Abstract
α-Tubulin detyrosination, largely catalyzed by vasohibins, is involved in many microtubule (MT)-related cellular events. In this study, we identified a core heterodimeric complex of human small vasohibin-binding protein (SVBP) and vasohibin 1 (VASH1) (hereafter denoted as SVBP-VASH1) that catalyzes the detyrosination of a peptide derived from C-terminus of α-tubulin. We further solved the crystal structures of the SVBP-VASH1 heterodimer alone and in complex with either an inhibitor or a mutant substrate peptide. Our structural research, complemented by biochemical and mutagenesis experiments, resulted in identification of the key residues for VASH1 binding to SVBP and α-tubulin substrate. Our in vivo experiments reveal that MT detyrosination in general, as well as the interactions between SVBP, VASH1, and α-tubulin, are critical for spindle function and accurate chromosome segregation during mitosis. Furthermore, we found that the phenotypes caused by the depletion of vasohibins were largely rescued upon co-depletion of kinesin13/MCAK, suggesting the coordination between the MT depolymerase and MT detyrosination during mitosis. Thus our work not only provides structural insights into the molecular mechanism of α-tubulin detyrosination catalyzed by SVBP-bound vasohibins, but also uncovers the key role of vasohibins-mediated MT detyrosination in spindle morphology and chromosome segregation during mitosis.
Subject terms: X-ray crystallography, Mitosis
Introduction
Microtubules (MTs) are essential components of cytoskeleton and play important roles in a wide variety of cellular events, such as maintaining cell shape, mediating cell motility, transporting materials, and driving chromosome segregation during cell division.1 MTs are assembled from α/β-tubulin heterodimers, which in the presence of GTP polymerize into a ~13-protofilament hollow tube.2 MT nucleation mainly begins at the MT organizing center (MTOC) and transitions between growth and shrinkage of MTs occur rapidly, accounting for dynamic nature of MTs.3,4
Different tubulin isotypes are encoded in human genome and many post-translational modifications (PTMs) are found to occur on tubulin,5,6 including acetylation, detyrosination/tyrosination, glycylation, and (poly)glutamylation, etc.7 A variety of combinations of α-tubulin and β-tubulin isotypes, together with the tubulin PTMs, constitute the known “tubulin code”, accounting for some of diverse cellular functions of MTs.7 Tubulin detyrosination occurs at the C-terminal tail of α-tubulin, which is typically translated containing a tyrosine as the terminal amino acid.8 This tyrosine can be cleaved by tubulin carboxypeptidases (TCPs) and tubulin detyrosination can be reversed by the action of tubulin tyrosine ligase (TTL),9,10 which can re-add tyrosine to the C-terminus of α-tubulin.11–13
The TCPs were only recently identified as the vasohibins-SVBP heterodimer.14,15 Mammalian vasohibins, including VASH1 and VASH2, belong to the transglutaminase-like cysteine protease superfamily with a predicted catalytic triad.16 VASH1 and VASH2 share all known motifs with an overall sequence identity ~52%,16 and display redundant functions, as evidenced by the observation that knockdown of vasohibins by shRNAs resulted in reduced level of detyrosinated MTs and aberrant neuron differentiation.14 VASH1 and VASH2 were previously reported to play roles in angiogenesis and VASH1 was also found to suppress tumor growth.17,18 SVBP, a small vasohibin-binding protein of 66 amino acids, was proposed as a chaperone of VASH1 in vascular endothelial cells.19
Since TCPs and TTL prefer polymerized tubulin and soluble tubulin subunits,11,15,20–22 respectively, tubulin detyrosination is unevenly distributed along the mitotic spindle, mainly decorating more stable MTs that point from the spindle pole towards the cell equator, while the shorter-living astral MTs, pointing towards the cell cortex, remain more tyrosinated.23,24 Recently, it was shown that this anisotropic distribution of MT detyrosination within the mitotic spindle influences the spatiotemporal regulation of two kinetochore-based motor proteins of opposite orientation: kinesin-7/CENP-E and dynein.25–28 Consequently, the antipodal preferences of CENP-E and dynein/dynactin complex for detyrosinated and tyrosinated MTs, respectively, control the directionality of chromosome movements during mitosis.29
In addition to spatiotemporal control of CENP-E and dynein, MT detyrosination was shown to affect several other proteins with important roles in mitosis, such as a centromere- and MT-plus-end tip-localized kinesin-13 protein family member MCAK,30,31 MT-plus-end tip tracking protein CLIP-170 and MT-severing protein fidgetin,28,32 as well as kinesin-mediated transport in neurons,33,34 dynein activity in neurons,27 localizations and functions of different MT-associated proteins (MAPs),28,35–40 and the motility of several kinesins in vitro.25,31,41,42
Although vasohibins are identified as tubulin TCPs in vivo, the molecular basis for α-tubulin detyrosination by SVBP-vasohibins heterodimer remains elusive owing to the low sequence identity between vasohibins and other peptidases with known structures. By presenting the structure of the core complex of SVBP-VASH1, we show here that VASH1, which contains an unexpected catalytic triad (Cys-His-Leu), is stabilized by SVBP via extensive intermolecular interactions. We also determined another two structures of SVBP-VASH1, one bound to a peptide mimic of α-tubulin C-terminus and the other bound to an epoxide-containing inhibitor epoY,14 unraveling how VASH1 recognizes the C-terminus of α-tubulin and how epoY irreversibly inhibits VASH1-mediated detyrosination.
Furthermore, by applying the knowledge gained from the solved structure, we generated mutants that could be tested in functional assays. Due to the established association between MT detyrosination and mitosis, we studied the role of SVBP-vasohibins in maintenance of mitotic spindle length and positioning, and its impact on chromosome segregation during cell division. We propose that vasohibins-mediated MT detyrosination regulates spindle morphology and positioning by affecting the activity of MT depolymerase MCAK on astral MTs.43 Overall, this study not only uncovers the molecular mechanism of α-tubulin detyrosination by SVBP-VASH1, but also provides insights for better understanding the role of vasohibins and vasohibins-mediated MT detyrosination in spindle function and mitosis.
Results
The core complex consisting of SVBP and VASH1 displays activity towards an α-tubulin peptide
Given that fragments of recombinant human VASH1 alone displayed extremely poor solubility during expression and were stable with human SVBP, we cloned, co-expressed, and purified human SVBP-VASH1 core complexes by using different fragments of VASH1 and SVBP to obtain functional SVBP-VASH1 complex in vitro (Supplementary information, Fig. S1). We found by mass spectrometry that heterodimers containing VASH170–306 and SVBP3–49 were able to catalyze the detyrosination of a heptapeptide with its sequence derived from the C-terminal sequence of TUBA1A (−6EEEGEEY0), which is numbered from −6 to 0 hereafter (Supplementary information, Figs. S2a–c and S3a, Table S1). Notably, SVBP3–49-VASH170–306 and SVBP1–66-VASH170–306 catalyze the detyrosination of almost 100 and 98% of the substrates, respectively, suggesting that both heterodimers have comparable activities (Supplementary information, Table S1). In addition, we performed the ITC (isothermal titration calorimetry) binding experiments and found that full length SVBP(SVBP1–66)and SVBP3–49 bind to VASH170–306 with Kds of 42 and 48 nM, respectively (Supplementary information, Fig. S3b), which are comparable with the one previously measured by SPR (surface plasmon resonance) for full length SVBP and VASH1 (Kd = 31 nM).19 Collectively, SVBP3–49 is sufficient to bind VASH1 for the detyrosination of the α-tubulin peptide (Supplementary information, Table S1).
Overall structure of SVBP-VASH1 and the non-canonical catalytic triad of VASH1
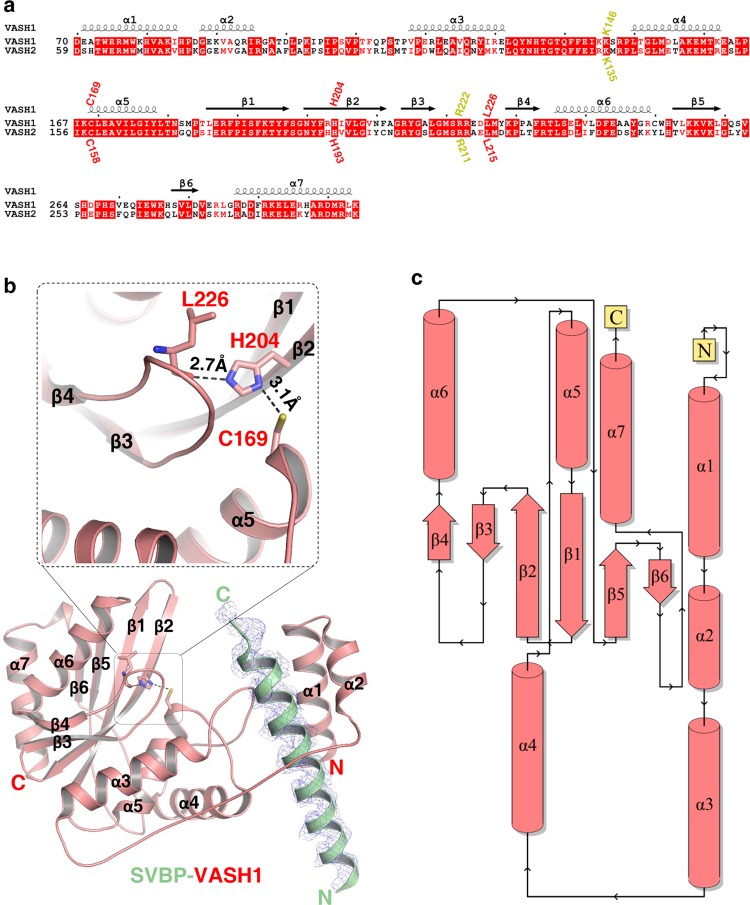
While every effort in our hand to crystallize the full length protein complex failed, we succeeded in crystallizing the core of SVBP-VASH1 heterodimer, consisting of VASH170–306 and SVBP1–66, and solved the structure at a resolution of 2.31 Å (Supplementary information, Table S2). There are two heterodimer molecules in the asymmetric unit. The two heterodimers are almost identical, with a root-mean-square deviation (R.M.S.D) of 0.14 Å over 261 Cα atoms. Residues 22–50 and 26–50 of SVBP are visible in two heterodimers, respectively, whereas the rest of SVBP residues are invisible probably owing to their internal flexibility (Fig. 1a, b).
Fig. 1.
Overall structure of the SVBP-VASH1 heterodimer. a Sequence alignment of human VASH1 (NP_055724.1) and VASH2 (NP_001129946.1), with secondary structures labeled at the top of sequences. Catalytic triad of VASH1 (Cys169, His204 and Leu226 of VASH1) and their corresponding residues in VASH2, are marked in red. Lys146 and Arg222 of VASH1 and their corresponding residues in VASH2, are marked in yellow. b Overall structure of the SVBP1–66-VASH170–306 heterodimer. SVBP and VASH1 are shown in green and red cartoon, respectively, with their secondary structures labeled. The catalytic residues of VASH1, including Cys169, His204, and Leu226, are indicated. The |Fo|–|Fc| omit map of SVBP22–50 is contoured at 2.7σ. Hydrogen bonds between His204 and Leu226 and between Cys169 and His204 are indicated by black dashes. c Topology diagram of VASH1 was provided by PDBsum,70 with secondary structures colored in red
Residues 70–303 of VASH1 is visible and adopts an α/β fold consisting of seven α-helices (α1–α7) and six β-strands (β1–β6) (Fig. 1a, b). β1–β6 constitute the core of VASH1 and are arranged into a six-stranded anti-parallel β-sheet (β4-β3-β2-β1-β5-β6) (Fig. 1b, c), with α3 and α5, and α6 and α7 packing against the β-sheet from each side to stabilize it. α1 and α2 form a small helix bundle and are connected to α3 via an extended loop (L23) (Fig. 1b). α4 buttresses α3 from the bottom and contacts the loop between β5 and β6 via its N-terminus (Fig. 1b).
Cys169 and His204 of VASH1, which were identified as two putative catalytic residues,16 are localized in α5 and β2, respectively (Fig. 1a, b). The thiol group of Cys169 is hydrogen bonded to the N1 atom of the His204 imidazole ring, with a bond length of 3.1 Å. His204 forms another hydrogen bond via its N3 atom with the main chain carbonyl group of Leu226, 2.7 Å distant, bearing a noncanonical catalytic triad consisting of Cys-His-Leu (Fig. 1b). Mutating either Cys169 or His204 to alanine, or mutating Cys169 to serine, makes the enzyme almost inactive, suggesting the essential roles of Cys169 and His204 of VASH1 in the detyrosination of the α-tubulin peptide (Supplementary information, Fig. S2d–f, Table S1). Notably, all three residues are conserved in VASH1 and VASH2 (Fig. 1b), suggesting the conservation of the catalytic triad in vasohibins.
We also solved a 1.95-Å resolution structure of the VASH1 C169A70–306 bound to SVBP1–66 (Supplementary information, Table S2) and found it to be almost identical to that of the wild-type heterodimer, with a R.M.S.D of 0.28 Å over 259 Cα atoms (Supplementary information, Fig. S4). Therefore, the mutant enzyme loses the enzymatic activity due to the lack of the catalytic cysteine side-chain thiol nucleophile. A search within the Dali database reveals that VASH1 adopts a peptidase fold similar to those of LapG (PDB id: 4FGP, Z-core: 7.3, R.M.S.D: 4.1 Å for 186 Cα atoms) and AVRPPH3 (PDB id: 1UKF, Z-core: 6.1, R.M.S.D: 2.7 Å for 188 Cα atoms). Specifically, configurations of the catalytic-triad residues for VASH1 are similar between VASH1 and AVRPPH3, despite their low sequence identity (~12%) (Supplementary information, Fig. S5).
SVBP stabilizes VASH1 via extensive intermolecular interactions
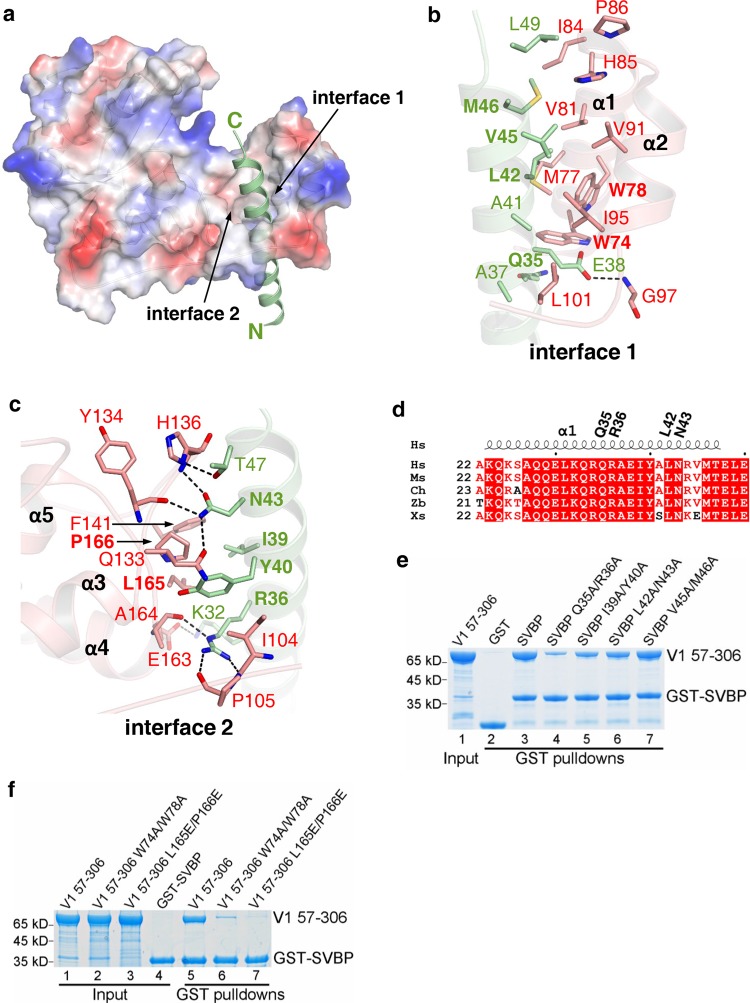
In the heterodimer structure, SVBP22–50 adopts an alpha-helical conformation and passes through a cavity formed by α1-α5 of VASH1, with its helix axis roughly parallel to that of α1 (Fig. 2a). Specifically, residues 32–49 of SVBP make intermolecular contacts along the helix axis with VASH1 via two faces. In one interface (interface 1), SVBP interacts with a region spanning α1-α2 of VASH1 primarily via hydrophobic interactions (Fig. 2b). Ala41, Leu42, Val45, Met46 and Leu49 of SVBP make hydrophobic interactions with Met77, Trp78, Val81, Ile84, His85, Pro86, Val91, and Ile95 of VASH1. Additionally, Ala37 and Ala41 of SVBP make hydrophobic interactions with Leu101 of VASH1 (Fig. 2b). The carboxyl group of SVBP Glu38 is hydrogen bonded to the main chain amide group of VASH1 Gly97 (Fig. 2b).
Fig. 2.
Detailed interactions between SVBP and VASH1. a The surface electrostatic potential of VASH170–306 bound to SVBP. Residues 22–50 of SVBP (green cartoon) are visible in the structure. b, c Residues 32–49 of SVBP interact with VASH1 via two interfaces. The VASH1 residues and SVBP residues that are involved in binding to each other are shown in red and green sticks, respectively. Mutated residues of SVBP and VASH1 are highlighted. d Sequence alignment of SVBP fragments (aa. 22–50) from different species, including homo sapiens (Hs, NP_955374.1), mouse (Ms, NP_077782.1), Gallus gallus (Ch, AADN04001018), Danio rerio (Zb, NP_001189361.1), and Xenopus laevis (Xs, XP_018083777.1). The secondary structures are labeled at the top of sequences. Gln35, Arg36, Leu42, and Asn43 of human SVBP are marked. e, f GST pull-down experiments, combined with mutagenesis experiments, to evaluate the roles of SVBP and VASH1 residues in forming the heterodimer. The levels of MBP-VASH157-306 in e, f are quantified in supplementary information, Fig. S6a, b, respectively
In the other interface (interface 2), SVBP interacts primarily via electrostatic interactions with the loop connecting α3 and α4 (L34) and with the loop connecting α4 and α5 (L45) (Fig. 2c). Lys32 of SVBP forms one salt bridge with the carboxyl group of VASH1 Glu163; Arg36 of SVBP forms three hydrogen bonds with the main chain amide groups of Ile104, Pro105, and Ala164 via its guanidino group; Asn43 of SVBP forms one salt bridge with VASH1 Gln133, and two main chain hydrogen bonds with Tyr134 and His136 of VASH1, respectively (Fig. 2c); the side chain hydroxyl group of SVBP Thr47 is hydrogen bonded to the N1 atom of the imidazole ring of VASH1 His136. In addition to above electrostatic interactions, Ile39 and Tyr40 of SVBP make hydrophobic contacts with Ile104, Phe141, Leu165, and Pro166 (Fig. 2c). Most of VASH1-binding residues in SVBP are conserved from Xenopus to human (Fig. 2d), suggesting the evolutionary conservation of the SVBP-VASH1 interactions.
To evaluate the roles of SVBP residues in binding to VASH1, we introduced different double mutations into SVBP and compared the binding affinities of these mutants to VASH1 with that of wild type by GST pull-down experiments (Fig. 2e). The binding experiments show that all mutants except SVBP V45A/M46A, display weaker VASH1 binding affinities. Specifically, Q35A/R36A greatly impairs the binding of VASH1, with the level of proteins pulled down dropping to ~30% (Supplementary information, Fig. S6a), while I39A/Y40A and L42A/N43A only weaken the binding moderately (40–60%, Supplementary information, Fig. S6a), suggesting the key role of SVBP Arg36 and VASH1 in maintaining intermolecular hydrogen-bonding interactions (Fig. 2c). Next, to evaluate the roles of VASH1 residues in binding to SVBP, we made some mutants containing double mutations and performed GST pull-down experiments to examine their SVBP-binding affinities. We found that W74A/W78A of VASH1 weakens the SVBP binding remarkably and L165E/P166E abolishes the binding (Fig. 2f; Supplementary information, Fig. S6b). All of above mutants display overall secondary structures similar to those of wild-type proteins, as indicated by the circular dichroism (CD) spectra (Supplementary information, Fig. S7). Taken together, mutagenesis and biochemical experiments further validate the SVBP-VASH1 interface.
The structure of SVBP-VASH1 bound to epoY
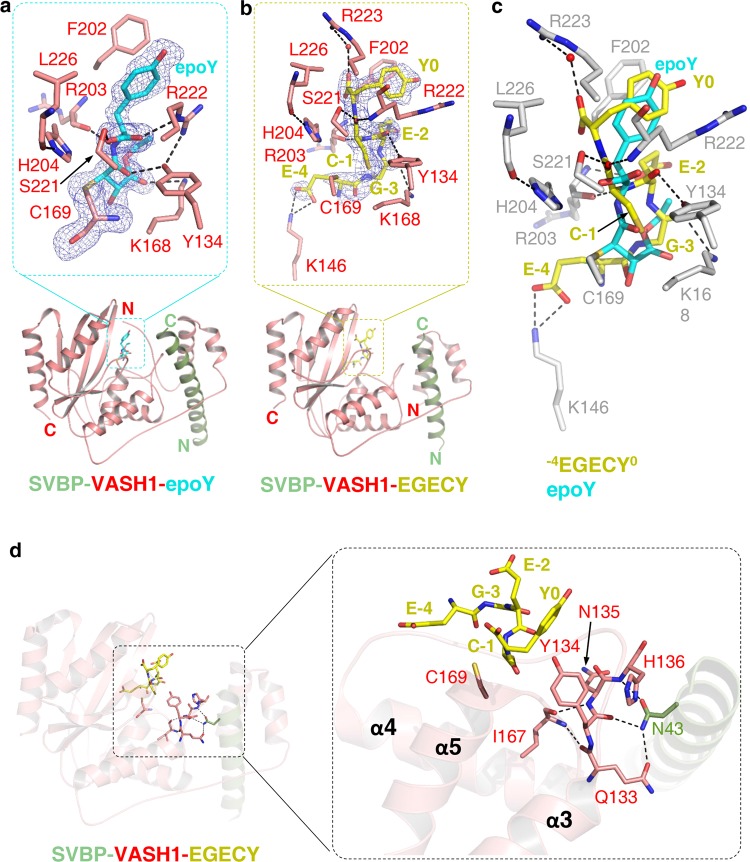
It has been reported that the activity of vasohibins is inhibited irreversibly by its potent inhibitor, epoY.14 To gain mechanistic insight into the inhibitory effect of epoY on VASH1, we solved the structure of SVBP1–52-VASH157–306 bound with epoY at a resolution of 1.62 Å. The R.M.S.D between the epoY-bound heterodimer and the apo one is 0.27 Å over 259 Cα atoms, suggesting that the SVBP-VASH1 conformation does not change remarkably upon binding to epoY.
In the ternary structure, epoY is positioned into an overall positively charged cleft (Fig. 3a; Supplementary information, Fig. S8a). The distance between the thiol group of VASH1 Cys169 and the O5 of epoY is 3.4 Å, which makes the structure represent a snapshot of the intermediate complex formed by VASH1 and α-tubulin, with the Tyr moiety of epoY mimicking the C-terminal Tyr of α-tubulin. Consistent with its role as an irreversible inhibitor, epoY masks Cys169 of VASH1 by forming a covalent bond with the thiol group of Cys169 via its C3 atom (Fig. 3a). Tyr of epoY is sandwiched between Phe202 and Arg222 of VASH1, making hydrophobic and cation–π interactions with Phe202 and Arg222, respectively (Fig. 3a). The two carbonyl groups of epoY Tyr form two hydrogen bonds with the side chain hydroxyl group of Ser221 and the guanidine group of Arg222, respectively. The side-chain conformation of Arg222 is stabilized by forming two hydrogen bonds with the terminal carbonyl group of epoY and the side chain hydroxyl group of Tyr134, respectively (Fig. 3a). Additional intermolecular hydrogen bonds are formed between Tyr134 and the O5 atom of epoY, between Arg203 and the main chain amide (N6) of epoY Tyr, and between Lys168 of VASH1 and the O20 atom of epoY (Fig. 3a; Supplementary information, Fig. S9a).
Fig. 3.
Molecular mechanism of tubulin detysosination by VASH1. a The structure of VASH157–306 (red) in complex with SVBP1–52 (green) and epoY (cyan). epoY and epoY-interacting residues of VASH1 are shown in sticks. The |Fo|–|Fc| difference electron density map of epoY is contoured at 3.0 σ and is shown as blue mesh with epoY deleted prior to one round of refinement. b The structure of VASH170–306 (red) in complex with SVBP3–49 (green) and −7GEEEGECY0 peptide (yellow). Peptide and peptide-interacting residues of VASH1 are shown in sticks. Only five residues of the peptide (−4EGECY0) are visible in the structure. The |Fo|–|Fc| difference electron density map of the peptide is contoured at 3.0 σ and is shown as blue mesh with the peptide deleted prior to one round of refinement. c Superposition of epoY-bound and −7GEEEGECY0-bound SVBP-VASH1 structures. epoY, −4EGECY0, and VASH1 residues involved in peptide binding are shown in cyan, yellow, and gray sticks, respectively. d Allosteric regulatory role of SVBP. SVBP, VASH1 and the peptide are colored in the same way as shown in b. The 133QYNH136 motif, Ile167 and Cys169 of VASH1, Asn43 of SVBP and peptide residues are shown in sticks.
The structure of SVBP-VASH1 bound to the peptide GEEEGECY
VASH1 was found to catalyze the detyrosination of a peptide derived from the C-terminus of TUBA1A, albeit with lower activity than it does on α-tubulin proteins in the context of MTs.15 Inspired by the finding that epoY is covalently bonded to Cys169, we synthesized a peptide (−7GEEEGECY0, numbered −7 to 0 hereafter) with its sequence derived from the C-terminus of TUBA1A (444GEEEGEEY451) except that Glu450 was replaced by cysteine. The mutant peptide could be catalyzed by SVBP3–49-VASH170–306, albeit with much lower catalytic efficiency (percentage of detyrosination: ~0.7%, Supplementary information, Table S1), suggesting that the peptide at least to some extent, mimics the substrate. We used the peptide for co-crystallization with the SVBP3–49-VASH170–306 heterodimer since the mutant peptide might be captured during crystallization before its detyrosination occurs.
Finally we succeeded in the co-crystallization of SVBP3-49-VASH170-306 with the −7GEEEGECY0 peptide and solved the structure at a 2.28 Å resolution (Fig. 3b; Supplementary information, Table S2). In the peptide-bound complex, the peptide occupies the cleft where epoY lies in, with its last five residues (−4EGECY0) visible in the density map (Fig. 3b; Supplementary information, Fig. S8b). Specifically, the distance between the thiol groups of Cys−1 and VASH1 Cys169 is 3 Å, suggesting that the thiol group of Cys169 is still available for catalyzing the peptide detyrosination. (Fig. 3b).
As expected, Tyr0 interacts with VASH1 in a way similar to that observed for Tyr of epoY, with its aromatic ring positioned between Phe202 and Arg222 and stacked with both residues; One of the main chain carbonyl group of Tyr0 forms one water-mediated hydrogen bond with the guanidino group of VASH1 Arg223 (Fig. 3b; Supplementary information, Fig. S9b). In addition to Tyr0, other peptide residues interact with VASH1 via intermolecular hydrogen-bonding interactions. The main chain carbonyl group of Cys−1 makes two hydrogen bonds with the side chain hydroxyl group of Ser221 and the main chain amide group of Arg222, respectively; the main chain amide group of Cys–1 and the side chain carboxyl group of Glu–2 are hydrogen bonded to the main chain carbonyl and amide groups of Arg203, respectively; the main chain carbonyl groups of Glu–2 and Gly–3 are hydrogen bonded to the side chain hydroxyl group of Tyr134 and the side chain of Lys168, respectively; Glu–4 forms salt bridges via its carboxyl groups with the side chain of Lys146 (Fig. 3b; Supplementary information, Fig. S9b).
The presented two ternary structures of SVBP-VASH1, one with epoY and the other with the −7GEEEGECY0 peptide, allow us to compare their binding modes in detail. After superimposing the two structures, we found that the two terminal tyrosines are partially overlapped (Fig. 3c). In addition, Cys−1 and Gly−3 of the peptide are fitted into the conformation of the inhibitor, while Glu−2 and Glu−4 of the peptide deviate from the inhibitor and make additional electrostatic interactions with the alkaline residues of VASH1 (Fig. 3c). Since mutant peptide is catalyzed by VASH1 with low efficiency, we could not exclude the possibility that mutant peptide might be accommodated differently than the wild-type sequence in the active site.
Next we decided to mutate the interface residues of VASH1, including Tyr134, Lys146, Ser221, and Arg222, to examine if those VASH1 residues involved in recognizing the mutant peptide also affect the detyrosination of wild-type peptide. We found by mass spectrometry that all five VASH1 mutants (Y134A, K146A, S221A, R222A, and K146A/R222A) displayed much weaker activity with the percentage of detyrosination ranging from 0.009% to 0.13%, Supplementary information, Fig. S2h–l, Table S1), suggesting the importance of those residues in peptide detyrosination.
The potential allosteric regulation of SVBP-VASH1
VASH1 was reported to require SVBP to achieve its full activiy,15 which prompted us to study how the binding of SVBP influences the activity of VASH1. We mutated the SVBP residues involved in binding to VASH1 and compared the activities of mutant heterodimers with that of the wild-type SVBP-VASH1 heterodimer by mass spectrometry. Strikingly, whereas Q35A/R36A, I39A/Y40A, and V45A/M46A of SVBP only slightly decrease the detyrosination activity of VASH1 (percentage of detyrosination: 20–29% versus 50%), L42A/N43A of SVBP greatly impairs the detyrosination activity of VASH1 (percentage of detyrosination: 1.6%) (Supplementary information, Fig. S10, Table S3), suggesting a regulatory role of L42 and/or N43 in VASH1-mediated detyrosination.
The severely impaired activity of VASH1 to bind with SVBP L42A/N43A mutant prompted us to analyze the structural roles of Leu42 and/or Asn43. In the peptide-bound structure, the side chain of SVBP Asn43 is hydrogen-bonded to the main chain groups of Tyr134 and His136 that reside in loop L34, and it also forms one salt bridge with the carboxyl group of VASH1 Gln133 (Fig. 3d). Loop L34 further interacts with loop L45 via two main chain hydrogen bonds, one between Gln133 and Ile167 and the other between Asn135 and Ile167. Above hydrogen-bonding interactions rigidify the 133QYNH136 motif, thereby positioning Tyr134 in a favorable position for efficient peptide detyrosination. Importance of VASH1 Tyr134 in peptide detyrosination is supported by the interaction between Tyr134 and the peptide (Fig. 3b), as well as evidenced by the impaired activity of Y134A mutant (Supplementary information, Table S1). It is noteworthy that 133QYNH136 is also conserved in VASH2 (Fig. 1a). Taken together, our structure analysis, supplemented by mass spectrometry and mutagenesis, suggests an allosteric role of SVBP in peptide detyrosination by vasohibins.
VASH1 and VASH2 equally contribute to tubulin detyrosination in human osteosarcoma cells
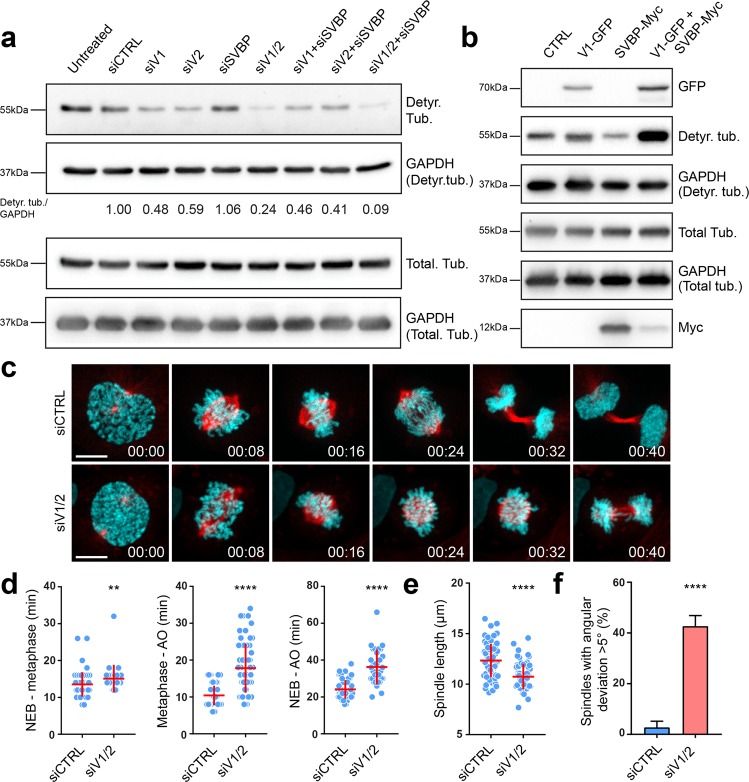
Since MT detyrosination was shown to be important for accurate chromosome segregation during cell division,25 we decided to further test the impact of vasohibins on mitosis, taking advantage of the new insights provided by the solved structure of SVBP-VASH1 complex. First, we tested the effect of RNAi-mediated depletion of vasohibins and SVBP on tubulin detyrosination levels in human osteosarcoma U2OS cells (Fig. 4a; Supplementary information, Fig. S11a–e). In contrast to an increase in tubulin detyrosination observed upon TTL knockdown (Supplementary information, Fig. S11d–f), co-depletion of VASH1 and VASH2 strongly decreased the level of tubulin detyrosination, which was clearly detected both by immunoblotting (Fig. 4a) and by immunofluorescence (Supplementary information, Fig. S11d, e). These data, together with a partial decrease in tubulin detyrosination obtained upon single VASH1 or VASH2 depletion (Fig. 4a), suggest redundant functions of VASH1 and VASH2 in U2OS cells. Overexpressing VASH1 and SVBP together additionally increased tubulin detyrosination compared to VASH1 overexpression alone (Fig. 4b), indicating that SVBP indeed stabilizes vasohibins and thus enhances their potency. However, we could not detect a strong decrease in tubulin detyrosination upon SVBP depletion (Fig. 4a; Supplementary information, Fig. S11a).
Fig. 4.
The impact of vasohibins and SVBP on tubulin detyrosination levels and mitosis. a Immunoblot analysis of detyrosinated tubulin levels upon RNAi-mediated knockdown of the components of vasohibin-SVBP complex. Protein extracts from U2OS cells transfected with target specific siRNAs against VASH1, VASH2 and SVBP were probed with α-tubulin antibody and an antibody that recognizes detyrosinated form of α-tubulin, with GAPDH serving as a loading control. Relative levels of detyrosinated α-tubulin normalized to GAPDH are indicated. b Effect of VASH1 and SVBP overexpression on detyrosinated tubulin levels. Immunoblot of cell lysates from U2OS cells transfected with plasmids expressing VASH1-GFP and/or SVBP-myc. The expression of VASH1 and SVBP were observed using anti-GFP and anti-myc antibodies, respectively. The levels of detyrosinated and total α-tubulin were assessed using the respective antibodies, with GAPDH serving as the loading control. c Spinning-disk confocal imaging of U2OS cells stably expressing H2B-GFP (cyan) and mCherry-α-tubulin (red) transfected with control and VASH1/2 siRNAs. Time in hour:min. Scale bar: 10 µm. d Duration of different phases of mitosis in control versus VASH1/2 siRNAs-treated cells. N (number of cells, number of independent experiments): NEB-metaphase—siCTRL (56, 3), siVASH1/2 (28, 5); Metaphase-AO—siCTRL (78, 3), siVASH1/2 (62, 5); NEB-AO—siCTRL (56, 3); siVASH1/2 (31, 5), NEB represents nuclear envelope breakdown, AO represents anaphase onset. e Quantification of the spindle length from live-cell imaging presented in c. N (number of cells, number of independent experiments): siCTRL (63, 3); siVASH1/2 (68, 5). f Percentage of cells with spindle positioning defect obtained from live-cell imaging presented in c. N (number of cells, number of independent experiments): siCTRL (88,3); siVASH1/2 (96, 5). P-values were calculated using Mann–Whitney U test. **p < 0.01, ****p < 0.0001
Vasohibins-mediated MT detyrosination regulates mitotic spindle length and positioning
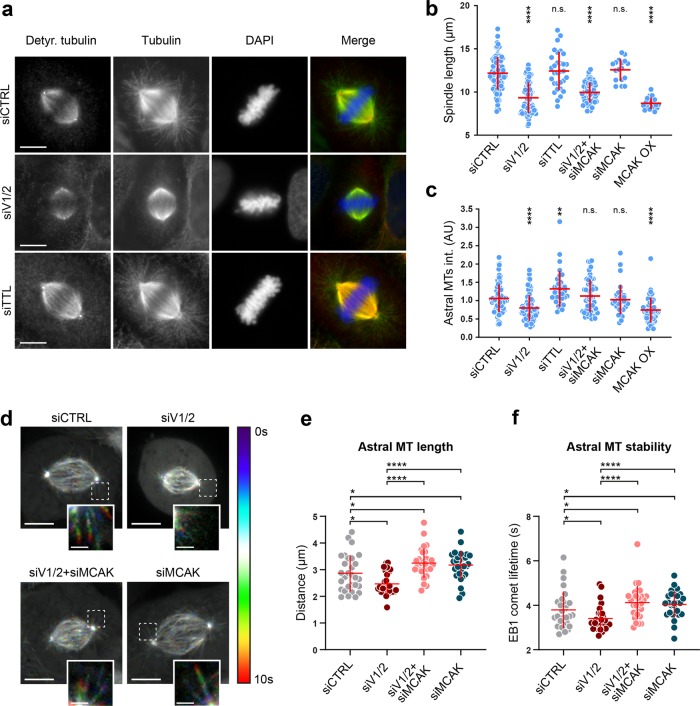
To test the potential role of vasohibins in mitosis, we used spinning-disk confocal live-cell imaging to monitor how vasohibins-depleted cells progress through mitosis. The cells with depleted vasohibins spent significantly longer time in mitosis (36.3 ± 9.5 min) compared to control cells (24.1 ± 5 min) (Fig. 4c, d). This delay was primarily a result of longer metaphase (17.8 ± 6.7 min in VASH1/2-depleted cells compared to 10.4 ± 2.6 min in controls), although the prometaphase was slightly prolonged as well (15.1 ± 3.8 in VASH1/2-depleted cells compared to 13.5 ± 3.3 in controls) (Fig. 4d). Strikingly, live-cell imaging also revealed a reduction in spindle size upon vasohibin depletion (10.7 ± 1.2 µm, compared to 12.3 ± 1.7 µm in controls) (Fig. 4e), as well as a strong increase in number of cells with tilted mitotic spindle in metaphase (42.7 ± 4.2% compared to 2.7 ± 2.5% in controls) (Fig. 4f), indicating that MT detyrosination plays an important role in accurate spindle positioning during cell division.
In order to investigate the impact of vasohibins on MT detyrosination within the mitotic spindle in more detail, we immunostained the mitotic spindles of U2OS cells using an antibody against detyrosinated tubulin. Compared to controls, mitotic cells that were co-depleted of VASH1 and VASH2 displayed reduced MT detyrosination along the mitotic spindles (Fig. 5a; Supplementary information, Fig. S12a, b). Notably, although VASH1/2 siRNAs showed a great efficiency in depletion of vasohibin protein levels (Supplementary information, Fig. S11b), as well as in downregulation of tubulin detyrosination in asynchronous cell population (Fig. 4a), significant MT detyrosination levels could still be detected in VASH1/2-depleted mitotic spindles (Fig. 5a; Supplementary information, Fig. S12a, b), further indicating the activity of other yet-unidentified TCP(s) in the cells. As expected, depletion of TTL led to an overall increase in detyrosination of spindle MTs (Fig. 5a; Supplementary information, Fig. S12b).
Fig. 5.
Vasohibins-mediated MT detyrosination regulates mitotic spindle length and positioning. a Representative images of mitotic spindles in control, VASH1/2- or TTL-depleted U2OS cells, immunostained with antibodies against detyrosinated and total α-tubulin. DNA was counterstained with DAPI. Detyrosinated tubulin in red, total α-tubulin in green, and DNA in blue in merged image. Scale bar: 10 µm. b Quantification of the spindle length from immunostained U2OS cells that were subjected to indicated treatments. c Astral MT intensities quantified as described in methods section. d Temporal color-coded projections of mitotic spindles of U2OS EB1-GFP cells treated with indicated siRNAs. Pronounced effect of the siRNA treatments on growing astral MTs shown in insets. Scale bar: 10 µm (Inset scale bar: 2 µm) e, f Astral MT length and stability quantified by manual tracking of EB1-GFP comet signal. N (number of cells, number of independent experiments): -spindle length: siCTRL (94, 6); siVASH1/2 (82, 5); siTTL (31, 2); siVASH1/2 + siMCAK (50, 3); siMCAK (32, 2); MCAK OX (49, 3); -astral MT intensity: siCTRL (106, 4); siVASH1/2 (73, 4); siTTL (33, 3); siVASH1/2 + siMCAK (51, 3); siMCAK (32, 2); MCAK OX (46, 3), -EB1-GFP comet tracking: siCTRL (29, 3); siVASH1/2 (27, 3); siVASH1/2 + siMCAK (29, 3); siMCAK (28, 3). P-values were calculated using Mann–Whitney U test. n.s. not significant, *p < 0.05, **p < 0.01, ****p < 0.0001
In addition to decreased MT detyrosination, our immunofluorescence experiments not only confirmed significant shortening of the spindles in VASH1/2-depleted cells (9.3 ± 1.8 µm, compared to 12.2 ± 1.9 µm in controls) (Fig. 5a, b; Supplementary information, Table S4), but also revealed a significant reduction in astral MTs that propagate from the spindle poles towards the cell cortex (Fig. 5a, c). TTL depletion-mediated increase in MT detyrosination did not have any effect on the spindle length (12.4 ± 2.2 µm, compared to 12.2 ± 1.9 µm in controls) (Fig. 5a, b; Supplementary information, Table S4).
Vasohibins-mediated MT detyrosination regulates spindle morphology and positioning via the spindle pole- and MT plus-end tips-based pool of MCAK
Observed reduction in the spindle size and number of astral MTs, as well as misoriented mitotic spindles, might be caused by dysregulated MT dynamics resulting from malfunctioning kinesin-13 protein family member MCAK44, which displayed increased MT depolymerizing activity on tyrosinated MTs.30,31 Thus, we examined whether the co-depletion of VASH1/2 and MCAK, could recover the spindle length and positioning, as well as astral MT population, which would rationalize the impact of VASH1/2 depletion on spindle morphology and positioning. MCAK co-depletion restored the tubulin detyrosination levels (Supplementary information, Figs. S12 and S13a), further implying the existence of additional TCPs that are able to detyrosinate MTs in the absence of vasohibins. Albeit vasohibins and MCAK co-depletion failed to fully recover the length of shorter spindles induced by depletion of vasohibins, we still observed a slight, but statistically significant spindle length recovery (10 ± 1.2 µm in VASH1/2 and MCAK co-depletion compared to 9.3 ± 1.8 µm in VASH1/2 depletion alone; p value = 0.0128) (Fig. 5b). Importantly, co-depletion of vasohibins and MCAK successfully recovered the amount of astral MTs in mitotic cells (Fig. 5c) and largely rescued the spindle misalignment phenotype (Supplementary information, Fig. S13b). In line with these results, when we transiently transfected the cells with mEmerald-MCAK, in order to overexpress MCAK and enhance its activity (Supplementary information, Fig. S13c), we observed shorter spindle lengths (8.7 ± 1.5 µm in MCAK OX compared to 12.2 ± 1.9 µm in controls) (Fig. 5b), as well as a reduction in amount of astral MTs (Fig. 5c; Supplementary information, Fig. S13c), which altogether mimicked the phenotype caused by vasohibin depletion.
Next, we focused on determining whether the observed cellular phenotypes caused by perturbations of vasohibins and MCAK could be explained by altered MT dynamics of the kinetochore-bound, interpolar or astral MTs. In order to answer that, we set up two experimental approaches. First, we measured MT turnover by photoactivation of mitotic spindles of U2OS cells stably expressing photoactivatable-GFP (PA-GFP) tubulin (Supplementary information, Fig. S14a). Neither single depletions, nor co-depletion of vasohibins and MCAK showed any significant effect on MT turnover of kinetochore- and interpolar (non-kinetochore) MTs in late prometaphase/metaphase compared to control cells (Supplementary information, Fig. S14b, c), excluding the possibility that shorter and tilted spindles were caused by a MT detyrosination-dependent increased activity of the centromere-based pool of MCAK. Next, we monitored MT dynamics of astral MTs in late prometaphase/metaphase using live-cell imaging of U2OS cells stably expressing EB1-GFP, as a marker for growing MT ends (Fig. 5d). In agreement with the immunofluorescence results, both the traversed distance of EB1-GFP comets, representing the astral MT length, as well as their half-life, representing the stability of astral MTs, were significantly reduced upon vasohibin depletion (Fig. 5e, f). Both MT length and stability were successfully recovered when MCAK was co-depleted along with vasohibins.
VASH1 catalytic triad-mutants and α-tubulin C-terminal tail-binding mutant failed to rescue spindle length and misorientation phenotypes caused by VASH1/2 depletion
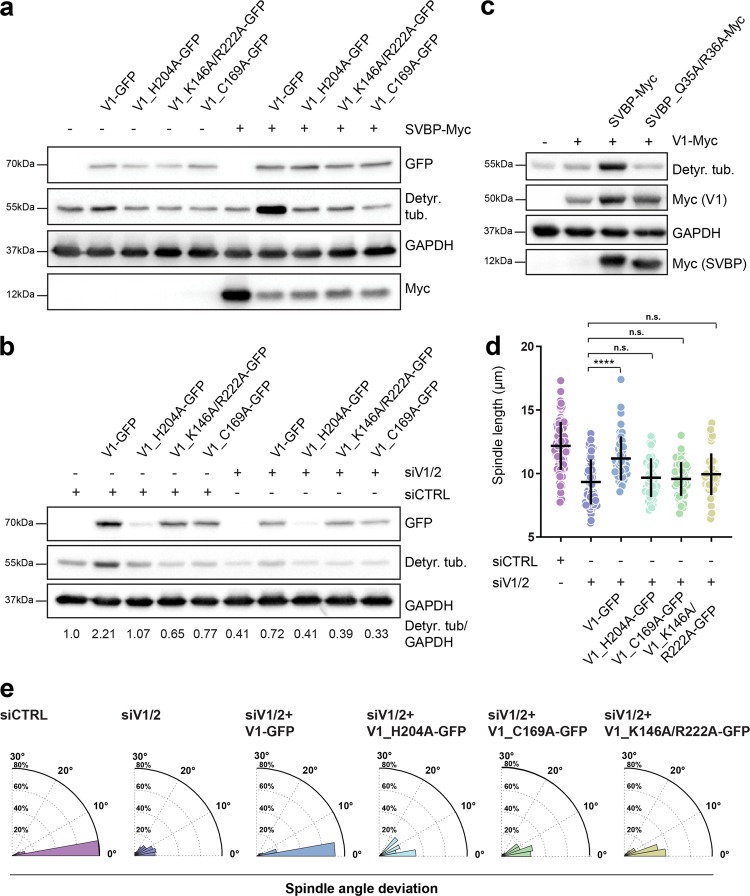
In order to further investigate the effect of vasohibin-SVBP protein complex on tubulin detyrosination, spindle morphology and positioning, we decided to assess the efficiency of the mutants linked to the structural features of SVBP-VASH1 complexes (Fig. 6). We generated three VASH1 mutants, two of which were mutated within the VASH1 catalytic triad (H204A and C169A) and one containing a double mutation that should affect the ability of VASH1 to efficiently bind the C-terminal tail of tubulin (K146A/R222A). While full-length wild-type VASH1 significantly increased tubulin detyrosination when it was overexpressed either alone or with SVBP, the mutants did not show any TCP activity even in the presence of SVBP (Fig. 6a). In addition, wild-type VASH1 successfully recovered VASH1/2 RNAi-induced reduction in tubulin detyrosination levels, whereas all three mutants failed to do so (Fig. 6b).
Fig. 6.
Analysis of the mutants linked to the structural features of SVBP-VASH1 complex. a Influence of VASH1 mutations on its activity in the presence and absence of SVBP. Immunoblot of lysates from cells expressing VASH1-GFP mutants with/without SVBP-myc. The activity of VASH1 was assessed with the antibody against detyrosinated tubulin pool. Expression levels of VASH1 and SVBP were observed using anti-GFP and anti-myc antibodies respectively with GAPDH as loading control. b Immunoblotting analysis of detyrosination levels of U2OS cells expressing GFP-tagged VASH1 mutants treated with control or VASH1/2 siRNAs. Expression of VASH1-GFP constructs was detected using anti-GFP antibody, with GAPDH serving as a loading control. Indicated are relative levels of detyrosinated α-tubulin normalized to GAPDH. c Effect of SVBP mutation on activity of SVBP-VASH1 complex was detected by immunoblotting. Levels of detyrosinated tubulin were analyzed in cells overexpressing VASH1-myc with or without overexpression of SVBP-myc constructs. Expression of VASH1 and SVBP constructs were monitored using an anti-myc antibody. d Quantification of the impact of VASH1 activity on preservation of proper spindle length in metaphase U2OS cells treated with control and VASH1/2 siRNAs in presence or absence of VASH1 constructs. N (number of cells, number of independent experiments): siCTRL (94, 6); siVASH1/2 (82, 5); V1 (49, 3); H204A (45, 3); C169A (45, 3); K146A/R222A (45, 3). P-values were calculated using Mann–Whitney U test. n.s.—not significant, ****p < 0.0001. e Polar distribution plots of spindle angle deviation of the same conditions as presented in d
Given that SVBP Q35A/R36A binds remarkably weaker to VASH1 (Fig. 2d), we introduced the double mutation and examined its effect on tubulin detyrosination in vivo. As predicted and in contrast to wild-type SVBP, the mutant failed to enhance the activity of VASH1, which was evidenced by the lack of strong increase in tubulin detyrosination levels (Fig. 6c), clearly endorsing the importance of these two residues for SVBP-VASH1 interaction, and indicating that the double mutation impairs the MT detyrosination by decreasing the amount of active SVBP-VASH1 heterodimer.
We further tested whether the VASH1 mutants could recover the effect of VASH1/2 depletion on spindle length and positioning. As it was the case with tubulin detyrosination levels, wild-type VASH1 was able to substantially recover the spindle length (Fig. 6d; Supplementary information, Table S4), and the spindle positioning in VASH1/2-depleted cells (Fig. 6e). In contrast, both catalytic-site mutants (H204A and C169A), as well as the tubulin-binding mutant (K146A/R222A) failed to recover either spindle length (Fig. 6d; Supplementary information, Table S4) or spindle misalignment phenotypes induced by vasohibin depletion (Fig. 6e).
Discussion
The identification of vasohibins as TCPs brought us a big step closer to completing the tyrosination/detyrosination cycle of α-tubulin. Vasohibins, together with TTL, regulate the dynamic tyrosination/detyrosination cycle of spindle MTs during mitosis.
Mechanism of tubulin recognition and detyrosination by VASH1
We identified that VASH170–306 and SVBP3–49 constitute the minimum region required for catalyzing the detyrosination of the α-tubulin peptide. By solving a series of crystal structures of SVBP-VASH1 that contain the minimum catalytic heterodimer, we reveal that VASH1 adopts a papain-like fold and further compare the catalytic pocket of VASH1 with that of AVRPPH3,45 a papain-like cysteine protease that displays the highest structural similarity to VASH1, as analyzed using the Dali database.
Superposition of the apo SVBP-VASH1 structure with that of AVRPPH3 clearly indicates that VASH1 and AVRPPH3 both adopt an open α/β fold and their catalytic residues overlap each other (Supplementary information, Fig. S5). Specifically, both peptidases possess a nucleophilic cysteine (Cys169 and Cys98, respectively) at the N-terminus of an α-helix and a histidine (His204 and His212, respectively) in a β-strand (Supplementary information, Fig. S5). Both Cys169 and His204 are critical for the activity of VASH1 since the histidine is proposed to increase the nucleophilicity of the Cys thiol group during catalysis.46
Despite above similarities, the two enzymes differ in several aspects: (i) AVRPPH3 has a canonical Cys-His-Asp catalytic triad, with its His212 hydrogen-bonded to the side chain carboxyl group of Asp227, whereas in VASH1, His204 is hydrogen-bonded to the main chain carbonyl group of Leu226 to generate a Cys-His-Leu triad (Fig. 1b). (ii) Asn93 of AVRPPH3 is the oxyanion hole residue to stabilize the intermediate, however, no such residue could be found in VASH1. Instead, Tyr134 occupies the corresponding position in VASH1 and forms an intramolecular hydrogen bond with Arg222 (Fig. 3a; Supplementary information, Fig. S5), thereby enhancing the cation–π stacking interaction between Arg222 and the tyrosine of epoY. (iii) AVRPPH3 does not require additional factors, whereas VASH1 requires SVBP for full activity and SVBP plays an allosteric role in peptide detyrosination (Fig. 3d; Supplementary information, Table S3).
Previous enzymatic experiments indicate that VASH1 catalyzes the detyrosination of an α-tubulin peptide only if a Tyr or a Phe is located at the C-terminus.15 By solving the inhibitor- and peptide-bound structures of SVBP-VASH1, we found some common features that might provide insights into substrate recognition. In both structures, the terminal tyrosine stacks with Arg222 of VASH1 via cation–π interactions and the terminal carboxyl group of Tyr is hydrogen-bonded to the guanidino group of an arginine (Fig. 3a–c). While replacing the Tyr with Ala would disrupt the stacking, the substitution of Phe for Tyr maintains the cation–π interaction, which well explains that VASH1 also proteolyzes the peptide with a Phe at its C-terminus.15 In addition to the tyrosine specific interaction, several alkaline residues of VASH1 recognize the acidic residues at C-terminus of α-tubulin via electrostatic interactions. Mutation of these VASH1 residues remarkably impairs the activity of VASH1.
Since vasohibins display higher activity toward tubulin molecules that have been already incorporated into MT, the fragments outside the core of the SVBP-VASH1 heterodimer might also be involved in recognizing full length α- or β-tubulin in the context of organized MT. In the peptide-bound structure, residues 70–306 of VASH1 constitute the minimum functional peptidase domain and the mutant peptide lies in a positively charged cleft of VASH1 (Supplementary information, Fig. S8b). The other surface of VASH1 outside of the cleft is still available for contacting with the globular region of α-tubulin. It is also possible that the N- and C-terminal flanking regions, including residues 1–69 and 307–365 of VASH1, respectively, and the N-terminal tail including residues 1–20 of SVBP might be required for contacting with the tubulin proteins in the assembled MT context given that vasohibins display higher activity towards MT tubulins.15 Further work is required to examine whether the SVBP-VASH1 dimer binds to MTs or soluble tubulin proteins with higher affinity.
Implication of vasohibin-mediated detyrosination in mitosis
Tubulin tyrosination-detyrosination cycle was shown to play an important role in various biological processes, such as neuronal differentiation, cardiomyocyte contraction and cell division.47 α-tubulin detyrosination by vasohibins might confer new ligand binding preference of MT for MT-associated proteins (MAPs). Therefore, we decided to apply the knowledge gained from the solved SVBP-VASH1 structures to the functional in vivo test of the SVBP-VASH1 complex, with an ultimate aim to further unveil the function of MT detyrosination in mitosis. RNAi-based depletion of vasohibins and SVBP in U2OS cells confirmed the role of VASH1/VASH2-SVBP complex in mediating microtubule detyrosination. Whereas the individual removals of VASH1 and VASH2 significantly reduced the levels of tubulin detyrosination, simultaneous knockdown of both proteins led to even stronger decrease (Fig. 4a). Although SVBP greatly improved the in vivo measured activity of VASH1, it did not seem to be essential for maintaining the activity of vasohibins-mediated MT detyrosiantion levels in U2OS cells (Fig. 4a, b). However, it is unclear whether SVBP-mediated stabilization of vasohibins is dispensable for its basal tubulin detyrosination activity in U2OS cells or it is associated to SVBP depletion efficiency.
The observed delay in cell division after depleting vasohibins-mediated detyrosination suggests that vasohibins play an important role in mediating cell cycle by impacting the chromosome segregation process (Fig. 4c). The impact of vasohibin depletion on chromosome congression during prometaphase that we presented here was milder than what was previously observed when MT detyrosination was depleted by either TTL-overexpression or using parthenolide,25 a drug known for its inhibitory effect on tubulin detyrosination.48 TTL overexpression- and parthenolide-mediated inhibition of accurate chromosome congression was explained by decreased affinity of CENP-E for tyrosinated MTs, which was demonstrated in in vitro motility assays.25 This milder effect on chromosome congression upon the depletion of vasohibins might be a consequence of the proven presence of additional, yet-unidentified, TCP(s) in the cells,15 the presence of which was also evident from the residual spindle detyrosination in vasohibins-depleted cells presented in this study. The largest effect of vasohibin depletion in mitotic cells was manifested by misoriented and shorter mitotic spindles, accompanied by a strong reduction of astral MTs, finally resulting in delayed chromosome segregation. The observed vasohibin depletion-induced phenotypes largely resembled the effects of over-expression of kinesin-13 protein family member MCAK (Fig. 5a–c; Supplementary information, Fig. S13b,c).
In line with earlier association between the activity of MCAK and (de)tyrosination status of MTs,30,31 and based on our data here, we propose that the MT depolymerase activity of MCAK43 is downregulated in mitosis by vasohibin-mediated MT detyrosination. We showed that co-depletion of MCAK together with vasohibins recovered the amount of astral MTs in mitosis and rescued the spindle misorientation phenotype (Fig. 5c; Supplementary information, Fig. S13b), uncovering MCAK as one of the main mediators of the observed mitotic phenotypes and an important reader of the tubulin code in mitosis. Furthermore, we demonstrated that neither the direct MCAK depletion, nor its potential indirect downregulation via vasohibin depletion-induced reduction of spindle detyrosination, affected the stability of either kinetochore-bound or interpolar MTs of late prometaphase/metaphase cells (Supplementary information, Fig. S14). However, the stability and growth of the astral MTs were reduced upon vasohibin depletion, both of which were successfully rescued by co-depleting MCAK (Fig. 5d–f). Together with our immunofluorescence results, this set of data strongly suggests that vasohibins-mediated MT detyrosination regulates spindle morphology and positioning via the spindle pole- and MT plus-end tips-based pool of MCAK,49 rather than via its centromere-localized fraction.50 Nevertheless, since MCAK co-depletion only partially rescued vasohibins depletion-induced shortening of the mitotic spindles (Fig. 5b), we cannot exclude the possibility that there are other mitotic proteins whose function depends on (de)tyrosination levels of MTs, and whose identity should be revealed by future research.
The presented binary and ternary complex structures uncover how the SVBP-VASH1 heterodimer recognizes the C-terminal tyrosine. The identified key residues of VASH1 involved in binding to SVBP or the α-tubulin peptide, also affect the detyrosination and function of spindle MTs in vivo, as evidenced by the detection of MT detyrosination levels, immunofluorescence-based analysis and live-cell imaging experiments (Fig. 6). Thus our study not only provides mechanistic insights into SVBP-VASH1 heterodimer-mediated detyrosination on α-tubulin, but it also reveals how the detyrosination of spindle MT impacts the spindle morphology and chromosome segregation during mitosis.
Tubulin detyrosination was recently revealed as a potent regulator of MT-dependent mechano-transduction in heart muscle, explaining the observed correlation between increased detyrosination and reduction in cardiac function in patients with hypertrophic cardiomyopathy.51 Experimental modification of the levels of MT detyrosination increased contractility in failing human cardiomyocytes, suggesting a new inotropic strategy for improving cardiac function.52 In addition to cardiomyocyte contraction,51–53 tubulin tyrosine cycle also plays an important role in nerve regeneration,54,55 and cell division.25 Finally, MT detyrosination was shown to be elevated in aggressive forms of neuroblastoma and breast cancers with poor prognosis,56,57 as well as in prostate cancer cells.58 Thus, design of the drugs that could modulate the activity of the enzymes involved will be of great benefit for treating health conditions, such as nerve damage, heart diseases and cancer. The solved structure of SVBP-VASH1 and its functional analysis presented here could be of great importance for achieving these goals.
Altogether, these data provide a structural insight into tubulin detyrosination by vasohibin-SVBP protein complex and reveal its previously overlooked role in the maintenance of mitotic spindle size and accurate spindle positioning during cell division, further emphasizing the importance of MT detyrosination in mitosis. This will provide a solid basis for future studies focusing on identification of the novel motor proteins and MAPs sensitive to MT (de)tyrosination levels, with the aim to elucidate the molecular mechanism underlying the impact of MT detyrosination on the properties of mitotic spindle.
Materials and methods
Cloning, mutation, protein expression and purification
Genes encoding full-length human SVBP and VASH1 were codon-optimized for expression in Escherichia coli (E. coli) and synthesized by Genscript. Genes corresponding to full length SVBP (SVBP1–66), truncated fragments of SVBP (SVBP3–49 and SVBP1–52) and VASH1 (VASH57–306 and VASH70–306) were amplified by polymerase chain reactions (PCR) using primers SVBP-F(ATAGGATCCATGGACCCGCCGGCGCGTAAG), SVBP-R(ATAAAGCTTTTACTCGCCCGGCGGCTG), SVBP1–52-R(ATAAAGCTTTTATTGCTGTTCCAGCTCGGTC), SVBP3–49-F (ATAGGATCCCCGCCGGCGCGTAAGGAG), SVBP3–49-R (ATAAAGCTTTTACAGCTCGGTCATAAC), VASH70–306-F(ATACATATGATGGATGAGGCGACCTGG), VASH57–306-F(ATACATATGGGTGTGCCGTTCTTTGTTAAC), and VASH70–306-R(ATACTCGAGTTAACCAATTTTCAGACG) (restriction sites are underlined), and cloned into upstream and downstream multiple cloning sites of a modified pCDFDuet-1 (Novagen), respectively. Plasmids were transformed into E. coli BL21 (DE3) and recombinant proteins were overexpressed at 16 °C for 20 h by adding IPTG to a final concentration of 0.4 mM. Cells were harvested at 3600 × g, 4 °C, and then were resuspended using a buffer containing 20 mM Tris-HCl (pH 7.8), 300 mM NaCl (suspension buffer) and were lysed by sonication. Lysate was centrifuged at 18000× g, 4 °C for 30 min and supernatant were collected. Recombinant SVBP/VASH1 complexes were purified by fast flow Ni-NTA column (GE healthcare) and eluted by 20 mM Tris-HCl (pH 7.8), 300 mM NaCl, 300 mM imidazole. Gel filtration and ion exchange experiments were employed for further purification. Gel filtration experiments were performed on a HiLoadTM 16/600 superdexTM 75 pg column (GE healthcare) with suspension buffer, the fraction containing SVBP/VASH1 complex was collected and dialyzed to ion exchange buffer I (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM DTT). Ion exchange experiments were performed on a HitrapTM SP HP (1 mL) column (GE healthcare) with ion exchange buffer I and ion exchange buffer II (20 mM Tris-HCl pH 7.4, 1 M NaCl, 1 mM DTT), fraction corresponding to the complex was collected and concentrated to 10 mg/mL with ion exchange buffer I. Seleno-Methionine (SeMet)-labeled protein was purified in the same way, except that cells were cultured in M9 medium supplied with 50mg/L Seleno-Methionine.
Site specific mutations were introduced using two reverse and complement primers containing the mutation codon as following: VASH1_Y134A-F (ATCCGTGAGCTGCAAGCTAACCACACCGGCACC), VASH1_Y134A-R (GGTGCCGGTGTGGTTAGCTTGCAGCTCACGGAT); VASH1_K146A-F(TTCTTTGAAATTAAGGCAAGCCGTCCGCTGACC), VASH1_K146A-R(GGTCAGCGGACGGCTTGCCTTAATTTCAAAGAA); VASH1_C169A-F(GCGCTGCCGATCAAGGCCCTGGAGGCGGTGATC), VASH1_C169A-R(GATCACCGCCTCCAGGGCCTTGATCGGCAGCGC); VASH1_H204A-F(GGCAACTATTTTCGTGCCATTGTGCTGGGTGTT), VASH1_H204A-R(AACACCCAGCACAATGGCACGAAAATAGTTGCC); VASH1_R222A-F (GCGCTGGGTATGAGCGCTCGTGAGGACCTGATG), VASH1_R222A-R(CATCAGGTCCTCACGAGCGCTCATACCCAGCGC). Mutated nucleotides are underlined. VASH1 mutants were co-expressed with SVBP3–49, and purified in the same way as wild-type proteins.
For GST pull-downs, VASH1, VASH1W74A/W78A, VASH1L165E/P166E were cloned into a modified RSFDuet-1 vector (Novagen) with an N-terminal His6-SUMO-MBP tag to get N-terminal MBP fusion proteins. The MBP-tagged proteins were purified in the same way as described above. Full-length SVBP and its mutants Q35A/R36A, I39A/Y40A, L42A/N43A, and V45A/M46A were cloned into the pGEX-6p-1 vector (GE Healthcare) to obtain N-terminal GST fusion proteins. The GST-tagged proteins were first purified using Glutathione Sepharose 4B beads (GE Healthcare) and further purified on a HiLoad 16/600 Superdex 200 size exclusion column. Primers used for mutagenesis are listed as follow: SVBP_Q35A/R36A-F(GAACTGAAACAACGTGCGGCTGCGGAGATCTACGCG), SVBP_Q35A/R36A-R(CGCGTAGATCTCCGCAGCCGCACGTTGTTTCAGTTC); SVBP_I39A/Y40A-F(CGTCAGCGTGCGGAGGCCGCCGCGCTGAACCGTGTT), SVBP_I39A/Y40A-R(AACACGGTTCAGCGCGGCGGCCTCCGCACGCTGACG); SVBP_L42A/N43A-F(GCGGAGATCTACGCGGCGGCCCGTGTTATGACCGAG), SVBP_L42A/N43A-R(CTCGGTCATAACACGGGCCGCCGCGTAGATCTCCGC); SVBP_V45A/M46A-F(TACGCGCTGAACCGTGCTGCGACCGAGCTGGAACAG), SVBP_V45A/M46A-R(CTGTTCCAGCTCGGTCGCAGCACGGTTCAGCGCGTA); VASH1_W74A/W78A-F(GGATGAGGCCACCGCGGAAAGGATGGCGAAACACG)
VASH1_W74A/W78A-R(CGTGTTTCGCCATCCTTTCCGCGGTGGCCTCATCC)
VASH1_L165E/P166E-F(ATGACCAAAGAGGCGGAGGAGATCAAGTGCCTGGAA), VASH1L165E/P166E-R(TTCCAGGCACTTGATCTCCTCCGCCTCTTTGGTCAT). Mutated nucleotides are underlined.
Crystallization, data collection and structure determination
All crystals were grown by using sitting drop vapor diffusion method at 18 °C. For crystallization, 1 μL sample was mixed with 1 µL crystallization buffer. SeMet-labeled SVBP3–49/VASH170–306 crystals were grown in buffer by mixing equal volume of protein complex and buffer containing 0.1 M sodium citrate tribasic dihydrate (pH 5.5), 16% polyethylene glycol (PEG) 8000, and 0.01 M Cadmium chloride hydrate. Apo SVBP3–49/VASH170–306 crystals were grown in the same condition. SVBP3–49/VASH1 C169A70–306 crystals were grown in buffer by mixing equal volume of protein complex and buffer containing 6% v/v Tacsimate (pH 6.0), 0.1 M MES monohydrate pH 6.0, 25% PEG 4000. For crystallization of SVBP/VASH1 with an peptide corresponding to E450C of TUBA1A (GEEEGECY), purified SVBP3–49/VASH170–306 was pre-incubated with the synthesized peptide (Genscript) at a molar ratio of 1:10 at 4 °C for 8 h, and crystals were grown in buffer by mixing equal volume of protein complex and buffer containing 0.1 M sodium citrate tribasic dihydrate pH 5.0, 18% PEG 20000. For crystallization of SVBP1–52/VASH157–306 with epoY, protein complex was incubated with the inhibitor epoY at a molar ratio of 1:5 at 4 °C for 30 min, crystals were grown in buffer by mixing equal volume of protein complex and buffer containing 20% PEG6000, 1 M LiCl, 0.1 M MES (pH 6.0). Before flash-freezing crystals in liquid nitrogen, all crystals were soaked shortly in cryo-protectant containing glycerol and precipitant to about 30% v/v.
The diffraction data were collected on beam line BL17U1, BL18U1, and BL19U1 at Shanghai Synchrotron Facility (SSRF).59 The statistic details about data collection were summarized in supplementary information, Table S2. Data sets were processed using HKL2000/3000,20,60 XDS.61 The initial model of SeMet SVBP-VASH1 was solved by CRANK2,62 built manually by COOT63 and refined by Phenix.64 Other structures were solved by Phenix by molecular replacement with the SeMet structure as the search model.
Glutathione S transferase pull down
For the pull-down of wild-type GST-SVBP and derived mutants Q35A/R36A, I39A/Y40A, L42A/N43A, and V45A/M46A with MBP-VASH1, 40 µL of Glutathione Sepharose beads (GE Healthcare) were suspended with 200 µL of binding buffer (20 mM Tris-HCl pH 7.5, 0.5 M NaCl), and 51.6 µg of GST-SVBP protein variants were added. Then 114 µg of MBP-VASH1c were added and incubated at 4 °C for 2 h. Beads were subsequently washed for three times with 1 mL of washing buffer (20 mM Tris-HCl pH 7.5, 0.5 M NaCl, and 0.5% Triton X-100) and then add 100 µL of 2× SDS-PAGE loading buffer. The GST pull-down of GST-SVBP with MBP-tagged VASH1, VASH1_W74A/W78A, and VASH1_L165E/P166E was performed in the same way.
Circular dichroism (CD)
Plasmids for expression of wild-type and mutant were prepared as described above. Proteins were purified as described above. Eluted proteins were dialyzed to buffer containing 20 mM NaH2PO4, 20 mM Na2HPO4 (pH 7.2), and then concentrated to 0.2 mg/mL. All CD experiments were performed on a ChirascanTM qCD spectrophotometer (Applied Photophysics Ltd.) over wave lengths ranging from 185 to 260 nm at 20 °C. Measurements were taken in a 1 mm path length quartz cuvette at a data pitch of 1 nm. The CD spectrum of the dialysis buffer was measured as control and subtracted. Two successive scans were recorded, and results were smoothed and averaged.
Isothermal titration calorimetry (ITC)
MBP-fused VASH157–306 protein and wild-type SVBP proteins and SVBP3–49 were purified using Ni-NTA column as described above, and further purified by gel-filtration using HiLoadTM 16/600 superdexTM 200 pg column and HiLoadTM 16/600 superdexTM 75 pg column (GE Healthcare), respectively. Phosphate-buffered saline (85 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) containing 1 mM EDTA was used for gel filtration and ITC experiments. ITC experiments were performed by titrating 2 μL of SVBP or SVBP3–49 (1 mM) into cell containing 50 μM MBP-VASH157–306 on MicroCal PEAQ ITC (Malvern Panalytical, UK) at 10 °C, with a spacing time of 120 s and a reference power of 10 μCal/s. Control experiments were performed by titrating SVBP or SVBP3–49 (1 mM) into buffer, and were subtracted during analysis, respectively. Binding isotherms were plotted, analyzed and fitted based on one-site binding model by MicroCal PEAQ-ITC Analysis software (Malvern Panalytical, UK). The dissociation constants were determined by two independent experiments (Mean ± SD).
Mass spectrometry
Detyrosination reactions were performed at 37 °C by mixing 4 µg synthesized peptides corresponding to carboxyl-terminus of alpha tubulin (Genscript) and 100 μg SVBP/VASH1 complexes into 50 µL buffer containing 20 mM Tris-HCl (pH 7.2), 50 mM NaCl, 1 mM DTT, for 3 h. Reactions were quenched at 75 °C for 10 min, and tubes were centrifuged at 15000 × g for 20 min. Supernatants were analyzed by LC-MS/MS. LC-MS/MS analyses were performed on QE-Plus (Thermo Scientific) connected to an Easy-nLC nanoflow HPLC (Proxeon Biosciences) with a self-packed 75 μm id × 15 cm C18 column in the data-dependent acquisition and positive ion mode at a flow rate of 300 nL/min. Spectra were manually inspected to confirm the fragment ions aligned with the assigned peptide sequences.
Cell culture, plasmids, RNAi and immuno blot analysis
Parental U2OS, U2OS cell lines stably expressing H2B-GFP and mCherry-α tubulin (gift from S. Geley, Innsbruck Medical University, Austria),65 photoactivatable GFP (PA-GFP) and mCherry-α tubulin (gift from R. Medema, NKI, Amsterdam, The Netherlands), and EB1-GFP66 (gift from P. Draber, IMG ASCR, Prague, Czech Republic) were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen) at 37 °C with 5% CO2.
Gateway compatible plasmids encoding human VASH1 (NM_014909.5) and SVBP (NM_199342.4) (kind gift from J. Nilsson, Center for Protein Research, Copenhagen, Denmark) were used to create expression vectors by LR recombination (Invitrogen) encoding C-terminal tags (3× myc or eGFP). Site specific mutations were introduced by overlap extension PCR using the following primers: VASH1_H204A-F – TCAGGGAACTACTTCCGCGCTATCGTGCTGGGGGTGAAC; VASH1_H204A-R – GTTCACCCCCAGCACGATAGCGCGGAAGTAGTTCCCTGA; VASH1_R222A-F – GGTGCGCTGGGCATGAGTGCGCGCGAGGACCTGATGTAC; VASH1_R222A-R – GTACATCAGGTCCTCGCGCGCACTCATGCCCAGCGCACC; VASH1_K146A-F – CAGTTCTTTGAAATTAAGGCCAGCAGGCCTCTGACAGGG; VASH1_K146A-R – CCCTGTCAGAGGCCTGCTGGCCTTAATTTCAAAGAACTG; SVBP_Q35A/R36A-F – CAGGAGCTCAAGCAGAGAGCTGCTGCAGAGATCTATGCCCTC; SVBP_Q35A/R36A-R – GAGGGCATAGATCTCTGCAGCAGCTCTCTGCTTGAGCTCCTG; SVBP_Y40A-F – CAGGAGCTCAAGCAGAGACAAAGAGCAGAGATCGCTGCCCTC AACAGAGTCATGACAGAA; SVBP_Y40A-R – TTCTGTCATGACTCTGTTGAGGGCAGCGATCTCTGCTCTTTGTCTCTGCTTGAGCTCCTG (Mutated nucleotides are underlined).
Plasmid encoding mEmerald-MCAK was used for MCAK expression (gift from Michael Davidson; Addgene plasmid #54161). For expression of the wild-type or mutant proteins, cells were transfected with 1–2 µg of the plasmids using GeneJuice (Merck) transfection reagent according to manufacturer’s protocol. For RNA interference (RNAi) experiments, cells were transfected at 30–50% confluency using Lipofectamine RNAiMAX (Thermo Fisher Scientific) with 50 nM of the following siRNAs for 24–72 h: VASH1-3′UTR (VASH1-A) 5′ CCAGACUAGGAUGCUUCUG 3′, VASH1-CDS (VASH1-B) 5′ CCAGACUAGGAUGCUUCUG 3′, VASH2-3’UTR (VASH2-A) 5′ GGAUAACCGUAACUGAAGU 3′, VASH2-3′UTR (VASH2-B) 5′ CCUUAUAGUGUAAUUCAGU 3′, SVBP-A 5′ CCUGCACGUAAAGAAAAAA 3′, SVBP-B 5′ GCAGAGAUCUAUGCCCUCA 3′, MCAK 5′ GAUCCAACGCAGUAAUGGU 3′, TTL 5’ GUGCACGUGAUCCAGAAAU 3′ Control siRNA 5′ UGGUUUACAUGUCGACUAA 3′. For combined knockdown of VASH1/2 VASH1-A and VASH2-A siRNAs were used.
Immunoblots were performed with the following primary antibodies: mouse anti-Myc (1:10000; Cell Signaling Technology 2276S), goat anti-GFP (1:1000; Rockland Immunochemicals Inc. 600-101-215), mouse anti α-tubulin (1:10000; Sigma-Aldrich T5168, B-5-1-2), rabbit anti-detyrosinated α-tubulin (1:40000; home-made), mouse anti-GAPDH (1:25000; Proteintech 60004-1-Ig), mouse anti-MCAK (1:1000; Abnova H00011004-M01), HRP-conjugated secondary antibodies (1: 10000; Jackson ImmunoResearch), and visualized using ECL system (Bio-Rad). Antibody against detyrosinated tubulin was generated at GenScript by immunizing rabbits with a short peptide (GEEEGEE) resembling the detyrosinated α-tubulin C-terminal tail similar to previously reported studies.24,67 The antibody was affinity purified using the same peptide and its specificity was further validated by ELISA as a part of the antibody generation process. Additionally, its specificity was tested by immunoblot and immunofluorescence analysis of cells treated with siVASH1/2 and siTTL.
Immunofluorescence microscopy
U2OS cells grown on glass coverslips were fixed by ice-cold methanol for 4 min at −20 °C and stained using the following primary antibodies: mouse anti-Myc (1:4000; Cell Signalling Technology 2276S), mouse anti α-tubulin (1:2000; Sigma-Aldrich T5168, B-5-1-2), rabbit anti-detyrosinated α-tubulin (1:200; Abcam ab48389 and 1:2000; home-made), rabbit anti-centrin (1:2000; provided by I. Cheeseman, Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA). Alexa Fluor 488, 568 and 647 (Invitrogen) were used as secondary antibodies (1:1000) and DAPI (1 µg/mL) as DNA counterstain. Images were acquired using Zeiss AxioObserver Z1 wide-field microscope (63× Plan-Apochromatic oil differential interference contrast objective lens, 1.4 NA) equipped with Metal halide arc lamp and a CoolSNAP HQ2 CCD camera operated by the Metamorph software (Molecular Devices, USA) unless stated otherwise.
For quantification of spindle length and orientation angles, the mitotic spindles were imaged using fifteen 0.5 µm thick Z-planes. The distance between the two spindle poles and number of z-slices between the peak intensities of centrin signal were measured using ImageJ (National Institute of Health, Bethesda, MD, USA) and used to quantify the three-dimensional distance between the spindle poles, as well as the corresponding shift in spindle orientation angle. Measurement of detyrosinated tubulin intensities at interphase and mitotic spindles were performed by quantifying the integrated densities from sum-projections of images along the z-plane and the intensities were normalized with total α-tubulin intensities. Astral MT intensities were quantified as (TI-SI-B)/B, where TI is the whole cell integrated density of total α-tubulin intensity, SI is integrated density of α-tubulin signal at the spindle and B is background signal. For comparison, the quantified intensities were normalized to the median of the control.
Live-cell imaging
Time-lapse imaging of U2OS-H2B-GFP/mCherry-α-tubulin cells and of U2OS EB1-GFP cells cultured in 35 mm glass-bottomed dishes (14 mm, No. 1.5, MatTek Corporation) was performed in a heated incubation chamber (37 °C) with controlled humidity and CO2 supply (5%), using a Plan-Apochromat DIC 63×/1.4NA oil objective mounted on an inverted Zeiss Axio Observer Z1 microscope (Marianas Imaging Workstation from Intelligent Imaging and Innovations Inc. (3i), Denver, CO, USA), equipped with a CSU-X1 spinning-disk confocal head (Yokogawa Corporation of America) and four laser lines (405 nm, 488 nm, 561 nm and 640 nm). Images were detected using an iXon Ultra 888 EM-CCD camera (Andor Technology). Fifteen 1 µm-separated z-planes were collected every 2 min for 2–3 h. The spindle length was quantified by measuring the three-dimensional distance between the spindle poles. For astral MT tracking, mitotic spindles of U2OS cells stably expressing plus-end tracking protein EB1 tagged with GFP were imaged every 500 ms for 2 min. About 5–10 EB1 comets per cell were tracked manually using ImageJ.
MT turnover was quantified using sum-projected kymographs68,69 of the decay of the corrected normalized photoactivated spindle intensities from late prometaphase/metaphase U2OS PA-GFP/mCherry-α tubulin cells. A transversal strip of PA-GFP tubulin was photoactivated over one half of a mitotic spindle with a 5 ms pulse by a 405 nm laser. Images were acquired every 5 s for 4 min over 3 z-slices of 1 µm thickness. Background-subtracted and normalized fluorescence of different cells was averaged at each time point and double exponential fitting was performed in GraphPad Prism, from which the half-lives were extracted. Background correction was performed by subtracting the background spindle signal intensity from the non-activated half-spindle. Photo-bleaching correction was done by normalization to the average curve obtained upon photo-conversion of taxol-treated spindles (1 µM).
Accession numbers
The coordinates and structure factors of SeMET SVBP1–66-VASH170–306, Apo-SVBP1–66/VASH170–306, SVBP3–49-VASH1_C169A70–306, SVBP3–49-VASH170–306 with the peptide GEEEGECY, and SVBP1–52-VASH157–306 with epoY have been deposited into Protein Data Bank (https://www.rcsb.org/) with accession number 6J91, 6J9H, 6J8N, 6J8F and 6J7B, respectively.
Supplementary information
Acknowledgements
We thank the staffs of Shanghai Synchrotron Radiation Facility at beam lines BL17U1, BL18U1, and BL19U1 for assistance in data collection and processing. This work was supported by the “Strategic Priority Research Program” of the Chinese Academy of Sciences (Grant No. XDB19000000) and also by National Natural Science Foundation of China Grants 31570737 and 31770806 (to C.X.), 31500601 (to S.L.), and 31501093 (to H.Y.) C.X. is also supported by the Major/Innovative Program of the Development Foundation of the Hefei Center for Physical Science and Technology (2018CXFX007) and the “Thousand Young Talent program”. H.Y. is supported by China Postdoctoral Science Foundation Grant (2015M5805470). The work was also supported by the Fundamental Research Funds for the Central Universities (WK2070080001). We would like to thank Martina Barisic for excellent technical support. We thank J. Nilsson, S. Geley, R. Medema, P. Draber and I. Cheeseman for sharing reagents and E. Vitiello for her help with the polar plots. We also thank Dr. Zhongliang Zhu for his assistance in processing the datasets of SeMet crystals.
Author contributions
S.L. and C.X. conceived the project; S.L. and N.W. performed structural biology and biochemical experiments with assistance from J.G., H.Y., G.W. and X.T.; G.R. and S.E. conducted the cell biology experiments; S.L., H.H., M.B. and C.X. wrote the manuscript and all authors contributed to editing the manuscript. H.H., M.B. and C.X. supervised the project.
Competing interests
The authors declare no competing interests.
Contributor Information
Hongda Huang, Email: huanghd@sustech.edu.cn.
Marin Barisic, Email: barisic@cancer.dk.
Chao Xu, Email: xuchaor@ustc.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0187-y.
References
- 1.Dogterom M, Koenderink GH. Actin-microtubule crosstalk in cell biology. Nat. Rev. Mol. Cell Biol. 2019;20:38–54. doi: 10.1038/s41580-018-0067-1. [DOI] [PubMed] [Google Scholar]
- 2.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat. Rev. Mol. Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 3.Brouhard GJ, Rice LM. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat. Rev. Mol. Cell Biol. 2018;19:451–463. doi: 10.1038/s41580-018-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 5.Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J. Cell Biol. 2014;206:461–472. doi: 10.1083/jcb.201406055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Y, Brady ST. Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol. 2015;25:125–136. doi: 10.1016/j.tcb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valenzuela P, et al. Nucleotide and corresponding amino acid sequences encoded by alpha and beta tubulin mRNAs. Nature. 1981;289:650–655. doi: 10.1038/289650a0. [DOI] [PubMed] [Google Scholar]
- 9.Argarana CE, Barra HS, Caputto R. Release of [14C]tyrosine from tubulinyl-[14C]tyrosine by brain extract Separation of a carboxypeptidase from tubulin-tyrosine ligase. Mol. Cell. Biochem. 1978;19:17–21. doi: 10.1007/BF00231230. [DOI] [PubMed] [Google Scholar]
- 10.Arce CA, Hallak ME, Rodriguez JA, Barra HS, Caputto R. Capability of tubulin and microtubules to incorporate and to release tyrosine and phenylalanine and the effect of the incorporation of these amino acids on tubulin assembly. J. Neurochem. 1978;31:205–210. doi: 10.1111/j.1471-4159.1978.tb12449.x. [DOI] [PubMed] [Google Scholar]
- 11.Arce CA, Rodriguez JA, Barra HS, Caputo R. Incorporation of L-tyrosine, L-phenylalanine and L-3,4-dihydroxyphenylalanine as single units into rat brain tubulin. Eur. J. Biochem. 1975;59:145–149. doi: 10.1111/j.1432-1033.1975.tb02435.x. [DOI] [PubMed] [Google Scholar]
- 12.Ersfeld K, et al. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 1993;120:725–732. doi: 10.1083/jcb.120.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raybin D, Flavin M. An enzyme tyrosylating alpha-tubulin and its role in microtubule assembly. Biochem. Biophys. Res. Commun. 1975;65:1088–1095. doi: 10.1016/S0006-291X(75)80497-9. [DOI] [PubMed] [Google Scholar]
- 14.Aillaud C, et al. Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science. 2017;358:1448–1453. doi: 10.1126/science.aao4165. [DOI] [PubMed] [Google Scholar]
- 15.Nieuwenhuis J, et al. Vasohibins encode tubulin detyrosinating activity. Science. 2017;358:1453–1456. doi: 10.1126/science.aao5676. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Pulido L, Ponting CP. Vasohibins: new transglutaminase-like cysteine proteases possessing a non-canonical Cys-His-Ser catalytic triad. Bioinformatics. 2016;32:1441–1445. doi: 10.1093/bioinformatics/btv761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosaka T, et al. Vasohibin-1 expression in endothelium of tumor blood vessels regulates angiogenesis. Am. J. Pathol. 2009;175:430–439. doi: 10.2353/ajpath.2009.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi Y, et al. The angiogenesis regulator vasohibin-1 inhibits ovarian cancer growth and peritoneal dissemination and prolongs host survival. Int. J. Oncol. 2015;47:2057–2063. doi: 10.3892/ijo.2015.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki Y, et al. Isolation of a small vasohibin-binding protein (SVBP) and its role in vasohibin secretion. J. Cell Sci. 2010;123:3094–3101. doi: 10.1242/jcs.067538. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Szyk A, Deaconescu AM, Piszczek G, Roll-Mecak A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 2011;18:1250–1258. doi: 10.1038/nsmb.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar N, Flavin M. Preferential action of a brain detyrosinolating carboxypeptidase on polymerized tubulin. J. Biol. Chem. 1981;256:7678–7686. [PubMed] [Google Scholar]
- 23.Gundersen GG, Bulinski JC. Distribution of tyrosinated and nontyrosinated alpha-tubulin during mitosis. J. Cell Biol. 1986;102:1118–1126. doi: 10.1083/jcb.102.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38:779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- 25.Barisic M, et al. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science. 2015;348:799–803. doi: 10.1126/science.aaa5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenney RJ, Huynh W, Vale RD, Sirajuddin M. Tyrosination of alpha-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J. 2016;35:1175–1185. doi: 10.15252/embj.201593071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nirschl JJ, Magiera MM, Lazarus JE, Janke C, Holzbaur EL. alpha-tubulin tyrosination and CLIP-170 phosphorylation regulate the initiation of dynein-driven transport in neurons. Cell Rep. 2016;14:2637–2652. doi: 10.1016/j.celrep.2016.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peris L, et al. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 2006;174:839–849. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barisic M, Maiato H. The tubulin code: a navigation system for chromosomes during mitosis. Trends Cell Biol. 2016;26:766–775. doi: 10.1016/j.tcb.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peris L, et al. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 2009;185:1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z, et al. Fidgetin regulates cultured astrocyte migration by severing tyrosinated microtubules at the leading edge. Mol. Biol. Cell. 2017;28:545–553. doi: 10.1091/mbc.e16-09-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 35.Bonnet C, et al. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J. Biol. Chem. 2001;276:12839–12848. doi: 10.1074/jbc.M011380200. [DOI] [PubMed] [Google Scholar]
- 36.Boucher D, Larcher JC, Gros F, Denoulet P. Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry. 1994;33:12471–12477. doi: 10.1021/bi00207a014. [DOI] [PubMed] [Google Scholar]
- 37.Ikegami K, et al. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J. Biol. Chem. 2006;281:30707–30716. doi: 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lacroix B, et al. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J. Cell Biol. 2010;189:945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larcher JC, Boucher D, Lazereg S, Gros F, Denoulet P. Interaction of kinesin motor domains with alpha- and beta-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J. Biol. Chem. 1996;271:22117–22124. doi: 10.1074/jbc.271.36.22117. [DOI] [PubMed] [Google Scholar]
- 40.Valenstein ML, Roll-Mecak A. Graded control of microtubule severing by tubulin glutamylation. Cell. 2016;164:911–921. doi: 10.1016/j.cell.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn S, et al. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J. Cell. Sci. 2008;121:1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- 42.Kaul N, Soppina V, Verhey KJ. Effects of alpha-tubulin K40 acetylation and detyrosination on kinesin-1 motility in a purified system. Biophy. J. 2014;106:2636–2643. doi: 10.1016/j.bpj.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/S0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 44.Bakhoum, S. F., Thompson, S. L., Manning, A. L. & Compton, D. A. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol.11, 27-35 (2009). [DOI] [PMC free article] [PubMed]
- 45.Zhu M, Shao F, Innes RW, Dixon JE, Xu Z. The crystal structure of pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site. Proc. Natl Acad. Sci. USA. 2004;101:302–307. doi: 10.1073/pnas.2036536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verma S, Dixit R, Pandey KC. Cysteine proteases: modes of activation and future prospects as pharmacological targets. Front. Pharmacol. 2016;7:107. doi: 10.3389/fphar.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nieuwenhuis Joppe, Brummelkamp Thijn R. The Tubulin Detyrosination Cycle: Function and Enzymes. Trends in Cell Biology. 2019;29(1):80–92. doi: 10.1016/j.tcb.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Fonrose X, et al. Parthenolide inhibits tubulin carboxypeptidase activity. Cancer Res. 2007;67:3371–3378. doi: 10.1158/0008-5472.CAN-06-3732. [DOI] [PubMed] [Google Scholar]
- 49.Moore AT, et al. MCAK associates with the tips of polymerizing microtubules. J. Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robison P, et al. Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science. 2016;352:aaf0659. doi: 10.1126/science.aaf0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen CY, et al. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat. Med. 2018;24:1225–1233. doi: 10.1038/s41591-018-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerr JP, et al. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat. Commun. 2015;6:8526. doi: 10.1038/ncomms9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gobrecht P, et al. Promotion of functional nerve regeneration by inhibition of microtubule detyrosination. J. Neurosci. 2016;36:3890–3902. doi: 10.1523/JNEUROSCI.4486-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song W, Cho Y, Watt D, Cavalli V. Tubulin-tyrosine ligase (TTL)-mediatedIncrease in tyrosinated alpha-tubulin in injured axons is required for retrograde injury signaling and axon regeneration. J. Biol. Chem. 2015;290:14765–14775. doi: 10.1074/jbc.M114.622753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kato C, et al. Low expression of human tubulin tyrosine ligase and suppressed tubulin tyrosination/detyrosination cycle are associated with impaired neuronal differentiation in neuroblastomas with poor prognosis. Int. J. Cancer. 2004;112:365–375. doi: 10.1002/ijc.20431. [DOI] [PubMed] [Google Scholar]
- 57.Mialhe A, et al. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 2001;61:5024–5027. [PubMed] [Google Scholar]
- 58.Soucek K, et al. Normal and prostate cancer cells display distinct molecular profiles of alpha-tubulin posttranslational modifications. Prostate. 2006;66:954–965. doi: 10.1002/pros.20416. [DOI] [PubMed] [Google Scholar]
- 59.Wang Q. S. et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Technol. 29, 68 (2018).
- 60.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 61.Kabsch W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pannu NS, et al. Recent advances in the CRANK software suite for experimental phasing. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:331–337. doi: 10.1107/S0907444910052224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 65.Barisic M, et al. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell. 2010;21:1968–1981. doi: 10.1091/mbc.e09-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vinopal S, et al. gamma-Tubulin 2 nucleates microtubules and is downregulated in mouse early embryogenesis. PLoS One. 2012;7:e29919. doi: 10.1371/journal.pone.0029919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paturle-Lafanechere L, et al. Accumulation of delta 2-tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. J. Cell Sci. 1994;107(Pt 6):1529–1543. doi: 10.1242/jcs.107.6.1529. [DOI] [PubMed] [Google Scholar]
- 68.Pereira AJ, Maiato H. Improved kymography tools and its applications to mitosis. Methods. 2010;51:214–219. doi: 10.1016/j.ymeth.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 69.Ferreira LT, Figueiredo AC, Orr B, Lopes D, Maiato H. Dissecting the role of the tubulin code in mitosis. Methods Cell Biol. 2018;144:33–74. doi: 10.1016/bs.mcb.2018.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laskowski RA, et al. PDBsum: a web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci. 1997;22:488–490. doi: 10.1016/S0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.