Abstract
Oxidation of methionine residues to methionine sulfoxide scavenges reactive species, thus protecting against oxidative stress. Reduction of the sulfoxide back to methionine by methionine sulfoxide reductases creates a cycle with catalytic efficiency. Protection by the methionine sulfoxide reductases is well documented in cultured cells, from microorganisms to mammals. However, knocking out one or two of the 4 mammalian reductases had little effect in mice that were not stressed. We hypothesized that the minimal effect is due to redundancy provided by the 4 reductases. We tested the hypothesis by creating a transgenic mouse lacking all 4 reductases and predicted that this mouse would be exceptionally sensitive to oxidative stress. The mutant mice were phenotypically normal at birth, exhibited normal post-natal growth, and were fertile. Surprisingly, rather than being more sensitive to oxidative stress, they were more resistant to both cardiac ischemia-reperfusion injury and to parenteral paraquat, a redox-cycling agent. Resistance was not a result of hormetic induction of the antioxidant transcription factor Nrf2 nor activation of Akt. The mechanism of protection may be novel.
Keywords: Methionine sulfoxide reductases, Oxidative stress, Oxidative defenses, Methionine-methionine sulfoxide signaling, Ischemia-reperfusion, Paraquat
Graphical Abstract
Introduction
Except for its role in initiation of protein synthesis, no special functions for methionine in proteins were generally appreciated. It is often assumed to be a generic hydrophobic amino acid, presumably replaceable by other hydrophobic amino acids such as valine. More recently, investigations from multiple laboratories have changed this view [1-8]. Methionine in proteins functions in antioxidant defense, protein structure, and redox sensing and regulation [9].
Methionine residues provide antioxidant defense through reversible oxidation and reduction. Oxidation of methionine creates a chiral center at the sulfur so that the methionine sulfoxide produced is a mixture of the S and R epimers [10]. Reduction of the sulfoxide back to methionine is catalyzed by the methionine sulfoxide reductases (MSR), enzymes found in almost all organisms from microbes to humans [11]. Recycling by the reductases allows the methionine residue to react again with oxidizing species, creating a system with catalytic efficiency in scavenging reactive species.
MSRA acts on the S epimer and MSRB on the R epimer. The A class was described some years ago and has been characterized in considerable detail [11]. The B class of reductases, some of which are selenoproteins in higher animals, was discovered more recently and has also been studied intensively [12, 13]. Mammals have 3 isoforms of the B class, and one of the A class. There is only one gene for MSRA, although it has two initiation sites [14]. Initiation at the first site generates a protein with a mitochondrial targeting sequence. The protein produced by initiation at the second site lacks the mitochondrial targeting sequence, is myristoylated, and is targeted to the cytosolic surface of late endosomes/lysosomes [15].
The importance of the reductases, particularly MSRA, is well established, at least in cultured cells. Knocking out MSRA caused increased susceptibility to oxidative stress in yeast [16] and bacteria [17-20]. Helicobacter pylori, a causative agent of gastric ulcers and carcinoma, requires MSRA for protection against oxidative stress, and MSRA appears to act through reduction of methionine sulfoxide in the bacterial catalase [20, 21]. Overexpression of MSRA conferred increased resistance to oxidative stress in Saccharomyces [22], PC-12 cells [23], Drosophila [24], Arabidopsis [25], human T cells [22], and to the mouse heart in a Langendorff model of ischemia reperfusion [26].
However, mice lacking MSRA have a normal lifespan but do exhibit an increased susceptibility to paraquat, a redox cycling compound that induces oxidative stress [27]. They also show an increased sensitivity to cisplatin [28], exhibit greater kidney damage and brain damage following ischemia-reperfusion [29, 30] as well as increased fibrosis in experimentally induced ureteral obstruction [31]. Liver damage by acetaminophen, a radical-mediated process, is exacerbated in MSRA knockout mice [32]. MSRA knockout mice are more susceptible to cardiomegaly and death induced by chronic oxidative stress [33] and to death from lipopolysaccharide challenge [34]. They also produce an arterial neointimal hyperplasia following injury to the common carotid artery [35]. MSRA knockout mice experience a progressive loss of hearing that is evident by 4 months of age ([36] and personal communication, Richard EM, Ahmed ZM, Gladyshev VN, Riazuddin S). Mice in which MSRB3 was knocked out are deaf [37], as are humans with a familial lack of MSRB3 [38]. While mice lacking MSRB1 have no recognized phenotypic changes, they do have increased levels of malondialdehyde, protein carbonylation, protein methionine sulfoxide, and oxidized glutathione in their livers and kidneys [39]. It is not known whether the 4 isoforms of MSR provide significant redundancy in protecting against oxidative stress or for cell signaling via reversible oxidation of methionines. We therefore constructed a quadruple knockout mouse lacking MSRA, MSRB1, MSRB2, and MSRB3. No methionine sulfoxide reductase activity was detected in tissues of the quadruple knockout animals, confirming that the 4 known isozymes are the only MSR in the mouse. The unexpected result in our studies was that when mice were subjected to severe oxidative stress (ischemia-reperfusion or paraquat), the quadruple knockout was distinctly more resistant than the wild-type mouse.
2. Materials and methods
2.1. Generation of quadruple knockout mouse MsrA−/−/MsrB1−/−/MsrB2−/−/MsrB3−/−
Zygotes were collected from MsrA−/−/MsrB1−/− double knockout mice (substrain C57BL/6J) whose generation was previously described [40]. Genomic sequences of MsrB2 or MsrB3 were submitted to the MIT’S Guide RNA Design Tool [41]. For each gene, 2 sgRNAs were selected for cutting the first coding exon, shortly after the translation initiation codon. They were:
MsrB2: 5’-CGACTGCTGCGAGCGCTGCG-3’
5’-GGCTCGGGGTTGCGCGGGAT-3’
MsrB3: 5’-CCGTGGCCCCGAGAGCCTGC-3’
5’-CACCAAGTGCAGCAGGTTGA-3’
The 4 sgRNAs (20 ng/ul each) and Cas9 mRNA (100 ng/ul) were co-injected into zygotes which were subsequently implanted into pseudopregnant foster mice. Offspring carrying the mutant alleles were identified by PCR amplification followed by sequencing using these primers:
MsrB2: Forward: 5’-AGAGGAGCAA GAGGATAAGT-3’
Reverse: 5’-ACTTATCCTCTTGCTCCTCT-3’
MsrB3: Forward: 5’-GAGCCTCGAT TGCACTCACA-3’
Reverse: 5’-GTCACTCAGAGGACGGTGAG-3’
The MsrB2−/− founder had disruption of MsrB2 by insertion of a fragment of about 200 base pairs from the knockout vector of MsrB1−/−. It was inserted into the sgRNA cutting site. The founder chosen for the MsrB3−/− line had a 7 base pair deletion (CCGAGAG). Founders were backcrossed with MsrA−/−/MsrB1−/−. Since the 4 Msr genes are on different chromosomes and the mutations segregated into different offspring, we generated triple mutants (MsrA−/−/MsrB1−/−/MsrB2−/− and MsrA−/−/MsrB1−/−/MsrB3−/−) first, and then cross bred them to produce the quadruple knockout.
The mice are being deposited in the Mutant Mouse Resource & Research Centers (MMRRC) from which qualified investigators can obtain them. The strain identification is RRID:MMRRC_065273-JAX [42].
2.2. Methionine sulfoxide reductase assay
With Western blotting, antisera against MSRA produced in house as well as product 14547-1-AP from Proteintech confirmed loss of MSRA in the quadruple knockout. Antiserum against MSRB3 (Proteintech 14251-1-AP) confirmed loss of MSRB3. However, we were unable to obtain antisera against MSRB1 and MSRB2 that possessed appropriate sensitivity and specificity. We evaluated Proteintech 15333-1-AP and 17629-1-AP against MSRB1 and MSRB2, Santa Cruz Biotechnologies sc-34848 against MSRB1, and Antibodies-online ABIN331357 against MSRB2. We therefore assayed liver extract for total MSR activity to confirm the genetic analysis of the quadruple knockout.
Reduction of the hexapeptide substrate, Pro-Met(O)-Ala-Ile-Lys-Lys, was assayed as described [43]. The substrate concentration was 50 μM, and the incubations were for 10 min at 37°. Extracted ion chromatograms for the substrate (M+H = 703.4171) and product (M+H = 687.4222) were obtained and integrated. The areas of the substrate and product were used to calculate activity. Freshly prepared liver homogenates were prepared and assayed from 4 each wild type and quadruple knockout male mice, ~11 months old. The homogenization buffer was 25 mM Hepes pH 7.8, 150 mM NaCl, 1% NP-40, 125 μM DTT, 1 mM phenylmethanesulfonyl fluoride, 1 mM Na3VO4, 4 mM NaF, Protein Inhibitor Cocktail, 1 tablet per 50 ml buffer (Sigma, 11836145001). The tissue:buffer ratio was 1:10. Homogenization was carried out on ice with 7 passes of a Teflon pestle tissue grinder. The homogenate was centrifuged at 21,000 g at 4° for 20 min. ~7 μg total protein of each lysate was used for activity measurement. The HPLC-Mass Spectrometry assays were performed twice (technical replicates) and the results averaged. The detection limit of the assay is 1 pmol/min/mg protein.
2.3. RNAseq and quantitative real-time reverse transcription-PCR (qRT-PCR)
RNAseq (Illumina) and data processing were performed in the core facilities of the National Heart, Lung, and Blood Institute. Experimental details and data have been deposited at the Gene Expression Omnibus (GEO) under accession number GSE130394. For qRT-PCR, 4 hearts from 3-6 months old female wild type and quadruple knockout mice were fast frozen in liquid nitrogen. RNA was extracted from frozen tissue with Trizol reagents (Thermo Fisher, # 15596026). cDNA was synthesized from 1 μg RNA of each sample with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher, # 4368814). qRT-PCR was performed on a LightCycler©96 Thermocycler (Roche) with iTaq Universal SYBR Green Supermix (Bio-Rad). 28S RNA was included on each plate as an internal standard.
2.4. Treadmill test
Exercise capacity was tested using a Columbus Instruments rodent treadmill (Model Eco-6M), set at a 10 degree incline. Total exercise time, distance, maximum speed, and work were recorded at the time of exhaustion which was defined as the moment the mouse was unable to continue running without repeatedly falling back onto the shock grid at the backend of the treadmill belt. The testing protocol was as follows: 10 minutes at 10 m/min, then 12 m/min for five minutes. At minute 15 the belt speed was increased to 15 m/min for three minutes and then increased 1.8 m/min every three minutes until the mouse became exhausted.
2.5. Dobutamine cardiac stress test
Six-month old mice were lightly anesthetized with isoflurane and placed on a heated platform with ECG leads and a rectal temperature probe to perform echocardiography exams before and after administering constant rate dobutamine infusions. Heart images were acquired using the Vevo2100 ultrasound system (VisualSonics, Toronto, Canada) with a 30 MHz ultrasound probe (VisualSonics, MS-400 transducer). After the baseline-scan, the mice received constant rate infusions of dobutamine (0.625 mg/ml in normal saline containing 5% dextrose), via the tail vein using an infusion syringe pump (Harvard Apparatus). The low dose infusion rate was 10 μg/kg/min. After the heart rate reached a steady state as determined by the ECG, the rate was increased to 40 μg/kg/min and the scans repeated.
2.6. Ischemia-reperfusion model
Male and female mice between 12 and 16 weeks of age were studied; cardiac hypertrophy was not observed in any mice. Hearts were Langendorff-perfused as described [44, 45]. After an equilibration period of 20 minutes, hearts were subjected to 25 minutes of no-flow ischemia followed by 90 minutes of reperfusion. Hearts receiving ischemic preconditioning were first equilibrated for 20 min, subjected to 4 cycles of 5 min of ischemia and 5 min of reperfusion, and then subjected to ischemia-reperfusion as above. Functional recovery and infarct size were assessed as described [26]. All values are presented as the mean ± standard error. Statistical significance was assessed by one way analysis of variance (ANOVA), followed by post hoc multiple comparison procedures using the Holm-Sidak method. A p value less than 0.05 was considered to be statistically significant.
2.7. Paraquat challenge
Paraquat (NN’-dimethyl-4,4’-bipyridilium) dichloride (Sigma), dissolved in sterile saline, was administered intraperitoneally to induce a global oxidative stress, and survival was followed for 10 days. Two separate experiments were carried out. In one, 8 wild type and 8 quadruple knockout mice received 40 mg/kg paraquat intraperitoneally. In the other, 10 mice of each genotype received 50 mg/kg paraquat intraperitoneally. Kaplan-Meier survival curves were constructed and evaluated with Prism software, version 7 (GraphPad, La Jolla, CA). Comparisons were made with the Mantel-Cox Log-Rank test. No statistically significant differences were observed between genotypes for the 40 and 50 mg/kg doses so the results for the two experiments were combined. Similar results were obtained in an experiment using 40 mg/kg body weight and performed 1 year apart by a different author.
2.8. Murine embryonic fibroblasts (MEF)
MEF were prepared with a modification of a published procedure [46]. MEF were obtained from 14-day old postcoitum embryos. They were cultured at 37° in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Invitrogen), penicillin, and streptomycin (Thermo Fisher #15070063). The atmosphere in the incubator was 3% O2, 5% CO2, and 92% N2. Cultures were split 1:3 on the fourth day. MEF were grown to confluency and then frozen in liquid nitrogen at a concentration of 106 cells/ml (passage #2). Subsequent passages were split 1:4, and medium was replaced twice a week. Cells were counted with a Countess™ automated cell counter (Invitrogen). Cells used in the cell viability experiments were 80-90% confluent and were from passages 3-6.
2.9. Cell viability assay
Cell viability assays followed the cleavage by mitochondrial dehydrogenases of stable water-soluble tetrazolium salt to a soluble formazan, a process that depends on the glycolytic production of NADPH in viable cells. The amount of formazan produced is directly proportional to the number of metabolically active cells in the culture and was quantified by measuring its absorbance at 440 nm.
MEF were seeded in a 96-well plate at 20,000 cells per well (200 μl) and allowed to attach to the bottom of the plate overnight, after which the culture medium was removed. To induce oxidative stress, one of the following reagents (freshly prepared in complete culture medium) was added to a well followed by incubation for the indicated time: paraquat (0-1000 μM, 24h), hydrogen peroxide (0-400 μM, 24h), potassium dichromate (0-100 μM, 6h), and cadmium chloride (0-16 μM, 24h). After incubation, medium was removed and replaced by fresh 180 μl complete culture medium, and the plates were incubated for another 3 h. Then 20 μl of water-soluble tetrazolium −1 reagent (Sigma , #11644807001) was added to each well. After 30 min incubation, absorbance was determined with a scanning multi-well spectrophotometer (InfiniteM200Pro, Tecan). Viability of treated cells was expressed as a percentage of untreated cells.
Paraquat (methyl viologen dichloride hydrate; #856177) and potassium dichromate (#207802) were obtained from Sigma-Aldrich. Hydrogen peroxide (30%) was Acros #UN2014. Cadmium chloride (#72242) was purchased from Merck (Rahway, NJ).
2.10. Nrf2 analysis
Heart and liver tissue (20 mg) were processed immediately after harvesting from 9 month old male mice. Nuclear and cytoplasmic fractionation was performed using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Scientific, # 78835). Immediately before use, a protease inhibitor cocktail (Millipore, # 539134) was added to CER-I and NER solutions at a dilution of 1:100. Tissues were washed with phosphate-buffered saline (PBS) (KD Medical, 15 mM NaCl, 10 mM KH2PO4, 56 mM Na2HPO4, pH 7.4) followed by centrifugation at 500g for 5 minutes. The supernatant was discarded, taking care to leave the tissue pellet as dry as possible. The tissue was homogenized in 200 μl of CER-I, using a Dounce homogenizer and incubated on ice for 10 minutes. Next, 11 μl of ice-cold CER-II was added and the sample was vortexed for 5 seconds and then incubated on ice for 2-3 minutes. The tube was centrifuged at ~16,000g for 10 minutes after which the supernatant, containing the cytoplasmic extract, was transferred to a clean pre-chilled tube. The insoluble pellet, containing nuclei, was suspended in 100 μl ice-cold NER and kept on ice for 40 minutes with vortexing for 15 seconds every minutes. The tube was then centrifuged at ~16,000g for 10 minutes. The supernatant, now containing the nuclear extract, was transferred to a clean pre-chilled tube. Extracts were then analyzed by immunoblotting.
Samples containing 100 μg protein were separated by SDS-PAGE (4-20%) and transferred to a nitrocellulose membrane. Membranes were incubated with Odyssey® blocking buffer (Licor, #927-40000) for 1 h at room temperature, followed by incubation with primary antibodies at 4° overnight. The blots were washed 3 times for 10 min with PBS supplemented with 0.1% Tween-20. The blots were then incubated with secondary antibodies labeled with IRDye near-infrared fluorescent dyes for 40 min and washed again 3 times for 10 min with PBS supplemented with 0.1% Tween-20. The blots were then scanned on an Odyssey imaging system (Licor Biosciences). Band intensities were quantitated with using Odyssey software version 2.1.
The primary antibodies and their dilutions were: anti-Nrf2 (AbCam, #ab31163, 1:2000), anti-GAPDH (Invitrogen, #MA5-15738, 1:2000), anti-histone H3 (Cell Signaling Technologies, D1H2 #4499, 1:2000). The secondary antibodies were: DyLight™ 680 labeled Goat anti-Mouse IgG (H+L) (KPL, #072-06-18-06, 1:5000) and DyLight™ 800 labeled Goat anti-Rabbit IgG (H+L) (KPL, #072-07-15-06, 1:5000).
2.11. Akt and phospho-Akt analysis
Heart tissue was taken from two wild type and two quadruple knockout male mice, ~11 months old. The tissue was homogenized and centrifuged as described above the reductase assay. The supernatant was subjected to SDS-PAGE, and proteins were transferred to a nitrocellulose membrane. The antibodies utilized to detect total and phospho-Ser473 were from Cell Signaling (#9272S and #9271S). A positive control for phospho-Akt was prepared by treating HEK293 cells with A-443654 (Cayman Biochemical Company, Cat# 16499), an Akt phosphorylation inducer [47]. An HEK293 cell lysate was prepared by sonication.
2.12. Animal study approval
All animals were treated humanely in accordance with the Guide for the Care and Use of Laboratory Animals [48]. The study, H-120R4, was approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute.
3.0. Results
3.1. The quadruple knockout mouse has no methionine sulfoxide reductase
The 4 methionine sulfoxide reductases are located on 4 different chromosomes in the mouse, facilitating generation of the quadruple knockout. We had previously generated a double knockout, MsrA−/−/MsrB1−/− [40], so that only MsrB2 and MsrB3 knockouts were required to eventually generate the quadruple knockout. CRISPR/Cas9 was used to target the first coding exon of MsrB2−/− and MsrB3−/−. sgRNA against both MSRB2 and MSRB3 were injected together, but no quadruple knockout was obtained. However, triple knockouts with either MsrB2−/− or MsrB3−/− were obtained. These triple knockout mice were bred to each other to generate the quadruple knockout, MsrA−/−/MsrB1−/−MsrB2−/−MsrB3−/−. The line is maintained by breeding quadruple knockouts to each other. Successful targeting was confirmed by DNA sequencing (Figure 1). The double knockout, MsrA−/−/MsrB1−/−, is on substrain C57BL/6J. The “J” designates a line carrying a mutation in the nicotinamide nucleotide translocator, and we confirmed that the quadruple knockout is also C57BL/6J (Supplemental Figure 1) [49].
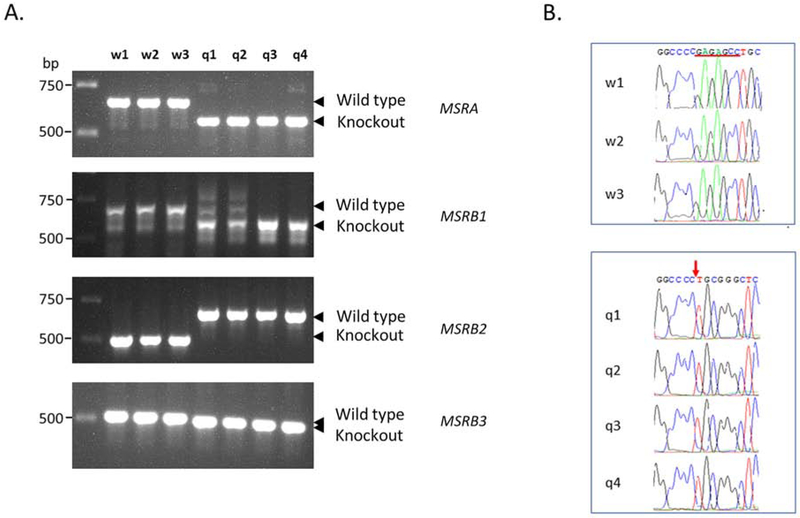
Figure 1. Genotyping of mice lacking the 4 known methionine sulfoxide reductases.
(A) Genomic DNA from tail tissue was analyzed by PCR. The product from the deletion in MSRA was 129 bp shorter than the wild type while the MSRB1 deletion was 182 bp shorter. Deletion of exons 2 and 3 in MSRB2 yielded a product that was 200 bp longer than the wild type. Only 7 bp were deleted in the MSRB3 knockout, and that could not reliably be established by PCR. Thus, the PCR products from the 3 wild type(w1, w2, w3) and 4 quadruple knockout mice (q1, q2, q3, q4) were subjected to DNA sequencing (B). The 7 nucleotides, GAGAGCC, are present in the wild type mice but missing in the mutants (red arrow).
Although there are only 4 identified methionine sulfoxide reductases in mammals, it is possible that other proteins may be able to reduce methionine sulfoxide. Liver is rich in the known reductases and many other enzymes, so we assayed the liver of the quadruple knockout mice for methionine sulfoxide reductase activity. No signal above background was detected (Figure 2). As expected, the mice are deaf – determined by their failure to respond to startle noises. The growth curves of the quadruple knockout matched those of wild-type mice (Figure 3).
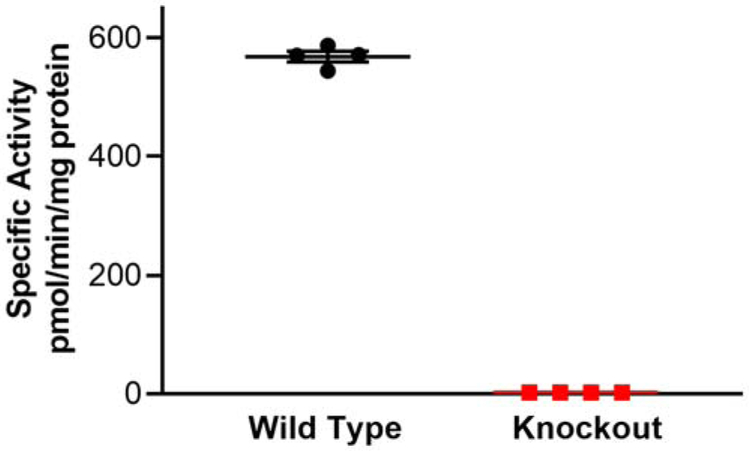
Figure 2. Liver methionine sulfoxide reductase activity assay.
The specific activity of methionine sulfoxide reductase in the knockout is less than 0.5% that of the wild type. The lines show the mean ± SD.
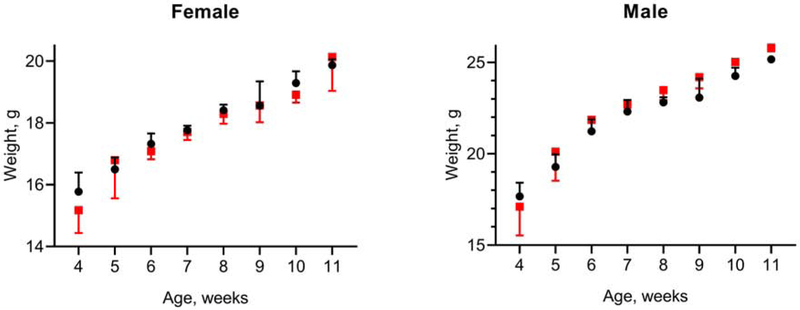
Figure 3. Growth curves.
Mice were weighed 3 times per week and these weights were averaged to yield the weekly weight plotted on the graph. Wild type, black circles; Quadruple knockout, red squares. The bars display the mean ± SD, up from mean for wild type and down for knockout. The number of animals in each group were: wild type male, 8; wild type female, 16; knockout male, 6; knockout female, 10.
3.2. Cardiac and muscular performance
Heart and skeletal muscle oxidative stress increases when stress-tested or exercised. We compared the running performance of wild type and quadruple knockout mice that were run to exhaustion on a treadmill. The knockout mice performed as well as their wild type counterparts (Figure 4). We then characterized cardiac performance, both at rest and during a dobutamine stress test. Since mice won’t voluntarily run on a treadmill to increase their heart rate as in a human cardiac stress test, dobutamine is used to do so. It is a β1-adrenergic agonist that increases heart rate as well as cardiac contractility and output. No significant differences were observed between wild-type and quadruple knockout mice (Table 1).
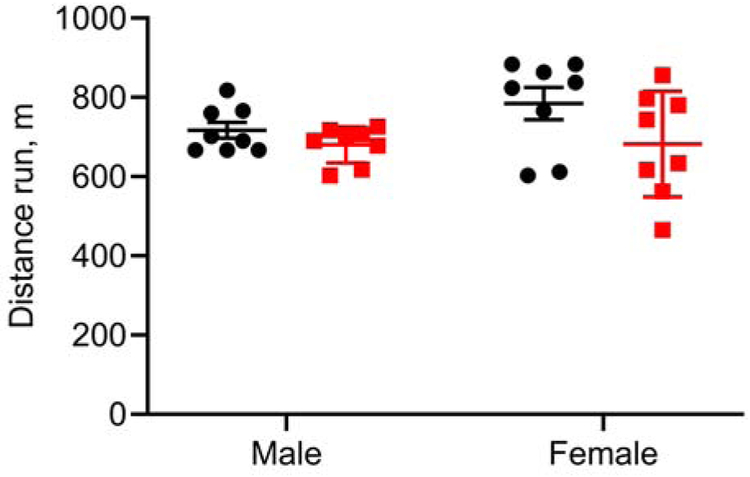
Figure 4. Treadmill running distance at exhaustion.
The results are plotted as the mean ± SD, wild type, black circles; quadruple knockout, red squares. Each group had 8 animals.
Table 1.
Cardiac parameters from dobutamine stress test
| Genotype and Sex |
Systolic volume of left ventricle (mm3) |
Diastolic volume of left ventricle (mm3) |
Systolic diameter of left ventricle (mm) |
Diastolic diameter of left ventricle (mm) |
Heart rate (beats per minute) |
Cardiac Output (ml/min) |
Ejection fraction (%) |
|---|---|---|---|---|---|---|---|
| Baseline | |||||||
| WT female | 21.7±4.4 | 58.0±8.2 | 2.5±0.2 | 3.7±0.2 | 505±43 | 18.4±3.3 | 63±2.9 |
| qKO female | 22.8±2.5 | 64±7.6 | 2.5±0.1 | 3.8±0.2 | 528±68 | 21.9±3.7 | 64±2.1 |
| WT male | 29±3.1 | 77±9.8 | 2.8±0.1 | 4.2±0.2 | 516±27 | 24.8±4.0 | 62.3±2.6 |
| qKO male | 26.5±3.2 | 70.5±5.2 | 2.7±0.1 | 4.0±0.1 | 548±40 | 24.2±3.1 | 62.4±2.4 |
| Low Dose dobutamine | |||||||
| WT female | 10±6.6 | 43±10.6 | 1.8±0.4 | 3.2±0.3 | 603±25 | 19.7±2.3 | 77.5±7.9 |
| qKO female | 16.2±5.1 | 56.1±10.0 | 2.2±0.3 | 3.6±0.3 | 594±44 | 23.7±3.5 | 71.5±5.2 |
| WT male | 15.1±5.7 | 59.8±11.3 | 2.1±0.4 | 3.7±0.3 | 607±15 | 27.1±3.9 | 75.6±6.0 |
| qKO male | 16.3±6.6 | 61.0±7.4 | 2.2±0.3 | 3.8±0.2 | 616±25 | 27.6±2.9 | 73.9±7.3 |
| High dose dobutamine | |||||||
| WT female | 6.7±4.2 | 39.4±9.0 | 1.5±0.3 | 3.1±0.3 | 617±32 | 20.2±3.1 | 84.0±5.7 |
| qKO female | 11.6±4.0 | 50.7±8.0 | 1.9±0.3 | 3.5±0.2 | 633±26 | 25.6±3.8 | 77.8±5.5 |
| WT male | 11.0±5.4 | 55.7±9.8 | 1.8±0.4 | 3.6±0.3 | 642±28 | 28.6±3.2 | 81.1±7.4 |
| qKO male | 10.8±5.4 | 56.5±6.8 | 1.8±0.3 | 3.7±0.2 | 639±28 | 29.1±1.5 | 81.4±7.1 |
The genotype abbreviation are WT, wild type and qKO, quadruple knockout. Each group had 8 animals in it. For each parameter, the wild type and quadruple knockout were compared, with the Bonferroni correction for the multiple comparisons. With the correction, there were no significant differences.
We previously showed that overexpression of the non-mitochondrial form of MSRA protected the heart from damage in a Langendorff model of ischemia-reperfusion. We thus anticipated that the quadruple knockout animal would suffer greater damage and loss of function when subjected to the same stress. Notably, the opposite was observed. Both male and female quadruple knockout were more resistant to cardiac ischemia-reperfusion. As there was no difference between males and females, the results were combined (Figure 5). Hemodynamic parameters for the mice used in this figure are in Table 2.
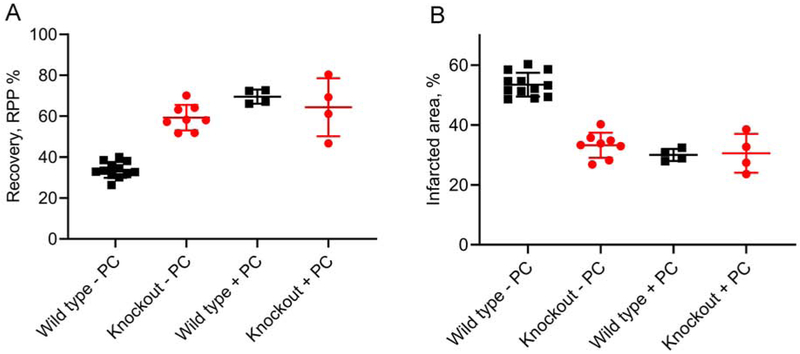
Figure 5. Effects of ischemia-reperfusion on the heart.
Isolated hearts were subjected to ischemia-reperfusion as described in Methods. (A) Recovery from reperfusion measured as the left ventricular rate-pressure product (RPP). (B) Infarcted area of the left ventricle. The groups are wild type without preconditioning (− PC), 6 female and 6 male; quadruple knockout without preconditioning, 4 male and 4 female; wild type with preconditioning (+PC), 2 male and 2 female; quadruple knockout with preconditioning, 2 male and 2 female. Without preconditioning, the quadruple knockout had a significantly higher RPP (p<0.0001) and lower infarct area (p<0.0001) than the wild type. With preconditioning, there was no significant difference in RPP (p=0.50) nor infarct area (p=0.88). Comparisons were made with the unpaired t-test. The line and bars show the mean and SD.
Table 2.
Hemodynamic parameters from Langendorff model of ischemia-reperfusion
| Pre-ischemic hemodynamic value | Post-ischemic hemodynamic value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | FR | LVDP | +dp/dt | −dp/dt | HR | FR | LVDP | +dp/dt | −dp/dt | ||
| Group | n | (bpm) | (ml/min) | (cmH2O) | (cmH2O/ms) | (bpm) | (ml/min) | (cmH2O) | (cmH2O/ms) | ||
| WT: I/R | 12 | 378±9 | 2.3±0.1 | 150±6 | 8.9±0.5 | −6.0±0.4 | 286±10 | 1.8±0.1 | 68±2 | 5.1±0.3 | −4.1±0.3 |
| qKO: I/R | 8 | 363±19 | 2.4±0.1 | 141±6 | 8.1±0.3 | −5.2±0.2 | 315±22 | 1.8±0.1 | 97±5 | 5.7±0.4 | −3.8±0.2 |
| WT: PC & I/R | 4 | 376±13 | 2.3±0.1 | 144±16 | 9.5±0.8 | −7.1±0.6 | 350±9 | 1.8±0.1 | 108±12 | 7.4±0.9 | −5.6±0.6 |
| qKO: PC & I/R | 8 | 352±8 | 2.4±0.1 | 127±7 | 8.5±0.5 | −6.6±0.3 | 302±19 | 1.8±0.1 | 97±8 | 6.5±0.4 | −5.4±0.2 |
The values are the mean ± SE. n, number of hearts; HR, heart rate (beats per min, bpm); FR, flow rate (ml/min); LVDP, left ventricular developed pressure (cmH2O); ±dp/dt, rates of pressure rise and fall (cmH2O/ms), respectively. The groups are: WT: I/R, Wild type subjected to ischemia-reperfusion without preconditioning; qKO: I/R, Quadruple knockout type subjected to ischemia-reperfusion without preconditioning; WT: PC & I/R, Wild type subjected to preconditioning and then ischemia-reperfusion; qKO: PC & I/R, Quadruple knockout type subjected to preconditioning and then ischemia-reperfusion.
If a heart is first exposed to a brief period of ischemia-reperfusion before the long period of ischemia-reperfusion, that heart is protected against damage and loss of function. This is a hormetic phenomenon, which in cardiac physiology is usually referred to as “preconditioning” [50]. We considered that the lack of methionine sulfoxide reductase activity in the quadruple knockout might mimic preconditioning, as do many other treatments, both ischemic and non-ischemic [51]. We therefore compared the effect of ischemic preconditioning in wild type and quadruple knockout mice (Figure 5). Preconditioning raised the level of protection of the wild type mouse to equal that of the quadruple knockout that was not preconditioned. Preconditioning of the quadruple knockout did not further increase protection, possibly because the mechanism of protection by preconditioning and knocking out the MSR is the same.
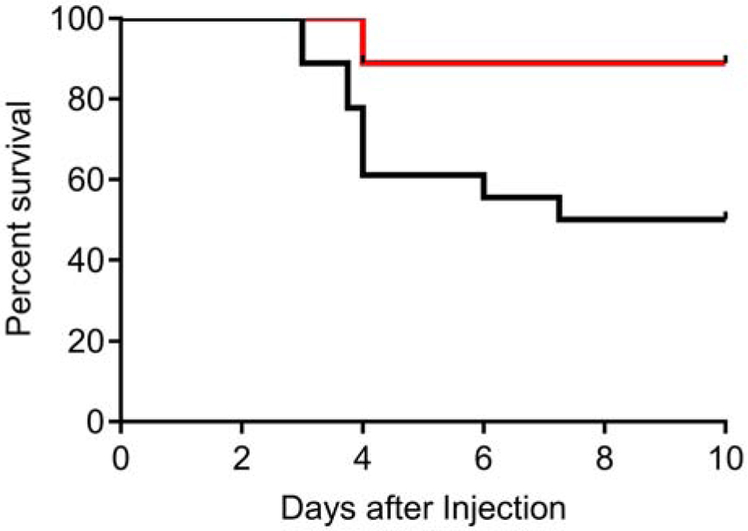
Paraquat is a redox cycling compound that induces severe, systemic oxidative stress, and animals lacking MSRA die faster when exposed to paraquat [52]. Quadruple knockout mice were expected to be distinctly more sensitive to paraquat than wild type mice, but again the opposite was observed. Quadruple knockout mice were reproducibly more resistant to paraquat than wild type controls (Figure 6).
Figure 6. Paraquat challenge.
Wild type (black line) and quadruple knockout (red line) survival plotted as a Kaplan Meier graph. The quadruple knockout mice were more resistant to killing by paraquat than the wild type mice (p=0.01, Mantel-Cox Log-Rank test).
3.3. Mechanism of increased resistance to oxidative stress
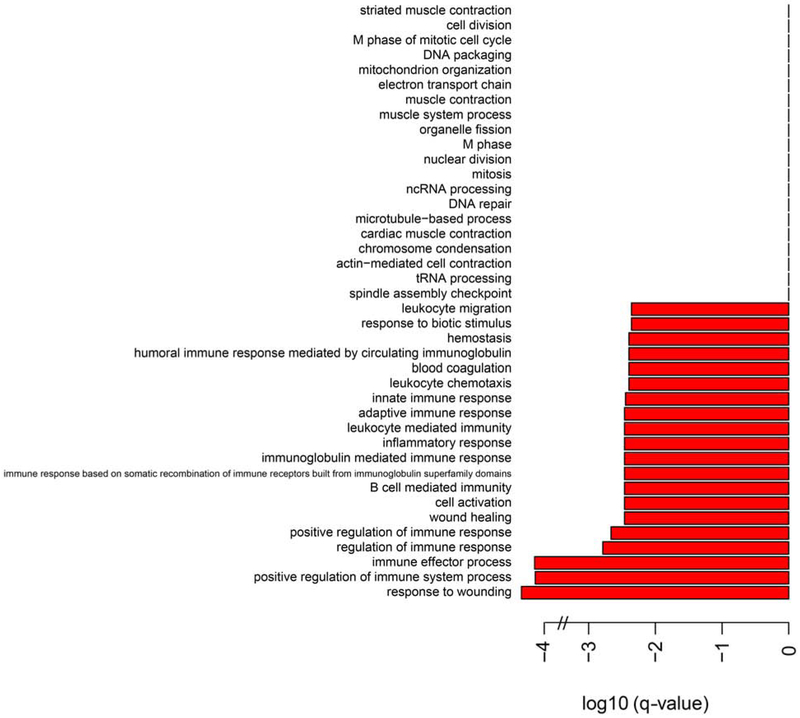
To initiate investigation of the mechanism of protection, we performed RNA-seq on the liver and heart of wild type and quadruple knockout animals (GEO Accession GSE130394). Analysis of the data sets at the Gene Ontology Consortium (GO) [53, 54] revealed a marked downregulation of inflammatory and immune responses in the heart, but not the liver (Figure 7).
Figure 7. Biological processes comparison of RNAseq results for the heart.
The categories are those of the Gene Ontology Consortium (GO) [53, 54].
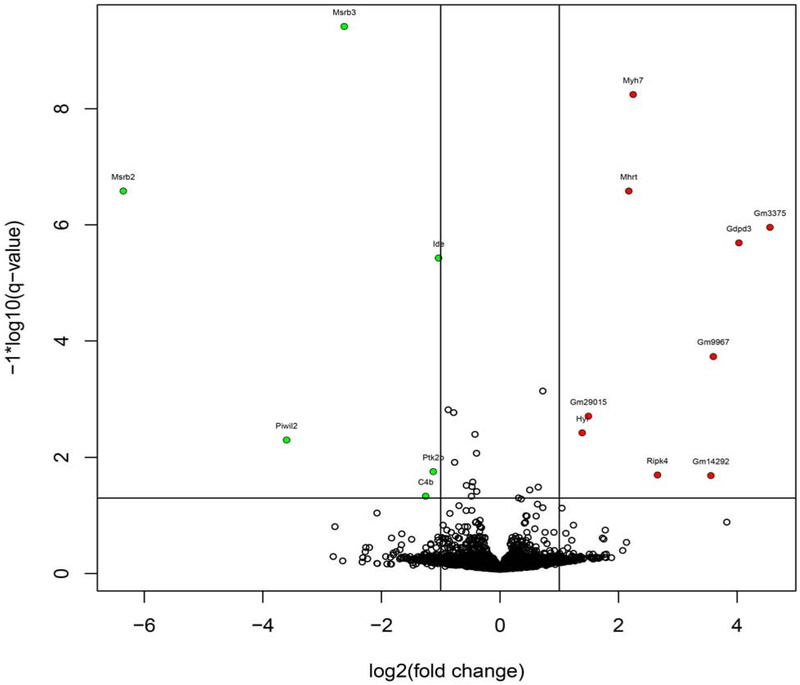
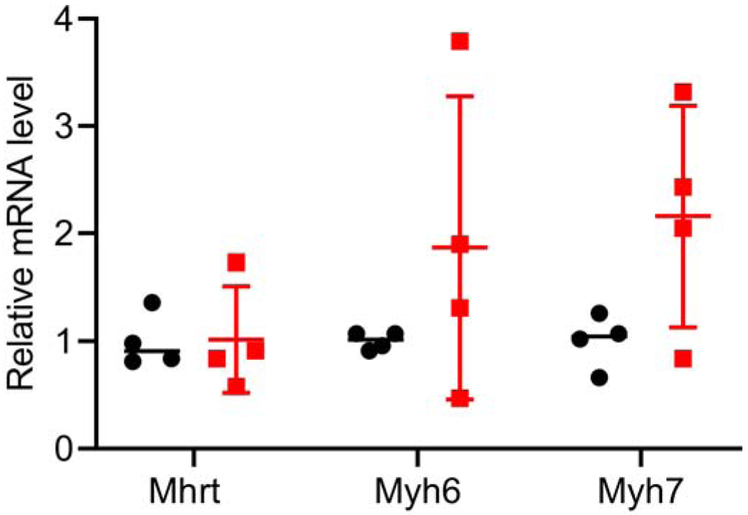
Volcano plots with q values comparing wild type and quadruple knockout mice are shown in Figure 8. MsrB2 and MsrB3 levels are, as expected, drastically reduced in the knockout. The counts for MsrA and MsrB1 were very low in both wild type and knockout. In the heart of the quadruple knockout, the level of Myh7, encoding the β-myosin heavy chain was elevated. Myh6, encoding the α-myosin heavy chain, was also elevated, albeit more modestly than Myh7. Mhrt, a long non-coding RNA whose DNA sequence is within the Myh7 gene, was also elevated. Elevated expression of the β-myosin heavy chain occurs in failing hearts and depresses cardiac function [55] and thus would not explain the increased stress resistance observed in the quadruple knockouts. Mhrt is a long noncoding RNA that protects the heart from pathological hypertrophy [56]. We performed qRT-PCR for Myh7, Mhrt, and Myh6. As shown in Figure 9, the RNA-seq results for the myosin heavy chains were confirmed, but there was no difference in the expression of Mhrt between wild type and quadruple knockout in the heart. In both liver and heart of the quadruple knockout, the level of Gm3375 is elevated in the quadruple knockout. It has been annotated as a processed pseudogene related to Psmb5, which codes for a β subunit of the proteasome. Gdpf3 (sometimes referred to as Gde7) levels are also elevated in the quadruple knockout. It codes for a protein that may be a calcium-dependent lysophospholipase D but has not been characterized in detail [57, 58].
Figure 8.
Volcano plot of RNAseq results for the heart.
Figure 9. qRT-PCR of heart Mhrt, Myh6, and Myh7.
The results are plotted as the mean ± SD. Four hearts of each genotype were analyzed. Wild type, black circles; quadruple knockout, red squares.
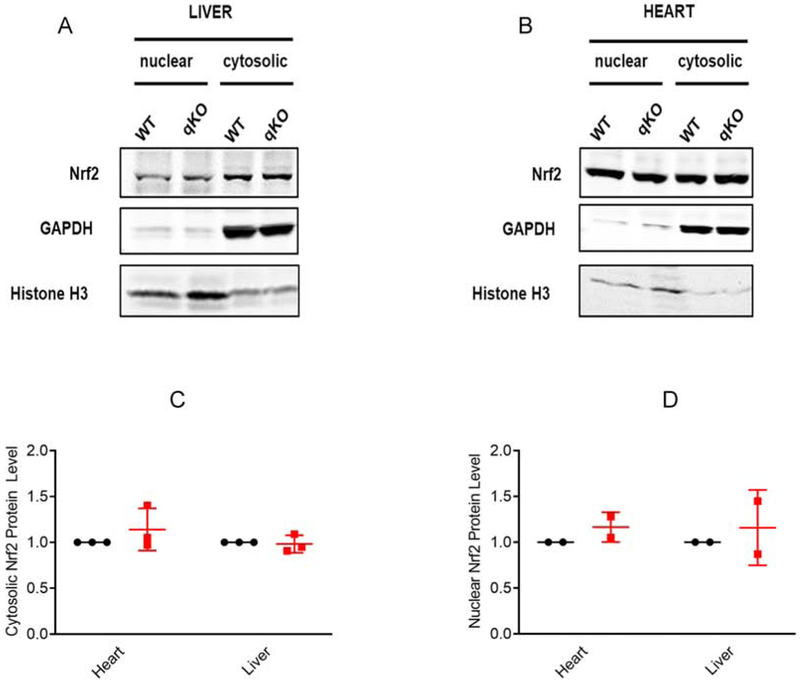
Nrf2 is a transcription factor that is the primary regulator of the antioxidant response [59]. Under oxidative stress as well as other triggers, both cytosolic and nuclear levels of Nrf2 are increased, and increased Nrf2 is cardioprotective [60]. Deletion of the MsrA gene in mice increased both cytosolic and nuclear Nrf2 which in turn increased the levels of reduced glutathione without a change in the level of oxidized glutathione [61]. Since we observed cardioprotection and resistance to paraquat in the quadruple knockout mouse, we measured Nrf2 in the heart and liver. We found no difference in cytosolic or nuclear levels of Nrf2 between the quadruple knockout and the wild type mice (Figure 10).
Figure 10. Cytosolic and nuclear Nrf2 protein levels.
Representative immunoblots showing expression of Nrf2 in the nuclear and cytosolic fractions of tissue lysates derived from the (A) liver and (B) heart of the wild type or quadruple knockout mice. GAPDH and histone H3 are shown as cytosolic and nuclear markers respectively. (C, D) Quantitation of the expression levels of Nrf2, wild type, black circles; quadruple knockout, red squares. The integrated intensities of each Nrf2 band were calculated as a ratio to the corresponding integrated intensities of the GAPDH or histone H3. The ratios were normalized by setting the wild type to 1.0. The bars display the mean ± SD from separate experiments (3 for the cytosolic and 2 for the nuclear). There were no significant differences (p≥0.3). This analysis was replicated by a different coauthor one year apart, with the same result. Uncropped blots are shown in Supplemental Figure 2.
Cardioprotection in the quadruple knockout without preconditioning was equivalent to that observed in the wild type mouse with preconditioning. The mechanism of cardioprotection by ischemic preconditioning has been studied in detail, and there is good evidence that activation of the pro-survival kinase Akt is required for protection [62, 63]. We therefore determined whether Akt was activated in the quadruple knockout and found that it was not (Figure 11).
Figure 11. Akt kinase activation.
Wild type (w1 and w2) and quadruple knockout (q1 and q2) heart extracts were analyzed for Akt activation by immunochemical detection of total Akt (upper panel) and of Akt phosphorylated on Ser473 (lower panel). A positive control from HEK293 cells was prepared as described in methods. The analysis was performed twice with the same results. Uncropped blots from the two experiments are in Supplemental Figure 3.
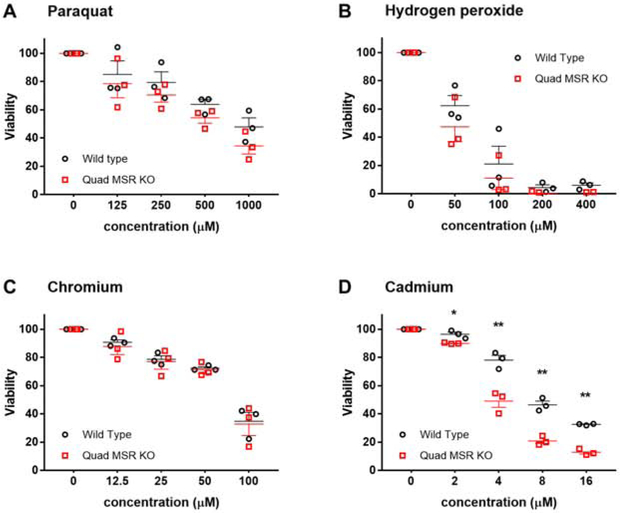
We next considered that elucidating the mechanism of protection might be facilitated by focusing on cultured cells rather than mouse tissues. Murine embryonic fibroblasts were prepared from wild type and quadruple knockout embryos and tested for susceptibility to oxidative stress (Figure 12). There was no difference in cell killing by paraquat, hydrogen peroxide, nor chromium, and quadruple knockout cells were slightly more sensitive to killing by cadmium.
Figure 12. Viability of MEF cells exposed to oxidative stress.
Viability of MEFs derived from wild type (black circles) versus quadruple knockout mice (red squares) in response to oxidative stress. Wild type and quadruple knockout fibroblasts were treated with paraquat, hydrogen peroxide, chromium or cadmium. Viability was averaged from 3 separate experiments performed on different days and normalized to 100% for the untreated controls. The only significant difference observed was a reduction in the viability of knockout cells exposed to cadmium (p = 0.02 at 2 μM and p ≤ 0.01 at 4-16 μM). The error bars with SEM are plotted in one direction to improve readability.
4. Discussion
The methionine sulfoxide reductases, particularly MSRA, provide defense against oxidative stress by reducing methionine sulfoxide residues in proteins back to methionine [64]. We successfully knocked out all 4 known methionine sulfoxide reductases as confirmed by gene analysis and enzymatic assay. MRB2 and MSRB3 were knocked out with CRISPR/Cas9. While it is still debatable whether an off-target effect of this technology is a major concern, recent whole genome sequencing studies have shown that off-target mutations are rare in germline-mutated mice [65, 66]. In addition, a recent large scale whole genome sequencing of 36 mice with 60x coverage also found that off-target mutations are rare [67], and the mice in this study were generated in the same core facility with the same experimental conditions that we employed. Specifically, we injected the Cas9 mRNA and guide RNAs into zygotes, in which they should be stable for only a few days. Some other off-target studies used somatic cell lines that constitutively expressed Cas9 and guide RNAs. Also, the original founder mice had been bred with unedited mice for several generations during quadruple knockout establishment and colony expansion. Therefore, rare off-target mutations, if any, were likely diluted out before the mice were used for experiments.
Knocking out all 4 known mammalian reductases was expected to render mice exquisitely sensitive to oxidative stress and provide an experimentally amendable system for identifying the targets of that stress. The most important finding of our studies is that the exact opposite occurred: Mice lacking methionine sulfoxide reductases are more resistant to the severe oxidative stress created by cardiac ischemia-reperfusion and by whole body redox cycling induced by paraquat. Moreover, resistance was not a result of hormetic induction of the antioxidant transcription factor Nrf2 nor activation of Akt.
A CRISPR screen in Jurkat cells led to the identification of 3 genes that are required for paraquat resistance [68]: POR, a cytochrome P450 reductase localized to the endoplasmic reticulum; ATP7A, a copper transporter localized mainly to the Golgi; and SLC45A4, a sugar transporter localized to the plasma membrane. A negative selection CRISPR screen identified two genes whose loss increased paraquat sensitivity: SOD1, the cytoplasmic copper-dependent superoxide dismutase, and SLC31A1, a plasma membrane bound copper importer. Mice lacking SHC1, which encodes the cytosolic adapter protein p66SHC, have a lengthened lifespan and are resistant to paraquat [69]. None of the RNA levels of these 6 genes differed between the wild-type and quadruple knockout in our RNAseq analyses of liver and heart.
With the relatively recent recognition that cyclic oxidation and reduction of methionines in proteins can function in cellular regulation [4-6, 9], we considered the possibility that the resistance of the quadruple knockout mouse to oxidative stress is a consequence of increased methionine sulfoxide in a specific protein. Unfortunately, a major technical limitation in the field is that there is no methodology for conveniently identifying and studying proteins subject to reversible methionine oxidation. As a consequence, to date, only one methionine-methionine sulfoxide cycle has been established to occur in vivo. Terman and coworkers identified an NADPH oxidoreductase, MICAL, that specifically oxidizes a methionine residue in actin, thereby inducing filament severing and decreasing actin polymerization [6]. Subsequently that group and Gladyshev’s showed that the oxidation was stereospecific and generated only the R-MetO [4, 5]. The modification was fully reversible by MSRB1, a cytosolic enzyme. Thus, oxidation of Met44 in actin by MICAL induces de-polymerization of actin and reduction of MetO44 by MSRB1 restores the ability to polymerize.
In summary, we report the novel finding that knocking out all 4 known mammalian reductases reduces cardiac ischemia-reperfusion injury and induces resistant to systemic paraquat, a redox cycling agent. This surprising result could be due to a hormetic upregulation of a common protective pathway induced by oxidative stress. We examined several candidates and found that it was not due to upregulation of the master antioxidant transcription factor Nrf2 nor to Akt signaling. Alternatively, the lack of reductases may increase the cellular level of the methionine sulfoxide form of one or more proteins that induce resistance. The activity of such proteins must be regulated by reversible oxidation and reduction of methionine sulfoxide, as has been demonstrated for the polymerization/depolymerization of actin [4-6]. Unfortunately, at present there are no tractable methods for identifying such proteins.
Supplementary Material
Highlights.
Mammals have 4 methionine sulfoxide reductases thought to provide oxidative defense
Knocking out one or two reductases in the mouse has little effect on phenotype
We thus produced a mouse line lacking all 4 reductases
This mouse line was predicted to be exquisitely sensitive to oxidative stress
Instead, the quadruple knockout is notably resistant to oxidative stress
Acknowledgments
We thank Vadim Gladyshev (Harvard Medical School) for his gift of antiserum against MSRB3. We also thank Elodie Richard (University of Maryland School of Medicine) for testing the mice and determining that the quadruple knockout mice are deaf. The treadmill and dobutamine stress experiments were performed in the Mouse Phenotyping Core Facility. The quadruple knockout mice were produced in the Transgenic Core Facility. RNAseq was performed in the DNA Sequencing and Genomics Core Facility, and the data produced were analyzed in the Bioinformatics and Computational Biology Core Facility. The methionine sulfoxide reductase assays were analyzed on the HPLC-Mass Spectrometer in the Biochemistry Core Facility.
Funding source
All of the core facilities are in the National Heart, Lung, and Blood Institute. The Institute’s Intramural Research Program provided funding under grants ZIA HL002066 (EM) and ZIA HL000225 (RLL).
Abbreviations:
- CRISPR
clustered regularly interspaced short palindromic repeats
- KO
knockout mouse
- MEF
murine embryonic fibroblasts
- MSR
methionine sulfoxide reductase
- PBS
phosphate-buffered saline
- qRT-PCR
quantitative real-time reverse transcription-PCR
- WT
wild-type mouse
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
All authors have declared no conflict of interest nor conflicting financial interests.
References
- [1].Levine RL; Mosoni L; Berlett BS; Stadtman ER Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U.S.A 93:15036–15040; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bender A; Hajieva P; Moosmann B Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc. Natl. Acad. Sci. U.S.A 105:16496–16501; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Valley CC; Cembran A; Perlmutter JD; Lewis AK; Labello NP; Gao J; Sachs JN The methionine-aromatic motif plays a unique role in stabilizing protein structure. J. Biol. Chem 287:34979–34991; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lee BC; Peterfi Z; Hoffmann FW; Moore RE; Kaya A; Avanesov A; Tarrago L; Zhou Y; Weerapana E; Fomenko DE; Hoffmann PR; Gladyshev VN MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol. Cell 51:397–404; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hung RJ; Spaeth CS; Yesilyurt HG; Terman JR SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat. Cell Biol 15:1445–1454; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hung R-J; Pak CW; Terman JR Direct redox regulation of F-actin assembly and disassembly by Mical. Science 334:1710–1713; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Netzer N; Goodenbour JM; David A; Dittmar KA; Jones RB; Schneider JR; Boone D; Eves EM; Rosner MR; Gibbs JS; Embry A; Dolan B; Das S; Hickman HD; Berglund P; Bennink JR; Yewdell JW; Pan T Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462:522–526; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee JY; Kim DG; Kim BG; Yang WS; Hong J; Kang T; Oh YS; Kim KR; Han BW; Hwang BJ; Kang BS; Kang MS; Kim MH; Kwon NH; Kim S Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J. Cell Sci 127:4234–4245; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lim JM; Kim G; Levine RL Methionine in proteins: It's not just for protein initiation anymore. Neurochem. Res 44:247–257; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Toennies G; Kolb JJ Methionine studies. J. Biol. Chem 128:399–405; 1939. [Google Scholar]
- [11].Weissbach H; Resnick L; Brot N Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim. Biophys. Acta 1703:203–212; 2005. [DOI] [PubMed] [Google Scholar]
- [12].Lee BC; Dikiy A; Kim HY; Gladyshev VN Functions and evolution of selenoprotein methionine sulfoxide reductases. Biochim. Biophys. Acta 1790:1471–1477; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Boschi-Muller S; Gand A; Branlant G The methionine sulfoxide reductases: Catalysis and substrate specificities. Arch. Biochem. Biophys 474:266–273; 2008. [DOI] [PubMed] [Google Scholar]
- [14].Kim G; Cole NB; Lim JC; Zhao H; Levine RL Dual sites of protein initiation control the localization and myristoylation of methionine sulfoxide reductase A. J. Biol. Chem 285:18085–18094; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lim JM; Lim JC; Kim G; Levine RL Myristoylated methionine sulfoxide reductase A is a late endosomal protein. J. Biol. Chem 293:7355–7366; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moskovitz J; Berlett BS; Poston JM; Stadtman ER The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc. Natl. Acad. Sci. U.S.A 94:9585–9589; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Moskovitz J; Rahman MA; Strassman J; Yancey SO; Kushner SR; Brot N; Weissbach H Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bacteriol 177:502–507; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Douglas T; Daniel DS; Parida BK; Jagannath C; Dhandayuthapani S Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J. Bacteriol 186:3590–3598; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].St John G; Brot N; Ruan J; Erdjument-Bromage H; Tempst P; Weissbach H; Nathan C Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. U.S.A 98:9901–9906; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mahawar M; Tran V; Sharp JS; Maier RJ Synergistic roles of Helicobacter pylori methionine sulfoxide reductase and GroEL in repairing oxidant-damaged catalase. J. Biol. Chem 286:19159–19169; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Benoit SL; Maier RJ Helicobacter catalase devoid of catalytic activity protects the bacterium against oxidative stress. J. Biol. Chem 291:23366–23373; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Moskovitz J; Flescher E; Berlett BS; Azare J; Poston JM; Stadtman ER Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc. Natl. Acad. Sci. U.S.A 95:14071–14075; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yermolaieva O; Xu R; Schinstock C; Brot N; Weissbach H; Heinemann SH; Hoshi T Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc. Natl. Acad. Sci. U.S.A 101:1159–1164; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ruan H; Tang XD; Chen ML; Joiner ML; Sun G; Brot N; Weissbach H; Heinemann SH; Iverson L; Wu CF; Hoshi T; Chen ML; Joiner MA; Heinemann SH High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. U.S.A 99:2748–2753; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Romero HM; Berlett BS; Jensen PJ; Pell EJ; Tien M Investigations into the role of the plastidial peptide methionine sulfoxide reductase in response to oxidative stress in Arabidopsis. Plant Physiol. 136:3784–3794; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao H; Sun J; Deschamps AM; Kim G; Liu C; Murphy E; Levine RL Myristoylated methionine sulfoxide reductase A protects the heart from ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol 301:H1513–H1518; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Salmon AB; Perez VI; Bokov A; Jernigan A; Kim G; Zhao H; Levine RL; Richardson A Lack of methionine sulfoxide reductase A in mice increases sensitivity to oxidative stress but does not diminish life span. FASEB J. 23:3601–3608; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Noh MR; Kim KY; Han SJ; Kim JI; Kim HY; Park KM Methionine sulfoxide reductase A deficiency exacerbates cisplatin-induced nephrotoxicity via increased mitochondrial damage and renal cell death. Antioxid. Redox Signal 27:727–741; 2017. [DOI] [PubMed] [Google Scholar]
- [29].Kim JI; Choi SH; Jung KJ; Lee E; Kim HY; Park KM Protective role of methionine sulfoxide reductase A against ischemia/reperfusion injury in mouse kidney and its involvement in the regulation of trans-sulfuration pathway. Antioxid. Redox Signal 18:2241–2250; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gu SX; Blokhin IO; Wilson KM; Dhanesha N; Doddapattar P; Grumbach IM; Chauhan AK; Lentz SR Protein methionine oxidation augments reperfusion injury in acute ischemic stroke. JCI Insight 1; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim JI; Noh MR; Kim KY; Jang HS; Kim HY; Park KM Methionine sulfoxide reductase A deficiency exacerbates progression of kidney fibrosis induced by unilateral ureteral obstruction. Free Radic. Biol. Med 89:201–208; 2015. [DOI] [PubMed] [Google Scholar]
- [32].Singh MP; Kim KY; Kim HY Methionine sulfoxide reductase A deficiency exacerbates acute liver injury induced by acetaminophen. Biochem. Biophys. Res. Commun 484:189–194; 2017. [DOI] [PubMed] [Google Scholar]
- [33].Erickson JR; Joiner ML; Guan X; Kutschke W; Yang J; Oddis CV; Bartlett RK; Lowe JS; O'Donnell SE; Aykin-Burns N; Zimmerman MC; Zimmerman K; Ham AJ; Weiss RM; Spitz DR; Shea MA; Colbran RJ; Mohler PJ; Anderson ME A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 133:462–474; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Singh MP; Kim KY; Kwak GH; Baek SH; Kim HY Methionine sulfoxide reductase A protects against lipopolysaccharide-induced septic shock via negative regulation of the proinflammatory responses. Arch. Biochem. Biophys 631:42–48; 2017. [DOI] [PubMed] [Google Scholar]
- [35].Klutho PJ; Pennington SM; Scott JA; Wilson KM; Gu SX; Doddapattar P; Xie L; Venema AN; Zhu LJ; Chauhan AK; Lentz SR; Grumbach IM Deletion of methionine sulfoxide reductase A does not affect atherothrombosis but promotes neointimal hyperplasia and extracellular signal-regulated kinase 1/2 signaling. Arteroscler. Thromb. Vasc. Biol 35:2594–2604; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Alqudah S; Chertoff M; Durham D; Moskovitz J; Staecker H; Peppi M Methionine sulfoxide reductase A knockout mice show progressive hearing loss and sensitivity to acoustic trauma. Audiol. Neurotol 23:20–31; 2018. [DOI] [PubMed] [Google Scholar]
- [37].Kwon TJ; Cho HJ; Kim UK; Lee E; Oh SK; Bok J; Bae YC; Yi JK; Lee JW; Ryoo ZY; Lee SH; Lee KY; Kim HY Methionine sulfoxide reductase B3 deficiency causes hearing loss due to stereocilia degeneration and apoptotic cell death in cochlear hair cells. Hum Mol Genet 23:1591–1601; 2014. [DOI] [PubMed] [Google Scholar]
- [38].Ahmed ZM; Yousaf R; Lee BC; Khan SN; Lee S; Lee K; Husnain T; Rehman AU; Bonneux S; Ansar M; Ahmad W; Leal SM; Gladyshev VN; Belyantseva IA; Van Camp G; Riazuddin S; Friedman TB; Riazuddin S Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am. J. Hum. Genet 88:19–29; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fomenko DE; Novoselov SV; Natarajan SK; Lee BC; Koc A; Carlson BA; Lee TH; Kim HY; Hatfield DL; Gladyshev VN MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J. Biol. Chem 284:5986–5993; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhao H; Kim G; Levine RL Methionine sulfoxide reductase contributes to meeting dietary methionine requirements. Arch. Biochem. Biophys 522:37–43; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. https://crispr.mit.edu.
- [42]. https://www.mmrrc.org/catalog/sds.php?mmrrc_id=65273.
- [43].Tarafdar S; Kim G; Levine RL Drosophila methionine sulfoxide reductase A (MSRA) lacks methionine oxidase activity. Free Radic. Biol. Med 131:154–161; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lin J; Steenbergen C; Murphy E; Sun J Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation 120:245–254; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sun J; Morgan M; Shen RF; Steenbergen C; Murphy E Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ. Res 101:1155–1163; 2007. [DOI] [PubMed] [Google Scholar]
- [46].Huang TT; Yasunami M; Carlson EJ; Gillespie AM; Reaume AG; Hoffman EK; Chan PH; Scott RW; Epstein CJ Superoxide-mediated cytotoxicity in superoxide dismutase-deficient fetal fibroblasts. Arch. Biochem. Biophys 344:424–432; 1997. [DOI] [PubMed] [Google Scholar]
- [47].Han EKH; Leverson JD; McGonigal T; Shah OJ; Woods KW; Hunter T; Giranda VL; Luo Y Akt inhibitor A-443654 induces rapid Akt Ser-473 phosphorylation independent of mTORC1 inhibition. Oncogene 26:5655; 2007. [DOI] [PubMed] [Google Scholar]
- [48].National Research Council Guide for the care and use of laboratory animals. Washington, DC: National Academies Press; 2011. [Google Scholar]
- [49].Freeman HC; Hugill A; Dear NT; Ashcroft FM; Cox RD Deletion of nicotinamide nucleotide transhydrogenase: a new quantitive trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes 55:2153–2156; 2006. [DOI] [PubMed] [Google Scholar]
- [50].Cohen MV; Baines CP; Downey JM Ischemic preconditioning: from adenosine receptor to KATP channel. Ann Rev Physiol 62:79–109; 2000. [DOI] [PubMed] [Google Scholar]
- [51].Wojcik B; Knapp M; Gorski J Non-ischemic heart preconditioning. J. Physiol. Pharmacol 69; 2018. [DOI] [PubMed] [Google Scholar]
- [52].Salmon AB; Perez VI; Bokov A; Jernigan A; Kim G; Zhao H; Levine RL; Richardson A Lack of methionine sulfoxide reductase A in mice increases sensitivity to oxidative stress but does not diminish life span. FASEB J. 23:3601–3608; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ashburner M; Ball CA; Blake JA; Botstein D; Butler H; Cherry JM; Davis AP; Dolinski K; Dwight SS; Eppig JT; Harris MA; Hill DP; Issel-Tarver L; Kasarskis A; Lewis S; Matese JC; Richardson JE; Ringwald M; Rubin GM; Sherlock G Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25:25–29; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47:D330–d338; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Krenz M; Robbins J Impact of beta-myosin heavy chain expression on cardiac function during stress. J. Am. Coll. Cardiol 44:2390–2397; 2004. [DOI] [PubMed] [Google Scholar]
- [56].Han P; Li W; Lin CH; Yang J; Shang C; Nuernberg ST; Jin KK; Xu W; Lin CY; Lin CJ; Xiong Y; Chien H; Zhou B; Ashley E; Bernstein D; Chen PS; Chen HV; Quertermous T; Chang CP A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514:102–106; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Corda D; Mosca MG; Ohshima N; Grauso L; Yanaka N; Mariggio S The emerging physiological roles of the glycerophosphodiesterase family. FEBS J. 281:998–1016; 2014. [DOI] [PubMed] [Google Scholar]
- [58].Rahman IAS; Tsuboi K; Hussain Z; Yamashita R; Okamoto Y; Uyama T; Yamazaki N; Tanaka T; Tokumura A; Ueda N Calcium-dependent generation of N-acylethanolamines and lysophosphatidic acids by glycerophosphodiesterase GDE7. Biochim Biophys Acta Mol Cell Biol Lipids 1861:1881–1892; 2016. [DOI] [PubMed] [Google Scholar]
- [59].Sies H; Berndt C; Jones DP Oxidative stress. Annu. Rev. Biochem 86:715–748; 2017. [DOI] [PubMed] [Google Scholar]
- [60].Chen QM; Maltagliati AJ Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genomics 50:77–97; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pennington SM; Klutho PR; Xie L; Broadhurst K; Koval OM; McCormick ML; Spitz DR; Grumbach IM Defective protein repair under methionine sulfoxide A deletion drives autophagy and ARE-dependent gene transcription. Redox Biol. 16:401–413; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hausenloy DJ; Tsang A; Mocanu MM; Yellon DM Ischemic preconditioning protects by activating prosurvival kinases at reperfusion. Am. J. Physiol. Heart Circ. Physiol 288:H971–976; 2005. [DOI] [PubMed] [Google Scholar]
- [63].Cohen MV; Downey JM Ischemic postconditioning: from receptor to end-effector. Antioxid. Redox Signal. 14:821–831; 2011. [DOI] [PubMed] [Google Scholar]
- [64].Stadtman ER; Moskovitz J; Berlett BS; Levine RL Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol. Cell Biochem 234-235:3–9; 2002. [PubMed] [Google Scholar]
- [65].Zhang XH; Tee LY; Wang XG; Huang QS; Yang SH Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 4:e264; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Iyer V; Shen B; Zhang W; Hodgkins A; Keane T; Huang X; Skarnes WC Off-target mutations are rare in Cas9-modified mice. Nat Methods 12:479; 2015. [DOI] [PubMed] [Google Scholar]
- [67].Willi M; Smith HE; Wang C; Liu C; Hennighausen L Mutation frequency is not increased in CRISPR-Cas9-edited mice. Nat. Methods 15:756–758; 2018. [DOI] [PubMed] [Google Scholar]
- [68].Reczek CR; Birsoy K; Kong H; Martínez-Reyes I; Wang T; Gao P; Sabatini DM; Chandel NS A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nature Chem. Biol 13:1274; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Migliaccio E; Giorgio M; Mele S; Pelicci G; Reboldi P; Pandolfi PP; Lanfrancone L; Pelicci PG The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature 402:309–313; 1999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.