Abstract
Bacterial colonization of the urogenital tract is limited by innate defenses, including the production of antimicrobial peptides (AMPs). Uropathogenic Escherichia coli (UPEC) resist AMP‐killing to cause a range of urinary tract infections (UTIs) including asymptomatic bacteriuria, cystitis, pyelonephritis, and sepsis. UPEC strains have high genomic diversity and encode numerous virulence factors that differentiate them from non‐UTI‐causing strains, including ompT. As OmpT homologs cleave and inactivate AMPs, we hypothesized that UPEC strains from patients with symptomatic UTIs have high OmpT protease activity. Therefore, we measured OmpT activity in 58 clinical E. coli isolates. While heterogeneous OmpT activities were observed, OmpT activity was significantly greater in UPEC strains isolated from patients with symptomatic infections. Unexpectedly, UPEC strains exhibiting the greatest protease activities harbored an additional ompT‐like gene called arlC (ompTp). The presence of two OmpT‐like proteases in some UPEC isolates led us to compare the substrate specificities of OmpT‐like proteases found in E. coli. While all three cleaved AMPs, cleavage efficiency varied on the basis of AMP size and secondary structure. Our findings suggest the presence of ArlC and OmpT in the same UPEC isolate may confer a fitness advantage by expanding the range of target substrates.
Keywords: antimicrobial peptides, ArlC, LL‐37, OmpP, OmpT, RNase 7, UPEC
Here, we investigated omptin protease‐mediated antimicrobial resistance among uropathogenic Escherichia coli clinical isolates. We found that clinical isolates with the highest omptin protease activity encode multiple omptin proteases, showing different substrate specificities. Our data suggest that the presence of multiple omptin family proteases in a single clinical isolate may provide a fitness advantage by expanding the range of natural antimicrobial peptides cleaved during urogenital tract colonization.
![]()
1. INTRODUCTION
Urinary tract infections (UTIs) are among the most common cause of bacterial infections requiring antibiotic treatment (Flores‐Mireles, Walker, Caparon, & Hultgren, 2015; Foxman, 2014; Hooton & Stamm, 1997). The majority of community‐acquired UTIs (70%–95%) and recurrent UTIs are caused by uropathogenic Escherichia coli (UPEC) (Flores‐Mireles et al., 2015; Nielubowicz & Mobley, 2010). The human gut acts as a reservoir for UPEC strains where they form part of the fecal flora (Kaper, Nataro, & Mobley, 2004; Moreno et al., 2006). Following colonization of the periurethral area, UPEC infect the urinary tract in an ascending manner, resulting in diseases ranging from asymptomatic bacteriuria (ABU), cystitis, pyelonephritis, and sepsis (Hooton, 2012). UPEC strains have high genomic diversity and encode numerous virulence factors that differentiate them from non‐UTI‐causing strains (Johnson, 1991; Lloyd, Rasko, & Mobley, 2007; Najafi, Hasanpour, Askary, Aziemzadeh, & Hashemi, 2018; Norinder, Koves, Yadav, Brauner, & Svanborg, 2012). These virulence factors contribute to disease progression allowing UPEC to colonize the uroepithelium, produce toxins, scavenge metabolites, and evade the host immune system (Schwab, Jobin, & Kurts, 2017; Terlizzi, Gribaudo, & Maffei, 2017).
Bacterial colonization is limited in the upper urogenital tract by several mechanisms including urine flow, chemical properties of urine, epithelial cell shedding, influx of immune cells including neutrophils upon bacterial stimulation, and secretion of soluble proteins and peptides by epithelial cells (Spencer, Schwaderer, Becknell, Watson, & Hains, 2014; Weichhart, Haidinger, Horl, & Saemann, 2008). Secreted proteins and antimicrobial peptides (AMPs) form part of the innate immune defenses of the urogenital tract and act through immunomodulation, indirect anticolonization activity, or direct bacterial killing (Kai‐Larsen et al., 2010; Zasloff, 2007). AMPs are small (12–50 amino acids), cationic, amphipathic peptides that exert bactericidal action by interacting with anionic bacterial membranes to form pores resulting in bacterial lysis (Jenssen, Hamill, & Hancock, 2006). Two types of AMPs are detected in the urogenital tract: defensins that form small disulfide bond‐stabilized ß‐sheets and the α‐helical cathelicidin LL‐37 (Chromek et al., 2006; Lehmann et al., 2002; Valore et al., 1998). In addition, the urogenital tract produces large structured antimicrobial proteins called ribonucleases (RNases) (Spencer et al., 2011, 2013). Human α‐defensin 5 (HD5), human ß‐defensins (hBD) 1 and 2, LL‐37, and RNase 7 are thought to prevent bacterial colonization as they are constitutively expressed in the urinary tract (Kjolvmark, Akesson, & Pahlman, 2017; Spencer et al., 2012). During UTIs, production of HD5, hBD2, LL‐37, and RNase 7 increases, suggesting an active role in bacterial clearance (Chromek & Brauner, 2008; Chromek et al., 2006; Nielsen et al., 2014; Spencer et al., 2012, 2013). Remarkably, increased cathelicidin expression and LL‐37 secretion are triggered a few minutes after bacteria encounter uroepithelial cells. This suggested role for AMPs in UTI immune defense is consistent with reports that UPEC strains are generally more resistant to AMPs than commensal E. coli strains that do not colonize the urogenital tract (Chromek et al., 2006).
Gram‐negative bacteria use several mechanisms to resist killing by AMPs, including capsules, efflux pumps, LPS modifications, and proteases (Gruenheid & Le Moual, 2012). Omptin proteases are found in the Gram‐negative outer bacterial membrane and have a conserved active site with features of both aspartate and serine proteases (Kramer et al., 2001; Vandeputte‐Rutten et al., 2001). With their active sites facing the extracellular environment, omptins contribute to virulence by cleaving a variety of proteins and peptides (Haiko, Suomalainen, Ojala, Lahteenmaki, & Korhonen, 2009). Both substrate specificity and amino acid identity are used to classify omptins into Pla‐like and OmpT‐like subfamilies. Pla readily cleaves the proenzyme plasminogen into active plasmin to promote bacterial dissemination during both bubonic and pneumonic plague (Lathem, Price, Miller, & Goldman, 2007; Sodeinde et al., 1992; Zimbler, Schroeder, Eddy, & Lathem, 2015). OmpT rapidly cleaves and inactivates AMPs, including LL‐37, protamine, and a synthetic peptide optimized to have maximum antibacterial activity called C18G (Brannon, Thomassin, Desloges, Gruenheid, & Le Moual, 2013; Stumpe, Schmid, Stephens, Georgiou, & Bakker, 1998; Thomassin, Brannon, Gibbs, Gruenheid, & Le Moual, 2012). OmpT‐mediated AMP inactivation is thought to support host colonization by some pathogenic E. coli strains (Thomassin, Brannon, Gibbs, et al., 2012). In addition to OmpT, two OmpT‐like proteases have been described in E. coli strains (Kaufmann, Stierhof, & Henning, 1994; McPhee et al., 2014; Zhuge et al., 2018); these genes, called ompP and arlC (ompTp), encode proteins that have approximately 74% amino acid identity to OmpT (GenBank accession numbers: AAC73666.1 (OmpT), BAA97899.1 (OmpP), ADR30001.1 (ArlC)). While the physiological substrates of OmpP and ArlC are unknown, OmpP has been shown to cleave the AMP protamine and ArlC is associated with AMP resistance (Hwang et al., 2007; McPhee et al., 2014).
The ompT gene is present in the genome of 85%–97% of UPEC clinical isolates and is used in epidemiological studies to identify virulent UPEC strains, yet its function across clinical isolates remains unclear (Foxman, Zhang, Palin, Tallman, & Marrs, 1995). As OmpT and OmpT‐like omptins play roles in resistance to host‐produced AMPs, we hypothesized that UPEC strains from patients with symptomatic UTIs have high OmpT protease activity. To test this hypothesis, we detected ompT and measured OmpT activity in a collection of 58 clinical E. coli isolates from groups of patients with infections of differing clinical severity (fecal, ABU, UTI [cystitis and pyelonephritis], and sepsis). Heterogeneous OmpT activity was observed, and in some isolates, high protease activity was correlated with the presence of an additional ompT‐like gene called arlC (ompTp). The presence of two OmpT‐like proteases in some UPEC isolates led us to compare the substrate specificity of the three E. coli omptins (OmpT, OmpP, and ArlC). We found that OmpT, OmpP, and ArlC all cleave AMPs, although cleavage efficiency of different AMP types varied. Our results suggest that the presence of multiple omptins allows UPEC to cleave at least two major subsets of AMPs encountered during infection.
2. MATERIALS AND METHODS
2.1. Bacterial strains and growth conditions
58 clinical E. coli isolates originating from patients diagnosed with extraintestinal infections or from the urine or stool of healthy individuals were obtained from the Manges collection. Included isolates were randomly selected from the E. coli category to ensure they were representative. Isolates were divided into four groups based on disease type. Fecal isolates (n = 12) were recovered from the feces of healthy subjects in Québec, Canada (2009–2010), ABU isolates (n = 10) were from patients with asymptomatic bacteriuria in California, USA (2005–2006) (Manges, Johnson, & Riley, 2004), UTI isolates (n = 24) were recovered from patients with cystitis in Québec, Canada (2005–2007) (Manges, Tabor, Tellis, Vincent, & Tellier, 2008) and cystitis or pyelonephritis in California, USA (1999–2000) (Larsen, Cosentino, Dietrich, & Riley, 2004), and sepsis isolates (n = 12) were from patients with sepsis in California, USA (2001–2003) (Manges, Perdreau‐Remington, Solberg, & Riley, 2006). Bacterial strains used in this study are listed in Table 1. Bacteria were routinely cultured in lysogeny broth (LB; 10% (w/v) tryptone, 5% (w/v) yeast extract, 10% (w/v) NaCl)) or in N‐minimal medium (50 mM Bis‐Tris, 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 0.5 mM KH2PO4, 0.1% casamino acids) adjusted to pH 7.5, supplemented with 1.4% glucose and 1 mM MgCl2 (UPEC isolates) or with 0.5% glucose and 1 mM MgCl2 (all other strains). Bacteria were cultured at 37°C with aeration (220 rpm).
Table 1.
Strains and plasmids used in this study
| Strains | Description | Source |
|---|---|---|
| XL1‐Blue | endA1 gyrA96(nalR) thi‐1 recA1 relA1 lac glnV44 F'[ ::Tn10 proAB + lacI q Δ(lacZ)M15] hsdR17(rK −mK +) | Stratagene |
| GMS 002A | O11:NM; coded as Fecal 1 | (Aslam et al., 2014) |
| GMS 003A | Coded as Fecal 2 | Manges strain collection |
| GMS 005A | Coded as Fecal 3 | Manges strain collection |
| GMS 006E | Coded as Fecal 4 | Manges strain collection |
| GMS 008A | Coded as Fecal 5 | Manges strain collection |
| GMS 009B | Coded as Fecal 6 | (Aslam et al., 2014) |
| GMS 010A | Coded as Fecal 7 | Manges strain collection |
| GMS 012A | Coded as Fecal 8 | Manges strain collection |
| GMS 015A | Coded as Fecal 9 | Manges strain collection |
| GMS 016D | Coded as Fecal 10 | Manges strain collection |
| GMS 017A | Coded as Fecal 11 | Manges strain collection |
| GMS 018A | Coded as Fecal 12 | Manges strain collection |
| 10001U001 | Coded as asymptomatic bacteriuria 1 | (Manges, Johnson, et al., 2004) |
| 10003U002 | Coded as asymptomatic bacteriuria 2 | (Manges, Johnson, et al., 2004) |
| 10004U001 | Coded as asymptomatic bacteriuria 3 | (Manges, Johnson, et al., 2004) |
| 10013U005 | Coded as asymptomatic bacteriuria 4 | (Manges, Johnson, et al., 2004) |
| 10014U005 | Coded as asymptomatic bacteriuria 5 | (Manges, Johnson, et al., 2004) |
| 10017U005 | Coded as asymptomatic bacteriuria 6 | (Manges, Johnson, et al., 2004) |
| 1,001006 | Coded as asymptomatic bacteriuria 7 | (Manges, Johnson, et al., 2004) |
| 10005004 | Coded as asymptomatic bacteriuria 8 | (Manges, Johnson, et al., 2004) |
| 10006001 | Coded as asymptomatic bacteriuria 9 | (Manges, Johnson, et al., 2004) |
| 10012007 | Coded as asymptomatic bacteriuria 10 | (Manges, Johnson, et al., 2004) |
| CLSC 36 | O1:H42; isolated from a patient with cystitis; coded as UTI 1 | (Manges et al., 2018) |
| MSHS 100 | O2:H7; isolated from a patient with cystitis; coded as UTI 2 | (Manges et al., 2018) |
| MSHS 1,070 | Isolated from a patient with cystitis; coded as UTI 3 | (Manges et al., 2018) |
| MSHS 233 | O9:H32; isolated from a patient with cystitis; coded as UTI 4 | (Manges et al., 2018) |
| MSHS 434 | O73:H18; isolated from a patient with cystitis; coded as UTI 5 | (Manges et al., 2018) |
| MSHS 472 | O82:NM; isolated from a patient with cystitis; coded as UTI 6 | (Manges et al., 2018) |
| MSHS 635 | Isolated from a patient with cystitis; coded as UTI 7 | (Manges et al., 2018) |
| MSHS 637 | Isolated from a patient with cystitis; coded as UTI 8 | (Manges et al., 2018) |
| MSHS 689 | Isolated from a patient with cystitis; coded as UTI 9 | (Manges et al., 2018) |
| MSHS 415 | O6:H1; isolated from a patient with cystitis; coded as UTI 10 | (Manges et al., 2018) |
| MSHS 133 | O24:NM; isolated from a patient with cystitis; coded as UTI 11 | (Manges et al., 2018) |
| MSHS 769 | O4:H5; isolated from a patient with cystitis; coded as UTI 12 | (Manges et al., 2018) |
| UTI PI 486 | O11:Neg; isolated from a patient with pyelonephritis; coded as UTI 13 | (Manges, Dietrich, et al., 2004) |
| UTI PI 141 | X19; isolated from a patient with pyelonephritis; coded as UTI 14 | (Manges, Dietrich, et al., 2004) |
| UTI PI 147 | Isolated from a patient with cystitis; coded as UTI 15 | (Manges, Dietrich, et al., 2004) |
| UTI PI 192 | Isolated from a patient with cystitis; coded as UTI 16 | (Manges, Dietrich, et al., 2004) |
| UTI PI 240 | Isolated from a patient with cystitis; coded as UTI 17 | (Manges, Dietrich, et al., 2004) |
| UTI PI 247 | Isolated from a patient with cystitis; coded as UTI 18 | (Manges, Dietrich, et al., 2004) |
| UTI PI 259 | Isolated from a patient with cystitis; coded as UTI 19 | (Manges, Dietrich, et al., 2004) |
| UTI PI 268 | Isolated from a patient with cystitis; coded as UTI 20 | (Manges, Dietrich, et al., 2004) |
| UTI PI 280 | Isolated from a patient with cystitis; coded as UTI 21 | (Manges, Dietrich, et al., 2004) |
| UTI PI 374 | O18; isolated from a patient with cystitis; coded as UTI 22 | (Manges, Dietrich, et al., 2004) |
| UTI PI 20 | Isolated from a patient with cystitis; coded as UTI 23 | (Manges, Dietrich, et al., 2004) |
| UTI PI 116 | Isolated from a patient with cystitis; coded as UTI 24 | (Manges, Dietrich, et al., 2004) |
| W26653 | O15; isolated from a patient with sepsis; coded as sepsis 1 | (Manges et al., 2006) |
| W55291 | O77; isolated from a patient with sepsis; coded as sepsis 2 | (Manges et al., 2006) |
| X19714 | O86; isolated from a patient with sepsis; coded as sepsis 3 | (Manges et al., 2006) |
| X37350 | O73; isolated from a patient with sepsis; coded as sepsis 4 | (Manges et al., 2006) |
| X47726 | O11; isolated from a patient with sepsis; coded as sepsis 5 | (Manges et al., 2006) |
| S49894 | O102; isolated from a patient with sepsis; coded as sepsis 6 | (Manges et al., 2006) |
| H15 | O153; isolated from a patient with sepsis; coded as sepsis 7 | (Manges et al., 2006) |
| F46700 | Isolated from a patient with sepsis; coded as sepsis 8 | (Manges et al., 2006) |
| F55268 | Isolated from a patient with sepsis; coded as sepsis 9 | (Manges et al., 2006) |
| M32569 | Isolated from a patient with sepsis; coded as sepsis 10 | (Manges et al., 2006) |
| M4026 | Isolated from a patient with sepsis; coded as sepsis 11 | (Manges et al., 2006) |
| M49611 | Isolated from a patient with sepsis; coded as sepsis 12 | (Manges et al., 2006) |
| CFT073 | Uropathogenic E. coli O6:K2:H1 | (Mobley et al., 1990) |
| CFT073∆ompT | Uropathogenic E. coli O6:K2:H1 ∆ompT | (Brannon et al., 2013) |
| BL21 | F– dcm ompT hsdSB (rB – mB –) gal | Novagen |
| BL21(pWSK129) | BL21(DE3) containing plasmid pWSK129 | This study |
| BL21(pompT) | BL21(DE3) expressing ompT from pWSKompT | This study |
| BL21(pompP) | BL21(DE3) expressing ompP from pWSKompP | This study |
| BL21(parlC) | BL21(DE3) expressing arlC from pWSKarlC | This study |
| BL21(ppla) | BL21(DE3) expressing pla from pWSKpla | This study |
| Plasmids | ||
| pWSK129 | Low‐copy‐number plasmid (KanR) | (Wang & Kushner, 1991) |
| pWSKarlC | arlC from Cys 6 cloned into pWSK129 | This study |
| pWSKpla | pla cloned into pWSK129 | This study |
| pWSKompT | ompT from isolate Cys 6 cloned into pWSK129 | This study |
| pWSKompP | ompP from XL1‐Blue cloned into pWSK129 | This study |
2.2. Multiplex PCR of UPEC virulence genes
Total DNA (genomic and large plasmid DNA) was isolated using the Puregene Yeast/Bact. kit (Qiagen). Phylogenetic groups were determined as described in Clermont, Bonacorsi, and Bingen (2000), using primer pairs listed in Table 2. To detect virulence genes present in the isolates, primer sequences were obtained from previous studies (Johnson & Stell, 2000) or designed de novo for this study (Table 2). Three multiplex PCR experiments were performed as follows: pool 1: hylA (1,177 bp), papAH (720 bp), fimH (508 bp), kpsMTIII (392 bp), and papEF (336 bp); pool 2: papC (200 bp), sfaS (240 bp), cnf1 (498 bp), fyuA (880 bp), iutA (300 bp), and kpsMTII (272 bp); pool 3: arlC (852 bp), ompT (670 bp), fimH (508 bp), and ompP (648 bp).
Table 2.
Primers used in this study
| Name | Sequence 5–3′a | Use | Source |
|---|---|---|---|
| iutA_f | GGCTGGACATCATGGGAACTGG | Multiplex PCR | (Johnson & Stell, 2000) |
| iutA_r | CGTCGGGAACGGGTAGAATCG | Multiplex PCR | (Johnson & Stell, 2000) |
| fimH_f | TGCAGAACGGATAAGCCGTGG | Multiplex PCR | (Johnson & Stell, 2000) |
| fimH_r | GCAGTCACCTGCCCTCCGGTA | Multiplex PCR | (Johnson & Stell, 2000) |
| papAH_f | ATGGCAGTGGTGTCTTTTGGTG | Multiplex PCR | (Johnson & Stell, 2000) |
| papAH_r | CGTCCCACCATACGTGCTCTTC | Multiplex PCR | (Johnson & Stell, 2000) |
| papC_f | GTGGCAGTATGAGTAATGACCGTTA | Multiplex PCR | (Johnson & Stell, 2000) |
| papC_r | ATATCCTTTCTGCAGGGATGCAATA | Multiplex PCR | (Johnson & Stell, 2000) |
| papEF_f | GCAACAGCAACGCTGGTTGCATCAT | Multiplex PCR | (Johnson & Stell, 2000) |
| papEF_r | AGAGAGAGCCACTCTTATACGGACA | Multiplex PCR | (Johnson & Stell, 2000) |
| sfaS_f | GTGGATACGACGATTAACTGTG | Multiplex PCR | (Johnson & Stell, 2000) |
| sfaS_r | CCGCCAGCATTCCCTGTATTC | Multiplex PCR | (Johnson & Stell, 2000) |
| fyuA_f | TGATTAACCCCGCGACGGGAA | Multiplex PCR | (Johnson & Stell, 2000) |
| fyuA_r | CGCAGTAGGCACGATGTTGTA | Multiplex PCR | (Johnson & Stell, 2000) |
| kpsMII_f | GCGCATTTGCTGATACTGTTG | Multiplex PCR | (Johnson & Stell, 2000) |
| kpsMII_r | CATCCAGACGATAAGCATGAGCA | Multiplex PCR | (Johnson & Stell, 2000) |
| kpsMIII_f | TCCTCTTGCTACTATTCCCCCT | Multiplex PCR | (Johnson & Stell, 2000) |
| kpsMIII_r | AGGCGTATCCATCCCTCCTAAC | Multiplex PCR | (Johnson & Stell, 2000) |
| cnf−1_f | AAGATGGAGTTTCCTATGCAGGAG | Multiplex PCR | (Johnson & Stell, 2000) |
| cnf−1_r | CATTCAGAGTCCTGCCCTCATTATT | Multiplex PCR | (Johnson & Stell, 2000) |
| hlyA_f | AACAAGGATAAGCACTGTTCTGGCT | Multiplex PCR | (Johnson & Stell, 2000) |
| hlyA_r | ACCATATAAGCGGTCATTCCCGTCA | Multiplex PCR | (Johnson & Stell, 2000) |
| ompT_mf | TTTGATGCCCCAGATATCTATCGG | Multiplex PCR | This study |
| ompT_mr | GGCTTTCCTGATATCCGGCCATG | Multiplex PCR | This study |
| arlC_mf | GATTCTTGCTACTGCACTCTCAGCTCC | Multiplex PCR | This study |
| arlC_mr | CTGGAGTACAGAGAAGTATCACC | Multiplex PCR | This study |
| ompP_mf | TGCTTCTGATTTCTTCGGCC | Multiplex PCR | This study |
| ompP_mr | GTAGTTTGTCTTACATAATGCTC | Multiplex PCR | This study |
| chuA_f | GACGAACCAACGGTCAGGAT | Phylogenetic typing | (Clermont et al., 2000) |
| chuA_r | TGCCGCCAGTACCAAAGACA | Phylogenetic typing | (Clermont et al., 2000) |
| yjaA_f | TGAAGTGTCAGGAGACGCTG | Phylogenetic typing | (Clermont et al., 2000) |
| yjaA_r | ATGGAGAATGCGTTCCTCAAC | Phylogenetic typing | (Clermont et al., 2000) |
| TSPE4.C2_f | GAGTAATGTCGGGGCATTCA | Phylogenetic typing | (Clermont et al., 2000) |
| TSPE4.C2_r | CGCGCCAACAAAGTATTACG | Phylogenetic typing | (Clermont et al., 2000) |
| ompT_cf | CATGTCTAGACCACGACTTAGAAGTTCCTAGAACG | Cloning | This study |
| ompT_cr | GCGAGCTCAAATCTGGTTAACTTCGTTAA | Cloning | This study |
| ompP_cf | GCATAGTCTAGATCCTGTAGTTGCGTCAGGCCCTCCA | Cloning | This study |
| ompP_cr | GCATAGCTGCAGTCCGGGTAATCCAGGTCCGCCACT | Cloning | This study |
| arlC_cf | CATGTCTAGACCCGGCATAAAGTGTCC | Cloning | This study |
| arlC_cr | CTAGGAGCTCATCGTTGAGCACATATAC | Cloning | This study |
| ompT_sf | ATGCGGGCGAAACTTCTGGGAATAG | Southern blot probe | This study |
| ompT_sr | TCCCAATTAATTGCACCTTTAATAATT | Southern blot probe | This study |
| arlC_sf | GATTCTTGCTACTGCACTCTCAGCTCC | Southern blot probe | This study |
| arlC_sr | CTAGGAGCTCATCGTTGAGCACATATAC | Southern blot probe | This study |
| rpoD_qf | GCTGGAAGAAGTGGGTAAAC | qPCR | This study |
| rpoD_qr | TAATCGTCCAGGAAGCTACG | qPCR | This study |
| ompT_qf | CAGCGGCTGGGTGGAAGCAT | qPCR | (Thomassin, Brannon, Gibbs, et al., 2012) |
| ompT_qr | ACCCGATTCCATGCGCCTTCA | qPCR | (Thomassin, Brannon, Gibbs, et al., 2012) |
| arlC_qf | AGGATCACCTATCGTAGCGATGT | qPCR | This study |
| arlC_qf | CGGTTCCATGTTCCTTCGACATAA | qPCR | This study |
Restriction sites are underlined.
2.3. Fluorescence resonance energy transfer (FRET) activity assay
The FRET substrate containing a dibasic motif (RK) in its center (2Abz‐SLGRKIQI‐K(Dnp)‐NH2) was purchased from Anachem. Bacteria were grown in N‐minimal medium to mid‐exponential phase and normalized to an OD595nm of 0.5. Bacterial cells were pelleted and resuspended in phosphate‐buffered saline (PBS). Bacteria (~2.25 × 107 CFU in 75 µl) were mixed in a 96‐well plate with 75 µl of the FRET substrate (final concentration 3 μM). Fluorescence (λ Ex 325 nm, λ Em 430 nm) was monitored for 1 hr at 25°C using a Biotek FLx800 plate reader. Data were normalized by subtracting the background fluorescence of the FRET substrate in PBS.
2.4. Plasmid construction
The ompT and arlC genes were PCR‐amplified from DNA isolated from the UPEC UTI clinical isolate 6, also called cystitis 6, using their respective primer pairs ompT_cf/ompT_cr and arlC_cf/arlC_cr (Table 2). PCR fragments were treated with XbaI and SacI and ligated into plasmid pWSK129 treated with the same enzymes, generating plasmids pWSKompT and pWSKarlC (Table 1). The ompP gene was PCR‐amplified from XL1‐Blue DNA using primer pair ompP_cf/ompP_cr. PCR products were treated with XbaI and PstI and ligated into pWSK129 treated with the same enzymes to generate plasmid pWSKompP. The pla gene under control of the croP promoter was subcloned from pYCpla (Brannon, Burk, et al., 2015) using XbaI and SacI and ligated into pWSK129 previously treated with the same enzymes, generating pWSKpla.
2.5. Southern blotting
Total DNA was isolated and treated with EcoRV. Southern blotting and hybridization were performed as previously described (Taylor, Ouimet, Wargachuk, & Marczynski, 2011) using Hybond‐XL membranes. Probes for ompT and arlC were PCR‐generated using primer pairs ompT_sf/ompT_sr and arlC_sf/arlC_sr, respectively (Table 2). Probes were radiolabeled with dATP [α‐32P] using the RadPrime kit (Invitrogen). The pWSKarlC plasmid was used as the positive control for the arlC probe.
2.6. Quantitative RT‐PCR
Quantitative RT‐PCR (qPCR) was performed as previously described (Thomassin, Brannon, Gibbs, et al., 2012). Briefly, bacterial strains were grown to an OD595nm of 0.5 in N‐minimal medium. Total RNA was isolated using TRIzol reagents (Invitrogen) and treated with TURBO DNase I (Ambion) to remove residual DNA. The absence of DNA was confirmed by qPCR using the primer pair rpoD_qf/rpoD_qr. RNA (100 ng) was reverse‐transcribed using Superscript II (Invitrogen) with 0.5 μg of random hexamer primers. A reaction mixture without Superscript II was also included and was used as the negative control. qPCRs were performed in a Rotor‐Gene 3,000 thermal cycler (Corbett Research) using the Maxima SYBR Green qPCR kit (Thermo Scientific), according to the manufacturer's instructions. Primers used are listed in Table 2. The relative expression levels were calculated by normalizing the threshold cycle (C T) of ompT and arlC transcripts to the C T of rpoD using the 2‐Δ C T method (Livak & Schmittgen, 2001).
2.7. Whole‐genome sequencing
Sequencing was performed on a PacBio platform (Pacific Biosciences). Genomic DNA samples were purified using the Gentra® Puregene® kit (Qiagen) and sheared to 20 kb using g‐tubes (Covaris). Libraries were prepared using the template preparation kit from Pacific Biosciences. A single SMRT cell was sequenced to generate datasets including unique subreads with a minimum length of 3 kb. Genome assemblies of sequence reads were generated using a combination of HGAP/Celera/Quiver following Pacific Biosciences recommendations. The complete chromosome and plasmid sequences were submitted to GenBank. The BioProject accession numbers are as follows: PRJNA551561 (cystitis 1), PRJNA551565 (cystitis 6), and PRJNA551566 (cystitis 11).
2.8. Preparation of whole‐cell lysates and outer membrane fractions
Bacteria were grown in N‐minimal medium until mid‐exponential phase and normalized to an OD595nm of 0.5. For whole‐cell lysate samples, bacterial cells were pelleted and resuspended in 1/10 volume of 2X ESB (Thomas et al., 2005). Outer membrane fractions were isolated as follows: bacterial cultures were centrifuged at 3, 600 g for 10 min, and pellets were resuspended in 1.5 ml low‐salt buffer (100 mM NaPi buffer [pH 7], 5 mM EDTA, and 10% glycerol). Samples were supplemented with 10 µl PMSF and sonicated. Samples were then centrifuged at 3,600 g for 10 min. Supernatants were collected and centrifuged at 100,000 g for 30 min at 4°C. Pellets were resuspended in 2 ml sarcosyl buffer (10 mM Tris [pH 7.5], 5 mM MgCl2, and 2% sarcosyl) and incubated for 30 min at 10°C. Samples were then centrifuged for 60 min at 100,000 g, and the pellet containing outer membranes was resuspended in buffer (20 mM Tris‐HCl pH 7.5 and 10% glycerol). Outer membrane samples were combined 1:1 with 2X ESB and boiled for 10 min prior to loading samples on an SDS‐PAGE gel.
2.9. Western blotting
Whole‐cell lysate and outer membrane fractions were resolved on a 10% SDS‐PAGE gel and transferred to a polyvinylidene fluoride membrane. Membranes were blocked for 1 hr in Tris‐buffered saline (TBS) supplemented with 5% skim milk, and OmpT was detected using the polyclonal anti‐CroP antibody as described in Thomassin, Brannon, Gibbs, et al. (2012). Membranes were washed extensively with TBS and incubated for 1 hr with a goat anti‐rabbit secondary antibody conjugated with HRP. Membranes were washed and developed using chemiluminescent HRP substrate.
2.10. Plasminogen activation assay
Bacteria were grown in N‐minimal medium to mid‐exponential phase and normalized to an OD595nm of 0.5. Bacterial cells were pelleted and resuspended in ½ volume of phosphate‐buffered saline (PBS; final 6 x108 CFU/mL). In a 96‐well plate, 178 μL of bacteria and 20 μL of 45 mM VLKpNA (Sigma‐Aldrich) were combined. Baseline assays were performed at OD405nm. After 5 min, 4 μg of plasminogen substrate was added and absorbance (405 nm) was measured every 10 min for 400 min at 37°C with agitation before every reading.
2.11. Proteolytic cleavage of AMPs
Bacteria were grown in N‐minimal medium to mid‐exponential phase, washed, and normalized to an OD595nm of 0.5 in PBS. Aliquots of bacteria (107 CFU) were combined 1:4 (v/v) with 2 μg/µL LL‐37, mCRAMP, C18G or Magainin II (BioChemia), or 1 μg/µL RNase 7 and incubated at room temperature for various time points. Bacteria were separated from peptide cleavage products by centrifugation, and supernatants were combined 1:1 with 2X ESB, then boiled and frozen at −20°C. Peptide cleavage products were resolved on 10%–20% Tris‐Tricine gels (Bio‐Rad), and RNase 7 samples were resolved on 20% SDS‐PAGE gels. Peptides were fixed in the gel by incubation in 20% (v/v) glutaraldehyde for 30 min; gels were rinsed with water and peptides stained for 1h with Coomassie blue G‐250 stain. Gels were destained in 20% (v/v) acetic acid.
2.12. Circular dichroism spectroscopy
Experiments were performed on a Jasco J‐810 spectropolarimeter (Easton, MD). AMPs (200 µg/ml in PBS) were placed in a quartz cuvette with a path length of 0.1 cm, and spectra were recorded from 260 to 195 nm. Samples were scanned three times at 20°C using a bandwidth of 1 nm, a time response of 2 s, and a scan rate of 100 nm/min. Spectra were corrected by subtracting the background spectrum of PBS, and values were converted from ellipticity to mean residue ellipticity (MRE; degree × cm2 × dmol−1).
2.13. Statistical analyses
Data were analyzed using GraphPad Prism software. Normality was verified using the D’Agostino–Pearson normality test. Fisher's exact test was performed to compare incidence of virulence genes within severity groups of UPEC clinical isolates. FRET activity was assessed using a two‐way ANOVA with Tukey's post hoc test. P value ≤ 0.05 was considered significantly different.
3. RESULTS
3.1. Phylogenetic and virulence profile of UPEC isolates
UPEC isolates from patients with different disease severities were obtained from the Manges collection (Manges et al., 2018, 2001, 2006; Manges, Dietrich, et al., 2004; Manges, Johnson, et al., 2004). Although UPEC strains are heterogeneous, clinical isolates from UTIs predominantly belong to E. coli phylogenetic groups B2 and D (Johnson, Delavari, Kuskowski, & Stell, 2001). To confirm that our isolates are generally representative of UPEC clinical strains, we determined the phylogenetic grouping of our 58 clinical isolates categorized into the fecal (n = 12), ABU (n = 10), UTI (cystitis and pyelonephritis, n = 24), and sepsis (n = 12) groups. Most isolates from the ABU and UTI groups associated with UTIs belong to the phylogenetic group B2 and, to a lesser extent, D (Table 3). In contrast, isolates from the sepsis group were predominantly from group D (Table 3). Finally, isolates from the fecal group had the most variable phylogenetic grouping with 5/12 isolates belonging to phylogenetic groups A and B1 (Table 3). Overall, this distribution is in agreement with previous reports, showing that UPEC strains mainly belong to E. coli phylogenetic groups B2 and D (Johnson et al., 2001).
Table 3.
Phylogenetic distribution of UPEC clinical isolates
| Phylogenetic groups | (B2 + D)/Total | ||||
|---|---|---|---|---|---|
| A | B1 | B2 | D | ||
| Fecal (n = 12) | 4 | 1 | 3 | 4 | 7/12 |
| ABU (n = 10) | 2 | 1 | 5 | 2 | 7/10 |
| UTI (n = 24) | 3 | 3 | 11 | 7 | 18/24 |
| Sepsis (n = 12) | 0 | 2 | 0 | 10 | 10/12 |
| Total (n = 58) | 9 | 7 | 19 | 23 | 42/58 |
The 58 isolates were further characterized using multiplex PCR to detect 12 recognized UPEC virulence genes (Table 4). Our data showed variations consistent with previous studies reporting that UPEC is a heterogeneous pathotype (Marschall et al., 2012; Maynard et al., 2004; Norinder et al., 2012; Poey, Albini, Saona, & Lavina, 2012). The fimH gene, involved in UPEC adherence, was present in all but 2 ABU isolates (Table 4). There was a difference in the distribution of virulence genes fyuA and ompT for which the incidence was significantly higher in symptomatic (i.e., UTI and sepsis) groups than asymptomatic (i.e., fecal and ABU) groups (Table 4). No other genes showed a significant difference in incidence between asymptomatic and symptomatic groups. In agreement with previous studies, we found that ompT is present in 89% of the UPEC isolates associated with symptomatic infections (Table 4).
Table 4.
Prevalence of virulence factors in UPEC clinical isolates
| Gene | Fecal (n = 12) | ABU (n = 10) | UTI (n = 24) | Sepsis (n = 12) | P valuea |
|---|---|---|---|---|---|
| iutA | 1 | 6 | 14 | 12 | 0.0541 |
| fimH | 12 | 8 | 24 | 12 | 0.5508 |
| papAH | 3 | 6 | 10 | 10 | 0.4173 |
| papC | 3 | 6 | 12 | 10 | 0.4263 |
| papEF | 4 | 7 | 12 | 10 | 0.4550 |
| sfaS | 1 | 1 | 4 | 0 | 1.0000 |
| fyuA | 9 | 7 | 23 | 11 | 0.0435 |
| kpsMTII | 7 | 7 | 14 | 8 | 1.0000 |
| kpsMTIII | 0 | 0 | 2 | 0 | 0.5203 |
| cnf1 | 4 | 4 | 8 | 0 | 0.3641 |
| hylA | 4 | 3 | 5 | 1 | 0.2078 |
| ompT | 7 | 7 | 22 | 10 | 0.0418 |
P value determined by Fisher's exact test, statistical significance (p ≤ 0.05) is indicated in bold.
3.2. Variability of omptin proteolytic activities among UPEC isolates
OmpT preferentially cleaves substrates between two consecutive basic residues (Dekker, Cox, Kramer, & Egmond, 2001; McCarter et al., 2004). Therefore, to assess OmpT proteolytic activity we measured cleavage of a FRET substrate (2Abz‐SLGRKIQI‐K(Dnp)‐NH2) that contains a dibasic motif in its center (Brannon, Burk, et al., 2015; Brannon et al., 2013; McPhee et al., 2014; Thomassin, Brannon, Gibbs, et al., 2012). Cleavage of the substrate by the 58 clinical E. coli isolates was monitored by measuring fluorescence emission over time and compared with substrate cleavage by the previously characterized reference UPEC strain CFT073 (Brannon et al., 2013). As shown in Figure 1a, omptin activity of the isolates was heterogeneous between groups. Isolates for which the ompT gene was not detected by PCR showed basal activity levels (red triangles in Figure 1a), whereas isolates harboring the ompT gene showed a wide range of omptin activity. The omptin activity of the isolates of the fecal group was significantly lower than that of the 2 symptomatic groups (UTI and sepsis) (Figure 1a). The mean activity of the isolates from the fecal group (0.75 ± 0.5) was lower than that of strain CFT073. In contrast, the activity means of the symptomatic groups (1.54 ± 0.66 and 1.71 ± 0.66) were higher than those of CFT073. Extensive variability in omptin activity was also observed within groups (Figure 1a). The UTI group exhibited the most heterogeneous omptin activity, and some isolates from the UTI group had threefold higher omptin activity than CFT073. Together, these results indicate that omptin activity is variable among fecal and UPEC clinical isolates.
Figure 1.

Omptin protease activity and distribution in clinical isolates. (a) Omptin activity was determined by monitoring fluorescence, indicative of FRET substrate cleavage, for 60 min. Data points indicate mean fold change in fluorescence of each isolate over the mean fold change in fluorescence of reference UPEC strain CFT073 from triplicate samples. Bars represent mean ± SD fold change in fluorescence for each group. Bacteria that contain the ompT gene are indicated by circles, and those that do not contain ompT are indicated by triangles. Indicated in green or purple are isolates that contain arlC. Statistical analysis was performed by one‐way ANOVA followed by Tukey's post hoc test using GraphPad Prism software (NS, not significant; *p ≤ 0.05; **p ≤ 0.01). (b) Multiplex PCR of arlC (852 bp), ompT (670 bp), and fimH (508 bp) from each of the clinical isolates. Amplification of fimH was used as a positive control. Numbers indicate isolate number for each group. Data are representative of at least three independent experiments
3.3. OmpT‐like proteases in UPEC
In addition to the chromosomally encoded ompT gene, plasmid‐borne ompT‐like genes ompP and arlC are present in several E. coli strains (Kaufmann et al., 1994; McPhee et al., 2014; Zhuge et al., 2018). These OmpT‐like proteins are approximately 74% identical to OmpT. To determine whether the presence of ompT‐like genes in some isolates may account for the heterogeneity of OmpT activity observed in Figure 1a, multiplex PCR screens were performed to detect ompT, ompP, and arlC. The ompP gene was not detected in any of the isolates (data not shown). In contrast, the arlC gene was present in 8 of the 58 isolates (Figure 1b). Strikingly, arlC was only present in symptomatic isolates, which was statistically significant according to Fisher's exact test (p = .0445). Most isolates harboring the arlC gene also contained ompT and generally had higher proteolytic activity (green circles, Figure 1a) than CFT073. This is consistent with the report that ArlC cleaves the FRET substrate (McPhee et al., 2014). Isolate 18 from the UTI group did not have ompT but harbored arlC (Figure 1b); this isolate exhibited moderate proteolytic activity (purple triangle, Figure 1a). Together, these data show that among commensal and clinical isolates, higher omptin activity is associated with symptomatic disease and isolates with the greatest omptin activity harbor both the ompT and arlC genes.
3.4. Variability of ompT and arlC expression among select UPEC cystitis isolates
To further understand omptin activity among UPEC isolates, we selected 12 isolates from the UTI group (Table 1) for further analysis because they have the most heterogeneous omptin activity. The presence of ompT genes in these isolates was confirmed by Southern blot analysis (Figure 2a). This analysis also indicated that two ompT genes may be present in isolates 7, 8, and 11. Consistent with the multiplex PCR results, arlC was detected in UTI isolates 1, 6, and 11 (Figure 2a). Next, qPCR was used to measure the expression levels of ompT and arlC. In agreement with our activity assay, ompT transcript levels were heterogeneous among these UTI isolates (Figure 2b and c). Only three isolates (2, 10, and 11) had similar expression levels to the reference strain CFT073, whereas all other isolates had higher ompT expression levels than the reference strain. As expected from the multiplex PCR screen and Southern blot, arlC expression was only detected in UTI 1, 6, and 11 isolates. UTI isolates 1 and 6, which showed the highest ompT and arlC expression levels, also had the highest omptin activity levels (Figure 2c). Although both ompT and arlC are present in UTI isolate 11, they have low expression levels, which is consistent with the low omptin activity observed (Figure 2c). These data indicate that heterogeneous omptin activity levels are associated with both the presence and the different expression levels of the ompT and arlC genes.
Figure 2.
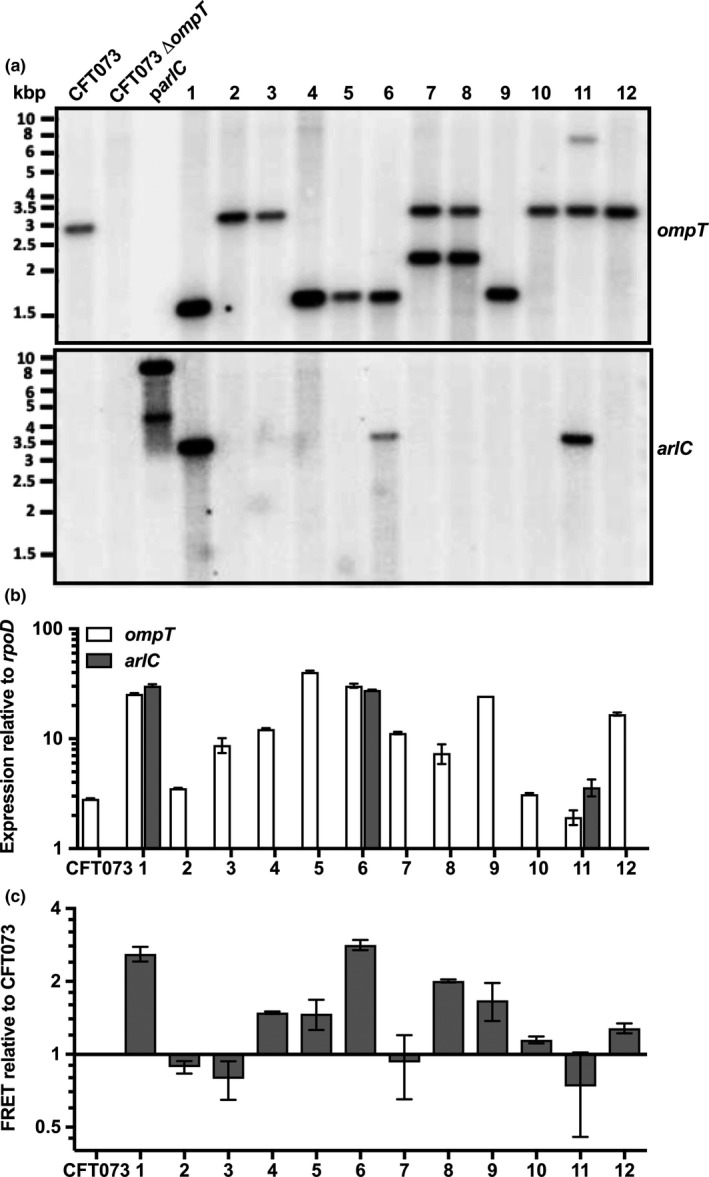
Presence and expression of ompT and arlC among select UTI isolates. (a) Southern blot of ompT and arlC from EcoRV‐treated total DNA isolated from 12 cystitis‐causing isolates, as well as control strains CFT073, CFT073∆ompT, and plasmid DNA from pWSKarlC. (b) Quantitative real‐time PCR (qRT‐PCR) of ompT and arlC from 12 clinical isolates causing cystitis, as well as from reference strain CFT073. Shown is mean ± SD of ompT or arlC expression relative to rpoD calculated using the 2−∆ C T method. Data are representative of three independent experiments. (c) Omptin activity of these cystitis clinical isolates was determined by monitoring cleavage of a synthetic FRET substrate for 60 min. Shown are mean ± SD change in fluorescence of each cystitis isolate over the change in fluorescence of reference stain CFT073 . Data are representative of at least three independent experiments
3.5. arlC is present on plasmids
To determine the genomic context of the ompT and arlC genes, isolates 1, 6, and 11 of the UTI group were sequenced on a PacBio platform. These isolates were then renamed cystitis 1, cystitis 6, and cystitis 11. Detailed descriptions of genomes and gene features are found in Appendix 1 (Figures A1a,b, A2a,b, Tables A1, A2, A3, A4, A5, A6, A7). In all three isolates, ompT was located within the bacterial chromosome and arlC was part of large plasmids (150‐195 kbp; Figures A1b and A2b). In addition, the ompT gene was invariably located downstream of nfrA and ybcH (Figure 3a). Some differences were noted in the genomic context of ompT among the clinical isolates. In cystitis 1 and cystitis 6, the envY gene, encoding a transcriptional regulator of porin synthesis, is inserted between ybcH and ompT (182 bp downstream of ybcH, 512 bp upstream of ompT). The appY gene, encoding a transcriptional activator, is located 249 bp downstream of the ompT gene in cystitis 1, whereas ymcE, encoding a putative cold shock gene, is located 186 bp downstream of ompT in cystitis 6. In cystitis 11, the ompT gene is located 657 bp downstream of ybcH and 272 bp upstream of ybcY; this is the same genomic context as that in UPEC strains CFT073, UTI89, 536, J96 ABU83972 and EPEC strain e2348/69, all of which were reported to have low omptin activity (Figure 3a; Brannon et al., 2013; Thomassin, Brannon, Gibbs, et al., 2012; Thomassin, Brannon, Kaiser, Gruenheid, & Le Moual, 2012)). These isolates all encode a functional ompT gene in their chromosomes in addition to a second truncated and, likely inactive, plasmid‐encoded ompT gene located adjacent to arlC (Figure 3a,b). For all isolates, the predicted amino acid sequence of ArlC is 100% identical to ArlC identified in adherent‐invasive E. coli (AIEC) strain NRG857c (McPhee et al., 2014). Although the three plasmids harboring arlC were different (Figure A2a and b), arlC was present in all cases as part of pathogenicity island PI‐6 previously reported to play a role in AMP resistance (Figure 3b (McPhee et al., 2014)).
Figure 3.

Genomic context of arlC and ompT. Schematic representation of the genomic contexts of the ompT (a) and arlC (b) genes in cystitis isolates 1, 6, and 11. Genomic contexts of ompT (a) and arlC (b) from respective reference strains CFT073 (a) and NRG857c (b) are included for comparison. Omptin genes are indicated in dark gray, light gray indicates genes located upstream and downstream of the omptin genes, stripes indicate pseudogenes, and black lines indicate intergenic space
3.6. Comparative analysis of OmpT, OmpP, and ArlC
With the unexpected detection of arlC among the UPEC clinical isolates, we hypothesized that the presence of a second or even a third omptin protease within a single species may provide an advantage by expanding the potential range of substrates cleaved. Therefore, we sought to compare the substrate specificities of these proteases. As OmpT undergoes autocleavage during purification (Kramer, Zandwijken, Egmond, & Dekker, 2000; Vandeputte‐Rutten et al., 2001) and mutagenesis of residues to stabilize the protein results in a significant decrease in FRET substrate cleavage ((Kramer et al., 2000); unpublished data Thomassin JL and Brannon JR), it was not possible to purify these proteases and directly compare their activities. Instead, we produced OmpT, OmpP, and ArlC in E. coli BL21, a laboratory strain that lacks omptin proteases. To test their production and correct localization in BL21, omptin proteins were detected by Western blot analysis from both whole cells and outer membrane preparations (Figure 4a). To determine whether the proteases were active in BL21, FRET substrate cleavage was monitored over time. As expected, BL21 with empty vector did not cleave the FRET substrate, whereas the three omptins readily cleaved the FRET substrate (Figure 4b). This demonstrates that when produced in BL21, ArlC, OmpP, and OmpT are found in the outer membrane and are proteolytically active.
Figure 4.

ArlC, OmpP, and OmpT are functional in BL21. (a) BL21 containing empty vector (ø) or plasmids encoding arlC, ompP, or ompT were grown until mid‐log phase and normalized to OD595 0.5. Proteins from whole‐cell preparations or isolated bacterial outer membranes were resolved by SDS‐PAGE and transferred to a PVDF membrane. Omptins were detected by Western blot using anti‐CroP polyclonal antibodies. (b) A synthetic FRET peptide containing a dibasic motif (RK) was incubated with BL21 (open circles, control) or BL21 expressing arlC (filled squares, ArlC), ompP (filled circles, OmpP), or ompT (filled triangles, OmpT). Peptide cleavage, indicated by increased fluorescence, was monitored over time. Data show the mean ± SD from triplicate samples and are representative of at least three independent experiments. (c) Plasmin activation by ArlC, OmpP, and OmpT. Glu‐plasminogen and VLKpNA (plasmin substrate) were incubated with BL21 (open circles, control), BL21(ppla) (open triangles, Pla), BL21(parlC) (filled squares, ArlC), BL21(pompP) (filled circles, OmpP), or BL21(pompT) (filled triangles, OmpT) strains. Absorbance at 405 nm was monitored over time. Data were normalized by subtracting initial absorbance from all values. Data represent mean ± SD and are representative of at least three independent experiments
Omptin proteases are generally subdivided into OmpT‐like or Pla‐like subfamilies. These subfamilies differ in their ability to cleave plasminogen to activate it into active plasmin, with Pla‐like omptins producing active plasmin more readily than OmpT‐like omptins (Haiko et al., 2009; Kukkonen et al., 2001). To verify that the three omptin proteases belong in the OmpT‐like subfamily, we tested their ability to cleave plasminogen into plasmin. Consistent with their presence in the outer membrane, all three omptins cleaved plasminogen to a greater extent than BL21 alone (Figure 4c). There was no difference in their ability to activate plasminogen. Compared with the positive control, Pla produced in BL21, the E. coli omptins converted significantly less plasminogen into plasmin. These data are consistent with previous publications (Brannon, Burk, et al., 2015; Kukkonen et al., 2001; McPhee et al., 2014) and suggest that all three omptins found in E. coli belong to the OmpT‐like subfamily of omptin proteases.
Omptin proteases belonging to the OmpT‐like subfamily have been associated with AMP cleavage (Le Sage et al., 2009; Stumpe et al., 1998; Thomassin, Brannon, Gibbs, et al., 2012). Previous work has shown that OmpT from EPEC, EHEC, and UPEC cleaves the human cathelicidin LL‐37. Although ArlC was shown to play a role in AMP resistance (McPhee et al., 2014), and OmpT and OmpP are reported to exhibit similar substrate specificities (Hwang et al., 2007; McCarter et al., 2004), their ability to cleave different AMPs has not been directly compared. Therefore, we investigated the ability of the E. coli omptins to cleave the synthetic cationic peptide C18G and various cathelicidins Magainin II (Xenopus laevis), mCRAMP (Mus musculus), and LL‐37 (Homo sapiens). As expected, AMPs incubated with BL21 did not show any degradation or cleavage products, indicating that BL21 does not contain intrinsic proteases that cleave these AMPs (Figure 5a). OmpT cleaved all peptides by the first time point tested (2 min C18G, 15 min mCRAMP, Magainin II, and LL‐37; Figure 5a). Similar to OmpT, OmpP readily cleaved C18G and Magainin II within 2 and 30 min, respectively. In contrast, OmpP only cleaved small amounts of mCRAMP after 60 min and did not appear to cleave LL‐37 (Figure 5a). ArlC cleaved mCRAMP, C18G, and Magainin II by the first time point tested (2 min C18G, 15 min mCRAMP, and Magainin II), but only a small amount of LL‐37 cleavage was observed after 60 min. Substrate properties, such as size and secondary structure, are known to influence omptin activity (Brannon, Thomassin, Gruenheid, & Le Moual, 2015; Hritonenko & Stathopoulos, 2007). Peptide secondary structure also influences omptin activity (Brannon, Thomassin, et al., 2015); therefore, we used circular dichroism spectroscopy to determine the secondary structure of these AMPs (Figure 5c). Under our experimental conditions, only LL‐37 is α‐helical, while mCRAMP, C18G, and Magainin II are unstructured (Figure 5c). While peptide structure did not affect OmpT activity, ArlC did not appear to cleave the only α‐helical AMP, LL‐37 (Figure 5b,c). Together, these findings suggest that OmpT, OmpP, and ArlC have differences in substrate specificities.
Figure 5.
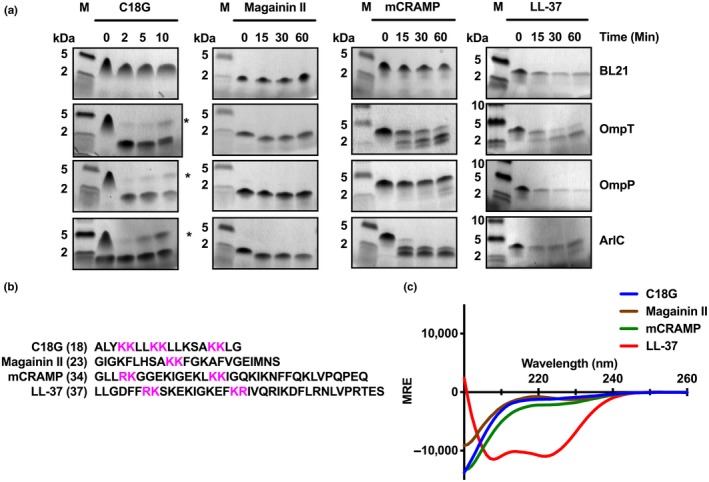
ArlC, OmpP, and OmpT cleave cathelicidins. (a) AMP cleavage assay. AMPs were incubated with BL21 alone or BL21 expressing arlC, ompP, or ompT for the indicated times. Resulting AMP cleavage products were separated by Tris‐Tricine SDS‐PAGE, fixed with glutaraldehyde, and visualized by Coomassie staining. M indicates molecular weight marker. Data are representative of three independent experiments. (b) Amino acid sequence of AMPs cleaved in (a) with dibasic motifs highlighted in magenta and sequence length indicated in parenthesis. (c) Far UV circular dichroism spectra (200–260 nm) of the indicated peptides measured in PBS. Data were normalized by subtracting spectra from PBS alone from the sample spectra. MRE indicates degree × cm2 × dmol−1
We previously reported that disulfide bonds present in defensins render them resistant to OmpT‐mediated proteolysis (Thomassin, Brannon, Kaiser, et al., 2012). Yet ArlC was shown to contribute to bacterial survival in the presence of human defensins (McPhee et al., 2014), suggesting that unlike OmpT, ArlC might cleave AMPs that are stabilized by disulfide bridges. RNase 7 contains four disulfide bridges and three dibasic sites (Figure 6a) and is abundant in the urinary tract (Spencer et al., 2011, 2013). The presence of dibasic sites suggests that RNase 7 might be an omptin substrate; therefore, we sought to investigate whether there was a difference in omptin‐mediated cleavage of this peptide. Under our experimental conditions, OmpT and OmpP did not cleave RNase 7 (Figure 6b). After a 60‐min incubation with ArlC, an RNase 7 cleavage product appeared, with more cleavage product appearing after 90 min. While cleavage appears limited, ArlC was the only OmpT‐like omptin able to cleave RNase 7. Taken together, these data indicate that ArlC, OmpP, and OmpT have different substrate specificities, suggesting that the presence of multiple omptin proteases in a single bacterial strain may enhance AMP resistance by increasing the range of substrates cleaved.
Figure 6.

ArlC cleaves RNase 7. (a) Pymol‐generated image of RNase 7 (Huang et al., 2007; PDB 2hky), peptide backbone is shown in blue, cysteines and disulfide bridges are in yellow, and dibasic sites are colored magenta. Numbers correspond to the following locations of the dibasic sites in the protein sequence: 1, residues 35 and 36; 2, residues 96 and 97; and 3, residues 111 and 112. (b) Proteolytic cleavage of RNase 7. RNase 7 was incubated with BL21 containing empty vector (ø) or BL21 expressing arlC, ompP, or ompT for 60 or 90 min. Cleavage products (arrows) were separated by SDS‐PAGE and visualized by Coomassie staining. M indicates molecular weight marker. Data are representative of three independent experiments
4. DISCUSSION
Detection of specific genes, including ompT, is often used to characterize virulent clinical UPEC isolates (Johnson et al., 2001; Najafi et al., 2018). Previous studies have suggested that OmpT from the UPEC strain CFT073 is involved in adhesion, invasion, and/or inactivation of AMPs (Brannon et al., 2013; He et al., 2015). While the presence of ompT is associated with virulent strains, its precise contribution remains unclear, as UPEC clinical isolates have highly variable genetic sequences (Schreiber et al., 2017). In addition, we previously observed large differences in OmpT protein activity due to differential ompT expression (Thomassin, Brannon, Gibbs, et al., 2012; Thomassin, Brannon, Kaiser, et al., 2012), suggesting that the presence of the ompT gene may not entirely correlate with its activity levels in different UPEC clinical isolates. In this study, we hypothesized that OmpT activity correlates with increased disease severity among UPEC clinical isolates. To test this hypothesis, we systematically measured omptin activity in 58 E. coli isolates representing colonization and a range of clinical outcomes. Increased omptin activity was correlated with clinical UPEC strains isolated from patients with symptomatic UTIs (UTI and sepsis groups).
Omptin activity was heterogeneous among the clinical isolates and could be related to differential ompT expression and the presence of a second OmpT‐like protease, arlC. For example, a 20‐fold difference in ompT expression was observed between isolates 5 and 11 of the UTI group (Figure 2b). This finding is not unprecedented, since it was previously shown that ompT expression was 32‐fold higher in EHEC than in EPEC (Thomassin, Brannon, Gibbs, et al., 2012). Differential ompT expression levels in EHEC and EPEC were attributed to differences in distal promoter sequences found more than 150 bp upstream of the ompT start codon (Thomassin, Brannon, Gibbs, et al., 2012). An EPEC‐like ompT distal promoter sequence and genomic context were also correlated with low OmpT activity in UPEC reference strains (Brannon et al., 2013). Therefore, it was not surprising that the EPEC‐like promoter in cystitis (UTI) isolate 11 resulted in low ompT expression and OmpT activity. The insertion of envY in the intergenic space between nfrA and ompT correlated with the increased ompT expression and OmpT activity levels observed in cystitis (UTI) isolates 1 and 6 (Figures 2b,c and 3a). These data further suggest that variations in distal promoter sequences are responsible for differential ompT expression and, in turn, proteolytic activity observed. It is also possible that in addition to differences in the promoter regions, transcription factors or post‐transcriptional factors regulating ompT expression are absent or differentially expressed in some isolates. In some cases, ompT expression levels did not correlate with proteolytic activity (Figure 2b,c). There are several possible explanations for this observation: (a) In some isolates, ompT might be subjected to additional post‐transcriptional controls, (b) truncated ompT genes present on some of the virulence plasmids contribute to the qPCR results, (c) the presence of different surface structures prevent the peptide from accessing the OmpT active site as described by Galvàn and colleagues (Galvan, Lasaro, & Schifferli, 2008), and (d) another explanation for heterogeneous omptin activity observed in this study could be attributed to the presence of a second plasmid‐encoded omptin, ArlC, in some isolates. The arlC gene was first identified as part of a large virulence plasmid of the AIEC strain NRG857c (McPhee et al., 2014). BLAST searches in the NCBI database revealed that arlC can also be found on plasmids harbored by various human ExPEC strains isolated from patients with meningitis and sepsis, as well as avian E. coli strains (Figure A2). Specifically, tBLASTn search of the nonredundant plasmid database identified arlC in 91 instances (Galata et al., 2018). The arlC gene is predominantly found in IncFIB (41/91) or IncFII (28/91) plasmids and less commonly in IncFIC(FII), IncQ1, IncN, or IncHI2 plasmids (13/91, 6/91, 2/91, 1/91, respectively). While we did not detect ompP in our study, ompP is present in some UPEC strains that were collected and sequenced by the Broad Institute (E.coli UTI Bacteremia Initiative, 2019). This opens the possibility that any combination of ompT‐like omptin may be present in a given UPEC strain.
Omptins belonging to the OmpT‐like subfamily are known to have subtle differences in substrate specificity (Brannon, Thomassin, et al., 2015; Hwang et al., 2007; McCarter et al., 2004). Studies using peptide libraries to compare OmpP and OmpT activity showed both omptins preferentially cleave substrates between two consecutive basic residues, but that OmpP appears to have a slight preference for Lys in the P and P’ sites (Hwang et al., 2007). In addition to subtle differences in amino acid motif preference, peptide size and secondary structure also impact substrate specificity (Brannon, Thomassin, et al., 2015; Haiko et al., 2009; Hritonenko & Stathopoulos, 2007). For example, AMP α‐helicity was shown to be a determining factor for proteolytic activity of the OmpT‐like omptin, CroP, from Citrobacter rodentium (Brannon, Thomassin, et al., 2015). While ArlC, OmpP, and OmpP all readily cleave small unstructured substrates, such as the FRET substrate and C18G, differences in cleavage efficiency were noted for larger or more structured AMPs. As all three proteases readily cleave the FRET substrate and C18G, the striking differences in ability to cleave Magainin II, mCRAMP, and LL‐37 are likely due to intrinsic differences between OmpT, OmpP, and ArlC. OmpP did not cleave Magainin II as efficiently as C18G and did not cleave larger substrates such as mCRAMP, LL‐37, and RNase 7 (Figures 4a, 5a,b, 6b). These findings suggest that larger peptides might be excluded from the OmpP active site. While OmpT and ArlC cleaved the FRET substrate, C18G, Magainin II, and mCRAMP relatively efficiently, there was a striking difference in LL‐37 and RNase 7 cleavage (Figures 4a, 5a, and 6b). Given the similarity in size of mCRAMP and LL‐37, and the ability of ArlC to cleave RNase 7, it is unlikely that the 3 amino acid size difference accounts for the marked difference in cleavage efficiency. It is possible that ArlC does not cleave α‐helical AMPs, but instead cleaves unstructured and disulfide bond‐stabilized peptides. While this possibility requires further study, it is supported by the finding that an arlC deletion strain is more susceptible to killing by human defensins (McPhee et al., 2014). Altogether, these findings suggest the presence ArlC and OmpT in the same UPEC isolate may confer a fitness advantage by expanding the spectrum of target substrates.
5. CONCLUSIONS
Here, we show that increased omptin activity is associated with UPEC strains causing symptomatic UTIs. Extensive heterogeneity of omptin activity among UPEC clinical isolates was due to variations in ompT expression and due to the presence of a plasmid‐encoded ompT‐like gene arlC. Our findings support current profiling practices of UPEC strains that include the ompT gene (Johnson & Stell, 2000), but suggest that additional screening for arlC should be considered as both genes were exclusively harbored in UPEC strains associated with symptomatic infections. Altogether, our findings suggest that the presence of two different omptins in a UPEC strain may provide an additional fitness advantage by expanding the range of AMPs cleaved during UTIs.
CONFLICT OF INTERESTS
The authors have declared that no conflict of interest exists.
AUTHOR CONTRIBUTIONS
AP, ID, JAT, JLT, JML, and JRB performed experiments. AM, GTM, HLM, ID, JDS, JLT, KD, and SG conceived and designed experiments. HLM, ID, JLT, and SG analyzed the data. HLM and ID wrote early drafts of the manuscript. JLT wrote and reviewed later drafts with support from all other authors.
ETHICS STATEMENT
None required.
DEDICATION
This publication is dedicated to Dr. Hervé Le Moual who passed away on 3 March 2018; he was a great mentor who always encouraged his trainees to follow their passions.
ACKNOWLEDGMENTS
This work was supported by the Canadian Institutes of Health Research (CIHR, MOP‐15551), the Natural Sciences and Engineering Research Council (NSERC, RGPIN‐217482, RGPIN‐2014‐05119), and the Fonds de Recherche Québec—Nature et Technologies (FRQNT 2013‐PR‐165926). ID was supported by the Fonds de Recherche Québec—Santé (FRQS). AP was supported by an Ontario‐Quebec Exchange Fellowship. JLT was supported by a Hugh Burke fellowship from the McGill Faculty of Medicine. JLT is supported by a NSERC postdoctoral fellowship and Pasteur‐Roux fellowship. Work performed by JAT and GTM was supported by the Canadian Institutes of Health Research (CIHR, MOP‐125998). We thank Dr. Mario Jacques and Mr. Frédéric Berthiaume (Faculté de Médecine Vétérinaire, Université de Montréal) for providing access to the Jasco J‐810 spectropolarimeter and technical assistance with CD experiments. We thank Dr. Selena Sagan for the gift of labeling reagents for Southern hybridization. We thank Drs. Olivera Francetic and Yannick Tremblay for helpful comments and suggestions. We gratefully acknowledge Mr. Gary Leveque from the McGill University and Genome Québec Innovation Center for his assistance with plasmid sequence assembly.
APPENDIX 1.
MATERIALS AND METHODS
Genome analysis
Sequenced genomes were annotated using PATRIC (Wattam et al., 2017). Isolate serotypes were determined using the online database SeroTypeFinder (Joensen, Tetzschner, Iguchi, Aarestrup, & Scheutz, 2015). Strain sequence type was determined using the MLST2 server (Larsen et al., 2012), and plasmids were typed using the pMLST 2.0 server (Carattoli et al., 2014). Pathogenicity islands were detected using IslandViewer 4 (Bertelli et al., 2017) and VRprofile 2 (Li et al., 2018). Antibiotic resistance genes were identified in PATRIC, IslandViewer 4, VRprofile 2, BLAST, and RGI 4.2.2 CARD 3.0.1 (Jia et al., 2017). Figures of genome alignments and plasmid alignments were generated using BRIGs software (Alikhan, Petty, Ben Zakour, & Beatson, 2011).
Figure A1.

Comparisons of cystitis (UTI) isolate genomes with reference strains. (a) Indicated cystitis isolates were used as subject sequences in multiple sequence alignments with the indicated UPEC strain genomes using BRIGs software. White fill indicates no homology. (b) Genes amplified by multiplex PCR (ompT, fimH, iutA, papA, papH, papC, papF, fyuA, kpsMTII, papE, sfaS, kpsMTIII, cnf‐1) were used as subject sequences for a multiple sequence alignment of the indicated UPEC strain genome using BRIGs software. Black fill indicates no homology
Figure A2.

Comparison of plasmid sequences containing pathogenicity island 6. (a) Plasmids from the indicated cystitis (UTI) isolates were used as subject sequences in multiple sequence alignments with the indicated plasmid containing pathogenicity island 6 from pO83 from E. coli NRG857c using BRIGs software. White fill indicates no homology. (b) Coding sequences for pathogenicity islands (PI‐) 1, 2, 3, 4, 5, and 6 from pO83 (indicated) were used as subject sequences for a multiple sequence alignment with the indicated plasmid nucleotide sequence using BRIGs software. Black fill indicates no homology
Table A1.
General features of sequenced cystitis (UTI) isolates
| Strain | Serotype | Pathotype | Origin/disease | Phylogenetic group | Sequence type | Chromosome | Plasmid | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Size (kb) | G + C content (%) | Size (kb) | G + C content (%) | Inc type | ||||||
| Cystitis 1 | O1:H42 | ExPEC | Homo sapiens/Cystitis | D | 648 | 5,083 | 50.5 | 195 | 49.5 | F18:A‐:B1 |
| Cystitis 6 | O82:H8 | ExPEC | Homo sapiens/Cystitis | B1 | 88 | 4,782 | 50.6 | 150 | 51.8 | F16:A‐:B1 |
| Cystitis 11 | O24:H4 | ExPEC | Homo sapiens/Cystitis | D | 48 | 4,946 | 50.7 | 157 | 49.8 | F18:A6:B42 |
Table A2.
Genome features of sequenced cystitis (UTI) isolates
| Strains | Cystitis 1 | Cystitis 6 | Cystitis 11 | ||||
|---|---|---|---|---|---|---|---|
| Chromosome | Plasmid | Chromosome | Plasmid | Chromosome | Plasmid | ||
| Number of genes | 5,148 | 262 | 4,904 | 236 | 4,865 | 235 | |
| rRNA | 22 | 0 | 22 | 0 | 22 | 0 | |
| tRNA | 88 | 0 | 91 | 0 | 89 | 0 | |
| Prophages | Complete | 4 | 1 | 0 | 0 | 6 | 0 |
| Incomplete | 3 | 1 | 3 | 1 | 1 | 1 | |
| Virulence factors | Number | 249 | 19 | 242 | 13 | 217 | 19 |
| % of genes | 4.8 | 7.2 | 4.9 | 5.5 | 4.5 | 8.1 | |
| Genomic islands | 12 | 4 | 11 | 5 | 15 | 5 | |
| Unique sequencesa | 73 | N/A | 34 | N/A | 100 | N/A | |
N/A not applicable.
Table A3.
Predicted genomic islands
| Isolate | Location | GIa | Start | Stop | Size | % GC | # VGb | #AMRc | tRNA | Features |
|---|---|---|---|---|---|---|---|---|---|---|
| Cystitis 1 | Chromosome | 1 | 108,501 | 129,938 | 21,438 bp | 47 | 6 | 0 | seC | GI‐like region‐1 |
| 2 | 147,071 | 185,155 | 38,085 bp | 49 | 10 | 0 | 0 | Prophage‐1 | ||
| 3 | 759,876 | 781,233 | 21,358 bp | 43 | 3 | 0 | 0 | GI‐like region‐2 | ||
| 4 | 1,067,617 | 1,083,797 | 16,181 bp | 37 | 0 | 0 | 0 | T3SS‐1 | ||
| 5 | 1,974,099 | 2,012,452 | 38,354 bp | 49 | 14 | 0 | 0 | GI‐like region‐3 | ||
| 6 | 2,496,955 | 2,544,448 | 47,494 bp | 49 | 0 | 0 | Arg | Prophage‐2 | ||
| 7 | 2,715,386 | 2,762,418 | 47,033 bp | 48 | 0 | 0 | 0 | Prophage‐3 | ||
| 8 | 2,924,030 | 2,960,151 | 36,122 bp | 46 | 3 | 0 | 0 | GI‐like region‐4 | ||
| 9 | 3,638,710 | 3,682,745 | 44,036 bp | 51 | 1 | 1 | 0 | Prophage‐4 | ||
| 10 | 3,972,222 | 3,989,123 | 16,902 bp | 56 | 0 | 0 | 0 | T6SS‐1 | ||
| 11 | 4,643,625 | 4,660,145 | 16,521 bp | 49 | 0 | 0 | 0 | Prophage‐5 | ||
| 12 | 4,802,101 | 4,835,389 | 33,289 bp | 51 | 1 | 0 | 0 | Prophage‐6 | ||
| Plasmid | 1 | 2 | 30,510 | 30,509 bp | 53 | 0 | 0 | 0 | T4SS‐1 | |
| 2 | 35,739 | 41,156 | 5,417 bp | 59 | 0 | 0 | 0 | Integron region | ||
| 3 | 72,375 | 80,367 | 7,992 bp | 56 | 0 | 1 | 0 | IS401 | ||
| 4 | 134,523 | 141,371 | 6,848 bp | 38 | 2 | 0 | 0 | PAI‐6 like region | ||
| Cystitis 6 | Chromosome | 1 | 9,045 | 26,763 | 17,719 bp | 40 | 0 | 0 | 0 | Prophage‐1 |
| 2 | 457,520 | 475,359 | 17,840 bp | 49 | 2 | 0 | 0 | GI‐like region‐1 | ||
| 3 | 897,443 | 919,427 | 21,985 bp | 46 | 0 | 0 | Arg | Prophage‐2 | ||
| 4 | 1,119,747 | 1,142,698 | 22,952 bp | 45 | 0 | 0 | Thr | Prophage‐3 | ||
| 5 | 1,951,044 | 1,969,169 | 18,126 bp | 47 | 1 | 0 | 0 | Prophage‐4 | ||
| 6 | 2,249,278 | 2,256,258 | 6,981 bp | 44 | 2 | 0 | 0 | Prophage‐5 | ||
| 7 | 2,316,877 | 2,325,663 | 8,787 bp | 53 | 0 | 0 | 0 | GI‐like region‐2 | ||
| 8 | 2,695,320 | 2,714,439 | 19,120 bp | 51 | 9 | 0 | 0 | GI‐like region‐3 | ||
| 9 | 2,728,674 | 2,734,357 | 5,684 bp | 52 | 1 | 0 | Leu | Prophage‐6 | ||
| 10 | 4,256,804 | 4,265,449 | 8,646 bp | 35 | 0 | 0 | 0 | T3SS‐1 | ||
| 11 | 4,460,470 | 4,468,593 | 8,124 bp | 48 | 0 | 0 | 0 | Prophage‐7 | ||
| Plasmid | 1 | 216 | 5,410 | 5,195 bp | 48 | 0 | 0 | 0 | T4SS‐1 | |
| 2 | 20,148 | 28,846 | 8,698 | 43 | 0 | 0 | 0 | IS1 | ||
| 3 | 24,163 | 149,472 | 24,163 bp | 54 | 0 | 0 | 0 | T4SS‐2 | ||
| 4 | 38,028 | 42,095 | 4,234 bp | 37 | 2 | 0 | 0 | PAI‐6 like region | ||
| 5 | 79,896 | 108,023 | 28,127 bp | 60 | 0 | 5 | 0 | Integron region | ||
| Cystitis 11 | Chromosome | 1 | 108,460 | 131,562 | 23,103 bp | 47 | 7 | 0 | seC | GI‐like region‐1 |
| 2 | 1,043,826 | 1,070,085 | 26,260 bp | 55 | 0 | 0 | 0 | T6SS‐1 | ||
| 3 | 1,855,773 | 1,873,433 | 17,661 bp | 37 | 6 | 1 | 0 | GI‐like region‐2 | ||
| 4 | 1,893,976 | 1,917,531 | 23,556 bp | 51 | 6 | 0 | 0 | GI‐like region‐3 | ||
| 5 | 1,969,787 | 2,013,948 | 44,162 bp | 51 | 0 | 0 | Gly | Prophage‐1 | ||
| 6 | 2,068,949 | 2,103,765 | 34,817 bp | 51 | 0 | 0 | 0 | Prophage‐2 | ||
| 7 | 2,436,309 | 2,488,552 | 52,244 bp | 49 | 0 | 0 | 0 | Prophage‐3 | ||
| 8 | 2,834,397 | 2,844,341 | 9,945 bp | 49 | 1 | 0 | 0 | Prophage‐4 | ||
| 9 | 3,120,160 | 3,147,806 | 27,647 bp | 51 | 1 | 2 | 0 | Prophage‐5 | ||
| 10 | 3,245,740 | 3,304,629 | 58,890 bp | 49 | 2 | 0 | 0 | Prophage‐6 | ||
| 11 | 3,370,608 | 3,382,255 | 11,648 bp | 48 | 0 | 0 | 0 | GI‐like region‐4 | ||
| 12 | 3,825,365 | 3,850,278 | 24,914 bp | 49 | 8 | 0 | 0 | GI‐like region‐5 | ||
| 13 | 4,115,143 | 4,151,037 | 35,895 bp | 51 | 3 | 1 | 0 | GI‐like region‐6 | ||
| 14 | 4,118,953 | 4,167,634 | 48,682 bp | 50 | 1 | 0 | Leu | Prophage‐7 | ||
| 15 | 4,402,939 | 443,880 | 40,942 bp | 43 | 7 | 0 | 0 | GI‐like region‐7 | ||
| Plasmid | 1 | 5,280 | 23,125 | 17,846 bp | 56 | 2 | 1 | 0 | Prophage‐1 | |
| 2 | 45,365 | 50,841 | 5,116 bp | 37 | 2 | 0 | 0 | PAI‐6 like region | ||
| 3 | 82,364 | 87,769 | 5,405 | 42 | 0 | 0 | 0 | IS2 and IS3 | ||
| 4 | 106,311 | 111,175 | 4,864 | 41 | 1 | 0 | 0 | IS1 | ||
| 5 | 125,191 | 157,658 | 32,468 bp | 53 | 0 | 0 | 0 | T4SS‐1 |
Genomic island.
VG indicates known virulence genes.
Antimicrobial resistance genes.
Table A4.
Antibiotic resistance genes in cystitis (UTI) isolates
| Isolate | Location | Gene | Function; resistance mechanism | Resistance to |
|---|---|---|---|---|
| Cystitis 1 | Chromosome | acrA | Subunit of an RND efflux pump; antibiotic efflux | Aminoglycosides |
| acrD | Part of an RND efflux pump; antibiotic efflux | Aminoglycosides | ||
| acrE | Part of AcrEF‐TolC efflux pump; antibiotic efflux | Fluoroquinolones, cephamycin, cephalosporin, penam | ||
| acrF | Part of AcrEF‐TolC efflux pump; antibiotic efflux | Fluoroquinolones, cephamycin, cephalosporin, penam | ||
| adeF | Membrane fusion protein of the multidrug efflux complex AdeFGH; antibiotic efflux | Fluoroquinolone, tetracycline | ||
| ampC | Enzymatic degradation of ß‐lactam rings; antibiotic inactivation | Broad and extended spectrum ß‐lactamases | ||
| cmeB | Inner membrane transporter in CmeABC RND efflux channel; antibiotic efflux | Cephalosporins, macrolides, fluoroquinolones, fusidic acid | ||
| cmeC | Outer membrane channel in CmeABC RND efflux channel; antibiotic efflux | Cephalosporins, macrolides, fluoroquinolones, fusidic acid | ||
| cyaA | Adenylate cyclase variant S352T; antibiotic target alteration | Fosfomycin | ||
| emrA | Membrane fusion protein in EmrAB‐TolC efflux pump complex; antibiotic efflux | Fluoroquinolone | ||
| emrB | Translocase in EmrAB‐TolC efflux pump complex; antibiotic efflux | Fluoroquinolone | ||
| emrD | Multidrug transporter that couples efflux of amphipathic compounds with proton import across the plasma membrane; antibiotic efflux | Detergents | ||
| emrE | Small multidrug resistance efflux; antibiotic efflux | Macrolides | ||
| emrY | Multidrug transport across the inner membrane; antibiotic efflux | Tetracycline | ||
| ermK | Membrane fusion protein that works with ErmY and TolC as part of a MFS efflux pump; antibiotic efflux | Tetracycline | ||
| ftsI | Sequence variant D350N, S357N of PBP3; antibiotic target alteration | Cephamycin, cephalosporin, penam, carbapenam, monobactam | ||
| glpT | Sequence variant E448K of the active importer GlpT; antibiotic target alteration | Fosfomycin | ||
| macA | Membrane fusion protein that acts with MacB and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides | ||
| macB | ABC transporter that acts with MacA and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides (14‐/15‐membered lactones) | ||
| marA | Regulates MDR efflux pump and regulates porin synthesis; reduced antibiotic permeability, antibiotic efflux | Tetracycline, penem, penam, carbapenem, cephamycin, cephalosporin, rifamycin, phenicol, monobactam, glycycline, fluoroquinolone, triclosan | ||
| marR | MarR variant G103S Y137H causes efflux pump overexpression; antibiotic target alteration, antibiotic efflux | Tetracyclines, penam, cephalosporins, glycycline, rifamycin, phenicol, triclosan, fluoroquinolones | ||
| mdfA | Multidrug efflux pump; antibiotic efflux | Tetracycline, benzalkonium chloride, rhodamine | ||
| mdtA | Membrane fusion protein RND efflux pump; antibiotic efflux | Aminocoumarins | ||
| mdtB | Transporter that forms multimeric complex with MdtC; antibiotic efflux | Aminocoumarins | ||
| mdtC | Transporter that forms multimeric complex with MdtB; antibiotic efflux | Aminocoumarins | ||
| mdtD | MFS transporter; antibiotic efflux | Aminocoumarins | ||
| mdtE | Membrane fusion protein that works with MdtF and TolC as part of a MFS efflux pump; antibiotic efflux | Penam, fluoroquinolones, macrolides | ||
| mdtF | Inner membrane transporter that works with MdtE and TolC as part of a MFS efflux pump; antibiotic efflux | Penam, fluoroquinolones, macrolides | ||
| mdtH | MFS transporter; antibiotic efflux | Fluoroquinolones | ||
| mdtM | MFS transporter; antibiotic efflux | Nucleosides, phenicol, lincosamides, fluoroquinolones, acridine dye | ||
| mdtN | Part of MdtNOP MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| mdtO | Part of MdtNOP MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| mdtP | Part of MdtNOP MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| msbA | Multidrug resistance transporter homolog; antibiotic efflux | Nitroimidazole | ||
| nfsA | Variant Y45C of major oxygen insensitive nitroreductase in Escherichia coli; antibiotic target alteration | Nitrofuran | ||
| pmrD | Histidine kinase involved in regulation of polymyxin resistance; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrF | Glycosyl transferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrH | UDP‐4‐amino‐4‐deoxy‐L‐arabinose‐oxoglutarate aminotransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrI | UDP‐4‐amino‐4‐deoxy‐L‐arabinose formyltransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrJ | Catalyzes deformylation of L‐Ara4‐formyL‐N; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrK | Undecaprenyl phosphate‐alpha‐4‐amino‐4‐deoxy‐L‐arabinosyltransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrL | Sucrose‐6 phosphate hydrolase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrM | Subunit of undecaprenyl phosphate‐alpha‐L‐Ara4N flippase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| sapA | Periplasmic solute binding protein; antibiotic efflux | Antimicrobial peptides | ||
| sapB | Permease subunit; antibiotic efflux | Antimicrobial peptides | ||
| sapC | Permease subunit; antibiotic efflux | Antimicrobial peptides | ||
| sapD | ATPase; antibiotic efflux | Antimicrobial peptides | ||
| sapF | ATPase; antibiotic efflux | Antimicrobial peptides | ||
| tufA | Sequence variant R234F of elongation factor Tu; antibiotic target alteration | Pulvomycin, elfamycin | ||
| Cystitis 6 | Chromosome | acrA | subunit of AcrAB‐TolC RND efflux pump; antibiotic efflux | Tetracycline, penam, cephalosporin, rifamycin, phenicol, glycycline, fluoroquinolone, triclosan |
| acrB | subunit of AcrAB‐TolC RND efflux pump; antibiotic efflux | Tetracycline, penam, cephalosporin, rifamycin, phenicol, glycycline, fluoroquinolone, triclosan | ||
| acrD | Part of an RND efflux pump; antibiotic efflux | Aminoglycosides | ||
| acrE | Part of AcrEF‐TolC RND efflux pump; antibiotic efflux | Cephamycin, cephalosporin, penam, fluoroquinolone | ||
| acrF | Part of AcrEF‐TolC RND efflux pump; antibiotic efflux | Cephamycin, cephalosporin, penam, fluoroquinolone | ||
| adeF | Membrane fusion protein of the AdeFGH RND efflux pump; antibiotic efflux | Tetracycline, fluoroquinolones | ||
| ampC | Enzymatic degradation of ß‐lactam rings; antibiotic inactivation | Broad and extended spectrum ß‐lactamases | ||
| bcr | Part of an efflux system; antibiotic efflux | Bicyclomycins | ||
| cmeB | Inner membrane transporter of the CmeABC RND efflux complex; antibiotic efflux | Macrolides, cephalosporins, fusidic acid, fluoroquinolones | ||
| emrA | Part of the EmrAB‐TolC MFS efflux pump; antibiotic efflux | Fluoroquinolones | ||
| emrB | Part of the EmrAB‐TolC MFS efflux pump; antibiotic efflux | Fluoroquinolones | ||
| emrK | Part of the EmKY‐TolC MFS efflux pump; antibiotic efflux | Tetracyclines | ||
| emrY | Part of the EmKY‐TolC MFS efflux pump; antibiotic efflux | Tetracyclines | ||
| ermK | Erm 23S rRNA methyltransferase; antibiotic target alteration | Lincosamides, macrolides, streptogramins | ||
| ftsI | Sequence variant D350N, S357N of PBP3; antibiotic target alteration | Cephamycin, cephalosporin, penam, carbapenam, monobactam | ||
| gyrA | Point mutation (S83L); antibiotic target modification | Fluoroquinolones, nybomycin | ||
| marA | Regulates MDR efflux pump and regulates porin synthesis; reduced antibiotic permeability, antibiotic efflux | Tetracycline, penem, penam, carbapenem, cephamycin, cephalosporin, rifamycin, phenicol, monobactam, glycycline, fluoroquinolone, triclosan | ||
| marR | Regulates marAB operon; antibiotic target alteration, antibiotic efflux | Tetracyclines, penam, cephalosporins, glycycline, rifamycin, phenicol, triclosan, fluoroquinolones | ||
| marR | MarR variant G103S Y137H causes efflux pump overexpression; antibiotic target alteration, antibiotic efflux | Tetracyclines, penam, cephalosporins, glycycline, rifamycin, phenicol, triclosan, fluoroquinolones | ||
| mdfA | Multidrug efflux pump; antibiotic efflux | Tetracycline, benzalkonium chloride, rhodamine | ||
| mdtA | Membrane fusion protein RND efflux pump; antibiotic efflux | Aminocoumarins | ||
| mdtB | Transporter that forms multimeric complex with MdtC; antibiotic efflux | Aminocoumarins | ||
| mdtC | Transporter that forms multimeric complex with MdtB; antibiotic efflux | Aminocoumarins | ||
| mdtE | Membrane fusion protein that works with MdtF and TolC as part of a MFS efflux pump; antibiotic efflux | Penam, fluoroquinolones, macrolides | ||
| mdtF | Inner membrane transporter that works with MdtE and TolC as part of a MFS efflux pump; antibiotic efflux | Penam, fluoroquinolones, macrolides | ||
| mdtH | MFS transporter; antibiotic efflux | Fluoroquinolones | ||
| mdtK | Part of a multidrug and toxic compounds extrusions transporter; antibiotic efflux | Norfloxacin, doxorubicin, acriflavine | ||
| mdtM | MFS transporter; antibiotic efflux | Nucleosides, phenicol, lincosamides, fluoroquinolones, acridine dye | ||
| mdtN | Part of MdtNOP MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| mdtO | Part of MdtNOP MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| mdtP | Part of MdtNOP MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| msbA | Multidrug resistance transporter homolog; antibiotic efflux | Nitroimidazole | ||
| nfsA | Variant Y45C of major oxygen insensitive nitroreductase in Escherichia coli; antibiotic target alteration | Nitrofuran | ||
| pmrD | Histidine kinase involved in regulation of polymyxin resistance; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrF | Glycosyl transferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrH | UDP‐4‐amino‐4‐deoxy‐L‐arabinose‐oxoglutarate aminotransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrI | UDP‐4‐amino‐4deoxy‐L‐arabinose formyltransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrJ | Catalyzes deformylation of L‐Ara4‐formyL‐N; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrK | Undecaprenyl phosphate‐alpha‐4‐amino‐4‐deoxy‐L‐arabinosyltransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrL | Sucrose‐6 phosphate hydrolase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrM | Subunit of undecaprenyl phosphate‐alpha‐L‐Ara4N flippase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| tufA | Sequence variant R234F of elongation factor Tu; antibiotic target alteration | Pulvomycin, elfamycin | ||
| Plasmid | cmeC | Outer membrane channel of the CmeABC RND antibiotic efflux pump; antibiotic efflux | Cephalosporin, macrolides, fluoroquinolones, fusidic acid | |
| macA | Membrane fusion protein that acts with MacB and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides | ||
| macB | ABC transporter that acts with MacA and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides (14‐/15‐membered lactones) | ||
| mdtH | MFS antibiotic efflux pump; antibiotic efflux | Fluoroquinolones | ||
| aadA | Aminoglycoside nucleotidyltransferase, antibiotic inactivation | Aminoglycosides | ||
| dfrA12 | Dihydrofolate reductase; antibiotic target replacement | Diaminopyrimidine | ||
| mphA | Macrolide 2’‐phosphotransferase; antibiotic inactivation | Macrolides; preferentially inactivates 14‐membered macrolides over 16‐membered macrolides | ||
| sul1 | Dihydropteroate synthase type‐2; antibiotic target replacement | Sulfonamides, sulfone | ||
| tetA | Tetracycline efflux protein TetA; antibiotic efflux | Tetracyclines, Glycylcycline | ||
| Cystitis 11 | Chromosome | acrA | subunit of AcrAB‐TolC RND efflux pump; antibiotic efflux | Tetracycline, penam, cephalosporin, rifamycin, phenicol, glycycline, fluoroquinolone, triclosan |
| acrB | subunit of AcrAB‐TolC RND efflux pump; antibiotic efflux | Tetracycline, penam, cephalosporin, rifamycin, phenicol, glycycline, fluoroquinolone, triclosan | ||
| acrD | RND antibiotic efflux pump; antibiotic efflux | Aminoglycosides | ||
| acrE | Membrane fusion protein of a RND efflux transporter; antibiotic efflux | Penam, cephamycin, cephalosporin, fluoroquinolones | ||
| acrF | Inner membrane transporter component of a RND efflux transporter; antibiotic efflux | Penam, cephamycin, cephalosporin, fluoroquinolones | ||
| adeF | Membrane fusion protein of the AdeFGH RND efflux pump; antibiotic efflux | Tetracycline, fluoroquinolones | ||
| ampC | Class C ß‐lactamase; antibiotic inactivation | Broad and extended spectrum cephalosporins | ||
| cmeB | Inner membrane transporter in CmeABC RND efflux channel; antibiotic efflux | Cephalosporins, macrolides, fluoroquinolones, fusidic acid | ||
| cmeC | Outer membrane channel in CmeABC RND efflux channel; antibiotic efflux | Cephalosporins, macrolides, fluoroquinolones, fusidic acid | ||
| cyaA | Adenylate cyclase variant S352T; antibiotic target alteration | Fosfomycin | ||
| emrA | Part of the EmrAB‐TolC MFS efflux pump; antibiotic efflux | Fluoroquinolones | ||
| emrB | Part of the EmrAB‐TolC MFS efflux pump; antibiotic efflux | Fluoroquinolones | ||
| emrD | Multidrug transporter that couples efflux of amphipathic compounds with proton import across the plasma membrane; antibiotic efflux | Detergents | ||
| emrE | Small MDR transporter; antibiotic efflux | Macrolides | ||
| emrK | Part of the EmKY‐TolC MFS efflux pump; antibiotic efflux | Tetracyclines | ||
| emrY | Part of the EmKY‐TolC MFS efflux pump; antibiotic efflux | Tetracyclines | ||
| emrY | MFS antibiotic efflux pump; antibiotic efflux | Tetracyclines | ||
| ermA | RNA methylase; antibiotic target alteration | Macrolides, streptogramins, lincosamides | ||
| ermB | RNA methylase; antibiotic target alteration | Macrolides, streptogramins, lincosamides | ||
| ermK | RNA methylase; antibiotic target alteration | Macrolides, streptogramins, lincosamides | ||
| ftsI | Sequence variant D350N, S357N of PBP3; antibiotic target alteration | Cephamycin, cephalosporin, penam, carbapenam, monobactam | ||
| glpT | Sequence variant E448K of the active importer GlpT; antibiotic target alteration | Fosfomycin | ||
| macA | Membrane fusion protein that acts with MacB and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides | ||
| macB | ABC transporter that acts with MacA and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides (14‐/15‐membered lactones) | ||
| marA | Global activator protein that induces MDR efflux and downregulates OmpF synthesis; reduced permeability to antibiotic, antibiotic efflux | Tetracycline, penem, penam, carbapenem, cephalosporin, rifamycin, phenicol, monobactam, glycycline, fluoroquinolone, triclosan | ||
| marR | MarR variant G103S Y137H causes efflux pump overexpression; antibiotic target alteration, antibiotic efflux | Tetracyclines, penam, cephalosporins, glycycline, rifamycin, phenicol, triclosan, fluoroquinolones | ||
| mdfA | Multidrug efflux pump; antibiotic efflux | Tetracycline, benzalkonium chloride, rhodamine | ||
| mdtA | Membrane fusion component of the MdtABC RND efflux pump; antibiotic efflux | Aminocoumarin resistance | ||
| mdtB | Transporter in the MdtABC RND efflux pump; antibiotic efflux | Aminocoumarin resistance | ||
| mdtC | Transporter in the MdtABC RND efflux pump; antibiotic efflux | Aminocoumarin resistance | ||
| mdtE | Membrane fusion protein of a RND efflux transporter; antibiotic efflux | Penam, macrolides, fluoroquinolones | ||
| mdtF | Multidrug inner membrane transporter of an RND efflux transporter; antibiotic efflux | Penam, macrolides, fluoroquinolones | ||
| mdtH | MFS transporter; antibiotic efflux | Fluoroquinolones | ||
| mdtM | MFS transporter; antibiotic efflux | Nucleosides, phenicol, lincosamides, fluoroquinolones, acridine dye | ||
| mdtN | Predicted inner membrane fusion protein of MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| mdtO | Uncharacterized component of MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| mdtP | Predicted outer membrane component of MFS efflux pump; antibiotic efflux | Nucleoside antibiotics, acridine dye | ||
| msbA | Multidrug resistance transporter homolog; antibiotic efflux | Nitroimidazole | ||
| nfsA | Variant Y45C of major oxygen insensitive nitroreductase in Escherichia coli; antibiotic target alteration | Nitrofuran | ||
| pmrD | Histidine kinase involved in regulation of polymyxin resistance; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrF | Glycosyl transferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrH | UDP‐4‐amino‐4‐deoxy‐L‐arabinose‐oxoglutarate aminotransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrI | UDP‐4‐amino‐4deoxy‐L‐arabinose formyltransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrJ | Catalyzes deformylation of L‐Ara4‐formyL‐N; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrK | Undecaprenyl phosphate‐alpha‐4‐amino‐4‐deoxy‐L‐arabinosyltransferase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrL | Sucrose‐6 phosphate hydrolase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| pmrM | Subunit of undecaprenyl phosphate‐alpha‐L‐Ara4N flippase; antibiotic target alteration | Polymyxins and peptide antibiotics | ||
| sapA | Periplasmic solute binding protein; antibiotic efflux | Antimicrobial peptides | ||
| sapB | Permease subunit; antibiotic efflux | Antimicrobial peptides | ||
| sapC | Permease subunit; antibiotic efflux | Antimicrobial peptides | ||
| sapD | ATPase; antibiotic efflux | Antimicrobial peptides | ||
| sapF | ATPase; antibiotic efflux | Antimicrobial peptides | ||
| tetA | Tetracycline efflux protein TetA; antibiotic efflux | Tetracyclines, Glycylcycline | ||
| tetB | Part of MFS efflux pump; antibiotic efflux | Tetracycline, doxycycline, minocycline | ||
| tetC | Part of MFS efflux pump; antibiotic efflux | Tetracycline | ||
| tufA | Sequence variant R234F of elongation factor Tu; antibiotic target alteration | Pulvomycin, elfamycin | ||
| Plasmid | cmeC | Outer membrane channel of the CmeABC RND antibiotic efflux pump; antibiotic efflux | Cephalosporin, macrolides, fluoroquinolones, fusidic acid | |
| macA | Membrane fusion protein that acts with MacB and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides | ||
| macB | ABC transporter that acts with MacA and TolC to form an ABC antibiotic efflux complex; antibiotic efflux | Macrolides (14‐/15‐membered lactones) |
Table A5.
Unique sequences in cystitis (UTI) isolate 1
| Lengtha | In GIb | Number of genes | Descriptionc |
|---|---|---|---|
| 6,492 | Y | 8 | Hypothetical protein, hypothetical protein, transcriptional regulator LacI family, PTS system IIA component, putative sugar phosphoesterase component IIB, putative integral membrane protein, transketolase N‐terminal section, transketolase C‐terminal section |
| 10,220 | Y/N/Y | Hypothetical protein, hypothetical protein, cobalt–zinc–cadmium resistance protein CzcA/ cation efflux system protein CusA, hypothetical protein, hypothetical protein, periplasmic lysozyme inhibitor of c‐type lysozyme, hypothetical protein, hypothetical protein, N‐acetylmannosamine‐6‐phosphate 2‐epimerase, PTS system/ maltose and glucose‐specific IIC component, RpiR family transcriptional regulator, putative exported protein, probable transposase | |
| 888 | Y | 1 | Hypothetical protein |
| 5,748 | Y | 3 | Mobile element protein, mobile element protein, AidA‐I adhesin‐like protein |
| 2,279 | Y | 2 | Beta‐1,4‐galactosyltransferase, O‐antigen ligase |
| 1,524 | Y | 2 | Hypothetical protein, hypothetical protein |
| 7,510 | Y/N | 7 | DsORF‐h1, hypothetical protein, core protein, hypothetical protein, hypothetical protein, core protein, hypothetical protein |
| 1,411 | N | 2 | Prophage Lp2 protein 6, hypothetical protein |
| 7,374 | Y | 7 | Integrase, hypothetical protein, tRNA‐dihydrouridine synthase, hypothetical protein, putative DNA‐binding protein, hypothetical protein, HigA (antitoxin to HigB) |
| 1,100 | Y | 2 | Hypothetical protein, hypothetical protein |
| 1,005 | Y | 1 | Transposase |
| 1,901 | Y | 2 | Hypothetical protein, hypothetical protein |
| 8,390 | Y | 6 | Type I restriction–modification system/DNA‐methyltransferase subunit M, anticodon nuclease, type I restriction–modification system/specificity subunit S, hypothetical protein, type I restriction–modification system/restriction subunit R, hypothetical protein |
| 1,534 | N | 2 | Hypothetical protein, hypothetical protein |
| 8,418 | N | 6 | Hypothetical protein, hypothetical protein, hypothetical protein, mobile element protein, beta‐1,3‐glucosyltransferase, UDP‐glucose 6‐dehydrogenase |
| 8,457 | Y | 8 | CRISPR‐associated helicase Cas3, hypothetical protein, CRISPR‐associated protein/Cse1 family, CRISPR‐associated protein/Cse2 family, CRISPR‐associated protein/ Cse4 family, CRISPR‐associated protein/ Cas5e family, CRISPR‐associated protein/Cse3 family, CRISPR‐associated protein Cas1 |
| 2,212 | N | 2 | Hypothetical protein, LPS glycosyltransferase |
| 1,960 | N | 4 | Minor fimbrial subunit StfE, minor fimbrial subunit StfF, minor fimbrial subunit StfG, uncharacterized protein YadU in stf fimbrial cluster |
| 970 | N | 1 | Uncharacterized protein YehA precursor |
| 6,783 | N | 2 | Membrane protein involved in the export of O‐antigen, UDP‐N‐acetylglucosamine 2‐epimerase |
| 5,126 | Y | 3 | Aspartate ammonia‐lyase, tripeptide aminopeptidase, anaerobic C4‐dicarboxylate transporter DcuC |
| 2,756 | Y | 4 | Hypothetical protein, isoaspartyl aminopeptidase, hypothetical protein, transposase |
| 4,757 | Y | 3 | Mg(2+)‐transport‐ATPase‐associated protein MgtC, inosine–uridine preferring nucleoside hydrolase, transporter/MFS superfamily |
| 7,477 | Y | 5 | Putative transcriptional regulator LysR‐type, aspartate racemase, anaerobic C4‐dicarboxylate transporter, aspartate ammonia‐lyase, Anaerobic C4‐dicarboxylate transporter DcuB |
| 8,357 | Y | 4 | Hypothetical protein, fumarate respiration transcriptional regulator DcuR, regulatory protein GntR, anaerobic C4‐dicarboxylate transporter |
| 1,562 | N | 1 | Flagellar hook‐associated protein FliD |
| 558 | N | 1 | Flagellar biosynthesis protein FliC |
| 814 | N | 1 | Hypothetical protein |
| 12,495 | Y | 6 | Type I restriction–modification system/DNA‐methyltransferase subunit M, type I restriction–modification system/ specificity subunit S, hypothetical protein, hypothetical protein, type I restriction–modification system/ restriction subunit R, Putative predicted metal‐dependent hydrolase |
| 2,441 | N | 3 | Hypothetical protein, deoxyguanosinetriphosphate triphosphohydrolase, dNTP triphosphohydrolase (putative) |
| 546 | Y | 1 | Hypothetical protein |
| 1,681 | Y | 1 | Hypothetical protein |
| 1,355 | Y | 2 | TolA protein, hypothetical protein |
| 528 | Y | 0 | Part of a phage tail fiber protein |
| 1,047 | Y | 1 | Hypothetical protein |
| 3,301 | N | 3 | VgrG protein, hypothetical protein, hypothetical protein |
| 1,052 | N | 3 | Phenylacetic acid degradation protein PaaY, phenylacetic acid degradation operon negative regulatory protein PaaX, transcriptional activator feaR |
| 936 | Y | 1 | Phage tail fiber protein |
| 17,933 | Y/N/Y | 20 | Phage tail length tape‐measure protein 1, phage tail length tape‐measure protein 1, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, Conserved hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, NAD+‐‐asparagine ADP‐ribosyltransferase, 62kDa structural protein, putative phage terminase, hypothetical protein, hypothetical protein, hypothetical protein, Chromosome (plasmid) partitioning protein ParB, Trk system potassium uptake protein TrkG |
| 1,982 | Y | 2 | Hypothetical protein, hypothetical protein |
| 3,502 | Y | 5 | Bacteriophage‐encoded homolog of DNA replication protein DnaC, hypothetical protein, hypothetical protein, hypothetical protein, Rac prophage repressor |
| 759 | Y | 1 | Superinfection exclusion protein B |
| 1,167 | Y | 3 | Kil protein, putative bacteriophage protein, phage protein |
| 729 | N | 1 | Exodeoxyribonuclease VIII |
| 3,389 | N | 6 | Exodeoxyribonuclease VIII, recombinational DNA repair protein RecT, hypothetical protein, phage protein, ydaQ protein, putative lambdoid prophage Rac integrase |
| 19,445 | Y | 15 | Integrase, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, unknown (no homologous in databases), DNA helicase, hypothetical protein, hypothetical protein, transposase, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, |
| 19,161 | Y | 14 | Hypothetical protein, hypothetical protein, site‐specific recombinase XerD, DNA repair protein RadC, hypothetical protein, hypothetical protein, transcriptional regulator/ (Cro/CI family), hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, type III restriction enzyme (res subunit), hypothetical protein |
| 9,113 | N | 5 | VgrG protein, hypothetical protein, Rhs family protein, hypothetical protein, Rhs family protein |
| 2,146 | Y | 2 | Hypothetical protein, hypothetical protein |
| 933 | Y | 1 | Hypothetical protein |
| 1,828 | N | 0 | |
| 7,085 | N | 5 | Protein ImpG/VasA, uncharacterized protein ImpH/VasB, type VI secretion lipoprotein/VasD, uncharacterized protein ImpJ/VasE, outer membrane protein ImpK/VasF (OmpA/MotB domain) |
| 3,584 | Y | 1 | IcmF‐related protein |
| 3,035 | Y | 2 | Secreted protein Hcp, hypothetical protein |
| 842 | N | 1 | Phosphoesterase |
| 1,367 | N | 1 | Ferric hydroxamate outer membrane receptor FhuA |
| 6,165 | N | 6 | Chaperone protein EcpD, outer membrane usher protein HtrE, fimbrial protein YadM, fimbrial protein YadL, fimbrial protein YadK, fimbrial protein YadC |
| 2,703 | N | 2 | Hypothetical protein, type I restriction–modification system/ restriction subunit R |
| 2,694 | N | 2 | Hypothetical protein, hypothetical protein |
| 5,659 | N | 1 | Adherence and invasion outer membrane protein (Inv, enhances Peyer's patches colonization) |
| 32,101 | N/Y | 11 | Transposase and inactivated derivative, hypothetical protein, hypothetical protein, putative DNA helicase, putative RNA helicase, type II restriction enzyme/methylase subunits, putative ATP‐dependent helicase, hypothetical protein, hypothetical protein, putative membrane protein, outer membrane protein and related peptidoglycan‐associated (lipo)proteins, hypothetical protein |
| 571 | Y | 1 | ORF25 |
| 9,482 | Y/N | 7 | Hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein |
| 563 | N | 1 | Hypothetical protein |
| 7,390 | N | 5 | IS/phage/ transposon‐related functions, hypothetical protein, hypothetical protein, core protein, core protein |
| 1,334 | N | 2 | Hypothetical protein, hypothetical protein |
| 831 | N | 1 | Phage tail fibers |
| 939 | Y | 1 | Hypothetical protein |
| 3,406 | Y | 3 | Hypothetical protein, hypothetical protein, DNA‐cytosine methyltransferase |
| 1,898 | N | 2 | Cox, C protein |
| 1,034 | N | 1 | Inner membrane protein |
| 5,750 | N | 6 | Putative cytoplasmic protein, hypothetical protein, putative membrane‐associated metal‐dependent hydrolase, hypothetical radical SAM family enzyme, hypothetical protein, putative hydrolase |
| 1,059 | N | 1 | Transposase |
| 636 | N | 1 | Hypothetical protein |
bp.
Y: yes, N: no; underline corresponds to the underlined gene description.
Italics indicates half the coding sequence is present; bold indicates internal deletion; underlined text corresponds to the underlined GI location.
Table A6.
Unique sequences in cystitis (UTI) isolate 6
| Lengtha | In GIb | Number of genes | Descriptionc |
|---|---|---|---|
| 17,937 | Y | 17 | Hypothetical protein, hypothetical protein, putative cytoplasmic protein USSDB7A, arylsulfatase regulator, radical SAM domain heme biosynthesis protein, radical SAM domain heme biosynthesis protein, His‐Xaa‐Ser repeat protein, hypothetical protein, hypothetical protein, chromosome (plasmid) partitioning protein ParB, recombinase, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, integrase |
| 3,686 | Y | 3 | Phage DNA transfer protein, Phage DNA transfer protein, regulatory protein |
| 2,006 | Y | 1 | Phage tail fibers |
| 8,809 | Y | 9 | Asparagine synthetase [glutamine‐hydrolyzing], asparagine synthetase [glutamine‐hydrolyzing], glycerol‐3‐phosphate cytidylyltransferase, membrane protein involved in the export of O‐ antigen, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, putative N‐acetylgalactosaminyl‐diphosphoundecaprenol glucuronosyltransferase |
| 12,940 | N/Y | 11 | Hypothetical protein, hypothetical protein, predicted ATP‐dependent endonuclease of the OLD family, ATP‐dependent DNA helicase UvrD/PcrA, hypothetical protein, hypothetical protein, hypothetical protein, exonuclease SbcC, hypothetical protein, hypothetical protein, hypothetical protein |
| 12,013 | N/Y | 14 | Hypothetical protein, hypothetical protein, probable monooxygenase, cysteinyl‐tRNA synthetase, Zinc uptake regulation protein ZUR, putative metal chaperone/ involved in Zn homeostasis/ GTPase of COG0523 family, C4‐type zinc finger protein(DksA/TraR family), GTP cyclohydrolase I, GTP cyclohydrolase I, carbonic anhydrase (gamma class), NADPH‐dependent preQ0 reductase, putative inner membrane protein, manganese ABC transporter/inner membrane permease protein SitD, manganese ABC transporter/inner membrane permease protein SitC |
| 16,136 | N/Y/N | 16 | Manganese ABC transporter/ATP‐binding protein SitB, manganese ABC transporter/periplasmic‐binding protein SitA, threonyl‐tRNA synthetase, peptidase, dihydroorotase, porphobilinogen synthase, hypothetical protein, FAD‐dependent oxidoreductase, hypothetical protein, putative secreted protein, Zinc ABC transporter/ periplasmic‐binding protein ZnuA, Zinc ABC transporter/ inner membrane permease protein ZnuB, Zinc ABC transporter/ inner membrane permease protein ZnuB, ABC transporter/ATP‐binding protein, putative phosphatase, putative phosphatase, |
| 3,203 | N | 2 | Flagellar hook‐associated protein FliD, flagellar biosynthesis protein FliC |
| 1,355 | Y | 2 | TolA protein, hypothetical protein |
| 1,368 | Y | 1 | Probable bacteriophage protein STY2043 |
| 897 | Y | 1 | Hypothetical protein |
| 1,056 | Y | 1 | Mobile element protein |
| 2,161 | Y | 1 | Bicyclomycin resistance protein |
| 2,877 | Y | 2 | Chromosome (plasmid) partitioning protein ParB, Trk system potassium uptake protein TrkG |
| 724 | Y | 0 | |
| 3,611 | Y | 5 | Phage antitermination protein, IS/ phage/transposon‐related functions, hypothetical bacteriophage protein, putative cytoplasmic protein, hypothetical protein |
| 1,010 | Y | 2 | Hypothetical protein, LygF |
| 3,314 | N | 4 | Bacteriophage‐encoded homolog of DNA replication protein DnaC, hypothetical protein, hypothetical protein, regulatory protein Cro of bacteriophage BP‐933W |
| 1,049 | N | 2 | Phage protein, IS/ phage/transposon‐related functions |
| 1,167 | N | 2 | Kil protein, phage protein, |
| 747 | N | 1 | Exodeoxyribonuclease VIII |
| 3,390 | N | 5 | Exodeoxyribonuclease VIII, recombinational DNA repair protein RecT, phage protein, ydaQ protein, putative lambdoid prophage Rac integrase |
| 940 | N | 4 | Transposase and inactivated derivative, hypothetical protein, transposase, transposase |
| 1,643 | N | 2 | Uncharacterized protein YcdU, uncharacterized protein YmdE |
| 3,808 | N | 3 | Hypothetical protein, hypothetical protein, phage integrase/ phage P4‐associated |
| 1,406 | N | 2 | Hypothetical protein, hypothetical protein/ unknown protein (putative secreted protein) |
| 505 | N | 0 | |
| 1,962 | Y | 3 | Hypothetical protein, hypothetical protein, unknown |
| 1,256 | N | 1 | Integrase |
| 3,163 | N | 3 | TRAP‐type transport system/small permease component/predicted N‐acetylneuraminate transporter, TRAP‐type C4‐dicarboxylate transport system/ large permease component, TRAP‐type C4‐dicarboxylate transport system/ periplasmic component |
| 12,866 | Y | 17 | Hypothetical protein, hypothetical protein, resolvase, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, diguanylate cyclase/phosphodiesterase (GGDEF & EAL domains) with PAS/PAC sensor(s), hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, integrase |
| 20,140 | Y | 27 | Hypothetical protein, hypothetical protein, DNA‐binding protein H‐NS, hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, GALNS arylsulfatase regulator (Fe‐S oxidoreductase), hypothetical protein, hypothetical protein, thiJ/pfpI family protein, hypothetical protein, NUDIX hydrolase, Transcriptional regulator (AraC family), integral membrane protein, hypothetical protein, transcriptional regulator (AlpA like), hypothetical protein, hypothetical protein, hypothetical protein, integrase/recombinase (XerC/CodV family), putative enzyme; integration, recombination (phage or prophage related), putative enzyme; integration, recombination (phage or prophage related), putative enzyme; integration, recombination (phage or prophage related), hypothetical protein, hypothetical protein, transposase and inactivated derivatives |
| 898 | Y | 1 | Hypothetical protein |
bp.
Y: yes, N: no; underline corresponds to the underlined gene description.
Underlined text corresponds to the underlined GI location.
Table A7.
Unique sequences in cystitis (UTI) isolate 11
| Lengtha | In GIb | Number of genes | Descriptionc |
|---|---|---|---|
| 1,154 | Y | 1 | Hypothetical protein |
| 1,329 | Y | 1 | Mobile element protein |
| 5,408 | Y | 6 | Hypothetical protein, hypothetical protein, DNA‐binding protein H‐NS homolog, hypothetical protein, dNTP triphosphohydrolase, hemolysin E |
| 6,492 | N/Y | 8 | Hypothetical protein, hypothetical protein, transcriptional regulator LacI family, PTS system IIA component, putative sugar phosphoesterase component IIB, putative integral membrane protein, transketolase N‐terminal section, transketolase C‐terminal section |
| 8,673 | Y/N | 10 | Cobalt–zinc–cadmium resistance protein CzcA/cation efflux system protein CusA, hypothetical protein, hypothetical protein, periplasmic lysozyme inhibitor of c‐type lysozyme, hypothetical protein, hypothetical protein, N‐acetylmannosamine‐6‐phosphate 2‐epimerase, PTS system maltose and glucose‐specific IIC component, RpiR family transcriptional regulator, putative exported protein |
| 1,336 | Y | 4 | Mobile element protein, probable transposase, transposase, hypothetical protein |
| 888 | Y | 1 | Hypothetical protein |
| 5,755 | Y | 3 | Mobile element protein, mobile element protein, AidA‐I adhesin‐like protein |
| 2,279 | Y | 2 | Beta‐1,4‐galactosyltransferase, O‐antigen ligase |
| 1,320 | N | 2 | Putative cytoplasmic protein, mobile element protein |
| 669 | N | 2 | Prevent host death protein/Phd antitoxin, death on curing protein/ Doc toxin |
| 1,312 | N | 2 | Mobile element protein, putative cytoplasmic protein |
| 1,324 | N | 1 | Mobile element protein |
| 1,329 | Y | 1 | Mobile element protein |
| 8,372 | Y | 8 | Hypothetical protein, glycerol kinase, hypothetical protein, ribokinase, hypothetical protein, ADP‐ribosylglycohydrolase, fatty acyl responsive regulator, possible GPH family transporter for arabinosides |
| 1,328 | N | 1 | Mobile element protein |
| 1,328 | N | 1 | Mobile element protein |
| 1,240 | N | 1 | Hypothetical protein |
| 1,369 | N | 2 | Hypothetical protein, hypothetical protein |
| 1,329 | N | 1 | Mobile element protein |
| 1,329 | N | 1 | Putative outer membrane protein |
| 817 | N | 1 | Uncharacterized protein YadU in stf fimbrial cluster |
| 1,443 | N | 1 | Molybdate metabolism regulator |
| 1,329 | Y | 1 | Mobile element protein |
| 970 | Y | 1 | Uncharacterized protein YehA precursor |
| 13,828 | Y | 9 | Putative glycosyltransferase, mobile element protein, mobile element protein, sialic acid biosynthesis protein NeuD/O‐acetyltransferase, N‐acetylneuraminate synthase, N‐acetylneuraminate cytidylyltransferase, UDP‐N‐acetylglucosamine 2‐epimerase, hypothetical protein, N‐acetylneuraminic acid synthase‐like protein |
| 1,683 | Y | 1 | Glycosyl transferase group 1 |
| 867 | N | 1 | Putative transcriptional regulator |
| 666 | N | 1 | Phage or prophage related |
| 759 | N | 1 | Bacteriophage‐encoded homolog of DNA replication protein DnaC |
| 652 | N | 0 | |
| 1,031 | Y | 2 | Hypothetical protein, hypothetical protein |
| 1,248 | Y | 1 | Phage tail fiber protein |
| 879 | Y | 1 | Mobile element protein |
| 1,564 | N | 1 | Flagellar hook‐associated protein FliD |
| 825 | N | 0 | |
| 1,812 | N | 1 | Hypothetical protein |
| 918 | N | 1 | Serine acetyltransferase |
| 1,196 | N | 1 | Hypothetical protein |
| 1,030 | N | 2 | Hypothetical protein, hypothetical protein |
| 3,151 | N | 6 | Putative DNA‐binding protein, hypothetical protein, Transcriptional regulator/XRE family, putative membrane protein, unknown protein encoded by prophage CP‐933T, putative integrase |
| 1,329 | N | 2 | Hypothetical protein, Flagellar biosynthesis protein FlhB |
| 1,329 | N | 1 | Mobile element protein |
| 1,329 | N | 1 | Mobile element protein |
| 1,321 | N | 2 | Excinuclease cho (excinuclease ABC alternative C subunit), mobile element protein |
| 546 | Y | 2 | Hypothetical protein, IS/phage/ transposon‐related functions |
| 1,680 | Y | 1 | Hypothetical protein |
| 1,328 | Y | 1 | Mobile element protein |
| 1,324 | N | 1 | Mobile element protein |
| 3,450 | N | 2 | Putative hydrolase, hypothetical protein |
| 9,840 | N | 8 | No significant similarities, hypothetical protein, hypothetical protein, Rhs family protein, VgrG protein, hypothetical protein, hypothetical protein, hypothetical protein |
| 1,329 | Y | 1 | Mobile element protein |
| 1,329 | N | 2 | Oxidoreductase (putative), mobile element protein |
| 1,121 | N | 2 | Hypothetical protein, hypothetical protein |
| 1,328 | Y | 1 | Mobile element protein |
| 1,329 | N | 1 | Mobile element protein |
| 1,311 | N | 2 | Putative cytoplasmic protein, mobile element protein |
| 984 | N | 0 | |
| 1,134 | N | 1 | Prophage Clp protease‐like protein |
| 1,291 | N | 2 | Hypothetical protein, hypothetical protein |
| 2,771 | N | 3 | Zinc binding domain/DNA primase/phage P4‐associated/replicative helicase RepA/ Pha, hypothetical protein, hypothetical protein |
| 1,170 | N | 2 | Putative ATPase component of ABC transporter with duplicated ATPase domain, L,D‐transpeptidase YbiS |
| 1,248 | N | 1 | Phage tail fiber protein |
| 2,030 | Y | 2 | Hypothetical protein, hypothetical protein |
| 2,095 | Y | 3 | Predicted transcriptional regulator, hypothetical protein, phage major capsid protein |
| 4,593 | N | 9 | Hypothetical protein, hypothetical protein, hypothetical protein, hypothetical protein, IS/ phage/ transposon‐related functions, hypothetical protein, hypothetical protein, phage DNA‐binding protein, site‐specific recombinase/phage integrase family |
| 705 | Y | 2 | Regulatory protein cro, phage repressor |
| 2,319 | Y | 3 | Hypothetical protein, phage antitermination protein N, hypothetical protein |
| 1,444 | Y | 2 | Eae protein, hypothetical protein |
| 1,328 | N | 1 | Mobile element protein |
| 1,653 | Y | 1 | Hypothetical protein |
| 1,394 | Y | 3 | No significant similarities, hypothetical protein, Rhs family protein |
| 6,494 | Y | 3 | Transposase, transposase, hypothetical protein |
| 2,136 | N | 3 | Hypothetical protein, ornithine decarboxylase, mobile element protein |
| 562 | N | 2 | Hypothetical protein, ABC‐type sugar transport system/periplasmic component |
| 1,329 | N | 1 | Mobile element protein |
| 614 | Y | 1 | Hypothetical protein |
| 1,328 | Y | 1 | Mobile element protein |
| 2,660 | Y | 2 | Mobile element protein, mobile element protein |
| 850 | N | 1 | Phosphoesterase |
| 1,367 | N | 1 | Ferric hydroxamate outer membrane receptor FhuA |
| 1,643 | Y | 1 | Hypothetical protein |
| 1,950 | Y | 1 | Hypothetical protein |
| 683 | N | 1 | Phage‐related protein |
| 1,505 | N | 1 | Hypothetical protein |
| 5,897 | N | 5 | Transcriptional regulator/RpiR family, pantothenate:Na + symporter, bona fide RidA/YjgF/TdcF/RutC subgroup, D‐aminoacylase, D‐serine deaminase |
| 8,320 | N | 5 | Hypothetical protein, hypothetical protein, hypothetical protein, type I restriction‐modification system/specificity subunit S, type I restriction–modification system/specificity subunit R |
| 571 | Y | 1 | ORF25 |
| 1,329 | N | 1 | Mobile element protein |
| 1,228 | N | 1 | Hypothetical protein |
| 1,402 | N | 1 | Hypothetical protein |
| 16,536 | Y/N | 11 | Conserved protein of unknown function, hypothetical protein, hypothetical protein, hypothetical protein, DNA sulfur modification protein DndE, DNA sulfur modification protein DndD, 3'‐phosphoadenosine 5'‐phosphosulfate sulfurtransferase DndC, DNA sulfur modification protein DndB, hypothetical protein, hypothetical protein, hypothetical protein |
| 9,146 | Y | 12 | Mobile element protein, LysR family transcriptional regulator YeiE, hypothetical protein, sodium/glutamate symport protein, hypothetical protein, hypothetical protein, hypothetical protein, transcriptional regulator/ ArsR family, transcriptional regulator/ TetR family, tetracycline efflux protein TetA, hypothetical protein, right origin‐binding protein, mobile element protein |
| 1,852 | Y | 2 | Transposase, hypothetical protein |
| 2,449 | Y | 3 | Hypothetical protein, hypothetical protein, hypothetical protein |
| 5,212 | Y | 5 | Hypothetical protein, hypothetical protein, hypothetical protein, conserved hypothetical protein, hypothetical protein |
| 1,154 | Y | 1 | Hypothetical protein |
| 3,592 | N | 3 | Mobile element protein, Hypothetical protein, Hypothetical protein |
| 1,329 | N | 1 | Mobile element protein |
| 1,153 | N | 1 | Possible exported protein |
bp.
Y: yes, N: no; underline corresponds to the underlined gene description.
Underlined text corresponds to the underlined GI location.
Desloges I, Taylor JA, Leclerc J‐M, et al. Identification and characterization of OmpT‐like proteases in uropathogenic Escherichia coli clinical isolates. MicrobiologyOpen. 2019;8:e915 10.1002/mbo3.915
Co‐author Hervé Le Moual is deceased.
DATA AVAILABILITY STATEMENT
Data related to plasmid and genome sequencing have been deposited to NCBI under BioProject accession numbers PRJNA551561 (cystitis 1), PRJNA551565 (cystitis 6), and PRJNA551566 (cystitis 11); BioSample accession numbers SAMN12158196 (cystitis 1), SAMN12158201 (cystitis 6), and SAMN12158203 (cystitis 11); GenBank accession numbers CP041299 (cystitis 1 plasmid), CP041300 (cystitis 1 chromosome), CP041301 (cystitis 6 plasmid), CP041302 (cystitis 6 chromosome), CP041303 (cystitis 11 plasmid), and CP041304 (cystitis 11 chromosome). Other datasets are available from the corresponding author upon reasonable request.
REFERENCES
- Alikhan, N. F. , Petty, N. K. , Ben Zakour, N. L. , & Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics, 12, 402 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam, M. , Toufeer, M. , Narvaez Bravo, C. , Lai, V. , Rempel, H. , Manges, A. , & Diarra, M. S. (2014). Characterization of extraintestinal pathogenic Escherichia coli isolated from retail poultry meats from Alberta, Canada. International Journal of Food Microbiology, 177, 49–56. 10.1016/j.ijfoodmicro.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Bertelli, C. , Laird, M. R. , Williams, K. P. , Lau, B. Y. , Hoad, G. , Winsor, G. L. , & Brinkman, F. S. L. (2017). IslandViewer 4: Expanded prediction of genomic islands for larger‐scale datasets. Nucleic Acids Research, 45(W1), W30–W35. 10.1093/nar/gkx343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon, J. R. , Burk, D. L. , Leclerc, J.‐M. , Thomassin, J.‐L. , Portt, A. , Berghuis, A. M. , … Le Moual, H. (2015). Inhibition of outer membrane proteases of the omptin family by aprotinin. Infection and Immunity, 83(6), 2300–2311. 10.1128/IAI.00136-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon, J. R. , Thomassin, J. L. , Desloges, I. , Gruenheid, S. , & Le Moual, H. (2013). Role of uropathogenic Escherichia coli OmpT in the resistance against human cathelicidin LL‐37. FEMS Microbiology Letters, 345(1), 64–71. 10.1111/1574-6968.12185 [DOI] [PubMed] [Google Scholar]
- Brannon, J. R. , Thomassin, J. L. , Gruenheid, S. , & Le Moual, H. (2015). Antimicrobial peptide conformation as a structural determinant of omptin protease specificity. Journal of Bacteriology, 197(22), 3583–3591. 10.1128/JB.00469-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli, A. , Zankari, E. , Garcia‐Fernandez, A. , Voldby Larsen, M. , Lund, O. , Villa, … H. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial Agents and Chemotherapy, 58(7), 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromek, M. , & Brauner, A. (2008). Antimicrobial mechanisms of the urinary tract. Journal of Molecular Medicine, 86(1), 37–47. 10.1007/s00109-007-0256-4 [DOI] [PubMed] [Google Scholar]
- Chromek, M. , Slamová, Z. , Bergman, P. , Kovács, L. , Podracká, L. , Ehrén, I. , … Brauner, A. (2006). The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nature Medicine, 12(6), 636–641. 10.1038/nm1407 [DOI] [PubMed] [Google Scholar]
- Clermont, O. , Bonacorsi, S. , & Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Applied and Environment Microbiology, 66(10), 4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, N. , Cox, R. C. , Kramer, R. A. , & Egmond, M. R. (2001). Substrate specificity of the integral membrane protease OmpT determined by spatially addressed peptide libraries. Biochemistry, 40(6), 1694–1701. [DOI] [PubMed] [Google Scholar]
- E. coli UTI Bacteremia initiative (2019).
- Flores‐Mireles, A. L. , Walker, J. N. , Caparon, M. , & Hultgren, S. J. (2015). Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nature Reviews Microbiology, 13(5), 269–284. 10.1038/nrmicro3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, B. (2014). Urinary tract infection syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infectious Disease Clinics of North America, 28(1), 1–13. 10.1016/j.idc.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Foxman, B. , Zhang, L. , Palin, K. , Tallman, P. , & Marrs, C. F. (1995). Bacterial virulence characteristics of Escherichia coli isolates from first‐time urinary tract infection. Journal of Infectious Diseases, 171(6), 1514–1521. 10.1093/infdis/171.6.1514 [DOI] [PubMed] [Google Scholar]
- Galata, V. , Backes, C. , Laczny, C. C. , Hemmrich‐Stanisak, G. , Li, H. , Smoot, L. , … Keller, A. (2018). Comparing genome versus proteome‐based identification of clinical bacterial isolates. Briefings in Bioinformatics, 19(3), 495–505. 10.1093/bib/bbw122 [DOI] [PubMed] [Google Scholar]
- Galvan, E. M. , Lasaro, M. A. , & Schifferli, D. M. (2008). Capsular antigen fraction 1 and Pla modulate the susceptibility of Yersinia pestis to pulmonary antimicrobial peptides such as cathelicidin. Infection and Immunity, 76(4), 1456–1464. 10.1128/IAI.01197-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheid, S. , & Le Moual, H. (2012). Resistance to antimicrobial peptides in Gram‐negative bacteria. FEMS Microbiology Letters, 330(2), 81–89. 10.1111/j.1574-6968.2012.02528.x [DOI] [PubMed] [Google Scholar]
- Haiko, J. , Suomalainen, M. , Ojala, T. , Lahteenmaki, K. , & Korhonen, T. K. (2009). Invited review: Breaking barriers–attack on innate immune defences by omptin surface proteases of enterobacterial pathogens. Innate Immun, 15(2), 67–80. 10.1177/1753425909102559 [DOI] [PubMed] [Google Scholar]
- He, X. L. , Wang, Q. , Peng, L. , Qu, Y.‐R. , Puthiyakunnon, S. , Liu, X.‐L. , … Huang, S.‐H. (2015). Role of uropathogenic Escherichia coli outer membrane protein T in pathogenesis of urinary tract infection. Pathogens and Disease, 73(3), 10.1093/femspd/ftv006 [DOI] [PubMed] [Google Scholar]
- Hooton, T. M. (2012). Clinical practice. Uncomplicated urinary tract infection. New England Journal of Medicine, 366(11), 1028–1037. 10.1056/NEJMcp1104429 [DOI] [PubMed] [Google Scholar]
- Hooton, T. M. , & Stamm, W. E. (1997). Diagnosis and treatment of uncomplicated urinary tract infection. Infectious Disease Clinics of North America, 11(3), 551–581. 10.1016/S0891-5520(05)70373-1 [DOI] [PubMed] [Google Scholar]
- Hritonenko, V. , & Stathopoulos, C. (2007). Omptin proteins: An expanding family of outer membrane proteases in Gram‐negative Enterobacteriaceae. Molecular Membrane Biology, 24(5–6), 395–406. 10.1080/09687680701443822 [DOI] [PubMed] [Google Scholar]
- Huang, Y.‐C. , Lin, Y.‐M. , Chang, T.‐W. , Wu, S.‐J. , Lee, Y.‐S. , Chang, M.‐T. , … Liao, Y.‐D. (2007). The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. Journal of Biological Chemistry, 282(7), 4626–4633. 10.1074/jbc.M607321200 [DOI] [PubMed] [Google Scholar]
- Hwang, B. Y. , Varadarajan, N. , Li, H. , Rodriguez, S. , Iverson, B. L. , & Georgiou, G. (2007). Substrate specificity of the Escherichia coli outer membrane protease OmpP. Journal of Bacteriology, 189(2), 522–530. 10.1128/JB.01493-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenssen, H. , Hamill, P. , & Hancock, R. E. (2006). Peptide antimicrobial agents. Clinical Microbiology Reviews, 19(3), 491–511. 10.1128/CMR.00056-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, B. , Raphenya, A. R. , Alcock, B. , Waglechner, N. , Guo, P. , Tsang, K. K. , … McArthur, A. G. (2017). CARD 2017: Expansion and model‐centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Research, 45(D1), D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen, K. G. , Tetzschner, A. M. , Iguchi, A. , Aarestrup, F. M. , & Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole‐genome sequencing data. Journal of Clinical Microbiology, 53(8), 2410–2426. 10.1128/JCM.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J. R. (1991). Virulence factors in Escherichia coli urinary tract infection. Clinical Microbiology Reviews, 4(1), 80–128. 10.1128/CMR.4.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J. R. , Delavari, P. , Kuskowski, M. , & Stell, A. L. (2001). Phylogenetic distribution of extraintestinal virulence‐associated traits in Escherichia coli . Journal of Infectious Diseases, 183(1), 78–88. 10.1086/317656 [DOI] [PubMed] [Google Scholar]
- Johnson, J. R. , & Stell, A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. Journal of Infectious Diseases, 181(1), 261–272. 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- Kai‐Larsen, Y. , Lüthje, P. , Chromek, M. , Peters, V. , Wang, X. , Holm, Å. , … Brauner, A. (2010). Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL‐37. PLoS Path, 6(7), e1001010 10.1371/journal.ppat.1001010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper, J. B. , Nataro, J. P. , & Mobley, H. L. (2004). Pathogenic Escherichia coli . Nature Reviews Microbiology, 2(2), 123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Kaufmann, A. , Stierhof, Y. D. , & Henning, U. (1994). New outer membrane‐associated protease of Escherichia coli K‐12. Journal of Bacteriology, 176(2), 359–367. 10.1128/jb.176.2.359-367.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjolvmark, C. , Akesson, P. , & Pahlman, L. I. (2017). Urine concentrations of human beta‐defensins and ribonuclease 7 in urinary tract infection and asymptomatic bacteriuria. Diagnostic Microbiology and Infectious Disease, 89(1), 58–60. 10.1016/j.diagmicrobio.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Kramer, R. A. , Vandeputte‐Rutten, L. , de Roon, G. J. , Gros, P. , Dekker, N. , & Egmond, M. R. (2001). Identification of essential acidic residues of outer membrane protease OmpT supports a novel active site. FEBS Letters, 505(3), 426–430. 10.1016/S0014-5793(01)02863-0 [DOI] [PubMed] [Google Scholar]
- Kramer, R. A. , Zandwijken, D. , Egmond, M. R. , & Dekker, N. (2000). In vitro folding, purification and characterization of Escherichia coli outer membrane protease ompT . European Journal of Biochemistry, 267(3), 885–893. 10.1046/j.1432-1327.2000.01073.x [DOI] [PubMed] [Google Scholar]
- Kukkonen, M. , Lahteenmaki, K. , Suomalainen, M. , Kalkkinen, N. , Emody, L. , Lang, H. , & Korhonen, T. K. (2001). Protein regions important for plasminogen activation and inactivation of alpha2‐antiplasmin in the surface protease Pla of Yersinia pestis . Molecular Microbiology, 40(5), 1097–1111. 10.1046/j.1365-2958.2001.02451.x [DOI] [PubMed] [Google Scholar]
- Larsen, M. V. , Cosentino, S. , Rasmussen, S. , Friis, C. , Hasman, H. , Marvig, R. L. , … Lund, O. (2012). Multilocus sequence typing of total‐genome‐sequenced bacteria. Journal of Clinical Microbiology, 50(4), 1355–1361. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem, W. W. , Price, P. A. , Miller, V. L. , & Goldman, W. E. (2007). A plasminogen‐activating protease specifically controls the development of primary pneumonic plague. Science, 315(5811), 509–513. 10.1126/science.1137195 [DOI] [PubMed] [Google Scholar]
- Le Sage, V. , Zhu, L. , Lepage, C. , Portt, A. , Viau, C. , Daigle, F. , … Le Moual, H. (2009). An outer membrane protease of the omptin family prevents activation of the Citrobacter rodentium PhoPQ two‐component system by antimicrobial peptides. Molecular Microbiology, 74(1), 98–111. 10.1111/j.1365-2958.2009.06854.x [DOI] [PubMed] [Google Scholar]
- Lehmann, J. , Retz, M. , Harder, J. , Krams, M. , Kellner, U. , Hartmann, J. , … Stöckle, M. (2002). Expression of human beta‐defensins 1 and 2 in kidneys with chronic bacterial infection. BMC Infectious Diseases, 2, 20 10.1186/1471-2334-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Tai, C. , Deng, Z. , Zhong, W. , He, Y. , & Ou, H. Y. (2018). VRprofile: Gene‐cluster‐detection‐based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Briefings in Bioinformatics, 19(4), 566–574. 10.1093/bib/bbw141 [DOI] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25(4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lloyd, A. L. , Rasko, D. A. , & Mobley, H. L. (2007). Defining genomic islands and uropathogen‐specific genes in uropathogenic Escherichia coli . Journal of Bacteriology, 189(9), 3532–3546. 10.1128/JB.01744-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges, A. R. , Dietrich, P. S. , & Riley, L. W. (2004). Multidrug‐resistant Escherichia coli clonal groups causing community‐acquired pyelonephritis. Clinical Infectious Diseases, 38(3), 329–334. 10.1086/380640 [DOI] [PubMed] [Google Scholar]
- Manges, A. R. , Johnson, J. R. , Foxman, B. , O'Bryan, T. T. , Fullerton, K. E. , & Riley, L. W. (2001). Widespread distribution of urinary tract infections caused by a multidrug‐resistant Escherichia coli clonal group. New England Journal of Medicine, 345(14), 1007–1013. 10.1056/NEJMoa011265 [DOI] [PubMed] [Google Scholar]
- Manges, A. R. , Johnson, J. R. , & Riley, L. W. (2004). Intestinal population dynamics of UTI‐causing Escherichia coli within heterosexual couples. Current Issues in Intestinal Microbiology, 5(2), 49–57. [PubMed] [Google Scholar]
- Manges, A. R. , Perdreau‐Remington, F. , Solberg, O. , & Riley, L. W. (2006). Multidrug‐resistant Escherichia coli clonal groups causing community‐acquired bloodstream infections. Journal of Infection, 53(1), 25–29. 10.1016/j.jinf.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Manges, A. R. , Tabor, H. , Tellis, P. , Vincent, C. , & Tellier, P. P. (2008). Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerging Infectious Diseases, 14(10), 1575–1583. 10.3201/eid1410.080102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall, J. , Zhang, L. , Foxman, B. , Warren, D. K. , Henderson, J. P. , & Program, C. D. C. P. E. (2012). Both host and pathogen factors predispose to Escherichia coli urinary‐source bacteremia in hospitalized patients. Clinical Infectious Diseases, 54(12), 1692–1698. 10.1093/cid/cis252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard, C. , Bekal, S. , Sanschagrin, F. , Levesque, R. C. , Brousseau, R. , Masson, L. , … Harel, J. (2004). Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. Journal of Clinical Microbiology, 42(12), 5444–5452. 10.1128/JCM.42.12.5444-5452.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter, J. D. , Stephens, D. , Shoemaker, K. , Rosenberg, S. , Kirsch, J. F. , & Georgiou, G. (2004). Substrate specificity of the Escherichia coli outer membrane protease OmpT. Journal of Bacteriology, 186(17), 5919–5925. 10.1128/JB.186.17.5919-5925.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee, J. B. , Small, C. L. , Reid‐Yu, S. A. , Brannon, J. R. , Le Moual, H. , & Coombes, B. K. (2014). Host defense peptide resistance contributes to colonization and maximal intestinal pathology by Crohn's disease‐associated adherent‐invasive Escherichia coli . Infection and Immunity, 82(8), 3383–3393. 10.1128/IAI.01888-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley, H. L. , Green, D. M. , Trifillis, A. L. , Johnson, D. E. , Chippendale, G. R. , Lockatell, C. V. , … Warren, J. W. (1990). Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: Role of hemolysin in some strains. Infection and Immunity, 58(5), 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, E. , Andreu, A. , Perez, T. , Sabate, M. , Johnson, J. R. , & Prats, G. (2006). Relationship between Escherichia coli strains causing urinary tract infection in women and the dominant faecal flora of the same hosts. Epidemiology and Infection, 134(5), 1015–1023. 10.1017/S0950268806005917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi, A. , Hasanpour, M. , Askary, A. , Aziemzadeh, M. , & Hashemi, N. (2018). Distribution of pathogenicity island markers and virulence factors in new phylogenetic groups of uropathogenic Escherichia coli isolates. Folia Microbiol (Praha), 63(3), 335–343. 10.1007/s12223-017-0570-3 [DOI] [PubMed] [Google Scholar]
- Nielsen, K. L. , Dynesen, P. , Larsen, P. , Jakobsen, L. , Andersen, P. S. , & Frimodt‐Moller, N. (2014). Role of urinary cathelicidin LL‐37 and human beta‐defensin 1 in uncomplicated Escherichia coli urinary tract infections. Infection and Immunity, 82(4), 1572–1578. 10.1128/IAI.01393-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielubowicz, G. R. , & Mobley, H. L. (2010). Host‐pathogen interactions in urinary tract infection. Nat Rev Urol, 7(8), 430–441. 10.1038/nrurol.2010.101 [DOI] [PubMed] [Google Scholar]
- Norinder, B. S. , Koves, B. , Yadav, M. , Brauner, A. , & Svanborg, C. (2012). Do Escherichia coli strains causing acute cystitis have a distinct virulence repertoire? Microbial Pathogenesis, 52(1), 10–16. 10.1016/j.micpath.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Poey, M. E. , Albini, M. , Saona, G. , & Lavina, M. (2012). Virulence profiles in uropathogenic Escherichia coli isolated from pregnant women and children with urinary tract abnormalities. Microbial Pathogenesis, 52(5), 292–301. 10.1016/j.micpath.2012.02.006 [DOI] [PubMed] [Google Scholar]
- Schreiber, H. L. T. , Conover, M. S. , Chou, W. C. , Hibbing, M. E. , Manson, A. L. , Dodson, K. W. , … Hultgren, S. J. (2017). Bacterial virulence phenotypes of Escherichia coli and host susceptibility determine risk for urinary tract infections. Science Translational Medicine, 9(382), 10.1126/scitranslmed.aaf1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, S. , Jobin, K. , & Kurts, C. (2017). Urinary tract infection: Recent insight into the evolutionary arms race between uropathogenic Escherichia coli and our immune system. Nephrology, Dialysis, Transplantation, 32(12), 1977–1983. 10.1093/ndt/gfx022 [DOI] [PubMed] [Google Scholar]
- Sodeinde, O. A. , Subrahmanyam, Y. V. , Stark, K. , Quan, T. , Bao, Y. , & Goguen, J. D. (1992). A surface protease and the invasive character of plague. Science, 258(5084), 1004–1007. [DOI] [PubMed] [Google Scholar]
- Spencer, J. D. , Hains, D. S. , Porter, E. , Bevins, C. L. , DiRosario, J. , Becknell, B. , … Schwaderer, A. L. (2012). Human alpha defensin 5 expression in the human kidney and urinary tract. PLoS ONE, 7(2), e31712 10.1371/journal.pone.0031712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, J. D. , Schwaderer, A. L. , Becknell, B. , Watson, J. , & Hains, D. S. (2014). The innate immune response during urinary tract infection and pyelonephritis. Pediatric Nephrology (Berlin, Germany), 29(7), 1139–1149. 10.1007/s00467-013-2513-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, J. D. , Schwaderer, A. L. , DiRosario, J. D. , McHugh, K. M. , McGillivary, G. , Justice, S. S. , … Hains, D. S. (2011). Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney International, 80(2), 174–180. 10.1038/ki.2011.109 [DOI] [PubMed] [Google Scholar]
- Spencer, J. D. , Schwaderer, A. L. , Wang, H. , Bartz, J. , Kline, J. , Eichler, T. , … Hains, D. S. (2013). Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney International, 83(4), 615–625. 10.1038/ki.2012.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe, S. , Schmid, R. , Stephens, D. L. , Georgiou, G. , & Bakker, E. P. (1998). Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli . Journal of Bacteriology, 180(15), 4002–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J. A. , Ouimet, M. C. , Wargachuk, R. , & Marczynski, G. T. (2011). The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Molecular Microbiology, 82(2), 312–326. 10.1111/j.1365-2958.2011.07785.x [DOI] [PubMed] [Google Scholar]
- Terlizzi, M. E. , Gribaudo, G. , & Maffei, M. E. (2017). UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non‐antibiotic antimicrobial strategies. Frontiers in Microbiology, 8, 1566 10.3389/fmicb.2017.01566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, N. A. , Deng, W. , Puente, J. L. , Frey, E. A. , Yip, C. K. , Strynadka, N. C. , & Finlay, B. B. (2005). CesT is a multi‐effector chaperone and recruitment factor required for the efficient type III secretion of both LEE‐ and non‐LEE‐encoded effectors of enteropathogenic Escherichia coli . Molecular Microbiology, 57(6), 1762–1779. 10.1111/j.1365-2958.2005.04802.x [DOI] [PubMed] [Google Scholar]
- Thomassin, J. L. , Brannon, J. R. , Gibbs, B. F. , Gruenheid, S. , & Le Moual, H. (2012). OmpT outer membrane proteases of enterohemorrhagic and enteropathogenic Escherichia coli contribute differently to the degradation of human LL‐37. Infection and Immunity, 80(2), 483–492. 10.1128/IAI.05674-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassin, J. L. , Brannon, J. R. , Kaiser, J. , Gruenheid, S. , & Le Moual, H. (2012). Enterohemorrhagic and enteropathogenic Escherichia coli evolved different strategies to resist antimicrobial peptides. Gut Microbes, 3(6), 556–561. 10.4161/gmic.21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valore, E. V. , Park, C. H. , Quayle, A. J. , Wiles, K. R. , McCray, P. B. Jr , & Ganz, T. (1998). Human beta‐defensin‐1: An antimicrobial peptide of urogenital tissues. J Clin Invest, 101(8), 1633–1642. 10.1172/JCI1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte‐Rutten, L. , Kramer, R. A. , Kroon, J. , Dekker, N. , Egmond, M. R. , & Gros, P. (2001). Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO Journal, 20(18), 5033–5039. 10.1093/emboj/20.18.5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. F. , & Kushner, S. R. (1991). Construction of versatile low‐copy‐number vectors for cloning, sequencing and gene expression in Escherichia coli . Gene, 100, 195–199. 10.1016/0378-1119(91)90366-J [DOI] [PubMed] [Google Scholar]
- Wattam, A. R. , Davis, J. J. , Assaf, R. , Boisvert, S. , Brettin, T. , Bun, C. , … Stevens, R. L. (2017). Improvements to PATRIC, the all‐bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Research, 45(D1), D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart, T. , Haidinger, M. , Horl, W. H. , & Saemann, M. D. (2008). Current concepts of molecular defence mechanisms operative during urinary tract infection. European Journal of Clinical Investigation, 38(Suppl 2), 29–38. 10.1111/j.1365-2362.2008.02006.x [DOI] [PubMed] [Google Scholar]
- Zasloff, M. (2007). Antimicrobial peptides, innate immunity, and the normally sterile urinary tract. Journal of the American Society of Nephrology, 18(11), 2810–2816. 10.1681/ASN.2007050611 [DOI] [PubMed] [Google Scholar]
- Zhuge, X. , Sun, Y. U. , Xue, F. , Tang, F. , Ren, J. , Li, D. , … Dai, J. (2018). A novel PhoP/PhoQ regulation pathway modulates the survival of extraintestinal pathogenic Escherichia coli in macrophages. Frontiers in Immunology, 9, 788 10.3389/fimmu.2018.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimbler, D. L. , Schroeder, J. A. , Eddy, J. L. , & Lathem, W. W. (2015). Early emergence of Yersinia pestis as a severe respiratory pathogen. Nature Communications, 6, 7487 10.1038/ncomms8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data related to plasmid and genome sequencing have been deposited to NCBI under BioProject accession numbers PRJNA551561 (cystitis 1), PRJNA551565 (cystitis 6), and PRJNA551566 (cystitis 11); BioSample accession numbers SAMN12158196 (cystitis 1), SAMN12158201 (cystitis 6), and SAMN12158203 (cystitis 11); GenBank accession numbers CP041299 (cystitis 1 plasmid), CP041300 (cystitis 1 chromosome), CP041301 (cystitis 6 plasmid), CP041302 (cystitis 6 chromosome), CP041303 (cystitis 11 plasmid), and CP041304 (cystitis 11 chromosome). Other datasets are available from the corresponding author upon reasonable request.


