Abstract
The National Institute of Environmental Health Sciences (NIEHS) Superfund Research Program (SRP) aims to support university-based multidisciplinary research on human health and environmental issues related to hazardous substances and pollutants. The Texas A&M Superfund Research Program comprehensively evaluates the complexities of hazardous chemical mixtures and their potential adverse health impacts due to exposure through a number of multi-disciplinary projects and cores. One of the essential components of the Texas A&M Superfund Research Center is the Data Science Core, which serves as the basis for translating the data produced by the multi-disciplinary research projects into useful knowledge for the community via data collection, quality control, analysis, and model generation. In this work, we demonstrate the Texas A&M Superfund Research Program computational platform, which houses and integrates large-scale, diverse datasets generated across the Center, provides basic visualization service to facilitate interpretation, monitors data quality, and finally implements a variety of state-of-the-art statistical analysis for model/tool development. The platform is aimed to facilitate effective integration and collaboration across the Center and acts as an enabler for the dissemination of comprehensive ad-hoc tools and models developed to address the environmental and health effects of chemical mixture exposure during environmental emergency-related contamination events.
Keywords: Data analytics, data integration, statistical analysis, collaborative networks
1. Introduction
The risk of chemical contamination and exposure to hazardous chemicals are elevated during and after natural catastrophic events (i.e., hurricanes) due to the increased mobility of many chemical toxicants. In such situations, the rapid and precise examination of potential sources and pathways of chemical contamination becomes essential: (i) for identifying their adverse health impacts and (ii) for delivering solutions to mitigate such adverse effects. To this end, Texas A&M Superfund Research Program (TAMU Superfund Research Center, 2018) aims to build both experimental and computational models, methods and tools through exposomics research and data analysis. The program extensively studies the health, economic and social impacts of hazardous complex chemical mixtures after environmental emergencies with Galveston Bay/Houston Ship Channel area being selected as a case study.
TAMU SRP is a cross-disciplinary program and has a tightly integrated structure which governs four main research projects (two environmental and two biomedical research projects). The two environmental projects, namely Project 1 and 2, focus on understanding dynamic exposure pathways under the conditions of environmental emergencies and designing novel broad-acting sorption materials for reducing bioavailability of contaminants. Project 3 and 4, being the two biomedical projects, are studying in vitro and in vivo hazard, kinetics and inter-individual variability of responses to chemical mixtures and developing in vitro multiplex single-cell assays to detect endocrine disruption potential of mixtures. Each of these projects utilizes various experimental methodologies for detecting, assessing, evaluating and characterizing the effects of complex chemical contaminants including Gas Chromatography-Mass Spectrometry (GC-MS), Ion Mobility-Mass Spectrometry (IM-MS), Inductively Coupled Plasma Mass Spectrometry (ICP-MS), Ultraviolet-Visible Spectroscopy (UV-Vis), high-throughput imaging and image analysis. Hence, these four projects generate large quantities of highly diverse datasets, where their maintenance and analysis require a systematic approach through the development of a computational platform.
In addition to the four main research projects, there are three research supporting cores within the TAMU SRP, one of which is the Data Science Core. The Data Science Core serves as the basis for translating the data produced by the four research projects into useful knowledge for the community via data collection, quality control, analysis, visualization and model generation. This Core functions as a hub that collects, processes and integrates the aforementioned diverse datasets over a computational platform to draw specific conclusions via supervised (i.e., regression, classification) and unsupervised (i.e., clustering) analysis. These techniques are widely used in process systems engineering (PSE) including process monitoring (Onel et al., 2018b) and grey-box optimization (Beykal et al., 2018). In this work, we present the TAMU SRP computational platform which aims to promote collaboration across the Center and facilitate dissemination of methods/data across all projects of the program as well as to the wider community. The computational platform is developed as an online tool that specifically uses a relational database for data storage as well as statistical and machine learning techniques to create decision support models, housing both novel computational methodologies and state-of the-art data analytics techniques. It establishes an accessible front-end interface for the application of the high-performance models and tools developed during collaborations with individual research projects (Onel et al., 2018a). The details of the computational platform are provided in the following sections where its integration and connection with one of the biomedical projects is demonstrated as a motivating example.
2. Computational Platform
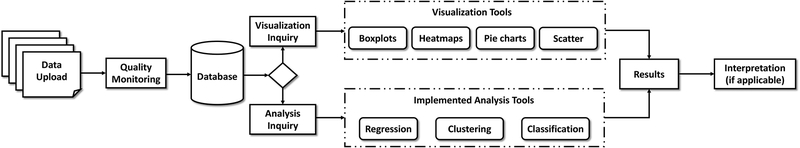
The online computational platform is developed in Python environment, whereas the backend functionalities utilize either R or Python environments. The relational database for storing and sharing data across the Center is based on SQLite. A flow diagram of the platform is shown in Figure 1. The computational platform is developed and implemented in two stages. The first stage entails the dissemination of datasets and methodologies across the Center for supplying a convenient environment for collaboration among all projects. The second stage enables access to the extracted knowledge, models, and tools with the scientific community, government and commercial stakeholders. In this work we will only present the developments of the first stage.
Figure 1.
Online computational platform flowchart.
The computational platform first requires a data upload by the user, which is further passed to an initial quality monitoring module. The quality monitoring module checks the dataset for any missing data and/or outliers. Missing data is handled two-fold: (i) Deletion of rows or columns that include missing data, (ii) imputation by k-nearest neighbor (k-NN) methodology (Ramaswamy et al., 2000). This pre-processed data is stored under the relational database for future reference. A summary of this first module is provided as a feedback to the user. Second, the user specifies a type of inquiry, namely visualization and analysis. Currently, four visualization techniques are implemented within the platform including, boxplots, heatmaps, pie charts and scatter plots. Guidelines for selecting the relevant visualization technique is provided online. The generated plots or maps are then displayed on the interface which can be downloaded by the user. Specifically, further interpretation of boxplots, containing the summary of statistics (i.e., median, interquartile range etc.) is provided along with the visuals. Next, the datasets can be analyzed via unsupervised or supervised techniques depending on the purpose of the study. For untargeted analysis, clustering with hierarchical, k-means, and deep learning techniques are utilized. For targeted analysis, where the output of certain experiments is known and used for training models, supervised learning approaches are chosen. Specifically, for the datasets with discrete type of output (or label), classification techniques are used. Current classification techniques include Support Vector Machines (SVM), Random Forest (RF) Algorithm, and logistic regression. Whereas if the output of the dataset is continuous, regression techniques are employed. Here, in addition to SVM and RF Algorithm, interpolation (i.e., Kriging, radial basis functions) and multivariate regression techniques (i.e., linear, quadratic) are employed. The analysis selection is guided by the collaboration between the Data Science Core personnel and individual research projects. Once the data analysis methodology is established for a specific type of data, custom tools are generated and implemented within the platform. This automates the workflow across the Center, minimizes repetitive efforts, thus increasing the overall efficiency.
It is important to note that the large (i.e., exposomics and imaging) datasets generated by the two biomedical research projects under TAMU SRP are in high dimensional space. This necessitates the use of dimensionality reduction techniques along with the aforementioned data analysis methodologies. To this end, numerous dimensionality reduction methodologies are implemented in the computational platform. These include Principal Component Analysis, Chi-squared test, built-in feature ranking algorithms of RF and in-house developed SVM-based feature selection algorithms (Onel et al., 2018b).
3. Motivating Example
Here, we present a motivating example from TAMU SRP Project 4 to showcase the use of the developed computational platform. This project focuses on understanding the hazardous effects of environmental contaminants and mixtures that may interfere with proper function of the human endocrine system, causing several adverse health effects (i.e., reproductive, developmental, metabolic etc.) due to modulations in hormone nuclear receptors’ action. Hence, Project 4 personnel develop single-cell high throughput microscopy experiments with associated image analysis and informatics, thus producing high dimensional imaging data to fingerprint the endocrine disruptor potential of chemicals and environmental mixtures, whereas Data Science Core personnel use the generated data to build predictive models that classifies and quantifies the endocrine disruptor potential. Below, the development of data-driven models that predict potential activity of chemicals on a prototypical target, the estrogen receptor (ER), and their use through the computational platform are described in detail.
In order to establish a framework, 45 known chemical compounds (agonists, antagonists and inactive for the ER) used by the United States Environmental Protection Agency (US EPA) are utilized to determine the effectors of ER action (Judson et al., 2015). The GFP-ERα:PRL-HeLa cell line, and its derivatives, is an engineered model that allows multi-parametric simultaneous measurements of many important features, including, ligand binding, DNA binding, chromatin remodeling and transcriptional output, required for the activation of Estrogen Receptors (ER) (Szafran et al., 2017). This high throughput microscopy assay is used to test the responses to the EPA 45 reference compounds as well as to the control agonist 17β-estradiol (E2) and antagonist 4-hydroxytamoxifen (4OHT). The effect of these test chemicals can broadly be classified three-fold: (i) agonist (which elicits a positive response of the ER signaling pathway – akin to E2), (ii) antagonist (mimicking a response like 4OHT), or (iii) inactive. By treating the cells with a six-point dose-response of these compounds, high throughput imaging data is generated and analyzed. This yields a data matrix of 180 (4 measurements for each 45 compounds) by 70 descriptors (features). Each descriptor considers various aspects of the ER pathway (i.e., Is the ER level changing? Does ER bind to DNA? How much chromatin remodeling happens? etc.). This dataset is later passed to the Data Science Core for further analysis and for modeling the ER disrupting potential of the tested compounds. The details on model generation and step-by-step use of the developed model within the computational platform are provided below.
Step 1 – Data Quality Monitoring:
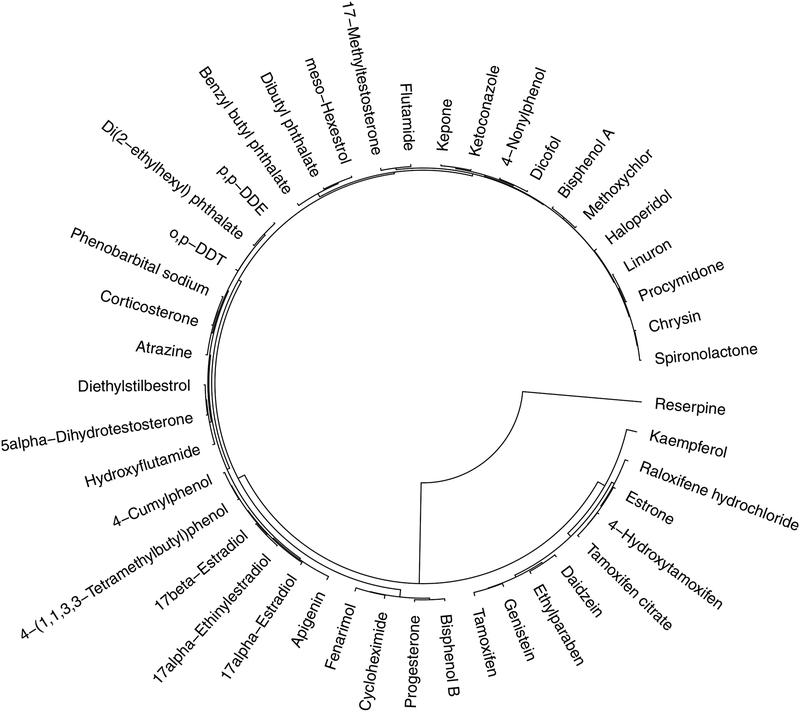
As an initial step, the quality of the received experimental data is inspected by identifying any potential missing data. In this case study, there are no missing data. The complete dataset is analyzed to detect any potential outliers via hierarchical clustering algorithm with complete linkage methodology and Euclidean distance metric. Identification and removal of outliers is essential in order to ensure accurate model development. The results reveal “Reserpine” as an outlier, which has been removed from further analysis (Figure 2).
Figure 2.
Outlier identification via hierarchical clustering. Reserpine is identified as outlier.
Step 2 – Normalization:
The goal is to classify the compounds based on agonist/antagonist activity. To achieve this, inactive compounds must be separated prior to normalization and model building. This is done by using a threshold for the cell population with a visible nuclear spot, signifying ER-DNA binding. Less than 10% of the cell population, that has a visible nuclear spot, are considered to be inactive and removed from further analysis. Then, the dataset is normalized in order to attain a consistent range per feature. Specifically, the order of magnitude of intensity related measurements significantly differ from measurements derived from nucleus shape. Therefore, normalization is performed by using Equation 1.
| (1) |
Step 3 – Predictive Modeling & Dimensionality Reduction:
Once the data is pre-processed, cleaned from outliers and normalized, various classification algorithms are applied to build predictive models. In this study, RF algorithm is employed for the classification of agonist/antagonist activity and for identifying important descriptors through the built-in feature ranking property (Breiman, 2001). Tuning is performed and optimal number of trees is identified as 500. Final model is then built with the optimal number of trees by using 5-fold cross-validation. The top 10 informative features achieved during modeling are also reported in Table 1. The model accuracy before dimensionality reduction is achieved as 90%, whereas the end-model, the reduced model that use the top 10 informative features, has 92% accuracy.
Table 1.
Top 10 informative features for agonist/antagonist classification.
| Rank | Measurement | Rank | Measurement |
|---|---|---|---|
| 1 | Nucleoplasm GFP Pixel Intensity Variance | 6 | Ratio of Nuclear Spot to Nucleoplasm GFP Intensity |
| 2 | Nuclear GFP Pixel Intensity Variance | 7 | Nuclear Spot GFP Pixel Intensity Variance |
| 3 | Nucleoplasm 90th Percentile GFP Pixel Intensity | 8 | Cytoplasm 75th Percentile GFP Pixel Intensity |
| 4 | Nuclear 90th Percentile GFP Pixel Intensity | 9 | Nuclear Spot 75th Percentile GFP Pixel Intensity |
| 5 | Nuclear Spot 90th Percentile GFP Pixel Intensity | 10 | Cytoplasm 90th Percentile GFP Pixel Intensity |
Step 4 – Automation of Analysis:
This end-model is then converted into an executable tool and implemented to the Python based environment of the computational platform.
4. Future directions
Development of the computational platform is an ongoing process. As new data are generated, corresponding tools and models are tailored, updated and incorporated in the platform. Current limitation of the platform is that the analysis only covers the Center projects and datasets. However, access to historical data provided by government agencies is crucial for comparative analysis in TAMU SRP (e.g., ToxCast and Tox21 initiatives). Therefore, additional features will be provided for access to the relevant public data repositories through integration. Finally, one of the main goals of the Data Science Core is to serve as a basis to facilitate TAMU SRP data analysis and understanding. Therefore, training for the use of generated models and tools within the computational platform will be provided in collaboration with the Research Translation and Training Cores.
5. Conclusions
In this study, development of a computational platform for the Texas A&M Superfund Research Center is presented. This platform provides on-demand, intuitive access to the custom-made data analysis tools and models developed for the environmental and biomedical projects within the Center. These analysis techniques are applicable to PSE problems. The ultimate goal is to establish an online data analytics services for rapid decision-making during environmental emergencies. This research is funded by U.S. National Institute of Health grant P42 ES027704 and Texas A&M Energy Institute.
References
- Szafran AT, Stossi F, Mancini MG, Walker CL, Mancini MA, 2017, Characterizing properties of non-estrogenic substituted bisphenol analogs using high throughput microscopy and image analysis, PloS one, 12(7), e0180141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beykal B, Boukouvala F, Floudas CA, Sorek N, Zalavadia H, Gildin E, 2018, Global Optimization of Grey-Box Computational Systems Using Surrogate Functions and Application to Highly Constrained Oil-Field Operations, Computers & Chemical Engineering, 114, 99–110. [Google Scholar]
- Breiman L, 2001, Random Forests, Machine Learning, 45, 1, 5–32. [Google Scholar]
- Onel M, Beykal B, Wang M, Grimm FA, Zhou L, Wright FA, Phillips TD, Rusyn I, Pistikopoulos EN, 2018a, Optimal Chemical Grouping and Sorbent Material Design by Data Analysis, Modeling and Dimensionality Reduction Techniques, Computer Aided Chemical Engineering, 43, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onel M, Kieslich CA, Guzman YA, Floudas CA, Pistikopoulos EN, 2018b, Big Data Approach to Batch Process Monitoring: Simultaneous Fault Detection and Diagnosis Using Nonlinear Support Vector Machine-based Feature Selection, Computers & Chemical Engineering, 115, 46–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia M, Huang R, Rotroff DM, Filer DL, Houck KA, Martin MT, Sipes N, Richard AM, Mansouri K, Setzer RW, Knudsen TB, Crofton KM, Thomas RS, 2015, Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor, Toxicological Sciences 148(1), 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAMU Superfund Research Center (2018). https://superfund.tamu.edu/ (accessed 9 November 2018).