Summary
The olive (Olea europaea L. subsp. europaea) is one of the oldest and most socio‐economically important cultivated perennial crop in the Mediterranean region. Yet, its origins are still under debate and the genetic bases of the phenotypic changes associated with its domestication are unknown. We generated RNA‐sequencing data for 68 wild and cultivated olive trees to study the genetic diversity and structure both at the transcription and sequence levels. To localize putative genes or expression pathways targeted by artificial selection during domestication, we employed a two‐step approach in which we identified differentially expressed genes and screened the transcriptome for signatures of selection. Our analyses support a major domestication event in the eastern part of the Mediterranean basin followed by dispersion towards the West and subsequent admixture with western wild olives. While we found large changes in gene expression when comparing cultivated and wild olives, we found no major signature of selection on coding variants and weak signals primarily affected transcription factors. Our results indicated that the domestication of olives resulted in only moderate genomic consequences and that the domestication syndrome is mainly related to changes in gene expression, consistent with its evolutionary history and life history traits.
Keywords: artificial selection, differential expression analysis, domestication, Olea europaea (olive tree), perennial crop, RNA‐sequencing, transcriptomics
Significance Statement
We revisited the evolutionary history of the iconic olive tree by reconstructing a model of domestication and inferring the genomic changes associated with its cultivation. Although more challenging than studying annual crops, further studies of the genomic of the domestication process in perennial should broaden our understanding of the genomic bases and dynamics of adaptation.
Introduction
The olive tree (Olea europaea L. subsp. europaea var. europaea) constitutes a cornerstone of Mediterranean culture by its multiple past and present uses and its omnipresence in traditional agrosystems (Loumou and Giourga, 2003). Historically, olives were restricted to the Mediterranean basin, but current cultivation includes also the Americas and Australia. By 2017, more than 10 million hectares were devoted to olive cultivation globally, with more than 90% in the Mediterranean area (FAO, 2018). Hundreds of clonally propagated cultivars have been described (Bartolini et al., 1998), but only a few are cultivated on a large scale, such as ‘Leccino’ or ‘Picual’. Today, the olive tree is one of the most important oil‐producing plant species, and demand for its oil is still increasing due to its nutritional quality (Kalua et al., 2007). Over 20 million tonnes of olives were harvested in 2017, and the production is in constant growth (FAO, 2018).
The domestication history of olives, however, remains unresolved and its origins are partly lost in the mists of time (Besnard et al., 2018). Olive domestication is a still ongoing process involving the selection and vegetative propagation of individuals of high agronomical value, but also the establishment of orchards with various cultivation practices. While archaeological remains indicate that Mediterranean wild olives (also called oleasters; var. sylvestris) were already used during the Palaeolithic (Kislev et al., 1992), cultivation and early domestication efforts are supposed to have started only about 8000–6000 years bp in the Levant, where the olive oil trade has been developed during the Bronze Age (Liphschitz et al., 1991; Galili et al., 1997; Terral et al., 2004; Kaniewski et al., 2012).
Botanical and genetic data attest that cultivated olives mostly derive from oleasters (Besnard et al., 2001b, 2013a, 2013b; Terral et al., 2004; Breton et al., 2006; Carrión et al., 2010; Díez et al., 2015). Oleasters are found all over the Mediterranean basin and are partitioned in two main gene pools in the East and the West, according to plastid and nuclear markers (Besnard et al., 2001b, 2013b; Breton et al., 2006; Díez et al., 2015). Likewise, the cultivated germplasm shows some degrees of geographic differentiation between the East, Central and West of the Mediterranean basin; yet, extensive admixture between these gene pools has occurred, especially in the western part of its distribution (Besnard et al., 2001a, 2013a; Breton et al., 2009, 2006; Díez et al., 2015; Lumaret et al., 2004). Whether these two gene pools were independently domesticated remains highly debated (Besnard et al., 2018). Some studies on a few dozens of genetic markers support a main domestication event in the East, followed by diffusion to the West where this genepool was introgressed by local oleaster populations (Besnard et al., 2013a,b; Khadari and El Bakkali, 2018). Other studies sustain at least two independent domestications in the East and Central Mediterranean basin (Breton et al., 2006; Díez et al., 2015).
Even less is known about the genetic bases underlying the domestication of olives. In many crops, domestication genes were recently identified through QTL mapping, genome‐wide association studies, candidate gene approaches and more recently population genomic screens for signatures of selection (Doebley et al., 2006; Meyer and Purugganan, 2013; Gepts, 2014). Despite examples of important protein‐altering mutations (e.g. Olsen and Purugganan, 2002; Olsen and Wendel, 2013), these studies highlight the predominant role of regulatory mutations affecting either coding (i.e. transcription factors) or non‐coding regions (i.e. cis‐ regulatory elements) (Meyer and Purugganan, 2013; Lemmon et al., 2014; Martínez‐Ainsworth and Tenaillon, 2016; Swinnen et al., 2016). For instance, the overexpression of the tb1 gene in maize is responsible for its apically dominant architecture (Doebley et al., 1995; Studer et al., 2011), and the rewiring of the expression of several genes in oilseed rape (Brassica napus), increased seed oil content (Liu et al., 2015). However, most of these findings stem from annuals, while the genetic architecture of perennial domestication remains elusive (Gaut et al., 2015). In olives, QTL analyses have identified molecular markers associated with fruit yield and flowering variables (Ben Sadok et al., 2013; Atienza et al., 2014), but the genetic basis of other phenotypic changes associated with domestication attested by the archaeobotanical record, such as an increase in fruit size or oil content (Galili et al., 1997), is still unknown.
Here we present a study based on transcriptome sequences of wild and cultivated olive trees that aims at clarifying the evolutionary history of olives and identifying the molecular changes associated with its domestication. Transcriptomic data at the population level are powerful to study both the changes in regulation, through the study of gene expression differential, as well as the change in protein‐coding genes, with no or minor biases when compared with genome‐wide data (Ray et al., 2011; Romiguier et al., 2014). It has previously been used successfully to study the domestication process in several crops, including African rice (Nabholz et al., 2014), common bean (Bellucci et al., 2014) and tomato (Koenig et al., 2013; Sauvage et al., 2017).
To date, genome assemblies of olives and oleasters are available (Barghini et al., 2014; Cruz et al., 2016; Unver et al., 2017), along with several transcriptome assemblies from different tissues of olives (Muñoz‐Mérida et al., 2013; Parra et al., 2013; Carmona et al., 2015; Iaria et al., 2016; Sarah et al., 2017). While transcriptomic studies have highlighted differences in gene expression in response to cold acclimation (de la O Leyva‐Pérez et al., 2015) or salt stress (Mousavi et al., 2019), neither the domestication history of olives nor the genetic architecture of its domestication syndrome have been studied with such data so far. We therefore generated transcriptomes of 39 cultivated accessions and 27 oleasters, which we used to refine the scenario of domestication and expansion. We further inferred the genomic changes associated with olive domestication using a two‐step strategy: we first contrasted transcriptome‐wide expression levels in wild and cultivated olives to identify changes in gene expression. Second, we scanned the transcriptomic sequences for signatures of selection to identify alleles potentially associated with domestication. The identification of regions under positive selection is methodologically challenging as selection and demography leave very similar signatures in the genome (Stephan, 2016). This is particularly problematic for perennial crops that have complex domestication history with possibly more than one origin and ongoing wild‐cultivated gene flows (Miller and Gross, 2011). We therefore made use of the demographic model inferred here to properly look for signatures of selection in the olive transcriptome.
Results
RNA‐sequencing and variant detection
RNA‐seq data were generated for 66 Mediterranean olive accessions and two accessions of O. e. cuspidata using an Illumina HiSeq2000 sequencer, resulting in a total of 2 billion reads of 100 bp with an average of 30 million reads per individual (Table S1). After removing low‐quality reads, trimming low‐quality bases and keeping only reads mapping to the reference transcriptome of var. ‘Arbequina’ (Sarah et al., 2017) with high quality (Methods S2), the average and median depth were 33.4× and 13.1× per accessions, respectively (Table S1). Variant calling led to 583 455 single nucleotide polymorphisms (SNPs) when the outgroup O. e. cuspidata was included, and to 536 341 SNPs when it was excluded. We eliminated four cultivated olive accessions identified as clones based on inferred relatedness (Figure S1) and therefore kept a total of 62 unique olive genotypes for downstream analyses. Variant calling on those samples resulted in 536 113 SNPs over 25 835 contigs.
Evolutionary history of olive tree domestication
Structure of nuclear and chloroplastic diversity
We examined population structure in our olive transcriptomic data using principal component analysis (PCAs) (Figure 1) and NGSadmix (Figure 2). A strong geographic structure between the West and East of the Mediterranean Basin was observed along with a subtler differentiation between wild and cultivated accessions (Figures 1, 2 and S2–S6). Eastern and western oleasters clustered into distinct groups in the admixture analysis (Figure 2; Appendix S1), and stretched the first principal component of the PCA (Figure 1). Many cultivars sampled in the East of the Mediterranean Basin could hardly be differentiated from eastern oleasters in the admixture analysis (Figure 2). They appeared distinct although close in the PCA with only one cultivated accession falling among the eastern oleasters (OGMed_019: cultivar Zard from Iran; Figure 1). The oleaster OS3 from Morocco clustered with cultivated accessions (Figure 1) and displayed a mixed ancestry (Figure 2). The distinction between western and eastern accessions was also supported by chloroplast lineages (Figure S7). Eastern and western oleasters displayed chlorotypes from lineages E1 and E2, respectively. Most cultivars carried a chlorotype from lineage E1, but some western cultivars carried chlorotypes from lineages E2 and E3, which had been previously reported to be western (Besnard et al., 2013b).
Figure 1.
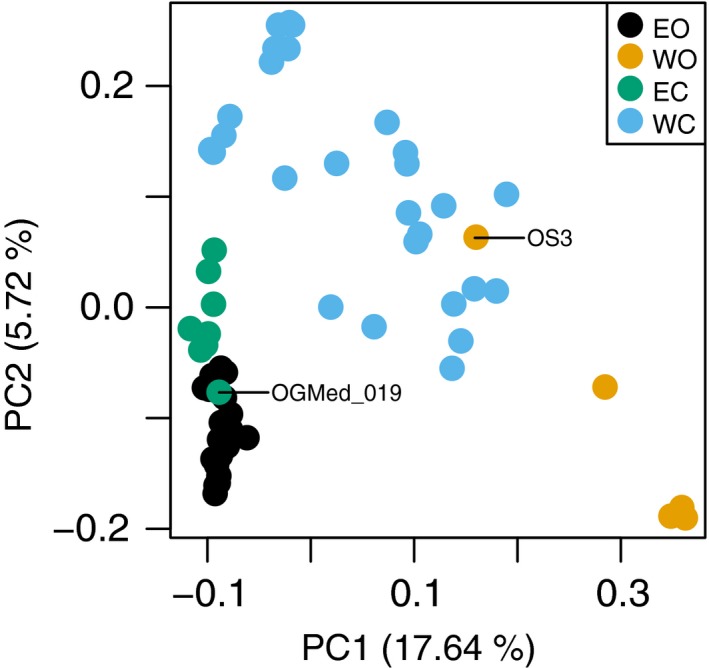
Principal component analysis plot (PCA) of the first two factors based on 536 341 SNPs genotyped in 62 olive accessions. Accessions are coloured according to the population they belong to as defined using their geographic origin, these results and the results from the admixture analysis and chloroplast genotyping (see text). EO, eastern oleasters; WO, western oleasters; EC, eastern cultivated olives; WC, western cultivated olives.
Figure 2.
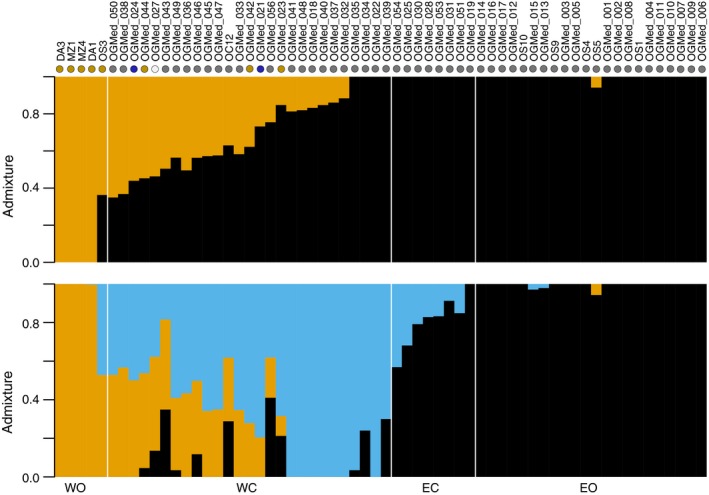
Admixture proportion in wild and cultivated olives (n = 62) with K = 2 and 3 inferred from 536 341 SNPs. On the top of the graph, coloured dots indicate the chlorotype of each accession. Grey: E1; brown: E2; blue: E3; white: L1 (Data S1). Accessions were split in four different populations: WC and EC for western and eastern oleasters, respectively, based on their geographic origin and WC and EC for western and eastern cultivars, respectively, based on PCA (Figure 1) and structure results (this figure) along with chloroplastic genotyping data (Data S1).
Based on these results, we split cultivated accessions into two populations: (1) a western cultivated population comprised of 27 accessions with <50% of ancestry from the eastern oleaster cluster in the admixture analysis with K = 3, and either chlorotypes from lineages E1, E2, E3 or L1 (Figure 2); and (2) an eastern cultivated population comprised of the remaining eight accessions that displayed ancestry mainly from the eastern oleaster clade (>50% with K = 3) and the eastern chlorotype lineage E1. This assignment matches the presumed geographic origin of cultivars (Table S1), with the eastern population including all cultivars from Iran, Syria, Lebanon, Egypt and Cyprus, and the western population all cultivars from Greece and further to the West. The only exception was accession OGMed_032 (cultivar Stanbouli), which was assigned to the western population while its presumed origin is Syria. But this misclassification was previously noted in an a microsatellite analysis (Haouane et al., 2011).
Genetic diversity
When grouped together, wild accessions displayed on average more diversity than cultivated accessions (Table 1), as predicted for crops (Gepts, 2004). We observed a loss of diversity of about 14% and a slight increase in Tajima's D in eastern cultivated accessions compared to eastern oleasters, compatible with a weak to moderate bottleneck during domestication. Western cultivated accessions displayed more variability than eastern cultivated accessions in all diversity estimators as well as a more positive Tajima's D than the other populations, likely due to its admixed origin (Figures 1 and 2).
Table 1.
Summary of population diversity statistics calculated on cultivated (1) and wild accessions (2) and on western and eastern subpopulations (a and b, respectively)
| Population | No. accessions | Site class | % π | % θW | Tajima's D | H E | H O | C |
|---|---|---|---|---|---|---|---|---|
| (1) Cultivated olives | 35 | Synonymous | 0.969 | 0.807 | 0.243 | 0.0110 | 0.00792 | 0.614 |
| Non‐synonymous | 0.374 | 0.339 | 0.160 | 0.0041 | 0.00263 | |||
| All sites | 0.576 | 0.490 | 0.423 | 0.00612 | 0.00395 | |||
| a) Western cultivated | 27 | Synonymous | 0.987 | 0.772 | 0.408 | 0.0121 | 0.00792 | 0.615 |
| Non‐synonymous | 0.380 | 0.317 | 0.411 | 0.00425 | 0.00263 | |||
| All sites | 0.582 | 0.463 | 0.693 | 0.00657 | 0.00395 | |||
| b) Eastern cultivated | 8 | Synonymous | 0.789 | 0.681 | 0.215 | 0.00920 | 0.00792 | 0.624 |
| Non‐synonymous | 0.297 | 0.268 | 0.224 | 0.00363 | 0.00263 | |||
| All sites | 0.448 | 0.392 | 0.434 | 0.00511 | 0.00395 | |||
| (2) Oleasters | 27 | Synonymous | 1.001 | 1.014 | −0.209 | 0.0114 | 0.00834 | 0.615 |
| Non‐synonymous | 0.385 | 0.448 | −0.494 | 0.00481 | 0.00263 | |||
| All sites | 0.589 | 0.621 | −0.270 | 0.00636 | 0.00398 | |||
| a) Western oleasters | 5 | Synonymous | 0.904 | 0.843 | 0.059 | 0.00936 | 0.00822 | 0.629 |
| Non‐synonymous | 0.335 | 0.322 | 0.017 | 0.00390 | 0.00263 | |||
| All sites | 0.497 | 0.468 | 0.197 | 0.00496 | 0.00394 | |||
| b) Eastern oleasters | 22 | Synonymous | 0.866 | 0.842 | −0.143 | 0.0106 | 0.00837 | 0.617 |
| Non‐synonymous | 0.332 | 0.368 | −0.393 | 0.00371 | 0.00263 | |||
| All sites | 0.508 | 0.512 | −0.178 | 0.00577 | 0.00398 |
π, Mean number of pairwise difference; θW, number of segregating sites; H E, expected heterozygosity; H O, observed heterozygosity; C, selective constraint.
The fixation index (F ST; Table 2) was the highest between western oleasters and eastern cultivated olives (21.8%). While the two oleaster populations were highly differentiated (16.7%), the two cultivated populations were much more similar (5.77%). Eastern cultivated olives appeared to be genetically very close to eastern oleasters (1.31%).
Table 2.
Pairwise genetic differentiation (F ST) and standard deviation between pairs of populations
| C | WO | WC | EC | EO | |
|---|---|---|---|---|---|
| O | 0.0243 ± 0.0559 | / | 0.0319 ± 0.0657 | 0.0107 ± 0.0838 | / |
| WO | 0.118 ± 0.239 | 0.000 | 0.116 ± 0.220 | 0.218 ± 0.307 | 0.167 ± 0.313 |
| WC | / | 0.116 ± 0.220 | 0.000 | 0.0559 ± 0.115 | 0.0577 ± 0.0883 |
| EC | / | 0.218 ± 0.307 | 0.0559 ± 0.115 | 0.000 | 0.0131 ± 0.0862 |
| EO | 0.0434 ± 0.0729 | 0.167 ± 0.313 | 0.0577 ± 0.0883 | 0.0131 ± 0.0862 | 0.000 |
C, cultivars (n = 35); EO, eastern oleasters (n = 22); EW, eastern cultivated olives (n = 8); O, oleasters (n = 27); WC, western cultivated olives (n = 27); WO, western oleasters (n = 5).
Inference of olive population splits and mixtures
We identified patterns of divergence and migration between the four olive populations with TreeMix, using the O. e. cuspidata individuals to root the tree (Figure 3). One migration event was required to explain >99.9% of the variance in the data (Figures S8 and S9) and results were consistent across five separate runs. The resulting tree confirms an initial domestication event in the East as eastern and western cultivars clustered together, separated from eastern oleasters by a very short branch only. However, the analysis also confirmed the admixed origin of western cultivars, which were estimated to have received 33.73% of their gene pool from western oleasters (Figure 3). To test for the robustness of this admixture signal, we repeated the analysis without the introgressed western oleaster (OS3), resulting in a 28.77% contribution of western oleasters to western cultivars. We further repeated the analysis by separating pure from admixed western cultivars based on the admixture analysis with K = 3. Both western cultivated populations clustered with the eastern oleasters and cultivars and we inferred a 40.60% contribution of western oleasters to the western cultivars (Figures S10 and S11).
Figure 3.
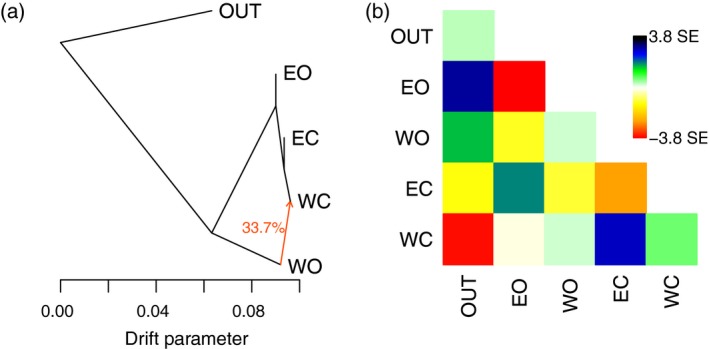
Patterns of divergence and migration between four olive populations with one migration event, as inferred by TreeMix. (a) Relationships between four populations; (b) Corresponding residuals of the model. OUT, O. e. cuspidata accessions; WO, western oleasters; EC, eastern cultivated accessions; WC, western cultivated accessions; EO, eastern oleasters.
Gene expression profiling
To avoid issues related to the independent construction of libraries (Conesa et al., 2016), we focused on the first sequencing batch, which contained data from all four populations studied here (Table S1). These data were generated by RNA‐sequencing both inflorescences and leaves.
Population structure and diversity in gene expression
We obtained expression profiles for each of the 19 accessions (Table S1) after filtering transcripts based on high intrapopulation variance and low read numbers, leading to 40 831 transcripts (90%). Hierarchical clustering on the expression profiles clustered the two known clones (OGMed_025 and OGMed_026) together, confirming the reliability of our expression analysis. We identified three major groups (Figure 4a). The first and most divergent group consisted of the western oleasters. The second group consisted of the eastern oleasters and eastern cultivars, which could not be separated into individual groups. The third groups consisted of western cultivars, which appeared intermediate between the other groups, in line with their admixed origin (Figures 1, 2, 3). Two samples, however, did not match this general pattern as the expression profile of both the western cultivar OGMed_018 and the introgressed western oleaster OS3 appeared nested within the eastern group. Although we observed a lower gene expression diversity in cultivars compared to oleasters, this difference was not significant (Student's t‐test, P = 0.056 and 0.093 for eastern and western accessions, respectively; Figure 4b).
Figure 4.
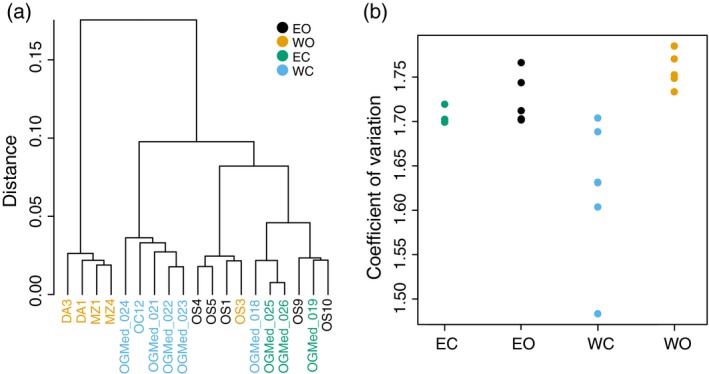
Gene expression data of 19 wild and cultivated olives over 40 831 transcripts. (a) Dendrogram of hierarchical clustering from gene expression data; (b) Gene expression diversity calculated as the level of gene expression variation for each sample. EO, eastern oleasters; WO, western oleasters; EC, eastern cultivars; WC, western cultivars.
Transcriptional differences in olives
Differentially expressed genes (DEGs) were identified in six pairwise comparisons (Table 3). Interestingly, the divergence in gene expression between western and eastern accessions (1280 DEGs) is comparable with that between oleasters and cultivated accessions (1269 DEGs). Additionally, the lowest number of DEGs among these six comparisons was found when comparing eastern oleasters to eastern cultivars, supporting a low differentiation between these two populations. Most of the DEGs identified between eastern oleasters and eastern cultivars were also divergent when comparing all wild and all cultivated olives (Figure 5). But only a fraction of the DEGs between western oleasters and western cultivated accessions were also divergent between all wild and all cultivated accessions.
Table 3.
Summary of the transcriptome‐wide differential of expression analysis performed on 40 831 transcripts profiled in 19 wild and cultivated olives
| Pairwise comparison | No. DEGs (total, upregulated, downregulated) | Enriched GO |
|---|---|---|
| Oleasters versus cultivated olives | 1269 |
GO:0000785: Chromatin; GO:0034728: Nucleosome organization; GO:0016837: Carbon‐oxygen lyase activity, acting on polysaccharides; GO:0006270: DNA replication initiation |
| 543 | / | |
| 726 |
GO:0000785: Chromatin; GO:0034728: Nucleosome organization; GO:0006270: DNA replication initiation; GO:0016837: Carbon‐oxygen lyase activity, acting on polysaccharides; GO:0006563: l‐serine metabolic process |
|
| Eastern oleasters versus eastern cultivated accessions | 566 | / |
| 233 | / | |
| 333 |
GO:0006270: DNA replication initiation; GO:0016837: Carbon‐oxygen lyase activity, acting on polysaccharides |
|
| Western oleasters versus western cultivated accessions | 749 | / |
| 257 | / | |
| 492 | / | |
| Western accessions versus eastern accessions (both wild and cultivated) | 1280 |
GO:0009648: Photoperiodism; GO:0009791: Post‐embryonic development; GO:0004486: Methylenetetrahydrofolate dehydrogenase activity; GO:0019238: Cyclohydrolase activity; |
| 433 | GO:0010557: Positive regulation of macromolecule biosynthetic process | |
| 847 |
GO:0009648: Photoperiodism; GO:0004486: Methylenetetrahydrofolate dehydrogenase activity; GO:0019238: Cyclohydrolase activity |
|
| Eastern oleasters versus western oleasters | 713 |
GO:0004486: Methylenetetrahydrofolate dehydrogenase activity; GO:0019238: Cyclohydrolase activity |
| 242 | / | |
| 471 |
GO:0004486: Methylenetetrahydrofolate dehydrogenase activity; GO:0019238: Cyclohydrolase activity |
|
| Eastern cultivated accessions versus western cultivated accessions | 571 | / |
| 195 | / | |
| 376 | / |
Figure 5.
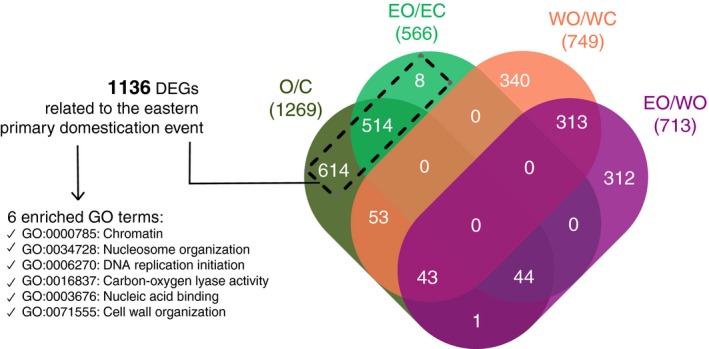
Overlap of differentially expressed genes (DEGs) identified in four differential expression analyses including a total of 19 wild and cultivated accessions. O/C: comparison between all oleasters and all cultivated accessions; EO/EC: comparison between eastern oleasters and eastern cultivars; WO/WC: comparison between western oleasters and western cultivars; EO/WO: comparison between eastern oleasters and western oleasters. Numbers in parentheses indicate the total number of DEGs for each of these four analyses. We identified 1136 genes differentially expressed in relation to the domestication process. These were DEGs either when comparing all oleasters and all cultivars or when comparing eastern oleasters and eastern cultivars or both but were removed from this list DEGs related to eastern/western geographic differentiation as identified by the analysis EO/WO and DEGs in relation to the divergence between western oleasters and western cultivars (WO/WC).
We identified primary domestication DEGs as genes differentially expressed between all wild and all cultivated accessions and/or eastern oleasters and eastern cultivated accessions, excluding genes differentially expressed when comparing eastern with western accessions (oleasters or cultivars) to correct for the high geographic differentiation in olives. Among these 1136 primary domestication DEGs, six Gene Ontology (GO) terms were significantly enriched (P < 0.05), of which four were related to transcriptional activity (Figure 5).
Detection of loci under selection in cultivated olives
No sign of relaxation of the selective constraint (C) on non‐synonymous sites was detected in domesticated olives with all four olive populations showing very similar values (Table 1).
To focus on the primary domestication event that took place in the East, and dispose of the effect of gene flows between western oleasters and the cultivated olive accessions, we first scanned the transcriptome for signatures of selection using eastern accessions only. Neither BayeScan nor PCAdapt detected outlier SNPs when comparing eastern oleasters with eastern cultivated accessions (Table 4 and Figure S12), nor did BayeScan identify outliers when including western cultivated accessions in the cultivated population.
Table 4.
Selection scans performed on the RNA‐seq olive dataset with BayeScan and PCAdapt
| Software | Population pairwise comparisons | Parameter | Threshold | No. outlier SNPs | No. contigs with outliers | No. enriched GO terms |
|---|---|---|---|---|---|---|
| BayeScan | Oleasters vs. cultivated | 100 | FDR = 0.05 | 30 | 22 | 0 |
| 1000 | FDR = 0.05 | 7 | 7 | 0 | ||
| Eastern oleasters vs. all cultivated | 100 | FDR = 0.05 | 0 | / | / | |
| 1000 | FDR = 0.05 | 0 | / | / | ||
| Eastern oleasters vs. eastern cultivated | 100 | FDR = 0.05 | 0 | / | / | |
| 1000 | FDR = 0.05 | 0 | / | / | ||
| Eastern accession vs. western accessions | 100 | FDR = 0.05 | 1 | 1 | 0 | |
| 1000 | FDR = 0.05 | 0 | / | / | ||
| Eastern cultivars vs. western cultivars | 100 | FDR = 0.05 | 0 | / | / | |
| 1000 | FDR = 0.05 | 0 | / | / | ||
| Eastern oleasters vs. western oleasters | 100 | FDR = 0.05 | 0 | / | / | |
| 1000 | FDR = 0.05 | 0 | / | / | ||
| PCAdapt | All olives | factors 1‐2 | FDR = 0.01 | 3211 | 1119 | 298 |
| All olives | factor 2 | top 1% | 3000 | 1649 | 97 | |
| Eastern olives | factor 1 | FDR = 0.01 | 0 | / | / |
The parameter column in the case of BayeScan indicates the value set for the prior odds. For PCAdapt, it indicates the principal component used to find outlier loci.
Under a model contrasting 27 wild against 35 cultivated olives, BayeScan identified 30 and seven SNPs as potentially under selection with a prior odds of 100 and 1000, respectively (Table 4 and Table S2; Figure S13). Nevertheless, these outliers could reflect the high differentiation between western cultivars and eastern oleasters that is not due to domestication but to their admixture with the highly differentiated western oleaster gene pool. To correct the candidate genes list, we therefore identified SNPs and contigs exhibiting large F ST between eastern and western accessions. Comparing all eastern with all western accessions, BayeScan identified a single outlier SNP, while when taking cultivars or oleasters only, the comparisons led to no outlier SNP. This single outlier did not overlap with the 30 and seven from the oleaster/cultivar comparison indicating that the oleaster/cultivar comparison could actually provide insights into the domestication event. Nevertheless, no enriched GO term was identified among these outliers SNPs (Table 4).
PCAdapt identified 3211 candidate SNPs that were atypically associated with both PC1 and PC2 (FDR < 0.01) when ran on all accessions together. PC1 captures the geographic partitioning of olive diversity while PC2 actually relates to the distinction between oleasters and cultivated olives (Figure 1). Therefore, to identify SNPs related to domestication, but not to the geographic differentiation, we focused on the top 1% hits associated to PC2 only. These 3000 SNPs were distributed over 1648 transcripts enriched for 97 GO terms (Figure S14). Enriched GO terms for cellular components were mostly related to chromosome organization, transcription complex as well as endomembrane structures (lipid bilayer) while enriched GO terms for molecular function were mostly related to protein synthesis (mRNA transcription and translation) and growth (growth factor binding, tubulin binding and cytoskeletal protein binding).
Ten of these outlier transcripts also contained an SNP identified by BayeScan and are therefore the strongest candidates for selection (Figure 6). Their sequence similarity to the nucleotide collection of NCBI was identified with blastn using default parameters (Zhang et al., 2000; Morgulis et al., 2008): three transcripts are proteins involved in transcriptional and translational activities and two are involved in cell cycle (Table S3).
Figure 6.
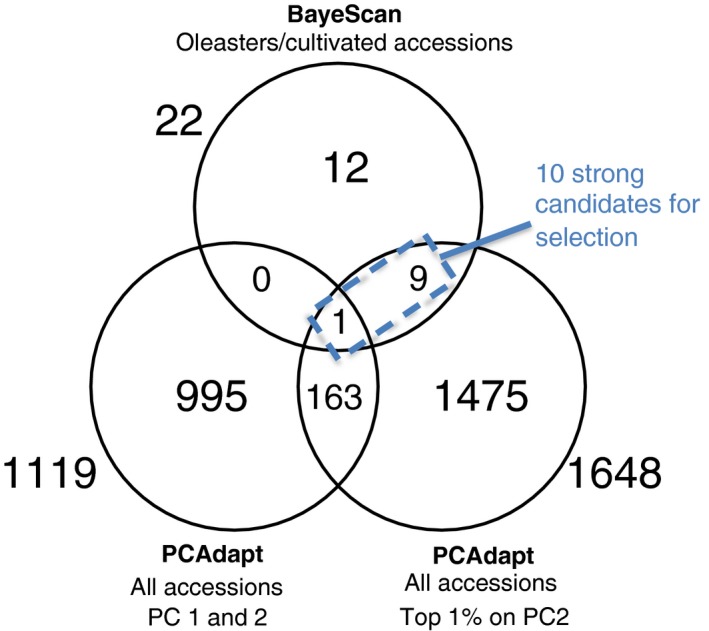
Overlap between selection scan analyses. The 10 contigs found as outliers in both BayeScan (all accessions) and PCAdapt (all accessions, top 1% on PC2) can be considered as strong candidates for selection.
Differentially expressed transcripts do not show more sign of selection
We investigated whether genes showing divergent expression in relation to the primary eastern domestication event were also identified as likely targets of selection. The intersection of the top 1% transcripts associated to PC2 in the PCAdapt analysis combining all samples (3000 SNPs on 1648 contigs) with the DEGs in all, eastern only and western only accessions (Table 3) led to, respectively, 73, 35 and 32 overlapped contigs. These numbers are, however, not different from what is expected by chance (hypergeometric test; P = 0.116). Additionally, when compared with non‐DEGs, they did not show a larger differentiation (as calculated using F ST between eastern wild and cultivated accessions, Wilcoxon rank sum test; P > 0.05) nor a decrease in nucleotide diversity, whether synonymous or non‐synonymous, in the cultivated accessions (Wilcoxon rank sum test; P > 0.05).
Discussion
Here, we studied the evolutionary history of the olive using transcriptomic data of both wild and cultivated accessions from all around the Mediterranean basin. Oleasters were carefully selected to represent genuinely wild olives not heavily impacted by human activities (Besnard and Rubio de Casas, 2016). Their wild origin was corroborated through their high diversity in cpDNA haplotypes and their genetic differentiation observed at the nuclear genome.
Insights into the domestication history of olives
East/West geographic structure in olives and admixture in the cultivated gene pool
Expression data, transcriptomic variants and chloroplastic data all confirmed the previously described high East/West differentiation observed in olives (Besnard et al., 2013a; Díez et al., 2015), in line with the hypothesis of a geographic structure older than the domestication process (Besnard et al., 2013b). The diversity of cultivated olives also displayed an East/West structure, but in contrast with oleasters, there is extensive admixture between these two gene pools, as already evidenced (Besnard et al., 2013b; Díez et al., 2015; Khadari and El Bakkali, 2018).
Based on few markers, previous investigations identified two cultivated populations (namely eastern and western) (Besnard et al., 2001b), or three (namely western, central and eastern populations, Haouane et al., 2011; Belaj et al., 2012; Díez et al., 2012, 2015; El Bakkali et al., 2013). Our SNP dataset identified two major groups: cultivated accessions sampled in the East of the Mediterranean as only marginally differentiated from eastern oleasters in terms of nuclear diversity, and those from the West and Central Mediterranean basin that had mixed ancestry and were difficult to be further differentiated into separate populations.
Support for a single primary domestication in olives
We re‐examined the debate of single versus multiple domestication event(s) in olives, and several lines of evidence corroborate the occurrence of a dominant event in the eastern part of the Mediterranean basin followed by human‐mediated spread towards the West and introgression by western oleasters. First, there was weak differentiation between eastern oleasters and eastern cultivated accessions observed in both expression and SNPs data along with a slight reduction in diversity observed in the domesticated gene pool. Agronomically interesting oleasters may therefore have been put into cultivation in this region through cloning directly or after only a few generations of (conscious or unconscious) selection. Conversely, we could not identify any cultivar unambiguously assigned to the western group as they are all at least admixed between the two gene pools. Second, most cultivars (82%), both eastern and western, carried a chlorotype that is restricted to the eastern part of the Mediterranean basin in oleasters. This situation indicates that, for a majority of olive cultivars, the maternal origin can be traced to the East of the distribution. Thirdly, in the analysis of population split and mixtures, the western cultivars grouped with eastern oleasters and cultivated accessions rather than grouped with western oleasters, even when the western cultivated population was split into two populations. Lastly, western cultivated accessions are also closer to eastern wild and cultivated olives than to western oleasters in term of expression patterns. We therefore argue in favour of a single domestication event in olive in the eastern region of the Mediterranean basin as previously proposed (Besnard et al., 2013a; Khadari and El Bakkali, 2018). But we do recognize a secondary contribution of western oleasters in the cultivated gene pool of the western and central Mediterranean basin. Western cultivars indeed showed up to 60% of admixture from the western oleasters while a minority displayed a chlorotype only found in western oleasters (see Besnard et al., 2013b for a large sampling of cultivars from this area). During the expansion of olive cultivation to the West, domesticated olives were therefore repeatedly crossed with western oleasters.
This model of domestication, with introgression from wild relatives leading to cultivars closely related to their secondary progenitor, has already been observed in other perennials such as grape (Myles et al., 2011), apple (Cornille et al., 2012) and date palm (Gros‐Balthazard et al., 2017; Flowers et al., 2019).
Based on our dataset, we did not retrieve the scenario of independent domestication in the central Mediterranean basin favoured by Díez et al. (2015) who, based on microsatellite data, suggested a separate olive domestication in the central Mediterranean basin. However, their conclusion might have been driven in part by eastern oleaster accessions that turned out to be feral rather than genuinely wild (Díez et al., 2015; Díez and Gaut, 2016). Also, such an additional domestication, if it existed, must have been rather localized as it did not leave any detectable trace in neither the western nor eastern cultivars analyzed here.
The consequence of domestication in the olive transcriptome
Reduction in nucleotide diversity but not in expression diversity
On average, cultivated olives retained 97% of the diversity found in oleasters. When only considering eastern populations, this number falls to 86% and 77% for nucleotide diversity (π) and the number of segregating sites (θW), respectively. This deficit is comparable with that found in other perennials (94.6% on average; Miller and Gross, 2011). For instance, genomic data indicated that cultivated grapes retained about 95% of the diversity found in its wild progenitor (Zhou et al., 2017). This limited loss of diversity in perennials is related to the low number of generations since domestication and ongoing gene flow between wild populations and domesticates (Miller and Gross, 2011; Gaut et al., 2015). Interestingly, the western cultivars showed the largest nucleotide diversity, in line with their mixed ancestry including both eastern cultivated accessions and western oleasters. This high level of genetic diversity following a secondary contact of the cultivated gene pool with other wild relative populations has already been described in olives (Díez et al., 2015) and in other crops such as the grape vine (Myles et al., 2011) or the date palm (Gros‐Balthazard et al., 2017; Flowers et al., 2019).
In contrast with the nucleotide diversity, however, we did not find a significant reduction in the diversity of gene expression. While such a reduction is reported for annual crops such as rice (5.1% reduction; Liu et al., 2019) or common bean (18% reduction; Bellucci et al., 2014), it might be a result of the generally weaker domestication syndrome in perennial plants. However, an important limitation of our result is the restricted number of samples used for the expression analysis, and further observations might provide a more robust outcome.
Altered gene expression in cultivated olives
We found evidence of an important transcriptome rewiring due to domestication. Indeed, many transcripts exhibited differential expression when comparing wild and cultivated olives, whether using the full dataset, eastern olives or western olives separately. The observed fraction of DEGs linked to olive domestication (1.39%) is comparable with that found in maize (2.18%; Swanson‐Wagner et al., 2012) or in common bean (0.7%; Bellucci et al., 2014).
We identified six GO terms that were enriched in DEGs related to domestication. Of those, four were associated with transcriptional activity. This result is consistent with previous findings underlying an over‐representation of transcription regulators among domestication genes (Doebley et al., 2006; Meyer and Purugganan, 2013; Olsen and Wendel, 2013). We did not identify any DEGs directly related to oil content or fruit/seed size, traits expected to have been impacted by domestication. However, we note that the expression data analyzed here were obtained from leaves and inflorescences rather than fruits. Nevertheless, we provide evidence of olive transcriptome reshaping by domestication, motivating the expression profiling of other tissues, especially the fruit, to further understand how the olive transcriptome was affected.
Lack of signatures of selection in the olive transcriptome
Estimates of the extent to which artificial selection has shaped crop genomes mostly stem from annuals and include ~7.6% for maize (Hufford et al., 2012), ~9% for the common bean (Bellucci et al., 2014) and ~7.3% in sunflower (Chapman et al., 2008). Estimates are lower for the perennial apple, for which regions selected during the initial phase of domestication or secondary introgression covered only 3.7% of the genome (Duan et al., 2017).
Here we report even weaker selection on protein‐coding genes in olives. Both statistical approaches used (PCAdapt and BayeScan) to contrast eastern oleasters and eastern cultivated accessions failed to identify candidate genes for domestication, and only 10 (0.04%) putative selected genes were detected when using the full dataset. While our estimates are based on transcriptomes rather than full genomes, the very weak selection inferred here is a likely result of olive life history. Indeed, olives are self‐incompatible with long generation times, and are extensively propagated clonally while still exchanging genes with oleasters (Rugini et al., 2016), all limiting the possibility to impose strong selection regimes. But we note that our selection scans were underpowered to detect selection on highly polygenic traits (Narum and Hess, 2011) that were proposed to contribute substantially to the domestication in other crops (Olsen and Wendel, 2013). In addition, and given the long generation time, selection during domestication was likely to act primarily on standing variation rather than on de novo mutations (Stetter et al., 2017), therefore leaving even softer signatures of selection that are harder to detect (Przeworski et al., 2005).
The olive domestication syndrome driven by gene expression changes
In contrast with the weak signature of selection we found on coding genes, there was a much higher proportion of changes in gene expression potentially associated with olive domestication. While we detected only 0.04% of putative genes under selection, we found that 1.39% of these same genes were differentially expressed in wild and cultivated eastern olives. Several non‐exclusive hypotheses may explain how the expression patterns were rewired during olive domestication without leaving strong signatures of selection in the transcriptome. First, gene expression may have changed as a result of drift, in relation to the demography of olives, rather than artificial selection. Second, and given the polygenic nature of gene regulation with a large numbers of transcription factors modulating the expression of single genes (Chen and Rajewsky, 2007), our data set was underpowered to detect selection signals. Third, mutations in cis‐regulatory regions (CREs) played a major role in changing expression patterns. CREs are known to be major targets of evolutionary changes, due to their weaker deleterious pleiotropic effects compared with mutations in coding sequences (Stern and Orgogozo, 2008), as was exemplified in annual crops (Lemmon et al., 2014; Wang et al., 2017; but see Rhoné et al., 2017). Yet, they were not captured by our transcriptomic data and, in case of rapid decay of linkage disequilibrium, signatures of selection could not extend up to the coding regions we sequenced. Lastly, epigenetic changes, maintained through vegetative propagation (Richards, 2011), can also contribute to gene expression changes associated with domestication as was recently shown for agronomically important traits such as fruit ripening in tomato (for review, see Gallusci et al., 2016; Piperno, 2017). Interestingly, siRNA involved in epigenetic processes were recently discovered to be involved in oil biosynthesis in olive (Unver et al., 2017).
Phenotypic differences between wild and cultivated olives may therefore have emerged from divergent gene regulation rather than changes in protein function through fixed difference in amino acid sequence of structural genes (aka gene coding for any RNA or protein product other than a regulatory factor). This finding is consistent with the evolutionary history and the life history of olives. Indeed, in perennials, there has been a limited number of generations since the onset of artificial selection (due to long generation time and clonal propagation) and previous experimental evolution studies showed that the response to artificial selection occurs primarily through gene expression changes rather than changes of allele frequencies at coding sites (Konczal et al., 2015). Additionally, it was shown in steelhead trout (Oncorhynchus mykiss) that the expression of hundreds of genes was heritably altered after just a single generation of domestication (Christie et al., 2016). The weak domestication syndrome observed in olives therefore probably arose following heritable changes of gene regulation.
Although more challenging than studying annual crops, further studies of the genomic of the domestication process in perennial should broaden our understanding of the genomic bases and dynamics of adaptation, especially to understand whether they are influenced by the life history and ecological traits of a species.
Experimental procedures
Plant material
We analyzed a total of 68 accessions of Olea europaea L., representing 27 oleasters [Olea europaea subsp. europaea var. sylvestris (Mill.) Lehr], 39 cultivated olive trees (Olea europaea subsp. europaea var. europaea), and two accessions of African olive [Olea europaea subsp. cuspidata (Wall. ex G.Don) Cif.] (Table S1).
Eastern and western oleasters were sampled to represent the two wild gene pools previously evidenced (Besnard et al., 2001b, 2013b; Breton et al., 2006). We sampled 17 oleasters in the area south of the Taurus mountains (South Turkey near the border with Syria), a region considered as very close to the primary centre of olive domestication (Besnard et al., 2013b) (Table S1). Five additional eastern oleasters were sampled in the CBNMed collection at Porquerolles Island, France: two from Western Turkey, and three from Northwestern Syria. Five western oleasters were sampled in three different locations in Morocco, with one of these conserved at the CBNMed. To minimize admixture with cultivated olives, most of these wild forms were sampled in natural areas far from olive agroecosystems. The sampled oleasters displayed smaller fruits characterized by less fleshy mesocarp than cultivated olives; yet this characteristic may also be found in feral individuals (Hannachi et al., 2008). Therefore, we characterized their chloroplastic (cpDNA) variation (Methods S1 and Data S1) as true oleasters are expected to harbour a higher diversity of plastid profiles with many haplotypes never observed in cultivated olives (Besnard et al., 2013b).
The 39 cultivated accessions were sampled in three germplasm collections (Table S1) and were chosen to maximize the Mediterranean diversity (El Bakkali et al., 2013). Three pairs of accessions (OGMed_025/OGMed_026 from Cyprus, OGMed_029/OGMed_046 from Lebanon and Italy, and OGMed_018/OGMed_055 from Morocco) sharing the same nuclear microsatellite profile (El Bakkali et al., 2013) were considered as clones and used to calibrate the relatedness and differential gene expression analyses.
RNA‐sequencing, read alignments and variant calling
We performed three sequencing runs in 2011, 2013 and 2015 to obtain a total of 68 Olea spp. transcriptomes (Table S1). Total RNA was extracted from leaves and/or inflorescence tissues. The construction of Illumina libraries and sequencing protocols are described in Sarah et al., (2017). The data obtained for the cultivar ‘Arbequina’ and on a set of 10 wild olive trees and for one accession O. e. cuspidata were previously used by Sarah et al. (2017) and Clément et al. (2017) (see Table S1).
We used a custom bioinformatic pipeline, following in large the standard Illumina pipeline, to process raw reads, obtain alignment files and generate variant call files (Figure S15 and Methods S2). In brief, raw Illumina reads were filtered based on quality before mapping on the annotated olive transcriptome assembly of var. ‘Arbequina’ (Sarah et al., 2017) with BWA (Li and Durbin, 2010). Keeping only high quality alignments, we performed local realignment near InDels using GATK (McKenna et al., 2010). Variants were called using GATK and filtered following different criteria (Methods S2).
Genetic structure and diversity in wild and cultivated olives
We first inferred relatedness between each pair of olive accessions (Methods S3). We estimated individual ancestries using NGSadmix (Skotte et al., 2013) on all olive accessions and on wild and cultivated accessions separately with a minimum minor allele frequency of 0.05. Input files were directly generated from alignment files with ANGSD v.0.911 (Korneliussen et al., 2014) using the genotype likelihood estimation as implemented in SAMtools (Li et al., 2009). A PCA was performed using genotype calls as input and considering a minimum minor allele frequency of 0.05 with the PCAdapt R package v.3.0.2 (Duforet‐Frebourg et al., 2014, 2015; Luu et al., 2017). We used TreeMix v.1.12 (Pickrell and Pritchard, 2012) to discriminate trees between wild and cultivated populations, and identify gene flows between them.
Genetic variation within O. europaea was assessed using different statistics. We calculated observed and expected heterozygosities (H O and H E), the mean number of pairwise differences π (Tajima, 1983), the Watterson estimator θW (Watterson, 1975), and Tajima's D (Tajima, 1989) based on site frequency spectra (SFSs) inferred with ANGSD v.0.911 (Korneliussen et al., 2014), as detailed in Methods S4. These statistics were calculated for all sites and on both synonymous and non‐synonymous sites annotated as described in Methods S5. In addition, the partitioning of diversity within and between populations, measured with F ST (Weir and Cockerham, 1984), was calculated using VCFtools (Danecek et al., 2011).
Gene expression in wild and cultivated olives
To avoid issues related to the independent construction of libraries (Conesa et al., 2016), we focused on the first sequencing run, as it contained data from all four populations studied here (Table S1). We therefore included five oleasters from both eastern and western Mediterranean basin, along with three and six accessions of eastern and western cultivated olives, respectively. Read counts (the number of read pairs aligned to each transcript) were calculated using the idxstats option of SAMtools. We excluded genes with very low expression levels (< 30 mapped reads across all 19 sequenced libraries) and high within‐group variance (> 100 000 in wild or cultivated populations). After data normalization using the DESeq2 package from Bioconductor (Anders and Huber, 2010; Love et al., 2014), we clustered gene expression hierarchically using hclust in R software (R Core Team, 2015). We inferred gene expression diversity for each sample by calculating the coefficient of variation (standard deviation divided by the mean) of the expression values (Bellucci et al., 2014). We tested for differences in gene expression diversity between populations using Student's t‐tests. We then identified DEGs with DESeq2 which fits a generalized linear model (GLM) modelling read counts per gene and samples using a negative binomial distribution. P‐values were computed using a Wald test (Love et al., 2014) and transformed into q‐values to correct for multiple testing using the R package qvalue (Storey, 2002; Bass et al., 2015). Genes were considered as differentially expressed if they showed a q‐value ≤ 0.01.
GO terms enriched in differentially expressed transcripts (functionally annotated by Sarah et al., 2017) were established with the R package goseq (Young et al., 2010), using a Wallenius distribution as null distribution and a false‐discovery threshold of 0.05 (Benjamini and Hochberg, 1995).
Genes under selection during domestication
We first measured the strength of the selective constraint C = 1 – (πNS/πS) (Keightley et al., 2011, also known as evolutionary constraint) from the diversity at non‐synonymous (πNS) and synonymous sites (πS), assuming the latter to be neutral.
To minimize false positives, we accounted for the demographic history of olives when identifying molecular signatures of selection using two complementary approaches (Lotterhos and Whitlock, 2014; Vatsiou et al., 2016). First, we used a Bayesian approach implemented in BayeScan v.2.1 (Foll and Gaggiotti, 2008) to detect SNPs with unusually high F ST values, which is a hallmark of divergent selection. This analysis was performed on different sets of populations and we ran BayeScan twice for each model with prior odds of the neutral model set to 100 and 1000, respectively. We used an initial burn‐in period of 5000 steps. After 20 pilot runs of 5000 iterations each, we sampled 5000 parameter combinations with a thinning interval of 10, resulting in a total of 100 000 iterations. The probability that a locus is under selection was estimated by calculating the Bayes factor of a model with selection over a neutral model and loci with a q‐value < 0.05 were considered as potential candidates for selection. Second, and because the presence of admixed individuals may impact the power of BayeScan (Luu et al., 2017), we also identified candidate SNPs using PCAdapt v2.2.1, a hierarchical Bayesian approach (Duforet‐Frebourg et al., 2014, 2015). PCAdapt first performs a PCA to infer population structure and then identifies loci that are atypically related to a specific principal component as measured by the latent factors. We corrected for multiple testing by transforming p‐values associated with each locus into q‐values (Storey, 2002) using the R package qvalue v.2.4.2 (Bass et al., 2015) and used a false‐discovery threshold of 0.01.
To gain a deeper biological insight into the candidate SNPs retrieved from these selection scans, we performed GO enrichment analysis using the R package SNP2GO version 1.0 (Szkiba et al., 2014) using default parameters. The lists of enriched GO terms were summarized and visualized using REVIGO (Supek et al., 2011).
Finally, we tested whether identified DEGs are more frequently identified as targets of selection than non‐DEGs by comparing their F ST and π values using Wilcoxon rank sum tests.
Accession numbers
The data were deposited in the Sequence Read Archive (SRA) under the Bioproject accession PRJNA525000.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
MG‐B, GB, SG and BK designed the research. MG‐B performed data analysis and interpretation and wrote the manuscript with the help of GB, DW, SG and BK. GB, SG YH, JL, SS, DW, SG and BK contributed to data analysis and interpretation. BK collected the material.
Supporting information
Figure S1. Heatmap displaying the relatedness between the 66 Mediterranean olive tree accessions.
Figure S2. Admixture proportions in 62 olive trees inferred from genotype likelihoods of 536 341 SNPs with NGSadmix for K = 4–6.
Figure S3. Admixture proportions inferred from genotype likelihoods with NGSadmix for K = 2–6.
Figure S4. Log‐likelihood estimates for different K values in the three admixture analyses.
Figure S5. Proportion of variance explained by the 10 first principal components of the PCA performed on 62 olive tree accessions.
Figure S6. Principal component analysis of 62 Olea europaea subsp. europaea.
Figure S7. Reduced median network of chlorotypes reconstructed with network v.5 using data from Data S1.
Figure S8. Fraction of variance in relatedness between four olive tree populations accounted for by phylogenetic models with zero through two migrations in TreeMix.
Figure S9. TreeMix relationships between oleasters and cultivars from all around the Mediterranean basin grouped in four populations.
Figure S10. TreeMix relationships between five olive tree populations.
Figure S11. Fraction of variance in relatedness between five olive tree populations accounted for by phylogenetic models with zero through two migrations in TreeMix.
Figure S12. Bayesian test for selection between 27 wild and 36 cultivated populations implemented in BayeScan.
Figure S13. Principal component analysis of SNPs data.
Figure S14. Visualizations of enriched GO terms from PCAdapt selection scan including all accessions.
Figure S15. Bioinformatic pipeline followed to process the raw reads into alignment files (final.bam files) and subsequently variant call format file (.vcf file) used in subsequent analyses.
Table S1. Summary of the 68 Olea europaea spp. accessions including their origin, the cultivar name when relevant and statistics related to sequencing (excel file).
Table S2. List of SNPs detected as outliers in BayeScan when comparing all oleasters and all cultivated accessions.
Table S3. Strong candidate transcripts for selection.
Methods S1. Plastid genome profiling.
Methods S2. Read alignments and variant calling.
Methods S3. Inference of relatedness among accessions.
Methods S4. Inference of site frequency spectra and calculation of diversity statistics.
Methods S5. Annotation of the transcriptome reference sites as synonymous or non‐synonymous.
Data S1. Chloroplastic microsatellite and CAPS profile of each individual analysed in the present study [according to the method developed by Besnard et al. (2011) (excel file).
Appendix S1. Detailed structure and diversity of oleaster populations.
Acknowledgements
This work was supported by Agropolis Fondation through the program OliveMed under the reference ID 1403‐026, «Investissements d'avenir» program (Labex Agro:ANR‐10‐LABX‐0001‐01). MGB has received the support of the EU in the framework of the Marie‐Curie FP7 COFUND People Programme, through the award of an AgreenSkills fellowship under grant agreement no. 267196. G.B. is member of the EDB laboratory supported by the excellence projects Labex CEBA (ANR‐10LABX‐25‐01) and Labex TULIP (ANR‐10LABX‐0041), managed by the French ANR.
We thank Sylvia Lochon‐Menseau, Daniel Bielmann, Bruno Bernazeau (CBNMed collection at Porquerolles, France), Hayat Zaher, Abdelmajid Moukhli, Sikaoui lhassane (Word Olive Germplasm Bank of Marrakech, Morocco), and the staff of the experimental field of the Platform "Terrains d'Expériences" of the LabEx CeMEB (Montpellier, France); Mohammed Ater, Melek Gurbuz, Mehmet Ulas and Mehmet Hakan for helping us in the wild olive sampling from in situ areas; Audrey Veber for helping with the RNA extraction and library preparation for sequencing; Christine Tollon for chloroplast DNA genotyping; Bertrand Pitollat, Stéphanie Sidibe‐Bocs and Gaëtan Droc for their support in the bioinformatic analyses; Yann Bourgeois, Athanasios Kousathanas, François Sabot, Baptiste Guitton, Jean‐Jacques Kelner, Rémy Tournebize, Yves Vigouroux, Olivier François and Eric Frichot for constructive discussions; Michael Blum, Keurcien Luu and Nicolas Duforet‐Frebourt for their help with PCAdapt software; Christopher Sauvage for providing with scripts for gene expression analysis and helpful discussions; Emeline Pobelle for graphic design and Philip P. Rodenbough for proofreading.
Contributor Information
Muriel Gros‐Balthazard, Email: muriel.grosb@gmail.com.
Bouchaib Khadari, Email: khadari@supagro.fr.
References
- Anders, S. and Huber, W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, 1344–1349. 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atienza, S.G. , de la Rosa, R. , León, L. , Martín, A. and Belaj, A. (2014) Identification of QTL for agronomic traits of importance for olive breeding. Mol. Breed. 34, 725–737. 10.1007/s11032-014-0070-y. [DOI] [Google Scholar]
- Barghini, E. , Natali, L. , Cossu, R.M. , Giordani, T. , Pindo, M. , Cattonaro, F. , Scalabrin, S. , Velasco, R. , Morgante, M. and Cavallini, A. (2014) The peculiar landscape of repetitive sequences in the olive (Olea europaea L.) genome. Genome Biol. Evol. 6, 776–791. 10.1093/gbe/evu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini, G. , Prevost, G. , Messeri, C. and Carignani, G. (1998) Olive germplasm: cultivars and world‐wide collections. Roma, Italy: FAO, 459 pp. [Google Scholar]
- Bass, A.J. , Storey, J.D. , Dabney, A. and Robinson, D. (2015) qvalue: Q‐value estimation for false discovery rate control. R package version 2.14.0, http://github.com/jdstorey/qvalue.
- Belaj, A. , Dominguez‐García, M. del C. , Atienza, S.G. et al (2012) Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes, 8, 365–378. 10.1007/s11295-011-0447-6 [DOI] [Google Scholar]
- Bellucci, E. , Bitocchi, E. , Ferrarini, A. et al (2014) Decreased nucleotide and expression diversity and modified coexpression patterns characterize domestication in the common bean. Plant Cell, 26, 1901–1912. 10.1105/tpc.114.124040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Sadok, I. , Celton, J.‐M. , Essalouh, L. , El Aabidine, A.Z. , Garcia, G. , Martinez, S. , Grati‐Kamoun, N. , Rebai, A. , Costes, E. and Khadari, B. (2013) QTL mapping of flowering and fruiting traits in olive. PLoS ONE, 8, e62831 10.1371/journal.pone.0062831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B, 57, 289–300. [Google Scholar]
- Besnard, G. and Rubio de Casas, R. (2016) Single vs multiple independent olive domestications: the jury is (still) out. New Phytol. 209, 466–470. 10.1111/nph.13518. [DOI] [PubMed] [Google Scholar]
- Besnard, G. , Baradat, P. and Bervillé, A. (2001a) Genetic relationships in the olive (Olea europaea L.) reflect multilocal selection of cultivars. Theor. Appl. Genet. 102, 251–258. [Google Scholar]
- Besnard, G. , Baradat, P. , Breton, C. , Khadari, B. and Bervillé, A. (2001b) Olive domestication from structure of oleasters and cultivars using nuclear RAPDs and mitochondrial RFLPs. Genet. Sel. Evol. 33, S251–S268. [Google Scholar]
- Besnard, G. , El Bakkali, A. , Haouane, H. , Baali‐Cherif, D. , Moukhli, A. and Khadari, B. (2013a) Population genetics of Mediterranean and Saharan olives: geographic patterns of differentiation and evidence for early generations of admixture. Ann. Bot. 112, 1293–1302. 10.1093/aob/mct196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard, G. , Hernández, P. , Khadari, B. , Dorado, G. and Savolainen, V. (2011) Genomic profiling of plastid DNA variation in the Mediterranean olive tree. BMC Plant Biol. 11, 80 10.1186/1471-2229-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard, G. , Khadari, B. , Navascués, M. et al (2013b) The complex history of the olive tree: from Late Quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proc. R. Soc. Ser. B 280, 20122833 10.1098/rspb.2012.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard, G. , Terral, J.‐F. and Cornille, A. (2018) On the origins and domestication of the olive: a review and perspectives. Ann. Bot. 121, 385–403. 10.1093/aob/mcx145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton, C. , Tersac, M. and Bervillé, A. (2006) Genetic diversity and gene flow between the wild olive (oleaster, Olea europaea L.) and the olive: several Plio‐Pleistocene refuge zones in the Mediterranean basin suggested by simple sequence repeats analysis. J. Biogeogr. 33, 1916–1928. 10.1111/j.1365-2699.2006.01544.x. [DOI] [Google Scholar]
- Breton, C. , Terral, J.F. , Pinatel, C. , Médail, F. , Bonhomme, F. and Bervillé, A. (2009) The origins of the domestication of the olive tree. C. R. Biol. 332, 1059–1064. 10.1016/j.crvi.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Carmona, R. , Zafra, A. , Seoane, P. et al (2015) ReprOlive: a database with linked data for the olive tree (Olea europaea L.) reproductive transcriptome. Front. Plant Sci. 6, 625 10.3389/fpls.2015.00625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión, Y. , Ntinou, M. and Badal, E. (2010) Olea europaea L. in the north Mediterranean Basin during the Pleniglacial and the Early–Middle Holocene. Quat. Sci. Rev. 29, 952–968. 10.1016/j.quascirev.2009.12.015. [DOI] [Google Scholar]
- Chapman, M.A. , Pashley, C.H. , Wenzler, J. , Hvala, J. , Tang, S. , Knapp, S.J. and Burke, J.M. (2008) A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus). Plant Cell, 20, 2931–2945. 10.1105/tpc.108.059808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. and Rajewsky, N. (2007) The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 8, 93–103. 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Christie, M.R. , Marine, M.L. , Fox, S.E. , French, R.A. and Blouin, M.S. (2016) A single generation of domestication heritably alters the expression of hundreds of genes. Nat. Commun. 7, 10676 10.1038/ncomms10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément, Y. , Gautier, S. , Holtz, Y. et al. (2017) Evolutionary forces affecting synonymous variations in plant genomes. PLoS Genet. 13, e1006799 10.1371/journal.pgen.1006799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa, A. , Madrigal, P. , Tarazona, S. et al (2016) A survey of best practices for RNA‐seq data analysis. Genome Biol. 17, 13 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team, R. (2015) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Cornille, A. , Gladieux, P. , Smulders, M.J. et al (2012) New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet. 8, e1002703 10.1371/journal.pgen.1002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, F. , Julca, I. , Gómez‐Garrido, J. et al (2016) Genome sequence of the olive tree Olea europaea . GigaScience, 5, 29 10.1186/s13742-016-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P. , Auton, A. , Abecasis, G. et al (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la O Leyva‐Pérez, M. , Valverde‐Corredor, A. , Valderrama, R. , Jiménez‐Ruiz, J. , Muñoz‐Mérida, A. , Trelles, O. , Barroso, J.B. , Mercado‐Blanco, J. and Luque, F. (2015) Early and delayed long‐term transcriptional changes and short‐term transient responses during cold acclimation in olive leaves. DNA Res. 22, 1–11. 10.1093/dnares/dsu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez, C.M. and Gaut, B.S. (2016) The jury may be out, but it is important that it deliberates: a response to Besnard and Rubio de Casas about olive domestication. New Phytol. 209, 471–473. 10.1111/nph.13780. [DOI] [PubMed] [Google Scholar]
- Díez, C.M. , Imperato, A. , Rallo, L. , Barranco, D. and Trujillo, I. (2012) Worldwide core collection of olive cultivars based on simple sequence repeat and morphological markers. Crop Sci. 52, 211–221. 10.2135/cropsci2011.02.0110. [DOI] [Google Scholar]
- Díez, C.M. , Trujillo, I. , Martinez‐Urdiroz, N. , Barranco, D. , Rallo, L. , Marfil, P. and Gaut, B.S. (2015) Olive domestication and diversification in the Mediterranean Basin. New Phytol. 206, 436–447. 10.1111/nph.13181. [DOI] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Gustus, C. (1995) teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics, 141, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J.F. , Gaut, B.S. and Smith, B.D. (2006) The molecular genetics of crop domestication. Cell, 127, 1309–1321. 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Duan, N. , Bai, Y. , Sun, H. et al (2017) Genome re‐sequencing reveals the history of apple and supports a two‐stage model for fruit enlargement. Nat. Commun. 8, 249 10.1038/s41467-017-00336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duforet‐Frebourg, N. , Bazin, E. and Blum, M.G.B. (2014) Genome scans for detecting footprints of local adaptation using a Bayesian factor model. Mol. Biol. Evol. 31, 2483–2495. 10.1093/molbev/msu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duforet‐Frebourg, N. , Luu, K. , Laval, G. , Bazin, E. and Blum, M.G.B. (2015) Detecting genomic signatures of natural selection with principal component analysis: application to the 1000 Genomes data. Mol. Biol. Evol. 33, 1082–1093. 10.1093/molbev/msv334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bakkali, A. , Haouane, H. , Moukhli, A. , Costes, E. , Van Damme, P. and Khadari, B. (2013) Construction of core collections suitable for association mapping to optimize use of Mediterranean olive (Olea europaea L.) genetic resources. PLoS ONE, 8, e61265 10.1371/journal.pone.0061265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2018) FAOSTAT [WWW Document]. United Nations, Rome, Italy: Food Agric. Organ; http://www.fao.org/faostat/en/#data/QC. [Google Scholar]
- Flowers, J.M. , Hazzouri, K.M. , Gros‐balthazard, M. et al (2019) Cross‐species hybridization and the origin of North African date palms. Proc. Natl Acad. Sci. USA, 116, 1651–1658. 10.1073/pnas.1817453116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll, M. and Gaggiotti, O. (2008) A genome‐scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics, 180, 977–993. 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili, E. , Stanley, D.J. , Sharvit, J. and Weinstein‐Evron, M. (1997) Evidence for earliest olive‐oil production in submerged settlements of the Carmel Coast. Israel. J. Archaeol. Sci. 24, 1141–1150. 10.1006/jasc.1997.0193. [DOI] [Google Scholar]
- Gallusci, P. , Hodgman, C. , Teyssier, E. and Seymour, G.B. (2016) DNA methylation and chromatin regulation during fleshy fruit development and ripening. Front. Plant Sci. 7, 807 10.3389/fpls.2016.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S. , Díez, C.M. and Morrell, P.L. (2015) Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 31, 709–719. 10.1016/j.tig.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Gepts, P. (2004) Plant domestication as a long‐term selection experiment. Plant Breed. Rev. 24, 1–44. [Google Scholar]
- Gepts, P. (2014) The contribution of genetic and genomic approaches to plant domestication studies. Curr. Opin. Plant Biol. 18, 51–59. 10.1016/j.pbi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Gros‐Balthazard, M. , Galimberti, M. , Kousathanas, A. et al (2017) The discovery of wild date palms in Oman reveals a complex domestication history involving centers in the middle East and Africa. Curr. Biol. 27, 2211–2218. 10.1016/j.cub.2017.06.045. [DOI] [PubMed] [Google Scholar]
- Hannachi, H. , Breton, C. , Msallem, M. , Ben El Hadj, S. , El Gazzah, M. and Bervillé, A. (2008) Are olive cultivars distinguishable from oleaster trees based on morphology of drupes and pits, oil composition and microsatellite polymorphisms?. Acta Bot. Gallica, 155, 531–545. 10.1080/12538078.2008.10516132. [DOI] [Google Scholar]
- Haouane, H. , El Bakkali, A. , Moukhli, A. , Tollon, C. , Santoni, S. , Oukabli, A. , El Modafar, C. and Khadari, B. (2011) Genetic structure and core collection of the World Olive Germplasm Bank of Marrakech: towards the optimised management and use of Mediterranean olive genetic resources. Genetica, 139, 1083–1094. 10.1007/s10709-011-9608-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford, M.B. , Xu, X. , van Heerwaarden, J. et al (2012) Comparative population genomics of maize domestication and improvement. Nat. Genet. 44, 808–811. 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaria, D. , Chiappetta, A. and Muzzalupo, I. (2016) De novo transcriptome sequencing of Olea europaea L. to identify genes involved in the development of the pollen tube. Sci. World J. 2016, 1–7. 10.1155/2016/4305252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalua, C.M. , Allen, M.S. , Bedgood, D.R. , Bishop, A.G. , Prenzler, P.D. and Robards, K. (2007) Olive oil volatile compounds, flavour development and quality: a critical review. Food Chem. 100, 273–286. 10.1016/j.foodchem.2005.09.059. [DOI] [Google Scholar]
- Kaniewski, D. , Van Campo, E. , Boiy, T. , Terral, J.‐F. , Khadari, B. and Besnard, G. (2012) Primary domestication and early uses of the emblematic olive tree: palaeobotanical, historical and molecular evidence from the Middle East. Biol. Rev. 87, 885–899. 10.1111/j.1469-185X.2012.00229.x. [DOI] [PubMed] [Google Scholar]
- Keightley, P.D. , Halligan, D.L. and Kirkpatrick, M. (2011) Inference of mutation parameters and selective constraint in mammalian coding sequences by Approximate Bayesian Computation. Genetics, 187, 1153–1161. 10.1534/genetics.110.124073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadari, B. and El Bakkali, A. (2018) Primary selection and secondary diversification: two key processes in the history of olive domestication. Int. J. Agron. 2018, 5607903. [Google Scholar]
- Kislev, M.E.E. , Nadel, D. and Carmi, I. (1992) Epipalaeolithic (19,000 BP) cereal and fruit diet at Ohalo II, Sea of Galilee. Israel. Rev. Palaeobot. Palynol. 73, 161–166. 10.1016/0034-6667(92)90054-K. [DOI] [Google Scholar]
- Koenig, D. , Jiménez‐Gómez, J.M. , Kimura, S. et al (2013) Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl Acad. Sci. USA, 110, E2655–E2662. 10.1073/pnas.1309606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konczal, M. , Babik, W. , Radwan, J. , Sadowska, E.T. and Koteja, P. (2015) Initial molecular‐level response to artificial selection for increased aerobic metabolism occurs primarily through changes in gene expression. Mol. Biol. Evol. 32, 1461–1473. 10.1093/molbev/msv038. [DOI] [PubMed] [Google Scholar]
- Korneliussen, T.S. , Albrechtsen, A. and Nielsen, R. (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinform. 15, 356 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, Z.H. , Bukowski, R. , Sun, Q. and Doebley, J.F. (2014) The role of cis regulatory evolution in maize domestication. PLoS Genet. 10, e10047 10.1371/journal.pgen.1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. and Durbin, R. (2010) Fast and accurate long‐read alignment with Burrows‐Wheeler transform. Bioinformatics, 26, 589–595. 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphschitz, N. , Gophna, R. , Hartman, M. and Biger, G. (1991) The beginning of olive (Olea europaea) cultivation in the Old World: a reassessment. J. Archaeol. Sci. 18, 441–453. 10.1016/0305-4403(91)90037-P. [DOI] [Google Scholar]
- Liu, F. , Xia, Y. , Wu, L. et al (2015) Enhanced seed oil content by overexpressing genes related to triacylglyceride synthesis. Gene, 557, 163–71. 10.1016/j.gene.2014.12.029. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Chen, L. , Zhang, S. et al (2019) Decrease of gene expression diversity during domestication of animals and plants. BMC Evol. Biol. 19, 19 10.1186/s12862-018-1340-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterhos, K.E. and Whitlock, M.C. (2014) Evaluation of demographic history and neutral parameterization on the performance of F ST outlier tests. Mol. Ecol. 23, 2178–2192. 10.1111/mec.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loumou, A. and Giourga, C. (2003) Olive groves: “The life and identity of the Mediterranean”. Agric. Human Values, 20, 87–95. 10.1023/A:1022444005336. [DOI] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumaret, R. , Ouazzani, N. , Michaud, H. , Vivier, G. , Deguilloux, M.‐F. and Di Giusto, F. (2004) Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin. Heredity, 92, 343–351. 10.1038/sj.hdy.6800430. [DOI] [PubMed] [Google Scholar]
- Luu, K. , Bazin, E. and Blum, M.G.B. (2017) PCAdapt: an R package to perform genome scans for selection based on principal component analysis. Mol. Ecol. Resour. 17, 67–77. 10.1111/1755-0998.12592. [DOI] [PubMed] [Google Scholar]
- Martínez‐Ainsworth, N.E. and Tenaillon, M.I. (2016) La domestication des plantes: une affaire de super‐héros et de masterminds. C. R. Biol. 339, 268–273. 10.1016/j.crvi.2016.05.005. [DOI] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res. 20, 1297–1303. 10.1101/gr.107524.110.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, R.S. and Purugganan, M.D. (2013) Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- Miller, A.J. and Gross, B.L. (2011) From forest to field: perennial fruit crop domestication. Am. J. Bot. 98, 1389–1414. 10.3732/ajb.1000522. [DOI] [PubMed] [Google Scholar]
- Morgulis, A. , Coulouris, G. , Raytselis, Y. , Madden, T.L. , Agarwala, R. and Schäffer, A.A. (2008) Database indexing for production MegaBLAST searches. Bioinformatics, 24, 1757–1764. 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi, S. , Regni, L. , Bocchini, M. et al (2019) Physiological, epigenetic and genetic regulation in some olive cultivars under salt stress. Sci. Rep. 9, 1093 10.1038/s41598-018-37496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz‐Mérida, A. , González‐Plaza, J.J. , Cañada, A. et al (2013) De novo assembly and functional annotation of the olive (Olea europaea) transcriptome. DNA Res. 20, 93–108. 10.1093/dnares/dss036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles, S. , Boyko, A.R. , Owens, C.L. et al (2011) Genetic structure and domestication history of the grape. Proc. Natl Acad. Sci. USA, 108, 3530–3535. 10.1073/pnas.1009363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz, B. , Sarah, G. , Sabot, F. , Ruiz, M. , Adam, H. , Nidelet, S. , Ghesquière, A. , Santoni, S. , David, J. and Glémin, S. (2014) Transcriptome population genomics reveals severe bottleneck and domestication cost in the African rice (O. glaberrima). Mol. Ecol. 23, 2210–2227. 10.1111/mec.12738. [DOI] [PubMed] [Google Scholar]
- Narum, S.R. and Hess, J.E. (2011) Comparison of F ST outlier tests for SNP loci under selection. Mol. Ecol. Resour. 11, 184–194. 10.1111/j.1755-0998.2011.02987.x. [DOI] [PubMed] [Google Scholar]
- Olsen, K.M. and Purugganan, M.D. (2002) Molecular evidence on the origin and evolution of glutinous rice. Genetics, 162, 941–950. 10.1007/s001220050477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K.M. and Wendel, J.F. (2013) A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 64, 47–70. 10.1146/annurev-arplant-050312-120048. [DOI] [PubMed] [Google Scholar]
- Parra, R. , Paredes, M.A. , Sanchez‐Calle, I.M. et al (2013) Comparative transcriptional profiling analysis of olive ripe‐fruit pericarp and abscission zone tissues shows expression differences and distinct patterns of transcriptional regulation. BMC Genom. 14, 866 10.1186/1471-2164-14-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell, J.K. and Pritchard, J.K. (2012) Inference of population splits and mixtures from genome‐wide allele frequency data. PLoS Genet. 8, e1002967 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, D.R. (2017) Assessing elements of an extended evolutionary synthesis for plant domestication and agricultural origin research. Proc. Natl Acad. Sci. USA, 114, 6429–6437. 10.1073/pnas.1703658114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski, M.P. , Coop, G. and Wall, J.D. (2005) The signature of positive selection on standing genetic variation. Evolution, 59, 2312–2323. [PubMed] [Google Scholar]
- Ray, A. , Pal, M.K. , Ghosh, M. and Sanyal, D. (2011) Psychodermatological disorders: an assessment of psychiatric morbidity. Int. Med. J. 18, 300–304. 10.1371/journal.pgen.1003457. [DOI] [Google Scholar]
- Rhoné, B. , Mariac, C. , Couderc, M. , Berthouly‐Salazar, C. , Ousseini, I.S. and Vigouroux, Y. (2017) No excess of CIS‐regulatory variation associated with intraspecific selection in wild pearl millet (Cenchrus americanus). Genome Biol. Evol. 9, 388–397. 10.1093/gbe/evx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E.J. (2011) Natural epigenetic variation in plant species: a view from the field. Curr. Opin. Plant Biol. 14, 204–209. 10.1016/j.pbi.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Romiguier, J. , Gayral, P. , Ballenghien, M. et al (2014) Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature, 515, 261–263. 10.1038/nature13685. [DOI] [PubMed] [Google Scholar]
- Rugini, E. , Baldoni, L. , Muleo, R. and Sebastiani, L. (2016) The Olive Tree Genome. Compendium of Plant Genomes: Springer International Publishing, AG. [Google Scholar]
- Sarah, G. , Homa, F. , Pointet, S. et al (2017) A large set of 26 new reference transcriptomes dedicated to comparative population genomics in crops and wild relatives. Mol. Ecol. Resour. 17, 565–580. 10.1111/1755-0998.12587. [DOI] [PubMed] [Google Scholar]
- Sauvage, C. , Rau, A. , Aichholz, C. , Chadoeuf, J. , Sarah, G. , Ruiz, M. , Santoni, S. , Causse, M. , David, J. and Glémin, S. (2017) Domestication rewired gene expression and nucleotide diversity patterns in tomato. Plant J. 91, 631–645. 10.1111/tpj.13592. [DOI] [PubMed] [Google Scholar]
- Skotte, L. , Korneliussen, T.S. and Albrechtsen, A. (2013) Estimating individual admixture proportions from next generation sequencing data. Genetics, 195, 693–702. 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W. (2016) Signatures of positive selection: from selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol. Ecol. 25, 79–88. 10.1111/mec.13288. [DOI] [PubMed] [Google Scholar]
- Stern, D.L. and Orgogozo, V. (2008) The loci of evolution: how predictable is genetic evolution? Evolution, 62, 2155–2177. 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter, M.G. , Gates, D.J. , Mei, W. and Ross‐Ibarra, J. (2017) How to make a domesticate. Curr. Biol. 17, R896–R900. 10.1016/j.cub.2017.06.048. [DOI] [PubMed] [Google Scholar]
- Storey, J.D. (2002) A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B 64, 479–498. 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Studer, A. , Zhao, Q. , Ross‐Ibarra, J. and Doebley, J. (2011) Identification of a functional transposon insertion in the maize domestication gene tb1 . Nat. Genet. 43, 1160–1163. 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek, F. , Bošnjak, M. , Škunca, N. and Šmuc, T. (2011) REVIGO summarizes and visualizes long lists of GO terms. PLoS ONE, 6, e21800 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson‐Wagner, R. , Briskine, R. , Schaefer, R. , Hufford, M.B. , Ross‐Ibarra, J. , Myers, C.L. , Tiffin, P. and Springer, N.M. (2012) Reshaping of the maize transcriptome by domestication. Proc. Natl Acad. Sci. USA, 109, 11878–11883. 10.1073/pnas.1201961109/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1201961109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinnen, G. , Goossens, A. and Pauwels, L. (2016) Lessons from domestication: targeting cis‐regulatory elements for crop improvement. Trends Plant Sci. 21, 506–515. 10.1016/j.tplants.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Szkiba, D. , Kapun, M. , von Haeseler, A. and Gallach, M. (2014) SNP2GO: functional analysis of genome‐wide association studies. Genetics, 197, 285–289. 10.1534/genetics.113.160341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics, 105, 437–460 https://doi.org/6628982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terral, J.F. , Alonso, N. , Capdevila, R.B.I. et al (2004) Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. J. Biogeogr. 31, 63–77. [Google Scholar]
- Unver, T. , Wu, Z. , Sterck, L. et al (2017) Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl Acad. Sci. USA, 114, E9413–E9422. 10.1073/pnas.1708621114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsiou, A.I. , Bazin, E. and Gaggiotti, O.E. (2016) Detection of selective sweeps in structured populations: a comparison of recent methods. Mol. Ecol. 25, 89–103. 10.1111/mec.13360. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Tu, L. , Lin, M. et al (2017) Asymmetric subgenome selection and cis‐regulatory divergence during cotton domestication. Nat. Genet. 49, 579–587. 10.1038/ng.3807. [DOI] [PubMed] [Google Scholar]
- Watterson, G.A. (1975) On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276. [DOI] [PubMed] [Google Scholar]
- Weir, B.S. and Cockerham, C.C. (1984) Estimating F‐statistics for the analysis of population‐structure. Evolution, 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- Young, M.D. , Wakefield, M.J. , Smyth, G.K. and Oshlack, A. (2010) Gene ontology analysis for RNA‐seq: accounting for selection bias. Genome Biol. 11, R14 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Schwartz, S. , Wagner, L. and Miller, W. (2000) A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214. 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Massonnet, M. , Sanjak, J.S. , Cantu, D. and Gaut, B.S. (2017) Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc. Natl Acad. Sci. USA, 114, 11715–11720. 10.1073/pnas.1709257114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Heatmap displaying the relatedness between the 66 Mediterranean olive tree accessions.
Figure S2. Admixture proportions in 62 olive trees inferred from genotype likelihoods of 536 341 SNPs with NGSadmix for K = 4–6.
Figure S3. Admixture proportions inferred from genotype likelihoods with NGSadmix for K = 2–6.
Figure S4. Log‐likelihood estimates for different K values in the three admixture analyses.
Figure S5. Proportion of variance explained by the 10 first principal components of the PCA performed on 62 olive tree accessions.
Figure S6. Principal component analysis of 62 Olea europaea subsp. europaea.
Figure S7. Reduced median network of chlorotypes reconstructed with network v.5 using data from Data S1.
Figure S8. Fraction of variance in relatedness between four olive tree populations accounted for by phylogenetic models with zero through two migrations in TreeMix.
Figure S9. TreeMix relationships between oleasters and cultivars from all around the Mediterranean basin grouped in four populations.
Figure S10. TreeMix relationships between five olive tree populations.
Figure S11. Fraction of variance in relatedness between five olive tree populations accounted for by phylogenetic models with zero through two migrations in TreeMix.
Figure S12. Bayesian test for selection between 27 wild and 36 cultivated populations implemented in BayeScan.
Figure S13. Principal component analysis of SNPs data.
Figure S14. Visualizations of enriched GO terms from PCAdapt selection scan including all accessions.
Figure S15. Bioinformatic pipeline followed to process the raw reads into alignment files (final.bam files) and subsequently variant call format file (.vcf file) used in subsequent analyses.
Table S1. Summary of the 68 Olea europaea spp. accessions including their origin, the cultivar name when relevant and statistics related to sequencing (excel file).
Table S2. List of SNPs detected as outliers in BayeScan when comparing all oleasters and all cultivated accessions.
Table S3. Strong candidate transcripts for selection.
Methods S1. Plastid genome profiling.
Methods S2. Read alignments and variant calling.
Methods S3. Inference of relatedness among accessions.
Methods S4. Inference of site frequency spectra and calculation of diversity statistics.
Methods S5. Annotation of the transcriptome reference sites as synonymous or non‐synonymous.
Data S1. Chloroplastic microsatellite and CAPS profile of each individual analysed in the present study [according to the method developed by Besnard et al. (2011) (excel file).
Appendix S1. Detailed structure and diversity of oleaster populations.


