolysin gene expression, a repressor function possessed by neither SinR nor SlrR alone. Although the exact nature of the SinR-SlrR complex is not known, it was predicted that the SinR-SlrR complex might be binding to pairs of inverted repeat DNA sequences; one constitutes SinR consensus sequences and the other constitutes SlrR consensus sequences [31]. Experiments are under progress in our lab to find the consensus sequences on which C. difficile SinR and SinR’ bind. These experiments may potentially lead to the identification of unique promoters targeted by the SinR-SinR’ complex.
Conclusion
In summary, the observed sinRR’ regulatory mechanism of toxin production, sporulation and motility is driven primarily by the inactivation of SinR through SinR’ using its Multimerization Domain. Even though the importance of SinR’-SinR interaction is evident, the mechanisms of SinR controlling toxin production, sporulation and motility remain elusive, warranting further study. Additionally, study of the functionality of the HTH domain within SinR’ may lead to undiscovered regulatory components and pathways of the sin locus.
Supplementary Material
Highlights.
Clostridioides difficile pathogenicity is controlled by the sporulation inhibition (sin) locus.
The pleiotropic regulator SinR acts as a regulator for sporulation, toxin production, and motility pathways.
Regulation of SinR is achieved by SinR’. The antagonistic relationship observed between SinR and SinR’ causes inactivation of SinR regulatory mechanisms.
SinR’ encodes an HTH DNA binding domain (1-63 aa) and Multimerization Domain (76-110 aa). SinR’ directly regulates SinR primarily through a protein-protein interaction with the Multimerization Domain.
Acknowledgement
We thank following investigators for sharing their lab resources: Nigel Minton, University of Nottingham, for the plasmid pMTL007C-E5; Robert Fagan for the vector pRPF185. We thank Thomas Williams for technical assistance throughout the study. RG is supported by NIAID (1R15AI122173, 1R03AI135762-01A1). Funds from the Johnson Cancer Center-KSU and a pilot project to RG from CBID-KU (1P20GM113117-01) also supported this work. A scholarship fund from the IDeA-P20 GM103418 supported YC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- [1].Martin JSH, Monaghan TM, Wilcox MH, Clostridium difficile infection: epidemiology, diagnosis and understanding transmission, Nat Rev Gastroenterol Hepatol. 13 (2016) 206–216. doi: 10.1038/nrgastro.2016.25. [DOI] [PubMed] [Google Scholar]
- [2].CDC Press Releases, CDC. (2016). https://www.cdc.gov/media/releases/2015/p0225-Clostridiumdifficile.html (accessed October 23, 2018).
- [3].Chandrasekaran R, Lacy DB, The role of toxins in Clostridium difficile infection, FEMS Microbiol. Rev. 41 (2017) 723–750. doi: 10.1093/femsre/fux048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ, Clostridium difficile infection, Nat Rev Dis Primers. 2 (2016) 16020. doi: 10.1038/nrdp.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Abt MC, McKenney PT, Pamer EG, Clostridium difficile colitis: pathogenesis and host defence, Nat. Rev. Microbiol. 14 (2016) 609–620. doi : 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Theriot CM, Bowman AA, Young VB, Antibiotic-Induced Alterations of the Gut Microbiota Alter Secondary Bile Acid Production and Allow for Clostridium difficile Spore Germination and Outgrowth in the Large Intestine, MSphere. 1 (2016). doi : 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Metabolism of bile salts in mice influences spore germination in Clostridium difficile. - PubMed - NCBI, (n.d.). https://www.ncbi.nlm.nih.gov/pubmed/20090901/ (accessed October 23, 2018). [DOI] [PMC free article] [PubMed]
- [8].Hunt JJ, Ballard JD, Variations in virulence and molecular biology among emerging strains of Clostridium difficile, Microbiol. Mol. Biol. Rev. 77 (2013) 567–581. doi: 10.1128/MMBR.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D, Clostridium difficile virulence factors: Insights into an anaerobic spore-forming pathogen, Gut Microbes. 5 (2014) 579–593. doi: 10.4161/19490976.2014.969632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK, Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response, Gut Microbes. 3 (2012) 121–134. doi: 10.4161/gmic.19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mani N, Dupuy B, Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor, Proc. Natl. Acad. Sci. U.S.A. 98 (2001) 5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R, The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD, J. Bacteriol. 195 (2013) 5174–5185. doi: 10.1128/JB.00501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons J-L, Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR, PLoS ONE. 8 (2013) e83748. doi: 10.1371/journal.pone.0083748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D, Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile, PLoS ONE. 8 (2013) e79666. doi: 10.1371/journal.pone.0079666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, Martin MJ, Goulding D, Duncan SH, Flint HJ, Dougan G, Choudhary JS, Lawley TD, Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism, BMC Genomics. 15 (2014) 160. doi: 10.1186/1471-2164-15-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Edwards AN, Tamayo R, McBride SM, A novel regulator controls Clostridium difficile sporulation, motility and toxin production, Mol. Microbiol. 100 (2016) 954–971. doi: 10.1111/mmi.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL, Repression of Clostridium difficile toxin gene expression by CodY, Mol. Microbiol. 66 (2007) 206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- [18].Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I, The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile, J. Bacteriol. 193 (2011) 3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nawrocki KL, Edwards AN, Daou N, Bouillaut L, McBride SM, CodY-Dependent Regulation of Sporulation in Clostridium difficile, J. Bacteriol. 198 (2016) 2113–2130. doi: 10.1128/JB.00220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R, A novel regulatory protein governing biofilm formation in Bacillus subtilis, Mol. Microbiol. 68 (2008) 1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kuroda A, Sekiguchi J, High-level transcription of the major Bacillus subtilis autolysin operon depends on expression of the sigma D gene and is affected by a sin (flaD) mutation, J. Bacteriol. 175 (1993) 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Barilla D, Caramori T, Galizzi A, Coupling of flagellin gene transcription to flagellar assembly in Bacillus subtilis, J. Bacteriol. 176 (1994) 4558–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Girinathan BP, Ou J, Dupuy B, Govind R, Pleiotropic roles of Clostridium difficile sin locus, PLoS Pathog. 14 (2018) e1006940. doi: 10.1371/journal.ppat.1006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Poquet I, Saujet L, Canette A, Monot M, Mihajlovic J, Ghigo J-M, Soutourina O, Briandet R, Martin-Verstraete I, Dupuy B, Clostridium difficile Biofilm: Remodeling Metabolism and Cell Surface to Build a Sparse and Heterogeneously Aggregated Architecture, Front Microbiol. 9 (2018) 2084. doi: 10.3389/fmicb.2018.02084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gaur NK, Cabane K, Smith I, Structure and expression of the Bacillus subtilis sin operon, J. Bacteriol. 170 (1988) 1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Voigt CA, Wolf DM, Arkin AP, The Bacillus subtilis sin operon: an evolvable network motif, Genetics. 169 (2005) 1187–1202. doi: 10.1534/genetics.104.031955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sheffield P, Garrard S, Derewenda Z, Over-coming expression and purification problems of RhoGDI using a family of “parallel” expression vectors, Protein Expr. Purif. 15 (1999) 34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- [28].Girinathan BP, Monot M, Boyle D, McAllister KN, Sorg JA, Dupuy B, Govind R, Effect of tcdR Mutation on Sporulation in the Epidemic Clostridium difficile Strain R20291, MSphere. 2 (2017). doi: 10.1128/mSphere.00383-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Putnam EE, Nock AM, Lawley TD, Shen A, SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins, J. Bacteriol. 195 (2013) 1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Newman JA, Rodrigues C, Lewis RJ, Molecular basis of the activity of SinR protein, the master regulator of biofilm formation in Bacillus subtilis, J. Biol. Chem. 288 (2013) 10766–10778. doi: 10.1074/jbc.M113.455592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chai Y, Norman T, Kolter R, Losick R, An epigenetic switch governing daughter cell separation in Bacillus subtilis, Genes Dev. 24 (2010) 754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R, Sticking together: building a biofilm the Bacillus subtilis way, Nat. Rev. Microbiol. 11 (2013) 157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW, Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium, Genome Biol. 10 (2009) R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Teng F, Murray BE, Weinstock GM, Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations, Plasmid. 39 (1998) 182–186. doi: 10.1006/plas.1998.1336. [DOI] [PubMed] [Google Scholar]
- [35].Fagan RP, Fairweather NF, Clostridium difficile has two parallel and essential Sec secretion systems, J. Biol. Chem. 286 (2011) 27483–27493. doi : 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.