SUMMARY
Forkhead box P2 (FOXP2) is a transcription factor expressed in the human brain that peaks during fetal development, and disruption in its ability to regulate downstream target genes leads to vulnerability to neurodevelopmental disorders. However, the mechanisms by which FOXP2 exerts regulatory control over targets during neuronal maturation have not been fully elucidated. Here, we use genome-wide chromatin accessibility assays and transcriptome-wide expression analyses in differentiating human neurons to show that FOXP2 represses proliferation-promoting genes in a DNA-binding-dependent manner. In contrast, FOXP2 and its cofactors, NFIA/B, activate neuronal maturation genes in a manner that does not require FOXP2 to interact with DNA directly. Moreover, comparisons with expression data from the developing human brain suggest that FOXP2/NFIA/B-dependent chromatin alterations drive maturation of excitatory cortical neurons. Thus, FOXP2 and its NFI cofactors may be specifically important for the development of cortical circuits underlying neurodevelopmental disorders.
INTRODUCTION
Mutations disrupting the ability of FOXP2 to bind to DNA, and thus regulate its target genes, cause heritable forms of childhood apraxia of speech together with deficits in grammatical function, linking FOXP2 to speech and language development (Lai et al., 2000, Lai et al., 2001, Lennon et al., 2007, MacDermot et al., 2005, Vargha-Khadem et al., 1995, Watkins et al., 2002a). Moreover, the first attention deficit/hyperactivity disorder (ADHD) genome-wide association study (GWAS) has identified a risk locus containing FOXP2 (Demontis et al., 2019), and many FoxP2 targets are disrupted in autism spectrum disorder (ASD) and schizophrenia (SCZ) (Vernes et al., 2008, Mukamel et al., 2011, Chen et al., 2016, Walker et al., 2012), suggesting that neurodevelopmental disorders occur upon perturbation of transcriptional networks governed by FOXP2. Indeed, genome-wide FoxP2 binding and expression microarray studies in mouse and human cells imply that FoxP2 regulates genes involved in neuron outgrowth and other neuronal functions (Vernes et al., 2007, Vernes et al., 2011, Spiteri et al., 2007), but the mechanism by which FoxP2 does so remains unclear.
Moreover, while FOXP2 is highly conserved among mammals, the hominin version of FOXP2 contains two amino acid changes that impact transcriptional activity and behaviors (Enard et al., 2002, Zhang et al., 2002, Enard et al., 2009, Konopka et al., 2009, Reimers-Kipping et al., 2011). Thus, human-specific transcriptional regulation mediated by FOXP2 during brain development may lead to the establishment of neural circuits necessary for normal cognitive development. Mouse models of Foxp2 loss-of-function and humanized FOXP2 have been highly valuable (Shu et al., 2005, French et al., 2007, Groszer et al., 2008, Fujita et al., 2008, French et al., 2018, Medvedeva et al., 2018, Enard et al., 2009, Reimers-Kipping et al., 2011); however; differences in brain development and structure between mouse and human, especially in the cortex (Lui et al., 2011), highlight the need to study transcriptional regulation by FOXP2 on a human genomic background. While promoter regions bound by FOXP2 in human fetal brain have been previously identified (Spiteri et al., 2007), genes regulated by FOXP2 during human neuronal differentiation have never been assayed at the transcriptome-wide level.
Here, using genome-wide chromatin accessibility assays and transcriptome-wide expression analyses in differentiating primary human neurons, we determined that FOXP2 binds DNA at its canonical binding motif to condense chromatin and repress genes involved in maintaining a non-neuronal state, while opening chromatin in conjunction with NFI transcription factors to activate genes that promote neuronal maturation. These FOXP2-regulated transcriptional programs are enriched for ASD and SCZ risk genes, suggesting a role for FOXP2 in psychiatric disease-relevant programs of early neuronal differentiation. Moreover, by comparing genes regulated by FOXP2 with in vivo expression data from human fetal brain tissue and single cells, we hypothesize that the co-regulation of activated genes by NFIA/B and FOXP2 may be especially important in the cortex, specifically in the maturation of excitatory neurons. Thus, we have uncovered a mechanism by which FOXP2 activates transcriptional targets that facilitate neuronal maturation in the developing human cortex and are at risk in cognitive diseases.
RESULTS
FOXP2 promotes mature neuronal gene expression profiles in hDNs
To uncover the mechanism by which FOXP2 regulates gene expression in human neurons, we manipulated FOXP2 expression in a human cellular system that recapitulates in vivo human brain development at the level of gene expression programs (Rosen et al., 2011, Konopka et al., 2012, Wexler et al., 2011, Palmer et al., 2001, Stein et al., 2014, de la Torre-Ubieta et al., 2018). We used human neural progenitors (hNPs) isolated from human fetal brain tissue that can be induced into post-mitotic neurons (human differentiated neurons or hDNs) and can be genetically modified using viruses.
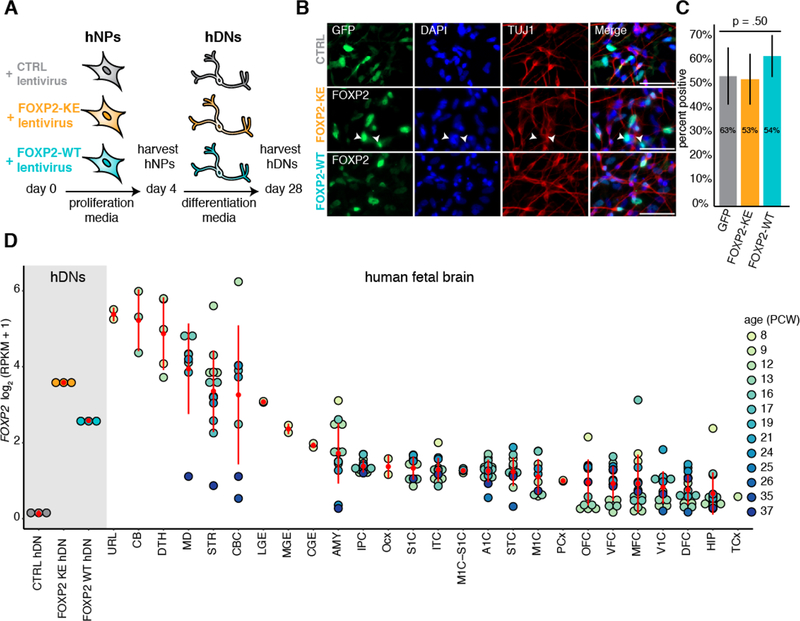
Because these cells do not endogenously express FOXP2, we controlled for exogenous expression of wild-type FOXP2 (FOXP2-WT) by expressing FOXP2 with a DNA-binding mutation (FOXP2-KE) or a control GFP construct (CTRL) (Figure 1A). FOXP2-KE contains the R553H point mutation in its DNA-binding domain which causes childhood apraxia of speech in the “KE” family (Lai et al., 2001), and electron mobility shift assays have shown that this altered form of FOXP2 is unable to bind to DNA (Vernes et al., 2006). This mutant has been shown to localize less efficiently to the nucleus (Vernes et al., 2006), and in line with this, immunocytochemistry of hDNs showed that while FOXP2-KE is mainly localized to the nucleus, it can be found in the cytoplasm (white arrow heads Figure 1B). Overall, 50–60% of the transduced cells were positive for FOXP2 or CTRL-GFP with no significant difference in the number of positive cells between conditions (Figure 1C).
Figure 1:
Expression of FOXP2-WT and –KE in hDNs
(A) Schematic representation of the human cell culture and RNA collection timeline. (B) Immunocytochemistry showing hDNs exogenously expressing CTRL-GFP, FOXP2-WT or –KE. White arrowheads indicate cells with cytoplasmic localization of FOXP2-KE. Scale bar represents 50 μm. (C) Bar plot showing the average percent of cells expressing GFP, FOXP2-WT, or FOXP2-KE. Four images from separate coverslips were quantified. Error bars represent standard deviation. The difference between conditions was assessed using a one-way ANOVA. (D) Dot plot showing the expression of FOXP2 in hDNs and in various human fetal brain regions (Brainspan, Zhu et al. 2018). Each dot represents an individual sample and in vivo samples are colored by age. Red dots represent the mean expression in each brain region, and red lines are the standard deviation. Occipital neocortex (Ocx), primary motor-sensory cortex (M1C-S1C), amygdaloid complex (AMY), medial ganglionic eminence (MGE), posterior (caudal) superior temporal cortex (STC), upper (rostral) rhombic lip (URL), caudal ganglionic eminence (CGE), dorsal thalamus (DTH), anterior (rostral) cingulate (medial prefrontal) cortex (MFC), dorsolateral prefrontal cortex (DFC), orbital frontal cortex (OFC), lateral ganglionic eminence (LGE), inferolateral temporal cortex (ITC), hippocampus (HIP), ventrolateral prefrontal cortex (VFC), parietal neocortex (PCx), temporal neocortex (TCx), primary auditory cortex (A1C), primary visual cortex (V1C), striatum (STR), primary motor cortex (M1C), posteroventral (inferior) parietal cortex (IPC), primary somatosensory cortex (S1C), cerebellum (CB), cerebellar cortex (CBC), mediodorsal nucleus of thalamus (MD)
We then carried out RNA-seq in proliferating and differentiated cells in the presence or absence of FOXP2. Across conditions, hDNs have reduced expression of neuroepithelial and immature neuron marker genes (i.e. NES, SOX2, TUBB3, and DCX), but increased expression of mature neuronal markers (SYP, RBFOX1, DLG, and MEF2C) and genes coding for many glutamate and GABBA receptors compared to hNPs (Figure S1A, Table S1A–C). Additionally, using Specific Expression Analysis (Xu et al., 2014), we found that genes activated during the differentiation program significantly overlap with genes enriched in multiple brain regions during fetal development including the thalamus, striatum, cortex, and cerebellum (Figure S1B, Table S1A). Thus, while hDNs express markers of mature neurons at higher levels than hNPs, the gene expression profile of these cells does not reflect that of any specific brain region during development.
In CTRL hDNs, FOXP2 expression is below 0.5 RPKM, our lower expression cutoff; however FOXP2 expression in FOXP2-KE and –WT hDNs falls within the range of endogenous FOXP2 levels in brain regions previously identified to contain a substantial proportion of FOXP2-expressing cells during human brain development including the cerebellum, thalamus, and striatum (Lai et al., 2003, Zhu et al., 2018)(Figure 1D). As FOXP2 is expressed in only a subset of neurons in the developing human cortex (Vernes et al., 2008, Lai et al., 2003), it is not surprising that FOXP2 transcript is diluted in bulk RNA-seq data from this tissue (Zhu et al., 2018).
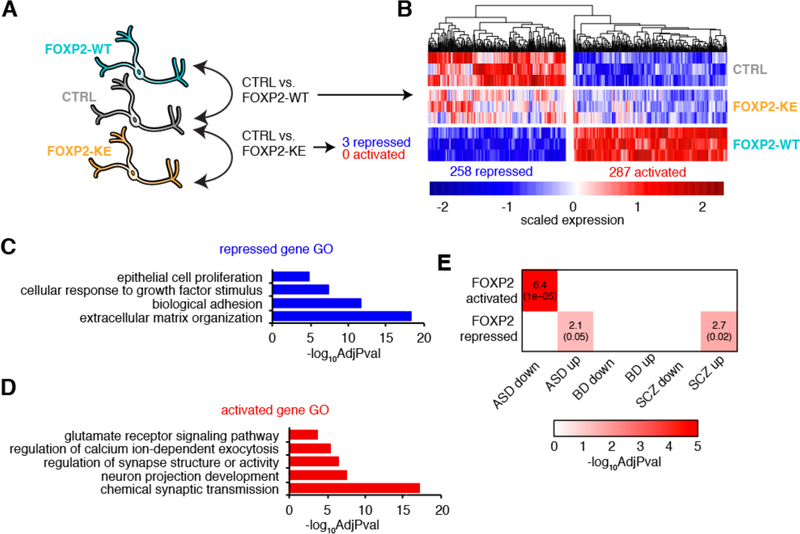
To identify genes regulated by FOXP2 in human neurons, we performed differential expression analysis comparing CTRL hDNs to either FOXP2-WT or -KE-expressing hDNs (Figure 2A). This revealed 258 genes repressed and 287 genes activated by FOXP2-WT (Figure 2B, Table S2A). Only three genes were significantly repressed and no genes were activated by FOXP2-KE (Figure 2A, Table S2B). This suggests that expression changes seen in FOXP2-WT hDNs are mediated by the FOXP2 protein and are not a consequence of forced expression. Gene ontology (GO) categories such as “cellular response to growth factor stimulus” and “extracellular matrix organization” are enriched for genes repressed by FOXP2-WT in hDNs (Figure 2C, Table S2C). Importantly, expression analyses of human and mouse cortical germinal zones suggest that growth factors and ECM components promote the proliferation of neural progenitors in mammals (Fietz et al., 2012). Activated genes are enriched for genes involved in neuron projection development, consistent with the role of murine Foxp2 in mediating neurite outgrowth (Enard et al., 2009, Vernes et al., 2011). Additionally, activated genes are enriched for genes that regulate synapse structure or activity or are involved in chemical synaptic transmission, including glutamate signaling and regulation of calcium ion-dependent exocytosis (Figure 2D, Table S2D). Moreover, using data from the PsychENCODE Consortium (Gandal et al., 2018) we found that genes downregulated in the brains of patients with ASD compared to control are enriched for genes activated by FOXP2, and genes upregulated in the brains of patients with ASD and SCZ are enriched for genes repressed by FOXP2 (Figure 2E, Table S1A). Taken together, these data suggest that FOXP2 promotes maturation of neurons while repressing a progenitor-like state that and disruptions in FOXP2-regulated neuronal maturation can lead to neurodevelopmental disorders.
Figure 2:
FOXP2 promotes mature neuronal gene expression profiles in hDNs
(A) A schematic showing the conditions compared for differential expression (DE) analysis. (B) A heatmap showing the relative expression of genes differentially expressed by FOXP2 across three replicates of each condition. (C) Summarized gene ontology terms enriched for genes repressed by FOXP2 (Benjamini-Hochberg adjusted p-value). (D) Summarized gene ontology terms enriched for genes activated by FOXP2 (Benjamini-Hochberg adjusted p-value). (E) Heat map showing the overlaps between FOXP2 DE genes and genes up- or downregulated in the brains of patients with neurodevelopment disorders versus unaffected controls as determined by Gandal et al. 2018. The number in the box is the percent overlap followed by the Benjamini-Hochberg adjusted p-value in parenthesis. Autism spectrum disorder (ASD); bipolar disorder (BD); schizophrenia (SCZ).
FOXP2 promotes mature neuronal gene expression via two distinct mechanisms
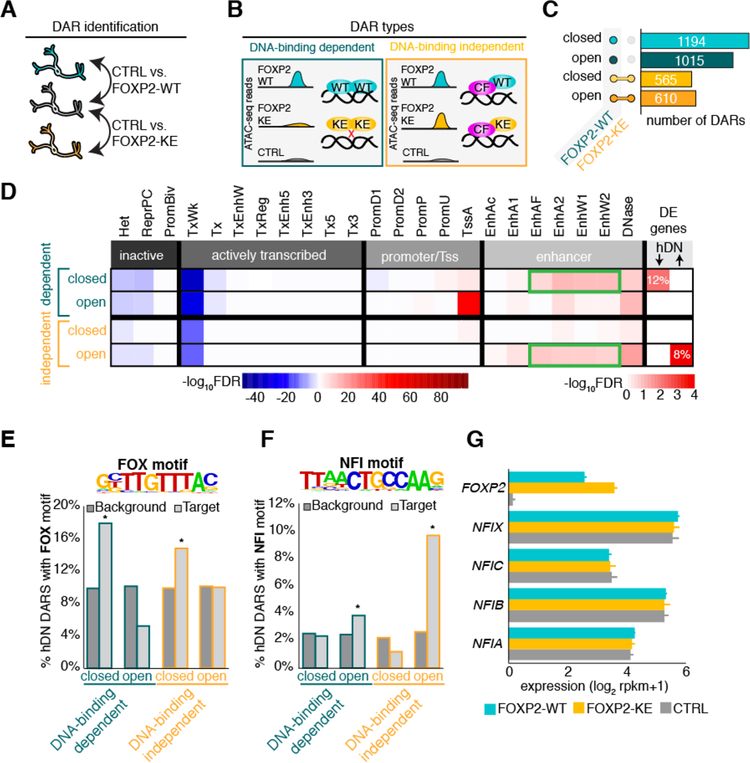
To uncover the mechanism by which FOXP2 promotes neuronal gene expression, we performed ATAC-seq (Buenrostro et al., 2015a) and identified differentially accessible regions of chromatin with expression of FOXP2 in differentiated cells. As in the RNA-seq experiment, we used cells expressing FOXP2-WT, -KE, and CTRL constructs. We hypothesized that alterations in chromatin structure could occur via two different mechanisms: 1) FOXP2 may open or close chromatin through direct DNA-binding, or 2) FOXP2 may change chromatin structure independently of its DNA-binding ability via interaction with a cofactor. Thus, while the R553H mutation of FOXP2-KE renders it unable to bind DNA and regulate gene expression, it may still be capable of altering chromatin through preserved protein-protein interactions. To identify both DNA-binding-dependent and -independent differentially accessible regions (DARs) of chromatin, we calculated the fold-difference between normalized reads in regions accessible in FOXP2-WT or -KE-expressing cells relative to CTRL cells in a pairwise manner (Figure 3A). Regions made differentially accessible with only FOXP2-WT expression were called DNA-binding-dependent DARs, while regions made differentially accessible if either FOXP2-WT or FOXP2-KE was expressed were called DNA-binding-independent DARs (Figure 3B, Table S3A). We found that DNA-binding-dependent DARs are more abundant than DNA-binding-independent DARs, and within each DAR type there are similar numbers of differentially open and closed regions, suggesting that there may be four different methods through which FOXP2 regulates transcription (Figure 3C).
Figure 3:
FOXP2 promotes mature neuronal gene expression via two distinct mechanisms
(A) Schematic showing pairwise comparison of ATAC-seq samples for identification of differentially accessible chromatin regions (DARs). (B) Examples of open DNA-binding dependent and independent DARs. FOXP2-WT (WT), FOXP2-KE (KE), hypothetical cofactor (CF). (C) Bar plot showing number of DARs open or closed compared to CTRL and the conditions in which they are present. DARs represented by teal and green bars occur only when FOXP2-WT is expressed (DNA-binding dependent), while DARs represented by yellow and orange bars co-occur with expression of FOPX2-WT or FOXP2-KE (DNA-binding independent). (D) Heat map representing the adjusted p-value of the overlaps between DARs and regions of the genome defined by 24 imputed chromatin states in human fetal brain(Roadmap Epigenomics et al., 2015, Ernst and Kellis, 2015) and the hypergeometric overlap between genes near DARs and FOXP2 DE genes. For the chromatin states, darker blue corresponds to −log10 Benjamini-Hochberg adjusted p-value of significantly under-enriched groups while red corresponds to significantly enriched groups. For the DE gene overlaps color corresponds to −log10 Benjamini-Hochberg adjusted p-value with red being most and white being least significant. Enhancer types enriched in DAR groups significantly overlapping with DE genes are outlined in green. (E) Top: Significantly enriched FOX motif identified in closed DNA-binding dependent DARs by de novo motif analysis. Bottom: Percentage of DARs with previously identified FOX motif compared to background regions. (F) Top: Significantly enriched NFI-half site identified in open DNA-binding independent DARs by de novo motif analysis. Bottom: Percent of DARs with previously identified NFI-half site compared to background regions. For E and F p-values were calculated using hypergeometric test (*p < 0.01). (G) Bar plot showing the average normalized expression of FOXP2 and NFIA-C and X in hDNs. Error bars represent the standard deviation.
To identify the functional significance of these DARs, we compared DAR locations to the locations of genomic regions with defined functional chromatin states imputed from epigenetic marks in human fetal brain (Roadmap Epigenomics et al., 2015, Ernst and Kellis, 2015) (Figure 3D, for abbreviation explanation Table S3B). DARs do not significantly overlap with inactive regions of chromatin, such as heterochromatin and regions repressed by the polycomb complex, suggesting FOXP2 alters chromatin structure in regulatory regions of the genome (Figure 3D). FOXP2 DARs are also not enriched in actively transcribed regions marked by H3K36me3, H4K20me1, and H3K79me2 (Ernst and Kellis, 2015), consistent with ATAC-seq data from human fetal brain (de la Torre-Ubieta et al., 2018) (Figure 3D). Interestingly, most DAR types were significantly enriched for enhancer regions, suggesting that FOXP2 might exert its effects through long-range interactions with promoter regions.
To distinguish how the DNA-binding-dependent and -independent DARs might influence gene expression, we compared genes with the closest transcriptional start sites to DARs in each group (Supplementary Table 3D–F) with FOXP2-WT differentially expressed (DE) genes (Figure 3D, Supplementary Table 1A). Genes near closed, DNA-binding-dependent DARs overlapped significantly with genes repressed by FOXP2 in hDNs, while genes near open, DNA-binding-independent DARs overlapped significantly with hDN FOXP2-activated genes (Figure 3D). Interestingly, the only DAR types that significantly overlapped with DE genes were those with the most significant enrichment for certain enhancer states (Figure 3D, outlined in green). These data suggest that FOXP2 directly binds DNA to facilitate chromatin compaction in enhancer regions that control target gene repression. Conversely, FOXP2 activates gene expression by opening chromatin in enhancer regions independently of direct DNA binding, perhaps in conjunction with a cofactor or cofactors.
To identify potential FOXP2 cofactors, we performed de novo motif analysis (Heinz et al., 2010) on closed DNA-binding-dependent and open DNA-binding-independent DARs. A Forkhead motif was among the most significantly enriched motifs identified in hDN closed DNA-binding-dependent DARs (p = 1×10−16), while hDN open DNA-binding-independent DARs were most significantly enriched with an NFI half-site motif (p = 1×10−16) (Figure 3E, F, Table S3C, S3F). Next, we performed a known motif enrichment analysis to compare the prevalence of the de novo identified motifs in all DAR types compared to background regions normalized for sequence bias (Heinz et al., 2010). The Forkhead motif was significantly enriched above background in hDN closed DNA-binding-dependent and -independent DARs (Figure 3E). Closed DNA-binding-dependent DARs likely represent areas of chromatin condensed by FOXP2 homodimers, while closed DNA-binding-independent DARs may be bound by heterodimers of FOXP2 and FOXP1 or FOXP4, which are both expressed in hDNs and are able to interact with FOXP2-KE, albeit with slightly decreased affinity (Estruch et al., 2016). As expected, open DNA-binding-independent DARs were highly enriched with NFI half-sites, and open DNA-binding-dependent DARs were enriched to a lesser extent (Figure 3F). NFI transcription factors (NFIA-C and X) are expressed in the developing human brain and have been shown to be important for various aspects of brain development (Betancourt et al., 2014, Heng et al., 2012, Martynoga et al., 2013). Importantly, all four NFI factors are expressed in hDNs, but none are activated by FOXP2-WT (Figure 3G), suggesting that the NFI motif is not present in differentially open regions simply because NFI transcription factors are increased in FOXP2-expressing hDNs. Taken together, these data suggest that FOXP2 binds its consensus sequence to close chromatin and repress gene expression, and to activate gene expression, FOXP2 may work with an NFI cofactor to open inaccessible chromatin.
FOXP2 and NFI are co-expressed in excitatory subplate neurons in the developing human cortex
As previously discussed, FOXP2 is expressed during human fetal brain development in the cerebellum, thalamus, deep layers of the cortex and striatum (Lai et al., 2003). To identify brain regions in which FOXP2 and any of the four NFI factors may co-regulate gene expression networks, we performed WGCNA (Langfelder and Horvath, 2008) with a subset of the Brainspan Developmental Transcriptome data (Zhu et al., 2018). We built a gene expression network using the transcriptomes of human fetal brains between 8 and 24 post-conception weeks collected across 26 brain regions and identified three modules containing NFI factors (Table S4A). Consistent with high expression levels in the developing mouse cortex (Chaudhry et al., 1997), the eigengenes of the modules containing NFIA/NFIB and NFIX are positively correlated only with cortical subregions (Figure 4A, Table S4B, C). Conversely, the eigengene of the NFIC module is negatively correlated with most cortical subregions and positively correlated with the amygdala, cerebellar cortex, and hippocampus (Figure 4A, Table S4B, C). While FOXP2 is expressed in both the cortex and the cerebellum, genes activated by FOXP2 in hDNs were only significantly enriched among genes in the modules containing NFIA/NFIB or NFIX (Figure 4A), suggesting that FOXP2 may interact with at least one of these factors to activate gene expression during human cortical development.
Figure 4:
FOXP2, NFIA, and NFIB are enriched in the human fetal subplate
(A) Heat maps showing the correlations of module eigengenes (ME) of the NFI factor-containing WGCNA modules built from Brainspan RNA-seq data (Zhu et al., 2018) to each brain region along with significant enrichments of each module with DE genes. For acronym meanings see Table S4B. Cortical regions are written in green. Asterisks represent a correlation with a p-value < 0.01. (B) Heat map showing the mean expression of FOXP2 and NFIA-C and X (Miller et al., 2014) scaled across developmental layer. Marginal zone (MZ), outer and inner cortical plate (CPo, CPi), subplate (SP), intermediate zone (IZ), outer and inner subventricular zone (SZo, SZi), ventricular zone (VZ). (C) T-distributed stochastic neighbor embedding (tSNE) clustering of the single-cell transcriptomes from Nowakowski et al. 2017 (CTX-RG, cortical derived radial glia; MGE-RG/PC, medial ganglionic eminence derived radial glia and progenitor cells; IN, interneuron; IPC/newborn-EN, intermediate progenitor cell/newborn excitatory neuron; ESP, excitatory subplate neuron; EPN excitatory projection neuron; OPC, oligodendrocyte progenitor cell). (D) Expression of FOXP2 and NFIA and NFIB in single-cells. Outlined clusters are significantly enriched for NFIA and/or NFIB expression (Wilcoxon rank sum test with a Bonferroni corrected p-value). (E) Pie charts showing the proportion of all FOXP2-expressing cells (left) or FOXP2-expressing ESP cluster cells (right) that also express NFIA, NFIB, or NFIA and NFIB.
In the developing human cortex, FOXP2 has enriched expression in the subplate (Miller et al., 2014), a transient region of the developing mammalian neocortex that contains some of earliest-born cortical neurons (Kostovic and Rakic, 1990, Hoerder-Suabedissen et al., 2009). Importantly, NFIA and NFIB, but not NFIC or NFIX, are enriched together with FOXP2 in this layer (Figure 4B), and may aid in the maturation of subplate neurons. To determine if FOXP2, NFIA, and NFIB are expressed in the same cells, we examined single-cell RNA-seq data from developing human fetal cortex (Nowakowski et al., 2017). We re-clustered the cells in this data set for visualization purposes, and labeled the clusters based on the cell-types identified by Nowakowski et al (Table S4D). As previously published, these cells can be clustered into distinct classes including radial glia collected from the cortex (CTX-RG), MGE derived radial glia and progenitor cells (MGE-RG/PC), interneurons (IN), and newborn excitatory neurons and intermediate progenitors (IPC/newborn-EN) (Figure 4C, Table S4D) (Nowakowski et al., 2017). Moreover, mature excitatory neurons can be separated into clusters of earlier-maturing neurons with a subplate gene signature (ESP) and later-maturing cortical projection neurons (EPN) (Figure 4C, Table S4D) (Nowakowski et al., 2017). In line with the enrichment of FOXP2 in the subplate layer in bulk RNA-seq data (Figure 4B) (Miller et al., 2014), FOXP2 is expressed in significantly more cells in the ESP cluster than in the EPN cluster (p = 3.29×10−09, Fisher’s exact test, Figure 4D). Moreover, while NFIA- and NFIB-expressing cells are found in all cell-type clusters, NFIA expression is significantly enriched in CTX-RG (FDR = 6.58×10−60), and expression of both NFIA and NFIB is enriched in IPC/newborn-ENs (FDR = 2.16×10−40 and 4.73×10−95) and ESPs (FDR = 9.31×10−88 and 4.33×10−179) suggesting a role for these transcription factors in the development of excitatory cortical neurons (Figure 4D). Additionally, 79% of all FOXP2-positive cells in this dataset also express NFIA and NFIB, and 100% of the FOXP2-positive ESP cluster cells are positive for both NFIA and NFIB (Figure 4E), supporting the potential role of FOXP2 and NFIA and/or NFIB working together to regulate subplate neuron maturation in vivo.
FOXP2 physically interacts with NFIA and NFIB
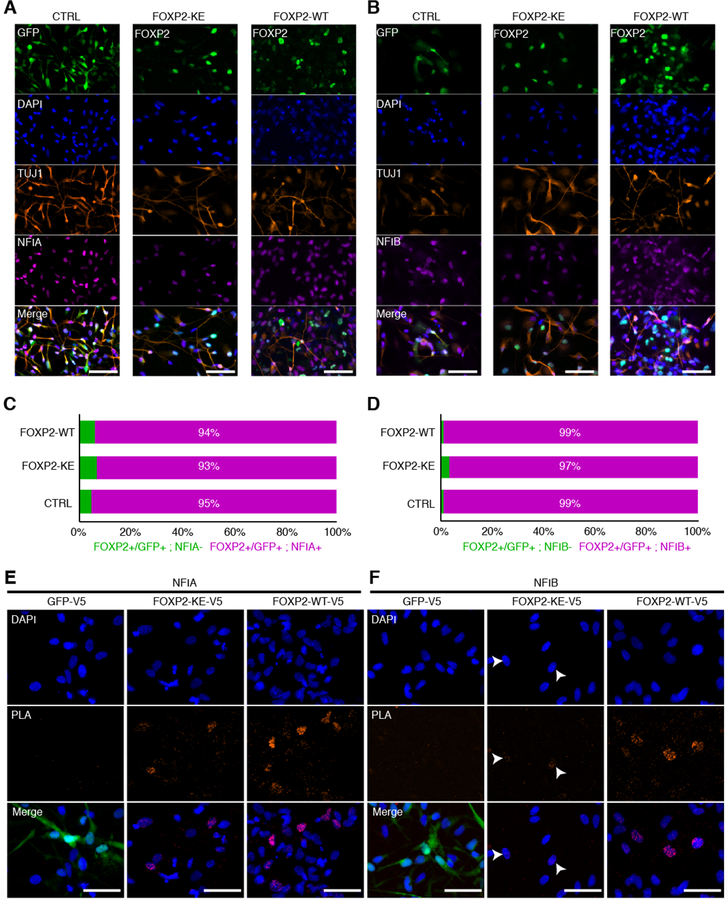
After identifying NFIA and/or NFIB as candidate FOXP2 cofactors based on motif enrichment in our ATAC-seq data and in vivo co-expression, we sought to determine if these transcription factors physically interact in hDNs. First, we confirmed that FOXP2 and NFIA or NFIB were co-expressed in hDNs via immunocytochemistry. Mirroring the co-expression we observed in human fetal cortex, more than 90% of FOXP2-WT, -KE, or GFP positive cells also expressed NFIA, and 97–99% expressed NFIB (Figure 5A–D). Next, we performed a proximity ligation assay (PLA) that generates a fluorescent signal when two proteins of interest are within 40 nm of one another, indicating physical interaction. We expressed V5-tagged FOXP2-WT, -KE, or GFP in hDNs and used antibodies against V5 and NFIA or NFIB to detect interactions between the tagged proteins and NFI transcription factors. As expected, only background PLA signal was detected between GFP and either NFIA or NFIB (Figure 5E, F). On the other hand, strong PLA signal was detected in FOXP2-WT expressing nuclei suggesting that FOXP2-WT-V5 interacts with both NFIA and NFIB (Figure 5E, F). PLA signal was enriched in FOXP2-KE nuclei when probed with NFIA antibody, but little, if any signal was detected with the NFIB antibody, suggesting that FOXP2-KE at most weakly interacts with NFIB (Figure 5F, white arrowheads). Taken together, NFIA and FOXP2 likely act as cofactors to open chromatin in a DNA-binding-independent manner.
Figure 5:
FOXP2 physically interacts with NFIA and NFIB
(A) Immunocytochemistry showing expression of TUJ1 and NFIA along with GFP, FOXP2-KE or FOXP2-WT in hDNs. Scale bars represent 100 μm. (B) Immunocytochemistry showing expression of TUJ1 and NFIB along with GFP, FOXP2-KE or FOXP2-WT in hDNs. Scale bars represent 100 μm. (C) Percentage of FOXP2 or GFP positive cells that also express NFIA calculated from one image per condition. (D) Percentage of FOXP2 or GFP positive cells that also express NFIB calculated from one image per condition. (E) Immunocytochemistry showing DAPI stained nuclei (blue) along with puncta representing V5 and NFIA antibodies within 40 nm of one another (red). GFP fluorescence is shown in the merged images of the CTRL condition. Scale bars represents 100 μm. (F) Immunocytochemistry showing DAPI stained nuclei (blue) along with puncta representing V5 and NFIB antibodies within 40 nm of one another (red). Arrowheads point to nuclei that may contain a small number of FOXP2-KE-V5/NFIB interactions. GFP fluorescence is shown in the merged images of the CTRL condition. Scale bars represent 100 μm.
Genes in FOXP2 driven networks are correlated with neuronal pseudodifferentiation
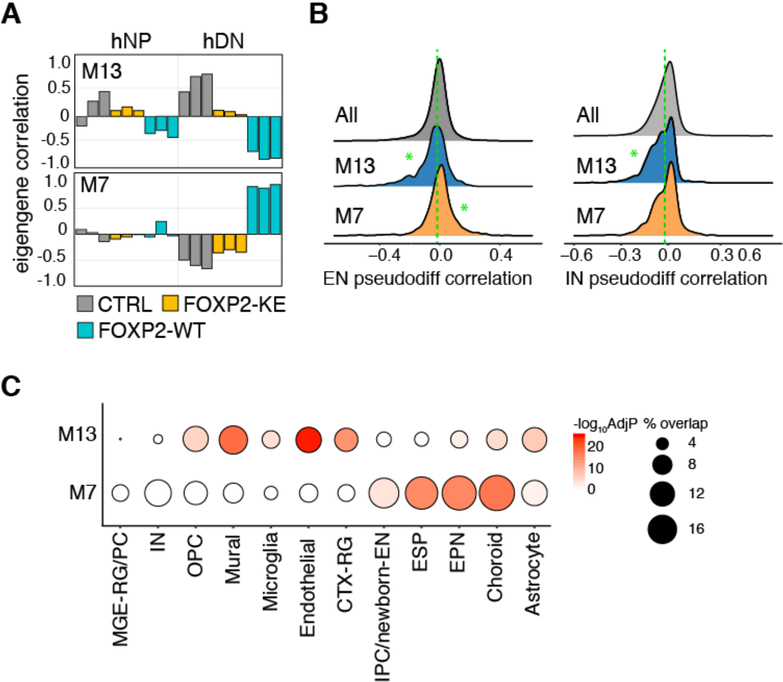
To determine the large-scale impact of FOXP2-regulated gene expression programs on neuronal maturation and brain development, we performed WGCNA using hNP and hDN transcriptomes and again integrated our data with single-cell RNA-seq data from developing human fetal cortex (Nowakowski et al., 2017) (Table S5A). We identified modules of interest in which the module eigengene is correlated with samples expressing FOXP2-WT (Table S5B). For example, the eigengene of module M13 is negatively correlated with hNPs and hDNs expressing FOXP2-WT and positively correlated with CTRL hNPs and hDNs, while the module eigengene of M7 is positively correlated with FOXP2-WT and negatively correlated with CTRL and FOXP2-KE in hDNs only (Figure 6A).
Figure 6:
Genes in FOXP2 driven networks are correlated with neuronal pseudodifferentiation
(A) Bar plots showing WGCNA modules with module eigengenes correlated with FOXP2-WT expressing samples. (B) Ridge plots showing the distribution of the correlations of each module member gene with EN- or IN-lineage derived pseudodifferentiation scores calculated by Nowakowski et al. 2017. The green dotted lines represent median correlation of genes not in M7 or M13. Significant differences between the correlations of M13 or M7 member genes and all other genes were calculated using Wilcoxon rank-sum tests (*p< 0.01). (C) Bubble plot showing the percent of M13 and M7 member genes that are cell-type enriched genes (bubble size) and the significance of the overlap calculated by the hypergeometric test and Benjamini-Hochberg corrected (color).
Nowakowski and colleagues used gene co-expression networks to quantify the level of maturity of each cell in their data set based on the cell’s unique transcriptome. The “pseudodifferentiation” scores of the cells fall on a continuum with mature neurons having a higher score than IPCs that, in turn, have higher scores than radial glia (Nowakowski et al., 2017). To assess the contribution of FOXP2-coordinated networks to cell-type-specific in vivo neuronal differentiation programs, we calculated the correlations of all genes expressed in hNPs or hDNs with pseudodifferentiation on the interneuron (IN) and excitatory cortical neuron (EN) lineages from human fetal brain data (Nowakowski et al., 2017). We then compared the correlations of M13 and M7 module genes with pseudodifferentiation to the pseudodifferentiation correlations of all other expressed genes (Figure 6B, Table S5A). We found that M13 genes are significantly more negatively correlated with pseudodifferentiation on both neuronal lineages than all expressed genes, while M7 genes are significantly more positively correlated with pseudodifferentiation than other expressed genes on the EN but not the IN lineage (Figure 6B). Moreover, M13 genes overlap significantly with genes enriched in several non-neuronal clusters as well as cortical radial glia, and while EPN, choroid, and astrocyte marker genes overlap with both clusters, only IPC/newborn-EN and mature ESP cluster markers are significantly enriched for M7 genes alone. Additionally, neither module is enriched for MGE derived progenitor or IN cluster makers (Figure 6C, Table S5C). Taken together, these data provide further evidence that FOXP2 positively regulates an excitatory neuronal maturation gene network while simultaneously negatively regulating transcriptional profiles that contribute to proliferating cell identity.
Chromatin accessibility modifications made by FOXP2 drive networks that promote neuron maturation
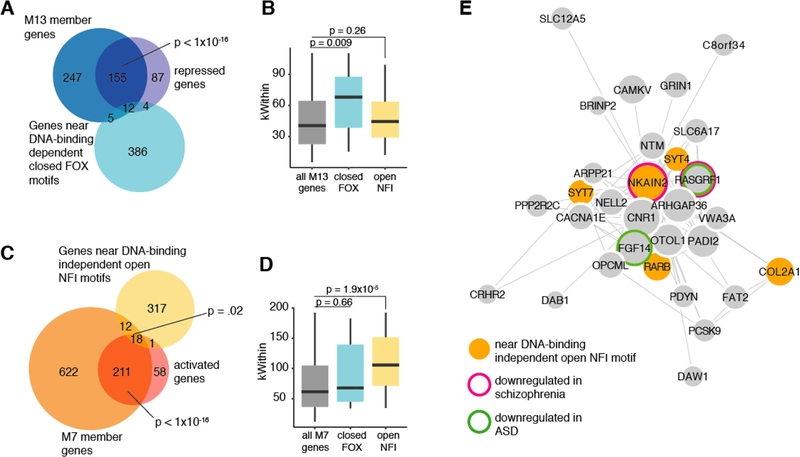
Next, we sought to determine whether changes in chromatin accessibility caused by FOXP2 expression contribute to these co-expression networks. M13 is significantly enriched with genes repressed by FOXP2 (Figure 7A). Moreover, the M13 genes near FOXP2 DNA-binding-dependent closed regions containing a FOX motif are more highly connected within the module than all M13 genes and M13 genes near DNA-binding-independent open regions with NFI motifs (Figure 7B). This suggests that FOXP2 DNA-binding-dependent chromatin condensation drives the repression of a network of genes inhibiting neuronal maturation. Moreover, M7 is significantly enriched for FOXP2-activated genes and genes near DNA-binding-independent open regions of chromatin containing an NFI motif (Figure 7C). M7 genes near DNA-binding-independent open regions of chromatin containing an NFI motif are significantly more highly connected within the module than all M7 genes and M7 genes near DNA-binding dependent closed regions with FOX motifs (Figure 7D), suggesting that FOXP2-dependent chromatin opening drives FOXP2-expressing excitatory neurons to a more mature state. Additionally, the most highly connected gene in M7, NKAIN2, is near a DNA-binding-independent open region of chromatin containing an NFI motif, is activated by FOXP2, and is expressed at lower levels in the brain tissue of schizophrenia patients than in non-affected individuals (Figure 7E) (Gandal et al., 2018), providing further evidence that the ability of FOXP2/NFI complexes to open chromatin is important for normal brain development.
Figure 7:
Chromatin accessibility modifications made by FOXP2 drive networks that promote neuron maturation
(A) Venn diagram showing the overlaps between M13 member genes, FOXP2 repressed genes, and genes near DNA-binding dependent closed FOX motifs. P-values calculated using the hypergeometric test. (B) Box plot comparing the median connectivity (kWithin) of all M13 module genes, M13 modules genes near DNA-binding dependent closed FOX motifs, and M13 modules genes near DNA-binding independent open NFI motifs. P-values calculated using the Wilcoxon rank-sum test. (C) Venn diagram showing the overlaps between M7 member genes, FOXP2 activated genes, and genes near DNA-binding independent open NFI motifs. P-values calculated using the hypergeometric test. (D) Box plot comparing the median connectivity (kWithin) of all M7 module genes, M7 modules genes near DNA-binding dependent closed FOX motifs, and M7 modules genes near DNA-binding independent open NFI motifs. P-values calculated using the Wilcoxon Rank Sum test. (E) Visualization of the top 100 weighted connections in M7. The length of the edges corresponds to the weight of the connection and the size of the nodes corresponds to the kwithin value.
Discussion
Uncovering transcriptional programs driving brain development is key to ultimately understanding and treating neuropsychiatric disorders. Mutations in the transcription factor FOXP2 have been linked to childhood apraxia of speech (Lai et al., 2001, Konopka and Roberts, 2016) and FOXP2 target genes have been implicated in ASD and schizophrenia (Lepp et al., 2013). We found that genes regulated by FOXP2 are enriched for genes with aberrant expression in the brains of patients with these disorders; however, the mechanisms by which FOXP2 regulates gene expression in human neurons during brain development remain unclear. Here, we show that exogenous expression of FOXP2 in human neurons leads to repression of genes expressed in non-neuronal and proliferating cells in the developing human cortex. Moreover, our analysis of differentially accessible chromatin in human neurons shows that FOXP2 binds to DNA at its canonical motif and closes chromatin near these repressed germinal genes.
FOXP2 can bind DNA as a homodimer or heterodimerize with other FOXP proteins, and has been shown to interact with NFAT, TBR1, and CTBP among other DNA-binding proteins (Wu et al., 2006, Deriziotis et al., 2014, Li et al., 2004). We identified NFIA and NFIB, which are essential for proper forebrain development (Betancourt et al., 2014, das Neves et al., 1999), as co-regulators of FOXP2-activated genes. Importantly, only NFIA robustly interacts with FOXP2-KE, implying that NFIA is likely to be responsible for opening chromatin together with FOXP2 in a manner that does not require FOXP2 to interact with DNA directly. Further studies will be necessary to uncover the mechanisms by which FOXP2-KE facilitates chromatin opening via NFIA without activating gene expression. One possible explanation is that the FOXP2/NFIA complex facilitates enhancer/promoter looping with NFIA binding the enhancer region directly and FOXP2 binding the promoter. NFIA alone may not be sufficient to bind these enhancer regions (as in the CTRL condition), but interaction with FOXP2-WT or -KE may allow NFIA within the FOXP2/NFIA complex to directly bind and open chromatin. However, gene activation may only be possible when FOXP2-WT is able to bind the corresponding promoter region. Thus, chromatin can be opened in a FOXP2 dependent manner at enhancer regions containing an NFI motif without gene activation.
Analysis of accessible chromatin regions across layers of the adult mouse visual cortex provides further evidence of the relationship between Foxp2, Nfia, and chromatin accessibility (Gray et al., 2017). Forkhead motifs were present in areas of chromatin less accessible in layer VI, where Foxp2 is expressed, compared with other cortical layers devoid of Foxp2; in contrast, enrichment of Nfi motifs was seen in regions of chromatin more accessible in layer VI, where Nfia is highly expressed (Gray et al., 2017). Moreover, Gray et al. suggest Nfia as a putative activator of Foxp2 based on the presence of an Nfi motif in a differentially accessible region near Foxp2. It is remarkable that our motif enrichment analysis in hDNs exogenously expressing FOXP2 also shows that Forkhead motifs are present in differentially closed chromatin but NFI motifs are present in differentially open chromatin. While we cannot confirm the regulation of FOXP2 by NFIA in our overexpression system, our study suggests that FOXP2 and NFIA physically interact at NFI motifs to open chromatin and promote neuron maturation.
While NFIA, -B, and -X are highly expressed in the neocortex during mouse development, only expression of NFIA and NFIB is correlated with the human subplate along with FOXP2. Additionally, all FOXP2-positive excitatory subplate neurons identified in developing human cortex using single-cell RNA-seq express NFIA and NFIB. Moreover, subplate neurons are among the first mature neurons in the developing cortex (Kostovic and Rakic, 1990, Hoerder-Suabedissen et al., 2009, Rakic, 1974). Thus, FOXP2 and NFIA and -B may aid in this early cortical maturation phase.
The subplate is an understudied part of the neocortex with potentially important evolutionary adaptations as the subplate is much larger, relative to overall neocortical size, in human and non-human primates than in rodents (Hoerder-Suabedissen and Molnar, 2015). Additionally, while FOXP2 is highly conserved among mammals, the hominin version of FOXP2 contains two amino acid changes that impact transcriptional activity and behaviors (Enard et al., 2002, Zhang et al., 2002, Enard et al., 2009, Konopka et al., 2009). While we show that FOXP2 activates gene networks present in both excitatory subplate neurons and later born excitatory neurons, it is tempting to speculate that FOXP2 modulates gene expression in the evolutionarily relevant subplate in a human-specific manner to facilitate speech and language abilities. For example, this brain region plays a crucial role in the patterning of cortical circuits by directing thalamocortical afferents to their proper targets (Molnar and Blakemore, 1995), and the glutamatergic input from the subplate to early layer IV neurons is critical for the maturation of thalamocortical and cortico-cortical synapses (Kanold et al., 2003, Kanold and Shatz, 2006). Thus, the subplate specifically supports cortico-basal ganglia-thalamocortical circuitry, which is abnormal in patients with childhood apraxia of speech caused by FOXP2 mutations (Vargha-Khadem et al., 1998, Watkins et al., 2002b, Liegeois et al., 2003, Belton et al., 2003), is critical for language (Konopka and Roberts, 2016), and is aberrant in patients with ASD and schizophrenia (Hutsler and Casanova, 2016, Peters et al., 2016). As no overt cortical abnormalities have been observed in mice with FOXP2 loss-of-function (Shu et al., 2005, French et al., 2007, Groszer et al., 2008, Fujita et al., 2008, French et al., 2018, Medvedeva et al., 2018), the role of FOXP2 and its NFI cofactors in the human subplate may be specifically important for the development of brain circuitry underlying language and other cognitive abilities.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Genevieve Konopka (genevieve.konopka@utsouthwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK 293T cells and lentivirus production
HEK 293T cells were plated in DMEM (#SH3024301, Thermo Fisher Scientific, Waltham, Massachusetts) with 1% Antibiotic-Antimycotic (#15240062, Thermo Fisher Scientific) and 10% FBS (#10437028, Thermo Fisher Scientific), kept at 37°C in 5% CO2, and passaged at least three times after thawing with the final passage 12–24 hours prior to transfection. For transfection of one 10 cm plate, viral packaging vectors VSV-G and psPAX2 (2μg each) and the lentiviral construct of interest (4 μg) were added to DMEM up to 250 μl. 32 μl of Fugene (#E2691, Fisher, Madison, WI) were then added to an additional 228 μl of DMEM, and the tube was flicked gently to mix. The plasmid solution was then added to the Fugene solution, the tube was flicked gently to mix and incubated for 30 minutes at room temperature. The 500 μl transfection mix was subsequently added dropwise, in a spiral motion to one 10 cm plate of HEK 293T cells at approximately 70–90% confluence and mixed. 12–24 hours later, the 293T cell media was replaced with proliferation media (Neurobasal A media (#10888–022, Thermo Fisher Scientific), 2% Antibiotic-Antimycotic, 10% BIT (#9500, STEMCELL Technologies, Vancouver, Canada), 1% GlutaMAX Supplement (#35050061, Thermo Fisher Scientific)). 12–24 hours after media replacement the viral supernatant was collected and centrifuged for 5 minutes at 1000 rpm. The supernatant was then filtered throμgh a 45 μm filter, and stored in small aliquots at −80 °C.
Human neural progenitor (hNP) culture
Female human neural progenitor cultures were obtained from Lonza (Line #OF4106, Product Code PT-2599, Lonza, Basel, Switzerland). Cells were stored in liquid nitrogen in Neurobasal A media with 10% DMSO until thawed and plated. Tissue culture plates were first coated for at least two hours at 37°C with 50 μg/mL Poly-L-ornithine hydrobromide (#P3655, Millipore Sigma, Burlington, Massachusetts) in PBS. The polyornithine solution was removed from the plate and replaced with 5 μg/mL fibronectin (#F1141, Millipore Sigma) in PBS for at least two hours at 37°C. Immediately before plating cells, the fibronectin solution was completely aspirated off of the plates and the remaining fibronectin film was allowed to dry for up to 5 minutes. Frozen cells were quickly thawed at 37°C and resuspended in 5 mL of proliferation media without growth factors (500 mL Neurobasal A media with 60 mL BIT serum substitute, 12 mL Antibiotic-Antimycotic and 7 mL Glutamax) to dilute the DMSO. The cells were pelleted by centrifugation for 5 minutes at 1000 rpm. The pellet was resuspended in 10 mL plating media comprised of 50% proliferation media and 50% conditioned media (proliferation media removed during half feedings and filtered through a 0.22 μm filter) with 5 ng/μL EGF (#2633, STEMCELL Technologies) and 5 ng/μL FGF (#2634, STEMCELL Technologies) added. The resuspended cells were then pipetted onto a coated 10 cm plate. Three times per week 50% of the media on the plate was removed, stored as conditioned media, and replaced with fresh proliferation media containing 10 ng/mL EGF and 10 ng/mL FGF. When the cells reached 80–100% confluency they were passaged to new, coated plates. Briefly, the media was removed from the plates and the cells were washed with 5 mL PBS. Next, 0.25% trypsin-EDTA (#25200114, Thermo Fisher Scientific) was added to cover the cells and immediately removed. After incubating the cells in the residual trypsin for approximately 1 minute, the trypsin was neutralized and cells were triturated and resuspended in 10 mL DMEM with 10% BSA or 10 mL conditioned media and pelleted by centrifugation for 5 minutes at 1000 rpm. The pellet was resuspended in plating media. One confluent plate was typically split into two to three new plates and kept at 37°C in 5% CO2. 24 hours later, lentivirus was added in a dropwise manner. Cells were harvested four days after transduction.
Human differentiating neuron (hDN) culture
Differentiation from hNPs into human differentiating neurons (hDNs) began four days after lentiviral transduction. Cells were kept at 37°C in 5% CO2. Tissue culture plates were first coated for at least two hours at 37°C with 50 μg/mL poly-L-ornithine hydrobromide in PBS. The polyornithine solution was removed from the plate and replaced with 5 μg/mL laminin (#354232, Corning, Corning, New York) in PBS for at least two hours at 37°C. Immediately before plating cells, the laminin solution was completely aspirated off of the plates but not allowed to dry before plating. hNPs were densely plated (3–5 million cells per 10 cm plate, 1million cells per well in a 6-well plate) in plating media as previously described (see Proliferating human neural progenitor (hNP) culture). After 48 hours, half of the media was removed from the plates and replaced with differentiation media (500 mL Neurobasal A media, 10 mL B27 without vitamin A (#12587010, Thermo Fisher Scientific), 12 mL Antibiotic-Antimycotic, 7 mL Glutamax, 10 ng/mL BDNF (#450–02, PeproTech, Rocky Hill, New Jersey), 10 ng/mL NT-3 (#450–03, PeproTech), 10 ng/mL retinoic acid (#R2625, Millipore Sigma), 10 μM forskolin (#344270, Millipore Sigma), and 10 mM KCl). Two days later, the media was completely removed, the cells were washed with PBS, and fresh differentiation media was added. For the first two weeks after initiation of differentiation, three times a week, 50% of the media was changed to fresh differentiation media with concentrations of growth factors adjusted to the total volume. After this initial two-week period, the cells were again washed with PBS and fresh differentiation media was added. For the following second two-week period, the cells were again fed three times per week with an adjusted 50% media change.
METHOD DETAILS
Plasmids
pLUGIP, a lentiviral expression vector with a Ubi-c promoter driven GFP-V5-IRES-Puromycin was used in the RNA-seq and ATAC-seq experiments as the control vector. The GFP in the vector was originally replaced with FOXP2-WT-3xFLAG (Konopka et al., 2009). Subsequently, site directed mutagenesis was used to remove the FLAG tag. For pLUGIP-FOXP2-KE a point mutation was made via site directed mutagenesis in untagged pLUGIP-FOXP2-WT construct to change R at position 553 to H in the DNA-binding domain of FOXP2. Subsequently, site directed mutagenesis was used to add a V5 tags to the GFP, FOXP2-WT and -KE constructs. Oligonucleotides were designed using Agilent’s Quikchange Primer Design tool. See key resources table for oligonucleotide sequences.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| goat anti-FOXP2 (N-terminal) | Santa Cruz | Cat#21069 |
| mouse-monoclonal anti-TUJ1 | BioLegend | Cat#801201 |
| rabbit-polyclonal anti-NFIA | Millipore Sigma | Cat#HPA008884 |
| rabbit-polyclonal anti-NFIB | Millipore Sigma | Cat#HPA003956 |
| mouse-monoclonal anti-V5-tag | Thermo Fisher Scientific | Cat#R960–25 |
| Alexa Fluor® 488 Donkey Anti-Goat IgG | Thermo Fisher | Cat # A-11055 |
| Alexa Fluor® 555 Donkey Anti-Mouse IgG | Thermo Fisher | Cat#A-31570 |
| Alexa Fluor® 647 Donkey Anti-Rabbit IgG | Thermo Fisher | Cat#711-605-152 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 500 mM EDTA, pH 8.0 | Sigma-Aldrich Corporation | Cat#E5134-500G |
| Actinomycin D | Sigma Aldrich | Cat#A1410 |
| Agencourt AMpure XP beads | Beckman Coulter | Cat# A63881 |
| Antibiotic-Antimycotic (100X), liquid | Thermo Fisher Scientific | Cat#15240062 |
| B27 without vitamin A | Life Technologies | Cat#12587010 |
| BDNF | Peprotech | Cat#450–02 |
| BIT | STEMCELL Technologies | Cat#9500 |
| Dimethyl sulfoxide (DMSO); filtered (for TC) | Sigma | Cat#D2438–50ML |
| DMEM | Fisher | Cat#SH3024301 |
| donkey serum | Millipore | Cat#S30–100ML |
| FBS | Thermo Fisher Scientific | Cat#10437028 |
| FGF | STEMCELL Technologies | Cat#2634 |
| fibronectin | Sigma | Cat# F1141–5MG |
| Forskolin, Coleus forskohlii | CAS 66575-29-9 | Calbiochem | 50MG | Millipore Sigma | Cat#344270–50MG |
| Fugene | Fisher | Cat#PR-E2691 |
| Glutamax | Thermo Fisher Scientific | Cat#35050–061 |
| IGEPAL CA-630 | Sigma | Cat#13021 |
| Laminin, Mouse | Corning | Cat#354232 |
| Methanol-free Formaldehyde Ampules | Fisher Scientific | Cat#PI28906 |
| Neurobasal A | Thermo Fisher Scientific | Cat#10888–022 |
| Neurotrophin-3 (NT-3) | Peprotech | Cat#450–03 |
| NEXTflex Poly(A) Beads | Bioo Scientific | Cat#512981 |
| Poly-L-ornithine hydrobromide | Sigma | Cat#P3655 |
| ProLong® Diamond Antifade Reagent with DAPI | Life Technologies | Cat#P36971 |
| Recombinant Human EGF | STEMCELL Technologies | Cat#2633 |
| Retinoic acid, =98% (HPLC) | Sigma Aldrich | Cat#R2625–50mg |
| Triton X-100 | Fisher | Cat#BP151–100 |
| Trizma Hydrochloride Solution, pH 8.0, 1 M | Sigma-Aldrich Corporation | Cat#T2694–100ML |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific | Cat#25200114 |
| 100x SYBR Green I | Invitrogen | Cat#S-7563 |
| NEBNext High-Fidelity 2x PCR Master Mix | New England Labs | Cat#M0541 |
| Critical Commercial Assays | ||
| TruSeq Stranded mRNA Library Prep | Illumina | Cat#20020595 |
| NEXTflex™ Barcoded Adapters | biooscientific | Cat#NOVA-514106 |
| Nextera DNA Library Preparation Kit | Illumina | Cat#FC-121–1030 |
| Qiagen MinElute Kit | Qiagen | Cat#28004 |
| KAPA Library Quantification Kit | Roche | Cat#07960140001 |
| Bioanalyzer RNA 6000 Nano chip | Agilent | Cat#5067–1511 |
| Bioanalyzer High Sensitivity DNA chip | Agilent | Cat#5067–1504 |
| Deposited Data | ||
| Expression Atlas of the Developing Human Brain | Miller et al. 2014 |
http://brainspan.org/ RRID:SCR_008083 |
| Layer Correlated Genes table | Miller et al. 2014 Supplemental table 3 | |
| consensus excludable ENCODE blacklist (lifted over to hg38) | ENCODE | http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/). |
| ATAC-seq mitochondrial blacklists hg19 (lifted over to hg38) | Buenrostro et al., 2015 | https://sites.google.com/site/atacseqpublic/atac-seq-analysis-methods/mitochondrialblacklists-1 |
| Gencode v24 (GRCh38.p5) | Gencode | https://www.gencodegenes.org/releases/24.html |
| Roadmap Epigenomics 25-state model | Kundaje et al., 2015 | http://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/imputed12marks/jointModel/final/ RRID:SCR_008924 |
| Human fetal brain single-cell RNA-seq expression matrix and cell meta data | Nowakowski et al., 2017 | https://cells.ucsc.edu/ |
| Human fetal brain single-cell RNA-seq cluster interpretations | Nowakowski et al., 2017, supplemental table 4 | |
| Brainspan Developmental Transcriptome Dataset (RNA-Seq Gencode v10 summarized to genes) | Zhu et al. 2018 | http://www.brainspan.org/static/download.html |
| Genes identified as up or downregulated in the frontal and temporal cerebral cortices of patients with ASD, bipolar disorder, or schizophrenia (DER-13_Disorder_DEX_Genes) | Gandal et al. 2018 | http://resource.psychencode.org/ |
| RNA-seq data | This paper | GSE111353 |
| ATAC-seq data | This paper | GSE111353 |
| Experimental Models: Cell Lines | ||
| normal human neural progenitor line Lot# OF4106 (female) | Lonza | Product Code: PT-2599 |
| Oligonucleotides | ||
| Add V5 to pLUGIP Forward primer: ACGTAAACGGCCACAAGTTC | This paper | N/A |
| Add V5 to pLUGIP Reverse primer: GTCGCGGCCGCTTTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCACCAGAACCACCCTTGTACAGCTCGTC | This paper | N/A |
| Remove 3xFLAG from pLUGIP FOXP2-WT-3xFLAG forward primer: ATGTTAAAGGAGCAGTATGG | This paper | N/A |
| Remove 3xFLAG from pLUGIP FOXP2-WT-3xFLAG reverse primer: TACCGGGCCCTATTCCAGATCTTCAGATAAAGGC | This paper | N/A |
| Make KE mutant in pLUGIP-FOXP2-WT-notag forward primer: ACTTGGAAGAATGCAGTACATCATAATCTTAGCCTGCAC | This paper | N/A |
| Make KE mutant in pLUGIP-FOXP2-WT-notag reverse primer: AGGCTCTTCTTCAATCTCTCTGTCG | This paper | N/A |
| Add V5 to pLUGIP FOXP2-WT or -KE-notag forward primer: ATGTTAAAGGAGCAGTATGG | This paper | N/A |
| Add V5 to pLUGIP FOXP2-WT or -KE-notag reverse primer: TACCGGGCCCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCACCAGAACCACCTTCCAGATCTTCAGATAAAGGC | This paper | N/A |
| Recombinant DNA | ||
| pLUGIP | ATTC | ATCC 10326372; http://www.signaling-gateway.org/data/plasmid/Plasmid.cgi?rq=s_atcc_id&atcc_id=10326372&barcode=&gene_name=&afcs_id=&orf_id=&keyword= |
| pLUGIP FOXP2-WT-3X-FLAG | Konopka et al., 2009 | N/A |
| pLUGIP FOXP2-WT-V5 | This paper | N/A |
| pLUGIP-GFP-V5 | This paper | N/A |
| pLUGIP FOXP2-KE-V5 | This paper | N/A |
| pLP/VSVG | Invitrogen | https://www.addgene.org/vector-database/6099/ |
| psPAX2 | Addgene | Cat#12260 |
| Software and Algorithms | ||
| Agilent’s Quikchange Primer Design tool | Agilent | http://www.genomics.agilent.com/primerDesignProgram.jsp;jsessionid=AJa3gAhGyUWhxzHPdkY9bCFhVpTwIy9tOo8xm64_2c2lOUvaOewx!-742858405 |
| Adobe Illustrator 18.0.0 | ||
| Picard 1.77 | The Broad Institute | http://broadinstitute.github.io/picard/ |
| bowtie 0.12.7 | Langmead et al., 2009 | |
| STAR 2.5.2b | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| homer v4.6 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| WGCNA 1.51 | Langfelder and Horvath, 2008 | http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/Rpackages/WGCNA/ |
| R 3.2.5 | R Core Team, 2016 | URL https://www.R-project.org/ |
| Toppgene | Chen et al., 2009 | https://toppgene.cchmc.org |
| Cytoscape 3.3.0 | Shannon et al., 2003 | http://www.cytoscape.org |
| Htseq version 0.9.1 | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.9.1/ |
| Trimmomatic 0.36 | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| DESeq2 1.10.1 | Love et al., 2014 | http://bioconductor.org/packages/DESeq2/ |
| MACS2 2.1.1.20160226 | Zhang et al., 2008 | http://liulab.dfci.harvard.edu/MACS/ |
| DiffBind 1.16.3 | Ross-Innes et al., 2012 | http://bioconductor.org/packages/DiffBind/ |
| pheatmap 1.0.8 | https://CRAN.R-project.org/package=pheatmap | |
| EdgeR 3.12.1 | Robinson et al., 2010 | http://bioconductor.org/packages/edgeR/ |
| biomaRt 2.26.1 | Durinck et al., 2009 | http://bioconductor.org/packages/biomaRt/ |
| Bedtools 2.25.0 | Quinlan et al., 2010 | http://bedtools.readthedocs.io/en/latest/ |
| ggridges 0.5.0 | https://github.com/clauswilke/ggridges | |
| Seurat 2.3.0 | Butler et al., 2018 | https://github.com/satijalab/seurat |
| REVIGO | Supek et al. 2011 | http://revigo.irb.hr/ |
| CSEA tool (SEA across brain regions and development) | Xu et al. 2014 | http://genetics.wustl.edu/jdlab/csea-tool-2/ |
RNA-collection
RNA from three technical replicates of approximately 1×106 Lonza line #OF4106 hNPs or hDNs expressing plugip-FOXP2-WT-V5, plugip-FOXP2-KE-V5, or plugip-eGFP-V5 was harvested using the Qiagen miRNeasy Mini Kit (217004, Qiagen, Hilden, Germany).
RNA-sequencing library prep
Prior to submission to the McDermott Sequencing Core at the University of Texas Southwestern Medical Center, RNA samples were randomized by ordering the samples based on a random list of numbers generated by the RAND() function in Excel. The samples were then relabeled in a way that de-identified them, and libraries were prepped based on the random order and not by sample type in order to reduce batch effect.
Libraries were prepared using the TruSeq Stranded mRNA Library Prep (#20020595, Illumina, San Diego, California) as per the manufacturer’s instructions by UTSW McDermott Next Generation Sequencing Core. The quality and concentration of the libraries was checked on an Agilent Bioanalyzer High Sensitivity DNA chip (#5067–1504, Agilent, Santa Clara, California). All samples were pooled and sequenced by the UTSW McDermott Next Generation Sequencing Core using Illumina’s NextSeq500. Reads are strand specific, single end, and 75bp long.
RNA-seq mapping, QC and expression quantification
Adapter removal and quality trimming was performed using Trimmomatic 0.36 (Bolger et al., 2014). Reads were aligned to the human hg38 (GRCh38) reference genome using STAR 2.5.2b (Dobin et al., 2013). Gencode v24 annotation was used as reference to build STAR indexes and alignment annotation. For each sample, a BAM file including mapped and unmapped reads with spanning splice junctions was produced. Secondary alignment and multi-mapped reads were further removed using in-house scripts. Only uniquely mapped reads were retained for further analyses. Quality control metrics were performed using RseqQC using the hg38 gene model provided (Wang et al., 2012). These steps include: number of reads after multiple-step filtering, ribosomal RNA reads depletion, and defining reads mapped to exons, UTRs, and intronic regions. Picard tool was implemented to refine the QC metrics (http://broadinstitute.github.io/picard/). The median number of reads per sample is ~22 million. Gene level expression was calculated using HTseq version 0.9.1 using intersection-strict mode by exon (Anders et al., 2015). Counts were calculated based on protein-coding genes annotation from the Gencode v24 annotation file.
RNA-seq differential expression analysis
Reads were normalized by RPKM, and DESeq2 1.10.1 (Love et al., 2014) was used for DE analysis. Genes were considered for DE analysis if RPKM was ≥ 0.5 in all replicates of at least one condition in the comparison. In all cases, genes were considered differentially expressed if the log2 fold change was greater than or equal to ± 0.3 and FDR was less than or equal to 0.05.
Conditions compared for DE analysis include the following:
CTRL hDNs vs CTR hNPs
FOXP2-KE hDNs vs CTR hNPs
FOXP2-WT hDNs vs CTR hNPs
FOXP2-WT hDNs vs CTR hDNs
FOXP2-KE hDNs vs CTRL hDNs
Gene ontology enrichments
ToppFun from the ToppGene suit (Chen et al., 2009) was used with default parameters to identify gene ontology enrichments among differentially expressed genes. The enriched biological process categories and the associated Benjamini-Hochberg adjusted p-values were entered into Revigo (Supek et al., 2011) to remove redundant terms (allowed similarity of 0.5, with homo sapiens GO term sizes). For repressed genes, only the top 350 enriched categories by Benjamini-Hochberg adjusted p-value were entered into Revigo. Four to five the remaining terms of interest were chosen for visualization.
Overlaps with neurodevelopmental disorder DE genes
Genes identified as up or downregulated in the frontal and temporal cerebral cortices of patients with ASD, bipoloar disorder, or schizophrenia were identified by Gandal et al. 2018 (Gandal et al., 2018) were downloaded from http://resource.psychencode.org/ (DER-13_Disorder_DEX_Genes). The hypergeometric overlap test was used to assess the significance of the overlaps between these genes and FOXP2 DE genes.
hNP and hDN WGCNA
Weighted gene coexpression network analysis was performed using the WGCNA R package (Langfelder and Horvath, 2008). Genes included in the analysis have RPKM ≥ 0.5 in all replicates of at least one condition. Before analysis, the batch effect between hNP and hDN data was removed using the removeBatchEffect function from the limma package (Ritchie et al., 2015) in order to identify modules correlated with FOXP2 expression and not only differentiation. The options used for the blockwiseModules function were: corType=“pearson”, maxBlockSize = 14000, networkType=“signed”, power=10, minModuleSize=50, TOMType = “signed”, TOMDenom = “mean”, deepSplit=2, verbose=5, mergeCutHeight=0.1, detectCutHeight = 0.999, reassignThreshold = 1e-6.
The 100 top weighted edges of M7 were selected for Cytoscape visualization (Shannon et al., 2003).
ATAC-seq
ATAC-seq was performed as per Buenrostro et al., Nature Methods, 2015 (Buenrostro et al., 2015a).
24 samples from hNPs were harvested for ATAC-seq: four technical replicates of Lonza line #OF4106 proliferating hNPs expressing FOXP2-WT, FOXP2-KE or GFP-CTRL, and four replicates each of four-week differentiating hNPs expressing FOXP2-WT, FOXP2-KE, or GFP-CTRL. These samples were collected in parallel with the samples for the RNA-seq experiment.
For each replicate from each condition, 1×106 hNPs were plated in one well of a six well plate. After 24 hours the cells were transduced with V5-tagged FOXP2-WT, V5-tagged FOXP2-KE, or V5-tagged GFP lentivirus. After four days or four weeks (for proliferating and differentiated cells, respectively) the cells were harvested by dissociating with 0.25% Trypsin-EDTA, centrifuged for 5 minutes at 1000 rpm, and resuspended in 10 mL Neurobasal-A Media. The cells were counted using a hemocytometer, and 50,000 cells per condition were isolated and spun down at 500 × g for 5 minutes at 4°C.
The cells were washed once with 50 μL cold 1x PBS and centrifuged again at 500 × g for 5 minutes at 4°C. The cell pellet was then re-suspended in cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and centrifuged immediately for 10 minutes at 500 × g at 4°C. The supernatant was discarded, and the remaining pellet was re-suspended in 50 μL transposition reaction mix from the Nextera DNA Library Preparation Kit (#FC-121–1030, Illumina, 25 μL 2x TD Buffer, 2.5 μL Tn5 Transposase, 22.5 μL nuclease free water) by gently pipetting. Following re-suspension the reaction was incubated at 37°C for 30 minutes in a thermal cycler. The transposed DNA was then cleaned up using a Qiagen MinElute Kit (#28004, Qiagen), and eluted in 10 μL Elution Buffer (10mM Tris buffer, pH 8).
After transposition and purification, 10 μL of DNA was added to 35 μL PCR Master Mix (9.7 μL nuclease free water, 0.3 μL 100x SYBR Green I (#S7563, Thermo Fisher Scientific), 25 μL NEBNext High-Fidelity 2x PCR Master Mix (#M054, New England Biolabs, Ipswich, Massachusetts). 2.5 μL of 25μM Customized Nextera PCR Primer 1, and 2.5 μL of 25μM Customized Nextera PCR Primer 2 (Buenrostro et al., 2015a) with a specific barcode for each sample were added to the DNA and PCR mix and amplified. Following PCR, the samples were again purified using a Qiagen MinElute Kit and eluted in 20 μL Elution Buffer.
The quality of the libraries was assessed using an Agilent 2100 Bioanalyzer with a High Sensitivity DNA Kit. A banding pattern consistent with nucleosome periodicity was observed; however, a large fraction of the fragments were larger than 1000 base pairs (bp), which can make Bioanalyzer quantification inaccurate. Therefore, the libraries were size selected using a 0.6:1 ratio of Agencourt AMpure XP beads (#A63881, Beckman Coulter, Brea, Califiornia) to DNA. This ratio allowed fragments larger than 1000 bp to bind to the beads while leaving fragments 1000 bp and smaller in the supernatant. The size-selected libraries were run on the Bioanalyzer in order to determine average fragment size (297–433 bp). These data along with the library concentration determined using the KAPA Library Quantification Kit (#07960140001, Roche, Basel, Switzerland) were used to calculate the amount of each library needed to make 300 μL of a 2 nM pool.
Sequencing was performed by the McDermott Sequencing Core at the University of Texas Southwestern Medical Center on an Illumina NextSeq500 sequencer. The library pool was sequenced three times generating paired-end, 75 bp reads.
ATAC-seq mapping and QC
The raw reads were mapped to hg38 using bowtie1 0.12.7 (Langmead et al., 2009) with the parameters -p 12 -y -v 2 --best --strata -m 3 -k 1 -X 2000 as per the Hardison ATAC-seq pipeline from ENCODE (https://www.encodeproject.org/pipelines/ENCPL035XIO/). Samtools (Li et al., 2009) was used to filter for MAPQ score greater than 10. Reads mapping to the mitochondria and unmapped contigs were removed. Reads mapping to a subset of high-signal regions generally unique to ATAC-seq, which were defined by the Greenleaf lab and are thought to represent mitochondrial homologues were also removed (Buenrostro et al., 2015a, Buenrostro et al., 2015b). Duplicates were removed from each replicate using Picard MarkDuplicates (http://broadinstitute.github.io/picard/) with REMOVE_DUPLICATES=TRUE. The final number of filtered reads used for analysis is between 3.62×107 and 6.69×107.
DAR identification
After the final bam files were generated, MACS2 callpeak was used to identify peaks with the following parameters: -f BAMPE --nomodel --nolambda --keep-dup all (Zhang et al., 2008). Peaks were filtered using the consensus excludable ENCODE blacklist (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeMapability/). Any peaks with a MACS2 fold change less than 6 were filtered out, resulting in a list of high confidence peaks for further analysis. Homer mergePeaks (Heinz et al., 2010) was then used to identify MACS2 peaks overlapping in at least three of four replicates of each condition.
The area 250 bp up- and downstream of the summits of these overlapping peaks was analyzed for differential accessibility using the bioconductor package DiffBind 1.16.3 using the option summits=250 in DBA count (Ross-Innes et al., 2012). FOXP2-KE and FOXP2-WT peaks were compared separately in a pairwise manner to CTRL-GFP peaks in hNPs and hDNs (hNP FOXP2-WT vs. hNP CTRL-GFP; hNP FOXP2-KE vs. hNP CTRL-GFP; hDN FOXP2-WT vs. hDN CTRL-GFP; hDN FOXP2-KE vs. hDN CTRL-GFP). Reads for each sample were normalized to their full library size, and EdgeR 3.12.1 (Robinson et al., 2010) was used to determine differential accessibility. Regions with an FDR < 0.05 were considered significantly differentially accessible regions (DARs).
DARs with fewer reads in the FOXP2-WT or -KE condition compared to CTRL are considered closed DARs, while those with more reads in the FOXP2-WT or -KE condition compared to CTRL are considered open DARs. DNA-binding independent DARs are regions that are significantly differentially accessible in the same direction (open or closed) with expression of both FOXP2-WT and FOXP2-KE compared to control in hNPs only or hDNs only. DNA-binding dependent DARs are only present with expression of FOXP2-WT and not FOXP2-KE in hNPs only or hDNs only.
DARs vs imputed chromatin states
A bed file (E081_25_imputed12marks_mnemonics.bed.gz) listing regions with imputed chromatin states defined by 12 regulatory marks from human fetal brain tissue was downloaded from the Roadmap Epigenomics Project web portal (http://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/imputed12marks/jointModel/final/) (Ernst and Kellis, 2015). These regions were lifted over from hg19 to hg38 with UCSC’s LiftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Region abbreviations can be found in Table S3B.
ZNF genes & repeats and Quiescent/Low regions were not included in the analysis. Bedtools Fisher function (Quinlan and Hall, 2010) was used to calculated the p-value of the overlap between DAR types and chromatin states and p-values were Benjamini-Hochberg corrected for multiple comparisons within DAR type. Left- and right-tail tests were included in the final heatmap. The sign on the −log10(FDR) was inverted in the final heat map if the left-tail test was significant to denote under enrichment.
DAR genes vs DE genes enrichment analysis
DNA-binding dependent and independent DARs were annotated to the gene with the nearest transcription start site using annoatePeaks.pl from Homer (Heinz et al., 2010) with a custom Homer formatted annotation file made from the Gencode v24 annotation file used for RNA-seq alignment. After identifying the genes nearest to DARs, genes near more than one type of DAR (hDN DNA-binding independent closed, hNP DNA-binding dependent open etc.) were excluded to make mutually exclusive DAR gene lists. These mutually exclusive lists were then overlapped with DE genes from RNA-seq. The significance of the enrichment for DAR genes that are also DE genes was evaluated with a hypergeometric test Benjamini-Hochberg adjusted for all comparisons. Overlaps deemed significant had an adjusted p-value < 0.05.
DAR motif enrichment analysis
Homer findMotifsGenome.pl program (Heinz et al., 2010) was used to identify de novo enriched motifs in hDN DNA-binding dependent closed and hDN DNA-binding independent open DARs using the DiffBind consensus peak set as background. Homer’s known motif enrichment algorithm (also part of findMotifsGenome.pl) was used to find the enrichment of de novo identified FOX (hDN DNA-binding dependent closed homer motif3 p= 1e-16) and NF1-halfsite motifs (hDN DNA-binding independent open homer motif1, p= 1e-16) in the other hDN DAR groups compared to the consensus background peak set.
Brainspan WGCNA
We downloaded the Brainspan Developmental Transcriptome Dataset (RNA-Seq Gencode v10 summarized to genes) from http://www.brainspan.org/static/download.html, and only used samples up to 24 post conception weeks and genes that were expressed in either hNPs or hDNs (see hNP and hDN WGCNA). We built the network using the following parameters for the blockwiseModules comand:
corType=“pearson”, maxBlockSize = 14000, networkType=“signed”, power=10, minModuleSize=50, nThreads=15, TOMType = “signed”, TOMDenom = “mean”, deepSplit=4, verbose=5, mergeCutHeight=0.1, detectCutHeight = 0.999, reassignThreshold = 1e-6, numericLabels=TRUE, saveTOMs=TRUE, pamStage=TRUE, pamRespectsDendro=TRUE, saveTOMFileBase=“TOM_SIGNED”
After identifying the module eigengenes, we used the moduleTraitCor and moduleTraitPvalue functions to define the relationships between module and the brain regions from which the samples were obtained.
Integration of single-cell RNA-seq data from Nowakowski et al. 2017
Cell-type definitions used in this paper are based on cluster interpretations from Nowakowski et al. 2017 as described in Table S4D. Nowakowski clusters were merged to make up larger cell-type groups.
TPM expression values and meta data for the data set were downloaded from https://cells.ucsc.edu/. TSNE visualization was performed after removing unknown clusters U1–4 using the Seurat R package (Butler et al., 2018) as follows:
matrix = read.delim(“matrix.tsv”) meta=read.delim(“meta.tsv”) nowa= CreateSeuratObject(raw.data= matrix) nowa = AddMetaData(nowa, meta) nowa= SetAllIdent(nowa, id=“WGCNAcluster”) nowa.noU= SubsetData (nowa, ident.remove = c(“U1”, “U2”, “U3”, “U4”, ““)) nowa.noU = NormalizeData(nowa.noU, normalization.method = “LogNormalize”, scale.factor = 10000) nowa.noU = FindVariableGenes(nowa.noU, mean.function = ExpMean, dispersion.function = LogVMR, x.low.cutoff = 0.0125, x.high.cutoff = 3, y.cutoff = 0.5) nowa.noU =ScaleData(nowa.noU, vars.to.regress = c(“nUMI”)) nowa.noU = RunPCA (nowa.noU, pc.genes = nowa.noU@var.genes, do.print = FALSE) nowa.noU = RunTSNE (nowa.noU, reduction.use = “pca”, dims.use = 1:20, dim.embed = 2)
After TSNE visualization, the cells were colored according to this paper’s cell-type definitions.
Cell-type markers were identified as follows where cell.type.ident represents the cell-type definitions used in this paper:
nowa= SetAllIdent(nowa, id=“cell.type.ident”) nowa.noU = FindAllMarkers(nowa.noU, only.pos = TRUE)
Pseudodifferentation values for each cell from Nowakowski et al. 2017 were obtained from the authors. EN and IN lineage values here are the same as “page” and “vage” distinctions in Nowakowski et al. 2017. Pearson correlation of gene expression with the pseudodifferentation was performed using the corr.test function in R. The ggridges R packages was used to visualize the distributions of the correlation.
Immunocytochemistry
Coverslips were fixed in PBS with 4% methanol-free formaldehyde (#PI28906, Fisher Scientific) for 15 mins, washed three times with TBS, permeabilized with TBS containing .05% Triton X-100 (#BP151–100, Thermo Fisher Scientific), washed three times with TBS, and blocked with TBS containing .02% Triton X-100 and 5–10% donkey serum (#S30–100ML, Millipore Sigma). Coverslips were incubated with antibodies and dilutions specific to each experiment in TBS containing .02% Triton X-100 and 5% donkey serum overnight at 4°C. Antibodies and dilutions used are as follows: goat-polyclonal anti-FOXP2 (N-16), (#sc-21069, Santa Cruz Biotechnology, Dallas, Texas), 1:1000; mouse-monoclonal anti-TUJ1 (#801201, BioLegend, San Diego, California), 1:1000; rabbit-polyclonal anti-NFIA (#HPA008884, Millipore Sigma), 1:500; rabbit-polyclonal anti-NFIB (#HPA003956, Millipore Sigma), 1:500; mouse-monoclonal anti-V5-tag (#R960–25,Thermo Fisher Scientific), 1:10,000. This was followed by incubation with 1:10,000 of the appropriate Alexa Fluor® IgG secondary antibodies (Thermo Fisher Scientific) in TBS containing .02% Triton X-100 and 5% donkey serum. Coverslips were mounted with ProLong® Diamond Antifade Reagent with DAPI (P36962, Thermo Fisher Scientific) and imaged using a Zeiss Observer.Z1 inverted microscope and ZEN 2011 software.
Proximity ligation
Proximity ligation was performed using the Duolink PLA Fluorescence reagents (#DUO92101, Millipore Sigma) as per the manufacturers’ instructions. Samples were probed with mouse-monoclonal anti-V5-tag (#R960–25, Thermo Fisher Scientific) at a 1:10,000 dilution and either rabbit-polyclonal anti-NFIA (#HPA008884, Millipore Sigma) at a 1:10,000 dilution or rabbit-polyclonal anti-NFIB (#HPA003956, Millipore Sigma) at a 1:10,000 dilution. Coverslips were imaged using a 60x objective on a Zeiss Observer.Z1 inverted microscope using ZEN 2011 software. The level of exposure in the red, PLA signal channel was kept consistent across conditions. Using FIJI, brightness and contrast were auto adjusted in the DAPI channel for visualization. Background was subtracted and brightness was adjusted in the red, PLA signal channel in order to visualize puncta above background signal.
QUANTIFICATION AND STATISTICAL ANALYSIS
Hypergeometric overlap tests
The hypergeometric overalp test function is as follows:
library(gmp)
enrich_pvalue <- function(N, A, B, k)
{
m <- A + k
n <- B + k
i <- k:min(m,n)
as.numeric(sum(chooseZ(m,i)*chooseZ(N-m,n-i))/chooseZ(N,n))
}
enrich_pvalue(N,A,B,k)
15585 was used for the background gene number for all hypergeometric overlap tests. This number represents genes with RPKM of 1 in 80% of the samples from at least one cortical region at one time in the Allen Human Brain Atlas (Hawrylycz et al., 2012), and has been previously used for this purpose (Parikshak et al., 2013).
Gene expression heat maps
Expression heat maps were made using the pheatmap package in R (https://CRAN.R-project.org/package=pheatmap). Expression was centered and scaled by column mean or by row mean as indicated in the figure legend. For heat maps showing the expression of human fetal cortical genes (Miller et al., 2014) the average expression of the gene of interest in samples corresponding to each layer was used.
Bar plots
All bar plots with error bars represent mean ± SD. The exact value of n and the appropriate statistical tests for each analysis are listed in the figure or figure legend. Significance was defined as p-value or Benjamini-Hochberg adjusted p-value (when appropriate) of <0.05.
Sample size estimation
No specific criteria were used for sample size estimation.
DATA AND SOFTWARE AVAILABILITY
The NCBI Gene Expression Omnibus (GEO) accession number for the RNA-seq and ATAC-seq data reported in this paper is GSE111353.
Supplementary Material
Table S1: Related to Figures 1 and S1 (A) Genes differentially expressed in CTRL hDNs vs CTRL hNPs. (B) Genes differentially expressed in FOXP2-KE hDNs vs CTRL hNPs. (C) Genes differentially expressed in FOXP2-WT hDNs vs CTRL hNPs.
Table S2: Related to Figure 2 (A) hDN DE genes (FOXP2-WT vs CTRL) and differential expression values from Gandal et. al. 2018. (B) hDN DE genes (FOXP2-KE vs CTRL). (C) Combined ToppGene and Revigo output for genes repressed by FOXP2-WT. (D) Combined ToppGene and Revigo output for genes activated by FOXP2-WT.
Table S3: Related to Figure 3 (A) Locations of all identified differentially accessible regions FDR < .05. (B) Imputed chromatin states defined by 12 regulatory marks from human fetal brain tissue by the Roadmap Epigenomics Project. (C) List of annotated hDN DNA-binding dependent closed DARs. (D) List of annotated hDN DNA-binding dependent open DARs. (E) List of annotated hDN DNA-binding independent closed DARs. (F) List of annotated hDN DNA-binding independent open DARs.
Table S4: Related to Figure 4 (A) Module member genes for Brainspan WGCNA. (B) Module eigengenes for Brainspan WGCNA. (C) Module eigengene:region correlations for Brainspan WGCNA. (D) Cell type-lables for Nowakowski et al. clusters.
Table S5: Related to Figure 6 (A) hNP/hDN WGCNA module member genes and correlations with pseudodifferentiation. (B) Module eigengenes for hNP/hDN WGCNA. (C) Single-cell cell-type enriched genes (reanalyzed from Nowakowski et. al. 2017).
ACKNOWLEDGMENTS
We thank Drs. Joseph S. Takahashi, Jane E. Johnson, and Taekyung Kim for critical reading of the manuscript. We thank Marissa Co for providing constructs and editing the manuscript. We thank Drs. Arnold Kriegstein and Tomasz Nowakowski for providing the pseudotime and pseudodifferentiation data. G.K. is a Jon Heighten Scholar in Autism Research at UT Southwestern. This work was supported by the James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition – Scholar Award and grants from the NIH (DC014702, DC016340, MH090238, MH102603, and MH107672) to G. K.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- ANDERS S, PYL PT & HUBER W 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics, 31, 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELTON E, SALMOND CH, WATKINS KE, VARGHA-KHADEM F & GADIAN DG 2003. Bilateral brain abnormalities associated with dominantly inherited verbal and orofacial dyspraxia. Hum Brain Mapp, 18, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BETANCOURT J, KATZMAN S & CHEN B 2014. Nuclear factor one B regulates neural stem cell differentiation and axonal projection of corticofugal neurons. J Comp Neurol, 522, 6–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOLGER AM, LOHSE M & USADEL B 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUENROSTRO JD, WU B, CHANG HY & GREENLEAF WJ 2015a. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol, 109, 21 29 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUENROSTRO JD, WU B, LITZENBURGER UM, RUFF D, GONZALES ML, SNYDER MP, CHANG HY & GREENLEAF WJ 2015b. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature, 523, 486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUTLER A, HOFFMAN P, SMIBERT P, PAPALEXI E & SATIJA R 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology, 36, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAUDHRY AZ, LYONS GE & GRONOSTAJSKI RM 1997. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn, 208, 313–25. [DOI] [PubMed] [Google Scholar]
- CHEN J, BARDES EE, ARONOW BJ & JEGGA AG 2009. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res, 37, W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN YC, KUO HY, BORNSCHEIN U, TAKAHASHI H, CHEN SY, LU KM, YANG HY, CHEN GM, LIN JR, LEE YH, CHOU YC, CHENG SJ, CHIEN CT, ENARD W, HEVERS W, PAABO S, GRAYBIEL AM & LIU FC 2016. Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nat Neurosci, 19, 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAS NEVES L, DUCHALA CS, TOLENTINO-SILVA F, HAXHIU MA, COLMENARES C, MACKLIN WB, CAMPBELL CE, BUTZ KG & GRONOSTAJSKI RM 1999. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci U S A, 96, 11946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE LA TORRE-UBIETA L, STEIN JL, WON H, OPLAND CK, LIANG D, LU D & GESCHWIND DH 2018. The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell, 172, 289–304 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEMONTIS D, WALTERS RK, MARTIN J, MATTHEISEN M, ALS TD, AGERBO E, BALDURSSON G, BELLIVEAU R, BYBJERG-GRAUHOLM J, BAEKVAD-HANSEN M, CERRATO F, CHAMBERT K, CHURCHHOUSE C, DUMONT A, ERIKSSON N, GANDAL M, GOLDSTEIN JI, GRASBY KL, GROVE J, GUDMUNDSSON OO, HANSEN CS, HAUBERG ME, HOLLEGAARD MV, HOWRIGAN DP, HUANG H, MALLER JB, MARTIN AR, MARTIN NG, MORAN J, PALLESEN J, PALMER DS, PEDERSEN CB, PEDERSEN MG, POTERBA T, POULSEN JB, RIPKE S, ROBINSON EB, SATTERSTROM FK, STEFANSSON H, STEVENS C, TURLEY P, WALTERS GB, WON H, WRIGHT MJ, CONSORTIUM AWGOTPG, EARLY L, GENETIC EPIDEMIOLOGY C, ANDME RESEARCH T, ANDREASSEN OA, ASHERSON P, BURTON CL, BOOMSMA DI, CORMAND B, DALSGAARD S, FRANKE B, GELERNTER J, GESCHWIND D, HAKONARSON H, HAAVIK J, KRANZLER HR, KUNTSI J, LANGLEY K, LESCH KP, MIDDELDORP C, REIF A, ROHDE LA, ROUSSOS P, SCHACHAR R, SKLAR P, SONUGA-BARKE EJS, SULLIVAN PF, THAPAR A, TUNG JY, WALDMAN ID, MEDLAND SE, STEFANSSON K, NORDENTOFT M, HOUGAARD DM, WERGE T, MORS O, MORTENSEN PB, DALY MJ, FARAONE SV, BORGLUM AD & NEALE BM 2019. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet, 51, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DERIZIOTIS P, O’ROAK BJ, GRAHAM SA, ESTRUCH SB, DIMITROPOULOU D, BERNIER RA, GERDTS J, SHENDURE J, EICHLER EE & FISHER SE 2014. De novo TBR1 mutations in sporadic autism disrupt protein functions. Nat Commun, 5, 4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOBIN A, DAVIS CA, SCHLESINGER F, DRENKOW J, ZALESKI C, JHA S, BATUT P, CHAISSON M & GINGERAS TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENARD W, GEHRE S, HAMMERSCHMIDT K, HOLTER SM, BLASS T, SOMEL M, BRUCKNER MK, SCHREIWEIS C, WINTER C, SOHR R, BECKER L, WIEBE V, NICKEL B, GIGER T, MULLER U, GROSZER M, ADLER T, AGUILAR A, BOLLE I, CALZADA-WACK J, DALKE C, EHRHARDT N, FAVOR J, FUCHS H, GAILUS-DURNER V, HANS W, HOLZLWIMMER G, JAVAHERI A, KALAYDJIEV S, KALLNIK M, KLING E, KUNDER S, MOSSBRUGGER I, NATON B, RACZ I, RATHKOLB B, ROZMAN J, SCHREWE A, BUSCH DH, GRAW J, IVANDIC B, KLINGENSPOR M, KLOPSTOCK T, OLLERT M, QUINTANILLA-MARTINEZ L, SCHULZ H, WOLF E, WURST W, ZIMMER A, FISHER SE, MORGENSTERN R, ARENDT T, DE ANGELIS MH, FISCHER J, SCHWARZ J & PAABO S 2009. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell, 137, 961–71. [DOI] [PubMed] [Google Scholar]
- ENARD W, PRZEWORSKI M, FISHER SE, LAI CS, WIEBE V, KITANO T, MONACO AP & PAABO S 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature, 418, 869–72. [DOI] [PubMed] [Google Scholar]
- ERNST J & KELLIS M 2015. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol, 33, 364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESTRUCH SB, GRAHAM SA, CHINNAPPA SM, DERIZIOTIS P & FISHER SE 2016. Functional characterization of rare FOXP2 variants in neurodevelopmental disorder. J Neurodev Disord, 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIETZ SA, LACHMANN R, BRANDL H, KIRCHER M, SAMUSIK N, SCHRODER R, LAKSHMANAPERUMAL N, HENRY I, VOGT J, RIEHN A, DISTLER W, NITSCH R, ENARD W, PAABO S & HUTTNER WB 2012. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc Natl Acad Sci U S A, 109, 11836–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRENCH CA, GROSZER M, PREECE C, COUPE AM, RAJEWSKY K & FISHER SE 2007. Generation of mice with a conditional Foxp2 null allele. Genesis, 45, 440–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRENCH CA, VINUEZA VELOZ MF, ZHOU K, PETER S, FISHER SE, COSTA RM & DE ZEEUW CI 2018. Differential effects of Foxp2 disruption in distinct motor circuits. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUJITA E, TANABE Y, SHIOTA A, UEDA M, SUWA K, MOMOI MY & MOMOI T 2008. Ultrasonic vocalization impairment of Foxp2 (R552H) knockin mice related to speech-language disorder and abnormality of Purkinje cells. Proc Natl Acad Sci U S A, 105, 3117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GANDAL MJ, ZHANG P, HADJIMICHAEL E, WALKER RL, CHEN C, LIU S, WON H, VAN BAKEL H, VARGHESE M, WANG Y, SHIEH AW, HANEY J, PARHAMI S, BELMONT J, KIM M, MORAN LOSADA P, KHAN Z, MLECZKO J, XIA Y, DAI R, WANG D, YANG YT, XU M, FISH K, HOF PR, WARRELL J, FITZGERALD D, WHITE K, JAFFE AE, PSYCH EC, PETERS MA, GERSTEIN M, LIU C, IAKOUCHEVA LM, PINTO D & GESCHWIND DH 2018. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRAY LT, YAO Z, NGUYEN TN, KIM TK, ZENG H & TASIC B 2017. Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROSZER M, KEAYS DA, DEACON RM, DE BONO JP, PRASAD-MULCARE S, GAUB S, BAUM MG, FRENCH CA, NICOD J, COVENTRY JA, ENARD W, FRAY M, BROWN SD, NOLAN PM, PAABO S, CHANNON KM, COSTA RM, EILERS J, EHRET G, RAWLINS JN & FISHER SE 2008. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr Biol, 18, 354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAWRYLYCZ MJ, LEIN ES, GUILLOZET-BONGAARTS AL, SHEN EH, NG L, MILLER JA, VAN DE LAGEMAAT LN, SMITH KA, EBBERT A, RILEY ZL, ABAJIAN C, BECKMANN CF, BERNARD A, BERTAGNOLLI D, BOE AF, CARTAGENA PM, CHAKRAVARTY MM, CHAPIN M, CHONG J, DALLEY RA, DAVID DALY B, DANG C, DATTA S, DEE N, DOLBEARE TA, FABER V, FENG D, FOWLER DR, GOLDY J, GREGOR BW, HARADON Z, HAYNOR DR, HOHMANN JG, HORVATH S, HOWARD RE, JEROMIN A, JOCHIM JM, KINNUNEN M, LAU C, LAZARZ ET, LEE C, LEMON TA, LI L, LI Y, MORRIS JA, OVERLY CC, PARKER PD, PARRY SE, REDING M, ROYALL JJ, SCHULKIN J, SEQUEIRA PA, SLAUGHTERBECK CR, SMITH SC, SODT AJ, SUNKIN SM, SWANSON BE, VAWTER MP, WILLIAMS D, WOHNOUTKA P, ZIELKE HR, GESCHWIND DH, HOF PR, SMITH SM, KOCH C, GRANT SGN & JONES AR 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEINZ S, BENNER C, SPANN N, BERTOLINO E, LIN YC, LASLO P, CHENG JX, MURRE C, SINGH H & GLASS CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell, 38, 576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENG YH, BARRY G, RICHARDS LJ & PIPER M 2012. Nuclear factor I genes regulate neuronal migration. Neurosignals, 20, 159–67. [DOI] [PubMed] [Google Scholar]
- HOERDER-SUABEDISSEN A & MOLNAR Z 2015. Development, evolution and pathology of neocortical subplate neurons. Nat Rev Neurosci, 16, 133–46. [DOI] [PubMed] [Google Scholar]
- HOERDER-SUABEDISSEN A, WANG WZ, LEE S, DAVIES KE, GOFFINET AM, RAKIC S, PARNAVELAS J, REIM K, NICOLIC M, PAULSEN O & MOLNAR Z 2009. Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb Cortex, 19, 1738–50. [DOI] [PubMed] [Google Scholar]
- HUTSLER JJ & CASANOVA MF 2016. Review: Cortical construction in autism spectrum disorder: columns, connectivity and the subplate. Neuropathol Appl Neurobiol, 42, 115–34. [DOI] [PubMed] [Google Scholar]
- KANOLD PO, KARA P, REID RC & SHATZ CJ 2003. Role of subplate neurons in functional maturation of visual cortical columns. Science, 301, 521–5. [DOI] [PubMed] [Google Scholar]
- KANOLD PO & SHATZ CJ 2006. Subplate neurons regulate maturation of cortical inhibition and outcome of ocular dominance plasticity. Neuron, 51, 627–38. [DOI] [PubMed] [Google Scholar]
- KONOPKA G, BOMAR JM, WINDEN K, COPPOLA G, JONSSON ZO, GAO F, PENG S, PREUSS TM, WOHLSCHLEGEL JA & GESCHWIND DH 2009. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature, 462, 213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONOPKA G & ROBERTS TF 2016. Insights into the Neural and Genetic Basis of Vocal Communication. Cell, 164, 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONOPKA G, WEXLER E, ROSEN E, MUKAMEL Z, OSBORN GE, CHEN L, LU D, GAO F, GAO K, LOWE JK & GESCHWIND DH 2012. Modeling the functional genomics of autism using human neurons. Mol Psychiatry, 17, 202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOSTOVIC I & RAKIC P 1990. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol, 297, 441–70. [DOI] [PubMed] [Google Scholar]
- LAI CS, FISHER SE, HURST JA, LEVY ER, HODGSON S, FOX M, JEREMIAH S, POVEY S, JAMISON DC, GREEN ED, VARGHA-KHADEM F & MONACO AP 2000. The SPCH1 region on human 7q31: genomic characterization of the critical interval and localization of translocations associated with speech and language disorder. Am J Hum Genet, 67, 357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI CS, FISHER SE, HURST JA, VARGHA-KHADEM F & MONACO AP 2001. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature, 413, 519–23. [DOI] [PubMed] [Google Scholar]
- LAI CS, GERRELLI D, MONACO AP, FISHER SE & COPP AJ 2003. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain, 126, 2455–62. [DOI] [PubMed] [Google Scholar]
- LANGFELDER P & HORVATH S 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANGMEAD B, TRAPNELL C, POP M & SALZBERG SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol, 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNON PA, COOPER ML, PEIFFER DA, GUNDERSON KL, PATEL A, PETERS S, CHEUNG SW & BACINO CA 2007. Deletion of 7q31.1 supports involvement of FOXP2 in language impairment: clinical report and review. Am J Med Genet A, 143A, 791–8. [DOI] [PubMed] [Google Scholar]
- LEPP S, ANDERSON A & KONOPKA G 2013. Connecting signaling pathways underlying communication to ASD vulnerability. Int Rev Neurobiol, 113, 97–133. [DOI] [PubMed] [Google Scholar]
- LI H, HANDSAKER B, WYSOKER A, FENNELL T, RUAN J, HOMER N, MARTH G, ABECASIS G, DURBIN R & GENOME PROJECT DATA PROCESSING, S. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI S, WEIDENFELD J & MORRISEY EE 2004. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol, 24, 809–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIEGEOIS F, BALDEWEG T, CONNELLY A, GADIAN DG, MISHKIN M & VARGHA-KHADEM F 2003. Language fMRI abnormalities associated with FOXP2 gene mutation. Nat Neurosci, 6, 1230–7. [DOI] [PubMed] [Google Scholar]
- LOVE MI, HUBER W & ANDERS S 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUI JH, HANSEN DV & KRIEGSTEIN AR 2011. Development and evolution of the human neocortex. Cell, 146, 18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACDERMOT KD, BONORA E, SYKES N, COUPE AM, LAI CS, VERNES SC, VARGHA-KHADEM F, MCKENZIE F, SMITH RL, MONACO AP & FISHER SE 2005. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet, 76, 1074–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTYNOGA B, MATEO JL, ZHOU B, ANDERSEN J, ACHIMASTOU A, URBAN N, VAN DEN BERG D, GEORGOPOULOU D, HADJUR S, WITTBRODT J, ETTWILLER L, PIPER M, GRONOSTAJSKI RM & GUILLEMOT F 2013. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev, 27, 1769–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEDVEDEVA VP, RIEGER MA, VIETH B, MOMBEREAU C, ZIEGENHAIN C, GHOSH T, CRESSANT A, ENARD W, GRANON S, DOUGHERTY JD & GROSZER M 2018. Altered social behavior in mice carrying a cortical Foxp2 deletion. Hum Mol Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER JA, DING SL, SUNKIN SM, SMITH KA, NG L, SZAFER A, EBBERT A, RILEY ZL, ROYALL JJ, AIONA K, ARNOLD JM, BENNET C, BERTAGNOLLI D, BROUNER K, BUTLER S, CALDEJON S, CAREY A, CUHACIYAN C, DALLEY RA, DEE N, DOLBEARE TA, FACER BA, FENG D, FLISS TP, GEE G, GOLDY J, GOURLEY L, GREGOR BW, GU G, HOWARD RE, JOCHIM JM, KUAN CL, LAU C, LEE CK, LEE F, LEMON TA, LESNAR P, MCMURRAY B, MASTAN N, MOSQUEDA N, NALUAI-CECCHINI T, NGO NK, NYHUS J, OLDRE A, OLSON E, PARENTE J, PARKER PD, PARRY SE, STEVENS A, PLETIKOS M, REDING M, ROLL K, SANDMAN D, SARREAL M, SHAPOURI S, SHAPOVALOVA NV, SHEN EH, SJOQUIST N, SLAUGHTERBECK CR, SMITH M, SODT AJ, WILLIAMS D, ZOLLEI L, FISCHL B, GERSTEIN MB, GESCHWIND DH, GLASS IA, HAWRYLYCZ MJ, HEVNER RF, HUANG H, JONES AR, KNOWLES JA, LEVITT P, PHILLIPS JW, SESTAN N, WOHNOUTKA P, DANG C, BERNARD A, HOHMANN JG & LEIN ES 2014. Transcriptional landscape of the prenatal human brain. Nature, 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOLNAR Z & BLAKEMORE C 1995. Guidance of thalamocortical innervation. Ciba Found Symp, 193, 127–49; discussion 192–9. [DOI] [PubMed] [Google Scholar]
- MUKAMEL Z, KONOPKA G, WEXLER E, OSBORN GE, DONG H, BERGMAN MY, LEVITT P & GESCHWIND DH 2011. Regulation of MET by FOXP2, genes implicated in higher cognitive dysfunction and autism risk. J Neurosci, 31, 11437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOWAKOWSKI TJ, BHADURI A, POLLEN AA, ALVARADO B, MOSTAJO-RADJI MA, DI LULLO E, HAEUSSLER M, SANDOVAL-ESPINOSA C, LIU SJ, VELMESHEV D, OUNADJELA JR, SHUGA J, WANG X, LIM DA, WEST JA, LEYRAT AA, KENT WJ & KRIEGSTEIN AR 2017. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science, 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALMER TD, SCHWARTZ PH, TAUPIN P, KASPAR B, STEIN SA & GAGE FH 2001. Cell culture. Progenitor cells from human brain after death. Nature, 411, 42–3. [DOI] [PubMed] [Google Scholar]
- PARIKSHAK NN, LUO R, ZHANG A, WON H, LOWE JK, CHANDRAN V, HORVATH S & GESCHWIND DH 2013. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell, 155, 1008–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERS SK, DUNLOP K & DOWNAR J 2016. Cortico-Striatal-Thalamic Loop Circuits of the Salience Network: A Central Pathway in Psychiatric Disease and Treatment. Front Syst Neurosci, 10, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QUINLAN AR & HALL IM 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAKIC P 1974. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science, 183, 425–7. [DOI] [PubMed] [Google Scholar]
- REIMERS-KIPPING S, HEVERS W, PAABO S & ENARD W 2011. Humanized Foxp2 specifically affects cortico-basal ganglia circuits. Neuroscience, 175, 75–84. [DOI] [PubMed] [Google Scholar]
- RITCHIE ME, PHIPSON B, WU D, HU Y, LAW CW, SHI W & SMYTH GK 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROADMAP EPIGENOMICS C, KUNDAJE A, MEULEMAN W, ERNST J, BILENKY M, YEN A, HERAVI-MOUSSAVI A, KHERADPOUR P, ZHANG Z, WANG J, ZILLER MJ, AMIN V, WHITAKER JW, SCHULTZ MD, WARD LD, SARKAR A, QUON G, SANDSTROM RS, EATON ML, WU YC, PFENNING AR, WANG X, CLAUSSNITZER M, LIU Y, COARFA C, HARRIS RA, SHORESH N, EPSTEIN CB, GJONESKA E, LEUNG D, XIE W, HAWKINS RD, LISTER R, HONG C, GASCARD P, MUNGALL AJ, MOORE R, CHUAH E, TAM A, CANFIELD TK, HANSEN RS, KAUL R, SABO PJ, BANSAL MS, CARLES A, DIXON JR, FARH KH, FEIZI S, KARLIC R, KIM AR, KULKARNI A, LI D, LOWDON R, ELLIOTT G, MERCER TR, NEPH SJ, ONUCHIC V, POLAK P, RAJAGOPAL N, RAY P, SALLARI RC, SIEBENTHALL KT, SINNOTT-ARMSTRONG NA, STEVENS M, THURMAN RE, WU J, ZHANG B, ZHOU X, BEAUDET AE, BOYER LA, DE JAGER PL, FARNHAM PJ, FISHER SJ, HAUSSLER D, JONES SJ, LI W, MARRA MA, MCMANUS MT, SUNYAEV S, THOMSON JA, TLSTY TD, TSAI LH, WANG W, WATERLAND RA, ZHANG MQ, CHADWICK LH, BERNSTEIN BE, COSTELLO JF, ECKER JR, HIRST M, MEISSNER A, MILOSAVLJEVIC A, REN B, STAMATOYANNOPOULOS JA, WANG T & KELLIS M 2015. Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON MD, MCCARTHY DJ & SMYTH GK 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSEN EY, WEXLER EM, VERSANO R, COPPOLA G, GAO F, WINDEN KD, OLDHAM MC, MARTENS LH, ZHOU P, FARESE RV JR. & GESCHWIND DH 2011. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron, 71, 1030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSS-INNES CS, STARK R, TESCHENDORFF AE, HOLMES KA, ALI HR, DUNNING MJ, BROWN GD, GOJIS O, ELLIS IO, GREEN AR, ALI S, CHIN SF, PALMIERI C, CALDAS C & CARROLL JS 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature, 481, 389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHANNON P, MARKIEL A, OZIER O, BALIGA NS, WANG JT, RAMAGE D, AMIN N, SCHWIKOWSKI B & IDEKER T 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 13, 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHU W, CHO JY, JIANG Y, ZHANG M, WEISZ D, ELDER GA, SCHMEIDLER J, DE GASPERI R, SOSA MA, RABIDOU D, SANTUCCI AC, PERL D, MORRISEY E & BUXBAUM JD 2005. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci U S A, 102, 9643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPITERI E, KONOPKA G, COPPOLA G, BOMAR J, OLDHAM M, OU J, VERNES SC, FISHER SE, REN B & GESCHWIND DH 2007. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet, 81, 1144–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEIN JL, DE LA TORRE-UBIETA L, TIAN Y, PARIKSHAK NN, HERNANDEZ IA, MARCHETTO MC, BAKER DK, LU D, HINMAN CR, LOWE JK, WEXLER EM, MUOTRI AR, GAGE FH, KOSIK KS & GESCHWIND DH 2014. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron, 83, 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUPEK F, BOSNJAK M, SKUNCA N & SMUC T 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARGHA-KHADEM F, WATKINS K, ALCOCK K, FLETCHER P & PASSINGHAM R 1995. Praxic and nonverbal cognitive deficits in a large family with a genetically transmitted speech and language disorder. Proc Natl Acad Sci U S A, 92, 930–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARGHA-KHADEM F, WATKINS KE, PRICE CJ, ASHBURNER J, ALCOCK KJ, CONNELLY A, FRACKOWIAK RS, FRISTON KJ, PEMBREY ME, MISHKIN M, GADIAN DG & PASSINGHAM RE 1998. Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci U S A, 95, 12695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VERNES SC, NEWBURY DF, ABRAHAMS BS, WINCHESTER L, NICOD J, GROSZER M, ALARCON M, OLIVER PL, DAVIES KE, GESCHWIND DH, MONACO AP & FISHER SE 2008. A functional genetic link between distinct developmental language disorders. N Engl J Med, 359, 2337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VERNES SC, NICOD J, ELAHI FM, COVENTRY JA, KENNY N, COUPE AM, BIRD LE, DAVIES KE & FISHER SE 2006. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet, 15, 3154–67. [DOI] [PubMed] [Google Scholar]
- VERNES SC, OLIVER PL, SPITERI E, LOCKSTONE HE, PULIYADI R, TAYLOR JM, HO J, MOMBEREAU C, BREWER A, LOWY E, NICOD J, GROSZER M, BABAN D, SAHGAL N, CAZIER JB, RAGOUSSIS J, DAVIES KE, GESCHWIND DH & FISHER SE 2011. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet, 7, e1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VERNES SC, SPITERI E, NICOD J, GROSZER M, TAYLOR JM, DAVIES KE, GESCHWIND DH & FISHER SE 2007. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet, 81, 1232–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALKER RM, HILL AE, NEWMAN AC, HAMILTON G, TORRANCE HS, ANDERSON SM, OGAWA F, DERIZIOTI P, NICOD J, VERNES SC, FISHER SE, THOMSON PA, PORTEOUS DJ & EVANS KL 2012. The DISC1 promoter: characterization and regulation by FOXP2. Hum Mol Genet, 21, 2862–72. [DOI] [PubMed] [Google Scholar]
- WANG L, WANG S & LI W 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics, 28, 2184–5. [DOI] [PubMed] [Google Scholar]
- WATKINS KE, DRONKERS NF & VARGHA-KHADEM F 2002a. Behavioural analysis of an inherited speech and language disorder: comparison with acquired aphasia. Brain, 125, 452–64. [DOI] [PubMed] [Google Scholar]
- WATKINS KE, VARGHA-KHADEM F, ASHBURNER J, PASSINGHAM RE, CONNELLY A, FRISTON KJ, FRACKOWIAK RS, MISHKIN M & GADIAN DG 2002b. MRI analysis of an inherited speech and language disorder: structural brain abnormalities. Brain, 125, 465–78. [DOI] [PubMed] [Google Scholar]
- WEXLER EM, ROSEN E, LU D, OSBORN GE, MARTIN E, RAYBOULD H & GESCHWIND DH 2011. Genome-wide analysis of a Wnt1-regulated transcriptional network implicates neurodegenerative pathways. Sci Signal, 4, ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU Y, BORDE M, HEISSMEYER V, FEUERER M, LAPAN AD, STROUD JC, BATES DL, GUO L, HAN A, ZIEGLER SF, MATHIS D, BENOIST C, CHEN L & RAO A 2006. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell, 126, 375–87. [DOI] [PubMed] [Google Scholar]
- XU X, WELLS AB, O’BRIEN DR, NEHORAI A & DOUGHERTY JD 2014. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci, 34, 1420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG J, WEBB DM & PODLAHA O 2002. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics, 162, 1825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y, LIU T, MEYER CA, EECKHOUTE J, JOHNSON DS, BERNSTEIN BE, NUSBAUM C, MYERS RM, BROWN M, LI W & LIU XS 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol, 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU Y, SOUSA AMM, GAO T, SKARICA M, LI M, SANTPERE G, ESTELLER-CUCALA P, JUAN D, FERRANDEZ-PERAL L, GULDEN FO, YANG M, MILLER DJ, MARQUES-BONET T, IMAMURA KAWASAWA Y, ZHAO H & SESTAN N 2018. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Related to Figures 1 and S1 (A) Genes differentially expressed in CTRL hDNs vs CTRL hNPs. (B) Genes differentially expressed in FOXP2-KE hDNs vs CTRL hNPs. (C) Genes differentially expressed in FOXP2-WT hDNs vs CTRL hNPs.
Table S2: Related to Figure 2 (A) hDN DE genes (FOXP2-WT vs CTRL) and differential expression values from Gandal et. al. 2018. (B) hDN DE genes (FOXP2-KE vs CTRL). (C) Combined ToppGene and Revigo output for genes repressed by FOXP2-WT. (D) Combined ToppGene and Revigo output for genes activated by FOXP2-WT.
Table S3: Related to Figure 3 (A) Locations of all identified differentially accessible regions FDR < .05. (B) Imputed chromatin states defined by 12 regulatory marks from human fetal brain tissue by the Roadmap Epigenomics Project. (C) List of annotated hDN DNA-binding dependent closed DARs. (D) List of annotated hDN DNA-binding dependent open DARs. (E) List of annotated hDN DNA-binding independent closed DARs. (F) List of annotated hDN DNA-binding independent open DARs.
Table S4: Related to Figure 4 (A) Module member genes for Brainspan WGCNA. (B) Module eigengenes for Brainspan WGCNA. (C) Module eigengene:region correlations for Brainspan WGCNA. (D) Cell type-lables for Nowakowski et al. clusters.
Table S5: Related to Figure 6 (A) hNP/hDN WGCNA module member genes and correlations with pseudodifferentiation. (B) Module eigengenes for hNP/hDN WGCNA. (C) Single-cell cell-type enriched genes (reanalyzed from Nowakowski et. al. 2017).
Data Availability Statement
The NCBI Gene Expression Omnibus (GEO) accession number for the RNA-seq and ATAC-seq data reported in this paper is GSE111353.