Supplemental Digital Content is available in the text.
Keywords: arteries, chromatin immunoprecipitation, cytokines, epigenetics, histone, inflammation
Objective:
Vascular inflammation underlies cardiovascular disease. Vascular smooth muscle cells (VSMCs) upregulate selective genes, including MMPs (matrix metalloproteinases) and proinflammatory cytokines upon local inflammation, which directly contribute to vascular disease and adverse clinical outcome. Identification of factors controlling VSMC responses to inflammation is therefore of considerable therapeutic importance. Here, we determine the role of Histone H3 lysine 9 di-methylation (H3K9me2), a repressive epigenetic mark that is reduced in atherosclerotic lesions, in regulating the VSMC inflammatory response.
Approach and Results:
We used VSMC-lineage tracing to reveal reduced H3K9me2 levels in VSMCs of arteries after injury and in atherosclerotic lesions compared with control vessels. Intriguingly, chromatin immunoprecipitation showed H3K9me2 enrichment at a subset of inflammation-responsive gene promoters, including MMP3, MMP9, MMP12, and IL6, in mouse and human VSMCs. Inhibition of G9A/GLP (G9A-like protein), the primary enzymes responsible for H3K9me2, significantly potentiated inflammation-induced gene induction in vitro and in vivo without altering NFκB (nuclear factor kappa-light-chain-enhancer of activated B cell) and MAPK (mitogen-activated protein kinase) signaling. Rather, reduced G9A/GLP activity enhanced inflammation-induced binding of transcription factors NFκB-p65 and cJUN to H3K9me2 target gene promoters MMP3 and IL6. Taken together, these results suggest that promoter-associated H3K9me2 directly attenuates the induction of target genes in response to inflammation in human VSMCs.
Conclusions:
This study implicates H3K9me2 in regulating the proinflammatory VSMC phenotype. Our findings suggest that reduced H3K9me2 in disease enhance binding of NFκB and AP-1 (activator protein-1) transcription factors at specific inflammation-responsive genes to augment proinflammatory stimuli in VSMC. Therefore, H3K9me2-regulation could be targeted clinically to limit expression of MMPs and IL6, which are induced in vascular disease.
Highlights.
The repressive epigenetic mark Histone H3 Lysine 9 Dimethylation (H3K9me2) is reduced in vascular smooth muscle cells on inflammation in cardiovascular disease models.
Loss of H3K9me2 in human and mouse vascular smooth muscle cells exacerbates inflammation-induced upregulation of disease-associated genes in vitro and in vivo, with functional consequences for matrix degradation and phenotypic switching.
H3K9me2 inhibits binding of NFκB (nuclear factor kappa-light-chain-enhancer of activated B cell) and AP-1 (activator protein-1) transcription factors to their cognate sequences within MMP (matrix metalloproteinase) and cytokine gene promoters.
See accompanying editorial on page 2199
Vascular smooth muscle cell (VSMC) accumulation is a hallmark of cardiovascular disease (CVD), including atherosclerosis and vascular remodeling after injury. In response to proinflammatory stimuli, VSMCs downregulate contractile proteins and upregulate selective gene sets, including matrix metalloproteinases, and proinflammatory cytokines (eg, IL [interleukin]-6), which directly contribute to the progression of vascular disease.1,2 Inflammation-responsive gene expression is controlled by the NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells) and AP-1 (activator protein 1) transcription factors, and dysregulation of NFκB and MAPK (mitogen-activated protein kinase)/AP-1 signaling is linked to the initiation and progression of vascular dysfunction.3 Indeed, directly reducing inflammation with Canakinumab, an IL1-β neutralizing antibody, significantly lowered cardiovascular events compared with placebo in patients4 although not in mice.5 Identification of mechanisms that regulate inflammation-associated changes in VSMC gene expression is therefore of considerable therapeutic importance.
MMPs (matrix metalloproteinases) comprise a family of proteases that degrade extracellular matrix components and mediate many structural changes associated with vascular disease.6,7 For example, animal atherosclerosis studies implicate MMP12 in plaque development and instability.8,9 MMP3 and MMP9 promote VSMC migration and neointima formation after carotid ligation in mice.10 In addition, increased MMP3, MMP9, and MMP12 activity is observed in vulnerable compared with stable human atherosclerotic lesions.11–14 The proinflammatory cytokine IL-6 also plays an important role in vascular disease progression. IL-6 attenuates VSMC contractility while promoting VSMC migration, proliferation, and vascular calcification.15–17 Furthermore, elevated serum IL-6 is associated with greater CVD risk.18–20
Gene expression is regulated by epigenetic pathways that modify histone proteins, resulting in either increased or reduced accessibility to the transcription machinery.21 Decreased levels of histone H3 lysine 9 di-methylation (H3K9me2) and histone H3 lysine 27 tri-methylation (H3K27me3) are observed in VSMCs derived from human atherosclerotic plaques compared with control arteries.22 However, how these mechanisms control inflammation-associated changes in VSMC gene expression are unclear. Interestingly, H3K9me2, a repressive mark associated with facultative heterochromatin23 and directly regulating the expression of inducible genes in other cells,24–28 affects the growth rate, migration, and contractility of pulmonary and airway smooth muscle cells.29,30 Yet, the functional importance and target genes of H3K9me2 in VSMCs remain largely unknown.
We demonstrate that global levels of H3K9me2 in VSMCs are reduced in atherosclerosis and in arteries undergoing injury-induced remodeling, concomitant with increased inflammation. H3K9me2 is deposited at a subset of inflammation-responsive VSMC genes, including MMP3, MMP9, MMP12, and IL6, which are strongly associated with CVD.11–14 Importantly, we show that H3K9me2 attenuates inflammation-associated upregulation of target genes by inhibiting NFκB/p65 and/or AP-1/cJUN transcription factor binding. These findings identify functional consequences of loss of H3K9me2 in vascular disease and suggest H3K9me2 as an important mechanism to prevent spurious induction of a proinflammatory state in VSMCs.
Methods
Disclosure Statement
The chromatin immunoprecipitation (ChIP)-seq data have been made publicly available at the Gene Expression Omnibus with the accession number GSE131212. All other supporting data are available within the article and in the online-only Data Supplement.
Animal Experiments
All experiments were approved by the United Kingdom Home Office (PPL70/7565 and P452C9545) and the local ethics committee. Wild-type and Apolipoprotein E-null (Apoe−/−) mice on a C57Bl/6 background were purchased from Charles River. Myh11-CreERt2 (Y-linked),31 Rosa26-Confetti, Rosa26-EYFP (enhanced yellow fluorescent protein), and Apoe−/− mice have been described previously.31,32 MYH11 is a marker of differentiated smooth muscle cells and the Myh11-CreERt2 transgene used here has been extensively tested for VSMC-specific expression in major arteries.31,33–36 Experimental animals (all males as the Myh11-CreERt2 transgene is Y-linked) received 10 intraperitoneal injections of 1 mg/mL tamoxifen between 4 and 8 weeks of age for lineage labeling. To inhibit G9A/GLP (G9A-like protein), lineage-labeled Myh11-CreERt2/Rosa26-EYFP+ males were treated with A366 delivered by osmotic pumps (30 mg/kg per day in 98:2 PEG 400/polysorbate 80), or vehicle alone, and either analyzed directly or subjected to ligation of the left common carotid artery as described previously.34 Surgery and tissue processing are described in the Methods in the online-only Data Supplement.
ChIP and Analysis
ChIP was performed as described in the Methods in the online-only Data Supplement. For genome-wide analysis, the DNA SMART ChIP-seq kit (Clontech, 634865) was used to generate Illumina-compatible sequencing libraries from 100 pg to 2 ng of DNA from 2 independent H3K9me2 ChIP experiments and associated input. Libraries were sequenced (Illumina NextSeq) using paired-end 75 bp reads. ChIP-seq reads were trimmed using Cutadapt v1.9,37 aligned to the mouse GRCm38 genome using Bowtie2 v2.2,38 and reads per gene promoter (within ±1 kb of the transcription start sites) quantified using SeqMonk v1.4 (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk). Genes showing fewer than 20 read counts in the input samples were removed from further analysis and the ratio of H3K9me2/input signal was computed. Of genes with H3K9me2/input ratios in the top 25th percentile, 63 genes were associated with arteriosclerosis according to the Cardiovascular Disease Portal39 (https://rgd.mcw.edu/rgdCuration/?module=portal&func=show&name=cardio).
VSMC Culture
Human aortic VSMCs (hVSMCs) were isolated as described40 from patients undergoing aortic valve replacement with ethics committee approval and used at passage 6 to 15. Primary mouse aortic VSMCs were derived from 8- to 12-week-old wild-type C57Bl/6 males and used at passage 4. VSMCs were treated with UNC0638 (UNC; 1 μM Tocris), small interfering RNA targeting G9A (sc-43777, Santa Cruz), control small interfering RNA (sc-37007, Santa Cruz), SP600125 (10 μM, Abcam), human recombinant IL-1α (2 ng/mL, Peprotech), and human recombinant TNF-α (tumor necrosis factor-α), 90 ng/mL, Peprotech as indicated and analyzed as described in the Methods in the online-only Data Supplement.
Statistics
Data are shown as mean±SEM, unless otherwise indicated. The number of animals per group and number of biologically independent experiments are indicated in Figure legends (at least 3). Independent in vitro experiment were done using isolates derived from different individuals for hVSMCs and independent primary cultures for murine VSMCs. Data were analyzed using Mann-Whitney U test, Kruskal-Wallis 1-way ANOVA with Dunn test to compare specific sample pairs, 1-sided exact Wilcoxon rank-sum tests or a linear model (described in the online-only Data Supplement) with P<0.05 as the threshold for considering results to be statistically significant.
Results
H3K9me2 Is Reduced Within VSMCs in Atherosclerosis and Arterial Remodeling, Concomitant With Inflammation
VSMCs downregulate lineage markers in atherosclerosis and after injury, making specific quantification of H3K9me2 in VSMCs difficult. We therefore used genetic lineage tracing to definitively identify VSMCs by crossing Myh11-CreERt2+ mice31 with recombination reporter alleles (Rosa26-Confetti32 or Rosa26-EYFP33). Tamoxifen treatment of these animals induces VSMC-specific recombination and stable fluorescent protein expression in 70% to 95% of VSMCs in Confetti34 and 40% to 60% in EYFP reporter mice.33,35 Importantly, the fluorescent lineage reporters are stably expressed, independent of the expression status of the Myh11-CreERt2 transgene, in medial VSMCs and in VSMC-derived neo-intimal and plaque cells.34
We first crossed Myh11-CreERt2/Rosa26-Confetti mice with Apoe−/− mice to assess H3K9me2 levels in atherosclerosis. Male mice received tamoxifen and were then fed an atherosclerosis-inducing high-fat diet (21% fat, 0.2% cholesterol) or a standard chow diet. Immunohistochemistry in carotid artery sections of animals after 3 to 4 months of high-fat diet revealed significantly lower H3K9me2 levels in Confetti+ cells within the media, core, and fibrous cap in atherosclerotic arteries compared with cells in control vessels (Figure 1A). Western blot analysis also indicated reduced H3K9me2 expression in aortas of mice fed a high-fat diet for 6 months compared with control aortas that had little or no visible plaque, and this was associated with increased levels of phosphorylated NFκB p65 (Figure 1B). This indicates that the previously reported reduced H3K9me2 levels in human atherosclerotic plaques compared with healthy controls22 is because of downregulation in VSMCs.
Figure 1.
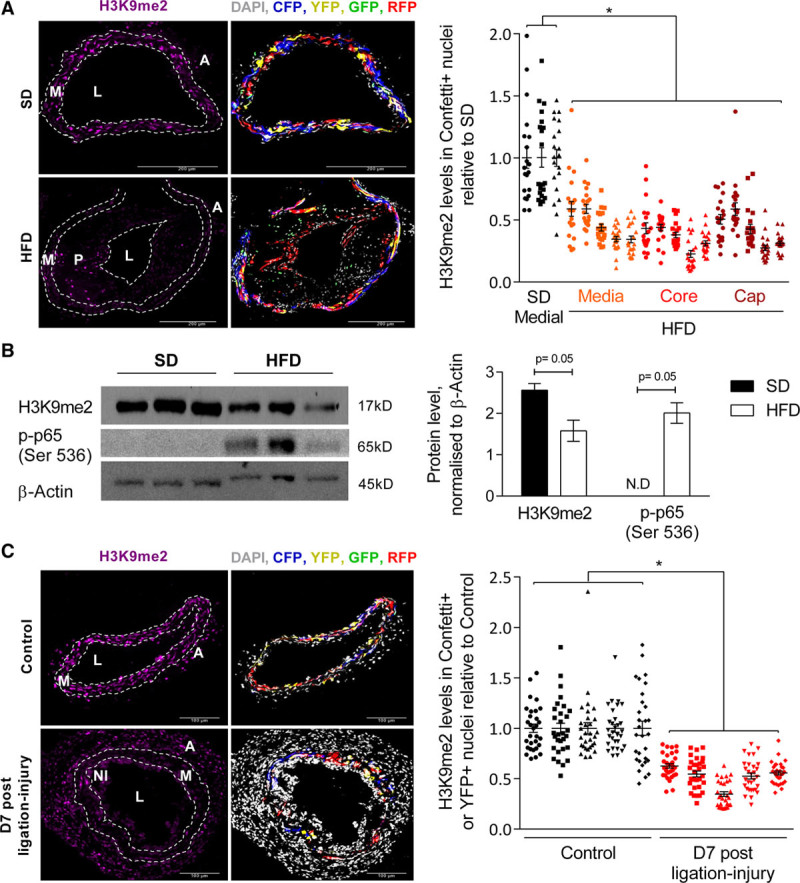
Histone H3 lysine 9 dimethylation (H3K9me2) is reduced in vascular smooth muscle cells (VSMCs) in atherosclerosis and vascular remodeling. A, Representative immunofluorescence images and quantification of H3K9me2 signal intensity in nuclei of Confetti+ cells within left common carotid arteries (LCCAs) from lineage-labeled Myh11-CreERt2+/Rosa26-Confetti+/Apoe−/− males fed a standard (SD, n=3 animals) or high-fat diet (HFD, n=5 animals). H3K9me2 quantification is displayed separately for medial, plaque core, and cap VSMCs in HFD mice. B, Western blot and quantification of H3K9me2 and p-p65 (Ser 536) in aortic media from SD and HFD animals, normalized to β-Actin; n=3 animals per group. N.D., not detected. P were calculated as described in the online-only Data Supplement. C, Representative immunofluorescence images and quantification of H3K9me2 signal intensity in nuclei of Confetti+ cells within LCCAs from ligated (7 days post-ligation) relative to no surgery control Myh11-CreERt2+/Rosa26-Confetti+ mice. n=5 animals per group. A and C, Signals for H3K9me2 (magenta), Confetti reporter proteins (red, blue, yellow, green) and DAPI (4′,6-diamidino-2-phenylindole; white) are shown. The dot plots show H3K9me2 intensity in individual nuclei (of Confetti+ cells) relative to the average H3K9me2 signal intensity in control animals (SD in A, nonligated control samples in C) analyzed in the same batch (batches are indicated with symbols). Mean (line) and SEM (error bars) are indicated. *P<0.05 (linear model, see online-only Data Supplement). A indicates adventitia; L, lumen; M, media; NI, neointima; and P, plaque.
To assess whether H3K9me2 regulates early events in the VSMC inflammatory response, we performed ligation of the left common carotid artery, which elicits reproducible vessel remodeling concomitant with changes in VSMC gene expression, including induction of IL6 and MMPs.41,42 Immunostaining revealed significantly decreased H3K9me2 levels in lineage-labeled VSMC nuclei in ligated compared with nonligated control arteries at 5, 7, and 28 days post-ligation (Figure 1C and Figure I in the online-only Data Supplement), suggesting that H3K9me2 may regulate the inflammatory response of VSMCs.
H3K9me2 Attenuates Inflammation-Induced MMP Gene Induction
The reduction of H3K9me2 in VSMCs concomitant with increased inflammation prompted us to examine local levels of H3K9me2 at promoters of arteriosclerosis-associated genes. Genome-wide mapping by ChIP-seq demonstrated that H3K9me2 levels at gene promoters correlated negatively with expression of the associated gene in ex vivo murine VSMCs (Figure II in the online-only Data Supplement), which is expected for a repressive epigenetic mark. We found that among the genes with high H3K9me2 levels within ±1kb of their transcription start site were 63 genes listed as arteriosclerosis-associated on the Cardiovascular Disease Portal39 (Table I in the online-only Data Supplement), including several inflammation-responsive MMPs, such as Mmp3, Mmp9, and Mmp12. ChIP-qPCR using cultured primary murine VSMCs confirmed abundant H3K9me2 levels at Mmp3, Mmp9, and Mmp12 as well as the positive control locus Magea2 compared with the negative control, Actb (Figure 2A). Surprisingly, despite increased mRNA expression of Mmp3, Mmp9, and Mmp12 after IL-1α treatment (Figure 2B), the levels of the repressive H3K9me2 modification at the Mmp3, Mmp9, and Mmp12 promoters were not reduced after IL-1α treatment (Figure 2A). The continued presence of this repressive epigenetic mark at target genes suggests that their activation in response to inflammation could be obstructed.
Figure 2.
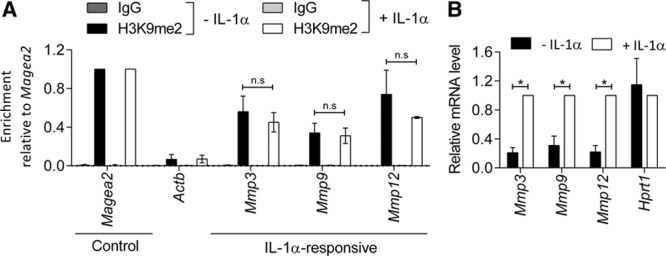
Histone H3 Lysine 9 Dimethylation (H3K9me2) is enriched at a subset of IL (interleukin)-1α−responsive gene promoters. A, Chromatin immunoprecipitation-qPCR analysis for H3K9me2 in untreated or IL-1α-treated murine vascular smooth muscle cells (mVSMCs; 6 h). H3K9me2 enrichment (relative to Magea2) at Actb (negative control) and the promoters of Mmp3, Mmp9, and Mmp12, compared with signal observed using negative control IgG (immunoglobulin). Graph show mean±SEM of 3 independent primary cultures. n.s., not significant (Kruskal-Wallis). B, RT-qPCR (reverse transcription with quantitative polymerase chain reaction) analysis of Mmp3, Mmp9, Mmp12, and Hprt1 in control and IL-1α-treated mVSMCs. Expression (mean±SEM in 6 independent primary cultures) is shown relative to IL-1α-treated mVSMCs, normalized to housekeeping genes (Hmbs and Hprt1). *P<0.05 (2-tailed Mann-Whitney U test).
To directly test the importance of H3K9me2-marking at MMP gene promoters, we pretreated cultured murine VSMCs with UNC, an inhibitor of the main H3K9 dimethyltransferases G9A/GLP, before IL-1α stimulation. UNC treatment significantly reduced H3K9me2 levels globally and at target gene promoters (Figure 3A and 3B). UNC did not affect basal MMP gene expression, but significantly potentiated IL-1α-mediated upregulation of Mmp3 (3.2-fold), Mmp9 (1.7-fold), and Mmp12 (7.1-fold) (Figure 3C). In contrast, neither IL-1α nor UNC treatment alone, or in combination, affected the expression of Mmp2, which is constitutively expressed by VSMCs.7 Importantly, UNC+IL-1α did not affect expression of Il6 and Ccl2 (Figure 3C), which showed no H3K9me2 enrichment (Figure 3B), demonstrating that the effect of UNC on MMP gene expression is not because of a general upregulation of the inflammatory response. This analysis suggests that H3K9me2 acts to specifically repress the induction of target genes in response to inflammatory signaling.
Figure 3.
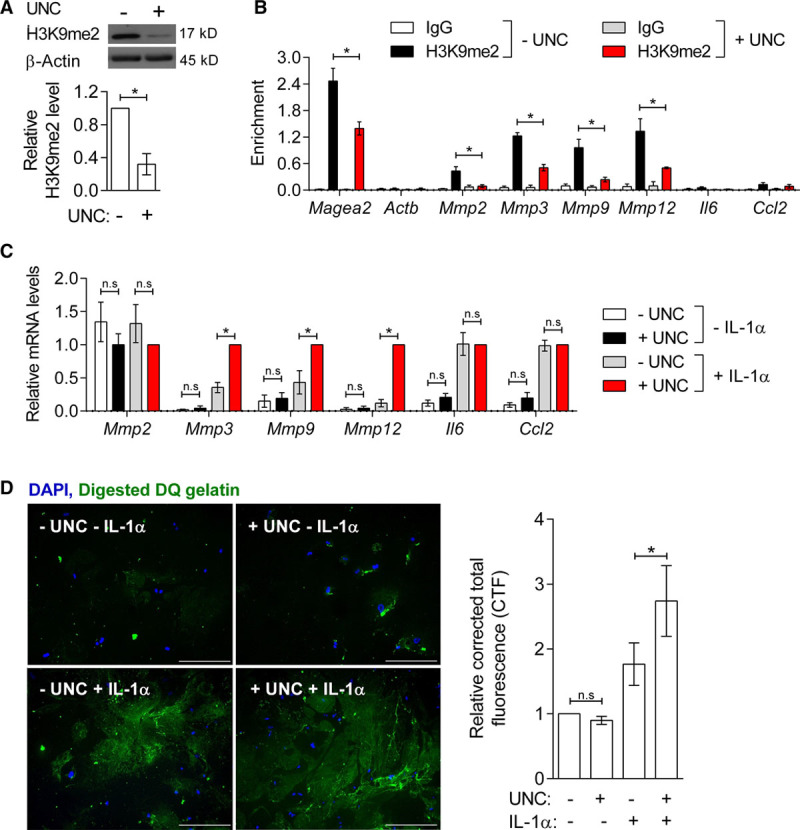
Inhibition of G9A/GLP (G9A-like protein) reduces Histone H3 Lysine 9 Dimethylation (H3K9me2) at MMP (matrix metalloproteinase) gene promoters and potentiates their upregulation in response to IL-1α. A, Representative western blot and quantification of H3K9me2 in UNC (UNC0638)-treated murine vascular smooth muscle cells (mVSMCs; 48 h) relative to controls (–UNC), normalized to β-actin. Graph shows mean±SEM of 4 independent primary cultures. *P<0.05 (Mann-Whitney U test). B, Chromatin immunoprecipitation-qPCR analysis in control and UNC-treated mVSMCs showing H3K9me2 enrichment (mean±SEM of 4 independent primary cultures) at control loci (Magea2, positive and Actb, negative) and the promoters of Mmp2, Mmp3, Mmp9, Mmp12, Il6, and Ccl2 compared with negative control IgG. *P<0.05 (Kruskal-Wallis). C, RT-qPCR (reverse transcription with quantitative polymerase chain reaction) analysis of Mmp2, Mmp3, Mmp9, Mmp12, Il6, and Ccl2 in control (white bars), UNC (black), IL-1α (gray), and IL-1α+UNC-treated (red) mVSMCs. Expression (mean±SEM of 4–5 independent primary cultures) is shown relative to cells treated with UNC+IL-1α (red bars) and normalized to housekeeping genes (Hmbs and Ywhaz). *P<0.05, n.s., not significant (Kruskal-Wallis). D, Representative images and quantification of dye-quenched (DQ)-gelatin digestion in control (top left), UNC (top right), IL-1α (lower left), and UNC+IL-1α-treated mVSMCs (lower right). Signals for digested DQ-gelatin (green) and DAPI (4′,6-diamidino-2-phenylindole)-stained nuclei (blue) are shown. Scale bars, 25 μm. Bar plot shows the corrected total fluorescence (CTF), relative to untreated cells (mean±SEM of 6 independent primary cultures). *P<0.05 (Kruskal-Wallis).
To assess whether MMP regulation by H3K9me2 could affect VSMC extracellular matrix remodeling, we examined whether G9A/GLP inhibition affects matrix degradation. We treated cultured murine VSMCs with IL-1α and UNC before incubating with dye-quenched gelatin, which is converted into fluorescent peptides upon digestion. Inhibition of G9A/GLP further increased IL-1α-induced gelatin digestion (1.6-fold) compared with VSMCs treated with IL-1α alone (Figure 3D), indicating that H3K9me2-regulation of MMP expression in VSMCs might affect vascular remodeling in disease.
VSMC-Specific Upregulation of Inflammation-Responsive H3K9me2-Target Genes After Carotid Ligation Is Enhanced by G9A/GLP Inhibition
We next assessed the effect of reducing H3K9me2 in vivo on injury-induced MMP expression following carotid artery ligation (Figure 4A). VSMC-lineage-labeled animals (Myh11-CreERt2/Rosa26-EYFP) were administered A366, a G9A/GLP inhibitor that displays similar specificity and efficacy but improved pharmacokinetics compared with UNC063843, 44. Two weeks of A366 treatment resulted in reproducible reduction of global H3K9me2 levels in VSMCs as revealed by immunostaining (45%, Figure 4B) and Western blotting (40%, Figure 4C), compared with vehicle-treated controls. Subsequent to A366 administration, the left common carotid artery was ligated and vessels were analyzed 7 days later (Figure 4A). We confirmed by ChIP that H3K9me2 levels were reduced at target gene promoters at the end of the protocol (Figure 4D). Interestingly, analysis of control animals revealed that H3K9me2 was also enriched at the Il6 promoter in VSMCs within tissue (Figure 4D, green bars), contrasting with observations in cultured cells (Figure 3B).
Figure 4.
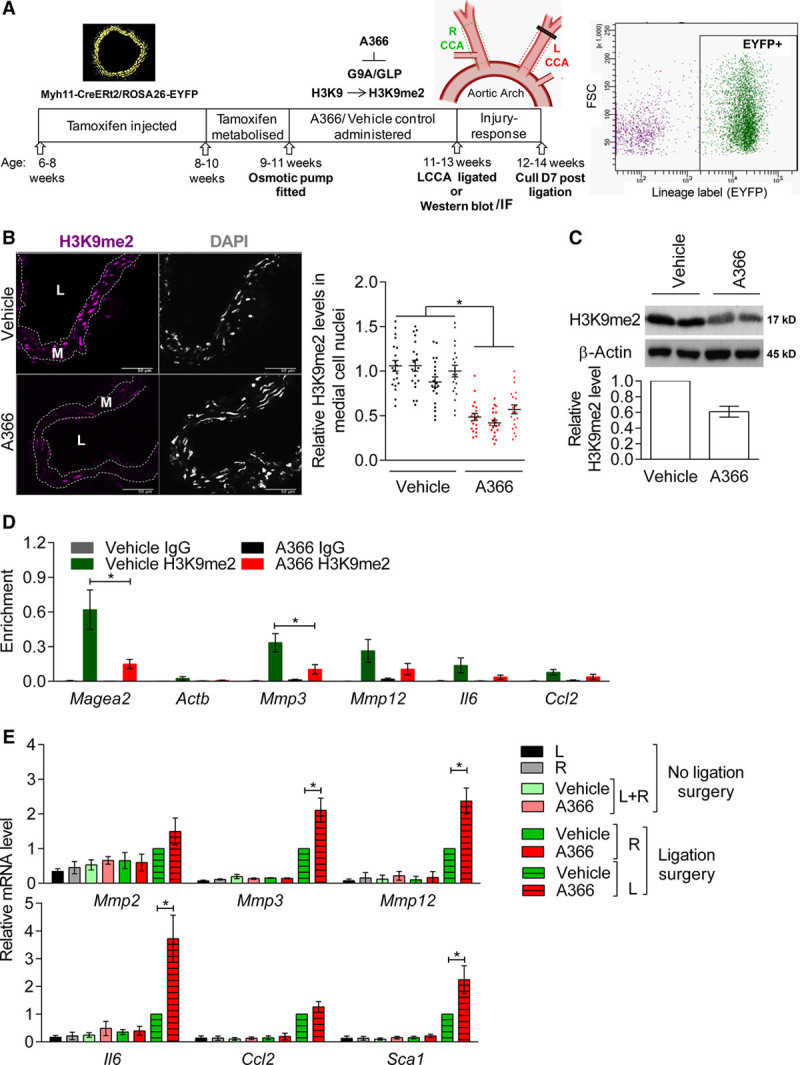
Vascular smooth muscle cell (VSMC)-specific upregulation of histone H3 lysine 9 dimethylation (H3K9me2)-target genes after carotid ligation is enhanced by G9A/GLP (G9A-like protein) inhibition. A, Schematic of experimental model. Lineage-labeled Myh11-CreERt2/ROSA26-EYFP (enhanced yellow fluorescent protein) mice were administered A366 (30 mg/kg per d) or vehicle control and subjected to ligation of the left common carotid artery (CCA). B, Representative immunofluorescence staining and quantification of H3K9me2 in left (L) CCA cryosections from mice administered vehicle control or A366 for 14 d. Signals for H3K9me2 (magenta) and DAPI (4′,6-diamidino-2-phenylindole; white) are shown. Scale bars, 50 μm. Dot plot shows H3K9me2 signal in individual medial cell nuclei, defined by their location between elastic lamina, relative to the average H3K9me2 signal in vehicle controls analyzed in the same batch (batches are indicated with symbols). Mean (line) and SEM (error bars) are indicated. n=4 animals for vehicle, n=3 animals for A366. *P<0.05 (linear model, see online-only Data Supplement). C, Representative western blot and quantification of H3K9me2 levels in aortic medial VSMCs of mice administered vehicle control or A366 for 14 days. Data (mean±SEM of 3 biological replicates) are shown relative to vehicle control, normalized to β-actin. D, Chromatin immunoprecipitation-qPCR analysis of the aortic medial layer (adventitial and endothelial cells removed) from mice 21 days after insertion of an osmotic pump delivering A366 or vehicle control. H3K9me2 enrichment at control loci (Magea2, positive and Actb, negative) and Mmp3, Mmp12, Il6, and Ccl2 promoters is shown alongside negative control IgG. Mean±SEM of 5 to 6 biological replicates. *P<0.05 (Kruskal-Wallis). E, RT-qPCR (reverse transcription with quantitative polymerase chain reaction) analysis of Mmp2, Mmp3, Mmp12, Il6, Ccl2, and Sca1 (Ly6a) in EYFP+ carotid VSMCs isolated by flow cytometry 7 d after carotid ligation. Mmp9 transcripts were not detected in any sample. Data (mean±SEM of 4 experiments) are shown relative to ligated LCCA from vehicle control (striped, green bars) and normalized to housekeeping genes (Hprt1 and Hmbs). *P<0.05 (Kruskal-Wallis). L indicates left, and R, right CCA.
To assess the specific effect of H3K9me2 on injury-induced gene regulation in VSMCs, we performed reverse transcription with quantitative polymerase chain reaction on lineage-labeled, EYFP+ VSMCs isolated by flow cytometry from ligated and control carotid arteries 7 days after surgery. Compared with vehicle-treated control mice, injury-induced expression of Mmp3, Mmp12, and Il6 was significantly increased in those treated with A366 (Figure 4E), correlating with reduced H3K9me2 at their promoters (Figure 4D). Interestingly, A366 also significantly increased expression of Sca1/Ly6a, a marker of phenotypically switched VSMCs,35 suggesting an increased VSMC response to injury compared with controls (Figure 4E). We confirmed that flow cytometry-purified VSMC cell populations did not express markers of adventitial fibroblasts (Pdgfra), endothelial cells (Cd31), or bone marrow-derived cells (Cd45), beyond background (Figure III in the online-only Data Supplement). Importantly, A366 did not affect expression of H3K9me2 target genes in uninjured vessels, the response of Ccl2 to ligation-injury, or the reduction of contractile VSMC gene expression after ligation (Figure 4E; Figure III in the online-only Data Supplement). Together, these results suggest that H3K9me2 attenuates induction of inflammation-responsive H3K9me2 target genes in VSMCs in vivo and inhibits VSMC phenotypic switching to a more proinflammatory state.
H3K9me2-Mediated Regulation of Inflammation-Associated VSMC Gene Expression Changes Is Conserved in Humans
ChIP analysis in primary hVSMC cultures revealed that enrichment of H3K9me2 at the MMP3, MMP9, and MMP12 gene promoters compared with the active ACTB control locus with and without IL-1α treatment was conserved in human (Figure 5A; Figure IV in the online-only Data Supplement). Similar to murine tissue VSMCs, IL6 was also a H3K9me2 target in hVSMCs, whereas this repressive epigenetic mark was not present at CCL2 and MMP2 promoters (Figure 5A). UNC treatment reduced global H3K9me2 levels 10-fold (Figure 5B) and potentiated the IL-1α response of H3K9me2 target genes at both the transcript and protein level (Figure 5C, D). This was most pronounced 6 hours after IL-1α stimulation for IL6, when expression peaked, whereas potentiation of MMP3, MMP9, and MMP12 transcript levels by UNC was also observed 24 hours after stimulation (Figure 5C).
Figure 5.
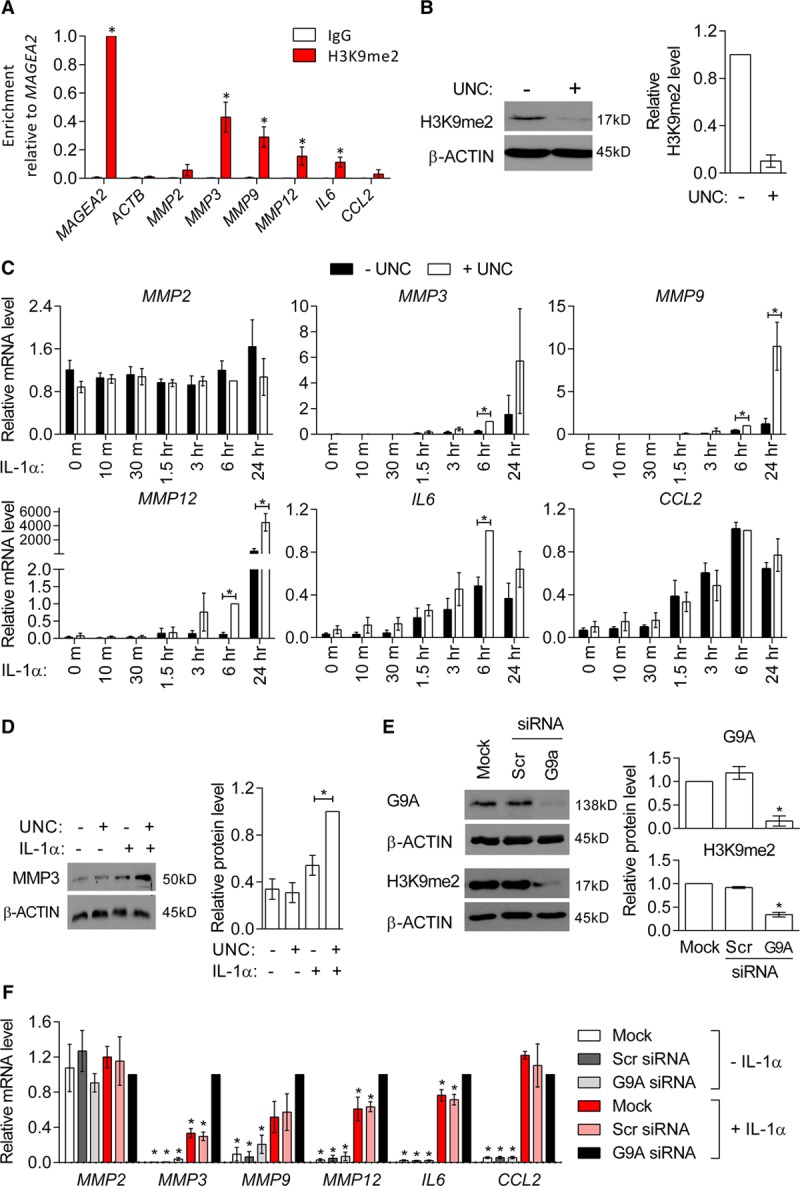
Histone H3 lysine 9 dimethylation (H3K9me2)-mediated regulation of inflammation-associated genes is conserved in human aortic VSMCs (hVSMCs). A, Chromatin immunoprecipitation-qPCR analysis in cultured hVSMCs, showing enrichment for H3K9me2 relative to MAGEA2 (positive control) at ACTB (negative control) and MMP2, MMP3, MMP9, IL6, and CCL2 promoters compared with signals for negative control IgG (immunoglobulin). Graph show mean±SEM of 6 experiments. *P<0.05 (Kruskal-Wallis, H3K9me2 enrichment compared with ACTB). B, Representative western blot and densitometric analysis of H3K9me2 levels in control (−UNC [UNC0638]) and UNC-treated (+UNC) cultured hVSMCs. Data (mean±SEM of 3 experiments) are shown relative to untreated cells and normalized to β-actin. C, RT-qPCR (reverse transcription with quantitative polymerase chain reaction) analysis of MMP2, MMP3, MMP9, MMP12, IL6, and CCL2 in control hVSMCs and after IL-1α treatment for 10, 30 min, 1.5, 3, 6, or 24 h, without (−UNC, black bars) and with prior UNC treatment (+UNC, white bars). Data are relative to cells treated with UNC+IL-1α for 6 h and normalized to housekeeping genes (HMBS and YWHAZ). Graph show mean±SEM of 4 to 10 experiments. *P<0.05 (Kruskal-Wallis). D, Representative western blot and densitomeric analysis of MMP3 in control, UNC, IL-1α, and UNC+IL-1α-treated cultured hVSMCs. Mean±SEM of 3 experiments is shown relative to cells treated with UNC+IL-1α and normalized to β-actin. *P<0.05 (Kruskal-Wallis). E, Representative western blot and densitometric analysis of G9A and H3K9me2 levels in mock, scrambled (Scr) siRNA, and G9A siRNA transfected hVSMCs. Data are shown relative to mock-transfected hVSMCs and normalized to β-actin (mean±SEM of 4 experiments). *P<0.05 (Kruskal-Wallis, compared with Scr). F, RT-qPCR analysis of MMP2, MMP3, MMP9, MMP12, IL6, and CCL2 in mock, Scr siRNA and G9A siRNA-transfected hVSMCs with and without IL-1α stimulation. Data (mean±SEM of 4 experiments) are relative to hVSMCs treated with G9A siRNA+IL-1α and normalized to housekeeping genes (HPRT1 and YWHAZ). *P<0.05 (Kruskal-Wallis, compared with G9A siRNA+IL-1α). Each experiment used hVSMC isolates derived from different individuals.
To ensure that the observed changes in gene expression were because of specific effects of UNC, we used siRNA to silence the G9A subunit of the heterodimeric G9A/GLP H3K9-methyltransferase complex. G9A knockdown resulted in > 7.5-fold reduction in G9A protein levels, which lead to a 2.7-fold reduction in global H3K9me2 levels compared with scrambled siRNA-transfected cells (Figure 5E). G9A knockdown alone did not influence basal transcript levels but enhanced IL-1α-induced MMP3, MMP9, MMP12 and IL6 expression (Figure 5F). Consistent with the effect of UNC, siRNA-mediated G9A knockdown did not affect the expression of MMP2 or the response of CCL2 to IL-1α (Figure 5F).
H3K9me2 Does Not Affect Inflammation-Induced Signal Transduction
In addition to their role as histone methyltransferases, G9A/GLP have nonhistone targets,45 which may influence protein stability,46 protein-protein interactions,47,48 and cellular signaling pathways.45,49 We therefore tested whether reducing G9A/GLP activity affects signaling cascades downstream of IL-1α (Figure VA in the online-only Data Supplement). Expression levels of the IL-1 receptor was modestly increased in IL-1α-treated hVSMCs but was not affected by UNC-mediated G9A/GLP inhibition (Figure VB in the online-only Data Supplement). Similar to IL-1α, TNF-α-mediated induction of H3K9me2 target genes MMP3, MMP9, MMP12, and IL6 was also potentiated by UNC, whereas MMP2 or CCL2 expression was not affected (Figure VC in the online-only Data Supplement). This suggests that UNC acts downstream of receptor binding and affects a factor that is common to TNF-α and IL-1α signaling such as NFκB and MAPK-mediated activation of the AP-1 transcription factor (Figure VA in the online-only Data Supplement). Binding of NFκB and AP-1 transcription factors to a number of MMP and proinflammatory cytokine promoters, including MMP350-52, MMP951, MMP12,51 and IL653, has been reported as necessary to induce their expression.
To investigate whether G9A/GLP regulates inflammation-induced NFκB activity, we analyzed regulation of the p65 subunit, which contains the transactivation domain necessary for gene induction.54 Compared with untreated control cells, p65 levels (Figure VI A in the online-only Data Supplement) were unchanged in hVSMCs treated with UNC±IL-1α. UNC treatment neither affected the nuclear-to-cytoplasmic ratio of p65 at basal conditions nor its nuclear translocation in response to IL-1α or TNF-α stimulation (Figure VI in the online-only Data Supplement). These results suggest that potentiation of IL-1α/TNF-α-induced IL6 and MMP expression by UNC is not because of increased upstream NFκB signaling.
Inflammatory cytokines induce phosphorylation of MAPKs at specific residues, which ultimately leads to phosphorylation of AP-1 subunits, such as cJUN.55–57 Western blotting showed that neither timing nor levels of phosphorylated JNK (cJUN N-terminal kinase), ERK (extracellular regulating kinase) 1/2, and p38 kinase (p38) were affected by UNC treatment before IL-1α stimulation (Figure VII in the online-only Data Supplement), suggesting that G9A/GLP inhibition does not influence MAPK activation.
H3K9me2 Attenuates NFκB and AP-1 Binding to Inflammation-Responsive Target Gene Promoters
Binding sites for NFκB and AP-1 and IL-1-responsive elements have been reported within 2 kb of the transcriptional start site of MMP3, IL6, and CCL2 (Figure 6A).50–53,58 To investigate whether H3K9me2 affects transcription factor binding at target gene promoters, we performed ChIP for p65 (NFκB) and cJUN (AP-1) in hVSMCs treated with IL-1α and UNC. Compared with the ACTB negative control locus, p65 was enriched at the IL6 and CCL2 gene promoters in untreated cells. At the IL6 locus, p65 binding was increased by IL-1α stimulation, and this was significantly enhanced by UNC pretreatment (Figure 6B). Increased p65 binding in UNC-treated cells correlated with reduced H3K9me2, suggesting that H3K9me2 at the IL6 locus prevents NFκB binding to its cognate sequence. In contrast, the enrichment of p65 at CCL2, which is not a H3K9me2 target (Figure 5A), was unaffected by inhibition of G9A/GLP (Figure 6B).
Figure 6.
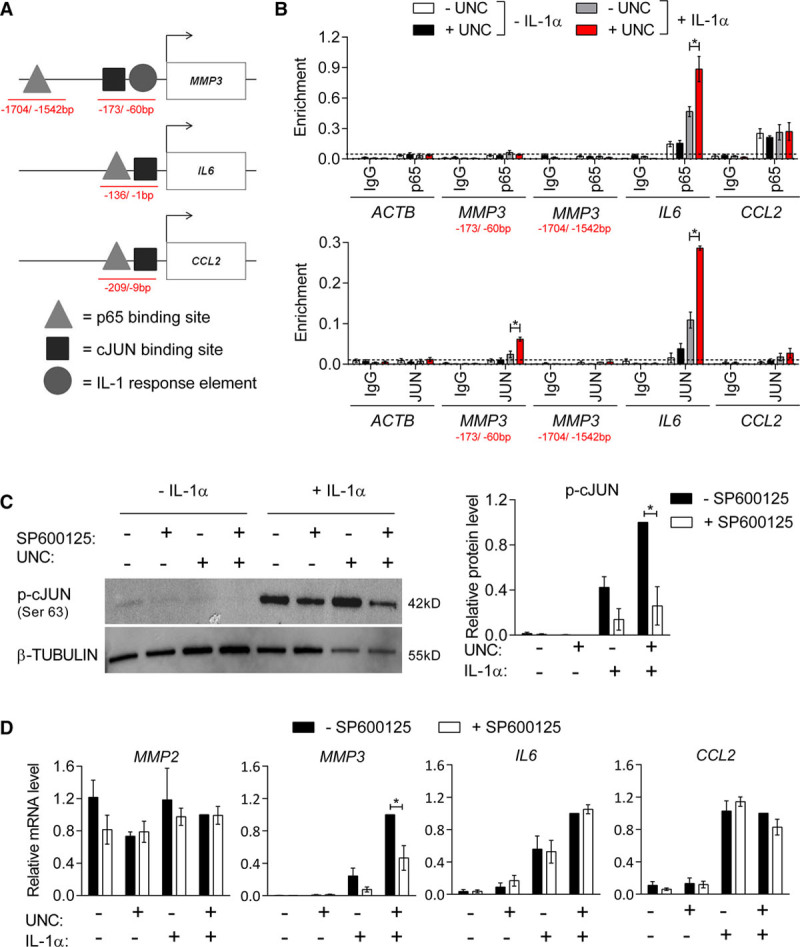
IL (interleukin)-1α-induced binding of p65 and cJUN at histone H3 lysine 9 dimethylation (H3K9me2)-target genes is enhanced by G9A/GLP (G9A-like protein) inhibition. A, Schematic indicating positions of reported p65 and cJUN binding sites and an IL-1-responsive element at the MMP3 (Borghaei et al, Clark et al, Quinones et al),41–43 IL6 (Gomard et al),44 and CCL2 (Sutcliffe et al)49 promoters. Red lines indicate chromatin immunoprecipitation (ChIP)-qPCR amplicons. B, ChIP-qPCR analysis of p65 and cJUN binding relative to input in untreated control (white bars), UNC (UNC0638; black), IL-1α (gray), and UNC+IL-1α-treated human aortic VSMCs (hVSMCs; red) showing enrichment levels at the reported binding sites within the MMP3, IL6, and CCL2 promoters and the ACTB negative control locus compared with negative control IgG (immunoglobulin). Background levels of p65 binding, indicated by a line, are based on enrichment at ACTB. Graph shows mean±SEM of 4 experiments. C, Representative western blot and densitometric analysis p-cJUN (Ser63) in untreated control hVSMCs and cells stimulated with IL-1α, without and with prior treatment with UNC and SP600125 for 48 h. Data (mean±SEM of 4 experiments) are relative to +UNC+IL-1α-SP600125 and normalized to β-TUBULIN levels. D, RT-qPCR (reverse transcription with quantitative polymerase chain reaction) analysis of MMP2, MMP3, IL6, and CCL2 in control and IL-1α-treated hVSMCs, without and with prior UNC and/or SP600125 treatment. Data (mean±SEM of 4 experiments) are relative to +UNC+IL-1α−SP600125 and normalized to the average of 2 housekeeping genes (HPRT1 and YWHAZ). Each experiment used hVSMCs derived from different individuals; *P<0.05 (Kruskal-Wallis).
Notably, p65 enrichment was not detected at the reported IL-1-responsive element in the proximal MMP3 promoter,51,52 nor at a reported NFκB site 1.5 kb upstream of the transcriptional start site,50 irrespective of cell stimulation (Figure 6B). In contrast, binding of the AP-1 subunit cJUN was detected at both IL6 and MMP3 promoters on IL-1α stimulation, and cJUN enrichment was significantly increased by UNC-mediated H3K9me2 depletion (Figure 6B). No cJUN binding was observed at the CCL2 promoter under any condition (Figure 6B).
To explore the impact of differences in transcription factor binding at the MMP3 and IL6 loci, we measured the sensitivity of IL-1α-induced transcription to inhibition of JNK (SP60012559), the primary MAPK responsible for cJUN phosphorylation at Ser63.60 Treatment with SP600125 significantly reduced IL-1α-induced p-cJUN levels (3.9-fold to 4.7-fold; Figure 6C). Interestingly, G9A/GLP inhibition caused a modest (2-fold) increase in p-cJUN in hVSMCs stimulated with IL-1α, despite not affecting the levels of phosphorylated MAPKs (Figure 6C; Figure VII in the online-only Data Supplement) or total cJUN (Figure VII in the online-only Data Supplement). Reduced levels of p-cJUN in SP600125-treated cells selectively affected MMP3 gene expression resulting in an attenuated induction by IL-1α, whereas IL-1α-induced IL6 and CCL2 expression was not affected by JNK inhibition (Figure 6D). These results suggest that JNK-mediated cJUN phosphorylation is necessary for IL-1α-induced expression of MMP3 but not required for activation of the NFκB target IL6.
Taken together, these findings suggest that H3K9me2 acts directly at the MMP3 and IL6 promoter to restrain IL-1α-induced upregulation by inhibiting NFκB (p65) and AP-1 (cJUN) transcription factor binding and demonstrates that inflammation-induced expression of MMP3, IL6, and CCL2 are regulated via different mechanisms.
Discussion
We here demonstrate an evolutionary conserved role of H3K9me2 in regulating the proinflammatory VSMC phenotype associated with vascular disease and identify a subset of inflammation-responsive genes, including MMP3, MMP9, MMP12, and IL6, as direct H3K9me2 targets in VSMCs. We show that loss of H3K9me2 increases binding of inflammation-induced transcription factors and increases expression of these MMP and proinflammatory VSMC genes in response to inflammatory stimuli both in vitro and in vivo. This functional role of H3K9me2 suggests that this epigenetic mark acts to restrain inflammation-induced, CVD-associated gene expression in VSMCs. It is therefore tempting to speculate that the reduced H3K9me2 levels we observed in atherosclerosis and after injury is required to elicit VSMC inflammatory activation and vascular remodeling. Increasing H3K9me2 levels by stimulating G9A/GLP activity or inhibiting H3K9me2-specific demethylases (eg, KDM3A) might therefore be a strategy for therapeutic targeting in CVD.
The global loss of H3K9me2 in VSMCs from atherosclerotic lesions and in arteries undergoing remodeling after vascular injury is consistent with the reported H3K9me2 reduction in atherosclerotic plaques from patients compared with healthy human arteries.22 Our analysis of lineage traced VSMCs supports the observation that αSMA-positive plaque cells have lower H3K9me2 levels61 and further indicates that this epigenetic mark is also reduced in VSMC-derived cells within the lesion core, that do not express contractile markers.
H3K9me2-mediated regulation of inflammation-induced genes in VSMCs might also explain the accelerated neointima formation following vascular injury previously detected in diabetic rats when H3K9me2 levels were reduced by overexpression of KDM3A, a H3K9me2-specific demethylase.62,63 VSMCs from diabetic patients also display reduced levels of H3K9me2 compared with nondiabetic controls, and KDM3A is induced by high insulin levels.62,64 Therefore, dysregulation of H3K9me2 might contribute to diabetes-associated vascular complications.
An inverse correlation between H3K9me2 occupancy at MMP and other inflammatory gene promoters and their expression has also been reported in other cell types.24–27 For example, H3K9me2 depletion at the Mmp9 promoter correlates with elevated Mmp9 expression in retinal endothelial cells from diabetic rats compared with controls.27 Similarly, solar-stimulated UV radiation induces loss of H3K9me2 at the MMP3 promoter in primary human dermal fibroblasts, correlating with enhanced MMP3 mRNA levels.25 In addition, genetic ablation or pharmacological inhibition of G9A/GLP stimulates the expression of inflammatory genes in mouse embryonic fibroblasts.24 Similar to our findings in VSMCs, reduction of H3K9me2 levels in fibroblasts potentiates poly(I:C)-induced upregulation of inflammatory genes.24 Moreover, G9A-deficient flies are hypersensitive to RNA virus infection and succumb faster to infection than wild-type controls.28 These observations suggest that G9A/GLP represents an evolutionarily conserved mechanism to control genes involved in processes that require tight and dynamic regulation—such as the inflammatory response—in a variety of cell types.
We demonstrate increased binding of AP-1 and NFκB specifically at H3K9me2 target genes on reduction of H3K9me2 levels following G9A/GLP inhibition (see Graphical Abstract). In contrast, inhibition of G9A/GLP did not affect IL-1α/TNF-α-induced activation of upstream signaling, including p65 activation and phosphorylation of p38, JNK, and ERK1/2. MAPKs can phosphorylate substrates once bound to chromatin.65 We propose that the 2-fold increase in p-cJUN in IL-1α-stimulated cells after G9A/GLP inhibition could be because of increased phosphorylation efficiency of chromatin-associated cJUN, which potentiates transactivation.66,67 Alternatively, G9A/GLP might regulate an as-yet-unidentified cJUN phosphatase.66 Regardless, our data demonstrate that key inflammation-induced genes have distinct regulatory mechanisms in VSMCs. Non-H3K9me2 targets such as CCL2 display transcription factor binding at basal levels resulting in rapid induction upon stimulation. In contrast, H3K9me2 target gene induction is delayed in a context-specific manner, which could depend on both specific transcription factor usage and locus-specific chromatin regulation.
In conclusion, our data implicate H3K9me2 as a functional epigenetic regulator of the VSMC inflammatory response. We propose that H3K9me2 acts to prevent spurious induction of CVD-associated proinflammatory genes, including MMPs and IL6. Differential transcriptional regulation of selective MMPs and other inflammation-induced genes could be leveraged for developing more specific drugs for future clinical use.
Acknowledgments
We thank the Cambridge Statistics Clinic (Department of Statistics) for advise on statistical analysis, Mikhail Spivakov for advise on chromatin immunoprecipitation-seq data analysis, Gregory Strachan and the Wellcome Trust-Medical Research Council, Institute of Metabolic Science, Metabolic Research Laboratories, Imaging core (Wellcome Trust Major Award [208363/Z/17/Z]) for technical assistance, the Cambridge National Institute for Health Research Biomedical Research Centre Cell Phenotyping Hub for cell sorting and all members of the Jørgensen group for helpful discussions.
Sources of Funding
J.L. Harman was funded by a British Heart Foundation (BHF) studentship (FS/15/38/31516); L. Dobnikar was funded by a BBSRC DTP studentship; J. Chappell was funded by the BHF Centre for Cardiovascular Research Excellence (RE/13/6/30180); B.G. Stokell was funded by a studentship from the Cantab Capital Institute for the Mathematics of Information. K. Foote was funded by the BHF PG/16/63/32307; A. Finigan and A. Uryga were funded by the BHF RG/13/14/30314. A.L. Taylor was funded by BHF studentship (FS/15/62/32032). M.R. Bennett and H.F. Jørgensen were supported by the BHF (CH/2000003), the Oxbridge BHF Centre for Regenerative Medicine (RM/13/3/30159, RM/17/2/33380), the BHF Centre for Cardiovascular Research Excellence (RE/13/6/30180), and the National Institute for Health Research Cambridge Biomedical Research Center.
Disclosures
None.
Supplementary Material
Nonstandard Abbreviations and Acronyms
- AP-1
- activator protein-1
- ChIP
- chromatin immunoprecipitation
- CVD
- cardiovascular disease
- GLP
- G9A-like protein
- H3K9me2
- histone H3 Lysine 9 di-methyl
- HFD
- high-fat diet
- hVSMC
- human aortic VSMC
- IL
- interleukin
- JNK
- cJUN N-terminal kinase
- LCCA
- left common carotid artery
- MAPK
- mitogen-activated protein kinase
- MMP
- matrix metalloproteinase
- mVSMC
- murine VSMC
- NFκB
- nuclear factor kappa-light-chain-enhancer of activated B cell
- TNF-α
- tumor necrosis factor-alpha
- UNC
- UNC0638
- VSMC
- vascular smooth muscle cell
For Sources of Funding and Disclosures, see page 2301.
The online-only Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.119.312765.
References
- 1.Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cells in atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim S, Park S. Role of vascular smooth muscle cell in the inflammation of atherosclerosis. BMB Rep. 2014;47:1–7. doi: 10.5483/BMBRep.2014.47.1.285. doi: 10.5483/bmbrep.2014.47.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chistiakov DA, Melnichenko AA, Grechko AV, Myasoedova VA, Orekhov AN. Potential of anti-inflammatory agents for treatment of atherosclerosis. Exp Mol Pathol. 2018;104:114–124. doi: 10.1016/j.yexmp.2018.01.008. doi: 10.1016/j.yexmp.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, et al. CANTOS Trial Group. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 5.Gomez D, Baylis RA, Durgin BG, Newman AAC, Alencar GF, Mahan S, St Hilaire C, Müller W, Waisman A, Francis SE, et al. Interleukin-1β has atheroprotective effects in advanced atherosclerotic lesions of mice. Nat Med. 2018;24:1418–1429. doi: 10.1038/s41591-018-0124-5. doi: 10.1038/s41591-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JL. Matrix metalloproteinases and their inhibitors in cardiovascular pathologies: current knowledge and clinical potential. Dove Medical Press Ltd. 2014;2014:21–36. [Google Scholar]
- 7.Galis ZS, Khatri JJ. Matrix metalloproteinases in vascular remodeling and atherogenesis: the good, the bad, and the ugly. Circ Res. 2002;90:251–262. [PubMed] [Google Scholar]
- 8.Johnson JL, George SJ, Newby AC, Jackson CL. Divergent effects of matrix metalloproteinases 3, 7, 9, and 12 on atherosclerotic plaque stability in mouse brachiocephalic arteries. Proc Natl Acad Sci USA. 2005;102:15575–15580. doi: 10.1073/pnas.0506201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang J, Liu E, Yu Y, Kitajima S, Koike T, Jin Y, Morimoto M, Hatakeyama K, Asada Y, Watanabe T, et al. Macrophage metalloelastase accelerates the progression of atherosclerosis in transgenic rabbits. Circulation. 2006;113:1993–2001. doi: 10.1161/CIRCULATIONAHA.105.596031. doi: 10.1161/CIRCULATIONAHA.105.596031. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JL, Dwivedi A, Somerville M, George SJ, Newby AC. Matrix metalloproteinase (MMP)-3 activates MMP-9 mediated vascular smooth muscle cell migration and neointima formation in mice. Arterioscler Thromb Vasc Biol. 2011;31:e35–e44. doi: 10.1161/ATVBAHA.111.225623. doi: 10.1161/ATVBAHA.111.225623. [DOI] [PubMed] [Google Scholar]
- 11.Halpert I, Sires UI, Roby JD, Potter-Perigo S, Wight TN, Shapiro SD, Welgus HG, Wickline SA, Parks WC. Matrilysin is expressed by lipid-laden macrophages at sites of potential rupture in atherosclerotic lesions and localizes to areas of versican deposition, a proteoglycan substrate for the enzyme. Proc Natl Acad Sci USA. 1996;93:9748–9753. doi: 10.1073/pnas.93.18.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas AC, Sala-Newby GB, Ismail Y, Johnson JL, Pasterkamp G, Newby AC. Genomics of foam cells and nonfoamy macrophages from rabbits identifies arginase-I as a differential regulator of nitric oxide production. Arterioscler Thromb Vasc Biol. 2007;27:571–577. doi: 10.1161/01.ATV.0000256470.23842.94. doi: 10.1161/01.ATV.0000256470.23842.94. [DOI] [PubMed] [Google Scholar]
- 13.Müller A, Krämer SD, Meletta R, Beck K, Selivanova SV, Rancic Z, Kaufmann PA, Vos B, Meding J, Stellfeld T, et al. Gene expression levels of matrix metalloproteinases in human atherosclerotic plaques and evaluation of radiolabeled inhibitors as imaging agents for plaque vulnerability. Nucl Med Biol. 2014;41:562–569. doi: 10.1016/j.nucmedbio.2014.04.085. doi: 10.1016/j.nucmedbio.2014.04.085. [DOI] [PubMed] [Google Scholar]
- 14.Galis ZS, Sukhova GK, Lark MW, Libby P. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest. 1994;94:2493–2503. doi: 10.1172/JCI117619. doi: 10.1172/JCI117619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurozumi A, Nakano K, Yamagata K, Okada Y, Nakayamada S, Tanaka Y. Il-6/stat3 pathway is critically involved in vascular calcification via histone modification of the runx2 promoter in vascular smooth muscle cells. Bone Abstracts. 2016;5:425. [Google Scholar]
- 16.Lee G-L, Wu J-Y, Tsai C-S, Lin C-Y, Tsai Y-T, Lin C-S, Wang Y-F, Yet S-F, Hsu Y-J, Kuo C-C. Tlr4-activated mapk-il-6 axis regulates vascular smooth muscle cell function. Int J Mo Sci. 2016;17:1394. doi: 10.3390/ijms17091394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe S, Mu W, Kahn A, Jing N, Li JH, Lan HY, Nakagawa T, Ohashi R, Johnson RJ. Role of JAK/STAT pathway in IL-6-induced activation of vascular smooth muscle cells. Am J Nephrol. 2004;24:387–392. doi: 10.1159/000079706. doi: 10.1159/000079706. [DOI] [PubMed] [Google Scholar]
- 18.Biasucci LM, Liuzzo G, Fantuzzi G, Caligiuri G, Rebuzzi AG, Ginnetti F, Dinarello CA, Maseri A. Increasing levels of interleukin (IL)-1Ra and IL-6 during the first 2 days of hospitalization in unstable angina are associated with increased risk of in-hospital coronary events. Circulation. 1999;99:2079–2084. doi: 10.1161/01.cir.99.16.2079. doi: 10.1161/01.cir.99.16.2079. [DOI] [PubMed] [Google Scholar]
- 19.Tousoulis D, Oikonomou E, Economou EK, Crea F, Kaski JC. Inflammatory cytokines in atherosclerosis: current therapeutic approaches. Eur Heart J. 2016;37:1723–1732. doi: 10.1093/eurheartj/ehv759. doi: 10.1093/eurheartj/ehv759. [DOI] [PubMed] [Google Scholar]
- 20.Anderson DR, Poterucha JT, Mikuls TR, Duryee MJ, Garvin RP, Klassen LW, Shurmur SW, Thiele GM. IL-6 and its receptors in coronary artery disease and acute myocardial infarction. Cytokine. 2013;62:395–400. doi: 10.1016/j.cyto.2013.03.020. doi: 10.1016/j.cyto.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 21.Delcuve GP, Rastegar M, Davie JR. Epigenetic control. J Cell Physiol. 2009;219:243–250. doi: 10.1002/jcp.21678. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- 22.Greißel A, Culmes M, Napieralski R, Wagner E, Gebhard H, Schmitt M, Zimmermann A, Eckstein HH, Zernecke A, Pelisek J. Alternation of histone and DNA methylation in human atherosclerotic carotid plaques. Thromb Haemost. 2015;114:390–402. doi: 10.1160/TH14-10-0852. doi: 10.1160/TH14-10-0852. [DOI] [PubMed] [Google Scholar]
- 23.Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, Chen MS, Rioja I, Parravicini V, Prinjha RK, Chandwani R, et al. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med. 2012;209:661–669. doi: 10.1084/jem.20112343. doi: 10.1084/jem.20112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gesumaria L, Matsui MS, Kluz T, Costa M. Solar-simulated ultraviolet radiation induces histone 3 methylation changes in the gene promoters of matrix metalloproteinases 1 and 3 in primary human dermal fibroblasts. Exp Dermatol. 2015;24:384–385. doi: 10.1111/exd.12675. doi: 10.1111/exd.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Yu Y, Liu F, Wei X, Wrobel JA, Gunawardena HP, Zhou L, Jin J, Chen X. A chromatin activity-based chemoproteomic approach reveals a transcriptional repressome for gene-specific silencing. Nat Commun. 2014;5:5733. doi: 10.1038/ncomms6733. doi: 10.1038/ncomms6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Q, Kowluru RA. Regulation of matrix metalloproteinase-9 by epigenetic modifications and the development of diabetic retinopathy. Diabetes. 2013;62:2559–2568. doi: 10.2337/db12-1141. doi: 10.2337/db12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkling SH, Bronkhorst AW, Kramer JM, Overheul GJ, Schenck A, Van Rij RP. The epigenetic regulator G9a mediates tolerance to RNA virus infection in Drosophila. PLoS Pathog. 2015;11:e1004692. doi: 10.1371/journal.ppat.1004692. doi: 10.1371/journal.ppat.1004692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Q, Lu Z, Singh D, Raj JU. BIX-01294 treatment blocks cell proliferation, migration and contractility in ovine foetal pulmonary arterial smooth muscle cells. Cell Prolif. 2012;45:335–344. doi: 10.1111/j.1365-2184.2012.00828.x. doi: 10.1111/j.1365-2184.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clifford RL, John AE, Brightling CE, Knox AJ. Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. J Immunol. 2012;189:819–831. doi: 10.4049/jimmunol.1103641. doi: 10.4049/jimmunol.1103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirth A, Benyó Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horváth B, Maser-Gluth C, Greiner E, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 32.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Methods. 2013;10:171–177. doi: 10.1038/nmeth.2332. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jørgensen HF. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res. 2016;119:1313–1323. doi: 10.1161/CIRCRESAHA.116.309799. doi: 10.1161/CIRCRESAHA.116.309799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jørgensen HF. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun. 2018;9:4567. doi: 10.1038/s41467-018-06891-x. doi: 10.1038/s41467-018-06891-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakraborty R, Saddouk FZ, Carrao AC, Krause DS, Greif DM, Martin KA. Promoters to study vascular smooth muscle. Arterioscler Thromb Vasc Biol. 2019;39:603–612. doi: 10.1161/ATVBAHA.119.312449. doi: 10.1161/ATVBAHA.119.312449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal [Online] 2011;17:1. doi: 10.14806/ej.17.1.200. [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimoyama M, De Pons J, Hayman GT, Laulederkind SJ, Liu W, Nigam R, Petri V, Smith JR, Tutaj M, Wang SJ, et al. The rat genome database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43(Database issue):D743–D750. doi: 10.1093/nar/gku1026. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah A, Gray K, Figg N, Finigan A, Starks L, Bennett M. Defective base excision repair of oxidative DNA damage in vascular smooth muscle cells promotes atherosclerosis. Circulation. 2018;138:1446–1462. doi: 10.1161/CIRCULATIONAHA.117.033249. doi: 10.1161/CIRCULATIONAHA.117.033249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herring BP, Hoggatt AM, Griffith SL, McClintick JN, Gallagher PJ. Inflammation and vascular smooth muscle cell dedifferentiation following carotid artery ligation. Physiol Genomics. 2017;49:115–126. doi: 10.1152/physiolgenomics.00095.2016. doi: 10.1152/physiolgenomics.00095.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali MS, Starke RM, Jabbour PM, Tjoumakaris SI, Gonzalez LF, Rosenwasser RH, Owens GK, Koch WJ, Greig NH, Dumont AS. TNF-α induces phenotypic modulation in cerebral vascular smooth muscle cells: implications for cerebral aneurysm pathology. J Cereb Blood Flow Metab. 2013;33:1564–1573. doi: 10.1038/jcbfm.2013.109. doi: 10.1038/jcbfm.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thienpont B, Aronsen JM, Robinson EL, Okkenhaug H, Loche E, Ferrini A, Brien P, Alkass K, Tomasso A, Agrawal A, et al. The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy. J Clin Invest. 2017;127:335–348. doi: 10.1172/JCI88353. doi: 10.1172/JCI88353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sweis RF, Pliushchev M, Brown PJ, Guo J, Li F, Maag D, Petros AM, Soni NB, Tse C, Vedadi M, et al. Discovery and development of potent and selective inhibitors of histone methyltransferase g9a. ACS Med Chem Lett. 2014;5:205–209. doi: 10.1021/ml400496h. doi: 10.1021/ml400496h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 46.Lee JY, Lee SH, Heo SH, Kim KS, Kim C, Kim DK, Ko JJ, Park KS. Novel function of lysine methyltransferase G9a in the regulation of sox2 protein stability. PLoS One. 2015;10:e0141118. doi: 10.1371/journal.pone.0141118. doi: 10.1371/journal.pone.0141118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ling BMT, Bharathy N, Chung T-K, Kok WK, Li S, Tan YH, Rao VK, Gopinadhan S, Sartorelli V, Walsh MJ, Taneja R. Lysine methyltransferase g9a methylates the transcription factor myod and regulates skeletal muscle differentiation. Proc Natl Acad Sci USA. 2012;109:841–846. doi: 10.1073/pnas.1111628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HJ, Kim JH, Kim JY, Seo SB, et al. Negative regulation of hypoxic responses via induced reptin methylation. Mol Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pless O, Kowenz-Leutz E, Knoblich M, Lausen J, Beyermann M, Walsh MJ, Leutz A. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-beta. J Biol Chem. 2008;283:26357–26363. doi: 10.1074/jbc.M802132200. doi: 10.1074/jbc.M802132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borghaei RC, Rawlings PL, Jr, Javadi M, Woloshin J. NF-kappaB binds to a polymorphic repressor element in the MMP-3 promoter. Biochem Biophys Res Commun. 2004;316:182–188. doi: 10.1016/j.bbrc.2004.02.030. doi: 10.1016/j.bbrc.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 51.Clark IM, Swingler TE, Sampieri CL, Edwards DR. The regulation of matrix metalloproteinases and their inhibitors. Int J Biochem Cell Biol. 2008;40:1362–1378. doi: 10.1016/j.biocel.2007.12.006. doi: 10.1016/j.biocel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Quinones S, Buttice G, Kurkinen M. Promoter elements in the transcriptional activation of the human stromelysin-1 gene by the inflammatory cytokine, interleukin 1. Biochem J. 1994;302(pt 2):471–477. doi: 10.1042/bj3020471. doi: 10.1042/bj3020471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomard T, Michaud HA, Tempé D, Thiolon K, Pelegrin M, Piechaczyk M. An NF-kappaB-dependent role for JunB in the induction of proinflammatory cytokines in LPS-activated bone marrow-derived dendritic cells. PLoS One. 2010;5:e9585. doi: 10.1371/journal.pone.0009585. doi: 10.1371/journal.pone.0009585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. 2015;89:867–882. doi: 10.1007/s00204-015-1472-2. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
- 56.Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600–604. doi: 10.3109/10799893.2015.1030412. doi: 10.3109/10799893.2015.1030412. [DOI] [PubMed] [Google Scholar]
- 57.Yang SH, Sharrocks AD, Whitmarsh AJ. MAP kinase signalling cascades and transcriptional regulation. Gene. 2013;513:1–13. doi: 10.1016/j.gene.2012.10.033. doi: 10.1016/j.gene.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 58.Sutcliffe AM, Clarke DL, Bradbury DA, Corbett LM, Patel JA, Knox AJ. Transcriptional regulation of monocyte chemotactic protein-1 release by endothelin-1 in human airway smooth muscle cells involves NF-kappaB and AP-1. Br J Pharmacol. 2009;157:436–450. doi: 10.1111/j.1476-5381.2009.00143.x. doi: 10.1111/j.1476-5381.2009.00143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han Z, Boyle DL, Chang L, Bennett B, Karin M, Yang L, Manning AM, Firestein GS. c-Jun N-terminal kinase is required for metalloproteinase expression and joint destruction in inflammatory arthritis. J Clin Invest. 2001;108:73–81. doi: 10.1172/JCI12466. doi: 10.1172/JCI12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morton S, Davis RJ, McLaren A, Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22:3876–3886. doi: 10.1093/emboj/cdg388. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greißel A, Culmes M, Burgkart R, Zimmermann A, Eckstein HH, Zernecke A, Pelisek J. Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc Pathol. 2016;25:79–86. doi: 10.1016/j.carpath.2015.11.001. doi: 10.1016/j.carpath.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Chen J, Zhang J, Yang J, Xu L, Hu Q, Xu C, Yang S, Jiang H. Histone demethylase KDM3a, a novel regulator of vascular smooth muscle cells, controls vascular neointimal hyperplasia in diabetic rats. Atherosclerosis. 2017;257:152–163. doi: 10.1016/j.atherosclerosis.2016.12.007. doi: 10.1016/j.atherosclerosis.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 63.Zhang BF, Jiang H, Chen J, Guo X, Hu Q, Yang S. KDM3A inhibition attenuates high concentration insulin-induced vascular smooth muscle cell injury by suppressing MAPK/NF-κB pathways. Int J Mol Med. 2018;41:1265–1274. doi: 10.3892/ijmm.2017.3351. doi: 10.3892/ijmm.2017.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone h3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein AM, Zaganjor E, Cobb MH. Chromatin-tethered MAPKs. Curr Opin Cell Biol. 2013;25:272–277. doi: 10.1016/j.ceb.2013.01.002. doi: 10.1016/j.ceb.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu J, Zhang J, Huang H, Li J, Yu Y, Jin H, Li Y, Deng X, Gao J, Zhao Q, et al. Crucial role of c-Jun phosphorylation at Ser63/73 mediated by PHLPP protein degradation in the cheliensisin a inhibition of cell transformation. Cancer Prev Res (Phila) 2014;7:1270–1281. doi: 10.1158/1940-6207.CAPR-14-0233. doi: 10.1158/1940-6207.CAPR-14-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pulverer BJ, Kyriakis JM, Avruch J, Nikolakaki E, Woodgett JR. Phosphorylation of c-jun mediated by MAP kinases. Nature. 1991;353:670–674. doi: 10.1038/353670a0. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]


