Abstract
Pharmacogenetic testing can help identify primary care patients at increased risk for medication toxicity, poor response or treatment failure and inform drug therapy. While testing availability is increasing, providers are unprepared to routinely use pharmacogenetic testing for clinical decision-making. Practice-based resources are needed to overcome implementation barriers for pharmacogenetic testing in primary care.The NHGRI’s IGNITE I Network (Implementing GeNomics In pracTicE; www.ignite-genomics.org) explored practice models, challenges and implementation barriers for clinical pharmacogenomics. Based on these experiences, we present a stepwise approach pharmacogenetic testing in primary care: patient identification; pharmacogenetic test ordering; interpretation and application of test results, and patient education. We present clinical factors to consider, test-ordering processes and resources, and provide guidance to apply test results and counsel patients. Practice-based resources such as this stepwise approach to clinical decision-making are important resources to equip primary care providers to use pharmacogenetic testing.
Keywords: : clinic, implementation, pharmacogenetics, pharmacogenomics, precision medicine, primary care
Pharmacogenetic testing can help predict variability in drug response to identify patients at risk for adverse effects or treatment failure with selected drugs. Although significant needs remain in building the clinical evidence base, pharmacogenetic testing is increasingly available across diverse practice settings to guide medication selection and dosing [1–4]. Much of the interest in pharmacogenetic and other genomic tests has been driven at the patient level by growth in the direct-to-consumer genetic testing marketing, which is expected to reach $6.36 billion dollars by 2028 [5]. This uptick in patient and provider interest in pharmacogenetic testing has been particularly notable in the primary care setting, where the majority of prescriptions in the USA are written [6]. Nearly 30% of primary care patients take a medication that is associated with genetic variability, with more than 300 medications having pharmacogenetic information in the US FDA-approved label [3,7,8]. One analysis found that 65% of medical home (mostly primary care) patients at one academic medical center were exposed to medications affected by pharmacogenetic variability over a 5-year period. Study authors concluded that implementation of pharmacogenetic testing could have potentially averted 383 severe adverse events [9]. It is anticipated that as precision medicine grows, the demand for primary care providers to apply pharmacogenetic principles in practice will rise sharply [10–13].
The primary care setting is a logical clinical entry point for pharmacogenetic testing. Primary care providers are an initial point of access into the healthcare system, focus on preventive care, treat a wide variety of illnesses, and communicate frequently with patients and other clinicians [14–17]. They are ideally positioned to identify patients who may need a pharmacogenetic test, determine prior testing, order necessary tests, and document and communicate test results to patients and other clinicians. Pharmacogenetic testing is also consistent with the longitudinal chronic care model employed in primary care that utilizes a patient- and community-centered approach [16,18,19]. Finally, incorporating pharmacogenetic testing into primary care is consistent with the evolution of this practice area, in which providers have long taken on new roles in specialized areas, such as HIV drug therapy and detection and management of certain cancers [20–22].
However, in spite of increasing accessibility of pharmacogenetic testing and its potential to guide the use of many medications commonly prescribed in primary care (e.g., selective serotonin reuptake inhibitors, codeine and tramadol), routine testing remains limited [8,11,12,23–25]. This lack of real-world adoption is driven, in large part, by clinical knowledge and evidence gaps in pharmacogenetics. In recent surveys, physician respondents agreed that genetic variability influences drug response, but fewer than 20% had ordered a pharmacogenetic test in the previous year. Less than 15% of physicians reported feeling informed about pharmacogenetic testing, with prescribers consistently expressing a desire for additional evidence and clinical guidance on applications and use of pharmacogenetics in practice [25–28]. Practical barriers have also slowed uptake of clinical pharmacogenetic testing, including poor access to systems to facilitate testing and return of results, electronic health record limitations, and sparse clinical resources [16,18,29–31]. Research in this area is ongoing and experience is increasing with strategies to help clinicians overcome these barriers, such as use of patient-centered and case-based provider education, clinical decision support, novel test reimbursement strategies and interdisciplinary collaboration [32].
However, it is our experience that clinical resources to help prescribers use pharmacogenetic and other precision medicine data in practice are not being developed quickly enough to keep pace with the rapidly growing commercial and direct-to-consumer genomic testing market. As we move into a future of increasing consumer and clinician access to precision medicine data, it is imperative that primary care clinicians and the healthcare system as whole are prepared to face this reality and use healthcare resources to apply pharmacogenetic data in an informed, efficient, and evidence-based manner that compliments routine clinical care. To achieve this goal, primary care clinicians and other frontline providers need clear clinical guidance to order pharmacogenetic tests, interpret results, and answer patients' questions in practice (e.g., from commercial clinical laboratories or DTC testing), while researchers continue to build the pharmacogenetic evidence base [26,33–35].
The National Human Genome Research Institute established and funded the IGNITE I Network in 2013 (Implementing GeNomics In pracTicE; https://gmkb.org/) to develop and test clinical models for using genomic information in diverse practice settings, including primary care [36]. All IGNITE I research sites actively investigated practice models, needs, challenges and pragmatic strategies to overcome implementation barriers in primary care settings. IGNITE I investigators explored methods to engage nongenetics specialists, specifically primary care providers, to manage pharmacogenetic tests and guide clinical decision-making. Our experiences have confirmed an urgent need for solutions to systems- and provider-level and challenges. While many barriers require systems-level solutions for education, reimbursement, point-of-care clinical decision support and increased capabilities for storage and use of pharmacogenetic data within electronic health records, in our experiences, provider-level guidance and resources are essential to support clinical adoption of genomic testing [16,18,19,26,29–31,33–35].
Herein, we present a stepwise process for ordering pharmacogenetic testing in primary care with clinical considerations that we have consistently seen emerge at each step. We hope that describing this process may provide a clinical scaffolding for prescribers who are interested in ordering pharmacogenetic tests but are unsure how to approach this process clinically. This process may also be of benefit for prescribers who are seeking a systematic approach to clinical pharmacogenetics to streamline provider education, use a consistent clinical approach to pharmacogenetic testing or improve efficiency and documentation of the testing and patient education process. Of note, in developing this stepwise process, we targeted an audience of clinicians who have already decided to adopt pharmacogenetic testing in their practice. We therefore do not address preimplementation planning steps (e.g., stakeholder engagement and provider education) or provide an extensive review of clinical utility literature for specific gene–drug pairs.
A stepwise approach to implementing pharmacogenetic testing
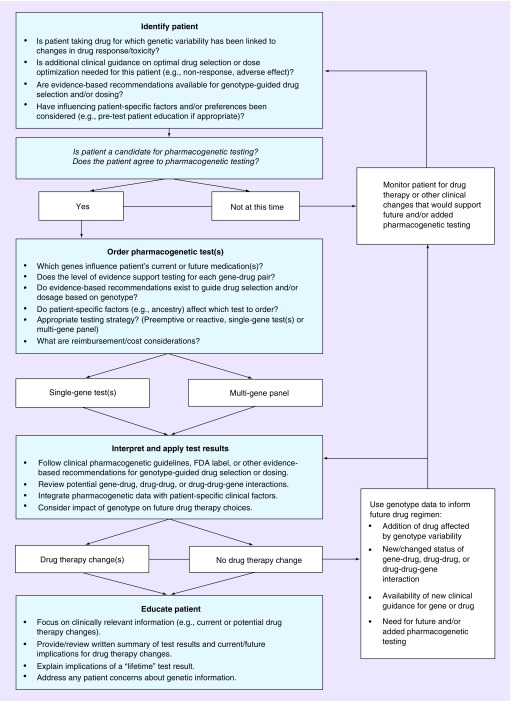
Although specific clinical needs and workflow vary among practices, we have observed the emergence of four primary steps and a number of clinical considerations that consistently arise with adoption of pharmacogenetic testing in the primary care setting. These steps include: patient Identification, test ordering, application of test results and patient education (Figure 1). This list of steps is not intended to be inclusive or prescriptive. Instead, providers can adapt the steps and considerations below to their individual practice structure, patient needs and clinical priorities.
Figure 1. . Clinical decision making process for pharmacogenetic testing.
Step 1: patient identification
When determining whether an individual patient may be a candidate for pharmacogenetic testing, providers should consider a patient’s medications, the level of evidence supporting a link between variability in a specific gene and clinically meaningful changes in drug effects or outcomes, the prevalence of specific genetic variants in individual patient subpopulations, and patient-specific factors. As with all tests, pharmacogenetic testing should be reserved for patients in whom test results will provide clinically meaningful information. This may include individuals currently taking or likely to need a drug for which genetic variability has been linked to drug response or toxicity and for which evidence-based clinical recommendations exist to guide drug dosing and/or selection based on genotype (Table 1). Within these agents, pharmacogenetic testing can be considered in patients who are not responding to or experiencing toxicity from an existing drug, patients taking multiple medications, those who have comorbid conditions that complicate drug therapy selection and/or dosing, or those who could otherwise benefit from pharmacogenetic data when initiating or changing treatment regimens.
Table 1. . Gene–drug pairs used in primary care with supporting clinical evidence.
| Medication(s) | Gene | CPIC guideline | PGx in US FDA label |
|---|---|---|---|
| Opioids/Analgesics | |||
| Codeine, tramadol | CYP2D6 | X | X |
| Celecoxib | CYP2C9 | X | |
| Selective serotonin reuptake inhibitors | |||
| Paroxetine, fluvoxamine | CYP2D6 | X | X |
| Citalopram, escitalopram | CYP2C19 | X | X |
| Sertraline | CYP2C19 | X | |
| Tricyclic antidepressants | |||
| Amitriptyline, imipramine |
CYP2C19 CYP2D6 |
X X |
X |
| Nortriptyline, desipramine | CYP2D6 | X | X |
| Doxepin, trimipramine |
CYP2C19 CYP2D6 |
X X |
X X |
| Other antidepressants | |||
| Venlafaxine, vortioxetine | CYP2D6 | X | |
| Proton pump inhibitors | |||
| Esomeprazole, lansoprazole, dexlansoprazole, omeprazole, pantoprazole, rabeprazole | CYP2C19 | X | |
| Cardiovascular medications | |||
| Clopidogrel | CYP2C19 | X | X |
| Simvastatin | SLCO1B1 | X | X |
| Warfarin |
CYP2C9 CYP4F2 VKORC1 |
X X X |
X X |
| Antipsychotics | |||
| Aripiprazole, risperidone, brexpiprazole | CYP2D6 | X | |
| Anticonvulsants | |||
| Carbamazepine |
HLA-A HLA-B |
X X |
X X |
| Oxcarbazepine | HLA-B | X | X |
| Phenytoin |
CYP2C9 HLA-B |
X X |
X X |
| Other medications | |||
| Allopurinol | HLA-B | X | |
| Atomoxetine | CYP2D6 | X | X |
| Ondansetron | CYP2D6 | X | X |
| Tamoxifen | CYP2D6 | X | |
Table includes medications with a Clinical Pharmacogenetics Implementation ‘A’ or ‘B’ evidence-level rating that are relevant to a primary care practice setting (source: www.cpicpgx.org).
CPIC: Clinical Pharmacogenetics Implementation Consortium; PGx: Pharmacogenetics.
Once a patient is identified as a candidate for pharmacogenetic testing, clinicians should consider having a brief discussion of the rationale, benefits and limitations of testing; implications for informing current and future drug therapy selection/changes; and potential testing strategies and reimbursement options to ensure joint clinician-patient decision-making. This discussion could be woven into traditional discussions in which they explain why they recommend specific medication choices, akin to discussing the use of renal function testing. There is no consensus on the need for an informed consent process for pharmacogenetic testing [37–40].
Step 2: pharmacogenetic test ordering
Providers should determine which pharmacogenetic tests should be ordered and identify the optimal testing method. In our experience, it can be helpful, at least initially, to approach this step from the perspective of ‘gene–drug’ pairs since the specific pharmacogenetic test(s) that is/are needed may differ for the same gene within varying patient populations, drugs or dosages, or even drug indications. The gene–drug pairs listed in Table 1 include medications commonly encountered in primary care that have the highest level of evidence across diverse patient populations and also have clinical guidance available to inform drug therapy changes. We recommend limiting genotype-based drug therapy changes to these and other gene–drug pairs with strong supporting evidence to maximize time and resource efficiency and provide the highest likelihood of a meaningful change in clinical outcomes. If a gene–drug pair is not included in this table and providers are unsure whether to order a pharmacogenetic test, we recommend consulting PharmGKB (www.pharmgkb.org/), an NIH-funded searchable online pharmacogenomics knowledgebase that has clinical evidence summaries to inform decision-making [41].
Pharmacogenetic tests are generally ordered as single-gene tests (e.g., CYP2D6 alone) or as part of a broader multigene panel [42]. There are advantages and disadvantages to these two strategies from clinical, practical and reimbursement perspectives [43,44]. Panel-based, multigene testing is often ‘preemptively’, in which test results for a broad number of genes are available in the medical record to be used at a later time when needed. Single gene tests are usually ordered ‘reactively’ (e.g., a CYP2D6 test to provide clinical insight in a patient who is experiencing a toxicity or a poor response to a specific drug). Therefore, panel-based testing allows providers to consider pharmacogenetic data before they prescribe a drug and use these data to guide future drug therapies and thus may offer certain cost and care advantages over single gene tests. Reactive single-gene test results are usually not available prior to initial drug use and may delay the time to treatment optimization. However, panel-based tests may be more challenging to order and/or access testing information. Panel-based test results are commonly available through standalone commercial laboratories, may require additional steps for test ordering and sample processing, and test results are usually reported in an external online portal or in a printed laboratory report that must be scanned and uploaded into the electronic health record. Single-gene tests are more likely to be offered by mainstream clinical laboratories and are usually ordered, processed and reported back to clinicians in the same manner as other laboratory tests.
Reimbursement for pharmacogenetic testing varies widely among insurance carriers. Single-gene tests ordered for specific clinical situations (e.g., a CYP2C19 test ordered in a postacute coronary syndrome patient taking clopidogrel) that have established current procedural terminology (CPT) codes are currently more likely to be covered by insurers than panel-based preemptive pharmacogenetic tests or those without CPT codes [45]. Commercial laboratories that offer panel-based testing may provide reimbursement support for patients through patient assistance programs, income-based sliding scale payment models and/or by helping individuals navigate the reimbursement process. This support is generally not provided by mainstream laboratory companies with single-gene testing. Clinicians should recommend patients contact their insurance provider directly to determine coverage if they are not confident a test or panel will be covered by insurance.
The IGNITE Network developed an interactive map of pharmacogenetic test reimbursement providing state-specific information coverage of genetic tests according to the Medicare Administrative Contractor, a list of pharmacogenetic tests determined to be medically necessary for Medicare coverage, and indications and noncovered indications for nonmedically necessary pharmacogenetic tests, available at: https://gmkb.org/ignite/. Information about which laboratories perform specific pharmacogenetic tests is freely available online through the National Center for Biotechnology Information’s Genetic Testing Registry (www.ncbi.nlm.nih.gov/gtr/) by searching for a targeted gene (e.g., CYP2C19).
Step 3: application of pharmacogenetic test results
Although turnaround time varies, pharmacogenetic test results are generally accessible within a few days to a couple of weeks after a test is ordered. Once results are available, they must be interpreted and integrated into evidence-based clinical recommendations for genotype-guided drug therapy changes (adding, discontinuing, changing medications or changing dosages) alongside individual patient-specific factors. The PharmGKB website (www.pharmgkb.org/) is an essential resource to help clinicians understand and interpret pharmacogenetic test results if additional information is needed.
In regards to clinical recommendations, most disease/treatment guidelines are silent on pharmacogenetic testing at this time, primarily due to the lack of randomized controlled trials definitively linking genotype-guided therapy to improved patient outcomes [46]. Until such data are available, multiple scientific groups have developed consensus recommendations based on the best available clinical evidence, which may include pharmacokinetic or pharmacodynamic studies, multisite pooled analyses, pragmatic clinical trials, and retrospective and/or observational data. The Clinical Pharmacogenetics Implementation Consortium (CPIC; www.cpicpgx.org), has developed more than 20 peer-reviewed, evidence-based guidelines with genotype-guided dosing recommendations for individual drugs and/or drug classes [47–50]. Guidelines are also available from other groups, including the Dutch Pharmacogenetics Working Group (www.pharmgkb.org/page/dpwg) and the European Pharmacogenomics Implementation Consortium (www.eu-pic.net). When CPIC guidelines are available, we recommend using them first to inform drug therapy changes as they provide the most updated, evidence-based recommendations for genotype-guided dosing or drug selection.
Additional pharmacogenetic recommendations are available in US FDA-approved labeling for more than 300 medications [3], although many of these are indicated to treat specialized conditions that may be encountered infrequently in primary care. Pharmacogenetic biomarker information is particularly helpful when a significant patient safety issue has been identified in drug development or through postmarketing adverse event reports. Drug labeling may include data supporting specific dose recommendations (e.g., eliglustat dosing based on CYP2D6 genotype), boxed warnings (e.g., life-threatening respiratory depression with codeine use in CYP2D6 ultrarapid metabolizers) and informational data (e.g., drug level may be affected by CYP2D6 variability or enzyme inhibition). A listing of drugs and labeling recommendations is available through the FDA’s Table of Pharmacogenomic Biomarkers (www.fda.gov/drugs/science-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling) and this information is curated on the PharmGKB website alongside clinical guideline recommendations. If guidelines or FDA labeling are not available for a specific gene–drug pair, PharmGKB also provides level-of-evidence ratings (levels 1 through 4, with level 1 representing the strongest evidence) for individual gene–drug pairs that can be found by searching for the gene or drug on the PharmGKB website.
Genotype-based recommendations may also be included in the laboratory report accompanying a pharmacogenetic test result. However, we strongly recommend clinicians review clinical guidelines (e.g., CPIC) and/or FDA-approved labeling prior to making drug therapy changes. This is especially true with commercial laboratories that test for multiple genes and/or drugs that have varying levels of evidence as well as for laboratory reports that display drug therapy recommendations in simplified proprietary formats (e.g., categorizing drugs as ‘safe’ or ‘unsafe’ without a clear evidentiary link). In our experience, lengthy commercial laboratory reports may ‘information overload’ and often fail to effectively communicate the degree of variability in supporting evidence among individual gene–drug pairs. Reports may also make clinical recommendations based on lower-quality evidence (e.g., conflicting studies, small sample sizes, preliminary results not yet confirmed or replicated) [13,42]. Although it may be necessary at times in practice to make drug therapy decisions based on the best available clinical evidence, we feel it essential that providers are aware of the level of evidence supporting specific clinical pharmacogenetic recommendations to better integrate this information with other patient-specific factors. When available, providers should also refer to point-of-care clinical decision support resources for pharmacogenetic testing, such as automated electronic health record alerts that fire at the time of prescribing, test order or return of results [51].
It is essential that clinicians integrate available genotype-guided dosing recommendations with other clinical and patient-specific factors that influence drug response for each individual. For example, CYP2D6 genotype data can help predict response and/or toxicity with selected opioids in patients with chronic pain but should be considered alongside other influencing factors such as patients’ previous response to opioids, concomitant medical conditions, and individual insurance and financial considerations. Consideration of the effects of drug–drug, drug–gene (e.g., genotype affects drug response) and drug–drug–gene interactions (e.g., genotype affects drug response and drug–drug interaction exists) is particularly important in pharmacogenetics since many common medications used in primary care may inhibit or induce CYP enzymes that are also affected by genetic variability (Table 2) [52,53].
Table 2. . Common medications that may inhibit or induce CYP2C19 or CYP2D6 enzyme activity.
| Inhibitor(s) | Inducer(s) | |
|---|---|---|
| CYP2C19 | Strong: – Fluconazole – Fluoxetine – Fluvoxamine Moderate: –– Weak: – Omeprazole – Voriconazole |
Strong: – Rifampin – Ritonavir Moderate: – Efavirenz – Phenytoin Weak: –– |
| CYP2D6 | Strong: – Bupropion – Fluoxetine – Paroxetine Moderate: – Cimetidine – Duloxetine – Fluvoxamine Weak: – Amiodarone – Celecoxib – Cimetidine – Desvenlafaxine – Escitalopram – Labetalol – Ritonavir – Sertraline |
None |
Source: FDA, drug development and drug interactions: table of substrates, inhibitors and inducers. Available at: www.fda.gov/Drugs/default.htm (accessed July 24, 2019).
Step 4: patient education
As with other laboratory tests, the primary goal when educating patients about pharmacogenetics is to ensure that they understand their test results and their significance for their treatment [40,54]. Patient understanding of pharmacogenetic test results can positively impact confidence in their medications and boost adherence to drug therapy regimens [55,56]. A key component to achieving this understanding is using effective communication strategies based on clinical education standards, including an individualized, patient-centered, structured and interactive approach [37,57–59]. When communicating pharmacogenetic information, providers should focus primarily on clinically relevant information, including current or potential future drug therapy changes based on the patient’s genetics [40,60]. It may be more productive to explain test results in terms of the clinical effects, such as having a slower or faster ability to activate or breakdown a specific drug, rather than to focus on specific genotype results [7,38,54].
Whenever possible, clinicians should provide patients written or online summaries (e.g., via patient portal) of pharmacogenetic test results that distill the most personally and clinically relevant information for patients [57–59]. These can reinforce patient understanding and serve as long-term documentation of lifetime test results that patients can maintain and share with other healthcare providers within or outside the ordering provider’s healthcare system [38,54,60]. These results should be simple, to the point and written at the appropriate health literacy level. Unfortunately, most currently available commercial lab reports for multigene panels do not meet this standard. An analysis of sample pharmacogenetic test reports from eight commercial laboratories revealed reports up to 27 pages in length, with an average of 14 pages for multigene panel reports, reinforcing patient education as an important need and immediate challenge with clinical pharmacogenetic testing [13].
The PCPs should anticipate areas of patient concern with pharmacogenetic testing and address them through education. Patients generally view pharmacogenetic testing positively and value its potential role in improving their medication regimen [38,61–64], but they have expressed concerns about potential disease implications or possible genetic discrimination with pharmacogenetic testing [38,62]. Providers can inform patients that the vast majority of clinical pharmacogenetic testing to assess variability in drug metabolizing enzymes has not been associated with disease risk [31,63]. Providers should also be conscious of the language and terminology they use to explain test results [54]. Terms such ‘mutant’, ‘defective’, or ‘abnormal’, may be stigmatizing; value-neutral, low health literacy terms such as ‘working’ or ‘non-working’ genes, ‘faster’ or ‘slower’ drug metabolism will likely be better received by patients [54,65]. Although the targeted nature of pharmacogenetic data mean, there are minimal disease-risk associations if patients have questions about how their genetic information is protected, educational resources about the Genetic Information Nondiscrimination Act of 2008 are available at www.GINAhelp.org.
Finally, as with other tests of genetic data, it is helpful for patients to understand the lifetime nature of pharmacogenetic test results. Patients should keep their written record of test results and share them with future providers as appropriate. Payers may not cover future repeat pharmacogenetic tests since results do not change over the patient’s lifetime. Patients should also be aware of the emerging nature of pharmacogenetic evidence for individual drug prescribing. As evidence develops or new data are published, patients may receive future communication about potential implications for drug therapy based on their current test results [56,58].
Conclusion
There is currently an imbalance between the potential benefits and demand for pharmacogenetic testing in primary care and its underuse to guide medication selection and dosing. Providers and patients value pharmacogenetic testing, but PCPs consistently report feeling unprepared to use pharmacogenetic data and cite a lack of point-of-care resources for applying test results in practice. Practice-based models and resources for pharmacogenetic testing are essential to equip prescribers for pharmacogenetics. In this manuscript, we present a practice-based stepwise approach to pharmacogenetic testing in primary care based on implementation experiences of the IGNITE Network in this manuscript. Synergy of practice-based resources such as this stepwise approach with systems-level solutions to technical, reimbursement, educational and other challenges will enable the healthcare system as a whole to integrate pharmacogenetic testing into patient care in a scalable, efficient manner.
Footnotes
Financial & competing interests disclosure
This work was supported by National Institutes of Health grants (grant numbers U01 HG007269, U01 GM074492, U01 HL105198 [KW Weitzel]; 5U01HG7278, 3U01HG00871-02S1, U01HG006380 [A Owusu-Obeng]; U01HG007253 [JC Denny]; U01HG007775 [TI Pollin]; 1U01HG007282-01 [RR Wu]; and National Center for Advancing Translational Sciences grants UL1TR000064 and UL1TR001427 [KW Weitzel]). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature 526(7573), 343–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans WE, McLeod HL. Pharmacogenomics – drug disposition, drug targets, and side effects. N. Engl. J. Med. 348(6), 538–549 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Administration FaD. Table of pharmacogenomic biomarkers in drug labeling (2019). www.fda.gov/Drugs/ScienceResearch/ucm572698.htm
- 4.Roden DM, Wilke RA, Kroemer HK, Stein CM. Pharmacogenomics: the genetics of variable drug responses. Circulation 123(15), 1661–1670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Global direct-to-consumer genetic testing market to reach $6.36 billion by 2028. PRNewswire (2019).. www.prnewswire.com/news-releases/global-direct-to-consumer-genetic-testing-market-to-reach-6-36-billion-by-2028--300853946.html
- 6.Soumerai SB, McLaughlin TJ, Avorn J. Improving drug prescribing in primary care: a critical analysis of the experimental literature. Milbank Q. 83(4), (2005). [PubMed] [Google Scholar]
- 7.Grice GR, Seaton TL, Woodland AM, McLeod HL. Defining the opportunity for pharmacogenetic intervention in primary care. Pharmacogenomics 7(1), 61–65 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Mills R, Voora D, Peyser B, Haga SB. Delivering pharmacogenetic testing in a primary care setting. Pharmgenomics Pers. Med. 6, 105–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schildcrout JS, Denny JC, Bowton E. et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin. Pharmacol. Ther. 92(2), 235–242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haga SB, Burke W, Ginsburg GS, Mills R, Agans R. Primary care physicians' knowledge of and experience with pharmacogenetic testing. Clin. Genet. 82(4), 388–394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson EA, Wilke RA. Integration of genomics in primary care. Am. J. Med. 128(11), 1251–1255 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Scott J, Trotter T. Primary care and genetics and genomics. Pediatrics 132(Suppl. 3), S231–S237 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Haga SB. Educating patients and providers through comprehensive pharmacogenetic test reports. Pharmacogenomics 18(11), 1047–1050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Primary health care: main terminology (2016). www.euro.who.int/en/health-topics/Health-systems/primary-health-care/main-terminology
- 15.Physicians AAoF. Primary care (2016). www.aafp.org/about/policies/all/primary-1care.html
- 16.Qureshi N, Modell B, Modell M. Timeline: raising the profile of genetics in primary care. Nat. Rev. Genet. 5(10), 783–790 (2004). [DOI] [PubMed] [Google Scholar]
- 17.David SP, Johnson SG, Berger AC. et al. Making personalized health care even more personalized: insights from activities of the IOM genomics roundtable. Ann. Fam. Med. 13(4), 373–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikat-Stevens NA, Larson IA, Tarini BA. Primary-care providers' perceived barriers to integration of genetics services: a systematic review of the literature. Genet. Med. 17(3), 169–176 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Paneque M, Turchetti D, Jackson L, Lunt P, Houwink E, Skirton H. A systematic review of interventions to provide genetics education for primary care. BMC Fam. Pract. 17, 89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abed Faghri NM, Boisvert CM, Faghri S. Understanding the expanding role of primary care physicians (PCPs) to primary psychiatric care physicians (PPCPs): enhancing the assessment and treatment of psychiatric conditions. Mental Health Fam. Med. 7(1), 17–25 (2010). [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin G, Berendsen A, Crawford SM. et al. The expanding role of primary care in cancer control. Lancet Oncol. 16(12), 1231–1272 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Goldschmidt RH, Chu C, Dong BJ. Initial management of patients with HIV Infection. Am. Fam. Physician 94(9), 708–716 (2016). [PubMed] [Google Scholar]
- 23.Lemke AA, Hutten Selkirk CG, Glaser NS. et al. Primary care physician experiences with integrated pharmacogenomic testing in a community health system. Per. Med. 14(5), 389–400 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Weitzel KW, Elsey AR, Langaee TY. et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am. J. Med. Genet. C Semin. Med. Genet. 166C(1), 56–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser D, Obeng AO, Fei K, Ramos MA, Horowitz CR. Views of primary care providers on testing patients for genetic risks for common chronic diseases. Health Aff. 37(5), 793–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansen Taber KA, Dickinson BD. Pharmacogenomic knowledge gaps and educational resource needs among physicians in selected specialties. Pharmgenomics Pers. Med. 7, 145–162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanek EJ, Sanders CL, Taber KA. et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin. Pharmacol. Ther. 91(3), 450–458 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Peterson JF, Field JR, Shi Y. et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. Pharmacogenomics J. 16(4), 393–398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll JC, Makuwaza T, Manca DP. et al. Primary care providers' experiences with and perceptions of personalized genomic medicine. Can. Fam. Physician 62(10), e626–e635 (2016). [PMC free article] [PubMed] [Google Scholar]
- 30.Houwink EJ, van Luijk SJ, Henneman L, van der Vleuten C, Jan Dinant G, Cornel MC. Genetic educational needs and the role of genetics in primary care: a focus group study with multiple perspectives. BMC Fam. Pract. 12, 5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abul-Husn NS, Owusu Obeng A, Sanderson SC, Gottesman O, Scott SA. Implementation and utilization of genetic testing in personalized medicine. Pharmacogenomics Pers. Med. 7, 227–240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cicali EJ, Weitzel KW, Elsey AR. et al. Challenges and lessons learned from clinical pharmacogenetic implementation of multiple gene–drug pairs across ambulatory care settings. Genet Med. 21(10), 2264–2274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramos E, Weissman SM. The dawn of consumer-directed testing. Am. J. Med. Genet. C Semin. Med. Genet. 178(1), 89–97 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Bartlett G, Zgheib N, Manamperi A. et al. Pharmacogenomics in primary care: a crucial entry point for global personalized medicine? Curr. Pharmacogenomics Pers. Med. 10(2), 101–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black EL, Hocum BT, Black KJ. The future: pharmacogenetics in primary care. Primary Care Rep. 20(10), 13 (2014). [Google Scholar]
- 36.Weitzel KW, Alexander M, Bernhardt BA. et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med. Genomics 9, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haga SB, Mills R. A review of consent practices and perspectives for pharmacogenetic testing. Pharmacogenomics 17(14), 1595–1605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haga SB, Mills R, Moaddeb J, Allen Lapointe N, Cho A, Ginsburg GS. Patient experiences with pharmacogenetic testing in a primary care setting. Pharmacogenomics 17(15), 1629–1636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose D, Russo J, Wykes T. Taking part in a pharmacogenetic clinical trial: assessment of trial participants understanding of information disclosed during the informed consent process. BMC Med. Ethics 14, 34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zierhut HA, Campbell CA, Mitchell AG, Lemke AA, Mills R, Bishop JR. Collaborative counseling considerations for pharmacogenomic tests. Pharmacotherapy 37(9), 990–999 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Whirl-Carrillo M, McDonagh EM, Hebert JM. et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92(4), 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haga SB, Kantor A. Horizon scan of clinical laboratories offering pharmacogenetic testing. Health Aff. 37(5), 717–723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haga SB, Moaddeb J. Comparison of delivery strategies for pharmacogenetic testing services. Pharmacogenetics Genomics 24(3), 139–145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vo TT, Bell GC, Owusu Obeng A, Hicks JK, Dunnenberger HM. Pharmacogenomics implementation: considerations for selecting a reference laboratory. Pharmacotherapy 37(9), 1014–1022 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Keeling NJ, Rosenthal MM, West-Strum D, Patel AS, Haidar CE, Hoffman JM. Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genetics Med., (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frigon MP, Blackburn MÈ, Dubois-Bouchard C. et al. Pharmacogenetic testing in primary care practice: opinions of physicians, pharmacists and patients. Pharmacogenomics 20(8), 589–598 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Hoffman JM, Dunnenberger HM, Kevin Hicks J. et al. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC). J. Am. Med. Inform. Assoc. 23(4), 796–801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crews KR, Gaedigk A, Dunnenberger HM. et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95(4), 376–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hicks JK, Bishop JR, Sangkuhl K. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 98(2), 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hicks JK, Sangkuhl K, Swen JJ. et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 102(1), 37–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Empey PE, Stevenson JM, Tuteja S. et al. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin. Pharmacol. Ther. 104(4), 664–674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verbeurgt P, Mamiya T, Oesterheld J. How common are drug and gene interactions? Prevalence in a sample of 1143 patients with CYP2C9, CYP2C19 and CYP2D6 genotyping. Pharmacogenomics 15(5), 655–665 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Bahar MA, Setiawan D, Hak E, Wilffert B. Pharmacogenetics of drug-drug interaction and drug–drug–gene interaction: a systematic review on CYP2C9, CYP2C19 and CYP2D6. Pharmacogenomics 18(7), 701–739 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Haga SB, Mills R, Bosworth H. Striking a balance in communicating pharmacogenetic test results: promoting comprehension and minimizing adverse psychological and behavioral response. Patient Educ. Couns. 97(1), 10–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charland SL, Agatep BC, Herrera V. et al. Providing patients with pharmacogenetic test results affects adherence to statin therapy: results of the additional KIF6 risk offers better adherence to statins (AKROBATS) trial. Pharmacogenomics J. 14(3), 272–280 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Sweet K, Hovick S, Sturm AC. et al. Counselees' perspectives of genomic counseling following online receipt of multiple actionable complex disease and pharmacogenomic results: a qualitative research study. J. Genet. Couns. 26(4), 738–751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trevena LJ, Davey HM, Barratt A, Butow P, Caldwell P. A systematic review on communicating with patients about evidence. J. Eval. Clin. Pract. 12(1), 13–23 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Friedman AJ, Cosby R, Boyko S, Hatton-Bauer J, Turnbull G. Effective teaching strategies and methods of delivery for patient education: a systematic review and practice guideline recommendations. J. Cancer Educ. 26(1), 12–21 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Marcus C. Strategies for improving the quality of verbal patient and family education: a review of the literature and creation of the EDUCATE model. Health Psychol. Behav. Med. 2(1), 482–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones LK, Kulchak Rahm A, Gionfriddo MR. et al. Developing pharmacogenomic reports: insights from patients and clinicians. Clin. Transl. Sci. 11(3), 289–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gibson ML, Hohmeier KC, Smith CT. Pharmacogenomics testing in a community pharmacy: patient perceptions and willingness-to-pay. Pharmacogenomics 18(3), 227–233 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Haga SB, O’Daniel JM, Tindall GM, Lipkus IR, Agans R. Survey of US public attitudes toward pharmacogenetic testing. Pharmacogenomics J. 12(3), 197–204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haga SB, Tindall G, O'Daniel JM. Public perspectives about pharmacogenetic testing and managing ancillary findings. Genet. Test. Mol. Biomarkers 16(3), 193–197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pulley JM, Denny JC, Peterson JF. et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther. 92(1), 87–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richards S, Aziz N, Bale S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17(5), 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]