Abstract
Primary germ layers have the potential to form all tissues in the mature organism, and their formation during gastrulation requires precise epigenetic modulation of both proximal and distal regulatory elements. Previous studies indicated that spatial and temporal patterns of gene expression in the gastrula predispose individual regions to distinct cell fates. However, the underlying epigenetic mechanisms remain largely unexplored. Here, we profile the spatiotemporal landscape of the epigenome and transcriptome of the mouse gastrula. We reveal the asynchronous dynamics of proximal chromatin states during germ layer formation as well as unique gastrula-specific epigenomic features of regulatory elements, which have strong usage turnover dynamics and clear germ layer-specific signatures. Importantly, we also find that enhancers around organogenetic genes, which are weakly expressed at the gastrulation stage, are frequently pre-marked by histone H3 lysine 27 acetylation (H3K27ac) in the gastrula. By using the transgenic mice and genome editing system, we demonstrate that a pre-marked enhancer, which is located in the intron of a brain-specific gene 2510009E07Rik, exhibits specific enhancer activity in the ectoderm and future brain tissue, and also executes important function during mouse neural differentiation. Taken together, our study provides the comprehensive epigenetic information for embryonic patterning during mouse gastrulation, demonstrates the importance of gastrula pre-marked enhancers in regulating the correct development of the mouse embryo, and thus broadens the current understanding of mammalian embryonic development and related diseases.
Subject terms: Epigenetic memory, Embryonic stem cells, Pluripotency, Epigenetics analysis
Introduction
During mammalian embryo development, one fertilized zygote will undergo multiple rounds of cell fate commitment to form diverse cell types.1 The gastrulation, which occurs at the early post-implantation stage, is one of the most crucial biological processes for the generation of the primary germ layers.2–4 During gastrulation, the pluripotent epiblast develops into the ectoderm, mesoderm, and endoderm, which set up the future body plan.3 Previous knowledge has revealed that regionalized cell distribution in the gastrula accounts for the formation of primary germ layers.2,4,5 Specifically, anterior epiblast cells develop into ectodermal cells, which give rise to the future nervous system and skin6; posterior epiblast cells develop into mesendoderm cells, which give rise to visceral organs7; while epiblast cells, which traverse the primitive streak and then migrate anteriorly, develop into mesodermal cells and mainly give rise to heart and blood cells.8
Multiple layers of molecular mechanisms including epigenetic regulation9 and signaling pathways10 are involved to orchestrate the precise embryonic development. Recently, a number of studies have determined the dynamics of epigenetic programming during different stages of the mouse embryo development. Epigenomic landscape studies have revealed the features of histone H3 lysine 4 tri-methylation (H3K4me3) and H3 lysine 27 tri-methylation (H3K27me3) re-establishment and the enrichment of H3K4me3 broad domain at key cell identity-related genes,11,12 determined the dynamics of chromatin accessibility,13 chromatin structures,14 and histone H3 lysine 9 tri-methylation (H3K9me3) reprogramming15–17 in pre- and early post-implantation embryos. Other studies also provided the global chromatin state landscape during late embryo development.18,19 These findings together highlight the importance of epigenetic regulations in early mouse embryonic patterning. However, the knowledge of epigenetic dynamics during mouse gastrulation particularly for the generation of spatially-restricted germ layers remains largely elusive.
Recently, we and others have reported the distinct distributions of H3K27me3 and DNA methylation at a range of development-related genes in intraembryonic and extraembryonic cells,20,21 thereby explaining the differential developmental plasticity of these two types of cells. Studies have also revealed that epigenetic decommissioning, priming, or activation of regulatory elements, especially enhancers, play critical roles in the regulation of stem cell fate by modulating target gene expression or by meditating the interactions between transcription factors (TFs).22,23 For example, epigenetic priming of enhancers by histone H3 lysine 4 mono-methylation (H3K4me1), which facilitates the binding of pioneer factors, signifies developmental competence during human endodermal lineage diversification.24 Histone H3 lysine 27 acetylation (H3K27ac)-marked cis-regulatory elements in pre-implantation embryos tend to reside near zygotic genome activation-related genes,12 while rapid and pervasive enhancer usage turnover happens during mouse organ development.18,19 However, there still remains a knowledge gap concerning the epigenetic basis for the correct development of the primary germ layers during gastrulation.
To determine the dynamics of epigenetic programming and to specify the epigenetic mechanisms of primary germ layers’ formation during mouse gastrulation, we collected sub-regions of the mouse gastrula for transcriptome and epigenome profiling. First, we revealed the asynchronous features of both proximal and distal chromatin state transitions among the three primary germ layers. Then, we identified that many key organogenetic gene-related enhancers are frequently pre-marked by H3K27ac at the gastrula stage. The epigenetic pre-patterning of organ development-related enhancers ensures the correct development of each germ layer towards a specific organ tissue. Finally, we uncovered the essential roles of an ectoderm- and brain-specific enhancer, Ect2, during mouse neural differentiation. Our study provides the first comprehensive epigenetic landscape of the mouse gastrula, reveals the in vivo epigenetic basis of primary germ layers’ generation and future organ development, and bridges the knowledge gap of epigenetic information between pre-implantation and organogenetic embryos.
Results
Epigenetic profiling of the mouse gastrula
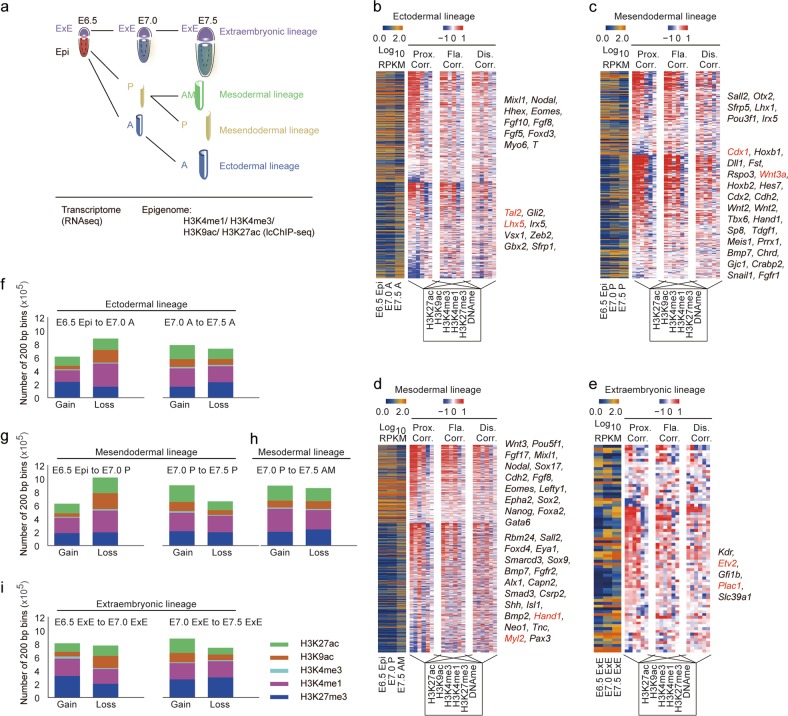
We have previously revealed the epigenetic dynamics of DNA methylation and H3K27me3 during mouse early embryo development from the pre-streak (embryonic day 6.5, E6.5), mid-streak (E7.0), and late-streak (E7.5) stages, on both epiblasts (Epi, anterior epiblast (A), posterior epiblast (P), as well as anterior mesoderm part (AM), were separated at corresponding stages) and extraembryonic (ExE) parts.20 Here, we investigated the dynamics of H3K4me1, H3K4me3, H3K9ac, and H3K27ac modifications as well as the transcriptome in the mouse gastrula (Fig. 1a). Dissected tissues from a single embryo of each stage were subjected to RNA-seq for transcriptome profiling. Pooled samples of corresponding tissues (~10,000 cells per sample) were subjected to limited cell number chromatin immunoprecipitation followed by sequencing (lcChIP-seq)17 (Fig. 1a). All data have been prepared with two highly reproducible biological replicates (Supplementary information, Fig. S1 and Table S1). Using this strategy, we were able to trace the transcriptomic and epigenomic trajectory of ectodermal, mesendodermal, mesodermal, and extraembryonic lineages.
Fig. 1.
Epigenomic profiling of the mouse gastrula. a An illustrative scheme describing the sample collection strategy for the study of mouse gastrula. b‒e Heatmap indicating gene expression dynamics (down-regulation (top), up-regulation (bottom)), the corresponding correlation with each epigenetic modification in proximal (Prox, TSS ± 2 kb), flanking (Fla, TSS ± 2–10 kb), and distal regions (Dis, TSS ± 10–100 kb) during the development of ectodermal (b), mesendodermal (c), mesodermal (d) and extraembryonic (e) lineages. Representative genes are listed on the right of each map. f‒i Bar plot showing the number of 200-bp bins defined by ChromHMM with gain or loss of selected modification during the developmental process of the ectodermal (f), mesendodermal (g), mesodermal (h) and extraembryonic (i) lineages. H3K27me3 ChIP-seq and DNA methylation data for the mouse gastrula are from our previous report (GSE104243)
We first analyzed gene expression dynamics during the development of each lineage (Fig. 1b–e; Supplementary information, Table S2) and found that expression of ectodermal genes (e.g., Tal2 and Lhx5), mesendodermal genes (e.g., Cdx1 and Wnt3a), mesodermal genes (e.g., Hand1 and Myl2), and placenta-related genes (e.g., Etv2 and Plac1) was specifically up-regulated during the development of anterior, posterior, anterior mesodermal, and extraembryonic tissues, respectively (Fig. 1b–e). The formation of the primitive streak and region-specific distribution of morphogen, such as BMP4 and Wnt3a in the gastrula,25 have been reported to be crucial for the development of mouse embryos and related cell fate commitment. We found that both epigenomic pattern and transcriptomic profile of the primitive streak marker gene T, and morphogen Bmp4 and Wnt3a accurately reflect the regional distribution of these factors in the gastrula (Supplementary information, Fig. S2a–c). These results robustly demonstrate the biological fidelity and validity of our sample collection strategy, which faithfully recapitulated the lineage diversification process during mouse gastrulation. We also analyzed the correlation between gene expression and epigenomic modifications at various ranges away from transcription start sites (TSSs) (Fig. 1b–e). As expected, epigenetic modifications in proximal regions (TSS ± 2 kb) were more highly correlated with gene expression dynamics than those in the distal regions. In addition, we observed a relatively high positive correlation between gene expression and representative enhancer-related modifications (H3K4me1/H3K27ac) at both flanking (±2–10 kb) and distal regions (±10–100 kb) in intraembryonic lineages (Fig. 1b–d). These observations suggest that the activities of distal enhancers play important regulatory roles in gastrulation.
To systematically analyze the global dynamics of epigenetic modifications in the mouse gastrula, we counted the number of modified regions, which was calculated as the sum-up of 200-bp bins with corresponding modification signals detected by ChromHMM. We found that histone modifications underwent extensive changes in both intra- and extra-embryonic lineages (Fig. 1f–i; Supplementary information, Fig. S2d), suggesting a dramatic genome-wide remodeling of the epigenomic landscape during gastrulation. In general, histone modifications of the three intraembryonic lineages exhibited similar patterns, with fewer gain than loss of modified regions from stages E6.5 to E7.0 and more gain than loss of modified regions from stages E7.0 to E7.5 (Fig. 1f–h). We also found that H3K4me1, but not H3K4me3, exhibited the most dramatic changes during gastrulation. Given the requirement for rapid and pervasive transcriptional remodeling during lineage specification at gastrulation stage26 (Fig. 1b–e), higher dynamics of H3K4me1 strongly suggests that enhancer-related regulatory mechanisms27 are widely involved in the regulation of the lineage commitment during gastrulation.
By combining our data with the published dataset of H3K4me3 and H3K27me3 in pre-implantation embryos,11 we detected a clear developmental trajectory from pre- to post-implantation stages (Supplementary information, Fig. S3a). Globally, the enrichment for H3K4me3 in the gastrula was highly different from that in the oocyte, but was similar to H3K4me3 marks in pre-implantation embryos (Supplementary information, Fig. S3b). Previous studies revealed that H3K4me3-marked broad domains are highly associated with the regulation of cell identity-related genes in pre-implantation embryos.11,12 However, the noticeable presence of H3K4me3 broad domains in pre-implantation embryos was substantially reduced in the gastrula where only medium H3K4me3 peaks distributed around the TSS regions (e.g., the Nanog promoter region) (Supplementary information, Fig. S3c, d).
Asynchronous cell fate commitment revealed by proximal chromatin state transition
To further illustrate the functions of histone modifications and related chromatin states for cell fate commitment during gastrulation, we trained a ten-state model28 consisting of five active states (active TSSs, flanking regions of active TSSs, bi-acetylated TSSs, bi-acetylated enhancers, and active enhancers), three bivalent states (poised enhancers, bivalent enhancers, and bivalent TSSs), and two repressive states (repressed Polycomb state and quiescent chromatin) based on the combinatorial histone modification patterns observed in all analyzed samples (Fig. 2a). We found that the TSS-related chromatin regions exhibited minimal levels of DNA methylation but maintained a high degree of evolutionary conservation of DNA sequences (Fig. 2a). By contrast, enhancer-related chromatin regions, especially poised enhancers, displayed high levels of DNA methylation and low evolutionary conservation (Fig. 2a). Previous studies revealed that bivalent enhancers show the broad distribution of DNA methylation only in pluripotent cells but not in primary tissues,29 therefore high levels of DNA methylation at bivalent enhancers in the gastrula indicate the high potential of epiblast cells for multi-lineage generation and future tissue development.
Fig. 2.
Proximal chromatin state transition reflects asynchronous cell fate commitment of intraembryonic lineages. a Average genomic coverage, histone mark probabilities, chromatin state definitions, genomic annotation overlap, average DNA methylation level and enrichment of non-coding mammalian conserved elements. b, d, f Bar plots showing the ratio of 200-bp bins with respective chromatin state changes along with gene expression changes during the development of each intraembryonic lineage at the promoter regions (ectodermal lineage (b); mesendodermal lineage (d); mesodermal lineage (f)). The promoter regions (TSS ± 2 kb) were divided into twenty 200-bp bins, and bins with the same chromatin state change were summed to calculate the percentage. Representative chromatin state changes are highlighted with rectangles and related genes are also listed. c, e, g Representative genome browser snapshots of H3K4me3, H3K27ac and H3K27me3 at the loci of representative genes (highlighted in red in b, d, f). H3K27me3 ChIP-seq and DNA methylation data for the mouse gastrula are from our previous report (GSE104243)
Next, we profiled the chromatin state dynamics of the proximal regions (TSS ± 2 kb) and specific histone modification changes during gastrulation. By profiling the relationship between chromatin state in the proximal region and gene expression, we found that the up- and down-regulation of gene expression during intraembryonic lineage development are likely controlled by the chromatin state transitions from bivalent to active and from active to bivalent, respectively (Fig. 2b–g). Surprisingly, we also found distinct patterns of chromatin state changes between the ectodermal lineage and the other two lineages. During ectoderm development, the bivalent-to-active transition, which is usually associated with gene up-regulation and stem cell fate specification,30 was more prominent from the E7.0 A to E7.5 A stages than that from the E6.5 Epi to E7.0 A stages (Fig. 2b, c). In contrast, during mesendoderm and mesoderm development, bivalent-to-active transition became extrusive from as early as the E6.5 Epi to E7.0 P stages (Fig. 2d–g). These results indicate that the cell fate of the anterior epiblast might be committed much later than the posterior part, reflecting the asynchronous fate commitment between ectodermal and mesendodermal lineages in the context of epigenetic regulation. The delayed commitment of ectodermal cells may explain the notion that the epiblast stem cells are developmentally similar to late-gastrula-stage ectoderm.31 Consistently, by analyzing the dynamics of H3K4me3 and H3K27me3 co-occupied domains, we also found the asynchronous feature between ectodermal and mesendodermal/mesodermal lineages (Supplementary information, Fig. S3e–g). Besides, we found that genes, such as Gsc and Mixl1, down-regulated in the mesendodermal lineage from E7.0 P to E7.5 P and in the mesodermal lineage from E7.0 P to E7.5 AM, were mainly regulated by active-to-bivalent transitions (Fig. 2d–g). These results indicate that mesendodermal/mesodermal cell fate specification occurs in the early stage of gastrulation and cell fate determination occurs thereafter.
Lineage-specific dynamics of enhancers drives embryonic patterning during mouse gastrulation
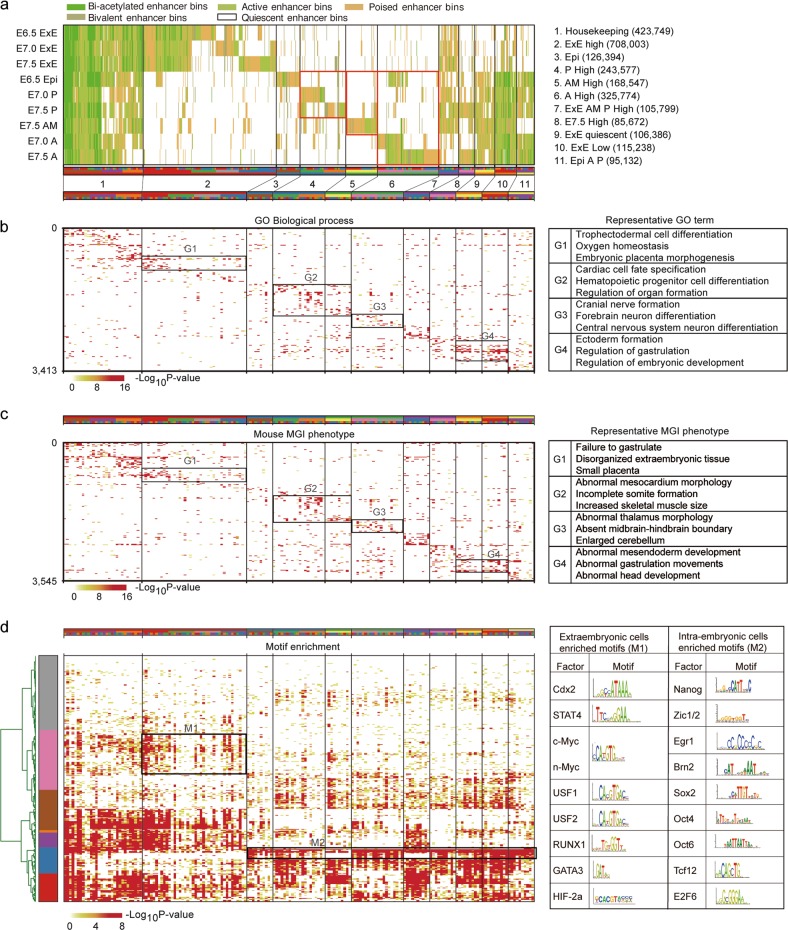
The tightly regulated activity of enhancers is crucial for stem cell differentiation,32 mammalian development,33 and disease pathology.34 Based on analyses of chromatin states (Fig. 2a), we profiled 2,504,271 bins with candidate enhancer activities in the mouse gastrula (Fig. 3a). We further clustered all enhancer bins into 11 clusters based on their distribution specificity among all samples (Fig. 3a). Using GREAT analysis, we found that lineage-specific enhancer bins were enriched near genes with coordinated biological functions and phenotypic changes (Fig. 3b, c; Supplementary information, Table S3). For example, genes around extraembryonic-specific group 1 enhancer bins (G1: cluster 2) are related to trophectodermal cell differentiation and placental development, and knockout of these genes leads to small placenta; genes neighboring posterior-specific group 2 enhancer bins (G2: clusters 4 and 5) are related to cardiac cell fate specification, hematopoietic progenitor cell specification and other processes in mesodermal development, and knockout of these genes leads to mesocardium morphology abnormality; genes neighboring anterior-specific group 3 enhancer bins (G3: cluster 6) are related to brain and neural development, and knockout of these genes leads to abnormal thalamus morphology; and genes neighboring intraembryonic-specific group 4 enhancers (G4: clusters 9 and 10) are involved in the regulation of gastrulation and embryonic development, and knockout of these genes leads to abnormalities in gastrulation (Fig. 3b, c). We also found the enrichment of TF-binding motifs in extraembryonic and intraembryonic enhancer bins (Fig. 3d; Supplementary information, Table S3). For example, binding motifs for Cdx2 and USF1/2, which are required for extraembryonic development,35,36 were enriched in extraembryonic-specific enhancer bins, and motifs for essential developmental TFs, such as Zic1/2,37 Oct438 and TCF12,39 were enriched in intraembryonic-specific enhancer bins (Fig. 3d).
Fig. 3.
Enhancer profiling of the mouse gastrula. a Enhancer modules based on k-means clustering of 2.5 million enhancer bins (200-bp) classified as bi-acetylated enhancer bins (dark green), active enhancer bins (green), poised enhancer bins (dark yellow) and bivalent enhancer bins (moss green) and quiescent enhancer bins across 9 mouse gastrula samples. All enhancer bins were grouped into 11 groups, 18 modules and 180 sub-modules for further functional annotation. The names of each group and module are shown. b‒d Gene ontology (GO) analysis (b), mouse phenotypes (c) and motif enrichment (d) of each sub-module for enhancer bin neighboring genes. Rectangles pinpoint enrichments for selected modules with representative functions, phenotypes, and motifs. Representative GO terms, phenotypes, and motifs are listed on the right
Furthermore, we analyzed the rapid and pervasive changes in enhancer activities during the development of intraembryonic lineages (Supplementary information, Table S4). We found that ectodermal cells in E7.5 A shared a large proportion of enhancers with E6.5 epiblast cells, whereas mesendodermal or mesodermal enhancers were predominantly generated at the E7.0 or E7.5 stage (Fig. 4a, f, g). In the ectodermal lineage, many ectodermal enhancer bins were maintained in the active state, such as enhancers residing near Otx2 (Fig. 4a). During the E6.5 Epi to E7.0 A stage, there was only a mild shift from the quiescent to active or poised enhancer bins, whereas during the E7.0 A to E7.5 A stage, a large number of enhancer bins shifted from quiescent to active, poised or bivalent states (Fig. 4a). Further analyses of enhancer-neighboring genes highlighted the importance of anterior-specific enhancers in ectoderm fate specification and brain development (Fig. 4a). For example, one of the enhancers, which we named as Ect1 here and is located upstream of the Tal2 TSS, exhibited anterior-specific H3K27ac enrichment in E7.5 A (Fig. 4a, b) and intensive interactions with nearby chromatin40 (Fig. 4c). Transgenic reporter assay showed that Ect1 was specifically activated in the anterior region of E7.5 embryos (Fig. 4d) and remained active until E9.5 in embryonic forebrain region (Fig. 4e).
Fig. 4.
Lineage-specific dynamics of enhancers drives embryonic patterning during gastrulation. a Alluvial plot depicting the global activity dynamics of enhancer bins during the development of ectodermal lineage. Each line represents a 200-bp bin defined based on the ChromHMM categories. Numbers of representative state enhancer bins are shown. The representative biological process terms identified using GREAT analysis are listed. Bar plots indicate the significance of corresponding terms. The representative genome browser snapshots are also shown. b, c Representative genome browser snapshots of H3K27ac in all samples (b) and chromatin interaction frequency heatmap at 10 kb resolution in mouse cortical neurons (c) at the range of Tal2 locus. The Tal2-related enhancer Ect1 (highlighted with a red rectangle, b) is located in the topologically associating domain (TAD). Chromatin conformation capture HiC data are from a published dataset (c).39 d, e Ect1 enhancer activity reporter expression (blue staining) in representative transgenic mouse gastrula (E7.5 stage, d) and organogenic embryo (E9.5 stage, e). A cryosection slide of the stained E7.5 embryo is also shown (d). All experiments were performed at least twice. f, g Alluvial plots depicting the global activity dynamics of enhancer bins during the development of mesendodermal (f) and mesodermal (g) lineages. Each line represents a 200-bp bin defined based on the ChromHMM categories. Numbers of representative state enhancer bins are shown. The representative biological process terms identified by GREAT analysis are listed. Bar plots indicate the significance of corresponding terms. The genome browser snapshots of representative genes are also shown
For the mesendodermal lineage, we found that most mesendodermal enhancer bins were in quiescent states in E6.5 Epi, and many enhancer bins in E7.0 P became silenced at E7.5 P stage (e.g., Fgf10- and Bmp10-related enhancers), and more than half of E7.5 P active or poised enhancer bins (such as enhancers near Mesp2 and Hand2 genes) were de novo generated (Fig. 4f). The difference in enhancer usage at E7.0 P and E7.5 P indicates that cells in these two stages likely execute distinct developmental functions during gastrulation. As expected, the enhancers with E7.0 P active/poised but E7.5 P quiescent state were usually related to heart growth and cyclin-dependent kinase activity, whereas enhancers with E7.0 P quiescent but E7.5 P active/poised state were usually responsible for nephron epithelium morphogenesis and muscle cell proliferation (Fig. 4f).
In the mesodermal lineage, we also found that most E7.5 AM enriched enhancer bins, which are usually located near genes responsible for heart development, were quiescent at E6.5 Epi and E7.0 P stages (Fig. 4g). These remarkable distinctions of enhancer usage turnover pattern further confirm the asynchronous commitment of different cell lineages in intraembryonic tissues (Fig. 2b–g) and emphasize the distinct functions of lineage-specific enhancers for gastrulation.
Identification of gastrula-specific enhancers
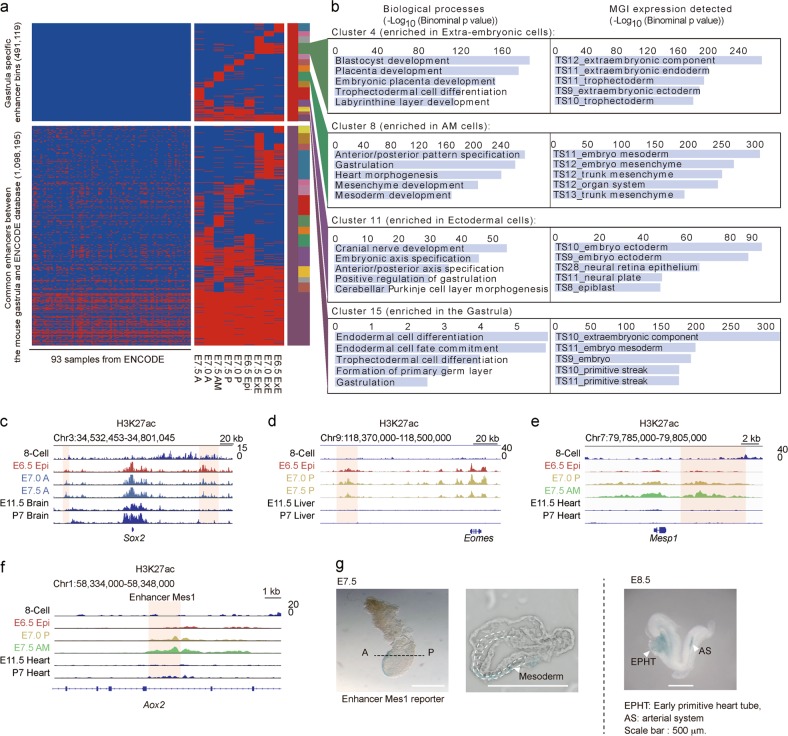
Next, we probed the existence of gastrula-specific enhancer bins by comparison with data from fetal and postnatal mouse tissue samples in the Encyclopedia of DNA Elements (ENCODE) project41 (Supplementary information, Table S5) and found 491,119 gastrula-specific enhancer bins (Fig. 5a). These gastrula-specific enhancer bins were enriched near genes that are related to embryonic pattern formation, gastrulation and primary germ layer specification, and are highly expressed at the gastrulation stage, such as Sox2, Eomes and Mesp1 (Fig. 5b–e). We also identified an enhancer, which we named as Mes1 and is located in the intron region of the Aox2 gene, exhibited enrichment of H3K27ac in E7.5 AM but not in E11.5 or P7 heart sample (Fig. 5f). Enhancer reporter assays further validated the activity of Mes1 in E7.5 anterior mesodermal cells as well as E8.5 early primitive heart tube and artery system (Fig. 5g), indicating a potential role of the Mes1 enhancer in mouse cardiac mesoderm progenitor cell development.
Fig. 5.
Identification of gastrula-specific enhancers. a Heatmap showing a large number of gastrula-specific enhancer bins based on comparison with the published dataset from ENCODE. The dataset used is also listed in Supplementary information, Table S4. b Representative biological process terms and expression stage information for selected enhancer bin clusters based on GREAT analyses. c‒e Representative genome browser snapshots of H3K27ac for gastrula-specific enhancers during development of the ectoderm (c), mesendoderm (d), and mesoderm (e). f Representative genome browser snapshot of H3K27ac for Mes1 enhancer in the gastrula and later embryonic samples. g Enhancer activity reporter expression (blue staining) in representative transgenic mouse gastrula (E7.5) and E8.5 stages. A cryosection slide of the stained E7.5 embryo is also shown. H3K27ac data of E11.5 and P7 (postnatal day 7) embryo samples are from a published dataset19
Vital organ development-related enhancers are epigenetically pre-marked by H3K27ac in the gastrula
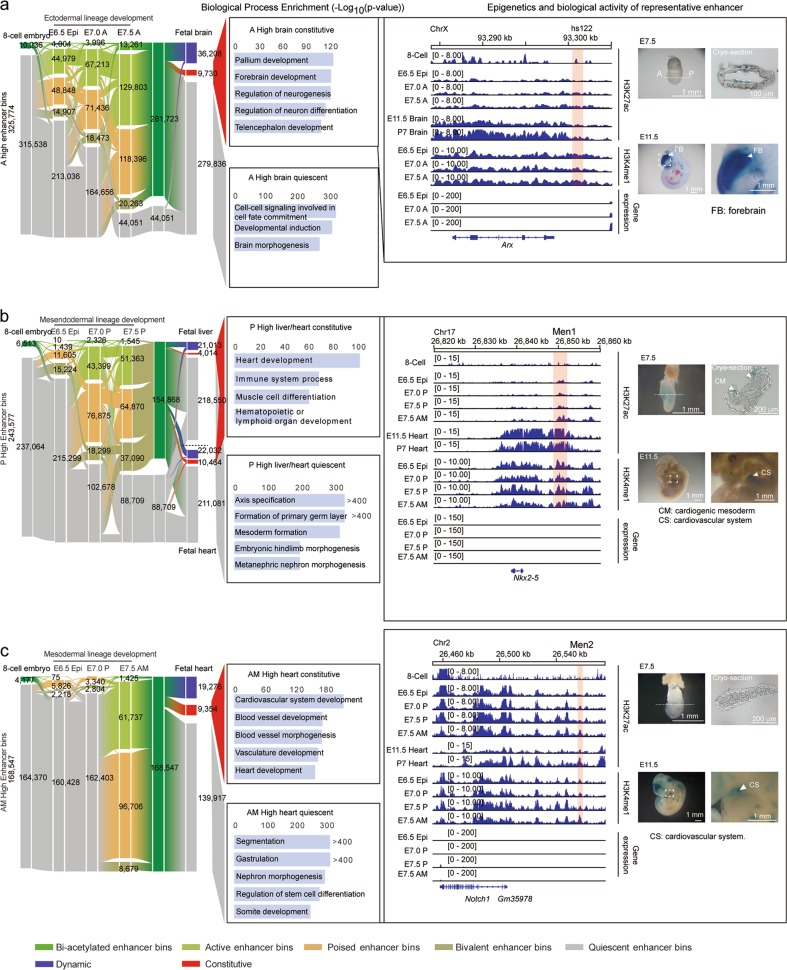
Given the important role of gastrulation in establishing the future body plan, we analyzed the dynamics of lineage-specific enhancers across early and late mouse development by comparing our dataset with the published H3K27ac datasets in the 8-cell embryo12 and in E11.5, E14.5, E17.5, P0, P7, P21, and P56 mouse brain, liver and heart samples19 (Supplementary information, Table S6). We found that most of the active enhancer bins in the gastrula were in quiescent states in the 8-cell embryo stage (Fig. 6a–c), whereas many gastrula enhancer bins were active in organogenic embryos with constitutive or dynamic patterns (Fig. 6a–c). Further analyses revealed that these enhancer bins were usually located near crucial organogenetic genes, which exhibited higher expression in later developmental stages (Fig. 6a–c; Supplementary information, Fig. S4a–h). For example, Arx, which plays important roles in forebrain development, has been reported to be regulated by hs122 and hs123 enhancers in the dorsal brain region.42 Double knockout of both enhancers, as well as single knockout of hs122 but not hs123, leads to structural and morphological defects in mouse brain.42 We found that, even though there was no apparent expression of Arx in the gastrula, the hs122 but not hs123 enhancer was already pre-marked by H3K27ac and exhibited enhancer activity in some ectodermal cells of the gastrula (Fig. 6a). This result indicates that biologically significant, organogenesis-related enhancers are preferentially pre-marked by H3K27ac in the gastrulation stage. Similarly, enhancers (we named as Men1, Men2, Men3, and Ect3) located nearby of Nkx2-5, Notch1, Lbh, or Pax6, which are responsible for heart/limb or brain development, exhibited epigenetic pre-marking and clear enhancer activities in future organs (Fig. 6b, c; Supplementary information, Fig. S4d, h). These data illustrate that the enhancers of genes essential for fetal organ development are often pre-marked by active epigenetic modifications in the gastrula, even though the corresponding genes are weakly expressed in this stage; and suggest that this region-specific pre-marking of enhancers in the gastrula may ensure the correct development of primary germ layers for organ development later in mouse embryogenesis.
Fig. 6.
Vital organ development-related enhancers are pre-marked by H3K27ac in the gastrula. a–c Alluvial plots revealing the global dynamics of enhancer bins during the development from the 8-cell stage embryo to primary germ layers in the gastrula and related future organs. Ectodermal lineage and fetal brain (a); mesendodermal lineage and fetal liver or fetal heart (b); mesodermal lineage and fetal heart (c). Numbers of enhancer bins for each state are shown. The related biological process terms of neighboring genes of selected enhancer clusters are shown in the middle panel. Genome browser snapshots of H3K27ac and H3K4me1 around representative genes are shown. Enhancer regions with relevant dynamics are highlighted with red rectangles and enhancer activity reporter expression (blue staining) for highlighted enhancer regions is also shown on the right. Enhancers for Nkx2-5 and Notch1 were named Men1 and Men2, respectively. H3K27ac data of E11.5 and P7 embryo samples are from a published dataset19
A brain activated enhancer with ectoderm specificity plays important roles in the neural differentiation process
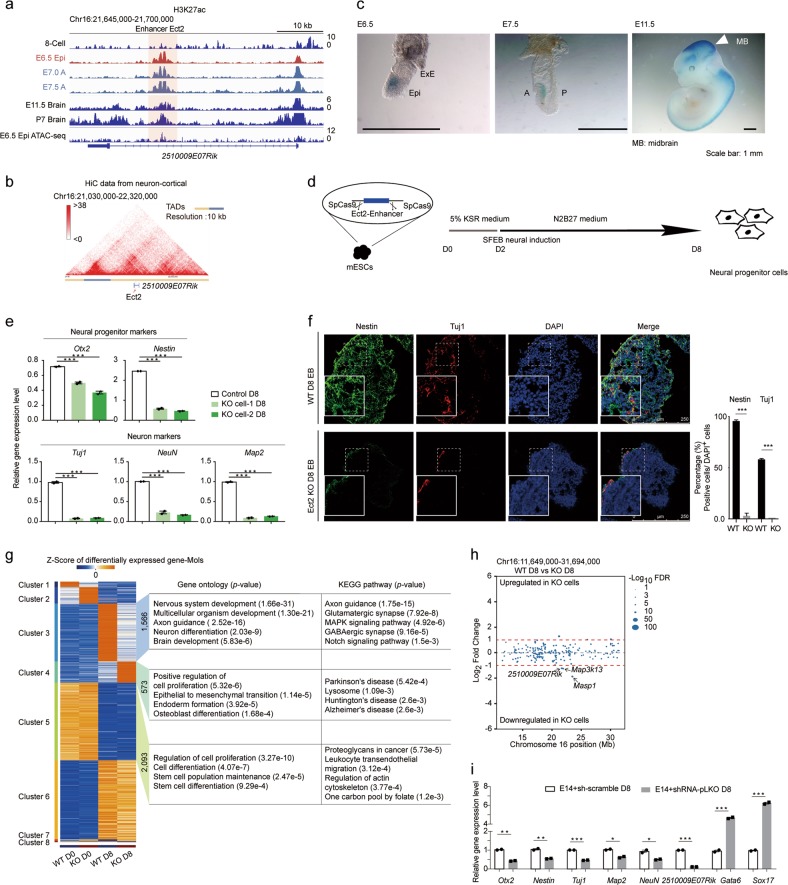
To further explore the biological significance of gastrula pre-marked enhancers, we identified a novel enhancer, which we named as Ect2 and is located in the intronic region of protein-coding gene 2510009E07Rik (Fig. 7a). The genomic region of Ect2 showed enrichment of ATAC-seq signal21 and strong ectoderm-specific enrichment of H3K27ac in the gastrula (Fig. 7a; Supplementary information, Fig. S5a). The region maintained the epigenetic pattern in brain tissues till postnatal stages (Fig. 7a), and also exhibited intensive interactions with nearby genomic regions that were located in the same topologically associated domain (TAD)40 (Fig. 7b). Moreover, enhancer reporter assays demonstrated that Ect2 exhibited strong enhancer activity in ectodermal cells of the gastrula and brain region-specific activity in E11.5 organogenetic embryos (Fig. 7c). This result strongly implies the potential role of Ect2 enhancer in regulating mouse brain development. To address this, we took advantage of the CRISPR-Cas9-based genome editing system to delete the Ect2 enhancer fragment in mouse embryonic stem cells (Fig. 7d). Multiple single cell-derived Ect2-knockout cell lines were generated, and independent knockout cell lines were further subjected to neural differentiation as previously described43 (Fig. 7d). We found that the expression of several neural progenitor markers (e.g., Otx2 and Nestin) and neuronal markers (e.g., Map2, Tuj1, and NeuN) was significantly compromised at both mRNA and protein levels compared to the wild-type cells after 8 days of differentiation (Fig. 7e, f). We also profiled the transcriptome of Ect2-knockout cells on day 0 and day 8 of differentiation, and found that the knockout of Ect2 specifically affected the transcriptomic state of the differentiated cells but not of pluripotent stem cells (Supplementary information, Fig. S5b, c). Analysis of differentially expressed genes (DEGs) revealed that knockout of Ect2 biased the cells towards endodermal fate and impaired neural differentiation (Fig. 7g; Supplementary information, Fig. S5d, and Table S7). To further identify targets of Ect2, which strongly interacts with neighboring chromatin (Fig. 7b), we profiled the expression changes of genes within 20 Mb of Ect2 between wild-type and Ect2-knockout cells (Fig. 7h). We observed significant down-regulation of 2510009E07Rik (P < 0.01), Map3k13 (P < 0.05), and Masp1 (P < 0.01) (Fig. 7h; Supplementary information, Fig. S5e). Considering the relatively low expression of Map3k13 and Masp1 and high expression of 2510009E07Rik during neural differentiation and in the central nervous system (Supplementary information, Fig. S5e–g), we inferred that 2510009E07Rik, a protein-coding gene with no reported function, might be a direct target of Ect2. Indeed, the expression level of 2510009E07Rik was down-regulated at the late stage of neural differentiation (Supplementary information, Fig. S5h), specific knockdown of 2510009E07Rik closely mimicked the transcriptomic state of Ect2-knockout cells (Fig. 7i). Moreover, re-expression of 2510009E07Rik in Ect2-knockout cells partially rescued the neural differentiation abnormalities (Supplementary information, Fig. S5i). Therefore, we concluded that the enhancer Ect2, which acts as an active enhancer in both ectoderm and fetal brain tissues, plays important roles in neural development by regulating the expression of 2510009E07Rik. These results clearly illustrate the importance of gastrula pre-activated enhancers in mammalian development.
Fig. 7.
A brain activated enhancer with ectoderm specificity plays important roles in neural differentiation. a A representative genome browser snapshot of H3K27ac enrichment and chromatin accessibility at the range of Ect2 enhancer from the 8-cell stage to P7 brain. H3K27ac data of E11.5 and P7 embryo samples are from a published dataset.19 Chromatin accessibility data of E6.5 epiblast is from a published dataset.21 b Interaction frequency heatmap at 10 kb resolution in mouse cortical neurons around the Ect2 enhancer. The Ect2 enhancer is located in the TAD. Chromatin conformation capture HiC data are from a published dataset.39 c Ect2 enhancer activity reporter expression in representative transgenic mouse gastrula (E6.5 and E7.5 stages) and organogenetic mouse embryo (E11.5) (blue staining). All experiments were performed at least twice. d Scheme showing the workflow for Ect2 enhancer functional validation. e Bar plots showing the relative expression levels of neural-related genes in both wild-type (WT) cells (control) and Ect2 enhancer knockout cells (KO cell-1 and KO cell-2) on day 8 of differentiation. Quantitative gene expression level was normalized to the expression level of Gapdh. Results are shown as means ± SEM. Student’s t-test, ***P < 0.001. f Immunocytochemistry analysis of Tuj1 and Nestin protein levels in mouse embryoid bodies on day 8. Quantification of the data is shown on the right. g Heatmap showing differentially expressed genes (DEGs) of WT cells and Ect2-knockout cells on day 0 and day 8 during mouse neural differentiation. Gene counts of each cluster, representative GO terms, and KEGG pathways are shown on the right. The color key from blue to red indicates low to high gene expression level. h Dot plot indicating RNA-seq results of all genes within a 20 Mb window around the Ect2 enhancer. Each dot represents one gene, and the size of each dot represents the significance for gene expression change. The red dotted line shows the threshold for DEGs. i Bar plot indicating gene expression pattern after the knockdown of the 2510009E07Rik gene in wild-type cells. Quantitative gene expression level was normalized to the expression level of Gapdh. Results are shown as means ± SEM. Student’s t test, *P < 0.05, **P < 0.01. All experiments were performed at least three times
Discussion
Lineage specification during mouse embryogenesis is one of the most important topics for biologists, due to the diversification of cell types comprising the whole body. The establishment of stem cells with different degrees of pluripotency44,45 and specific primary germ layer progenitor cells46–49 has greatly accelerated the understanding of these processes. However, the molecular mechanisms of lineage specification in vivo remain poorly understood. With the advent of high-throughput sequencing technologies, many studies have been performed to profile the epigenetic landscape of early mammalian embryos.11–14,50,51 However, the epigenetic profile for histone modifications and its underlying biological significance during mouse primary germ layer formation remain largely unknown. In this study, by using limited cell number epigenome profiling technologies and careful embryo dissection, we present a comprehensive and lineage-specific epigenomic landscape of the mouse gastrula from the pre-streak to the late-streak stages. By applying multiple bioinformatics and experimental analyses, we reveal the asynchronous features of cell fate commitment in the gastrula and identify the existence of gastrula-specific enhancers. We further determine and validate the epigenetic basis for the primary germ layers’ formation and related organ development through the discovery of epigenetically pre-marked enhancers in the gastrula. Taken together, our study provides a comprehensive interpretation of epigenetic regulatory mechanisms in mouse embryogenesis and greatly broadens the current understanding of mammalian development.
Chromatin state transition has been reported to be fully involved in cell fate commitment.52 Here, we comprehensively profile both TSS proximal and distal chromatin state transitions in the gastrula. We reveal the lineage-specific dynamics of chromatin state transitions in different primary germ layers. Our analyses of proximal chromatin state transition reveal that the bivalent-to-active transition has already become prominent from stage E6.5 to stage E7.0 in mesendodermal and mesodermal lineages, while this becomes dominant from stage E7.0 to stage E7.5 in ectoderm lineage. Further investigation identifies that the chromatin state transition pattern is highly related to lineage development-related genes (Fig. 2b–f), suggesting the occurrence of lineage commitment. The study of distal regulatory elements shows that E7.5 ectoderm cells (E7.5 A) still share a large number of enhancers with E6.5 Epi cells (Fig. 4a), indicating similar enhancer signature between ectoderm cells and E6.5 epiblast cells; in contrast, most enhancers in mesendodermal and mesodermal cells are de novo generated at the late gastrula stages (E7.0-E7.5) (Fig. 4f, g), suggesting that distinct enhancer usage patterns exist between mesendodermal/mesodermal cells and E6.5 epiblast cells. Bioinformatic analyses reveal a strong correlation between these enhancers and lineage development. Thus, the temporal distinction of chromatin state transitions at both TSS proximal and distal regions between mesendoderm/mesoderm and ectoderm strongly indicates that cell fate commitment during gastrulation is asynchronous. These findings not only support the current notion that epiblast stem cells are more similar to ectoderm,31 but also address the discrepancy in molecular mechanisms for cell fate commitment, which requires further exploration.
Developmental competence is a key concept in mammalian development, describing that lineage intermediate cells possess the ability to respond to inductive signals with precise timing.24 The developmental competence established in primary germ layers has been reported to be crucial for future body development.53 Studies have revealed the important roles of the interaction among extrinsic signals, pioneer factors and chromatin states in mediating the formation of developmental competence and related mammalian development.54 For example, enhancers primed with H3K4me1 mediate the formation of developmental competence and further facilitate the binding of pioneer factors in endodermal progenitor cells.24 However, it remained elusive whether tissue-specific enhancers are also epigenetically pre-marked in vivo. Here, we profile the enhancer turnover continuum during mouse embryogenesis by combining our data with published dataset.12,19 Surprisingly, we find that enhancers, residing near key organ development-related genes, are frequently pre-marked by H3K27ac in the gastrula (such as Men1, Men2, Men3, Ect2, and Ect3) (Fig. 6a–c). Similarly, studies based on DNA methylation profiling of the post-implantation embryos have also revealed that a number of developmental gene-related enhancers in E14.5 embryos and somatic tissues are hypo-methylated or lowly methylated in the mouse gastrula.51,55 These results indicate that the competence for organ development has been established in the gastrula through epigenetically pre-marking. This may propose a general principle for epigenetic regulation of developmental competence in mouse embryo development. In the future, further exploration of the interactions among extrinsic signals, pioneer factors, DNA methylation, and chromatin states at these pre-marked enhancers will provide more comprehensive explanations for developmental competence.
A recent study reported the accessibility of the regulatory genome before gastrulation of the Drosophila embryo by using a single-cell combinatorial indexing assay for transposase accessible chromatin with sequencing (sci-ATAC-seq).56 In this study, we have reasonably used an embryo dissection strategy15,21,51 to profile the epigenomic landscape of the mouse gastrula based on current knowledge of mouse embryo pattern and current histone modification profiling technologies. In the future, with continued improvement in technologies,57 such as the uliCUT&RUN technology,58 epigenomic profiling of histone modifications using single cells from the gastrula will provide more thorough explanations not only for the formation of primary germ layers but also for the heterogeneities that exist at this stage.
Materials and methods
Animal use, embryo staging and collection
All animal experiments were performed under the guidelines of the Animal Ethical Committee of the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. Plugged female mice were picked after mating and counted as embryonic day 0.5 (E0.5). Mice were euthanized when embryos developed into nominal day 6.5, 7.0 or 7.5. Embryos were removed from the implantation site as described previously.59 Embryos were then transferred to petri dishes with phosphate buffered saline (PBS). The removal of decidua and Reichert’s membrane was performed using sharpened surgeon tweezers. The developmental stage and morphology of embryos were reconfirmed according to the Theiler stage methodology. The acquired embryos were then dissected into essential parts for the following experiments.
Transcript preparation, quantitative real-time PCR, cDNA sequencing library construction and sequencing
For in vitro culture cells, bulk cells were collected, and RNAs were extracted using Trizol (Invitrogen, 15596018) and precipitated using isopropanol. RNAs were then reverse transcribed into cDNA by using FastQuant RT Super Mix (Tiangen, KR108). For embryo samples, cDNA pools were prepared following a previous protocol.60 Quantitative real-time PCR was performed using diluted acquired cDNA with Stormstar SYBR green qPCR master mix (DBI Bioscience, DBI-143). cDNA sequencing libraries were prepared by using NEBNext Ultra II RNA Library Prep kit (NEB, E7775L) or Nextera XT DNA Library Preparation Kit (FC-131-1024) according to the manufacturer’s instructions. Paired-end 125 bp sequencing was performed on a Hiseq2500 platform at the Berry Genomics Co., Ltd (Beijing, China) and Novogene Company (Beijing, China).
Embryo ChIP sample preparation and ChIP library construction
Essential parts of embryos were collected and pooled to reach a scale of 10,000 cells per sample. The basic workflow for sample collection and chromatin immunoprecipitation (ChIP) was as previously reported.20 Noticeably, for the experiment of H3K9ac and H3K27ac, 20 mM sodium butyrate was added to inhibit histone deacetylase, and in all experiments, proteinase inhibitor cocktail (Roche, 11697498001) and PMSF were added to inhibit protein degradation. In sum, we performed the ChIP assay for histone modifications with specific antibodies against H3K4me3 (Abcam, 8580), H3K4me1 (Abcam, 8895), H3K27ac (Active Motif, 39133), and H3K9ac (Millipore, 06-942). ChIP libraries were prepared using NEBNext Ultra DNA library preparation kit (NEB, E7370) according to the manufacturer’s instructions.
Sequencing data quality control
All the de-multiplexed sequencing reads were first cleaned to remove any artificial sequences, such as sequencing adapters introduced during the experimental process, and reads with >10% low-quality bases were also discarded.
RNA-seq data processing
For the RNA-seq data, the annotation of the transcriptome was defined by combining the UCSC mm10 RefSeq genes and the GENCODE vM5 lncRNAs. Then the cleaned reads were mapped using TopHat (version 2.0.9)61 with default parameters. HTSeq62 was used to count the number of reads mapped to each annotated gene based on the mapping results. These results were further used for calculation of DEGs using DESeq263 package in Bio-conductor. Furthermore, for the quantification of RNA expression, the gene expression levels (RPKMs, reads per kilobase per million reads) of each sample were calculated based on the number of reads for each gene counted by HTSeq:
where gen for genes and s for samples.
Genes with variation above 1 among all samples were selected and the ComBat function in sva package of Bio-conductor64 was used to remove the systematic error introduced by different sequencing batches. The normalized RPKMs after removing batch effects were used for the subsequent analyses, including clustering and combinational analyses with ChIP-seq results.
ChIP-seq data processing
The whole ChIP-seq analysis pipeline was available in https://github.com/kundajelab/chipseq_pipeline. In brief, for the ChIP-seq data, the trimmed reads were mapped to the mouse genome (mm10 assembly) using BWA aligner (version 0.7.5a-r405)65 with the options “-i 15 -q 10 -t 4” and the density tracks and peaks were called by MACS2(v2.10)66 based on the mapping results. The density track of interval i in each sample was further normalized by the “macs2 bdgcmp” command with the control sample.
Especially, for all biological replicates, the peaks were called using the Irreproducibility Discovery Rate(IDR) framework developed by Kundaje et al. (https://sites.google.com/site/anshulkundaje/projects/idr)67 and the densities were calculated using the merged results for samples that passed the IDR framework.
For the broad domain analysis of H3K4me3, the MACS2(v2.10) was again used to call the broad peaks using the options:
Then based on the broad peaks called by MACS2, we classified the H3K4me3-marked domains into four groups based on the segment lengths at their promoter regions.11 To specify, promoters overlapping with segments > 5000 bp were defined as broad H3K4me3 domain genes; those overlapping with 1000 bp < segments ≤ 5000 bp were defined as median H3K4me3 domain genes; those overlapping with 200 bp < segments ≤ 1000 bp were defined as narrow H3K4me3 domain genes; those overlapping with segments ≤ 200 bp were defined as null H3K4me3 domain genes. Stage-specific broad domain genes were defined as the broad domains that only occurred at that specific developmental stage.
For the H3K4me3 and H3K27me3 domain analysis, we called the H3K4me3 and H3K27me3 co-occupied domains across all the samples using the R package chromstaR in a joint multivariate analysis, and the parameters were set as: bin size = 1,000 bp; step size = 200 bp; mode = combinatorial. The state transitions along the developmental lineages were then plotted through alluvial diagrams in R package ggalluvial.
The DNA methylation analysis pipeline
The trimmed reads were mapped to mm10 mouse genome following our modified bisulfite sequencing pipeline.68 PCR duplicates were removed using samtools and DNA methylation level of every cytosine was calculated based on our previous publication.68 The pairwise comparisons of consecutive samples on a single CpG level were calculated using a b-binomial model and the b difference distribution.
The relationship between the epigenetic modifications and gene expression
The epigenetic modifications that we took into consideration include H3K27ac, H3K9ac, H3K4me3, H3K4me1, H3K27me3, and DNA methylation. The epigenetic signals of the histone markers for the ±2 kb TSS region, ±2–10 kb from TSS and ±10–100 kb from TSS were calculated by averaging the signal density around the promoter region, and DNA methylation level was calculated by averaging the methylation level of the single bases with the sequencing depth above 3 at respective ranges from TSS.
Then we examined the relationship between epigenetic modification and gene expression for each gene and each marker. In principle, the Pearson correlation value calculated using the average epigenetic track density and the gene expression level for a given gene in different samples were used to represent this relationship. However, Pearson correlation will usually be influenced when the expression level of genes or the epigenetic track density is stable between different stages. For example, if the RPKM and DNA methylation level of ±2 kb around TSS for a given gene are like below in three continuous stages:
the Pearson correlation will be 1.0 in this case. However, as the RPKM will present a high level variation when the mean RPKM is low, a 0.1 difference between two continuous stages may not be large enough to get rid of the effect of technical errors. The same thing also influenced the DNA methylation levels. Therefore, in order to get a more accurate index for this relationship, the random jitter with Gaussian distribution was introduced into the input vector and the Pearson correlation was calculated in a bootstrap strategy. The average and standard deviation of the TSS signal for the Gaussian distribution for each epigenetic marker m was calculated using TSS signal in all samples and all genes:
However, as the standard deviation of RPKM would change with the different average level of RPKM, a k-nearest neighbor strategy was used to estimate the standard deviation of RPKM. We first calculated the average expression level and the standard deviation for each gene among all samples:
Then all (μRPKM,gen, σRPKM,gen) pairs for different genes were ordered according to μRPKM,gen. For a given gene, the average for the Gaussian distribution of the jitter is μRPKM,gen and the standard deviation is the average value of the standard deviation for the 20 genes nearest to the corresponding average RPKM value.
Then the recalibrated Pearson correlation using the bootstrap strategy is:
where
ChromHMM analysis
Chromatin states were identified and characterized using ChromHMM(v1.11).69 The alignment files of H3K27ac, H3K9ac, H3K4me3, H3K4me1 and H3K27me3 modifications across 9 samples were binned into 200-bp bins using the BinarizeBam command, with the input alignment file as the control. Next, we trained the model with 8, 10, 12, 15 and 18 emission states at 200-bp resolution and default parameters using the LearnModel command. The 15-state result was selected, and we then merged states that shared similar patterns with other states and finally obtained 10 states (Fig. 2a). The enrichment level of non-coding mammalian-conserved elements (GERP) for each state was annotated using the files downloaded from http://mendel.stanford.edu/SidowLab/downloads/gerp/mm9.GERP_scores.tar.gz and then lifted over to mm10. To describe the chromatin states in a more intuitive way, the emission matrix for the 9 states (except Quiescent state) was clustered using R command:
Clustering tree was then cut into three sub-trees. These three resulting sub-trees were then concluded as repressive state, bivalent state and active state (Fig. 2a), which were further used in the following analyses.
Dynamics of enhancers
We analyzed the dynamics of enhancers using the results of ChromHMM. Four states among the 10 states were assumed to imply enhancers, including bivalent enhancer bins, poised enhancer bins, active enhancer bins and bi-acetylated enhancer bins. The matrix used 1 for “Bivalent enhancer bin”, 2 for “Poised enhancer bin”, 3 for “Active enhancer bin” and 4 for “Bi-acetylated enhancer bin”, with 9 samples for columns and 2.5 million 200-bp tiles for rows, which have at least one enhancer bin that existed using the segment results of ChromHMM (Fig. 3a). We used a two-step K-means clustering strategy to cluster this matrix. First, the 2.5 million of 200-bp tiles were clustered into 18 modules with the parameters:
by using the mllib package of pyspark engine(v1.6.2) and reordered the position of the 18 modules manually. Second, for each cluster, we used K-means (scikit-learn v0.18.1) with parameter (k = 10, random_state = 0) to divide 1 group into 10 sub-modules, and then determine the order for the 10 sub-modules by clustering the center of the resulting 10 sub-modules using ward distance (scipy v0.18.1). Finally, the regions of the resulting 18 × 10 = 180 sub-modules were extracted and used for the enrichment analyses of proximity to gene members of a catalog of gene sets (Biological Process (BP) of Gene Ontology (GO), phenotypes of Mouse Genotype Informatics (MGI) and MGI expressed detection) by GREAT.70 The GREAT web API was used to automatically submit region descriptions and retrieve results for subsequent parsing. We restricted ourselves to the interpretation of results with an enrichment ratio of at least 2 and showed the results of binomial FDR P-values in the correspondent figure. Details of enhancer dynamics during the development of each lineage in the gastrula have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE98101.
Identification of gastrula-specific enhancer bins in the mouse gastrula
The 0/1 binarized 200-bp intervals generated by ChromHMM for samples in the gastrula were compared with H3K27ac narrow peak file downloaded from ENCODE (See Supplementary information, Table S5). The downloaded H3K27ac peak intervals were merged using “bedtools multiinter” command and then joined with our data using “bedtools intersect”. If there were any peaks in ENCODE dataset that were intersected with a 200-bp interval in the gastrula sample, it would be considered as “common H3K27ac peak” for this interval, while gastrula-specific enhancer bins were defined as H3K27ac peaks in the gastrula samples that did not overlap with any peaks in the ENCODE data (Fig. 5a).
Mouse enhancer activity reporter construction
Hsp68 minimal promoter driving LacZ reporter plasmid (pHsp68-LacZ plasmid) was kindly given by Prof. Bin Zhou (Shanghai Institute of Biochemistry and Cell Biology) as a gift. The plasmid was firstly linearized by HindIII (FastDigest, FD0504) at 37 °C for 2 h and purified (Qiagen, 28704). Enhancer fragments were cloned from mouse genomic DNA by using KOD neo polymerase system (Toyobo, KFX-201), and then ligated to linearized pHsp68-LacZ plasmid by seamless cloning system (Beyotime, D7010M). Acquired plasmid was further digested with NotI (FastDigest, FD0596) and XhoI (FastDigest, FD0694) followed by purification to obtain linearized fragment for transgenic mouse generation. The primers used here were included in Supplementary information, Table S8.
Transgenic mouse acquisition and LacZ staining
Generally, we used zygote pronuclear microinjection to produce LacZ transgenic mice. Egg donor female mice (3-week-old C57BL/6) were superovulated by injecting PMSG (pregnant mare’s serum gonadotropin) and HCG (human chorionic gonadotropin) prior to mating. Zygotes were harvested about 18 h after mating. Purified DNA was diluted with injection buffer (10 mM Tris, pH 7.5, 0.1 mM EDTA) to a final concentration of 3–4 ng/μL and used for pronuclear injections of PN4 stage zygotes with a FemtoJet Microinjection System (Eppendorf). The injected embryos were cultured to the 2-cell stage in KSOM medium with amino acids at 37 °C under 5% CO2, and then transferred to the oviduct of pseudo-pregnant ICR females and marked as 0.5 dpc. Embryos were collected at the appropriate stage for LacZ staining. Lac staining was performed as previously reported.71
CRISPR-Cas9-mediated enhancer fragment deletion
Small guided RNAs were designed by using the online tool Chop-chop (http://chopchop.cbu.uib.no/). The plasmid preparation scheme and knockout cell line generation was performed as previously reported.72 The potential off-target sites were also tested based on the website’s indication.
shRNA design
shRNAs for knocking down the expression of 2510009E07Rik were designed by using online tool GPP Web Portal (https://portals.broadinstitute.org/gpp/public/). Replicated shRNAs were selected. Scramble sequence for each shRNA was designed using online tool Create Scrambled Sequence (https://www.genscript.com/tools/create-scrambled-sequence).
Mouse embryonic stem cell culture and neural differentiation
Mouse embryonic stem cell E14Tg2A was cultured in standard medium supplemented with 3 μM ChIR99021, 1 μM PD0325901 and 10 ng/mL mouse leukemia inhibitory factor (mLIF, Millipore). The neural differentiation was performed as previously reported.43
Data availability
The gene expression and histone modification data are deposited in NCBI GEO under the accession number GSE98101.
Supplementary information
Acknowledgements
We thank Drs Patrick Tam, Chi-chung Hui, Bing Zhu and Dangsheng Li for helpful advice and discussion. We thank Dr Bin Zhou for the generous gift of the pHsp68-LacZ plasmid. This work was supported in part by the National Key Basic Research and Development Program of China (2018YFA0800100, 2018YFA0108000, 2018YFA0107200, 2017YFA0102700, 2014CB964804, 2018YFA0107601 and 2015CB964500), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16020501 and XDA16020404), the Genome Tagging Project, and the National Natural Science Foundation of China (31900454, 31501178, 31430058, 31571513, 31630043, 31871456, 91519314, 31661143042, 81561138005, 31625018, 81521002, 31530048, 81672117 and 31730062), and Shanghai Municipal Commission for Science and Technology (17JC1400900 and 17411954900).
Author contributions
Y. Qiao, J.L., F.T., and N.J. conceived and supervised the project. X.Y., J.L., Y.C., Y. Qian, S.F., F.Y., H.X., and Y.H. performed the experiments. B.H., J.D., Y.H., and R.W. performed the computational analyses. X.Y., B.H., Y. Qiao, G.P., F.T, and N.J wrote the paper with the help from all other authors.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xianfa Yang, Boqiang Hu, Jiaoyang Liao, Yunbo Qiao
Contributor Information
Yunbo Qiao, Email: ybqiao@gzhu.edu.cn.
Jinsong Li, Email: jsli@sibcb.ac.cn.
Fuchou Tang, Email: tangfuchou@pku.edu.cn.
Naihe Jing, Email: njing@sibcb.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0234-8.
References
- 1.Wolpert, L., Tickle, C. & Martinez-Arias, A. Principles of Development. Fifth edn (Oxford, UK: Oxford University Press, 2015).
- 2.Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell. Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 3.Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- 4.Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- 5.Takaoka K, Hamada H. Cell fate decisions and axis determination in the early mouse embryo. Development. 2012;139:3–14. doi: 10.1242/dev.060095. [DOI] [PubMed] [Google Scholar]
- 6.Benitah SA, Frye M. Stem cells in ectodermal development. J. Mol. Med. 2012;90:783–790. doi: 10.1007/s00109-012-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tada S, et al. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 8.Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 9.Atlasi Y, Stunnenberg HG. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017;18:643–658. doi: 10.1038/nrg.2017.57. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Peng G, Jing N. TGF-beta signaling pathway in early mouse development and embryonic stem cells. Acta. Biochim. Biophys. Sin. 2018;50:68–73. doi: 10.1093/abbs/gmx120. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 12.Dahl JA, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 14.Ke Y, et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell. 2017;170:367–381. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Wang C, et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell. Biol. 2018;20:620–631. doi: 10.1038/s41556-018-0093-4. [DOI] [PubMed] [Google Scholar]
- 16.Zylicz JJ, et al. G9a regulates temporal preimplantation developmental program and lineage segregation in blastocyst. Elife. 2018;7:pii: e33361. doi: 10.7554/eLife.33361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zylicz JJ, et al. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife. 2015;4:pii: e09571. doi: 10.7554/eLife.09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorkin, D. U. et al. Systematic mapping of chromatin state landscapes during mouse development. BioRxiv 2017. 10.1101/166652
- 19.Nord AS, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, et al. Silencing of developmental genes by H3K27me3 and DNA methylation reflects the discrepant plasticity of embryonic and extraembryonic lineages. Cell. Res. 2018;28:593–596. doi: 10.1038/s41422-018-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith ZD, et al. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature. 2017;549:543–547. doi: 10.1038/nature23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam RC, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong CT, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–430. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang A, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- 26.Peng G, et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell. 2016;36:681–697. doi: 10.1016/j.devcel.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlet J, et al. Bivalent regions of cytosine methylation and H3K27 acetylation suggest an active role for DNA methylation at enhancers. Mol. Cell. 2016;62:422–431. doi: 10.1016/j.molcel.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Gobbi M, et al. Generation of bivalent chromatin domains during cell fate decisions. Epigenetics Chromatin. 2011;4:9. doi: 10.1186/1756-8935-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima Y, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Whyte WA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterwalder M, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243. doi: 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen H, et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016;165:331–342. doi: 10.1016/j.cell.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strumpf D, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 36.Jiang B, Mendelson CR. USF1 and USF2 mediate inhibition of human trophoblast differentiation and CYP19 gene expression by Mash-2 and hypoxia. Mol. Cell. Biol. 2003;23:6117–6128. doi: 10.1128/MCB.23.17.6117-6128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai T, et al. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev. Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- 38.Wu G, Scholer HR. Role of Oct4 in the early embryo development. Cell. Regen. 2014;3:7. doi: 10.1186/2045-9769-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mesman S, Smidt MP. Tcf12 is involved in early cell-fate determination and subset specification of midbrain dopamine neurons. Front. Mol. Neurosci. 2017;10:353. doi: 10.3389/fnmol.2017.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonev B, et al. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–572. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickel DE, et al. Ultraconserved enhancers are required for normal development. Cell. 2018;172:491–499. doi: 10.1016/j.cell.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang K, et al. Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development. 2010;137:2095–2105. doi: 10.1242/dev.049494. [DOI] [PubMed] [Google Scholar]
- 44.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144:365–373. doi: 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kubo A, et al. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 47.Cheng X, et al. Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell. 2012;10:371–384. doi: 10.1016/j.stem.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar N, et al. Generation of an expandable intermediate mesoderm restricted progenitor cell line from human pluripotent stem cells. Elife. 2015;4:pii: e08413. doi: 10.7554/eLife.08413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu C, et al. Suppressing nodal signaling activity predisposes ectodermal differentiation of epiblast stem cells. Stem Cell Rep. 2018;11:43–57. doi: 10.1016/j.stemcr.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du Z, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 2018;50:96–105. doi: 10.1038/s41588-017-0003-x. [DOI] [PubMed] [Google Scholar]
- 52.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossant., J. & Tam., P. P. L. Mouse Development: Patterning, Morphogenesis, and Organogenesis. (San Diego, USA: Gulf Professional Publishing, 2002).
- 54.Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 55.Bogdanovic O, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat. Genet. 2016;48:417–426. doi: 10.1038/ng.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cusanovich DA, et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 2018;555:538–542. doi: 10.1038/nature25981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hainer SJ, Boskovic A, McCannell KN, Rando OJ, Fazzio TG. Profiling of pluripotency factors in single cells and early embryos. Cell. 2019;177:1319–1329. doi: 10.1016/j.cell.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Behringer, R. R., Gertsenstein, M., Nagy, K. V. & Nagy, A. Manipulating the mouse embryo: a laboratory manual. (New York, USA: Cold Spring Harbor Laboratory Press, 2014).
- 60.Chen J, et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017;12:566–580. doi: 10.1038/nprot.2017.003. [DOI] [PubMed] [Google Scholar]
- 61.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome. Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo H, et al. Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat. Protoc. 2015;10:645–659. doi: 10.1038/nprot.2015.039. [DOI] [PubMed] [Google Scholar]
- 69.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser - a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene expression and histone modification data are deposited in NCBI GEO under the accession number GSE98101.