Summary
CD8+ T cell responses to hepatotropic viruses like HBV range from dysfunction to differentiation into effector cells, but the mechanisms underlying these distinct outcomes remain poorly understood. Here we show that priming by Kupffer cells –not natural targets of HBV – leads to differentiation into effector cells that form dense, extravascular clusters of rather immotile cells scattered throughout the liver. By contrast, priming by hepatocytes – natural targets of HBV - leads to local activation and proliferation but lack of differentiation into effector cells; these cells form loose, intravascular clusters of motile cells that coalesce around portal tracts. Transcriptomic and chromatin accessibility analyses unveil unique features of these dysfunctional CD8+ T cells, with limited overlap with those of exhausted or tolerant T cells; accordingly, CD8+ T cells primed by hepatocytes cannot be rescued by anti-PD-L1 treatment, but instead respond to IL-2. These findings suggest new immunotherapeutic strategies against chronic HBV infection.
Priming of circulating naïve CD8+ T cells in non-lymphoid organs is hindered by the endothelial barrier limiting antigen (Ag) recognition on epithelial cells. The liver is an exception: slow blood flow1, presence of endothelial fenestrations and absence of a basement membrane allow CD8+ T cells to sense MHC-Ag complexes on hepatocytes2,3. Liver priming is thought to result in T cell unresponsiveness or dysfunction4,5 but the underlying mechanisms, particularly in the context of HBV pathogenesis, are incompletely understood. HBV is a noncytopathic virus replicating in hepatocytes and causing acute or chronic infections6,7. Infection outcome is mainly determined by the kinetics, breadth, vigour and effector functions of HBV-specific CD8+ T cell responses6. Chronic HBV infection is typically acquired at birth or in early childhood8 and proceeds from an initial “immune tolerant” phase (characterized by high viremia and no liver inflammation) to an “immune active” phase (in which viremia is lower and liver inflammation is present)8,9. HBV-specific CD8+ T cells in young immune tolerant patients are considered akin to exhausted T cells characterizing the immune active phase10, as well as to other infection- or cancer-related conditions of immune dysfunction, although a detailed characterization is lacking11.
Spatiotemporal dynamics of naïve CD8+ T cells undergoing intrahepatic priming
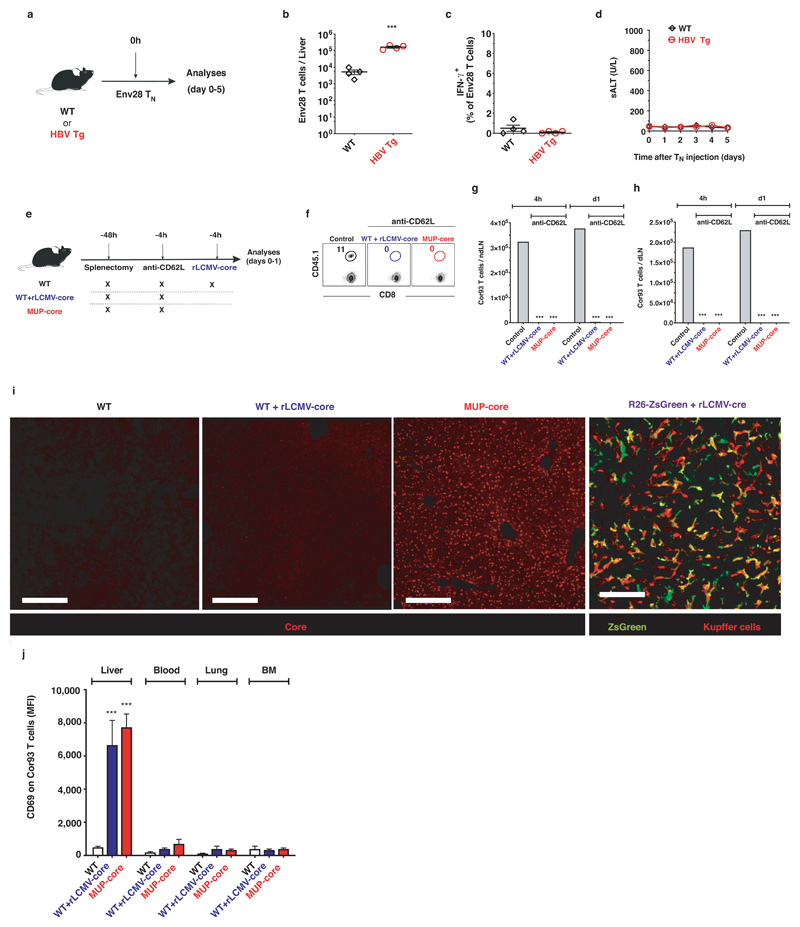
To study the immune mechanisms of early HBV unresponsiveness, we initially analysed HBV-specific CD8+ T cells undergoing priming in a non-inflamed liver. In accordance to previous data12, envelope-specific naïve CD8+ TCR transgenic T cells (Env28 TN)12 adoptively transferred into HBV replication-competent transgenic mice expressing all viral proteins in the hepatocyte13 proliferated but failed to develop IFN-γ-producing or cytolytic capacities (Extended Data Fig. 1a-d). As an effective CD8+ T cell response is induced in immunocompetent individuals exposed to HBV in adulthood14, it remains to be determined whether this is due to cross-priming events in secondary lymphoid organs or whether the liver itself is capable of supporting full effector differentiation.
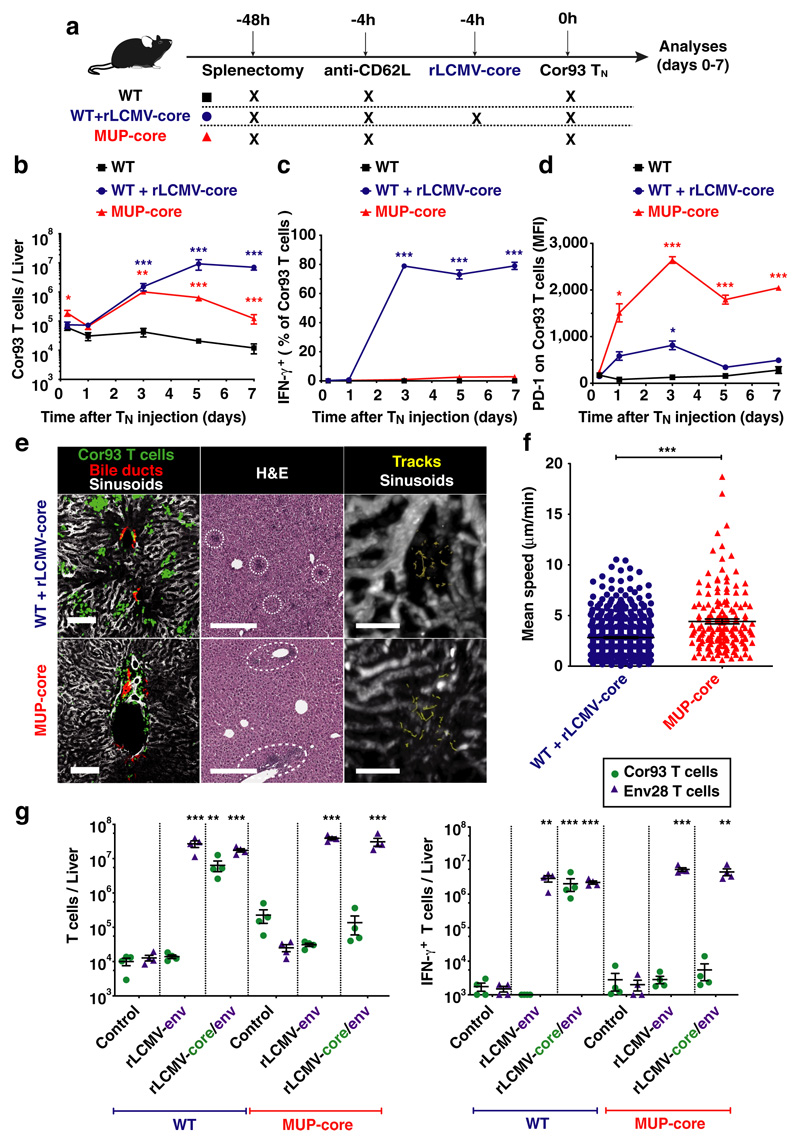
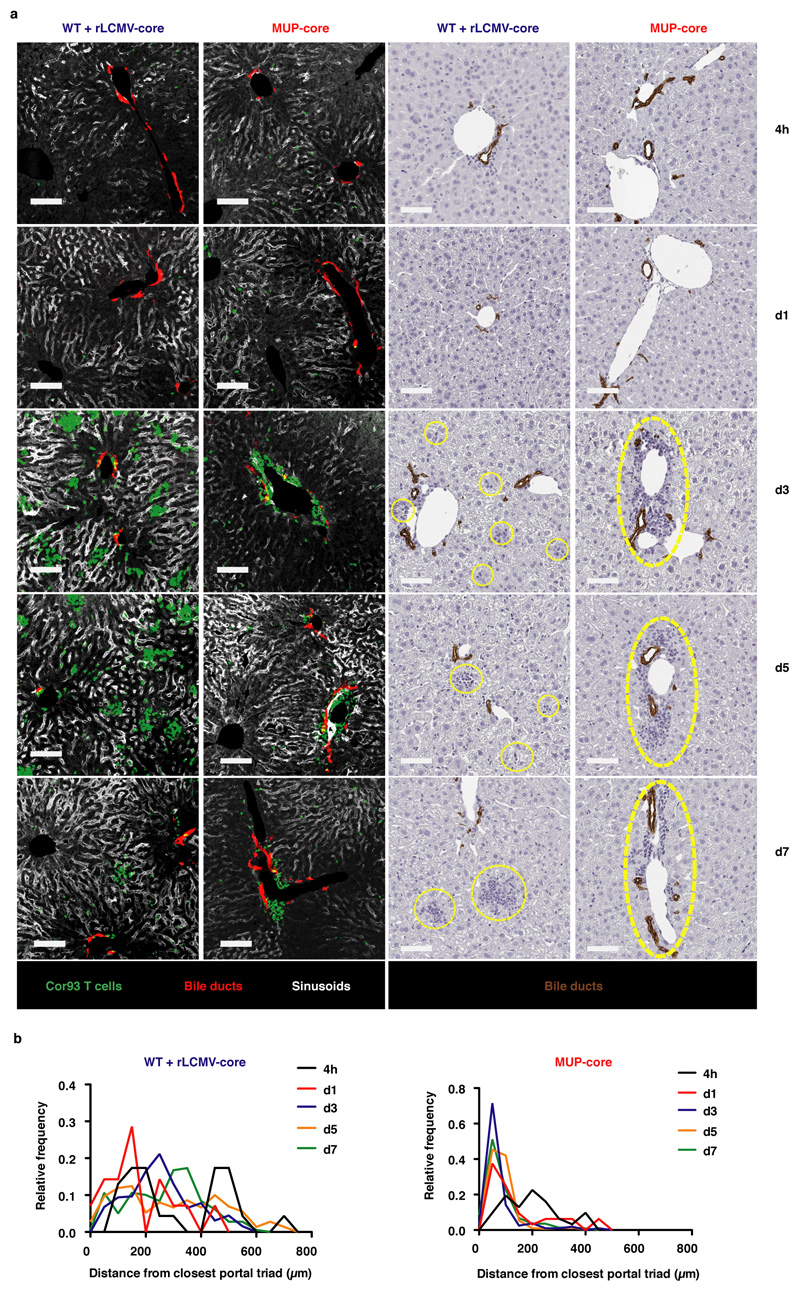
Using a system whereby T cell priming is restricted to the liver (Fig. 1a and Extended Data Fig. 1f-h), we injected naïve CD8+ TCR transgenic T cells specific for the core protein of HBV (Cor93 TN)12 into MUP-core transgenic mice15, which exclusively express a non-secretable version of the HBV core protein in 100% of hepatocytes (Extended Data Fig. 1i). Two additional groups of mice served as controls (Fig. 1a): i) WT mice; and ii) WT mice that are transduced with recombinant replication-defective, lymphocytic choriomeningitis virus (LCMV)-based vectors16 targeting a non-secretable version of the HBV core protein (rLCMV-core) to Kupffer cells (KCs) and hepatic dendritic cells (DCs) that are not naturally infected by HBV (Extended Data Fig. 1i). Ag recognition was restricted to hepatocytes in MUP-core mice or to KCs and hepatic DCs in rLCMV-transduced WT mice, as Cor93 TN isolated 1 hour after transfer up-regulated CD69 (a proxy for Ag recognition) in the liver but not in the blood, lung and bone marrow (Extended Data Fig. 1j). We then characterized the fate and function of naïve CD8+ T cells undergoing intrahepatic priming. HBV-specific naïve CD8+ T cells recognizing Ag in the liver underwent local activation (Extended Data Fig. 1j) and proliferation, so that by day 3 after transfer we could recover ~30-fold more intrahepatic Cor93 T cells in Ag-expressing mice than control animals (Fig. 1b). Whereas Ag recognition on KCs and hepatic DCs yielded bona fide effector cells endowed with IFN-γ-producing (Fig. 1c) and cytolytic abilities, Ag recognition on hepatocytes led to the generation of dysfunctional cells that produce little or no IFN-γ upon in vitro peptide re-stimulation (Fig. 1c), did not develop cytotoxic activity and instead up-regulated the inhibitory receptor PD-1 (Fig. 1d). Taken together, these results indicate that, depending upon the nature of the Ag-presenting cell, the liver can support the development of either functional or dysfunctional CD8+ T cells. Spatiotemporal analyses of rLCMV-core transduced mice revealed T cell clusters scattered throughout the liver lobule (Fig. 1e and Extended Data Fig. 2a, b) in a pattern that is reminiscent to that observed during acute self-limited HBV infection17. By contrast, CD8+ T cells formed clusters that coalesced around portal tracts in MUP-core mice (Fig. 1e and Extended Data Fig. 2a, b), a situation that is similar to chronic HBV infection18. These periportal clusters occurred in spite of the fact that the core protein is uniformly expressed in all hepatocytes15 (Extended Data Fig. 1i) and that in the first few hours after transfer CD8+ T cells recognize Ag on hepatocytes that can be distant from portal tracts (Extended Data Fig. 2). Multiphoton intravital imaging of the liver uncovered that clusters formed in WT mice transduced with rLCMV-core are dense, extravascular and composed of largely immotile cells; by contrast clusters formed in MUP-core mice are looser, intravascular and composed of more motile cells (Fig. 1e, f and Supplementary Videos 1, 2). By day 5-7, clusters in WT mice transduced with rLCMV-core start to disaggregate as cells move out from the liver, whereas clusters in MUP-core mice remain in place, possibly reflecting Ag persistence (Supplementary Video 3).
Fig. 1. Spatiotemporal dynamics of naïve CD8+ T cells undergoing intrahepatic priming.
(a) Schematic representation of the experimental setup. (b) Absolute numbers of Cor93 T cells in the livers of indicated mice at indicated time points. (c) Frequency of IFN-γ-producing Cor93 T cells in the livers of indicated mice at indicated time points. (d) MFI of PD-1 expression on Cor93 T cells in the livers of indicated mice. (e) (Left panels) Representative confocal immunofluorescence micrographs of liver sections from WT + rLCMV-core (upper panels) or from MUP-core mice (lower panels) three days after Cor93 TN transfer. Distribution of Cor93 T cells (green) relative to portal tracts (red). Sinusoids are in white. Scale bars represent 100 μm. (Middle panels) H&E staining of liver sections from the same mice, where dotted lines denote leukocyte clusters. Scale bars represent 300 μm. (Right panels) Snapshots from representative intravital multiphoton microscopy movies of the same mice. Cor93 T cells tracks are in yellow and blood vessels are in white. Scale bars represent 40 μm. (f) Mean speed of Cor93 T cells in the livers of indicated mice. (g) Cor93 and Env28 naïve CD8+ T cells were co-transferred into splenectomized and anti-CD62L-treated C57BL/6 x Balb/c F1 (WT) or MUP-core x Balb/c F1 (MUP-core) recipients. When indicated, mice were injected with rLCMV-env or rLCMV-core/env. Livers were collected and analysed five days after T cell transfer. Total numbers (left) and numbers of IFN-γ-producing (right) Cor93 and Env28 T cells in the livers of indicated mice.
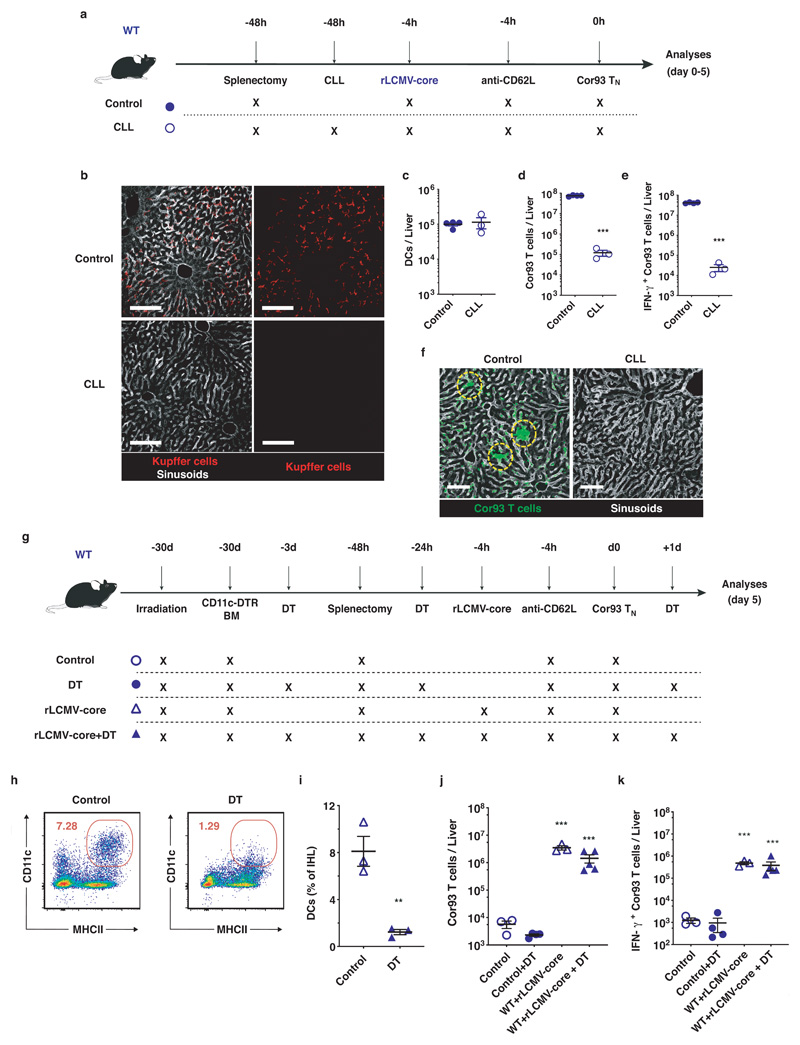
The notion that the liver is capable of supporting full effector differentiation is not without precedent4,19,20 but stands in contrast to the immunological dogma that T cell priming occurs exclusively in secondary lymphoid organs. As rLCMV targets both KCs and hepatic DCs, we next investigated which of these two cell populations support intrahepatic priming of naïve CD8+ T cells. To this end, WT mice were injected with clodronate liposomes (CLL) that effectively deplete KCs while sparing hepatic DCs21 (Extended Data Fig. 3a-c). As shown in Extended Data Fig. 3d-f, KC depletion abolished TN expansion and effector differentiation. We then depleted hepatic DCs through diphtheria toxin injection in WT mice reconstituted with CD11c-DTR bone marrow, but this treatment did not affect the capacity of rLCMV-core to efficiently prime and promote effector differentiation of Cor93-specific CD8+ T cells (Extended Data Fig. 3g-k). Together, the data indicate that KCs – but not hepatic DCs – promote effective CD8+ T cell priming upon rLCMV injection.
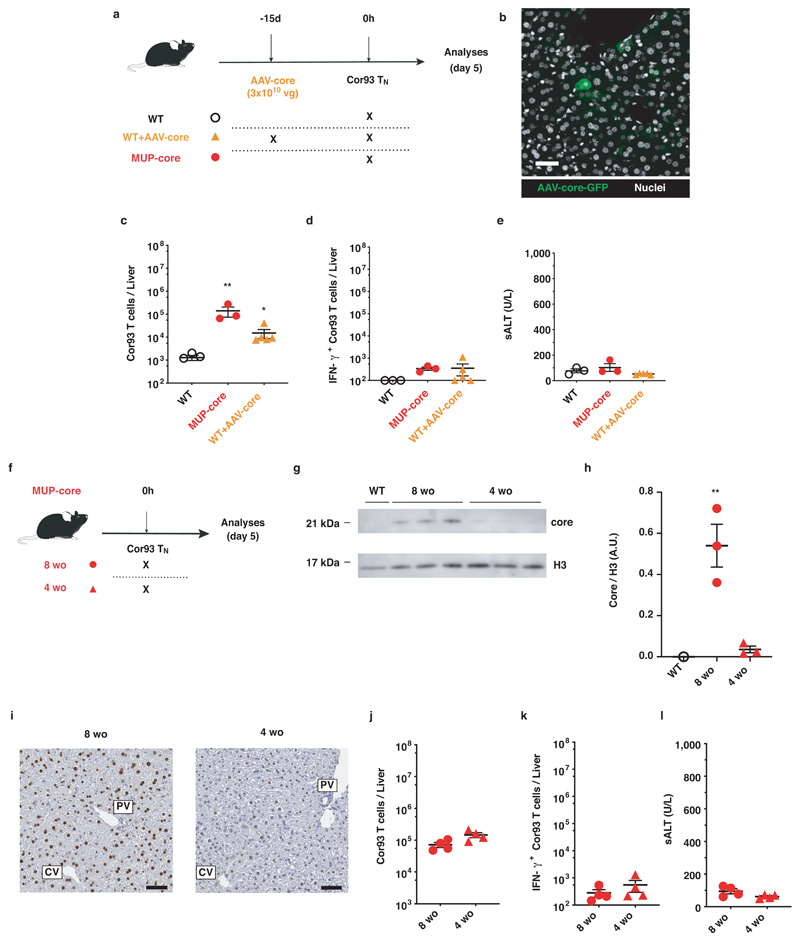
We next evaluated the fate of naïve T cells that are primed within livers expressing low levels of HBV core Ag. First, we transferred Cor93 TN into WT mice previously injected with a low dose of a hepatotropic adeno-associated viral vector (AAV) encoding the HBV core protein. This dose (transducing <5% of hepatocytes) supported Cor93 CD8+ T cell proliferation but not effector differentiation (Extended Data Fig. 4). Second, we transferred Cor93 TN into 3-4 week-old MUP-core mice, which express only trace amounts of this protein per hepatocyte (note that core protein expression in these animals is developmentally regulated, reaching plateaus at 6-8 week of age15). As shown in Extended Data Fig. 4, reducing the amount of expressed Ag by >15 folds within individual hepatocytes did not affect the differentiation of intrahepatically primed CD8+ T cells. Altogether, these experiments indicate that low expression of hepatocellular core Ag is per se not sufficient to induce effector differentiation.
Finally, we investigated the fate of intrahepatic TN primed by Ag presented by both hepatocytes as well as KCs and DCs by transferring Env28 and Cor93 TN into WT and MUP-core mice transduced with rLCMV vectors encoding either the HBV envelope protein alone (rLCMV-env) or both the HBV core and envelope proteins (rLCMV-core/env). As expected, in WT mice TN expanded and differentiated into IFN-γ-secreting cells only when cognate Ag is present (Fig. 1g). In MUP-core mice, injection of rLCMV-env allowed for Env28 (but not Cor93) TN expansion and effector differentiation, indicating that i) innate immune signal carried by rLCMV vectors are not sufficient to overcome Cor93 T cell dysfunction and ii) dysfunctional Cor93 T cells do not produce soluble or membrane-bound mediators that inhibit Env28 T cell effector differentiation (Fig. 1g). Finally, injection of rLCMV-core/env to MUP-core mice led to Env28 (but not Cor93) TN expansion and effector differentiation, indicating that – when Ag is presented by both hepatocytes as well as KCs and hepatic DCs – hepatocellular Ag presentation is dominant in inducing immune dysfunction (Fig. 1g).
Genomic landscape of CD8+ T cells undergoing intrahepatic priming
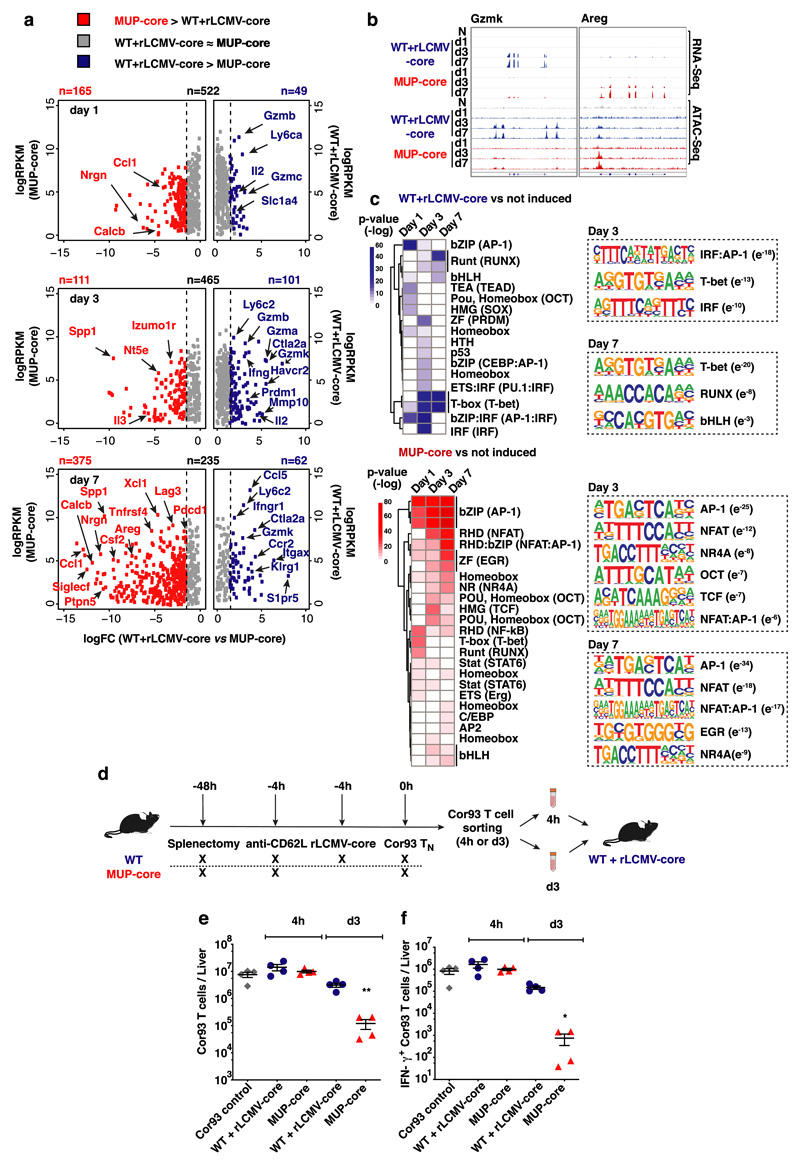
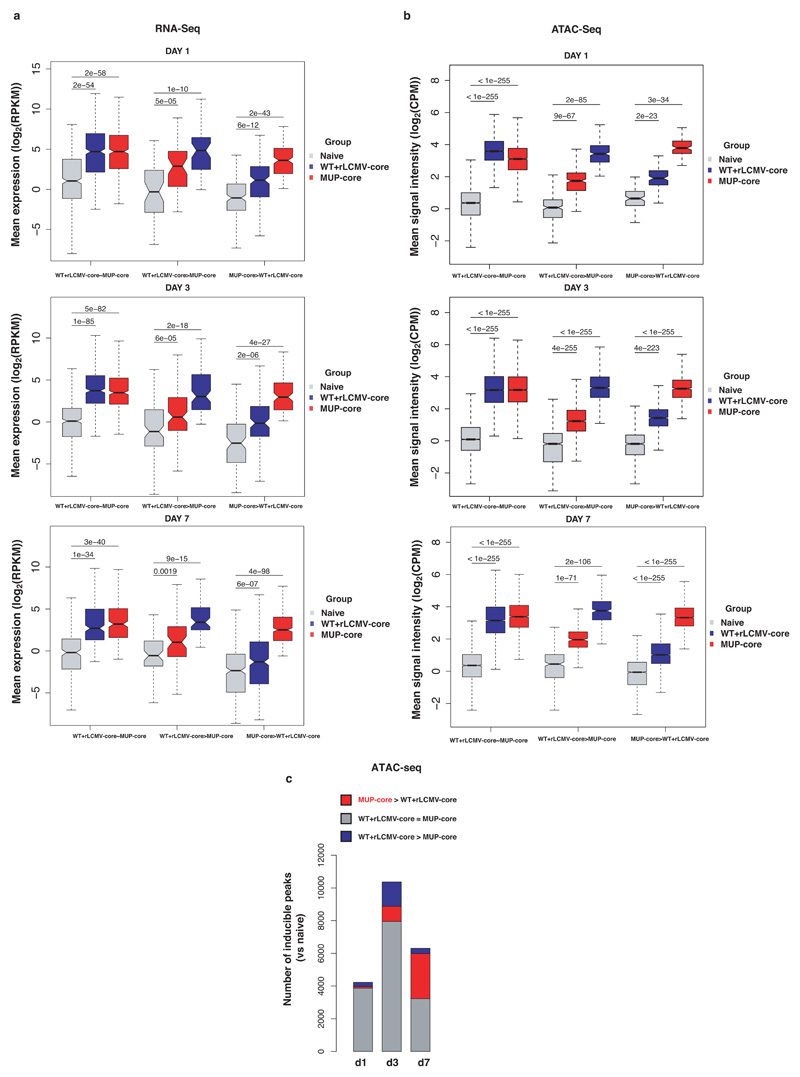
To unveil molecular determinants of this immune dysfunction, we performed transcriptomic (RNA-Seq) and chromatin accessibility (ATAC-Seq) analyses of Cor93 CD8+ T cells isolated from the livers of MUP-core or control rLCMV-core-transduced WT mice at day 1, 3 and 7 after transfer. We observed a broad and progressive transcriptional divergence in intrahepatic Cor93 CD8+ T cells sorted from the two groups of mice (Fig. 2a, Extended Data Fig. 5a and Supplementary Table 1, 2). Hepatic CD8+ T cells from rLCMV-core-transduced WT mice, but not those from MUP-core mice, upregulated canonical genes of the T cell effector program such as Gzma, Gzmb and Ifng 22,23. By contrast, CD8+ T cells isolated from the livers of MUP-core mice upregulated transcripts encoding for a different set of cytokines and chemokines (Ccl1, Csf2, Xcl1), growth factors and hormones (Areg, Calcb), inhibitory molecules (Pdcd1, Lag3, Havcr2) or surface markers (Siglecf) (Fig. 2a, b and Supplementary Table 2). CD8+ T cells from rLCMV-core-transduced WT mice or MUP-core mice had distinct chromatin accessibility profiles at days 3 and 7 after transfer (Extended Data Fig. 5b, c and Supplementary Table 3). Motif enrichment analysis on differentially induced (versus naïve CD8+ T cells) ATAC-Seq peaks revealed an over-representation of binding sites for transcription factor (TF) families involved in effector T cell differentiation, such as IRF, IRF:AP-1 and T-bet at day 3, as well as T-bet, RUNX and bHLH at day 7 in CD8+ T cells from rLCMV-core-transduced WT mice24–27. By contrast, CD8+ T cells from MUP-core mice were enriched in binding sites for AP-1, NFAT, NFAT:AP-1 as well as for NR4A (recently associated to CD8+ T cell dysfunction28,29), OCT, TCF and EGR (Fig. 2c, Supplementary Table 4).
Fig. 2. Transcriptomic and chromatin accessibility analyses of CD8+ T cells undergoing intrahepatic priming.
(a) Scatter plot showing the level (y axis) and the difference in expression (x axis) of inducible genes in the dataset (versus Cor93 TN) in the indicated conditions. Genes expressed at higher levels in WT + rLCMV-core or MUP-core mice are shown in blue or red, respectively. (b) Integrative Genomics Viewer (IGV)40 snapshots showing RNA-seq and ATAC-seq data at Gzmk and Areg loci, selected as representative genes with differential expression in WT + rLCMV-core or MUP-core mice, respectively. (c) Left panels. Heatmap showing the enrichment of DNA motifs (HOMER)41 within the top 200 inducible (versus Cor93 TN) and differential ATAC-Seq peaks in WT + rLCMV-core (blue) or MUP-core mice (red). A set of 3899 non-inducible ATAC-seq peaks was used as background. Right panels. Selected enriched motifs and putative cognate transcription factors in ATAC-Seq peaks from WT + rLCMV-core (top) or MUP-core (bottom) mice. Values between brackets indicates p-value. (d) Schematic representation of the experimental setup. (e-f) Total numbers (e) and numbers of IFN-γ-producing (f) Cor93 T cells in the livers of the indicated mice.
Our genomic analyses indicated that Ag recognition on KCs can support priming and differentiation into effector CD8+ T cells similar to those recovered from secondary lymphoid organs (Supplementary Table 5). By contrast, Ag recognition on hepatocytes initiates a defective differentiation program with progressive accumulation of chromatin and transcriptional landscape alterations that ultimately result in a dysregulated T cell phenotype.
We next looked at the plasticity of the dysfunctional Cor93 T cells recovered from MUP-core livers. When Cor93 T cells were sorted from MUP-core livers 4 hours after injection and then transferred into rLCMV-core-transduced WT mice, they were fully capable of expanding and differentiating into effector cells (Fig. 2d-f). By contrast, Cor93 T cells isolated from MUP-core livers at day 3 (a time point in which chromatin alterations are evident, Fig. 2c and Extended Data Fig. 5) and transferred into rLCMV-core-injected WT mice were significantly impaired in their ability to expand and differentiate into IFN-γ-producing cells (Fig. 2d-f). These data indicate that 3 days of hepatocellular Ag exposure are sufficient to render cells partially refractory to effector differentiation stimuli.
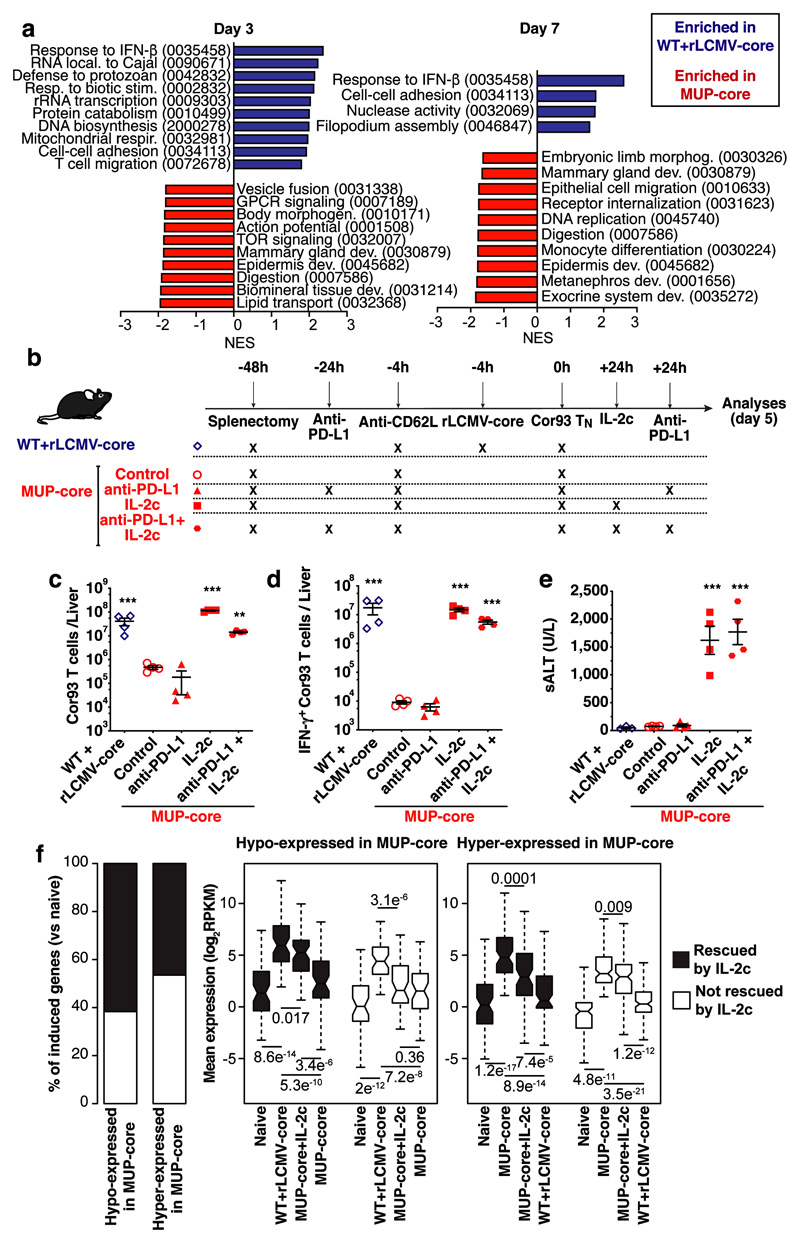
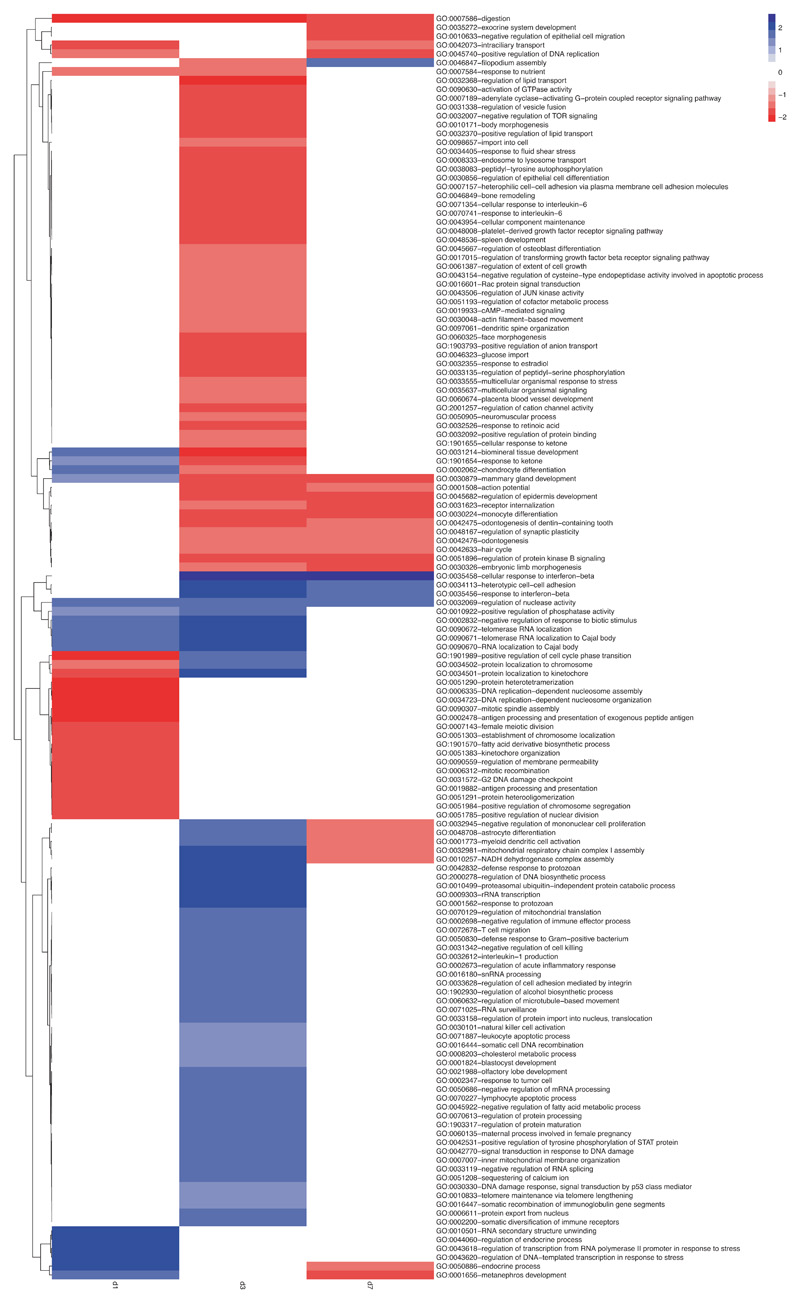
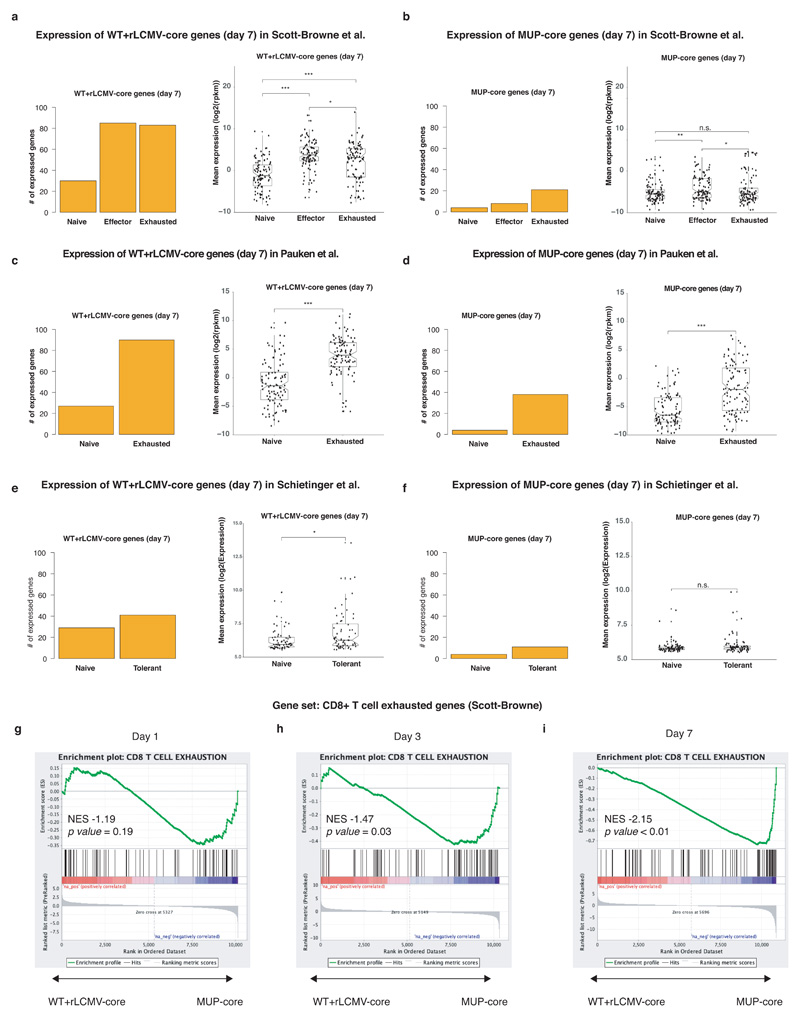
Gene Set Enrichment Analysis (GSEA) identified distinct sets of gene ontology (GO) categories in the transcriptomes of CD8+ T cells from the two groups. Genes with higher expression in CD8+ T cells from rLCMV-core-transduced WT mice were enriched in GO categories linked to effector immune responses such as response to type I interferon, cell proliferation, T cell migration and cell-cell adhesion. By contrast, CD8+ T cells from MUP-core livers failed to express genes linked to effector T cell responses beyond day 1, and instead expressed genes belonging to GO categories linked to tissue development and organ remodelling, cell differentiation and cell-matrix interaction (Fig. 3a, Extended Data Fig. 6, Supplementary Table 6). The transcriptional program of hepatic CD8+ T cells isolated from MUP-core mice at day 7 after transfer was not obviously overlapping with that of other known dysfunctional CD8+ T cell fates, as selectively represented genes in these cells were poorly expressed in reference transcriptomic datasets generated on splenic LCMV-specific exhausted CD8+ T cells30,31 or on tolerant self-Ag-specific CD8+ T cells32 (Extended Data Fig. 7, Supplementary Table 7, 8). An exhaustion-like signature30, however, was progressively enriched in the transcriptome of CD8+ T cells from MUP-core mice at day 3 and day 7 after transfer, as determined by GSEA (Extended Data Fig. 7). These data indicate that, while priming by hepatocytes initiates a unique dysfunctional program, hepatocellular Ag persistence gradually triggers an additional exhaustion profile.
Fig. 3. Intrahepatically-primed, dysfunctional CD8+ T cells can be rescued by IL-2, but not by anti-PD-L1 Abs.
(a) Normalized enrichment score (NES) of selected GO categories enriched within genes expressed at higher levels in WT + rLCMV-core (blue) or from MUP-core (red) mice at the indicated time points. GO categories were identified by GSEA42 and grouped by similarity with REVIGO43. (b) Schematic representation of the experimental setup. (c-d) Total numbers (c) and numbers of IFN-γ-producing (d) Cor93 T cells in the livers of the indicated mice. (e) ALT levels in the sera of the indicated mice. (f) Left panel. Stacked bar plot showing the effect of IL-2c on genes induced at day 5 (versus naïve) in Cor93 T cells from WT + rLCMV-core or MUP-core mice. Genes hypo-expressed or hyper-expressed in MUP-core mice as compared to WT + rLCMV-core mice are shown separately. Right panels. Box plots showing expression levels of hypo-expressed (left) or hyper-expressed (right) genes at day 5 in the indicated conditions. Genes the expression of which is rescued or not rescued in MUP-core+IL2c mice are shown in black or white, respectively.
Dysfunctional CD8+ T cells can be rescued by IL-2 but not by anti-PD-L1 Abs
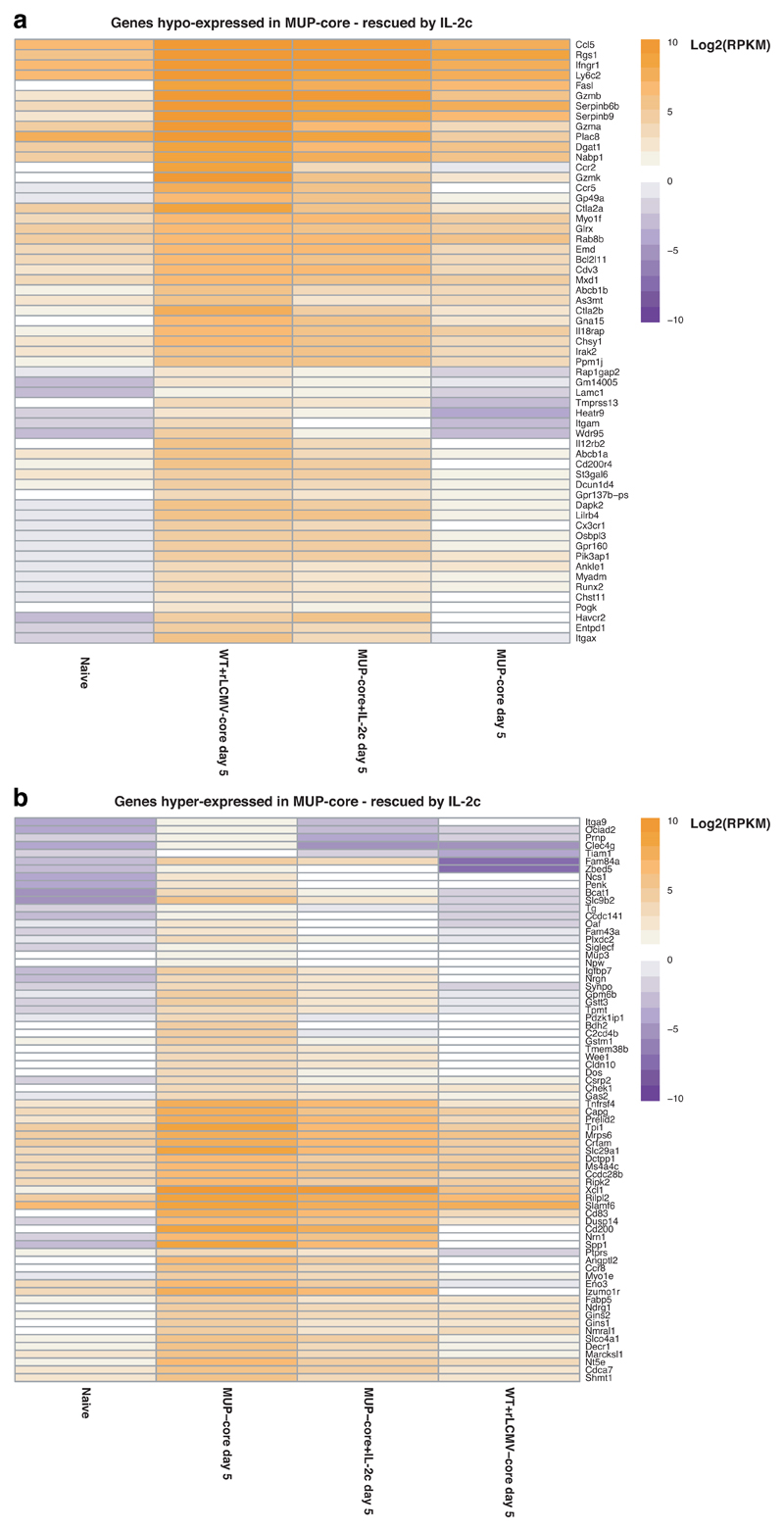
Among the genes that are differentially expressed (Fig. 2a), we focused our attention on two known regulators of T cell function: Pdcd1 and Il233–36. Pdcd1 was found to be hyper-expressed on hepatic Cor93 CD8+ T cells sorted from MUP-core mice (Fig. 2a), whereas Il2 was found to be induced in the livers of rLCMV-core-transduced WT mice as well as hyper-expressed on Cor93 CD8+ T cells sorted from the livers of rLCMV-core-transduced WT mice (Fig. 2a). We assessed the functional consequences of these findings by treating Cor93 TN-injected MUP-core mice with anti-PD-L1 blocking Abs, with recombinant IL-2 coupled with anti-IL-2 Abs (IL-2c)37, or with a combination of both (Fig. 3b). IL-2c administration promoted expansion and differentiation of Cor93 T cells into IFN-γ producing, cytotoxic effector cells (Fig. 3c-e), whereas anti-PD-L1 treatment either failed to do so when given alone or did not show a synergistic effect when given in combination with IL-2c (Fig. 3c-e). IL-2c administration 1 day after transfer of Cor93 TN into MUP-core mice substantially rescued the transcriptional program of dysfunctional CD8+ T cells, as measured by RNA-seq at day 5 (Fig. 3f, Extended Data Fig. 8, Supplementary Table 9). More than half of the genes with defective expression (hypo-expressed genes) in hepatic CD8+ T cells from MUP-core mice were upregulated in IL-2c-treated MUP-core mice, often reaching expression levels comparable to those detected in WT mice injected with rLCMV-core. Similarly, a comparable fraction of genes with higher expression (hyper-expressed genes) in hepatic CD8+ T cells from MUP-core mice were downregulated by IL-2c treatment (Fig. 3f, Extended Data Fig. 8 and Supplementary Table 9).
To assess the specificity of our treatment37,38, we co-transferred Ag-specific (Cor93) and irrelevant (Env28) TN into MUP-core mice 24 hours prior to IL-2c administration. Cor93 and Env28 TN were also transferred into control WT mice previously injected with rLCMV-core/env. IL-2c improved the capacity of Ag-specific Cor93 T cells to expand, differentiate into IFN-γ-producing cells and accumulate in clusters scattered throughout the liver lobules, but it had no effect on irrelevant Env28 TN (data not shown).
Therapeutic potential of IL-2 treatment for T cell restoration during chronic HBV infection
Next we tested the effect of IL-2 treatment in HBV replication-competent transgenic mice that were neither splenectomized nor treated with anti-CD62L blocking Abs. IL-2c administration promoted differentiation of Cor93 T cells into IFN-γ producing, cytotoxic effector cells that accumulated in clusters scattered throughout the liver lobules and exerted potent antiviral activity (Extended Data Fig. 9).
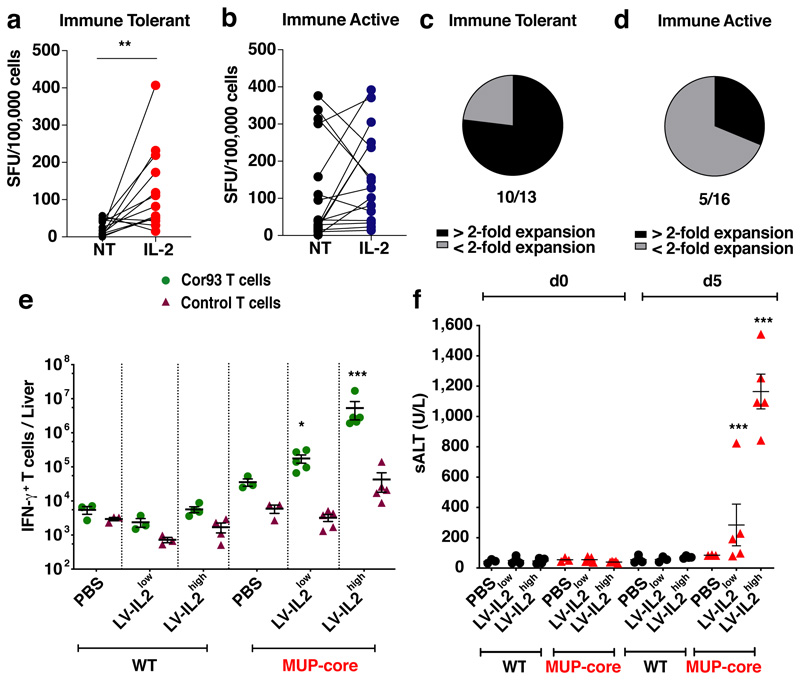
We then hypothesized that HBV-specific T cells present in “immune tolerant” (IT) patients have a different functional behaviour than those present in “immune active” (IA) patients and might be more closely related to T cells primed by hepatocytes in the mouse models. Peripheral T cells from 13 IT and 16 IA patients (Supplementary Table S10) were stimulated with overlapping HBV peptides in the presence or absence of recombinant human IL-2 and the frequency of HBV-specific T cells was determined by IFN-γ ELISpot assay. Only very low frequencies of IFN-γ-secreting cells were detected in IT patients in the absence of IL-2 (mean = 24 SFU/105 cells, Fig. 4a); IL-2 addition, however, significantly augmented their frequency in 10 out of the 13 patients (mean = 122 SFU/105 cells; Fig. 4a, c). By contrast, HBV-specific T cells from IA patients did not require and could not be boosted by IL-2 during their expansion and their frequency was similar to that of IT patients in the presence of IL-2 (Fig. 4b, d). The data suggest that HBV-specific T cells from IT patients, but not from IA patients, resemble hepatocellularly primed murine CD8+ T cells in that they can expand and secrete IFN-γ only upon IL-2 treatment. Whether IL-2 exerts an even greater effect on HBV-specific T cell restoration if administered directly to IT patients (where KCs could cross-present hepatocellular Ags) remains to be determined.
Fig. 4. Therapeutic potential of IL-2 treatment for T cell restoration during chronic HBV infection.
(a-d) HBV-specific T cell frequency from 13 Immune Tolerant (IT, a) and 16 Immune Active (IA, b) chronic HBV patients (Supplementary Table 10) cultured with or without IL-2. The percentage of IT (c) and IA patients (d) for which the HBV-specific T cell expansion increased by more than 2-fold with addition of IL-2 during the culture are shown in black. (e) Absolute numbers of IFN-γ-producing T cells in the livers of the indicated mice. (f) Serum ALT levels at day 0 and day 5 in the same mice.
To test the clinical potential of IL-2 in a system that may limit its systemic toxicity34, we generated third-generation, self-inactivating lentiviral vectors (LV.ET.mIL2.142T) that allow selective hepatocellular expression of murine IL-239. We injected WT or MUP-core mice with 2.5 x 108 (LV-IL2low) or 5 x 108 (LV-IL2high) transducing units (TU)/mouse, 7 days prior to Cor93 or control TN injection. As shown in Fig. 4e, f, lentiviral-mediated hepatic expression of IL-2, even at a dose that transduces less than 10% of hepatocytes in vivo, increased the capacity of Cor93 (but not control) T cells to expand and differentiate into IFN-γ-producing cells endowed with cytolytic capacities.
Discussion
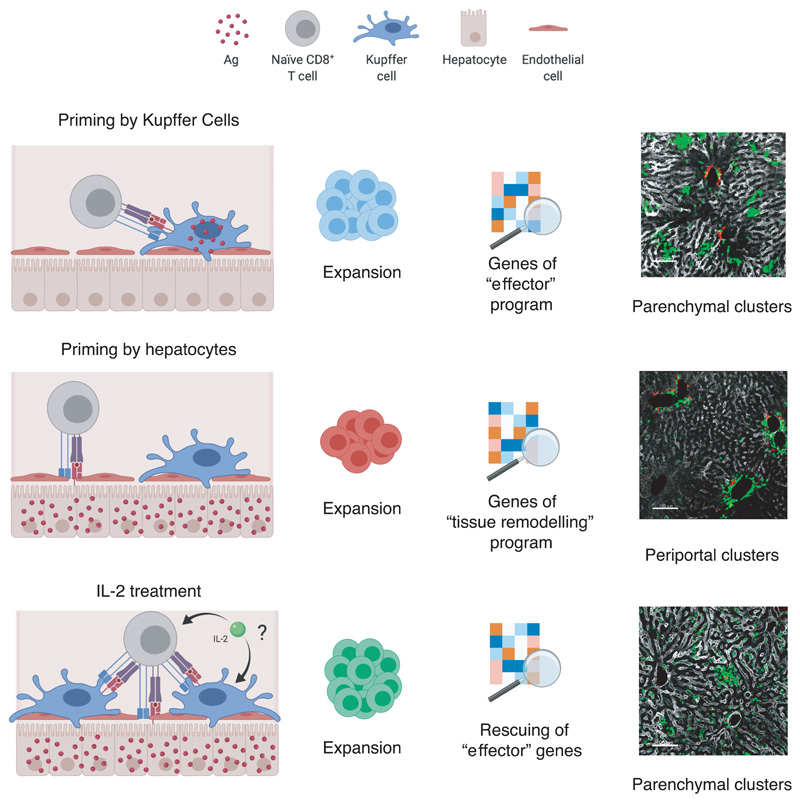
We have delineated the spatiotemporal dynamics, molecular programs and functional consequences of naïve CD8+ T cells undergoing intrahepatic priming (Extended Data Fig. 10). We showed that hepatocellular presentation leads to a CD8+ T cell dysfunction that is distinct from T cell alterations reported in other viral infections and cancer and, as such, is not readily responsive to anti-PD-L1 treatment. As immune checkpoint inhibitors are beginning to be tested in patients persistently infected with HBV, the results reported here should help interpreting the outcome of those studies and eventually inform the design of modified trials in selected cohorts of patients. Our data identify IL-2 as a potent immunotherapeutic capable of rescuing CD8+ T cells rendered dysfunctional by hepatocellular priming. Thus, IL-2-based strategies should be considered for the treatment of chronic HBV infection.
Methods
Mice
C57BL/6, CD45.1 (inbred C57BL/6), Balb/c, Thy1.1 (CBy.PL(B6)-Thya/ScrJ), β-actin-GFP [C57BL/6-Tg(CAG-EGFP)1Osb/J], β-actin-DsRed [B6.Cg-Tg(CAG-DsRed*MST)1Nagy/J], Tap1-deficient (B6.129S2-Tap1tm1Arp/J), TCR-I [B6.Cg-Tg(TcraY1, TcrbY1)416Tev/J], CD11c-DTR [B6.FVB-1700016L2RikTg(Itgax-DTR/EGFP)57Lan/J] mice were purchased from Charles River or The Jackson Laboratory. MHC-II-/- mice were obtained through the Swiss Immunological Mutant Mouse Repository (Zurich, Switzerland). MUP-core transgenic mice (lineage MUP-core 50 [MC50], inbred C57BL/6, H-2b), that express the HBV core protein in 100% of the hepatocytes under the transcriptional control of the mouse major urinary protein (MUP) promoter, have been previously described15. HBV replication-competent transgenic mice (lineage 1.3.32, inbred C57BL/6, H-2b), that express all of the HBV antigens and replicate HBV in the liver at high levels without any evidence of cytopathology, have been previously described13. In indicated experiments, MUP-core and HBV replication-competent transgenic mice were used as C57BL/6 x Balb/c H-2bxd F1 hybrids. Cor93 TCR transgenic mice (lineage BC10.3, inbred CD45.1), in which > 98% of the splenic CD8+ T cells recognize a Kb-restricted epitope located between residues 93–100 in the HBV core protein (MGLKFRQL), have been previously described12. Env28 TCR transgenic mice (lineage 6C2.36, inbred Thy1.1 Balb/c), in which ∼83% of the splenic CD8+ T cells recognize a Ld-restricted epitope located between residues 28–39 of HBsAg (IPQSLDSWWTSL), have been previously described12. For imaging experiments Cor93 and TCR-I transgenic mice were bred against both β-actin-GFP and β-actin-DsRed mice, while Env28 transgenic mice were bred against β-actin-DsRed mice that were previously back-crossed more than 10 generations against Balb/c. Bone marrow (BM) chimeras were generated by irradiation of MUP-core or C57BL/6 mice with one dose of 900 rad and reconstitution with the indicated BM; mice were allowed to reconstitute for at least 8 weeks before use. In some experiments, to achieve full reconstitution of Kupffer cells from donor-derived BM, mice were injected with 200 μl of clodronate-containing liposomes 28 and 31 days after BM injection. Mice were housed under specific pathogen-free conditions and used at 8-10 weeks of age. In all experiments, mice were matched for age, sex and (for the 1.3.32 animals) serum HBeAg levels before experimental manipulations. In selected experiments, 1.3.32 mice were matched for serum HBV DNA levels before experimental manipulations. All experimental animal procedures were approved by the Institutional Animal Committee of the San Raffaele Scientific Institute and are compliant with all relevant ethical regulations.
Viruses and Viral Vectors
Replication-incompetent lymphocytic choriomeningitis virus (LCMV)-based vectors encoding for HBV core protein, HBV envelope protein, HBV core and envelope proteins, or Cre recombinase (termed rLCMV-core, rLCMV-env, rLCMV-core/env and rLCMV-cre, respectively) were generated, grown and titrated as described16. Mice were injected i.v. with 2.5 x 105 infectious units of the indicated rLCMV vector 4 hours prior CD8+ T cells injection.
Adeno-associated viruses expressing GFP and HBV core protein (AAV-core-GFP) have already been described3. Mice were injected with 3 x 1010 or 3 x 1011 viral genomes (vg) of AAV-core-GFP 15 days prior to further experimental manipulation.
Third-generation, self-inactivating lentiviral vectors (LV.ET.mIL2.142T) that allow expression of murine IL-2 exclusively in hepatocytes thanks to the presence of a synthetic hepatocyte-specific promoter/enhancer as well as specific microRNA 142 target sequences that suppress expression in hematopoietic-lineage cells39 were generated, produced and titrated as described44. Briefly, the gene-synthesized murine interleukin 2 (mIL-2) cDNA was cloned into the previously described transfer vector pCCLsin.cPPT.ET.GFP.142T44 by standard cloning techniques. Third-generation LVs were produced by calcium phosphate transient transfection of 293T cells of the transfer vector, the packaging plasmid pMDLg/p.RRE, pCMV.REV, the vesicular stomatitis virus glycoprotein G (VSV-G) envelope plasmid pMD2.G and the pAdvantage plasmid (Promega), as previously described44. For integrase-defective lentiviral vector (IDLV) production, the pMDLg/p.RRE.D64Vint packaging with a mutant integrase was used instead of pMDLg/p.RRE, as described45. Briefly, 9x106 293T cells were seeded 24 hours before transfection in 15-cm dishes. Two hours before transfection culture medium was replaced with fresh medium. For each dish, a solution containing a mix of the selected transfer plasmid, the packaging plasmids pMDLg/pRRE and pCMV.REV, pMD2.G and the pAdvantage plasmid was prepared using 35, 12.5, 6.25, 9 and 15 μg of plasmid DNA, respectively. A 0.1X TE solution (10 mM Tris-HCl, 1 mM EDTA pH 8.0 in dH20) and water (1:2) was added to the DNA mix to 1,250 μL of final volume. The solution was left on a spinning wheel for 20-30 minutes, then 125 μl of 2.5M CaCl2 were added. Right before transfection, a precipitate was formed by adding 1,250 μL of 2X HBS (281 mM NaCl, 100 mM HEPES, 1.5 mM Na2HPO4, pH 7.12) while the solution was kept in agitation on a vortex. The precipitate was immediately added to the culture medium and left on cells for 14-16 hours and after that the culture medium was changed. Supernatant was collected 30 hours after medium change and passed through a 0.22 μm filter (Millipore). Filtered supernatant was transferred into sterile 25 x 89 mm polyallomer tubes (Beckman) and centrifuged at 20,000g for 120 min at 20°C (Beckman Optima XL-100K Ultracentrifuge). Vector pellet was dissolved in the appropriate volume of PBS to allow a 500X concentration. For LV titration, 105 293T cells were transduced with serial vector dilutions in the presence of polybrene (16 μg/ml). Genomic DNA (gDNA) was extracted 14 days after transduction. gDNA was extracted by using Maxwell 16 Cell DNA Purification Kit (Promega) according to manufacturer’s instructions. Vector copies per diploid genome (vector copy number, VCN) were quantified by quantitative PCR (qPCR) starting from 100 ng of template gDNA using primers (HIV sense: 5’-TACTGACGCTCTCGCACC-3’; HIV antisense: 5’-TCTCGACGCAGGACTCG-3’) and a probe (FAM 5’-ATCTCTCTCCTTCTAGCCTC-3’) designed to amplify the primer binding site region of LV. Endogenous DNA amount was quantified by a primers/probe set designed to amplify the human telomerase gene (Telo sense: 5’-GGCACACGTGGCTTTTCG-3’; Telo antisense: 5’-GGTGAACCTCGTAAGTTTATGCAA-3’; Telo probe: VIC 5’-TCAGGACGTCGAGTGGACACGGTG-3’ TAMRA). Copies per genome were calculated by the formula = [ng LV/ng endogenous DNA] x [n° of LV integrations in the standard curve]. The standard curve was generated by using a CEM cell line stably carrying 4 vector integrants, which were previously determined by Southern blot and FISH analysis. All reactions were carried out in duplicate or triplicate in an ABI Prism 7900HT or Viia7 Real Time PCR thermal cycler (Applied Biosystems). Each qPCR run carried an internal control generated by using a CEM cell line stably carrying 1 vector integrant, which were previously determied by Southern blot and FISH analysis. Titre is expressed as transducing units293T (TU)/mL and calculated using the formula TU/mL = [VCNx105x1/dilution factor]. IDLV titre was determined on 293T cells 3 days after transduction using an ad hoc qPCR, which selectively amplifies the reverse transcribed vector genome (both integrated and non-integrated) discriminating it from plasmid carried over from the transient transfection (RT-LV; ΔU3 sense: 5’-TCACTCCCAACGAAGACAAGATC-3’, gag antisense: 5’ GAGTCCTGCGTCGAGAGAG-3’). Vector particles were measured by HIV-1 Gag p24 antigen immunocapture assay (Perkin Elmer) according to manufacturer’s instructions. Vector infectivity was calculated as the ratio between titre and particles. Vector administration was carried out by tail vein injection in mice at 2.5-10x108 TU/mouse, 7 days prior to T cell injection.
All infectious work was performed in designated BSL-2 or BSL-3 workspaces, in accordance with institutional guidelines.
Naïve T Cell Isolation, Adoptive Transfer and In Vivo Treatments
CD8+ T cells from the spleens of Cor93, Env28, TCR-I transgenic mice were purified by negative immunomagnetic sorting (Miltenyi Biotec). Mice were adoptively transferred with 2-5 x 106, 2 x 105 or 2 x 104 CD8+ T cells. In selected experiments, mice were splenectomized and treated with 200 μg of anti-CD62L mAb (clone MEL-14, BioXcell) 48 hours and 4 hours prior to cell injection, respectively. Splenectomy was performed according to standard procedures46. In selected experiments, CD4+ T cells were depleted by injecting i.v. 200 μg of anti-CD4 Ab (clone GK1.5, BioXcell) 3 days and 1 day prior to T cell transfer. In selected experiment, mice were treated with 200 μg of anti-PD-L1 (Clone 10F.9G2, BioXcell) 1 day before and 1 day and 3 days after T cell transfer. In some experiments, Tregs were depleted by injecting i.p. 200 μg of purified anti-CD25 mAbs (clone PC61, BioXcell) 8 days prior to T cell transfer. In selected experiments, WT or MUP-core mice were lethally irradiated and reconstituted with BM from CD11c-DTR mice; dendritic cells were subsequently depleted by injecting i.p. 25 ng/g of diphtheria toxin (Sigma) 3 days and 1 day prior to T cell transfer. In indicated experiments, Kupffer cells were depleted by intravenous injection of clodronate-containing liposomes 2 days prior to T cell injection, as described21. IL-2/anti-IL-2 complexes (IL-2c) were prepared by mixing 1.5 μg of rIL-2 (BioLegend) with 50 μg anti-IL-2 mAb (clone S4B6-1, BioXcell) per mouse, as previously described37. Mice were injected with IL-2c i.p. one day after T cell transfer, unless otherwise indicated.
Cell Isolation and Flow Cytometry
Single-cell suspensions of livers, spleens, lymph nodes, bone marrow, lung and blood were generated as described47,48. Kupffer cell isolation was performed as described3,49. All flow cytometry stainings of surface-expressed and intracellular molecules were performed as described3. Cell viability was assessed by staining with Viobility™ 405/520 fixable dye (Miltenyi). Abs used included: anti-CD3 (clone: 145-2C11, Cat#562286, BD Biosciences), anti-CD11b (clone: M1/70, Cat#101239), anti-CD19 (clone: 1D3, Cat#562291 BD Biosciences), anti-CD25 (clone: PC61, Cat#102015), anti-CD31 (clone: 390, Cat#102427), anti-CD45 (clone: 30-F11, Cat#564279 BD Biosciences), anti-CD49b (clone: DX5, Cat#562453 BD Biosciences), anti-CD64 (clone: X54-5/7.1, Cat#139311), anti-F4/80 (clone: BM8, Cat#123117), anti-I-A/I-E (clone: M5/114.15.2, Cat#107622), anti-TIM4 (polyclonal, Cat#orb103599 Biorbyt), anti-CD69 (clone: H1.2F3, Cat#104517), anti-CD45.1 (clone: A20, Cat#110716), anti-IFN-γ (clone: XMG1.2, Cat#557735 BD Biosciences), anti-CD4 (clone: RM4-5, Cat#553048 BD Biosciences), anti-CD11c (clone: N418, Cat#117308), anti-I-Ab (clone: AF6-120.1, Cat#116420), anti-PD-1 (clone: J43, Cat#17-9985 eBioscience), anti-NK1.1 (clone: PK136, Cat# 108706), anti-NKp46 (clone: 29A1.4, Cat# 137623), anti-Stat5 pY694 (clone: 47, Cat# 560117 BD Biosciences), anti-Foxp3 (clone FJK-16s, Cat# 12-5773-80 eBioscience). All Abs were purchased from BioLegend, unless otherwise indicated. Recombinant dimeric H-2Ld:Ig and H-2Kb:Ig fusion proteins (BD Biosciences) complexed with peptides derived from HBsAg (Env28-39) or from HBcAg (Cor93-100), respectively, were prepared according to the manufacturer’s instructions. Dimer staining was performed as described47. Flow cytometry staining for phosphorylated STAT5 was performed using Phosflow™ Perm Buffer III (Cat# 558050, BD Bioscience), following the manufacturer’s instructions. Flow cytometry staining for Foxp3 was performed using Foxp3 / Transcription Factor Staining Buffer Set (Cat# 00-5523-00, eBioscience), following the manufacturer’s instructions.
All flow cytometry analyses were performed in FACS buffer containing PBS with 2 mM EDTA and 2% FBS on a FACS CANTO or LSRII (BD Biosciences) and analysed with FlowJo software (Treestar).
Cell Sorting
Single-cell suspensions from spleens and livers were stained with Viobility 405/520 fixable dye (Miltenyi), with PB-conjugated anti-CD8α (clone 53-6.7) and PE-conjugated anti-CD45.1 Abs. Live CD8+ CD45.1+ cells were sorted on a MoFlo Legacy (Beckman Coulter) cell sorter in a buffer containing PBS with 2% FBS. Cells were always at least 98% pure (data not shown).
RNA Purification and RNA-seq Library Preparation
Total RNA was purified from 8,000-300,000 sorted cells with the ReliaPrep RNA Cell Miniprep System (Promega). Sequencing libraries were generated using the Smart-seq2 method50. Briefly, 5 ng of RNA were retrotranscribed and cDNA was amplified using 15 PCR cycles and purified with AMPure XP beads (Beckman Coulter). After purification, the concentration was determined using Qubit 3.0 (Life Technologies) and the size distribution was assessed using Agilent 4200 TapeStation system. Then, the tagmentation reaction was performed starting from 0.5 ng of cDNA for 30 minutes at 55°C and the enrichment PCR was carried out using 12 cycles. Libraries were then purified with AMPure XP beads, quantified using Qubit 3.0 and single-end sequenced (75 bp) on an Illumina NextSeq 500.
RNA-Seq Data Processing and Analysis
Reads were generated on a NextSeq 500 (Illumina) instrument following the manufacturer’s recommendations. Single end reads (75bp) were aligned to the mm10 reference genome using STAR51 aligner. featureCounts function from Rsubread package52 was used to compute reads over RefSeq Mus musculus transcriptome, with option minMQS set to 255. Further analyses were performed with edgeR R package53. Pearson’s correlation was computed for each couple of samples on log transformed RPKM. Read counts were normalized with the Trimmed Mean of M-values (TMM) method54 using calcNormFactors function and dispersion was estimated with the estimateDisp function. Differential expression across different conditions was evaluated fitting a negative binomial generalized linear model on the dataset with glmQLFit function and then performing a quasi-likelihood (QL) F-test with glmQLFTest function. Batch information was included in the design as covariate.
Differential Gene Expression Analysis
Hepatic CD8+ T cells from WT mice injected with rLCMV-core versus MUP-core mice
Genes with an RPKM (Reads Per Kilobase per Million) value higher than 1 in at least two samples in the datasets were retained. We first defined inducible genes, namely those genes with log2FCRPKM >2.5 and FDR<0.01 relative to naïve T cells in at least one condition or time point. For each comparison, only genes with an RPKM value higher than 1 in at least two samples in the comparison were selected. For each time point, induced genes were classified as expressed at higher levels in the WT + rLCMV-core condition setting FDR < 0.1 and log2FCRPKM > 1.5 (WT + rLCMV-core vs MUP-core) as cut-offs. Genes with an FDR < 0.1 and a log2FCRPKM < -1.5 in the WT + rLCMV-core vs MUP-core comparison were classified as expressed at higher levels in MUP-core. The remaining genes were defined as non-differentially expressed between WT + rLCMV-core and MUP-core.
Hepatic or splenic CD8+ T cells from WT mice injected with rLCMV-core versus Cor93 TN
We first defined as expressed genes those having CPM > 1 in at least two samples in the dataset. Induced genes were defined using as cut-offs a log2FCRPKM > 2.5 and FDR < 0.01 relative to naïve T cells in at least one condition or time point. For each comparison, only genes with an RPKM value higher than 1 in at least two samples in the comparison were selected.
Hepatic CD8+ T cells from WT mice injected with rLCMV-core versus MUP-core mice with or without IL-2c treatment
We first defined as expressed genes those having CPM > 1 in at least two samples in the dataset. Induced genes were defined using as cut-offs a log2FCRPKM > 2.5 and FDR < 0.01 relative to naïve T cells in at least one condition. For each comparison, only genes with an RPKM value higher than 1 in at least two samples in the comparison were selected. Induced genes were then classified as expressed at higher levels in the WT + rLCMV-core condition (hypo-expressed in MUP-core at day 5) setting log2FCRPKM > 1.5 and FDR < 0.01 (WT + rLCMV-core vs MUP-core) as cut-off. Genes with a log2FCRPKM < -1.5 and FDR < 0.01 in the WT + rLCMV-core vs MUP-core comparison were classified as expressed at higher levels (hyper-expressed in MUP-core at day 5) in MUP-core. We then classified genes hypo-expressed in MUP-core as rescued if they displayed log2FCRPKM > 1 and FDR < 0.01 in the IL-2c-treated MUP-core versus MUP-core comparison. Conversely, genes hyper-expressed in MUP-core were defined as rescued if displaying log2FCRPKM < -1 and FDR < 0.01 in the IL-2c-treated MUP-core versus MUP-core comparison. The remaining genes were classified as not rescued.
Gene Ontology (GO) Analyses
For each time point, we ranked expressed genes by decreasing order of log2FCRPKM values in the WT+rLCMV-core versus MUP-core comparison. We then performed Gene set enrichment analysis (GSEA)42 on each of these ranked lists using the clusterProfiler R package55 and the Gene Sets contained in the Biological Processes ontology from the org.Mm.eg.db database. GO categories with q-value < 0.1 were retained and aggregated using REVIGO43 (similarity score=0.7), yielding 143 seed GO categories showing enrichment in WT+rLCMV-core or in MUP-core in at least one time point.
Gene Expression Analysis in Published Datasets
RNA-seq/SRA data were downloaded from the Gene Expression Omnibus repository (GEO) and converted to the FastQ format. Reads were then aligned against the whole Mus musculus mm10 genome build using STAR aligner (v 2.6.0a) with default options, generating BAM files. Read counts for all expressed genes (Ensembl annotation v94; GENCODE M19) were obtained using featureCounts (Rsubread v 3.7). Features with < 1 CPM were filtered out. The resulting count matrix was then normalized using the normalization factors generated by the upperquartile method56 implemented in edgeR Bioconductor package. Hierarchical cluster analysis was performed on RPKM values. The similarity of the samples was measured using the Pearson correlation coefficient and the complete-linkage was used as the distance measure of the agglomerative hierarchical clustering. For Illumina BeadChip data, the normalized expression matrix was downloaded from the GEO repository. Genes whose expression level corresponded to the 65th percentile of the distribution of the log2 expression values were considered to be expressed.
ATAC-seq
ATAC (Assay for Transposase Accessible Chromatin)-seq was performed as described57 with slight modifications. Briefly, 8,000-50,000 cells per sample were sorted and centrifuged at 1,600 rpm for 5 minutes. Then, the transposition reaction was performed using digitonin 1% (Promega), Tn5 transposase and TD Buffer (Illumina) for 45 minutes at 37°C. Immediately following transposition, the reaction was stopped using a solution of 900 mM NaCl and 300 mM EDTA, 5% SDS and Proteinase K (Sigma-Aldrich) for 30 minutes at 40°C. Transposed DNA fragments were purified using AMPure XP beads (Beckman Coulter), barcoded with dual indexes (Illumina Nextera) and PCR amplified with KAPA HiFi PCR Kit (KAPA Biosystems). Then, the concentration of the library was determined using Qubit 3.0 (Life Technologies) and the size distribution was assessed using Agilent 4200 TapeStation system. Libraries were single-end sequenced (75 bp) on an Illumina NextSeq 500.
ATAC-seq Data Processing and Analysis
Reads were generated on NextSeq 500 (Illumina) instrument following manufacturer’s recommendations. Single end reads (75bp) were aligned to the mm10 reference genome using BWA58 aligner. Bam files were processed using samtools59 and BEDTools60 suits: reads with a mapping quality lower then 15 or duplicated were discarded. Moreover, unassigned reads and reads mapped on chromosomes Y and M were removed. MACS261 callpeak function with parameters -g mm --nomodel --shift -100 --extsize 200 was used for peak calling. For each sample peaks with a q-value lower than 1e-10 were selected. Peaks from all samples that passed filter were then merged with mergeBed function form BEDTools, resulting in 72884 regions. Reads counts were computed on this set of regions using coverageBed function from BEDTools. The set of 72884 regions was annotated using ChIPpeakAnno R package62. Each region was associated to the gene with the closest TSS. Further analyses were performed with edgeR R package. Pearson’s correlation was computed for each pair of samples on log transformed CPM. As previously described for RNA-seq data, read counts were normalized with the TMM method using calcNormFactors function and dispersion was estimated with the estimateDisp function. Differences in peaks intensities across different conditions were evaluated fitting a negative binomial generalized linear model on the dataset with glmQLFit function and then performing a quasi-likelihood (QL) F-test with glmQLFTest function. Batch information was included in the design as covariate.
Definition of Induced and Differentially Induced ATAC-seq peaks
We first defined inducible peaks, namely those regions with log2FCCPM>2.5 and FDR<0.001 relative to naïve T cells in at least one condition or time point. For each time point, induced peaks were classified as induced at higher levels in the WT + rLCMV-core condition setting FDR < 0.1 and log2FCCPM > 1.5 (WT + rLCMV-core vs MUP-core) as cut-offs. Peaks with an FDR < 0.1 and a log2FCCPM < -1.5 in the WT + rLCMV-core vs MUP-core comparison were classified as induced at higher levels in MUP-core. The remaining peaks were defined as non-differentially induced between WT + rLCMV-core and MUP-core.
Motif Enrichment Analysis
Enrichment analysis of known motifs was performed with HOMER41 using findMotifsGenome.pl script. For each time point we ranked ATAC-seq peaks according to log2FCCPM values in the WT + rLCMV-core versus MUP-core comparison and selected the 200 regions showing highest or lowest log2FCCPM values. These sets of differentially induced regions were compared to a background composed by a set of 3899 regions with unchanged intensities (FDR > 0.1 and abs(log2FC) < 0.5) between both MUP-core and WT + rLCMV-core versus naïve in all time points.
Purification of viral nucleic acids from serum
Twenty μl of serum were incubated for 2 hours at 37°C with 180 μl IsoHi Buffer (150 mM NaCl, 0.5% NP40, 10 mM Tris pH 7.4), 5 mM CaCl2, 5 mM MgCl2, 1U DNaseI (Life Technologies), 5U Micrococcal Nuclease (Life Technologies). The digestion was stopped by the addition of 20 mM EDTA pH 8.0 and viral nucleic acid purification performed with the QIAmp MiniElute Virus Spin Kit (Qiagen, Cat #57704), according to the manufacturer’s instructions.
RT-qPCR
Total RNA was extracted from frozen livers using ReliaPrep™ RNA Tissue Miniprep System (Promega), according to the manufacturer’s instructions, as described63, genomic DNA contamination was removed using Ambion® TURBO DNA-freeTM DNase. 1 μg of total RNA was reverse transcribed with Superscript IV Vilo (Life Technologies) prior to qPCR analysis for mouse il2 (TaqMan Mm00434256, Life Technologies), ifng (TaqMan Mm01168134, Life Technologies), HBV core (forward TACCGCCTCAGCTCTGTATC, reverse CTTCCAAATTAACACCCACCC, probe TCACCTCACCATACTGCACTCAGGCAA). Reactions were run and analysed on Quant Studio 5 instrument (Life Technologies). For viremia quantification, a standard curve was drawn using plasmid DNA. All experiments were performed in triplicate and normalized to the reference gene GAPDH.
Western blot analysis
Western blot analysis on frozen liver homogenates or on KCs was performed exactly as described64. Primary Abs include anti-STAT5 and anti-pSTAT5 (Tyr694) (rabbit; Cell Signaling #8215), anti-HBcAg (polyclonal, Dako), β-Actin (polyclonal; Abcam #ab228001) and H3 (polyclonal; abcam #ab1791). Secondary Ab include horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch). Reactive proteins were visualized using a Clarity Western ECL substrate kit (Bio-Rad), and exposure was performed using UVItec (Cambridge MINI HD, Eppendorf). Images were acquired by NineAlliance software. Band quantification was performed with ImageJ software on 16-bit images and normalized on the matching housekeeping protein as a loading control. Each lane corresponds to a different mouse.
Southern blot analysis
Southern blot analysis on total DNA isolated from frozen livers (left lobe) was performed exactly as described63.
Confocal Immunofluorescence Histology and Histochemistry
Confocal microscopy analysis of livers was performed as described3. The following primary Abs were used for staining: anti-F4/80 (BM8, Invitrogen), anti-cytokeratin 7 (EPR17078, Abcam), anti-Lyve-1 (NB600-1008, Novus Biological), anti-HBcAg (polyclonal, Dako). The following secondary Abs were used for staining: Alexa Fluor 488-, Alexa Fluor 514-, Alexa Fluor 568-, or Alexa Fluor 647-conjugated anti-rabbit or anti-rat IgG (Life Technologies). Images were acquired on an inverted Leica microscope (TCS STED CW SP8, Leica Microsystems) with a motorized stage for tiled imaging. To minimize fluorophore spectral spillover, we used the Leica sequential laser excitation and detection modality. The bleed-through among sequential fluorophore emission was removed applying simple compensation correction algorithms to the acquired images. The semiautomatic surface-rendering module in Imaris (Bitplane) was used to create 3D volumetric surface objects corresponding either to individual cells or to the liver vascular system. Signal thresholds were determined using the Imaris Surface Creation module, which provides automatic threshold. T cells were tracked manually for single cell distance from the centre of each bile duct (CK7+) using Fiji.
For H&E and HBcAg immunohistochemistry, livers were perfused with PBS, harvested in Zn-formalin and transferred into 70% ethanol 24 hours later. Tissue was then processed, embedded in paraffin and stained as previously described3. Bright-field images were acquired through an Aperio Scanscope System CS2 microscope and an ImageScope program (Leica Biosystem) following the manufacturer’s instructions.
Intravital Multiphoton Microscopy
Liver intravital multiphoton microscopy was performed as described3,65. Liver sinusoids were visualized by injecting nontargeted Quantum Dots 655 (Invitrogen) i.v. during image acquisition. Images were acquired with a LaVision BioTec TriMScope II coupled to a Nikon Ti-U inverted microscope enclosed in a custom-built environmental chamber (Life Imaging Services) that was maintained at 37-38°C with heated air. Continuous body temperature monitoring through a rectal probe was performed to ensure that a narrow range of 37–38°C was maintained at all times. Fluorescence excitation was provided by two tuneable femtosecond (fs)- pulsed Ti:Sa lasers (680–1080 nm, 120 fs pulse-width, 80 MHz repetition rate, Ultra II, Coherent), an Optical Parametric Oscillator (1000–1600 nm, 200 fs pulse-width, 80 MHz repetition rate, Chameleon Compact OPO, Coherent). The setup includes four non-descanned photomultiplier tubes (Hamamatsu H7422-40 GaAsP High Sensitivity PMTs and Hamamatsu H7422-50 GaAsP High Sensitivity red-extended PMT from Hamamatsu Photonics K.K.), a 25X, 1.05 NA, 2 mm working distance, water-immersion multiphoton objective (Olympus). For 4D analysis of cell migration, stacks of 7-15 square xy sections (512 × 512 pixel) sampled with 4 μm z spacing were acquired every 5-32 s for up to 2 hours, to provide image volumes that were 40 μm in depth and with an xy field of view variable between 100×100 μm2 and 450×450 μm2. Sequences of image stacks were transformed into volume-rendered, 4D time-lapse movies with Imaris (Bitplane). The 3D positions of the cell centroids were segmented by semi-automated cell tracking algorithm of Imaris. The semiautomatic surface-rendering module in Imaris (Bitplane) was used to create 3D volumetric surface objects corresponding either to individual cells or to the liver vascular system. Signal thresholds were determined using the Imaris Surface Creation module, which provides automatic threshold.
Biochemical Analyses
The extent of hepatocellular injury was monitored by measuring sALT activity at multiple time points after treatment, as previously described3.
Patients and Study Approval
A total of 34 patients with chronic HBV infection (HBsAg+) were included. The patients were subdivided into the disease categories Immune Tolerant (IT), Immune Active (IA) based on their clinical history (see legend to Supplementary Table 10). Briefly, the 13 IT patients had no history of hepatitis (normal ALT) and are all positive for HBeAg. The 16 IA patients (5 HBeAg+, 12 HBeAg-) have or had previously signs of hepatic inflammation (ALT > 40 IU/L), six of them are currently or were previously treated with nucleoside analogues. Supplementary Table 10 summarizes the available clinical and virological parameters. Blood donors were recruited from the viral hepatitis clinic at The Royal London Hospital. Written informed consent was obtained from all subjects. The study was conducted in accordance with the Declaration of Helsinki and approved by the Barts and the London NHS Trust local ethics review board and the NRES Committee London–Research Ethics Committee (reference 10/H0715/39) and by the Singapore National Healthcare Group ethical review board (DSRB 2008/00293).
Clinical and Virological Parameters
On recruitment to the study, viral serology and HBV DNA levels were tested. HBsAg, HBeAg and anti-HBe levels were measured with a chemiluminescent microparticle immunoassay (CMIA; Architect Assay, Abbott Diagnostics). HBV DNA levels in serum were quantified by real-time PCR (COBAS AmpliPrep/COBAS TaqMan HBV test v2.0; Roche Molecular Diagnostics) and HBV genotyping was performed by restriction fragment length polymorphism analysis of a pre-S amplicon, as described66.
HBV Peptide Library
Three libraries of 311-313 15-mer peptides overlapping by 10 amino acids were used to identify HBV-specific T cells. The peptides covered the entire sequence of HBV genotypes B, C, and D (GenBank AF121243, AF 112063, AF 21241, respectively) and were purchased from Mimotopes. The purity of the peptides was above 80%, and their composition was confirmed by mass spectrometry analysis. Peptides were pooled as previously described67. The peptide libraries were matched to the HBV genotype of each patient as indicated in Supplementary Table 10. For patients infected with HBV genotype A or E, the peptide library of genotype D was used.
PBMC Isolation and T cell culture
PBMCs were isolated from peripheral blood by Ficoll gradient and cryopreserved. Cells were thawed, and T cell lines were generated as follows: 20% of PBMCs were pulsed with 10 μg/ml of the overlapping HBV peptides for 1 hour at 37°C, subsequently washed, and cocultured with the remaining cells in AIM-V medium (Gibco; Thermo Fisher Scientific) supplemented with 2% AB human serum (Gibco; Thermo Fisher Scientific). T cell lines were cultured for 10 days, with or without the presence of 20 U/ml of recombinant IL-2 (R&D Systems).
ELISpot Assays
ELISpot assays for the detection of IFN-γ–producing cells were performed on in vitro expanded T cell lines using HBV peptides pooled into the following mixtures: X, core, envelope 1 (env 1), env 2, polymerase 1 (pol 1), pol 2, pol 3, pol 4. T cell lines were incubated overnight at 37°C with pools of HBV peptides (1 μg/ml), where final DMSO concentrations did not exceed 0.2%. Medium was supplemented as before with or without 20 U/ml of recombinant IL-2. IFN-γ ELISpot assays (Millipore) were performed as previously described67.
Statistical Analyses
Results are expressed as mean ± SEM. All statistical analyses were performed in Prism (GraphPad Software). Statistical tests, sample size and p values used for Figures:
-
Fig. 1. (b) n (4h and d3) = 4 (WT), 7 (WT + rLCMV-core and MUP-core); n (d5) = 3 (WT), 13 (WT + rLCMV-core and MUP-core); n (d7) = 4 (WT), 6 (WT + rLCMV-core), 10 (MUP-core). (c) n (4h-d3-d7) = 3; n (d5) = 3 (WT), 6 (WT + rLCMV-core), 7 (MUP-core); (d) n = 3 (f) n = 613 tracks (WT+rLCMV) and 156 tracks (MUP-core) (g) n = 4.
Data are representative of at least 3 independent experiments. Results are expressed as mean ± SEM. p-values were calculated using two-tailed t-test or one-way ANOVA with Bonferroni post-test. * p value < 0.05, ** p value < 0.01, *** p value < 0.001
-
Fig. 2. a) Differential gene expression was evaluated fitting a negative binomial generalized linear model on the dataset and then performing a quasi-likelihood (QL) F-test. The Benjamini-Hochberg procedure was applied in order to correct for multiple tests. Sample size: naïve (n=2), WT+rLCMV-core (n=3), MUP-core (day1 and day3, n=2; day7, n=3). c) Motif enrichment was calculated using cumulative binomial distributions. Sample size: naïve (n=2), WT+rLCMV-core (day 1 and day 7, n=2; day 3, n=3), MUP-core (day1 and day3, n=2; day7, n=3).
(e-f) n = 4.
Results are expressed as mean ± SEM. Means among groups were compared with one-way ANOVA with Bonferroni post-test.
Data are representative of at least 3 independent experiments. ** p value < 0.01.
-
Fig. 3. (c-d) n = 4. Results are expressed as mean ± SEM. p-values were calculated using one-way ANOVA with Bonferroni post-test. (e) n = 3 (WT+rLCMV) or 4 (all other groups). Results are expressed as mean ± SEM. p-values were calculated using one-way ANOVA with Bonferroni post-test. (f) For each boxplot the median (horizontal line), the upper quartile and the lower quartile (dashed lines) outside the interquartile range are reported. Results of two-sided Wilcoxon Rank Sum Test are shown for the indicated comparison. Sample size: naïve (n=2), WT+rLCMV-core (n=3), MUP-core (day1 and day3, n=2; day7, n=3)
Data are representative of at least 2 independent experiments. ** p value < 0.01, *** p value < 0.001
-
Fig. 4. (a-d) T cells from 13 Immune Tolerant (IT) and 16 Immune Active (IA) chronic HBV patients (Supplementary Table 10). Samples were compared using Wilcoxon matched-pairs signed rank test. (e-f) n = 3 (WT + PBS, WT+LV-IL-2low and MUP-core+PBS), 4 (WT+LV-IL-2high) or 5 (MUP-core+LV-IL-2low and MUP-core+LV-IL-2high). Results are expressed as mean ± SEM. Means among groups were compared with one-way ANOVA with Bonferroni post-test.
Data are representative of at least 3 independent experiments. * p value < 0.05, ** p value < 0.01, *** p value < 0.001
Extended Data
Extended Data Fig. 1. Naïve CD8+ T cells that recognize hepatocellular Ag are activated locally and expand, but fail to develop effector function.
(a) Schematic representation of the experimental setup. 5 x 106 Env28 TN were transferred into C57BL/6 x Balb/c F1 (WT) or HBV replication-competent transgenic (HBV Tg, C57BL/6 x Balb/c F1) recipients. Livers were collected and analysed five days after Env28 TN transfer and sera from the same mice were collected every day from day 0 to day 5 after Env28 TN transfer. (b-c) Absolute numbers (b) and frequency of IFNγ-producing (c) Env28 T cells in the livers of the indicated mice. (d) ALT levels detected in the sera of the indicated mice at the indicated time points. n = 4. Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test (e) Schematic representation of the experimental setup. 5 x 106 Cor93 TN were transferred into C57BL/6 (WT) or MUP-core recipients. Mice were splenectomized and treated with anti-CD62L 48 hours and 4 hours prior to cell transfer, respectively. Untreated WT mice that received 5 x 106 Cor93 TN were used as controls. Where indicated, mice were injected with 2.5 x 105 infectious units of non-replicating rLCMV-core 4 hours prior to Cor93 TN transfer. Liver-draining lymph nodes68 (dLN) and non-draining inguinal lymph nodes (ndLN) were collected at four hours and one day after Cor93 TN. (f) Representative flow cytometry plot at four hours upon Cor93 TN transfer. Numbers indicate the percentage of cells within the indicated gate. (g-h) Quantification of the absolute numbers of cells recovered from the ndLN (g) and dLN (h) of the indicated mice four hours and one day upon Cor93 TN transfer. n = 3. Results are expressed as mean ± SEM. Means among groups were compared with one-way ANOVA with Bonferroni post-test (i) Confocal immunofluorescence micrographs of liver sections from WT mice (WT), rLCMV-core-injected WT mice (WT + rLCMV-core), MUP-core mice and R26-ZsGreen mice injected with 2.5 x 105 infectious units of non-replicating rLCMV-cre (R26-ZsGreen + rLCMV-cre). Scale bars represent 100 μm. Note that, because HBV core protein did not accumulate at detectable levels in KCs and hepatic dendritic cells [DCs] upon rLCMV-core injection, we confirmed the tropism of this vector by injecting rLCMV-cre into R26-ZsGreen mice – these mice express the fluorescent protein ZsGreen upon Cre-mediated recombination. (j) Mean Fluorescent Intensity (MFI) of CD69 expression on Cor93 T cells in the liver, blood, lung and bone marrow of the indicated mice four hours after Cor93 TN transfer. n = 4. Results are expressed as mean ± SEM. Means among groups were compared with one-way ANOVA with Bonferroni post-test.
Data are representative of at least 3 independent experiments. *** p value < 0.001. Mouse drawings were adapted from reference 69.
Extended Data Fig. 2. Spatiotemporal dynamics of naïve CD8+ T cells upon intrahepatic priming.
5 x 106 fluorescent Cor93 TN were transferred into MUP-core or rLCMV-core-injected WT recipients. Mice were splenectomized and treated with anti-CD62L 48 hours and 4 hours prior to Cor93 TN transfer, respectively. (a) (left panels) Confocal immunofluorescence micrographs of liver sections from the indicated mice at the indicated timepoints upon Cor93 TN transfer, showing the distribution of Cor93 T cells (green) relative to portal tracts (highlighted by anti-cytokeratin 7 Ab-mediated staining of bile ducts in red). Sinusoids are highlighted by anti-Lyve-1+ Abs (white). Scale bars represent 100 μm. (right panels) Immunohistochemical micrographs of liver sections from the indicated mice at the indicated timepoints upon Cor93 TN transfer, showing the distribution of leukocyte infiltrates relative to portal tracts (highlighted by anti-cytokeratin 7 Ab-mediated staining of bile ducts in brown). Scale bars represent 100 μm. (b) Distribution of the distances (μm) of each Cor93 T cell from the centre of the closest portal triad at the indicated timepoints. n = 3 mice.
Data are representative of at least 3 independent experiments.
Extended Data Fig. 3. Kupffer cells, but not dendritic cells, promote CD8+ T cell effector differentiation upon rLCMV injection.
(a) Schematic representation of the experimental setup. 5 x 106 Cor93 TN were transferred into C57BL/6 (WT) recipients. Mice were splenectomized and treated with anti-CD62L 48 hours and 4 hours prior to cell transfer, respectively and injected with 2.5 x 105 infectious units of non-replicating rLCMV-core 4 hours prior to Cor93 TN transfer. Where indicated, mice were treated with clodronate liposomes (CLL) 48 hours prior to Cor93 TN transfer. (b) Confocal microscopy of liver sections from control mice (left panels) and clodronate liposomes-treated mice (right panels). Kupffer cells are depicted in red in all panels, while sinusoids are depicted in grey only in the first and third panel. Scale bars represent 100 μm. (c) Absolute numbers of CD11c+ MHC-IIhigh dendritic cells (DCs) in the livers of the indicated mice. (d-e) Absolute numbers of total (d) and of IFN-γ producing (e) Cor93 T cells in the livers of the indicated mice five days after Cor93 TN transfer. n = 4 (Control), 3 (CLL). Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test. (f) Confocal immunofluorescence micrographs of liver sections from the indicated mice five days after Cor93 TN transfer. Scale bars represent 100 μm. (g) Schematic representation of the experimental setup. WT mice were lethally irradiated and reconstituted with CD11c-DTR bone marrow (BM). 1 x 106 Cor93 TN were transferred into recipients. Mice were injected with 2.5 x 105 infectious units of non-replicating rLCMV-core 4 hours prior to Cor93 TN transfer. Indicated mice were treated with 400 ng of diphtheria toxin three days before, one day before and one day after T cell transfer. Livers were collected and analysed five days after Cor93 TN transfer. (h) Representative flow cytometry plots in the liver of control (left) or DT-treated (right) mice. (i) CD11c+ MHC-II+ DCs (expressed as percentage of the total intrahepatic leukocyte population, IHL) in the livers of the indicated mice. n = 3. Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test. (j-k) Absolute numbers of total (j) and of IFN-γ-producing (k) Cor93 T cells in the livers of the indicated mice five days after Cor93 TN transfer. n = 3 (Control and WT + rLCMV-core), 4 (Control + DT), 5 (WT + rLCMV-core + DT). Results are expressed as mean ± SEM. Means among groups were compared with one-way ANOVA with Bonferroni post-test.
Data are representative of 3 independent experiments. ** p value < 0.01, *** p value < 0.001. Mouse drawings were adapted from reference 69.
Extended Data Fig. 4. A strong reduction in the levels of hepatocellular core Ag expression is per se not sufficient to induce effector differentiation.
(a) Schematic representation of the experimental setup. 1 x 106 Cor93 TN were transferred into C57BL/6 (WT) or MUP-core recipients. Indicated WT mice were injected with 3 x 1010 viral genomes of AAV-core 15 days prior to Cor93 TN transfer. Livers were collected and analysed five days after Cor93 TN transfer. (b) Representative confocal immunofluorescence micrographs of a liver section from an AAV-core injected mouse 15 days after virus injection. Transduced hepatocytes are depicted in green and nuclei in grey. Scale bar represents 50 μm. n = 3 mice. (c-e) Absolute numbers of total (c) and of IFN-γ-producing (d) Cor93 T cells in the livers of the indicated mice five days after Cor93 TN transfer. (e) ALT levels detected in the sera of the indicated mice. n = 3 (WT and MUP-core), 5 (AAV-core). Results are expressed as mean ± SEM. Means among groups were compared with one-way ANOVA with Bonferroni post-test. (f) Schematic representation of the experimental setup. 1 x 106 Cor93 TN were transferred into 8-week-old (8 wo) or 4-week-old (4 wo) MUP-core mice. Livers were collected and analysed five days after Cor93 TN transfer. (g) HBcAg expression in the livers of the indicated mice was analysed by Western Blotting (WB). (h) Quantification of the WB shown in C. Core expression, normalized to the housekeeping nuclear protein H3, is expressed as arbitrary units (A.U.). n=1 (WT), 3 (MUP-core 8wo and MUP-core 4wo). Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test. (i) Immunohistochemical micrographs of liver sections from the indicated mice, showing core Ag expression (brown). Scale bars represent 50 μm. PV, portal vein; CV, central vein. n = 3 mice. (j-k) Absolute numbers of total (j) and of IFN-γ-producing (k) Cor93 T cells in the livers of the indicated mice five days after Cor93 TN transfer. n = 4. Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test. (l) ALT levels detected in the sera of the indicated mice. n = 4. Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test.
Data are representative of 2 independent experiments. * p value < 0.05, ** p value < 0.01. Mouse drawings were adapted from reference 69.
Extended Data Fig. 5. Genomic landscape of naïve CD8+ T cells undergoing intrahepatic priming.
(a) Box plots showing expression levels (log2RPKM) in the indicated experimental condition of genes belonging to the categories described in Fig. 2A. Results of two-sided Mann-Whitney U test are shown for the indicated comparisons. The midline of the box plot represents the median; lower and upper limits of the box plot represent the first and third quartile, respectively. Whiskers extend up to 1,5 times the interquartile range from the top and the bottom of the box plot. Sample size: naïve (n = 2), WT+rLCMV-core (n = 3), MUP-core (day1 and day3, n = 2; day7, n = 3). (b) Box plots showing ATAC-seq signal intensity (log2CPM) in the indicated experimental condition of peaks belonging to the categories described in Fig. 2C. Results of two-sided Mann-Whitney U test are shown for the indicated comparisons. The midline of the box plot represents the median; lower and upper limits of the box plot represent the first and third quartile, respectively. Whiskers extend up to 1,5 times the interquartile range from the top and the bottom of the box plot. Sample size: naïve (n = 2), WT+rLCMV-core (day 1 and day 7, n = 2; day 3, n = 3), MUP-core (day1 and day3, n = 2; day7, n = 3). (c) Bar plot showing the number of inducible ATAC-seq peaks (logFCCPM > 2.5, FDR < 0.001 versus Cor93 TN) in the indicated conditions. ATAC-seq peaks with higher intensity signal in Cor93 T cells from WT + rLCMV-core (logFCCPM > 1.5, FDR < 0.1) or from MUP-core mice (logFCCPM < -1.5, FDR < 0.1) are shown in blue and red, respectively. Differences in peak signal intensities were evaluated fitting a negative binomial generalized linear model on the dataset and then performing a quasi-likelihood (QL) F-test. The Benjamini-Hochberg procedure was applied in order to correct for multiple tests. Sample size: naïve (n=2), WT+rLCMV-core (day 1 and day 7, n = 2; day 3, n = 3), MUP-core (day1 and day3, n = 2; day7, n = 3).
Extended Data Fig. 6. Gene ontology analysis of intrahepatically primed CD8+ T cells.
Heatmap showing the NES value associated to the seed GO categories (identified by REVIGO) found enriched in the indicated time points by GSEA. Colour legends indicate NES, with positive values (in blue) reflecting enrichment of GOs in hepatic CD8+ T cells isolated from WT mice injected with rLCMV-core mice, and negative values (in red) reflecting enrichment of GOs in hepatic CD8+ T cells isolated from MUP-core mice.
Extended Data Fig. 7. While priming by hepatocytes initiates a unique dysfunctional program, hepatocellular Ag persistence may gradually trigger an additional exhaustion signature.
(a) Number (left panel) of top 100 genes from Cor93 T cells recovered from WT + rLCMV-core livers reaching log2RPKM>1 in the indicated conditions in RNA-seq data from splenic LCMV-specific effector or exhausted CD8+ T cells (ref.30). Box plot (right panel) showing the expression levels of top 100 genes from Cor93 T cells recovered from WT + rLCMV-core livers in the indicated conditions in RNA-seq data from splenic LCMV-specific effector or exhausted CD8+ T cells (ref.30) [two-tailed Wilcoxon Rank Sum Test: (*) p-value ≤ 0.05; (**) p-value ≤ 0.01; (***) p-value ≤ 0.001; (n.s.) Not Significant]. For each boxplot the median (horizontal line), the upper quartile and the lower quartile (straight lines) outside the interquartile range are reported. Dots represent the expression distribution of the set of 100-genes. Sample size: naïve (n = 2), effector (n = 2), exhausted (n = 2). (b) Number (left panel) of top 100 genes from Cor93 T cells recovered from MUP-core mice reaching log2RPKM>1 in the indicated conditions in RNA-seq data from splenic LCMV-specific effector or exhausted CD8+ T cells (ref.30). Box plot (right panel) showing the expression levels of top 100 genes from Cor93 T cells recovered from MUP-core mice in the indicated conditions in RNA-seq data from ref30 [two-tailed Wilcoxon Rank Sum Test: (*) p-value ≤ 0.05; (**) p-value ≤ 0.01; (***) p-value ≤ 0.001; (n.s.) Not Significant)]. For each boxplot the median (horizontal line), the upper quartile and the lower quartile (straight lines) outside the interquartile range are reported. Dots represent the expression distribution of the set of 100-genes. Sample size: naïve (n = 2), effector (n = 2), exhausted (n = 2). (c) Number (left panel) of top 100 genes in Cor93 T cells isolated from WT + rLCMV-core livers reaching log2RPKM>1 in the indicated conditions in RNA-seq data from splenic LCMV-specific exhausted CD8+ T cells from ref.31. Box plot (right panel) showing the expression levels of top 100 genes in Cor93 T cells isolated from WT + rLCMV-core livers in the indicated conditions in RNA-seq data from splenic LCMV-specific exhausted CD8+ T cells from ref.31 [two-tailed Wilcoxon Rank Sum Test: (*) p-value ≤ 0.05; (**) p-value ≤ 0.01; (***) p-value ≤ 0.001; (n.s.) Not Significant)]. For each boxplot the median (horizontal line), the upper quartile and the lower quartile (straight lines) outside the interquartile range are reported. Dots represent the expression distribution of the set of 100-genes. Sample size: naïve (n = 2), exhausted (n = 3). (d) Number (left panel) of top 100 genes from Cor93 T cells recovered from MUP-core mice reaching log2RPKM>1 in the indicated conditions in RNA-seq data from splenic LCMV-specific exhausted CD8+ T cells from ref.31. Box plot (right panel) showing the expression levels of top 100 genes from Cor93 T cells recovered from MUP-core mice in the indicated conditions in RNA-seq data from splenic LCMV-specific exhausted CD8+ T cells from ref.31 [two-tailed Wilcoxon Rank Sum Test: (*) p-value ≤ 0.05; (**) p-value ≤ 0.01; (***) p-value ≤ 0.001; (n.s.) Not Significant)]. For each boxplot the median (horizontal line), the upper quartile and the lower quartile (straight lines) outside the interquartile range are reported. Dots represent the expression distribution of the set of 100-genes. Sample size: naïve (n = 2), exhausted (n = 3). (e) Number (left panel) of top 100 genes in Cor93 T cells isolated from WT + rLCMV-core livers expressed (log2(normalized data) > 65th percentile of the full distribution) in the indicated conditions in microarray data from tolerant self-Ag-specific CD8+ T cells from ref.32. Box plot (right panel) showing the expression levels of genes retrieved in the dataset among the top 100 genes in Cor93 T cells isolated from WT + rLCMV-core livers in the indicated conditions in microarray data from tolerant self-Ag-specific CD8+ T cells from ref.32. Only genes for which microarray probes were retrieved were kept for these analyses. [two-tailed Wilcoxon Rank Sum Test: (*) p-value ≤ 0.05; (**) p-value ≤ 0.01; (***) p-value ≤ 0.001; (n.s.) Not Significant)]. For each boxplot the median (horizontal line), the upper quartile and the lower quartile (straight lines) outside the interquartile range are reported. Dots represent the expression distribution of the set of 100-genes. Sample size: naïve (n = 3), tolerant (n = 3). (f) Number (left panel) of top 100 genes from Cor93 T cells recovered from MUP-core mice expressed (log2(normalized data) > 65th percentile of the full distribution) in the indicated conditions in microarray data from tolerant self-Ag-specific CD8+ T cells from ref.32. Box plot (right panel) showing the expression levels of genes retrieved in the dataset among the top 100 genes from Cor93 T cells recovered from MUP-core mice in the indicated conditions in microarray data from tolerant self-Ag-specific CD8+ T cells from ref.32. Only genes for which microarray probes were retrieved were kept for these analyses. [two-tailed Wilcoxon Rank Sum Test: (*) p-value ≤ 0.05; (**) p-value ≤ 0.01; (***) p-value ≤ 0.001; (n.s.) Not Significant)]. For each boxplot the median (horizontal line), the upper quartile and the lower quartile (straight lines) outside the interquartile range are reported. Dots represent the expression distribution of the set of 100-genes. Sample size: naïve (n = 3), tolerant (n = 3). (g-i) Enrichment plot showing the results of a GSEAPreanked analysis (Kolmogorow-Smirnov statistics) performed on genes expressed in CD8+ T cells from WT + rLCMV-core or MUP-core mice (gene lists ranked by logFCRPKM) and using as gene set a curated list of genes induced in exhausted CD8+ T cells (n = 2) but not in effector CD8+ T cells (n = 2) as compared to naïve cells (n = 2) (ref.30). NES and FDR values are reported for each time point.
Extended Data Fig. 8. IL-2c substantially rescues the transcriptional program of dysfunctional CD8+ T cells.
(a-b) Heatmap showing expression values (log2RPKM) of genes hypo-expressed (a) or hyper-expressed (b) in Cor93 CD8+ T cells from MUP-core livers at day 5, which are rescued by IL-2c.
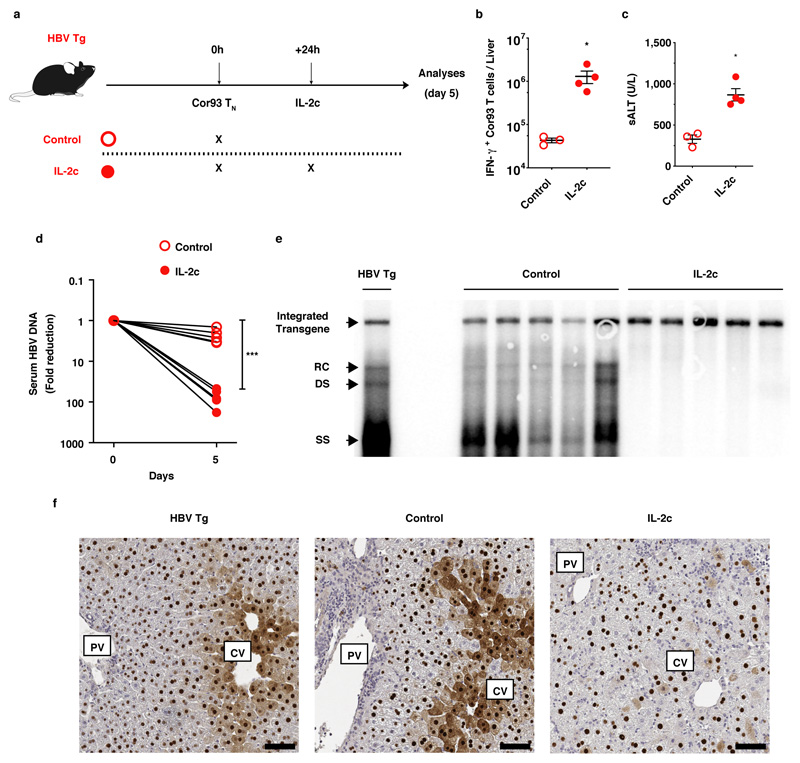
Extended Data Fig. 9. Therapeutic restoration of intrahepatically-primed, dysfunctional CD8+ T cells by IL-2.
(a) Schematic representation of the experimental setup. 1 x 106 Cor93 TN were transferred into HBV replication-competent transgenic (HBV Tg) mice. Indicated HBV Tg mice received IL-2c treatment one day after CD8+ T cell transfer. Livers were collected and analysed five days after Cor93 TN transfer. Sera were collected right before and five days after Cor93 TN transfer. (b) Absolute numbers of IFN-γ-producing Cor93 T cells in the livers of the indicated mice. n = 3 (Control), 4 (IL-2c). Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test. (c) ALT levels detected in the sera of the indicated mice. n = 3 (Control), 4 (IL-2c). Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test. (d) HBV DNA quantification (expressed as fold reduction over pre-treatment levels) in sera of the indicated mice prior to and five days after Cor93 TN transfer. n = 5. Results are expressed as mean ± SEM. Means among groups were compared with two-tailed t test. (e) HBV DNA quantification by Southern Blot analysis in the liver of the indicated mice. Bands corresponding to the expected size of the integrated transgene, relaxed circular (RC), double-stranded linear (DS), and single-stranded (SS) HBV DNAs are indicated. n = 5. (f) Representative immunohistochemical micrographs of liver sections from the indicated mice showing HBV core Ag expression (brown). Scale bars represent 100 μm. PV, portal vein; CV, central vein. n = 5 mice.
Data are representative of at least 2 independent experiments. * p value < 0.05, *** p value < 0.001.
Mouse drawings were adapted from reference 69.
Extended Data Fig. 10. Graphical abstract summarizing the paper’s main findings.
(Top panels) Priming by Kupffer cells – that are not natural targets of HBV – leads to differentiation into bona fide effector cells that form dense, extravascular clusters of rather immotile cells scattered throughout the liver. (Middle panels) Priming by hepatocytes – the natural targets of HBV - leads to local activation and proliferation but lack of differentiation into effector cells; these dysfunctional cells express a unique set of genes including some belonging to GO categories linked to tissue remodelling and they form loose, intravascular clusters of motile cells that coalesce around portal tracts. (Bottom panels) CD8+ T cells primed by hepatocytes can be rescued by IL-2 treatment.
Supplementary Material
Supplementary Information is available in the online version of this paper.
Acknowledgments
We thank F.V. Chisari for critical comments and suggestions, and for providing transgenic mouse lineages 1.3.32, MUP-core 50, Cor93 and Env28 TCR transgenic mice that were produced in his laboratory at The Scripps Research Institute in La Jolla, CA, USA; A. Fiocchi, M. Freschi, M. Mainetti, M. Raso and G. Sitia for technical support; M. Silva for secretarial assistance; E. Lugli, A. Mondino, L. Pace, R. Pardi, S. Trifari for critical reading of the manuscript and the members of the Iannacone laboratory for helpful discussions. Confocal immunofluorescence histology was carried out at Alembic, an advanced microscopy laboratory established by the San Raffaele Scientific Institute and the Vita-Salute San Raffaele University. Flow cytometry was carried out at FRACTAL, a flow cytometry resource and advanced cytometry technical applications laboratory established by the San Raffaele Scientific Institute. We would like to acknowledge the PhD program in Basic and Applied Immunology and Oncology at Vita-Salute San Raffaele University, as G.D.S., F.C., V.F. and V.B. conducted this study as partial fulfilment of their PhD in Molecular Medicine within that program. M.I. is supported by European Research Council (ERC) Consolidator Grant 725038, Italian Association for Cancer Research (AIRC) Grant 19891 and 22737, Italian Ministry of Health (MoH) Grant RF-2018-12365801, Lombardy Foundation for Biomedical Research (FRRB) Grant 2015-0010, the European Molecular Biology Organization Young Investigator Program, and a Career Development from the Giovanni Armenise-Harvard Foundation; A.P.B. is the recipient of EMBO Long-Term Fellowship ALTF 694-2016; F.M. is the recipient of Marie Curie Intra-European Fellowship (IEF) for Career Development SEP-210371319; F.A. is the recipient of a Fondazione Umberto Veronesi postdoctoral fellowship; M.K. is supported by the Italian Ministry of Education grant SIR-RBSI14BAO5; L.G.G. is supported by the Italian MoH grant RF-2013-02355209 and the Lombardy Open Innovation grant 229452; R.O. is supported by ERC Starting Grant 759532, Italian Telethon Foundation SR-Tiget Grant Award F04, Italian MoH grant GR-2016-02362156, AIRC MFAG 20247, Cariplo Foundation Grant 2015-0990 and the EU Infect-ERA 126.
Footnotes
Author contributions A.P.B and G.D.S. designed and performed experiments, analysed data, performed the statistical analyses, prepared the figures and edited the paper; P.D.L., F.M., P.Z., V.F., E.B., L.G., M.K., F.A. performed experiments and analysed data; F.C. generated RNA-seq and ATAC-seq data with help from V.B; G.B. and E.L. analysed RNA-seq and ATAC-seq data; R.O. supervised F.C., G.B., E.L. and V.B and prepared figures; N.L.B, K.K., P.T.F.K. and A.B. performed experiments on HBV-infected patients, analysed data, prepared the figures and edited the manuscript; C.B. and F.G. analysed the expression of genes involved in IL-2 sensing on KCs; A.C. and L.N. generated the lentiviral vectors encoding IL-2; G.G.-A. generated recombinant adeno-associated viruses; W.V.B. and D.D.P. generated rLCMV vectors; M.K., R.O. and L.G.G. provided funding, conceptual advice and edited the manuscript; M.I. designed and coordinated the study, provided funding, analysed the data, and wrote the paper.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare competing financial interest: M.I., L.G.G., R.O., A.C. and L.N. are inventors on patents filed, owned and managed by Telethon Foundation and San Raffaele Scientific Institute on LV technology related to the work presented in this manuscript. Readers are welcome to comment on the online version of the paper.
Data and materials availability
The RNA-seq and ATAC-seq data on sorted hepatic CD8+ T cells have been deposited in the ArrayExpress database under the accession code E-MTAB-7462 and E-MTAB-7461, respectively. All data is available in the main text or the supplementary materials.
References
- 1.Sironi L, et al. In vivo flow mapping in complex vessel networks by single image correlation. Sci Rep. 2014;4:7341. doi: 10.1038/srep07341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren A, et al. T lymphocytes interact with hepatocytes through fenestrations in murine liver sinusoidal endothelial cells. Hepatology. 2006;44:1182–1190. doi: 10.1002/hep.21378. [DOI] [PubMed] [Google Scholar]
- 3.Guidotti LG, et al. Immunosurveillance of the liver by intravascular effector CD8(+) T cells. Cell. 2015;161:486–500. doi: 10.1016/j.cell.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Wong YC, Tay SS, McCaughan GW, Bowen DG, Bertolino P. Immune outcomes in the liver: Is CD8 T cell fate determined by the environment? J Hepatol. 2015;63:1005–1014. doi: 10.1016/j.jhep.2015.05.033. [DOI] [PubMed] [Google Scholar]
- 5.Holz LE, et al. Naïve CD8 T cell activation by liver bone marrow-derived cells leads to a ‘neglected’ IL-2low Bimhigh phenotype, poor CTL function and cell death. J Hepatol. 2012;57:830–836. doi: 10.1016/j.jhep.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 7.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy PTF, Litwin S, Dolman GE, Bertoletti A, Mason WS. Immune Tolerant Chronic Hepatitis B: The Unrecognized Risks. Viruses. 2017;9:96. doi: 10.3390/v9050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Association for the Study of the Liver Electronic address: easloffice@easloffice.eu European Association For The Study Of The Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. Journal of hepatology. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 10.Fisicaro P, et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat Med. 2017;23:327–336. doi: 10.1038/nm.4275. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy PTF, et al. Preserved T-cell function in children and young adults with immune-tolerant chronic hepatitis B. 2012;143:637–645. doi: 10.1053/j.gastro.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Isogawa M, Chung J, Murata Y, Kakimi K, Chisari FV. CD40 activation rescues antiviral CD8+ T cells from PD-1-mediated exhaustion. PLoS Pathog. 2013;9:e1003490. doi: 10.1371/journal.ppat.1003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertoletti A, Ferrari C. Adaptive immunity in HBV infection. J Hepatol. 2016;64:S71–S83. doi: 10.1016/j.jhep.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Guidotti LG, Martinez V, Loh YT, Rogler CE, Chisari FV. Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice. J Virol. 1994;68:5469–5475. doi: 10.1128/jvi.68.9.5469-5475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flatz L, et al. Development of replication-defective lymphocytic choriomeningitis virus vectors for the induction of potent CD8+ T cell immunity. Nat Med. 2010;16:339–345. doi: 10.1038/nm.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guidotti LG, et al. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 18.Ishak K, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 19.Klein I, Crispe IN. Complete differentiation of CD8+ T cells activated locally within the transplanted liver. J Exp Med. 2006;203:437–447. doi: 10.1084/jem.20051775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Böttcher JP, et al. IL-6 trans-Signaling-Dependent Rapid Development of Cytotoxic CD8+ T Cell Function. Cell Reports. 2014;8:1318–1327. doi: 10.1016/j.celrep.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Sitia G, et al. Kupffer cells hasten resolution of liver immunopathology in mouse models of viral hepatitis. PLoS Pathog. 2011;7:e1002061. doi: 10.1371/journal.ppat.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Best JA, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez CX, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med. 2015;212:2041–2056. doi: 10.1084/jem.20150186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Intlekofer AM, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 26.Cruz-Guilloty F, et al. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurachi M, et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol. 2014;15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 2019;6:1. doi: 10.1038/s41586-019-0985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature. 2019;15:1. doi: 10.1038/s41586-019-0979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott-Browne JP, et al. Dynamic Changes in Chromatin Accessibility Occur in CD8(+) T Cells Responding to Viral Infection. Immunity. 2016;45:1327–1340. doi: 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:aaf2807–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schietinger A, Delrow JJ, Basom RS, Blattman JN, Greenberg PD. Rescued tolerant CD8 T cells are preprogrammed to reestablish the tolerant state. Science. 2012;335:723–727. doi: 10.1126/science.1214277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 34.Spolski R, Li P, Leonard WJ. Biology and regulation of IL-2: from molecular mechanisms to human therapy. Nat Rev Immunol. 2018 doi: 10.1038/s41577-018-0046-y. [DOI] [PubMed] [Google Scholar]
- 35.Manske K, et al. Outcome of anti-viral immunity in the liver is shaped by the level of antigen expressed in infected hepatocytes. Hepatology. 2018 doi: 10.1002/hep.30080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolksdorf F, et al. The PDL1-inducible GTPase Arl4d controls T effector function by limiting IL-2 production. Sci Rep. 2018;8 doi: 10.1038/s41598-018-34522-4. 16123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 38.Kamimura D, Bevan MJ. Naive CD8+ T cells differentiate into protective memory-like cells after IL-2 anti IL-2 complex treatment in vivo. J Exp Med. 2007;204:1803–1812. doi: 10.1084/jem.20070543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 40.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantore A, et al. Liver-directed lentiviral gene therapy in a dog model of hemophilia B. Sci Transl Med. 2015;7:277ra28–277ra28. doi: 10.1126/scitranslmed.aaa1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mátrai J, et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeves JP, Reeves PA, Chin LT. Survival surgery: removal of the spleen or thymus. Curr Protoc Immunol. 2001;Chapter 1:Unit 1.10. doi: 10.1002/0471142735.im0110s02. [DOI] [PubMed] [Google Scholar]
- 47.Iannacone M, et al. Platelets mediate cytotoxic T lymphocyte-induced liver damage. Nat Med. 2005;11:1167–1169. doi: 10.1038/nm1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tonti E, et al. Bisphosphonates target B cells to enhance humoral immune responses. Cell Reports. 2013;5:323–330. doi: 10.1016/j.celrep.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li P-Z, Li J-Z, Li M, Gong J-P, He K. An efficient method to isolate and culture mouse Kupffer cells. Immunol Lett. 2014;158:52–56. doi: 10.1016/j.imlet.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108–e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21.29.1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fioravanti J, et al. Effector CD8(+) T cell-derived interleukin-10 enhances acute liver immunopathology. J Hepatol. 2017;67:543–548. doi: 10.1016/j.jhep.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zordan P, et al. Tuberous sclerosis complex–associated CNS abnormalities depend on hyperactivation of mTORC1 and Akt. J Clin Invest. 2018;128:1688–1706. doi: 10.1172/JCI96342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benechet AP, Ganzer L, Iannacone M. Intravital Microscopy Analysis of Hepatic T Cell Dynamics. Methods Mol Biol. 2017;1514:49–61. doi: 10.1007/978-1-4939-6548-9_4. [DOI] [PubMed] [Google Scholar]
- 66.Lindh M, Gonzalez JE, Norkrans G, Horal P. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J Virol Methods. 1998;72:163–174. doi: 10.1016/s0166-0934(98)00026-3. [DOI] [PubMed] [Google Scholar]
- 67.Tan AT, et al. Host ethnicity and virus genotype shape the hepatitis B virus-specific T-cell repertoire. J Virol. 2008;82:10986–10997. doi: 10.1128/JVI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barbier L, et al. Two lymph nodes draining the mouse liver are the preferential site of DC migration and T cell activation. J Hepatol. 2012;57:352–358. doi: 10.1016/j.jhep.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 69.Thierry GR, et al. The conduit system exports locally secreted IgM from lymph nodes. J Exp Med. 2018;245 doi: 10.1084/jem.20180344. jem.20180344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.