Abstract
Burkholderia bacteria are multifaceted organisms that are ecologically and metabolically diverse. The Burkholderia genus has gained prominence because it includes human pathogens; however, many strains are nonpathogenic and have desirable characteristics such as beneficial plant associations and degradation of pollutants. The diversity of the Burkholderia genus is reflected within the large genomes that feature multiple replicons. Burkholderia genomes encode a plethora of natural products with potential therapeutic relevance and biotechnological applications. This review highlights Burkholderia as an emerging source of natural products. An overview of the taxonomy of the Burkholderia genus, which is currently being revised, is provided. We then present a curated compilation of natural products isolated from Burkholderia sensu lato and analyze their characteristics in terms of biosynthetic class, discovery method, and bioactivity. Finally, we describe and discuss genome characteristics, and highlight the biosynthesis of a select number of natural products that are encoded in unusual biosynthetic gene clusters. The availability of >1,000 Burkholderia genomes in public databases provides an opportunity to realize the genetic potential of this underexplored taxon for natural product discovery.
Graphical Abstract

Bacterial natural products are a valuable source of bioactive compounds with applications in medicine and agriculture.1 Although members of the Actinomycetales order have been the major focus of natural product drug discovery programs over the last decades,2 it has become evident, particularly since the rise of whole genome sequencing, that neglected bacterial taxa are also promising sources of natural products.3 One of the emerging, gifted producers of natural products that have attracted scientific attention is Burkholderia, a heterogeneous and ecologically diverse group of gram-negative bacteria belonging to the Proteobacteria phylum, β-Proteobacteria class, and Burkholderiales order (Figure 1).3–5 Because of the heterogeneity of the Burkholderia genus, its taxonomy is currently being revised.6–13 The term Burkholderia is used below to mean Burkholderia sensu lato.
Figure 1.
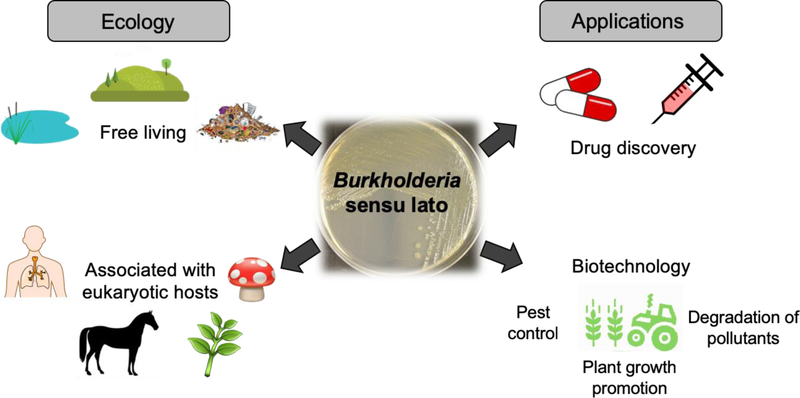
Overview of Burkholderia sensu lato, its ecological roles and potential applications. Members of Burkholderia sensu lato occupy diverse ecological niches ranging from pristine soil and aquatic environments to contaminated landfill, and they can be free-living or associated with a wide set of eukaryotic hosts, from fungi to humans. Host associations can be harmful (e.g. human and animal pathogens that include biological warfare agents) or beneficial (e.g. endosymbionts that promote plant growth). Ecological niche diversity translates into diverse natural products that mediate host interactions, that are beneficial for adaptation and survival, and that may be harnessed for biotechnological applications and drug discovery.
Burkholderia are found in a wide range of terrestrial and aquatic niches, as free-living organisms or in association with eukaryotic hosts such as humans, animals, plants, and fungi (Figure 1).14,15 The interaction between Burkholderia and their hosts can be either beneficial or harmful.16,17 For instance, a group of ~20 closely related species referred to as Burkholderia cepacia complex (Bcc) include opportunistic, human pathogens that can cause lung infections in immunocompromised individuals and are of particular concern to cystic fibrosis patients.18–20 Moreover, B. pseudomallei and B. mallei are the causative agents of melioidosis and glanders, respectively, and are listed as potential bioweapons by the Center for Disease Control.21–23 The genus also includes plant pathogens of agricultural importance, such as B. glumae that causes rice rot.24 More complex host associations involving three players have also been reported, including an interesting association that was described between a rice pathogenic fungus (Rhizopus sp.) and a Burkholderia species that lives inside of the fungus (endosymbiont) and produces the polyketide precursor of rhizoxin, a phytotoxin that binds β-tubulin of rice cells causing cell cycle arrest.25–31
In contrast to pathogenicity, members of Burkholderia sensu lato are also known to promote growth of plants and protect plants from pests (Figure 1).15,17 For instance, Burkholderia ambifaria and Burkholderia caribensis are presumably diazotrophic strains that promote growth of the grain crop amaranth.32 Burkholderia rinojensis, a soil-dwelling bacterium, was shown to exhibit activity against arthropod pests, making them desirable as a potential alternative to the use of pesticides.33
There are also free-living species that have biotechnological potential, including the production of commercially relevant hydrolytic enzymes that can degrade natural and synthetic pollutants (Figure 1). For instance, Burkholderia xenovorans (now Paraburkholderia xenovorans)12 and Burkholderia jiangsuensis (now Caballeronia jiangsuensis)8 have the ability to break down environmental contaminants, polychlorinated biphenyl (PCB) and methyl parathion, respectively.34,35
Regardless if free-living or in association with eukaryotic hosts, Burkholderia are known to produce a variety of natural products that are beneficial for adaptation and survival, that mediate host interactions, and that may be applied for therapeutic and biotechnological purposes (Figure 1). In this review, we start by providing an overview of the taxonomy of the Burkholderia genus, which is currently being revised, followed by a curated list of natural products isolated from Burkholderia and an analysis of their characteristics in terms of biosynthetic class, bioactivity, and discovery method. We then discuss genome characteristics and the biosynthesis of a select number of natural products that are encoded in unusual biosynthetic gene clusters (BGCs).
Taxonomy and Ecological Diversity of Burkholderia Bacteria
The first Burkholderia isolates were reported in 1950 by W.H. Burkholder as the causative agent of an onion bulb rot termed “sour skin”.36 At that time, the onion isolates were named Pseudomonas cepacia. Due to the emergence of DNA-DNA hybridization and rRNA gene sequencing tools, several bacteria previously characterized as Pseudomonas were reclassified and then transferred into the then newly proposed genus Burkholderia.37 In addition, Burkholderia picketti and Burkholderia solanacearum were transferred into another newly proposed genus, Ralstonia.38
The identification of newly discovered Burkholderia species has not been straightforward. A complicating issue is that Burkholderia isolates may not be phenotypically distinguishable, despite being genetically distinct. A term used in the literature to signify strains that are phenotypically indistinguishable but that are distinct at the DNA level is “genomovar”.18,39 Thus, the use of commercial phenotypic assays is not a feasible option for proper classification of organisms in this genus. Molecular methods that employ not only 16S rRNA analysis but also other conserved genes are essential for identification of Burkholderia genomovars.18,39
Burkholderia included >100 species as of 2015, representing a large, heterogeneous and taxonomically controversial group of bacteria.6–12,15,40 In fact, phylogenetic analyses performed by different research groups indicated that Burkholderia is polyphyletic.6,10–12 Due to this heterogeneity, a split of the genus Burkholderia was proposed.11,12,40 However, 16S rRNA phylogenetic trees or even conventional multilocus sequence analysis using a small number of genes initially provided limited support for the new lineages within Burkholderia.6,9,12,40 Nevertheless, recent whole genome sequence data followed by maximum-likelihood phylogeny using 106 concatenated orthologous genes10 provided strong support for separating Burkholderia sensu lato into Burkholderia sensu stricto, Paraburkholderia, Caballeronia, and Robbsia.12,13,41–43 Further phylogenomic analyses supported inclusion of the fungal endosymbionts Paraburkholderia rhizoxinica (basonym Burkholderia rhizoxinica) and Paraburkholderia endofungorum (basonym Burkholderia endofungorum) in the recently proposed genus Mycetohabitans.44 In addition, the new genus Trinickia was also proposed, including the nodulating Paraburkholderia symbiotica, the plant pathogen Paraburkholderia caryophylli, and the soil bacterium Paraburkholderia soli.44
Burkholderia sensu stricto includes members such as those pathogenic to humans (e.g. members of the Bcc),18–20 animals (e.g. B. pseudomallei and B. mallei),21–23 and plants (e.g. B. glumae, B. gladioli and B. plantarii),45 along with soil, low-virulence or nonpathogenic isolates (e.g. B. thailandensis E264).46 Paraburkholderia consists of diverse species including free-living organisms capable of fixing nitrogen (e.g. P. caballeronis),47 plant symbionts capable of fixing nitrogen (e.g. P. nodosa),48 and free-living, environmental species involved in the degradation of pollutants such as PCB (e.g. P. xenovorans).12,49 Caballeronia currently includes environmental species and no nitrogen-fixing species. An example is C. jiangsuensis that can degrade the organophosphate and extremely hazardous pesticide methyl parathion.8,35
The phylogenetic clusters of the Burkholderia sensu lato lineages do not strictly distinguish between pathogenic and nonpathogenic/beneficial strains. Although Burkholderia sensu stricto consists primarily of pathogenic strains, some nonpathogenic/beneficial strains fall within this group as well. For instance, although the Bcc clade includes pathogenic species such as B. cenocepacia, an opportunistic pathogen to immunocompromised individuals and cystic fibrosis patients, it also includes B. vietnamiensis and B. ambifaria which have been used for promoting plant growth and biocontrol, respectively.18,19 Within the B. pseudomallei clade, B. mallei and B. pseudomallei are identified as bioterrorism agents by the Centers for Disease Control and Prevention, while B. thailandensis is a low virulence strain.23,46 Due to their phylogenetic relationship, B. thailandensis is studied as a model, nonpathogenic species to gain insight into the pathogenic behavior of strains in the B. pseudomallei group. Finally, members of the Paraburkholderia and Caballeronia genera contain mostly nonpathogenic species.8,12,13 Although Paraburkholderia and Caballeronia species have been isolated from clinical samples, pathogenicity has not been unambiguously demonstrated.10,13,50,51
For more details on the taxonomy of Burkholderia, the reader is referred to recent reviews by Depoorter et al.,9 Beukes et al.,10 and Estrada de los Santos et al.11
Undeniably, bacteria from the Burkholderia sensu lato group have attracted attention of the scientific community. The revisions within the genus reflect the diversity of the microorganisms populating this group of bacteria and the challenges in physically distinguishing isolates within this group solely on phenotype and 16S rRNA gene sequences. Apart from the complicated taxonomy and reflecting its ecological versatility, the Burkholderia sensu lato group has emerged as a promising source of natural products.
Natural Products Isolated from Burkholderia Bacteria
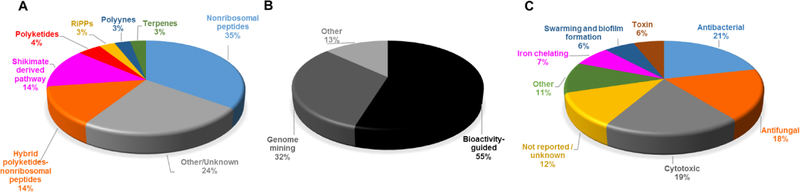
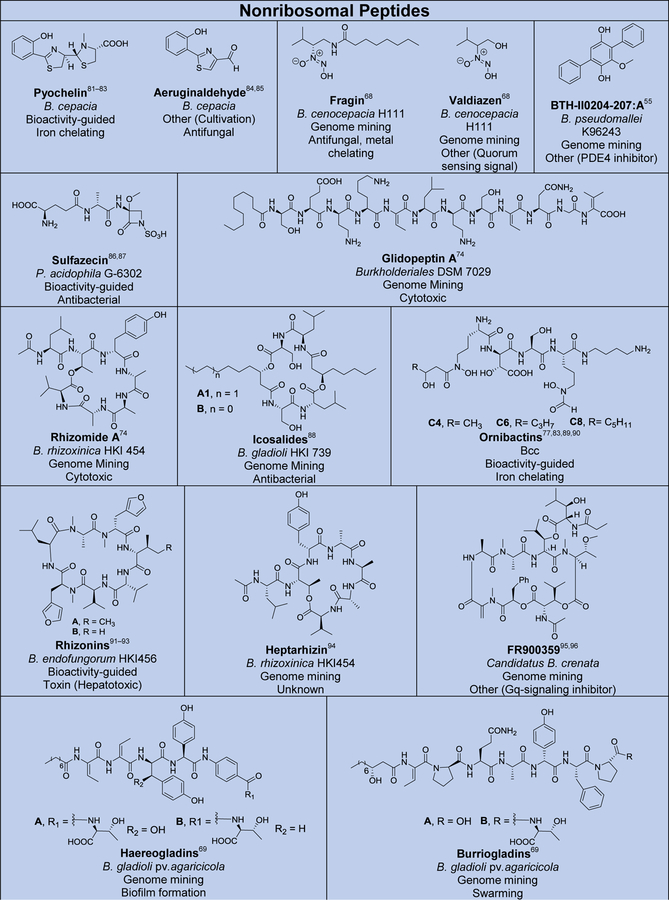
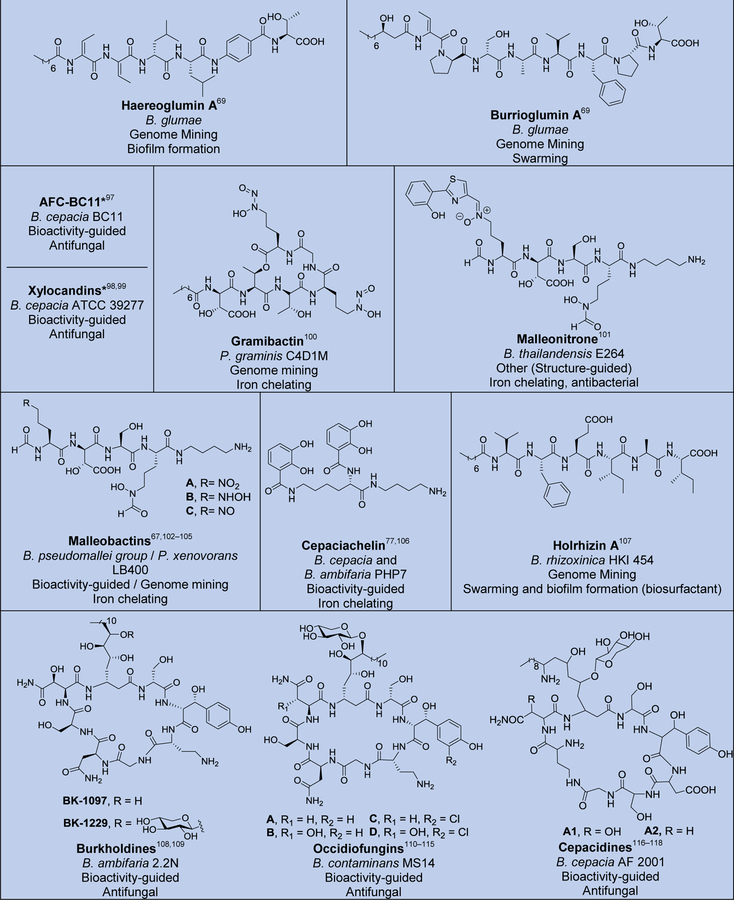
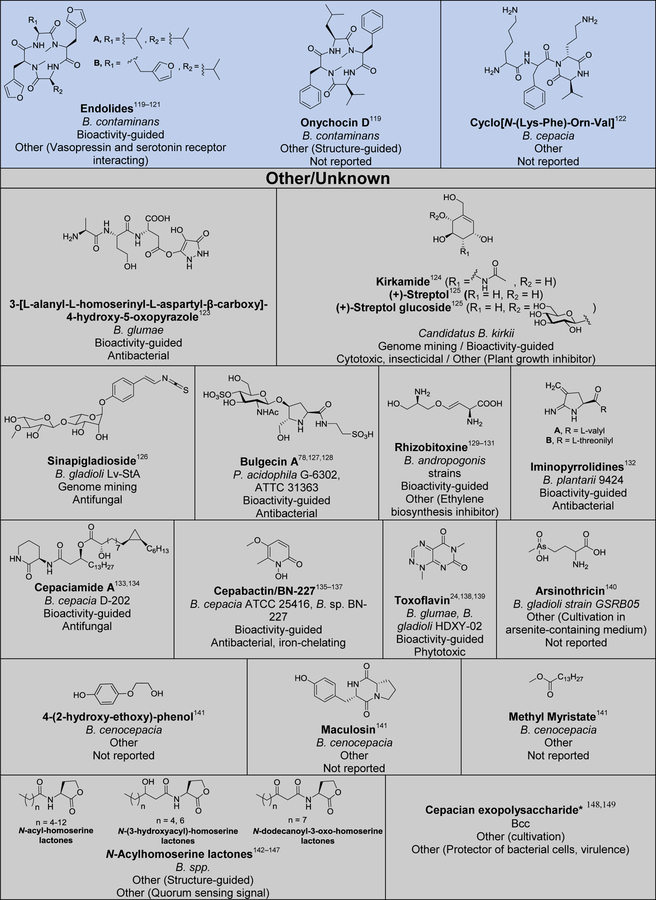
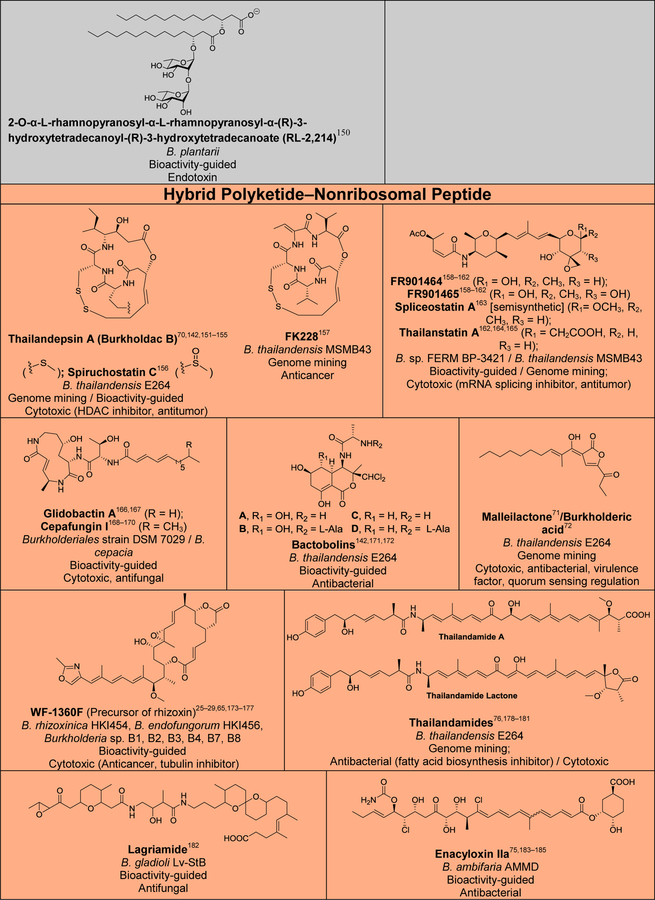
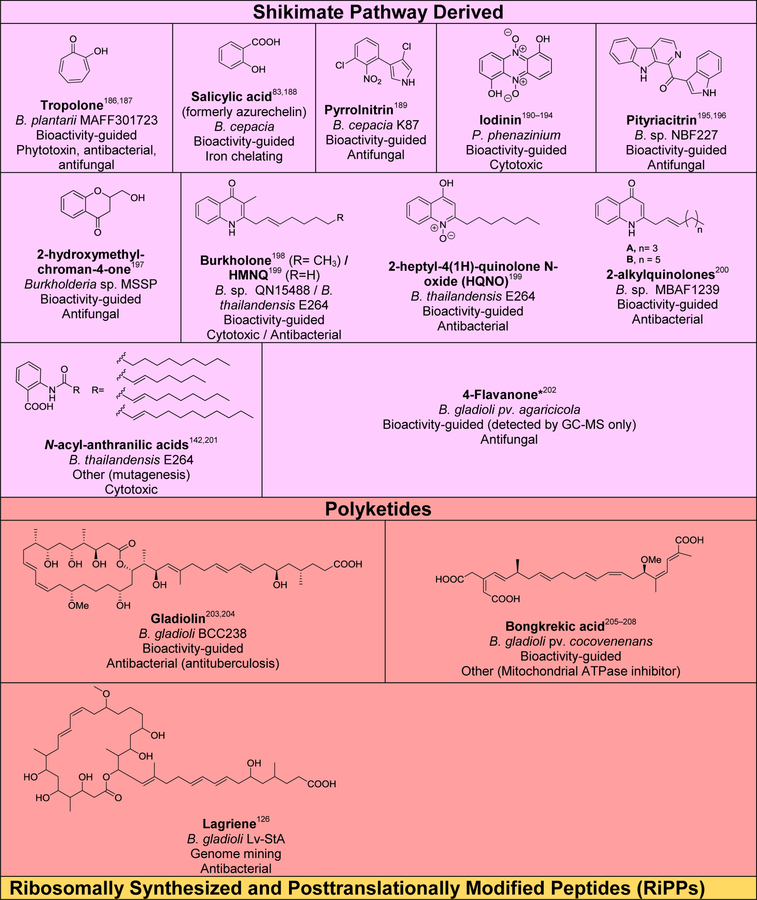
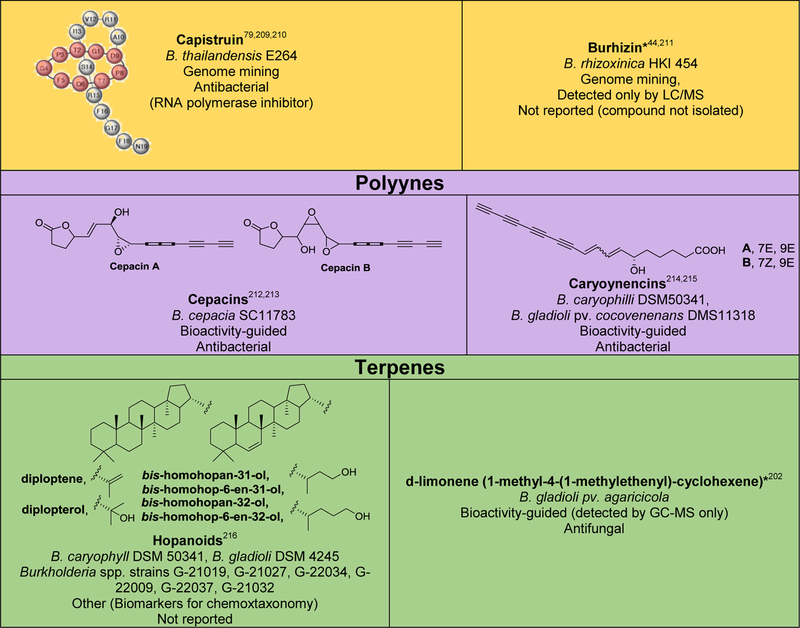
Although several reviews have been published on specific aspects of Burkholderia natural products, such as natural products discovered via genome mining,5 siderophores and lipopeptides,52 nonribosomal peptides and polyketides,53 antibiotics from neglected bacteria,3 and antibiotics from Gram-negative bacteria,54 a comprehensive account of Burholderia as a source of natural products is outstanding. In order to compile information regarding compounds isolated, their reported bioactivities, and the method of isolation, we searched Web of Science and SciFinder using following search terms “[Burkholderia or Paraburkholderia or Caballeronia or Robbsia] and [natural product or secondary metabolite or isolated compound]”. Based on these searches, 66 structural classes were identified. We defined a structural class as compounds known or expected to be encoded in the same (or very similar) gene cluster. It is important to note that Burkholderia strains isolated before the early 1990s were misclassified as Pseudomonas. Although we were able to identify a small number of such cases, 66 compound classes is likely an underestimation. An analysis of biosynthetic class, reported bioactivities, and the method of identification is presented in Figure 2. The structures of the analyzed compounds are depicted in Figure 3.
Figure 2.
Analysis of natural products isolated from Burkholderia sensu lato. (A) Pie chart depicting biosynthetic class. Compounds belonging to the same structural class (defined as known or expected to be encoded in the same or very similar BGC) were counted as one. The 66 structural classes (corresponding to 66 cells in Figure 3) were then classified into seven biosynthetic classes as shown. Compounds that did not belong to any of the seven classes or for which the biosynthesis was unknown were classified as “other/unknown”. (B) Natural product identification method. “Other” includes structure-guided isolation. (C) Reported bioactivity. If a compound displayed more than one bioactivity, they were categorized as follows: Cytotoxic compounds that had more than one bioactivity were counted as “cytotoxic” only. Antitumor and anticancer compounds were also included under cytotoxic. Compounds that had antifungal and antibacterial activity were added to one of the two categories based on highest displayed potency. Activities that did not fit within the depicted groups were designated as “other”, which includes phosphodiesterase 4 inhibitor, Gq-signaling inhibitor, vasopressin and serotonin receptor interacting, plant growth inhibitor, ethylene biosynthesis inhibitor, virulence, and quorum sensing signal.
Figure 3.
Natural products isolated from Burkholderia sensu lato. Compounds are grouped and color-coded based on biosynthetic class as in Figure 2A. The bacterial source (B., Burkholderia, P., Paraburkholderia), discovery method, reported bioactivity, and references are indicated. For the “other/unknown” category, the biosynthesis is either not yet elucidated or the biosynthetic class does not belong in the categories depicted. In cases where many congeners of a compound class have been isolated, only representative examples are shown. Note that B. rhizoxinica and B. endofungorum as the reported sources of rhizomide A, heptarhizin, holrhizin A, WF-1360F, burhizin, and rhizonins were later revised as P. rhizoxinica and P. endofungorum and most recently transferred to the new genus Mycetohabitans. *Denotes that chemical structure was not fully elucidated or that the compound was detected by mass spectrometry only, in which cases we opted to not show the proposed structure with the exception of N-acylhomoserine lactones.
As it can be seen from Figures 2 and 3, and Table S1, known Burkholderia natural products are diverse in terms of biosynthetic class and structure. The biosynthetic class with the largest representation is that of nonribosomal peptides. Hybrid polyketide-nonribosomal peptides also appear to be common, with trans-AT type I polyketides appearing more often than cis-AT, which is the opposite of what is seen with the more extensively studied actinomycetes, in which trans-AT type I polyketides are rather rare.
In terms of biological activity, examples of reported bioactivity of therapeutic potential include antibacterial, antifungal, and cytotoxic. More unusual activities such as phosphodiesterase 4 (PDE4) inhibition and Gq-signaling inhibition were also reported. PDE4 inhibitors have anti-inflammatory and neuroprotective effects,55–57 whereas inhibition of Gq signaling is being investigated for asthma treatment.58 Moreover, activities involved in ecological functions and virulence such as swarming, biofilm formation, iron acquisition, and quorum sensing have also been reported.
Based on our dataset, the percentage of natural products coming from genome mining (32%) seems rather high. Factors that may contribute to this observation include a) our dataset is potentially biased towards compounds that were discovered after the early 1990s since Burkholderia strains were misclassified as Pseudomonas before then and even after; and b) Burkholderia have been neglected as a source of natural products and have only more recently been studied, a timing that coincides with the genomics era.
The main advantage of bioactivity-guided isolation is clearly to identify compounds that show a desired bioactivity. Disadvantages include a) not all gene clusters are actively transcribed to levels that allow detection in bioassays, and b) bioactivity-guided isolation requires cultivation of the native producer. On one hand, genome mining offers the chance to overcome some of these challenges to a) discover compounds encoded in gene clusters that are poorly expressed under given laboratory growth conditions, and b) discover compounds from uncultivated bacteria and metagenomics data sets. On the other hand, a drawback of genome mining is that biological activity can often not be predicted, although examples of target-directed or resistance-gene-directed genome mining have been described.59–62 Another drawback of genome mining is that some compounds may be missed if biosynthesis is unknown and consequently the BGC is not picked up by automated software tools,63 in which case bioactivity-guided isolation has the upper hand. Therefore bioactivity-guided isolation and genome mining are complementary approaches.
In order to enable genome mining of Burkholderia, genetic engineering techniques are important. If the gene cluster is actively transcribed under the used culture condition, gene deletion can help with compound identification through comparative metabolite analysis of mutant and wild-type strains. Gene knockouts can also aid biosynthesis investigations. Moreover, if the gene cluster is not transcribed to levels that enable compound detection, promoter exchange can be used to activate gene expression.
Many examples of genetic engineering of Burkholderia have been reported. We highlight some examples here to illustrate the different methods used. Gene knockouts via traditional homologous recombination have been performed in several Burkholderia strains for biosynthesis investigations of e.g. spliceostatins,64 rhizoxin,65 malleobactin,66,67 and fragin.68 The construction of gene knockouts using the Red/ET recombination method enabled confirmation of the BGCs encoding for the production of haereogladin and burriogladin in Burkholderia gladioli pv. agaricicola.69 The Flp-FRT recombination system was employed to generate mutants for the investigation of thailandepsin.70 Promoter exchange with a rhamnose inducible promoter71 and with the constitutive PthaA promoter72 were used to activate the malleilactone/burkholderic acid pathway in B. thailandensis E264. Additionally, the constitutive PS7 promoter and the E. coli PBAD L-arabinose inducible promoter were used to increase expression of a cytochrome P450 gene and improve production of thailanstatin A.73 The discovery and exploitation of cloned, native recombinase genes enabled the activation of previously silent BGCs in Burkholderiales strain DSM7029, resulting in the isolation of glidopeptin.74 Finally, transposon mutagenesis was applied in investigations of the enacyloxin,75 thailandamide,76 ornibactin,77 and bulgecin78 BGCs.
In addition to genetic engineering of native producers, heterologous expression has also been used for natural product discovery from Burkholderia. Examples include heterologous expression of BGCs encoding the lasso peptide capistruin,79 and the polyketide-nonribosomal peptide glidobactin80 in E. coli, and of the nonribosomal peptide BTH-II0204–207:A in P. aeruginosa.55
This section highlighted Burkholderia bacteria as a promising source of natural products of diverse structures and bioactivities. The next section will introduce Burkholderia genomes as an untapped source of yet more natural products.
The Versatile Burkholderia Genome – An Opportunity to Mine for Natural Products
With the growing interest in Burkholderia there are numerous sources of publicly available genomes. In order to provide an user-friendly database of annotated Burkholderia genomes to the public, the Brinkman lab at Simon Fraser University launched the Burkholderia Genome Database (see http://www.burkholderia.com), currently featuring 208 complete genomes and 1,603 draft genomes, compiled from many databases (including those from NCBI) in one platform.217
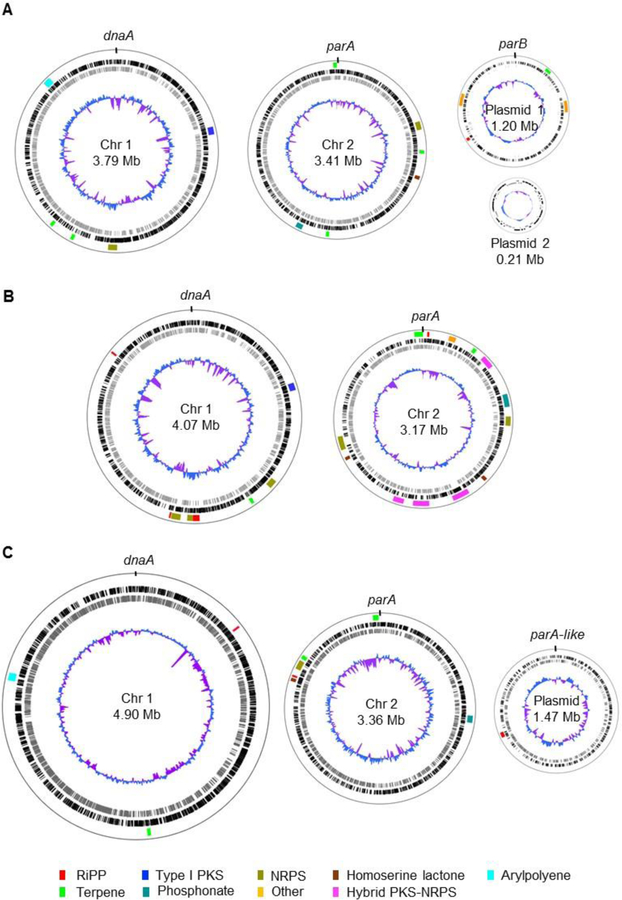
The versatility and diversity of Burkholderia species are attributed to their large and complex genomes. The average genome size is ~7.5 Mb, ranking among the top 5% of bacterial genomes.5,218 The genomes of Burkholderia bacteria usually feature two circular chromosomes (Figure 4), with the exception of the endosymbiont B. rhizoxinica which has a small genome of 3.75 Mb and only one chromosome.218,219 Moreover, Burkholderia can have none or up to six plasmids (Table S2).220 Burkholderia genomes contain a large number of insertion sequences (IS), which can lead to genomic rearrangements, replicon fusion, mobilization of DNA elements, and recruitment of foreign genes.221,222 Additionally, conserved DNA regions may have distinct distributions within different genomes. This adaptable genetic make-up is believed to play a role in the evolution of many biological functions.222
Figure 4.
Genome maps of representative Burkholderia species highlighting their multi-replicon nature and the distribution of biosynthetic gene clusters. (A) Burkholderia cepacia ATCC 25416 (accession codes: NZ_CP012981, NZ_CP012982, NZ_CP012983 and NZ_CP012984 for chromosomes 1 and 2 and plasmids 1 and 2, respectively). Megaplasmid 1 has also been referred to as chromosome 3. Plasmid 2 was named pBC25416. (B) Burkholderia pseudomallei K96243 (accession codes: NC_006350 and NC_006351 for chromosomes 1 and 2, respectively). (C) Paraburkholderia xenovorans LB400 (accession codes: NC_007951, NC_007952 and NC_007953 for chromosomes 1 and 2 and plasmid). The megaplasmid has also been termed chromosome 3. In all cases, chromosome 1 is oriented to dnaA and chromosome 2 to parA. Plasmids are oriented to parB or parA-like proteins. The location of biosynthetic gene clusters for natural products is indicated and color-coded by biosynthetic class as shown (lane 1 from the outside in). Predicted open reading frames (ORFs) on the leading (black) and lagging (gray) strands are shown on lanes 2 and 3, respectively. A normalized plot of guanosine + cytosine (G+C) content (blue/purple) is depicted in lane 4.
The large genome sizes of most Burkholderia sensu lato genomes combined with their plasticity translate into remarkable phenotypic diversity, including the production of a diverse array of natural products. As more Burkholderia genomes become available, there has been a collective realization that this bacterial group is a promising and relatively untapped source of natural products with therapeutic and biotechnological relevance.3,5,9,52–54
The number of putative natural product BGCs varies depending on the species and their ecological niches and do not necessarily correlate with genome size. For example, an antiSMASH223,224 analysis of B. cepacia ATCC 25416 (genome size of 8.6 Mb) yielded 15 putative BGCs (1.7 BGCs per Mb), that of B. pseudomallei K96243 (genome size of 7.25 Mb) yielded 21 putative BGCs (2.9 BGCs per Mb), and that of P. xenovorans LB400 (genome size of 9.7 Mb) yielded only ten putative BGCs, that is 1.0 BGC per Mb (Figure 4). We were first intrigued by the paucity of BGCs in P. xenovorans LB400 genome despite its large size. It turns out that although the large P. xenovorans LB400 genome does not encode the biosynthesis of many secondary metabolites, it does encode a remarkable capacity to degrade aromatic compounds as evidenced by the presence of 31 aromatic catabolic pathways, reflecting its ecological niche.34 Thus, it seems that the P. xenovorans LB400 genome dedicates more coding capacity to catabolic rather than anabolic pathways. Our next question was whether other Paraburkholderia strains follow the same trend of large genomes with relatively fewer natural product BGCs. To answer that question, we compiled data from 45 genomes available in the antiSMASH database (Table S2). The average number of BGCs per Mb of genome is 1.4 and the range is 0.8 to 2.2. Thus, while there are certainly other Paraburkholderia that have a low ratio of BGCs to genome size, Paraburkholderia can also be a rich source of BGCs with ratios of up to 2.2.
Furthermore, a skewed distribution of BGCs has been observed, with chromosome 1 containing lesser and chromosome 2 and plasmids containing more BGCs per base pair on average. The opposite is true for essential genes and genes involved in primary metabolism, which tend to be more concentrated on chromosome 1.34,225,226 This skewed concentration of BGCs is reminiscent of other BGC-rich genomes such as those of actinomycetes. However, Burkholderia tend to segregate essential and non-essential functions between different chromosomes, whereas Streptomyces species, for instance, have only one linear chromosome containing a core region around the origin of replication that is rich in essential and housekeeping genes, and two “arms” encoding mostly secondary metabolism.227
One interesting observation is that at least one phosphonate BGC is observed in most Burkholderia sensu lato genomes (Figure 4). According to an analysis by Yu et al., ~5% of sequenced bacterial genomes encode phosphonate biosynthesis.228 Yet this percentage is greatly skewed in Burkholderia in which over 90% of Burkholderia genomes appear to feature at least one phosphonate BGC, with selected strains containing as many as four. Burkholderia strains have been shown to solubilize phosphate salts present in soil, presumably via the production of organic acids.229 Phosphate solubilization increases its bioavailability, promoting the growth of plants,229 and perhaps facilitating uptake and phosphonate production by Burkholderia themselves.
The abundance of BGCs encoded in Burkholderia sensu lato genomes provides remarkable opportunities for natural product discovery.
Biosynthesis of Selected Burkholderia Natural Products Highlighting Unusual Features
One of the rewards of connecting natural products to their BGCs is the ability to gain insight into how natural products are biosynthesized and into the enzymes that have evolved to catalyze complex chemical reactions. One of the most striking examples of complex enzymology is arguably that of modular, type I polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs). Here, we start by briefly describing canonical PKSs and NRPSs. We then highlight unusual features of the biosynthesis of three, selected natural products produced by Burkholderia via PKS and NRPS catalysis.
NRPS and PKS megaenzymes are distinctively arranged in an assembly line fashion.230–232 A set of catalytic domains grouped into modules govern the incorporation of each monomer through sequential condensation. PKSs utilize a wide range of starter and extender units.233,234 The most common starter unit is acetyl-CoA and the most typical extender units are malonyl-CoA and methylmalonyl-CoA. Minimally, a PKS loading module contains two domains, an acyl-transferase (AT), and an acyl-carrier protein (ACP)/thiolation (T) domain for monomer selection and activation, respectively, whereas PKS extension modules also contain a β-ketosynthase (KS) domain to catalyze decarboxylative Claisen-type condensation.235–237 A common, alternative loading module organization consists of a mutated KS domain (KSQ, active site C to Q mutation, condensation-incompetent), an AT and an ACP. In this case, the starter AT selects malonyl-CoA or methylmalonyl-CoA and the KSQ serves to decarboxylate the dicarboxylic acids to yield the acetyl and propionyl starter units, respectively.238 Auxiliary domains can also be present to catalyze reduction of the β-carbonyl, these are ketoreductase (KR), dehydratase (DH) and enoylreductase (ER). Notably, in addition to the cis-AT type PKS described above, an evolutionarily distinct trans-AT type PKS was discovered in which the AT function is not covalently fused with the PKS but is rather provided in trans.239 trans-AT PKSs were only relatively recently discovered because they are rare in actinomycetes, but they are now known to be present in various bacterial groups. In fact, about 38% of PKSs found in sequenced bacterial genomes are of the trans-AT type.239
The organization of NRPSs is analogous to that of cis-AT PKSs. NRPSs utilize both proteinogenic and nonproteinogenic amino acids as monomers. The adenylation (A) domain is analogous to the AT in selecting monomers for activation. Here, amino acids are activated using ATP and transferred to the T domain or peptidyl carrier protein (PCP) in which a thioester is formed. The condensation (C) domain is analogous to KS in catalyzing monomer condensation, in this case via amide bond formation.240 At the end of the assembly line, thioesterase (TE) domains are responsible for product release which can happen by hydrolysis or cyclization. Some PKSs and NRPSs contain a reductive terminal domain for product offloading.241
For both PKSs and NRPSs, the loading of each substrate usually follows co-linearity in which the number and order of modules reflect the number and order of monomers in the final product. Although there are exceptions to this co-linearity rule – including skipping, stuttering and inactive domains – the structure of the final product can be roughly predicted based on the domain organization of the modular enzymes. Several bioinformatics tools have been launched to aid BGC detection and annotation, including attempts at structure prediction of PKS and NRPS products.223,242
The following examples highlight unconventional characteristics of PKSs and/or NRPSs from Burkholderia. Our criteria for selecting the three examples presented below were a) relatively well-characterized pathway, b) unusual biosynthesis, and c) interesting structural motifs.
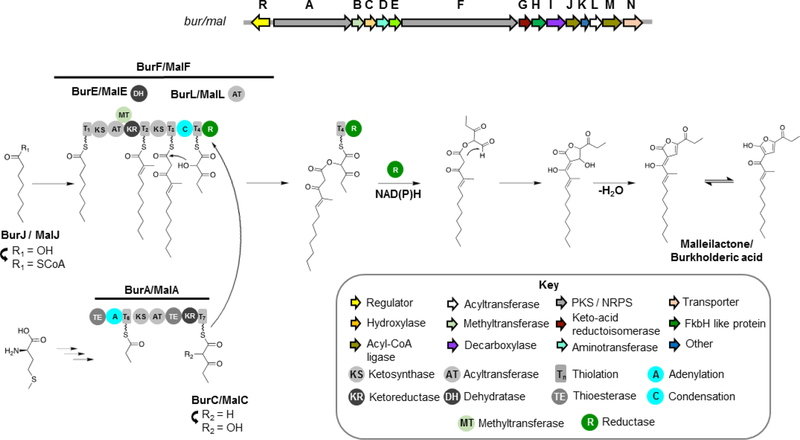
Malleilactone/burkholderic acid.
Malleilactone and burkholderic acid are tautomers that were independently reported by the Brady71 and Hertweck72 groups, respectively. The cognate orphan BGC (mal, and bur, respectively) was found to be conserved within the genomes of three of the B. pseudomallei group species, i.e. B. mallei, B. pseudomallei, and B. thailandensis. Disruption of the mal BGC was shown to attenuate virulence in animal models, suggesting a role for these compounds in pathogenesis.71
The hybrid mal/bur PKS-NRPS features unconventional domains and module organization, from which the structure of the natural product could not be accurately predicted based on precedent literature. The mal/bur BGC showcases a PKS-NRPS system that generates a furan-containing compound from two polyketide chains. Noncanonical features of the PKS-NRPS include the presence of two TE domains at unusual locations within the PKS-NRPS (not terminal), a dehydratase protein that acts in trans rather than being part of a module, mixed cis-AT and trans-AT architecture, and the incorporation of a propionate unit derived from methionine (Figure 5).
Figure 5.
Biosynthesis of malleilactone and burkholderic acid by B. pseudomallei, B. mallei and B. thailandensis. The model shown is according to studies in B. thailandensis E264 described in references71,72
Stable isotope labeling studies showed that acetate is incorporated into the whole backbone of burkholderic acid except for the presumed propionate starter (C15-C17) unit of BurA, which was surprisingly shown to be derived from methionine.72 Accordingly, the biosynthesis hypothesis put forth is that the propionyl starter unit loaded into BurA is derived from transamination, decarboxylation, and subsequent desulfurization of methionine, which is unprecedented for polyketide biosynthesis.72 Next, the AT domain in BurA shows specificity for malonyl-CoA and thus this extender unit is proposed to further undergo an α-hydroxylation, supported by the presence of hydroxylase BurC. An alternative hypothesis would be that hydroxymalonyl-ACP is the extender unit here based on the presence of a FkbH-like protein in the gene cluster, since the glyceryl transferase/phosphatase FkbH has been shown to be involved in the biosynthesis of hydroxymalonyl-ACP.71 However, feeding studies with 13C-labelled acetate support malonyl-CoA as extender unit.72 The two TE domains in BurA show sequence similarity to type II thioesterases which are involved in proof-reading rather than offloading.71
The other polyketide precursor is biosynthesized by BurF. The short chain fatty acid caprylic acid would be activated by CoA ligase BurJ and loaded as the starter unit of BurF. The first extender module likely incorporates malonyl-CoA followed by α-methylation as proposed by Franke et al.,72 given the presence of a methyltransferase domain in this module. An unusual feature of this module is that the DH domain is provided in trans by BurE rather than being fused with the PKS-NRPS. The next extender module would incorporate malonyl-CoA via trans AT BurL. The product of BurA is then loaded into BurF and the two polyketide precursors are condensed via a condensation (C) domain present in the final NRPS module, to yield an ester bond in contrast to the canonical amide bond. Release of the final product would be catalyzed by the terminal reductase (R) domain in BurF, which would catalyze reductive cleavage to form an aldehyde and subsequent intramolecular cyclization to yield the lactone/hydroxyfuran moiety.71,72
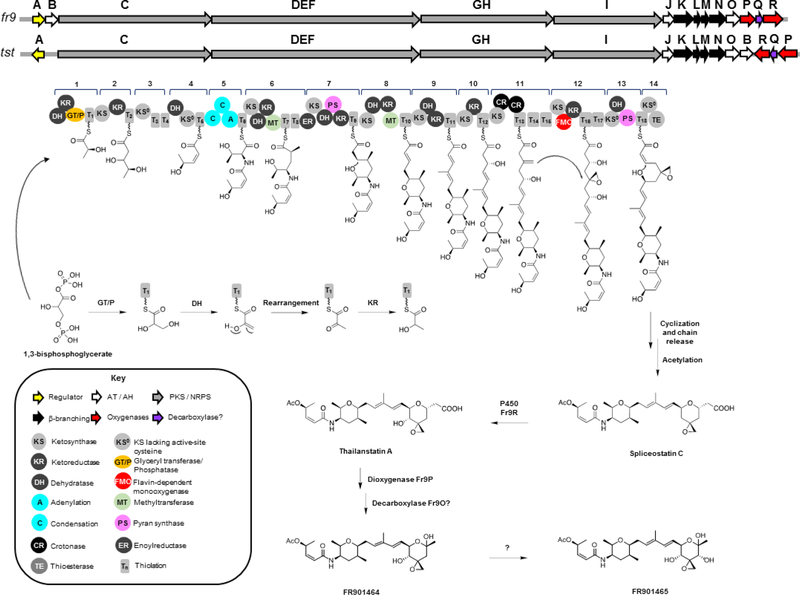
Spliceostatins/FR01464/thailanstatins.
FR901464 and FR901465 (Figure 6) were first isolated from Burkholderia sp. FERM BP-3421 (formerly Pseudonomas sp. no. 2663).158–160 Later, a stabilized, methyl ketal derivative of FR901464 was generated by semi-synthesis and shown to target the spliceosome; the compound was thus termed spliceostatin A.163An analog of FR901464 containing a terminal carboxylic acid instead of the hemiketal was later isolated from B. thailandensis MSMB43 and named thailanstatin A.164 Concomitantly, compounds with a terminal carboxylic acid were also isolated from strain FERM BP-3421.162 In fact, both strains were shown to produce hemiketal and carboxylic acid analogs, albeit in different ratios.64 Due to its promising activity as splicing modulator and much improved stability, thailanstatin A and derivatives were evaluated as antibody drug conjugates in preclinical studies.243
Figure 6.
Biosynthesis of spliceostatins by Burkholderia sp. FERM BP-3421 and B. thailandensis MSMB43. The model shown is according to several lines of evidence described in references64,161,244,245 Module numbering is according to the current convention for trans-AT PKSs.239
To make the discussion below easier to follow we will refer to this class of compounds as spliceostatins. Biosynthesis of spliceostatins is encoded in hybrid trans-AT PKS-NRPS gene clusters in both strains. Biosynthesis of FR901464 was first proposed by Zhang161 and Liu164 and later revised by Eustáquio et al.64 The biosynthesis of spliceostatins involves the fr9 and tst loci in B. sp. FERM BP-3421 and in B. thailandensis MSMB43, respectively, which was confirmed by construction of knockout mutants and biochemical studies.64,161,164
The spliceostatin BGC (fr9/tst) exemplifies the distinctiveness of trans-AT PKSs, which in addition to having stand-alone ATs, also feature unusual domain orders, unique domains, split modules (meaning that they are divided between two proteins), non-elongating modules, and other functions provided in trans in addition to the AT.239 Spliceostatin biosynthesis starts with loading of the unusual starter unit 1,3-bisphosphoglycerate (1,3-BPG).161 In vitro studies using recombinant glyceryl transferase/phosphatase (GT/P) and T domains from the starter module support glyceryl loading to the T domain as catalyzed by GT/P.161 As such, loading of 1,3-BPG is analogous to what has been proposed for bryostatin biosynthesis.246 After 1,3-BPG loading to T and dephosphorylation, the DH in the starter module was shown to catalyze dehydration followed by ketoreduction by KR to yield L-lactyl-S-T (Figure 6).244
Another interesting feature in spliceostatin biosynthesis is the cis-double bond. The corresponding modules 2–4 have following domain organization within the C-terminal end of Fr9C: KS-KR-T-KS0-T-T-TE/DH-KS0-T. He et al.245 provided biochemical evidence showing that the domain initially predicted to be a TE actually functions as dehydratase DH catalyzing the formation of the cis-double bond. Additionally, the second KS0 was shown to function as a gatekeeper ensuring that only the intermediate containing the cis-double bond is transferred down the assembly line.245
According to the biosynthesis model, product hydrolysis catalyzed by TE and acetylation leads to the free carboxylic acid compound termed spliceostatin C. The function of the cytochrome P450 Fr9R as a 4-hydroxylase is corroborated by gene knockout studies. As evidenced by genetic and biochemical investigations, the Fe(II)/α-ketoglutarate-dependent dioxygenase Fr9P catalyzes hydroxylation at C-1, which followed by decarboxylation leads to hemiketal FR901464.64 Decarboxylation of the β-hydroxyacid generated by Fr9P appears to be enzyme-catalyzed, based on studies using a cell-free lysate from strain FERM BP-3421. However, the decarboxylase remains to be experimentally identified. The hydroxylase responsible for formation of FR901465 has yet to be identified as well.
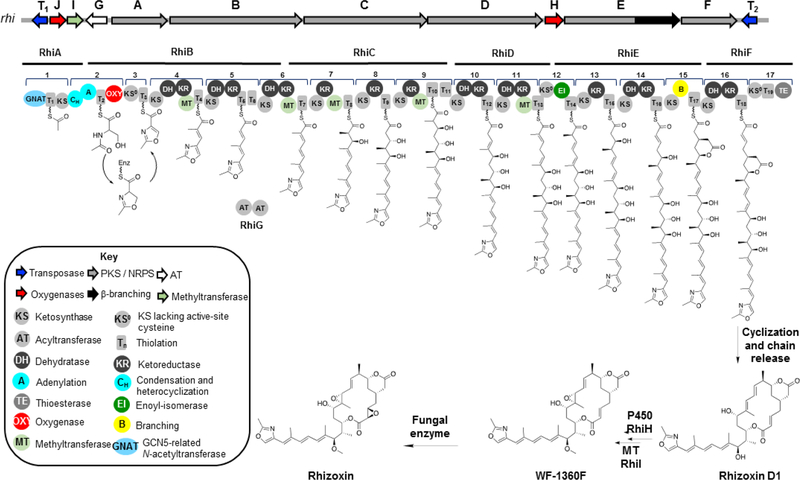
Rhizoxin.
Rhizoxin is an important natural product from both agricultural and pharmaceutical perspectives. While rhizoxin is a phytotoxin that causes rice seedling blight leading to significant crop losses, it has also been investigated in human clinical trials for cancer treatment due to its microtubule inhibitory effects.29,173–177 Rhizoxin was originally isolated from rice pathogenic fungi of the Rhizopus genus but later shown to be encoded in the genome of the endosymbiont B. rhizoxinica (later referred to as P. rhizoxinica and now Mycetohabitans rhizoxinica).8,25,44,65,247 Interestingly, biosynthesis of the bisepoxide rhizoxin was shown to involve both the bacterial rhi BGC for production of the monoepoxide precursor WF-1360F and a fungal enzyme that catalyzes the final epoxidation leading to rhizoxin (Figure 7).26
Figure 7.
Biosynthesis of rhizoxin. The model shown is according to several lines of evidence provided by references26,65,248,251–253
The intricate biosynthesis of rhizoxin was elucidated by the Hertweck group. The hybrid trans-AT PKS-NRPS consists of one PKS loading module containing the unusual GCN5-related N-acetyltransferase (GNAT), one NRPS module for extension with serine, heterocyclization and oxidation to the methyloxazoline ring, eleven PKS modules for linear extension with malonate, one module for double-bond isomerization (#12), and one beta-branching module (#15, Figure 7). Experimental evidence for gene cluster assignment was initially provided by disruption of the trans AT gene rhiG via homologous recombination.65
The PKS extension modules correlate well with the backbone structure of rhizoxin except for the 9,11-diene moiety and the δ-lactone. The formation of alkenes is typically predicted between acetate building blocks and, consequently, a 8,10-diene would be expected. Kusebauch et al. showed through in vivo mutagenesis experiments followed by structure elucidation of accumulated intermediates that the diene shift happens sequentially.248 The DH in module 10 appears to catalyze double bond formation and isomerization concomitantly, whereas the second shift occurs in a step-wise manner in which module 11 catalyzes the formation of an α-β-double bond as usual, and the downstream module 12 composed of KS0-DH*-ACP catalyzes the β-γ-double bond shift. The noncanonical DH* contains active site mutations that would make it catalytically inactive for dehydration but enable double bond migration. This unusual DH* domain (in fact, an enoyl isomerase, EI) is also observed in other PKS modules that encode double bond shifts such as in bacillaene biosynthesis.249 X-ray crystallography studies of such an enoyl isomerase domain revealed a catalytic histidine shuttles a proton between the γ- and the α-positions of the α-β-unsaturared intermediate.250
Arguably, the most exciting feature of rhizoxin structure and biosynthesis is the β-branched δ-lactone that branches off at C-5. The δ-lactone is not only required for biological activity, but elucidation of its biosynthesis revealed an unprecedented strategy for polyketide β-branching.248,251,252 The rhi gene cluster contains no genes that show sequence similarity to the mevalonate-like pathway genes as seen e.g. in the spliceostatin BGC presented in the previous section. Given that the corresponding module 11 is expected to introduce a double bond – rather than the carbonyl that is the substrate for mevalonate pathway-like β-branching –, the authors proposed a Michael-type addition mechanism (Figure 7).65,253 Based on TE domain inactivation and structure elucidation of the accumulated intermediates, Kusebauch et al. indeed showed that the substrate for β-branching contains a double bond.248 These analyses also demonstrated that the product of the β-branching module bears the δ-lactone. Subsequent biochemical and X-ray crystallography studies of the β-branching module showed that both the KS and the novel B domain are required for β-branching, but while the KS has catalytic function, the B domain, which shows a double hot dog fold characteristic of dehydratases, appears to have a structural role.251,252
Conclusion
Bacteria belonging to Burkholderia sensu lato are a promising source of natural products. In the past few decades, the Burkholderia genus has gained increasing scientific attention due to its impact on human health, on agriculture, and the potential pharmaceutical and biotechnological applications of its natural products. Given its heterogeneity, a proposal to split the Burkholderia genus was put forth. The current taxonomic revisions within the genus reveal the diversity of the isolates populating this group of bacteria and the challenges in physically distinguishing isolates within this group solely on phenotype and 16S rRNA gene sequences. In addition to the complicated taxonomy and diverse ecological niches, the Burkholderia sensu lato group also features genomes with unique characteristics including multiple replicons. A skewed distribution of function between replicons has been observed, where the larger chromosome 1, termed the “core chromosome”, encodes core cellular functions such as translation machinery, whereas the smaller chromosome 2 is biased towards secondary metabolism and has been termed the “life-style-determining chromosome”. Plasmids, if present, encode specialized, strain-specific functions and have been referred to as “individuality replicons”.34
Natural products that have been discovered from Burkholderia are diverse in terms of structure and bioactivity (Figures 2 and 3). Burkholderia genomes provide exciting opportunities for genome mining towards the discovery of yet more natural products. Unlike more extensively studied natural product producers, many of the BGCs found in Burkholderia harbor characteristics not seen in canonical PKS or NRPS systems. Unconventional biosynthesis translates into structurally and functionally diverse natural products of potential therapeutic and biotechnological relevance.
Supplementary Material
ACKNOWLEDGMENTS
We thank anonymous reviewers for their constructive criticism that helped improve this manuscript. Financial support for this work was provided by the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), under grant KL2TR002002 (to A.S.E.), by a UIC Provost Graduate Award and the Charles Wesley Petranek Memorial Scholarship (to S.K.), and by startup funds from the Department of Medicinal Chemistry and Pharmacognosy and the Center for Biomolecular Sciences, University of Illinois at Chicago (to A.S.E.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supporting Information.
The following files are available free of charge.
Table S1 (Natural products isolated from Burkholderia sensu lato), and Table S2 (Natural product BGCs detected in Paraburkholderia genomes). (PDF)
References
- (1).Cragg GM; Newman DJ Biochim. Biophys. Acta, Gen. Subj 2013, 1830, 3670–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Genilloud O Nat. Prod. Rep 2017, 34, 1203–1232. [DOI] [PubMed] [Google Scholar]
- (3).Pidot SJ; Coyne S; Kloss F; Hertweck C Int. J. Med. Microbiol 2014, 304, 14–22. [DOI] [PubMed] [Google Scholar]
- (4).Baltz RH J. Ind. Microbiol. Biotechnol 2017, 44, 573–588. [DOI] [PubMed] [Google Scholar]
- (5).Liu X; Cheng Y-Q J. Ind. Microbiol. Biotechnol 2014, 41, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Estrada-de los Santos P; Vinuesa P; Martínez-Aguilar L; Hirsch AM; Caballero-Mellado J Curr. Microbiol 2013, 67, 51–60. [DOI] [PubMed] [Google Scholar]
- (7).Vandamme P; Peeters C Antonie Van Leeuwenhoek 2014, 106, 57–65. [DOI] [PubMed] [Google Scholar]
- (8).Dobritsa AP; Samadpour M Int. J. Syst. Evol. Microbiol 2016, 66, 2836–2846. [DOI] [PubMed] [Google Scholar]
- (9).Depoorter E; Bull MJ; Peeters C; Coenye T; Vandamme P; Mahenthiralingam E Appl. Microbiol. Biotechnol 2016, 100, 5215–5229. [DOI] [PubMed] [Google Scholar]
- (10).Beukes CW; Palmer M; Manyaka P; Chan WY; Avontuur JR; van Zyl E; Huntemann M; Clum A; Pillay M; Palaniappan K; Varghese N; Mikhailova N; Stamatis D; Reddy TBK; Daum C; Shapiro N; Markowitz V; Ivanova N; Kyrpides N; Woyke T; Blom J; Whitman WB; Venter SN; Steenkamp ET Front. Microbiol 2017, 8, 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Estrada-de los Santos P; Rojas-Rojas FU; Tapia-García EY; Vásquez-Murrieta MS; Hirsch AM Ann. Microbiol 2016, 66, 1303–1314. [Google Scholar]
- (12).Sawana A; Adeolu M; Gupta RS Front. Genet 2014, 5, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dobritsa AP; Linardopoulou EV; Samadpour M Int. J. Syst. Evol. Microbiol 2017, 67, 3846–3853. [DOI] [PubMed] [Google Scholar]
- (14).Coenye T; Vandamme P Environ. Microbiol 2003, 5, 719–729. [DOI] [PubMed] [Google Scholar]
- (15).Compant S; Nowak J; Coenye T; Clément C; Ait Barka E FEMS Microbiol. Rev 2008, 32, 607–626. [DOI] [PubMed] [Google Scholar]
- (16).Eberl L; Vandamme P F1000Research 2016, 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Suárez-Moreno ZR; Caballero-Mellado J; Coutinho BG; Mendonça-Previato L; James EK; Venturi V Microb. Ecol 2012, 63, 249–266. [DOI] [PubMed] [Google Scholar]
- (18).Parke JL; Gurian-Sherman D Annu. Rev. Phytopathol 2001, 39, 225–258. [DOI] [PubMed] [Google Scholar]
- (19).Mahenthiralingam E; Baldwin A; Dowson CG J. Appl. Microbiol 2008, 104, 1539–1551. [DOI] [PubMed] [Google Scholar]
- (20).LiPuma JJ J. Nematol 2003, 35, 212–217. [PMC free article] [PubMed] [Google Scholar]
- (21).Cheng AC; Currie BJ Clin. Microbiol. Rev 2005, 18, 383–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Whitlock GC; Mark Estes D; Torres AG FEMS Microbiol. Lett 2007, 277, 115–122. [DOI] [PubMed] [Google Scholar]
- (23).Larsen JC; Johnson NH Mil. Med 2009, 174, 647–651. [PubMed] [Google Scholar]
- (24).Jeong Y; Kim J; Kim S; Kang Y; Nagamatsu T; Hwang I Plant Dis. 2003, 87, 890–895. [DOI] [PubMed] [Google Scholar]
- (25).Partida-Martinez LP; Hertweck C Nature 2005, 437, 884–888. [DOI] [PubMed] [Google Scholar]
- (26).Scherlach K; Busch B; Lackner G; Paszkowski U; Hertweck C Angew. Chemie Int. Ed 2012, 51, 9615–9618. [DOI] [PubMed] [Google Scholar]
- (27).Scherlach K; Partida-Martinez LP; Dahse H-M; Hertweck CJ Am. Chem. Soc 2006, 128, 11529–11536. [DOI] [PubMed] [Google Scholar]
- (28).Scherlach K; Brendel N; Ishida K; Dahse H-M; Hertweck C Org. Biomol. Chem 2012, 10, 5756–5759. [DOI] [PubMed] [Google Scholar]
- (29).Takahashi M; Iwasaki S; Kobayashi H; Okuda S; Murai T; Sato Y Biochim. Biophys. Acta 1987, 926, 215–223. [DOI] [PubMed] [Google Scholar]
- (30).Koga-Ban Y; Niki T; Nagamura Y; Sasaki T; Minobe Y DNA Res. 1995, 2, 21–26. [DOI] [PubMed] [Google Scholar]
- (31).Schmitt I; Partida-Martinez LP; Winkler R; Voigt K; Einax E; Dölz F; Telle S; Wöstemeyer J; Hertweck C ISME J. 2008, 2, 632–641. [DOI] [PubMed] [Google Scholar]
- (32).Parra-Cota FI; Peña-Cabriales JJ; de los Santos-Villalobos S; Martínez-Gallardo NA; Délano-Frier JP PLoS One 2014, 9, e88094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cordova-Kreylos AL; Fernandez LE; Koivunen M; Yang A; Flor-Weiler L; Marrone PG Appl. Environ. Microbiol 2013, 79, 7669–7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chain PSG; Denef VJ; Konstantinidis KT; Vergez LM; Agullo L; Reyes VL; Hauser L; Cordova M; Gomez L; Gonzalez M; Land M; Lao V; Larimer F; LiPuma JJ; Mahenthiralingam E; Malfatti SA; Marx CJ; Parnell JJ; Ramette A; Richardson P; Seeger M; Smith D; Spilker T; Sul WJ; Tsoi TV; Ulrich LE Zhulin IB; Tiedje JM Proc. Natl. Acad. Sci. U. S. A 2006, 103, 15280–15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Liu X-Y; Li C-X; Luo X-J; Lai Q-L; Xu J-H Int. J. Syst. Evol. Microbiol 2014, 64, 3247–3253. [DOI] [PubMed] [Google Scholar]
- (36).Burkholder WH Phytopathology 1950, 40, 115–117. [Google Scholar]
- (37).Yabuuchi E; Kosako Y; Oyaizu H; Yano I; Hotta H; Hashimoto Y; Ezaki T; Arakawa M Microbiol. Immunol 1992, 36, 1251–1275. [DOI] [PubMed] [Google Scholar]
- (38).Yabuuchi E; Kosako Y; Yano I; Hotta H; Nishiuchi Y Microbiol. Immunol 1995, 39, 897–904. [DOI] [PubMed] [Google Scholar]
- (39).Mahenthiralingam E; Urban TA; Goldberg JB Nat. Rev. Microbiol 2005, 3, 144–156. [DOI] [PubMed] [Google Scholar]
- (40).Gyaneshwar P; Hirsch AM; Moulin L; Chen W-M; Elliott GN; Bontemps C; Estrada-de los Santos P; Gross E; dos Reis FB; Sprent JI; Young PW; James EK Mol. Plant-Microbe Interact 2011, 24, 1276–1288. [DOI] [PubMed] [Google Scholar]
- (41).Garrity GM; Oren A Int. J. Syst. Evol. Microbiol 2015, 65, 2017–2025. [DOI] [PubMed] [Google Scholar]
- (42).Oren A; Garrity GM Int. J. Syst. Evol. Microbiol 2017, 67, 4291–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Lopes-Santos L; Castro DBA; Ferreira-Tonin M; Corrêa DBA; Weir BS; Park D; Ottoboni LMM; Neto JR; Destéfano SAL Antonie Van Leeuwenhoek 2017, 110, 727–736. [DOI] [PubMed] [Google Scholar]
- (44).Estrada-de los Santos P; Palmer M; Chávez-Ramírez B; Beukes C; Steenkamp E; Briscoe L; Khan N; Maluk M; Lafos M; Humm E; Arrabit M; Crook M; Gross E; Simon MF; dos Reis Junior FB; Whitman WB; Shapiro N; Poole PS; Hirsch AM; Venter SN; James EK Genes (Basel). 2018, 9, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Mannaa M; Park I; Seo Y-S Int. J. Mol. Sci 2018, 20, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Haraga A; West TE; Brittnacher MJ; Skerrett SJ; Miller SI Infect. Immun 2008, 76, 5402–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Rojas-Rojas FU; Tapia-García EY; Maymon M; Humm E; Huntemann M; Clum A; Pillay M; Palaniappan K; Varghese N; Mikhailova N; Stamatis D; Reddy TBK; Markowitz V; Ivanova N; Kyrpides N; Woyke T; Shapiro N; Hirsch AM; Estrada-de los Santos P Stand. Genomic Sci 2017, 12, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Dall’Agnol RF; Plotegher F; Souza RC; Mendes IC; dos Reis Junior FB; Béna G; Moulin L; Hungria M FEMS Microbiol. Ecol 2016, 92, fiw108. [DOI] [PubMed] [Google Scholar]
- (49).Goris J Int. J. Syst. Evol. Microbiol 2004, 54, 1677–1681. [DOI] [PubMed] [Google Scholar]
- (50).Peeters C; Meier-Kolthoff JP; Verheyde B; De Brandt E; Cooper VS; Vandamme P Front. Microbiol 2016, 7, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Deris ZZ; Van Rostenberghe H; Habsah H; Noraida R; Tan GC; Chan YY; Rosliza AR; Ravichandran M Int. J. Infect. Dis 2010, 14, e73–e74. [DOI] [PubMed] [Google Scholar]
- (52).Esmaeel Q; Pupin M; Kieu NP; Chataigné G; Béchet M; Deravel J; Krier F; Höfte M; Jacques P; Leclère V Microbiologyopen 2016, 5, 512–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Esmaeel Q; Pupin M; Jacques P; Leclère V Environ. Sci. Pollut. Res 2017, 25, 29794–29807. [DOI] [PubMed] [Google Scholar]
- (54).Masschelein J; Jenner M; Challis GL Nat. Prod. Rep 2017, 34, 712–783. [DOI] [PubMed] [Google Scholar]
- (55).Biggins JB; Liu X; Feng Z; Brady SF J. Am. Chem. Soc 2011, 133, 1638–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Omori K; Kotera J Circ. Res 2007, 100, 309–327. [DOI] [PubMed] [Google Scholar]
- (57).Press NJ; Banner KH Prog. Med. Chem 2009, 47, 37–74. [DOI] [PubMed] [Google Scholar]
- (58).Matthey M; Roberts R; Seidinger A; Simon A; Schröder R; Kuschak M; Annala S; König GM; Müller CE; Hall IP; Kostenis E; Fleischmann BK; Wenzel D; Sci. Transl. Med 2017, 9, eaag2288. [DOI] [PubMed] [Google Scholar]
- (59).Tang X; Li J; Millán-Aguiñaga N; Zhang JJ; O’Neill EC; Ugalde JA; Jensen PR; Mantovani SM; Moore BS ACS Chem. Biol 2015, 10, 2841–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Yan Y; Liu Q; Zang X; Yuan S; Bat-Erdene U; Nguyen C; Gan J; Zhou J; Jacobsen SE; Tang Y Nature 2018, 559, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Yeh H-H; Ahuja M; Chiang Y-M; Oakley CE; Moore S; Yoon O; Hajovsky H; Bok J-W; Keller NP; Wang CCC; Oakley BR ACS Chem. Biol 2016, 11, 2275–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Panter F; Krug D; Baumann S; Müller R Chem. Sci 2018, 9, 4898–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Braesel J; Lee J-H; Arnould B; Murphy BT; Eustáquio AS J. Nat. Prod 2019, 82, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Eustáquio AS; Janso JE; Ratnayake AS; O’Donnell CJ; Koehn FE Proc. Natl. Acad. Sci. U. S. A 2014, 111, E3376–E3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Partida-Martinez LP; Hertweck C ChemBioChem 2007, 8, 41–45. [DOI] [PubMed] [Google Scholar]
- (66).Alice AF; López CS; Lowe CA; Ledesma MA; Crosa JH J. Bacteriol 2006, 188, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Franke J; Ishida K; Ishida-Ito M; Hertweck C Angew. Chemie Int. Ed 2013, 52, 8271–8275. [DOI] [PubMed] [Google Scholar]
- (68).Jenul C; Sieber S; Daeppen C; Mathew A; Lardi M; Pessi G; Hoepfner D; Neuburger M; Linden A; Gademann K; Eberl L Nat. Commun 2018, 9, 1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Thongkongkaew T; Ding W; Bratovanov E; Oueis E; García-Altares M; Zaburannyi N; Harmrolfs K; Zhang Y; Scherlach K; Müller R; Hertweck C ACS Chem. Biol 2018, 13, 1370–1379. [DOI] [PubMed] [Google Scholar]
- (70).Wang C; Henkes LM; Doughty LB; He M; Wang D; Meyer-Almes F-J; Cheng Y-QJ Nat. Prod 2011, 74, 2031–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Biggins JB; Ternei MA; Brady SF J. Am. Chem. Soc 2012, 134, 13192–13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Franke J; Ishida K; Hertweck C Angew. Chemie Int. Ed 2012, 51, 11611–11615. [DOI] [PubMed] [Google Scholar]
- (73).Eustáquio AS; Chang L-P; Steele GL; ÓDonnell CJ; Koehn FE Metab. Eng 2016, 33, 67–75. [DOI] [PubMed] [Google Scholar]
- (74).Wang X; Zhou H; Chen H; Jing X; Zheng W; Li R; Sun T; Liu J; Fu J; Huo L; Li YZ; Shen Y; Ding X; Müller R; Bian X; Zhang Y Proc. Natl. Acad. Sci. U. S. A 2018, 115, E4255–E4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Mahenthiralingam E; Song L; Sass A; White J; Wilmot C; Marchbank A; Boaisha O; Paine J; Knight D; Challis GL Chem. Biol 2011, 18, 665–677. [DOI] [PubMed] [Google Scholar]
- (76).Wozniak CE; Lin Z; Schmidt EW; Hughes KT; Liou TG Antimicrob. Agents Chemother 2018, 62, 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Agnoli K; Lowe CA; Farmer KL; Husnain SI; Thomas MS J. Bacteriol 2006, 188, 3631–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Horsman ME; Marous DR; Li R; Oliver RA; Byun B; Emrich SJ; Boggess B; Townsend CA; Mobashery S ACS Chem. Biol 2017, 12, 2552–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Knappe TA; Linne U; Zirah S; Rebuffat S; Xie X; Marahiel MA J. Am. Chem. Soc 2008, 130, 11446–11454. [DOI] [PubMed] [Google Scholar]
- (80).Bian X; Huang F; Wang H; Klefisch T; Müller R; Zhang Y ChemBioChem 2014, 15, 2221–2224. [DOI] [PubMed] [Google Scholar]
- (81).Sokol PA J. Clin. Microbiol 1986, 23, 560–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Quadri LEN; Keating TA; Patel HM; Walsh CT Biochemistry 1999, 38, 14941–14954. [DOI] [PubMed] [Google Scholar]
- (83).Darling P; Chan M; Cox AD; Sokol PA Infect. Immun 1998, 66, 874–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Bukovits GJ; Mohr N; Budzikiewicz H Zeitschrift für Naturforsch. B 1982, 37, 877–880. [Google Scholar]
- (85).Ye L; Cornelis P; Guillemyn K; Ballet S; Hammerich O Nat. Prod. Commun 2014, 9, 789–794. [PubMed] [Google Scholar]
- (86).Asai M; Haibara K; Muroi M; Kintaka K; Kishi T J. Antibiot. (Tokyo) 1981, 34, 621–627. [DOI] [PubMed] [Google Scholar]
- (87).Li R; Oliver RA; Townsend CA Cell Chem. Biol 2017, 24, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Dose B; Niehs SP; Scherlach K; Flórez LV; Kaltenpoth M; Hertweck C ACS Chem. Biol 2018, 13, 2414–2420. [DOI] [PubMed] [Google Scholar]
- (89).Stephan H; Freund S; Meyer J-M; Winkelmann G; Jung G Liebigs Ann. der Chemie 1993, 1993, 43–48. [Google Scholar]
- (90).Stephan H; Freund S; Beck W; Jung G; Meyer J-M; Winkelmann G Biometals 1993, 6, 93–100. [DOI] [PubMed] [Google Scholar]
- (91).Partida-Martinez LP; Flores de Looss C; Ishida K; Ishida M; Roth M; Buder K; Hertweck C Appl. Environ. Microbiol 2007, 73, 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Steyn PS; Tuinman AA; van Heerden FR; van Rooyen PH; Wessels PL; Rabie CJ J. Chem. Soc. Chem. Commun 1983, 136, 47–49. [Google Scholar]
- (93).Wilson T; Rabie CJ; Fincham JE; Steyn PS; Schipper MA Food Chem. Toxicol 1984, 22, 275–281. [DOI] [PubMed] [Google Scholar]
- (94).Niehs SP; Dose B; Scherlach K; Roth M; Hertweck C ChemBioChem 2018, 19, 2167–2172. [DOI] [PubMed] [Google Scholar]
- (95).Crüsemann M; Reher R; Schamari I; Brachmann AO; Ohbayashi T; Kuschak M; Malfacini D; Seidinger A; Pinto-Carbó M; Richarz R; Reuter T; Kehraus S; Hallab A; Attwood M; Schiöth HB; Mergaert P; Kikuchi Y; Schäberle TF; Kostenis E; Wenzel D; Müller CE; Piel J; Carlier A; Eberl L; König GM Angew. Chemie Int. Ed 2018, 57, 836–840. [DOI] [PubMed] [Google Scholar]
- (96).Fujioka M; Koda S; Morimoto Y; Biemann KJ Org. Chem 1988, 53, 2820–2825. [Google Scholar]
- (97).Kang Y; Carlson R; Tharpe W; Schell MA Appl. Environ. Microbiol 1998, 64, 3939–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Meyers E; Bisacchi GS; Dean L; Liu WC; Minassian B; Slusarchyk DS; Sykes RB; Tanaka SK; Trejo W J. Antibiot. (Tokyo) 1987, 40, 1515–1519. [DOI] [PubMed] [Google Scholar]
- (99).Bisacchi GS; Hockstein DR; Koster WH; Parker WL; Rathnum ML; Unger SE J. Antibiot. (Tokyo) 1987, 40, 1520–1529. [DOI] [PubMed] [Google Scholar]
- (100).Hermenau R; Ishida K; Gama S; Hoffmann B; Pfeifer-Leeg M; Plass W; Mohr JF; Wichard T; Saluz H-P; Hertweck C Nat. Chem. Biol 2018, 14, 841–843. [DOI] [PubMed] [Google Scholar]
- (101).Trottmann F; Franke J; Ishida K; García-Altares M; Hertweck C Angew. Chemie Int. Ed 2019, 58, 200–204. [DOI] [PubMed] [Google Scholar]
- (102).Franke J; Ishida K; Hertweck C Chem. Eur. J 2015, 21, 8010–8014. [DOI] [PubMed] [Google Scholar]
- (103).Yang HM; Chaowagul W; Sokol PA Infect. Immun 1991, 59, 776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Vargas-Straube MJ; Cámara B; Tello M; Montero-Silva F; Cárdenas F; Seeger M PLoS One 2016, 11, e0151273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Franke J; Ishida K; Hertweck CJ Am. Chem. Soc 2014, 136, 5599–5602. [DOI] [PubMed] [Google Scholar]
- (106).Barelmann I; Meyer J-M; Taraz KBH Z Naturforsch 1996, 51C, 627–630. [Google Scholar]
- (107).Niehs SP; Scherlach K; Hertweck C Org. Biomol. Chem 2018, 16, 8345–8352. [DOI] [PubMed] [Google Scholar]
- (108).Tawfik KA; Jeffs P; Bray B; Dubay G; Falkinham JO; Mesbah M; Youssef D; Khalifa S; Schmidt EW Org. Lett 2010, 12, 664–666. [DOI] [PubMed] [Google Scholar]
- (109).Lin Z; Falkinham JO; Tawfik KA; Jeffs P; Bray B; Dubay G; Cox JE; Schmidt EW J. Nat. Prod 2012, 75, 1518–1523. [DOI] [PubMed] [Google Scholar]
- (110).Gu G; Smith L; Liu A; Lu S-E Appl. Environ. Microbiol 2011, 77, 6189–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Ravichandran A; Geng M; Hull KG; Li J; Romo D; Lu S-E; Albee A; Nutter C; Gordon DM; Ghannoum MA; Lockless SW; Smith L Antimicrob. Agents Chemother 2019, 63, e01585–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Lu S-E; Novak J; Austin FW; Gu G; Ellis D; Kirk M; Wilson-Stanford S; Tonelli M; Smith L Biochemistry 2009, 48, 8312–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Ellis D; Gosai J; Emrick C; Heintz R; Romans L; Gordon D; Lu S-E; Austin F; Smith L Antimicrob. Agents Chemother 2012, 56, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Chen K-C; Ravichandran A; Guerrero A; Deng P; Baird SM; Smith L; Lu S-E Appl. Environ. Microbiol 2013, 79, 2899–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Gu G; Wang N; Chaney N; Smith L; Lu S-E FEMS Microbiol. Lett 2009, 297, 54–60. [DOI] [PubMed] [Google Scholar]
- (116).Lee CH; Kim S; Hyun B; Suh JW; Yon C; Kim C; Lim Y; Kim CJ Antibiot. (Tokyo) 1994, 47, 1402–1405. [DOI] [PubMed] [Google Scholar]
- (117).Lim Y; Suh J-W; Kim S; Hyun B; Kim C; Lee C J. Antibiot. (Tokyo) 1994, 47, 1406–1416. [DOI] [PubMed] [Google Scholar]
- (118).Lee C-H; Suh JW; Cho Y-HJ Microbiol. Biotechnol 1999, 9, 672–674. [Google Scholar]
- (119).Almeida C; Silva Pereira C; Gonzalez-Menendez V; Bills G; Pascual J; Sánchez-Hidalgo M; Kehraus S; Genilloud O Appl. Environ. Microbiol 2018, 84, e00660–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Almeida C; Maddah F. El; Kehraus S; Schnakenburg G; König GM Org. Lett 2016, 18, 528–531. [DOI] [PubMed] [Google Scholar]
- (121).El Maddah F; Kehraus S; Nazir M; Almeida C; König GM J. Nat. Prod 2016, 79, 2838–2845. [DOI] [PubMed] [Google Scholar]
- (122).Pérez-Picaso L; Rios MY; Hernández AN; Martínez J Magn. Reson. Chem 2006, 44, 959–961. [DOI] [PubMed] [Google Scholar]
- (123).Mitchell RE; Greenwood DR; Sarojini V Phytochemistry 2008, 69, 2704–2707. [DOI] [PubMed] [Google Scholar]
- (124).Sieber S; Carlier A; Neuburger M; Grabenweger G; Eberl L; Gademann K Angew. Chemie Int. Ed 2015, 54, 7968–7970. [DOI] [PubMed] [Google Scholar]
- (125).Hsiao C-C; Sieber S; Georgiou A; Bailly A; Emmanouilidou D; Carlier A; Eberl L; Gademann K Chem. Eur. J 2019, 25, 1722–1726. [DOI] [PubMed] [Google Scholar]
- (126).Flórez LV; Scherlach K; Gaube P; Ross C; Sitte E; Hermes C; Rodrigues A; Hertweck C; Kaltenpoth M Nat. Commun 2017, 8, 15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Tomoshige S; Dik DA; Akabane-Nakata M; Madukoma CS; Fisher JF; Shrout JD; Mobashery S ACS Infect. Dis 2018, 4, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Shinagawa S; Kasahara F; Wada Y; Harada S; Asai M Tetrahedron 1984, 40, 3465–3470. [Google Scholar]
- (129).Mitchell RE; Frey EJ Physiol. Mol. Plant Pathol 1988, 32, 335–341. [Google Scholar]
- (130).Mitchell RE; Coddington JM Phytochemistry 1991, 30, 1809–1814. [Google Scholar]
- (131).Yasuta T; Satoh S; Minamisawa K Appl. Environ. Microbiol 1999, 65, 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Mitchell RE; Teh KL Org. Biomol. Chem 2005, 3, 3540–3543. [DOI] [PubMed] [Google Scholar]
- (133).Jiao Y; Yoshihara T; Ishikuri S; Uchino H; Ichihara A Tetrahedron Lett. 1996, 37, 1039–1042. [Google Scholar]
- (134).Toshima H; Maru K; Saito M; Ichihara A Tetrahedron 1999, 55, 5793–5808. [Google Scholar]
- (135).Meyer J-M; Hohnadel D; Hallé F Microbiology 1989, 135, 1479–1487. [DOI] [PubMed] [Google Scholar]
- (136).Itoh J; Miyadoh S; Takahasi S; Amano S; Ezaki N; Yamada Y J. Antibiot. (Tokyo) 1979, 32, 1089–1095. [DOI] [PubMed] [Google Scholar]
- (137).Itoh J; Amano S; Ogawa Y; Kodama Y; Ezaki N; Yamada Y J. Antibiot. (Tokyo) 1980, 33, 377–382. [DOI] [PubMed] [Google Scholar]
- (138).Suzuki F; Sawada H; Azegami K; Tsuchiya K J. Gen. Plant Pathol 2004, 70, 97–107. [Google Scholar]
- (139).Li X; Li Y; Wang R; Wang Q; Lu L Appl. Environ. Microbiol 2019, 85, e00106–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Kuramata M; Sakakibara F; Kataoka R; Yamazaki K; Baba K; Ishizaka M; Hiradate S; Kamo T; Ishikawa S Environ. Chem 2016, 13, 723–731. [Google Scholar]
- (141).Yap A-C; Chan K-G; Choo Y-M Sains Malaysiana 2016, 45, 1073–1077. [Google Scholar]
- (142).Mao D; Bushin LB; Moon K; Wu Y; Seyedsayamdost MR Proc. Natl. Acad. Sci. U. S. A 2017, 114, E2920–E2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Duerkop BA; Herman JP; Ulrich RL; Churchill MEA; Greenberg EP J. Bacteriol 2008, 190, 5137–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Duerkop BA; Ulrich RL; Greenberg EP J. Bacteriol 2007, 189, 5034–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Poonguzhali S; Madhaiyan M; Sa T Res. Microbiol 2007, 158, 287–294. [DOI] [PubMed] [Google Scholar]
- (146).Suárez-Moreno ZR; Caballero-Mellado J; Venturi V Microbiology 2008, 154, 2048–2059. [DOI] [PubMed] [Google Scholar]
- (147).Gotschlich A; Huber B; Geisenberger O; Tögl A; Steidle A; Riedel K; Hill P; Tümmler B; Vandamme P; Middleton B; Camara M; Williams P; Hardman A; Eberl L Syst. Appl. Microbiol 2001, 24, 1–14. [DOI] [PubMed] [Google Scholar]
- (148).Cescutti P; Foschiatti M; Furlanis L; Lagatolla C; Rizzo R Carbohydr. Res 2010, 345, 1455–1460. [DOI] [PubMed] [Google Scholar]
- (149).Ferreira AS; Leitão JH; Silva IN; Pinheiro PF; Sousa SA; Ramos CG; Moreira LM Appl. Environ. Microbiol 2010, 76, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Andrä J; Rademann J; Howe J; Koch MHJ; Heine H; Zähringer U; Brandenburg K Biol. Chem 2006, 387, 301–310. [DOI] [PubMed] [Google Scholar]
- (151).Wang C; Flemming CJ; Cheng Y-Q Med. Chem. Commun 2012, 3, 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Jang S; Janssen A; Aburjania Z; Robers MB; Harrison A; Dammalapati A; Cheng Y-Q; Chen H; Jaskula-Sztul R Oncotarget 2017, 8, 70828–70840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Biggins JB; Gleber CD; Brady SF Org. Lett 2011, 13, 1536–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Wilson AJ; Cheng Y-Q; Khabele DJ Ovarian Res. 2012, 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Weinlander E; Somnay Y; Harrison AD; Wang C; Cheng Y-Q; Jaskula-Sztul R; Yu X-M; Chen HJ Surg. Res 2014, 190, 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Klausmeyer P; Shipley SM; Zuck KM; McCloud TG J. Nat. Prod 2011, 74, 2039–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Liu X; Xie F; Doughty LB; Wang Q; Zhang L; Liu X; Cheng Y-Q Synth. Syst. Biotechnol 2018, 3, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Nakajima H; Sato B; Fujita T; Takase S; Terano H; Okuhara M J. Antibiot. (Tokyo) 1996, 49, 1196–1203. [DOI] [PubMed] [Google Scholar]
- (159).Nakajima H; Hori Y; Terano H; Okuhara M; Manda T; Matsumoto S; Shimomura K J. Antibiot. (Tokyo) 1996, 49, 1204–1211. [DOI] [PubMed] [Google Scholar]
- (160).Nakajima H; Takase S; Terano H; Tanaka H J. Antibiot. (Tokyo) 1997, 50, 96–99. [DOI] [PubMed] [Google Scholar]
- (161).Zhang F; He H-Y; Tang M-C; Tang Y-M; Zhou Q; Tang G-L J. Am. Chem. Soc 2011, 133, 2452–2462. [DOI] [PubMed] [Google Scholar]
- (162).He H; Ratnayake AS; Janso JE; He M; Yang HY; Loganzo F; Shor B; O’Donnell CJ; Koehn FE J. Nat. Prod 2014, 77, 1864–1870. [DOI] [PubMed] [Google Scholar]
- (163).Kaida D; Motoyoshi H; Tashiro E; Nojima T; Hagiwara M; Ishigami K; Watanabe H; Kitahara T; Yoshida T; Nakajima H; et al. Nat. Chem. Biol 2007, 3, 576–583. [DOI] [PubMed] [Google Scholar]
- (164).Liu X; Biswas S; Berg MG; Antapli CM; Xie F; Wang Q; Tang M-C; Tang G-L; Zhang L; Dreyfuss G; Cheng Y-QJ Nat. Prod 2013, 76, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Ghosh AK; Veitschegger AM; Nie S; Relitti N; MacRae AJ; Jurica MS J. Org. Chem 2018, 83, 5187–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Oka M; Nishiyama Y; Ohta S; Kamei H; Konishi M; Miyaki T; Oki T; Kawaguchi H J. Antibiot. (Tokyo) 1988, 41, 1331–1337. [DOI] [PubMed] [Google Scholar]
- (167).Schellenberg B; Bigler L; Dudler R Environ. Microbiol 2007, 9, 1640–1650. [DOI] [PubMed] [Google Scholar]
- (168).Shoji J; Hinoo H; Kato T; Hattori T; Hirooka K; Tawara K; Shiratori O; Terui Y J. Antibiot. (Tokyo) 1990, 43, 783–787. [DOI] [PubMed] [Google Scholar]
- (169).Terui Y; Nishikawa J; Hinoo H; Kato T; Shoji J J. Antibiot. (Tokyo) 1990, 43, 788–795. [DOI] [PubMed] [Google Scholar]
- (170).Krahn D; Ottmann C; Kaiser M Nat. Prod. Rep 2011, 28, 1854–1867. [DOI] [PubMed] [Google Scholar]
- (171).Kondo S; Horiuchi Y; Hamada M; Takeuchi T; Umezawa H J. Antibiot. (Tokyo) 1979, 32, 1069–1071. [DOI] [PubMed] [Google Scholar]
- (172).Seyedsayamdost MR; Chandler JR; Blodgett JAV; Lima PS; Duerkop BA; Oinuma K-I; Greenberg EP; Clardy J Org. Lett 2010, 12, 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Iwasaki S; Kobayashi H; Furukawa J; Namikoshi M; Okuda S; Sato Z; Matsuda I; Noda T J. Antibiot. (Tokyo) 1984, 37, 354–362. [DOI] [PubMed] [Google Scholar]
- (174).McLeod HL; Murray LS; Wanders J; Setanoians A; Graham MA; Pavlidis N; Heinrich B; ten Bokkel Huinink WW; Wagener DJ; Aamdal S; Verweij J Br. J. Cancer 1996, 74, 1944–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Fox BW Ann. Oncol 1992, 3, 707–709. [DOI] [PubMed] [Google Scholar]
- (176).Bissett D; Graham MA; Setanoians A; Chadwick GA; Wilson P; Koier I; Henrar R; Schwartsmann G; Cassidy J; Kaye SB Cancer Res. 1992, 52, 2894–2898. [PubMed] [Google Scholar]
- (177).Tsuruo T; Oh-hara T; Iida H; Tsukagoshi S; Sato Z; Matsuda I; Iwasaki S; Okuda S; Shimizu F; Sasagawa K Cancer Res. 1986, 46, 381–385. [PubMed] [Google Scholar]
- (178).Ishida K; Lincke T; Hertweck C Angew. Chemie Int. Ed 2012, 51, 5470–5474. [DOI] [PubMed] [Google Scholar]
- (179).Wu Y; Seyedsayamdost MR Biochemistry 2018, 57, 4247–4251. [DOI] [PubMed] [Google Scholar]
- (180).Nguyen T; Ishida K; Jenke-Kodama H; Dittmann E; Gurgui C; Hochmuth T; Taudien S; Platzer M; Hertweck C; Piel J Nat. Biotechnol 2008, 26, 225–233. [DOI] [PubMed] [Google Scholar]
- (181).Ishida K; Lincke T; Behnken S; Hertweck C J. Am. Chem. Soc 2010, 132, 13966–13968. [DOI] [PubMed] [Google Scholar]
- (182).Flórez LV; Scherlach K; Miller IJ; Rodrigues A; Kwan JC; Hertweck C; Kaltenpoth M Nat. Commun 2018, 9, 2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (183).Parmeggiani A; Krab IM; Watanabe T; Nielsen RC; Dahlberg C; Nyborg J; Nissen P J. Biol. Chem 2006, 281, 2893–2900. [DOI] [PubMed] [Google Scholar]
- (184).Cetin R; Krab IM; Anborgh PH; Cool RH; Watanabe T; Sugiyama T; Izaki K; Parmeggiani A EMBO J. 1996, 15, 2604–2611. [PMC free article] [PubMed] [Google Scholar]
- (185).Ross C; Opel V; Scherlach K; Hertweck C Mycoses 2014, 57, 48–55. [DOI] [PubMed] [Google Scholar]
- (186).Azegami K; Nishiyama K; Watanabe Y; Suzuki T; Yoshida M; Nose K; Toda S Ann. Phytopath. Soc. Japan 1985, 51, 315–317. [Google Scholar]
- (187).Wang M; Tachibana S; Murai Y; Li L; Lau SYL; Cao M; Zhu G; Hashimoto M; Hashidoko Y Sci. Rep 2016, 6, 22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Sokol PA; Lewis CJ; Dennis JJ J. Med. Microbiol 1992, 36, 184–189. [DOI] [PubMed] [Google Scholar]
- (189).Sultan MZ; Park K; Lee SY; Park JK; Varughese T; Moon S-S J. Antibiot. (Tokyo) 2008, 61, 420–425. [DOI] [PubMed] [Google Scholar]
- (190).Bell SC; Turner JM Biochem. Soc. Trans 1973, 1, 751–753. [Google Scholar]
- (191).Byng GS; Turner JM Biochem. J 1977, 164, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Hollstein U; McCamey DA J. Org. Chem 1973, 38, 3415–3417. [DOI] [PubMed] [Google Scholar]
- (193).Byng GS; Turner JM J. Gen. Microbiol 1976, 97, 57–62. [DOI] [PubMed] [Google Scholar]
- (194).Viktorsson EÖ; Melling Grøthe B; Aesoy R; Sabir M; Snellingen S; Prandina A; Høgmoen Åstrand OA; Bonge-Hansen T; Døskeland SO; Herfindal L; Rongved P Bioorg. Med. Chem 2017, 25, 2285–2293. [DOI] [PubMed] [Google Scholar]
- (195).Xu T; Shi L; Zhang Y; Wang K; Yang Z; Ke S Eur. J. Med. Chem 2019, 168, 293–300. [DOI] [PubMed] [Google Scholar]
- (196).Zuther K; Mayser P; Hettwer U; Wu W; Spiteller P; Kindler BLJ; Karlovsky P; Basse CW; Schirawski J Mol. Microbiol 2008, 68, 152–172. [DOI] [PubMed] [Google Scholar]
- (197).Kang JG; Shin SY; Kim MJ; Bajpai V; Maheshwari DK; Kang SC J. Antibiot. (Tokyo) 2004, 57, 726–731. [DOI] [PubMed] [Google Scholar]
- (198).Mori T; Yamashita T; Furihata K; Nagai K; Suzuki K; Hayakawa Y; Shin-ya K J. Antibiot. (Tokyo) 2007, 60, 713–716. [DOI] [PubMed] [Google Scholar]
- (199).Wu Y; Seyedsayamdost MR Cell Chem. Biol 2017, 24, 1437–1444.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Li D; Oku N; Hasada A; Shimizu M; Igarashi Y Beilstein J. Org. Chem 2018, 14, 1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Park WJ; Ma E Arch Pharm Res 2012, 35, 1379–1386. [DOI] [PubMed] [Google Scholar]
- (202).Elshafie HS; Bufo SA; Racioppi R, Camele I Int. J. Drug Discov 2013, 5, 181–184. [Google Scholar]
- (203).Song L; Jenner M; Masschelein J; Jones C; Bull MJ; Harris SR; Hartkoorn RC; Vocat A; Romero-Canelon I; Coupland P; Webster G; Dunn M; Weiser R; Paisey C; Cole ST; Parkhill J; Mahenthiralingam E; Challis GL J. Am. Chem. Soc 2017, 139, 7974–7981. [DOI] [PubMed] [Google Scholar]
- (204).Perry C; Sargeant JR; Song L; Challis GL Tetrahedron 2018, 74, 5150–5155. [Google Scholar]
- (205).Hu WJ; Chen XM; Meng HD; Meng ZH Biomed. Environ. Sci 1989, 2, 65–71. [PubMed] [Google Scholar]
- (206).Rohm B; Scherlach K; Hertweck C Org. Biomol. Chem 2010, 8, 1520–1522. [DOI] [PubMed] [Google Scholar]
- (207).de Bruijn J; Frost DJ; Nugteren DH; Gaudemer A; Lijmbach GWM; Cox HC; Berends W Tetrahedron 1973, 29, 1541–1547. [Google Scholar]
- (208).Moebius N; Ross C; Scherlach K; Rohm B; Roth M; Hertweck C Chem. Biol 2012, 19, 1164–1174. [DOI] [PubMed] [Google Scholar]
- (209).Knappe TA; Linne U; Robbel L; Marahiel MA Chem. Biol 2009, 16, 1290–1298. [DOI] [PubMed] [Google Scholar]
- (210).Kuznedelov K; Semenova E; Knappe TA; Mukhamedyarov D; Srivastava A; Chatterjee S; Ebright RH; Marahiel MA; Severinov KJ Mol. Biol 2011, 412, 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Hegemann JD; Zimmermann M; Zhu S; Klug D; Marahiel MA Biopolymers 2013, 100, 527–542. [DOI] [PubMed] [Google Scholar]
- (212).Parker WL; Rathnum ML; Seiner V; Trejo WH; Principe PA; Sykes RB J. Antibiot. (Tokyo) 1984, 37, 431–440. [DOI] [PubMed] [Google Scholar]
- (213).Mullins AJ; Murray JAH; Bull MJ; Jenner M; Jones C; Webster G; Green AE; Neill DR; Connor TR; Parkhill J; Challis GL; Mahenthiralingam E Nat. Microbiol 2019, 4, 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Kusumi T, Ohtani I, Nishiyama K, and Kakisawa H Tetrahedron Lett. 1987, 28, 3981–3984. [Google Scholar]
- (215).Ross C; Scherlach K; Kloss F; Hertweck C Angew. Chemie Int. Ed 2014, 53, 7794–7798. [DOI] [PubMed] [Google Scholar]
- (216).Cvejic JH; Putra SR; El-Beltagy A; Hattori R; Hattori T; Rohmer M FEMS Microbiol. Lett 2000, 183, 295–299. [DOI] [PubMed] [Google Scholar]
- (217).Winsor GL; Khaira B; Van Rossum T; Lo R; Whiteside MD; Brinkman FSL Bioinformatics 2008, 24, 2803–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (218).Mahenthiralingam E; Drevinek P In Burkholderia: Molecular Microbiology and Genomics; Coenye T; Vandamme P, Eds.; Horizontal Bioscience: Gent, 2007; pp 53–79. [Google Scholar]
- (219).Lackner G; Moebius N; Partida-Martinez LP; Boland S; Hertweck C BMC Genomics. 2011, 12, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Deng P; Wang X; Baird SM; Showmaker KC; Smith L; Peterson DG; Lu S Microbiologyopen 2016, 5, 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Cheng HP; Lessie TG J. Bacteriol 1994, 176, 4034–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).Lessie TG; Hendrickson W; Manning BD; Devereux R FEMS Microbiol. Lett 1996, 144, 117–128. [DOI] [PubMed] [Google Scholar]
- (223).Blin K; Wolf T; Chevrette MG; Lu X; Schwalen CJ; Kautsar SA; Suarez Duran HG; de los Santos ELC; Kim HU; Nave M; Dickschat JS; Mitchell DA; Shelest E; Breitling R; Takano E; Lee SY; Weber T; Medema MH Nucleic Acids Res. 2017, 45, W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Blin K; Medema MH; Kottmann R; Lee SY; Weber T Nucleic Acids Res. 2017, 45, D555–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Holden MTG; Titball RW; Peacock SJ; Cerdeño-Tarraga AM; Atkins T; Crossman LC; Pitt T; Churcher C; Mungall K; Bentley SD; Sebaihia M; Thomson NR; Bason N; Beacham IR; Brooks K; Brown KA; Brown NF; Challis GL; Cherevach I; Chillingworth T; Cronin A; Crossett B; Davis P; DeShazer D; Feltwell T; Fraser A; Hance Z; Hauser H; Holroyd S; Jagels K; Keith KE; Maddison M; Moule S; Price C; Quail MA; Rabbinowitsch E; Rutherford K; Sanders M; Simmonds M; Songsivilai S; Stevens K; Tumapa S; Vesaratchavest M; Whitehead S; Yeats C; Barrell BG; Oyston PC; Parkhill J Proc. Natl. Acad. Sci. U. S. A 2004, 101, 14240–14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Rodley PD; Römling U; Tümmler B Mol. Microbiol 1995, 17, 57–67. [DOI] [PubMed] [Google Scholar]
- (227).Hopwood DA Annu. Rev. Genet 2006, 40, 1–23. [DOI] [PubMed] [Google Scholar]
- (228).Yu X; Doroghazi JR; Janga SC; Zhang JK; Circello B; Griffin BM; Labeda DP; Metcalf WW Proc. Natl. Acad. Sci. U. S. A 2013, 110, 20759–20764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Pande A; Pandey P; Mehra S; Singh M; Kaushik SJ Genet. Eng. Biotechnol 2017, 15, 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Walsh CT Science 2004, 303, 1805–1810. [DOI] [PubMed] [Google Scholar]
- (231).Fischbach MA; Walsh CT Chem. Rev 2006, 106, 3468–3496. [DOI] [PubMed] [Google Scholar]
- (232).Hertweck C Angew. Chemie Int. Ed 2009, 48, 4688–4716. [DOI] [PubMed] [Google Scholar]
- (233).Chan YA; Podevels AM; Kevany BM; Thomas MG Nat. Prod. Rep 2009, 26, 90–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Moore Bradley S; Hertweck C Nat. Prod. Rep 2002, 19, 70–99. [DOI] [PubMed] [Google Scholar]
- (235).Khosla C; Herschlag D; Cane DE; Walsh CT Biochemistry 2014, 53, 2875–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Robbins T; Liu Y-C; Cane DE; Khosla C Curr. Opin. Struct. Biol 2016, 41, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Kornfuehrer T; Eustáquio AS Med. Chem. Commun 2019, Advance Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Bisang C; Long PF; Cortés J; Westcott J; Crosby J; Matharu A-L; Cox RJ; Simpson TJ; Staunton J; Leadlay PF Nature 1999, 401, 502–505. [DOI] [PubMed] [Google Scholar]
- (239).Helfrich EJN; Piel J Nat. Prod. Rep 2016, 33, 231–316. [DOI] [PubMed] [Google Scholar]
- (240).Süssmuth RD; Mainz A Angew. Chemie Int. Ed 2017, 56, 3770–3821. [DOI] [PubMed] [Google Scholar]
- (241).Du L; Lou L Nat. Prod. Rep 2010, 27, 255–278. [DOI] [PubMed] [Google Scholar]
- (242).Boddy CN J. Ind. Microbiol. Biotechnol 2014, 41, 443–450. [DOI] [PubMed] [Google Scholar]
- (243).Puthenveetil S; Loganzo F; He H; Dirico K; Green M; Teske J; Musto S; Clark T; Rago B; Koehn F; Veneziale R; Falahaptisheh H; Han X; Barletta F; Lucas J; Subramanyam C; O’Donnell CJ; Tumey LN; Sapra P; Gerber HP; Ma D; Graziani EI Bioconjug. Chem 2016, 27, 1880–1888. [DOI] [PubMed] [Google Scholar]
- (244).He H-Y; Yuan H; Tang M-C; Tang G-L Angew. Chemie Int. Ed 2014, 53, 11315–11319. [DOI] [PubMed] [Google Scholar]
- (245).He H-Y; Tang M-C; Zhang F; Tang G-L J. Am. Chem. Soc 2014, 136, 4488–4491. [DOI] [PubMed] [Google Scholar]
- (246).Hildebrand M; Waggoner LE; Liu H; Sudek S; Allen S; Anderson C; Sherman DH; Haygood M Chem. Biol 2004, 11, 1543–1552. [DOI] [PubMed] [Google Scholar]
- (247).Partida-Martinez LP; Groth I; Schmitt I; Richter W; Roth M; Hertweck C Int. J. Syst. Evol. Microbiol 2007, 57, 2583–2590. [DOI] [PubMed] [Google Scholar]
- (248).Kusebauch B; Busch B; Scherlach K; Roth M; Hertweck C Angew. Chemie Int. Ed 2010, 49, 1460–1464. [DOI] [PubMed] [Google Scholar]
- (249).Moldenhauer J; Götz DCG; Albert CR; Bischof SK; Schneider K; Süssmuth RD; Engeser M; Gross H; Bringmann G; Piel J Angew. Chemie Int. Ed 2010, 49, 1465–1467. [DOI] [PubMed] [Google Scholar]
- (250).Gay DC; Spear PJ; Keatinge-Clay AT ACS Chem. Biol 2014, 9, 2374–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (251).Bretschneider T; Heim JB; Heine D; Winkler R; Busch B; Kusebauch B; Stehle T; Zocher G; Hertweck C Nature 2013, 502, 124–128. [DOI] [PubMed] [Google Scholar]
- (252).Sundaram S; Heine D; Hertweck C Nat. Chem. Biol 2015, 11, 949–951. [DOI] [PubMed] [Google Scholar]
- (253).Kusebauch B; Busch B; Scherlach K; Roth M; Hertweck C Angew. Chemie Int. Ed 2009, 48, 5001–5004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.