Abstract
We report the results of two fully-automated structure prediction pipelines, “Zhang-Server” and “QUARK”, in CASP13. The pipelines were built upon the C-I-TASSER and C-QUARK programs, which in turn are based on I-TASSER and QUARK but with three new modules: (1) a novel multiple sequence alignment (MSA) generation protocol to construct deep sequence-profiles for contact prediction; (2) an improved meta-method, NeBcon, which combines multiple contact predictors, including ResPRE that predicts contact-maps by coupling precision-matrices with deep residual convolutional neural-networks; and (3) an optimized contact potential to guide structure assembly simulations. For 50 CASP13 FM domains that lacked homologous templates, average TM-scores of the first models produced by C-I-TASSER and C-QUARK were 28% and 56% higher than those constructed by I-TASSER and QUARK respectively. For the first time, contact-map predictions demonstrated usefulness on TBM domains with close homologous templates, where TM-scores of C-I-TASSER models were significantly higher than those of I-TASSER models with a p-value <0.05. Detailed data analyses showed that the success of C-I-TASSER and C-QUARK was mainly due to the increased accuracy of deep-learning-based contact-maps, as well as the careful balance between sequence-based contact restraints, threading templates, and generic knowledge-based potentials. Nevertheless, challenges still remain for predicting quaternary structure of multi-domain proteins, due to the difficulties in domain partitioning and domain reassembly. In addition, contact prediction in terminal regions was often unsatisfactory due to the sparsity of MSAs. Development of new contact-based domain partitioning and assembly methods and training contact models on sparse MSAs may help address these issues.
Keywords: CASP13, protein structure prediction, ab initio folding, contact prediction, deep multiple sequence alignment, deep convolutional neural networks
INTRODUCTION
Depending on the availability of homologous templates in the PDB library, the computational methods for protein structure prediction can be generally categorized into two categories:1 template-based modeling (TBM) or template-free (FM) modeling. In TBM,2,3 structural models are constructed based on the template structures identified by threading the query sequence through the PDB,4–6 while FM builds models from scratch without using global template structures.7,8 In previous Critical Assessment of Protein Structure Prediction (CASP) experiments, we developed and tested two pipelines, “Zhang-Server” and “QUARK”, in the TBM and FM categories, respectively. From its initial implementation in CASP7,9 “Zhang-Server” has been mainly based on the Iterative Threading ASSEmbly Refinement (I-TASSER) method.10–12 On the other hand, “QUARK”, which was introduced in CASP9, has been based on the QUARK13,14 program, which was designed to fold proteins by assembling continuously-distributed fragments from unrelated protein structures under the guidance of a knowledge-based energy function.15 In the last decade, we have been constantly working on extending the capabilities of these pipelines by introducing new modules in each of the previous CASP experiments. In CASP12, for instance, we included contact information, derived from a meta-method, NeBcon16, into the “QUARK” pipeline, which greatly improved the accuracy of the FM target models. However, the modeling accuracy was often limited by low contact prediction precision. While the precision of contact prediction algorithms has recently improved dramatically, which in turn has helped enhance the modeling accuracy, careful optimization was required to further improve the final 3D model quality by appropriately integrating the contact prediction information into the structure assembly simulations.
In CASP13, we improved the “Zhang-Server” and “QUARK” pipelines by introducing three new modules into both frameworks. First, an iterative multiple sequence alignment (MSA) construction program, DeepMSA, was developed which builds deep MSAs with more effective sequence homologs and thus helped improve the precision of contact-map prediction. Second, the capability of NeBcon for accurate contact-map prediction was extended by integrating nine state-of-the-art contact prediction programs, including several deep-learning-based methods, such as ResPRE17, which is a newly developed contact-map predictor that couples coevolution-derived precision matrices with deep residual convolutional neural networks.18 Finally, a novel contact potential term was developed and carefully trained to balance its contribution with the other energy terms, including the threading-based distance restraints and the inherent knowledge/physics-based potentials, in order to guide the structure assembly simulations to fold target proteins. Due to the addition of these contact-based features, the classical I-TASSER and QUARK pipelines have been renamed to C-I-TASSER and C-QUARK, which stand for contact-guided I-TASSER and contact-guided QUARK, respectively.
Our C-I-TASSER and C-QUARK servers are available at https://zhanglab.ccmb.med.umich.edu/C-I-TASSER/ and https://zhanglab.ccmb.med.umich.edu/C-QUARK/.
METHODS
Overview of the “Zhang-Server” and “QUARK” pipelines used in CASP13
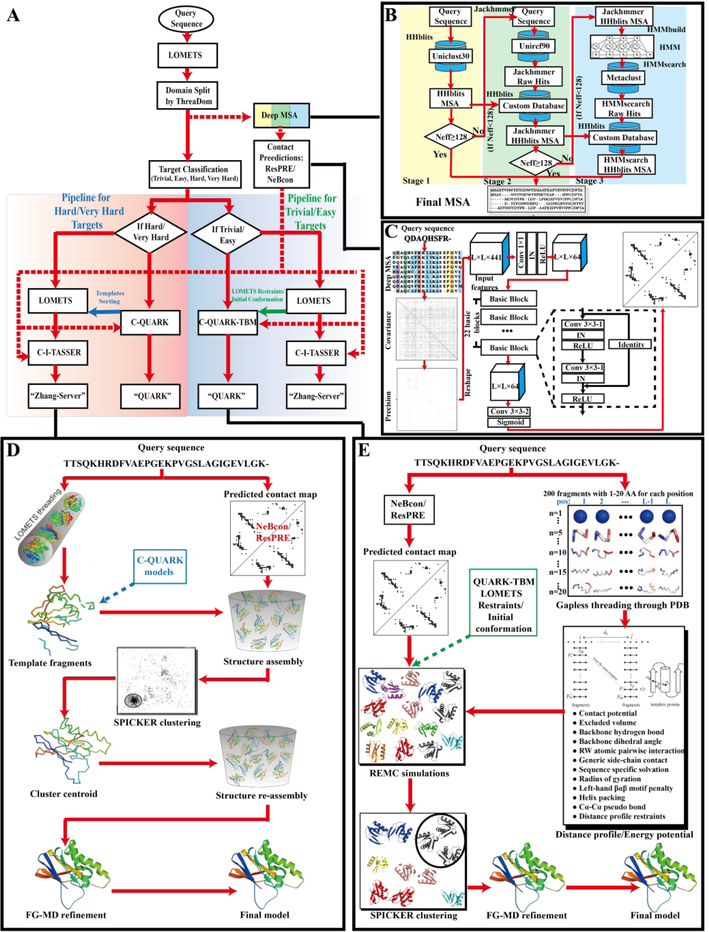
The protein structure prediction procedures used by the “Zhang-Server” and “QUARK” pipelines during CASP13 are depicted in Figure 1A. Starting from a query sequence, ThreaDom19 was first used to split the full-length sequence into multiple domains. Then, the DeepMSA program was used to generate deep multiple sequence alignments for each domain and full-length sequence by iteratively searching multiple sequence databases using several different methods including HHblits20, Jackhmmer, and HMMsearch21. The resulting MSAs were used to predict contact-maps for each domain and for the full-length sequences. Meanwhile, templates were identified for each domain by threading them through LOMETS22, where the domains were classified as “Trivial”, “Easy”, “Hard” or “Very Hard”23 based on the quality and number of threading alignments.
Figure 1.
(A) Automated “Zhang-Server” and “QUARK” protein structure prediction pipelines used in CASP13, including (B) MSA generation, (C) ResPRE Contact prediction, (D) the C-I-TASSER pipeline, and (E) the C-QUARK pipeline.
For “Trivial”/ “Easy” targets, “Zhang-Server” generally predicted structural models using the C-I-TASSER pipeline with LOMETS templates and contacts from ten predictors including NeBcon, while for “Hard”/ “Very Hard” targets, the LOMETS templates were reordered based on their similarity to the models produced by the “QUARK” pipeline. For “Hard”/ “Very Hard” targets, “QUARK” used the C-QUARK pipeline to generate structure models, while for “Trivial”/ “Easy” targets, it collected spatial restraints and the initial conformations from the LOMETS templates and used them to guide the C-QUARK folding simulations. All the procedures were fully automated. The “Zhang” human group used essentially the same pipeline as our “Zhang-Server” group, except that the initial conformations and distance restraints for certain targets came from other groups’ server models.
For multi-domain proteins, after the generation of the initial domain models by either C-I-TASSER or C-QUARK, they were assembled to form a full-length structural model. For that purpose, first, the whole-chain structure of each target was modeled by C-I-TASSER in order to obtain a full-length model that provided a reference orientation to assemble the individual domain models. Each individual domain model was then docked together using a quick Metropolis Monte Carlo simulation, which was guided by the following force field:
| (1) |
Here, di,i is the Cα-Cα distance between residue i of an individual domain and the corresponding residue i in the whole-chain reference model, L is the length of the protein, and di,j is the Cα-Cα distance between residue i of the first domain and residue j of the second domain. In the simulation, we considered those distances di,j that were smaller than dcut = 3.7 Å, which means residue i in the first domain potentially clashed with residue j in the second domain. The first term of Eq 1 was designed to superpose the individual domains to the whole chain reference model, and the second term was a penalty score to reduce the number of Cα atom clashes. The decoy with the lowest assembly energy was selected as the final full-length model. Finally, fragment-guided molecular dynamics-based refinement (FG-MD24) was applied to remove steric clashes between the domains in the assembled full-length structures. Below, we outline each part of the “Zhang-Server” and “QUARK” pipelines in greater detail.
Deep multiple sequence alignment generation by DeepMSA
Starting from an input sequence, an MSA was generated using the newly developed DeepMSA program, which uses a maximum of three stages (Figure 1B). In stage 1 of DeepMSA, HHblits from the HH-suite package is used to search the query sequence against the UniClust3025 database to generate the first-stage MSA. If the number of effective sequences (Neff, Eq 2) generated by stage 1 is less than 128, stage 2 will be performed. During stage 2, Jackhmmer, from the HMMER package, is used to search the query sequence against the UniRef9026 database to extract full-length sequences (hits). These hits are then converted into a custom HHblits-formatted database, and HHblits is used to search this custom database by jump-starting it from the first-stage MSA in order to generate the second-stage MSA. If the Neff of the second-stage MSA is still less than 128, stage 3 will be performed. In stage 3, the second-stage MSA is converted into a Hidden Markov Model (HMM) by hmmbuild from the HMMER package. This HMM is then searched against the Metaclust27 metagenomics sequence database by hmmsearch, which is also from the HMMER package, to extract full-length hits. Similar to stage 2, hits from hmmsearch are converted into a custom HHblits-formatted database. The second-stage MSA is used to jump-start an HHblits search against this new custom HHblits database to get the third-stage MSA, which is considered as the final MSA.
In DeepMSA, the depth of a multiple sequence alignment (MSA) is measured by the normalized number of effective sequence (Neff):
| (2) |
where L is the length of the query protein, N is the number of sequences in the MSA, and Sm,n is the sequence identity between the mth and nth sequences. I[Sm,n ≥ 0.8] is equal to 1 if Sm,n ≥ 0.8, or zero otherwise. Therefore, the Neff value is essentially equal to the number of non-redundant sequences (sequence identity<0.8) in the MSA normalized by the query length. Here, the sequence identity cut-off of 0.8 was determined in a previous study28. As mentioned above, in our pipeline, an MSA was considered to have a sufficient number of effective sequences if the Neff value reached the cutoff of 128. This is because we found on a benchmark dataset of 614 proteins that further sequence database search takes much more time without yielding more accurate contact prediction results when Neff ≥ 128.
Contact-map prediction
In CASP13, we retrained NeBcon16 (Figure S1) to improve its long-range contact prediction precision by integrating nine state-of-the-art contact prediction methods into the program, namely, ResPRE17, DeepPLM, Deepcontact29, DNCON230, DeepCov31, MetaPSICOV232, CCMpred33, GREMLIN34 and FreeContact35.
Among the component predictors, ResPRE (Figure 1C) is a newly developed deep-learning-based method to predict the contact-map (with Cβ-Cβ distance <8 Å) of a query sequence by coupling evolutionary precision matrices with deep residual neural networks36. In brief, given the MSA obtained for a query sequence, ResPRE first calculates the covariance between every pair of residue types at every pair of positions. A precision matrix is then generated by ridge estimation of the inverse covariance matrix in order to weed out translational noise in the covariance matrix. The estimated precision matrix is represented by an L × L × 21 × 21 array of evolutionary couplings between all pairs of residues in the protein. For each residue pair, the 21 × 21 coupling matrix is considered as the feature, and the full L × L × 21 × 21 precision matrix is fed directly into the deep residual network (ResNet)18. The ResNet is composed of a set of 22 residual blocks that add the identity map of the input to the output of the feedforward neural networks. Gradients can flow smoothly from deeper to shallower layers by adding such an identity shortcut, which makes the training of extremely deep neural networks possible. ResPRE was trained using the Adam37 method under the supervision of binary cross entropy loss and is implemented in PyTorch38. DeepPLM is another in-house deep-learning-based contact prediction approach that has the same deep-learning architecture as ResPRE, except it uses different features that are generated by CCMpred.
Following contact-map prediction by its component predictors, a Naïve Bayes classifier (NBC) is used by NeBcon to combine the confidence scores of the predicted contacts to obtain posterior probabilities for the contacts. A single hidden layer fully-connected neural network with 350 hidden units is further applied to refine the contact-map derived from the NBC model by training on the posterior probabilities together with additional sequence-based features, such as amino acid composition, Shannon entropy, residue separation, predicted solvent accessibility and secondary structure (Figure S1).
Contact energy potential
The predicted contact information from NeBcon and the individual predictors were used as restraints, together with the other energy terms in C-QUARK and C-I-TASSER, to guide the structural assembly simulations in the two pipelines. For a residue pair (i and j) that was predicted to be in contact, the following optimized contact potential was used to draw them together:
| (3) |
Here, Uij is the depth of the potential and the value of Uij between residue pair i and j is calculated by
| (4) |
where m is the index of each individual contact predictor. In total, ten predictors (NeBcon and its nine component predictors) are included here. (CScoreij)m is the confidence score of the predicted contact between residue pair i and j for the mth predictor, and is the confidence score cut-off, which corresponds to an average contact prediction precision of 0.5 for the mth predictor at range R (short, medium and long) in the protein training set.
dij is the Cβ-Cβ distance between residue i and j in the simulation decoys and D = 8 Å + db, where db is the well width of the first sine function term and 80-D is the well width of the second sine function term. The well width (db) is a crucial parameter to determine the rate at which residues that are predicted to be in contact are drawn together, and it was tuned based on the length of the training proteins. For instance, the width of the first well (db) is typically narrow, e.g. 6Å, when the length of the target is relatively small, e.g. < 200. However, the well width increases to 8Å when the length is between 200 and 250 residues, and the well width is fixed to 10Å for proteins with length >250 residues. The selection of a wider well for large proteins facilitates transitioning the residue pairs predicted to form contacts, which are generally farther apart from each other in larger proteins, to the well smoothly and bringing the residues pairs within 8Å quickly.
C-I-TASSER pipeline
The “Zhang-Server” pipeline in CASP13 was based on our new protein folding approach, C-I-TASSER, which is shown in Figure 1D. In C-I-TASSER, starting from the sequence of each domain or the full-length protein, the target is first threaded through a representative template library from the PDB using LOMETS. Fragments are extracted from the continuously aligned regions of the template structures and assembled into full-length structural models using a modified REMC simulation procedure39. A composite force field, which combines the distance restraints obtained from the threading templates with the sequence-based contact restraints (Eq. 3) and the inherent knowledge-based energy terms, is used to guide the structural assembly simulations.
Following the first round of simulations, the decoys from the simulation trajectories are clustered by SPICKER40. Then the cluster centroids are aligned against the structures in the PDB library using TM-align41. Spatial restraints extracted from the TM-align templates are used during the second round of REMC simulations to re-assemble the intermediate structural models. The re-assembled structures are further refined by FG-MD to generate the final structure models.
For each of these models, we obtain different rankings from seven Model Quality Assurance Programs (MQAP42) based on the C-score11, structural consensus (the average TM-score of the target model to all other models), the contact satisfaction between the model contacts and predicted contacts, and four statistical energy functions (RWplus43, GOAP44, DOPE45 and ROTAS46). The final model rankings are determined by ascending order of overall MQAP score. Lastly, the residue level quality of these models is estimated by ResQ47. The ResQ algorithm extracts the following features for each model: (1) coverage and structural variation compared to the LOMETS templates, (2) consistency between the solvent accessibility (and secondary structure) of the model residues and those predicted from the sequence, (3) structural variation between the decoys obtained from the REMC simulations, and (4) the deviation of the final model structures from structures identified by TM-align structural alignment search of the models through the PDB database. These features are used by Support Vector Regression to predict the deviation of each residue position in the models from the native residue positions.
C-QUARK pipeline
The “QUARK” pipeline in CASP13 was based on a new version of the ab initio protein structure prediction program C-QUARK, which is outlined in Figure 1E. In C-QUARK, starting from the sequence of each domain or full-length protein, the query is first threaded through a non-redundant PDB structure library by gapless threading to generate position-specific fragment structures. The scoring function for the gapless threading is comprised of profile-profile, secondary structure, solvent accessibility, and torsion angle matches between the target and the templates. A histogram of distances dij for each residue pair (i and j) of the target is derived from the top 200 fragments at the ith and jth positions, if the fragments are from the same PDB structure. Histograms that have a peak at a position of dij < 9Å are converted into distance profiles for the residue pair13,14.
In addition to obtaining distance profiles from the fragments, C-QUARK predicts contacts between the residues using NeBcon and its component predictors. The contact potential (Eq. 3) together with the fragment-derived distance profile restraints and the inherent knowledge-based energy function are used as the composite C-QUARK energy function to guide the REMC simulation. Following the REMC simulation, the resulting decoy conformations from the simulation trajectories are clustered by SPICKER to identify cluster centroids, which correspond to low free-energy states. The cluster centroids from the five largest clusters are refined by FG-MD to obtain five final models. The models from the corresponding clusters are ranked based on the size of the SPICKER clusters they were selected from, where the models from the more populated clusters are ranked higher. Finally, the residue-level quality is predicted by ResQ.
RESULTS AND DISCUSSIONS
121 domains from 80 protein chains were assessed in CASP13. Based on the modeling difficulty and template availability, the CASP13 assessors classified 111 domains into 32 FM targets, 13 FM/TBM targets, 44 TBM-easy targets and 22 TBM-hard targets, while the 10 remaining domains were defined as “not evaluated” by the CASP13 assessors. Here, we manually classified these 10 “not evaluated” proteins based on the TM-score48,49 between the first template identified by LOMETS and the experimental structure. The target was defined as FM, FM/TBM, TBM-hard, or TBM-easy, if the TM-score of the first LOMETS template belonged to the interval (0, 0.3], (0.3, 0.5], (0.5, 0.8], or (0.8, 1.0], respectively. Using this rule, 5 of them (T0974s2-D1, T0999-D1, T1000-D1, T1004-D3 and T1011-D2) were classified as TBM-easy targets and the remaining 5 (T0960-D1, T0960-D4, T0963-D1, T0963-D4 and T0980s2-D1) as FM targets. In the following analysis, we treat TBM-easy and TBM-hard targets as TBM targets, while FM and FM/TBM targets are treated as FM targets. The official CASP domain definitions were used to assess the results. Since the “Zhang” human group used essentially the same pipeline as our “Zhang-Server” group, the following discussion mainly focuses on the results obtained by the “Zhang-Server” and “QUARK” pipelines.
Impact of sequence-based contact prediction on “Zhang-Server” and “QUARK” modeling results
In previous CASP experiments, the “Zhang-Server” group was built on the classic I-TASSER pipeline. However, during CASP13, “Zhang-Server” utilized a new approach, namely, C-I-TASSER, which takes advantage of the strengths of both template-based modeling and sequence-based contact-maps derived from deep-learning approaches. Additionally, while “QUARK” started to use sequence-based contact-maps in CASP12, it used a former version of NeBcon16 that combined contacts mainly from co-evolution-based predictors that did not use deep-learning.
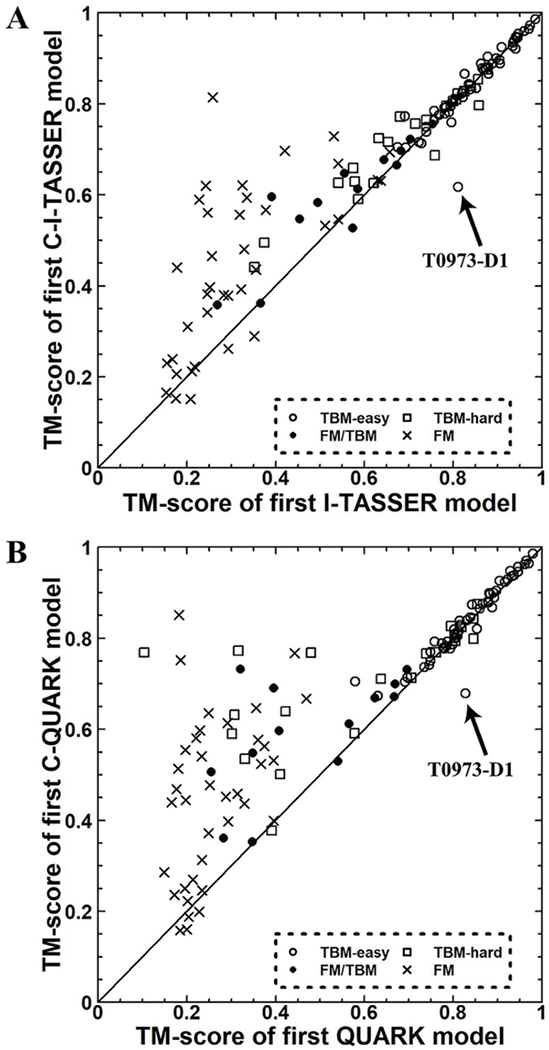
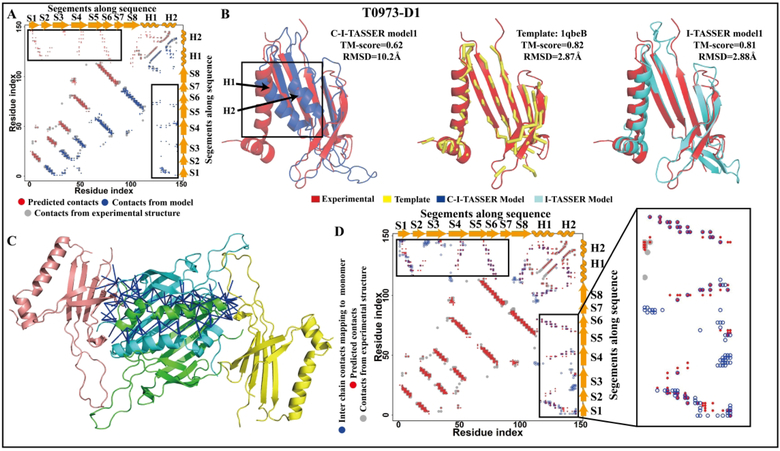
To examine the effect of implementing the new deep-learning-based contact predictors into C-I-TASSER and C-QUARK, following CASP13, we ran I-TASSER and QUARK for each CASP target using the same domain partitions and the same set of templates used by C-I-TASSER and C-QUARK (C-QUARK and QUARK use templates for “Trivial” and “Easy” targets) in CASP13. The results of this head-to-head comparison are shown in Figure 2. For the 50 FM targets, the average TM-score48,49 of the first models generated by C-I-TASSER was 0.487, which was 28% greater than that of I-TASSER’s first models (p-value=2.2E-07). In particular, 27 FM targets were foldable by C-I-TASSER, which was 80% higher than the number of targets that were foldable by I-TASSER (15 targets were foldable) (see Table S1 for details). In addition, 27 of the 50 FM targets were successfully folded by C-QUARK, while QUARK could only fold 6 of the targets. The average TM-score of C-QUARK’s first models was 0.49 for the 50 FM targets, which was 56% higher (p-value=2.14E-10) than the average TM-score for the models generated by QUARK (see Table S2 for details). For the 71 TBM targets, the first models of 47 (52) targets obtained by C-I-TASSER (C-QUARK) were better than the corresponding I-TASSER (QUARK) models. The average TM-score of the C-I-TASSER (C-QUARK) first models was 0.806 (0.803), which was 1% (5%) better than the average TM-score for the I-TASSER (QUARK) models with a p-value of 3.5E-02 (2.2E-03). However, interestingly, there were several TBM targets for which the first models produced by I-TASSER were considerably better than the C-I-TASSER models. Among them, T0973-D1 (TBM-easy) was a single domain protein that consisted of 146 residues, where the experimental structure for the T0973-D1 domain had 128 solved residues with two disordered regions (residue 60–65 and 84–95). It was an α+β protein that contained 8 β-strands (S1–8) and 2 α-helices (H1–2). From the experimental structure-derived contact-map (Figure 3A), no contacts appeared between the helix regions and strands 1–7. This implied that the N-terminus should be separated from the C-terminus, as shown in Figure 3B. The first template identified by LOMETS was 1qbeB, which had a high TM-score (0.82) to the experimental structure. Like the experimental structure, the N-terminus and C-terminus of this template were separated and did not form any contacts. After the I-TASSER simulation, the first I-TASSER model had a high TM-score of 0.81 to the experimental structure, similar to the template. During the C-I-TASSER simulation, however, the C-terminus was brought close to the N-terminus, resulting in the formation of new contacts in the model (Figure 3A blue points). This is because a set of false positive contacts were predicted between H1–2 and S1–7 (Figure 3A, red points), and, hence, the contact-based potential brought these two regions together during the structure assembly simulation. The first C-QUARK model for this target was also worse than the first model generated by QUARK (TM-score=0.828 vs 0.678) due to the same reason that the C-I-TASSER model was worse than the I-TASSER model.
Figure 2.
(A) Results of the head-to-head comparison between the first models produced by C-I-TASSER and I-TASSER. (B) Results of the head-to-head comparison between the first models produced by C-QUARK and QUARK.
Figure 3.
(A) and (B) The contact-map and structures (superposing the first “Zhang-Server” model (blue) with the experimental structure (red), superposing the first LOMETS template (yellow) with the experimental structure (red), and superposing the first I-TASSER model (cyan) with the experimental structure (red)) for target T0973-D1. The bad terminal contacts and the corresponding wrong structural model are highlighted with the black box. The red, blue and grey points on the contact-map correspond to the contacts predicted by NeBcon, extracted from the “Zhang-Server” model, or extracted from the experimental structure, respectively. (C) The homo-oligomeric T0973-D1 complex which was constructed using 2vf9 as a reference. The center copy of T0973-D1 is shown in green and inter-chain contacts are shown by the blue sticks. (D) The contact-map which maps inter-chain contacts to one monomer contact map. The terminal contacts and the corresponding inter-chain contacts are highlighted with the black box. The red, blue and grey points on the contact-map correspond to the contacts predicted by NeBcon, mapped from the inter-chain contacts, or extracted from the experimental structure, respectively.
Interestingly, we found that the false positive contacts between the N-terminus and C-terminus came from co-evolution between inter-chain residues of a homo-oligomer complex structure. T0973-D1 was a viral coat protein that formed homo-oligomer complexes. Since the biological assembly for the full homo-oligomer is not yet available, to check where the false positive contacts came from, we used a template-based approach to assemble the native T0973-D1 into a complex. The closest structure template of T0973-D1 was another viral coat protein from bacteriophage PRR1 (PDB 2vf9) with a TM-score of 0.80 and a sequence identity of 24% as determined by TM-align structure alignment. The biological assembly of 2vf9 is a homo-oligomer that contains 180 chains. Among these chains, we used one chain as the central chain, and chose all chains in the biological assembly that had more than one contact to the C-terminus of this central chain, which resulted in four monomers in total. TM-align was then used to separately superpose four copies of T0973-D1 to the four chains of 2vf9 to construct the predicted complex structure (Figure 3C). Using this predicted complex, we calculated the inter-chain contacts (Blue sticks in Figure 3C) between the C-terminus of the center chain (Green chain in Figure 3C) and every other chain. From the complex structure, there were 144 inter-chain contacts formed between H1–2 (C-terminus) of the center copy of T0973-D1 and S1–7 of the neighboring copies of T0973-D1. Furthermore, we tried to map the inter-chain contacts (Blue circle) into one monomer contact map, which is shown in Figure 3D. The co-evolutionary relationship should exist between residues from H1–2 of the center copy and residues from S1–7 of the neighboring copies, but a co-evolution-based contact prediction method cannot classify it as inter-chain co-evolution or intra-chain co-evolution if the complex is a homo-oligomer. NeBcon predicted 121 false positive contacts in this area, and 54% of them overlapped with the inter-chain contacts. Since most of the component programs in NeBcon are co-evolution-based methods or deep-learning methods that utilize co-evolutionary features, it is not strange that the false positive contacts predicted by co-evolution, propagate to the deep-learning-based contact maps, and eventually affect the quality of the final models. This is consistent with a previous study that showed that over 30% of false positive predicted contacts by co-evolution are actually inter-chain contacts50.
This case demonstrates that false positive contacts may lead to incorrect local folding even though high quality templates are identified, and that there are methodology-level limitations in current co-evolution-based contact prediction methods when dealing with homo-oligomer complexes. Although deep-learning contact-maps can help significantly improve the quality of C-I-TASSER and C-QUARK models in most cases, the inclusion of incorrect contacts can negatively impact the model quality. Increasing the contact prediction accuracy and improving the simulation strategy in order to selectively incorporate correct contacts are the keys to address this issue.
Model quality depends on the interplay between contact prediction and threading templates
Despite the fact that the new contact prediction potential/algorithms greatly improve the overall quality of the C-I-TASSER and C-QUARK predictions, template-based information is still a very important factor for protein folding, especially for TBM targets. Therefore, balancing the contributions from templates and predicted contacts is one of the key procedures that need to be considered. Since “QUARK” only used templates when a target was defined as “Trivial” or “Easy”, this section will mainly focus on “Zhang-Server” (C-I-TASSER) in order to examine the effectiveness of combining information from templates and predicted contacts.
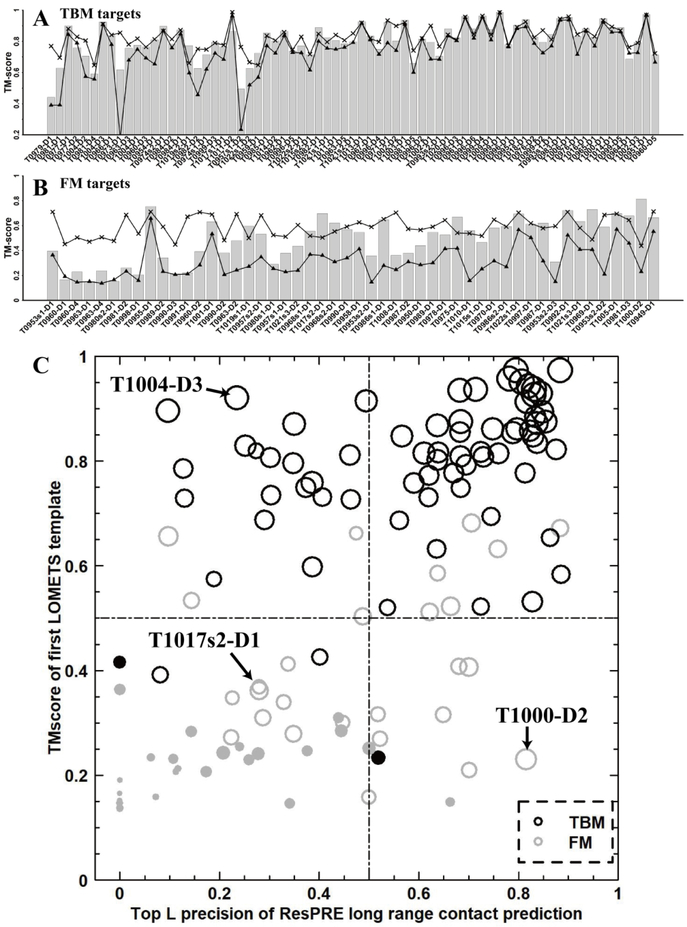
In Figure 4A and 4B, we present the TM-scores for the “Zhang-Server” first models, first LOMETS templates and the best available templates for the 121 CASP domains. The TM-scores of the first “Zhang-Server” models are shown by the bar plot, while the TM-scores of the first templates identified by LOMETS are shown by the triangle marks. Additionally, the TM-scores of the best available templates, which were identified by searching the experimental structures through our template library using TM-align41, are shown as cross marks. For a fair comparison with the LOMETS results, we excluded the templates that were released after the CASP server submission deadline for each target (see detailed information in Table S3). For 36 out of the 71 TBM targets (Figure 4A), the quality of the first template identified by LOMETS was very close (ΔTM-score<0.05) to the best available template in our template library. For the remaining 35 cases, although the first template identified by LOMETS was far from the best available template, 21 of the targets could still be folded by C-I-TASSER close to or even better than the best available template in the template library (ΔTM-score>−0.05). For 38 out of the 71 TBM targets, the first LOMETS template was very close to the “Zhang-Server” first model (ΔTM-score>−0.05), which demonstrates that templates are still an important component for folding TBM targets. For 42 out of the 50 FM targets (Figure 4B), high quality templates with TM-scores to the experimental structure >0.5 existed in the template library, but LOMETS could not detect them. This indicates that threading still requires improvement, especially for FM targets. With the help of contact information, for 15 out of the 50 FM targets, C-I-TASSER produced better structural models than the best available templates detected by TM-align, even though LOMETS failed to identify the best templates. This shows that the complementarity between predicted contacts and threading templates may help improve protein structure prediction.
Figure 4.
TM-scores of the first models built by “Zhang-Server” (bar), TM-align templates (crosses), and the first LOMETS templates (triangles) for the 71 TBM targets (A) and 50 FM targets (B), where the targets are sorted by the precision of the top L long-range predicted contacts. (C) The relationship between folding success and TM-scores of the templates identified by LOMETS and the top L long-range contact precision by NeBcon. An empty circle means that the target is foldable (TM-score≥0.5 for the first “Zhang-Server” model), while a solid point means the target is not foldable (TM-score<0.5). The point size is proportional to the value of the TM-score. The 71 TBM targets are shown in black and the 50 FM targets are shown in grey.
Figure 4C provides a closer look at the impact of contact and template quality on the predicted structural models, where the x-axis represents the precision of long-range top L predicted contacts and the y-axis represents the quality of the first templates identified by LOMETS. The figure can be divided into 4 sub-regions: (1) the top left sub-region represents targets with poor contact prediction quality (top L long-range precision ≤0.5) but good threading templates (TM-score >0.5 to the experimental structure48), (2) the top right sub-region represents targets with both high quality contact prediction and threading templates, (3) the bottom right sub-region represents targets with high quality contact prediction but poor threading templates, and (4) the last sub-region represents the most challenging targets with both low quality contact prediction and threading templates. Since the y-axis represents the quality of the first templates identified by LOMETS, it can mainly reflect whether a target should be a TBM or FM target. We found that 94% of the TBM targets were located in the upper two sub-regions, while 80% of the FM targets were located in the lower two sub-regions. For 10 of the FM targets that fell into the upper two sub-regions, 7 of them were FM/TBM targets. Thus, it is not surprising that they had relatively better templates. We observed that overall 80% (=96/121) of the CASP13 targets were foldable by “Zhang-Server”, where 100% of the targets from sub-regions 1, 100% of the targets from sub-region 2, 78% of the targets from sub-region 3, and only 34% of the targets from sub-region 4 were foldable. Below and in Figure 5, we examine in greater detail three representative examples from three of the sub-regions (1, 3 and 4) to analyze the advantages and disadvantages of integrating predicted sequence-based contacts with template information.
Figure 5.
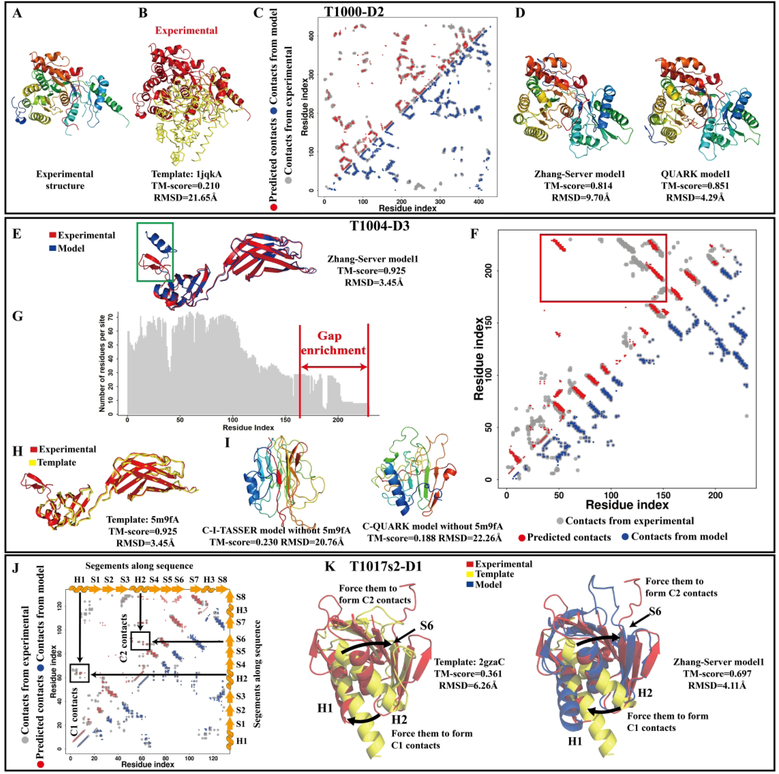
Case studies for the advantages and disadvantages of integrating predicted contacts with template information. The top panel from (A) to (D) shows the analyses for target T1000-D2, including the experimental structure (A), superposition of the experimental structure (red) and the first LOMETS template (yellow) (B), contact-map (C) and “Zhang-Server” and “QUARK” models (D). The middle panel from (E) to (I) shows the analyses for target T1004-D3, including the superposition of the experimental structure (red) and the “Zhang-Server” model (blue) (E), contact-map (F), number of effective residues in each sequence position of the deep MSA (G), superposition of the experimental structure (red) and the first LOMETS template (yellow) (H) and models generated by the control test which removed template 5m9fA (I). The false positive contacts between the terminal regions are highlighted using the red box, while the incorrect secondary structure prediction is shown using the green box in the model. The bottom panel from (J) to (K) shows the analyses for target T1017s2-D1, including the contact-map (J) and structure superposition of the “Zhang-Server” model (blue) and first LOMETS template (yellow) with the experimental structure (red). The red points in the contact-map are the predicted contacts by NeBcon, the grey points are contacts derived from the experimental structure, and the blue points are the contacts calculated from the first model built by “Zhang-Server” in (C) (F) and (J).
T1000-D2
T1000 was a two-domain protein with 523 residues, where the second domain (residues 93 to 523), T1000-D2, was an FM target. It was an α/β protein with 10 helices and 11 β-strands that formed a very complex topology (Figure 5A). The first template (PDB ID: 1jqkA) identified by LOMETS had a low TM-score of 0.21 (Figure 5B), indicating that it had a completely different fold than T1000-D2. Furthermore, the best template (PDB ID: 3lq2A) identified by TM-align also had a different fold than the experimental structure of T1000-D2 (TM-score=0.438), indicating that this target was very difficult to model solely based on information from threading templates. To demonstrate this, after CASP13, we also predicted the structure of this target using I-TASSER, with the same set of templates utilized in CASP13. The first model produced by I-TASSER only had a TM-score of 0.258, illustrating that it failed to produce a reasonable prediction. However, the contact prediction accuracy was very high for this target, where the precision of the top L long-range predicted contacts by NeBcon was 0.82 (Figure 5C). As a result, the C-I-TASSER model achieved a very high TM-score of 0.814. Similarly, due to the accurate prediction of long-range contacts, C-QUARK folded this target with a slightly better TM-score of 0.851 (Figure 5D). This target demonstrates the importance of contacts in template-based modeling, especially when good templates are not identified.
T1004-D3
T1004 was a protein with 458 residues and consisted of three continuous domains, where the third domain defined by the assessors, T1004-D3, was a β-protein that contained 230 residues (Figure 5E). Although it was a TBM-easy target, most of the long-range contacts between the C-terminus and other areas of the protein were not correctly predicted (Figure 5F), where the precision of the long-range contact prediction was only 0.235. The low contact prediction performance was mainly because of the poor quality of the MSA, where the last 50 positions in the C-terminus contained 62–94% gaps (Figure 5G). While the contact prediction quality was low, LOMETS successfully detected a very good template (PDB ID: 5m9fA) for this target with a TM-score to the experimental structure of 0.909 (Figure 5H). As a result, C-I-TASSER folded this target correctly, where the final structural model had a TM-score of 0.925. Since LOMETS identified a very reliable template for this domain, it was defined as a trivial target, so “QUARK” utilized the C-QUARK-TBM approach, where the initial conformation and distance restraints were taken from the LOMETS templates and introduced into the simulation. Like “Zhang-Server”, the first model produced by “QUARK” was also very high quality (TM-score=0.926). To investigate the influence of template quality on T1004-D3 modeling, we ran two control tests for this target. For the first test, we excluded the template 5m9fA, while we input all the other templates (all of them had TM-scores <0.5 to the experimental structure) and the predicted contacts used in CASP13 for this target into C-I-TASSER. For the second test, we ran the C-QUARK ab initio folding approach without using any information from templates, but we used the same predicted contacts in the simulation. The goal of these two tests was to remove the influence of the high-quality templates, and primarily use the inherent potentials and predicted contacts to guide the simulations. Without information from the high-quality templates, C-I-TASSER only produced a model for this target with a TM-score of 0.23, and the C-QUARK model also had a low TM-score of 0.188 (Figure 5I). These data suggest that identification of good templates is an important component for protein structure prediction when predicted contacts are not accurate, especially for large β-proteins, such as this target, that are difficult to fold through pure ab initio simulations51.
From this target, we further note that the N-terminus of the first model built by C-I-TASSER formed a short helix (highlighted by the green box in Figure 5E), while the correct local topology was a beta-pair. The wrong prediction of the local topology stemmed from incorrect secondary structure prediction by PSIPRED52 and PSSpred53 in that region. This example demonstrates that although incorrect secondary structure prediction may not have a great influence on folding the global topology, it may affect the local quality of the model.
T1017s2-D1
T1017s2 was an FM target with 240 residues, where its first domain, T1017s2-D1, spanned residues 2 through 129. T1017s2-D1 was an α+β protein that consisted of 3 alpha helices (H1–3) and 8 beta strands (S1–8). For this target, both the quality of the LOMETS threading templates and the NeBcon contact prediction were poor, where the TM-score of the best template detected by LOMETS was 0.36 and the top L long-range contact prediction precision was 0.28 (Figure 5J). Despite these facts, “Zhang-Server” still folded this target with a high TM-score of 0.697, which was mainly due to the complementarity between the templates and predicted contacts. Specifically, LOMETS detected template 2gzaC (Figure 5K), where most of its segments were correctly aligned (H1, H2, S3, S5, S6, S7 and S8). However, the orientation of H2 deviated from the native structure, which resulted in the low threading TM-score. NeBcon predicted that helix H2 formed C1 contacts with helix H1 and C2 contacts with strand S6. During the simulation, these contacts helped change the orientation of H2 to the correct direction and produced a high-quality structural model. The most challenging targets for “Zhang-Server” are located in sub-region 4, since neither good templates nor accurate contact prediction could be obtained. The successful folding of T1017s2-D1 and the other 11 foldable targets from sub-region 4 demonstrate that the appropriate coupling of both predicted contacts and templates may help minimize the weaknesses of each individual feature. This further highlights the importance of the complementarity between templates and predicted contacts as well as the optimized inherent knowledge-based energy function.
Better contact prediction improves FM target folding
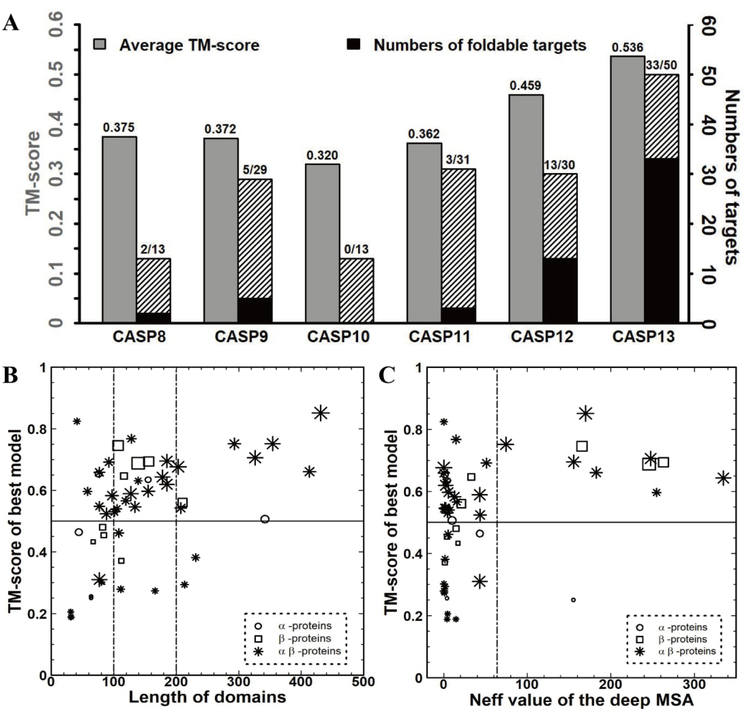
Since predicted residue contacts were introduced in protein structure prediction, a significant improvement can be observed in the folding performance for FM targets. To highlight this impact, in Figure 6A we summarize the folding performance for FM targets based on the best models submitted by either “Zhang-Server” or “QUARK” since CASP8. Here, a new contact prediction module that implemented the previous version of NeBcon without using deep-learning-based predictors16 was incorporated into “Zhang-Server” and “QUARK” during CASP12. This resulted in an average TM-score of 0.459 for the 30 FM targets in CASP12, which was at least 22.4% better than the results of the former CASP experiments. In CASP13, the inclusion of residue-residue contact prediction by deep-learning-based methods in NeBcon further improved the folding performance for FM targets, where the average TM-score of the best models was 16.8% better than that of CASP12 and 42.9% better than CASP8–11. Additionally, 43% of the FM targets were foldable (TM-score>0.5) in CASP12, which was around 2-fold more than the number of foldable FM targets in CASP8–11, where predicted residue-residue contacts were not used. It is remarkable that, 66% of the FM targets were foldable in CASP13, which is the first time that the number of foldable FM targets was over half of the total FM targets.
Figure 6.
(A) The folding performance for FM targets based on the best models that were submitted by “Zhang-Server” or “QUARK” from CASP8 to CASP13, the grey bar represents the average TM-score of the best “Zhang-Server” model, the black bar indicates the numbers of foldable targets (TM-score≥0.5) and the hatched bar depicts the number of non-foldable targets. (B) The TM-scores of the best “Zhang-Server” or “QUARK” models for the 50 FM targets in CASP13 versus the domain lengths. (C) The TM-scores of the best “Zhang-Server” or “QUARK” models for the 50 FM targets in CASP13 versus the Neff value of the deep MSA. The size of the points in (B) and (C) is proportional to the precision of the top L long-range contact prediction by NeBcon.
Figure 6B shows the TM-scores of the best “Zhang-Server” or “QUARK” models for the 50 FM targets in CASP13 versus the target lengths. Among the 33 foldable targets, 3 targets were α-proteins, 5 targets were β-proteins, and 25 targets were αβ (α/β or α+β) proteins. In CASP13, 9 FM targets with lengths greater than 200 residues were correctly folded, while in CASP12, only 2 FM targets whose size was greater than 200 residues were correctly folded. There is strong evidence that more accurate contact prediction can help fold larger proteins, where the Pearson Correlation coefficient was 0.674 between the model TM-score and top L long-range contact prediction precision.
The high accuracy of predicted contacts used in “Zhang-Server” and “QUARK” is attributed to the newly added deep-learning-based contact prediction methods into NeBcon. Table S4 shows the performance of NeBcon and its nine component contact prediction methods. The accuracies of the deep-learning-based methods (ResPRE, DeepPLM, Deepcontact, DNCON2, DeepCOV and MetaPSICOV2) that were newly added to NeBcon are significantly better than those of the co-evolution-based methods used in the former version of NeBcon16. Among all of the individual deep-learning-based methods, ResPRE has the best performance, followed by another in-house program, DeepPLM. For long-range top L contact prediction in particular, ResPRE is at least 20% better than the external contact predictors. Therefore, the high performance of NeBcon can be mainly attributed to the contributions from ResPRE and DeepPLM.
Besides the newly incorporated deep-learning-based approaches, the new MSA construction method is another factor that improves contact prediction performance as well as structure prediction. For the 50 FM targets, we compared the performance of contact prediction derived from NeBcon with HHblits MSAs as the input to the performance of NeBcon with DeepMSA MSAs as the input (Table S5). HHblits is the default MSA construction method for many contact prediction programs. The results show that the usage of deep MSAs increased NeBcon’s long-range top L precision by 18%. The reason that DeepMSA improved NeBcon’s performance is mainly due to the detection of more homologous and effective sequences compared to other very popular tools (HHblits and PSI-BLAST). In particular, for the 50 FM targets (Figure S2), on average, DeepMSA detected 1,936 sequences (or Neff=60), which was 5 times more sequences (or 4-fold higher Neff) than HHblits identified and 10 times more sequences (or 9 times higher Neff) than PSI-BLAST detected.
To further investigate the impact of MSA quality on protein structure prediction, Figure 6C shows the Neff value of the MSA input into NeBcon and the TM-score of the best “Zhang-Server” or “QUARK” model for the 50 FM targets. Almost all of the targets (except T0963-D4, which was close to a long loop structure without any long-range contacts) that had an MSA with a Neff value greater than 64 were foldable, and most targets that were not foldable had MSAs with a low number of effective sequences (low Neff) (Table S6). The targets with lower Neff values were the most challenging FM targets to fold. A future direction to solve the low Neff protein folding problem is to develop novel contact predictors trained on targets with few or no sequence homologs.
Domain partitioning and assembly affects the modeling quality
CASP13 contained 21 targets with multiple domains, following the CASP assessment. In the “Zhang-Server” and “QUARK” pipelines, the query sequences were first split into domains by ThreaDom19 and each domain was individually folded. The final full-length models were then assembled from the individual domain models using the full-length C-I-TASSER models as the reference templates (Figure 1). Table S7 lists a comparison of the ThreaDom predictions and actual domain splits based on the experimental structures. Here, the normalized domain overlap score54 (NDO-score) and the domain boundary distance score55 (DBD-score) implemented in the former CASP assessment were utilized to assess the domain boundary prediction accuracy. The NDO-score evaluates the overlap between the predicted domain regions and the true domain regions, while the DBD-score is defined as the distance between the predicted domain boundaries and the true domain boundaries along the sequence. All of the linker regions between the domains were considered as the true boundaries. On average, the NDO-score was 0.739 and the DBD-score was 0.400 for the 21 multi-domain targets, which indicates that domain partitioning remains a problem, although for T0982, T0989, T1014 and T1021s3, ThreaDom had nearly perfect domain partitions (NDO-score>0.94 and DBD-score=1.00). Moreover, the accuracy of discontinuous domain partitioning was still lower than that of continuous domain partitioning. This is demonstrated by the fact that for discontinuous domain boundary prediction, the average NDO-score and DBD-score of ThreaDom were 0.613 and 0.235, respectively, which were much lower than those for the continuous domain proteins (0.789 and 0.467 for NDO-score and DBD-score, respectively).
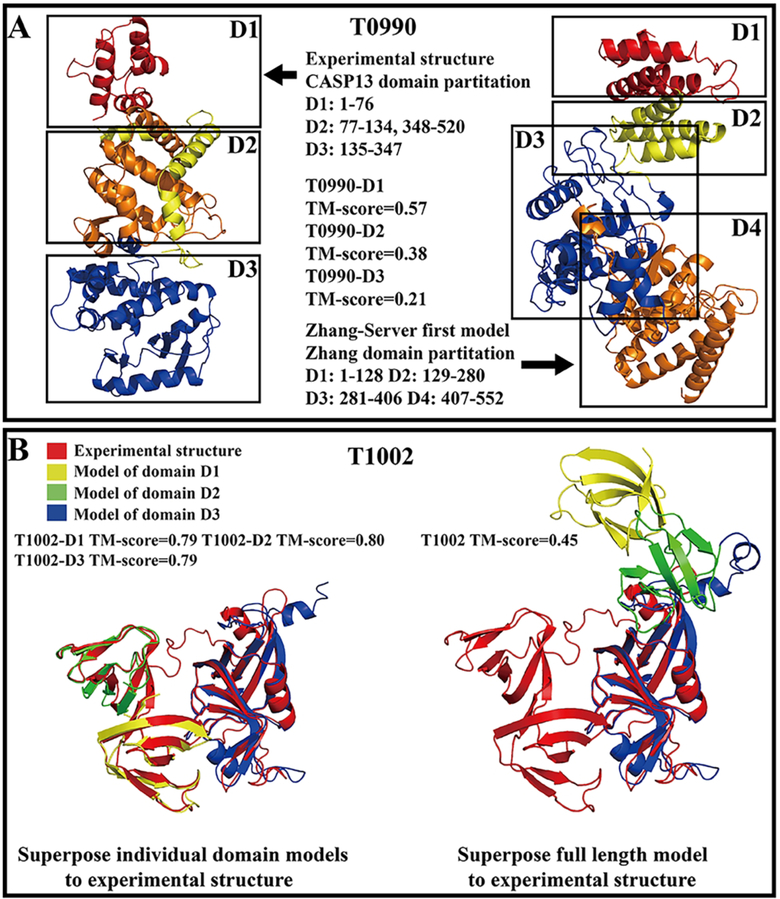
Incorrect domain partitioning can significantly impact the quality of both the individual domain and full-chain models. Figure 7A shows an example from T0990, which was a three-domain α-protein (T0990-D1, T0990-D2 and T0990-D3) that consisted of 552 residues, where the first and third domains were continuous (D1: 1–76; D3: 135–347) and the second domain was discontinuous (D2: 77–134, 348–520). ThreaDom predicted this target to be a protein with four continuous domains (D1: 1–136; D2: 128–289; D3: 280–406; D4: 403–552) and the prediction had a low NDO-score of 0.41. For the first “Zhang-Server” model, our predicted first domain covered the entire experimental domain, resulting in a relatively high TM-score (0.57) compared to the other two domains, as the domain boundaries of the other two domains were predicted completely inaccurately. Therefore, the other two domains were folded improperly as their models had TM-scores of 0.38 and 0.21 for T0990-D2 and T0990-D3, respectively.
Figure 7.
(A) The structures of different domains for target T0990. Different colors represent different domains or different parts of discontinuous domains. (B) The structures of different domains for target T1002. The red, yellow, green, and blue structures correspond to the experimental structure and the first C-I-TASSER model of the T1002-D1 domain, T1002-D2 domain, and T1002-D3 domain, respectively.
Figure 7B presents an example from T1002, which was a three-domain β-fold protein (T1002-D1: 1–59; T1002-D2: 60–118 and T1002-D3: 127–270) that consisted of 270 residues. All three domains were TBM-easy targets, and “Zhang-Server” folded the individual domains successfully with TM-scores of 0.79, 0.80 and 0.79. However, since a low quality full-length reference model was constructed (the best full-length template identified by LOMETS only had a TM-score=0.388), the three domains were assembled together in an incorrect orientation, resulting in a low TM-score (0.45) for the full-length model. This same problem also occurred for two of the three FM-sp targets (T0984-D0, T1000-D0 and T1002-D0) which were multi-domain FM targets but were assessed based on the full-chain model quality. Although “Zhang-Server” constructed correct folds with TM-scores >0.782 for all the individual domains of the three FM-sp targets, only T0984-D0 had a correct domain orientation with a TM-score of 0.86 due to the correct reference model from LOMETS (TM-score=0.83), while the domain orientations for the other two targets were completely wrong (with an overall TM-score=0.680 for T1000, and 0.450 for T1002, see Table S7).
In summary, both the domain partitioning and domain assembly procedures in the current pipeline depend on the threading results. Therefore, the template quality has a crucial impact on the performance of modeling multi-domain proteins. However, the quality of the templates detected by LOMETS threading was much worse for FM targets than for TBM targets. One way to improve the quality of the detected templates might be to use DeepMSA to construct sequence profiles for use by LOMETS to improve the profile quality for FM targets. In our analysis, we found that DeepMSA detected more sequences than PSI-BLAST, which is the current sequence profile construction method used in LOMETS. Additionally, beyond the threading-based approaches, predicted contacts could also be used to partition multi-domain proteins or assemble the domain models together. Since the current deep-learning-based predicted contacts can give a clear overall topology for a protein, inter-domain contact/distance prediction might be helpful for partitioning multi-domain proteins or assembling domain models.
CONCLUSION
In CASP13, we tested two improved pipelines, C-I-TASSER and C-QUARK, which participated as “Zhang-Server” and “QUARK”, respectively. Here, C-I-TASSER and C-QUARK are based on the well-established I-TASSER and QUARK frameworks with several new developments. These developments include the incorporation of a deep MSA generation method, deep-learning-based contact prediction, and a newly optimized contact-based potential. The most notable takeaway from CASP13 is that the interplay of deep MSA generation methods and deep-learning-based contact prediction can help improve the accuracy of protein structure prediction for both FM and TBM targets. In particular, the deep MSA generation method helps to extract a higher number of effective sequences and produces more accurate evolutionary coupling information for contact prediction that in turn helps improve the performance of structure prediction. Moreover, the addition of a novel deep-learning-based contact prediction method, ResPRE, into the NeBcon pipeline greatly improves the accuracy of contact prediction. Another notable takeaway is that properly balancing the components of the energy function is vital for accurate structure prediction. In other words, contact restraints should be carefully optimized and implemented into the folding simulations. Hence, in the new pipelines, a new contact potential term, used as a restraint to guide the structural assembly simulations, was fine-tuned to balance the contributions from the predicted contacts and the other energy terms in the C-I-TASSER and C-QUARK energy functions. Due to the high accuracy of contact prediction and the advantageous interplay of template and contact information, the accuracy of the final models from C-I-TASSER and C-QUARK were significantly better than the models generated by the original I-TASSER and QUARK pipelines, especially for FM targets.
Despite the success, there are still significant challenges to be overcome in the current pipelines. One of the major problems comes from the incorrectly predicted contacts between the N- and C-terminal protein regions, which guide the simulation in the wrong directions for a few of the CASP targets, such as T0973-D1, T1021s2-D1 and T0964-D1. The low accuracy of contact prediction in the terminal regions is mainly due to MSAs with many gaps in these regions, as the accuracy of contact-map prediction and FM target modeling is highly influenced by the number of effective sequences in the MSA. This can be clearly seen by the fact that the MSA Neff values for most failed targets were less than 64. A future direction to solve the low Neff protein folding problem might be to develop novel contact predictors trained on targets with low or no sequence homologs. Furthermore, the detection of correct templates is crucial to improve the performance of template-based structure prediction. Note that even for FM targets, we found that there existed distant-homology templates with the same fold as the query protein in the PDB, even though our current threading methods were unable to detect them, which is consistent with the former findings on the completeness of the PDB library56. One reason could be the poor quality of the MSAs used to build the profiles. Therefore, the new deep MSA and profile construction approaches may also improve the threading alignments, thereby enhancing the modeling performance for both FM and TBM targets.
Another significant challenge is domain partitioning and assembly for modeling multi-domain proteins. The current pipelines highly rely on the threading programs in two aspects: ThreaDom uses the threading template alignments to guide the domain boundary predictions and the full-chain reference models are constructed based on the full-chain threading templates. However, threading programs often have difficulty identifying distantly homologous templates, which impacts both aspects of multi-domain structure prediction. One way to address these issues is to utilize the deep-learning-derived contact-maps to guide the domain splitting processes. Meanwhile, in addition to the homologous templates, the structure-based analogous templates can also be utilized to guide the domain assembly. Our preliminary data demonstrates encouraging results along these lines, which should be used to improve future multi-domain structure prediction.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr. Xiaogen Zhou for insightful discussions on the domain partitioning and assembly problem, and Dr. Jungkap Park and Dr. Kazuhiro Saitou for technical assistance with the Rotas program. The I-TASSER and QUARK servers use the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation [ACI-1548562]. This work was supported by the National Institutes of Health [GM083107, GM116960, and AI134678]; and National Science Foundation [DBI1564756].
REFERENCES
- 1.Zhang Y Progress and challenges in protein structure prediction. Current opinion in structural biology. 2008;18(3):342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kryshtafovych A, Monastyrskyy B, Fidelis K, Moult J, Schwede T, Tramontano A. Evaluation of the template-based modeling in CASP12. Proteins. 2018;86 Suppl 1:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunbrack R Template-based modeling assessment in CASP11. Paper presented at: 11th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction2014; Riviera Maya, Mexico. [Google Scholar]
- 4.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. [DOI] [PubMed] [Google Scholar]
- 5.Soding J Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. [DOI] [PubMed] [Google Scholar]
- 6.Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72(2):547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinch LN, Li W, Monastyrskyy B, Kryshtafovych A, Grishin NV. Evaluation of free modeling targets in CASP11 and ROLL. Proteins. 2016;84 Suppl 1:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abriata LA, Tamo GE, Monastyrskyy B, Kryshtafovych A, Dal Peraro M. Assessment of hard target modeling in CASP12 reveals an emerging role of alignment-based contact prediction methods. Proteins. 2018;86 Suppl 1:97–112. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(S8):108–117. [DOI] [PubMed] [Google Scholar]
- 10.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015;12(1):7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu D, Zhang Y. Toward optimal fragment generations for ab initio protein structure assembly. Proteins: Structure, Function, and Bioinformatics. 2013;81(2):229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins: Structure, Function, and Bioinformatics. 2012;80(7):1715–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu D, Zhang J, Roy A, Zhang Y. Automated protein structure modeling in CASP9 by I-TASSER pipeline combined with QUARK-based ab initio folding and FG-MD-based structure refinement. Proteins. 2011;79 (Suppl 10):147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Mortuza SM, Wang Y, Shen H-B, Zhang Y. NeBcon: protein contact map prediction using neural network training coupled with naïve Bayes classifiers. Bioinformatics. 2017;33(15):2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Hu J, Zhang C, Yu D-J, Zhang Y. ResPRE: high-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016(770–778). [Google Scholar]
- 19.Xu D, Wang Y, Zhang Y, Xue Z. ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics. 2013;29(13):i247–i256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nature Methods. 2011;9:173. [DOI] [PubMed] [Google Scholar]
- 21.Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics. 2010;11(1):431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucleic Acids Research. 2007;35(10):3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y Interplay of I-TASSER and QUARK for template-based and ab initio protein structure prediction in CASP10. Proteins: Structure, Function, and Bioinformatics. 2014;82(S2):175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Liang Y, Zhang Y. Atomic-Level Protein Structure Refinement Using Fragment-Guided Molecular Dynamics Conformation Sampling. Structure. 2011;19(12):1784–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galiez C, Mirdita M, Söding J, von den Driesch L, Steinegger M, Martin MJ. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Research. 2016;45(D1):D170–D176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.the UniProt C, Suzek BE, Wu CH, Huang H, McGarvey PB, Wang Y. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2014;31(6):926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinegger M, Söding J. Clustering huge protein sequence sets in linear time. Nature Communications. 2018;9(1):2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovchinnikov S, Park H, Varghese N, et al. Protein structure determination using metagenome sequence data. Science. 2017;355(6322):294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Palmedo P, Ye Q, Berger B, Peng J. Enhancing Evolutionary Couplings with Deep Convolutional Neural Networks. Cell Systems. 2018;6(1):65–74.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikari B, Hou J, Cheng J. DNCON2: improved protein contact prediction using two-level deep convolutional neural networks. Bioinformatics. 2017;34(9):1466–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kandathil SM, Jones DT. High precision in protein contact prediction using fully convolutional neural networks and minimal sequence features. Bioinformatics. 2018;34(19):3308–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchan DWA, Jones DT. Improved protein contact predictions with the MetaPSICOV2 server in CASP12. Proteins: Structure, Function, and Bioinformatics. 2018;86(S1):78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söding J, Gruber M, Seemayer S. CCMpred—fast and precise prediction of protein residue–residue contacts from correlated mutations. Bioinformatics. 2014;30(21):3128–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue–residue contact predictions in a sequence- and structure-rich era. Proceedings of the National Academy of Sciences. 2013;110(39):15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaján L, Hopf TA, Kalaš M, Marks DS, Rost B. FreeContact: fast and free software for protein contact prediction from residue co-evolution. BMC Bioinformatics. 2014;15(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Li JH, Chengxin Zhang, Dong-Jun Yu, Yang Zhang. ResPRE: high-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics. 2019;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014. [Google Scholar]
- 38.Paszke A, Gross S, Chintala S, et al. Automatic differentiation in pytorch. 2017. [Google Scholar]
- 39.Zhang Y, Kihara D, Skolnick J. Local energy landscape flattening: Parallel hyperbolic Monte Carlo sampling of protein folding. Proteins. 2002;48:192–201. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Skolnick J. SPICKER: A clustering approach to identify near-native protein folds. Journal of Computational Chemistry. 2004;25(6):865–871. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Yang J, He B, et al. Integration of QUARK and I-TASSER for Ab Initio Protein Structure Prediction in CASP11. Proteins: Structure, Function, and Bioinformatics. 2016;84(S1):76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Zhang Y. A Novel Side-Chain Orientation Dependent Potential Derived from Random-Walk Reference State for Protein Fold Selection and Structure Prediction. PLOS ONE. 2010;5(10):e15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H, Skolnick J. GOAP: A Generalized Orientation-Dependent, All-Atom Statistical Potential for Protein Structure Prediction. Biophysical Journal. 2011;101(8):2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen M-y, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Science. 2006;15(11):2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J, Saitou K. ROTAS: a rotamer-dependent, atomic statistical potential for assessment and prediction of protein structures. BMC Bioinformatics. 2014;15(1):307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Wang Y, Zhang Y. ResQ: An Approach to Unified Estimation of B-Factor and Residue-Specific Error in Protein Structure Prediction. Journal of Molecular Biology. 2016;428(4):693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26(7):889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins: Structure, Function, and Bioinformatics. 2004;57(4):702–710. [DOI] [PubMed] [Google Scholar]
- 50.Anishchenko I, Ovchinnikov S, Kamisetty H, Baker D. Origins of coevolution between residues distant in protein 3D structures. Proceedings of the National Academy of Sciences. 2017;114(34):9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinch LN, Li W, Monastyrskyy B, Kryshtafovych A, Grishin NV. Assessment of CASP11 contact-assisted predictions. Proteins: Structure, Function, and Bioinformatics. 2016;84(S1):164–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices11Edited by G. Von Heijne. Journal of Molecular Biology. 1999;292(2):195–202. [DOI] [PubMed] [Google Scholar]
- 53.Yan R, Xu D, Yang J, Walker S, Zhang Y. A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Scientific Reports. 2013;3:2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tai C-H, Lee W-J, Vincent JJ, Lee B. Evaluation of domain prediction in CASP6. Proteins: Structure, Function, and Bioinformatics. 2005;61(S7):183–192. [DOI] [PubMed] [Google Scholar]
- 55.Tress M, Cheng J, Baldi P, et al. Assessment of predictions submitted for the CASP7 domain prediction category. Proteins: Structure, Function, and Bioinformatics. 2007;69(S8):137–151. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(4):1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.