A repetitive basic-acidic motif in U1-70K is critical for its phase separation and aggregation.
Abstract
Liquid-liquid phase separation (LLPS) facilitates the formation of functional membraneless organelles and recent reports have linked this phenomenon to protein aggregation in neurodegenerative diseases. Understanding the mechanism of LLPS and its regulation thus promises to shed light on the pathogenesis of these conditions. The RNA-binding protein U1-70K, which aggregates in brains of Alzheimer’s disease patients, is considered a potential target for Alzheimer’s therapy. Here, we report that two fragments in the low-complexity (LC) domain of U1-70K can undergo LLPS. We have demonstrated that the repetitive basic-acidic motifs in these fragments induce nucleotide-independent phase separation and initiate aggregation in vitro. We also have confirmed that LLPS and aggregation occur in vivo and that the content of ampholytic motifs in a protein domain determines the transition between droplets and aggregation, providing insights into the mechanism underlying the formation of diverse assembly states.
INTRODUCTION
Liquid-liquid phase separation (LLPS) is a process in which proteins or protein-RNA complexes spontaneously segregate to form coexisting high- and low-concentration phases. LLPS is considered to play important roles in membraneless organelle formation (1). Such transient organelles have multiple cellular functions, including P-granules that enrich intracellular RNA binding protein during transcription (2), large cytoplasmic RNA-protein assembly in stress granules (3), and transcription factors condensing in Cajal bodies (4). Recent studies reported that LLPS was observed in several neurodegenerative disease–associated proteins, such as FUS, TDP-43, and tau (5–7), whereas it was widely accepted that the fibrillar assemblies of amyloidogenic proteins are central to the pathogenesis of neurodegenerative diseases including amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease (AD) (8–10). LLPS may initiate the aggregation and potentially lead to pathological assembly in disease (6, 11–13). It is thus hypothesized that LLPS may regulate the balance between intracellular organization and pathogenesis. However, specific molecular interactions that drive phase separation and the mechanism that determines the dichotomy between physiology and pathology remain largely unclear.
In most cases, LLPS is driven by multivalent, weak interactions involving low-complexity (LC), intrinsically disordered regions, which contain biased amino acid compositions or repetitive sequences (5, 14). Arginine-rich domains are believed to mediate LLPS via electrostatic interactions between guanidine and inorganic counteranions or RNA (15, 16). For example, RGG/RG motifs are common among proteins that can undergo LLPS, such as FUS, EWS, and hnRNPA1 (17). Tyrosine-containing repeats in the FUS and other hnRNPs (heterogeneous nuclear ribonucleoproteins) can also form hydrophobic and π interactions that are crucial in promoting hydrogel formation (14, 18). Those motifs are important in phase separation and aggregation via various weak interactions like cation-π and π-π interaction (19). However, it remains unclear what is the sequence determinant of the threshold concentration for initial separation and the aggregation status of the final phase.
Currently, most literature on LLPS focuses on RNA binding proteins involved in ALS, and we believe that proteins related to other neurodegenerative diseases may share a similar mechanism. Recent studies showed that an RNA binding protein, U1 small nuclear ribonucleoprotein 70 kDa (U1-70K), aggregates and associates exclusively with tau in the cortex of AD patients (20). Later, the Seyfried group demonstrated that the AD brain homogenate could induce the aggregation of U1-70K from both healthy brain homogenate and recombinantly expressed protein, indicating that U1-70K aggregation may contribute to AD pathogenesis (21). The same group further demonstrated that the highly repetitive pattern of basic-acidic residues in U1-70K is critical for aggregation, localization, and interaction with tau (22).
We thus hypothesize that the basic-acidic motifs in the LC domains may facilitate intermolecular interactions and phase separation, and report here the LLPS of U1-70K LC domain as well as the mechanism and its conversion from LLPS to aggregates.
RESULTS
Both LC1 and LC2 of U1-70K can undergo LLPS
On the basis of in silico predictions (fig. S1) (23–25) and previous studies (21, 22, 26), the LC domains of U1-70K were divided into two fragments: LC1 (231–308) and LC2 (317–407) (Fig. 1A). We examined their behavior of LLPS in response to different regulatory factors.
Fig. 1. Sequences and LLPS of LC1 and LC2 of U1-70K.
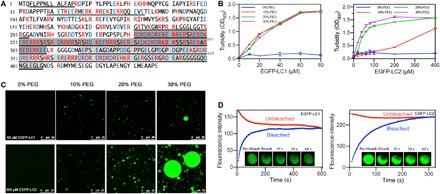
(A) U1-70K amino acid sequence with basic residues in red and acidic residues in blue. Disordered sequences by predictions were underlined, and LC domains were highlighted in gray. (B) Turbidity demonstrated phase separation of LC1 and LC2. (C) Confocal microscopy showed droplet formation of 60 μM LC1 and 200 μM LC2. (D) FRAP of the droplets formed by LC1 and LC2.
LC1 and LC2 with enhanced green fluorescent protein (EGFP) fusion on their N termini were expressed in Escherichia coli and purified by Ni–nitrilotriacetic acid (NTA) affinity chromatography. Purified proteins were mixed with polyethylene glycol (PEG 8000) at different ratios, which mimics the crowded environment in cells (27). Phase separation was evaluated by turbidity and droplet formation under a confocal microscope (Fig. 1, B and C).
As expected, both LC1 and LC2 formed dynamic liquid phases. The solution turbidity increased as protein or PEG concentration increased, indicating phase separation. The droplets merged and enlarged as more PEG was added, demonstrating stronger phase separation under more crowded environments. Furthermore, fluorescence recovery after photobleaching (FRAP) suggested that those droplets are in dynamic liquid phase, not gels or aggregation. A “half-bleach” method was used, where almost half of the droplet area was bleached and the fluorescence intensity was monitored over time to observe the redistribution of the proteins (Fig. 1D). For both LC1 and LC2, the bleached droplets got fully recovered, indicating their liquid status.
Nonetheless, LC1 and LC2 showed a quantitative difference in phase separation behavior. LC1 underwent phase separation under lower concentration of PEG than LC2 and formed precipitates at a protein concentration higher than 80 μM. In contrast, LC2 formed stable droplets at concentrations up to 400 μM. This difference is consistent with previous reports that the LC1 has a higher tendency to aggregate (22). This may be due to different numbers of ampholytic motifs within the two domains, as LC1 contains more ordered basic-acidic residual repeats.
The phase separation of U1-70K was affected by pH
To test whether the electrostatic interaction in ampholytic motifs drives phase separation, we changed buffer pH to alter charges on the proteins and tested their phase separation behavior (Fig. 2). LC1 and LC2 still underwent phase separation at altered pH, as indicated by increased turbidity. Higher concentrations of LC1 or LC2 were required to reach the same turbidity at high pH than low pH, suggesting that acidic conditions promoted phase separation, presumably due to increased positive charges at low pH. Electrostatic interaction is thus critical for phase separation.
Fig. 2. LLPS of LC1 and LC2 under various pH.
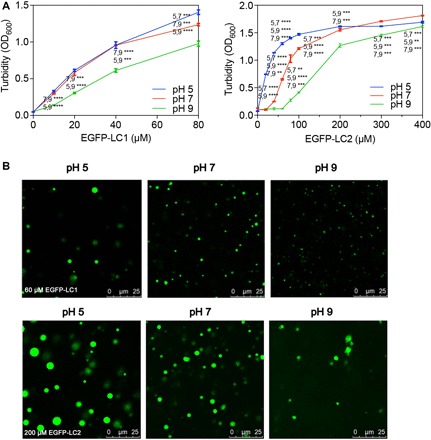
(A) pH affects the progress of LLPS occurring as concentrations of LC1 or LC2 increasing. (B) Droplet morphology of LC1 (60 μM) or LC2 (200 μM) changes under pH 5, 7, or 9. Multiple t tests, n = 3; **P < 0.0332, ***P < 0.0021, and ****P < 0.0001.
As turbidity is affected by the size, density, and shapes of the droplets, further examination of fluorescent images was conducted to dig more details. Statistical analysis of droplet sizes and densities was performed based on the images acquired (fig. S2 and table S1). LC1 formed smaller but more droplets at higher pH. LC2 formed larger droplets at pH 5 than at pH 7, but at pH 9, it formed irregular droplets and the density dropped markedly. Fluorescence of these irregular droplets recovered over time in FRAP (fig. S3), suggesting that they are liquid droplets. The irregular shape may be due to weaker interactions at this pH. The different response to pH change may arise from the difference in amino acid composition. LC1 consisted of only six different amino acids, with 56.4% of basic residues (R/K) and 29.5% of acidic residues (D/E). This domain is highly ampholytic and the intermolecular electrostatic interaction would dominate phase separation behavior. In contrast, amino acid composition in LC2 is more diverse, with 27.5% basic residues, 30.0% acidic residues, and 27.5% hydrophobic residues (A/G/L). The differences in composition result in different pI (isoelectric point) of these two proteins: 11.77 for LC1 and 5.30 for LC2. From pH 5 to pH 9, although the quantity of charges may vary, LC1 stayed positively charged, and its phase separation only differed in droplet size and density. However, at pH 9, LC2 could barely remain as droplets, as the dominant charges changed to negative from positive at pH 5. The results suggested that positive charges from the basic residues are more important in mediating phase separation.
Free charges in solution regulate LLPS of LC1 and LC2
The contribution of opposite charges should be considered separately instead of the total positive and negative charges or net charge. To test the role of both acidic and basic residues on phase separation of LC domains, we added free acidic (Asp), basic (Arg), and neutral (Ala) amino acids to the solution of LC1 and LC2, respectively, and inspected the phase separation (Fig. 3). Both Arg and Asp substantially decreased the turbidity of LC1, while Ala had little effect (Fig. 3A). Confocal microscopy further confirmed the changes (Fig. 3B, fig. S2, and table S1). With Arg or Asp added, the formation of the droplets was disrupted, as the droplets became irregularly shaped and highly variable in size. FRAP was used to examine whether the irregular “droplets” were liquid phase (fig. S3). When Asp was added to LC1, the fluorescence of irregular entities was not able to recover, indicating that they were solid aggregates rather than liquid phase. In contrast, droplets formed by LC1 mixed with Arg remained liquid. Neutral Ala did not change the morphology of droplets. The results indicated that both cations and anions can interact with LC1 and interfere with its phase separation. The Asp anion had stronger effect, which is consistent with our conclusion that positive charges in the protein are more critical for LLPS.
Fig. 3. LLPS of LC1 and LC2 with Arg, Ala, or Asp doped.
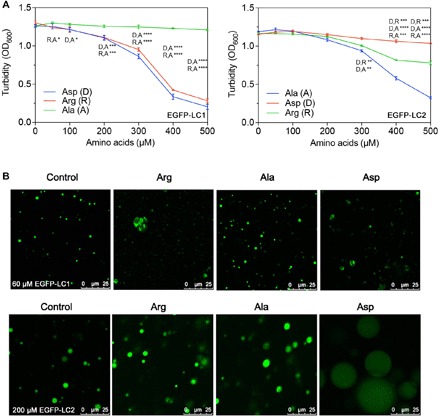
(A) Turbidity changes with increasing concentrations of free amino doped into LC1 (60 μM) or LC2 (200 μM). (B) Droplet morphology of LC1 or LC2 changes with the free amino acids. Multiple t tests, n = 3; *P < 0.1234, **P < 0.0332, ***P < 0.0021, and ****P < 0.0001.
LC2 behaved differently when free amino acids were added. As increasing amount of amino acids was doped into the samples, Arg had little effect on the turbidity, while Ala and Asp decrease the turbidity, the latter with much stronger effect. Under confocal microscopy, LC2 with Arg did not show significant difference compared with LC2 alone. With Ala and Asp added, LC2 formed larger but dimmer droplets, indicating that LC2 interacted with these amino acids through its abundant basic residues via cation-anion and potential cation-π interaction. Although LC2 contains a smaller fraction of basic residues, free negative charges still have a strong effect on its phase separation. This is consistent with our observation that basic residues are more critical for phase separation.
An alternative explanation may be the screen effect from the high concentration of ions. Thus, we repeated phase separation experiments with 500 mM NaCl (fig. S4). High salt had different effect from the charged amino acids. Although we cannot fully rule out screen effect, the change in LLPS caused by adding free amino acids was mainly due to the interaction between the protein and free charges in solution.
Peptide models reveal a charge-driven mechanism of LLPS and aggregation
To further study the effect of charges, we tested phase separation behavior of an array of peptides derived from LC domains. We synthesized a representative fragment containing 19 amino acids from U1-70K LC domain with repetitive basic-acidic residues, termed RD19, and peptide variants with acidic or basic residues mutated to Ala (AD19 and RA19; Fig. 4A). These peptides were mixed with PEG to examine their properties of phase separation. Both RD19 and RA19 showed increased turbidity and droplet formation, while AD19 remained one-phase solution (Fig. 4B). Under confocal microscope, RD19 formed regular, highly circular, and stable droplets but RA19 produced irregular droplets (Fig. 4C). These observations suggest that basic residues are essential for phase separation. A lack of acidic residues may not prevent phase separation but will destabilize the droplets.
Fig. 4. Peptide model of charge-driven LLPS.

(A) Sequence of RD19 and its mutants. (B) Turbidity increased as peptide concentrations increased, indicating that LLPS occurred. (C) Microscopy images of the peptide. RD19 formed nice circular droplets; RA19 formed irregular droplets, while no droplets were observed with AD19. Multiple t tests, n = 3; **P < 0.0332, ***P < 0.0021, and ****P < 0.0001.
The peptide of RD19 provided a simplified model of this family of repetitive basic-acidic proteins. We tested whether it could further aggregate after LLPS. The peptide was incubated for 3 days and observed under transmission electron microscope (TEM) (fig. S5). RD19 formed short but well-aligned aggregates with interlaced structures. Usually, the charge-driven peptides or proteins aggregate to disordered form (28). We were wondering whether any secondary structure, like β sheet, resulted in the morphology of the aggregations. Thus, we obtained circular dichroism (CD) of RD19, both freshly made and incubated for 24 hours. The spectrum both showed typical random coil structure and did not change over time. The results strongly suggested that the repetitive anion-cation motif is sufficient to form aggregates with ordered structures.
So far, all the results confirmed that both LC domains can undergo LLPS, and the main driving force is electrostatic interactions. Although LC1 and LC2 share similar compositions and mechanism of LLPS, their behaviors are distinct. The basic and acidic residues constitute over 80% of LC1 so that the interactions between the molecules are dominated by charges, resulting in strong tendency to phase separate and aggregation. The diversity of residues in LC2 grants it more diverse interactions, potentially including hydrophobic effect and interactions with counterions. In both cases, cations in the domains contributed more interactions during the LLPS.
LLPS and aggregation of the LC domains was observed in cells
In vitro assays showed that both LC domains can undergo LLPS. To test whether a similar pattern applies in vivo, we studied the LLPS of LC domains under physiological conditions. The sequences of LC1, LC2, LC, and full-length U1-70K were cloned into the pCDNA3.1-EGFP vector and transfected with Lipo8000 to Neuro2a (N2a) cells to express the corresponding proteins. After 24 hours, cells were imaged with a confocal microscope (Fig. 5A). Highly circular droplet-like entities were observed in cells, indicating that LLPS occurred with all the four proteins. LC1, LC, and full-length U1-70K were only localized in the nucleus, and the droplets were colocalized with condensed nuclear bodies, whereas LC2 was distributed all over the cells and the droplets were not correlated with the condensed parts in nucleus. The difference in localization of the two segments drew our attention because the LC domain of U1-70K is outside of the RNA recognition motif and should not directly interact with RNA (21). When we titrated different amounts of polyU, no significant difference was observed (fig. S6). Unlike other RNA binding domains with abundant basic residues, the LC domain of U1-70K contains evenly distributed acidic and basic residues, weakening its interaction with large homo-charged molecules like RNA. The localization of LC1 should thus result from interactions with other nuclear proteins, which is consistent with the literature that LC1 facilities nuclear localization and phase separation of U1-70K (22).
Fig. 5. LLPS and aggregation of U1-70K and the LC fragments in cells.
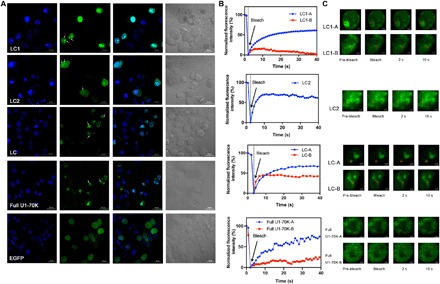
(A) Confocal microscopy of EGFP-LC1, EGFP-LC2, EGFP-LC, and full-length U1-70K in N2a cells. Nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI) in blue. EGFP alone was expressed as control. Arrows showed droplet-like entities in cells. (B) FRAP results of EGFP- LC1, EGFP-LC2, EGFP-LC, and full-length U1-70K in N2a cells. The fluorescence intensities of each area marked by red circles in (C) were plotted versus time. The intensity was normalized with the pre-bleached as 100% and first time point after bleach as 0%. (C) Fluorescence images during FRAP.
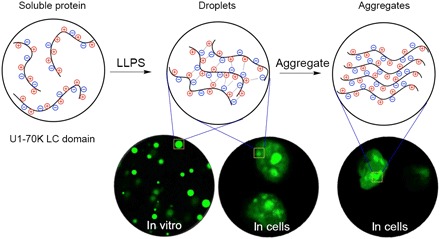
In addition, large irregular aggregate-like entities were found in cells with LC1, LC, and full-length U1-70K transformed. The findings suggested that liquid phase and solid aggregates can coexist in vivo and that the aggregates may be the next stage of liquid-phase droplets. LC1-mediated phase separation dominated the behavior of LC and the full-length protein.
To further characterize the droplets and aggregates and explore the potential conversion process, we performed FRAP on living cells overexpressing LC1, LC2, LC, and full-length U1-70K. For LC1, LC, and full-length protein, we selected both highly circular droplets and irregular aggregate-like particulates and performed photobleaching. For LC2, only droplets were bleached because no aggregates were observed. Fluorescence recovery over time was recorded (Fig. 5, B and C). The protein assemblies with different morphology demonstrated distinct behavior in fluorescence recovery. The highly circular droplets formed by all four species, shown as LC1-A, LC2, LC-A, and full U1-70K-A in Fig. 5B, recovered more than 60% after bleaching, indicating liquid-like properties. However, when we bleached those irregular aggregates labeled as “-B” areas, less recovery was observed. Especially for LC1-B, only less than 15% fluorescence was recovered, suggesting that fabric aggregations were formed. For LC-B and full U1-70K-B, some recovery was observed. Compared with LC1, the aggregation structure of LC was more like a mixture of droplets and aggregates. In addition, we have observed similar assemblies with full-length U1-70K—some with good mobility showing more droplet properties and others having less mobility, which contained more aggregates. This heterogeneity in behavior suggested that the assembly may be undergoing conversion from droplets to aggregates. It would be the first observation that this kind of repetitive ampholytic proteins aggregates via LLPS.
As structural details of the aggregates in cells were beyond the resolution limits of optical microscope, we examined the aggregates of LC1 and LC2 in vitro with TEM (fig. S7). Both aggregates are well aligned, showing great similarity with RD19. Given that the LC domain barely has any secondary structure, we concluded that the aligned, interlaced structures resulted from regular packing of repetitive charged motifs. To capture the transition states of the conversion from droplets to aggregates, we incubated the phase separation samples at 37°C for 12 hours and prepared the TEM samples. Aggregates grown inside of the droplets were observed (fig. S7), confirming that the conversion did occur. On the basis of these observations, we propose a model for the assembly of proteins rich in ampholytic motifs, in which LLPS occurs first and then the liquid droplets convert to solid-like aggregates (Fig. 6).
Fig. 6. Cartoon presenting the mechanism of the process from soluble proteins to phase separation and finally to fibric aggregations by the repetitive basic-acidic motif.
DISCUSSION
A number of models of LLPS that were proposed as proteins with different properties were reported to undergo LLPS, driven by various interactions, including π-π, cation-π, dipole-dipole, and cation-anion interactions (1). A common feature of phase separating proteins is multivalent motifs, such as the Phe-Gly repeats in DEAD-box helicase 4 (DDX4), in which the cation-π and π-stacking interactions contribute to its LLPS (19). Secondary structures and domains could also drive LLPS. For example, proteins like TDP-43 can phase separate through helix-helix interactions in its LCD and oligomerization domain (29, 30). Many of the phase separation processes require multiple proteins or motifs, especially those driven by electrostatic interactions. Arg is rich in many RNA binding proteins to interact with RNA, represented by FUS (31). LAF-1 existing in P granule contains separate acidic and basic motifs on C and N termini (32).
We have found a new amino acid pattern that induces LLPS by electrostatic interactions. Unlike the known charge-driven LLPS patterns listed above, in which opposite charges are distributed unevenly, usually in different molecules or domains, the LC domains of U1-70K contain evenly distributed basic and acidic residues, resulting in a strong self-assembling tendency to undergo LLPS. In the past few years, several groups have tried to predict the conformation and phase separation properties of polyampholytes (28, 33, 34). These investigators have demonstrated that the tendency for LLPS is affected both by the composition of charged residues and by sequence patterns. The properties of the polyampholytic proteins can be predicted by several parameters, such as fraction of charged residues (FCR) and charge patterning parameter κ. The sequences of LC1, LC2, and LC were analyzed with CIDER tool by the Pappu laboratory (table S3) (33). All three proteins are strong polyampholytes with low κ, which indicates that they have strictly alternating sequence. For comparison, LC1 has higher FCR, which results in higher tendency to self-assemble than LC2. LC2 is more hydrophobic, having more complicated interactions, as shown in the experiments. Combining theoretical predictions with analysis of the effects of pH and free charged amino acids on the phase separation of LC domains, we demonstrated how this alternately charged pattern drives LLPS and revealed that cations play a more critical role during the process.
These observations also gave us insights into the in vivo behavior of U1-70K under physiological and pathological conditions. The LC domain is sufficient for LLPS and may promote the formation of functional nuclear granules for RNA processing. Furthermore, strong interaction from the ampholytic motifs may promote assembly of pathological aggregations. As positive charges in the LC domain dominate in self-assembly, negatively charged molecules may co-aggregate under pathological conditions. Consistent with this notion, tau protein was found to co-aggregate with U1-70K when it is overphosphorylated in AD (35, 36).
Moreover, we observed for the first time the conversion of U1-70K LC domains from liquid phase to aggregation in cells. We found that the fraction of ampholytic motifs in a protein domain may determine its assembly states—LC1 with almost exclusively ampholytic motif forms both droplets and aggregations, whereas LC2 with a more diverse sequence pattern can only undergo LLPS. A higher fraction of ampholytic motifs results in stronger association and promote the conversion from liquid phase into aggregates. The mechanism we found explains the self-assembly behavior of proteins with ampholytic motifs and would facilitate the understanding of the conversion between functional granules and pathological aggregations. In addition to AD, increasing reports have suggested that LLPS is implicated in multiple neurodegenerative diseases. Understanding the mechanism of the conversion from LLPS to fabric aggregations would offer a new route for therapy.
MATERIALS AND METHODS
Materials and reagents
The DNA template was a gift from J. Han (Xiamen University, China). The primers were purchased from BGI, China. DNA polymerase KOD FX (TOYOBO Life Science Department Inc.) was used for polymerase chain reaction (PCR). The gel-purified kit was purchased from Axygen, China. All the restriction endonucleases were from New England Biolabs (NEB), USA. The plasmids of pET-28a-EGFP and pCDNA3.1-EGFP were purchased from Miao Ling Plasmid, China. Amplified plasmids were purified using the Plasmid Mini Kit I (100) from OMEGA BIO-TEK. The Transetta (DE3) Chemically Competent Cell was purchased from TransGen Biotech, China. Lysogeny broth (LB) was purchased from Beijing Aoboxing Biotech Co. Ltd., China. N2a cells were gifts from Y. Chen (Tsinghua University, China). The Dulbecco’s modified Eagle’s medium (DMEM) 1× with glucose (4.5 g/liter) and MEM Alpha 1× were from Corning, USA. Fetal bovine serum (FBS) was purchased from Thermo Fisher Scientific. Penicillin/streptomycin was purchased from General Electric. All other chemical reagents, unless otherwise specified, were purchased from Sigma-Aldrich.
Construction and expression of EGFP-LC1 and EGFP-LC2
Three DNA fragments (LC1, LC2, and LC) were amplified using four primers (table S2). The PCR fragment was gel-purified, digested by restriction endonucleases Bam HI and Xho I, and cloned into the pET-28a-EGFP vector (Bam HI and Xho I digested). The EGFP-LC1, EGFP-LC2, and GST-LC2 proteins were expressed in Transetta (DE3) E. coli cells. For protein expression, E. coli cells were cultured in standard LB medium at 37°C until the OD600 reached 0.9 to 1.0 and induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were incubated overnight at 16°C for protein expression. Then, cells were harvested by centrifugation at 4000 rpm for 18 min. EGFP-LC1 and EGFP-LC2 were purified by Ni-NTA metal affinity column. Particularly, we used 1 M NaCl in the buffers during the purification of EGFP-LC1. EGFP-LC1 and EGFP-LC2 were desalted into 200 mM potassium phosphate buffer by ÄKTA pure (General Electric, USA) with desalting column. GST-LC2 was cleaved by PreScission Protease (a gift from Y. Feng, Beijing University of Chemical and Technology, China) to obtain tag-free LC2, desalted into 200 mM potassium phosphate buffer.
Construction plasmid of pCDNA3.1-LC1-EGFP, pCDNA3.1-LC2-EGFP, pCDNA3.1-LC-EGFP, and pCDNA3.1 full-length U1-70K-EGFP for eukaryotic expression
Three DNA fragments (LC1, LC2, and LC) and full-length U1-70K were amplified using six primers (table S2). LC1 was cloned with U1-Nhe231f and U1-Sac308rnostop; LC2 was cloned with U1-Nhe317f and U1-Sac407rnostop; LC was cloned with U1-Nhe231f and U1-Sac407rnostop; full-length U1-70K was cloned with U1-Nhe1f and U1-Sac1314rnostop. The PCR fragment was gel-purified, digested by restriction endonucleases (Nhe I and Sac II), and cloned into the pCDNA3.1-EGFP vector (Nhe I and Sac II digested). The EGFP-fused proteins were expressed by N2a cells.
N2a cells were cultured in complete media [44.5% DMEM 1× with glucose (4.5 g/liter), 44.5% MEM Alpha 1×, 10% FBS, and 1% penicillin/streptomycin]. N2a cells were plated in 24-well plate with microscope coverglass (φ14 mm, CITOGLAS, China) on the bottom and were allowed to grow for 12 hours. The plasmids of pCDNA3.1-LC1-EGFP, pCDNA3.1-LC2-EGFP, or pCDNA3.1-LC-EGFP were transfected with the Lipo8000 Transfection Reagent (Beyotime Biotechnology, China) into the cells for protein expression. After 24 hours, 4% paraformaldehyde was used to fix the cells and cell nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI) (Beyotime Biotechnology, China) for 30 min at 37°C. The Antifade Polyvinylpyrrolidone Mounting Medium (Beyotime Biotechnology, China) was applied to the samples and mounted to the slide glass (CITOGLAS, China).
Sample preparation for phase separation
EGFP-LC1 and EGFP-LC2 were purified by Ni-NTA column and ÄKTA pure. EGFP-LC1/EGFP-LC2 was dissolved in potassium phosphate buffer (pH 7) to a concentration of 160 μM/800 μM as stock and diluted with potassium phosphate buffer (pH 7) in the assay. Then, EGFP-LC1/EGFP-LC2 was mixed with 20, 40, and 60% PEG 8000 solution with a volume ratio of 1:1 at room temperature to obtain various concentrations.
Phase separation with arginine, alanine, or aspartic acids
Purified EGFP-LC1/EGFP-LC2 was dissolved in potassium phosphate buffer (pH 7) to a concentration of 240 μM/800 μM as stock and diluted with potassium phosphate buffer (pH 7) in the assay. Arginine, alanine, or aspartic acids were dissolved in potassium phosphate buffer (pH 7) to a concentration of 2, 1.5, and 2 M as stock, respectively. EGFP-LC1/EGFP-LC2 solution was mixed with amino acid solution first to obtain various concentrations of R, A, and D. Then, 40% PEG 8000 was added to the mixture to a volume ratio of 1:1. For LC2, 500 mM NaCl was added instead of amino acids for comparison.
Phase separation at different pH
Purified EGFP-LC1 and EGFP-LC2 were desalted into potassium phosphate buffer of pH 5, pH 7, or pH 9. EGFP-LC1/EGFP-LC2 solution was then concentrated into 320 μM/800 μM as stock and diluted with corresponding buffer in the assay. Then, EGFP-LC1/EGFP-LC2 solution was mixed with 40% PEG 8000 solution in a volume ratio of 1:1 at room temperature to obtain various concentrations.
Phase separation with polyU
Stock EGFP-LC1/EGFP-LC2 was prepared as above. Polyuridylic acid potassium salt form Sigma-Aldrich was dissolved in potassium phosphate buffer and mixed with the proteins at mass ratios of 0:1, 0.5:1, 1:1, 2:1, 3:1 (polyU:protein). Then, 40% PEG 8000 in a volume ratio of 1:1 was added.
Turbidity assay
Proteins and peptides were prepared as described above. Turbidity was measured by absorption at 600 nm in 384-well plates using a SpectraMax M2 microplate reader. All samples were examined in triplicate (n = 3).
Confocal microscopy of droplets in vitro
Confocal microscopy was performed with an inverted Leica SP8 microscope, equipped with lasers for 405- and 488-nm excitation. Images were acquired using a 20× objective. The following fluorescence setting was used for detection of EGFP: 498 to 533 nm. The images were analyzed with Fiji (ImageJ) for droplet size and density by analyzing particle function.
FRAP of droplets in vitro
Samples containing droplets were applied to an eight-well Lab-Tek chambered coverglass (Thermo Fisher Scientific). Experiments were performed on an inverted microscope (LSM 780, Observer.Z1; Carl Zeiss, Oberkochen, Germany) equipped with a confocal spinning disk unit (CSU-X1; Yokogawa, Tokyo, Japan) and a Zeiss 63× oil immersion lens. Half-bleach was performed at pixel resolutions of x = 64 nm and y = 64 nm. Droplets of an approximate diameter of 3.5 to 5 μm were selected, and approximately half of the area was bleached for 20 ns with 35 to 45% of maximum laser power of a 488-nm laser [1 AU (Airy Unit)]. The recovery was recorded at a rate of 2 s per frame for 650 s.
Transmission electron microscopy
For aggregate observation, all the protein or peptides were incubated with shaking at 37°C for 3 days. For the examination of aggregation in droplets, the phase separation samples prepared above were incubated at 37°C for 12 hours. A total of 8 μl of solution was pipetted onto a carbon-coated copper grid, rested there for 2 min, and then stained with a 2% sodium phosphotungstic acid aqueous solution (pH 6.5) for 1 min. After the samples were dried, images of all samples were recorded on an HT7700 electron microscope (Hitachi, Japan) at 100 kV.
CD spectroscopy
The CD spectroscopy of peptides was recorded on a Jasco J-810 spectropolarimeter (Jasco Co., Japan). RD19 was dissolved in water and incubated at 37°C and 200 rpm. Before recording the spectrum, the solution was mixed by gently tapping its Eppendorf tube container five times. The CD spectra were collected at a bandwidth of 1 nm with a standard 4-s response time. The scanning mode was continuous with a scanning speed of 50 nm/min. Four scans were averaged for each sample. Molar residual ellipticity was calculated using ellipticity in millidegrees (θ), path length in centimeters (l), molecular weight in grams per mole (M), peptide concentration in milligrams per milliliter (c), and the number of residues (nr) in the following equation: [θ] = (θ × M)/(10 × c × l × nr).
Transfection of EGFP fused LC1, LC2, and LC in eukaryotic cells
N2a cells were cultured in complete media and plated in 24-well plate with microscope coverglass. After 12-hour growth, the plasmids of pCDNA3.1-LC1-EGFP, pCDNA3.1-LC2-EGFP, or pCDNA3.1-LC-EGFP were transfected with Lipo8000. The transfected cells were cultured for another 24 hours to allow the proteins to express.
Cell imaging and FRAP
Cells growing on coverglasses were fixed with 4% paraformaldehyde, and the nucleus was stained with DAPI. The coverglasses were mounted to the slide glass with antifade polyvinylpyrrolidone mounting medium. Images were acquired by Zeiss LSM 780 with 63× oil immersion lens. Fluorescence signals were obtained with excitation/emission at 405/498 nm for DAPI and 488/546 nm for EGFP.
FRAP was performed on an LSM 780 inverted microscope from Carl Zeiss with 63× oil immersion lens. The transfected N2a cells were cultured in glass petri dish (φ20 mm). Before FRAP, the culture medium was exchanged to Opti-MEM. The droplets in cells were bleached for 8 to 10 s with 10 to 20% of maximum laser power of a 488-nm laser (1 AU). The recovery was recorded for 20 to 30 time points after bleaching (30 to 40 s). The fluorescent intensity of bleached area over time was calculated by Zen.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (91853116 and 21672019), National Key Research and Development Program (2017YFF0205402), the Fundamental Research Funds for the Central Universities (XK1802-8), and Research projects on biomedical transformation of China-Japan Friendship Hospital (nos. PYBZ1812 and PYBZ1815). Author contributions: S.X. and S.-Z.L. designed the experiments, analyzed the data, and wrote the manuscript. R.G. and F.H. performed the experiments and analyzed the data. Y.L. and Y.W. helped construct the plasmids and protein expression. S.-Z.L., S.X., and T.T. supervised the whole project. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/11/eaax5349/DC1
Fig. S1. Disordered motif of U1-70K predicted by different models.
Fig. S2. Statistics of average sizes of droplets of LC1 and LC2.
Fig. S3. FRAP of the irregular droplets.
Fig. S4. Images showing effects of high salt (500 mM NaCl) to LC2 LLPS.
Fig. S5. Characterization of aggregation and secondary structure of RD19.
Fig. S6. The effects of RNA to the LLPS of LC1 and LC2.
Fig. S7. TEM images of aggregates formed by LC1 and LC2 in solution and in droplets.
Table S1. Densities of droplets of LC1 and LC2 under various pH values or with different amino acids added.
Table S2. Primers used for gene construction.
Table S3. Parameters of LC1, LC2, and LC predicted by CIDAR.
REFERENCES AND NOTES
- 1.Boeynaems S., Alberti S., Fawzi N. L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., Tompa P., Fuxreiter M., Protein phase separation: A new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brangwynne C. P., Eckmann C. R., Courson D. S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A. A., Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Kedersha N., Ivanov P., Anderson P., Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 38, 494–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handwerger K. E., Cordero J. A., Gall J. G., Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell 16, 202–211 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., Taylor J. P., Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel A., Lee H. O., Jawerth L., Maharana S., Jahnel M., Hein M. Y., Stoynov S., Mahamid J., Saha S., Franzmann T. M., Pozniakovski A., Poser I., Maghelli N., Royer L. A., Weigert M., Myers E. W., Grill S., Drechsel D., Hyman A. A., Alberti S., A Liquid-to-solid phase transition of the als protein fus accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Ambadipudi S., Biernat J., Riedel D., Mandelkow E., Zweckstetter M., Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8, 275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Da Cruz S., Cleveland D. W., Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 21, 904–919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C. X., Tan W., Whittle C., Qiu L., Cao L., Akbarian S., Xu Z., The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLOS ONE 5, e15878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goedert M., Spillantini M. G., Propagation of Tau aggregates. Mol. Brain 10, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K. M., Bennett R. E., Dujardin S., Laskowski P. R., MacKenzie D., Kamath T., Commins C., Vanderburg C., Roe A. D., Fan Z., Molliex A. M., Hernandez-Vega A., Muller D., Hyman A. A., Mandelkow E., Taylor J. P., Hyman B. T., Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guenther E. L., Cao Q., Trinh H., Lu J., Sawaya M. R., Cascio D., Boyer D. R., Rodriguez J. A., Hughes M. P., Eisenberg D. S., Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol. 25, 463–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun F., Chen L., Wei P., Chai M., Ding X., Xu L., Luo S.-Z., Dimerization and structural stability of amyloid precursor proteins affected by the membrane microenvironments. J. Chem. Inf. Model. 57, 1375–1387 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Kato M., Han T. W., Xie S., Shi K., du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., Eisenberg D., Tycko R., McKnight S. L., Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varadi M., Zsolyomi F., Guharoy M., Tompa P., Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLOS ONE 10, e0139731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A., Guharoy M., De Decker M., Jaspers T., Ryan V. H., Janke A. M., Baatsen P., Vercruysse T., Kolaitis R. M., Daelemans D., Taylor J. P., Kedersha N., Anderson P., Impens F., Sobott F., Schymkowitz J., Rousseau F., Fawzi N. L., Robberecht W., Van Damme P., Tompa P., Van Den Bosch L., Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong P. A., Vernon R. M., Forman-Kay J. D., RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Zhou X.-R., Cao Y., Zhang Q., Tian X. B., Dong H., Chen L., Luo S.-Z., Self-assembly nanostructure controlled sustained release, activity and stability of peptide drugs. Int. J. Pharm. 528, 723–731 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., Baldwin A. J., Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai B., Hales C. M., Chen P.-C., Gozal Y., Dammer E. B., Fritz J. J., Wang X., Xia Q., Duong D. M., Street C., Cantero G., Cheng D., Jones D. R., Wu Z., Li Y., Diner I., Heilman C. J., Rees H. D., Wu H., Lin L., Szulwach K. E., Gearing M., Mufson E. J., Bennett D. A., Montine T. J., Seyfried N. T., Wingo T. S., Sun Y. E., Jin P., Hanfelt J., Willcock D. M., Levey A., Lah J. J., Peng J., U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 110, 16562–16567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diner I., Hales C. M., Bishof I., Rabenold L., Duong D. M., Yi H., Laur O., Gearing M., Troncoso J., Thambisetty M., Lah J. J., Levey A. I., Seyfried N. T., Aggregation properties of the small nuclear ribonucleoprotein U1-70K in Alzheimer disease. J. Biol. Chem. 289, 35296–35313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishof I., Dammer E. B., Duong D. M., Kundinger S. R., Gearing M., Lah J. J., Levey A. I., Seyfried N. T., RNA-binding proteins with basic-acidic dipeptide (BAD) domains self-assemble and aggregate in Alzheimer’s disease. J. Biol. Chem. 293, 11047–11066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchan D. W. A., Minneci F., Nugent T. C. O., Bryson K., Jones D. T., Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D. T., Cozzetto D., DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wootton J. C., Non-globular domains in protein sequences: Automated segmentation using complexity measures. Comput. Chem. 18, 269–285 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Sun F., Xu L., Chen P., Wei P., Qu J., Chen J., Luo S.-Z., Insights into the packing switching of the EphA2 transmembrane domain by molecular dynamic simulations. J. Phys. Chem. B 119, 7816–7824 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Baskir J. N., Hatton T. A., Suter U. W., Protein partitioning in two-phase aqueous polymer systems. Biotechnol. Bioeng. 34, 541–558 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Priftis D., Leon L., Song Z., Perry S. L., Margossian K. O., Tropnikova A., Cheng J., Tirrell M., Self-assembly of α-helical polypeptides driven by complex coacervation. Angew. Chem. Int. Ed. Engl. 54, 11128–11132 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Conicella A. E., Zerze G. H., Mittal J., Fawzi N. L., ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afroz T., Hock E.-M., Ernst P., Foglieni C., Jambeau M., Gilhespy L. A. B., Laferriere F., Maniecka Z., Plückthun A., Mittl P., Paganetti P., Allain F. H. T., Polymenidou M., Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M., Ruepp M. D., Simons M., Niessing D., Madl T., Dormann D., Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C.-H., Eckmann C. R., Myong S., Brangwynne C. P., The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das R. K., Pappu R. V., Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 110, 13392–13397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das S., Eisen A., Lin Y. H., Chan H. S., A lattice model of charge-pattern-dependent polyampholyte phase separation. J. Phys. Chem. B 122, 5418–5431 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Alonso A. D., Grundke-Iqbal I., Barra H. S., Iqbal K., Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: Sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc. Natl. Acad. Sci. U.S.A. 94, 298–303 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson G. V., Stoothoff W. H., Tau phosphorylation in neuronal cell function and dysfunction. J. Cell Sci. 117, 5721–5729 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/11/eaax5349/DC1
Fig. S1. Disordered motif of U1-70K predicted by different models.
Fig. S2. Statistics of average sizes of droplets of LC1 and LC2.
Fig. S3. FRAP of the irregular droplets.
Fig. S4. Images showing effects of high salt (500 mM NaCl) to LC2 LLPS.
Fig. S5. Characterization of aggregation and secondary structure of RD19.
Fig. S6. The effects of RNA to the LLPS of LC1 and LC2.
Fig. S7. TEM images of aggregates formed by LC1 and LC2 in solution and in droplets.
Table S1. Densities of droplets of LC1 and LC2 under various pH values or with different amino acids added.
Table S2. Primers used for gene construction.
Table S3. Parameters of LC1, LC2, and LC predicted by CIDAR.


