Summary
In order to survive, bacteria must adapt to multiple fluctuations in their environment, including coping with changes in metal concentrations. Many metals are essential for viability, since they act as cofactors of indispensable enzymes. But on the other hand, they are potentially toxic because they generate reactive oxygen species or displace other metals from proteins, turning them inactive. This dual effect of metals forces cells to maintain homeostasis using a variety of systems to import and export them. These systems are usually inducible, and their expression is regulated by metal sensors and signal‐transduction mechanisms, one of which is mediated by extracytoplasmic function (ECF) sigma factors. In this review, we have focused on the metal‐responsive ECF sigma factors, several of which are activated by iron depletion (FecI, FpvI and PvdS), while others are activated by excess of metals such as nickel and cobalt (CnrH), copper (CarQ and CorE) or cadmium and zinc (CorE2). We focus particularly on their physiological roles, mechanisms of action and signal transduction pathways.
Bacteria use ECF sigma factors to maintain metal homeostasis. They regulate the expression of genes either to import metals under conditions of metal depletion or to reduce toxicity when metal concentrations are excessive. These adaptive mechanisms can be complex, consisting of several proteins with different cell locations or very simple, consisting of only the sigma factor capable of sensing the metal and triggering the response.

Metal ion homeostasis: beneficial functions and toxicity
Due to their exposure to a variable environment, bacteria have developed several adaptive mechanisms to rapidly respond to changes in their habitat and thereby increase their chances of survival. For instance, bacteria must adapt to the presence of metals, as they are required as cofactors of many enzymes. Iron is thought to be one of the first metals used as a cofactor for enzymatic reactions, and nowadays is an essential metal for almost all living organisms, with a few exceptions such as Lactobacilli and Borrelia (Posey and Gherardini, 2000). Iron‐containing proteins are not only excellent electron carriers (81% of the oxidoreductases use this metal to transfer electrons) (Waldron et al., 2009), but they also participate in enzyme catalysis and regulate gene expression as they function as sensors for environmental or intracellular signals (Lill, 2009). Additionally, iron plays a central role in host–pathogen interplay, and the fight between host cells and intercellular or intracellular pathogens for this essential metal will influence the outcome of infectious diseases in favor of either the host or the pathogenic invaders (Nairz et al., 2010). When the atmosphere became oxygenated, iron became insoluble and the preference of many enzymes shifted to more bioavailable metals, such as copper and zinc, which also became essential cofactors for many organisms (Dupont et al., 2010; Festa and Thiele, 2011). Other metals such as nickel are essential for a limited number of proteins, including glyoxalases, ureases, hydrogenases and some superoxide dismutases (Hausinger and Zamble, 2007; Boer et al., 2014), while cobalt has no clear physiological role except as a component of several cobalamins and as a cofactor of a few noncorrin‐cobalt‐containing enzymes (Kobayashi and Shimizu, 1999; Barras and Fontecave, 2011). In contrast, cadmium is not required for any biological function so its presence in the cell, even at low concentrations, is considered toxic (Moulis, 2010).
Nevertheless, all of the abovementioned metals are potentially toxic, and high concentrations can lead to the generation of hydroxyl radicals through Fenton or Fenton‐like reactions and singlet oxygen, damage to DNA and cell membranes by generation or enhancement of oxidative stress, inhibition of enzymes with histidine or cysteine residues in their active sites, and replacement of other metal cofactors on several metalloproteins (Waldron et al., 2009; Moraleda‐Muñoz et al., 2010a; 2010b; Macomber and Hausinger, 2011; Rensing and McDevitt, 2013; Chandrangsu et al., 2017; Cheng et al., 2018; Pérez et al., 2018; Grosse et al., 2019). This capacity to replace other metals is especially important in the case of copper, since it occupies the highest position in the Irving–Williams series (Mn2+ < Fe2+ < Co2+ < Ni2+ < Cu2+> Zn2+) and can potentially substitute any other biological metal used as a cofactor inside the cell (Irving and Williams, 1953). It is therefore critical for cells to maintain metal homeostasis both to ensure an adequate supply for their metal requirements and to avoid and alleviate the toxicity of metals.
Metal ion transport
Bacteria have developed a very diverse set of mechanisms for acquiring metals, which are often scarce in their natural habitats, and for exporting and detoxifying them when they reach high concentrations. Although iron is abundant in natural terrestrial and aquatic niches, under physiological conditions free iron levels are frequently very low. The ferrous form (Fe2+) is only soluble in anoxic environments and the presence of oxygen prompts its rapid oxidation to the ferric form (Fe3+), which is poorly soluble. Because of this, bacteria have developed complex strategies to acquire this important metal, including the reduction of ferric to ferrous ions, uptake of iron in the form of exogenous iron chelators such as Fe3+‐citrate (Silva et al., 2009) or siderophores (compounds that complex the ferric ions) such as pyoverdine (Koster et al., 1994), and direct acquisition of iron from other organisms' iron‐binding proteins, such as iron‐ and heme‐carrier proteins and hemophores (Wandersman and Delepelaire, 2004).
Because of their high solubility, most metal homeostasis mechanisms for cobalt, nickel, copper, zinc and cadmium are focused on their detoxification rather than on their acquisition. These mechanisms include exporting metals by P1B‐type ATPases, transenvelope CBA transporters and cation diffusion facilitators, metal oxidation to a less toxic form (as with multicopper oxidases, which turn Cu+ into Cu2+), or binding metals to metallochaperones (Sánchez‐Sutil et al., 2007; Moraleda‐Muñoz et al., 2010a; 2010b; Rensing and McDevitt, 2013; Pérez et al., 2018; Grosse et al., 2019).
These adaptive mechanisms are regulated by several transcription factors and signaling systems that have been traditionally classified in the four pillars of bacterial signal‐transduction mechanisms: one‐component systems, two‐component systems, extracytoplasmic function (ECF) sigma factors and serine‐threonine protein kinases (Staroń et al., 2009; Muñoz‐Dorado et al., 2012).
Regulation of metal ion homeostasis by ECF sigma factors
ECF sigma factors represent group IV of the σ70 family of sigma factors, which are directly involved in the transcription process by recognizing the −10 and −35 promoter sequences and, together with the core RNA polymerase (RNAP) enzyme, are responsible for initiating the transcription of the genes they regulate (Helmann, 2002; Gruber and Gross, 2003; Mooney et al., 2005; Lee et al., 2013). This promoter recognition will only happen after their specific stimulus has been detected (Nies, 2004; Staroń et al., 2009; Mascher, 2013; Pinto and Mascher, 2016). ECF sigma factors are smaller than other sigma factors, and they only contain the σ2 and σ4 domains (Lonetto et al., 1994; Helmann, 2002). Canonical ECF sigma factors are regulated by anti‐sigma factors. Anti‐sigma factors are usually membrane proteins with a high affinity for their cognate sigma factor, so in the absence of the specific stimulus, the sigma factor is sequestered by the anti‐sigma factor. Anti‐sigma factors often act as the sensor part of this signal‐transduction system, and upon detection of the stimulus, they release the sigma factors. ECF sigma factors and anti‐sigma factors are usually co‐transcribed to ensure that no sigma factor is released in the absence of an appropriate stimulus (Missiakas and Raina, 1998; Ho and Ellermeier, 2012; Muñoz‐Dorado et al., 2012). However, not all ECF sigma factors function in the same manner. A phylogenetic classification of the ECF sigma factors into 94 different groups has shed some light on the diversity of mechanisms regulating their activity and the stimuli detected (Staroń et al., 2009; Mascher, 2013; Pinto and Mascher, 2016).
Metals such as iron, nickel, cobalt, copper, zinc and cadmium activate a number of ECF sigma factors to trigger the specific response in the bacteria to keep metal homeostasis. In this review we will discuss the signaling mechanisms of the best characterized ECF sigma factors involved in the regulation of metal homeostasis genes, covering the iron uptake regulator systems FecI, FpvI and PvdS, the cobalt and nickel resistance regulator system CnrH, the copper‐responsive regulator CarQ involved in the biosynthesis of carotenoids, and the CorE‐like sigma factors involved in copper, zinc and cadmium resistance.
Iron starvation‐responsive ECF sigma factors
In most cases, iron acquisition genes (normally forming operons) are only expressed under conditions of metal deficiency. In the presence of iron, these operons are repressed by the regulator Fur, which forms complexes with ferrous iron and binds to conserved sites of DNA termed Fur boxes, preventing the transcription of iron‐uptake genes (Fig. 1A) (Baichoo and Helmann, 2002; Cornelis et al., 2009; Fillat, 2014). Under iron‐depletion, the regulator does not bind the metal, thus preventing Fur from binding to DNA and transcription of the genes involved in iron acquisition often occurs under the control of transcriptional activators. Several iron supply systems are positively regulated by ECF sigma factors that are induced by the availability of specific iron sources (Sexton et al., 1996; Leoni et al., 2000; Biville et al., 2004; Lindeberg et al., 2008; Thakur et al., 2013; Chevalier et al., 2018; Lang et al., 2018).
Figure 1.
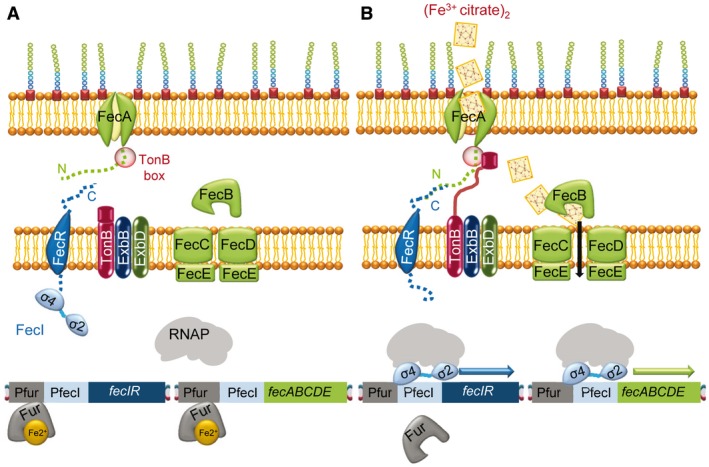
The E. coli CSS Fe3+‐citrate transport is regulated by the FecR/FecI system. A. In the absence of Fe3+‐citrate and in the presence of Fe2+‐Fur, the repressor binds to the region upstream of the operons fecIR and fecABCDE. Moreover, the FecR anti‐sigma factor sequesters the FecI ECF sigma factor by interaction of the N‐terminal region of the anti‐sigma factor and the σ4 domain of the ECF sigma factor, preventing transcription of these two operons. B. Signaling pathway in the presence of Fe3+‐citrate and under low iron availability. The Fur repressor is not bound to Fe2+ and cannot bind DNA. The outer membrane protein receptor FecA suffers conformational changes, interacts with TonB through the TonB box domain, and allows the transport of the substrate to the periplasm, and through FecB and the FecCDE transporter to the cytoplasm. FecA changes also allow interaction between the FecA N‐terminal region and the C‐terminal region of the anti‐sigma factor FecR, releasing the ECF sigma factor FecI. This sigma factor can now up‐regulate the transcription of fecIR and fecABCDE after recruiting the core RNAP.
Most iron starvation‐responsive ECF sigma factors are regulated by cell surface signaling (CSS) that transmits the input signal from a substrate‐bound outer membrane protein (OMP) across the periplasmic space to a cytoplasmic membrane‐spanning anti‐sigma factor, releasing the cytoplasmic ECF sigma factor that transcriptionally regulates the expression of the stimulus‐responsive genes (Braun et al., 2006; Brooks and Buchanan, 2008; Llamas et al., 2014). CSS was first described in the transport of ferripyoverdine in Pseudomonas putida WCS358 (Koster et al., 1994) and in the Fe3+‐citrate transport system in E. coli (Härle et al., 1995). These iron starvation‐responsive ECF sigma factors belong to groups ECF05‐ECF09 (Staroń et al., 2009; Mascher, 2013; Pinto and Mascher, 2016), and the best characterized FecI‐like sigma factors, FecI from Escherichia coli and PvdS and FpvI from Pseudomonas aeruginosa, are described below.
The Escherichia coli Fec signaling pathway
The Fe3+‐citrate system is regulated by the anti‐sigma/ECF sigma factor pair FecR/FecI via CSS (Angerer et al., 1995; Härle et al., 1995; Mahren and Braun, 2003), which in the presence of iron is repressed by Fur‐Fe2+ (Fig. 1A). Under conditions of iron depletion and presence of Fe3+‐citrate, Fur does not bind the DNA. The OMP receptor protein FecA has a dual role: it transports the Fe3+‐citrate complexes across the outer membrane and is involved in signaling the extracellular presence of these complexes to the genetic machinery in the cytoplasm (Brooks and Buchanan, 2008). The C‐terminal portion of FecA forms a beta barrel (with twenty‐two anti‐parallel beta‐strands) that spans the outer membrane, modeling a pore that is occluded by a plug domain that prevents the unspecific diffusion of large molecules. The plug must undergo conformational changes to allow the opening of the pore to facilitate transport. Fe3+‐citrate is first adsorbed from the medium by aromatic residues located in the external pocket of FecA and from there it is transferred to its high‐affinity binding site, which is formed mainly of several arginine residues that bind the negatively charged ferric citrate (Ferguson et al., 2002). Binding of Fe3+‐citrate to FecA provokes structural changes in two extracellular loops involved in the iron complex transport (Ferguson et al., 2002; Yue et al., 2003) and in a small loop in the plug domain that seems to play a role in the transmission of the signal to TonB (Buchanan et al., 1999; Brooks and Buchanan, 2008). FecA interacts with TonB through the TonB box domain (which also changes its conformation by Fe3+‐citrate binding) and it is able to transport iron through the periplasmic FecB protein and the ABC transporter FecCDE (Fig. 1B) to the cytoplasm (Staudenmaier et al., 1989). In the cytoplasm, a ferric reductase catalyzes the release of Fe2+ from the citrate complexes (Miethke et al., 2011), although other enzymes may also be implicated in this process (Miethke and Marahiel, 2007).
FecA belongs to the group of TonB‐dependent transporters (TBDT) (Noinaj et al., 2010), and receives energy from the complex TonB‐ExbB‐ExbD, which is anchored to the cytoplasmic membrane and extends into the periplasm. Additionally, the N‐terminal domain of FecA interacts with the C‐terminal domain of the inner membrane anti‐sigma factor FecR, which then releases FecI to recruit the core RNAP and binds to the promoters to initiate transcription of the clusters fecIR and fecABCDE to accelerate the uptake of Fe3+‐citrate (Fig. 1B) (Van Hove et al., 1990; Wriedt et al., 1995; Mahren and Braun, 2003).
The pair FecR/FecI does not function as a canonical anti‐sigma/sigma pair because FecR is also required for full FecI activity, probably by inducing the binding of FecI to the core RNAP (Mahren and Braun, 2003). The positive role of FecR on FecI could also be due to the fact that the ECF sigma factor might be unstable in the absence of FecR, and under these conditions, it would be more susceptible to be degraded by proteolysis, probably by the protease RseP (Braun et al., 2006).
Other FecI homologues, such as FiuR/FiuI, FoxR/FoxI, HasS/HasI and FemR/FemI, are present in many bacterial species belonging to the Proteobacteria phylum. Most of them are often clustered with genes coding for a FecA‐like OMP and a putative anti‐sigma factor containing a FecR‐like domain (Sexton et al., 1996; Biville et al., 2004; Braun et al., 2006; Brooks and Buchanan, 2008; Thakur, et al., 2013; Llamas et al., 2014; Chevalier et al., 2018; Lang et al., 2018). The great variety of associated domains, organized in almost 200 different architectures according to the Pfam database (El‐Gebali et al., 2019), seems to indicate that this FecIR system might be a generalized signal‐transduction mechanism used to regulate the entry of different types of products (Staroń et al., 2009; Karlsson et al., 2011; Mascher, 2013; Pinto and Mascher, 2016) such as complex polysaccharides in the gut environment, as suggested for FecIR‐like systems in Bacteroidetes (Xu et al., 2004).
Mechanism of action of Pseudomonas aeruginosa PvdS and FpvI
The P. aeruginosa PAO1 genome encodes 19 ECF sigma factors, 14 of which are regulated by iron starvation and are involved in the expression of TBDTs for siderophores, heme or iron citrate uptake (Llamas et al., 2008; Chevalier et al., 2018; Otero‐Asman et al., 2019). The best‐studied system in this bacterium is the pyoverdine CSS system (Ravel and Cornelis, 2003; Visca et al., 2007). Although the pyoverdine signaling pathway is similar to the Fe3+‐citrate system described above, it differs in several aspects. First, it responds to the endogenously produced siderophore pyoverdine complexed with Fe3+, as opposed to an exogenous source as in the case of Fe3+‐citrate; second, one anti‐sigma factor controls two sigma factors (Beare et al., 2003); and third, genes involved in this system are not adjacent in the genome, and even the sigma and anti‐sigma factors are not co‐transcribed (Beare et al., 2003; Llamas et al., 2014).
This CSS consists of the ferripyoverdine receptor FpvA (energized by the complex TonB‐ExbB‐ExbD), the anti‐sigma factor FpvR and the two ECF sigma factors FpvI and PvdS, which remain sequestered in the membrane by FpvR in the absence of ferripyoverdine (Fig. 2A). In this cascade, the interaction of ferric siderophore with the FpvA binding pocket, formed by at least 14 amino acid (most of them aromatic residues), transmits a signal to TonB that facilitates the import of ferripyoverdine (Schalk et al., 1999; Schalk et al., 2002; Schalk et al., 2004; Cobessi et al., 2005; Wirth et al., 2007). The conformational changes in FpvA also result in the proteolytic cleavage of FpvR by the RseP/MucP protease (Visca et al., 2007). The subsequent liberation of FpvI and PvdS allows the expression of the target regulons (Fig. 2B). The FpvI regulon includes the fpvA gene (Beare et al., 2003), some genes involved in pyoverdine biosynthesis (pvdA, pvdIJD and fpvGHIJ), and other genes such as the heme‐uptake transporter hasR or the small RNA gene prrF1 (Ravel and Cornelis, 2003; Schulz et al., 2015). PvdS controls the transcription of a regulon of about 80 genes (Ochsner et al., 2002; Schulz et al., 2015), including genes involved in pyoverdine biosynthesis, secretion and utilization, other virulence genes such as those encoding exotoxin A, the extracellular protease PrpL, and several type III secretion systems that function as toxins, such as ExoT and ExoS (Wilderman et al., 2001; Gaines et al., 2007). Moreover, PvdS regulates the expression of the sigma factor PA14_21540 (PA3285), the ferrochalatase hemH and other genes involved in cellular functions unrelated to iron uptake (for a review see Chevalier et al., 2018).
Figure 2.
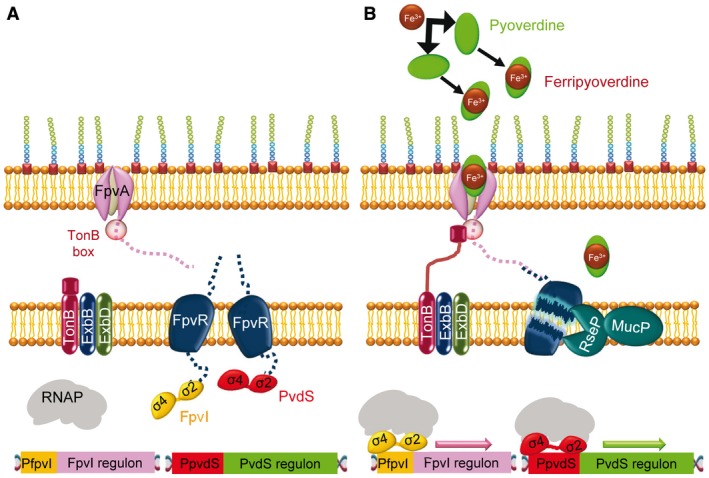
The P. aeruginosa ferripyoverdine CSS is regulated by FpvR/FpvI/PvdS. A. In the absence of ferripyoverdine, the anti‐sigma factor FpvR sequesters the sigma factors FpvI and PvdS, preventing them from binding to the core RNAP and DNA. The expression of both ECF sigma factors is also repressed by Fur (in a similar way to that shown for FecI in Fig. 1 and not shown here). In these conditions, the target regulons for each sigma factor are not transcribed. B. When pyoverdine binds to Fe3+ to form ferrypioverdine, a signal is transmitted to FpvA and TonB, enabling the import of ferripyoverdine and the proteolysis of FpvR by the RseP/MucP protease, liberating FpvI and PvdS and allowing expression of the two regulons.
As happens with most iron‐starvation ECF sigma factors, pvdS (and fpvI) is primarily controlled by Fur (not shown in Fig. 2), but its expression is very complex and it is modulated by several environmental signals and different factors, such as regulators related to the response to oxidative stress or sulfur homeostasis. Moreover, pvdS is also under the control of another FecI‐like ECF sigma factor, FecI2 (Llamas et al., 2014; Chevalier et al., 2018). The complexity of the activation mechanism of these ECF sigma factors, and the cross‐talk between them and other regulatory elements, such as two‐component systems, have been recently reviewed by Chevalier et al. (2018).
Nickel‐ and cobalt‐responsive ECF sigma factors
A co‐founder of the ECF sigma factor family (Lonetto et al., 1994), CnrH (group ECF20), controls nickel and cobalt resistance in Cupriavidus metallidurans CH34, an aerobic β‐proteobacterium that prevails in heavy metal‐rich environments.
The activity of CnrH is regulated by a complex of two transmembrane (TM) proteins: the metal‐sensor CnrX and the anti‐sigma factor CnrY. At the resting state, CnrH is sequestered at the membrane by CnrY (Fig. 3A), whereas binding of nickel or cobalt to CnrX frees CnrH from CnrY, allowing the expression of genes coding for proteins that extrude metals to the periplasm and the exterior (Fig. 3B) (Grass et al., 2005; Trepreau et al., 2011; Maillard et al., 2014; Trepreau et al., 2014).
Figure 3.
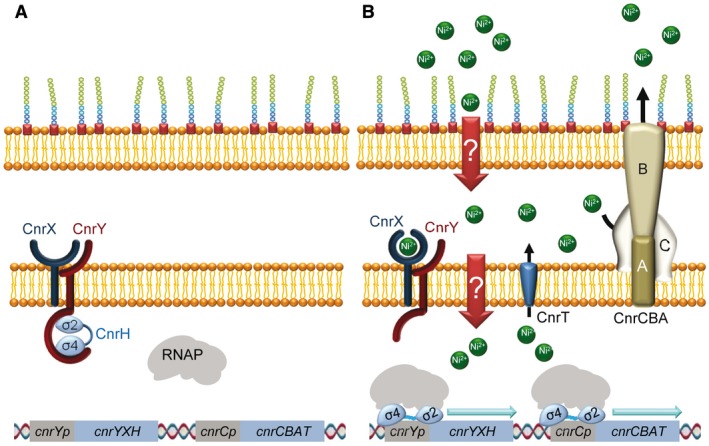
Mechanism of action of the C. metallidurans CnrH sigma factor. A. In the absence of nickel, CnrH is sequestered at the inner membrane by the protein complex CnrYX, where CnrX is an inner membrane protein that possesses a periplasmic metal sensor domain and CnrY is a transmembrane anti‐sigma factor. In this condition, CnrY wraps around CnrH and blocks the sites where the beta subunit of the RNAP binds. B. Nickel‐binding to CnrX results in a modification of the interaction between CnrX and CnrY that provokes a conformational change in CnrY, releasing CnrH to initiate transcription from the promoters cnrYp and cnrCp. Transcription from these promoters leads to synthesis of CnrH, CnrYX, the transenvelope complex CnrCBA and the exporter of cytoplasmic nickel ions CnrT.
The sensor CnrX is a membrane‐anchored dimeric protein with a C‐terminal periplasmic metal‐sensor domain. This portion binds Ni2+ or Co2+ and discriminates against the other transition metal cations, displaying subtle allosteric modifications depending on the nature of the metal ion (Trepreau et al., 2011; 2014). The affinity of CnrX for Ni2+ is 10‐ to 100‐fold higher than its affinity for Co2+, which is consistent with the expression observed in the genes under control of CnrH (Grass et al., 2000; 2005; Monchy et al., 2007; Trepreau et al., 2011; Maillard et al., 2015). These affinities are also in good agreement with the trend in the Irving‐Williams series. Accordingly, the complex CnrYXH plays a minor role in the resistance of C. metallidurans to Co2+ (Nies et al., 2006). At the resting state of this complex, zinc ions bind CnrX in a 3N2O coordination sphere (formed by His42, His46, Glu63 and His119). However, since nickel and cobalt exhibit a stronger affinity for CnrX, these ions displace the zinc ions at the metal binding site. The antagonistic effect of zinc on nickel and cobalt may be explained by the ability of nickel and cobalt to recruit the only methionine (Met123) of the sensor domain of CnrX as an additional ligand. This residue lies at the bottom of the cavity that harbors the metal ion and is central to the CnrX protomer architecture. Recruitment of Met123 to the coordination sphere of the metal results in a dramatic change in the geometry of the metal‐binding site that remodels the four‐helix bundle where this residue is located and elicits the biological response (Trepreau et al., 2011; 2014; Maillard et al, 2015).
After metal sensing in the periplasm, signal propagation proceeds through a modulation of the CnrX–CnrY interaction. On the periplasmic side, the C‐terminal portion of the anti‐sigma factor CnrY is docked in the hydrophobic cavity of the CnrX dimer facing the membrane. Within the membrane, interactions between the TM helices would be sensitive to any movement depending on the metal status of CnrX, affecting the interaction between the CnrX and CnrY TM domains (Fig. 3) (Trepreau et al., 2011). CnrY is a single‐pass TM protein with a 45 amino acid cytoplasmic domain that generates two helices that embrace the σ2 and σ4 domains of CnrH in a closed conformation so that the σ4 domain is buried against the −10 interaction surface of σ2, blocking the CnrH RNAP‐binding determinants (Grass et al., 2005; Maillard et al., 2014; Paget, 2015). CnrY belongs to class II of anti‐sigma domains (Grass et al., 2005; Maillard et al., 2014). Members of this class of anti‐sigma factors carry out their function via a short N‐terminal cytoplasmic domain that displays helical propensity but no canonical structure on its own (Staroń et al., 2009; Campagne et al., 2012; 2015; Huang et al., 2015).
Genes regulated by the CnrYXH complex are part of the cobalt‐nickel resistance (cnr) determinant cnrYXHCBAT borne in the megaplasmid pMOL28 of C. metallidurans CH34, consisting of two operons: cnrCBAT and cnrYXH. cnrCBAT is under control of the cnrCp promoter and encodes the transenvelope heavy‐metal efflux pump complex CnrCBA and the inner membrane exporter CnrT. At high concentrations of nickel, this metal is removed by CnrT from the cytoplasm to the periplasm, and from there to the exterior by CnrCBA (Fig. 3). On the other hand, cnrYXH is under the control of the cnrYp promoter and encodes the signal‐transduction system that regulates the expression of cnrCBAT (Liesegang et al., 1993; Grass et al., 2000; 2005; Monchy et al., 2007).
In addition to CnrH, the adaptive mechanism to nickel and cobalt in C. metallidurans is also regulated by two other ECF sigma factors, RpoE and RpoP. However, they do not respond to metals and their role covers the maintenance of cell wall integrity and the repair of nickel‐ and cobalt‐mediated damage (Grass et al., 2000; 2005; 2019; Tibazarwa et al., 2000).
Copper‐, cadmium‐ and zinc‐responsive ECF sigma factors
In most bacteria the expression of copper, cadmium and zinc resistance mechanisms is under control of one‐ and two‐component systems. But in the case of the myxobacterium Myxococcus xanthus three ECF sigma factors have been found to be involved in the adaptive mechanism to toxic concentrations of these metals. One of them is CarQ, an ECF sigma factor that responds to light and copper and regulates the biosynthesis of carotenoids. The others are members of the group ECF44, which respond to copper, zinc and/or cadmium and are known as CorE‐like sigma factors.
Mechanism of action of CarQ
M. xanthus is able to synthesize carotenoids in response to light and copper (Pérez et al., 2018) to quench singlet oxygen and other reactive oxygen species that are generated by these two environmental stimuli (Ziegelhoffer and Donohue, 2009). Genes involved in the synthesis of carotenoids are located in the operon carB, which contains six genes (crtE, crtIa, crtB, crtD, crtC and orf6), and the gene crtIb (Fig. 4). Expression of these seven genes is directly or indirectly regulated by the ECF sigma factor CarQ, which in the dark and in the absence of copper is sequestered at the membrane by the anti‐sigma factor CarR (Fig. 4A) (Gorham et al., 1996). Copper and light act at different levels to inactivate CarR and release CarQ. Light is indirectly sensed through CarF, which mediates signaling by the singlet oxygen generated via photoexcited protoporphyrin IX (Fontes et al., 2003; Elías‐Arnanz et al., 2011; Galbis‐Martínez et al., 2012). In this condition, CarF acts as an anti‐anti‐sigma factor to inactivate CarR by an unknown mechanism and release CarQ (Fig. 4B). In contrast, copper does not require CarF to release CarQ (Fig. 4B) (Moraleda‐Muñoz et al., 2005), although the mechanism by which CarR is inactivated by this metal remains to be elucidated. Once CarQ is released, the signal transduction pathway triggered is the same for both stimuli, which eventually up‐regulates the expression of carB (which was repressed by CarA and CarH, both encoded in the carA operon, consisting of five genes named crtYc, crtYd, orf9, carA and carH) and crtIb (directly regulated by CarQ) to synthesize carotenoids (see Fig. 4B for details) (Moraleda‐Muñoz et al., 2005; Elías‐Arnanz et al., 2011; Pérez et al., 2018).
Figure 4.
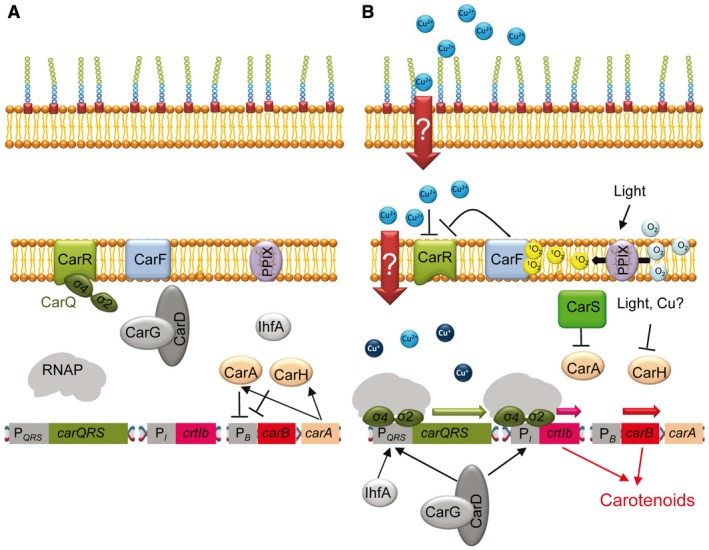
Mechanism of action of the M. xanthus CarQ sigma factor. A. In the absence of light and copper, the ECF sigma factor CarQ is sequestered at the membrane by the anti‐sigma factor CarR. Genes involved in carotenogenesis (located in the gene crtIb and the operon carB) are not expressed because crtIb is regulated by CarQ and the operon carB is repressed by CarA and CarH, encoded in the carA operon. B. Light is sensed via CarF, which is activated by singlet oxygen generated by photoexcited protoporphyrin IX (PPIX). In this condition, CarF functions as an anti‐anti‐sigma factor, inactivating CarR and releasing CarQ. Copper does not require CarF to inactivate CarR. Free CarQ by any of the two stimuli can bind to two promoters, PI, with the participation of CarD‐CarG, to express the gene crtIb, and PQRS, in conjunction with CarD‐CarG and IhfA, to express the operon carQRS. The operon carB can now be expressed after eliminating the repression by CarA and CarH. The repressor CarA is inactivated by CarS, which is encoded in the operon carQRS. CarH is a photoreceptor that is directly inactivated by light. Although it is not known how copper inactivates CarH, most likely this metal displaces cobalt in the vitamin B12 used as a cofactor by this repressor, thus allowing transcription of carB. Once carB and crtIb are expressed, carotenoids are synthesized. Arrows indicate positive regulation and blunt‐ended lines indicate negative regulation.
Although the CarR/CarQ pair mainly functions as a canonical anti‐sigma/sigma pair, one peculiarity is that CarQ requires additional proteins to bind to the promoters: IhfA and the CarD‐CarG pair to bind to PcarQRS (PQRS in Fig. 4), and only CarD‐CarG to bind to PcrtIb (PI in Fig. 4). IhfA is the α subunit of the integration host factor, which seems to be an essential architectural element of the appropriated macromolecular complex at the carQRS promoter (Moreno et al., 2001). CarD is a DNA‐binding architectural factor with similarities to the eukaryotic high mobility group A proteins (Padmanabhan et al., 2001). CarD always functions in conjunction with CarG, a zinc‐binding protein that regulates gene expression without binding to DNA. Instead, CarG interacts with the N‐terminal domain of CarD to function as a transcriptional regulatory unit (Peñalver‐Mellado et al., 2006). Interestingly, the CarD‐CarG complex is also required for proper activity by other M. xanthus ECF sigma factors, none of which are known to respond to metals (Abellón‐Ruiz et al., 2014).
Mechanism of action of the CorE‐like sigma factors
CorE‐like sigma factors (group ECF44) represent the best understood sigma factors regulated by C‐terminal extensions (Mascher, 2013; Pinto and Mascher, 2016). This mechanism provides a mode of action independent of an anti‐sigma factor, where the sensor domain responsible for triggering the response is part of the DNA‐binding sigma factor (see Pinto et al., 2019b for an overview on ECF sigma factors with regulatory extensions). Therefore, only one protein participates in this adaptive mechanism, whose activity is directly modulated by metals.
Even though only two members of the CorE‐like sigma factors have been characterized, 67 sigma factors have been predicted to fall within this group, with most of them encoded in a metal‐related genetic environment (Marcos‐Torres et al., 2016). These metal‐sensing ECF sigma factors can be identified because they contain two highly conserved regions: a CxC motif, located between the σ2 and σ4 domains, and a C‐terminal cysteine rich domain (CRD). Cysteines present in both regions are predicted to coordinate the metal, thus determining its activation state (Gómez‐Santos et al., 2011a; Marcos‐Torres et al., 2016; Pérez et al., 2018).
Both characterized CorE‐like sigma factors belong to the model myxobacterium M. xanthus: the copper‐regulated ECF sigma factor CorE (Gómez‐Santos et al., 2011a) and its homologue CorE2, which is regulated by cadmium and zinc (Marcos‐Torres et al., 2016; Pérez et al., 2018).
Although CorE and CorE2 are highly similar in sequence, they exhibit significant differences. CorE is involved in the immediate response to copper in M. xanthus, regulating the expression of the corE gene itself, genes for the P1B‐type ATPases CopA and CopB, and the multicopper oxidase CuoB (Sánchez‐Sutil et al., 2007; Moraleda‐Muñoz et al., 2010b; Gómez‐Santos et al., 2011a). Genes regulated by CorE exhibit a characteristic expression profile, in which expression levels rapidly increase after copper addition, reaching a peak at 2 h. Thereafter, expression rapidly decreases to basal levels in spite of the fact that copper is still present in the medium (Gómez‐Santos et al., 2011a). The use of chelators of Cu+, reducing agents and metals that mimic Cu+ and Cu2+ has revealed that this quick on/off molecular switch is caused by the ability of CorE to distinguish between the two oxidation states of copper, with Cu2+ acting as an activator and Cu+ as an inactivator (Fig. 5) (Gómez‐Santos et al., 2011a). Therefore, the resting state of CorE would be in the absence of copper (Fig. 5A) and in the presence of Cu+ (Fig. 5C), while it will be activated only in the presence of Cu2+ (Fig. 5B), which explains the typical expression profile of the genes regulated by CorE.
Figure 5.
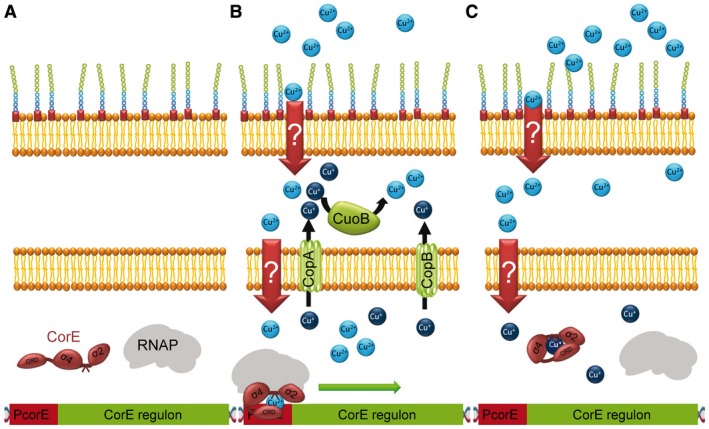
Mechanism of action of the M. xanthus CorE sigma factor. A. In the absence of copper, CorE remains inactive. B. When copper enters the cell, CorE is activated by binding this metal in its divalent oxidation state, initiating the transcription of genes involved in the immediate response to this metal. The immediate response includes the P1B‐type ATPases CopA and CopB, which will extrude copper to the periplasm, and the multicopper oxidase CuoB, which will oxidize Cu+ to Cu2+ in the periplasm. C. Due to the strongly reducing environment of the cytoplasm, Cu2+ will be quickly reduced to Cu+, inactivating the ECF sigma factor (presumably via a conformational change), and stopping the immediate response even when copper is still present in the medium.
In contrast, CorE2 is activated only in the presence of cadmium and zinc, and genes regulated by this sigma factor, among them a cation diffusion facilitator and a hypothetical protein with a glyoxal oxidase domain (Marcos‐Torres et al., 2016; Pérez et al., 2018), exhibit a different expression profile, in which expression rapidly increases after the metal addition, reaching a maximum that remains nearly constant thereafter (Marcos‐Torres et al., 2016). The difference between the expression profile exhibited by genes regulated by CorE2 and that of genes regulated by CorE seems to be related to the fact that cadmium and zinc are divalent metals with only one oxidation state. Therefore, the resting state of CorE2 will be in the absence of cadmium and zinc, while it will be active when bound to either of these metals (similar to the situation depicted in Fig. 5A and B, respectively, for CorE).
CorE and CorE2 share features that may be common to all members of the group ECF44 that differ from the canonical ECF sigma factors. One of those common traits is that they are not completely auto‐regulated. Even though there is some auto‐regulation in the case of corE expression, genes for both regulators have metal‐independent expression patterns (Gómez‐Santos et al., 2011a; Marcos‐Torres et al., 2016). Another common feature of CorE‐like sigma factors is the absence of an anti‐sigma factor that modulates their activity. Instead, the activation and inactivation of these sigma factors is controlled by the CxC motif and the CRD located at the carboxyl terminus. These two motifs are reminiscent of those present in the ZAS domains of the zinc‐binding anti‐sigma factors responsible for redox sensing (Jung et al., 2011; Devkota et al., 2017). However, neither the CRD nor CxC regions act as anti‐sigma factors, since deletion of the CRD or substitution of any cysteine in the CxC motif results in inactivation (instead of activation) of the sigma factor, despite keeping the σ2 and σ4 domains present in all ECF sigma factors (Gómez‐Santos et al., 2011a; Marcos‐Torres et al., 2016). Therefore, they are essential for activity.
Although the CRD domains of CorE and CorE2 are very similar, investigations into the role of each cysteine in both sigma factors have revealed that only one (Cys189 in CorE and 178 in CorE2) plays the same role in both regulators (Gómez‐Santos et al., 2011a; Marcos‐Torres et al., 2016). Interestingly, only one residue of each CRD seems to be the determinant for the metal specificity. Thus, the metal affinity of CorE and CorE2 (copper for CorE, and cadmium and zinc for CorE2) could be exchanged just by mutating a single amino acid of their CRD to the one found in the paralogous regulator (CorE Ala185 into a Cys and CorE2 Cys174 into an Ala) (Marcos‐Torres et al., 2016).
Whereas the CxC motif is strictly conserved in all ECF44 sigma factors, the length and cysteine distribution of the CRD vary between the different members. Its composition ranges from 21 to 50 residues and it may contain other metal‐binding residues such as methionines, aspartates and histidines. In view of the shift in metal recognition discussed previously, the differences in the CRDs of uncharacterized sigma factors in this group suggest that they may sense other metals (Marcos‐Torres et al., 2016).
Concluding remarks and future perspectives
Here we have reviewed the mechanisms developed by bacteria to adapt to fluctuations in metal concentrations that are mediated by ECF sigma factors. Interestingly, metal responsive ECF sigma factors share few common traits apart from participating in signal‐transduction mechanisms that are activated by the availability of metals and from regulating the expression of genes that modulate metal homeostasis.
Although metal‐responsive ECF sigma factors are small proteins that contain two conserved domains, they also exhibit sufficient differences in their sequences to be phylogenetically classified in different groups (Staroń et al., 2009; Mascher, 2013; Pinto and Mascher, 2016). Moreover, while the activity of most of these sigma factors is regulated by an anti‐sigma factor, CorE‐like sigma factors function in a different manner, as they are not sequestered by an anti‐sigma factor and require the presence of a metal (even in a specific redox state) to become activated. However, the most striking differences in the mechanism of action of these regulators are found in the proteins that function as metal sensors in each signal transduction pathway. Although all these sensor proteins can bind metals, their sequences do not exhibit significant similarities. In the pathways of the iron starvation‐responsive ECF sigma factors, the sensor is a TBDT located in the outer membrane. Both TBDTs, FecA and FpvA, bind iron, which is complexed with citrate and pyoverdine, respectively. The residues involved in binding these complexes are different in the two proteins due to the different chemical natures of these chelators. However, both TBDTs share a common domain structure consisting of a 22‐stranded β‐barrel with an inserted plug domain (Ferguson et al., 2002; Yue et al., 2003; Cobessi et al., 2005). Although the rest of sensors seem to directly bind specific metals, they do not exhibit sequence similarities, either. Moreover, in the CnrH pathway, the sensor is an inner membrane protein (CnrX) that interacts with the anti‐sigma factor CnrY. In the carotenogenesis pathway, copper seems to directly inactivate the inner membrane anti‐sigma factor CarR. And in the CorE‐like sigma factors, these loner cytoplasmic proteins directly bind the metal, bypassing the usual multiprotein complexes displayed by the other signaling mechanisms. These differences between sensor domains may be related to the type of metal that each protein recognizes, although the location of the protein sensor may also play a crucial role. For instance, it is known that proteins mainly use cysteines to bind copper in reducing intracellular compartments (cytoplasm), whereas they use methionines and histidines in the oxidizing compartments (periplasm) and the extracellular milieu (Frausto da Silva and Williams, 1991; Davis and O'Halloran, 2008; Sánchez‐Sutil et al., 2016). One intriguing question is why these metal sensor proteins are located at different locations. In the case of iron uptake, a location at the OM may be expected, since the same protein senses and transports a metal under conditions of iron depletion. However, an obvious reason is not found for metal sensors involved in metal detoxification, since some are located in the inner membrane (CnrX and CarQ) while others are located the cytoplasm (CorE and CorE2), in spite of the fact that CarQ and CorE respond to the same metal and in the same oxidation state (Cu2+).
Metal‐responsive ECF sigma factors may also have biotechnological applications. For instance, due to the specificity of several ECF sigma factors, bacteria could be designed to function as metal biosensors. Moreover, metal‐responsive promoters recognized by ECF sigma factors could be used to engineer plasmids that allow cheap heterologous gene expression, in a similar manner to how they are already available using promoters that are activated by two‐component systems that respond to copper (Gómez‐Santos et al., 2011b) or to non‐metal‐responsive ECF sigma factors (Pinto et al., 2019a).
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
This work has been supported by the Spanish Government, grant BFU2016‐75425‐P to Aurelio Moraleda‐Muñoz (70% funded by FEDER).
References
- Abellón‐Ruiz, J. , Bernal‐Bernal, D. , Abellan, M. , Fontes, M. , Padmanabhan, S. , Murillo, F.J. and Elías‐Arnanz, M. (2014) The CarD/CarG regulatory complex is required for the action of several members of the large set of Myxococcus xanthus extracytoplasmic function σ factors. Environmental Microbiology, 16, 2475–2490. [DOI] [PubMed] [Google Scholar]
- Angerer, A. , Enz, S. , Ochs, M. and Braun, V. (1995) Transcriptional regulation of ferric citrate transport in Escherichia coli K‐12. FecI belongs to a new subfamily of σ70‐type factors that respond to extracytoplasmic stimuli. Molecular Microbiology, 18, 163–174. [DOI] [PubMed] [Google Scholar]
- Baichoo, N. and Helmann, J.D. (2002) Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. Journal of Bacteriology, 184, 5826–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras, F. and Fontecave, M. (2011) Cobalt stress in Escherichia coli and Salmonella enterica: molecular bases for toxicity and resistance. Metallomics, 3, 1130–1134. [DOI] [PubMed] [Google Scholar]
- Beare, P.A. , For, R.J. , Martin, L.W. and Lamont, I.L. (2003) Siderophore‐mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Molecular Microbiology, 47, 195–207. [DOI] [PubMed] [Google Scholar]
- Biville, F. , Cwerman, H. , Létoffé, S. , Rossi, M.S. , Drouet, V. , Ghigo, J.M. , et al. (2004) Haemophore‐mediated signalling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in haem acquisition. Molecular Microbiology, 53, 1267–1277. [DOI] [PubMed] [Google Scholar]
- Boer, J.L. , Mulroone, S.B. and Hausinger, R.P. (2014) Nickel‐dependent metalloenzymes. Archives of Biochemistry and Biophysics, 544, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, V. , Mahren, S. and Sauter, A. (2006) Gene regulation by transmembrane signaling. BioMetals, 19, 103–113. [DOI] [PubMed] [Google Scholar]
- Brooks, B.E. and Buchanan, S.K. (2008) Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochimica et Biophysica Acta, 1778, 1930–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan, S.K. , Smith, B.S. , Venkatramani, L. , Xia, D. , Esser, L. , Palnitkar, M. , et al. (1999) Crystal structure of the outer membrane active transporter FepA from Escherichia coli . Nature Structural Biology, 6, 56–63. [DOI] [PubMed] [Google Scholar]
- Campagne, S. , Damberger, F.F. , Kaczmarczyk, A. , Francez‐Charlot, A. , Allain, F.H. and Vorholt, J.A. (2012) Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria. Proccedings of the National Academy of Sciences of the United States of America, 109, E1405–E1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne, S. , Allain, F.H. and Vorholt, J.A. (2015) Extra Cytoplasmic Function sigma factors, recent structural insights into promoter recognition and regulation. Current Opinion in Structural Biology, 30, 71–78. [DOI] [PubMed] [Google Scholar]
- Chandrangsu, P. , Rensing, C. and Helmann, J.D. (2017) Metal homeostasis and resistance in bacteria. Nature Reviews Microbiology, 15, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Yang, R. , Lyu, M. , Wang, S. , Liu, X. , Wen, Y. , et al. (2018) IdeR, a DtxR family iron response regulator, controls iron homeostasis, morphological differentiation, secondary metabolism, and the oxidative stress response in Streptomyces avermitilis . Applied and Environmental Microbiology, 84, e01503–01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier, S. , Bouffartigues, E. , Bazire, A. , Tahrioui, A. , Duchesne, R. , Tortuel, D. , et al. (2018) Extracytoplasmic function sigma factors in Pseudomonas aeruginosa . Biochimica et Biophysica Acta, 1862, 706–721. [DOI] [PubMed] [Google Scholar]
- Cobessi, D. , Celia, H. , Folschweiller, N. , Schalk, I.J. , Abdallah, M.A. and Pattus, F. (2005) The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. Journal of Molecular Biology, 347, 121–134. [DOI] [PubMed] [Google Scholar]
- Cornelis, P. , Matthijs, S. and Van Oeffelen, L. (2009) Iron uptake regulation in Pseudomonas aeruginosa . BioMetals, 22, 15–22. [DOI] [PubMed] [Google Scholar]
- Davis, A.V. and O'Halloran, T.V. (2008) A place for thioether chemistry in cellular copper ion recognition and trafficking. Nature Chemical Biology, 4, 148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota, S.R. , Kwon, E. , Ha, S.C. , Chang, H.W. and Kim, D.Y. (2017) Structural insights into the regulation of Bacillus subtilis SigW activity by anti‐sigma RsiW. PLoS ONE, 12, e0174284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, C.L. , Butcher, A. , Valas, R.E. , Bourne, P.E. and Caetano‐Anolles, G. (2010) History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proccedings of the National Academy of Sciences of the United States of America, 107, 10567–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Gebali, S. , Mistry, J. , Bateman, A. , Eddy, S.R. , Luciani, A. , Potter, S.C. , et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Research, 47, D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elías‐Arnanz, M. , Padmanabhan, S. and Murillo, F.J. (2011) Light‐dependent gene regulation in nonphototrophic bacteria. Current Opinion in Microbiology, 14, 128–135. [DOI] [PubMed] [Google Scholar]
- Ferguson, A.D. , Chakraborty, R. , Smith, B.S. , Esser, L. , van der Helm, D. and Deisenhofer, J. (2002) Structural basis of gating by the outer membrane transporter FecA. Science, 295, 1715–1719. [DOI] [PubMed] [Google Scholar]
- Festa, R.A. and Thiele, D.J. (2011) Copper: An essential metal in biology. Current Biology, 21, R877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillat, M. (2014) The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Archives of Biochemistry and Biophysics, 546, 41–52. [DOI] [PubMed] [Google Scholar]
- Fontes, M. , Galbis‐Martínez, L. and Murillo, F.J. (2003) A novel regulatory gene for light‐induced carotenoid synthesis in the bacterium Myxococcus xanthus . Molecular Microbiology, 47, 561–571. [DOI] [PubMed] [Google Scholar]
- Frausto da Silva, J.J.R. and Williams, R.J.P. (1991) The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. New York: Oxford University Press. [Google Scholar]
- Gaines, J.M. , Carty, N.L. , Tiburzi, F. , Davinic, M. , Visca, P. , Colmer‐Hamood, J.A. , et al. (2007) Regulation of the Pseudomonas aeruginosa toxA, regA and ptxR genes by the iron‐starvation sigma factor PvdS under reduced levels of oxygen. Microbiology, 153, 4219–4233. [DOI] [PubMed] [Google Scholar]
- Galbis‐Martínez, M. , Padmanabhan, S. , Murillo, F.J. and Elías‐Arnanz, M. (2012) CarF mediates signaling by singlet oxygen, generated via photoexcited protoporphyrin IX, in Myxococcus xanthus light‐induced carotenogenesis. Journal of Bacteriology, 194, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Santos, N. , Pérez, J. , Sánchez‐Sutil, M.C. , Moraleda‐Muñoz, A. and Muñoz‐Dorado, J. (2011a) CorE from Myxococcus xanthus is a copper‐dependent RNA polymerase sigma factor. PLoS Genetics, 7, e1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Santos, N. , Treuner‐Lange, A. , Moraleda‐Muñoz, A. , García‐Bravo, E. , García‐Hernández, R. , Martínez‐Cayuela, M. , et al. (2011b) A comprehensive set of integrative plasmid vectors for copper inducible gene expression in Myxococcus xanthus . Applied and Environmental Microbiology, 78, 2515–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham, H.C. , McGowan, S.J. , Robson, P.R. and Hodgson, D.A. (1996) Light‐induced carotenogenesis in Myxococcus xanthus: light‐dependent membrane sequestration of ECF sigma factor CarQ by anti‐sigma factor CarR. Molecular Microbiology, 19, 171–186. [DOI] [PubMed] [Google Scholar]
- Grass, G. , Grosse, C. and Nies, D.H. (2000) Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. strain CH34. Journal of Bacteriology, 182, 1390–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass, G. , Fricke, B. and Nies, D.H. (2005) Control of expression of a periplasmic nickel efflux pump by periplasmic nickel concentrations. BioMetals, 18, 437–448. [DOI] [PubMed] [Google Scholar]
- Grosse, C. , Poehlein, A. , Blank, K. , Schwarzenberger, C. , Schleuder, G. , Herzberg, M. and Nies, D.H. (2019) The third pillar of metal homeostasis in Cupriavidus metallidurans CH34: preferences are controlled by extracytoplasmic function sigma factors. Metallomics, 11, 291–316. [DOI] [PubMed] [Google Scholar]
- Gruber, T.M. and Gross, C.A. (2003) Multiple sigma subunits and the partitioning of bacterial transcription space. Annual Review of Microbiology, 57, 441–466. [DOI] [PubMed] [Google Scholar]
- Härle, C. , Kim, I. , Angerer, A. and Braun, V. (1995) Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO Journal, 14, 1430–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausinger, R.P. and Zamble, D.B. (2007) Microbial physiology of nickel and cobalt In: Nies D.H. and Silver S. (Eds.) Molecular Microbiology of Heavy Metals. Berlin: Springer‐Verlag, pp. 287–320. [Google Scholar]
- Helmann, J.D. (2002) The extracytoplasmic function (ECF) sigma factors. Advances in Microbial Physiology, 46, 47–110. [DOI] [PubMed] [Google Scholar]
- Ho, T.D. and Ellermeier, C.D. (2012) Extra cytoplasmic function σ factor activation. Current Opinion in Microbiology, 15, 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Pinto, D. , Fritz, G. and Mascher, T. (2015) Environmental sensing in Actinobacteria: a comprehensive survey on the signaling capacity of this phylum. Journal of Bacteriology, 197, 2517–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving, H. and Williams, R.J.P. (1953) The stability of transition metal complexes. Journal of the Chemical Society, 3192–3210. [Google Scholar]
- Jung, Y.G. , Cho, Y.B. , Kim, M.S. , Yoo, J.S. , Hong, S.H. and Roe, J.H. (2011) Determinants of redox sensitivity in RsrA, a zinc‐containing anti‐sigma factor for regulating thiol oxidative stress response. Nucleic Acids Research, 39, 7586–7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, F.H. , Ussery, D.W. , Nielsen, J. and Nookaew, I. (2011) A closer look at Bacteroides: phylogenetic relationship and genomic implications of a life in the human gut. Microbial Ecology, 61, 473–485. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M. and Shimizu, S. (1999) Cobalt proteins. European Journal of Biochemistry, 261, 1–9. [DOI] [PubMed] [Google Scholar]
- Koster, M. , van Klompenburg, W. , Bitter, W. , Leong, J. and Weisbeek, P. (1994) Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO Journal, 13, 2805–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, C. , Barnett, M.J. , Fisher, R.F. , Smith, L.S. , Diodati, M.E. and Long, S.R. (2018) Most Sinorhizobium meliloti extracytoplasmic function sigma factors control accessory functions. mSphere, 3, e00454‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.H. , Nam, K.H. and Helmann, J.D. (2013) A mutation of the RNA polymerase β' subunit (rpoC) confers cephalosporin resistance in Bacillus subtilis . Antimicrobial Agents and Chemotherapy, 57, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni, L. , Orsi, N. , de Lorenzo, V. and Visca, P. (2000) Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa . Journal of Bacteriology, 182, 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesegang, H. , Lemke, K. , Siddiqui, R.A. and Schlegel, H.G. (1993) Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. Journal of Bacteriology, 175, 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill, R. (2009) Function and biogenesis of iron‐sulphur proteins. Nature, 460, 831–838. [DOI] [PubMed] [Google Scholar]
- Lindeberg, M. , Myers, C.R. , Collmer, A. and Schneider, D.J. (2008) Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Molecular Plant‐Microbe Interactions, 21, 685–700. [DOI] [PubMed] [Google Scholar]
- Llamas, M.A. , Mooij, M.J. , Sparrius, M. , Vandenbroucke‐Grauls, C.M. , Ratledge, C. and Bitte, W. (2008) Characterization of five novel Pseudomonas aeruginosa cell‐surface signaling systems. Molecular Microbiology, 67, 458–472. [DOI] [PubMed] [Google Scholar]
- Llamas, M.A. , Imperi, F. , Visca, P. and Lamont, I.L. (2014) Cell‐surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiology Reviews, 38, 569–597. [DOI] [PubMed] [Google Scholar]
- Lonetto, M.A. , Brown, K.L. , Rudd, K.E. and Buttner, M.J. (1994) Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proccedings of the National Academy of Sciences of the United States of America, 91, 7573–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber, L. and Hausinger, R.P. (2011) Mechanisms of nickel toxicity in microorganisms. Metallomics, 3, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahren, S. and Braun, V. (2003) The FecI extracytoplasmic‐function sigma factor of Escherichia coli interacts with the subunit of RNA polymerase. Journal of Bacteriology, 185, 1796–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard, A.P. , Girard, E. , Ziani, W. , Petit‐Härtlein, I. , Kahn, R. and Covès, J. (2014) The crystal structure of the anti‐σ factor CnrY in complex with the σ factor CnrH shows a new structural class of anti‐σ factors targeting extracytoplasmic function σ factors. Journal of Molecular Biology, 426, 2313–2327. [DOI] [PubMed] [Google Scholar]
- Maillard, A.P. , Künnemann, S. , Grosse, C. , Volbeda, A. , Schleuder, G. , Petit‐Härtlein, I. , et al. (2015) Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding. Metallomics, 7, 622–631. [DOI] [PubMed] [Google Scholar]
- Marcos‐Torres, F.J. , Pérez, J. , Gómez‐Santos, N. , Moraleda‐Muñoz, A. and Muñoz‐Dorado, J. (2016) In depth analysis of the mechanism of action of metal‐dependent sigma factors: characterization of CorE2 from Myxococcus xanthus . Nucleic Acids Research, 44, 5571–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher, T. (2013) Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Current Opinion in Microbiology, 16, 148–155. [DOI] [PubMed] [Google Scholar]
- Miethke, M. and Marahiel, M.A. (2007) Siderophore‐based iron acquisition and pathogen control. Microbiology and Molecular Biology Reviews, 71, 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke, M. , Hou, J. and Marahiel, M.A. (2011) The siderophore‐interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli . Biochemistry, 50, 10951–10964. [DOI] [PubMed] [Google Scholar]
- Missiakas, D. and Raina, S. (1998) The extracytoplasmic function sigma factors: role and regulation. Molecular Microbiology, 28, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Monchy, S. , Benotmane, M.A. , Janssen, P. , Vallaeys, T. , Taghavi, S. , van der Lelie, D. and Mergeay, M. (2007) Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. Journal of Bacteriology, 189, 7417–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney, R.A. , Darst, S.A. and Landick, R. (2005) Sigma and RNA polymerase: an on‐again, off‐again relationship? Molecular Cell, 20, 335–345. [DOI] [PubMed] [Google Scholar]
- Moraleda‐Muñoz, A. , Pérez, J. , Fontes, M. , Murillo, F.J. and Muñoz‐Dorado, J. (2005) Copper induction of carotenoid synthesis in the bacterium Myxococcus xanthus . Molecular Microbiology, 56, 1159–1168. [DOI] [PubMed] [Google Scholar]
- Moraleda‐Muñoz, A. , Pérez, J. , Extremera, A.L. and Muñoz‐Dorado, J. (2010a) Differential regulation of six heavy metal efflux systems in the response of Myxococcus xanthus to copper. Applied and Environmental Microbiology, 76, 6069–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraleda‐Muñoz, A. , Pérez, J. , Extremera, A.L. and Muñoz‐Dorado, J. (2010b) Expression and physiological role of three Myxococcus xanthus copper‐dependent P1B‐type ATPases during bacterial growth and development. Applied and Environmental Microbiology, 76, 6077–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, A.J. , Fontes, M. and Murillo, F.J. (2001) ihfA gene of the bacterium Myxococcus xanthus and its role in activation of carotenoid genes by blue light. Journal of Bacteriology, 183, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulis, J.M. (2010) Cellular mechanisms of cadmium toxicity related to the homeostasis of essential metals. BioMetals, 23, 877–896. [DOI] [PubMed] [Google Scholar]
- Muñoz‐Dorado, J. , Gómez‐Santos, N. and Pérez, J. (2012) A novel mechanism of bacterial adaptation mediated by copper‐dependent RNA polymerase σ factors. Transcription, 3, 63–67. [DOI] [PubMed] [Google Scholar]
- Nairz, M. , Schroll, A. , Sonnweber, T. and Weiss, W. (2010) The struggle for iron ‐ a metal at the host‐pathogen interface. Cellular Microbiology, 12, 1691–1702. [DOI] [PubMed] [Google Scholar]
- Nies, D.H. (2004) Incidence and function of sigma factors in Ralstonia metallidurans and other bacteria. Archives of Microbiology, 181, 255–268. [DOI] [PubMed] [Google Scholar]
- Nies, D.H. , Rehbein, G. , Hoffmann, T. , Baumann, C. and Grosse, C. (2006) Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. Journal of Molecular Microbiology and Biotechnology, 11, 82–93. [DOI] [PubMed] [Google Scholar]
- Noinaj, N. , Guillier, M. , Barnard, T.J. and Buchanan, S.K. (2010) TonB‐dependent transporters: regulation, structure, and function. Annual Review of Microbiology, 64, 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner, U.A. , Wilderman, P.J. , Vasil, A.I. and Vasil, M.L. (2002) GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Molecular Microbiology, 45, 1277–1287. [DOI] [PubMed] [Google Scholar]
- Otero‐Asman, J. , Wettstadt, S. , Bernal, P. , and Llamas, M.A. (2019). Diversity of extracytoplasmic function sigma factor‐dependent signalling in Pseudomonas . Molecular Microbiology. 10.1111/mmi.14331. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, S. , Elías‐Arnanz, M. , Carpio, E. , Aparicio, P. and Murillo, F.J. (2001) Domain architecture of a high mobility group A‐type bacterial transcriptional factor. Journal of Biological Chemistry, 276, 41566–41575. [DOI] [PubMed] [Google Scholar]
- Paget, M.S. (2015) Bacterial sigma factors and anti‐sigma factors: structure, function and distribution. Biomolecules, 5, 1245–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalver‐Mellado, M. , García‐Heras, F. , Padmanabhan, S. , García‐Moreno, D. , Murillo, F.J. and Elías‐Arnanz, M. (2006) Recruitment of a novel zinc‐bound transcriptional factor by a bacterial HMGA‐type protein is required for regulating multiple processes in Myxococcus xanthus . Molecular Microbiology, 61, 910–926. [DOI] [PubMed] [Google Scholar]
- Pérez, J. , Muñoz‐Dorado, J. and Moraleda‐Muñoz, A. (2018) The complex global response to copper in the multicellular bacterium Myxococcus xanthus . Metallomics, 10, 876–886. [DOI] [PubMed] [Google Scholar]
- Pinto, D. and Mascher, T. (2016) The ECF classification: a phylogenetic reflection of the regulatory diversity in the extracytoplasmic function sigma factor protein family In: Bruijn F.J. (Ed.) Stress and Environmental Control of Gene Expression in Bacteria. Hoboken, NJ: Wiley‐Blackwell, pp. 64–96. [Google Scholar]
- Pinto, D. , Dürr, F. , Froriep, F. , Araújo, D. , Liu, Q. and Mascher, T. (2019a) Extracytoplasmic function σ factors can be implemented as robust heterologous genetic switches in Bacillus subtilis . iScience, 13, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, D. , Qiang, L. and Mascher, T. (2019b). ECF “sigma” factors with regulatory extensions: the one‐component systems of the “sigma” universe. Molecular Microbiology. 10.1111/mmi.14323. [DOI] [PubMed] [Google Scholar]
- Posey, J.E. and Gherardini, F.C. (2000) Lack of a role for iron in the Lyme disease pathogen. Science, 288, 1651–1653. [DOI] [PubMed] [Google Scholar]
- Ravel, J. and Cornelis, P. (2003) Genomics of pyoverdine‐mediated iron uptake in pseudomonads. Trends in Microbiology, 11, 195–200. [DOI] [PubMed] [Google Scholar]
- Rensing, C. and McDevitt, S.F. (2013) The copper metallome in prokaryotic cells. Metal Ions in Life Sciences, 12, 417–450. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Sutil, M.C. , Gómez‐Santos, N. , Moraleda‐Muñoz, A. , Martins, L.O. , Pérez, J. and Muñoz‐Dorado, J. (2007) Differential expression of the three multicopper oxidases from Myxococcus xanthus . Journal of Bacteriology, 189, 4887–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Sutil, M.C. , Marcos‐Torres, F.J. , Pérez, J. , Ruiz‐González, M. , García‐Bravo, E. , Martínez‐Cayuela, M. , et al. (2016) Dissection of the sensor domain of the copper‐responsive histidine kinase CorS from Myxococcus xanthus . Environmental Microbiology Reports, 8, 363–370. [DOI] [PubMed] [Google Scholar]
- Schalk, I.J. , Kyslik, P. , Prome, D. , Van Dorssealer, A. , Poole, K. , Abdallah, M.A. and Pattus, F. (1999) Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron‐free ligand: implication for siderophore‐mediated iron transport. Biochemistry, 38, 9357–9365. [DOI] [PubMed] [Google Scholar]
- Schalk, I.J. , Abdallah, M.A. and Pattus, F. (2002) A new mechanism for membrane iron transport in Pseudomonas aeruginosa . Biochemical Society Transactions, 30, 702–705. [DOI] [PubMed] [Google Scholar]
- Schalk, I.J. , Yue, W.W. and Buchanan, S.K. (2004) Recognition of iron‐free siderophores by TonB‐dependent iron siderophores. Molecular Microbiology, 54, 14–22. [DOI] [PubMed] [Google Scholar]
- Schulz, S. , Eckweiler, D. , Bielecka, A. , Nicolai, T. , Franke, R. , Dotsch, A. , et al (2015) Elucidation of sigma factor associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function‐specific crosstalk. PLoS Pathogens, 11, e1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton, R. , Gill, P.R. Jr , Dowling, D.N. and O'Gara, F. (1996) Transcriptional regulation of the iron‐responsive sigma factor gene pbrA . Molecular and General Genetics, 250, 50–58. [DOI] [PubMed] [Google Scholar]
- Silva, A.M. , Kong, X. , Parkin, M.C. , Cammack, R. and Hider, R.C. (2009) Iron (III) citrate speciation in aqueous solution. Dalton Transaction, 28, 8616–8625. [DOI] [PubMed] [Google Scholar]
- Staroń, A. , Sofia, H.J. , Dietrich, S. , Ulrich, L.E. , Liesegang, H. and Mascher, T. (2009) The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Molecular Microbiology, 74, 557–581. [DOI] [PubMed] [Google Scholar]
- Staudenmaier, H. , van Hove, B. , Yaraghi, Z. and Braun, V. (1989) Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic‐binding‐protein‐dependent transport mechanism for iron(III) dicitrate in Escherichia coli . Journal of Bacteriology, 171, 2626–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, P.B. , Vaughn‐Diaz, V.L. , Greenwald, J.W. and Gross, D.C. (2013) Characterization of five ECF sigma factors in the genome of Pseudomonas syringae pv. syringae B728a. PLoS ONE, 8, e58846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibazarwa, C. , Wuertz, S. , Mergeay, M. , Wyns, L. and van Der Lelie, D. (2000) Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. Journal of Bacteriology, 182, 1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepreau, J. , Girard, E. , Maillard, A.P. , de Rosny, E. , Petit‐Haertlein, I. , Kahn, R. and Covès, J. (2011) Structural basis for metal sensing by CnrX. Journal of Molecular Biology, 408, 766–779. [DOI] [PubMed] [Google Scholar]
- Trepreau, J. , Grosse, C. , Mouesca, J.M. , Sarret, G. , Girard, E. , Petit‐Haertlein, I. , et al. (2014) Metal sensing and signal transduction by CnrX from Cupriavidus metallidurans CH34: role of the only methionine assessed by a functional, spectroscopic, and theoretical study. Metallomics, 6, 263–273. [DOI] [PubMed] [Google Scholar]
- Van Hove, B. , Staudenmaier, H. and Braun, V. (1990) Novel two‐component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K‐12. Journal of Bacteriology, 172, 6749–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visca, P. , Imperi, F. and Lamont, I.L. (2007) Pyoverdine siderophores: from biogenesis to biosignificance. Trends in Microbiology, 15, 22–30. [DOI] [PubMed] [Google Scholar]
- Waldron, K.J. , Rutherford, J.C. , Ford, D. and Robinson, N.J. (2009) Metalloproteins and metal sensing. Nature, 460, 823–830. [DOI] [PubMed] [Google Scholar]
- Wandersman, C. and Delepelaire, P. (2004) Bacterial iron sources: from siderophores to hemophores. Annual Reviews in Microbiology, 58, 611–647. [DOI] [PubMed] [Google Scholar]
- Wilderman, P.J. , Vasil, A.I. , Johnson, Z. , Wilson, M.J. , Cunliffe, H.E. , Lamont, I.L. and Vasil, M.L. (2001) Characterization of an endoprotease (PrpL) encoded by a PvdS‐regulated gene in Pseudomonas aeruginosa . Infection and Immunity, 69, 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth, C. , Meyer‐Klaucke, W. , Pattus, F. and Cobessi, D. (2007) From the periplasmic signaling domain to the extracellular face of an outer membrane signal transducer of Pseudomonas aeruginosa: crystal structure of the ferric pyoverdine outer membrane receptor. Journal of Molecular Biology, 368, 398–406. [DOI] [PubMed] [Google Scholar]
- Wriedt, K. , Angerer, A. and Braun, V. (1995) Transcriptional regulation from the cell surface: conformational changes in the transmembrane protein FecR lead to altered transcription of the ferric citrate transport genes in Escherichia coli . Journal of Bacteriology, 177, 3320–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Chiang, H.C. , Bjursell, M.K. and Gordon, J.I. (2004) Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends in Microbiology, 12, 21–28. [DOI] [PubMed] [Google Scholar]
- Yue, W.W. , Grizot, S. and Buchanan, S.K. (2003) Structural evidence for iron‐free citrate and ferric citrate binding to the TonB‐dependent outer membrane transporter FecA. Journal of Molecular Biology, 332, 353–368. [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer, E.C. and Donohue, T.J. (2009) Bacterial responses to photooxidative stress. Nature Review Microbiology, 7, 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]


