Abstract
Choline kinase catalyzes the conversion of choline to phosphocholine (PC) by transferring a phosphate group from adenosine triphosphate (ATP) as the first step in the biosynthetic pathway for the membrane phospholipid phosphatidylcholine, an essential pathway in the Leishmania parasitic protozoan. Commonly used methods for kinetically quantifying the enzyme include a radioisotope assay utilizing labeled choline and a coupled spectrophotometric assay with multiple enzymes and substrates that indirectly measures choline kinase activity. When testing potential inhibitors with the coupled assay, results can cast doubt on whether choline kinase is being inhibited or one of the coupled enzymes. Therefore, 31P NMR spectroscopy was used to quantitatively measure the formation of the key product, phosphocholine, and to evaluate choline kinase activity. Interrogation of 31P NMR spectroscopy offers a number of benefits. Since this isotope is 100% abundant and has a relatively large gyromagnetic ratio, it is considered one of the more sensitive nuclides. As such, the need for costly isotopic enriched phosphorous is not required and detection of the 31P signal is possible even at relatively low concentrations. The enzymatic activity of Leishmania infantum choline kinase was able to be directly measured via integration of the 31P resonance associated with the phosphocholine product (δ = 3.94 ppm). These initial studies reveal that a 31P NMR spectroscopic-based assay could be used for testing substrate or transition state analogs as competitive inhibitors of Leishmania choline kinase that may prevent phosphatidylcholine synthesis in the parasite.
Keywords: TBC, 31P NMR, Choline, Kinase, Leishmaniasis, Enzyme, Assay
TBC
1. Introduction
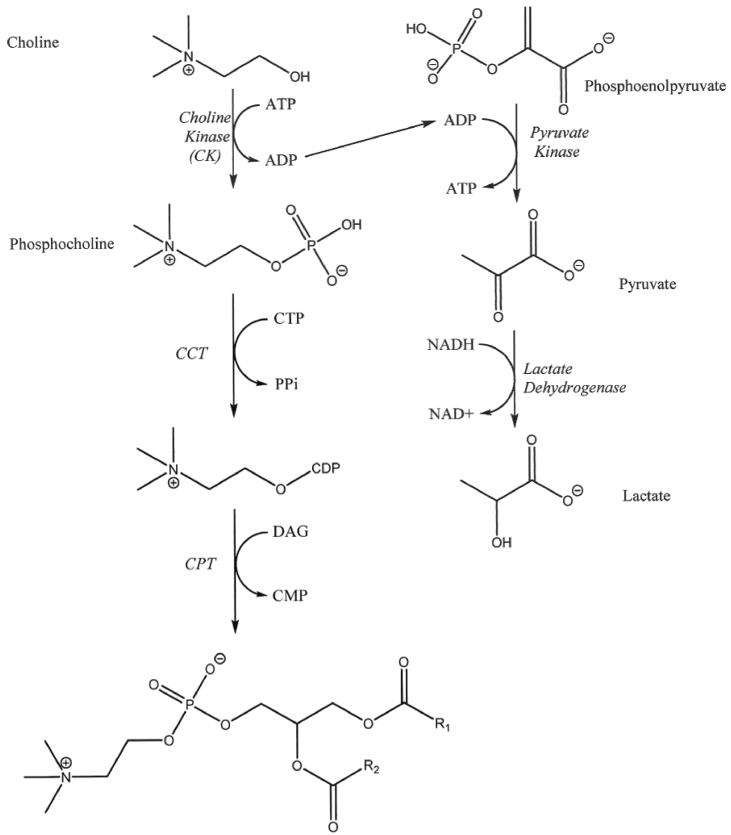
Choline kinase (EC 2.7.1.32) is an enzyme that transfers a phosphate from adenosine 5’-triphosphate (ATP) to choline, typically for synthesis of molecules that are precursors to membrane phospholipids [1, 2, 3]. Choline kinase is the first enzyme of the CDP-choline biosynthetic pathway for phosphatidylcholine (PC) synthesis, the major phospholipid in mammalian cell membranes [4, 5]. In mammals, PC can be synthesized via the CDP-choline pathway by the sequential catalytic action of three enzymes: choline kinase produces phosphocholine from choline and ATP, phosphocholine is converted to CDP-choline by CTP:phosphocholine cytidylyltransferase, and choline phosphotransferase synthesizes PC from CDP-choline and diacylglycerol (Fig. 1). In addition to serving as a membrane constituent, PC plays a role in signal transduction pathways and regulation of lipid second messengers, including fatty acids and diacylglycerols [6, 7].
Fig. 1.
CDP-choline pathway and choline kinase coupled enzyme assay. Synthesis of phosphatidylcholine is accomplished by the sequential action of three enzymes, choline kinase (CK), which converts choline to phosphocholine, CTP:phosphocholine cytidylyltransferase (CCT), which produces CDP-choline from phosphocholine, and choline phosphotransferase (CPT), the enzyme that synthesizes phosphatidylcholine from CDP-choline and diacylglycerol (DAG). The coupled assay is one of the traditional methods used to determine choline kinase activity. In this assay, the ADP product from choline kinase catalysis serves as a substrate for pyruvate kinase to generate pyruvate, which is then reduced by lactate dehydrogenase to lactate by transferring a hydride from NADH. The absorbance is monitored at 340 nm and an absorbance decrease is associated with NADH oxidation to NAD+. ATP is adenosine 5’-triphosphate, ADP is adenosine 5’-diphosphate, CTP is cytidine 5’-triphosphate, PPi is pyrophosphate, and CMP is cytidine 5’-monophosphate.
A search of the protein database (www.ncbi.nlm.nih.gov/protein) reveals choline kinase is found in mammals, bacteria, protozoans, archaea, and plant cells. Choline kinases from mammals and the nematode Caenorhabditis elegans have been characterized [8, 9] as has the enzyme from Trypanosoma brucei [10]. The gene encoding choline kinase of the malaria parasite Plasmodium falciparum was cloned and the enzyme characterized [11]. More recently, a gene encoding a choline kinase isoform from Leishmania infantum was cloned and the recombinant enzyme expressed, purified, and characterized [12].
Leishmania are a single cell parasitic protozoan that cause the neglected tropical disease leishmaniasis. Symptoms of infection include fever, weight loss, and an enlarged abdomen due to swollen spleen and liver. Affected individuals also routinely suffer from low white blood cell (leukopenia), red blood cell (anemia) and platelet (thrombocytopenia) counts. Identification of in vitro inhibitors that target phospholipid synthesis in parasitic protozoans would provide additional potential pharmaceutical agents to target leishmaniasis. Leishmania membrane lipids include phospholipids, sterols, and sphingolipids and are involved in cellular roles including protein anchoring, infection, and cell death. Approximately 70% of leishmanial lipids are phospholipids, comprised of 30–40% phosphatidylcholine (PC), 10% phosphatidylethanolamine (PE) and 10% phosphatidylinositol (PI) [13]. In order to assess the effectiveness of potential inhibitory compounds, a simple, inexpensive, and effective enzyme assay that directly measures choline kinase activity is needed. In the past, a radioisotope-dependent choline kinase enzyme assay, described by Gee and Kent, was often utilized [9]. To determine enzyme activity the radiolabeled product [14C]phosphocholine is separated from the substrate [14C]choline by ascending paper chromatography. Unfortunately, the radioactive substrate is expensive and generates undesirable radioactive waste. In addition, recovery and measurement of [14C]phosphocholine often results in poor reproducibility and greater standard error from replicates than desired. An alternative, non-radioactive, enzyme assay method also used for choline kinase is a coupled spectrophotometric method originally described by Wittenberg and Kornberg [14, 15] that requires the addition of two other enzymes. The first, pyruvate kinase, converts the ADP produced by choline kinase to ATP while simultaneously producing pyruvate from phosphoenolpyruvate. Therefore, phosphoenolpyruvate is also included as one of the assay components. The second additional enzyme, lactate dehydrogenase, then converts the pyruvate, produced by pyruvate kinase, to lactate along with the oxidation of NADH to NAD+. This, therefore, requires the addition of NADH to the assay mix. Finally, the activity is assessed by measuring the change in absorbance at 340 nm due to oxidation of NADH and activity is calculated using the absorptivity value for NADH (Fig. 1). Disadvantages of this assay include the expense of extra enzymes and substrates and the requirement of a buffer system and experimental conditions to optimize activity of all three enzymes. Also, the indirect measurement of choline kinase activity can result in significant variability in replicate measurements. In addition, when testing the effectiveness of a potential choline kinase inhibitor, any decrease in activity may be the result of any one of the three enzymes in the coupled assay being inhibited. Therefore, the major goal of this research was to develop a method utilizing nuclear magnetic resonance (NMR) spectroscopy to measure the concentration of phosphocholine produced by choline kinase. This NMR spectroscopic technique provides a way to directly quantify the choline kinase enzyme activity and is an alternative to the current radioisotope method or the indirect coupled choline kinase enzyme assay. In order to accomplish this goal, 31P NMR spectroscopy was used to investigate the conversion of choline to phosphocholine by purified, recombinant Leishmania infantum choline kinase.
2. Materials and methods
2.1. Protein purification
The recombinant 6x His-tagged Leishmania infantum choline kinase was expressed in ArcticExpress E. coli (Agilent) and purified as described previously [12]. In summary, protein expression was induced by addition of 1 mM IPTG to one liter cultures and induced cells were harvested after growth at 10 °C for 24 hours. Cells were resuspended in 20 mL of lysis buffer (50 mM Tris-Cl, 100 mM NaCl, pH 7.5) containing 1X protease inhibitor cocktail (Sigma) and lysed in a French Hydraulic press (Spectronic Instruments) at 10,000 psi. Following centrifugation in a Beckman JA-20 rotor for 40 min at 4 °C and 9800 g, the supernatant was applied to a 1 mL column of Ni-NTA resin (Qiagen) at 4 °C for metal-affinity chromatography and washed in succession with 50 mL lysis buffer and 50 mL lysis buffer containing 15 mM imidazole. Recombinant His-tagged protein was eluted by washing with 20 mL of lysis buffer containing 150 mM imidazole. Fractions (1 mL each) were collected and protein purity was evaluated by SDS-PAGE [16]. Protein concentration was determined using the method of Bradford [17] with a BSA standard curve and the Bio-Rad reagent following manufacturer's instructions.
2.2. Choline kinase enzyme assay
Choline kinase assay tubes contained a total volume of 1000 μL with 50 mM Tris-Cl, 100 mM sodium chloride, 10 mM magnesium chloride, 10 mM adenosine 5’-triphosphate (ATP), and 10 mM choline chloride, pH 7.5. Assay mixtures contained 90%/10% H2O/D2O, needed for NMR lock. The enzymatic reaction was initiated by the addition of ATP, incubated at 37 °C for the desired time, and terminated by placing the reaction tubes in a boiling water bath for 5 minutes. Once the reaction was stopped, the tubes were centrifuged for 2 minutes at 13,000 x g to pellet the denatured protein. Finally, 500 μL of the 1000 μL reaction was pipetted into a 5 mm NMR tube and 162 MHz 31P NMR data were collected.
2.3. NMR and sample preparation
All 162 MHz 31P{1H}NMR spectroscopic data were collected using a 400 MHz Bruker Avance III NMR spectrometer with broadband probe and the following instrument parameters used: TD = 16384, NS = 500, SW = 50 ppm and the acquisition time (AQ) = 1.01 sec. The pre-scan delay (d1) was set to 2.0 sec. The pulse program used was a routine 1D pulse sequence using a 30° flip angle. Phase cycling was performed using the AQ_mod: Digital Quadrature Detection (DQD). With this setting the digitization mode was also set to digital as required. The time domain FID obtained was multiplied by an exponential function ( with LB = 4.0) prior to Fourier transformation to improve S/N ratio. Digital resolution was also improved by doubling the digital data points in the frequency domain spectrum (i.e., zero filling). All spectra were phased accordingly and the baseline corrected prior to the integration of the 31P resonances. To ensure the complete integration, all peak integrals began and ended 0.25 ppm downfield and upfield, respectively from where the resonance meets the baseline. All samples were interrogated using the same acquisition parameters, which is necessary to ensure that the measured integrated peak areas correlate with the initial concentrations used in the kinetic assays. 85% H3PO4 was used as a chemical shift reference.
2.4. Determination of substrate and product chemical shift
Prior to performing the choline kinase assay, the 31P chemical shifts for the ATP, ADP and phosphocholine were measured under the same aqueous conditions that will be used in the assay and to ensure that little or no overlap of the phosphorous resonances is observed. The ability to measure accurately the integrated areas associated with the 31P peaks is important to obtaining the concentrations of all species in solution as choline reacts with ATP to form phosphocholine. All chemical shifts were referenced to an 85% phosphoric acid solution at δ = 0.0 ppm for this acid. This was accomplished by placing a sealed capillary containing the phosphoric acid into the NMR tube prior to collecting the 31P NMR spectral data.
2.5. Preparation of phosphocholine and ATP standard curves
A total of nine NMR samples were prepared in triplicate (27 samples) with a mixture of phosphocholine and ATP ranging in concentration from 0.2 mM to 25 mM. NMR spectral data were obtained for each sample using the NMR parameters discussed in Section 2.3. The integrated areas of the 31P resonances associated with the β-phosphate in ATP and the phosphocholine were plotted versus their respective concentrations in solution. The β-phosphate peak is shifted far up-field from the other phosphate resonances (including those resonances associated with ADP) and is the best choice for monitoring the change in ATP concentration during the choline kinase assay. These standard curves were used to determine the concentration of each species in solution throughout the enzyme-catalyzed reaction.
2.6. Assessment of choline analogs as potential choline kinase inhibitors
Choline analogs were purchased from Sigma. Analogs used were betaine hydrochloride, 3-bromopropyl trimethylammonium bromide, trimethylamine hydrochloride, 3-acrylamidopropyl trimethylammonium chloride, 3-chloro-2-hydroxypropyl trimethylammonium chloride, 2-aminoethyl trimethylammonium chloride, 3-carboxypropyl trimethylammonium chloride, chlorocholine chloride, trimethylammonium chloride, hexamethonium bromide, and 5-bromopentyl trimethylammonium bromide. Choline kinase was assayed in the presence of 5 mM of each inhibitor at either 10 mM or 1 mM choline.
3. Results and discussion
3.1. The 31P chemical shifts and the correlation of integrated signal area to concentration of phosphocholine
Use of 31P NMR spectroscopy as a means to explore choline kinase activity is feasible because the resonances associated with the substrate (ATP) and products (ADP and phosphocholine) show little or no overlap in the spectral data. This was confirmed by interrogating a NMR sample containing a mixture of ATP, ADP and phosphocholine (concentration of each was 10 mM) prepared in 90%/10% H2O/D2O with 50 mM Tris-Cl pH 7.5, 100 mM NaCl and 10 mM MgCl2, the same conditions used in the choline kinase assay. This control sample also contained a sealed capillary filled with 85% H3PO4, a compound which is commonly used as a chemical shift reference in phosphorous NMR. Fig. 2 shows the 31P NMR spectrum of this solution with the capillary tube present. The peaks associated with the α, β, and γ phosphates of ATP can be found at δ = -10.09, -18.57, and -4.81ppm, respectively while that for the phosphocholine is at δ = 3.94 ppm. The peaks associated with the α- and β-phosphates of ADP can be found at δ = -9.54 and δ = -5.53 ppm respectively. With good separation of peaks observed in the NMR data, the ability to correlate the integrated peak areas for all phosphates with the concentrations of substrate and products is straightforward.
Fig. 2.
162 MHz 31P{1H}-NMR spectrum of a sample containing a mixture of 10 mM ATP, 10 mM ADP and 10 mM phosphocholine in 90:10 H2O:D2O buffered solution (D2O is needed for NMR lock). See text for exact composition of the aqueous solution. A sealed capillary tube containing 85% phosphoric acid was placed in the NMR sample tube. The phosphoric acid resonance is used as a reference in 31P NMR and is assigned a δ = 0.0 ppm. A closer look at the phosphate resonances associated with ATP and ADP indicate that 2JPP coupling between adjacent phosphorous nuclei is observed. The doublets observed for the α- and γ-phosphates and the triplet for the β-phosphate of ATP all give a 2JPP = 16.0 Hz. The doublets observed for the α- and β-phosphates of ADP all give 2JPP = 19.1 Hz.
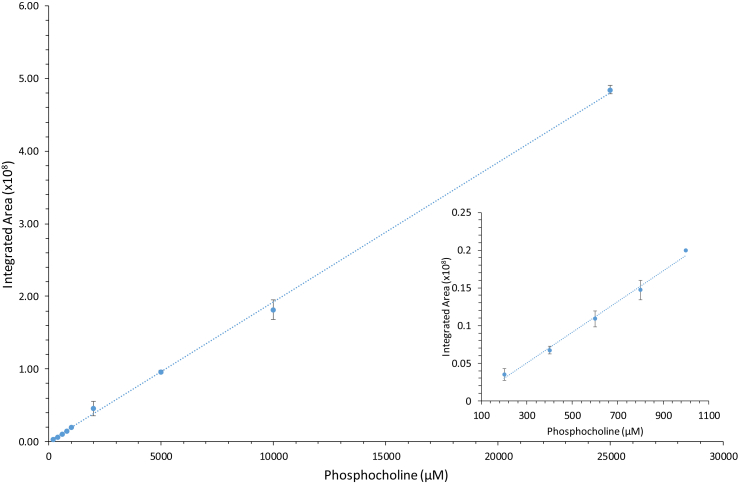
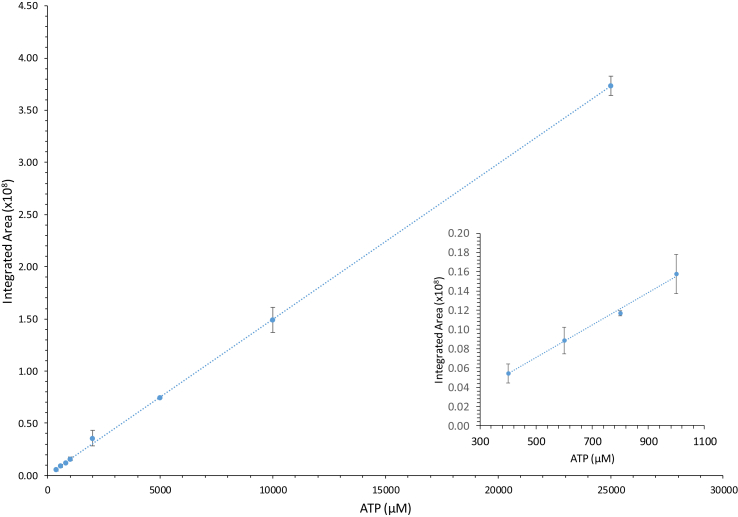
In order to accurately measure the [ATP] and [phosphocholine] in solution throughout the course of the choline kinase catalyzed reaction, standard curves were generated to show the linear relationship between the integrated areas of the phosphate resonances and the known concentrations of these two species. Fig. 3 and Fig. 4 contain these standardized curves for phosphocholine and ATP over concentrations ranging from 0.2 mM and 25 mM. In the case of ATP, the β-phosphate peak (δ = -18.57 ppm) was integrated and this area was plotted against the [ATP].
Fig. 3.
Preparation of a phosphocholine standard curve. A plot of the integrated peak area of the phosphate resonance for phosphocholine as a function of known concentration. The inset plots show the region of lower concentration. Data points are plotted as the average of three independent integrated areas determined for each peak and the standard deviation in these averages is indicated by the error bars. Regression analysis yielded a linear equation of y = 0.000193x-0.00328 with R2 = 0.9991.
Fig. 4.
Preparation of an ATP standard curve. A plot of the integrated peak area of the β-phosphate resonance for ATP as a function of known concentration. The inset plots show the region of lower concentration. Data points are plotted as the average of three independent integrated areas determined for each peak and the standard deviation in these averages is indicated by the error bars. Regression analysis yielded a linear equation of y = 0.000149x+0.00887 with R2 = 0.9997.
3.2. Utilization of 31P NMR spectroscopy to evaluate catalysis by Leishmania infantum choline kinase
The efficiency by which choline kinase produces phosphocholine from the reaction between choline and ATP was measured at varying concentrations of the enzyme while the concentration of the two substrates was held constant, as was the required Mg2+ cofactor needed for enzyme activity. As expected, when no choline kinase is present in solution only those resonances associated with the triphosphate of ATP are present in the NMR spectrum, no measurable amount of phosphocholine is observed indicating the reaction is essentially nonexistent without enzyme (Fig. 5). However, upon addition of purified, recombinant Leishmania infantum choline kinase, the production of phosphocholine after only 15 minutes is clearly observed in the NMR spectrum by the presence of a new peak at δ = 3.94 ppm (Fig. 5). Also seen in the NMR spectrum are the two phosphate resonances associated with the other product ADP found at δ = -5.53 and -9.54 ppm while those for the ATP have decreased in intensity. Additional experiments were performed at higher choline kinase concentrations, while maintaining the same concentration of substrates, and it is clear from the three top NMR spectra in Fig. 5 that the amount of phosphocholine produced after 15 minutes has increased with more enzyme present. Fig. 6 shows the linear growth in phosphocholine as a function of choline kinase concentration when the assay was performed over a fixed time period at each of the enzyme concentrations.
Fig. 5.
162 MHz 31P{1H}-NMR spectra collected after the choline kinase assay was performed with varying amount of enzyme present in solution. Enzyme concentrations are displayed on the right side of each spectrum and each assay was for 15 minutes at 37 °C and pH 7.5 and contained 10 mM ATP and 10 mM choline chloride.
Fig. 6.
A plot of phosphocholine concentration (μM) versus choline kinase protein concentration plotted as a function of protein concentration. Enzyme concentrations were 0 μg/mL, 20 μg/mL, 30 μg/mL, and 50 μg/mL. The substrates ATP and choline were each held constant at 10 mM. Data points are the average of three independent determinations with standard deviation indicated by the error bars. Regression analysis yielded a linear equation of y = 139.13x-158.53 with R2 = 0.9938.
In addition, the rate of phosphocholine formation was determined at a fixed concentration of choline kinase (35 μg/mL) by interrogating samples via 31P NMR from assays that were incubated over time. Fig. 7 shows four NMR spectra of samples obtained from the choline kinase assay, which were incubated with enzyme at four different times. After one minute the presence of phosphocholine in solution is clearly seen in the NMR spectrum. Over a period of 35 minutes the steady growth in phosphocholine and ADP is observed (Fig. 7). Once the area under the phosphocholine peaks was determined through integration, the phosphocholine standard curve generated (Fig. 3) was used to determine the [phosphocholine] in solution as a function of time. Fig. 8 shows the production of phosphocholine over this period of time; the rate is found to be 36.7 μM/min.
Fig. 7.
162 MHz 31P{1H}-NMR spectra of choline kinase assays incubated for various times with a fixed concentration of CK (35 μg/mL). Spectra illustrate the growth of the signals associated with the products ADP and phosphocholine.
Fig. 8.
Production of phosphocholine by choline kinase using the 31P NMR assay. The concentration of enzyme was held constant at 35 μg/mL and ATP and choline were each 10 mM. The integrated area was converted to concentration of phosphocholine or ATP and plotted versus time. Data points are the average of three independent determinations with standard error indicated by the error bars. Regression analysis yielded a linear equation of y = 36.67x+276.34 with R2 = 0.9569.
3.3. Calculation of specific activity of Leishmania infantum choline kinase
The data in Fig. 5, phosphocholine produced as a function of choline kinase concentration, were used to calculate the specific activity value in units of μmoles phosphocholine produced per minute per milligram of enzyme (μmol/min/mg). The concentration of the phosphocholine was calculated by integrating the phosphocholine signal at 3.94 ppm. The production of phosphocholine was determined at three concentrations of choline kinase (20 μg/mL, 30 μg/mL, and 50 μg/mL). For each enzyme concentration, the phosphocholine peak was integrated and the corresponding μmoles of phosphocholine determined from the phosphocholine standard curve (Fig. 3). By taking into account the assay time (15 minutes) and the mg of enzyme used in each assay, the μmoles of phosphocholine produced per minute per mg of enzyme was calculated. After calculating the individual specific activity at each enzyme concentration, the three values were averaged, and the standard error determined. The average specific activity for purified, recombinant Leishmania infantum choline kinase was calculated to be 8.74 ± 0.89 μmoles/min/mg protein. Pulido et al [12] reported a Vmax value of 3.52 μmoles/min/mg using the coupled enzyme assay and specific activity values of approximately 6.5 μmoles/min/mg using the radioisotope assay for a pH activity study. Therefore, the specific activity values for Leishmania infantum choline kinase obtained in this study using the 31P NMR assay compare favorably to previous values obtained using the coupled assay and the radioisotope assay with the same recombinant enzyme.
3.4. Assessment of choline analogs as potential choline kinase inhibitors
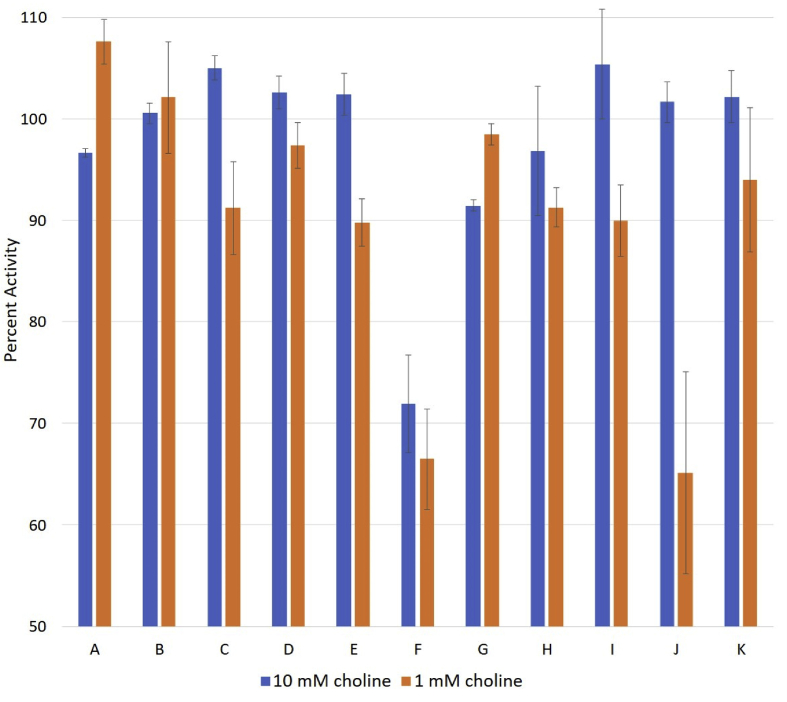
The 31P NMR choline kinase assay developed in the present work was used to assess whether L. infantum choline kinase was inhibited by a variety of choline analogs (Fig. 9). The production of phosphocholine was determined in the presence of 5 mM of each choline analog and compared to phosphocholine production in the absence of a choline analog. The concentration of choline substrate was either 10 mM or 1 mM. If any of the choline analogs were to inhibit the activity of choline kinase, they would be expected to be competitive in nature. Therefore, a decrease in choline kinase activity would be more likely to be observed when the choline substrate is present at the lower 1 mM concentration. The greatest inhibition was seen in the presence of either 2-aminoethyl trimethylammonium ion (analog F) or hexamethonium ion (analog J), which both decreased activity approximately 35% in the presence of 1 mM choline (Fig. 10).
Fig. 9.
Chemical structures of choline analogs tested as potential inhibitors of L. infantum choline kinase. Analogs are denoted as A–K.
Fig. 10.
Investigation of choline analogs as potential inhibitors of L. infantum choline kinase. Production of phosphocholine by choline kinase at either 10 mM or 1 mM choline was assessed in the presence of 5 mM of each choline analog. Activity in the presence of each analog is plotted as the percent activity in the absence of analog. Values are the average of three determinations with standard error indicated by the error bars. Analogs are denoted as A-K and correspond to the structures shown in Fig. 9.
3.5. Summary and conclusions
Using 31P NMR as a means to assess the activity of choline kinase is a new approach to analyze catalysis by this specific enzyme of phospholipid biosynthesis, however, 31P NMR has been used for decades to characterize a variety of phosphate-utilizing enzymes. In theory, any enzyme that uses a phosphate-containing molecule as a substrate or produces a phosphorylated product is a candidate to be studied via 31P NMR. As an example of the versatility of the technique, in the 1980s Rosch [18] reported the usefulness of 31P NMR for studying phosphoryl transferases such as adenylate kinase, creatine kinase, pyruvate kinase, arginine kinase, 3-phosphoglycerate kinase, and hexokinase. In addition, phosphoric ester hydrolases such as alkaline phosphatase and glycogen phosphorylase were also investigated. More recently, Richter et al [19] determined the specificity of ADP-dependent glucokinase for D-glucose by incubating the enzyme with a variety of potential sugar substrates and showing the lack of signals corresponding to the product AMP or the phosphorylated sugar. In this current study simple one-dimensional 31P NMR was employed to assess the catalytic activity of choline kinase in vitro. However, 31P NMR has also been applied to in vivo studies of enzymes, including in cancer cells [20, 21]. Others have even measured the 31P NMR signal of bound substrates and products to elucidate enzyme mechanisms [22, 23].
The principle motivation for development of a 31P NMR-based choline kinase assay was to replace the radioisotope assay and the coupled assay, methods used previously to characterize choline kinase. The financial burden of the radioisotope assay is a significant barrier to productivity when a large number of choline kinase assays are being conducted. In addition, undesirable radioactive waste is generated, subjecting research programs to additional costs and regulatory requirements. In the coupled enzyme assay the reduction in absorbance at 340 nm correlated to the oxidation of NADH to NAD+ by lactate dehydrogenase is actually measured instead of a direct measurement of choline kinase activity. In addition, optimizing activity of three enzymes (choline kinase, pyruvate kinase, and lactate dehydrogenase) is a difficult balancing act. In contrast, the 31P NMR spectroscopic assay described in this report provides an inexpensive, direct determination of product formation and is potentially invaluable for screening compounds as potential inhibitors of choline kinase. The possibility also exists that the 31P NMR spectroscopic assay methodology presented here could be useful as a basis for the development of new methods for assaying additional enzymes that catalyze phosphorylation reactions.
Declarations
Author contribution statement
Jacob A. Walker: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Joshua D. Friesen: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Steven J. Peters, Jon A. Friesen: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Marjorie A. Jones: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.McMaster C.R. From yeast to humans – roles of the Kennedy pathway for phosphatidylcholine synthesis. FEBS Lett. 2018:1256–1272. doi: 10.1002/1873-3468.12919. [DOI] [PubMed] [Google Scholar]
- 2.Wu G., Vance D.E. Choline kinase and its function. Biochem. Cell Biol. 2010;88:559–564. doi: 10.1139/O09-160. [DOI] [PubMed] [Google Scholar]
- 3.Li Z., Vance D.E. Phosphatidylcholine and choline homeostasis. J. Lipid Res. 2008;49:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Kent C. CTP:Phosphocholine cytidylyltransferase. Biochem. Biophys. Acta. 1997;1348:79–90. doi: 10.1016/s0005-2760(97)00112-4. [DOI] [PubMed] [Google Scholar]
- 5.Fagone P., Jackowski S. Phosphatidylcholine and the CDP-choline cycle. Biochim. Biophys. Acta. 2013;1831:523–532. doi: 10.1016/j.bbalip.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornell R. Chemical cross-linking reveals a dimeric structure for CTP: phosphocholine cytidylyltransferase. J. Biol. Chem. 1989;264:9077–9082. [PubMed] [Google Scholar]
- 7.Kalmar G.B., Kay R.J., Lachance A., Aebersold R., Cornell R.B. Cloning and expression of rat liver CTP: phosphocholine cytidylyltransferase: an amphipathic protein that controls phosphatidylcholine synthesis. Proc. Natl. Acad. Sci. 1990;87:6029–6033. doi: 10.1073/pnas.87.16.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoyama C., Liao H., Ishidate K. Structure and function of choline kinase isoforms in mammalian cells. Prog. Lipid Res. 2004;43:266–281. doi: 10.1016/j.plipres.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Gee P., Kent C. Multiple isoforms of choline kinase from Caenorhabditis elegans: cloning, expression, purification, and characterization. Biochim. Biophys. Acta. 2003;1648:33–42. doi: 10.1016/s1570-9639(03)00106-7. [DOI] [PubMed] [Google Scholar]
- 10.Gibellini F., Hunter W.N., Smith T.K. Biochemical characterization of the initial steps of the Kennedy pathway in Trypanosoma brucei: the ethanolamine and choline kinases. Biochem. J. 2008;415:135–144. doi: 10.1042/BJ20080435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choubey V., Maity P., Guha M., Kumar S., Srivastava K., Puri S.K., Bandyopadhyay U. Inhibition of Plasmodium falciparum choline kinase by hexadecyltrimethylammonium bromide: a possible antimalarial mechanism. Antimicrob. Agents Chemother. 2007;51:696–706. doi: 10.1128/AAC.00919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulido S.A., Nguyen V.H., Alzate J.F., Cedeño D.L., Makurath M.A., Ríos-Vásquez A., Duque-Benítez S.M., Jones M.A., Robledo S.M., Friesen J.A. Insights into the phosphatidylcholine and phosphatidylethanolamine biosynthetic pathways in Leishmania parasites and characterization of a choline kinase from Leishmania infantum. Comp. Biochem. Physiol. B. 2017;213:45–54. doi: 10.1016/j.cbpb.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K., Beverley S.M. Phospholipid and sphingolipid metabolism in Leishmania. Mol. Biochem. Parasitol. 2010;170:55–64. doi: 10.1016/j.molbiopara.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittenberg J., Kornberg A. Choline phosphokinase. J. Biol. Chem. 1953;202:431–444. [PubMed] [Google Scholar]
- 15.Major L.L., Denton H., Smith T.K. Coupled enzyme activity and thermal shift screening of the Maybridge role of 3 fragment library against Trypanosoma brucei choline kinase: a genetically validated drug target. In: El-Shemy H.A., editor. Drug Discovery. InTech; 2013 Jan. Chapter 14. [PubMed] [Google Scholar]
- 16.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Rosch P. NMR-studies of phosphoryl transferring enzymes. Prog. NMR Spectr. 1986;18:123–169. [Google Scholar]
- 19.Richter J.P., Goroncy A.K., Ronimus R.S., Sutherland-Smith A.J. The structural and functional characterization of mammalian ADP-dependent glucokinase. J. Biol. Chem. 2016;291:3694–3704. doi: 10.1074/jbc.M115.679902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brindle K.M. NMR methods for measuring enzyme kinetics in vivo. Prog. NMR Spectr. 1988;20:257–293. [Google Scholar]
- 21.Daly P.F., Lyon R.C., Faustino P.J., Cohen J.S. Phospholipid metabolism in cancer cells monitored by 31P NMR spectroscopy. J. Biol. Chem. 1987;262:14875–14878. [PubMed] [Google Scholar]
- 22.Vasavada K.V., Kaplan J.I., Nageswara Rao B.D. Analysis of 31P NMR spectra of enzyme-bound reactants and products of adenylate kinase using density matrix theory of chemical exchange. Biochemistry. 1984;23:961–968. doi: 10.1021/bi00300a025. [DOI] [PubMed] [Google Scholar]
- 23.Liu F.L., Fromm H.J. 31P nuclear magnetic resonance spectroscopy studies of substrate and product binding to fructose-1,6-bisphosphatase. J. Biol. Chem. 1991;266:11774–11778. [PubMed] [Google Scholar]