Significance
PP1, a metal-dependent enzyme responsible for >50% of all dephosphorylation reactions, forms distinct holoenzymes with >200 regulatory proteins, which create specificity for its substrates. Intriguingly, one of its most ancient regulators, SDS22, is not only an essential gene but also is both a PP1 inhibitor and an activator. How such divergent functions are mediated by the same regulator is unknown. Here, we discovered that SDS22 binds a unique, metal-deficient conformation of PP1, which renders PP1 inactive. Furthermore, once the complex forms, it does not permanently dissociate until PP1 binds a M1 metal. Thus, SDS22 is a "PP1 trap," providing a pool of PP1 poised for the rapid formation of new holoenzymes in response to dynamically changing cellular events.
Keywords: protein phosphatase 1 (PP1), SDS22, inhibitor, crystal structure, metal binding
Abstract
The metalloenzyme protein phosphatase 1 (PP1), which is responsible for ≥50% of all dephosphorylation reactions, is regulated by scores of regulatory proteins, including the highly conserved SDS22 protein. SDS22 has numerous diverse functions, surprisingly acting as both a PP1 inhibitor and as an activator. Here, we integrate cellular, biophysical, and crystallographic studies to address this conundrum. We discovered that SDS22 selectively binds a unique conformation of PP1 that contains a single metal (M2) at its active site, i.e., SDS22 traps metal-deficient inactive PP1. Furthermore, we showed that SDS22 dissociation is accompanied by a second metal (M1) being loaded into PP1, as free metal cannot dissociate the complex and M1-deficient mutants remain constitutively trapped by SDS22. Together, our findings reveal that M1 metal loading and loss are essential for PP1 regulation in cells, which has broad implications for PP1 maturation, activity, and holoenzyme subunit exchange.
Protein phosphatase 1 (PP1; Glc7 in Saccharomyces cerevisiae) is the most widely expressed and abundant Ser/Thr phosphoprotein phosphatase (PPP) (1). PP1 is a single-domain enzyme that is exceptionally well conserved from fungi to humans in both sequence and function. Thus, human PP1 rescues the lethality caused by deletion of GLC7 in S. cerevisiae (2). The active sites of all PPPs bind 2 metal ions [M1 and M2; both Mn2+ when PP1 is expressed in bacteria (3, 4) Zn2+ and Fe2+ when isolated from eukaryotic cells/tissue (5, 6); the identity of the metal that occupies the M1 versus M2 site is determined in this work]. While both metals coordinate the phosphorylated residue, the M1 metal activates a water molecule that ultimately enables the hydrolysis of the phosphate group from substrates (Figs. 1 A and B, Top) (4, 7). Although the specificity of PPPs appears low, PP1 is nevertheless able to dephosphorylate its targets with high specificity (8). This specificity is achieved via its interaction with >200 known regulatory proteins that bind directly to PP1, targeting it to specific cellular locations and substrates (9–11). Biochemical and structural studies have revealed that the majority of these regulators bind PP1 via one or more short linear motifs (SLiMs; ∼4–8 residues). SLiMs are commonly found within intrinsically disordered regions (IDRs) and function to mediate protein:protein interactions. Most PP1 regulators bind PP1 using a primary PP1 SLiM, the RVxF motif (12–14), and various combinations of additional SLiMs; these multivalent SLiM interactions enhance regulator binding to PP1 through avidity (12, 15). Furthermore, the binding of regulators via SLiM interactions does not alter the conformation of PP1, nor does it influence active site metal binding (16–19). Rather, the SLiM binding pockets on PP1 are “preformed,” ready to bind any regulator. However, even with these advances, little is understood about how PP1 holoenzymes form and how the various regulators are exchanged in the cell.
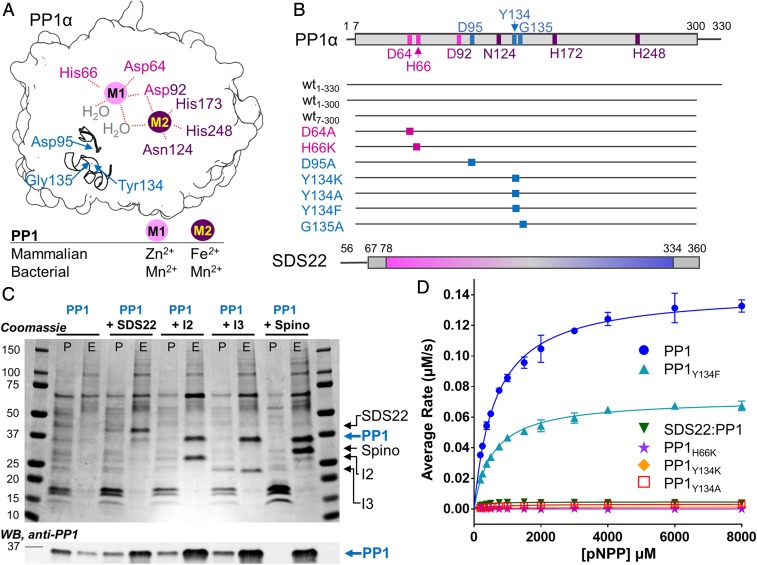
Fig. 1.
SDS22 is not a PP1 chaperone. (A) Cartoon of PP1 highlighting the M1 (pink sphere) and M2 (purple sphere) metals and their corresponding coordinating residues (labels colored; Asp92 is shared between M1 and M2) at the catalytic site. Two secondary structural elements (Arg96 loop and the Tyr134 loop) are shown with the location of key residues indicated. The M1 and M2 metals present in mammalian (whose binding positions are identified in this work) and bacterial PP1 (3) are indicated. (B) Schematic of the constructs examined in this study. (C) SDS/PAGE (Upper) and Western Blot (anti-PP1; Lower) of pull-downs with PP1 expressed either alone or with SDS22, Inhibitor-2 (I-2), Inhibitor-3 (I-3), or spinophilin (Spino). The pellet (P) and elution (E) fractions are shown. (D) pNPP activity assay of metal-loaded PP1 (blue; Mn2+/Mn2+ for M1/M2), PP1Y134F (cyan; Mn2+/Mn2+); SDS22:PP1 (dark green; -/Mn2+; "-" indicates no metal), PP1H66K (purple; -/Mn2+); PP1Y134K (orange; Mn2+/Mn2+); PP1Y134A (red; Mn2+/Mn2+). Error bar = SD, n = 3–6.
SDS22 (suppressor of Dis2 mutant 2; PPP1R7) is an essential PP1 regulator that is conserved from yeast to humans (Fig. 1B) (20). As observed for PP1, human SDS22 rescues the lethality of an SDS22 deletion in yeast (21), suggesting both structural and functional conservation. It consists of 2 domains: an N-terminal IDR (residues 1–77; no RVxF motif) and a folded leucine-rich repeat (LRR) domain (residues 78–360). This makes SDS22 unusual among PP1 regulators, in that SDS22 is a primarily folded protein that lacks the RVxF SLiM that is present in nearly all known PP1 regulators. Data suggest that SDS22 binds PP1 via its folded domain (22). Although the SDS22:PP1 complex has been suggested to play key roles in processes as diverse as chromosome segregation (23), cell-shape regulation (24), and sperm motility (25), the functional consequences of SDS22 binding to PP1 are still not clearly understood. Mutations in yeast SDS22 disrupt normal Glc7 nuclear localization, suggesting a positive role in nuclear targeting of Glc7 (26–29). Loss of Sds22 function leads to Glc7 aggregation, which leads to the hypothesis that Sds22 with I-3 (Ypi1 in S. cerevisiae) prevents Glc7 misfolding (26–29). Finally, SDS22, which is highly expressed in cells, was recently hypothesized to function as a PP1 maturation factor via an unknown mechanism and, thus, is critical for PP1 biogenesis (30). Taken together, the lack of molecular insight into this essential complex has limited our advances in understanding the biological functions of SDS22 and the SDS22:PP1 holoenzyme.
Here, we combined genetic, cellular, and molecular techniques to understand how SDS22 interacts with PP1 and how this interaction influences PP1 activity. We show that SDS22 prevents aggregation of PP1. However, unlike what would be predicted for a classical chaperone, the crystal structure of the SDS22:PP1 complex shows that SDS22 exclusively binds and traps a previously undescribed conformation of PP1, one in which the M1 metal is missing from the active site. Using surface plasmon resonance (SPR), we show that SDS22 does not induce the loss of the metal but instead binds selectively to M1-free PP1. Consistent with this, PP1 variants that fail to bind metal at the M1 site associate constitutively with SDS22 both in vitro and in cells. Our structure also shows that SDS22 binding inhibits PP1 activity by both blocking its C-terminal substrate binding groove and displacing residues critical for substrate recruitment. Critically, this is also the only structure of PP1 purified from cultured mammalian cells, and it shows no conformational differences from bacterially expressed PP1 (either alone or as a holoenzyme), allowing for the confident molecular interpretation of cellular data. Taken together, our data reveal the critical role for M1 metal binding in PP1 activation and provide fundamental insights into the mechanisms by which SDS22 inhibits and stabilizes PP1 prior to holoenzyme formation.
Results
SDS22 and Other PP1 Regulatory Proteins Prevent PP1 Aggregation.
In mammalian and yeast cells, SDS22 and I-3/Ypi1 associate with newly synthesized PP1/Glc7 and together are hypothesized to play an essential role in PP1 biogenesis (28, 30). Furthermore, defects in yeast Sds22 and I-3/Ypi1 result in aggregation of newly synthesized PP1/Glc7 (29). These data suggest that SDS22 plays a key role in the PP1 maturation process. However, folded, active PP1 is readily expressed in Escherichia coli in the absence of these PP1 regulators. Furthermore, PP1 coexpressed in bacteria with PP1-specific regulators (i.e., MYPT1; ref. 19) or purified from bacteria and incubated with purified PP1-specific regulators (17, 18, 31) readily forms active functional PP1 holoenzymes. Thus, SDS22 and I-3 are not PP1-specific chaperones required for PP1 folding, activity, or holoenzyme formation; rather, these data suggest that SDS22 has an alternative function, such as preventing the aggregation of newly synthesized PP1. To test this model, we assessed the requirements for the formation of soluble human PP1 in mammalian cells. We used transient expression experiments in HEK293F cells followed by PP1 pull-down, monitoring PP1 in both the pellet (insoluble) and the eluate (soluble) fractions to determine if SDS22 facilitates PP1 solubility. Although transient expression of PP1 alone is not detectable by SDS/PAGE, immunoblotting with a PP1-specific antibody shows it is present in both the pellet and the eluate fractions with the majority present in the pellet. In contrast, cotransfection of both PP1 and SDS22 results in robust soluble expression of both proteins with PP1 and SDS22 detected in the eluate using either SDS/PAGE or immunoblot (Fig. 1C). Comparatively little PP1 is present in the pellet. Thus, coexpression with SDS22 enhances the solubility of PP1. To test if this behavior is specific to SDS22, we measured the ability of 2 proteins that inhibit PP1 activity in vitro, I-2 (32) and I-3 (33) as well as a well-studied targeting protein, spinophilin (34), to facilitate the soluble expression of PP1 (Fig. 1C). Cotransfection of each of the 3 regulatory proteins results in a robust increase in the yield of soluble PP1, even higher than that obtained for SDS22. Together, these data show that not only SDS22, but also several other PP1 regulators, stabilize PP1 by preventing its aggregation.
SDS22 Is a Potent Inhibitor of PP1.
It is well established that SDS22 potently inhibits the PP1-mediated dephosphorylation of the canonical PP1 substrate glycogen phosphorylase (phos a) (22). However, how this inhibition is achieved is unknown. We and others have shown that the inability of PP1 to dephosphorylate phos a can be achieved via 2 distinct mechanisms: (1) the loss of PP1 catalytic activity or (2) the inability of phos a to bind the PP1 C-terminal substrate binding groove, a docking event that is essential for its PP1-mediated dephosphorylation (18, 35). To distinguish between these possibilities for SDS22-bound PP1, we measured the activity of PP1 in the presence and absence of SDS22 using paranitrophenylphosphate (pNPP) as a substrate. pNPP only requires access to the PP1 active site for dephosphorylation, so its dephosphorylation cannot be inhibited by blocking the substrate-binding groove. The data show that PP1 is unable to dephosphorylate pNPP when bound to SDS22 (Fig. 1D). Thus, SDS22 inhibits PP1 by either blocking or altering the conformation of the PP1 active site.
SDS22 Interacts with PP1 over an Extensive Interface.
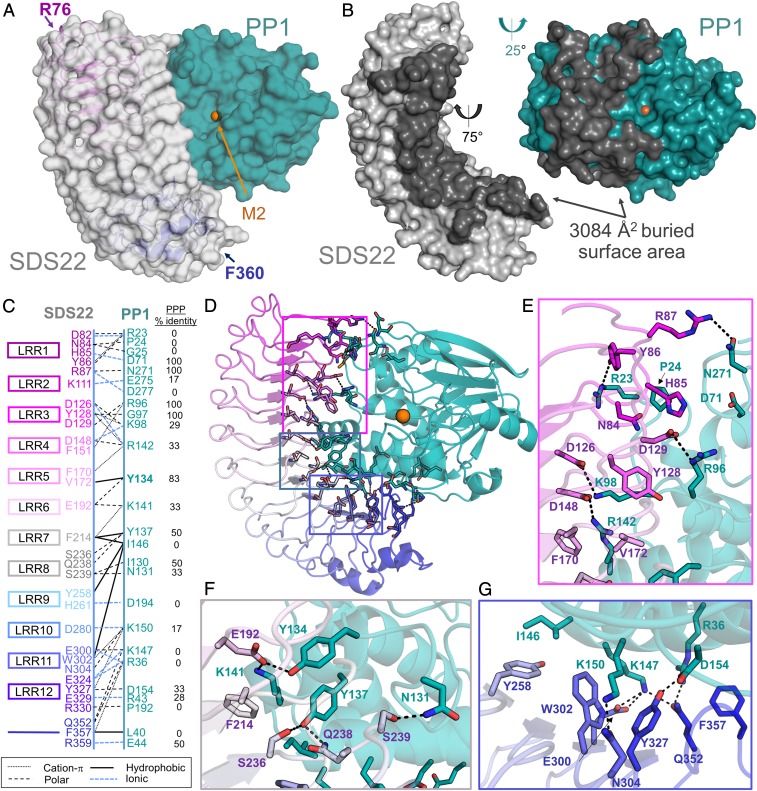
To understand how SDS22 binds and inhibits PP1, we determined the crystal structure of the coexpressed mammalian SDS22:PP1 complex (SDS2256–360:PP1α1–300 coexpressed in HEK293F; 2.7 Å resolution). The molecular replacement solution identified 2 SDS22:PP1 complexes in the asymmetric unit. The 2 complexes are essentially identical (root mean square deviation [rmsd] of 0.28 Å for SDS22 and 0.44 Å for PP1), and the final model includes residues 1–298 of PP1 and 76–360 of SDS22 (SI Appendix, Table S1 and Fig. 2A). No electron density was observed for SDS22 residues 56–75, consistent with previous data demonstrating that these residues are both dispensable for PP1 binding (22) and less well ordered than the Leu-rich repeats (LRRs) (36, 37). The SDS22:PP1 interaction surface is extensive, burying more than 3084 Å2 of solvent accessible surface area (Fig. 2B). SDS22 binds PP1 via its entire LRR concave face with between 1 and 4 residues from each of the 12 LRRs contributing to PP1 binding (Fig. 2 C and D). Conversely, PP1 binds SDS22 exclusively via its left lobe, extending from the C-terminal substrate binding groove to the left surface of the active site and the hydrophobic substrate binding groove to the bottom of PP1 (Fig. 2B). The interface is composed of 57 residues (31 residues, SDS22; 26 residues, PP1; Fig. 2 C–G). The majority of the interactions are electrostatic and/or polar, consistent with the high concentration of charged amino acids at the interface (15 Glu/Asp and 13 Lys/Arg; Fig. 2 E–G). However, hydrophobic and π-stacking interactions also contribute to complex stability. A sequence alignment of the PPP family catalytic subunits (PP1, PP2A, PP2B/calcineurin, PP4, PP5, PP6, and PP7; Fig. 2C and SI Appendix, Fig. S1A) shows that the majority of the PP1 residues that define the SDS22:PP1 interface are unique to PP1, explaining why SDS22 is specific for PP1.
Fig. 2.
SDS22 and PP1 interact via an extensive surface. (A) The SDS22:PP1 complex is shown as a transparent surface (SDS22, gray; PP1, teal). The LRR repeats of SDS22 are colored from magenta (LRR1) to blue (LRR12) and shown as a cartoon with the first and last SDS22 residues indicated. The bound M2 metal (orange sphere) is the location of the PP1 catalytic site. (B) SDS22 (gray) and PP1 (teal) from the SDS22:PP1 complex separated and rotated as indicated to illustrate the binding interface between the 2 proteins (dark gray). (C) Schematic of SDS22 (Left) and PP1 (Right) residues that form polar, ionic, hydrophobic, or cation-π interactions. SDS22 residue names are colored according to the LRR repeat they are a part of. (D) SDS22:PP1 complex shown as a cartoon and colored as in A. The residues that comprise the interface are shown as sticks. Colored boxed areas correspond to the region highlighted in E (magenta), F (gray) and G (blue). (E) Residues that define the SDS22:PP1 interface, focused on LRRs 1–5; polar/ionic interactions are indicated by dashed black lines. (F) The same as E but focusing on the interactions with LRRs 6–8. (G) The same as E but focusing on the interactions with LRRs 9–12.
SDS22 Stabilizes an Atypical Conformation of PP1.
The structures of free and PP1-bound SDS22 are essentially identical with a rmsd of 0.4 Å (SI Appendix, Fig. S1 B and C). This is consistent with the observation that LRR proteins have well-defined curvatures that are determined solely by the number of residues per LRR. In contrast, the rmsd between free and SDS22-bound PP1 is much higher at ∼1 Å (Fig. 3A). This is unprecedented for PP1 as its free and holoenzyme-bound conformations are largely identical for all structures determined to date (16, 18, 31). One of the most significant differences between free and SDS22-bound PP1 is that, in the SDS22:PP1 complex, PP1 contains only an M2 ion bound at the M2 metal-binding site (Fe2+, as determined by anomalous scattering; SI Appendix, Fig. S2 A–E). The M1 metal site is empty (Fig. 3A). The lack of metal at the M1 site is accompanied by unique conformational changes at the active site and adjacent secondary structural elements, including the Arg96 loop, the β12-β13 loop, and the Tyr134 loop (127CASINRIYG135). In particular, Tyr134 and Arg132 move by more than 12 Å between the metal-loaded and SDS22-bound PP1 conformations (Fig. 3 B and C).
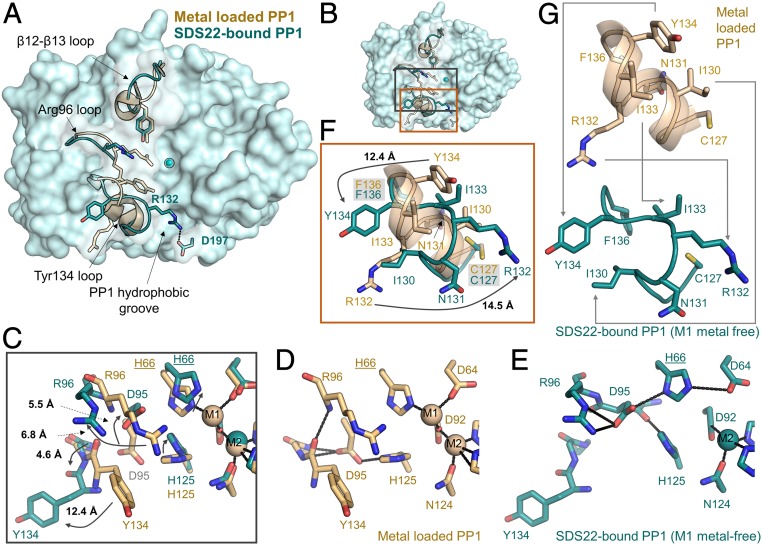
Fig. 3.
SDS22 stabilizes a unique conformation of PP1. (A) Overlay of metal-loaded PP1 (PDBID 4MOV; beige) and SDS22-bound PP1 (teal). The loops that are most different between the 2 conformations are shown as cartoons and labeled, with some of the key residues shown as sticks. The M2 metal is shown as a sphere. The location of the PP1 hydrophobic groove is also indicated. Arg132 in SDS22-bound PP1 lies across the PP1 hydrophobic groove [the binding pocket for PP1-specific toxins, such a microcystin-LR (MC)] to form a salt bridge with Asp197. (B) Overlay of the active sites of metal-loaded (beige) and SDS22-bound (teal) PP1 with the Arg96 and Tyr134 loop residues that change conformation shown as sticks; the bound M2 metal in SDS22-bound PP1 is shown as a sphere. (C) Closeup of B, black box. The arrows highlight the rotations of the residues that change conformation between the metal-loaded (beige) and the SDS22-bound (teal) states with the distances between the states indicated. (D) The same as C except showing only the metal-loaded PP1 conformation. The polar interactions between different residues are shown as black dashed lines. (E) The same as C except showing only the SDS22-bound state. The polar interactions formed between residues are indicated by dashed lines. (F) Closeup of B, orange box. The Tyr134 loop residues that change conformation as a consequence of SDS22 binding are indicated with arrows with the distances indicated. (G) The same as F except the metal-loaded PP1 (beige, Upper) and SDS22-bound (teal, Lower) states are shown separately with the conformational changes in residues between the 2 states indicated by arrows.
A detailed comparison of metal-loaded versus SDS22-bound (M1 metal-free) PP1 shows that the observed conformational changes are coupled. Specifically, the absence of the metal at the M1 position (normally coordinated by Asp64, His66, and Asp92) allows Asp64 to rotate upward and form a salt bridge with His66. As a consequence, His125 is now able to fill the M1 pocket (Fig. 3 C–E). Because His125 forms a salt bridge with Asp95 in metal-loaded PP1 (Fig. 3D), this rotation of His125 causes Asp95 to rotate upward by nearly 90° and point into the catalytic pocket (Fig. 3E). These changes have 2 consequences for the adjacent secondary structural elements (Fig. 3 C–E). First, the hydrogen bonds between Asp95 and amide hydrogens of Gly135 and Phe136 in metal-loaded PP1 are lost. Second, Arg96, which coordinates phosphorylated substrates, rotates out of the substrate binding pocket and forms a salt bridge with Asp95. The latter change not only disrupts a π-stacking interaction between Arg96 and Tyr134, but also breaks a main chain hydrogen bond between the Arg96 amide hydrogen and the Tyr134 carbonyl. In the absence of these stabilizing interactions, the Tyr134 loop (127CASINRIYG135) changes conformation from a short 2 turn α-helix in metal-loaded PP1 to an extended kinked loop in the SDS22-bound state (Fig. 3 B, F, and G). As a consequence, Tyr134, which is normally oriented below the catalytic pocket, rotates by nearly 180° to point away from the PP1 surface with the Tyr134 hydroxyl moving by more than 12 Å. Simultaneously, Arg132, which is normally at the bottom outer surface of PP1, rotates by nearly 180° in the opposite direction, resulting in the formation of a salt bridge with Asp197 that blocks access to the PP1 hydrophobic groove.
SDS22 Binds Exclusively to Metal-Deficient PP1.
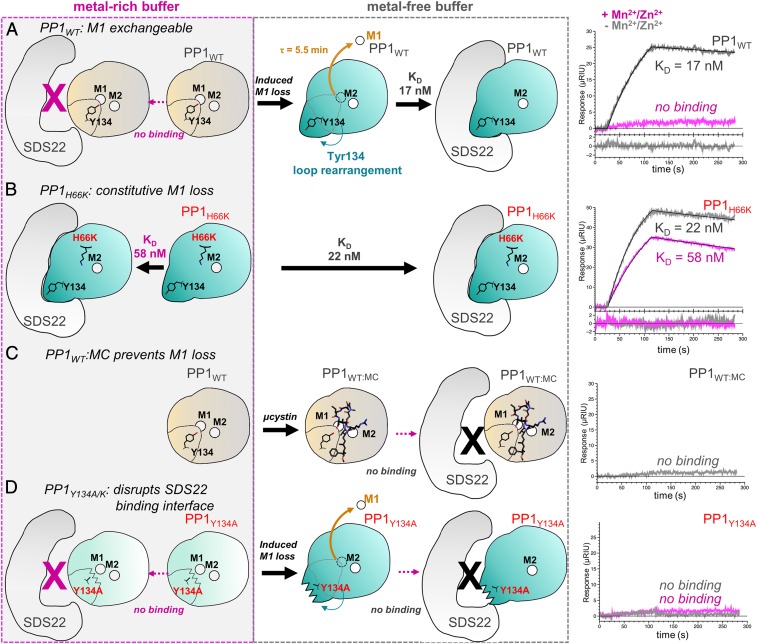
Although the observed conformational changes in PP1 are clearly coupled to the lack of metal, it was unclear if binding of SDS22 causes ejection of metal from the active site, or, alternatively, if SDS22 selectively binds M1 metal-free PP1 (e.g., SDS22 binds to PP1 before the M1 metal has been loaded, as might occur during PP1 biogenesis, or binds after metal-loaded PP1 has lost its M1 metal, as a mechanism of protein maintenance). To distinguish between these possibilities, we used SPR with SDS22 purified from HEK293F cells and active metal-loaded PP1 purified from E. coli in the presence of Mn2+. The data show that SDS22 does not bind PP1 when metal is present in the buffer (either Mn2+ or its native M1 metal Zn2+ Figs. 1A and 4A, Table 1, and SI Appendix, Fig. S3). However, once PP1 is equilibrated in metal-free buffer, binding is readily observed, with the maximum signal observed after ∼40 min [SI Appendix, Fig. S4 A and B; it is well established that Mn2+ readily dissociates from PP1 when Mn2+ is not present in the buffer (6, 38); SDS22 binding decreases at 70 min, which we interpret as M2 metal loss]. These data show that SDS22 binding does not eject the M1 metal from the PP1 active site, but rather that SDS22 selectively binds PP1 that is either completely metal deficient (neither M1 nor M2 is bound) or, as the structure shows, bound to only M2. To confirm that PP1 is folded under these conditions, we measured the binding of the PP1 regulator nuclear inhibitor of PP1 (NIPP1)158–216 to PP1 in metal-rich and metal-free buffers. NIPP1 binds to PP1 in both conditions with the same affinities (SI Appendix, Table S2 and Fig. S5), confirming that PP1 is folded and competent to bind regulators distal to the active site.
Fig. 4.
SDS22 binds exclusively to metal-deficient PP1 and requires Tyr134 for productive binding. (A) SPR experiments between SDS22 and WT PP1 (PP1WT) performed either in metal-rich buffer (Left; magenta dashed box) or in metal-free buffer (Middle; gray dashed box). The SPR sensorgrams for SDS22 and PP1 in metal-rich (magenta) and metal-free (gray) buffer are shown at the right. Cartoons (Left and Middle) illustrate SPR results. Metal-loaded PP1 conformation indicated by a "closed" Tyr134 loop (light beige with closed Tyr134 loop; typically contains both the M1 and the M2 metals). The SDS22-bound conformation is shown in teal with an "open" Tyr134 loop. The SPR data show that the SDS22-bound conformation is accessible only after the M1 metal is lost (half-life of metal loss determined using SPR, SI Appendix, Fig. S4). (B) The same as A except with PP1H66K, which does not bind M1. (C) The same as A, except with PP1 bound to the cyanobacterial toxin MC, which binds the PP1 hydrophobic pocket. (D) The same as A except with PP1Y134A.
Table 1.
SPR-binding kinetics for the association of SDS22 with PP1 variants
| PP1 variant | kon (M−1 s−1)* | koff (s−1)* | KD (nM)* | χ2* | n |
| No metal† | |||||
| PP1α 7–330 | 1.98 × 104 ± 2.93 × 103 | 3.25 × 10−4 ± 7.0 × 10−5 | 16.9 ± 5.1 | 0.21 ± 0.11 | 6 |
| PP1γ 7–323 | 1.74 × 104 ± 6.30 × 103 | 4.21 × 10−4 ± 8.2 × 10−5 | 25.2 ± 4.5 | 0.31 ± 0.34 | 3 |
| PP1α 1–300 | 2.33 × 104 ± 4.13 × 103 | 7.15 × 10−4 ± 2.5 × 10−4 | 30.4 ± 9.6 | 0.16 ± 0.08 | 6 |
| PP1α 7–300 | 2.03 × 104 ± 5.40 × 103 | 5.57 × 10−4 ± 1.5 × 10−4 | 29.6 ± 11.8 | 0.30 ± 0.20 | 4 |
| PP1α 7–300 Y134A | No binding | — | — | — | 4 |
| PP1α 7–300 Y134F | 2.13 × 104 ± 4.80 × 103 | 5.12 × 10−3 ± 8.3 × 10−4 | 257 ± 98 | 0.15 ± 0.07 | 5 |
| PP1α 7–300 Y134K | No binding | — | — | — | 4 |
| PP1α 7–300: MC | No binding | — | — | — | 4 |
| PP1α 7–300 H66K | 2.84 × 104 ± 1.99 × 103 | 6.12 × 10−4 ± 2.3 × 10−5 | 21.6 ± 1.3 | 0.53 ± 0.13 | 3 |
| with Mn2+ | |||||
| PP1α 7–330 | No binding | — | — | — | 4 |
| PP1γ 7–323 | No binding | — | — | — | 4 |
| PP1α 7–300 | No binding | — | — | — | 4 |
| PP1α 7–300 Y134A | No binding | — | — | — | 4 |
| PP1α 7–300 Y134F | No binding | — | — | — | 5 |
| PP1α 7–300 Y134K | No binding | — | — | — | 4 |
| PP1α 7–300 H66K | 1.84 × 104 ± 9.5 × 102 | 1.07 × 10−3 ± 3.1 × 10−5 | 58.4 ± 4.0 | 0.22 ± 0.02 | 3 |
| PP1α 7–330 | No binding | — | — | — | 4 |
| with Zn2+ | |||||
| PP1α 7–330 | No binding | — | — | — | 3 |
Values are represented as the mean ± SD.
No metal included in the SPR ligand buffer.
To assess whether the reconstituted SDS22:PP1 complex detected by SPR (SDS22 purified from HEK293F cells and PP1 purified from E. coli BL21 [DE3]) is identical to that obtained by coexpression in mammalian cells, we determined the crystal structure of the reconstituted complex to a resolution of 2.7 Å. The structure of the reconstituted complex is identical to that obtained using the complex purified from coexpression in mammalian cells. That is, no metal is present at the M1 site, and the conformational changes observed in PP1 are identical between the 2 SDS22:PP1 complexes (SI Appendix, Fig. S2 D and E). Critically, these structures also show that mammalian and bacterially expressed PP1 adopt identical conformations. Thus, the structure of PP1 is identical, independent of the expression system and independent of the identity of the metal at the active site.
Using SPR, we then tested if the reintroduction of metal would destabilize the SDS22:PP1 complex, resulting in SDS22 dissociation (SI Appendix, Fig. S6). No changes in the rate of dissociation of SDS22 from PP1 occur in the presence of metal, demonstrating that the presence of metal alone is not sufficient to destabilize the complex. The SPR experiments showed that the interaction between SDS22 and metal-deficient PP1 is tight, with a KD of 16.9 ± 5.1 nM (Fig. 4A and Table 1). It is also PP1 isoform-independent and requires only the SDS22 and PP1 folded domains (the PP1 and SDS22 IDR residues do not enhance binding; SI Appendix, Fig. S3 and Table 1). However, while the KD of SDS22 for PP1 is similar to that observed for other PP1 regulators (e.g., spinophilin, PP1 nuclear targeting subunit, NIPP1), the interaction is highly atypical in that the on- and especially the off-rates (kon, koff) are unusually slow. Together, these data confirm that SDS22 stabilizes and traps the inactive metal-deficient conformation of PP1.
The structures of the coexpressed and reconstituted SDS22:PP1 complexes, coupled with the SPR data, strongly suggest that the 127CASINRIYG135 loop is free to change its conformation only when the M1 site is empty. Thus, we reasoned that destabilizing or stabilizing M1 metal coordination should enhance or destabilize SDS22 binding, respectively. To test this, 2 PP1 M1 metal coordination variants were generated, H66K and D64A. Although H66K is folded (D64A does not fold; SI Appendix, Table S3), it is not active (Fig. 1D), suggesting that the M1 metal is not bound. We confirmed this using X-ray crystallography, which showed that the M1 site in PP1H66K is empty (SI Appendix, Fig. S7). The affinity of H66K for SDS22 was then measured using SPR. As predicted, strong binding was observed, even in the presence of metal (KD with metal, KD 58 ± 4 nM; without metal, 22 ± 1 nM; Fig. 4B and Table 1). Next, we used a potent inhibitor of PP1, the cyanobacterial toxin MC to stabilize bacterially expressed metal-loaded wild-type (WT) PP1. Because MC binds PP1 at the active site and hydrophobic groove, MC binding should not only prevent the loss of the M1 metal from metal-loaded WT PP1, but also should inhibit the 127CASINRIYG135 loop from adopting the SDS22-bound conformation. As predicted, SPR showed that PP1:MC does not bind SDS22 either in the presence or in the absence of metal (Fig. 4C and Table 1). Finally, we tested if Mn2+ competes with SDS22 for PP1 binding. In this SPR experiment, the M1 metal is removed by incubating PP1 in metal-free buffer after which SDS22 is added either in the absence or in the presence of Mn2+. The data show that there is a 3.7-fold reduction of SDS22 binding in the presence of Mn2+ (SI Appendix, Fig. S8A), demonstrating that Mn2+ competes with SDS22 for PP1 binding.
Tyr134 Is Essential for SDS22 Binding and PP1 Activity.
PP1 residue Tyr134 defines the center of the SDS22:PP1 interface, suggesting it is critical for SDS22 complex formation. To test this, we used SPR to measure the affinity of SDS22 for 3 PP1 variants, Y134F, Y134A, and Y134K (Fig. 4D, Table 1, and SI Appendix, Fig. S3). Weakened binding was observed with Y134F (∼9-fold weaker), consistent with the observation that the Tyr134 hydroxyl hydrogen bonds with SDS22 residue Glu192 (Fig. 2F). In contrast, no binding was observed to either Y134A or Y134K, confirming that, despite the extensive interface between SDS22 and PP1, Tyr134 is essential for SDS22 binding (both PP1 Y134A and Y134K bind to NIPP1 and exhibit similar unfolding temperatures as WT-PP1 [SI Appendix, Table S2 and S3]). We also assayed the activity of the Y134F/A/K variants. Y134F is active, consistent with the observation that calcineurin has a Phe residue at the same position. In contrast, Y134A and Y134K are inactive (Fig. 1D). To test if the lack of activity of the latter 2 variants is due to the loss of the M1-bound metal, we determined the crystal structures of these variants (SI Appendix, Fig. S8 B and C). Structures of the variants (PP1Y134K, PP1Y134K:MC, and PP1Y134A:MC) show metals bound at both the M1 and the M2 sites. Thus, the loss of activity is not due to either the lack of metal or a global change in PP1 conformation but, instead, is due solely to the substitution of the Tyr residue. In WT metal-loaded PP1, Arg96 and Tyr134 form a strong π-stacking interaction that orients the Arg guanidinium side chain to optimally position phosphorylated substrates at the active site (SI Appendix, Fig. S8D). In the absence of this π-stacking interaction, the Arg96 side chain adopts other conformations that, based on the activity assay results, do not appear conducive to dephosphorylation (SI Appendix, Fig. S8E). Our PP1Y134K variant structure shows that this is the case. The electron density for the Arg96 side chain in the PP1Y134K structure is weaker than that observed in the WT PP1 structure and, furthermore, is consistent with a conformation of Arg96 that will not result in productive substrate binding and dephosphorylation. Thus, our data show that Tyr134, via its ability to optimally position the substrate coordinating residue Arg96, is essential for PP1 activity.
In Vivo, Sds22 Traps PP1 Variants That Lack Metals at the M1 Site.
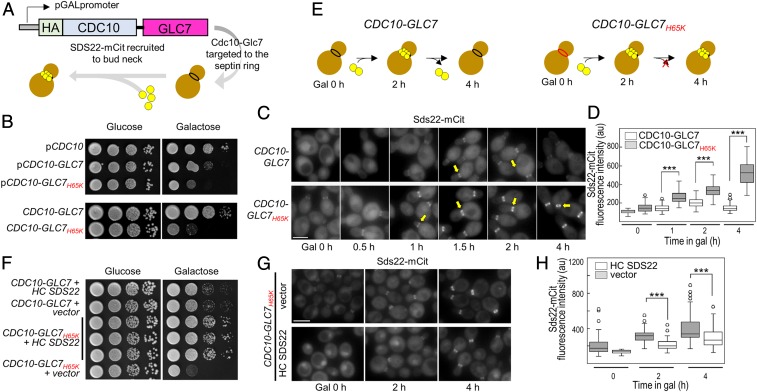
To understand how the interaction between SDS22 and PP1 manifests in cells, we developed an assay to monitor Sds22 binding to Glc7 (PP1) in live budding yeast cells (SI Appendix, Fig. S9 A–C). Briefly, GLC7 is fused to the CDC10 gene, which encodes a septin protein that targets Cdc10-Glc7 to the bud neck, a narrow constriction between the mother cell and the growing bud that is supported by a collar of septin filaments (39, 40). Because septin filaments are stable from late G1 through late anaphase (41, 42), a Cdc10 fusion can provide a platform for monitoring protein:protein interactions in living cells by fluorescence microscopy. Here, Cdc10-Glc7 is targeted to the bud neck, and fluorescently tagged Glc7 regulators, e.g., SDS22, are observed to associate with the bud neck when they bind Cdc10-Glc7 (Fig. 5A). To ensure that the Cdc10-Glc7 fusion is functional, we showed that it complements the glycogen deficiency of a glc7-1 mutant (SI Appendix, Fig. S9D).
Fig. 5.
In yeast, SDS22 and PP1 form a constitutive complex when the M1-metal coordination site is disrupted. (A) Model of the system: HA-CDC10-GLC7 chimera was placed under the transcriptional control of the GAL1 promoter. A flexible linker was introduced between the 3′ end of CDC10 and the 5′ end of GLC7 coding sequences to allow the fused proteins to adopt their native conformations. Upon induction, mCit-tagged Glc7 regulators (i.e., Sds22) are visualized at the bud neck. (B, Top) A WT strain (KT2762) was transformed with the designated plasmids. Transformants were serially diluted onto selective media lacking tryptophan, containing either glucose or galactose, and plates were imaged after incubation for 40–44 h at 24 °C. (B, Bottom): CDC10-GLC7WT (KT4038) and CDC10-GLC7H65K (KT2947) cells were serially diluted as in the Top (mammalian and bacterial PP1 residue numbers differ by −1). (C) Sds22-mCit was imaged in CDC10-GLC7 (KT4022 × KT4023) and CDC10-GLC7H65K (KT4020 × KT4021) cells grown in galactose for the indicated times. (Scale bar, 5 µm.) (D) Quantitative analysis of Sds22-mCit fluorescence in cells imaged in C. P values were calculated by two-tailed t test (***P < 0.001). (E) Cartoon model illustrating transient Sds22 association at the bud neck with Cdc10-Glc7 (Left) and the constitutive association of Sds22 with Cdc10-Glc7H65K (Right). (F) A WT strain (KT2764) was cotransformed with plasmids as indicated. Transformants were serially diluted onto selective media for both plasmids containing either glucose or galactose, and plates were imaged after incubation for 40–44 h at 24 °C. Three different transformants expressing CDC10-GLC7H65K and overexpressing SDS22 are shown (rows 3–5). (G) A Sds22-mCit strain (KT2764) was cotransformed with a plasmid containing CDC10-GLC7H65K and either the empty vector (Top) or a plasmid containing SDS22 that replicates to high copy number (HC SDS22; Bottom). Transformants were grown to logarithmic phase in selective media for both plasmids, and Sds22-mCit was imaged after addition of galactose for the indicated times. (Scale bar, 5 µm.) (H) Quantitative analysis of Sds22-mCit fluorescence at the bud neck in strains imaged in G (n = 100). P values were calculated by two-tailed t test (***P < 0.001).
Next, we tested if the Glc7 interface and metal-deficient variants (interface, Glc7Y133A and Glc7K146A/K149A; metal-deficient, Glc7H65K; residue numbering in mammalian and yeast PP1 differs by −1) can recruit SDS22. Induction of interface variants Cdc10-Glc7Y133A and Cdc10-Glc7K146A/K149A does not affect cell growth, demonstrating that these mutations are recessive (a WT GLC7 allele is present at its normal chromosomal position). We then tested the ability of Cdc10-Glc7, Cdc10-Glc7Y133A, and Cdc10-Glc7K146A/K149A to target Sds22-mCitrine (Sds22-mCit) to the bud neck. After induction of Cdc10-Glc7 for 2 h, robust Sds22-mCit fluorescence is visible at the bud neck, demonstrating Glc7-Sds22 binding (SI Appendix, Fig. S10 A–C). In contrast, only low levels of fluorescence are visible with the interface variants Cdc10-Glc7Y133A and Glc7K146A/K149A (SI Appendix, Fig. S10 A–C). These data are consistent with our PP1Y134A SPR data (Fig. 4D) and previously published PP1K147A/K150A pull-down data (37) which also show that these variants fail to bind SDS22. Together, these data show that, both in vivo and in vitro, Lys147/Lys150 and Tyr134 are critical for SDS22 binding.
We then tested if the metal-deficient variant can bind and recruit SDS22. In contrast to the interface variants, induction of Cdc10-Glc7H65K reduces cell growth (Fig. 5B). Because our strains have a WT GLC7 allele at its normal chromosomal position, we tested whether Cdc10 fused to the Glc7H65K variant interferes with the localization of endogenous Glc7-mCit following Cdc10-Glc7 induction. Strikingly, within 4 h of inducing Cdc10-Glc7H65K, cells exhibit a reduced nuclear/cytoplasmic ratio of Glc7-mCit and the formation of bright Glc7 foci (SI Appendix, Fig. S11 A–C). Thus, Glc7H65K behaves as a dominant-negative mutant (Fig. 5B). Next, we tested the ability of Cdc10-Glc7 and Cdc10-Glc7H65K to target Sds22-mCit to the bud neck. After induction of Cdc10-Glc7 for 1 h, Sds22-mCit fluorescence is clearly visible at the bud neck (Fig. 5C). Sds22-mCit fluorescence levels continue to increase and then plateau at ∼2 h following induction (Fig. 5 C and D), even though total Cdc10-Glc7 protein levels continue to rise (SI Appendix, Fig. S11D). In contrast, Cdc10-Glc7H65K induction results in significantly higher levels of Sds22-mCit at the bud neck as compared to WT Cdc10-Glc7 induction (Fig. 5 C and D) with ∼3-fold higher fluorescence intensity relative to WT Cdc10-Glc7 ∼4 h post induction. Immunoblot analysis shows that Cdc10-Glc7, Cdc10-Glc7H65K, and Sds22-mCit levels are similar over this time course, ruling out the possibility that the H65K mutation increases the expression or stability of either Cdc10-Glc7H65K or Sds22-mCit (SI Appendix, Fig. S11D). Together, these data show that, in cells, Sds22 binds newly synthesized (2 h after induction) WT Cdc10-Glc7 and, subsequently, dissociates (Fig. 5 E, Left). In contrast, once Sds22 binds Cdc10-Glc7H65K, it cannot dissociate and instead forms a constitutive complex (Fig. 5 E, Right).
The inability of Sds22 to dissociate from Cdc10-Glc7H65K, coupled with the observed growth arrest of cells expressing Cdc10-Glc7H65K, suggests that the dominant-negative effect of Cdc10-Glc7H65K is due to sequestration of Sds22. Thus, increased expression of Sds22 should rescue WT-Glc7 dysregulation and growth arrest. To test this, we transformed Cdc10-Glc7H65K-expressing cells with a high copy number plasmid containing untagged SDS22. As expected, the growth rates of the transformants show that the high copy SDS22 plasmid completely suppresses the growth defect of Cdc10-Glc7H65K (Fig. 5F). Consistent with this, Sds22-mCit fluorescence intensity at the bud neck is lower in Cdc10-Glc7H65K cells overexpressing SDS22 (Fig. 5 G and H). Importantly, Cdc10-Glc7H65K-expressing cells with high copy Sds22 exhibit significantly reduced formation of Glc7 foci as compared to the empty vector, while simultaneously showing a higher nuclear to cytoplasmic Glc7-mCit fluorescence ratio (SI Appendix, Fig. S11 E–G). These data confirm that the dominant negative effects of Cdc10-Glc7H65K on Glc7 localization and cell growth and division result from sequestration of Sds22 due to irreversible binding to the metal-deficient Glc7 variant (SI Appendix, Fig. S11), leading to inadequate concentrations of Sds22 available for endogenous Glc7.
Discussion
The activity of PP1, a metal-dependent enzyme that controls ≥50% of all human dephosphorylation reactions, is tightly controlled by ≥200 regulatory proteins, which allow it to become a highly specific holoenzyme (9, 12, 14). SDS22 is a highly conserved PP1-binding protein, but its function in PP1 maturation and regulation is not well understood. SDS22 has been described as a PP1 inhibitor, a PP1-specific chaperone, and a PP1 nuclear targeting protein (20, 24, 29, 30). These contradictory models of SDS22 function have posed a long-standing conundrum.
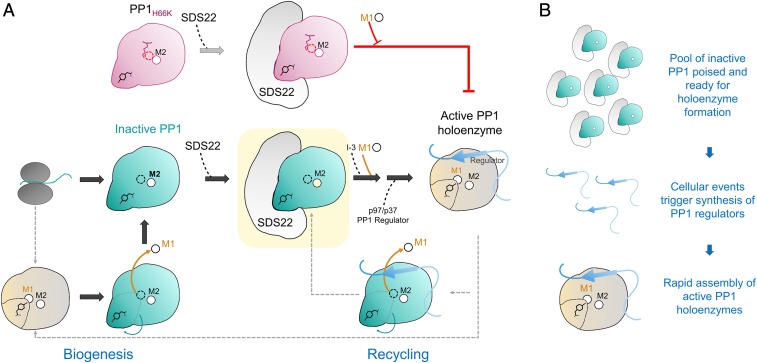
Our data resolve this conundrum. Our results show that SDS22 binds PP1 in a completely unexpected manner, namely, it selectively binds PP1 lacking a metal in the M1 site. Furthermore, we show that dissociation of PP1 from SDS22 strictly requires that a metal ion binds the M1 site since a PP1 variant (PP1H66K; Glc7H65K in yeast) that is unable to bind metal at M1 remains constitutively bound to SDS22 in vitro and in cells. Together, these data have important implications for the function of SDS22 in PP1 biology (Fig. 6A). First, the extensive interface between SDS22 and PP1 strongly suggests that SDS22 is unlikely to bind nascent PP1. Rather, SDS22 binds selectively to folded, yet M1 metal-deficient PP1. This interaction is a key step in the PP1 maturation process (i.e., before the M1 metal is loaded), but the possibility remains that SDS22 also binds mature PP1 from which the M1 metal has been lost (Fig. 4A). Second, when PP1 cannot bind a metal at the M1 site (PP1H66K and Glc7H65K in yeast), SDS22 binding is constitutive. In cells, the interaction of the SDS22:PP1 complex with I-3 and, subsequently, with the AAA+ ATPase p97/Cdc48 is required for PP1:SDS22 dissociation (28, 30). However, our data now show that this model is incomplete. Via a currently unknown mechanism, triple complex formation and subsequent disassociation are also accompanied by M1 metal loading into the PP1 active site.
Fig. 6.
SDS22:PP1 provides a pool of inactive PP1 capable of rapidly assembling into active, functional PP1 holoenzymes. (A) Nascent PP1 folds and contains either 1 (M2) or 2 (M1 and M2) metals. Once folded, SDS22 then selectively binds and traps M1-metal-deficient PP1, stabilizing a unique conformation of PP1 that is catalytically inactive (all steps discovered in this study shown with large block arrows). The subsequent association of the SDS22:PP1 complex with I-3, metal (M1), and p37/p97, via unknown mechanisms, ultimately results in the dissociation of SDS22 from metal-loaded PP1 and, finally, PP1 holoenzyme formation. Critically, our data show that, if metal cannot be loaded into the M1 position of PP1 (i.e., as is case for the H66K variant, pink), PP1 holoenzyme formation does not occur and, instead, SDS22 remains constitutively bound to M1-metal-deficient PP1. Thus, M1-metal loading is strictly required for p37/p97 to effectively dissociate SDS22 from PP1. (B) SDS22 functions to bind and maintain PP1 in an inactive state, providing a pool of PP1 that is poised and ready for holoenzyme assembly (Top). As a result of cellular signaling events, distinct PP1 regulators are rapidly synthesized (Middle). These regulators (with M1, I-3, p97/p37; see A), via a mechanism not fully understood, then assemble into metal-loaded, active PP1 holoenzymes.
As has been previously observed (6), our data confirm that Mn2+ is readily exchanged from PP1 in solution (SI Appendix, Fig. S4A). Even in the presence of Mn2+, long incubation times of Mn2+-loaded PP1 and SDS22 reproducibly result in the formation of the SDS22:PP1 complex because a fraction of the metal-deficient PP1 is irreversibly trapped by SDS22. This suggests that metal association at the M1 site could be a mechanism by which PP1 activity is regulated in cells. However, the M1 metal in mammalian expressed PP1 is not Mn2+ but Zn2+. Following the long-established Irving–Williams series, which reports on the affinity of metal ions to ligands [Mn2+ < Fe2+ < Co2+ < Ni2+ < Cu2+ > Zn2+ (43)], Zn2+ is expected to have a considerably higher affinity for PP1 than Mn2+. As a consequence, it is likely that the majority of PP1 in the cell is fully loaded with metals (M1, Zn2+; M2, Fe2+). Despite this, there are conditions in which the M1 zinc might be lost (i.e., metal starvation, thermal, or other stresses). Also, the M1 metal may be ejected by an, as yet, unknown mechanism. Either would render PP1 capable of reassociating with SDS22. This will be an important area of investigation for future studies.
Finally, our observation that the reconstituted SDS22:PP1 complex, i.e., that obtained after the M1 metal is ejected from PP1, is structurally identical to the complex obtained via coexpression of SDS22 and PP1 in mammalian cells has multiple implications for PP1 biology and biogenesis. First, the conformation of PP1 is independent of the metal bound at the M2 site. This is because, as we show, bacterially expressed PP1 contains Mn2+, while mammalian expressed PP1 contains Fe2+ at the M2 position. Yet, despite the different metals at this site, the structures of the reconstituted and coexpressed SDS22:PP1 complex are identical. Thus, any differences observed in the catalytic activities of bacterial versus mammalian (typically isolated from rabbit muscle) PP1 are not due to a difference in PP1 conformation, but instead due only to the nature of the metals themselves. Second, SDS22 cannot bind unfolded PP1 and thus, it is unlikely to bind PP1 solely during the maturation process. Rather, SDS22 is fully capable of trapping any PP1 from which the M1 metal has been lost. These results, coupled with the observation that fully active PP1 is readily produced in E. coli in the absence of SDS22, show that SDS22 is a PP1-specific regulator that functions to stabilize/trap a metal-deficient inactive conformation of PP1.
Trapping of metal-deficient PP1 by SDS22 is achieved via an atypical conformation of PP1 in which the 127CASINRIYG135 loop is completely reorganized. Previous docking-based models of the SDS22:PP1 complex (37) did not predict the unprecedented changes in the conformation of SDS22-bound PP1. The change in the 127CASINRIYG135 loop allows Tyr134, which is normally adjacent to the PP1 active site, to shift by more than 12 Å and bind the center of the SDS22 interface. As a consequence, SDS22 traps PP1. Interestingly, some of these now experimentally detected changes were predicted by computational methods ∼20 y ago (44).
Why might it be necessary for PP1 to be trapped in an inactive conformation by SDS22? The most likely explanation is that it provides a "pool" of inactive PP1 poised for holoenzyme formation, while simultaneously preventing unregulated PP1 activity in the cell (Fig. 6B). PP1 is directed to its substrates via its interactions with >200 distinct regulatory proteins. In response to cellular signals, these distinct holoenzymes must be rapidly assembled. We posit that SDS22:PP1 complexes function as a kind of PP1 "bank," keeping a pool of PP1 that is ready for holoenzyme formation yet completely inactive, preventing it from catalyzing unwanted dephosphorylation reactions. In this way, the cell only needs to synthesize regulators in response to incoming cellular signals (the PP1 is ready for holoenzyme formation via its association with SDS22). Then, via a process that involves p97/CDC48 but whose molecular details are still unknown, the regulator associates with PP1, and the M1 metal is loaded, resulting in a fully active and specific PP1 holoenzyme. Taken together, our studies of the SDS22:PP1 complex have significantly transformed our understanding of PP1 regulation. Our findings illustrate the central and defining role of metals in PP1 regulation and activity, revealing that both the loading and the loss of metal at the M1 site in PP1 are likely major mechanisms by which PP1 activity, via SDS22, is regulated in cells.
Materials and Methods
All buffers, proteins, DNA, and yeast strains (SI Appendix, Tables S4 and S5) are described in the SI Appendix (36, 45–47). SPR experiments performed using a 4-channel Reichert 4SPR instrument fitted with autosampler and degassing pump (SI Appendix). Crystallization and structure determination were performed as described (SI Appendix).
Supplementary Material
Acknowledgments
We are grateful to Ms. Makayla M. Freitas for help in PP1 Y134A protein expression and purification. We thank Malcolm Goldsby, Adam Sipes, and Charles Richardson for their help with plasmid construction. This work was supported by Grant R01GM134683 from the National Institute of General Medicine to W.P., and Grant R01GM098482 from the National Institute of General Medicine to R.P. Portions of this work were also supported by the Department of Biochemistry and Molecular Biology, the Feist-Weiller Cancer Center, and the Research Core facility at Louisiana State University Health Sciences Center (LSUHSC)-Shreveport. R.R. was supported by an Ike Muslow Predoctoral Fellowship awarded by the School of Graduate Studies, LSUHSC-Shreveport. This research used beamline 12-2 at the Stanford Synchrotron Radiation Lightsource. Use of the Stanford Synchrotron Radiation Lightsource (SSRL), Stanford Linear Accelerator Center (SLAC) is supported by the US Department of Energy, Office of Science and Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program was supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including Grant P41GM103393). Work at the FMX (Frontier Microfocusing Macromolecular Crystallography; 17-ID-2) beamline was supported by the National Institute of Health, National Institute of General Medical Sciences (Grant P41GM111244), and by the DOE Office of Biological and Environmental Research (Grant KP1605010). The National Synchrotron Light Source II at Brookhaven National Laboratory was supported by the DOE Office of Basic Energy Sciences under Contract DE-SC0012704 (KC0401040).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.V.O. is a guest editor invited by the Editorial Board.
Data deposition: Structures reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org/pdb/ (accession nos. 6OBN, 6OBP, 6OBQ, 6OBR, 6OBS, and 6OBU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908718116/-/DCSupplemental.
References
- 1.Brautigan D. L., Shenolikar S., Protein serine/threonine phosphatases: Keys to unlocking regulators and substrates. Annu. Rev. Biochem. 87, 921–964 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gibbons J. A., Kozubowski L., Tatchell K., Shenolikar S., Expression of human protein phosphatase-1 in Saccharomyces cerevisiae highlights the role of phosphatase isoforms in regulating eukaryotic functions. J. Biol. Chem. 282, 21838–21847 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Dancheck B., et al. , Molecular investigations of the structure and function of the protein phosphatase 1-spinophilin-inhibitor 2 heterotrimeric complex. Biochemistry 50, 1238–1246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salvi F., et al. , Effects of stably incorporated iron on protein phosphatase-1 structure and activity. FEBS Lett. 592, 4028–4038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan S. C. B., Graves D. J., Inactivation and reactivation of phosphoprotein phosphatase. Mol. Cell. Biochem. 42, 21–29 (1982). [DOI] [PubMed] [Google Scholar]
- 6.Chu Y., Lee E. Y., Schlender K. K., Activation of protein phosphatase 1. Formation of a metalloenzyme. J. Biol. Chem. 271, 2574–2577 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Goldberg J., et al. , Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376, 745–753 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Kumar G. S., et al. , Identification of the substrate recruitment mechanism of the muscle glycogen protein phosphatase 1 holoenzyme. Sci. Adv. 4, eaau6044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bollen M., Peti W., Ragusa M. J., Beullens M., The extended PP1 toolkit: Designed to create specificity. Trends Biochem. Sci. 35, 450–458 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choy M. S., Page R., Peti W., Regulation of protein phosphatase 1 by intrinsically disordered proteins. Biochem. Soc. Trans. 40, 969–974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peti W., Nairn A. C., Page R., Structural basis for protein phosphatase 1 regulation and specificity. FEBS J. 280, 596–611 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choy M. S., et al. , Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc. Natl. Acad. Sci. U.S.A. 111, 4097–4102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egloff M. P., et al. , Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16, 1876–1887 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrickx A., et al. , Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16, 365–371 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Grigoriu S., et al. , The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin. PLoS Biol. 11, e1001492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajaj R., Bollen M., Peti W., Page R., KNL1 binding to PP1 and microtubules is mutually exclusive. Structure 26, 1327–1336.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choy M. S., et al. , Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep. 11, 1885–1891 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ragusa M. J., et al. , Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat. Struct. Mol. Biol. 17, 459–464 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terrak M., Kerff F., Langsetmo K., Tao T., Dominguez R., Structural basis of protein phosphatase 1 regulation. Nature 429, 780–784 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Ohkura H., Yanagida M., S. pombe gene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell 64, 149–157 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Kachroo A. H., et al. , Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348, 921–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ceulemans H., et al. , Binding of the concave surface of the Sds22 superhelix to the alpha 4/alpha 5/alpha 6-triangle of protein phosphatase-1. J. Biol. Chem. 277, 47331–47337 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Duan H., et al. , Phosphorylation of PP1 regulator Sds22 by PLK1 ensures accurate chromosome segregation. J. Biol. Chem. 291, 21123–21136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues N. T. L., et al. , Kinetochore-localized PP1-Sds22 couples chromosome segregation to polar relaxation. Nature 524, 489–492 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Goswami S., et al. , Regulators of the protein phosphatase PP1γ2, PPP1R2, PPP1R7, and PPP1R11 are involved in epididymal sperm maturation. J. Cell. Physiol. 234, 3105–3118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bharucha J. P., Larson J. R., Gao L., Daves L. K., Tatchell K., Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol. Biol. Cell 19, 1032–1045 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Y.-L., Chen R.-H., The AAA-ATPase Cdc48 and cofactor Shp1 promote chromosome bi-orientation by balancing Aurora B activity. J. Cell Sci. 123, 2025–2034 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Cheng Y.-L., Chen R.-H., Assembly and quality control of the protein phosphatase 1 holoenzyme involves the Cdc48-Shp1 chaperone. J. Cell Sci. 128, 1180–1192 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Peggie M. W., et al. , Essential functions of Sds22p in chromosome stability and nuclear localization of PP1. J. Cell Sci. 115, 195–206 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Weith M., et al. , Ubiquitin-independent disassembly by a p97 AAA-ATPase complex drives PP1 holoenzyme formation. Mol. Cell 72, 766–777.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Kumar G. S., et al. , The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism. eLife 5, e16539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang F. L., Glinsmann W. H., Separation and characterization of two phosphorylase phosphatase inhibitors from rabbit skeletal muscle. Eur. J. Biochem. 70, 419–426 (1976). [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Zhang L., Zhao S., Lee E. Y., Identification and characterization of the human HCG V gene product as a novel inhibitor of protein phosphatase-1. Biochemistry 37, 16728–16734 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Hsieh-Wilson L. C., Allen P. B., Watanabe T., Nairn A. C., Greengard P., Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry 38, 4365–4373 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Zhang J., Zhang Z., Brew K., Lee E. Y., Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry 35, 6276–6282 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Choy M. S., Bolik-Coulon N., Archuleta T. L., Peti W., Page R., The structure of SDS22 provides insights into the mechanism of heterodimer formation with PP1. Acta Crystallogr. F Struct. Biol. Commun. 74, 817–824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heroes E., et al. , Structure-Guided exploration of SDS22 interactions with protein phosphatase PP1 and the splicing factor BCLAF1. Structure 27, 507–518.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapinos L. E., Song B., Sigel H., Metal ion-coordinating properties of imidazole and derivatives in aqueous solution: Interrelation between complex stability and ligand basicity. Inorg. Chim. Acta 280, 50–56 (1998). [Google Scholar]
- 39.Marquardt J., Chen X., Bi E., Architecture, remodeling, and functions of the septin cytoskeleton. Cytoskeleton 76, 7–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh Y., Bi E., Septin structure and function in yeast and beyond. Trends Cell Biol. 21, 141–148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caviston J. P., Longtine M., Pringle J. R., Bi E., The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobbelaere J., Gentry M. S., Hallberg R. L., Barral Y., Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345–357 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Irving H., Williams J. P., Order of stability of metal complexes. Nature 162, 746–747 (1949). [Google Scholar]
- 44.Wozniak E., Ołdziej S., Ciarkowski J., Molecular modeling of the catalytic domain of serine/threonine phosphatase-1 with the Zn2+ and Mn2+ di-nuclear ion centers in the active site. Comput. Chem. 24, 381–390 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Peti W., Page R., Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr. Purif. 51, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- 46.O’Connell N., et al. , The molecular basis for substrate specificity of the nuclear NIPP1:PP1 holoenzyme. Structure 20, 1746–1756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravindran R., Polk P., Robinson L. C., Tatchell K., New ubiquitin-dependent mechanisms regulating the Aurora B-protein phosphatase 1 balance in Saccharomyces cerevisiae. J. Cell Sci. 131, jcs217620 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.