Significance
How to ensure effectors are loaded efficiently is a key question to understanding protein secretion systems. Our study demonstrates that the type VI secretion system (T6SS) of Vibrio cholerae requires the physical presence of effectors as a prerequisite for assembling a functional secretion apparatus, representing a distinct posttranslational energy-saving strategy that may be conserved among the diverse T6SS systems in gram-negative bacteria.
Keywords: protein secretion, interspecies interaction, effector, synthetic biology
Abstract
The type VI secretion system (T6SS) is a lethal yet energetically costly weapon in gram-negative bacteria. Through contraction of a long sheath, the T6SS ejects a few copies of effectors accompanied by hundreds of structural carrier proteins per delivery. The few ejected effectors, however, dictate T6SS functions. It remains elusive how the T6SS ensures effector loading and avoids futile ejection. Here, by systemically mutating the active sites of 3 Vibrio cholerae effectors, TseL, VasX, and VgrG3, we show that the physical presence but not their activities is crucial for T6SS assembly. We constructed catalytic mutants of TseL and VgrG3 and truncated VasX mutants. These mutations abolished the killing of the effector-cognate immunity mutants. We determined that the VasX-mediated antimicrobial activity is solely dependent on the C-terminal colicin domain. Removal of the colicin domain abolished VasX secretion and reduced T6SS assembly, while deletion of the colicin internal loop abolished its toxicity but had little effect on secretion and assembly. The triple effector-inactive mutant maintains an active T6SS that is capable of delivering chimeric VgrG, PAAR, and TseL proteins fused with a cargo nuclease, indicating effector activities are not required for T6SS assembly or penetration into the cytosol of recipient cells. Therefore, by recruiting effectors as critical components for T6SS assembly, it represents an effective onboard checking mechanism that ensures effectors are loaded in place to prevent futile secretion. Our study also demonstrates a detoxified secretion platform by inactivating native effector activities that could translocate engineered cargo proteins via multiple routes.
Recent progress in microbiome studies has greatly expanded our traditional view of microbes in human health from infectious disease-focused to almost every aspect ranging from mental health to autoimmune diseases (1, 2). It is important not only to identify what microbes are present but also to understand how they exist and interact in complex communities. Of the diverse tools microbes have evolved for interaction, the type VI protein secretion system (T6SS) is one with wide distribution and diverse functions in microbial competition and host–microbe interactions in gram-negative commensals as well as pathogens (3–6).
Vibrio cholerae is a waterborne pathogen that causes cholera, an acute infectious disease from which patients develop severe diarrhea and dehydration (7). Cholera is a global public health threat with an estimated ∼3 million annual cases and ∼1.3 billion people at risk in 69 cholera-endemic countries (8). In addition to the key virulence factors cholera toxin and toxin-coregulated pili, V. cholerae has also acquired the T6SS that functions as a molecular weapon to inject antibacterial and antieukaryotic effectors into target cells through direct contact (4, 5, 9, 10). The T6SS is activated during infection in the host and facilitates V. cholerae colonization by outcompeting host commensal microbiota (11–13).
Belonging to the contractile injection systems (14), the T6SS consists of an intracellular double-tubular structure that ejects the inner needle outward through contraction of the outer sheath (15–17). The needle and the sheath are made of hundreds of stacks of Hcp and VipA/VipB (TssB/TssC) proteins, respectively (15–17). The tip of the needle carries a sharpening trimeric VgrG spike and a cone-shaped PAAR proposed for piercing through target cells (18, 19). V. cholerae encodes 3 VgrG proteins, of which VgrG1 and VgrG3 carry extended functional domains and display actin cross-linking and cell wall-degrading activities, respectively (9, 18, 20). VgrG1 and VgrG2 also serve as carrier proteins for their respective antibacterial effectors, TseL and VasX, both targeting the membrane (21, 22). TseL carries a lipase domain, and VasX carries an N-terminal predicted Pleckstrin-homology (PH) domain for interacting with phosphorylated membrane lipids and a C-terminal predicted colicin domain (9, 23–25). Whether the C-terminal colicin domain is solely responsible for the antibacterial activity is not known. Each antibacterial effector has a cognate immunity protein to confer self-protection. In addition, chaperone proteins are needed for VgrG-mediated delivery of TseL and VasX (21, 22). V. cholerae possesses another cell wall-targeting effector TseH (26). However, its role as a secreted effector is unclear since TseH has no effect in killing Escherichia coli or V. cholerae mutant lacking its cognate immunity gene tsiH (26).
As a protein delivery system, the diverse physiological functions of T6SS largely depend on its secreted effector proteins in different organisms (10, 18, 27–30). In addition, effectors also contribute to T6SS assembly because deletion of effector genes ΔvgrG3/ΔvasX and ΔtseL/ΔvgrG3/ΔvasX abolished Hcp secretion and sheath assembly in V. cholerae (9, 23, 31). In contrast to the evolved VgrG, Hcp, and PAAR effectors (18, 19, 32), it is less intuitive to envision how dedicated effectors without conserved T6SS structural domains, such as TseL and VasX, could affect the T6SS assembly. Indeed, deletion of all known effectors in Agrobacterium tumefaciens and Acinetobacter baylyi ADP1 has little effect on T6SS secretion (33, 34). It is not known whether the T6SS requirement for effectors is dependent on their activities or physical structures. For example, the T6SS of Acinetobacter requires a membrane-associated peptidoglycan hydrolase TagX to facilitate formation of the transenvelope TssJLM complex of the T6SS (35). Since V. cholerae lacks a TagX homolog, how the T6SS of V. cholerae assembles its transenvelope complex across the cell wall remains elusive. Considering that both TseL and VgrG3 are capable of reaching the periplasm when expressed in the cytosol (36), could V. cholerae effectors fulfill such TagX-like function through their membrane/cell wall-targeting activities?
To address this question, here we constructed catalytically inactive effector mutants to abolish their functions while minimizing perturbation to their structures in V. cholerae. We report that T6SS does not require effector activities for T6SS assembly or cytosol-to-cytosol penetration. In addition, the effector-inactive T6SS could deliver engineered VgrG, PAAR, and effector carrying a C-terminal cargo nuclease, demonstrating that PAAR and effectors are also delivered into the cytosol by the T6SS and can serve as carriers. Therefore, we conclude that the T6SS of V. cholerae requires the physical presence but not the antimicrobial activities of effectors. This represents an effective energy-conserving strategy to prevent futile secretion of structural proteins.
Results
Construction of Chromosomal Effector-Inactive Mutants.
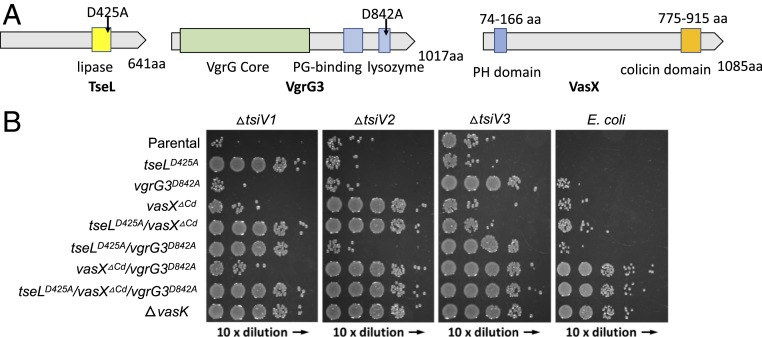
To determine how effectors contribute to T6SS assembly in V. cholerae, we decided to construct combinatorial deletion mutants and functionally inactive mutants of 3 known antibacterial effectors, VgrG3, TseL, and VasX (9) (Fig. 1A). To monitor T6SS assembly, we constructed all mutants in a fluorescence-labeled sheath VipA-mCherry2 V52 strain that maintains wild-type level T6SS activities (17, 37). Since we have previously identified active sites of TseL and VgrG3 (9), we constructed chromosomally encoded inactive mutants, TseLD425A and VgrG3D842A, accordingly. Unlike TseL and VgrG3 possessing enzymatic activities, VasX carries a C-terminal colicin domain without a catalytic pocket and it is unknown whether the observed antibacterial activity of VasX is solely dependent on this colicin domain (Fig. 1A). To inactivate VasX, we constructed a mutant VasXΔCd by deleting the colicin domain.
Fig. 1.
Chromosomal inactivation of effectors. (A) Predicted domains and mutation sites in V. cholerae effectors. (B) Bacterial competition assays. Killer strains are indicated on the left and prey strains on the top. A tetracycline-resistant strain CC114 is used as E. coli prey. Representative data from 3 independent assays are shown.
We then tested catalytic mutants competing with their respective immunity gene mutants and E. coli to compare their antibacterial activities (Fig. 1B). Competition assays show that tseLD425A, vgrG D842A, and vasXΔCd individual mutants failed to outcompete their specific immunity mutants but effectively killed E. coli and V. cholerae mutants lacking other immunity protection. Interestingly, E. coli could be killed by double-effector mutants of tseLD425A/vasXΔCd or tseLD425A/vgrG3D842A but not by vgrG3D842A/vasXΔCd. The vgrG3D842A/vasXΔCd double mutant could kill the TseL-specific ΔtsiV1 mutant, indicative of an active T6SS capable of delivering TseL. As expected, the triple mutant exhibited no difference from the ΔvasK mutant in all competition assays, indicating the loss of T6SS-mediated toxicity.
VgrG3 and TseL Enzymatic Activities Are Not Required for T6SS Assembly.
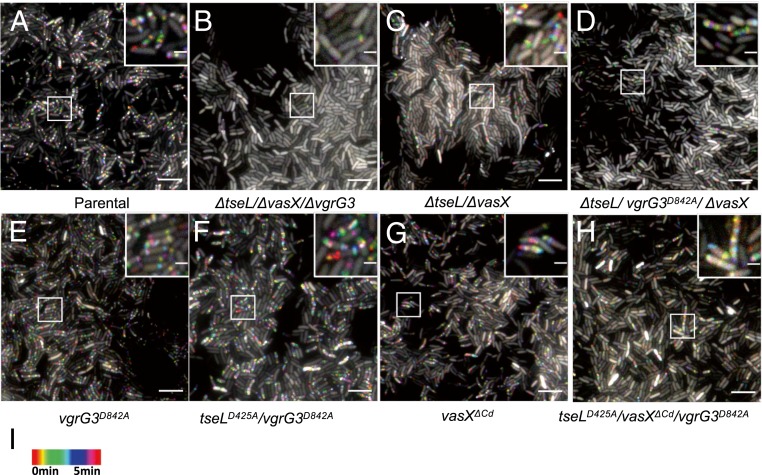
As VgrG3 is a T6SS structural protein, we then investigated whether the loss of VgrG3 function is responsible for abolishing T6SS activities in the triple effector deletion ΔtseL/ΔvgrG3/ΔvasX mutant (9). As expected, fluorescence microscopy imaging of T6SS sheath formation reveals that the T6SS was actively assembled in wild-type cells but not in the triple effector deletion mutant ΔtseL/ΔvasX/ΔvgrG3 (Fig. 2 A and B). Deletion of ΔtseL/ΔvasX significantly reduced T6SS assembly activities to about 25% of wild-type level (Fig. 2C and SI Appendix, Fig. S1). Catalytic inactivation of VgrG3 (VgrG3D842A) slightly reduced T6SS assembly in the ΔtseL/ΔvasX mutant and had little effect in wild type (Fig. 2 D and E and SI Appendix, Fig. S1). We then tested the chromosomally inactivated tseLD425A/vgrG3D842A mutant (Fig. 2F). This mutant exhibited wild-type-level T6SS activities as the vgrG3D842A mutant, indicating the catalytic activities of TseL and VgrG3 are not required for T6SS transenvelope complex assembly despite their capability of periplasmic translocation (36).
Fig. 2.
Fluorescence microscopy of T6SS assembly in effector mutants. (A–H) Time-lapse imaging of VipA-mCherry2 signal was captured every 10 s for 5 min and temporally color-coded. Representative images are shown with a close-up of a selected region as an inset. A 40- × 40-μm representative field of cells with a 3× magnified 5- × 5-μm inset (marked by box) is shown. Genotypes are indicated at the bottom. (Scale bars: large field of view, 5 µm and insets, 1 µm.) (I) Color scale used to temporally color-code the VipA-mCherry2 signal.
Deletion of the Colicin Domain of VasX Reduces T6SS Assembly.
VasX deletion has been shown to reduce T6SS assembly but the mechanism is elusive (31, 38). We then compared T6SS assembly in mutants lacking the colicin domain. The vasXΔCd mutant and the triple tseLD425A/vgrG3D842A/vasXΔCd mutant both show about 50% reduced T6SS assemblies relative to the wild type and the double tseLD425A/vgrG3D842A mutant, respectively (Fig. 2 G and H). These data suggest that the colicin domain-mediated toxicity is critical for T6SS assembly or, alternatively, the deletion of colicin domain causes structural deformation of VasX affecting T6SS assembly. Nonetheless, the active T6SS in those mutants suggests effector activities are not required for T6SS assembly.
Construction of a Colicin-Inactive and Secreted VasX Mutant.
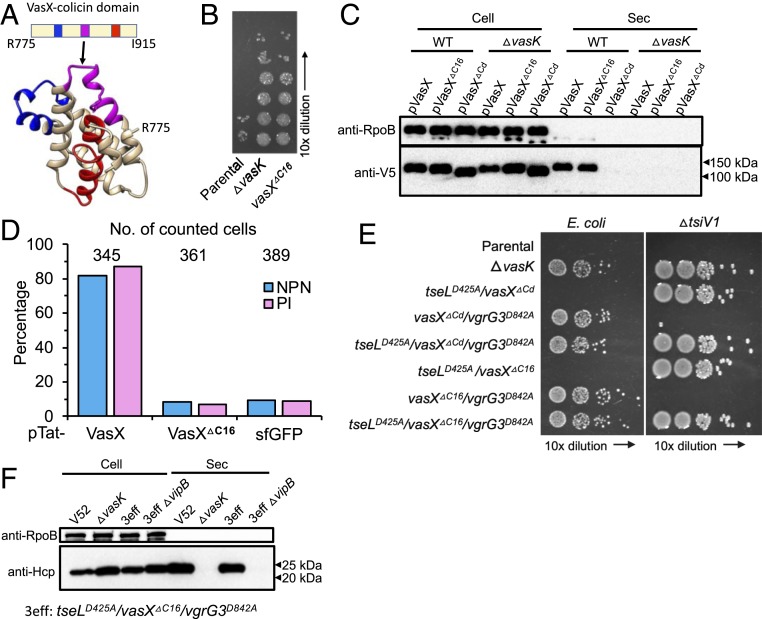
Protein sequence analysis using the HMMER program predicted 3 transmembrane regions within in the colicin domain (Fig. 3A). To further test whether the colicin-mediated activity is critical for T6SS assembly, we deleted the second transmembrane loop within the colicin domain to inactivate VasX toxicity while minimizing structural changes. The resultant VasXΔC16 mutant (lacking A852 to F867) showed no toxicity against the VasX immunity mutant ΔtsiV2 similar to the ΔvasK mutant, suggesting VasXΔC16 is inactivated (Fig. 3B).
Fig. 3.
Deletion of a colicin loop abolishes VasX toxicity but maintains secretion. (A) Predicted structural model of the colicin domain based on template 3FEW, with 13% identity and 84.3% confidence. Three predicted transmembrane regions within the colicin domain are highlighted in different colors. The loop within the second transmembrane region (pink) between A852 and F867 is deleted to result in VasXΔC16. (B) Competition assay using the ΔtsiV2 mutant as prey. (C) Secretion of VasX and its derivative mutants. Secretion of 3xV5 epitope-tagged VasX and its derivative mutants was detected using anti-V5 antiserum. (D) Membrane permeability and live–dead staining using NPN and PI fluorescence dyes. VasX and VasXΔC16 were induced in the periplasm using a Tat signal. Cells exhibiting NPN-positive and PI-positive are likely dead cells or cells with damaged membranes. Induction of sfGFP serves as a background control. See also SI Appendix, Fig. S2 for images. (E) Bacterial competition assay. Killers are indicated on the left and prey on the top. E. coli CC114 was used to test the overall antibacterial toxicity and ΔtsiV1 was used to specifically test TseL secretion in the activity mutant defective in both VasX and VgrG3. (F) Western blot analysis of Hcp secretion as an indicator of T6SS activity. Comparable secretion was found between wild-type V52 and the tseLD425A/vasXΔC16/vgrG3D842A mutant. In C and F, the RNA polymerase beta-subunit RpoB serves as a loading and cell lysis control.
To confirm that the loss of killing of the vasXΔC16 mutant is due to VasX inactivation but not failed delivery, we performed secretion assays expressing plasmid-borne VasX and its inactive mutants in wild type and the ΔvasK mutant. While no secretion of VasXΔCd was detected, wild-type VasX and VasXΔC16 were secreted at comparable levels (Fig. 3C), indicating VasXΔC16 but not VasXΔCd can be recognized as a T6SS substrate similar to wild-type VasX.
Although VasX has been shown to cause increased membrane permeability (23), whether this depends solely on its C-terminal activity or also requires it N-terminal lipid-binding PH domain has not been determined. We then compared the membrane permeability and cell death using the fluorescent probes 1-N-phenylnapthylamine (NPN) and propidium iodide (PI) in cells periplasmically expressing VasX and its VasXΔC16 using the twin-arginine translocation signal sequence (Tat) (26, 39). NPN interacts with membrane phospholipids and PI intercalates DNA, respectively, but both are not membrane-permeable except in damaged cells. Indeed, expression of VasX showed considerable cell damage, while the vasXΔC16 mutation reduced both NPN- and PI-positive cells to background levels, supporting that the C-terminal colicin domain dictates VasX toxicity (Fig. 3D and SI Appendix, Fig. S2).
The Triple Effector-Inactive Mutant Actively Assembles Sheath Structures.
We next constructed VasXΔC16 in combination with other effector mutations and tested their activities in bacterial competition assays (Fig. 3E). The triple effector mutant tseLD425A/vasXΔC16/vgrG3D842A exhibited no difference from the ΔvasK mutant in competition assays, indicating abolished T6SS toxicity. Notably, the vasXΔC16/vgrG3D842A mutant efficiently killed the ΔtsiV1 prey, suggesting TseL is delivered, but it failed to kill E. coli. Consistent results were also obtained using the vasXΔCd/vgrG3D842A mutant. These data suggest that TseL might have evolved to selectively target V. cholerae but not E. coli.
To test whether the tseLD425A/vasXΔC16/vgrG3D842A is T6SS-active, we monitored sheath assembly by microscopy and Hcp secretion by Western blot analysis (Fig. 3F and SI Appendix, Fig. S1). Indeed, T6SS was actively assembled and secretion of Hcp was equivalent to wild-type levels in the tseLD425A/vasXΔC16/vgrG3D842A mutant.
Distribution of VasX in Gram-Negative Bacteria.
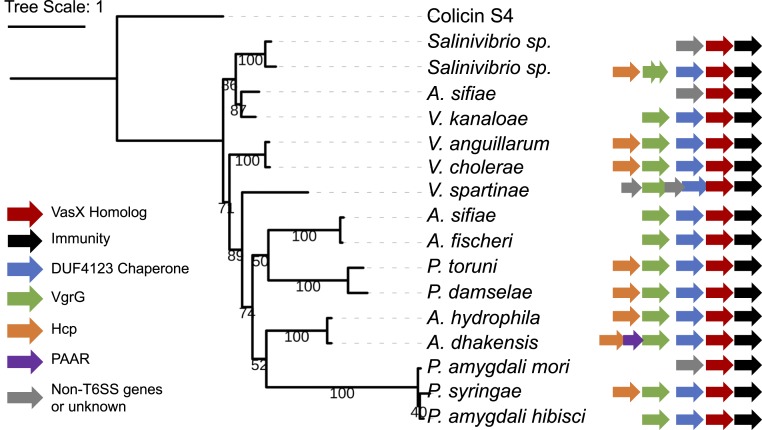
Our data highlight the importance of effector presence for T6SS assembly. Since TseL is known to belong to a large lipase family (25), we next investigated the distribution of VasX using HMMER to search for homologs of VasX in the UniProtKB database (40). By visually inspecting the pairwise hit positions and setting a stringent cutoff E-value of e-51, we reduced the number of 1,026 hits to a smaller set of 198 hits. Neighbor-joining clustering of the identified homologs demonstrates that the majority of VasX homologs are found in the families of Vibrionaceae, Aeromonadaceae, and Pseudomonadaceae with representative species including the bioluminescent Aliivibrio fischeri and Photobacterium damselae, Aeromonas hydrophila, and plant pathogens Pseudomonas syringae and Pseudomonas amygdali (SI Appendix, Fig. S3). We constructed a maximum-likelihood phylogenetic tree of 16 representative homologs using the E. coli colicin S4 protein as the outgroup and compared their gene clusters (Fig. 4). Thirteen VasX homologs are encoded in clusters containing upstream genes coding for the conserved Tec chaperone (DUF4123) (21) and VgrG proteins. Two VasX homologs were found in the strain Salinivibrio sp. IB872, suggesting a duplication event. In Vibrio spartinae, a gene of unknown function is inserted between vgrG and its downstream chaperone gene, which is reminiscent of the chaperone–cochaperone pair upstream of an H2-T6SS–dependent effector TseT in Pseudomonas aeruginosa (41). Comparison of A. hydrophila and its closely related Aeromonas dhakensis reveals a PAAR gene between hcp and vgrG in A. dhakensis. The copresence of vgrG-tec-vasX suggests that VasX secretion might be also tightly linked to T6SS assembly in those Vibrio, Pseudomonas, and other distantly related species.
Fig. 4.
Maximum likelihood phylogeny of VasX homologs and their gene clusters. Sixteen representative VasX homologs were aligned by MUSCLE and phylogeny (bootstrap = 100) was generated by PhyML using the MPI Bioinformatics Toolkit with default settings. Bootstrap value for each branch is indicated. E. coli colicin S4 protein, the structural template used for VasX modeling, serves as an outgroup. Gene clusters are retrieved from National Center for Biotechnology Information and curated. The size of each gene is not to scale.
Depth of T6SS Penetration Is Independent of Effector Activities.
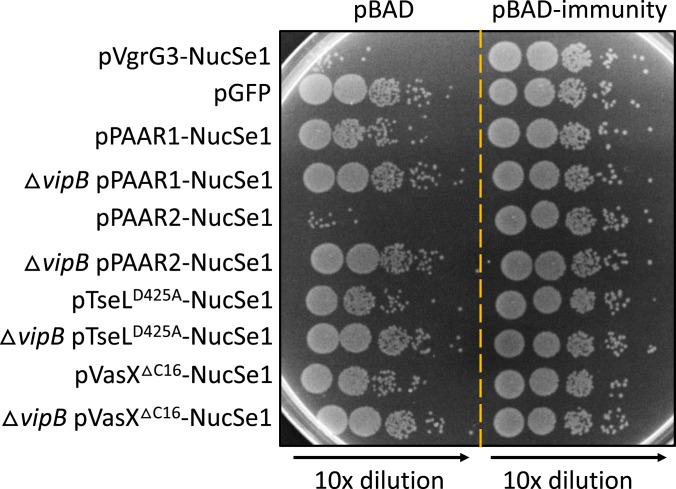
Finally, we asked whether effector activities are required for T6SS penetration into the cytosol of recipient cells. We employed a recently constructed chimeric VgrG3-NucSe1 nuclease in which the native lysozyme domain of VgrG3 is replaced with a nuclease domain NucSe1 (36). Bacterial competition assays show that the tseLD425A/vasXΔC16/vgrG3D842A mutant could deliver VgrG3-NucSe1 and kill prey cells except those expressing the specific immunity protein to NucSe1 (Fig. 5 and SI Appendix, Fig. S4). Therefore, the T6SS penetration into the cytosol is independent of effector activity but driven by the physical force of sheath contraction.
Fig. 5.
Cytosol-to-cytosol delivery of chimeric cargo by the triple effector-inactive T6SS. Prey and killer cells are derivatives of the same parental tseLD425A/vasXΔC16/vgrG3D842A mutant except for carrying different plasmids or the T6SS null ΔvipB deletion as indicated. Survival of prey was compared by serial dilutions. Delivery of chimeric NucSe1 proteins would reduce the survival of prey with the empty vector but not the ones expressing the NucSe1-specific immunity protein. See SI Appendix, Fig. S4 for comparison with statistics.
We next tested whether the NucSe1 cargo could also be delivered by the tip protein PAAR and effectors as fusion proteins. In contrast to VgrG and Hcp proteins (31), cytosol-to-cytosol delivery for the 2 PAAR proteins and membrane-targeting effectors TseL and VasX in V. cholerae has not been demonstrated. Using competition assays, we found that chimeric PAAR1(VCA0105), PAAR2 (VCA0284), and TseLD425A could deliver NucSe1, while VasXΔC16 showed a minor effect (Fig. 5 and SI Appendix, Fig. S4). These results not only indicate that T6SS-delivered PAAR proteins and TseL can reach the cytosol but also demonstrate the potential of using the tseLD425A/vasXΔC16/vgrG3D842A mutant as a detoxified delivery platform to translocate cargo proteins into recipient cells via multiple routes.
Discussion
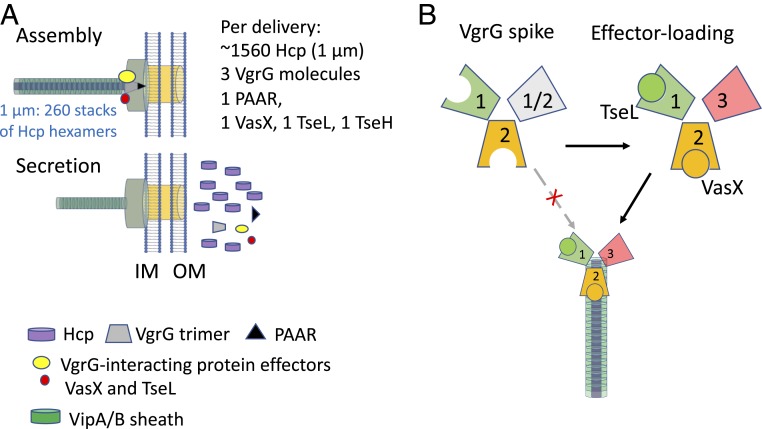
T6SS assembly and secretion is energetically costly. Assembling a 1-µm-long sheath needle (average length in V. cholerae) requires 260 rings of hexameric VipA/B-Hcp subunits (16). Assuming sheath contraction ejects the whole Hcp needle out, each contraction will cost 1,560 molecules of Hcp to accompany the delivery of a few molecules of effectors (Fig. 6A). Therefore, how could cells prevent futile delivery of T6SS, that is, T6SS secretion without any effectors loaded? Our results demonstrate an onboard checking mechanism by which the physical presence of effectors is required for T6SS assembly in V. cholerae, thereby ensuring effectors are loaded for each T6SS ejection (Fig. 6B).
Fig. 6.
Model for effector-dependent T6SS assembly that prevents from wasteful secretion of structural proteins. (A) T6SS secretes a large number of structural proteins with a small number of effectors per delivery. (B) VgrG spike lacking effectors, especially VasX and VgrG3, cannot support sheath-tube assembly, indicating each secretion would secrete at least 1 or 2 copies of VasX or VgrG3.
In addition to Hcp and effectors, each delivery also secretes a heterotrimeric VgrG spike complex (18, 42). This is evidenced by the fact that lacking VgrG1 or VgrG3 individually has little effect on T6SS functions but lacking both VgrG1 and VgrG3 severely reduces T6SS formation; by contrast, deletion of VgrG2 completely abolishes T6SS activities (18, 31, 43). Therefore, each delivery is expected to translocate at least 1 molecule of VgrG2 with 1 or 2 molecules of VgrG1/VgrG3 or both. Accordingly, considering that VasX secretion is VgrG2-dependent and TseL is VgrG1-dependent and that deletion of vasX and vgrG3 severely impairs assembly (9), it is likely that each secretion event delivers 1 or 2 VgrG2-VasX with VgrG3 and/or with VgrG1-TseL (Fig. 6B). There are 2 PAAR proteins encoded by V. cholerae. Although PAAR proteins are critical for the T6SS in A. baylyi, they are dispensable in V. cholerae (19). The carrier for TseH secretion is currently unclear, but it is likely dependent on its upstream gene product PAAR VCA0284.
How effectors contribute to T6SS assembly remains to be elucidated. Considering the VgrG dependence of TseL and VasX (9, 23), it is plausible that the presence of effector–VgrG interaction provides a more stable VgrG-trimeric complex that is required for the sheath-needle assembly (18, 42, 44). Indeed, when we used a noncontractile sheath construct (VipA-N3) to image assembly in the triple effector deletion mutant, long sheaths were observed suggesting effectors are not required for assembling the highly stable noncontractile sheath (SI Appendix, Fig. S5). When T6SS-delivered proteins enter the recipient cells, the interacting VgrG–effector complex should disassociate quickly to find their destined targets. Therefore, the intermolecular interaction is expected to be weak rather than strong. Stabilization of VgrG–effector complex may also require chaperone proteins carrying conserved domains (DUF4123, DUF1795, and DUF2169) that have now been identified in many T6SS species (41, 45–48). Because chaperones are not cosecreted, the delivered VgrG complex decorated with effectors might be quickly destabilized after entering recipient cells. This hypothesis can be tested by tracking the fate of delivered protein complex directly through imaging. However, this is a considerable technical challenge since it would require superresolution live-cell microscopy with millisecond temporal resolution (49).
VasX is a transkingdom effector targeting both eukaryotic amoeba and bacterial cells (9, 23, 24). Here we show that its C-terminal colicin domain is responsible for its antibacterial activity. Interestingly, despite that the domain is only 13% of the full-length protein at the C-terminal end, deletion of this domain abolishes VasX secretion, highlighting its critical role in T6SS assembly. Notably, a recent study quantifying T6SS protein levels reports that VasX is highly unstable and prone to degradation (38). To inactivate the colicin toxicity and maintain secretion, we removed 1 of the 3 predicted transmembrane regions within the colicin domain. The resulting VasXΔC16 mutant was indeed inactivated and secreted, and in contrast to domain deletion this mutation has little effect on T6SS assembly (Fig. 2C). Considering the wide distribution of VasX-like effectors, this demonstration of inactivating colicin-like effectors with minimal structural effect would be informative to study other VasX homologs.
Not all species require effectors for T6SS assembly, nor do all effectors contribute to assembly equally. Indeed, combinatorial effector deletions in A. tumefaciens and A. baylyi have little effect on T6SS secretion (33, 34). This might result from differences in effector secretion routes since there are multiple routes of delivery for T6SS in different organisms (41). Both TseL and VasX rely on the VgrG spike as carrier for delivery, while other effectors are carried inside the Hcp tubule for delivery, as exemplified by the Tse1–4 effectors in P. aeruginosa (50). Considering the essential role of VgrG in T6SS assembly (18, 44), it is conceivable that VgrG-interacting effectors are more likely to modulate assembly. Of the 3 antibacterial effectors TseL, VasX, and VgrG3, lacking both VasX and VgrG3 abolishes T6SS secretion while TseL seems to contribute but is not critical for T6SS assembly (9). In addition, while VasX or VgrG3 alone showed strong E. coli killing activities, TseL alone did not (Figs. 1B and 3E).
Since VasX homologs can be found in a variety of species, the requirement for VasX or VgrG3 in T6SS assembly may represent a conserved resource-saving strategy to ensure the most effective toxins are delivered. This resembles an onboard checking strategy distinct from known mechanisms that conserve resources through transcriptional regulation of T6SS genes (51, 52) or through secretion of negative regulators (53, 54). The latter refers to the type III secretion system and the flagellum system that commonly produce a negative regulator/substrate, whose accumulation under noninducing conditions represses expression but translocation relieves the repression upon assembly of a functional apparatus (53, 55, 56). Our findings thus provide important additional insights on how protein secretion systems balance fitness demand and energy cost by promoting efficiency during evolution.
Materials and Methods
Bacterial Strains and Plasmids.
V. cholerae and E. coli strains and plasmids are listed in SI Appendix, Table S1. Chromosomal deletions and site-directed mutations in V. cholerae were constructed using homologous recombination (57). Cultures were grown in lysogeny broth (LB) medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, and 0.5% [wt/vol] NaCl) aerobically. Antibiotics were as follows: ampicillin (100 µg/mL), streptomycin (100 µg/mL), chloramphenicol (25 µg/mL for E. coli, 2.5 µg/mL for V52), and kanamycin (50 µg/mL). Expression vectors were constructed as previously described (31). All constructs were verified by sequencing.
Bacterial Killing Assay.
The killing assay was performed as previously described (9, 58). Briefly, killer and prey cultures were grown to exponential (optical density at 600 nm [OD600] = 1) and stationary phase (OD600 = 2), respectively. Cells were centrifuged at 10,000 × g for 2 min and the pellets were washed in fresh LB. Killer and prey cells were gently mixed at a ratio of 5:1 and coincubated at 37 °C on an LB plate for 3 h. Cells were then transferred from the plate to fresh LB. Survival of prey cells was quantified by serial dilution in LB and plating on appropriate antibiotic LB plates.
NPN and PI Staining of VasX-Expressing Cells.
Cells were grown overnight in LB medium supplemented with 0.3% (wt/vol) glucose and 20 µg/mL gentamicin, diluted 1:100, and subcultured for 3 h in LB medium supplemented with 20 µg/mL gentamicin. Cells were grown to OD600 of ∼0.8 to 1 and induced with 0.5 mM isopropyl-β-d-thiogalactoside for 30 to 45 min, resuspended in 0.5× PBS to OD600 = 10, stained with 1 μg/mL PI and 0.5 mM NPN fluorescent probe, and spotted on 1% agarose pads for imaging.
Protein Secretion Assay.
Protein secretion was performed as previously described (9). Briefly, cultures (5 mL) were grown in LB at 30 °C to OD600 = 1 and collected by centrifugation. Pellets were resuspended in fresh LB. For induced expression of genes cloned to pBAD vectors, 0.01% (wt/vol) l-arabinose was added (59). Resuspended cells were grown at 30 °C for 1 h and centrifuged at 2,500 × g for 8 min at room temperature. Cell pellets were treated with SDS-loading dye and used as whole-cell samples. The supernatants were transferred to centrifuge tubes, precipitated in 20% (vol/vol) trichloroacetic acid at −20 °C for 30 min, and centrifuged at 15,000 × g for 30 min at 4 °C. The pellets were washed twice with 100% acetone by centrifugation, air-dried, and resuspended in SDS-loading dye. The samples of whole cells and secretion were boiled for 10 min prior to SDS/PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) analysis.
Western Blot Analysis.
Proteins were resolved on 12% SDS/PAGE gels and transferred to a poly(vinylidene difluoride) membrane (Bio-Rad). The membrane was blocked with 5% (wt/vol) nonfat milk in TBST buffer (50 mM Tris, 150 mM NaCl, and 0.05% Tween 20, pH 7.6) for 1 h at room temperature, incubated with primary antibodies for 1 h at room temperature, washed 3 times in TBST buffer, and incubated with an HRP-conjugated secondary antibody (Cell Signaling) for 1 h followed by detection using ECL solution (Bio-Rad). Monoclonal antibodies to the V5 epitope tag and RpoB, the beta subunit of RNA polymerase, were purchased from Abclonal and Biolegend, respectively. The polyclonal antibody to Hcp was custom-made by Shanghai Youlong Biotech.
Fluorescence Microscopy.
V. cholerae overnight cultures were diluted 1:100 in fresh LB medium and grown at 37 °C for 3 h to an OD600 of ∼1. The cells were concentrated 10 times in 0.5× PBS buffer, spotted onto an agarose pad, and covered with a coverslip. The images were obtained using a Nikon Ti-E inverted motorized microscope equipped with Perfect Focus System and CFI Plan Apochromat Lambda 100× oil objective. Intensilight C-HGFIE (Nikon), ET-GFP (Chroma 49002), and ET-mCherry (Chroma 49008) filter sets were used for fluorescence excitation and filtration. The images were captured at 10-s intervals for 5 min using an ANDOR Clara camera (DR 328G-C01-SIL), pixel size 60 nm and NIS-Elements AR 4.40 software. Fiji software was used for all image analysis and manipulation (60). Images from a time series were normalized to the same mean intensity as previously described (37).
Bioinformatic Analysis.
For the structural prediction of VasX, we used the Phyre2 web server (61). The predicted structural model was visualized using the Chimera program (62). HMMER was used to identify protein homologs (63). All identified homologs were manually inspected for the hit positions. Jalview was used for sequence alignment and removal of redundant sequences with 100% identity (64). Neighbor-joining clustering was performed using The EMBL-EBI online tools (65). Maximum-likelihood phylogeny was constructed with 100 bootstrap using PhyML and the MPI Bioinformatics Toolkit (66, 67), and the phylogenetic tree visualization was generated using the Interactive Tree Of Life (iTOL) server (68).
Supplementary Material
Acknowledgments
This work was supported by funding from National Key R&D Program of China (2018YFA0901200), National Natural Science Foundation of China (31770082), National Institute of Allergy and Infectious Diseases Grant AI-01845 (to J.J.M.), the Canadian Institutes of Health Research, and the Natural Sciences and Engineering Research Council of Canada (T.G.D). We thank Xinran Zhang, Steve Hersch, and Le Tang for technical assistance.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914202116/-/DCSupplemental.
References
- 1.Blaser M. J., The past and future biology of the human microbiome in an age of extinctions. Cell 172, 1173–1177 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Hsiao E. Y., et al. , Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho B. T., Dong T. G., Mekalanos J. J., A view to a kill: The bacterial type VI secretion system. Cell Host Microbe 15, 9–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pukatzki S., et al. , Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mougous J. D., et al. , A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wexler A. G., et al. , Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. U.S.A. 113, 3639–3644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson E. J., Harris J. B., Morris J. G. Jr, Calderwood S. B., Camilli A., Cholera transmission: The host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7, 693–702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali M., Nelson A. R., Lopez A. L., Sack D. A., Updated global burden of cholera in endemic countries. PLoS Negl. Trop. Dis. 9, e0003832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong T. G., Ho B. T., Yoder-Himes D. R., Mekalanos J. J., Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hood R. D., et al. , A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao W., Caro F., Robins W., Mekalanos J. J., Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359, 210–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y., Waldor M. K., Mekalanos J. J., Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y., Ho B. T., Mekalanos J. J., Tracking Vibrio cholerae cell-cell interactions during infection reveals bacterial population dynamics within intestinal microenvironments. Cell Host Microbe 23, 274–281.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor N. M. I., van Raaij M. J., Leiman P. G., Contractile injection systems of bacteriophages and related systems. Mol. Microbiol. 108, 6–15 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Basler M., Type VI secretion system: Secretion by a contractile nanomachine. Philos. Trans. R Soc. B Biol. Sci. 370, 20150021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J., et al. , Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat. Microbiol. 2, 1507–1512 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J., Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J., Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shneider M. M., et al. , PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks T. M., Unterweger D., Bachmann V., Kostiuk B., Pukatzki S., Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J. Biol. Chem. 288, 7618–7625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang X., et al. , Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc. Natl. Acad. Sci. U.S.A. 112, 9106–9111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unterweger D., et al. , Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 34, 2198–2210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyata S. T., Unterweger D., Rudko S. P., Pukatzki S., Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog. 9, e1003752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata S. T., Kitaoka M., Brooks T. M., McAuley S. B., Pukatzki S., Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect. Immun. 79, 2941–2949 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell A. B., et al. , Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altindis E., Dong T., Catalano C., Mekalanos J., Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. MBio 6, e00075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell A. B., Peterson S. B., Mougous J. D., Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cianfanelli F. R., Monlezun L., Coulthurst S. J., Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Trunk K., et al. , The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong M., et al. , Microbial herd protection mediated by antagonistic interaction in polymicrobial communities. Appl. Environ. Microbiol. 82, 6881–6888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vettiger A., Basler M., Type VI secretion system substrates are transferred and reused among sister cells. Cell 167, 99–110.e12 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Ma J., et al. , The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence 8, 1189–1202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L.-S. S., Hachani A., Lin J.-S. S., Filloux A., Lai E.-M. M., Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ringel P. D., Hu D., Basler M., The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep. 21, 3927–3940 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Weber B. S., et al. , Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. MBio 7, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho B. T., Fu Y., Dong T. G., Mekalanos J. J., Vibrio cholerae type 6 secretion system effector trafficking in target bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 114, 9427–9432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Basler M., Ho B. T., Mekalanos J. J., Tit-for-tat: Type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin L., Lezan E., Schmidt A., Basler M., Abundance of bacterial Type VI secretion system components measured by targeted proteomics. Nat. Commun. 10, 2584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer T., Berks B. C., The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10, 483–496 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Potter S. C., et al. , HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burkinshaw B. J., et al. , A type VI secretion system effector delivery mechanism dependent on PAAR and a chaperone-co-chaperone complex. Nat. Microbiol. 3, 632–640 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Leiman P. G., et al. , Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng J., Ho B., Mekalanos J. J., Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6, e23876 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renault M. G., et al. , The gp27-like hub of VgrG serves as adaptor to promote hcp tube Assembly. J. Mol. Biol. 430, 3143–3156 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Alcoforado Diniz J., Coulthurst S. J., Intraspecies competition in Serratia marcescens is mediated by type VI-secreted rhs effectors and a conserved effector-associated accessory protein. J. Bacteriol. 197, 2350–2360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cianfanelli F. R., et al. , VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 12, e1005735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitney J. C., et al. , An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bondage D. D., Lin J.-S., Ma L.-S., Kuo C.-H., Lai E.-M., VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vettiger A., Winter J., Lin L., Basler M., The type VI secretion system sheath assembles at the end distal from the membrane anchor. Nat. Commun. 8, 16088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitney J. C., et al. , Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng J., Shin O. S., Cameron D. E., Mekalanos J. J., Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 21128–21133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borgeaud S., Metzger L. C., Scrignari T., Blokesch M., The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347, 63–67 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Rietsch A., Vallet-Gely I., Dove S. L., Mekalanos J. J., ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 102, 8006–8011 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hauser A. R., The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 7, 654–665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller V. L., Connections between transcriptional regulation and type III secretion? Curr. Opin. Microbiol. 5, 211–215 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Darwin K. H., Miller V. L., Type III secretion chaperone-dependent regulation: Activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20, 1850–1862 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller V. L., Mekalanos J. J., A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacIntyre D. L., Miyata S. T., Kitaoka M., Pukatzki S., The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U.S.A. 107, 19520–19524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guzman L. M., Belin D., Carson M. J., Beckwith J., Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Söding J., Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madeira F., et al. , The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guindon S., et al. , New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Zimmermann L., et al. , A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Letunic I., Bork P., Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.