Abstract
Purpose of review:
Emerging evidence has shown that epigenetic derangements might drive and promote tumorigenesis in various types of malignancies and is prevalent in both B cell and T cell lymphomas. The purpose of this review is to explain how the epigenetic derangements result in a chromatin-remodeled state in lymphoma and contribute to the biology and clinical features of these tumors.
Relevant findings:
Studies have explored on the functional role of epigenetic derangements in chromatin remodeling and lymphomagenesis. For example, the haploinsufficiency of CREBBP facilitates malignant transformation in mice and directly implicates the importance to re-establish the physiologic acetylation level. New findings identified 4 prominent DLBCL subtypes, including EZB-GC-DLBCL subtype that enriched in mutations of CREBBP, EP300, KMT2D and SWI/SNF complex genes. EZB subtype has a worse prognosis than other GCB-tumors. Moreover, the action of the histone modifiers as well as chromatin remodeling factors (e.g. SWI/SNF complex) cooperate to influence the chromatin state resulting in transcription repression. Drugs that alter the epigenetic landscape have been approved in T cell lymphoma. In line with this finding, epigenetic lesions in histone modifiers have recently been uncovered in this disease, further confirming the vulnerability to the therapies targeting epigenetic derangements.
Summary:
Modulating the chromatin state by epigenetic-modifying agents provides precision-medicine opportunities to patients with lymphomas that depend on this biology.
Keywords: Chromatin, lymphoma, epigenetics
Introduction
DNA, carrying our genetic code, is packaged in the nucleus around histone proteins H1, H2A, H2B, H3, and H4. This tight complex is known as chromatin. Chromatin structure is regulated by two general classes of complexes: those that covalently modify histone tails and those that remodel nucleosomes in an ATP-dependent manner. Together, these classes cooperate to dynamically regulate the structure of chromatin, and thus have essential roles in the control of gene expression, DNA repair, cell division and apoptosis. Not surprisingly, these processes are often mutated in cancer. Diseases characterized by a chromatin remodeled state often have inherent resistance to chemotherapy and therefore tend to be highly aggressive.
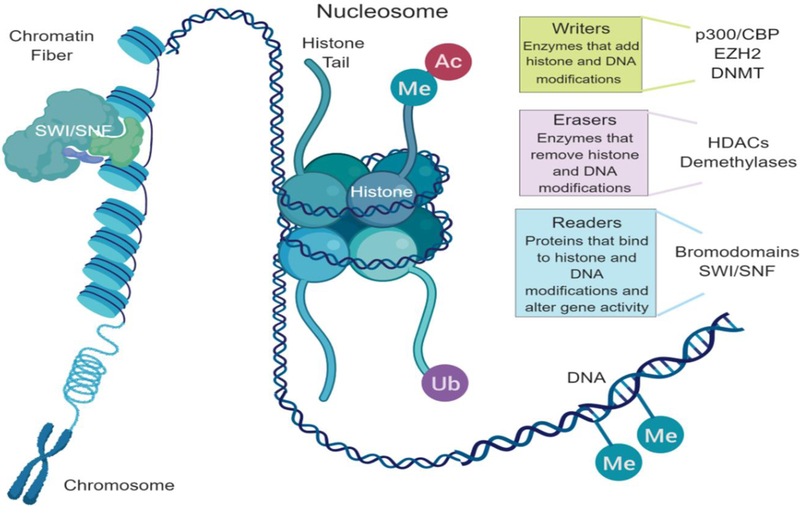
Epigenetic regulation of the chromatin state and transcription is a dynamic process coordinated by the interplay of multiple complexes working in concert. Three main groups of enzymes and proteins control these processes: writers, erasers, and readers (Fig 1). Writers include enzymes that add acetyl groups such as histone acetyltranserases (HATs) and methyl groups such as histone and DNA methyltransferases. Acetylation of histone leads to an open, active chromatin state. HAT enzymes that are known to influence the chromatin state in the context of lymphoma are p300 and CBP. New efforts to target HATs are underway. Methylation of histone or DNA can have either an activating or repressive role, depending on the location of the modification. The histone methyltransferase EZH2 is commonly mutated in lymphoma. This derangement contributes to lymphomagenesis by enforcing trimethylation of histone. Drugs targeting EZH2 are now in clinical development. Erasers are enzymes that remove post-translational marks on histone and DNA. Histone deacetylases (HDACs) are a large family of erasers that remove acetyl groups from histone. Targeting of HDACs with inhibitors leads to increased acetylation on histone. HDAC inhibitor drugs have demonstrated successful clinical application in T-cell lymphomas. Readers are proteins that recognize and bind to specific histone acetylation marks. This recognition and binding contributes to chromatin remodeling by co-activating the signal carried by the lysine state. There is a large family of these readers which are called bromodomains. Bromodomains are contained within HAT enzymes and members of the SWI/SNF complex to name a few. Bromodomain inhibitors are being studied across a broad range of cancers including lymphomas and have shown promise.
Figure 1:
Epigenetic regulation of the chromatin state. Three main groups of enzymes and proteins control these processes: writers, erasers, and readers. Me=Fmethylation, Ac=acetylation, Ub= ubiquitination, SWI/SNF=switch/sucrose non-fermentable.
Old and new insights into these derangements and their interplay, have opened the possibility of directing treatment toward epigenetic processes. By modulating the chromatin state, there is an opportunity to restore normal physiological epigenetic controls and thus reverse malignant phenotypes. Herein, we will discuss epigenetic controls of the nucleosome that lead to a chromatin remodeled state and methods for targeting specific derangements driving lymphoma.
Histone lysine methylation
EZH2
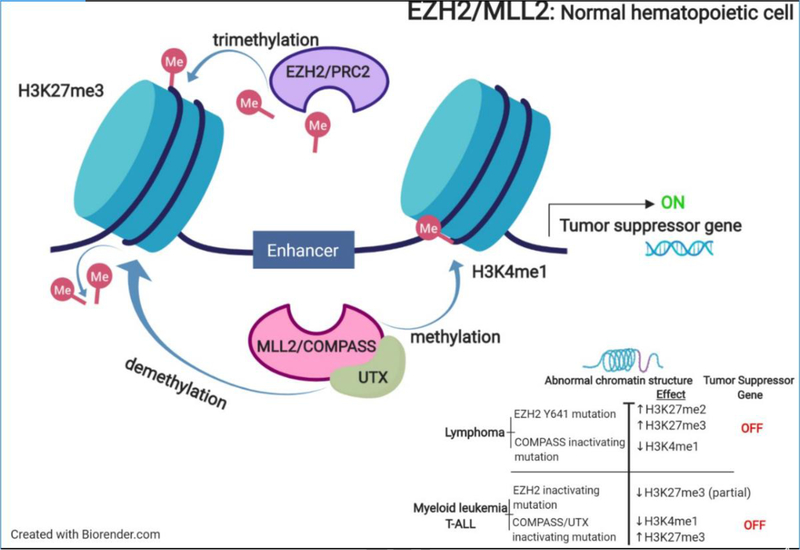
Chromatin modifying gene (CMG) mutations have led to altered effects of key enzymes driving lymphoma. Many of these enzymes are being studied as therapeutic targets in follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL), including CBP, EZH2, and KMT2D (alias, ALR/MLL2). EZH2 (Enhancer of Zeste Homolog 2) has a multifaceted role as a catalytic histone methyltransferase of histone H3 on lysine 27 (H3K27). It is the catalytic subunit of the polycomb repressive complex 2 (PRC2). The PRC2 contains multiple subunits including EZH2, EED and SUZ12, and together they play an essential role in transcriptional regulation. There are two PRC2 complexes containing either EZH2 or EZH1 [1]. PRC2 catalyzes trimethylation of H3K27 (H3K27me3) that lead to chromatin silencing, suppressing the expression of genes regulating cell proliferation and differentiation in the normal physiologic state (Figure 2). EZH2 activity is upregulated during the germinal center (GC) reaction, where B-cells undergo somatic hypermutation; this allows DNA damage to go unchecked and prevents programmed cell death. Abnormal, hyperactive PRC2 activity as identified in lymphomas has been attributed to trimethylation of histone [2]. This enforces a condensed chromatin state that prohibits normal transcriptional processes from occurring. Alterations in EZH2 at Ala677 and Tyr641 has been recognized in both FL and DLBCL [3]. As a subunit of the PRC2 complex, EED brings the N-terminal activation loop of EZH2 proximal to the catalytic SET domain; together these cofactors enable PRC2 basal activity and further contribute to transcriptional repression [4]. In fact, many histone methyltransferases, like MLL2 and EZH2, have been linked to the development of lymphomas, predominantly GC-derived lymphomas, including DLBCL and FL. Several EZH2 inhibitors have been tested in preclinical models, including GSK 343, EPZ-6438, ZLD10A, DZNep, and CPI 1205, as a way of targeting aberrant histone methylation. Results conclude that EZH2 inhibitors show strong antitumor effects by suppressing the overall methylation of H3K27 and cause an anti-proliferation effect in a concentration and time-dependent manner in DLBCL cell lines [3]. The clinical development of EZH2 inhibitors for the treatment of lymphoma has revealed that EHZ2 is a bonafide target. A phase 2 study of tazemetostat in patients with FL demonstrated an objective response rate in 74% (29/39) of relapsed/refractory patients harboring EZH2 mutations. A complete response was achieved in 10% of the responders. In contrast, the response rate in the wild type group was only 34%. Stable disease was achieved in both groups at a rate of 26% and 30% respectively. Tazemetostat was very well tolerated and therefore has a potential role in the treatment of FL [5].
Figure 2:
Interplay between EZH2 and MLL2 in normal hematopoietic cells
EZH2 has been found to lead to a chromatin-condensed state even in the absence of its mutational status. In T-cell lymphomas (TCL) such as Adult T-cell leukemia/lymphoma (ATLL), there is immunohistochemistry evidence demonstrating a correlation between increased activity of EZH2 and increased levels of H3K27me3. Additionally, EZH2 upregulation is facilitated by MYC‐induced suppression of its regulatory microRNAs (miRNAs) [6]. Both miRNAs, miR-26a and miR-548, are repressed by MYC, which contributes to lymphoma cell survival by silencing EZH2 [7]. Furthermore, MYC regulation of miRNA expression is further supported by miRNA array data showing that EZH2 inhibition, induced expression of a number of MYC-regulated miRNAs [7].
With that in mind, in TCL cell lines the EZH2 inhibitor, DZNep in combination with daunoblastine inhibited cell growth and increased apoptosis by decreasing EZH2 protein expression levels [8]. Moreover, the inhibitor DS-3201b, targeting EZH1 and EZH2, has shown anti-tumor activity in 80% of TCL patients with an overall response rate of about 53% [9]. Taken together, the in vitro and clinical data demonstrate the effectiveness of the inhibiting EZH2 in TCL, which may be a therapeutic target in these diseases.
In addition, the EZH2 protein has been demonstrated to be overexpressed in mantle cell lymphoma (MCL). This has led to abnormal methylation patterns on H3K27me3, which is thought to be linked to proliferation of MCL cell lines [10]. This suggests that MCL cell lines may be susceptible to EZH2 inhibition despite not possessing the mutations in EZH2. Recent studies have demonstrated that treatment with GSK 343 inhibited binding between EZH2 and its cofactor EED in MCL cell lines. To analyze the outcome of EED inhibition alongside EZH2, an immunoprecipitation was performed on transfected cells overexpressed with EED and EZH2, which revealed that it caused an increase in cell proliferation and resulted in increased H3K27 trimethylation [10]. Exposure to the EZH2 inhibitor, GSK 343, significantly inhibited cell growth (more than 90%) at 20uM concentration in the MCL cell lines Mino, Granta and Z138 [10]. Together these outcomes propose that wild type EZH2 is an epigenetic target for MCL. Although, EZH2 inhibition has been correlated with a greater cytotoxic impact on mutated EZH2 in the setting of DLBCL and FL, this example seen with MCL indicates that lymphomas with wild-type EZH2 may also have sensitivity to EZH2 inhibitor agents [10].
In our laboratory, we studied the impact of combining EZH2 inhibition and pan-HDAC inhibition. Our hypothesis was that dual epigenetic targeting would induce greater modulation in the histone state and therefore lead to increased programmed cell death. The combination exhibited synergy in lymphoma cell lines with both EZH2 mutations as well as wild type EZH2 cell lines with overexpression of the enzyme. The combination of EZH2 and HDAC inhibitors led to both modification of acetylation and methylation of H3K27. Additionally the combination disrupts the PRC2 complex possibly due to acetylation of the RBAP subunit [11]. With combined increased histone acetylation induced by HDAC inhibition and reduced histone methylation induced by EZH2 inhibition, chromatin are likely less compact and more accessible allowing for the expression of differentiation factors and tumor suppressors.
MLL2
Chromatin modifying genes play an essential part in modifying normal B-cell differentiation programs and inhibiting germinal center exit [12]. The mutated KMT2D (MLL2) gene is frequently found in FL (approximately 72% of cases) and in a lower frequency found in DLBCL (approximately 24% of cases) [12]. Among the mixed lineage leukemia (MLL) family of proteins, KMT2D has a role in the SET1 methyltransferases through the methylation of histone 3 lysine 4 (H3K4).
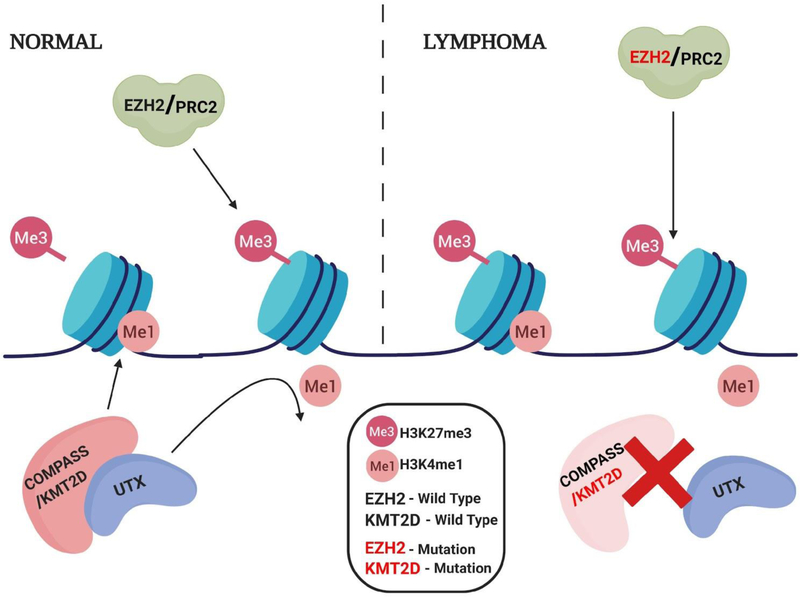
KMT2D also functions as a scaffolding protein. It forms a complex with H3K27 demethylase UTX. The mutations of KMT2D, which are frameshift or stop codons are deleterious to protein abundance and function. This results in the loss of KMT2D, and subsequently leads to a reduction of the KMT2D/KMT2C-UTX complex. This KMT2D-UTX complex is responsible for the removal of tri-methylation of H3K27. Because KMT2D is part of the complex, the loss of KMT2D leads to the accumulation of H3K27me3 (Figure 3) [12]. Like EZH2, KMT2D protein’s function is responsible for cell differentiation and embryonic development. KMT2D gene deletion increased levels of sister chromatid exchange, gross chromosomal aberrations, and micronuclei [13, 14]. This is an example of opposing effects of histone methylation on chromatin status.
Figure 3:
As part of the COMPASS complex, KMT2D has a role as a SET1 methyltransferase leading to the monomethylation of H3K4, which opposes H3k27me3. UTX, part of the KMT2D/COMPASS complex, is a demthylase responsible for the demethylation of H3K4. Removal of the H3K4 methyl group, allows for trimethylation of H3K27me3 by the EZH2/PRC2 complex.
The first gene in this family, KMT2A (alias, MLL1) was initially discovered because of its translocation (11q23 with more than 80 different partners) in a majority of lymphoid and myeloid leukemias arising in infants [15]. These translocations generate fusion proteins that aberrantly activate oncogenes such as HOXA9 and EV11 [16, 17]. Many of the MLL translocation partners such as ENL, AF9, AFF1, AFF4 are found in a super elongation complex (SEC) with elongation factors eleven-nineteen Lys-rich leukaemia (ELL) and the RNA polymerase II elongation factor P-TEFb [18]. MLL2KMT2D expression is controlled in part by PTEFbP-TEFb following the misrecruitment of SEC to MLL target genes, perturbing transcription elongation checkpoints at these loci, and resulting in leukemic pathogenesis. Accordingly, in preclinical models, CDK9/PTEFbP inhibitors have shown efficacy in MLL rearranged models of AML [19]. Further evaluation in lymphomas with MLL2 mutations may be merited.
DOT1L
Disruptor of telomeric silencing 1-like (DOT1L) is the only known H3 lysine 79 (H3K79) methyltransferase in mammals. Genetic silencing of DOT1L, leading to complete loss of H3K79 methylation, which affects mitotic spindle formation and cell cycle progression. Genome-wide analysis has demonstrated H3K79 methylation is a marker of active euchromatin and highly correlated to transcriptional activity [20, 21]. The efficacy of DOT1L inhibition has been reported in AML preclinical models. The DOT1L containing complex is present as a frequent translocation partner of MLL fusions in AML, which have aberrant H3K79 methylation patterns. Pinometostat (EPZ-5676), a DOT1L inhibitor, is currently being evaluated in clinical trials in AML. It demonstrated sensitivity in AML models harboring recurrent mutations of IDH1/2 and DNMT3A. Given these mutations are also frequent in T cell lymphomas, it would be interesting to determine the vulnerability of TCL to the DOT1L inhibition. Thus far, pinometostat has been shown to decrease cell viability in PTEN-inactivated TCL preclinical models [22, 23].
Histone acetylation
Histone Acetyltransferases (HATs)
CBP/p300
The acetylation of histone lysine residues is a dynamic process regulated by the balance between the activity of HATs and histone deacetylases (HDACs) [24]. Acetylation mediated by HATs leads to an open chromatin structure and recruitment of bromodomain protein “readers” to induce an open chromatin state and subsequent transcriptional activation [25, 26]. Besides altering histone acetylation patterns, HATs also acetylate and affect the activity of non-histone substrates that directly regulate transcription, including a diverse array of transcription factors [27]. Choudhary and colleagues discovered 1750 proteins in addition to histones are modified by lysine acetylation in leukemia cells, indicating that HATs play widespread roles in regulating cellular activities [28].
Among 17 HATs in four different HAT families, CBP and p300 are the best-characterized HATs in relation to lymphomas. Several studies have demonstrated that HAT proteins CBP and p300 act as tumor suppressors in B cell lymphomas. CBP and p300 function as co-activators of transcription factors and acetylation proteins relevant to lymphomagenesis, such as p53, NF-kB, BCL6 and Hsp90 [29–32]. Inhibition of BCL6 induce p300 expression and acetyltransferase activity, which is lethal to DLBCL cells [31].
Multiple findings suggest that histone acetylation has a key role in lymphoid malignancies and particularly in germinal center-derived tumors. Monoallelic inactivating mutations in CREBBP harbor frequently in 39% in a cohort of 134 DLBLCs and 33%, 68% respectively in another 2 different studies of follicular lymphoma (FL) as well as in 17% of ocular marginal zone lymphoma [33, 34]. Less commonly, somatic mutations truncating or disrupting the HAT domain of p300 were identified in 20% of ATLLs [35], 10% in 2 different studies of DLBCLs [31, 34] and 9% in a cohort of above 46 FLs [34]. CREBBP mutations are relatively rare in PTCL and other T-lymphoid malignancies compared to B cell lymphomas [36–39]. CREBBP mutations cluster within the substrate-binding pocket and decrease the affinity for acetyl-CoA, resulting in a reduction of acetylation in the H3K18 and H3K27 residue [40]. The reduced histone acetylation changes the electrical charge of the core histone, and thereby transforms chromatin structure to a resting closed conformation and less accessible to transcriptional factors. Pasqualucci and colleagues discovered that mutated CBP led to the impaired ability to acetylate BCL6, a transcriptional repressor and master regulator of the germinal center. Acetylation of BCL6 abrogates its effects, whereas the lack of acetylation allows for transcriptional repression imposed by BCL6 [34]. Activated BCL6 can recruit PRC1-like BCOR complexes and SMRT/HDAC3 complexes in B cell promoters to most effectively repress transcription and deacetylate H3K27 resulting in a repressed chromatin environment [41]. In addition, mutations led to impaired acetylation of p53, which compromised its effects as a tumor suppressor. Mutations of CREBBP may be related to chemotherapy resistance. CREBBP mutations were identified in 18% of relapsed pediatric B-acute lymphoblastic leukemia patients but only half of these were present at diagnosis, indicating these mutations may be induced by chemotherapy possibly contributing to chemo-resistance [40]. Although GC-specific loss of Crebbp was insufficient to initiate malignant transformation, compound Crebbp-haploinsufficient/BCL2-transgenic mice, mimicking the genetics of FL and DLBCL, develop clonal lymphomas recapitulating the features of the human diseases. It further suggests CREBBP loss contribute to lymphomagenesis indicate itself as a haplosufficient tumor suppressor in GC-B cells [42]. In FL patients, 94% of CREBBP mutations were identified in progenitor cells indicating they are early event in the genomic evolution of FL. CREBBP mutant FLs have significantly lower expression of HLA class II and thus less number of infiltrating T cell compared to wildtype tumors, suggesting CREBBP mutations play a role in promoting immune invasion in FL [43].
Loss of function of HAT genes in germinal center derived B-cell lymphomas shifts the balance toward a condensed chromatin state and transcriptional repression of differentiation genes. HAT mutations are monoallelic suggesting that their activity could be restored if the wild type allele’s function can be enhanced in some way, providing a strong rationale to induce HAT activity through therapeutic approaches. HAT activators have been reported [44, 45], but their therapeutic application has been limited by their lack of solubility and membrane permeability. We recently reported a HAT activator [46], YF2 induces p300-mediated acetylation of histone and p53 and demonstrates selective cytotoxic effect in EP300-mutated DLBCLs. We observed strong synergistic effect when combining HAT activator with HDAC inhibitor in vitro and in vivo [46]. Conversely, CBP/p300 has been detected in several oncogenic fusions and act as co-activator for leukemogenic proteins in leukemia. In line of this, a CBP/p300 bromodomain inhibitor, I-CBP112 impairs aberrant self-renewal of leukemic cells [47]. More recently, a second p300/CBP inhibitor, A-485 targeting the catalytic domain of p300/CBP demonstrated potent activity in a subset of multiple myeloma, acute myeloid leukemia and non-Hodgkin’s lymphoma cell lines [48]. However, it is unclear if the killing effect of the compounds is associated with HAT mutation status and the long-term effect of the treatment. Taken together, these preclinical data point towards the potential therapeutic advantage to targeting HAT enzymes in hematologic malignancies.
MYST family (TIP60, KAT6A)
Among the genes coding for the MYST family of KATs (KAT5-KAT8), the acetyltransferases implicated in lymphoma includes TIP60 (alias, KAT5) and KAT6A (alias, MYST4, MOZ). Tip60 is a haplo-insufficient tumor suppressor which suffers mono-allelic loss and reduced expression in a small fraction of FLs and DLBCLs. The key function of Tip60 is to protect cells from genomic instability and to suppress potential transforming events which can lead to cancer. Tip60 participates in two pathways, chromatin modeling at DNA-strand double breaks (DSBs) via NuA4-Tip60 complex and acetylation and activation of the ATM kinase. The NuA4-Tip60 complex acetylates histones H2AX and H4 at DSBs, modifying the chromatin structure to facilitate DSB repair. In line with it, the haplo-insufficiency of Tip60 accelerates the tumor development in Eμ-myc mice and abrogates a Myc-induced DNA-damage response [49].
KAT6A has an essential role in normal haematopoietic stem cells [50–52]. Chromosomal translocation of this gene with CREBBP has been identified in multiple cancer types, indicating it is the oncogene [53, 54]. Loss of one allele of KAT6A prolongs the median survival of mice with MYC-induced lymphoma from 103 to 413 days [55], indicating that the progression of lymphoma is highly dependent on KAT6A. A KAT6A inhibitor, WM-1119 demonstrated significant anti-tumor efficacy in a xenograft model of lymphoma [56]. Although inhibition of KAT6A may provide a therapeutic benefit in lymphoma, careful study will be required to determine if KAT6A contributes to lymphomagenesis via aberrant chromatin acetylation state.
GCN5 family (PCAF)
Histone acetyltransferases PCAF belonging to the GCN5 family which regulates various epigenetic events for transcriptional regulation through alterations in the chromatin structure. Kikuchi and colleagues revealed that Bcl-6 and Pax5 were downregulated in PCAF-deficient immature B cell line DT40 cells. PCAF-deficiency caused remarkable decrease in acetylation levels of both H3K9 and H3K14 residues within chromatin surrounding the 5’-flanking regions of Bcl-6 and Pax5 genes in vivo [57]. It suggests that PCAF is essential in epigenetic regulation in transcription factors for normal development of B cells.
HDACs
Human HDACs are categorized into four major groups based on sequence homology to yeast proteins and function: class I (HDAC1, class I (HDAC1, HDAC2, HDAC3 and HDAC8); class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10), class III (NAD-dependent protein deacetylase sirtuin 1 (SIRT1)–SIRT7); and class IV (HDAC11) [58].[58]. HDACs generally act as transcriptional corepressors by deacetylating nucleosomal histones, which can lead to chromosomal condensation and the exclusion of transcriptional activating complexes [59, 60]. In addition, large HDAC-containing repressor complexes can localize to specific gene loci and exclude activating molecules from interacting with transcriptional factors.
Mutations in genes encoding HDACs have not been found in any B- and T- cell lymphoid malignancies. However, non-mutant HDACs have been reported to be dysregulated (usually increased) in various types of leukemia and lymphomas [61]. HDACs tend to be recruited by oncoproteins to support repressive malignant gene expression. For example, multiple studies demonstrated that the inhibition of HDAC induce apoptosis in cancer cells by modulating the expression of c-myc and BCL2 family proteins [62–65]. Several HDAC inhibitors (vorinostat, romidepsin, panobinostat, belinostat) are currently approved by the FDA for relapsed T cell lymphomas or in combination for multiple myeloma [66–70]. Despite approval in T cell lymphoma, single agent HDAC inhibitors have demonstrated limited activity in relapsed DLBCL. Our previous work has demonstrated that the combination of niacinamide, a sirtuin inhibitor, and HDAC inhibitors (romidepsin, vorinostat, belinostat, or panobinostat) leads to synergistic cytotoxicity in GC-DLBCL but not ABC-DLBCL [71]. Following exposure to romidepsin and niacinamide, mouse lymphoid tissue demonstrated acetylation of both Bcl6 and p53 as well as modulation of downstream targets BLIMP1 and p21. This proof-of-principle study demonstrates a potential role of combined HDAC inhibition therapy in B-cell lymphomas [71]. In addition, it suggests that the cumulative epigenetic derangements of GC-DLBCL may be amenable to targeted therapy through modulation of the acetylation state.
Given the conversely biological effect of HDAC and HAT, it is intriguing to determine if the presence of HAT mutations can predict the clinical activity of HDAC inhibitors. In B-cell lymphoma, HDAC inhibitors have been shown to rescue deficits in histone acetylation induced by EP300/CREBBP mutations [72]. Although in other studies, the sensitivity to romidepsin was not associated with HAT mutation status in a panel of lymphoma cell lines [11]. The HDAC inhibitor chidamide has shown favorable efficacy in PTCL-NOS patients bearing EP300/CREBBP mutations [39], indicating such mutations may alter the protein function on chromatin state regulation, sensitizing PTCL-NOS patients to HDAC inhibitors. PTCL-NOS commonly displays mutations in genes involved in histone methylation and acetylation. The presence of histone modifier gene mutations was associated with decreased progression-free survival in a cohort of 125 PTCL-NOS patients. PTCL-NOS cells were shown to experience growth inhibition when treated with a HDAC inhibitor, chidamide, and a DNA methyltransferase (DNMT) inhibitor decitabine, both in vitro and in vivo [39]. Dual therapy enhanced the interaction of KMT2D with the transcription factor PU.1, consequently inactivating the MAPK, which tends to be constitutively activated in TCL. PU.1 interact with DNMTs to control hematopoesis and suppress leukemia, via the histone modifier KMT2D. In addition, clinical response to HDAC inhibitors is strongly associated with a concurrent gain in chromatin accessibility in CTCL. HDAC inhibitors causes distinct chromatin responses in leukemic and host CD4+ T cells, reprogramming host T cells toward normalcy [73].
BET
Bromodomains are protein motifs that recognize and bind to acetylated lysine moieties located on histone tails. BETs consist of two amino-terminal tandem bromodomains and an extra-terminal (non-bromodomain) region. The BET family includes include BRD2, BRD3, BRD4, and bromodomain testis-specific protein. Upon binding to the acetylated lysine residues on histones, BETs promote gene transcription either directly by recruiting and stabilizing transcription effectors or indirectly by interacting with gene super-enhancers [74]. Therefore, BETs contribute to the tumor development and progression by activating and potentiating the expression of key oncogenes.
Although BET mutations or translocations are rare, the overexpression of BETs are common [75]. Consequently, BET inhibition has been demonstrated to be effective in preclinical studies in different types of cancers, including breast, ovarian, neuroendocrine, glioma, sarcoma and hematological malignancies [76–80]. In B cell lymphomas, BRD4 was found to preferentially bind in the proximity of critical lymphoma-related oncogenes, such as c-Myc and CD79B, and enhancers essential for B-cell fate determination, such as PAX5 and IRF8 [81]. BETs activate the MYC oncogene and the BCL2 anti-apoptotic signaling pathways. The BET inhibitor, JQ1 results in a decreased expression of MYC in myeloma cells [82]. JQ1 has also been shown to prolong the survival duration of xenograft models of MYC-driven lymphoma, including those that were resistant to etoposide and those carrying TP53 deletions [83]. In patients with double-hit DLBCL, resistance to venetoclax can be overcome by administration of the BET inhibitor CPT-203, likely due to downregulation of BCL2-like-protein (BFL1) [84]. High concentration of BRD4 have been observed in the site of super-enhancers known to regulate genes involved in B-cell receptor signaling pathways as well as that of genes with oncogenic effects in CLLs. BET inhibition was further shown to downregulate the expression of those genes and reduce tumor burden in vivo [75]. BET proteolysis-targeting chimaeras (PROTACs) are hetero-bifunctional compounds which induce the profound deletions of BET proteins by promoting their ubiquitylation via through E3 ligases. They have been shown to work more effectively than BET inhibitor (OTX015) in ibrutinib-resistant mantle cell lymphoma [85]. In the context of T cell lymphomas, BET inhibition by JQ1 resulted in the downregulation of c-myc expression, induced cell cycle arrest in vitro and inhibited tumor growth in vivo [86].[86]. OTX015, an oral BRD2/3/4 inhibitor, induced cell cycle arrest in 5/8 ALCL cell lines. ALK status had no impact on likelihood of response. OTX015 was found to suppress the transcription of the MYC gene in 4/4 cell lines [87]. Moreover, several other BET inhibitors were found synergistically induced cell death in combination with HDAC or BCL2 inhibition in 4 CTCL cell lines. MYC and BCL2 expression were repressed when CTCL cells were treated with JF1 and HDAC inhibitors, indicating cooperative activities at the level of epigenetic regulation [86].
Higher order chromatin structure
SWI/SNF
Among the families of ATP-dependent chromatin remodelers, SWI/SNF chromatin remodeling complex (also known as BAF complex) was originally described (in yeast) as the complex is critical for cellular responses to mating type switching (SWI) or sucrose fermentation (SNF) [88, 89]. SWI/SNF complex containing more than 15 subunits [90–92] utilizes the energy from ATP to remodel chromatin by shuffling nucleosomes along the DNA [93].[93]. It is capable in regulating the accessibility of DNA to other proteins involved in replication or repair as well as allowing for the activation or suppression of gene expression [94].
Specific inactivating mutations in subunits of SWI/SNF chromatin remodeling complexes occur at a high frequency in several types of cancer [93, 95–98], including SNF5 (alias, SMARCB1, INI1 and BAF47), ARID1A (alias, BAF250A and SMARCF1), BRM/SWI2-related gene 1 (BRG1; alias, SMARCA4) and BCL7A/B/C subunits. Mutations of these genes are largely mutually exclusive and are commonly small deletions that introduce stop codon or frameshift resulting in loss of function in protein [99]. In detail, loss of SNF5 was first described in rhabdoid sarcomas [95, 96] and the list of SNF5-deficient tumors has since expanded to include several types of cancers [93, 98]. Notably, conditional knockout of SNF5 in mice results in mature CD8+ T cell lymphoma or rare rhabdoid tumors with a median onset of only 11 weeks, indicating SNF5 is a bona fide tumor suppressor [100]. In addition, ARID1A is the most commonly mutated subunit. In the context of Burkitt lymphoma, truncated mutations in ARID1A were observed in 17% of 29 cases [101]. KrysiakI and colleagues identified recurrent mutations in 9 SWI/SNF complex genes affecting 32 (30.5%) patients in cohort of 105 follicular lymphoma patients at mutation frequencies ranging from 0.95% to 19.4% [102]. Among these was ARID1A (5.7%), a modifier gene in the m7-FLIPI associated with longer failure-free survival [103].
SMARCA4 mutation were observed significantly more frequently in endemic Burkitt lymphomas harboring EBV type 2 latency and those without EBV than tumors with EBV type 1 latency, whereas ARID1A was mutated in roughly equivalent levels in either categorization [99]. BCL7A-MYC translocation has been previously reported in a Burkitt lymphoma cell line [104]. Overexpression of BCL-7A was associated with germinal center-DLBCL.
In a cohort of 24 mantle cell lymphoma patients, chromosome 9p21.1-p24.3 loss and/or mutations in components of the SWI-SNF chromatin-remodeling complex, i.e. SMARCA2, ARID2, SMARCA4, were present in all patients who were primarily refractory to ibrutinib plus venetoclax treatment and two-thirds of patients with relapsed disease [105]. Interestingly, SMARCA4-deficient and ARID1A-mutant cancers are particularly vulnerable when the other components of SWI/SNF complex are inhibited [106, 107]. This implication points to the rationale of combinatorial therapeutic approaches targeting these components in lymphoma and other disease harboring these mutations.
EZB-GC-DLBCL was recently defined as a distinct subtype of DLBCL, which has a worse prognosis than its former designation of GC-DLBCL [108]. The signature includes discrete genetic signatures including BCL2 translocation and mutations of EZH2, CREBBP, EP300, TNFRSF14, KMT2D and MEF2B [108]. Mutations of two SWI/SNF complex genes, ARID1A and BCL7A, were enriched in this subtype of patients.
Inactivating mutations of members of the SWI/SNF complex lead to unfettered activity of EZH2 and the PRC2 complex. Both disruption of the PRC2 complex and inhibition of the EZH2 SET domain have been demonstrated to lead to cellular demise in cell lines harboring mutations in the SWI/SNF complex [109]. Consistently, tumors carrying SWI/SNF inactivating mutations are particularly vulnerable to genetic and pharmacological inhibition of PRC2 [110, 111]. This has been demonstrated in epitheloid sarcomas, where patients with mutations in SMARCB1 have an enriched response to EZH2 inhibitors [112].
Linker histone genes
Linker histones bind to DNA at the entry and exit sites of the nucleosome, and are generally required to achieve chromatin condensation in the nucleus. Mutations in linker histone family genes (HIST1H1 B-E) are identified as recurrent mutated genes in follicular lymphoma and less frequent in DLBCL [113, 114]. The most commonly mutated linker histone, HIST1H1E can be methylated at lysine 26 by EZH2 to facilitate the formation of transcriptionally inactive heterochromatin. Linker histone can also recruit DNA methyltransferases and recruitment DNMT3B is impaired by somatic mutations [113]. Inactivating somatic mutations of linker histone genes may lead to greater accessibility of chromatin, which is in opposite effect of heterochromatin formation by activating EZH2 mutations and loss-of-function KMT2D or CREBBP mutations. However, acetylation of HIST1H1E at lysine 34 by GCN5 can also lead to recruitment of TAF1 and promote transcriptional activation, meaning that loss-of-function mutations may have a transcriptionally repressive effect. Given that each of the linker histone family genes may have unique biological function in gene transcription [115], future studies will be required to understand the precise role of these mutations in lymphomas.
Conclusion
Recurrent mutations leading to a chromatin remodeled state have been identified as pathogenic in lymphoma. These mutations can occur across multiple enzymes mechanisms affecting the control of chromatin condensation. As such, there is likely not one Achilles heal in these diseases. New therapies targeting histone methyltransferes, deacetylases, and bromodomains have successfully targeted each of these individual derangements. Clinical therapeutic success will likely be best achieved with combined targeting of these epigenetic derangements.
Table 1.
Frequency of chromatin modifying and organizing gene mutations in B-cell and T-cell lymphomas. Data from genome, exome and transcriptome sequencing studies of FL [43, 113, 114, 116, 117, 119], BL [99, 120], DLBCL [34, 117, 118, 121, 122] and TCL [35, 123] with sufficient data quality are summarized.
| Epigenetic modifers | Function | Mutation frequency | Mutation type | Protein activity | Histone state (as a consequence of mutations) | |||
|---|---|---|---|---|---|---|---|---|
| FL, % [43, 114, 116] | GCB-like, % | BL, % | TCL, % | |||||
| EZH2 | H3K27 methyltransferase | 25 | 15–20 [117] | 2 | ATLL, 3 CTCL, 0 | Gain of function | active | H3K27me3 ↑ |
| KMT2D | H3K4 methyltransferase | 72 | 25 [118] | 2 | CTCL, 2 | Loss of function | inactive | H3K4me1/me2↓ |
| CREBBP | lysine acetyltransferase | 65 | 32 [34] | 6 | CTCL, 5 | Loss of function | inactive | H3K27ac ↓ H3K18ac ↓ |
| EP300 | lysine acetyltransferase | 15 | 10 [34] | 0 | ATLL, 20 [35] | Loss of function | inactive | H3K27ac ↓ H3K18ac ↓ |
| ARID1A | SWI/SNF component | 11 | 10 | 7 | CTCL, 5 | Loss of function | inactive | |
| SMARCA 4 | SWI/SNF component | 1 | 10 | 21 | CTCL, 5 | Loss of function | inactive | |
| HIST1H1E | linker histone | 14 | 17 | 0 | 0 | Loss of function | inactive | |
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest: Dr. Amengual declares that she has received a grant from Appia Pharmaceuticals and has been paid to speak by Epizyme. Dr. Yuxuan and Dr. Rosario have no conflicts to disclose.
Human and Animal Rights and Informed Consent: This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- [1].Honma D, Kanno O, Watanabe J et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci 2017; 108:2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yang CY, Wang S. Allosteric Inactivation of Polycomb Repressive Complex 2 (PRC2) by Inhibiting Its Adapter Protein: Embryonic Ectodomain Development (EED). J Med Chem 2017; 60:2212–2214. [DOI] [PubMed] [Google Scholar]
- [3].Song X, Zhang L, Gao T et al. Selective inhibition of EZH2 by ZLD10A blocks H3K27 methylation and kills mutant lymphoma cells proliferation. Biomed Pharmacother 2016; 81:288–294. [DOI] [PubMed] [Google Scholar]
- [4].Qi W, Zhao K, Gu J et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat Chem Biol 2017; 13:381–388. [DOI] [PubMed] [Google Scholar]
- [5].Morschhauser F, Tilly H, Chaidos A et al. INTERIM UPDATE FROM A PHASE 2 MULTICENTER STUDY OF TAZEMETOSTAT, AN EZH2 INHIBITOR, IN PATIENTS WITH RELAPSED OR REFRACTORY FOLLICULAR LYMPHOMA. Hematological Oncology 2019; 37:154–156. [Google Scholar]; • The most recent updated report showed the promising efficacy of EZH2 inhibitor in follicular lymphoma
- [6].Chng WJ, Yan J, Li B et al. NON-CANONICAL ROLE OF EZH2 IN NATURAL KILLER / T-CELL LYMPHOMA. Hematological Oncology 2017; 35:121–122. [Google Scholar]
- [7].Zhao X, Zhang X, Tao J. MYC and MiR: vicious circle. Oncotarget 2013; 4:2168–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].D’Angelo V, Iannotta A, Ramaglia M et al. EZH2 is increased in paediatric T-cell acute lymphoblastic leukemia and is a suitable molecular target in combination treatment approaches. J Exp Clin Cancer Res 2015; 34:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maruyama D, Tobinai K, Makita S et al. First-in-Human Study of the EZH1/2 Dual Inhibitor DS-3201b in Patients with Relapsed or Refractory Non-Hodgkin Lymphomas — Preliminary Results. Blood 2017; 130:4070–4070. [Google Scholar]
- [10].Gupta M, Demosthenous C, Stenson MJ, Price-Troska T. Oncogenic Role of Chromatin Modifier Polycomb Repressive Complex-2 in Mantle Cell Lymphoma. Blood 2018; 132. [Google Scholar]
- [11].Lue JK, Prabhu SA, Liu Y et al. Precision Targeting with EZH2 and HDAC Inhibitors in Epigenetically Dysregulated Lymphomas. Clin Cancer Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This is the first manuscript to demonstrate the synergistic effects by dual targeting EZH2 and HDAC in B cell lymphoma preclinical models.
- [12].Green MR. Chromatin modifying gene mutations in follicular lymphoma. Blood 2018; 131:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pruitt K Molecular and Cellular Changes During Cancer Progression Resulting From Genetic and Epigenetic Alterations. Prog Mol Biol Transl Sci 2016; 144:3–47. [DOI] [PubMed] [Google Scholar]
- [14].Xin CQ, Wang C, Wang YC et al. Identification of novel KMT2D mutations in two Chinese children with Kabuki syndrome: a case report and systematic literature review. Bmc Med Genet 2018; 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 2007; 7:823–833. [DOI] [PubMed] [Google Scholar]
- [16].Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 2010; 10:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Arai S, Yoshimi A, Shimabe M et al. Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood 2011; 117:6304–6314. [DOI] [PubMed] [Google Scholar]
- [18].Shilatifard A The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012; 81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marschalek R Mixed lineage leukemia: roles in human malignancies and potential therapy. FEBS J 2010; 277:1822–1831. [DOI] [PubMed] [Google Scholar]
- [20].van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 2002; 109:745–756. [DOI] [PubMed] [Google Scholar]
- [21].Ng HH, Ciccone DN, Morshead KB et al. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proceedings of the National Academy of Sciences 2003; 100:1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gallipoli P, Huntly BJP. Novel epigenetic therapies in hematological malignancies: Current status and beyond. Semin Cancer Biol 2018; 51:198–210. [DOI] [PubMed] [Google Scholar]
- [23].Stein EM, Garcia-Manero G, Rizzieri DA et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 2018; 131:2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 2006; 23:289–296. [DOI] [PubMed] [Google Scholar]
- [25].Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem 2001; 70:81–120. [DOI] [PubMed] [Google Scholar]
- [26].Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 2000; 64:435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene 2005; 363:15–23. [DOI] [PubMed] [Google Scholar]
- [28].Choudhary C, Kumar C, Gnad F et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- [29].Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997; 90:595–606. [DOI] [PubMed] [Google Scholar]
- [30].Rothgiesser KM, Fey M, Hottiger MO. Acetylation of p65 at lysine 314 is important for late NF-kappaB-dependent gene expression. BMC Genomics 2010; 11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cerchietti LC, Hatzi K, Caldas-Lopes E et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest 2010; 120:4569–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The first paper reported inactive p300 mutation in B-cell lymphoma patients and demonstrated re-activating p300 is toxic to lymphoma cells.
- [32].Yang Y, Rao R, Shen J et al. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res 2008; 68:4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jung H, Yoo HY, Lee SH et al. The mutational landscape of ocular marginal zone lymphoma identifies frequent alterations in TNFAIP3 followed by mutations in TBL1XR1 and CREBBP. Oncotarget 2017; 8:17038–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pasqualucci L, Dominguez-Sola D, Chiarenza A et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011; 471:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This manuscript reported for the first time inactivating mutation of CREBBP in DLBCL and follicular lymphoma patients and confirmed the presence of EP300 mutations in DLBCL. It also demonstrated mutant CBP is unable to acetylate p53.
- [35].Shah UA, Chung EY, Giricz O et al. North American ATLL has a distinct mutational and transcriptional profile and responds to epigenetic therapies. Blood 2018; 132:1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Schatz JH, Horwitz SM, Teruya-Feldstein J et al. Targeted mutational profiling of peripheral T-cell lymphoma not otherwise specified highlights new mechanisms in a heterogeneous pathogenesis. Leukemia 2015; 29:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhu X, He F, Zeng H et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet 2014; 46:287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jiang L, Gu ZH, Yan ZX et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet 2015; 47:1061–1066. [DOI] [PubMed] [Google Scholar]
- [39].Ji MM, Huang YH, Huang JY et al. Histone modifier gene mutations in peripheral T-cell lymphoma not otherwise specified. Haematologica 2018; 103:679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mullighan CG, Zhang J, Kasper LH et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011; 471:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hatzi K, Jiang Y, Huang C et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep 2013; 4:578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang J, Vlasevska S, Wells VA et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer Discov 2017; 7:322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Green MR, Kihira S, Liu CL et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A 2015; 112:E1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. The Journal of biological chemistry 2003; 278:19134–19140. [DOI] [PubMed] [Google Scholar]
- [45].Mantelingu K, Kishore AH, Balasubramanyam K et al. Activation of p300 histone acetyltransferase by small molecules altering enzyme structure: probed by surface-enhanced Raman spectroscopy. The journal of physical chemistry. B 2007; 111:4527–4534. [DOI] [PubMed] [Google Scholar]
- [46].Liu Y, Fiorito J, Gonzalez Y et al. Development of First-in-Class Histone Acetyltransferase (HAT) Activators for Precision Targeting of Epigenetic Derangements in Lymphoma. Blood 2018; 132:37–37. [Google Scholar]
- [47].Picaud S, Fedorov O, Thanasopoulou A et al. Generation of a Selective Small Molecule Inhibitor of the CBP/p300 Bromodomain for Leukemia Therapy. Cancer Res 2015; 75:5106–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lasko LM, Jakob CG, Edalji RP et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017; 550:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gorrini C, Squatrito M, Luise C et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 2007; 448:1063–1067. [DOI] [PubMed] [Google Scholar]
- [50].Katsumoto T, Aikawa Y, Iwama A et al. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev 2006; 20:1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Thomas T, Corcoran LM, Gugasyan R et al. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev 2006; 20:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sheikh BN, Yang Y, Schreuder J et al. MOZ (KAT6A) is essential for the maintenance of classically defined adult hematopoietic stem cells. Blood 2016; 128:2307–2318. [DOI] [PubMed] [Google Scholar]
- [53].Borrow J, Stanton VP, Andresen JM Jr. et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet 1996; 14:33–41. [DOI] [PubMed] [Google Scholar]
- [54].Huang F, Abmayr SM, Workman JL. Regulation of KAT6 Acetyltransferases and Their Roles in Cell Cycle Progression, Stem Cell Maintenance, and Human Disease. Mol Cell Biol 2016; 36:1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sheikh BN, Lee SC, El-Saafin F et al. MOZ regulates B-cell progenitors and, consequently, Moz haploinsufficiency dramatically retards MYC-induced lymphoma development. Blood 2015; 125:1910–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Baell JB, Leaver DJ, Hermans SJ et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 2018; 560:253–257. [DOI] [PubMed] [Google Scholar]
- [57].Kikuchi H, Nakayama M, Kuribayashi F et al. Histone acetyltransferase PCAF is involved in transactivation of Bcl-6 and Pax5 genes in immature B cells. Biochem Biophys Res Commun 2015; 467:509–513. [DOI] [PubMed] [Google Scholar]
- [58].McClure JJ, Li X, Chou CJ. Advances and Challenges of HDAC Inhibitors in Cancer Therapeutics. Adv Cancer Res 2018; 138:183–211. [DOI] [PubMed] [Google Scholar]
- [59].Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009; 10:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 2008; 9:206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Haery L, Thompson RC, Gilmore TD. Histone acetyltransferases and histone deacetylases in B- and T-cell development, physiology and malignancy. Genes Cancer 2015; 6:184–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chen S, Dai Y, Pei XY, Grant S. Bim upregulation by histone deacetylase inhibitors mediates interactions with the Bcl-2 antagonist ABT-737: evidence for distinct roles for Bcl-2, Bcl-xL, and Mcl-1. Mol Cell Biol 2009; 29:6149–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhao Y, Tan J, Zhuang L et al. Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc Natl Acad Sci U S A 2005; 102:16090–16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu Y, Mondello P, Erazo T et al. NOXA genetic amplification or pharmacologic induction primes lymphoma cells to BCL2 inhibitor-induced cell death. Proc Natl Acad Sci U S A 2018; 115:12034–12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gui CY, Ngo L, Xu WS et al. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A 2004; 101:1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Olsen EA, Kim YH, Kuzel TM et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol 2007; 25:3109–3115. [DOI] [PubMed] [Google Scholar]
- [67].Whittaker SJ, Demierre MF, Kim EJ et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol 2010; 28:4485–4491. [DOI] [PubMed] [Google Scholar]
- [68].Coiffier B, Pro B, Prince HM et al. Results from a pivotal, open-label, phase II study of romidepsin in relapsed or refractory peripheral T-cell lymphoma after prior systemic therapy. J Clin Oncol 2012; 30:631–636. [DOI] [PubMed] [Google Scholar]
- [69].O’Connor OA, Horwitz S, Masszi T et al. Belinostat in Patients With Relapsed or Refractory Peripheral T-Cell Lymphoma: Results of the Pivotal Phase II BELIEF (CLN-19) Study. J Clin Oncol 2015; 33:2492–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].San-Miguel JF, Hungria VT, Yoon SS et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol 2014; 15:1195–1206. [DOI] [PubMed] [Google Scholar]
- [71].Amengual JE, Clark-Garvey S, Kalac M et al. Sirtuin and pan-class I/II deacetylase (DAC) inhibition is synergistic in preclinical models and clinical studies of lymphoma. Blood 2013; 122:2104–2113. [DOI] [PubMed] [Google Scholar]
- [72].Jiang Y, Ortega-Molina A, Geng H et al. CREBBP Inactivation Promotes the Development of HDAC3-Dependent Lymphomas. Cancer Discov 2017; 7:38–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Qu K, Zaba LC, Satpathy AT et al. Chromatin Accessibility Landscape of Cutaneous T Cell Lymphoma and Dynamic Response to HDAC Inhibitors. Cancer Cell 2017; 32:27–41 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The manuscript demonstrated clinical response to HDAC inhibitors is strongly associated with chromatin accessibility by ATAC-seq.
- [74].Wang CY, Filippakopoulos P. Beating the odds: BETs in disease. Trends Biochem Sci 2015; 40:468–479. [DOI] [PubMed] [Google Scholar]
- [75].Ozer HG, El-Gamal D, Powell B et al. BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor. Cancer Discov 2018; 8:458–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Vazquez R, Riveiro ME, Astorgues-Xerri L et al. The bromodomain inhibitor OTX015 (MK-8628) exerts anti-tumor activity in triple-negative breast cancer models as single agent and in combination with everolimus. Oncotarget 2017; 8:7598–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wang L, Matkar S, Xie G et al. BRD4 inhibitor IBET upregulates p27kip/cip protein stability in neuroendocrine tumor cells. Cancer Biol Ther 2017; 18:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yokoyama Y, Zhu H, Lee JH et al. BET Inhibitors Suppress ALDH Activity by Targeting ALDH1A1 Super-Enhancer in Ovarian Cancer. Cancer Res 2016; 76:6320–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Piunti A, Hashizume R, Morgan MA et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 2017; 23:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gryder BE, Yohe ME, Chou HC et al. PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov 2017; 7:884–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Chapuy B, McKeown MR, Lin CY et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 2013; 24:777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Loven J, Hoke HA, Lin CY et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Hogg SJ, Newbold A, Vervoort SJ et al. BET Inhibition Induces Apoptosis in Aggressive B-Cell Lymphoma via Epigenetic Regulation of BCL-2 Family Members. Mol Cancer Ther 2016; 15:2030–2041. [DOI] [PubMed] [Google Scholar]
- [84].Esteve-Arenys A, Valero JG, Chamorro-Jorganes A et al. The BET bromodomain inhibitor CPI203 overcomes resistance to ABT-199 (venetoclax) by downregulation of BFL-1/A1 in in vitro and in vivo models of MYC+/BCL2+ double hit lymphoma. Oncogene 2018; 37:1830–1844. [DOI] [PubMed] [Google Scholar]
- [85].Sun B, Fiskus W, Qian Y et al. BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia 2018; 32:343–352. [DOI] [PubMed] [Google Scholar]
- [86].Kim SR, Lewis JM, Cyrenne BM et al. BET inhibition in advanced cutaneous T cell lymphoma is synergistically potentiated by BCL2 inhibition or HDAC inhibition. Oncotarget 2018; 9:29193–29207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Boi M, Todaro M, Vurchio V et al. Therapeutic efficacy of the bromodomain inhibitor OTX015/MK-8628 in ALK-positive anaplastic large cell lymphoma: an alternative modality to overcome resistant phenotypes. Oncotarget 2016; 7:79637–79653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol 1984; 178:853–868. [DOI] [PubMed] [Google Scholar]
- [89].Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 1984; 108:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nair SS, Kumar R. Chromatin remodeling in Cancer: A Gateway to regulate gene Transcription. Molecular Oncology 2012; 6:611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Clapier CR, Cairns BR. The Biology of Chromatin Remodeling Complexes. Annual Review of Biochemistry 2009; 78:273–304. [DOI] [PubMed] [Google Scholar]
- [92].Mathur R, Roberts CWM. SWI/SNF (BAF) Complexes: Guardians of the Epigenome. Annual Review of Cancer Biology 2018; 2:413–427. [Google Scholar]
- [93].Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer 2004; 4:133–142. [DOI] [PubMed] [Google Scholar]
- [94].Kwon H, Imbalzano AN, Khavari PA et al. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 1994; 370:477–481. [DOI] [PubMed] [Google Scholar]
- [95].Shain AH, Pollack JR. The Spectrum of SWI/SNF Mutations, Ubiquitous in Human Cancers. PLOS ONE 2013; 8:e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lu C, Allis CD. SWI/SNF complex in cancer. Nature genetics 2017; 49:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Medina PP, Romero OA, Kohno T et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Hum Mutat 2008; 29:617–622. [DOI] [PubMed] [Google Scholar]
- [98].Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene 2009; 28:1653–1668. [DOI] [PubMed] [Google Scholar]
- [99].Kaymaz Y, Oduor CI, Yu H et al. Comprehensive Transcriptome and Mutational Profiling of Endemic Burkitt Lymphoma Reveals EBV Type-Specific Differences. Mol Cancer Res 2017; 15:563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2002; 2:415–425. [DOI] [PubMed] [Google Scholar]
- [101].Giulino-Roth L, Wang K, MacDonald TY et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood 2012; 120:5181–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Krysiak K, Gomez F, White BS et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood 2017; 129:473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Pastore A, Jurinovic V, Kridel R et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol 2015; 16:1111–1122. [DOI] [PubMed] [Google Scholar]
- [104].Zani VJ, Asou N, Jadayel D et al. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood 1996; 87:3124–3134. [PubMed] [Google Scholar]
- [105].Agarwal R, Chan YC, Tam CS et al. Dynamic molecular monitoring reveals that SWI-SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. Nat Med 2019; 25:119–129. [DOI] [PubMed] [Google Scholar]
- [106].Helming KC, Wang X, Wilson BG et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med 2014; 20:251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Hoffman GR, Rahal R, Buxton F et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci U S A 2014; 111:3128–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Schmitz R, Wright GW, Huang DW et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. The New England journal of medicine 2018; 378:1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••The study identified 4 prominent genetic subtypes of DLBCL based on shared genomic abnormalities.
- [109].Kim KH, Kim W, Howard TP et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nature medicine 2015; 21:1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wilson BG, Wang X, Shen X et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010; 18:316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bitler BG, Aird KM, Garipov A et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 2015; 21:231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Gounder MM, Stacchiotti S, Schöffski P et al. Phase 2 multicenter study of the EZH2 inhibitor tazemetostat in adults with INI1 negative epithelioid sarcoma (). Journal of Clinical Oncology 2017; 35:11058–11058. [Google Scholar]
- [113].Li H, Kaminski MS, Li Y et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood 2014; 123:1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Okosun J, Bodor C, Wang J et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 2014; 46:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep 2015; 16:1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Pasqualucci L, Khiabanian H, Fangazio M et al. Genetics of follicular lymphoma transformation. Cell Rep 2014; 6:130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Morin RD, Johnson NA, Severson TM et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics 2010; 42:181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This article reported for the first time the recurrent mutation in EZH2 in large cohorts of DLBCL and follicular lymphoma patients
- [118].Zhang J, Dominguez-Sola D, Hussein S et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015; 21:1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This manuscript demonstrated the functional consequences of tumor suppressor KMT2D mutations to lymphomagenesis.
- [119].Green MR, Gentles AJ, Nair RV et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood 2013; 121:1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Love C, Sun Z, Jima D et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 2012; 44:1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Morin RD, Mendez-Lago M, Mungall AJ et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011; 476:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Morin RD, Mungall K, Pleasance E et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 2013; 122:1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].da Silva Almeida AC, Abate F, Khiabanian H et al. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat Genet 2015; 47:1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]