ABSTRACT
A repertoire of sophisticated genetic tools has significantly enhanced studies of Methanosarcina genera, yet the lack of multiple positive selectable markers has limited the types of genetic experiments that can be performed. In this study, we report the development of an additional positive selection system for Methanosarcina that utilizes the antibiotic nourseothricin and the Streptomyces rochei streptothricin acetyltransferase (sat) gene, which may be broadly applicable to other groups of methanogenic archaea. Nourseothricin was found to inhibit growth of four different methanogen species at concentrations ≤300 μg/ml in liquid or on solid media. Selection of nourseothricin resistant transformants was possible in two genetically tractable Methanosarcina species, M. acetivorans and M. barkeri, using the sat gene as a positive selectable marker. Additionally, the sat marker was useful for constructing a gene deletion mutant strain of M. acetivorans, emphasizing its utility as a second positive selectable marker for genetic analyses of Methanosarcina genera. Interestingly, two human gut-associated methanogens Methanobrevibacter smithii and Methanomassillicoccus luminyensis were more sensitive to nourseothricin than either Methanosarcina species, suggesting the nourseothricin-sat gene pair may provide a robust positive selection system for development of genetic tools in these and other methanogens.
Keywords: Methanosarcina; selectable marker; streptothricin acetyltransferase; nourseothricin; Methanobrevibacter smithii, Methanomassillicoccus luminyensis
The streptothricin acetyltransferase (sat) gene from Streptomyces rochei can be used as a positive selectable marker in Methanosarcina species.
INTRODUCTION
Sophisticated genetic tools available for Methanosarcina genera have enabled studies investigating the physiology, genetics and metabolism of this versatile and environmentally important group of methane-producing archaea. In particular, the discovery that Methanosarcina can be efficiently transformed using liposomes provided the foundation for development of the genetic tools used to study these organisms, including those for constructing in-frame chromosomal gene deletions, tightly-regulated gene expression, transposon mutagenesis and most recently Cas9-mediated genome editing (Kohler and Metcalf 2012; Nayak and Metcalf 2017). Application of these tools requires the use of a selectable marker, in these examples an antibiotic resistance marker, to separate cells that acquire desired genetic changes from those that have escaped transformation. The puromycin acetyltransferase (pac) gene from Streptomyces alboniger was the first selectable marker developed for methanogens and has been used in both Methanosarcina and Methanococcus species (Gernhardt et al.1990; Kohler and Metcalf 2012). Additionally, the aph(3’)-II gene confers neomycin resistance in M. mazei providing this species with a second positive selectable marker (Mondorf, Deppenmeier and Welte 2012); however, in our experience this marker has not been useful for either Methanosarcina acetivorans or Methanosarcina barkeri, leaving these species with the pac gene as the sole positive selectable marker for genetic manipulation. Finally, a mutant version of the isoleucyl-tRNA synthetase gene (ileS) from M. barkeri was developed as a selectable marker conferring resistance to pseudomonic acid (Mupirocin) in Methanosarcina (Boccazzi, Zhang and Metcalf 2000). This marker is not commonly used due to higher cost, the large size of the ileS gene and the potential for recombination between mutant M. barkeri ileS allele and the nearly identical ileS genes found in all Methanosarcina species.
Although Cas9-mediated genome editing has streamlined the time-consuming process of constructing mutants of M. acetivorans, it has not eliminated the need for a second positive selectable marker to assist certain genetic experiments. For example, a second positive selectable marker would allow the complementation of genes disrupted with pac, such as M. acetivorans mini-mariner transposon mutants (Zhang et al.2000). Secondly, while the Cas9 system allows preliminary assessment of gene essentiality in M. acetivorans (Nayak et al.2017), additional validation is required to confirm the results. To exemplify one such validation experiment, the Cas9 system could be used to attempt deletion of a suspected essential gene in an M. acetivorans strain containing an ectopic, tetracycline-inducible second copy of the gene. Results from this type of experiment support essential gene hypotheses if the native gene can be deleted only when expression of the second copy is induced. Since M. acetivorans Cas9 plasmids encode pac, a second positive selectable marker gene would be required to construct the host strain containing the tetracycline-regulable gene. Development of additional selectable markers for Methanosarcina would also benefit researchers who are not using the Cas9 system. For example, a second marker gene could be used to introduce additional mutations in existing Methanosarcina mutants, which maintain pac from their initial construction. Finally, development of a second positive selectable marker for Methanosarcina could facilitate the development of genetic tools for other methanogens, just as the pac selection was first developed in Methanococcus and later used in Methanosarcina (Gernhardt et al.1990; Metcalf et al.1997).
Methanogenic archaea are not susceptible to many of the antibiotic compounds that inhibit bacteria and eukaryotes; thus, it has been difficult to find selective agent/resistance gene pairs for use in genetic experiments (Hilpert et al.1981). Given its utility in organisms from both Bacteria and Eukarya, we suspected the nourseothricin antibiotic/streptothricin acetyltransferase (sat) gene pair might comprise a useful positive selection for methanogens. Nourseothricin (a mixture of streptothricins C, D, E and F) is a commercially available antibiotic that induces mRNA miscoding during protein translation in both Bacteria and Eukarya (Haupt et al.1980). Moreover, the streptothricin acetyltransferase (sat) gene from the producing organism Streptomyces rochei has been shown to confer nourseothricin resistance in selected bacteria, fungi, plants and protozoa (Haupt, Hubener and Thrum 1978; Horinouchi et al.1987; Joshi et al.1995; Jelenska et al.2000; Reuss et al.2004). Here, we demonstrate that this is also the case for Methanosarcina. Additionally, we show that at least two other phylogenetically distinct human gut-associated methanogens, Methanomassiliicoccus luminyensis and Methanobrevibacter smithii, are inhibited by nourseothricin at low concentrations in both liquid and on solid media suggesting that nourseothricin and the sat marker gene may provide a stringent selection system in these methanogens as well.
MATERIALS AND METHODS
Strains, media and growth conditions
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. All manipulations of methanogens were carried out under strictly anoxic conditions in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI). Methanosarcina strains used in this study are listed in Supplemental Table I (Supporting Information). All derivatives of M. acetivorans C2A and M. barkeri Fusaro were grown in single-cell morphology (Sowers, Boone and Gunsalus 1993) at 37°C in bicarbonate-buffered high-salt (HS) liquid medium containing either 50 mM trimethylamine (TMA) or 125 mM methanol as growth substrates in Balch tubes with a N2/CO2 (80/20) headspace. Methanobrevibacter smithii DSM 861 was grown in liquid MSM medium containing 0.225 g/l K2HPO4, 0.225 g/l KH2PO4, 0.20 g/l (NH4)2SO4, 0.45 g/l NaCl, 0.0975 g/l MgSO4 x 7H20, 0.006 g/l CaCl2 x 2H2O, 0.2% yeast extract, 0.2% tryptone, 0.25% acetate, 0.0002% FeSO4, 0.01% resazurin, 2.8 g/l NaHCO3, 1 g/l NH4Cl, 0.50 g/l cysteine, 0.4 mM Na2S x 9H20, 2% clarified rumen fluid, and 1% each of trace element and vitamin solutions (Metcalf et al.1997). MSM medium was made anaerobic by boiling under N2/CO2 (80/20) for 10 minutes and cooling under N2/CO2 (80/20) before it was moved into an anaerobic glove chamber, dispensed into Balch tubes, autoclaved and pressurized to 135.5 kPa with H2/CO2 (80/20) for growth. Methanomassiliicoccus luminyensis DSM 25720 was grown in liquid MLM medium containing 0.225 g/l KH2PO4, 0.40 g/l NaCl, 0.40 g/l MgSO4 x 7H20, 0.05 g/l CaCl2 x 2H2O, 0.1% yeast extract, 0.1% tryptone, 0.25% acetate, 0.20% formate, 0.0002% FeSO4, 0.01% resazurin, 4.0 g/l NaHCO3, 0.40 g/l NH4Cl, 0.50 g/l cysteine, 0.50 g/l Na2S x 9H20, 2.5 mM each of the fatty acids valeric acid, isovaleric acid, 2-methylbutyric acid, and isobutyric acid, 125 mM methanol, 2% clarified rumen fluid, and 1% each of trace element and vitamin solutions (Metcalf et al.1997). MLM medium was made anaerobic by sparging under N2/CO2 (80/20) for 1 hour before it was moved into an anaerobic glove chamber, dispensed into Balch tubes, autoclaved and pressurized to 135.5 kPa with H2/CO2 (80/20) for growth. Rumen fluid was kindly donated by the Dairy Cattle Research Unit, University of Illinois at Urbana-Champaign. Rumen fluid was clarified by filtration through cheese cloth, centrifugation at 10 000 rpm for 30 minutes at 4°C, and filtration using a 0.2 μm Nalgene Filtration Apparatus (Thermo-Fisher Scientific, Waltham, MA). Growth on media solidified with 1.6% agar was performed as previously described (Metcalf et al.1996) except that M. acetivorans, Met. luminyensis and Me. smithii cells were spread directly on solid media plates, while M. barkeri cells were plated after suspension in high-salt medium solidified with 0.5% agar (high-salt top agar). Solid media plates were incubated in 3.5 L anaerobic jars (Oxoid) under a N2/CO2/H2S (80/20/10%) atmosphere for M. acetivorans or H2/CO2/H2S (80/20/10%) atmosphere for M. barkeri, Met. luminyensis and Me. smithii. Anaerobic jars were incubated inside an anerobic chamber. Puromycin dihydrochloride (Research Products International, Mt. Prospect, IL) and 2-bromoethanesulfonate (BES) were added to final concentrations of 2 μg/ml or 0.4 mM, respectively, from sterile, anaerobic stock solutions. Nourseothricin sulfate (GoldBio, St. Louis, MO) was added at concentrations indicated in the text from a sterile, anaerobic stock solution. Streptomyces rochei (ATCC 10 739) was grown at 30°C in liquid ATCC 172 medium substituted with tryptone instead of N-Z amine type A or on mannitol soy flour agar (Hobbs et al.1989). For plasmid construction and maintenance, the Escherichia coli DH5α/λpir and WM4489, a DH10B derivative designed to allow control the copy number oriV-based plasmids (Kim et al.2012), were used. E. coli strains were grown in LB broth at 37°C with 100 μg/ml ampicillin or 20 μg/ml chloramphenicol, as appropriate. Rhamnose was added to a final concentration of 10 mM to increase plasmid copy number of oriV plasmids in WM4489 hosts as needed.
Minimal inhibitory concentration determination
Liquid minimal inhibitory concentrations (MIC)s were determined as follows: saturated liquid cultures (OD600nm = ∼1.0 for Me. smithii, Met. luminyensis and ∼2.5 for Methanosarcina) were used to inoculate liquid media containing different concentrations of nourseothricin at a 1:100 final dilution of cells. For solid media MIC determination, saturated liquid cultures were spun down and concentrated 50x by resuspension in an appropriate volume of spent liquid media. A total of 20 μl of the cell suspension (∼1 × 109 CFU) were spotted on solid media plates containing different concentrations of nourseothricin and streaked for isolated colonies using sterile sticks. Growth on liquid and solid media containing the different concentrations of nourseothricin was monitored over time and compared with growth on media lacking nourseothricin for MIC determination.
Transformation methods
Escherichia coli strains were transformed via electroporation using an E. coli Pulser (Bio-Rad, Des Plaines, IL) as recommended. Liposome-mediated transformations of M. acetivorans and M. barkeri were performed as previously described (Metcalf et al.1997). All Methanosarcina transformants were single colony purified on solid media containing puromycin or nourseothricin before PCR verification and phenotypic analysis.
Isolation of genomic DNA from S. rochei
Streptomyces rochei spore suspension stocks were prepared as follows: 5 ml of sterile 0.001% Triton X-100 solution was added to a confluent lawn of S. rochei after growth and sporulation on mannitol soy flour agar (Hobbs et al.1989) plates. The spores were gently resuspended by scraping with a sterile glass slide, collected, vortexed for 1 minute and filtered using a sterile glass cotton-plugged syringe. This filtrate was centrifuged at 5000 rpm for 15 minutes and spores were resuspended in 1 ml of 0.001% Triton X-100 solution. On ice, resuspended spores were mixed with DMSO to a final concentration of 7%, then stored at −80°C. Streptomyces rochei genomic DNA was extracted by a salting out procedure as follows. A 30 ml culture of S. rochei mycelia was grown after inoculation from a spore suspension stock. Mycelia were spun down at 5000 rpm for 15 minutes, resuspended in SET buffer (20 mM Tris HCl pH 7.5, 75 mM NaCl, 25 mM EDTA) and incubated with 1 mg/ml final concentration of lysozyme at 37°C for 1 hour. After cell wall digestion, Proteinase K (Invitrogen) and SDS were added to 500 μg/ml and 1%, respectively, and the sample was incubated at 55°C for 2 hours. NaCl was added to 1.25 M and the sample was allowed to cool to room temperature before the addition of 5 ml chloroform and incubation at 20°C for 30 minutes. The sample was centrifuged at 4000 x g for 15 minutes at 20°C and the supernatant was transferred to a clean tube. The volume of supernatant was measured and 0.6 volume of 100% isopropanol was added to precipitate genomic DNA. DNA was carefully spooled onto a sterile, sealed Pasteur pipet, rinsed with 5 ml of 70% ethanol, air dried and dissolved in 2 ml of sterile deionized water.
Molecular methods
All primers used in this study are described in Supplemental Table II (Supporting Information). All reagents and enzymes used for molecular work were purchased from New England Biolabs (Ipswich, MA) unless otherwise specified. Primers were ordered from Integrated DNA Technologies (Coralville, IA) and all PCR reactions needed for plasmid construction used Phusion High-Fidelity DNA Polymerase. Plasmids were constructed using the NEBuilder HiFi DNA Assembly Master Mix as recommended. The NEBuilder Assembly Tool (v2.0.8) and Geneious (v9.1.8) were used to design plasmid constructs in silico. All plasmids were verified using restriction digestion and DNA sequencing (UIUC Core Sequencing Facility). To construct plasmid pKF05, pJK031A was digested with NdeI and HindIII and the plasmid backbone was gel purified (Omega E.Z.N.A. Gel Extraction Kit). The sat gene was amplified using pJK031A-sat gene forward and reverse primers and S. rochei genomic DNA. Purified pJK031A plasmid backbone and sat were assembled to produce pKF05. For Methanosarcina transformations, pKF05 was purified using the Zyppy Plasmid Miniprep DNA Isolation Kit (Zymo Research, Irvine, CA) and quantified via Nanodrop (Thermo-Fisher Scientific, Waltham, MA) or estimated by visual inspection of ethidium bromide stained agarose gels. Plasmid pKF010 was constructed in a step-wise manner. First, the pac gene in pJK301 was replaced with sat. To do this, the entirety of pJK301 (excluding the pac gene) was PCR amplified with primers pJK301 F and R and assembled with sat, which was PCR amplified using primers pJK301-sat F and R to make plasmid pKF08. Both pJK301 and sat PCR products were treated with DpnI to remove template DNA prior to assembly. The 2 kilobase sequence upstream of the M. acetivorans C2A ssuC gene was PCR amplified with ssuC upstream F and R primers and assembled with HindIII-digested pKF08 to make plasmid pKF09. The 2 kilobase sequence downstream of the M. acetivorans C2A ssuC gene was PCR amplified with ssuC downstream F and R primers and assembled with NotI-digested pKF09 to make plasmid pKF010. For transformation of M. acetivorans, pKF010 was purified using the Omega E.Z.N.A. Plasmid DNA Maxi Kit (Omega Bio-Tek, Norcross, GA) and digested with PvuII and XhoI. The linear sat gene-containing fragment of interest (∼6.1 kilobases) was gel purified (Omega E.Z.N.A. Gel Extraction Kit) and quantified via Nanodrop and agarose gel estimations. OneTaq 2X Master Mix with Standard or GC Buffer was used for PCR verification of Methanosarcina transformants and mutants.
Genome sequencing and analysis
Genomic DNA was extracted from four independent spontaneous nourseothricin resistant isolates, as well as the parental strain of Me. smithii as previously described (Boccazzi, Zhang and Metcalf 2000) except cells were enzymatically lysed using purified Me. smithii pseudomurein endoisopeptidase (MsPei) encoded by msm_1691 using methods described for Methanobrevibacter ruminantium PeiR (Leahy et al.2010). DNA libraries were prepared with the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA) and quantified via fluorometry (Qubit). All libraries were sequenced on one lane of an Illumina MiSeq v2 at the Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign using a 500 cycles v2 sequencing kit (Illumina, San Diego, CA). Trimmed, paired-end 250 nucleotide reads were mapped to the Me. smithii reference genome (NC_0 09515.1) using default parameters for breseq v0.25d (Deatherage and Barrick 2014). Mutations unique to the spontaneous nourseothricin resistant mutants were identified by comparing the mutations found in the parent strain with those found in each mutant.
RESULTS
Growth of at least four different methanogens is inhibited by nourseothricin
To test whether nourseothricin was an effective selective agent for methanogens, we determined the MIC in both liquid and on solid media for M. acetivorans, M. barkeri, Met. luminyensis, and Me. smithii. Growth of M. acetivorans and M. barkeri was completely inhibited by 300 μg/ml or 200 μg/ml nourseothricin, respectively, on solid media and by 200 μg/ml nourseothricin in liquid media. Growth of Met. luminyensis and Me. smithii was completely inhibited on solid media at 50 or 100 μg/ml, respectively, while the MIC in liquid media was 6.25 and 5 μg/ml, respectively.
Mutations in three different genes give rise to nourseothricin resistance in Me. smithii
Spontaneous nourseothricin resistant mutants were never observed when M. acetivorans, M. barkeri or Met. luminyensis were plated on solid media with nourseothricin; however, after an extended 1 month incubation period, spontaneous Me. smithii nourseothricin resistant mutants could be obtained that arose at frequencies between 6 × 10−8 and 11 × 10−8. To identify the basis of resistance in these mutants, we resequenced the genomes of four independent mutants, as well as the parental strain. The mutants examined carried missense mutations in msm_1096, encoding the TrkA-like component of a putative potassium transporter, or in msm_1318 or msm_1319 genes, which encode two hypothetical proteins with putative membrane-spanning domains (Table 1). Orthologs of msm_1096, msm_1318 or msm_1319 were not found in M. acetivorans, M. barkeri or Met. luminyensis, which may explain why no spontaneous nourseothricin resistant mutants were observed for these three strains throughout the course of this work.
Table 1.
Methanobrevibacter smithii spontaneous nourseothricin resistant mutants.
| Nourseothricin resistant mutant | Mutant gene | Gene annotation | Nucleotide change | Protein variant |
|---|---|---|---|---|
| 1 | msm_1319 | Hypothetical protein | A236C | Q79P |
| 2 | msm_1318 | Hypothetical protein | G43A | G15S |
| 3 | msm_1096 | Potassium transporter TrkA | C412A | L138I |
| 4 | msm_1319 | Hypothetical protein | C173A | A58D |
Expression of the S. rochei streptothricin acetyltransferase (sat) gene in Methanosarcina allows selection of nourseothricin resistant transformants
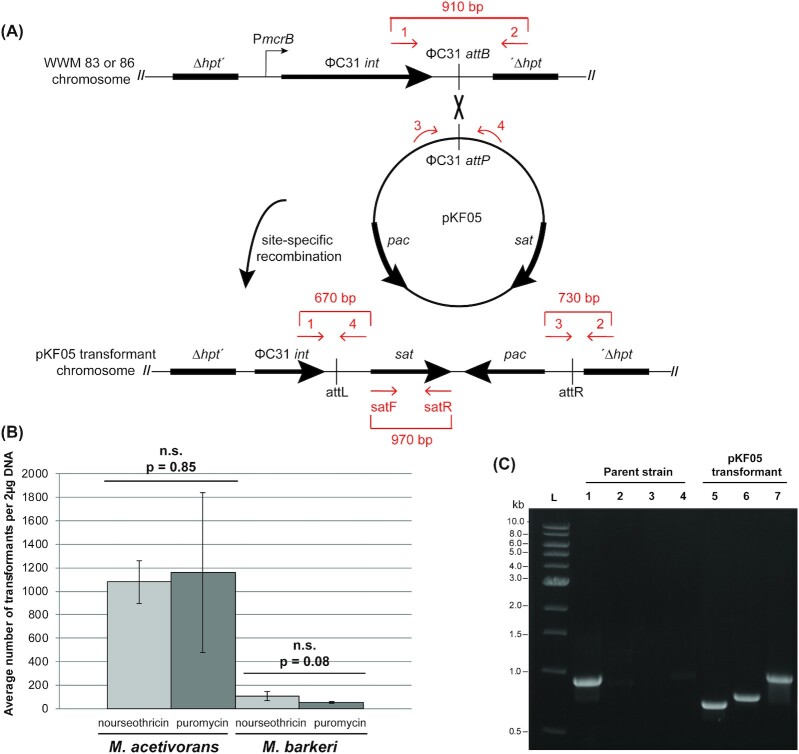
To test the utility of sat as a positive selectable marker for Methanosarcina, plasmid pKF05 was constructed to allow expression of sat from S. rochei under the control of the tetracycline-regulable Methanosarcina PmcrB(tetO1) promoter. Note that the base vector (pJK031A) also encodes the established puromycin resistance gene (pac) as a positive selectable marker and this marker was retained in pKF05 (Supplemental Figure 1, Supporting Information). Plasmid pKF05 cannot replicate independently in Methanosarcina, but can be integrated into the chromosome of strains that express the integrase gene from the Streptomyces bacteriophage ΦC31 (Guss et al.2008) (Fig. 1A). Importantly, because the Methanosarcina strains used for transformation do not encode the TetR repressor protein, expression of sat from PmcrB(tetO1) is constitutive. Separate selections for either puromycin-resistant or nourseothricin-resistant transformants produced similar numbers of colonies in M. acetivorans, Fig. 1B. Qualitatively similar results were obtained in M. barkeri; however, as previously observed, the transformation efficiency was lower in this host (Metcalf et al.1997). Growth of M. acetivorans and M. barkeri pKF05 transformants was similar to strains lacking pKF05, suggesting that expression of sat has minimal impact on either host.
Figure 1.
The streptothricin acetyltransferase (sat) gene is a useful positive selectable marker for Methanosarcina transformations. (A) A schematic of Streptomyces ΦC31 integrase-mediated site-specific recombination of pKF05 into M. acetivorans (WWM83) or M. barkeri (WWM86) chromosomes. ΦC31 integrase-mediated chromosomal integration of pKF05 introduces the selectable markers pac and sat for selection of Methanosarcina transformants. Primers used to verify pKF05 transformants are represented by the red arrows in the schematic. (B) Comparison of pKF05 transformation efficiency under puromycin or nourseothricin selection in M. acetivorans and M. barkeri. Triplicate transformation experiments with pKF05 were completed with M. acetivorans (WWM83) and M. barkeri (WWM86). Transformants were selected on both nourseothricin and puromycin plates. The number of colonies that arose was counted to determine the transformation efficiency: 1157 ± 681 and 1078 ± 182 transformants on puromycin and nourseothricin, respectively, for M. acetivorans and 55 ± 7 and 106 ± 37 on puromycin and nourseothricin, respectively, for M. barkeri. Error bars represent the standard deviation of three independent transformations and P-values were calculated using two-tailed unpaired t-tests (α = 0.01). Importantly, no colonies were observed for controls lacking DNA. (C) PCR verification of Methanosarcina pKF05 transformants. pKF05 transformants produced 670, 730, or 970 bp PCR products from reactions containing primers 1 and 4 (Lane 5), 2 and 3 (Lane 6) or satF and satR (Lane 7), respectively, while the parent strain only produced a 910 bp PCR product from reactions containing primers 1 and 2 (Lane 1) but no products with the other three primer pairs (Lanes 2–4).
Methanosarcina acetivorans and M. barkeri transformants were verified to have pKF05 integrated into the chromosome at the expected site using a previously described PCR assay and phenotypic analysis (Guss et al.2008) (Fig. 1A). We also used sat specific primers to verify the presence of this gene in selected transformants (Fig. 1A). A total of 7 independent puromycin and nourseothricin resistant M. acetivorans and M. barkeri pKF05 transformants produced the PCR products expected for pKF05 transformants (as shown for a single representative in Fig. 1C) regardless of the selection antibiotic or Methanosarcina strain used (data not shown). Furthermore, all puromycin resistant transformants were nourseothricin resistant and vice versa. Taken together these data show sat is a useful positive selectable marker analogous to pac for selection of Methanosarcina transformants when expressed in single copy under the control of a strong, constitutive Methanosarcina promoter.
sat is a useful positive selectable marker for constructing gene deletions in M. acetivorans
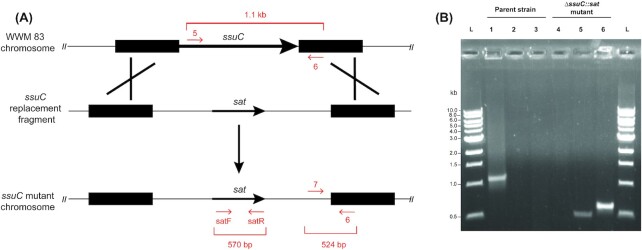
To further examine the utility of sat as a selectable marker for making chromosomal deletions in M. acetivorans we constructed pKF010 (Supplemental Figure 1, Supporting Information). This plasmid is designed to replace the M. acetivorans ssuC gene with our sat marker. The ssuC gene encodes the permease component of a putative coenzyme M ABC transporter and is required for the uptake of the methanogenesis inhibitor BES (Guss et al.2005). Transformants that successfully deleted ssuC, and thus incorporated sat into the chromosome, were therefore expected to be both BES and nourseothricin resistant (Fig. 2A). To test this, M. acetivorans was transformed with a linear DNA fragment from pKF010 containing 2 kilobase regions homologous to the upstream and downstream regions of ssuC flanking DNA containing sat and transformants were selected on nourseothricin-containing solid media plates (Fig. 2A).
Figure 2.
sat can be used as a positive selectable marker for the construction of gene deletion mutants in M. acetivorans. (A) A schematic of M. acetivorans ΔssuC mutant generation. A linear DNA fragment containing sat flanked by 2 kilobase regions homologous to the upstream and downstream regions of ssuC was transformed into M. acetivorans. Incorporation of sat-containing DNA at the ssuC locus gives rise to mutants which are both nourseothricin and BES resistant. Primers used to verify ΔssuC::sat mutants are represented by the red arrows in the schematic. (B) PCR analysis of M. acetivorans ΔssuC mutants. Lanes 1–3 represent PCR reactions containing M. acetivorans (WWM 83; Parent strain) DNA with primers 5 and 6 (Lane 1), 6 and 7 (Lane 2) or satF and satR (Lane 3). Lanes 4–6 represent PCR reactions containing DNA from a single M. acetivorans nourseothricin resistant ssuC mutant (ΔssuC::sat) with primers 5 and 6 (Lane 4), 6 and 7 (Lane 5) or satF and satR (Lane 6). A total of 15 out of 15 independent mutants produced identical PCR products to those shown for the mutant in this figure.
Methanosarcina acetivorans ΔssuC mutants were verified using PCR and phenotypic analyses. PCR analysis was carried out with primer pairs specific for the wild-type or mutant ssuC locus (Fig. 2A). As shown in Fig. 2B, both the wild-type M. acetivorans parent strain and ΔssuC::sat mutant strain produced the expected PCR amplicons. Identical results were obtained when 14 additional independent nourseothricin-resistant transformants were analyzed with the described primer sets (data not shown). These transformants were also phenotypically verified to be both nourseothricin and BES resistant as expected.
DISCUSSION
The results presented here demonstrate that the S. rochei sat gene, when expressed from a strong, constitutive promoter, confers nourseothricin resistance to both M. acetivorans and M. barkeri. As with the S. alboniger pac gene, functional expression of sat in Methanosarcina did not require codon optimization. The numbers of transformants selected using pac or sat were shown to be essentially equivalent across triplicate transformation experiments indicating that the two selections work equally well. Additionally, the sat gene was shown to be useful for generating a gene deletion in M. acetivorans. Together these data establish sat as an additional positive selectable marker for genetically tractable Methanosarcina species, enabling a variety of genetic experiments. It is worth noting that the sat cassette in these plasmids also confers nourseothricin resistance in E. coli, which simplifies construction of mutagenic plasmids.
The growth of two human-gut associated methanogens, Met. luminyensis and Me. smithii, is inhibited by nourseothricin at lower concentrations than Methanosarcina. Given its utility in bacteria, fungi, protozoa, plants and now Methanosarcina, it is likely that sat may be a useful positive selectable marker to aid the development of genetic tools for methanogens like Met. luminyensis and Me. smithii. Spontaneous nourseothricin resistant Me. smithii mutants had mutations in one of three genes hypothesized to encode cellular membrane-associated proteins: msm_1096, msm_1318 or msm_1319. Interestingly, msm_1096 shares homology with TrkA, a component of the bacterial TrkAH potassium transporter complex, which was shown to be required for antibiotic resistance in certain bacteria (Chen et al.2004). These results suggest mutations in these putative membrane spanning proteins/transporters block the inhibitory effects of nourseothricin in Me. smithii, either by preventing nourseothricin entry or by promoting export of the antibiotic. The fact that msm_1096, msm_1318 or msm_1319 homologs are absent in Methanosarcina and Methanomassillicoccus, coupled with our inability to isolate spontaneous resistant mutants in these strains, suggests that nourseothricin entry or export is significantly different between these distantly related methanogens.
Although advanced genetic tools such as Cas9-mediated genome editing are established for M. acetivorans, the sat marker can now aid experiments to further test essential gene hypotheses generated using these tools and benefit researchers using traditional genetic tools to study Methanosarcina species. As the repertoire of genetic tools is expanded for methanogenic archaea, the nourseothricin-sat gene pair will likely become a positive selection system routinely used for genetic analyses of Methanosarcina and the establishment of genetic tools for additional methanogen groups.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr. Mary Elizabeth Metcalf and Dr. Kou-San Ju for technical assistance.
FUNDING
This work was supported by a grant from the National Institute of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID), R21AI122019 to W.W.M.
Conflict of Interest . None declared.
REFERENCES
- Boccazzi P, Zhang JK, Metcalf WW. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri fusaro. J Bacteriol. 2000;182: 2611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Chuang YC, Chang CCet al.. A K+ uptake protein, TrkA, is required for serum, protamine, and polymyxin B resistance in Vibrio vulnificus. Infect Immun. 2004;72:629–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Meth Mol Biol. 2014;1151:165–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernhardt P, Possot O, Foglino Met al.. Construction of an integration vector for use in the Archaebacterium methanococcus-voltae and expression of a eubacterial resistance gene. Mol Gen Genet. 1990;221:273–9. [DOI] [PubMed] [Google Scholar]
- Guss AM, Mukhopadhyay B, Zhang JKet al.. Genetic analysis of mch mutants in two Methanosarcina species demonstrates multiple roles for the methanopterin-dependent C-1 oxidation/reduction pathway and differences in H(2) metabolism between closely related species. Mol Microbiol. 2005;55:1671–80. [DOI] [PubMed] [Google Scholar]
- Guss AM, Rother M, Zhang JKet al.. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea. 2008;2:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt I, Hubener R, Thrum H. Streptothricin F, an inhibitor of protein synthesis with miscoding activity. J Antibiot. 1978;31:1137–42. [DOI] [PubMed] [Google Scholar]
- Haupt I, Jonak J, Rychlik Iet al.. Action of streptothricin-F on ribosomal functions. J Antibiot. 1980;33:636–41. [DOI] [PubMed] [Google Scholar]
- Hilpert R, Winter J, Hammes Wet al.. The sensitivity of archaebacteria to antibiotics. Zentralblatt für Bakteriologie Mikrobiologie und Hygiene: I. Abt. Originale C: Allgemeine, angewandte und ökologische Mikrobiologie. ScienceDirect. 1981;2:11–20. [Google Scholar]
- Hobbs G, Frazer CM, Gardner DCJet al.. Dispersed growth of streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–7. [Google Scholar]
- Horinouchi S, Furuya K, Nishiyama Met al.. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J Bacteriol. 1987;169:1929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Tietze E, Tempe Jet al.. Streptothricin resistance as a novel selectable marker for transgenic plant cells. Plant Cell Rep. 2000;19:298–303. [DOI] [PubMed] [Google Scholar]
- Joshi PB, Webb JR, Davies JEet al.. The gene encoding streptothricin acetyltransferase (Sat) as a selectable marker for leishmania expression vectors. Gene. 1995;156:145–9. [DOI] [PubMed] [Google Scholar]
- Kim SY, Ju KS, Metcalf WWet al.. Different biosynthetic pathways to fosfomycin in Pseudomonas syringae andStreptomycesspecies. Antimicrob Agents Chemother. 2012;56:4175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler PR, Metcalf WW. Genetic manipulation of Methanosarcinaspp. Front Microbiol. 2012;3:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy SC, Kelly WJ, Altermann Eet al.. The genome sequence of the rumen methanogen Methanobrevibacter ruminantiumreveals new possibilities for controlling ruminant methane emissions. PLoS One. 2010;5:e8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Zhang JK, Shi Xet al.. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol. 1996;178: 5797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Zhang JK, Apolinario Eet al.. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A. 1997;94:2626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondorf S, Deppenmeier U, Welte C. A novel inducible protein production system and neomycin resistance as selection marker for Methanosarcina mazei. Archaea. 2012;2012: 973743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DD, Metcalf WW. Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc Natl Acad Sci U S A. 2017;114:2976–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak DD, Mahanta N, Mitchell DAet al.. Post-translational thioamidation of methyl-coenzyme M reductase, a key enzyme in methanogenic and methanotrophic Archaea. Elife. 2017;6:e29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter Ret al.. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–27. [DOI] [PubMed] [Google Scholar]
- Sowers KR, Boone JE, Gunsalus RP. Disaggregation of Methanosarcina spp and growth as single cells at elevated osmolarity. Appl Environ Microbiol. 1993;59:3832–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JK, Pritchett MA, Lampe DJet al.. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivoransC2A using a modified version of the insect mariner-family transposable element Himar1. Proc Natl Acad Sci U S A. 2000;97:9665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.