Abstract
The translocation of proteins across membranes is a fundamental cellular function. Bacteria have evolved a striking array of pathways for delivering proteins into or across cytoplasmic membranes and, when present, outer membranes. Translocated proteins can form part of the membrane landscape, reside in the periplasmic space situated between the inner and outer membranes of Gram-negative bacteria, deposit on the cell surface, or be released to the extracellular milieu or injected directly into target cells. One protein translocation system, the general secretory pathway (GSP), is conserved in all domains of life. A second, the twin-arginine translocation (Tat) pathway, is also phylogenetically distributed among most bacteria and plant chloroplasts. While all cell types have evolved additional systems dedicated to the translocation of protein cargoes, the number of such systems in bacteria is now known to exceed nine. These dedicated protein translocation systems, which include the types 1 through 9 secretion systems (T1SSs - T9SSs), the chaperone-usher (CU) pathway, and type IV pilus (T4P) system, are the subject of this review. Most of these systems were originally identified and have been extensively characterized in Gram-negative or diderm (two-membrane) species. It is now known that several of these systems also have been adapted to function in Gram-positive or monoderm (single-membrane) species, and at least one pathway (T7SS) is found only in monoderms. This review briefly summarizes the distinctive mechanistic and structural features of each dedicated pathway, as well as the shared properties, that together account for the broad biological diversity of protein translocation in bacteria.
Keywords: protein translocation, pilus, pathogenesis, traffic ATPases
Introduction
Among bacteria, the translocation of proteins across the cell envelope is fundamental for survival, niche establishment, and pathogenesis. In recent years, bacterial protein translocation has received considerable attention in the context of human medicine, as it has become increasingly evident that pathogens deliver proteins to the cell surface or into eukaryotic target cells as a major armament for invasion and infection [1,2]. Bacteria deploy the highly conserved general secretory pathway (GSP) and twin arginine translocation (TAT) systems to deliver proteins across the inner or cytoplasmic membrane (CM) [3,4]. Bacteria also have evolved many specialized translocation systems to deposit proteins on their cell surfaces, or deliver them to the extracellular milieu or into target cells [5,6]. Collectively, these systems accommodate a highly diverse repertoire of protein substrates, ranging from small to large, hydrophilic to hydrophobic, monomeric or multimeric, unfolded or folded, unmodified or posttranslationally modified, or even covalently attached to DNA. Gram-negative bacteria, whose cell envelopes consist of two membranes with intervening periplasm and cell wall, have evolved more than nine specialized translocation systems dedicated to protein trafficking to the cell surface or beyond. Gram-positive bacteria, whose envelopes are comprised of a single cytoplasmic membrane and a thick cell wall, have appropriated several of the systems functioning in Gram-negative species, and also have evolved their own specialized systems.
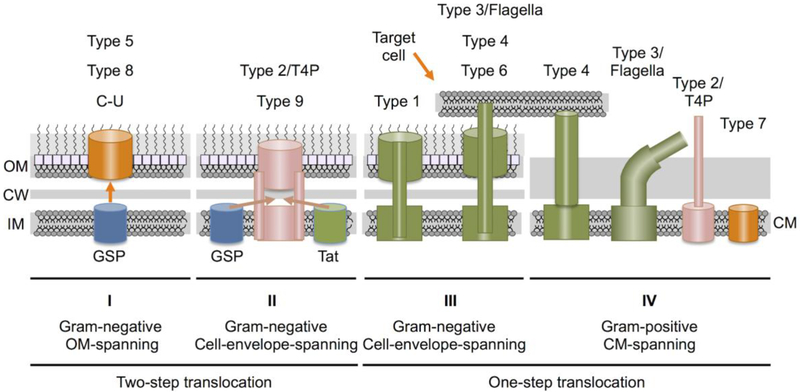
A full accounting of dedicated protein translocation in bacteria is beyond the scope of this review. Instead, here, I will highlight the broad biological diversity of protein translocation by describing system-specific features as well as shared properties. We can envision bacterial protein translocation as a rich tapestry, the dominant patterns corresponding to each system and the interconnecting threads representing the common mechanistic or biological functions. Over the past decade or so, exciting details of this tapestry have been unveiled through many state-of-the art approaches, including most recently the use of high-resolution cryoelectron tomography to visualize bacterial translocation ‘nanomachines’ in the native context of the bacterial cell envelope [7]. For purposes of discussion, these systems are grouped into four Classes (see Fig. 1 & Table 1). The first three Classes function in Gram-negative (diderm) species and the fourth in Gram-positive (monoderm) species.
Fig. 1. Bacteria have evolved more than nine distinct pathways for delivering proteins across their cell envelopes.
Systems are grouped into one of four different Classes based on their location within the cell envelope and mechanism of transfer. Class I: These systems assemble at the outer membrane (OM) of Gram-negative bacteria and mediate substrate transfer across the OM. Class I systems fall into the broad category of two-step translocation systems because substrates are delivered to the cell surface in two-steps, first across the inner membrane (IM) via the general secretory pathway (GSP) and second across the outer membrane (OM) via the Class I machine. Class II: These systems assemble across the entire Gram-negative cell envelope but mediate substrate transfer only across the OM. Class II systems are also designated as two-step translocation systems because substrates are delivered across the IM via the GSP or twin-arginine-translocation (Tat) pathways, and across the OM by the Class II system. Class III: These systems assemble across the entire Gram-negative cell envelope and function as one-step translocation systems by recruiting substrates from the cytoplasm and delivering the cargoes to the cell surface through channels that span the IM, periplasm and OM. A subset of Class III machines deliver substrates directly into bacterial or eukaryotic target cells. Class IV: These systems assemble across the cytoplasmic membranes (CM) of Gram-positive bacteria. They generally function as one-step translocation systems by delivering substrates across the CM, although specialized features are also postulated to be necessary for translocation across the thick cell wall, or Mycobacterial spp. mycolic acid layer. Systems assigned to each Class are listed at top. C-U, chaperone-usher pathway; T4P, type 4 pilus assembly pathway.
Table 1.
Properties of Dedicated Protein Translocation Systems in Bacteria.
| System | Distribution | Translocated substrates | Translocation requirements | Machine properties | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Types | Destination(s) | Function | Secretion signal(s) | Additional requirements | OM channel | IM channel | Requirements for biogenesis | Energetics | ||
| Class I: Two-step translocation systems—assemble only at the OM and translocate substrates from the periplasm across the OM | ||||||||||
| Type 5 (Auto transporters) | Gram-negative; mainly studied in Proteobacteia | Monomric/trimeric passenger domains; two-partner systems | Cell surface; extracellular milieu | Adhesin; toxins; proteases; receptor-binding proteins | N-termina Sec; Bam complex targeting signals | Autoproteolytic release from cell surface for some substrates | Hybrid anchor domain/BamA β-barrel channel | GSP | SurA/Skp/DegP chaperones, Bam complex | Charge interaction; vectorial folding TamA/B complex for subset of T5SSs |
| C-U pilus assembly | Gram-negative; Enterobacteriaceae | Type I/Pap pili CFA pili; CS pili; Afa/Dr fibers; F1 capsular antigen | Cell surface | Adhesion; Aggregation; Biofilms; IBCs; Activation of host cell pathways | N-terminal Sec; signal; Chaperone-pilin binding to usher | Donor strand complementation(DSC) for chaperone-pilin-contacts; Donor strand exchange(DSE) for pilin-pilin interactions | Usher: 24-strandei β-barrel with substrate binding/& pilus assembly domains | GSP | Bam-dependent Insertion of usher into OM; Dedicated chaperone binds pilins | Chaperone-pilin/usher binding affinities; Pilus subunit-subunit interaction affinities; entropy-based diffusion; evidence for a role of the TamA/B complex |
| Type 8(Curii) | Gram-negative; Proteobacteria, Bacteriodetes | Curli amyloid fibers | Cell surface; extracellular milieu | Adhesion biofilms; Host colonization; Innate response activation | N-terminal Sec signal; Chaperone-pilin binding to CsgG translocon | Charge-based chaperone-pilin and pilin-pilin interactions | CsgG: 36-stranded β-barrel; CsgE gate CsgB/F curli nucleator | GSP | LOL sorting/Bam-independent insertion of OM translocon; dedicated chaperone binds pilins | Peptide diffusion; entropy free-energy gradient |
| Class II: Two-step translocation systems—assemble across entire cell envelope but translocate substrates from the periplasm across the OM | ||||||||||
| Type 2 | Gram-negative; Proteobacteria | Monomeric, multimeric proteins | Cell surface via lipid or other attachment; Extracellular milieu | Toxins; lipases, other lytic enzymes; biofilm matrix components | N-terminal Sec or Tat; signal for binding to pseudopilus | Substrate folding in the periplasm; Recruitment to pseudopilus tip for extrusion through OM secretin channel | Secretin: 15 copies of GspD::GspS complexes | GSP or Tat | LOL-sorting & Bam-independent OM insertion; Secretin assembles first and stabilizes IM platform & GspE ATPase; Pseudopilus assembles for substrate extrusion | GspE ATPase drives pseuodopilus assembly |
| T4P | Gram-negative & -positive; Archaea | Dynamic type 4 pili; some T4Ps export exoproteins | Cell surface | Attachment; Bio-films; Twitching motility; Archaeal T4Ps can function as flagella | N-termina Sec for insertion of pilin into IM; Signals for pilin extrusion from and reinsertion into IM | T4P assembly on IM/CM platform; Pilus extension through G-negative secretin channel or G -positive cell wall | T2SS GspD secretin | GSP for insertion of pilins into IM | IM platform and GspE/PilT ATPases for pilus assembly/retraction | GspE/PilaT ATPases & PMF for Pilus extension/retraction; PilT-independent retraction in some systems |
| Type 9 | Gram-negative; Bacteriodetes | Monomeric proteins, including very large (~670 kDa SprB) adhesins | Cell surface; Extracellular milieu | Adhesion; Polymer degradation; Biofilms; Gliding motility: SprB adhesin moves rapidly between cell poles along closed helical loop | N-terminal Sec;Conserved CTDs for recruitment to T9SS in periplasm | Cell surface anchoring to acidic LPS by 'sortase-like' mechanism | SprA/sov: 36-strand β-barrel; PorV complex cleaves CTDs and attaches substrates to LPS | GSP | SprA/sov channel regulated by PorN,P,K,V; OM channel physically linked to IM-spanning PorM/L complex | IM PMF; secretion coupled to gliding motility |
| Class III: One-step translocation systems—span Gram-negative cell envelope and translocate substrates without a periplasmic intermediate | ||||||||||
| Type 1 | Gram-negative | Monomeric unfolded proteins | Cell surface; Extracellular milieu | Adhesin; Proteases; Lipases; Heme-binding | C-terminal signal forbinding to MFP/ATPase complex | Lap/Iba substrates are released by environmentally-induced, post-translocation cleavage | TolC-like channel/tunnel | ATPase/MFP complex | Substrate and ATP-binding induced channel activation and recruitment of TolC | ABC ATPase; Ca2+-mediated protein folding to prevent backsliding |
| Type 3: Injectisomes and flagella | Gram-negative; Gram-positive flagella | Monomeric unfolded proteins | Eukaryotic cell Flagellum assembly | Effector translocation disrupts various eukaryotic cell pathways and physiological processes; Flagellar-based motility | N-terminal peptide signal or 5′ RNA signal | Ordered contacts with sorting platform; ATPase-mediated unfolding; Target-cell-contact mediated machine activation | Secretion channel through which needle complex extrudes; L-/P-rings for Flagella assembly | ATPase/sorting platform/Export apparatus | IM complex and OM secretin complex form and join together as the needle complex (NC). NC is a scaffold for substrate sorting platform and injection needle | InvC-Like ATPase for substrate unfolding; PMF for translocation |
| Type 4 | Gram-negative and -positive; Archaea | Monomeric unfolded proteins; Multimeric A/B5 toxin, Single-stranded DNA-relaxase intermediates of MGEs | Bacterial or eukaryotic cells; Extracellular milieu; Surface-displayed conjugative pili | Conjugative DNA transfer; Effector translocation disrupts eukaryotic cell pathways and physiological processes; interbacterial toxin transmission; pilus-mediated adherence and biofilm development | C-terminal charged or hydrophobic, or internal motifs; Two-step translocation for PT export by B. pertussis Ptl system | Substrate docking & target-cell-contact mediated machine activation; Some systems elaborate conjugative pili | α-Helical OM pore connected to barrel/disc-shaped OMC | IMC:IM platform; VirB4, VirD4 ± VirB11 ATPases | LOL-sorting/Bam-independent insertion of OMC into OM; OMC assembles & stabilizes IMC and VirB4; VirB11 and VirD4 dock stably or transiently with IMC | ATPase- & substrate-induced conformational changes & PMF |
| Type 6 | Gram-negative | Monomeric proteins; may be covalently bound to machine components | Bacterial or eukaryotic cells | Effectors modulate eukaryotic cell processes; anti-bacterial and anti-eukaryotic cell toxins | Signals for effector docking with sheath, tube, or spike proteins; Covalent binding of effectors with these machine subunits | Target-cell-contact mediated machine activation | TssL/J/M membrane complex | TssL/J/M membrane complex/Baseplate | Sheath/tube assembly & contraction | PMF for sheath/tube contraction; ClpV ATPase-mediated recycling of sheath components |
| Class IV: One-step translocation systems-span Gram-positive cytoplasmic membrane and translocate substrates to cell surface or beyond | ||||||||||
| Type 7 | Gram-positive Mycobacteria, Actinobacteria, Firmicutes | Monomeric proteins; Homo- and hetero-dimers; WxG1OO & PPE/PE proteins | Cell surface; Extracellular milieu | Membrane permeabilization; Virulence; DNA conjugation; sliding motility; other? | C-terminal signals mediate binding to EccC ATPase | Homo- or hetero-dimer formation for some substrates | Two-step mechanism postulated for translocation across mycobacterial mycolic membrane | EccB/C/D/E membrane complex; VirD4- like EccC has 3 ATPase domains | Machine assembly & translocation mechanism unknown | EccC ATPase-mediated translocation |
1. Class I: Two-step translocation systems - Assemble only at the OM and translocate substrates from the periplasm across the OM.
Systems designated as Class I are phylogenetically widely distributed among Gram-negative bacteria, but are best characterized in members of the phylum Proteobacteria. These systems assemble only at the OM. The overall mechanism of translocation is designated as two-step because substrates are first delivered across the IM via the GSP to the periplasm and then they are recruited by the Class I system for translocation across the OM. Class I systems rely on energy sources other than ATP hydrolysis or the electrochemical gradient that are associated with inner membrane (IM) to drive substrate transfer. Class I systems include the T5SSs (autotransporters), T8SSs (curli), and the C-U (chaperone-usher pilus/fimbriae) pathway (Fig. 2). Functions of translocated substrates vary widely, but often are associated with surface display of adhesive proteins or pilus organelles of importance for cellular aggregation, establishment of biofilms, or colonization of mammalian host tissues.
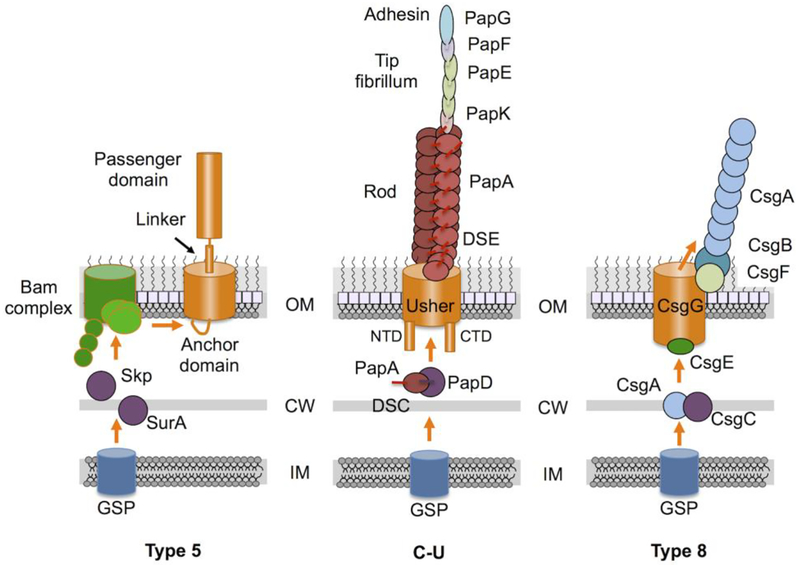
Fig. 2. Schematics of Class I assembly pathways.
Type 5 secretion systems (autotransporters) rely on the GSP for delivery across the IM and then are recruited to the Bam complex for OM insertion and surface display. C-U (Chaperone - Usher) systems as exemplified by the Pap pilus assembly pathway. Pilin subunits bind a periplasmic chaperone, e.g., PapD, by donor-strand complementation (DSC) and are delivered to the OM usher for delivery across the OM and pilus assembly by donor-strand exchange (DSE). Type 8 secretion systems are responsible for assembly of curli amyloid fibers.
1.1. Type 5 secretion systems (T5SSs).
T5SSs are also termed ‘autotransporters’ to reflect an early view that they mediate the delivery of proteins or domains across the OM independently of other cellular factors [8]. T5SSs are characteristically composed of a single polypeptide consisting of an N-terminal signal sequence (Sec) for translocation through the GSP, an extracellular or ‘passenger’ domain, a linker domain, and a ‘translocator’ or ‘anchor’ domain. Some T5SSs additionally possess a protease domain that cleaves off the passenger domain upon delivery to the cell surface. T5SSs have been subgrouped into 5 types based on structural variations in the passenger and/or anchor domains; subtype 5b systems are termed two-partner secretion systems because separate genes encode the passenger and anchor domains as distinct polypeptides [8–10]. Surface-exposed passenger domains exhibit a range of protease, adhesin, toxin or receptor binding activities, which play important roles in bacterial attachment, biofilm development, and infection [11].
The anchor domain folds as a β-barrel, which originally was thought to form the OM channel responsible for extrusion of the passenger domain to the cell exterior, hence, the term autotransporter [12,13]. It is now evident that maturation of T5SSs is considerably more complex, and involves a number of cellular factors required for insertion of β-barrel proteins in the OM (Fig. 2). These factors include periplasmic chaperones, e.g., SurA, Skp, DegP, and the β-barrel assembly machine (Bam complex) [8,14–17]. In a new model for autotransporter translocation [8], substrates are first delivered through the GSP to the periplasm where they bind periplasmic chaperones. The anchor domain is recruited by the Bam complex, which initiates - but does not complete - insertion of the anchor domain into the OM. As a result, a hybrid channel is formed, composed of the anchor domain and the β-barrel of BamA in an open conformation. Next, the passenger domain binds the Bam complex and is threaded through the hybrid β-barrel - BamA channel. Once the passenger domain is extruded to the cell surface, surface-exposed basic or large polar residues stimulate release of the anchor from BamA and closing of the β-barrel [18]. Finally, the passenger domain folds and either remains tethered to the β-barrel anchor domain or is proteolytically cleaved and released into the milieu [8].
It is postulated that directionality of autotransporter translocation is driven by charge-based interactions that help promote insertion of the anchor domain into the OM [18]. Vectorial folding of secreted regions might also ‘tug’ the passenger domain through the hybrid OM channel and block reentry into the channel [19–21]. Interestingly, recent work showed that TamA and its partner protein TamB facilitates the assembly of a subset of autotransporters [17,22]. TamA, like BamA, is member of the Omp85 superfamily, but its interaction partner, TamB extends across the periplasm and through the IM [23]. Through its contacts with the IM, the Tam complex might provide a source of mechanical energy for insertion of the autotransporter anchor domain into the OM [24].
1.2. Chaperone-usher (C-U) pilus biogenesis systems.
The C-U pathway mediates assembly of surface structures termed pili or fimbriae that adhere to eukaryotic cell targets [25]. Pilus-mediated attachment contributes to the formation of biofilms as well as intracellular biofilm-like communities (IBCs) that are highly resistant to antibiotics [26,27]. C-U pili can also activate host cell pathways leading to actin cytoskeletal rearrangements and subsequent bacterial invasion into the host cells [28]. Attached CU pili often must withstand strong shear forces, a property conferred by a combination of their rigidity and flexibility [28,29].
The best-characterized C-U pili are type 1 pili elaborated by different species of the Enterobacteriaceae, and the Pap pili elaborated by uropathogenic E. coli (UPEC) [27]. These pili are configured as a helical rod consisting of hundreds of pilus subunits, and a flexible tip fibrillum composed of several subunits in one or a few copies that mediate target cell attachment (Fig. 2) [30]. Assembly initiates by delivery of pilus subunits via the GSP to the periplasm, where the pilus subunits fold and form disulfide bridges, then interact with a dedicated chaperone (e.g., FimC, PapD) [31,32]. C-U pilus subunits typically are composed of an N-terminal flexible extension of ~10–15 residues in length and an incomplete immunoglobulin (Ig)-like pilin domain composed only of six β-strands. Because Ig-folded proteins typically contain seven β-strands, the lack of the seventh β-strand confers instability. Pilus subunits are stabilized through a specific interaction with the chaperone that involves donation of a β-strand by the chaperone subunit into the groove in the pilus that is normally occupied by the seventh β -strand. This interaction, termed donor strand complementation (DSC), stabilizes the pilus subunit, facilitates pilus subunit folding and inhibits premature pilus-pilus interactions [32,33].
The chaperone-pilus complex is recruited to and binds the OM usher, a 24-stranded β-barrel pore composed of an N-terminal domain (NTD), two C-terminal domains (CTD1 and CTD2), and a plug domain [34,35]. The usher catalyzes the exchange of the chaperone-subunit for subunit-subunit interactions in a process termed donor strand exchange (DSE) [36–38]. This involves displacement of the β-strand donated by the chaperone by the N-terminal extension of an incoming pilus, resulting in release of the chaperone and formation of a pilus subunit-subunit interaction. As pilus subunits are added to the base of the pilus fiber, the growing pilus is extruded through the β-barrel channel of the usher. The periplasmic NTD and CTDs of the usher orchestrate pilus assembly, a process that has been visualized by a series of high-resolution structures and modeled states [25,39–41]. In the absence of IM energy sources, ordered assembly is attributed to differences in affinities of chaperone-pilus complexes for the usher NTD, periplasmic concentrations of pilus subunits, and pilus subunit-subunit interactions that drive pilus fiber assembly and formation of the flexible helical rod at the cell surface [42].
The C-U pathway elaborates other types of pili than the well-characterized Pap and Type I. These include rigid pili termed Colonization Factor Antigen (CFA) or Coli Surface antigen (CS). Additionally, Afa/Dr pili made by pathogenic E. coli strains and the F1 capsular antigen of Yersinia pestis assemble as thin, flexible fibers that in some cases form amorphous, capsular-like or “afimbrial” structures [43–45]. As shown for T5SSs, insertion of the usher β-barrel relies on the Bam complex, but there is also evidence for involvement of the Tam complex [46].
1.3. Type 8 secretion systems (T8SSs).
T8SSs mediate assembly of amyloid fibers termed curli [47]. These systems are primarily found in Proteobacteria and Bacteroidetes [48]. Curli fibers are abundantly produced on the cell surface where they form dense mats. Their main functions are to promote aggregation, and in biofilm settings they function as adhesive and structural scaffolds for the microbial community [49,50]. Curli have also been implicated in host colonization, innate response activation, and cell invasion [51,52].
Curli are coiled fibers of ~2 – 5 nm in thickness that form via a nucleation-dependent self-assembly process [53]. The distinctive feature of curli is that, upon export of the CsgA curli subunits, curli pili form that adopt an amyloid fold as their native structure [54]. In E. coli, a dedicated chaperone, CsgC, binds the curli structural subunit, CsgA, upon delivery via the GSP to the periplasm (Fig. 2) [55]. CsgC apparently does not bind unfolded CsgA, but instead acts at the growth poles of curli fibers through charge-based interactions that block CsgA nucleation. Chaperone - curli complexes are delivered to the OM translocon, which is considerably larger than the β-barrel channels associated with the T5SS and C-U pathways. The translocon is composed of nine copies of the 262-kDa lipoprotein, CsgG, arranged as a 36-stranded OM-spanning β-barrel [56]. In the periplasm, the proximal end of CsgG forms a cage-like vestibule with an inner diameter of 35 Å [57]. A small subunit, CsgE, docks at the base of the vestibule to regulate substrate entry through charge-based interactions [58]. The size of the CsgG channel dictates that the CsgA pilin subunit is secreted in a largely extended conformation. CsgG can also export folded nonnative substrates, with size constraints delimited by channel dimensions [59].
Upon translocation, CsgA interacts with the nucleator protein, CsgB. CsgB resembles CsgA in sequence and structure, but a C-terminal repeat sequence anchors CsgB to the cell surface in proximity to CsgG. At this location and together with contributions by CsgF, CsgB nucleates assembly of CsgA into a β-sheet-rich amyloid fiber [60,61]. The transition from the extended preassembly to amyloid states may provide a driving force for directional translocation. A local high concentration and conformational confinement of curli subunits in the CsgG vestibule might also raise an entropic free-energy gradient over the translocation channel to ensure forward diffusion across the channel [57]. Interestingly, despite its β-barrel configuration, the CsgG channel assembles independently of the Bam or Tam complexes [62]. Rather, an N-terminal lipid motif targets CsgG to the lipoprotein sorting (LOL) pathway for delivery to the OM. CsgG subunits then oligomerize, which is thought to induce conformational changes that drive integration of amphipathic segments of the protein into the OM [62].
2. Class II: Two-step translocation systems - Span two membranes but translocate proteins across the OM.
Systems designated as Class II also recruit protein substrates from the periplasm for translocation across the OM, but are distinguished from the Class I systems by the fact that they span the entire cell envelope. This enables these systems to utilize IM energy sources such as ATP hydrolysis and the proton motive force (PMF) to drive translocation. Class II systems include the T2SSs (originally termed the terminal branch of the GSP) and the very recently described T9SSs. Type 4 pilus systems (T4Ps) are included in this Class because of their closely shared ancestry with the T2SSs.
2.1. Type 2 secretion systems (T2SSs) and type IV pilus assembly systems (T4Ps).
The T2SSs are large, envelope-spanning complexes composed of ~12–15 components [63]. The T2SSs are prevalent in the Proteobacteria, where they are deployed by both pathogenic and nonpathogenic species for release of various monomeric and multimeric proteins to the extracellular milieu. T2SS substrates include toxins, lipases and other lytic enzymes, and matrix proteins that contribute to establishment of robust biofilms in environmental or pathogenic settings [64]. Most substrates are released from the cell, but some such as pullulanase and heat-labile A/B5 enterotoxin (LT) remain tethered to the cell surface [63,65,66].
The main components of T2SSs include: i) a cytoplasmic ATPase (GspE) that powers substrate translocation, ii) an IM platform comprised of GspC and several other integral IM subunits, iii) a ‘pseudopilus’ comprised of GspG that dynamically extends and retracts, and iv) the secretin complex, a ~150 Å × ~195 Å cylindrical structure composed of 15 copies each of the GspD secretin and GspS lipoprotein [63,67–71]. Reminiscent of the curli channel, GspS - GspD building blocks are thought to be sorted via the LOL pathway to the OM where the secretin oligomerizes and inserts into the OM independently of the Bam complex [62,72,73].
GspG pilin monomers integrate into the IM as a pool for use to build the pseudopilus (Fig. 3) [63]. Upon delivery to the periplasm, T2SS substrates localize as soluble monomeric or multimeric proteins or are tethered via lipidation at the periplasmic face of the IM [66]. Protein folding ensues, although the folded structures vary considerably and a common translocation signal conferring recognition by the T2SS machinery is not yet defined. Upon binding of the substrate to the pseudopilus tip, the growing pseudopilus extrudes it through the secretin channel to the cell surface via proposed ‘piston’- or ‘Archimedes screw’-like actions [63]. The GspE ATPase is thought to induce structural transitions in the IM platform required for pseudopilus extension and, possibly, retraction [63,74].
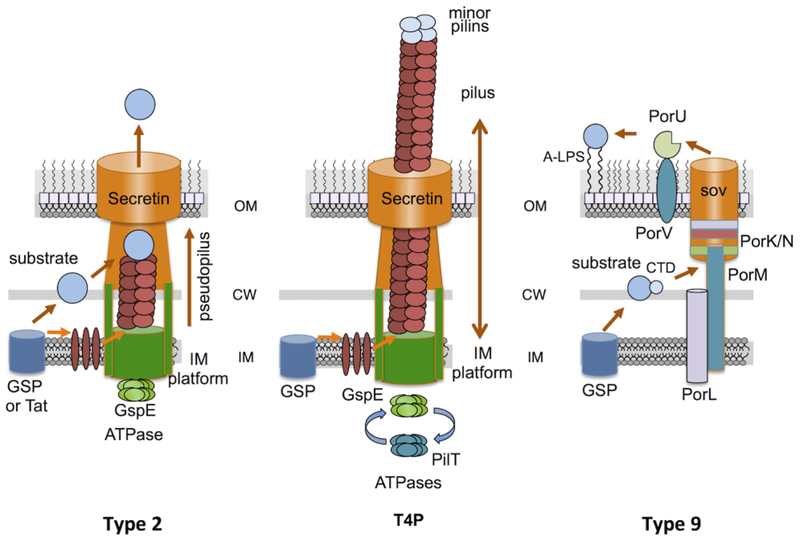
Fig. 3. Schematics of Class II assembly pathways.
Type 2 secretion systems elaborate ‘pseudopili’ on an IM platform. They recruit substrates delivered across the IM via the GSP or Tat systems to the tip of the pseudopilus, which might act as a piston to export the substrate through the secretin channel to the cell surface. T4Ps are phylogenetically and functionally related to the T2SSs, except they build pili that dynamically extend and retract through the coordinated actions of the GspE and PilT ATPases. Type 9 secretion systems recruit substrates from the periplasm through binding of conserved C-terminal domains (CTDs). Some substrates are covalently bound to the cell surface via a ‘sortase’ like mechanism.
T4P assembly systems are closely phylogenetically related to T2SSs, and, indeed, many structural and mechanistic similarities exist [75]. The main difference is that T4P systems elaborate a bona fide pilus that dynamically extends several microns from the cell surface and then retracts (Fig. 3) [76]. Extension and retraction generate considerable mechanical force, which enables the T4P to pull bacteria along abiotic and biotic surfaces resulting in a jerky trajectory of motility termed twitching motility [77]. T4P retraction can also pull extracellularly-bound molecules such as DNA and bacteriophages into the periplasm to initiate DNA transformation and phage infection [77,78]. T4Ps also mediate adherence of bacteria, resulting in formation of microcolonies or biofilms [77].
T4Ps are further distinguished from the T2SSs in their use of a second ATPase, e.g., PilT, which is responsible for pilus retraction [76]. Pilus extension/retraction dynamics remain poorly understood, but appear to involve opposite rotary motions of the GspE and PilT ATPases during cycles of ATP binding and hydrolysis [76]. The underlying mechanism of T4P retraction is complicated by the fact that some T4Ps, such as those found in Vibrio cholerae, Neisseria gonorrhoeae. and Caulobacter crescentus, lack a retraction ATPase but are still proficient in retraction [78–81]. T4Ps also assemble in Gram-positive bacteria, where they group into three functional subtypes that are not mutually exclusive: i) Pil (pilin), ii) Com (competence), and iii) Tad (tight adherence) (Fig. 1) [82]. T4Ps functioning in Gram-positive species also appear to retract independently of a PilT-like retraction ATPase [82]. Interestingly, T4Ps can also function as secretion systems by exporting exoproteins that are presumptively docked at the pilus tip [83]. The broad phylogenetic distribution and functional versatility of T4Ps is further exemplified by the fact that they are present in many archaeal species. In archaea, T4Ps assemble as adhesive pili or novel surface structures, and even as flagella termed ‘archaella’ that are capable of generating rotational forces enabling swimming motility [84,85].
2.2. Type 9 secretion systems (T9SSs).
The recently described T9SSs are found in the phylum Bacteriodetes, where they function both in protein secretion and a type of motility termed gliding motility [86,87]. Unlike substrates of the T2SSs whose translocation signals are not well-defined, substrates of T9SSs possess 70- to 100-residue C-terminal domains (CTDs) that are required for translocation across the OM [88]. CTDs have Ig-like folds that are thought to mediate binding of the substrate with components of the T9SS [89]. Screens for the presence of conserved CTDs have yielded estimates from a few dozen to over 200 T9SS substrates in different Bacteroides spp. [87]. Secreted substrates aid in growth of Bacteriodes in the environment or mammalian host by promoting polymer degradation, adhesion, virulence, and biofilm formation [87]. T9SS-mediated gliding motility is coupled to secretion and involves the rapid movement of cell surface adhesins [90]. Cells glide on their long axis at speeds of ~2 μm/s, powered by the IM PMF [91].
T9SS-mediated secretion and gliding motility require about ten proteins, although species-specific differences exist in subunit copy numbers and some machine subunits also appear to be involved in secretion of only a subset of T9SS substrates (Fig. 3) [87]. The secretion machinery consists of two IM-spanning subunits, PorL and PorM (an alternative Gld nomenclature is used for gliding motility systems), which likely are involved in coupling IM energy derived from the PMF to the OM-associated secretion or motility complex. The large ~270-kDa protein, SprA (also termed sov) adopts a 36-strand β-barrel whose postulated OM channel activity is regulated by other OM proteins including PorN, P, K, and V. An internal pore of ~70 Å, which could allow transit of folded or partially folded proteins, is blocked on its periplasmic side by a plug protein and on its external side by PorV [92]. Several Por proteins interact with PorV and this complex is thought to modify secreted proteins, e.g., by removing the CTD, and attaching them to the cell surface [93]. This ‘attachment complex’ is envisioned to function analogously to the sortases of Gram-positive species by covalently anchoring C-termini of secreted proteins to surface lipopolysaccharide, which accounts for observations that many T9SS substrates are C-terminally lipid-modified [93,94].
The mechanism responsible for gliding motility is not deciphered, but involves the rapid movement of a large, ~670-kDa highly-repetitive surface adhesin SprB, or alternative motility adhesins such as RemA, from pole to pole [95,96]. SprB follows a closed helical loop, leading to a model that the T9SS motility machinery functions as a PMF-dependent rotary motor that propels SprB or other motility adhesins along their helical tracks [91,97]. When the adhesins attach to a surface, the motors pushing against them result in rotation and forward movement of the cell in a screw-like manner [87].
3. Class III: One-step translocation systems - Span Gram-negative cell envelope and translocate substrates without a periplasmic intermediate.
The systems designated as Class III assemble as channels that span the IM, periplasm, and OM through which substrates are conveyed in one step from the cytoplasm to the cell surface. They rely on one or more associated ATPases or the PMF to drive translocation. Class III systems include the T1SSs (specialized ATP-binding cassette or ABC transporters), T3SSs (ancestrally related to flagella), T4SSs (ancestrally related to conjugation machines), and T6SSs (ancestrally related to bacteriophage injection systems) (Fig. 4). These systems function primarily to deliver ‘effector’ proteins to the cell surface or into other bacteria or eukaryotic cells to kill or modulate physiological processes of the target cells.
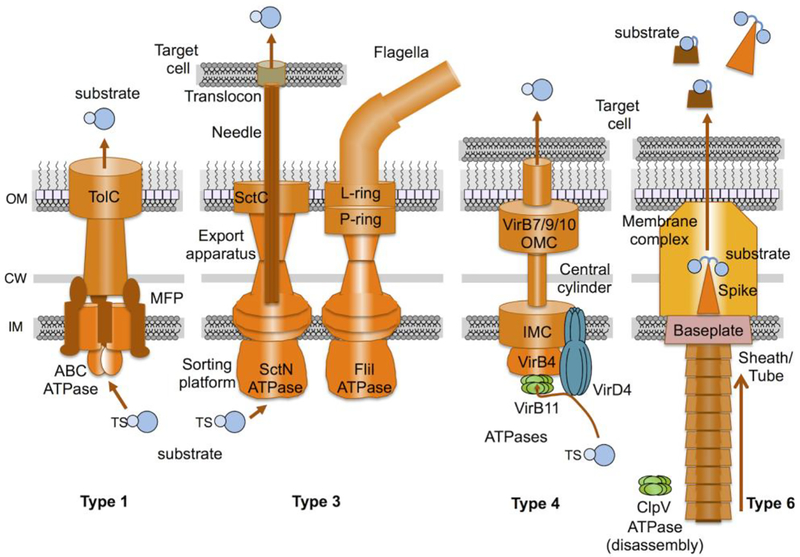
Fig. 4. Schematics of Class III assembly pathways.
Type 1 secretion systems are related to the ATP-binding cassette (ABC) transporter superfamily. They recruit substrates carrying C-terminal translocation signals (TSs) to the ATPase/MFP complex, and deliver substrates to the cell surface through the TolC channel/tunnel or ‘chunnel’. Type 3 systems consist of ‘injectisomes’ and phylogenetically-related flagella. Injectisomes elaborate injection needles for delivery of substrates into mammalian cell targets or pili for delivery to plant cell targets. Flagellar systems elaborate dynamic flagella for cellular chemotaxis. Substrates of injectisomes are recruited by recognition of N-terminal TSs or 5’ RNA structures with or without the aid of a chaperone. The SctN ATPase mediates unfolding of substrates prior to translocation through the needle. Type 4 systems are related to DNA conjugation machines. They elaborate cell envelope-spanning channels and deliver DNA or protein substrates via a contact-dependent mechanism into bacterial or eukaryotic target cells. Substrates are recognized by one or more TSs located at the C terminus or internally, with or without the aid of chaperones or adaptor proteins. Three ATPases (VirD4, VirB4, VirB11) are required for substrate translocation in most Gram-negative systems. Type 6 systems are bacteriophage contractile tail-like structures that are inverted for extrusion of substrates. Substrates associate covalently or noncovalently with the spike complex or the Hcp tube. The ClpV ATPase disassembles the sheath/tube complex after contraction and substrate ejection.
3.1. Type 1 secretion systems (T1SSs).
The T1SSs are phylogenetically and functionally related to the ABC transporter superfamily, which import or export small molecules [98]. T1SS substrates can range in size, up to several hundred kDa, and are either released to the milieu or affixed to the cell surface. T1SS substrates include proteases, lipases, adhesins, and heme-binding proteins [99,100]. Substrates are usually defined by the presence of several blocks of nonapeptide-binding sequences, termed RTX (repeats in toxins) that have a consensus GGxGxDxUx sequence, where x can be any amino acid and U is a large hydrophobic amino acid [101,102]. RTX motifs specifically bind Ca2+ and are implicated in post-translocation folding [103]. The RTX domain is located proximal to the C-terminal secretion signal. There is still debate whether the secretion signal consists of a specific linear code, a structural motif, or a combination of the two [99].
T1SSs are composed of an ABC transporter ATPase, a membrane fusion protein (MFP), and a TolC-like protein (Fig. 4) [99,100]. The ATPase functions as a dimer and catalyzes ATP hydrolysis to provide the energy for substrate transport. The ATPase interacts with the MFP in a 2::3 (or possibly 2::6) stoichiometric ratio to coordinate recognition of secretion substrates [104,105]. Upon binding of substrate, the ATPase/MFP complex recruits TolC, which inserts at the OM as a β-barrel channel and extends as a long α-helical ‘tunnel’ through the periplasm. The activated MFP/ATPase/TolC complex thus transiently forms a transenvelope channel/tunnel or ‘chunnel’ [104,106,107]. The unfolded C terminus of the substrate enters the channel first, and once portions of the secreted substrate reach the cell surface, Ca2+ binding induces folding, which prevents backsliding of the protein into the translocation channel[108].
Two T1SS substrates, the LapA and IBA adhesins, recently were shown to carry N-terminal “retention modules” (RMs) that anchor the adhesins to the cell surface by stalling further translocation [109,110]. This leaves a stalled short stub in the periplasm, apparently stuck in TolC, and a fully translocated, functional adhesin in the extracellular space. Upon sensing of an environmental signal, which in the case of LapA corresponds to conditions unfavorable for biofilm formation, the adhesin is released to the milieu by proteolytic removal of the RM [99,111]. The existence of a transient RM/TolC periplasmic intermediate, coupled with environmentally-induced toxin release, has been interpreted as a form of two-step, as opposed to one-step, translocation [111].
3.2. Type 3 secretion systems (T3SSs).
Studies have established that a conserved T3SS machinery contributes to the assembly of two phylogenetically-related but structurally and functionally distinct organelles, the ‘injectisome’ and the ‘flagellum’ [6,112,113]. Hence, the T3SS superfamily consists of systems dedicated to the translocation of effector proteins to eukaryotic cells and a motility/chemotaxis apparatus (Fig. 4). Injectisomes play important roles during pathogenesis through their capacity to deliver effector proteins directly into eukaryotic target cells [114]. These translocated effectors modulate a wide variety of physiological processes in the eukaryotic target cell to the benefit of the invading bacteria [112]. T3SSs of certain species, e.g., Yersinia and Pseudomonas, translocate only a few effectors, whereas those deployed by other species, e.g., Shigella, enterohemorrhagic E. coli, translocate several dozen effectors. Flagella also function as dedicated protein translocation systems, but the principal substrates are components of the flagellum itself that are translocated during biogenesis [113].
Effectors are recognized by cognate injectisomes in part by their translocation signals (TSs), which consist of short N-terminal motifs [115], but there is also evidence that substrate signals can alternatively consist of RNA secondary structures encoded by the 5’ ends of effector genes [116,117]. Many T3SS effectors also interact with chaperones, which maintain the effectors in an unfolded state and can guide them to the base of the T3SS [118]. Various internal signals also dictate a temporal or hierarchical order to the translocation of effectors during the infection process [119]. As with injectisome trafficking, protein translocation during assembly of the flagellum proceeds through a combination of chaperone-independent and -dependent recruitment of machine subunits and spatiotemporal control of substrate trafficking by various internal signals acting at the transcriptional and posttranscriptional levels [113].
At least eight subunits of injectisomes are highly conserved with components of the flagellar apparatus [112,113]. Another ten to twenty proteins are needed for injectisome assembly or function. Through single particle cryoelectron microscopy and in situ CryoET approaches, recent studies have defined the architectures of injectisomes and flagella at high resolutions (Fig. 4) [120–123]. In general, T3SSs consist of a cytoplasmic base (termed the sorting platform for injectisomes), the cell-envelope-spanning export apparatus, and either the extracellular needle complex or flagellum (Fig. 4) [114]. Flagella additionally are composed of stator and other components at the IM and OM that enable their rotary motion in response to environmental signals [113].
By reference to a universal nomenclature adopted for injectisomes [112], the sorting platform adopts a cage-like complex composed of cytoplasmic proteins SctQ, K, L, and N [124,125]. It engages and unfolds substrates through the action of a member of the AAA+ ATPase superfamily, SctN [126], and it specifies a temporal order for substrate transfer through the export channel. The sorting platform abuts the base of the export apparatus, which is composed of five IM proteins (SctV,R, S, T, U) that form a channel through which substrates are conveyed to the needle complex [127,128]. The needle complex extends through an OM secretin complex, which is similar to that described above for the T2SSs but distinct from the L- and P-rings comprising the OM-spanning substructure of flagella [70,113,129]. The needle has an inner hollow core of ~25 Å, a diameter wide enough to permit translocation of unfolded effectors [130]. A tip complex located at the distal end of the needle, senses contact with host cells and regulates secretion of effectors [131]. Upon contact with the host cell, the tip complex is thought to undergo conformational changes that are transduced to the secretion machine [114]. The first proteins delivered through the T3SS nanomachine are the translocator proteins SctB and SctE, which are deposited on eukaryotic cell membranes and form a pore or translocon [122]. Effectors are then delivered in temporal order to their final destinations in the eukaryotic cell cytosol.
3.3. Type 4 secretion systems (T4SSs).
T4SSs are unique among the dedicated protein translocation systems in that a large subfamily functions to deliver DNA substrates intercellularly by a process termed conjugation. Conjugation systems are present in nearly all bacterial and some archaeal species, where they mediate the intra- and inter-species transfer of mobile genetic elements (MGEs) and their cargoes of antibiotic resistance and virulence genes and other fitness traits [132]. Despite their prominent roles in MGE transfer, conjugation machines in fact function as protein translocation systems through their ability to recognize and translocate relaxase proteins and, coincidentally, covalently-attached DNA substrates [133,134]. Relaxases coordinate DNA transfer by processing DNA substrates at their origin-of-transfer (oriT) sequences into their single-stranded transfer intermediates (relaxase-T-strand). They additionally carry translocation signals for recruitment to the T4SS, and they ‘pilot’ the attached T-strand through the transfer channel into recipient cells by a mechanism requiring direct cell-to-cell contact [135]. Another large subfamily of T4SSs, the ‘effector translocators’ are deployed by pathogenic bacteria to deliver effector proteins to eukaryotic cells during the course of infection [136,137]. Recent studies have shown that some T4SSs also have evolved for niche establishment through delivery of toxic proteins to neighboring bacteria [138,139]. Other T4SSs, designated uptake or release systems, acquire DNA substrates from the milieu or release DNA or protein substrates into the milieu [136]. Finally, T4SSs can elaborate ‘conjugative pili’, which function as attachment devices to promote adherence, biofilm development, and colonization [140,141].
In Gram-negative species, T4SSs are built from a minimum of 12 ‘signature’ subunits (VirB1 - VirB11, VirD4) [142]. T4SSs are composed of two large subassemblies, an outer membrane complex (OMC) and an inner membrane complex (IMC), which are connected by a thin flexible stalk or a central cylinder (Fig. 4) [143–147]. The minimal OMCs are composed of three signature subunits (VirB7, VirB9, VirB10) that assemble as a barrel-shaped structure of ~185 × 185 Å in length and width [144,148]. Compositionally larger T4SSs elaborate OMCs exceeding 250 Å in width [146,147]. An α-helical domain of the highly conserved VirB10-like subunit appears to form the OM channel [144,149]. Reminiscent of the curli channel and secretin complexes, studies suggest that the lipoprotein VirB7 directs OMC building blocks to the LOL pathway for sorting to the OM where oligomerization and channel formation proceed independently of the Bam complex [150–152]. The IMCs are composed of an IM platform and two or three ATPases (VirB4, VirB11, VirD4) [145–147,153]. Among the T4SSs whose structures have been solved, VirB4 stably associates with the IMC but for unknown reasons it adopts two very different architectures in different systems. In one configuration, VirB4 assembles as two side-by-side hexameric barrels [145], and in the other as a hexamer of dimers surrounding the entrance to a central channel [146,147]. VirD4, a member of the SpoIIIE/FtsK ATPase superfamily, functions as the receptor for T4SS substrates [154–156]. VirB11, a member of the AAA ATPase superfamily, might function in unfolding of secretion substrates prior to translocation, although it is not present in all systems [157]. Interestingly, while only inferred for VirD4 subunits at this time, a recent study established that VirB11 subunits cycle on and off the cognate T4SS as a function of ATP binding and hydrolysis [146].
Most T4SSs are thought to translocate substrates in one step across the entire cell envelope via a route that was described originally by use of a modified chromatin-immunoprecipitation assay, and more recently via a channel visualized by in situ CryoET [146,147,156]. Interestingly, however, in Bordetella pertussis, the Ptl T4SS relies instead on a two-step translocation pathway to export pertussis toxin (PT) [158]. PT is a member of the A/B5 toxin superfamily, whose A and B subunits are first delivered across the IM via the Sec system and then assemble as the holotoxin in the periplasm prior to export via the Ptl system across the OM [159]. Other A/B5 toxins are typically exported by T2SSs, drawing interesting mechanistic parallels between the types 2 and 4 systems [63]. T4SSs also function in many different Gram-positive species [160]. These systems lack the OMC and conjugative pilus, and instead require large multidomain cell wall hydrolases for assembly across the thick PG layer [161,162]. In Gram-positive species, T4SSs rely on cell surface adhesins as opposed to conjugative pili to mediate attachment with recipient cells [163].
3.4. Type 6 secretion systems (T6SSs).
T6SSs belong to the versatile family of contractile injection systems (CISs), which characteristically deliver effectors into target cells using a spring-like mechanism [164,165]. T6SSs contribute to the virulence of bacterial pathogens by delivering effector proteins to host cells, and also by injecting toxic substrates into neighboring bacteria to gain a competitive advantage for niche establishment in the host [166,167]. T6SSs have a broad repertoire of secretion substrates, which include effectors that act on the eukaryotic cell cytoskeleton, peptidoglycan hydrolases acting on other bacteria, and DNases, phospholipases and other toxins that can target all cell types [168–170]. Toxic effectors are often coproduced with immunity proteins that interact with and block the activity of the effector in the T6SS-carrying donor cell, thereby preventing self-intoxication [167,171]. Although T6SSs are typically depicted as intercellular injection systems, there is also evidence that these systems can function to deliver effectors into the extracellular milieu. Many exported effectors target bacterial or eukaryotic cells, but recent studies have also identified exported effectors that bind manganese, zinc or vesicles for iron scavenging from the environment to provide metals to the bacterial cell [172–174]. In Proteus mirabilis, there is also evidence for T6SS-mediated coordination of swarming behavior and collective movement through export of proteins (Ids) that are involved in self-recognition and territorial behavior [175].
T6SSs use a mechanism reminiscent of contractile-tail bacteriophages, but inverted, to translocate effectors across the cell envelope and into target cells (Fig. 4) [164]. They are assembled from ~20 proteins, many of which are related to components of bacteriophage contractile tails. The main components include a sheath (TssBC) that envelopes a tube or needle (Hcp) [176]. The tube possesses a spike complex at its tip (VgrG) that pierces the membrane of the target cell [177,178]. The tube/sheath complex assembles on a platform termed the baseplate, a large multisubunit complex (VgrG/TssK/E/F/G) that anchors the tube/sheath complex to membrane complex and also orients the tube/sheath relative to the cell envelope [179,180]. The baseplate connects to a membrane complex composed of TssJ/M/L, which spans the entire cell envelope and forms the channel through which the tube passes upon sheath contraction [178,181].
T6SS biogenesis starts with the assembly of the membrane complex in the cell envelope and the baseplate in the cytoplasm. Upon docking of the baseplate with the membrane complex, the inner tube and sheath are coordinately assembled in the bacterial cytoplasm [164,182]. A stimulus, presumptively transduced by contact with a target cell, triggers a rapid contraction (<2–5 msec) of the sheath that initiates at the baseplate [183]. This results in expulsion of the tube through the baseplate and membrane complex and, ultimately, through the membrane of a bacterial or eukaryotic target cell [184]. After contraction, the sheath is disassembled by a dedicated AAA+ ATPase termed ClpV [185]. T6SS effectors are injected into target cells by virtue of their association with the VgrG spike complex or tube or sheath components. Effectors can be covalently or noncovalently linked with spike or tube structures [170,186]. When associated with tube components, the effectors are likely embedded into the tube lumen, whose fixed dimensions limits the sizes of effectors conveyed via this mechanism [187].
4. Class IV: One-step translocation systems - Span Gram-positive cytoplasmic membrane and translocate substrates to cell surface or beyond.
Gram-positive species deploy the conserved GSP and Tat pathways, as well as specialized systems for delivery of protein substrates across their cytoplasmic membranes. The latter include some of the organelles discussed above, e.g., flagella, T4SSs, T4Ps, adapted for assembly across the single cytoplasmic membrane and thick cell wall. Another translocation pathway, the T7SS, is unique to Gram-positive species. The T7SSs have been studied mainly in the mycobacteria, but are broadly distributed among many species of the phyla Actinobacteria and Firmicutes [188].
4.1. Type 7 secretion systems (T7SSs).
All T7SSs have two features in common: i) they are composed of a transmembrane protein of the FtsK/SpoIIIE/VirD4 ATPase family, and ii) they secrete small proteins of approximately 100 amino acids with conserved Trp-X-Gly (WXG) motifs [189,190]. Other features of T7SSs are quite diverse, suggestive of a long evolutionary process involving gene duplication, mutation, and horizontal gene transfer [188]. Little is known about the biological functions of secreted substrates, but one property of interest is that they can be secreted as folded homo- or heterodimers. For example, two well-characterized WXG substrates in Mycobacterium tuberculosis, EsxA and EsxB, interact to form a heterodimer that is secreted as a folded four-helix-bundle [191,192]. Also intriguingly, monomeric or dimeric substrates, e.g., EspA/C, EsbB, on EsxA/B, can be co-dependent on each other for secretion [193]. EsxB has a flexible C-terminal tail with a translocation signal that mediates binding to the FtsK/SpoIIIE ATPase subunit, EccC. This and other T7SS TSs consist of a helix-turn-helix domain followed by a characteristic Tyr-X-X-X-D/E motif; these signals may or may not reside at the C terminus of the translocated substrate [194,195]. EsxB and other small Esp/Esx subunits exhibit structural features similar to those of secretion chaperones associated with T3SSs, which has led to the proposal that dedicated chaperones facilitate folding of partner substrates and provide the necessary signals for cotranslocation of chaperone/substrate dimers through the T7SS [196,197].
In M. tuberculosis, the ESX-1 system is often used as a model for T7SSs [188,197]. Its components include three polytopic cytoplasmic membrane proteins (EccB, EccD, EccE), two subunits (EccCa, EccCb) that together comprise the FtsK/SpoIIIE-like ATPase, and an ATPase (EccA) of the AAA+ superfamily [188,197]. Members of the FtsK/SpoIIIE ATPase superfamily typically have a single nucleotide-binding domain, but interestingly EccC has three successive ATP binding domains [198]. Although the T7SS channel has not yet been visualized, its capacity to accommodate folded monomeric or dimeric substrates suggests it must be quite large and also tightly gated. Finally, in mycobacteria, all Ecc subunits required for secretion are thought to assemble as the CM translocon, leaving unanswered the question of how substrates are translocated through the mycolic acid layer, which is formally equivalent to the OM of Gram-negative bacteria [188].
T7SSs are not restricted to protein secretion. For example, in Mycobacterium smegmatis, an Esx-1-like system is involved in a special form of conjugation that involves the exchange of different large chromosomal DNA fragments between donor and recipient strains [199,200]. Curiously, conjugation requires production of the ESX-1 system in the recipient cell, not the donor cell, and, in fact, donor strains with an inactive ESX-1 system are hyperconjugative. There is also evidence that Esx systems function in zinc or iron metal ion uptake [201–203]. Finally, in Mycobacteria marinum, an ESX-1-like system is implicated in biofilm development and a form of motility termed sliding motility [204]. Overall, the diverse roles of T7SSs appear to be linked to survival of nonpathogenic bacteria in different environments, and of pathogenic bacteria in the harsh conditions encountered in the host.
5. Bacterial protein translocation: A rich tapestry of mechanism, structure, and function.
Bacteria have evolved a rich tapestry of pathways for moving proteins across their cell envelopes. We are making rapid progress in illuminating the dominant patterns of the tapestry representing each system, as evidenced most strikingly by recent successes in solving high-resolution structures of protein translocation nanomachines in situ. We are also identifying an increasing number of threads that tie the different systems to each other. On one level, the connecting threads can be depicted as the features inherent among all protein translocation systems required for: i) recruitment of specific substrate repertoires, ii) assembly of membrane or cell envelope-spanning channels or pores, and iii) energizing substrate transfer (Table 1). On another level, the connecting threads represent overlapping mechanistic properties that blur the distinctions of the different systems (Table 1). Most Class I systems, for example, assemble as OM channels or pores for substrate translocation across the OM. Some Class I systems, however, require the envelope-spanning Tam complex for assembly, thus linking Class I systems with the cell-envelope-spanning Class II systems. Most Class III systems characteristically deliver substrates in one-step across the entire cell envelope. However, several Class III systems have now been shown to recruit their substrates from the periplasm or transiently retain a portion of the secretion substrate in the periplasm, reminiscent of the two-step translocation pathways of Class II systems. As a final example, while several Class III systems, e.g., T3SSs, T4SSs, T6SSs, typically convey their substrates directly into target cells, some members of these system are now known to export proteins to the cell surface or extracellular milieu. These Class III ‘injection’ systems thus function instead like exporters, reminiscent of T1SSs, T2SSs, T7SSs, and T8SSs.
On a third level, the connecting threads link systems that have evolved to couple protein translocation to diverse biological processes such as attachment, motility, or import (Table 1). For attachment and biofilm formation, many systems, e.g., T1SSs, T2SSs, T4SSs, T5SSs, T9SSs, evolved the common strategy of elaborating large adhesins that extend appreciably away from the cell surface. Other systems, e.g., T3SSs, T4SSs, T4Ps, T8SSs, elaborate long pili/fimbriae or needles to promote attachment at a distance. For motility, many systems elaborate highly dynamic appendages: i) T3SSs mediate flagellar-based motility, ii) T4Ps mediate twitching and social motility, and in archaea these systems can promote flagellar-based motility, iii) T6SSs regulate swarming motility and group behavior, iv) T7SSs are involved in sliding motility, and v) T9SSs confer gliding motility. Even the dynamic conjugative pili of T4SSs impart a limited form of motility in the form of donor and recipient cells being drawn together during retraction of conjugative pili. Finally, several systems, e.g., T2Ps, T4SSs, T7SSs, have evolved the capacity to import or exchange DNA between cells, while other systems, e.g., T1SSs, T2SSs, T6SSs, T7SSs, can directly import metals or other small molecules or indirectly contribute to nutrient acquisition from the milieu.
Clearly, as our studies proceed, we are continuing to shed light on both the elegance and complexity of this rich tapestry of bacterial protein translocation.
Acknowledgements.
This work was supported by the National Institute of General Medical Sciences (Grants R01GM48746, R35GM131892)
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Green ER, Mecsas J (2016) Bacterial secretion systems: An overview. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerlach RG, Hensel M (2007) Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol 297: 401–415. [DOI] [PubMed] [Google Scholar]
- 3.Berks BC (2015) The twin-arginine protein translocation pathway. Annu Rev Biochem 84: 843–864. [DOI] [PubMed] [Google Scholar]
- 4.Tsirigotaki A, De Geyter J, Sostaric N, Economou A, Karamanou S (2017) Protein export through the bacterial Sec pathway. Nat Rev Microbiol 15: 21–36. [DOI] [PubMed] [Google Scholar]
- 5.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, et al. (2015) Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13: 343–359. [DOI] [PubMed] [Google Scholar]
- 6.Galan JE, Waksman G (2018) Protein-injection machines in bacteria. Cell 172: 1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oikonomou CM, Jensen GJ (2019) Electron cryotomography of bacterial secretion systems. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0019-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein HD (2019) Type V secretion in Gram-negative bacteria. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan E, Chauhan N, Udatha DB, Leo JC, Linke D (2016) Type V secretion systems in bacteria. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0009-2015. [DOI] [PubMed] [Google Scholar]
- 10.Nash ZM, Cotter PA (2019) Bordetella filamentous hemagglutinin, a model for the two-partner secretion pathway. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0024-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson IR, Nataro JP (2001) Virulence functions of autotransporter proteins. Infect Immun 69: 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, et al. (2004) Structure of the translocator domain of a bacterial autotransporter. EMBO J 23: 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Berg B (2010) Crystal structure of a full-length autotransporter. J Mol Biol 396: 627–633. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, et al. (2009) Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol 191: 6571–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ieva R, Bernstein HD (2009) Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci U S A 106: 19120–19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlova O, Peterson JH, Ieva R, Bernstein HD (2013) Mechanistic link between beta barrel assembly and the initiation of autotransporter secretion. Proc Natl Acad Sci U S A 110: E938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albenne C, Ieva R (2017) Job contenders: roles of the beta-barrel assembly machinery and the translocation and assembly module in autotransporter secretion. Mol Microbiol 106: 505–517. [DOI] [PubMed] [Google Scholar]
- 18.Kang’ethe W, Bernstein HD (2013) Charge-dependent secretion of an intrinsically disordered protein via the autotransporter pathway. Proc Natl Acad Sci U S A 110: E4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velarde JJ, Nataro JP (2004) Hydrophobic residues of the autotransporter EspP linker domain are important for outer membrane translocation of its passenger. J Biol Chem 279: 31495–31504. [DOI] [PubMed] [Google Scholar]
- 20.Baclayon M, Ulsen P, Mouhib H, Shabestari MH, Verzijden T, et al. (2016) Mechanical unfolding of an autotransporter passenger protein reveals the secretion starting point and processive transport intermediates. ACS Nano 10: 5710–5719. [DOI] [PubMed] [Google Scholar]
- 21.Besingi RN, Chaney JL, Clark PL (2013) An alternative outer membrane secretion mechanism for an autotransporter protein lacking a C-terminal stable core. Mol Microbiol 90: 1028–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen HH, Leyton DL, Shiota T, Belousoff MJ, Noinaj N, et al. (2014) Reconstitution of a nanomachine driving the assembly of proteins into bacterial outer membranes. Nat Commun 5: 5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stubenrauch CJ, Lithgow T (2019) The TAM: A translocation and assembly module of the beta-barrel assembly machinery in bacterial outer membranes. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0036-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bamert RS, Lundquist K, Hwang H, Webb CT, Shiota T, et al. (2017) Structural basis for substrate selection by the translocation and assembly module of the beta-barrel assembly machinery. Mol Microbiol 106: 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hospenthal MK, Waksman G (2019) The remarkable biomechanical properties of the Type 1 chaperone-usher pilus: A structural and molecular perspective. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0010-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright KJ, Seed PC, Hultgren SJ (2007) Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol 9: 2230–2241. [DOI] [PubMed] [Google Scholar]
- 27.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ (2015) Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13: 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Psonis JJ, Thanassi DG (2019) Therapeutic approaches targeting the assembly and function of chaperone-usher pili. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0033-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas WE, Nilsson LM, Forero M, Sokurenko EV, Vogel V (2004) Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated Escherichia coli. Mol Microbiol 53: 1545–1557. [DOI] [PubMed] [Google Scholar]
- 30.Hung CS, Bouckaert J, Hung D, Pinkner J, Widberg C, et al. (2002) Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol Microbiol 44: 903–915. [DOI] [PubMed] [Google Scholar]
- 31.Jacob-Dubuisson F, Striker R, Hultgren SJ (1994) Chaperone-assisted self-assembly of pili independent of cellular energy. J Biol Chem 269: 12447–12455. [PubMed] [Google Scholar]
- 32.Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, et al. (1999) Structural basis of chaperone function and pilus biogenesis. Science 285: 1058–1061. [DOI] [PubMed] [Google Scholar]
- 33.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, et al. (1999) X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285: 1061–1066. [DOI] [PubMed] [Google Scholar]
- 34.Nishiyama M, Horst R, Eidam O, Herrmann T, Ignatov O, et al. (2005) Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J 24: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remaut H, Tang C, Henderson NS, Pinkner JS, Wang T, et al. (2008) Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133: 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, et al. (2006) Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol Cell 22: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen WJ, Phan G, Waksman G (2012) Pilus biogenesis at the outer membrane of Gram-negative bacterial pathogens. Curr Opin Struct Biol 10.1016/j.sbi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Morrissey B, Leney AC, Toste Rego A, Phan G, Allen WJ, et al. (2012) The role of chaperone-subunit usher domain interactions in the mechanism of bacterial pilus biogenesis revealed by ESI-MS. Molec Cell Proteom: MCP 11: M111 015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Smith BS, Chen LX, Baxter RH, Deisenhofer J (2009) Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc Natl Acad Sci U S A 106: 7403–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phan G, Remaut H, Wang T, Allen WJ, Pirker KF, et al. (2011) Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature 474: 49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werneburg GT, Henderson NS, Portnoy EB, Sarowar S, Hultgren SJ, et al. (2015) The pilus usher controls protein interactions via domain masking and is functional as an oligomer. Nat Struct Mol Biol 22: 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Ng TW, Dodson KW, So SS, Bayle KM, et al. (2010) The differential affinity of the usher for chaperone-subunit complexes is required for assembly of complete pili. Mol Microbiol 76: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labigne-Roussel A, Schmidt MA, Walz W, Falkow S (1985) Genetic organization of the afimbrial adhesin operon and nucleotide sequence from a uropathogenic Escherichia coli gene encoding an afimbrial adhesin. J Bacteriol 162: 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaastra W, Svennerholm AM (1996) Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 4: 444–452. [DOI] [PubMed] [Google Scholar]
- 45.Titball RW, Howells AM, Oyston PC, Williamson ED (1997) Expression of the Yersinia pestis capsular antigen (F1 antigen) on the surface of an aroA mutant of Salmonella typhimurium induces high levels of protection against plague. Infect Immun 65: 1926–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stubenrauch C, Belousoff MJ, Hay ID, Shen HH, Lillington J, et al. (2016) Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nat Microbiol 1: 16064. [DOI] [PubMed] [Google Scholar]
- 47.Bhoite S, van Gerven N, Chapman MR, Remaut H (2019) Curli biogenesis: bacterial amyloid assembly by the Type VIII secretion pathway. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0037-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dueholm MS, Albertsen M, Otzen D, Nielsen PH (2012) Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One 7: e51274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S (2005) Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol 49: 875–884. [DOI] [PubMed] [Google Scholar]
- 50.Hufnagel DA, Depas WH, Chapman MR (2015) The biology of the Escherichia coli extracellular matrix. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0014-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, et al. (2005) CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol 58: 289–304. [DOI] [PubMed] [Google Scholar]
- 52.Van Gerven N, Van der Verren SE, Reiter DM, Remaut H (2018) The role of functional amyloids in bacterial virulence. J Mol Biol 430: 3657–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, et al. (2013) Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio 4: e00645–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, et al. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295: 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans ML, Chorell E, Taylor JD, Aden J, Gotheson A, et al. (2015) The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Mol Cell 57: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR (2006) Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol 59: 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goyal P, Krasteva PV, Van Gerven N, Gubellini F, Van den Broeck I, et al. (2014) Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516: 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, et al. (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol 81: 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Gerven N, Goyal P, Vandenbussche G, De Kerpel M, Jonckheere W, et al. (2014) Secretion and functional display of fusion proteins through the curli biogenesis pathway. Mol Microbiol 91: 1022–1035. [DOI] [PubMed] [Google Scholar]
- 60.Hammer ND, McGuffie BA, Zhou Y, Badtke MP, Reinke AA, et al. (2012) The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation. J Mol Biol 422: 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nenninger AA, Robinson LS, Hultgren SJ (2009) Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A 106: 900–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunstan RA, Hay ID, Wilksch JJ, Schittenhelm RB, Purcell AW, et al. (2015) Assembly of the secretion pores GspD, Wza and CsgG into bacterial outer membranes does not require the Omp85 proteins BamA or TamA. Mol Microbiol 97: 616–629. [DOI] [PubMed] [Google Scholar]
- 63.Korotkov KV, Sandkvist M (2019) Architecture, function, and substrates of the Type II secretion system. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0034-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cianciotto NP, White RC (2017) Expanding role of Type II secretion in bacterial pathogenesis and beyond. Infect Immun 85 pii: e00014–17. doi: 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferrandez Y, Condemine G (2008) Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol Microbiol 69: 1349–1357. [DOI] [PubMed] [Google Scholar]
- 66.East A, Mechaly AE, Huysmans GHM, Bernarde C, Tello-Manigne D, et al. (2016) Structural basis of pullulanase bembrane binding and secretion revealed by X-ray crystallography, molecular dynamics and biochemical analysis. Structure 24: 92–104. [DOI] [PubMed] [Google Scholar]
- 67.Sauvonnet N, Vignon G, Pugsley AP, Gounon P (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. EMBO J 19: 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Korotkov KV, Sandkvist M, Hol WG (2012) The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol 10: 336–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hay ID, Belousoff MJ, Lithgow T (2017) Structural Basis of Type 2 secretion system engagement between the inner and outer bacterial membranes. MBio 8. doi: 10.1128/mBio.01344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Majewski DD, Worrall LJ, Strynadka NC (2018) Secretins revealed: structural insights into the giant gated outer membrane portals of bacteria. Curr Opin Struct Biol 51: 61–72. [DOI] [PubMed] [Google Scholar]
- 71.Lopez-Castilla A, Thomassin JL, Bardiaux B, Zheng W, Nivaskumar M, et al. (2017) Structure of the calcium-dependent type 2 secretion pseudopilus. Nat Microbiol 2: 1686–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, et al. (1999) Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci U S A 96: 8173–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoang HH, Nickerson NN, Lee VT, Kazimirova A, Chami M, et al. (2011) Outer membrane targeting of Pseudomonas aeruginosa proteins shows variable dependence on the components of Bam and Lol machineries. MBio 2. doi: 10.1128/mBio.00246-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camberg JL, Sandkvist M (2005) Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J Bacteriol 187: 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sandkvist M (2001) Biology of type II secretion. Mol Microbiol 40: 271–283. [DOI] [PubMed] [Google Scholar]
- 76.McCallum M, Burrows LL, Howell PL (2019) The dynamic structures of the type IV pilus. Microbiol Spectr 7 oi: 10.1128/microbiolspec.PSIB-0006-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Craig L, Forest KT, Maier B (2019) Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol 10.1038/s41579-019-0195-4. [DOI] [PubMed] [Google Scholar]
- 78.Ellison CK, Dalia TN, Vidal Ceballos A, Wang JC, Biais N, et al. (2018) Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat Microbiol 3: 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clausen M, Jakovljevic V, Sogaard-Andersen L, Maier B (2009) High-force generation is a conserved property of type IV pilus systems. J Bacteriol 191: 4633–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, et al. (2017) Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zollner R, Cronenberg T, Maier B (2019) Motor properties of PilT-independent type 4 pilus retraction in gonococci. J Bacteriol 10.1128/JB.00778-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muschiol S, Aschtgen MS, Nannapaneni P, Henriques-Normark B (2019) Gram-positive type IV pili and competence. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0011-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kirn TJ, Bose N, Taylor RK (2003) Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol 49: 81–92. [DOI] [PubMed] [Google Scholar]
- 84.Albers SV, Szabo Z, Driessen AJ (2006) Protein secretion in the archaea: multiple paths towards a unique cell surface. Nat Rev Microbiol 4: 537–547. [DOI] [PubMed] [Google Scholar]
- 85.Chaudhury P, Quax TEF, Albers SV (2018) Versatile cell surface structures of archaea. Mol Microbiol 107: 298–311. [DOI] [PubMed] [Google Scholar]
- 86.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, et al. (2010) A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107: 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McBride MJ (2019) Bacteroidetes gliding motility and the type IX secretion system. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0002-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, et al. (2006) The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol 188: 6376–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Diego I, Ksiazek M, Mizgalska D, Koneru L, Golik P, et al. (2016) The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain. Sci Rep 6: 23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnston JJ, Shrivastava A, McBride MJ (2018) Untangling Flavobacterium johnsoniae gliding motility and protein secretion. J Bacteriol 200 pii: e00362–17. doi: 10.1128/JB.00362-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K (2013) Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110: 11145–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lauber F, Deme JC, Lea SM, Berks BC (2018) Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Glew MD, Veith PD, Chen D, Gorasia DG, Peng B, et al. (2017) PorV is an outer membrane shuttle protein for the type IX secretion system. Sci Rep 7: 8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Veith PD, Glew MD, Gorasia DG, Reynolds EC (2017) Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol 106: 35–53. [DOI] [PubMed] [Google Scholar]
- 95.Nelson SS, Bollampalli S, McBride MJ (2008) SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190: 2851–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ (2012) Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J Bacteriol 194: 3678–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shrivastava A, Berg HC (2015) Towards a model for Flavobacterium gliding. Curr Opin Microbiol 28: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Higgins CF, Hiles ID, Salmond GP, Gill DR, Downie JA, et al. (1986) A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323: 448–450. [DOI] [PubMed] [Google Scholar]
- 99.Holland IB, Peherstorfer S, Kanonenberg K, Lenders M, Reimann S, et al. (2016) Type I protein secretion-deceptively simple yet with a wide range of mechanistic variability across the family. EcoSal Plus 7. doi: 10.1128/ecosalplus.ESP-0019-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spitz O, Erenburg IN, Beer T, Kanonenberg K, Holland IB, et al. (2019) Type I secretion systems - one mechanism for all? Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0003-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Letoffe S, Wandersman C (1992) Secretion of CyaA-PrtB and HlyA-PrtB fusion proteins in Escherichia coli: involvement of the glycine-rich repeat domain of Erwinia chrysanthemi protease B. J Bacteriol 174: 4920–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lecher J, Schwarz CK, Stoldt M, Smits SH, Willbold D, et al. (2012) An RTX transporter tethers its unfolded substrate during secretion via a unique N-terminal domain. Structure 20: 1778–1787. [DOI] [PubMed] [Google Scholar]
- 103.Sanchez-Magraner L, Viguera AR, Garcia-Pacios M, Garcillan MP, Arrondo JL, et al. (2007) The calcium-binding C-terminal domain of Escherichia coli alpha-hemolysin is a major determinant in the surface-active properties of the protein. J Biol Chem 282: 11827–11835. [DOI] [PubMed] [Google Scholar]
- 104.Thanabalu T, Koronakis E, Hughes C, Koronakis V (1998) Substrate-induced assembly of a contiguous channel for protein export from E.coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J 17: 6487–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Du D, Wang Z, James NR, Voss JE, Klimont E, et al. (2014) Structure of the AcrAB-TolC multidrug efflux pump. Nature 509: 512–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C (2000) Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405: 914–919. [DOI] [PubMed] [Google Scholar]
- 107.Balakrishnan L, Hughes C, Koronakis V (2001) Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J Mol Biol 313: 501–510. [DOI] [PubMed] [Google Scholar]
- 108.Lenders MH, Weidtkamp-Peters S, Kleinschrodt D, Jaeger KE, Smits SH, et al. (2015) Directionality of substrate translocation of the hemolysin A type I secretion system. Sci Rep 5: 12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smith TJ, Font ME, Kelly CM, Sondermann H, O’Toole GA (2018) An N-terminal retention module anchors the giant adhesin LapA of Pseudomonas fluorescens at the cell surface: a novel subfamily of type I secretion systems. J Bacteriol 200. doi: 10.1128/JB.00734-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guo S, Stevens CA, Vance TDR, Olijve LLC, Graham LA, et al. (2017) Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice. Sci Adv 3: e1701440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smith TJ, Sondermann H, O’Toole GA (2018) Type 1 does the two-Step: Type 1 secretion substrates with a functional periplasmic intermediate. J Bacteriol 200. doi: 10.1128/JB.00168-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, et al. (2017) Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol 15: 323–337. [DOI] [PubMed] [Google Scholar]
- 113.Diepold A, Armitage JP (2015) Type III secretion systems: the bacterial flagellum and the injectisome. Philos Trans R Soc Lond B Biol Sci 370 pii: 20150020. doi: 10.1098/rstb.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lara-Tejero M, Galan JE (2019) The injectisome, a complex nanomachine for protein injection into mammalian cells. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0039-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McDermott JE, Corrigan A, Peterson E, Oehmen C, Niemann G, et al. (2011) Computational prediction of type III and IV secreted effectors in gram-negative bacteria. Infect Immun 79: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Anderson DM, Schneewind O (1997) A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278: 1140–1143. [DOI] [PubMed] [Google Scholar]
- 117.Niemann GS, Brown RN, Mushamiri IT, Nguyen NT, Taiwo R, et al. (2013) RNA type III secretion signals that require Hfq. J Bacteriol 195: 2119–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee SH, Galan JE (2004) Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol Microbiol 51: 483–495. [DOI] [PubMed] [Google Scholar]
- 119.Deng W, Yu HB, Li Y, Finlay BB (2015) SepD/SepL-dependent secretion signals of the type III secretion system translocator proteins in enteropathogenic Escherichia coli. J Bacteriol 197: 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Radics J, Konigsmaier L, Marlovits TC (2014) Structure of a pathogenic type 3 secretion system in action. Nat Struct Mol Biol 21: 82–87. [DOI] [PubMed] [Google Scholar]
- 121.Hu B, Lara-Tejero M, Kong Q, Galan JE, Liu J (2017) In situ molecular architecture of the Salmonella type III secretion machine. Cell 168: 1065–1074 e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park D, Lara-Tejero M, Waxham MN, Li W, Hu B, et al. (2018) Visualization of the type III secretion mediated Salmonella-host cell interface using cryo-electron tomography. Elife 7. doi: 10.7554/eLife.39514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu S, Schniederberend M, Zhitnitsky D, Jain R, Galan JE, et al. (2019) In situ structures of polar and lateral flagella revealed by cryo-electron tomography. J Bacteriol 201 pii: e00117–19. doi: 10.1128/JB.00117-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lara-Tejero M, Kato J, Wagner S, Liu X, Galan JE (2011) A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331: 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, et al. (2015) Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A 112: 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Akeda Y, Galan JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911–915. [DOI] [PubMed] [Google Scholar]
- 127.Wagner S, Konigsmaier L, Lara-Tejero M, Lefebre M, Marlovits TC, et al. (2010) Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci U S A 107: 17745–17750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuhlen L, Abrusci P, Johnson S, Gault J, Deme J, et al. (2018) Structure of the core of the type III secretion system export apparatus. Nat Struct Mol Biol 25: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schraidt O, Marlovits TC (2011) Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331: 1192–1195. [DOI] [PubMed] [Google Scholar]
- 130.Hu J, Worrall LJ, Hong C, Vuckovic M, Atkinson CE, et al. (2018) Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat Commun 9: 3840. doi: 10.1038/s41467-018-06298-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Epler CR, Dickenson NE, Bullitt E, Picking WL (2012) Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri. J Mol Biol 420: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li YG, Hu B, Christie PJ (2019) Biological and structural diversity of type IV secretion systems. Microbiol Spectr 7. doi: 10.1128/microbiolspec.PSIB-0012-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lang S, Zechner EL (2012) General requirements for protein secretion by the F-like conjugation system R1. Plasmid 10.1016/j.plasmid.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Redzej A, Ilangovan A, Lang S, Gruber CJ, Topf M, et al. (2013) Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Molec Microbiol 89: 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de la Cruz F, Frost LS, Meyer RJ, Zechner EL (2010) Conjugative DNA metabolism in Gram-negative bacteria. FEMS Micro Rev 34: 18–40. [DOI] [PubMed] [Google Scholar]
- 136.Cascales E, Christie PJ (2003) The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gonzalez-Rivera C, Bhatty M, Christie PJ (2016) Mechanism and function of type IV secretion during infection of the human host. Microbiol Spectr 4. doi: 10.1128/microbiolspec.VMBF-0024-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, et al. (2015) Bacterial killing via a type IV secretion system. Nat Commun 6: 6453. doi: 10.1038/ncomms7453 [DOI] [PubMed] [Google Scholar]
- 139.Nas MY, White RC, DuMont AL, Lopez AE, Cianciotto NP (2019) Stenotrophomonas maltophilia encodes a VirB/D4 type IV secretion system that modulates apoptosis in human cells and promotes competition against heterologous bacteria including Pseudomonas aeruginosa. Infect Immun 10.1128/IAI.00457-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ghigo JM (2001) Natural conjugative plasmids induce bacterial biofilm development. Nature 412: 442–445. [DOI] [PubMed] [Google Scholar]
- 141.Arutyunov D, Frost LS (2013) F conjugation: back to the beginning. Plasmid 70: 18–32. [DOI] [PubMed] [Google Scholar]
- 142.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59: 451–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, et al. (2009) Structure of a type IV secretion system core complex. Science 323: 266–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, et al. (2009) Structure of the outer membrane complex of a type IV secretion system. Nature 462: 1011–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, et al. (2014) Structure of a type IV secretion system. Nature 508: 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chetrit D, Hu B, Christie PJ, Roy CR, Liu J (2018) A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat Microbiol 3: 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu B, Khara P, Song L, Lin AS, Frick-Cheng AE, et al. (2019) In situ molecular architecture of the Helicobacter pylori Cag type IV secretion System. MBio 10. doi: 10.1128/mBio.00849-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sgro GG, Costa TR, Cenens W, Souza DP, Cassago A, et al. (2018) CryoEM structure of the core complex of a bacterial killing type IV secretion system. Nat Microbiol 3(12):1429–1440. doi: 10.1038/s41564-018-0262-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, et al. (2009) Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol Microbiol 71: 779–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Spudich GM, Fernandez D, Zhou XR, Christie PJ (1996) Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci U S A 93: 7512–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fernandez D, Spudich GM, Zhou XR, Christie PJ (1996) The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol 178: 3168–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gonzalez-Rivera C, Khara P, Awad D, Patel R, Li YG, et al. (2019) Two pKM101-encoded proteins, the pilus-tip protein TraC and Pep, assemble on the Escherichia coli cell surface as adhesins required for efficient conjugative DNA transfer. Mol Microbiol 111: 96–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Atmakuri K, Cascales E, Christie PJ (2004) Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54: 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cabezon E, Sastre JI, de la Cruz F (1997) Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet 254: 400–406. [DOI] [PubMed] [Google Scholar]
- 155.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, et al. (2001) The bacterial conjugation protein TrwB resembles ring helicases and F1- ATPase. Nature 409: 637–641. [DOI] [PubMed] [Google Scholar]
- 156.Cascales E, Christie PJ (2004) Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304: 1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bhatty M, Laverde Gomez JA, Christie PJ (2013) The expanding bacterial type IV secretion lexicon. Res Microbiol 164: 620–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Burns DL (2003) Type IV transporters of pathogenic bacteria. Curr Opin Microbiol 6: 29–34. [DOI] [PubMed] [Google Scholar]
- 159.Locht C, Coutte L, Mielcarek N (2011) The ins and outs of pertussis toxin. The FEBS journal 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- 160.Grohmann E, Christie PJ, Waksman G, Backert S (2018) Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol 107: 455–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Arends K, Celik EK, Probst I, Goessweiner-Mohr N, Fercher C, et al. (2013) TraG encoded by the pIP501 type IV secretion system is a two domain peptidoglycan degrading enzyme essential for conjugative transfer. J Bacteriol 195(19):4436–44. doi: 10.1128/JB.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Laverde Gomez JA, Bhatty M, Christie PJ (2014) PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone-responsive plasmid pCF10. J Bacteriol 196: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bhatty M, Cruz MR, Frank KL, Gomez JA, Andrade F, et al. (2015) Enterococcus faecalis pCF10-encodes surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol 95: 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, et al. (2014) Architecture and assembly of the type VI secretion system. Biochim Biophys Acta 1843: 1664–1673. [DOI] [PubMed] [Google Scholar]
- 165.Taylor NMI, van Raaij MJ, Leiman PG (2018) Contractile injection systems of bacteriophages and related systems. Mol Microbiol 108: 6–15. [DOI] [PubMed] [Google Scholar]
- 166.Alcoforado Diniz J, Liu YC, Coulthurst SJ (2015) Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol 17: 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Coyne MJ, Comstock LE (2019) Type VI secretion systems and the gut microbiota. Microbiol Spectr 7 10.1128/microbiolspec.PSIB-0009-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, et al. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475: 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, et al. (2016) Salmonella typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113: E5044–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Durand E, Cambillau C, Cascales E, Journet L (2014) VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol 22: 498–507. [DOI] [PubMed] [Google Scholar]
- 171.Russell AB, Peterson SB, Mougous JD (2014) Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang T, Si M, Song Y, Zhu W, Gao F, et al. (2015) Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog 11: e1005020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Si M, Zhao C, Burkinshaw B, Zhang B, Wei D, et al. (2017) Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci U S A 114: E2233–E2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lin J, Zhang W, Cheng J, Yang X, Zhu K, et al. (2017) A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8: 14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Saak CC, Gibbs KA (2016) The self-identity protein IdsD Is communicated between cells in swarming Proteus mirabilis colonies. J Bacteriol 198: 3278–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cascales E (2008) The type VI secretion toolkit. EMBO Rep 9: 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, et al. (2013) PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500: 350–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E (2015) The type VI secretion TssEFGK-VgrG phage-like baseplate Is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet 11: e1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, et al. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106: 4154–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, et al. (2018) Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3: 1404–1416. [DOI] [PubMed] [Google Scholar]
- 181.Rapisarda C, Cherrak Y, Kooger R, Schmidt V, Pellarin R, et al. (2019) In situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. EMBO J 38. 38(10). pii: e100886. doi: 10.15252/embj.2018100886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, et al. (2015) Structure of the type VI secretion system contractile sheath. Cell 160: 952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang J, Brackmann M, Castano-Diez D, Kudryashev M, Goldie KN, et al. (2017) Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat Microbiol 2: 1507–1512. [DOI] [PubMed] [Google Scholar]
- 184.Brackmann M, Wang J, Basler M (2018) Type VI secretion system sheath inter-subunit interactions modulate its contraction. EMBO Rep 19: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, et al. (2013) ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87: 1013–1028. [DOI] [PubMed] [Google Scholar]
- 186.Unterweger D, Kostiuk B, Pukatzki S (2017) Adaptor proteins of type VI secretion system effectors. Trends Microbiol 25: 8–10. [DOI] [PubMed] [Google Scholar]
- 187.Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, et al. (2013) Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51: 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Groschel MI, Sayes F, Simeone R, Majlessi L, Brosch R (2016) ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol 14: 677–691. [DOI] [PubMed] [Google Scholar]
- 189.Pallen MJ (2002) The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends Microbiol 10: 209–212. [DOI] [PubMed] [Google Scholar]
- 190.Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, et al. (2005) Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J 24: 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Brodin P, de Jonge MI, Majlessi L, Leclerc C, Nilges M, et al. (2005) Functional analysis of early secreted antigenic target-6, the dominant T-cell antigen of Mycobacterium tuberculosis, reveals key residues involved in secretion, complex formation, virulence, and immunogenicity. J Biol Chem 280: 33953–33959. [DOI] [PubMed] [Google Scholar]
- 192.Poulsen C, Panjikar S, Holton SJ, Wilmanns M, Song YH (2014) WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern. PLoS One 9: e89313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, et al. (2005) Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A 102: 10676–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Champion PA, Stanley SA, Champion MM, Brown EJ, Cox JS (2006) C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313: 1632–1636. [DOI] [PubMed] [Google Scholar]
- 195.Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, et al. (2012) General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci U S A 109: 11342–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ekiert DC, Cox JS (2014) Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A 111: 14758–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Houben EN, Korotkov KV, Bitter W (2014) Take five - Type VII secretion systems of Mycobacteria. Biochim Biophys Acta 1843: 1707–1716. [DOI] [PubMed] [Google Scholar]
- 198.Burton B, Dubnau D (2010) Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol 2: a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Coros A, Callahan B, Battaglioli E, Derbyshire KM (2008) The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol 69: 794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Derbyshire KM, Gray TA (2014) Distributive conjugal transfer: New insights into horizontal gene transfer and genetic exchange in mycobacteria. Microbiol Spectr 2: doi: 10.1128/microbiolspec.MGM2-0022-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Serafini A, Pisu D, Palu G, Rodriguez GM, Manganelli R (2013) The ESX-3 secretion system is necessary for iron and zinc homeostasis in Mycobacterium tuberculosis. PLoS One 8: e78351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, et al. (2009) Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A 106: 18792–18797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Siegrist MS, Steigedal M, Ahmad R, Mehra A, Dragset MS, et al. (2014) Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio 5: e01073–01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lai LY, Lin TL, Chen YY, Hsieh PF, Wang JT (2018) Role of the Mycobacterium marinum ESX-1 secretion system in sliding motility and biofilm formation. Front Microbiol 9: 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]