Abstract
Spinocerebellar ataxia type 3 (SCA3), or Machado-Joseph disease (MJD), is a progressive neurodegenerative disorder caused by a CAG expansion in the ATXN3 gene. The expanded CAG repeat is translated into a prolonged polyglutamine repeat in the ataxin-3 protein and accumulates within inclusions, acquiring toxic properties, which results in degeneration of the cerebellum and brain stem. In the current study, a non-allele-specific ATXN3 silencing approach was investigated using artificial microRNAs engineered to target various regions of the ATXN3 gene (miATXN3). The miATXN3 candidates were screened in vitro based on their silencing efficacy on a luciferase (Luc) reporter co-expressing ATXN3. The three best miATXN3 candidates were further tested for target engagement and potential off-target activity in induced pluripotent stem cells (iPSCs) differentiated into frontal brain-like neurons and in a SCA3 knockin mouse model. Besides a strong reduction of ATXN3 mRNA and protein, small RNA sequencing revealed efficient guide strand processing without passenger strands being produced. We used different methods to predict alteration of off-target genes upon AAV5-miATXN3 treatment and found no evidence for unwanted effects. Furthermore, we demonstrated in a large animal model, the minipig, that intrathecal delivery of AAV5 can transduce the main areas affected in SCA3 patients. These results proved a strong basis to move forward to investigate distribution, efficacy, and safety of AAV5-miATXN3 in large animals.
Introduction
Spinocerebellar ataxia type 3 (SCA3), or Machado-Joseph disease (MJD), is the most common spinocerebellar ataxia worldwide and the second most common polyglutamine (polyQ) disease after Huntington’s disease (HD).1, 2, 3, 4, 5 Similar to the other polyQ disorders, SCA3 is inherited in an autosomal dominant manner, which is progressively neurodegenerative and ultimately fatal. SCA3 is caused by an expanded stretch of CAG triplets in the coding region of the ATXN3 gene.6 Healthy individuals have up to 44 CAG repeats, while affected individuals have between 52 and 86 glutamine repeats.6, 7, 8 Repeat ranges from 45 to 51 are associated with incomplete penetrance of the disease. There is a clear correlation between CAG repeat size and age of onset, although CAG repeat length only accounts for approximately 50% of the total variability in age of onset,9 Age at onset of SCA3 is highly variable but most commonly in the second to fifth decade, with an average age at onset of 40 years.10 The CAG expansion has full penetration, as patients harboring the mutation will inevitably develop the disease and have a 50% chance to pass it on to their offspring.
The ATXN3 transcript is alternatively spliced and produces different isoforms of the ataxin-3 protein.10,11 The most abundant protein isoform contains an N-terminal Josephin domain that has a deubiquitinase activity and a C-terminal region that has three ubiquitin-interacting motifs (UIMs), implicating a role of ataxin-3 in the ubiquitin-proteasome pathway.1,12 The expanded CAG repeat in the ATXN3 gene leads to formation of an expanded polyQ tract in the C-terminal region of the ataxin-3 protein. This mutated ataxin-3 protein causes toxic gain of function and leads to formation of neuronal aggregates, which is a hallmark of polyQ diseases.13 Despite extensive research, the mechanisms leading to the observed neurodegeneration in SCA3 patients have not been completely elucidated. Ataxin-3 is normally ubiquitously found throughout the cell and can translocate from the cytoplasm to the nucleus and vice versa.14 In neurons, ataxin-3 is predominantly expressed in the cytoplasm while the mutated protein mainly accumulates in the nucleus and acquires toxic properties.13, 14, 15, 16, 17. Formation of neuronal aggregates comprising the mutated ataxin-3 protein is a typical neuropathological hallmark of the disease. Besides protein toxicity, RNA toxicity may also contribute to pathogenicity of the disease,18 as the expanded CAG repeat, and CUG-containing RNA molecules, can form RNA foci, which colocalize with RNA binding proteins and sequester their functions. For example, colocalization of CAG- and CUG-containing RNA foci with the muscleblind-like 1 (MBNL1) splicing factor in nuclei of both muscle cells and neurons resulted in inactivation of MBNL1, leading to dysregulation of alternative splicing.18, 19, 20, 21, 22
Neuropathological studies have detected widespread neuronal loss in the cerebellum, thalamus, midbrain, and spinal cord of SCA3 patients.17 Although widespread pathology is reported in later disease stage of SCA3 patients, the general consensus is that the main neuropathology in SCA3 patients is located in the cerebellum and brain stem.23 The main clinical symptom observed in SCA3 patients is progressive ataxia, affecting balance, gait, and speech.18 Other frequently described symptoms include pyramidal signs, progressive external ophthalmoplegia, dysarthria, dysphagia, rigidity, distal muscle atrophies, and double vision.2 Most of the patients die due to pulmonary complications, usually within 6–29 years after onset, and up to now there is no disease-modifying treatment available.24,25
From a therapeutic standpoint, an advantage of monogenetic disorders such as SCA3 is that reducing expression of the responsible gene should result in alleviation of mutant RNA and protein toxicity.18 Silencing approaches by RNAi or antisense oligonucleotides (ASOs) are attractive to achieve silencing of the mutant ataxin-3. The silencing can be allele specific, silencing only the mutant ataxin-3, or non-allele specific, silencing both wild-type and mutant alleles. Both approaches demonstrated that neuropathology in SCA3 rodents can be improved.26, 27, 28
In the current study, we investigated a non-allele-specific RNAi-based gene therapy for SCA3 patients with a potentially long-lived therapeutic effect. The therapeutic product is an adeno-associated virus (AAV) vector expressing a microRNA that binds ATXN3 mRNA, leading to its degradation via the RNA-induced silencing complex (RISC). Non-allele-specific silencing of ataxin-3 by RNAi has been tested in wild-type and SCA3 rodent models and demonstrated improvement of the observed neuropathology.27 This approach was well tolerated despite the concomitant reduction of the wild-type ataxin-3. We engineered artificial microRNAs (miATXN3) to target various exons within the ATXN3 mRNA. The miATXN3 candidates were incorporated in the primary miR-451 scaffold, which has been extensively studied by us and proved to be safe in human neurons, rodents, pigs, and non-human primates.29, 30, 31 By targeting both wild-type and mutant ataxin-3 we are aiming for a therapy to treat the whole SCA3 patient population. Because miR-451 has been shown to lack passenger activity due to dicer-independent processing, this significantly reduces the risk for off-target effects.32,33 A preselection for the most efficient miATXN3 candidates was performed in vitro on a luciferase (Luc) reporter. The best candidates were incorporated in AAV serotype 5 (AAV5), and their efficacies were tested in human-derived induced pluripotent stem cell (iPSC)-neurons and in a knockin SCA3 mouse model. We observed strong reductions of ATXN3 mRNA and mutant ataxin-3 protein, suggesting that our AAV-microRNA-based approach could have therapeutic benefits in SCA3 human patients. In addition, we investigated the intrathecal delivery of AAV5 in a minipig and confirmed that the main areas affected in SCA3 patients are transduced at a therapeutically relevant dose. These data demonstrate that AAV-based RNAi gene therapy has potential for use in SCA3 treatment by lowering of the mutant ataxin-3 protein in the CNS.
Results
Design of Artificial miATXN3 Constructs and Prescreening on Luc Reporter
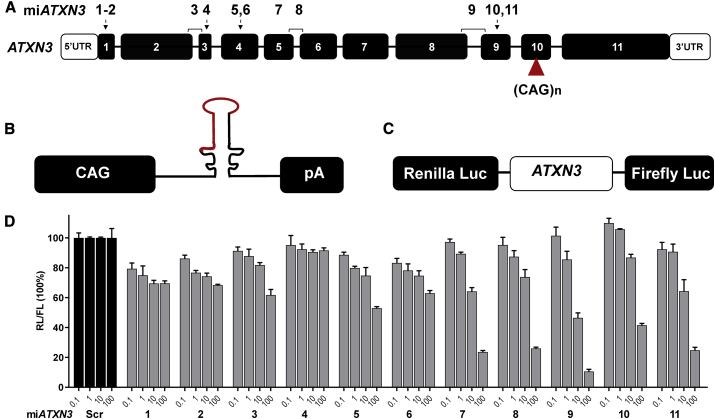
Silencing of mutant ATXN3 mRNA should result in alleviation of toxicity caused by the mutant ataxin-3 protein. Using this rationale, we engineered artificial microRNAs complementary to various regions of the ATXN3 gene aiming at a knockdown of ATXN3 mRNA. Different regions of the human ATXN3 gene were selected to design anti-ATXN3 microRNAs (miATXN3) constructs (Figure 1A). All miATXN3 constructs were designed to specifically target the human ATXN3 transcripts with high to full conservation for non-human primates, mouse and rat ATXN3. The miATXN3 sequences were embedded in the engineered pre-miR-451 scaffold and expressed by the ubiquitous CAG promoter that consists of the cytomegalovirus immediate-early enhancer fused to chicken β-actin promoter (Figure 1B). This promoter has been broadly used in CNS indications, showing stable and high transgene expression.34 To test the silencing efficacy of the miATXN3 constructs, we designed a Luc reporter bearing complementary ATXN3 target mRNA fused to the renilla luciferase (RL) gene (Figure 1C). The firefly luciferase (FL) gene was independently expressed from RL in the same reporter to correct for transfection efficiency. We co-transfected the miATXN3 constructs with the Luc reporter and prescreened for the best candidates. miATXN3_7, miATXN3_9, and miATXN3_11 were selected for further testing, as these candidates showed knockdown on the Luc reporter at lower concentrations and reached up to 80%–90% knockdown efficiencies at higher concentrations (Figure 1D).
Figure 1.
Design and Screening of Engineered miATXN3 Constructs
(A) Schematic representation of the human ATXN3 gene and miATXN3 binding sites. The ATXN3 gene (NG_008198.2) consists of 11 exons shown by the numbered black boxes. The white boxes represent the 5′ and 3′ UTRs. The CAG expansion in exon 10 is depicted by a red triangle. The position of the miATXN3 candidates are shown on top of the exons and indicated with numbers 1–11. miATXN3_3, miATXN3_8, and miATXN3_9 are exon spanning. (B) Schematic representation of the miATXN3 constructs. Each construct was expressed by the CAG promotor, followed by the primary miATXN3 sequence in the miR-451 scaffold, and a human growth hormone polyadenylation (hGH poly(A)) signal. (C) Schematic representation of the Luc reporter. The whole sequence of the ATXN3 mRNA (NM_004993.5) was cloned downstream of the RL gene. In addition, FL was co-expressed from the vector as an internal control. (D) Dose-dependent knockdown of ATXN3 Luc reporter by miATXN3 constructs. HEK293T cells were co-transfected with 50 ng of the Luc reporter and 0.1, 1, 10, and 100 ng of the miATXN3 constructs. RL and FL were measured 2 days post-transfection, and RL was normalized to FL expression. Scrambled microRNA (miScr) served as a negative control and was set at 100%.
Strong Silencing of Endogenous ATXN3 and Ataxin-3 Protein by miATXN3
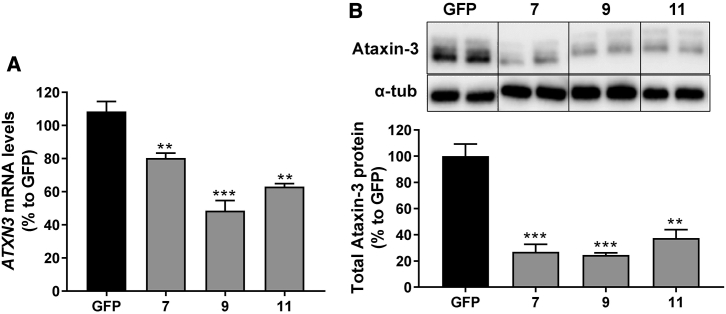
We next investigated whether the selected miATXN3 candidates were capable of reducing the endogenous levels of total ATXN3 mRNA and protein in HEK293T cells. Cells were transfected with the miATXN3_7, miATXN3_9 or miATXN3_11 and the endogenous levels of ATXN3 mRNA was determined 2 days post-transfection by qRT-PCR. A significant reduction was observed by all three miATXN3 constructs (Figure 2A). The most effective candidate was miATXN3_9 with a silencing efficacy of 52%. Consistently, silencing of the ATXN3 mRNA also resulted in significant reduction of ataxin-3 protein with up to 75% reduction achieved with miATXN3_9 (Figure 2B). Thus, all three selected candidates can reduce ATXN3 mRNA and protein in transfected cells, with miATXN3_9 being the most potent.
Figure 2.
Silencing of ATXN3 mRNA and Protein in HEK293T Cells
(A) Endogenous knockdown ATXN3 mRNA by the selected miATXN3 candidates. qRT-PCR ATXN3 mRNA was performed on RNA from HEK293T cells that were transfected with 250 ng of miATXN3_7, miATXN3_9, or miATXN3_11 for 3 days. mRNA input levels were normalized to GAPDH mRNA. Cells transfected with a GFP construct served as negative control, which was was set at 100%. (B) Silencing of total ataxin-3 protein. HEK293T cells were transfected as describe in (A), and protein expression was determined by western blot. α-Tubulin was included as internal control. Western blot intensity bands of ataxin-3 were quantitated and the knockdown was calculated relative to GFP. Data were analyzed using a multiple comparison one-way ANOVA to determine statistically significances of cells treated the miATXN3 constructs. The p values are listed in the graph as follows: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Each graph represents the mean values with SD (n = 3).
AAV5-miATXN3 Is Highly Effective in Human iPSC-Neurons
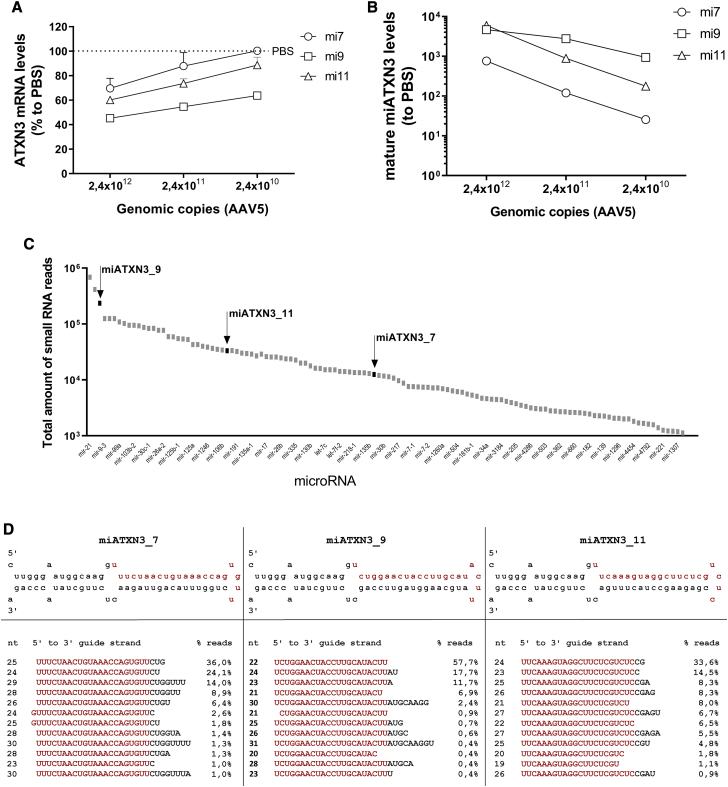
To further confirm the silencing of ATXN3 in the context of a gene therapy for SCA3, we incorporated all three candidates in AAV5. Subsequently, increasing doses of AAV5-miATXN3 were tested in iPSC-neurons. Human iPSCs were differentiated into frontal brain-like neurons, which represent mainly neurons of the frontal cortex. About 90% of these neurons expressed β-tubulin III, indicating a successful differentiation.35 The cells were transduced with AAV5-miATXN3_7, AAV5-miATXN3_9, or AAV5-miATXN3_11 for 2 weeks. As shown previously, at this time point, about 80% of cells are GFP positive.35 Following miATXN3 transductions, the silencing efficacy of ATXN3 mRNA was determined. ATXN3 mRNA was reduced up to 65% in a dose-dependent manner by all three miATXN3 candidates (Figure 3A). Consistent with the knockdown of the Luc reporter, AAV5-miATXN3_9 had the strongest silencing efficacy in iPSC-neurons. The mature miATXN3 guide strands levels were also determined by a small RNA TaqMan assay. All three mature miATXN3s were expressed in the cell in a dose-dependent manner, suggesting a successful transduction by AAV5-miATXN3 and processing into a mature microRNA (Figure 3B).
Figure 3.
Silencing of ATXN3 mRNA by AAV5-miC in Human iPSC-Neurons
(A) Dose-dependent silencing of ATXN3 in transduced iPSC-neurons. Frontal brain-like neurons were transduced with 2.4 × 1012, 2.4 × 1011, and 2.4 × 1010 gc of miATXN3_7, miATXN3_9, or miATXN3_11 incorporated into AAV5. RNA was isolated 2 weeks post-transduction and ATXN3 mRNA levels was determined by qRT-PCR. mRNA input was normalized to GAPDH and set relative to PBS-treated cells. (B) Levels of mature miATXN3 guide strands in transduced cells. Performed as described in (A). Expression of the mature miATXN3_7, miATXN3_9, and miATXN3_11 was determined by small RNA TaqMan. MicroRNA input levels were normalized to U6 small nuclear RNA and set relative to PBS-treated cells. (C) Relative miATXN3 expression levels in transduced cells by small RNA sequencing. Frontal brain-like neurons were transduced with 2.4 × 1012 gc of miATXN3_7, miATXN3_9, or miATXN3_11 incorporated in AAV5. Small RNA sequencing was performed 2 weeks post-transduction. The total amounts of small RNA reads corresponding to the three lead miATXN3 candidates are shown by the black arrows. The total amount of reads from other natural expressed endogenous microRNAs are shown in gray. (D) Processing of miATXN3 in iPSC-neurons by small RNA sequencing. Frontal brain-like neurons were transduced as described in (C). The secondary miATXN3 structure based on miRbase prediction is shown on the first row, including their predicted 22-nt guide strands shown in red. The sequence distributions of the different guide strand length (nt) mapping to miATXN3_7, miATXN3_9, and miATXN3_11 pre-microRNA sequences in the miR451 scaffold were calculated as percentages (% reads).
No Saturation of Endogenous RNAi Machinery by miATXN3
As microRNA overexpression can lead to toxicity, we determined the overall expression of the miATXN3 candidates in human-transduced iPSC-neurons relative to other endogenous microRNAs. Small transcriptome analysis was performed on total RNAs isolated from frontal brain-like neurons that were transduced with AAV5-miATXN3_7, AAV5-miATXN3_9, or AAV5-miATXN3_11 for 2 weeks. We then compared the total amount of reads corresponding to the mature guide strand sequences of miATXN3_7, miATXN3_9, and miATXN3_11 to the total amount of reads of other naturally expressed microRNAs found in the cell (Figure 3C). All three miATXN3 candidates were expressed at normal levels and within the expression range of other natural microRNAs in the cell. Additionally, none of the endogenously expressed miRNAs was found differentially expressed between miATXN3_7- and miATXN3_9-treated neurons (Figure S1), despite a nearly 10-fold difference in expression of these two constructs. It can thus be concluded that the microRNA biogenesis system was not overloaded by the relatively high levels of miATXN3 expression.
miATXN3 Is Processed Exclusively into Guide Strands in Human iPSC-Neurons
The miATXN3 candidates were incorporated in the natural human precursor miR-451 scaffold. miR-451 is processed in the non-canonical pathway, first by Drosha to generate a precursor microRNA (pre-miATXN3). The pre-miATXN3 is then transported by exportin 5 (EXP5) into the cytoplasm for further processing by Argonaute 2 (AGO2) and poly(A)-specific ribonuclease (PARN).32,36, 37, 38, 39 Ago2 cleaves the 3′ arm of pre-miR-451 by its slicer activity and generates an ∼30-nt intermediate that is further trimmed by PARN to generate a mature miATXN3 of ∼22 nt. The trimming is not essential for efficiency, and mature guide strands longer than the mature length can still be functional.39 However, mature microRNAs that are too long could increase the risk of binding to off-target genes. The processing was investigated by small transcriptome analysis, and all three miATXN3 candidates resulted exclusively in guide strands between 19 and 30 nt long, with no passenger strands detected (Figure 3D). However, the individual lengths of mature guide strands that were generated varied between the three miATXN3 candidates. The majority of miATXN3_7 guide strands were between 2 and 7 nt longer at the 3′ end than the in silico 22-nt prediction. miATXN3_9 was processed into primarily 22-nt guide strands, as was predicted on mfold. The processing of miATXN3_11 mainly generated guide strands between 1 and 4 extra nt at the 3′ end than predicted. Thus, Drosha cleavage sites at the 5′ end of the mature guide strand was precise for all of the miATXN3 candidates, but the trimming at the 3′ ends by PARN was different and resulted in a variety of mature lengths.
Overall, no passenger strands were detected by the miATXN3 candidates, and the processing of miATXN3_9 was the closest to our prediction.
Diffused AAV5 Transduction of Cerebellum and Brain Stem upon Administration in the Cisterna Magna or DCN of SCA3 Knockin Mice
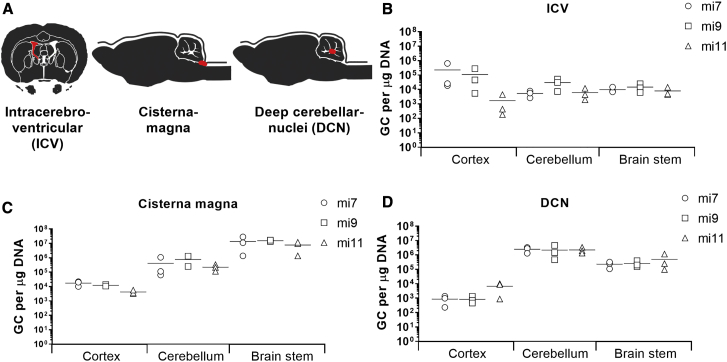
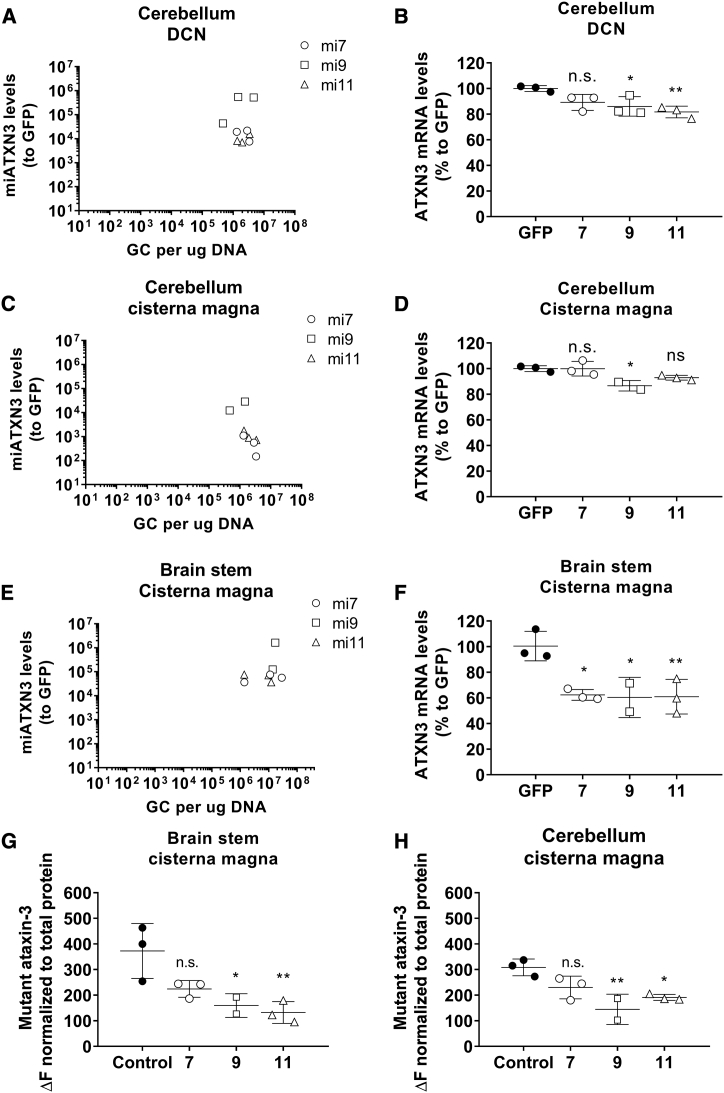
The primary pathology in SCA3 is degeneration of the cerebellum and brain stem. Thus, for an AAV-based gene therapy for SCA3, diffused transduction of both brain structures is required in order to prevent neuronal dysfunction caused by mutant ataxin-3 protein. To determine the most effective delivery route needed for transduction of the cerebellum and brain stem, a study was conducted in a SCA3 knockin mouse model. The SCA3 knockin mouse model was generated using zinc finger technology by cutting the murine (CAG)6 and subsequent homologous recombination with a (CAACAGCAG)48 donor vector with interrupted repeat. This mouse model was characterized to express a mutant ataxin-3 protein with a 304 glutamine repeat (E.H. et al., unpublished data). The expansion of the polyQ tract did not affect the fertility or life expectancy of the mice. At 4.5 months, the time point of sacrifice, the knockin mice were in a pre-symptomatic stage with no differences in body weight or motor phenotype compared to their wild-type littermates. Three routes of injection were explored: (1) intracerebroventricular(ly) (i.c.v.), (2) intracisterna magna, or (3) bilateral into the deep cerebellar nuclei (DCN) (Figure 4A). Injections were performed with AAV5-miATXN3_7, AAV5-miATXN3_9, or AAV5-miATXN3_11, whereas AAV5-GFP was taken along as control. The animals were sacrificed 6 weeks post-surgery, and qPCR of the transgene was performed on the miATXN3 or GFP to determine the biodistribution in the cortex, cerebellum, and brain stem. The i.c.v. administration resulted in a relative low vector copy distribution to all three analyzed tissues. Some transduction was observed only in the cortex (Figure 4B). Administration into the cisterna magna resulted in low transduction of the cortex but strong transduction of the brain stem and cerebellum (Figure 4C). The highest transduction was detected in the brain stem with up to 2.9 × 107 genome copies (gc)/μg tissue DNA. One mouse that received AAV5-miATXN3_9 in the cisterna magna was excluded because no genomic copies were detected, suggesting that the injection failed. Direct injection into the DCN also resulted in relatively high transduction of the cerebellum and the brain stem. Compared to cisterna magna administration, DCN injection resulted in better transduction of the cerebellum and less transduction of the brain stem (Figure 4D). Up to 4.6 × 106 gc/μg tissue DNA were detected in the cerebellum. Based on the current observation, we concluded that administration into the cisterna magna resulted in the highest combined transduction of both cerebellum and brain stem of mice.
Figure 4.
Vector Copy Distribution of AAV5 in SCA3 Knockin Mice
(A) Schematic representation of the routes of administration. Three-month-old mice (n = 3) were injected i.c.v., or in the cisterna magna, or DCN with 2.43 × 1013 gc/mL AAV5-miATXN3_7, AAV5-miATXN3_9, or AAV5-miATXN3_11. 10 μL of AAV5 was injected either i.c.v. or in the cisterna magna, and 2 μL was injected bilaterally in the DCN. The injection sites are depicted in red. All mice were sacrificed 6 weeks after surgeries. (B–D) Vector copy distribution in cortex, cerebellum, and brain stem of mice injected i.c.v. (B), or in the cisterna magna (C) or DCN (D) with AAV5-miATXN3. DNA was isolated from the cortex, cerebellum, and brain stem tissues and qRT-PCR was performed to determine the vector copy distribution. The genome copies per μg of DNA were calculated for each brain region using a standard curve. No genomic copies were detected in untreated mice.
Significant Reduction of ATXN3 mRNA in SCA3 Knockin Mice Brain
Having established distribution of AAV5 upon different routes of injection, we further investigated the efficacy of the AAV5-miATXN3 candidates to reduce the total ATXN3 mRNA in the transduced tissues. Direct injection into the DCN showed highest expression of mature miATXN3 in the cerebellum (Figure 5A). miATXN3_9 had the highest microRNA expression, consistent with the observation in vitro. The microRNA expression correlated well with a mild (∼15%–20%) but significant reduction of ATXN3 mRNA by miATXN3_9 and miATXN3_11 in the cerebellum (Figure 5B). Administration to the cisterna magna resulted into lower mature microRNA expression in the cerebellum as compared to DCN injection (Figure 5C). Nevertheless, miATXN3_9 was best expressed and resulted in significant lowering (∼15%) of ATXN3 mRNA in the cerebellum (Figure 5D). The highest microRNA expression and silencing efficacy from all three delivery routes were observed in the brain stem after administration in the cisterna magna (Figures 5E and 5F). Expression of the miATXN3 candidates were high in the brain stem, and all led to a strong reduction of ATXN3 mRNA of about 40%. Both AAV5-miATXN3_9 and AAV5-miATXN3_11 had comparable efficacies in the brain stem, although the significance of AAV5-miATXN3_9 was smaller because one mouse was excluded. Overall, we concluded that administration of AAV5-miATXN3_9 to the cisterna magna resulted in ATXN3 mRNA reduction in both cerebellum and brain stem, which are the main areas affected in SCA3 patients.
Figure 5.
Silencing of Mutant Ataxin-3 in SCA3 Knockin Mice
(A) Expression of mature miATXN3 guide strands in the cerebellum after DCN administration. Total RNA was isolated from the cerebellum for small RNA TaqMan. MicroRNA input levels were normalized to U6 small nuclear RNA and set relative to AAV-GFP-treated mice. (B) Lowering of total ATXN3 mRNA in cerebellum of DCN-injected mice. Total RNA was isolated from cerebellum and qRT-PCR was performed to detect the mouse wild-type ATXN3 mRNA. RNA input levels were normalized to GAPDH and set relative to AAV-GFP treated mice. (C) Expression of mature miATXN3 guide strands in the brain stem after cisterna magna administration. Performed as described in (A). (D) Lowering of total ATXN3 mRNA in cerebellum of cisterna magna injected mice. Performed as described in (B). (E) Expression of mature miATXN3 guide strands in the brain stem after cisterna magna administration. Performed as described in (A). (F) Lowering of total ATXN3 mRNA in brain stem of cisterna magna-injected mice. Performed as described in (B). (G) Reduction of mutant ataxin-3 protein in the brain stem after cisterna magna delivery. TR-FRET immunoassay was performed on tissue homogenates to specifically detect the mutant ataxin-3 protein containing more than 37 glutamine repeats (no detection of wild-type mouse ataxin-3). (H) Reduction of mutant ataxin-3 protein in the cerebellum after cisterna magna delivery.
Reduction of Mutant Ataxin-3 Protein in Brain Stem and Cerebellum of Mice
Heterozygous SCA3 knockin mice showed reduced body weight in male mice at 48 weeks in life. Additionally, male and female SCA3 knockin mice developed an ataxic gait at 18 months of age. Neuropathologically, accumulations of mutant ataxin-3 and protein aggregates were found in cerebellum, DCN, and pons of heterozygous SCA3 knockin mice starting at the age of 3 months, with higher amounts at later age.
The polyQ expansion within the mutant ataxin-3 protein causes toxic gain of function, leading to the formation of neuronal intranuclear inclusions, neuronal dysfunction, and degeneration.15,26 Decreasing the levels of the mutant proteins leads to therapeutic benefit in several preclinical models.18,26, 27, 28,40 We used a time-resolved fluorescence energy transfer (TR-FRET) immunoassay to measure the levels of mutant ataxin-3 in cerebellum and brain stem homogenates of treated mice.41 We found a strong lowering of up to 64% of the mutant ataxin-3 protein in the cerebellum and brain stem homogenates of mice receiving AAV5-miATXN3_9 and AAV5-miATXN3_11 in the cisterna magna (Figures 5G and 5H). The reduction of the mutant ataxin-3 protein in the cerebellum and brain stem of mice confirmed the feasibility of an RNAi-based gene therapy approach for lowering of mutated ataxin-3 protein in the affected brain regions of patients.
Prediction of Off-Target Genes Due to miATXN3 Treatment
We next investigated whether miATXN3 treatment results in major alterations in RNA expression profile in SCA3 knockin mouse brain and human-derived iPSC-neurons. Although the miATXN3 candidates were designed to specifically target ATXN3 mRNA transcript, complementarity with other transcripts might result in off-target lowering of other genes. The off-target activity of AAV5-miATXN3_7, AAV5-miATXN3_9, and AAV5-miATXN3_11 was predicted using BLAST to search for transcripts with (partial) complementarity with the guide strand using the Homo sapiens reference RNA sequence database (refseq_rna [taxid 9606]). The BLAST results were subsequently compared to RNA sequencing expression values that were performed on total RNA from human-derived frontal brain-like neurons transduced with the three AAV5-miATXN3 candidates and the formulation buffer (control). As expected, all three candidates showed 100% coverage to ATXN3 mRNA, and RNA sequencing data revealed a significant downregulation of ATXN3 mRNA (Table S2). Some other genes showed partial coverage to ATXN3 mRNA, and minor alterations (<1.5-fold) were found in those genes by RNA sequencing.
MicroRNAs can act by binding along the entire mRNA, but studies have shown that the 3′ UTR of transcripts is the most frequently targeted site for microRNAs.42 Further analysis was performed on all three miATXN3 candidates using siSPOTR to predict the binding of their predicted seed sequences to the 3′ UTR of human target transcripts.43 The transcript probability of off-target score (tPOTS) was calculated by siSPOTR based on the number and type of seed matches (8-mer, 7-mer-M8, 7-mer-1A, and 6-mer) found in each transcript.43 The top 20 genes with the highest tPOTS were compared to the gene expression values obtained by RNA sequencing in human-derived frontal brain-like neurons treated with AAV5-miATXN3_7, AAV5-miATXN3_9, AAV5-miATXN3_11, or control (Tables S3–S5). We did not find a correlation between tPOTS and gene expression. In most cases, only minor changes in gene expression (<1.5-fold) were accompanied with the miATXN3-mediated reduction of ATXN3. In frontal brain-like neurons treated with AAV5-miATXN3_7, the expression of all 20 genes was less than 1.5-fold. For AAV5-miATXN3_9-treated cells, only two genes showed a higher alteration in expression. Prostaglandin-endoperoxide synthase 1 (PTGS1) was found downregulated (−3.4-fold change) and formin 1 (FMN1) was upregulated (2.1-fold change). In AAV5-miATXN3_11-treated cells, Tudor domain containing 6 (TDRD6) was found downregulated (−1.6-fold change) and lysyl oxidase (LOX) was upregulated (1.8-fold change).
The 3′ UTR off-target analysis was also performed on RNA derived from brain stem of SCA3 knockin mice. We selected mice injected with AAV5-miATXN3_9 in the cisterna magna, as we observed the strongest knockdown using this route of administration. Consistent with the human cells, no correlation was found between tPOTS values, and the expression of two genes was slightly over the 1.5-fold threshold (Table S6). Overall, we found no major alterations in gene expression after treatment with the three miATXN3 candidates, suggesting that the risks for off-target effects are limited.
Widespread AAV5-GFP Transduction in Minipig Brain after Intrathecal Administration
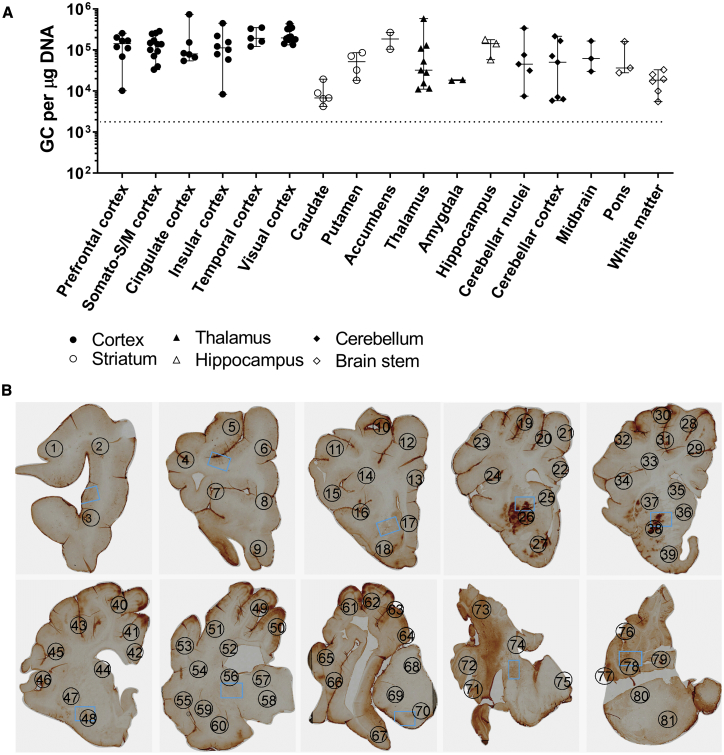
The translation of preclinical studies performed in rodent models to the clinic is a challenging process, particularly due to the relatively small brain and spinal cord sizes of rodents. Successful delivery of AAV to reach the target tissues in humans is likely dependent on brain size and structure. Therefore, we further investigated the delivery of AAV5 in minipig in order to better predict the CNS biodistribution of AAV5. The brain stem and cerebellum are the primary affected regions in SCA3 patients, and transduction of these brain regions is required for an ATXN3-based lowering gene therapy. Because cisterna magna injection in minipig has a slightly increased risk for spinal trauma, we investigated the biodistribution of AAV5 upon intrathecal injection in the lumbar region of the spinal cord. The biodistribution of AAV5 was determined by vector DNA genome copies and immunohistochemistry for the transgene (GFP). We found a relatively equal distribution of the vector DNA across the whole brain, including the cerebellum and brain stem (Figure 6A). The cerebellar cortex, DCN, and brain stem were transduced at therapeutically relevant doses of ∼105 gc/μg DNA. To visualize AAV5 distribution in the brain, GFP immunohistochemical analysis was performed, and the widespread GFP expression in the minipig brain confirmed an equal distribution of AAV5 (Figure 6B). Overall, the current data suggest that intrathecal administration of AAV5 could be sufficient for an AAV-based ATXN3-lowering gene therapy.
Figure 6.
AAV5 Biodistribution in Brain of Minipig upon Intrathecal Administration
(A) Vector copy distribution in brain of a 7-month-old Libechov minipig. The minipig was injected with 5 mL of AAV5-GFP (4 × 1013 gc/mL) into the lumbar region. The minipig was sacrificed 4 weeks post-injection and DNA was isolated from several punches of the brain. qRT-PCR was performed to determine the vector copy distribution. The genomic copies per μg of DNA were calculated for each brain region using a standard curve. Dotted lines represent lower limit of quantification. (B) Widespread GFP expression in minipig brain transduced with AAV5-GFP. Brains were fixed and serially cut in 5-μm sections. GFP staining was performed to visualize GFP expression throughout the brain. Different areas of the brain were numbered according to an atlas of the Libechov minipig brain (Cense): (1) prefrontal cortex (PFC) (lateral), (2) PFC (lateral), (3) PFC (ventral), (4) primary motor cortex (M1), (5) M1, (6) cingulate cortex, (7) PFC (lateral), (8) PFC (medial), (9) PFC (ventral), (10) motor cortex, (11) primary somatosensory cortex (S1) (dorsal), (12) frontal lobe white matter (WM), (13) cingulate cortex, (14) centrum semioval WM, (15) S1 (ventral), (16) insular cortex, (17) PFC (medial), (18) olfactory cortex, (19) M1/S1, (20) M1, (21) M1, (22) cingulate cortex, (23) S1 (dorsal), (24) insular cortex, (25) caudate (frontal), (26) putamen, (27) nucleus accumbens, (28) M1, (29) cingulate cortex, (30) occipital lobe (visual cortex), (31) occipital lobe (visual cortex), (32) S1 (ventral), (33) centrum semioval WM, (34) insular lobe, (35) caudate, (36) caudate, (37) putamen, (38) putamen, (39) nucleus accumbens, (40) occipital lobe (visual cortex), (41) cingulate cortex, (42) cingulate cortex, (43) S1 (ventral), (44) caudate, (45) insular cortex, (46) insular cortex, (47) putamen, (48) globus pallidus, (49) visual cortex, (50) cingulate cortex, (51) visual cortex, (52) centrum semiovale WM, (53) temporal cortex, (54) temporal lobe WM, (55) insular cortex, (56) caudate, (57) thalamus anterior, (58) thalamus ventral, (59) putamen, (60) globus pallidus, (61) visual cortex, (62) visual cortex, (63) visual cortex, (64) occipital lobe, (65) temporal cortex, (66) insular cortex, (67) hippocampus, (68) thalamus (medial), (69) thalamus (lateral), (70) thalamus (ventral), (71) temporal cortex, (72) occipital cortex (visual cortex), (73) cerebellum lobe, (74) cerebellum lobe, (75) reticular formation (pons), (76) cerebellum lobe, (77) cerebellum lobe, (78) cerebellar nuclei, (79) medulla oblongata, (80) cerebellar peduncle, (81) reticular formation (pons).
Discussion
In the current study we focused on the development of a microRNA-based gene therapy that could benefit the entire human SCA3 patient population. We aimed for a non-allele selective reduction of human ATXN3 mRNA, as this approach has been tested in several rodent preclinical models and showed no overt symptoms or toxicity.27,44 Furthermore, ataxin-3 knockout mice have no major abnormalities, suggesting that lowering of ataxin-3 in the context of SCA3 is safe and effective.44 An additional advantage of this approach is that the entire human patient population can be treated. Allele-specific approaches for downregulation of only the mutant ataxin-3 have also been tested. For example, allele-specific silencing was achieved using short hairpin RNAs (shRNAs) directed against a SNP unique to the mutant ataxin-3 transcript, which seems to be present in more than 70% of SCA3 patients. The SNP-specific shRNA was able to specifically silence mutant ataxin-3 and was found to be neuroprotective in SCA3 mouse and rat models.45. Single-stranded silencing RNAs (ssRNAs), ASOs, and peptide nucleic acids (PNAs) directly targeting the expanded CAG repeat also resulted in translational blockage of only the mutant ataxin-3.46, 47, 48, 49 The latter approach is very interesting because it may be used to treat multiple polyQ diseases but knockdown of several other endogenous CAG repeat-containing transcripts, such as huntingtin (HTT), TATA-binding protein (TBP), and other ATXNs, might be detrimental.
Several miATXN3s were designed in the natural human miR-451 scaffold to target different regions within exons of ATXN3 mRNA by using the knowledge and technologies developed for a Huntington’s disease RNAi gene therapy.29, 30, 31 A pre-screening of the miATXN3 candidates was performed on a Luc reporter construct, and miATXN3_7, miATXN3_9, and miATXN3_11 showed the highest silencing efficacy. All three candidates also successfully reduced the endogenous ATXN3 mRNA and protein levels in cells, and the strongest efficacy was consistently obtained with miATXN3_9.
For clinical development, it is important to establish the fidelity of cellular processing of the microRNA in the target human tissue and select the candidates with the least probability for off-target effects. For this reason, we analyzed the microRNA processing patterns in iPSC-neurons transduced with the three selected AAV5-miATXN3 candidates by small RNA sequencing. We found high expression of all three miATXN3s in transduced iPSC-neurons, but they were not overexpressed, even after transducing the cells with a very high concentration of AAV-miATXN3. Furthermore, no dysregulation in endogenously expressed microRNA molecules was observed, strongly limiting the possibility of off-target effects due to the saturation of the endogenous RNAi pathway. We detected a typical variability in the read length (19–30 nt) occurring when the miR-451 precursor is intracellularly processed. This observation is consistent with expected processing mechanism of miR-451, which escapes Dicer cleavage in the cytoplasm and is instead processed by Ago2. Ago2 cleavage generates a 30-nt guide strand that is further trimmed by PARN, leading to the observed variability in length. The processing and expression varied per miATXN3, with miATXN3_9 being most abundantly expressed. The higher expression could be a result of a more efficient processing because the thermodynamic stability at the loop of miATXN3_9 is lower, compared to miATXN3_7 and miATXN3_11. miATXN3_9 has an A-U nucleotide at the loop, and thus less energy may be needed to break this A-U bond by PARN. miATXN3_7 and miATXN3_11 both have a G-C nucleotide. The efficient processing and high expression of miATXN3_9 is likely to be responsible for its strong silencing efficacy. For miATXN3_9 the most abundant reads were, as predicted in silico, 22 nt long and showed the exact miATNX3 guide strand sequence. The second most abundant reads had either 1 or 2 extra nt at the 3′ end, and no passenger strands were detected, indicating that off-target effects due to a microRNA-like effect of the passenger strand can be excluded. The absence of a passenger strand associated with the miR-451 precursor also confirms our previous findings in the human neuronal cells and animal models that this scaffold does not produce a passenger strand.30,50
The success of an RNAi-based gene therapy for SCA3 is also dependent on the delivery method to reach the affected brain regions. AAV vectors are of particular interest due to their high safety profile, and different AAV serotypes have proven to be stable and safe, each with different tropism for a wide range of tissues.51, 52, 53, 54 AAVs can be delivered to the CNS either by systemic administration, direct intraparenchymal administration, or in the cerebrospinal fluid (CSF). Systemic AAV infusion has been used in preclinical models and in clinical studies and some serotypes were identified that can cross the blood-brain barrier.55,56 Based on our own experience, we anticipate that intravenous (i.v.) infusion of AAV would not be sufficient to achieve silencing of ATXN3 in the cerebellum and brain stem with the current technology. Direct injection of AAV in the parenchyma of brain is more invasive, but studies in rodents, minipig, and non-human primates consistently showed very strong transduction of the deeper brain structures and vector spread to the adjacent brain areas.29,57 The target regions in SCA3 patients are the cerebellum and brain stem and they are the primary affected areas in the early stages of the disease. Therefore, direct intraparenchymal administration would not result in spread of the vector to those areas and a CSF-mediated delivery would be necessary. In SCA3 mouse models, i.c.v. injection of ASOs resulted into sufficient reduction of the mutant ataxin-3 in the cerebellum and brain stem.58 MicroRNAs delivered by AAV directly in the cerebellum (DCN) also reduced mutant ataxin-3 in the cerebellum, while the effect was not evaluated in the brain stem.28,59 We investigated the transduction of cerebellum and brain stem of mice injected with AAV5-miATXN3 in the CSF via i.c.v. injection or cisterna magna, or directly to the cerebellum via DCN injection. Upon i.c.v. injection, the transduction of the cerebellum and brain stem of mice was low and not sufficient to expect a therapeutic benefit. In contrast, intracisterna magna administration resulted in transduction of the brain stem and cerebellum, with the highest transduction in the brain stem. DCN injection also transduced both brain regions, but transduction was the highest in the cerebellum. However, a single delivery to the cisterna magna resulted in the most optimal transduction of cerebellum and brain stem combined. Consistently, lowering of ATXN3 mRNA and the mutant ataxin-3 protein was observed in both cerebellum and brain stem of SCA3 knockin mice injected with AAV5-miATXN3_9 and AAV5-miATXN3_11 in the cisterna magna. The efficacy of both miATXN3 candidates was comparable, despite that AAV5-miATXN3_9 was the most effective candidate in our in vitro screening. A logical explanation is that while the miATXN3_11 target sequence had 100% homology with the human and mouse ATXN3 gene, miATXN3_9 had 1 nt mismatch with the mouse ATXN3. This means that the efficacy of miATXN3_9 seen could be underestimated in the mouse model used in this study, as mismatches interfere with the binding efficacy of a microRNA. Nevertheless, we showed that miATXN3_9 and miATXN3_11 are highly effective by mediating a strong knockdown of the mutant ataxin-3 protein in the cerebellum and brain stem after a single administration in the cisterna magna. Previous studies using compounds that use the RNAi or RNase H machineries showed that a reduction of about 37% up to 80% of the mutant ataxin-3 protein is sufficient to reverse molecular phenotypes associated with mutant ataxin-3 gain of toxicity in SCA3.27,28,45 We showed that we can reach 53.1% mutant ataxin-3 protein lowering in the cerebellum and 64% in the brain stem, which is within the desired therapeutic range. To further predict potential off-target genes, we examined the impact of AAV5-miATXN3 treatment on the overall gene expression profile. In silico analysis was performed using BLAST to search for off-target genes with partial guide-strand complementarity to human and mice microRNA, and the siSPOTR tool was used to predict potential off-target effects related to the miATXN3 seed sequence. The data obtained by BLAST and siSPOTR were compared to an unbiased RNA sequencing to look at differential changes in gene expression upon treatment. We selected an exclusion criterion of −1.5- to 1.5-fold change, based on observations from previous studies in large animals that most of the genes within this range do not show significant changes when these data are revalidated with other applications such as qPCR. Two genes (PTGS1 and FMN) were differentially expressed upon AAV5-miATXN3_9 treatment. PTGS1 encodes the enzyme cyclooxygenase (COX), which is involved in conversion of arachinodate to prostaglandin. PTGS1 is also downregulated by nonsteroidal anti-inflammatory drugs such as aspirin and ibuprofen, suggesting its downregulation is tolerable in humans.60 Interestingly, ibuprofen was found to have neuroprotective properties in SCA3 mouse model by increasing levels of neural progenitor proliferation and synaptic markers. Whether PTGS1 downregulation by ibuprofen may play a role is unclear and may be worth investigating. If this is the case, a combination therapy based on PTGS1 lowering and ATXN3 downregulation could add to therapeutic benefit. FMN belongs to the formin family of proteins and is involved in actin nucleation.61 Upregulation of FMN has not been linked to diseases. Similarly, two genes (TDRD6 and LOX1) were slightly modulated upon AAV5-miATXN3-11 treatment. TDRD6 encodes a protein that is specific to the male germline and essential for chromatoid body structure.62 TDRD6 is usually only expressed in mid prophase I spermatocytes. Thus, its downregulation in the brain is not expected to be relevant. LOX1 is a member of the lysyl oxidase family and has a role in crosslinking of collagens and elastin.63 Its role in disease is unclear, as it has been reported to enhance metastasis of certain cancers, but it was also shown to have tumor suppressor function.64, 65, 66, 67 Overall, dysregulation in one of these genes was not directly linked to any known diseases, increasing our confidence that AAV5-miATXN3-mediated silencing of ATXN3 mRNA may have limited risk for off-target effects. However, further investigation will be needed across multiple human cellular systems and larger animal models transduced with therapeutically relevant doses of AAV5-miATXN3 to better predict gene dysregulation and the effects after treatment with AAV5-miATXN3 in humans.
We demonstrated thus far that AAV5-miATXN3 can successfully lower the mutant ATXN3 mRNA and protein in iPSC-neurons and SCA3 knockin mice. One major challenge that remains is the translation of AAV5 delivery results acquired from the mouse model to humans. Multiple neuronal systems are affected in SCA3 patients, and degeneration of neurons is especially observed in the cerebellum, brain stem, basal ganglia, some cranial nerves, and the spinal cord.1,17,68 In the cerebellum, neurodegeneration is observed in dentate nucleus. The cerebellar cortex seems less affected, but loss of granule and Purkinje cells has been reported in the cerebellar vermis.1,69, 70, 71, 72 Main areas affected in the brain stem include the vestibular, pontine, and motor nuclei.1,70,73 Other regions such as the cerebral cortex, autonomic ganglia, striatum, substantia nigra, nerve motor nuclei, Clarke’s column nuclei, and the anterior horn of the spinal cord are also affected.1,45,71,74 We investigated AAV5 brain distribution upon intra-CSF delivery in the minipig with a brain and spinal cord size that closely resembles the situation in humans. Next to widespread vector distribution and GFP transgene expression, successful transduction of the structures mostly affected in SCA3 patients was observed. The current data increase of our confidence that mutant ataxin-3 protein can be modulated in these brain regions. Moving forward, further studies in large animal models are needed to determine the silencing efficacy and tolerability of AAV5-miATXN3.
In summary, we presented the development of a gene therapy for SCA3 patients based on non-allele-specific lowering of ATXN3 mRNA and protein by AAV5-miATXN3. We demonstrated the efficacy in cellular models and in a SCA3 knockin mouse model. In addition, we demonstrated that administration of AAV5 to the cisterna magna of mice can be successfully translated to intrathecal administration of a minipig to simultaneously transduce the cerebellum and brain stem, and potentially lower ATXN3 mRNA and mutant ataxin-3 protein in these brain regions. The current preclinical data support the feasibility for an AAV-based microRNA gene therapy, and further studies in animals with a larger brain would be required to investigate the translation of the current observations in the small mouse brain to a larger brain size closer to that of humans.
Materials and Methods
DNA Constructs
To generate the miATXN3 vectors, we searched for sequences on the ATXN3 gene that were mostly conserved between humans, non-human primates, and rodents. The sequences were incorporated into the cellular miR-451 scaffold of humans. 200-nt 5′ and 3′ flanking regions were included with EcoRV and BamHI restriction sites, and the mfold program (http://unafold.rna.albany.edu/?q=mfold) was used to determine whether the miATXN3 candidates are folded correctly into their secondary structures. The complete sequences were ordered from GeneArt gene synthesis (Invitrogen). These constructs were subsequently cloned into an expression vector containing the cytomegalovirus (CMV) immediate-early enhancer fused to chicken β-actin (CAG) promoter (Inovio, Plymouth Meeting, PA, USA) using the EcoRV and BamHI sites. For generation of the Luc reporter, the complete ATXN3 mRNA (NM_004993.5) sequence was synthesized at GeneArt gene synthesis and cloned in the 3′ UTR of the RL gene of the psiCHECK-2 vector (Promega, Madison, WI, USA). The FL gene was also expressed in this vector and served as internal control.
Culture and Transfections of HEK293T Cells
HEK293T cells were maintained in DMEM (Invitrogen) containing 10% fetal calf serum (Greiner, Kremsmünster, Austria), 100 U/mL penicillin, and 100 U/mL streptomycin (Thermo Fisher, Waltham, MA, USA) at 37°C and 5% CO2. With regard to transfections, for all assays, cells were seeded in 24-well plates at a density of 0.1 × 106 cells/well in DMEM. Transfections in HEK293T cells were performed 1 day after plating with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
Luciferase Assays
HEK293T cells were co-transfected with the miATXN3 expression constructs and the Luc reporter. The cells were lysed at 48 h post-transfection in 100 μL of 1× passive lysis buffer (Promega) by gentle rocking for 15 min at room temperature. The cell lysates were centrifuged for 5 min at 4,000 rpm to get rid of cell debris, and 10 μL of the supernatant was used to measure FL and RL activities with the Dual-Luc reporter assay system (Promega). Relative Luc activity was calculated as the ratio between RL and FL activities.
Culture of iPSC-Neurons
Frontal brain-like neurons were generated as described previously.35 Control iPSCs (ND42245) derived from fibroblasts were ordered from the Coriell Biorepository and were cultured on Matrigel (Corning)-coated six-well plates in mTeSR1 (STEMCELL). For embryoid body-based neural induction, iPSCs were seeded on AggreWell800 plates and cultured in STEMdiff neural induction medium (STEMCELL) for 5 days with daily medium changes. Embryoid bodies were harvested and plated on six-well plates coated with poly-d-lysine (Sigma-Aldrich) and laminin (Sigma-Aldrich) in STEMdiff neural induction medium for 7 days with daily medium changes. Rosettes were selected with rosette selection medium and plated on poly-d-lysine- and laminin-coated six-well plates in STEMdiff neural induction medium for 24 h. For differentiation into frontal brain-like neurons, STEMdiff neural induction medium was replaced for STEMdiff neuron differentiation medium (STEMCELL) and neuroprogenitor cells were differentiated for 5 days. The neuroprogenitor cells were then plated on poly-d-lysine- and laminin-coated plates in STEMdiff neuron maturation medium (STEMCELL) for 1 week. The mature frontal brain-like neurons were stored in liquid nitrogen in neuroprogenitor freezing medium (STEMCELL).
Western Blot
Western blot was performed to detect wild-type ataxin-3 protein expression in transfected HEK293T cells. In brief, cells were lysed using radioimmunoprecipitation assay (RIPA) lysis buffer solution, containing Tris-HCl (pH 8.0), NaCl, 1% IGEPAL CA-630, 0.5% deoxycholate (DOC), and cOmplete protease inhibitor and extracted by centrifugation. Equal amounts of tissue (30 μg) were loaded through a 10% SDS-PAGE gel. The gel was transferred to a Bio-Rad nitrocellulose membrane using a Bio-Rad turbo transfer system, running for 30 min at 90 V. Membranes were blocked for 1 h in 3% milk in TBS containing 0.1% Tween 20 (TBST) and incubated overnight with primary antibody mouse anti-ATXN3 (Abcam, ab61392, 1:1,000) and mouse anti-α-tubulin (Abcam, ab13533, 1:1,000) at 4°C. Secondary antibody incubation was with horseradish peroxidase (HRP)-conjugated rabbit anti-mouse (DAKO, 1:5,000) for 1 h at room temperature.
AAV5 Vector Production
AAV5 encoding miATXN3_7, miATXN3_9, and miATXN3_11 were produced by a baculovirus-based AAV production system as described previously.30 Briefly, the miATXN3 cassettes were obtained by digestion with restriction enzymes HindIII and PvuI and cloned in a uniQure transfer plasmid in order to generate an entry plasmid. The presence of the two inverted terminal repeats (ITRs) was confirmed by restriction digestion with SmaI. The ITR-CAG-miC cassettes were inserted in a recombinant baculovirus vector by homologous recombination in Spodoptera frugiperda (Sf) Sf9 cells, and clones were selected and screened by plague purification and PCR. The recombinant baculovirus containing the ITR-CAG-miC cassettes were further amplified until passage 6 in Sf+ cells and screened for the best production and stability by PCR and qRT-PCR. To generate AAV5, Sf+ cells were triple infected with three different recombinant baculoviruses expressing the ITR-CAG-miATXN3, the replicon enzyme, and the capsid protein. The cells were lysed 72 h after the triple infection, and the crude lysate was treated with 50 U/mL Benzonase (Merck, Darmstadt, Germany) for 1 h at 37°C. AAV5 was purified on an AVB Sepharose column (GE Healthcare, Little Chalfont, UK) using an AKTA purification system (GE Healthcare). The final titer concentration was determined by qRT-PCR with primers amplifying a 95-bp fragment from the CAG promoter region.
Transduction of iPSC-Neurons
Mature frontal brain-like neurons were plated in 500 μL of STEMdiff neuron maturation medium (STEMCELL) at 0.3 × 106 cells/well in 24-well plates. 1 week after plating, the cells were transduced with 100, 10, or 1 μL of 2.4 × 1013 gc/mL AAV5-miATXN3_7, AAV5-miATXN3_9, AAV5-miATXN3_11, or AAV5-GFP. The cells were incubated with AAV for 2 weeks and the media were changed twice a week.
Intrathecal Delivery of AAV5 and Tissue Collection in Minipig
All experiments were carried out according to the guidelines for the care and use of experimental animals and approved by the State Veterinary. A male 7-month-old Libechov minipig was selected at the Institute of Animal Physiology and Genetics in Libechov (Czech Republic). General anesthesia was induced by intramuscular application of tiletamine (4 mg/kg), zolazepam (4 mg/kg Zoletil 100; Virbac), ketamine (5 mg/kg Narketan 10; Chassot), and xylazine (1 mg/kg Rometar 2%; Spofa) mixture (TKX mixture) followed by intravenous ear cannulation and intubation. Artificial ventilation and isoflurane anesthesia were used during the rest of the procedure. For intrathecal administration, polyethylene tubing (PE10, 427401, Intramedic Clay Adams) attached to a 5-mL syringe passed through a spinal needle (Spinocan; 4501195-13; 1.1 × 88 mm; B. Braun Melsungen) was used. After tubing insertion into the intrathecal space, we collected approximately 1–2 mL of CSF to ensure precise localization of the tubing, and 5 mL of AAV5-GFP (4 × 1013 gc/mL) was then delivered slowly (over approximately 5 min) by hand injection into the lumbar region of the minipig.
Four weeks after treatment, the minipig was sacrificed under deep anesthesia using TKX mixture combined with propofol 1% (Fresenius) intravenous injection (10 mg/kg) followed by whole-body perfusion. The animal was heparinized and then transcardially perfused with 20 L of ice-cold PBS. Once removed from the skull, the brain was coronally sliced into 3- to 4-mm blocks from frontal to occipital. The left hemisphere was used for biomolecular analyses, where 102 brain punches were taken and stored at −80°C. The right hemisphere was fixed by 4% paraformaldehyde (PFA) overnight and then transferred to 30% sucrose with sodium azide for cryosectioning.
DNA Isolation from Minipig Brain Tissue
DNA isolation from brain tissue punches was performed using the DNeasy blood and tissue kit (QIAGEN, Germany). Primers specific for the CAG promoter sequence were used to measure the vector genome copies (gcs) by SYBR Green Fast qPCR (Thermo Fisher Scientific). The amount of vector DNA was calculated based on a plasmid standard curve. Results were reported as gc/mg genomic DNA.
Immunohistochemistry on Minipig Brain Slices
PFA-fixed brain slices were cut in 20-μm sections (10–15 sections/slice), which were spread on a microscopic slide and air dried. For immunostaining analysis, the sections on glass were antigen retrieved by treating in citrate buffer (pH 6) at 90°C for 30 min. Subsequently, endogenous peroxidase activity was blocked with a solution of 0.3% hydrogen peroxide in methanol for 20 min, and the brain sections were immunostained using the rabbit primary antibody anti-GFP (1:1,000, ab6556; Abcam). Sections were then treated with a biotinylated donkey anti-rabbit secondary antibody (1:400, RPN 1004V; GE Healthcare) followed by an avidin-peroxidase complex (1:400, A3151; Sigma-Aldrich). The avidin-peroxidase complex was visualized by incubation with solution containing a dissolved 3,3′-diaminobenzidine tablet (4170; Kementec Diagnostics). The sections were dehydrated and mounted with DePeX (Sigma). Images were acquired using a histological scanner (VS120-5 Virtual Slide Microscope fluorescence; Olympus).
SCA3 Mice and Treatment Groups
All procedures were approved by the local ethics committee at the Regierungspraesidium Tuebingen and performed in accordance with the German Animal Welfare Act and the guidelines of the Federation of European Laboratory Animal Science Associations based on the European Union legislation (directive 2010/63/EU). All efforts were made to minimize the suffering and the number of animals used.
Heterozygous SCA3 knockin animals expressing 304 glutamines within the murine ataxin-3 polyQ locus were generated (E.H. et al., unpublished data). No SCA3 phenotype was observed at the time of sacrifice (4.5 months). Increasing amounts of ataxin-3 and ubiquitin-positive aggregate formation were detectable in whole-brain homogenates and in hippocampal regions of older (18 months of age) mice. Consistently, significant reduction in body weight and mild motor deficits were observed (E.H. et al., unpublished data). All mice were maintained in a controlled facility in a 12-h light/12-h dark cycle with free access to food and water. Three animals (two males and one female) were recruited to each treatment group (Table S1).
Viral Delivery by Stereotaxic Injections and Tissue Collection in Mice
Animals were injected at 3 months of age. For each injection, the animal was initially anesthetized with 5% isoflurane in an oxygen-air mixture in the induction box. After induction, the animal was injected intraperitoneally with a combination of fentanyl (0.05 mg/kg), midazolam (5 mg/kg), and medetomidine (0.5 mg/kg) and transferred to the stereotaxic frame. Isoflurane was then delivered via the facemask and reduced to 1.5%–2.0% depending on the weight and responses of the animal. Eye ointment was given to the animal to prevent the eyes from drying during surgery. The body temperature of the animal was monitored and maintained at 37 ± 0.5°C through a rectal probe and a negative feedback-controlled heat pad (Harvard Apparatus, USA). The depth of anesthesia was checked by the lack of pedal withdrawal to a firmly pinched hind toe or foot. The head was shaved and disinfected, and local anesthetic agent (2% lidocaine, 5 mg/kg) was injected subcutaneously. For i.c.v. and DCN injections, bregma and lambda were used for alignments and locating the injection sites after a midline skin incision. A small skull hole was drilled to expose the dura over the injection site. Craniotomy (0.5–0.8 mm in diameter) was performed using a microscope or magnifying glass and a pneumatic drill. In the case of cisterna magna injections, skin incision was performed at the neck region. Dura mater was exposed by blunt dissection of the subcutaneous neck muscles. The animal received bilateral DCN (anteroposterior [AP], −6.5 mm; mediolateral [ML], ±1 mm; dorsoventral [DV], −2.8 mm [relative to bregma]), i.c.v. (AP, +0.3 mm; ML, +1 mm; DV, −3 mm [relative to bregma]), or cisterna magna injection according to the assigned treatment group. 2.0 μL per hemisphere (bilateral DCN) or 10 μL (i.c.v. and cisterna magna) of AAV5-miATXN3_7, AAV5-miATXN3_9, AAV5-miATXN3_11, or AAV5-GFP was delivered at a rate of 0.25 μL/min using a microinjection pump (UMP3-1, WPI) with a 10-μL Hamilton syringe and a 32-gauge needle. The AAVs were titer matched at 2.4 × 1013 gc/mL. After the injection, the needle remained at the injection site for 5 min before withdrawal. The animal was then sutured and given carprofen (5 mg/kg) subcutaneously as an analgesic after surgery. For recovery from anesthesia, the animal was given a mixture of naloxone (1.2 mg/kg), flumazenil (0.5 mg/kg), and atipamezole (2.5 mg/kg) subcutaneously and transferred to a warm (37°C) environment with access to food and water. The operated animal was returned to its home cage after recovery from anesthesia. All animals were given carprofen (5 mg/kg) subcutaneously every 24 h and provided with antibiotic (enrofloxacin, 10 mg/kg)-containing water until the third and seventh day post-surgery, respectively. Health and other conditions of all animals were closely monitored throughout the project duration using score sheets.
All treated animals were euthanized 6 weeks after viral delivery. For the untreated group, animals were sacrificed at 4.5 months of age. The animals were given saturated carbon dioxide in the home cage until death. Tissues were dissected and snap frozen in liquid nitrogen and stored at −80°C for subsequent analyses.
RNA Isolation, qRT-PCR, and MicroRNA TaqMan Assay
For all RNA isolation, cells and tissues were lysed in 300 μL of TRIzol, and RNA isolation was performed using the Direct-zol kit (R2061, Zymo Research) according to the manufacturer’s protocol. Genomic DNA contamination was removed by DNase treatment using recombinant shrimp dsDNase (Themo Fisher Scientific). First-strand cDNA was reverse transcribed using random hexamer primers with the Dynamo kit (Finnzymes, Espoo, Finland). Real-time PCR amplification was performed with a customized TaqMan for human ATXN3 (assay ID Hs01026440_g1) or mouse ATXN3 (assay ID Mm00804702_m1) on an ABI 7000 or ABI 7500 (Applied Biosystems, Foster City, CA, USA). The mRNA expression levels were normalized to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; assay ID Hs02758991_g1) or mouse GAPDH (assay ID Mm99999915_g10) as an internal control. A custom TaqMan small RNA assay design tool (Thermo Fisher Scientific) was used to design microRNA TaqMan for the mature miATXN3_7 (assay ID CTCE3VJ), miATXN3_9 (assay ID CTEPRZE), and miATXN3_11 (assay ID CTFVKKC). The microRNA expression levels were normalized to U6 snRNA (assay ID 001973) as an internal control. All reverse transcription reactions and TaqMan for small RNAs were performed according to the manufacturers’ protocols.
Next-Generation Sequencing and Data Analysis of Small RNAs
Small RNA sequencing libraries for the Illumina sequencing platform were generated using high-quality total RNA as input and the NEXTflex small RNA sequencing kit (Bioo Scientific, Austin, TX, USA). Briefly, the small RNA species were subjected to ligation with 3′ and 5′ RNA adapters, first-strand reverse transcription, and PCR amplification. Sample-specific barcodes were introduced in the PCR step. The PCR products were separated on Tris-borate-EDTA-PAGE electrophoresis, and the expected band around 30 bp was recovered for each sample. The resulting sequencing libraries were quantified on a BioAnalyzer (Agilent, Santa Clara, CA, USA). The libraries were multiplexed, clustered, and sequenced on an Illumina HiSeq 2000 (TruSeq v3 chemistry) with a single-read 36-cycle sequencing protocol and indexing. The sequencing run was analyzed with the Illumina CASAVA pipeline (v1.8.2), with demultiplexing based on sample-specific barcodes. The raw sequencing data produced were processed by removing the sequence reads that were of too low quality (only “passing filter” reads were selected). In total, between 15 and 35 million reads per sample were generated. Next-generation sequencing (NGS) small RNA raw datasets were analyzed using the CLC Genomics Workbench 8 (QIAGEN). The reads were adaptor trimmed and aligned against the corresponding reference sequence. Reads that were shorter than 10 nt, longer than 45 nt, or represented less than 10 times were excluded from the analysis. Illumina TruSeq small RNA adaptor (5′ -TGGAATTCTCGGGTGCCAAGG-3′) was removed and 3′ trimmed, allowing for both internal and end matches. Extracted reads were then annotated using miRbase 22.1 (http://www.mirbase.org/), including reference sequences of the pre-miATXN3 constructs with a maximum of 3 nt mismatches allowed. The percentages of reads based on the total number of reads matching the reference sequence were calculated. Differential miRNA expression analysis for the three treatment groups was performed using the “Empirical Analysis of differential gene expression” function in CLC Genomics. Significance was evaluated relative to the data dispersion with total count filter cutoff of 5.0. Expression changes were considered significant when the false discovery rate (FDR) < 0.05. miATXN3_7-treated cells were used as the reference condition given the relatively low expression of this construct.
RNA Sequencing and Transcriptome Analysis
For human transcriptome-related off-target analysis, total RNA from healthy human-derived frontal brain-like neurons was treated with the AAV5-miATXN3 candidates or formulation buffer as control. For analysis in mice, total RNA was isolated from brain stem of SCA3 knockin mice transduced with miATXN3_9 or the formulation buffer. Total RNA was reverse transcribed and single-end sequence reads were generated using the Illumina HiSeq 2500 system (BaseClear, Leiden, the Netherlands). FASTQ sequence files were generated using the Illumina Casava pipeline version 1.8.3. Initial quality assessment was based on data passing the Illumina chastity filtering. Subsequently, reads containing PhiX control signal were removed using an in-house filtering protocol. In addition, reads containing (partial) adapters were clipped (up to a minimum read length of 50 bp). The second quality assessment was based on the remaining reads using the FASTQC quality control tool version 0.10.0. The analysis of the weighted proportions fold change and p value scores between the conditions were calculated using Baggerley’s beta-binomial test. The fold change in gene expression was calculated for each comparison (AAV5-miATXN3_7 versus control, AAV5-miATXN3_9 versus control, and AAV5-miATXN3_11 versus control).
TR-FRET Assay
The TR-FRET assay was performed as described before.41 Brain tissues were lysed by sonication in 10× v/w of ice-cold lysis buffer (PBS + 1% Triton X-100 + 1× protease inhibitor cocktail [Roche]). Anti-ataxin-3 clone 1H9 (MAB5360, Millipore) was labeled with donor Lumi4-Tb-fluorophore (Cisbio). Anti-ployglutamine expansion marker clone 5TF1-1C2 (MAB1574, Millipore) was labeled with D2 acceptor fluorophore (Cisbio). The combination of 1H9 and 5TF1-1C allow detection of more than 37 glutamine repeats. After optimization of antibody titers and incubation conditions, quantification of mutant ataxin-3 levels was performed in low-volume polystyrene 384 microtiter plates (Greiner Bio-One) with 5 μL of sample volume and addition of 1 μL of antibody solution (50 mM NaHPO4 + 400 mM NaF + 0.1% BSA + 0.05% Tween 20 + 1 ng/mL 1H9-Tb + 10 ng/mL 1C2-D2). Plates were then incubated at 4°C for 20 h and analyzed by time-resolved fluorescence at 620 nm and 665 nm on an EnVision multilabel reader (PerkinElmer).
Statistical Analysis
Data were analyzed using the one-way ANOVA or Student’s t test to determine statistically significances between control and treated cells. A two-way ANOVA was used to determine statistically significances between multiple treated groups. The p values were either listed or represented as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Author Contributions
Conceptualization, M.M.E., R.M., P.K., S.v.D.; Methodology, R.M., J.H.-S., H.P.N., J.S.-S., J.K., J.J., S.J., Z.E., J.M.; Investigation, R.M., M.S.G., J.S.-S., S.K., J.H.-S., E.H.; Writing – Original Draft, R.M., M.M.E.; Writing – Review & Editing, R.M., M.S.G., J.S.-S., J.H.-S., L.J.T., E.H., P.K., H.P.N., S.v.D., M.M.E.; Funding Acquisition, M.M.E., H.P.N., P.K.
Acknowledgments
The authors would like to thank Jorien Witjas, Mark van Veen, Maroeska Oudshoorn-Dickmann, Tamar Grevelink, and Erich Ehlert for technical assistance and Ellen Broug and Eileen Sawyer for reviewing this manuscript. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 2012-305121 “Integrated European -omics research project for diagnosis and therapy in rare neuromuscular and neurodegenerative diseases (NEUROMICS).” This work was also supported by the National Sustainability Program I, project number LO1609 (Czech Ministry of Education, Youth and Sports), and RVO: 67985904.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.10.008.
Supplemental Information
References
- 1.Matos C., de Almeida L.P., Nóbrega C. Machado-Joseph disease/spinocerebellar ataxia type 3: lessons from disease pathogenesis and clues into therapy. J. Neurochem. 2019;148:8–28. doi: 10.1111/jnc.14541. [DOI] [PubMed] [Google Scholar]
- 2.Coutinho P., Andrade C. Autosomal dominant system degeneration in Portuguese families of the Azores Islands. A new genetic disorder involving cerebellar, pyramidal, extrapyramidal and spinal cord motor functions. Neurology. 1978;28:703–709. doi: 10.1212/wnl.28.7.703. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg R.N. Machado-Joseph disease: an autosomal dominant motor system degeneration. Mov. Disord. 1992;7:193–203. doi: 10.1002/mds.870070302. [DOI] [PubMed] [Google Scholar]
- 4.Ranum L.P., Lundgren J.K., Schut L.J., Ahrens M.J., Perlman S., Aita J., Bird T.D., Gomez C., Orr H.T. Spinocerebellar ataxia type 1 and Machado-Joseph disease: incidence of CAG expansions among adult-onset ataxia patients from 311 families with dominant, recessive, or sporadic ataxia. Am. J. Hum. Genet. 1995;57:603–608. [PMC free article] [PubMed] [Google Scholar]
- 5.Schöls L., Bauer P., Schmidt T., Schulte T., Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- 6.Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 7.Maciel P., Costa M.C., Ferro A., Rousseau M., Santos C.S., Gaspar C., Barros J., Rouleau G.A., Coutinho P., Sequeiros J. Improvement in the molecular diagnosis of Machado-Joseph disease. Arch. Neurol. 2001;58:1821–1827. doi: 10.1001/archneur.58.11.1821. [DOI] [PubMed] [Google Scholar]
- 8.Cummings C.J., Zoghbi H.Y. Trinucleotide repeats: mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- 9.Maciel P., Gaspar C., DeStefano A.L., Silveira I., Coutinho P., Radvany J., Dawson D.M., Sudarsky L., Guimarães J., Loureiro J.E. Correlation between CAG repeat length and clinical features in Machado-Joseph disease. Am. J. Hum. Genet. 1995;57:54–61. [PMC free article] [PubMed] [Google Scholar]
- 10.Bettencourt C., Lima M. Machado-Joseph disease: from first descriptions to new perspectives. Orphanet J. Rare Dis. 2011;6:35. doi: 10.1186/1750-1172-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goto J., Watanabe M., Ichikawa Y., Yee S.B., Ihara N., Endo K., Igarashi S., Takiyama Y., Gaspar C., Maciel P. Machado-Joseph disease gene products carrying different carboxyl termini. Neurosci. Res. 1997;28:373–377. doi: 10.1016/s0168-0102(97)00056-4. [DOI] [PubMed] [Google Scholar]
- 12.Ichikawa Y., Goto J., Hattori M., Toyoda A., Ishii K., Jeong S.Y., Hashida H., Masuda N., Ogata K., Kasai F. The genomic structure and expression of MJD, the Machado-Joseph disease gene. J. Hum. Genet. 2001;46:413–422. doi: 10.1007/s100380170060. [DOI] [PubMed] [Google Scholar]
- 13.Paulson H.L., Perez M.K., Trottier Y., Trojanowski J.Q., Subramony S.H., Das S.S., Vig P., Mandel J.L., Fischbeck K.H., Pittman R.N. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 14.Macedo-Ribeiro S., Cortes L., Maciel P., Carvalho A.L. Nucleocytoplasmic shuttling activity of ataxin-3. PLoS ONE. 2009;4:e5834. doi: 10.1371/journal.pone.0005834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt T., Landwehrmeyer G.B., Schmitt I., Trottier Y., Auburger G., Laccone F., Klockgether T., Völpel M., Epplen J.T., Schöls L., Riess O. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol. 1998;8:669–679. doi: 10.1111/j.1750-3639.1998.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trottier Y., Cancel G., An-Gourfinkel I., Lutz Y., Weber C., Brice A., Hirsch E., Mandel J.L. Heterogeneous intracellular localization and expression of ataxin-3. Neurobiol. Dis. 1998;5:335–347. doi: 10.1006/nbdi.1998.0208. [DOI] [PubMed] [Google Scholar]
- 17.Riess O., Rüb U., Pastore A., Bauer P., Schöls L. SCA3: neurological features, pathogenesis and animal models. Cerebellum. 2008;7:125–137. doi: 10.1007/s12311-008-0013-4. [DOI] [PubMed] [Google Scholar]
- 18.Evers M.M., Toonen L.J.A., van Roon-Mom W.M.C. Ataxin-3 protein and RNA toxicity in spinocerebellar ataxia type 3: current insights and emerging therapeutic strategies. Mol. Neurobiol. 2014;49:1513–1531. doi: 10.1007/s12035-013-8596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L.C., Chen K.Y., Pan H., Wu C.C., Chen P.H., Liao Y.T., Li C., Huang M.L., Hsiao K.M. Muscleblind participates in RNA toxicity of expanded CAG and CUG repeats in Caenorhabditis elegans. Cell. Mol. Life Sci. 2011;68:1255–1267. doi: 10.1007/s00018-010-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fardaei M., Larkin K., Brook J.D., Hamshere M.G. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang H., Mankodi A., Swanson M.S., Moxley R.T., Thornton C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 23.Eichler L., Bellenberg B., Hahn H.K., Köster O., Schöls L., Lukas C. Quantitative assessment of brain stem and cerebellar atrophy in spinocerebellar ataxia types 3 and 6: impact on clinical status. AJNR Am. J. Neuroradiol. 2011;32:890–897. doi: 10.3174/ajnr.A2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudarsky L., Corwin L., Dawson D.M. Machado-Joseph disease in New England: clinical description and distinction from the olivopontocerebellar atrophies. Mov. Disord. 1992;7:204–208. doi: 10.1002/mds.870070303. [DOI] [PubMed] [Google Scholar]
- 25.Sequeiros J., Coutinho P. Epidemiology and clinical aspects of Machado-Joseph disease. Adv. Neurol. 1993;61:139–153. [PubMed] [Google Scholar]
- 26.Nóbrega C., Nascimento-Ferreira I., Onofre I., Albuquerque D., Hirai H., Déglon N., de Almeida L.P. Silencing mutant ataxin-3 rescues motor deficits and neuropathology in Machado-Joseph disease transgenic mice. PLoS ONE. 2013;8:e52396. doi: 10.1371/journal.pone.0052396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alves S., Nascimento-Ferreira I., Dufour N., Hassig R., Auregan G., Nóbrega C., Brouillet E., Hantraye P., Pedroso de Lima M.C., Déglon N., de Almeida L.P. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum. Mol. Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez-Lebrón E., Costa Mdo.C., Luna-Cancalon K., Peron T.M., Fischer S., Boudreau R.L., Davidson B.L., Paulson H.L. Silencing mutant ATXN3 expression resolves molecular phenotypes in SCA3 transgenic mice. Mol. Ther. 2013;21:1909–1918. doi: 10.1038/mt.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evers M.M., Miniarikova J., Juhas S., Vallès A., Bohuslavova B., Juhasova J., Skalnikova H.K., Vodicka P., Valekova I., Brouwers C. AAV5-miHTT gene therapy demonstrates broad distribution and strong human mutant huntingtin lowering in a Huntington’s Disease minipig model. Mol. Ther. 2018;26:2163–2177. doi: 10.1016/j.ymthe.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, characterization, and lead selection of therapeutic miRNAs targeting huntingtin for development of gene therapy for Huntington’s disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miniarikova J., Evers M.M., Konstantinova P. Translation of microRNA-based huntingtin-lowering therapies from preclinical studies to the clinic. Mol. Ther. 2018;26:947–962. doi: 10.1016/j.ymthe.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera-Carrillo E., Berkhout B. Dicer-independent processing of small RNA duplexes: mechanistic insights and applications. Nucleic Acids Res. 2017;45:10369–10379. doi: 10.1093/nar/gkx779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keskin S., Brouwers C.C., Sogorb-Gonzalez M., Martier R., Depla J.A., Vallès A., van Deventer S.J., Konstantinova P., Evers M.M. AAV5-miHTT lowers huntingtin mRNA and protein without off-target effects in patient-derived neuronal cultures and astrocytes. Mol. Ther. Methods Clin. Dev. 2019;15:275–284. doi: 10.1016/j.omtm.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein R.L., Hamby M.E., Gong Y., Hirko A.C., Wang S., Hughes J.A., King M.A., Meyer E.M. Dose and promoter effects of adeno-associated viral vector for green fluorescent protein expression in the rat brain. Exp. Neurol. 2002;176:66–74. doi: 10.1006/exnr.2002.7942. [DOI] [PubMed] [Google Scholar]
- 35.Martier R., Liefhebber J.M., García-Osta A., Miniarikova J., Cuadrado-Tejedor M., Espelosin M., Ursua S., Petry H., van Deventer S.J., Evers M.M., Konstantinova P. Targeting RNA-mediated toxicity in C9orf72 ALS and/or FTD by RNAi-based gene therapy. Mol. Ther. Nucleic Acids. 2019;16:26–37. doi: 10.1016/j.omtn.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J.-S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O’Carroll D., Lai E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cifuentes D., Xue H., Taylor D.W., Patnode. H., Mishima. Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheloufi S., Dos Santos C.O., Chong M.M.W., Hannon G.J. A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bezprozvanny I., Klockgether T. Therapeutic prospects for spinocerebellar ataxia type 2 and 3. Drugs Future. 2009;34:991–999. doi: 10.1358/dof.2009.034.12.1443434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen H.P., Hübener J., Weber J.J., Grueninger S., Riess O., Weiss A. Cerebellar soluble mutant ataxin-3 level decreases during disease progression in spinocerebellar ataxia type 3 mice. PLoS ONE. 2013;8:e62043. doi: 10.1371/journal.pone.0062043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plass M., Rasmussen S.H., Krogh A. Highly accessible AU-rich regions in 3′ untranslated regions are hotspots for binding of regulatory factors. PLoS Comput. Biol. 2017;13:e1005460. doi: 10.1371/journal.pcbi.1005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boudreau R.L., Spengler R.M., Hylock R.H., Kusenda B.J., Davis H.A., Eichmann D.A., Davidson B.L. siSPOTR: a tool for designing highly specific and potent siRNAs for human and mouse. Nucleic Acids Res. 2013;41:e9. doi: 10.1093/nar/gks797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt I., Linden M., Khazneh H., Evert B.O., Breuer P., Klockgether T., Wuellner U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem. Biophys. Res. Commun. 2007;362:734–739. doi: 10.1016/j.bbrc.2007.08.062. [DOI] [PubMed] [Google Scholar]
- 45.Alves S., Nascimento-Ferreira I., Auregan G., Hassig R., Dufour N., Brouillet E., Pedroso de Lima M.C., Hantraye P., Pereira de Almeida L., Déglon N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS ONE. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evers M.M., Pepers B.A., van Deutekom J.C.T., Mulders S.A.M., den Dunnen J.T., Aartsma-Rus A., van Ommen G.J., van Roon-Mom W.M. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS ONE. 2011;6:e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I., Corey D.R. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J., Yu D., Aiba Y., Pendergraff H., Swayze E.E., Lima W.F., Hu J., Prakash T.P., Corey D.R. ss-siRNAs allele selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucleic Acids Res. 2013;41:9570–9583. doi: 10.1093/nar/gkt693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datson N.A., González-Barriga A., Kourkouta E., Weij R., van de Giessen J., Mulders S., Kontkanen O., Heikkinen T., Lehtimäki K., van Deutekom J.C. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS ONE. 2017;12:e0171127. doi: 10.1371/journal.pone.0171127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martier R., Liefhebber J.M., Miniarikova J., van der Zon T., Snapper J., Kolder I., Petry H., van Deventer S.J., Evers M.M., Konstantinova P. Artificial microRNAs targeting C9orf72 can reduce accumulation of intra-nuclear transcripts in ALS and FTD patients. Mol. Ther. Nucleic Acids. 2019;14:593–608. doi: 10.1016/j.omtn.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colella P., Ronzitti G., Mingozzi F. Emerging Issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 2017;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saraiva J., Nobre R.J., Pereira de Almeida L. Gene therapy for the CNS using AAVs: the impact of systemic delivery by AAV9. J. Control. Release. 2016;241:94–109. doi: 10.1016/j.jconrel.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Hocquemiller M., Giersch L., Audrain M., Parker S., Cartier N. Adeno-associated virus-based gene therapy for CNS diseases. Hum. Gene Ther. 2016;27:478–496. doi: 10.1089/hum.2016.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 55.Foust K.D., Nurre E., Montgomery C.L., Hernandez A., Chan C.M., Kaspar B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 57.Samaranch L., Blits B., San Sebastian W., Hadaczek P., Bringas J., Sudhakar V., Macayan M., Pivirotto P.J., Petry H., Bankiewicz K.S. MR-guided parenchymal delivery of adeno-associated viral vector serotype 5 in non-human primate brain. Gene Ther. 2017;24:253–261. doi: 10.1038/gt.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toonen L.J.A., Rigo F., van Attikum H., van Roon-Mom W.M.C. Antisense oligonucleotide-mediated removal of the polyglutamine repeat in spinocerebellar ataxia type 3 mice. Mol. Ther. Nucleic Acids. 2017;8:232–242. doi: 10.1016/j.omtn.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa Mdo.C., Luna-Cancalon K., Fischer S., Ashraf N.S., Ouyang M., Dharia R.M., Martin-Fishman L., Yang Y., Shakkottai V.G., Davidson B.L. Toward RNAi therapy for the polyglutamine disease Machado-Joseph disease. Mol. Ther. 2013;21:1898–1908. doi: 10.1038/mt.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dean B., Gibbons A., Gogos A., Udawela M., Thomas E., Scarr E. Studies on prostaglandin-endoperoxide synthase 1: lower levels in schizophrenia and after treatment with antipsychotic drugs in conjunction with aspirin. Int. J. Neuropsychopharmacol. 2018;21:216–225. doi: 10.1093/ijnp/pyx092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou F., Leder P., Martin S.S. Formin-1 protein associates with microtubules through a peptide domain encoded by exon-2. Exp. Cell Res. 2006;312:1119–1126. doi: 10.1016/j.yexcr.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 62.Akpınar M., Lesche M., Fanourgakis G., Fu J., Anastassiadis K., Dahl A., Jessberger R. TDRD6 mediates early steps of spliceosome maturation in primary spermatocytes. PLoS Genet. 2017;13:e1006660. doi: 10.1371/journal.pgen.1006660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T.H., Hsia S.M., Shieh T.M. Lysyl oxidase and the tumor microenvironment. Int. J. Mol. Sci. 2016;18:E62. doi: 10.3390/ijms18010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Erler J.T., Bennewith K.L., Nicolau M., Dornhöfer N., Kong C., Le Q.T., Chi J.T., Jeffrey S.S., Giaccia A.J. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 65.Cox T.R., Gartland A., Erler J.T. Lysyl oxidase, a targetable secreted molecule involved in cancer metastasis. Cancer Res. 2016;76:188–192. doi: 10.1158/0008-5472.CAN-15-2306. [DOI] [PubMed] [Google Scholar]
- 66.Kaneda A., Wakazono K., Tsukamoto T., Watanabe N., Yagi Y., Tatematsu M., Kaminishi M., Sugimura T., Ushijima T. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 2004;64:6410–6415. doi: 10.1158/0008-5472.CAN-04-1543. [DOI] [PubMed] [Google Scholar]
- 67.Ozdener G.B., Bais M.V., Trackman P.C. Determination of cell uptake pathways for tumor inhibitor lysyl oxidase propeptide. Mol. Oncol. 2016;10:1–23. doi: 10.1016/j.molonc.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scherzed W., Brunt E.R., Heinsen H., de Vos R.A., Seidel K., Bürk K., Schöls L., Auburger G., Del Turco D., Deller T. Pathoanatomy of cerebellar degeneration in spinocerebellar ataxia type 2 (SCA2) and type 3 (SCA3) Cerebellum. 2012;11:749–760. doi: 10.1007/s12311-011-0340-8. [DOI] [PubMed] [Google Scholar]
- 69.Sudarsky L., Coutinho P. Machado-Joseph disease. Clin. Neurosci. 1995;3:17–22. [PubMed] [Google Scholar]
- 70.Dürr A., Stevanin G., Cancel G., Duyckaerts C., Abbas N., Didierjean O., Chneiweiss H., Benomar A., Lyon-Caen O., Julien J. Spinocerebellar ataxia 3 and Machado-Joseph disease: clinical, molecular, and neuropathological features. Ann. Neurol. 1996;39:490–499. doi: 10.1002/ana.410390411. [DOI] [PubMed] [Google Scholar]
- 71.Yamada M., Sato T., Tsuji S., Takahashi H. CAG repeat disorder models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:71–86. doi: 10.1007/s00401-007-0287-5. [DOI] [PubMed] [Google Scholar]
- 72.Muñoz E., Rey M.J., Milà M., Cardozo A., Ribalta T., Tolosa E., Ferrer I. Intranuclear inclusions, neuronal loss and CAG mosaicism in two patients with Machado-Joseph disease. J. Neurol. Sci. 2002;200:19–25. doi: 10.1016/s0022-510x(02)00110-7. [DOI] [PubMed] [Google Scholar]
- 73.Rüb U., Brunt E.R., Deller T. New insights into the pathoanatomy of spinocerebellar ataxia type 3 (Machado-Joseph disease) Curr. Opin. Neurol. 2008;21:111–116. doi: 10.1097/WCO.0b013e3282f7673d. [DOI] [PubMed] [Google Scholar]
- 74.Paulson H.L., Das S.S., Crino P.B., Perez M.K., Patel S.C., Gotsdiner D., Fischbeck K.H., Pittman R.N. Machado-Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann. Neurol. 1997;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.