Abstract
Since 2012, the interest for TMEM165 increased due to its implication in a rare genetic human disease named TMEM165-CDG (Congenital Disorder(s) of Glycosylation). TMEM165 is a Golgi localized protein, highly conserved through evolution and belonging to the uncharacterized protein family 0016 (UPF0016). Although the precise function of TMEM165 in glycosylation is still controversial, our results highly suggest that TMEM165 would act as a Golgi Ca2+/Mn2+ transporter regulating both Ca2+ and Mn2+ Golgi homeostasis, the latter is required as a major cofactor of many Golgi glycosylation enzymes. Strikingly, we recently demonstrated that besides its role in regulating Golgi Mn2+ homeostasis and consequently Golgi glycosylation, TMEM165 is sensitive to high manganese exposure. Members of the UPF0016 family contain two particularly highly conserved consensus motifs E-φ-G-D-[KR]-[TS] predicted to be involved in the ion transport function of UPF0016 members. We investigate the contribution of these two specific motifs in the function of TMEM165 in Golgi glycosylation and in its Mn2+ sensitivity.
Our results show the crucial importance of these two conserved motifs and underline the contribution of some specific amino acids in both Golgi glycosylation and Mn2+ sensitivity.
Keywords: TMEM165, CDG, UPF0016, Golgi glycosylation, Mn2+
1. Introduction
Congenital Disorders of Glycosylation (CDG) are a rapidly expanding family of genetic diseases. The first patient cases were reported 38 years ago; today more than hundred different CDG have been reported [1]. The frequency of most CDG is unknown but they are probably underestimated. The genetic transmission is mostly autosomal recessive [1]. Congenital disorders of protein glycosylation are classified in two groups. CDG-I are assembly defects in the cytosol and the endoplasmic reticulum (ER), while CDG-II are defects in glycan remodeling in the Golgi [2]. They are multisystem disorders with a broad spectrum of severity and mostly comprising neurological involvement.
In 2012, a new CDG called TMEM165-CDG or CDG-IIk (OMIM #614727) has been described [3]. To date, a dozen of TMEM165-CDG patients have been worldwide diagnosed with a common semiology. The most severe phenotypes present a growth retardation resistant to human growth hormone, associated with a psychomotor disability, microcephaly, facial hypoplasia, hypotonia, seizures and hepatosplenomegaly with increased serum transaminases [4]. Some patients also harbor cardiac defects [5] but the pathognomonic signs remain bone and cartilage dysplasia with early and severe osteoporosis. All TMEM165-CDG present a strong defect in the Golgi glycosylation characterized by hypogalactosylation of total serum N-glycoproteins [3].
This CDG is due to a deficiency in TMEM165 protein, also named TPARL [3], a 324 amino-acids transmembrane protein member of the UPF family 0016 (Uncharacterized Protein Family 0016; Pfam PF01169). This protein is mainly localized in Golgi membranes [3,6], predominantly in the trans-Golgi subcompartment. Similarly to other UPF0016 family members, TMEM165 is highly conserved throughout evolution (919 different species in prokaryotes and 405 species in eukaryotes) [7].
The cellular and molecular functions of UPF0016 family members remain controversial. Gdt1p (Grc1 dependent translation factor 1), the yeast ortholog of TMEM165 in Saccharomyces cerevisiae was initially postulated to be a Ca2+/H+ exchanger [8]. Recent results however question the nature of the exchanged ions. Unexpectedly, PAM71 (Photosynthesis Affected Mutant 71), the Arabidopsis thaliana plant ortholog ofTMEM165 has been shown to function as a Ca2+/Mn2+ cation antiport transporter localized in the thylakoid membranes system and crucial for the regulation of chloroplastic Mn2+ homeostasis [9]. In addition, we recently demonstrated that the Golgi glycosylation defect due to TMEM165 deficiency also results from a defect in Golgi Mn2+ homeostasis [10]. Very importantly, a slight Mn2+ supplementation is sufficient to suppress the observed Golgi glycosylation defect in both deficient yeasts and human cells [11]. Furthermore, our recent studies suggested the function of Gdt1p as a Ca2+/Mn2+ cation antiport transporter [12]. In agreement with these results, Thines and collaborators have recently demonstrated that the yeast protein Gdt1p transports Mn2+ ions and thereby regulates manganese homeostasis in the Golgi [13].
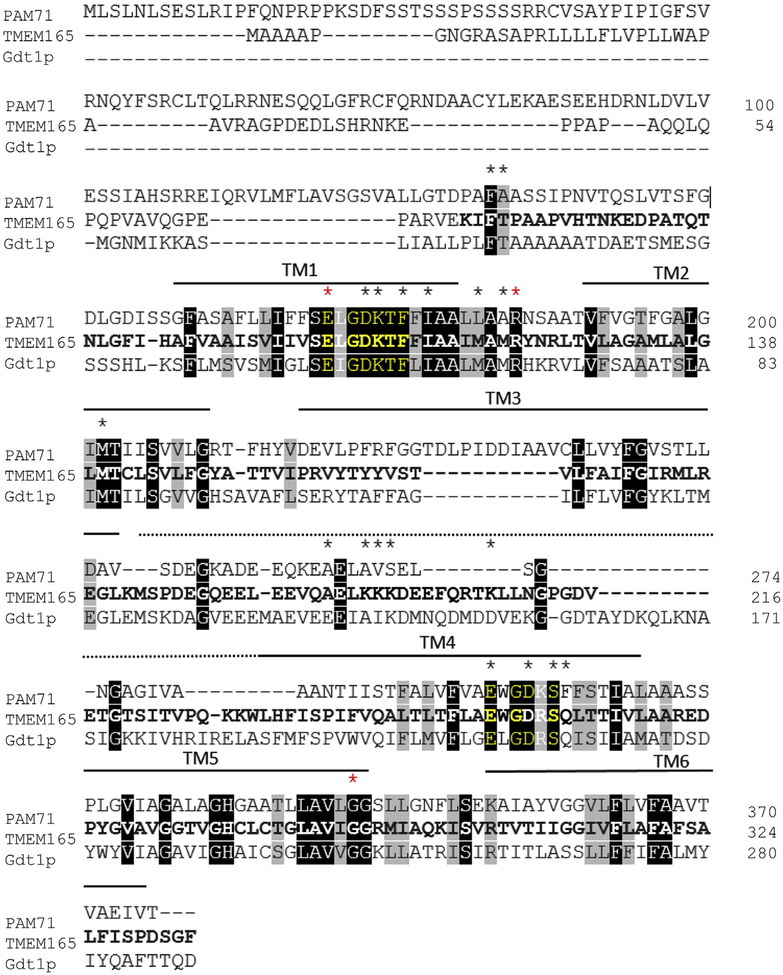
Protein sequence alignments between PAM71, TMEM165 and Gdt1p underline highly conserved amino acids (Fig. 1) [12]. Two patterns of highly conserved successive amino acid sequences emerge from this alignment: E-x-G-D-K-[TF] and E-x-G-D-R-[SQ]. These motifs are enshrined in the first and fourth transmembrane protein domains (TM1 and TM 4) (Fig. 1).
Fig. 1. Protein sequence alignment of TMEM165 and its orthologs PAM71 from Arabidopsis thaliana and Gdt1p from Saccharomyces cerevisiae.
The sequences were found in Uniprot database (www.uniprot.org) and the protein sequence alignment was generated using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo). Black boxes indicate the amino acid residues that are identical whereas gray boxes show the homologous amino acid residues. The black asterisks indicate the position of the generated mutated amino acids. The red asterisks indicate the mutated amino acids found in TMEM165-CDG patients’ proteins. The bold characters correspond to the amino acid residues that are found conserved in the mammalian TMEM165 sequence (SwissProt Database) using the Cobalt-NCBI multiple alignment tool (NCBI). Conserved domains (motif 1 and 2) are highlighted in yellow. Black horizontal bars on the top of the sequences indicate the amino acids within the predictive transmembrane domains (TMHMM v2.0 server tool). The dotted line indicates the cytosolic central loop.
In this paper, we particularly explored the contribution of these two highly conserved motifs in the function of TMEM165 in Golgi glycosylation and also in its Mn2+ sensitivity.
2. MATERIAL and METHODS
2.1. Sequence alignment
Uniprot accession codes are: Homo sapiens TMEM165_HUMAN, Arabidopsis thaliana PAM71_ARATH and Saccharomyces cerevisiae GDT1_YEAST.
2.2. Cell culture, transfection and treatment
Control and KO TMEM165 HEK293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Lonza, Basel, Switzerland), at 37 °C in humidity-saturated 5% CO2 atmosphere. Transfections were performed using 4 μl of Lipofectamine 2000® (Thermo Scientific) for 0.5 μg of plasmid for each well of 6 wells-plate at 70% confluence in 1 ml DMEM medium. Transfections were stopped after 5 h. Wells were split 24 h after transfection and treated 48 h post-transfection. Manganese (II) chloride tetrahydrate, from Riedel-de-Haën (Seelze, Germany) treatment 500 μM was pursued for 4 and 8 h. Chloroquine (ICN Biomedicals) 100 μM was added 1 h before manganese as a pretreatment.
2.3. Constructs, vector engineering and mutagenesis
Mutated TMEM165 plasmids were generated and supplied by e-Zyvec (Lille, France).
2.4. Antibodies and other reagents
Anti-TMEM165 and anti–β-actin antibodies were purchased from MilliporeSigma (Burlington, MA, USA), anti-LAMP2 antibody from Santa Cruz Biotechnology (Dallas, TX, USA) and anti-GM130 antibody from BD Biosciences. Polyclonal goat anti-rabbit IgG and goat anti-mouse IgG Horse Radish Peroxidase-conjugated were from Dako (Denmark).
2.5. Immunofluorescence staining
Cells were seeded on coverslips for 12–24 h, washed once in Dulbecco’s Phosphate Buffer Saline (DPBS, Lonza) containing Calcium and Manganese and fixed with 4% paraformaldehyde (PAF) in PBS (Phosphate Buffer Saline) pH 7.3, for 30 min at room temperature. Coverslips were subsequently washed three times with PBS. Cells were permeabilized with 0.5% Triton X-100 in PBS for 15 min then washed three times with PBS. Coverslips were then saturated for 1 h in blocking buffer [0.2% gelatin, 2% Bovine Serum Albumin (BSA), 2% FBS (Lonza) in PBS], followed by the incubation for 1 h with primary antibody diluted at 1:100 in blocking buffer, except for GPP130 that was diluted at 1:300. After washing with PBS, cells were incubated for 1 h with Alexa 488- or Alexa 568- secondary antibody (Life Technologies) diluted at 1:600 in blocking buffer. After three washes with PBS, nuclei were labeled with DAPI 1:300 for 15 min and then coverslips were mounted on glass slides with Mowiol. Fluorescence was detected through an inverted Zeiss LSM700 confocal microscope. Acquisitions were done with ZEN pro 2.1 software (Zeiss, Oberkochen, Germany).
2.6. Image analyses
Immunofluorescent images were edited using imageJ software (http://imagej.nih.gov/ij) developed by Fiji©.
2.7. Western blotting
Cells were collected in PBS after 2 washes and centrifuged at 6000 rpm for 10 min. Cells were lysed in RIPA buffer [Tris/HCl 50 mM pH 7.9, NaCl 12 0 mM, NP40 0.5%, EDTA 1 mM, Na3VO4 1 mM, NaF 5 mM] supplemented with a protease inhibitors mix (Roche Diagnostics, Penzberg, Germany) by a 30 min centrifugation at 14 000 rpm. Concentration of extracted proteins was determined with the Micro BCA™ Protein Assay Reagent kit (Thermo Fisher Scientific, Waltham, MA USA). For LAMP2 study only samples were preheated 10 min at 95 °C. 10 or 20 μg of total proteins of each sample were dissolved in reducing NuPage® Sample buffer and resolved by MOPS 4–12% Bis–Tris gel (Thermo Fisher Scientific, Waltham, MA USA) and then transferred with iBlot 2 Dry Blotting System (Thermo Fisher Scientific, Waltham, MA USA). Nitrocellulose membranes were blocked 1 h in TBS (Tris Buffer Saline) containing 0.05% Tween 20 5% (w/v) non-fat dried milk for at least 1 h at room temperature, then incubated 1 h with primary antibodies (used at a dilution of 1:2000 for TMEM165, 1: 20 000 for β Actin) and overnight for LAMP2 primary antibody 1:20 000. After three 5 min-TBS-T washes, membranes were incubated with respective secondary antibodies for 1 h (1:10 000 dilution for polyclonal goat anti-rabbit IgG and 1:20 000 for goat anti-mouse IgG Horse Radish Peroxidase-conjugated).
Signal was detected using chemiluminescence reagent Pierce™ Pico Plus Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA USA) on imaging film (GE Healthcare, Buckinghamshire, UK) or Camera Fusion® (Vilber Lourmat) and its software.
3. Results
3.1. Functionality of TMEM165 mutants in Golgi glycosylation
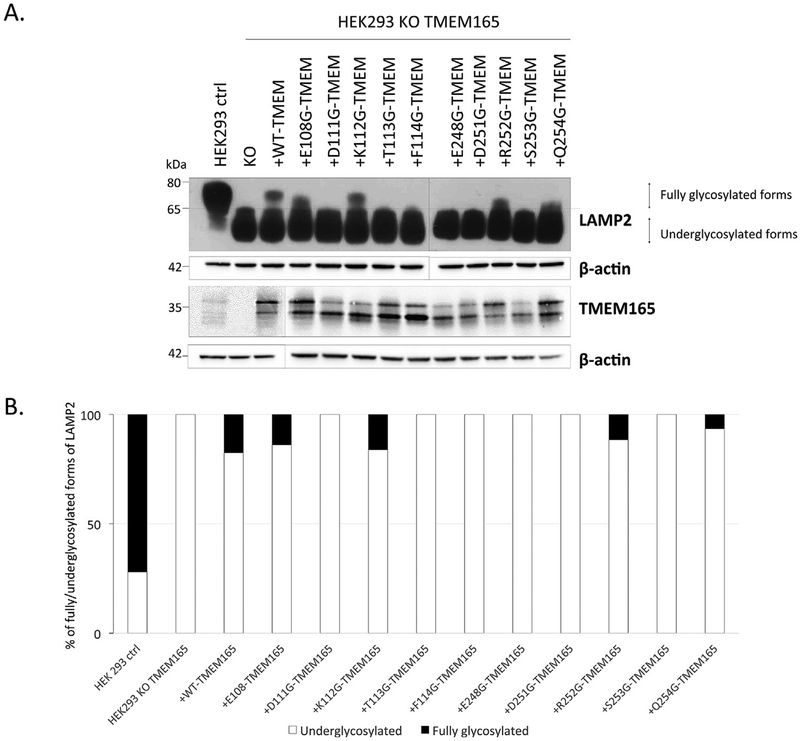
The ability for TMEM165 to rescue LAMP2 glycosylation defect in TMEM165 KO HEK293T cells was used to investigate the involvement each amino acid of the two highly conserved sequences. To explore this, 10 different mutations in the most conserved amino acids that lay in the two signature-motifs were generated (Fig. 1). The wild type (wt-) and mutated forms of TMEM165 were then transiently expressed in TMEM165 KO HEK293T cells and the Golgi glycosylation of LAMP2 was followed by Western blot experiments as previously described [14] (Fig. 2A). Compared to untransfected cells (KO), the expression of the wt-TMEM165 rescued fully glycosylated forms of LAMP2 similar to control cells. Even though the mutated forms of TMEM165 transfection gave heterogeneous results (Fig. 2A), only 3 mutants showed a partial restoration of LAMP2 glycosylation: E108G-, K112G-, R252G-TMEM165, E108G and R252G giving the mildest restorations. Interestingly among all our mutants, 6 of the conserved E-x-GDKT/E-x-GDRSQ motifs are unable to restore LAMP2 glycosylation (Fig. 2B). The mutants (D111G-, T113G-, F114G-, E248G-, D251G-, S253G-, Q254G) were found unable to restore LAMP2 glycosylation (Fig. 2B). This result is characteristic from these two motifs as most of the TMEM165 mutated forms, except G304R (patient mutation), were able to rescue LAMP2 glycosylation (Supp. Fig. 1).
Fig. 2. LAMP2 glycosylation profile after TMEM165 mutants transfection.
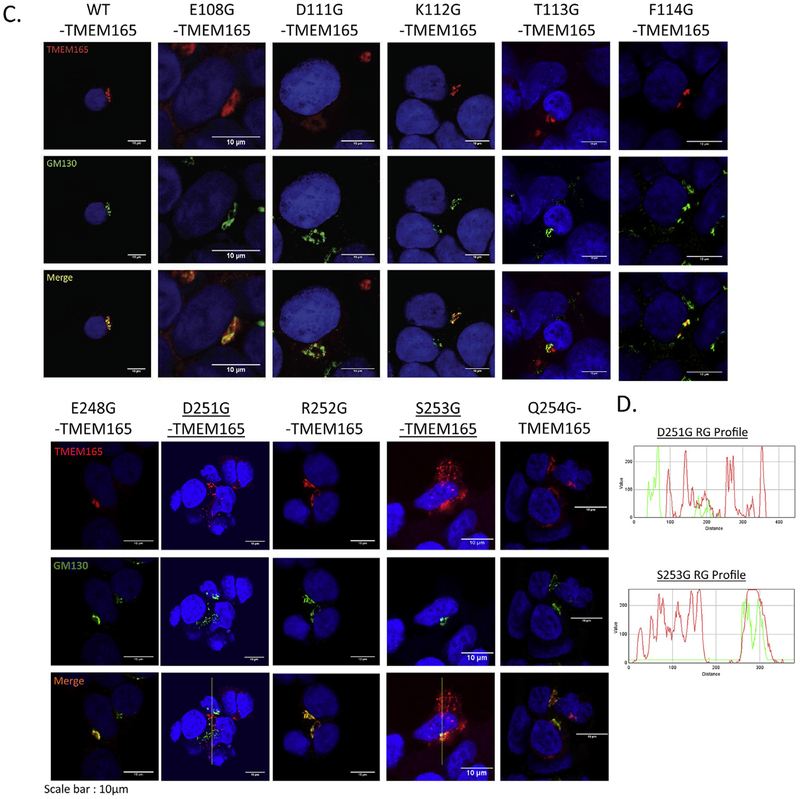
HEK293T KO TMEM165 cells were transfected with empty-vector, wild-type or TMEM165 constructs. Total cells lysates were obtained, subjected to SDS-PAGE, Western blot was performed with the respective antibodies. A. LAMP2 and TMEM165 profiles obtained 24 h after transfection. B. Ratio of fully glysosylated forms of LAMP2 (percentage of fully glysosylated forms versus the total LAMP2). C. Immunofluorescence analysis of the expression and localization of TMEM165 in transfected cells with mutated forms of TMEM165 in conserved amino-acids. (GM130 = Golgi marker) D. Illustration of red and green fluorescence merge with RGB Profiler (ImageJ Fiji®).
To assess these results, the expression level among the mutated forms of TMEM165 was then investigated by Western blot experiments. Although the TMEM165 profile is found heterogeneous with two major bands, there was no major difference in TMEM165 expression level (Fig. 2A). Altogether these results emphasize the importance of some specific amino acids of the two conserved motifs in TMEM165 function in Golgi glycosylation.
3.2. Subcellular localization of TMEM165 mutants
The functionality of TMEM165 mutants in Golgi glycosylation depends of the TMEM165 mutants’ expression but also on their subcellular Golgi localization. To reinforce the above results, the Golgi localization of the mutated forms of TMEM165 was then investigated by immunofluorescence and confocal microscopy experiments.
Most of the mutated forms of TMEM165 displayed a Golgi localization as observed by colocalization experiments using the GM130 Golgi marker (Fig. 2C and D). Very interestingly, the D251G- and S253G-TMEM165 mutants, did not entirely colocalize with GM130 as vesicular structures localized throughout the cytoplasm could be detected. To further assess the subcellular localization of these mutants, immunofluorescence staining using the lysosomal/endosomal intracellular marker LAMP2 was performed. A partial colocalization was observed with LAMP2 demonstrating the differential subcellular localization for these mutated forms (Supplementary Figs. 2A and B). For these mutants, it is likely that the observed lack of Golgi glycosylation restoration results from a subcellular mislocalization.
3.3. Sensitivity of TMEM165 mutants to manganese exposure
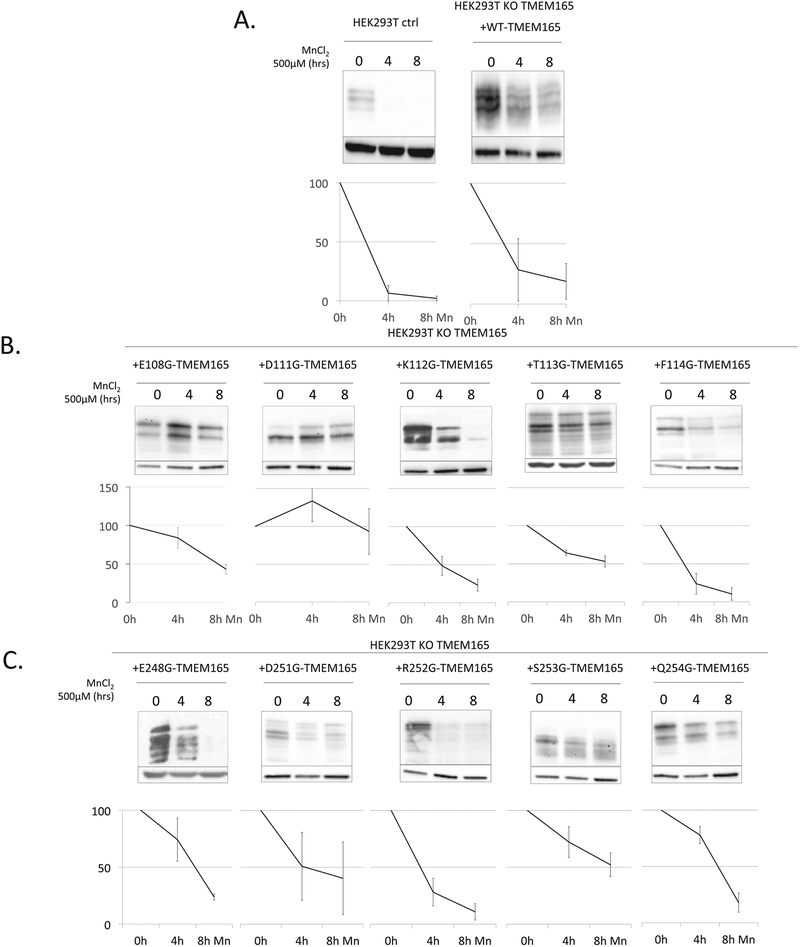
We recently highlighted that, when exposed to high manganese concentration, TMEM165 was efficiently targeted to lysosomes for degradation [15]. As for the glycosylation study, we investigated the Mn2+ sensitivity of these different mutants. To assess this point, the wild-type and mutated forms of TMEM165 were transiently transfected in KO cells. The impact of high Mn2+ concentration supplementation on the stability and subcellular localization was investigated during a 4 and 8 h time course by Western blot (Fig. 3) and immunofluorescence experiments (data not shown). Diagrams under each mutant’s Western blot describe the quantification of the remaining TMEM165 after 4 and 8 h of Mn2+ treatment. As previously published [15] we observed that TMEM165 in normal HEK293T cells is highly sensitive to manganese, with a complete loss of this protein after 8 h treatment (Fig. 3A). Same observation is made after transfection of the wild-type form of TMEM165 in HEK293T KO TMEM165 cells with a loss over 75% of TMEM165 expression after 4 h manganese treatment (Fig. 3A).
Fig. 3. Sensitivity of TMEM165 mutants to manganese exposure.
TMEM165 expression in cells transfected with transfected cells forms of TMEM165 in the conserved sequences with or without manganese (N ≥ 3) A. In control cells and transfected cells with WT-TMEM165. B. In the cytosolic E-x-G-D-K-[TF] motif. C. In the luminal E-x-G-D-R-[SQ] motif. Relative quantification of TMEM165 degradation at 4 and 8 h manganese treatment below each respective Western blot.
Concerning the mutated forms of TMEM165, 5 are found partially resistant, E108G, D111G, T113G, D251G and S253G. At the opposite, K112G, F114G, E248G, R252G and Q254G are sensitive to manganese treatment. These results were confirmed by immunofluorescence confocal microscopy (data not shown) and demonstrate the crucial importance of specific amino acids in the differential Mn induced sensitivity of TMEM165.
3.4. The functional mutants are targeted and degraded into lysosomes
We recently established that the Mn2+ induced degradation of TMEM165 was inhibited by chloroquine treatment [15]. To assess whether the mutated forms of TMEM165 fall under the same regulation, the stability of wt- and mutated forms of TMEM165 were analyzed by Western blot and immunofluorescence after Mn2+ exposure, in the presence or the absence of chloroquine. The degradation of every mutated forms of TMEM165 was completely blocked by chloroquine (data not shown). The molecular mechanism by which TMEM165 is sent to lysosomes following Mn2+ exposure is currently unknown. Monoubiquitination is known to be a very efficient mechanism to target proteins for lysosomal degradation. It appears that the cytosolic loop of TMEM165 contains 4 lysine residues K198, K199, K200 and K208 that could be involved in the Mn2+ induced lysosomal targeting. In order to investigate the role of these lysines, TMEM165 mutants (K198R, K199R, K200R, K208R and K198–K200R) were generated and analyzed by Western blot and immunofluorescence after Mn2+ exposure, in the presence or in the absence of chloroquine (Supp. Figure 3). We observed that after Mn2+ exposure, the lysine mutants of TMEM165 were localized in the Golgi and were degraded similarly to what is observed for wt-TMEM165. Altogether, these results indicate that the lysine residues of the cytosolic loop are not involved in the expression, neither in the Golgi localization nor in its Mn2+-induced degradation of TMEM165.
4. Discussion
Although the precise molecular and cellular functions of TMEM165 are still under debate, its functional role in Golgi glycosylation is now clearly established. The link between TMEM165 and cellular/Golgi Mn2+ homeostasis maintenance is shown by (i) the alteration of GPP130 Mn2+ induced degradation in TMEM165 depleted cells, (ii) the restoration of Golgi glycosylation by Mn2+ supplementation [11], and (iii) the TMEM165 Mn2+ sensitivity [15]. It is now highly suspected that TMEM165 functions as a Golgi Ca2+/Mn2+ transporter regulating both Ca2+/Mn2+ Golgi homeostasis. As observed in yeasts, the Golgi glycosylation defect due to a lack of TMEM165 would result in an alteration of the Golgi Mn2+ homeostasis crucial for the activities of Golgi glycosyltransferases using UDP-sugars as donors [11]. TMEM165 is a member of the UPF0016 family characterized by two highly conserved consensus motifs E-φ-G-D-[KR]-[TS]. Our previous results showed that the E-φ-G-D-K-T motif (motif 1) was facing the cytosol while the E-φ-G-D-R-S (motif 2) was exposed to the Golgi luminal side and hence are predicted to be involved in the transport function of UPF0016 members [15]. In this paper we wanted to further understand the contribution of these two highly conserved motifs in the role of TMEM165 in Golgi glycosylation and also in its sensitivity to high Mn2+ concentration.
Our results first emphasized that some of the mutated forms of TMEM165 are unable to rescue Golgi glycosylation. The mutation of the amino acid E108G does not seem to strongly affect the function of TMEM165 in Golgi glycosylation as a slight restoration of LAMP2 glycosylation is observed. At the opposite, the E248G mutation (second motif) cannot rescue Golgi glycosylation. Interestingly, the polar amino acids (T113 and S253) are found crucial for the function of TMEM165 in Golgi glycosylation while basic amino acids (K112 and R252) are dispensable. We hypothesize that these polar amino acids, via post-translational modifications, play a crucial role in the regulation of TMEM165 functionality.
As proposed for yeasts, it is most likely that amino acids of the two conserved motifs constitute the cation binding sites of TMEM165. In such hypothesis, mutations in specific amino acids of the two conserved motifs alter the transport function of TMEM165 by impairing cation affinity or pocket conformation changes.
The other particularity of TMEM165 is its sensitivity to high Mn2+ concentrations. We recently demonstrated that following high Mn2+ exposure, TMEM165 was targeted to lysosomes for its degradation [15]. The targeting molecular mechanism is unclear but recent investigations propose the requirement of Sortilin in the Mn2+ induced degradation of TMEM165 [16]. The Mn2+ sensitivity of the mutated forms of TMEM165 was evaluated. As pointed out for the glycosylation, most of the generated mutated forms of TMEM165 are resistant to Mn2+ exposure and only few are sensitive. Our results demonstrate that the acidic amino acids (E and D) of the first conserved motif are crucial in conferring the Mn2+ sensitivity to TMEM165. The two resistant mutants D251G and S253G of the second motif are insensitive to manganese presumably due to their mislocalization. Another interesting observation deals with the T113G that is also clearly found resistant to Mn exposure while correctly Golgi localized. The roles of these amino acids in the Mn2+ induced degradation mechanism/Golgi subcellular localization are not clear but one can imagine that they are part of a regulatory mechanism that delicately governs the Golgi ion homeostasis.
In conclusion, this paper highlights the importance of the two very conserved regions for the functionality of TMEM165 in Golgi glycosylation, its subcellular Golgi localization and Mn2+ sensitivity.
Supplementary Material
Acknowledgements
This work was supported by grants from Agence Nationale de la Recherche (SOLV-CDG project to F.F.), and EURO-CDG-2 that has received a funding from the European Union’s Horizon 2020 research and innovation program under the ERA-NET Cofund action N° 643578. We are also indebted to Dr Dominique Legrand for the Research Federation FRABio (Univ. Lille, CNRS, FR 3688, FRABio, Biochimie Structurale et Fonctionnelle des Assemblages Biomoléculaires) for providing the scientific and technical environment conducive to achieving this work. We thank the BioImaging Center of Lille, especially Christian Slomianny and Elodie Richard, for the use of the Leica LSM700.
Abbreviations
- CQ
Chloroquine
- CDG
Congenital Disorder(s) of Glycosylation
- GPP130
Golgi Phosphoprotein 4
- ER
Endoplasmic Reticulum
- LAMP2
Lysosomal-associated membrane protein 2
- Mn
Manganese
- Mn2+
Manganese, ion (2+)
- MnCl2
Manganese (II) chloride tetrahydratex
- TMEM165
Transmembrane Protein 165
- SPCA1
Secretory-Pathway Ca2+-ATPase 1
- PAM71
Photosynthesis Affected Mutant 71
- UPF
Uncharacterized Protein Family
Footnotes
Competing interests
The author(s) declare no competing interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biochi.2019.07.016.
References
- [1].Jaeken J, Péanne R, What is new in CDG? J. Inherit. Metab. Dis 40 (2017) 569–586, 10.1007/s10545-017-0050-6. [DOI] [PubMed] [Google Scholar]
- [2].Aebi M, Helenius A, Schenk B, Barone R, Fiumara A, Berger EG, Hennet T, Imbach T, Stutz A, Bjursell C, Uller A, Wahlström JG, Briones P, Cardo E, Clayton P, Winchester B, Cormier-Dalre V, de Lonlay P, Cuer M, Dupré T, Seta N, de Koning T, Dorland L, de Loos F, Kupers L, Carbohydrate-deficient glycoprotein syndromes become congenital disorders of glycosylation: an updated nomenclature for CDG. First International Workshop on CDGS, Glycoconj. J 16 (1999) 669–671. [DOI] [PubMed] [Google Scholar]
- [3].Foulquier F, Amyere M, Jaeken J, Zeevaert R, Schollen E, Race V, Bammens R, Morelle W, Rosnoblet C, Legrand D, Demaegd D, Buist N, Cheillan D, Guffon N, Morsomme P, Annaert W, Freeze HH, Van Schaftingen E, Vikkula M, Matthijs G, TMEM165 deficiency causes a congenital disorder of glycosylation, Am. J. Hum. Genet 91 (2012) 15–26, 10.1016/j.ajhg.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Marques-da-Silva D, Francisco R, Webster D, dos Reis Ferreira V, Jaeken J, Pulinilkunnil T, Cardiac complications of congenital disorders of glycosylation (CDG): a systematic review of the literature, J. Inherit. Metab. Dis 40 (2017) 657–672, 10.1007/s10545-017-0066-y. [DOI] [PubMed] [Google Scholar]
- [5].Marques-da-Silva D, dos Reis Ferreira V, Monticelli M, Janeiro P, Videira PA, Witters P, Jaeken J, Cassiman D, Liver involvement in congenital disorders of glycosylation (CDG). A systematic review of the literature, J. Inherit. Metab. Dis 40 (2017) 195–207, 10.1007/s10545-016-0012-4. [DOI] [PubMed] [Google Scholar]
- [6].Rosnoblet C, Legrand D, Demaegd D, Hacine-Gherbi H, de Bettignies G, Bammens R, Borrego C, Duvet S, Morsomme P, Matthijs G, Foulquier F, Impact of disease-causing mutations on TMEM165 subcellular localization, a recently identified protein involved in CDG-II, Hum. Mol. Genet 22 (2013) 2914–2928, 10.1093/hmg/ddt146. [DOI] [PubMed] [Google Scholar]
- [7].Demaegd D, Colinet A-S, Deschamps A, Morsomme P, Molecular evolution of a novel family of putative calcium transporters, PLoS One 9 (2014) e100851, 10.1371/journal.pone.0100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Demaegd D, Foulquier F, Colinet A-S, Gremillon L, Legrand D, Mariot P, Peiter E, Van Schaftingen E, Matthijs G, Morsomme P, Newly characterized Golgi-localized family of proteins is involved in calcium and pH homeostasis in yeast and human cells, Proc. Natl. Acad. Sci 110 (2013) 6859–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schneider A, Steinberger I, Herdean A, Gandini C, Eisenhut M, Kurz S, Morper A, Hoecker N, Rühle T, Labs M, Flügge UI, Geimer S, Schmidt SB, Husted S, Weber APM, Spetea C, Leister D, The evolutionarily conserved protein PHOTOSYNTHESIS AFFECTED MUTANT71 is required for efficient manganese uptake at the thylakoid membrane in Arabidopsis, Plant Cell 2015 (2016) 00812, 10.1105/tpc.15.00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Potelle S, Morelle W, Dulary E, Duvet S, Vicogne D, Spriet C, Krzewinski-Recchi M-A, Morsomme P, Jaeken J, Matthijs G, De Bettignies G, Foulquier F, Glycosylation abnormalities in Gdt1p/TMEM165 deficient cells result from a defect in Golgi manganese homeostasis, Hum. Mol. Genet 25 (2016) 1489–1500, 10.1093/hmg/ddw026. [DOI] [PubMed] [Google Scholar]
- [11].Morelle W, Potelle S, Witters P, Wong S, Climer L, Lupashin V, Matthijs G, Gadomski T, Jaeken J, Cassiman D, Morava E, Foulquier F, Galactose supplementation in patients with TMEM165-CDG rescues the glycosylation defects, J. Clin. Endocrinol. Metab 102 (2017) 1375–1386, 10.1210/jc.2016-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dulary E, Potelle S, Legrand D, Foulquier F, TMEM165 deficiencies in Congenital Disorders of Glycosylation type II (CDG-II): clues and evidences for roles of the protein in Golgi functions and ion homeostasis, Tissue Cell 49 (2017) 150–156, 10.1016/j.tice.2016.06.006. [DOI] [PubMed] [Google Scholar]
- [13].Thines L, Deschamps A, Sengottaiyan P, Savel O, Stribny J, Morsomme P, The yeast protein Gdt1p transports Mn2+ ions and thereby regulates manganese homeostasis in the Golgi, J. Biol. Chem 293 (2018) 8048–8055, 10.1074/jbc.RA118.002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Houdou M, Lebredonchel E, Garat A, Duvet S, Legrand D, Decool V, Klein A, Ouzzine M, Gasnier B, Potelle S, Foulquier F, Involvement of thapsigargin- and cyclopiazonic acid–sensitive pumps in the rescue of TMEM165-associated glycosylation defects by Mn2+, FASEB J. 33 (2019) 2669–2679, 10.1096/fj.201800387R. [DOI] [PubMed] [Google Scholar]
- [15].Potelle S, Dulary E, Climer L, Duvet S, Morelle W, Vicogne D, Lebredonchel E, Houdou M, Spriet C, Krzewinski-Recchi M-A, Peanne R, Klein A, de Bettignies G, Morsomme P, Matthijs G, Marquardt T, Lupashin V, Foulquier F, Manganese-induced turnover of TMEM165, Biochem. J 474 (2017) 1481–1493, 10.1042/BCJ20160910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Venkat S, Linstedt AD, Manganese-induced trafficking and turnover of GPP130 is mediated by sortilin, Mol. Biol. Cell (2017), 10.1091/mbc.E17-05-0326 mbc.E17-05-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.